Abstract
Terminal restriction fragment (TRF) analysis of 16S rRNA genes is an increasingly popular method for rapid comparison of microbial communities, but analysis of the data is still in a developmental stage. We assessed the phylogenetic resolution and reproducibility of TRF profiles in order to evaluate the limitations of the method, and we developed an essential analysis technique to improve the interpretation of TRF data. The theoretical phylogenetic resolution of TRF profiles was determined based on the specificity of TRFs predicted from 3,908 16S rRNA gene sequences. With sequences from the Proteobacteria or gram-positive division, as much as 73% of the TRFs were phylogenetically specific (representing strains from at most two genera). However, the fraction decreased when sequences from the two divisions were combined. The data show that phylogenetic inference will be most effective if TRF profiles represent only a single bacterial division or smaller group. The analytical precision of the TRF method was assessed by comparing nine replicate profiles of a single soil DNA sample. Despite meticulous care in producing the replicates, numerous small, irreproducible peaks were observed. As many as 85% of the 169 distinct TRFs found among the profiles were irreproducible (i.e., not present in all nine replicates). Substantial variation also occurred in the height of synonymous peaks. To make comparisons of microbial communities more reliable, we developed an analytical procedure that reduces variation and extracts a reproducible subset of data from replicate TRF profiles. The procedure can also be used with other DNA fingerprinting techniques for microbial communities or microbial genomes.
Comparative analysis of complex microbial communities in natural environments has been hampered by the lack of effective ways to rapidly measure community diversity, composition, and structure. The shortcomings of methods that rely on cultivation are well known, and although DNA-based, culture-independent techniques have provided new ways to examine the microbial world, the methods that are effective in community analysis are still quite limited in number and scope (23). Terminal restriction fragment (length polymorphism (T-RFLP or TRF) analysis is currently one of the most powerful methods in microbial ecology for rapidly comparing the diversity of bacterial DNA sequences amplified by PCR from environmental samples (19, 23). The method relies on variation in the position of restriction sites among sequences and determination of the length of fluorescently labeled TRFs by high-resolution gel electrophoresis on automated DNA sequencers (1, 16). The method's many strengths include speed and high sample throughput, which enables replicated experiments with statistical analysis to be conducted. Highly precise fragment length determination is achieved by use of an automated DNA sequencer with internal size standards in every profile and provides numerical data of exceptional resolution. In theory, data from the method can also be compared with data predicted from rapidly expanding sequence databases in order to infer the potential composition of a sample.
The TRF method has been used successfully for differentiation of bacterial communities in marine samples (20), the digestive tracts of fish (24), soil samples (3, 6, 7, 21), and enrichment cultures over time (4, 5, 14) and for differentiation of wastewater treatment plant sludge, laboratory bioreactor, aquifer sand, groundwater, and termite gut communities (16). It has also been compared with denaturing gradient gel electrophoresis (DGGE) and 16S rRNA gene (rDNA) cloning for its effectiveness and consistency in differentiating microbial communities (7, 20, 23). Given the increasing popularity of the TRF method as a tool for community analysis, widespread use of the method in field ecology studies is anticipated. However, while the method is easy to perform, analysis of the data generated is still in a developmental stage, with several technical and theoretical issues yet to be addressed. At this juncture, a thorough understanding of the strengths and current limitations of the method is essential so that we can correctly assess the value of TRF analysis, improve its capabilities, and interpret the results properly.
The TRF method could conceivably be used in three ways for analysis of microbial communities. TRF profiles could be used for differentiation of communities, for comparison of the relative phylotype richness and structure of communities, and for identifying specific organisms in a community. The method's robust ability to differentiate microbial communities has been validated (7, 20). However, for highly complex soil bacterial communities, the method has been shown to be ineffective in assessing relative phylotype richness and structure (7). In the present study, we extended the work of previous studies (16, 19) by providing a more thorough and detailed evaluation of the capacity for inferring the phylogenetic composition of a community from TRF profiles. This was done in part by examining the phylogenetic relationship of every group of 16S rDNA sequences that yielded the same TRF size. We also addressed two critical questions in data analysis: how reproducible are TRF profiles, and how can TRF data be analyzed such that samples of equal size are compared? In theory, replicate profiles (i.e., aliquots of a single restriction digest) should be identical, but in practice, we found a substantial amount of variability that could lead to erroneous conclusions if unreplicated profiles were used to compare microbial communities in different environmental samples. Here we present strategies for increasing the utility of TRF profiles for phylogenetic inference and present a data analysis method that improves the reliability of TRF data for differentiation of communities. The analysis techniques can also be used with other methods (e.g., DGGE and amplified fragment length polymorphism) that generate DNA profiles of microbial communities or microbial genomes.
MATERIALS AND METHODS
Prediction of TRFs from 16S rDNA sequences.
