Abstract
Neural stemness is suggested to be the ground state of tumorigenicity and pluripotent differentiation potential. However, the relationship between these cell properties is unclear. Here, by disrupting the neural regulatory network in neural stem and cancer cells and by serial transplantation of cancer cells, we show that tumorigenicity and pluripotent differentiation potential are coupled cell properties unified by neural stemness. We show that loss of neural stemness via inhibition of SETDB1, an oncoprotein with enriched expression in embryonic neural cells during vertebrate embryogenesis, led to neuronal differentiation with reduced tumorigenicity and pluripotent differentiation potential in neural stem and cancer cells, whereas enhancement of neural stemness by SETDB1 overexpression caused the opposite effects. SETDB1 maintains a regulatory network comprising proteins involved in developmental programs and basic cellular functional machineries, including epigenetic modifications (EZH2), ribosome biogenesis (RPS3), translation initiation (EIF4G), and spliceosome assembly (SF3B1); all of these proteins are enriched in embryonic neural cells and play active roles in cancers. In addition, SETDB1 represses the transcription of genes promoting differentiation and cell cycle and growth arrest. Serial transplantation of cancer cells showed that neural stemness, tumorigenicity, and pluripotent differentiation potential were simultaneously enhanced; these effects were accompanied by increased expression of proteins involved in developmental programs and basic machineries, including SETDB1 and the abovementioned proteins, as well as by increased alternative splicing events. These results indicate that basic machineries work together to define a highly proliferative state with pluripotent differentiation potential and also suggest that neural stemness unifies tumorigenicity and differentiation potential.
Keywords: neural stemness, cancer cell, tumorigenicity, alternative splicing, cell differentiation, pluripotency, ribosome, translation initiation factor, SETDB1, serial transplantation
Abbreviations: % FP, percent false positive; ChIP, chromatin immunoprecipitation; ESCs, embryonic stem cells; FBS, fetal bovine serum; IF, immunofluorescence; mESCs, mouse embryonic stem cells; MT, Myc-tag; NSCs, neural stem cells; primNSCs, primitive NSCs
Neural stemness has usually been considered to be a type of tissue stemness. However, it features some unique properties that are not displayed by other types of tissue stem cells. Neural stem cells (NSCs) exhibit tumorigenicity, because similar to cancer cells, NSCs are capable of tumor formation when transplanted into immunodeficient mice (1, 2). NSCs have pluripotent differentiation potential, which means that, in addition to the capability of differentiation into cell types of the nervous system, they can also be induced to differentiate into different nonneural cell types (1, 2, 3, 4). Although embryonic stem cells (ESCs) have both tumorigenic and pluripotent differentiation potential, their intrinsic property is neural stemness because their default fate is primitive NSCs (primNSCs), according to the “neural default model” of pluripotent embryonic cells (1, 4, 5, 6, 7). In somatic cells, blocking endogenous factors also leads to gain of neural stemness and tumorigenicity at the expense of the original cell identity (2, 8, 9, 10). The association among neural stemness, tumorigenicity, and pluripotent differentiation potential is supported by the fact that most genes promoting tumorigenesis and pluripotency are neural stemness genes or genes with localized or at least with enriched expression in embryonic neural cells (1, 11). The unique significance of neural stemness should be derived from the evolutionary advantage of neural genes and the neural-biased state in the last common unicellular ancestor of metazoans (1, 2, 12). We previously suggested that neural stemness represents the ground state of tumorigenicity and pluripotent differentiation potential and that tumorigenesis is a process of gradual loss of the original cell identity and restoration of the neural ground state in somatic cells (1, 13).
The neural ground state is also reflected by the observation that genes involved in basic physiological functions in cells, such as the cell cycle, ribosome biogenesis, proteasome or spliceosome assembly, and protein translation, are mostly enriched in neural cells during vertebrate embryogenesis. The genes involved in the basic machineries usually had a unicellular origin during evolution (1, 2). The cell cycle is tightly linked to cell fate decisions. Pluripotent stem cells and NSCs have a shorter cell cycle, and differentiated cells have a longer cell cycle (14), in agreement with the differential expression of cell cycle genes in neural and nonneural cells during embryogenesis (1). Cells with a short cell cycle need high protein production and homeostasis. Accordingly, genes encoding ribosomal and proteasomal proteins show dominant expression in embryonic neural cells during embryogenesis (1), suggesting the high activity of protein synthesis and turnover in embryonic neural cells. Recently, we showed that this is concertedly regulated and required for the maintenance of neural stemness in both NSCs and cancer cells (15). The spliceosome is responsible for alternative splicing, a mechanism contributing to phenotypic novelty during evolution and to cell differentiation, lineage determination, and tissue or organ formation during embryogenesis (16, 17, 18). The enriched expression of genes involved in spliceosome assembly and hence alternative splicing is consistent with the enrichment of long genes comprising more exons/introns in both embryonic neural cells and cancer cells (1, 2, 19). Moreover, genes involved in developmental programs, such as those involved in epigenetic modification, pluripotency, or promotion of pluripotency, are also neural stemness genes or genes with specific or enriched expression in embryonic neural cells (1 and references therein). These genes work concertedly together to define a cellular state with high proliferation and pluripotent differentiation potential, both of which are features of tumorigenic cells. SETDB1 is an epigenetic modification factor responsible for trimethylation of lysine 9 of histone H3 (H3K9me3) and mediates transcriptional repression. SETDB1 is required for early embryogenesis, because gene knockout in mice causes embryonic lethality before E7.5 (20), suggesting that it is critical during embryogenesis. Its gene is primarily transcribed in neural precursor cells during vertebrate embryogenesis, and brain-specific gene ablation causes severe brain developmental defects (11, 21). Accordingly, SETDB1 is upregulated in most cancers and promotes their initiation and progression (22, 23). Serial transplantation (or in vivo passaging) of cancer cells in immunodeficient mice is an approach that mimics cancer progression (24). In the present study, by disruption of neural regulatory network via alteration of SETDB1/Setdb1 activity in cancer cells and NSCs and serial transplantation of cancer cells, we show that neural stemness is a cell property unifying tumorigenicity and pluripotent differentiation potential.
Results
Setdb1 is required for the maintenance of neural stemness and differentiation potential of NSCs
When cultured in defined NSC-specific serum-free medium, mouse ESCs change into primNSCs (7, 25), which form free-floating neurospheres in the medium (Fig. 1A). Treatment of the neurospheres with retinoic acid (RA), a reagent that induces neuronal differentiation in NSCs, induced a neuronal differentiation phenotype (Fig. 1A). RA treatment led to a decrease in the expression of the neural stemness markers Sox1 and Msi1 in differentiated cells. In addition, Myc, Hes1, Fgfr1, Vim, and Cdh2, which are expressed specifically or dominantly in embryonic neural cells during vertebrate embryogenesis and promote or are upregulated during tumorigenesis, exhibited reduced expression in response to RA treatment (Fig. 1B). In contrast, the neuronal markers Map2 and Tubb3 were upregulated, supporting the neuronal differentiation effect (Fig. 1B). We observed decreased expression of the epigenetic modification factors Setdb1 and Ezh2 in cells treated with RA (Fig. 1B). Ezh2 mediates trimethylation of lysine 27 of histone H3 (H3K27me3) and hence transcriptional repression and is required for early embryogenesis, maintaining or promoting neural stemness (26, 27, 28). Intriguingly, the expression levels of proteins involved in translation initiation (Eif4g), ribosome biogenesis (Rps3 and Rpl26), and alternative splicing (Srsf3 and Sf3b1) were simultaneously reduced (Fig. 1B). This result suggests that loss of neural stemness leads to simultaneously decreases in the levels of proteins composing machineries for basic cellular physiological functions and developmental programs.
Figure 1.
Neuronal differentiation of primNSCs induced by RA treatment and by knockdown of Setdb1.A and B, neuronal differentiation phenotype in ESC-derived primNSCs that were treated with RA at an indicated dose and cultured in NSC-specific serum-free medium for a period as indicated (A), and IB detection of protein expression in control (DMSO) and treated (RA) cells (B). C and D, neuronal differentiation phenotype in ESC-derived primNSCs induced by knockdown of Setdb1 and cultured in serum-free medium (C), and IF detection of protein expression in control (shCtrl) and knockdown (shSetdb1) cells (D). E–G, neuronal differentiation phenotype in NE-4C cells induced by knockdown of Setdb1 and cultured in serum-free medium (E), and IF (F) and IB (G) detection of protein expression in control (shCtrl) and knockdown (shSetdb1) cells. H and I, enrichment analysis on GO and KEGG pathway terms for upregulated (H) and downregulated (I) genes identified in a transcriptome profiling on control and knockdown cells in (E). In (D) and (F), cell nuclei were counterstained with DAPI. In (F), Map2 and Tubb3 were simultaneously detected with cells on the same coverslips, either control (shCtrl) or knockdown cells (shSetdb1). ESCs, embryonic stem cells; IF, immunofluorescence; IB, immunoblotting; NSCs, neural stem cells; primNSCs, primitive NSCs; RA, retinoic acid.