TRF data were obtained primarily from version 7.1 (v7.1) (18) of the Ribosomal Database Project (RDP), with key updates based on the RDP v8.0 (17). A set of 2,156 aligned small-subunit (SSU) rRNA sequences were obtained from the RDP v7.1 (18) for analysis. The sequences represented bacteria from the gram-positive and Proteobacteria divisions only and matched the 3′-terminal 10 nucleotides of PCR primer 8-27f (pA; 5′-AGAGTTTGATCCTGGCTCAG [9] according to the criterion that the sequences had 100% identity with the 3′-terminal 3 nucleotides of primer 8-27f and one or no mismatches with the remaining 7 nucleotides. From the set of 2,156 sequences, 129 sequences with suspicious sequence gaps were eliminated. Suspicious gaps were defined as gaps of three or more nucleotides that were phylogenetically random (i.e., appearing in the sequence of one strain of a species but no other strains of the same species) or that occurred in regions of the SSU rRNA gene known to be highly conserved among bacteria. To maximize the number of sequences available for TRF analysis, the final set of 2,027 sequences was not matched with a reverse PCR primer. Many partial sequences that can be used for prediction of TRFs are too short to be matched against both forward and reverse PCR primers. TRFs were predicted for the enzymes HaeIII, BstUI, HhaI, MspI, and RsaI by identifying restriction site positions with the program Patscan (22). The TAP-TRFLP function from the RDP was used to update a subset of the data based on the new release (v8.0) (17) of the RDP in order to demonstrate the impact of the larger sequence database on TRF analysis.
Soil sample.
Soil was collected in 1994 from a site in the Coconino National Forest, Arizona (15). The site is a pinyon pine-juniper woodland with light sandy loam soil (11), and the interspaces (areas between widely spaced trees) are sparsely covered with grass and forb species. A composite soil sample for the interspaces was collected by combining 10 subsamples collected at a depth of 10 to 15 cm from different locations (15).
DNA extraction.
DNA was extracted from the soil sample using a four-step procedure that included three cycles of freeze-thaw, incubation at 70°C with sodium dodecyl sulfate, bead mill homogenization, and ethanol precipitation as described previously (15). Precipitated DNA was cleaned by phenol-chloroform extraction. The DNA was stored frozen at −20°C, then further purified by passage through Sephadex G-200 spin columns, and precipitated with ethanol for use in this study.
TRF profiles.
16S rDNA for TRF analysis was amplified with primer 8-27f fluorescently labeled with 6′-carboxyfluorescein (ABI, Perkin-Elmer, Foster City, Calif.) and with primer 1507-1492r (5′-TACCTTGTTACGACTT [25]). Each 50-μl reaction mixture contained 30 mM Tris (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 50 μM each deoxynucleoside triphosphate, 50 pmol of each primer, and 0.75 U of Taq polymerase (AmpliTaq LD; Perkin-Elmer). Cycling conditions were as follows: 2 min of denaturation at 94°C, 35 cycles of 30 s at 50°C, 1 min at 72°C, and 10 s at 94°C, and a final cycle of annealing at 55°C for 1 min and extension at 72°C for 5 min. Three independent 50-μl PCRs were performed for each sample; the products were combined and purified with a Qiaquick PCR cleanup kit (Qiagen, Inc., Chatsworth, Calif.). Approximately 50 ng of purified 16S rDNA was digested in a 20-μl reaction volume with 5 U of RsaI for 3 h. Following restriction digestion, the DNA was passed through a Sephadex G-200 Centrisep column (Princeton Separations Inc., Princeton, N.J.) for purification.
Nine replicate TRF profiles were obtained from the digested DNA by loading three aliquots of the digested DNA on each of three separate polyacrylamide gels. Replication at this level was performed to measure the degree of variation in TRF profiles arising solely as a result of experimental error during electrophoresis of digested DNA samples. For each gel, three 1-μl aliquots of the digested DNA were dried, suspended in 1.75 μl of loading buffer (0.25 μl of Genescan 2500 TAMRA size standard [ABI], 1.25 μl of deionized formamide, and 0.25 μl of a 3% [wt/vol] blue dextran–25 mM EDTA solution), denatured at 94°C for 2 min, and placed immediately on ice for 2 min. The aliquots were electrophoresed in denaturing 4% polyacrylamide gels with an ABI 377 DNA sequencer. Between runs, the stock of digested DNA was stored frozen at −20°C. Reagents for polyacrylamide gel electrophoresis were purchased from Bio-Rad (Hercules, Calif.). Terminal restriction fragment sizes between 94 and 827 bp with peak heights of ≥25 fluorescence units were determined using Genescan analytical software v2.02 (ABI).
Analysis of TRF profiles.
A five-step analysis procedure for comparison of TRF profiles was performed as follows, using S+ v3.2 (MathSoft, Inc., Seattle, Wash.).
(i) Alignment of replicate profiles.
A clustering algorithm was used to identify synonymous fragment sizes in replicate profiles and to align the profiles. Genescan analysis software calculates DNA fragment sizes to 1/100 of a base pair. The resulting values cannot simply be rounded up or down to the nearest integer value because in replicate profiles the measured value of a fragment size may float sometimes above and sometimes below the median of two integers (e.g., 133.38 and 133.53 bp). In this example, comparison of numerical values rounded to the nearest integer would incorrectly suggest the presence of two fragments of distinct sizes. Clustering values that fit within empirically determined margins of error circumvents this problem. The error in determining fragment sizes with our ABI 377 automated DNA sequencer was less than 0.5 bp, and typically the error was less than 0.2 bp. Therefore, TRFs that differed by less than 0.5 bp in different profiles were considered identical and were clustered. All fragments within a cluster were assigned the average of the sizes within the cluster. In some cases, distinct peaks differing by 0.5 bp or less occur in a single profile and can be reproducibly resolved, suggesting the presence of at least two DNA fragments that either differ in sequence composition or differ in length but migrate close together. In an attempt to avoid clustering such fragments among a set of profiles, the maximum number of fragments assigned to a cluster was limited to the number of profiles being aligned. Thus, as soon as a cluster is filled with the maximum number of fragments with the smallest differences in measured lengths, a new cluster is created.
(ii) Standardization of DNA quantity between replicate profiles.
The sum of all peak heights of ≥25 fluorescence units (i.e., the total fluorescence; 25 fluorescence units is the baseline noise threshold) in each replicate profile was calculated as an indication of the total DNA quantity represented by each profile. DNA quantity was standardized between replicate profiles to the smallest quantity by proportionally reducing the height of each peak in larger profiles. To accomplish this, the proportion of the smallest DNA quantity (i.e., total fluorescence) and a larger DNA quantity was calculated and used as a correction factor to adjust each peak height in the profile representing the larger DNA quantity. For example, given two profiles with total fluorescence values of 24,000 and 40,000, respectively, each peak in the latter profile would be multiplied by a correction factor of 0.6 (i.e., the quotient of 24,000/40,000). This procedure often eliminated peaks from larger profiles by reducing some peak heights below the baseline noise threshold (25 fluorescence units). Therefore, after adjustment of a profile, the new sum of peak heights of ≥25 fluorescence units was calculated, and the standardization procedure was repeated until, by iteration, the DNA quantity (i.e., total fluorescence) of the larger profile was equal to the quantity of the smaller profile.
In rare cases, the total DNA quantity represented by a larger profile cannot be made exactly equal to the quantity of the smaller profile. In these cases, the total DNA quantity of the larger profile fluctuates between a value above and a value below the quantity of the smaller profile in successive iterations of the standardization routine. This occurs when one or more peaks fall below the noise threshold (25 fluorescence units) in one iteration, resulting in a total DNA quantity less than that in the smaller profile, and then rise above the threshold in the next iteration, resulting in a total quantity greater than that in the smaller profile. In these cases, the average of the two iterations is calculated in order to make the larger profile as close to the small profile as possible.
(iii) Creation of a derivative, reproducible sample profile.
For each sample, a derivative profile containing only the most conservative and reliable TRF information was created by identifying the subset of TRFs that appeared in all replicate profiles of a sample. Irreproducible TRFs (i.e., fragments observed in less than 100% of the replicate profiles of a sample) were discarded. The average peak height (abundance) of each reproducible TRF from a sample was calculated from the peak heights observed in individual replicates. The resulting list of TRFs and the average height of each TRF were used as the derivative sample profile.
(iv) Standardization of DNA quantity between different environmental samples.
To compare different samples, the derivative profiles for a set of samples were standardized as described in step iii for replicate profiles. In brief, the sum of all peak heights of ≥25 fluorescence units in each derivative sample profile was calculated as an indication of the total DNA quantity represented by the profile. The DNA quantities for a set of samples were then standardized using the iterative standardization procedure described above.
(v) Alignment of standardized, derivative sample profiles.
Following standardization, derivative sample profiles were aligned as described in step i for replicate profiles. The average size of TRFs in each alignment cluster was calculated to produce a single, composite list of the TRF sizes found among all samples. By comparison of each average sample profile with the composite list, a binary vector was constructed for each sample representing the presence or absence of the TRFs in the composite list.
(vi) Comparison of binary sample profiles.
The Jaccard coefficient was used as a measure of similarity of binary vectors, and a matrix of pairwise comparisons was constructed (13). The Jaccard coefficient was used for the binary data because it describes the similarity of each sample pair based only on TRFs that are present in one or both samples (in other words, TRFs that are not present in either of two samples being compared do not contribute to the similarity of two samples). Agglomerative hierarchical clustering was performed using the similarity matrix of Jaccard coefficients and the unweighted pair-group average method and was displayed as dendrograms (13).
RESULTS AND DISCUSSION
Inference of phylogenetic composition of natural samples.
The ability to use a TRF profile to infer the phylogenetic composition of a sample depends greatly on two factors: the phylogenetic resolution of TRFs (i.e., the similarity of all organisms that can produce a given TRF size) and the number and quality of reference sequences available for comparative analysis. In previous studies, the capacity for TRFs to discriminate among sequences has been summarized either by reporting the maximum number of sequences predicted to generate the same TRF size (i.e., the maximum redundancy) (16, 19) or by reporting the fraction of sequences represented by the five most redundant TRFs (19). Both measures are partial indications of the amount of skew in the distribution of sequences among a set of predicted TRF sizes. However, these measures provide information for only one or a few TRFs (the least informative TRFs) in a distribution and provide no general information about the phylogenetic relationships between sequences that yield the same TRF size. As a result, one might mistakenly conclude that an enzyme which yields a set of predicted TRFs with relatively low redundancy might be useful for inferring some of the phylogenetic composition of a community.
In this study, we have attempted to provide more detailed information for assessing the use of TRF profiles for phylogenetic inference. Toward this end, we evaluated not only the frequency distribution of sequences among predicted TRF sizes as others have done but also the phylogenetic information that could be derived from each predicted TRF size. Fragment sizes that represented strains from three or fewer species were counted as species-specific TRFs, and fragment sizes that represented strains from two or fewer genera were counted as genus-specific TRFs. These arbitrary criteria for describing phylogenetic specificity were chosen as a compromise. For any given enzyme, extremely few TRFs are truly specific for a single species (i.e., representing numerous strains of a single species) or members of a single genus. Therefore, TRF specificity was evaluated by using groups that are larger than a single species or genus but are small enough to be informative for comparative community diversity.