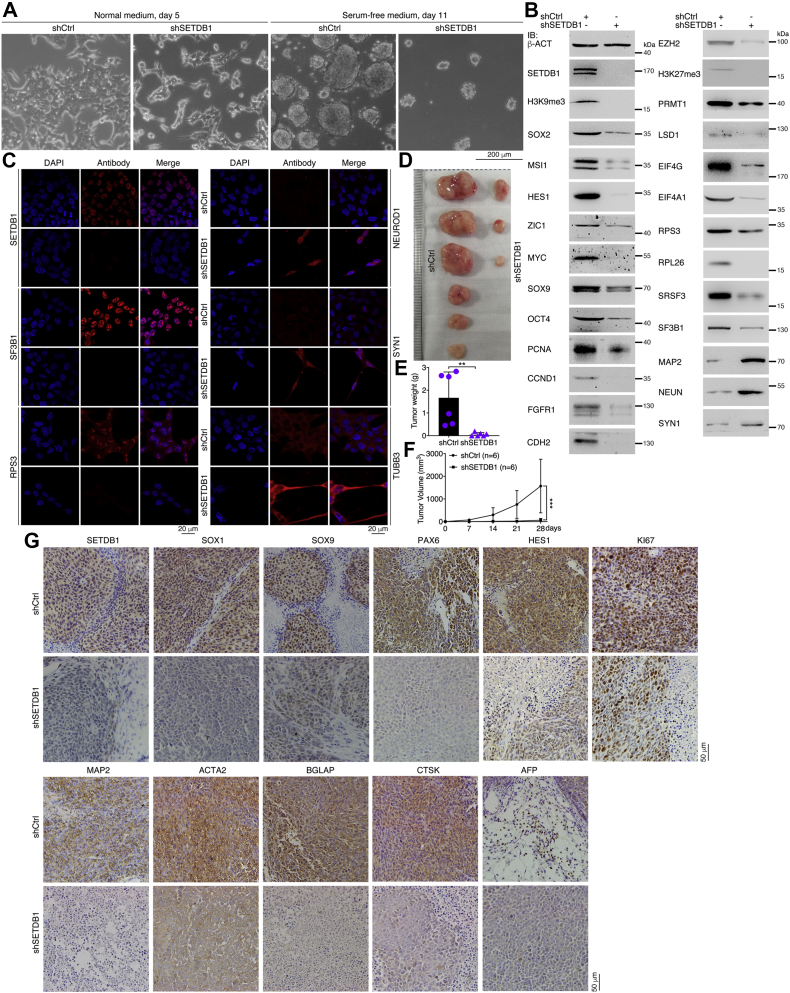
Neuronal differentiation is accompanied by downregulation of Setdb1, and we next examined how Setdb1 functions in NSCs. Control primNSCs infected with lentivirus generated from empty vector (shCtrl) formed neurospheres in serum-free medium, and knockdown of Setdb1 with a validated shRNA (shSetdb1) caused failure of neurosphere formation. The resulting cells assumed a neuronal differentiation phenotype (Fig. 1C). The shRNA efficiently blocked Setdb1 expression in cells. Correspondingly, immunofluorescence (IF) staining showed that Sox1 and Msi1 were repressed but Map2 and Tubb3 were stimulated in response to Setdb1 knockdown (Fig. 1D), suggesting an essential role of Setdb1 in maintaining stemness in primNSCs.
We next explored whether this finding holds true for different NSCs. NE-4C cells, an NSC line derived from cerebral vesicles of mouse E9 embryos, formed neurospheres in serum-free medium. When Setdb1 was knocked down, the cells exhibited a neuronal differentiation effect (Fig. 1E). IF staining revealed that Sox1 and Msi1 were repressed when Setdb1 was lost, whereas the expression of the neuronal proteins Map2, Tubb3, and Nefl was strongly induced (Fig. 1F). Detection of the expression of more proteins demonstrated that in addition to Sox1 and Msi1, neural stemness proteins and proteins with gene expression localized to or enriched in embryonic neural cells—that is, Hes1, Zic1, Cdh2, Fgfr1, and Pcna—were also downregulated (Fig. 1G). In contrast, Neun and Map2 were upregulated (Fig. 1G). Setdb1 knockdown caused decreased expression of the epigenetic factors Ezh2, Prmt1, and Lsd1 (Fig. 1G), which maintain neural stemness in NSCs and cancer cells or confer neural stemness on differentiated cells (15, 27, 28, 29, 30). Moreover, decreased expression of proteins involved in translation initiation (Eif4g and Eif4a1), ribosome biogenesis (Rps3 and Rpl26), and alternative splicing (Srsf3 and Sf3b1) was observed (Fig. 1G).