The phylogenetic specificities of TRFs predicted for five different enzymes are summarized in Table 1. The sequences represented 370 named genera and 1,288 named species. The importance of strategically choosing restriction enzymes to give an optimal distribution of TRFs has been discussed previously (16, 19); however, the data in the present study provide more detailed indications of the phylogenetic specificity that can be achieved with different enzymes. The data also demonstrate that combining two enzymes in a single digest does not significantly improve the utility of TRF profiles. For example, the total number of TRFs predicted from Proteobacteria sequences is not significantly increased by double digestion compared to the results achieved with single enzyme digests. Whereas 148 RsaI TRFs (between 94 and 827 bp) were predicted from Proteobacteria sequences, only 9 and 17 additional TRFs were produced by combining either HhaI or MspI with RsaI, respectively. Combining RsaI with HaeIII resulted in a decrease in the number of TRFs in the analysis range (94 to 827 bp) compared to the number that could be achieved with RsaI alone. The data also show that fewer phylogenetically informative TRFs are obtained from double digestion than by combining results from separate single enzyme digests. Whereas 116 species or genus-specific TRFs occurred in the profile derived from MspI and RsaI double digestion of the Proteobacteria sequences, a total of 217 informative TRFs were obtained by combining the data from separate MspI and RsaI digests. These data demonstrate that use of combinations of single enzyme digests will typically be the best strategy for general profiling of bacterial communities and for phylogenetic inference.
TABLE 1.
Phylogenetic resolution of primer 8-27f TRFs predicted for different enzymes
| Enzyme(s) | Divisiona | No. of TRFs | TRFs between 94 and 827 bpc | % Species-specific TRFsd | % Species- or genus-specific TRFsd | % Species- or genus-specific binse |
|---|---|---|---|---|---|---|
| HaeIII | G+, Proteo | 228 (2,025b) | 182 (1,442b) | 46 (11f) | 50 (15f) | 19 (7f) |
| BstUI | G+, Proteo | 358 (2,015) | 290 (1,706) | 63 (15) | 70 (24) | 42 (11) |
| HhaI | G+, Proteo | 406 (2,021) | 294 (1,607) | 62 (15) | 70 (21) | 37 (6) |
| MspI | G+, Proteo | 311 (2,007) | 276 (1,836) | 61 (13) | 70 (16) | 34 (9) |
| RsaI | G+, Proteo | 354 (2,015) | 266 (1,489) | 68 (23) | 70 (31) | 43 (15) |
| HaeIII | G+ | 183 (1,089) | 164 (927) | 61 (27) | 64 (32) | 22 (8) |
| BstUI | G+ | 197 (1,090) | 179 (1,036) | 65 (23) | 69 (28) | 50 (18) |
| HhaI | G+ | 305 (1,089) | 226 (901) | 68 (30) | 76 (42) | 46 (21) |
| MspI | G+ | 220 (1,081) | 200 (1,011) | 66 (28) | 72 (38) | 44 (19) |
| RsaI | G+ | 244 (1,090) | 185 (819) | 75 (30) | 77 (33) | 59 (24) |
| HaeIII + RsaI | G+ | 217 (1,086) | 194 (888) | 69 (35) | 74 (45) | NDg |
| BstUI + RsaI | G+ | 212 (1,088) | 184 (880) | 70 (31) | 74 (39) | ND |
| HhaI + RsaI | G+ | 286 (1,086) | 251 (867) | 73 (42) | 81 (61) | ND |
| MspI + RsaI | G+ | 228 (1,079) | 202 (860) | 70 (36) | 50 (76) | ND |
| HaeIII | Proteo | 133 (936) | 97 (515) | 63 (23) | 70 (34) | 36 (17) |
| BstUI | Proteo | 129 (935) | 120 (886) | 61 (13) | 66 (18) | 44 (10) |
| HhaI | Proteo | 176 (926) | 128 (705) | 67 (22) | 72 (28) | 47 (14) |
| MspI | Proteo | 165 (926) | 141 (825) | 67 (20) | 73 (30) | 40 (16) |
| RsaI | Proteo | 200 (925) | 148 (670) | 73 (29) | 77 (36) | 53 (23) |
| HaeIII + RsaI | Proteo | 146 (936) | 122 (519) | 70 (32) | 75 (42) | ND |
| BstUI + RsaI | Proteo | 142 (935) | 125 (866) | 61 (14) | 66 (21) | ND |
| HhaI + RsaI | Proteo | 186 (932) | 157 (755) | 69 (24) | 77 (33) | ND |
| MspI + RsaI | Proteo | 190 (926) | 165 (809) | 67 (26) | 70 (35) | ND |
G+, gram positive; Proteo, Proteobacteria.
Number of sequences represented by the TRFs.
The range was derived from the Genescan 2500 size standard (ABI) that was used for determination of fragment sizes in gel electrophoresis.
Only the specificity of TRFs in the analysis range (94 to 827 bp) was examined. A species-specific TRF was defined as a TRF size representing strains from three or fewer species, and each TRF size representing strains from two or fewer genera was counted as a genus-specific TRF. The listed values are percentages of the numbers of TRFs between 94 and 827 bp (from column 4).
Each bin consisted of a TRF size of ±1 bp.
Percentages of sequences (from the totals in column 4) that were represented by phylogenetically specific TRFs.
ND, not determined.
As shown in Table 1, the fraction of phylogenetically informative TRFs that are theoretically possible in a TRF profile can often be quite high. For example, with the enzymes HhaI, MspI, and RsaI, the fraction of species-specific TRFs predicted from gram-positive and Proteobacteria sequences combined ranged from 61 to 68%. For each enzyme, the fraction of phylogenetically informative TRFs was increased to 70% by including genus-specific TRFs, demonstrating that the criteria used to define phylogenetic specificity can alter the perceived utility of TRF profiles for phylogenetic inference. Relaxing the definition of specificity to include TRFs that represent members of more than two genera but belong to the same phylogenetic subgroup or assemblage would increase further the percentage of informative TRFs (data not shown), although the utility of this type of information in community analysis is questionable.
The fraction of phylogenetically informative TRFs was highest when TRFs were predicted from sequences representing a single bacterial division. For example, 77% of RsaI TRFs were species or genus specific when predicted from Proteobacteria sequences only or from gram-positive sequences only. Combining the two divisions decreased the fraction of phylogenetically specific TRFs to 70%. The total number of informative TRFs that could be derived from the sequences also decreased. A total of 257 informative TRFs could be derived by analyzing the divisions separately, while only 192 occurred when the divisions were combined. The extent of division level diversity observed thus far in natural environments is typically much higher than two divisions. For example, 9 divisions were identified in an anaerobic digestor (10), an average of 16 divisions were identified in soil from two different sites in Arizona (J. Dunbar, S. M. Barns, J. A. Davis, G. Fisher, and C. R. Kuske, unpublished data), and 25 divisions were detected in a Yellowstone hot spring sample (12). Thus, the average phylogenetic resolution of TRFs in general profiles of natural environments may be so low that phylogenetic inference of community composition is not feasible.
In practice, the number of phylogenetically informative TRFs that are possible in a profile will be lower than the number predicted from sequence databases. Since the migration of fragments in polyacrylamide gels is influenced to some extent by sequence composition, discrepancies may occur between observed and predicted fragment sizes. Discrepancies have been noted between predicted and observed fragment sizes (6, 16, 20). Due to the inaccuracy of fragment size measurements from gels, observed fragment sizes must be matched with a bin of predicted, contiguous fragment sizes. Thus, although each individual fragment size in a bin of predicted TRFs may be phylogenetically specific, the range of organisms represented by all TRFs in the bin may be quite broad. Applying a bin size of ±1 bp (i.e., each bin contains three consecutive fragment sizes such as 350, 351, and 352 bp) to the TRFs predicted from either the Proteobacteria sequences or gram-positive sequences reduced the fraction of species- or genus-specific RsaI TRFs from approximately 77% (Table 1) to 53 or 59%, respectively. These data underscore the importance of amplifying sequence mixtures from single bacterial divisions or smaller groups, since even more substantial reductions in bin specificity occur when TRFs are derived simultaneously from multiple divisions.
The predicted fraction of phylogenetically informative TRFs and TRF bins may decrease even further as the variety of sequences available for analysis increases. For example, the fraction of phylogenetically informative TRFs in profiles predicted from 1,007 Proteobacteria or gram-positive division sequences (analyzed separately) from the RDP v6.0 decreased from an average of 80 to 77% after adding 482 sequences from the RDP v7.1 (1,489 sequences analyzed in total). With the RDP v8.0, only 71% of TRFs predicted from 3,908 aligned Proteobacteria or gram-positive division sequences were phylogenetically specific. The fraction of informative TRFs from these two bacterial divisions is clearly decreasing as the number and variety of reference sequences increase. For TRF profiles obtained using universal 16S rDNA primers, even greater impacts on predicted TRF specificity can be anticipated as the variety of sequences in the eubacterial domain expands.
If phylogenetic inference from TRF profiles is confined (as it should be) to profiles created with primers specific for a single division or smaller group, the fraction of bacterial communities that could be studied is presently quite small. The RDP v8.0 recognizes 30 major eubacterial divisions but is severely limited in coverage of these divisions. For example, the aligned sequences that reasonably match primer 8-27f represent 20 of the 30 major divisions, but the Proteobacteria, gram-positive, and Flexibacter-Cytophaga-Bacteroides divisions account for 90% (45, 39, and 7%, respectively) of the sequences, while the remaining 17 divisions are represented by an average of 26 sequences each (Table 2). Natural environments may often be dominated by microbes belonging to divisions other than the Proteobacteria and gram-positive bacteria. For example, 16S rDNA sequences representing members of the Acidobacterium division were the most common sequences amplified from four different soils (including the one used in this study) from Arizona (8, 15; Dunbar et al., unpublished). Members of the Acidobacterium division accounted for 49% of 766 16S rDNA sequences obtained from the four Arizona soils, while Proteobacteria and gram-positive sequences accounted for only 17 and 6%, respectively (Dunbar et al., unpublished). Primers for amplification of 16S rDNA sequences specifically from members of the Acidobacterium division are available (2). However, attempts to infer the composition of this fraction (the largest fraction) of the community are clearly constrained by the limited supply of reference sequences currently available in sequence databases. Although this situation will certainly improve over time, at present the use of 16S rDNA clone libraries in conjunction with the TRF method will provide the most reliable phylogenetic information from microbial communities (4, 5, 14, 19, 24).
TABLE 2.