Transcriptome analysis showed that Setdb1 knockdown caused upregulation of 2619 genes and downregulation of 2457 genes in NE-4C cells (Table S1). The upregulated genes were primarily involved in events associated with neuronal differentiation (Fig. 1H). Notably, among the upregulated genes were the Hox genes, which are critical for patterning of the anterior-posterior body plan and specification of segment identity of tissues, including neural tissue, during embryogenesis. Some genes that are involved in cell cycle or growth arrest (Bcl6, Cdkn1a, and Eif2ak3), DNA repair, and apoptosis (Gadd45a, Gadd45b, and Gadd45g) were also upregulated (Table S1). The downregulated genes were associated with RNA and amino acid metabolism (Fig. 1I). This change might be related to the decrease in the expression of proteins involved in alternative splicing, ribosome biogenesis, and protein translation in knockdown cells. Among the downregulated genes were Aspm, Cenpk, Enc1, Lin28a, Mcm8, Nucks1, Pou5f1, and Utf1 (Table S1), which regulate pluripotency and neural stemness, transcription, or cell cycle. Taken together, these results indicate that inhibition of Setdb1 in NSCs causes neuronal differentiation and corresponding changes in regulatory networks.
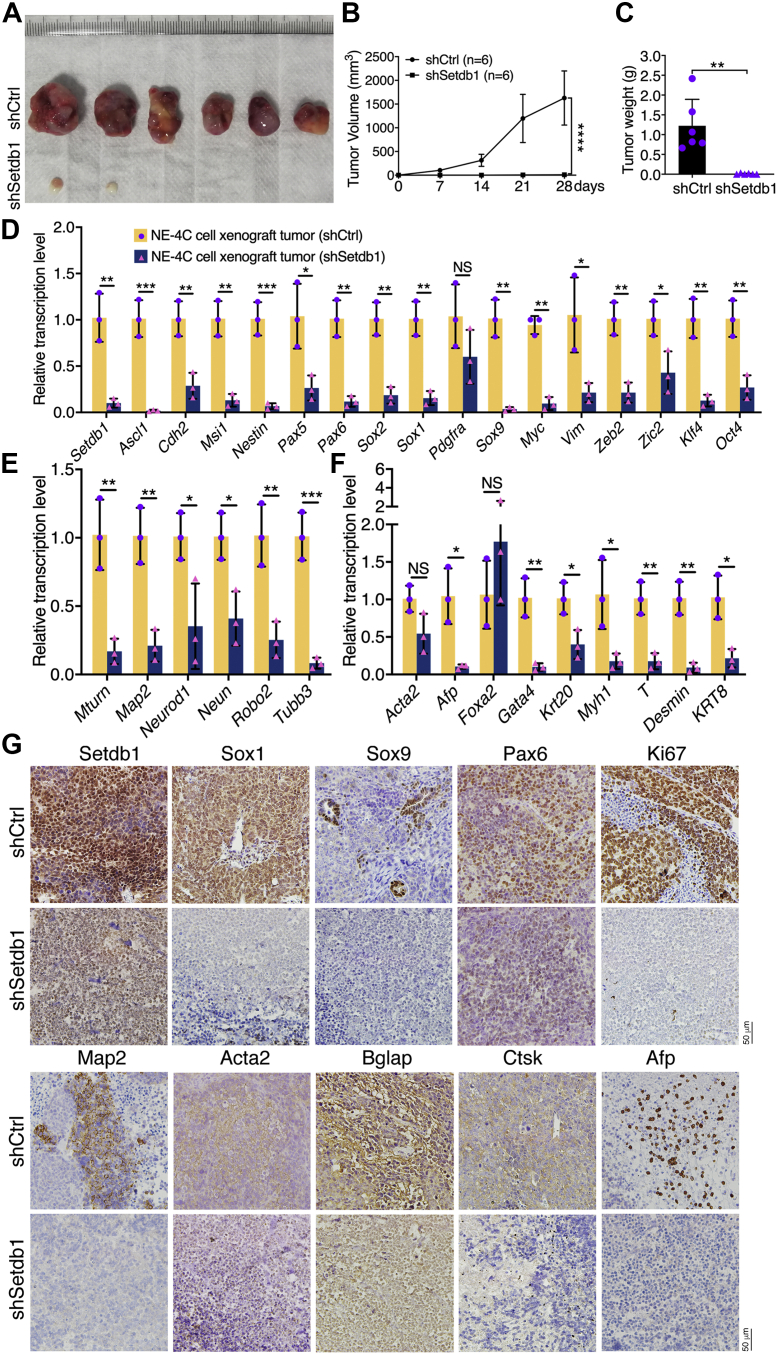
Subsequently, we observed that Setdb1 knockdown strongly inhibited cell invasion and migration (Fig. S1A) and reduced the colony-formation ability (Fig. S1, B and C). NSCs, such as NE-4C cells, are capable of forming xenograft tumors that contain cell types of all three germ layers (2). As reported, NE-4C cells formed xenograft tumors in all six immunodeficient nude mice, whereas knockdown cells formed smaller tumors in only two of the six mice (Fig. 2, A–C and Table S2), indicating the lower tumorigenicity of knockdown cells. The expression of neural stemness genes or genes with enriched expression in embryonic neural cells during vertebrate embryogenesis was much lower in xenograft tumors formed by knockdown cells than in those formed by control cells (Fig. 2D). The same trends in differential expression were observed for genes representing neuronal differentiation (Fig. 2E) and genes representing mesodermal or endodermal tissue differentiation (Fig. 2F). Immunohistochemical