Division-level distribution of aligned RDP v8.0 sequences that have primer sites matching primer 8-27f
| Divisiona |
No. of sequencesb |
|---|---|
| Proteobacteria | 2,076 |
| Gram-positive bacteria | 1,832 |
| Flexibacter-Cytophaga-Bacteroides | 304 |
| Cyanobacteriac | 99 |
| Spirochetes | 75 |
| Planctomyces | 48 |
| Fibrobacter and Acidobacterium | 41 |
| Nitrospina subdivision | 37 |
| Green nonsulfur bacteria | 25 |
| Leptospirillum-Nitrospira | 22 |
| Thermophilic oxygen reducers | 19 |
| Fusobacteria | 19 |
| Green sulfur bacteria | 15 |
| Thermotogales | 11 |
| Anaerobaculum thermoterrenum group | 10 |
| Environmental clone WCHB1-31 group | 10 |
| Prosthecobacter group | 6 |
| Flexistipes sinusarabici assemblage | 6 |
| Coprothermobacter proteolyticus group | 1 |
| Environmental clone OPB45 | 1 |
Obtained from the RDP (18).
The RDP v8.0 contains 16,277 aligned sequences, of which 4,657 match primer 8-27f. The remaining sequences either are too short to contain the primer site or contain mismatches, with the primer exceeding the specified limits. Ten divisions are not represented.
The number of sequences reported does not include sequences from chloroplasts and cyanelles.
Reproducibility of TRF profiles.
In previous studies, either TRF profiles of different samples were not replicated or the data and analysis methods were not described in sufficient detail to permit evaluation of the reproducibility of TRF profiles (6, 16, 19, 20, 24). Thus, the general quality of TRF profiles as reliable fingerprints of microbial communities is unknown. We examined the reproducibility of replicate TRF profiles by comparing the total quantity of DNA represented by different profiles, the number of TRFs in each profile, and the height (abundance) of individual TRFs in each profile. The quantity of DNA represented by each of nine replicate RsaI TRF profiles was determined by calculating the sum of peak heights in each profile. Although the profiles were produced by using aliquots of a single RsaI digest of 16S rDNA, the total fluorescence (representational DNA quantity) measured from each TRF profile by summing the heights of individual peaks varied by a factor of 2 and ranged from 14,845 to 35,207 fluorescent units. Similar variation was observed with the sum of peak areas for each profile (data not shown), demonstrating that choice of variable (height versus area) was unimportant in assessing variation in total fluorescence between replicates. Even when extreme care is taken to handle DNA samples uniformly, variation in the total fluorescence between replicates can arise routinely from small pipetting errors when aliquots are withdrawn from a restriction digest and loaded on a gel. Variation in the relative abundance (fluorescence) of individual fragments can also be introduced during use of Genescan analysis software to track gel lanes and extract fragment abundance data (peak area and peak height data) from each lane. A similar degree of variation probably occurs in other DNA fingerprinting methods but has not been examined. Variation among replicate TRF profiles (in comparison to profiles from other methods) is especially apparent because of the uniquely high detection sensitivity, fragment resolution, precision in fragment sizing, and numerical data obtained with the TRF method.
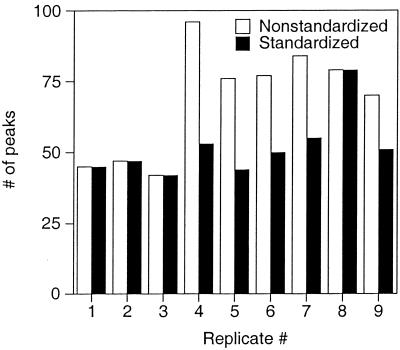
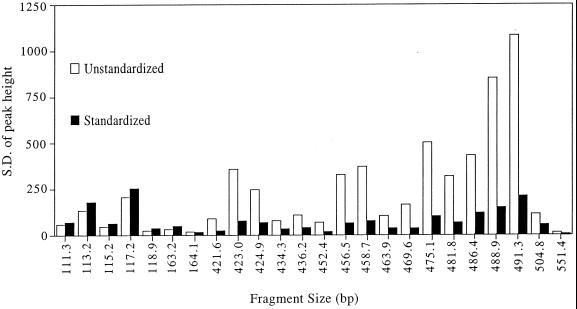
To reduce artifacts arising from variation in DNA quantity between replicate profiles, a procedure was developed to standardize DNA quantities after data collection. Effects of the standardization procedure on the number and average height of peaks in replicate profiles are shown in Fig. 1 and 2. Prior to standardization, the number of TRFs ranged from 42 to 96 (median = 76 [Fig. 1]), with an average variation in peak number (i.e., the average of pairwise comparisons with the smallest profile having only 42 TRFs) of 71%. Following standardization, the number of TRFs in the replicate profiles ranged from 42 to 79 (median = 50), with an average variation of 26%. Figure 2 shows the standard deviation (SD) of the average height of 24 separate TRFs consistently detected in nine replicate profiles. The average SD prior to standardization was 336 (range = 13 to 1,400), whereas after standardization it was 77 (range = 4 to 254). On average, the SD of each peak height was reduced from 24% of the mean peak height to 19% of the mean. For replicate profiles run on the same gel, standardization reduced the SD of each peak height from an average of 13% of the mean (a value similar to that reported by others [21]) to 7% of the mean. These data demonstrate that although the standardization procedure does not eliminate all of the variation between replicate profiles, it significantly reduces variation both in the number of TRFs observed in each profile and in the height of individual peaks among replicate profiles.
FIG. 1.
Effects of fluorescence standardization on variation in the number of TRFs observed in each of nine replicate RsaI TRF profiles of 16S rDNA amplified from a single soil DNA sample.
FIG. 2.
Effects of fluorescence standardization on variation in the height of TRFs observed in nine replicate RsaI TRF profiles of 16S rDNA amplified from a soil DNA sample. Each bar represents the SD of the mean height of a given peak.
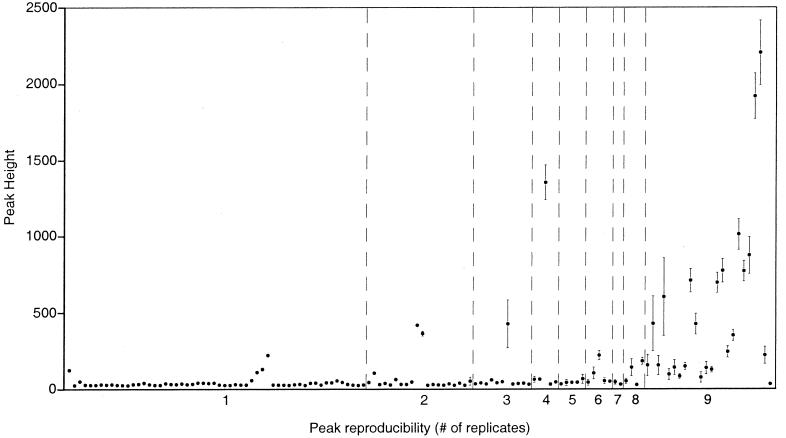
Reproducibility of the nine replicate profiles is illustrated in Fig. 3. Although a total of 169 distinct TRF sizes were observed among the nine profiles combined, only 24 TRF sizes were consistently detected in all profiles. Standardization of the DNA quantities represented by each profile reduced the total number of distinct TRFs from 169 to 132 but did not alter the number of TRFs that were consistently detected. This degree of noise was unexpected and arose mostly from the presence of small peaks, 90% of which had fluorescence values (after standardization) between 25 and 67 units. The 24 reproducible peaks had relatively high fluorescence values (median = 299) compared to the irreproducible peaks (median = 35), with all but one reproducible peak having a fluorescence value of 77 units or greater. The small, irreproducible peaks arise from unknown components of the DNA samples loaded on the sequencing gels, not from background noise from gel components. Blank gel lanes containing size standards but no sample DNA have maximum noise spikes of approximately 15 fluorescence units, which is significantly less than the height of the irreproducible peaks routinely observed in our sample profiles. Since the set of reproducible peaks in a profile cannot be clearly (or reliably) distinguished from the set of irreproducible peaks by a simple height threshold, the reproducible peaks must be identified by comparison of at least two or three replicate profiles.
FIG. 3.
Composite profile showing the average abundance of all the TRFs (169 in total) observed among nine replicate, standardized RsaI TRF profiles of 16S rDNA amplified from a soil DNA sample. The values along the x axis indicate the number of replicates in which each TRF was observed. Error bars are SDs of the mean height of each TRF.
Reproducibility of TRF profiles has been rigorously examined in one other published study. Osborn et al. (21) carefully and systematically examined the influence of many different experimental factors (including, for example, template concentration, PCR cycle number, and different brands of Taq polymerase) on the reproducibility of TRF profiles. In contrast to our results, the authors found that replicate profiles from the same sample DNA were almost identical, and only one or two irreproducible peaks were detected. The dramatic difference in reproducibility between studies arises largely from the use of different baseline noise thresholds (i.e., the value defining which fluorescence data collected by an ABI sequencer will be retained for user analysis and which fluorescence data will be discarded as noise). Osborn et al. (21) used a threshold value of 100 fluorescence units, whereas we used 25 fluorescence units. Applying a threshold of 100 to our data reduces the number of irreproducible peaks from 169 (prestandardization) to 13. However, the higher threshold also eliminated 4 of the 24 reproducible peaks. Although use of a high threshold value can eliminate much of the noise in TRF profiles, the threshold value required to eliminate a standard percentage of noise from a set of profiles cannot be predicted a priori and can vary from one sample to the next (data not shown). For these reasons, we prefer the use of the lowest possible threshold and comparison of replicate profiles to distinguish irreproducible data from reproducible data.
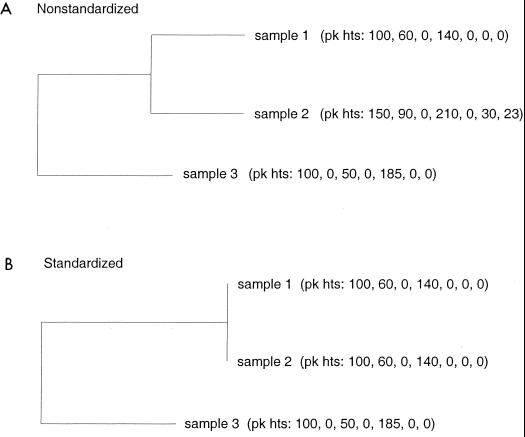
Identification of reproducible TRFs between replicates is required to obtain a reliable and representative TRF profile for a sample. After reliable profiles for a set of samples are obtained, standardization of sample quantities is necessary to enable comparisons that are based on samples of equal size. Even if care is taken to standardize DNA sample quantities prior to TRF analysis, the small errors that can accumulate during processing of samples for TRF analysis can result in different quantities of DNA being represented in TRF profiles. This problem is overcome by applying a procedure that can standardize sample quantities after data collection. Figure 4 shows the effects of standardization on three theoretical samples. When sample sizes are unequal, similarity relationships among samples can be severely distorted. Samples 1 and 2 in Fig. 4A have identical TRF profiles except that sample 2 has two additional TRFs because a larger quantity of DNA was examined. Standardization of sample size eliminates spurious comparisons and can reveal more accurate relationships among samples (Fig. 4B). Although standardization of sample size may not always alter the general topology of similarity dendrograms, it can routinely alter the branch lengths of such trees. Of course, standardization of sample sizes must be used sensibly. If one sample in a set of samples being compared is represented by an extremely low quantity of DNA compared to the other samples, standardization of sample quantities across the set may result in the loss of a large amount of data and distortion of sample relationships. Samples that are represented by large DNA quantities and that are different from one another may appear to be similar if much of the information in their profiles is lost during standardization to a low DNA quantity. In this circumstance, standardization of sample pairs during pairwise comparisons may be more appropriate than standardization of DNA quantities across an entire sample set.