staining showed that the expression level of Setdb1 was lower in sections of the tumors formed by knockdown cells than in tumors formed by control cells (Fig. 2G), further indicating the knockdown effect of Setdb1. Accordingly, the neural stemness markers Sox1, Sox9, and Pax6 and the proliferation marker Ki67 showed much weaker expression in tumors formed by knockdown cells (Fig. 2G). In tumors formed by control cells, there was significant neuronal differentiation, as shown by Map2 expression; however, Map2 expression was barely detectable in tumors formed by knockdown cells (Fig. 2G). Similarly, the mesodermal and endodermal tissue differentiation markers Acta2, Bglap, Ctsk, and Afp were detected strongly in tumors formed by control cells but were detected weakly or not detected in tumors formed by knockdown cells (Fig. 2G). These results demonstrate that loss of neural stemness in NE-4C cells via Setdb1 knockdown is accompanied by reduced tumorigenicity and pluripotent differentiation potential.
Figure 2.
Difference in xenograft tumor formation and differentiation potential between control and knockdown NE-4C cells.A–C, tumor formation of control (shCtrl) and knockdown (shSetdb1) cells in each six injected mice (A) and difference in tumor volume (B) and weight (C) between the two groups. Significance of difference in tumor volume (B) between two groups of mice was calculated using two-way ANOVA-Bonferroni/Dunn test. Significance of difference in tumor weight (C) was calculated using unpaired Student’s t test. Data are shown as mean ± SD. ∗∗p < 0.01, ∗∗∗∗p < 0.0001. D–F, RT-qPCR detection of differential expression of genes representing neural stemness or enriched in embryonic neural cells (D), neuronal differentiation (E), and mesodermal and endodermal tissue differentiation (F) between tumors of control NE-4C cells and of knockdown cells. Significance in transcription change was calculated based on experiments in triplicate using unpaired Student’s t test. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NS, not significant. G, IHC detection of cell/tissue markers in sections of tumors derived from control and knockdown cells. Objective magnification: 20 × . IHC, immunohistochemical; RT-qPCR, Reverse transcriptase-quantitative PCR.