FIG. 4.
Dendrograms showing the similarity of three theoretical samples before and after standardization of the sample profiles. The aligned TRF profile for each sample is shown in parentheses and lists the peak heights for seven theoretical TRFs.
The significance of the TRF method for microbial ecologists is indicated in part by the integration of TRF profile analysis programs into the RDP. With such centralized support, widespread use of the TRF method in microbial ecology studies is anticipated. The limitations and technical details of the method should be clearly understood in order to strengthen the method where possible and to wisely interpret data generated by the method. Toward this end, we identified limitations underlying the use of TRF profiles and have presented procedures that reduce the impact of these limitations. Application of the analytical procedures outlined in this study should strengthen the use of the TRF method for inferring the phylogenetic composition of environmental samples and for differentiation of microbial communities. The analytical techniques presented here are a necessary first step toward incorporating peak height as an additional parameter for profile comparisons. The data analysis procedures can also be applied to profiles created by other methods such as DGGE, rapid amplified polymorphic DNA, community restriction fragment polymorphism, or amplified fragment length polymorphism analysis.
ACKNOWLEDGMENTS
This research was supported by grants from the DOE Chemical and Biological Nonproliferation program, DOE Program for Ecosystem Research, and the Los Alamos National Laboratory.
REFERENCES
- 1.Avaniss-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques. 1994;17:144–149. [PubMed] [Google Scholar]
- 2.Barns S M, Takala S L, Kuske C R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol. 1999;65:1731–1737. doi: 10.1128/aem.65.4.1731-1737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce K D. Analysis of mer gene subclasses within bacterial communities in soils and sediments resolved by fluorescent-PCR-restriction fragment length polymorphism profiling. Appl Environ Microbiol. 1997;63:4914–4919. doi: 10.1128/aem.63.12.4914-4919.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin K J, Lukow T, Conrad R. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl Environ Microbiol. 1999;65:2341–2349. doi: 10.1128/aem.65.6.2341-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin K J, Lukow T, Stubner S, Conrad R. Structure and function of the methanogenic archaeal community in stable cellulose-degrading enrichment cultures at two different temperatures (15° and 30°C) FEMS Microbiol Ecol. 1999;30:313–326. doi: 10.1111/j.1574-6941.1999.tb00659.x. [DOI] [PubMed] [Google Scholar]
- 6.Clement B G, Kehl L E, DeBord K L, Kitts C L. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Methods. 1998;31:135–142. [Google Scholar]
- 7.Dunbar J, Ticknor L O, Kuske C R. Assessment of microbial diversity in two southwestern U.S. soils by terminal restriction fragment analysis. Appl Environ Microbiol. 2000;66:2943–2950. doi: 10.1128/aem.66.7.2943-2950.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunbar J, Takala S, Barns S M, Davis J A, Kuske C R. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microbiol. 1999;65:1662–1669. doi: 10.1128/aem.65.4.1662-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards U, Rogall T, Blocker H, Emde M, Bottger E C. Isolation and direct complete determination of entire genes. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godon J, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendricks D M. Arizona soils. Tucson, Ariz: University of Arizona Press; 1985. [Google Scholar]
- 12.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufman L, Rousseeuw P J. Finding groups in data: an introduction to cluster analysis. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 14.Knight V K, Kerkhof L J, Häggblom M M. Community analyses of sulfidogenic 2-bromophenol dehalogenating and phenol-degrading microbial consortia. FEMS Microbiol Ecol. 1999;29:137–147. [Google Scholar]
- 15.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maidak B L, Cole J R, Parker C T, Garrity G M, Larsen N, Bing L, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh T L. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr Opin Microbiol. 1999;2:323–327. doi: 10.1016/S1369-5274(99)80056-3. [DOI] [PubMed] [Google Scholar]
- 20.Moeseneder M M, Arrieta J M, Muyzer G, Winter C, Herndl G J. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:3518–3525. doi: 10.1128/aem.65.8.3518-3525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osborn A M, Moore E R B, Timmis K N. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- 22.Overbeek R. Scan for matches. Argonne, Ill: Argonne National Laboratory; 1996. [Google Scholar]
- 23.Tiedje J M, Asuming-Brempong S, Nusslein K, Marsh T L, Flynn S J. Opening the black box of soil microbial diversity. Appl Soil Ecol. 1999;13:109–122. [Google Scholar]
- 24.van der Maarel M J, Artz R R, Haanstra R, Forney L J. Association of marine Archaea with the digestive tracts of two marine fish species. Appl Environ Microbiol. 1998;64:2894–2898. doi: 10.1128/aem.64.8.2894-2898.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson K H, Blitchington R B, Green R C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]