Loss of neural stemness via SETDB1 knockdown in cancer cells leads to reduced tumorigenicity and pluripotent differentiation potential
We then investigated whether SETDB1 loss also causes a similar effect in cancer cells. The colorectal cancer cell line HCT116 exhibits neural stemness because it expresses neural stemness markers, forms neurosphere-like structures in NSC-specific serum-free medium, and has pluripotent differentiation potential (2). In normal culture medium, knockdown of SETDB1 using a validated shRNA (shSETDB1) led to a significant phenotypic change in HCT116 cells. The cells grew long neurite-like processes, suggesting an effect of neuronal differentiation (Fig. 3A). When cultured in serum-free medium, control cells formed free-floating spherical structures, similar to NSCs. However, knockdown cells could not form such structures efficiently (Fig. 3A), an indication of reduced neural stemness. Immunoblotting showed efficient SETDB1 knockdown in cells, which was accompanied by a reduction in the level of H3K9me3. The cells expressed a series of neural stemness markers and proteins that are primarily expressed in embryonic neural cells during embryogenesis, including SOX2, MSI1, HES1, ZIC1, MYC, SOX9, OCT4, PCNA, CCND1, FGFR1, CDH2, and the epigenetic factors EZH2, PRMT1, and LSD1, which promote cancers or are upregulated in cancer cells. All were repressed in knockdown cells (Fig. 3B). Reduced expression of CCND1 and PCNA indicates reduced cell proliferation. Additionally, EIF4G, EIF4A1, RPS3, RPL26, SRSF3, and SF3B1 were downregulated (Fig. 3B). The upregulated proteins were MAP2, NEUN, and SYN1 (Fig. 3B). In addition, IF staining detected reduced expression of SF3B1 and RPS3 but increased expression of the neuronal proteins NEUROD1, SYN1, and TUBB3 in knockdown cells (Fig. 3C). Transcriptome profiling revealed that blocking SETDB1 in HCT116 cells downregulated the transcription of genes that were significantly linked with biological processes of DNA replication and cell cycle, as well as the corresponding cellular components, cellular functions, and pathways (Fig. S2A and Table S3). The upregulated genes were associated with endoplasmic reticulum to Golgi vesicle-mediated transport and processes of unfolded and topologically incorrect proteins (Fig. S2B and Table S3). The upregulated genes in NE-4C knockdown cells, such as HOX genes (HOXA1, HOXB9), BCL6, CDKN1A, EIF2AK3, GADD45A, GADD45B, and GADD45G, were also upregulated in HCT116 knockdown cells, and the downregulated genes ASPM, CENPK, ENC1, MCM8, and NUCKS1 were downregulated in HCT116 knockdown cells (Table S3). Therefore, blocking SETDB1 in HCT116 cells caused concerted downregulation of basic cellular functional machineries (e.g., ribosome biogenesis, spliceosome assembly, translation), proteins involved in developmental programs, gene regulatory networks of cell proliferation, and the phenotype of neuron-like differentiation.
Figure 3.
Influence of reduced neural stemness via SETDB1 knockdown on the property of HCT116 cells.A, phenotypic change in HCT116 cells after SETDB1 knockdown and cultured in normal and serum-free medium in a period as indicated. B and C, IB (B) and IF (C) detection of protein expression in control and knockdown cells. In (C), cell nuclei were counterstained with DAPI. D–F, tumor formation of control and knockdown cells in each six injected mice (D) and difference in tumor weight (E) and volume (F) between the two groups. Significance of difference in tumor weight (E) was calculated using unpaired Student’s t test. Significance of difference in tumor volume (F) was calculated using two-way ANOVA-Bonferroni/Dunn test. Data are shown as mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001. G, IHC detection of cell/tissue markers in sections of tumors derived from control and knockdown cells. Objective magnification: 20 × . IF, immunofluorescence; IHC, immunohistochemical; IB, immunoblotting.
Accordingly, the invasion, migration, and colony formation capabilities were compromised in HCT116 knockdown cells (Fig. S2, C–E). These cells showed a much weaker xenograft tumor-formation ability (Fig. 3, D–F and Table S2). Tumors formed by control cells contained cells with high expression of SOX1, SOX9, PAX6, and HES1 and the proliferation marker KI67. However, in tumors formed by knockdown cells, these markers exhibited decreased expression (Fig. 3G), suggesting that these tumors contained cells with weaker neural stemness and proliferation ability. Control tumors had cells showing high expression of MAP2, ACTA2, BGLAP, or CTSK, indicating neuronal differentiation and mesodermal differentiation. Detection of AFP in scattered cells indicates endodermal tissue differentiation. Tumors formed by knockdown cells exhibited lower expression of these markers (Fig. 3G). In summary, knockdown of SETDB1 in HCT116 cells led to reduced neural stemness and consequently compromised tumorigenicity and differentiation potential, similar to the effect observed in NE-4C cells.
Enhanced neural stemness by SETDB1 overexpression is accompanied by increased tumorigenicity and pluripotent differentiation potential
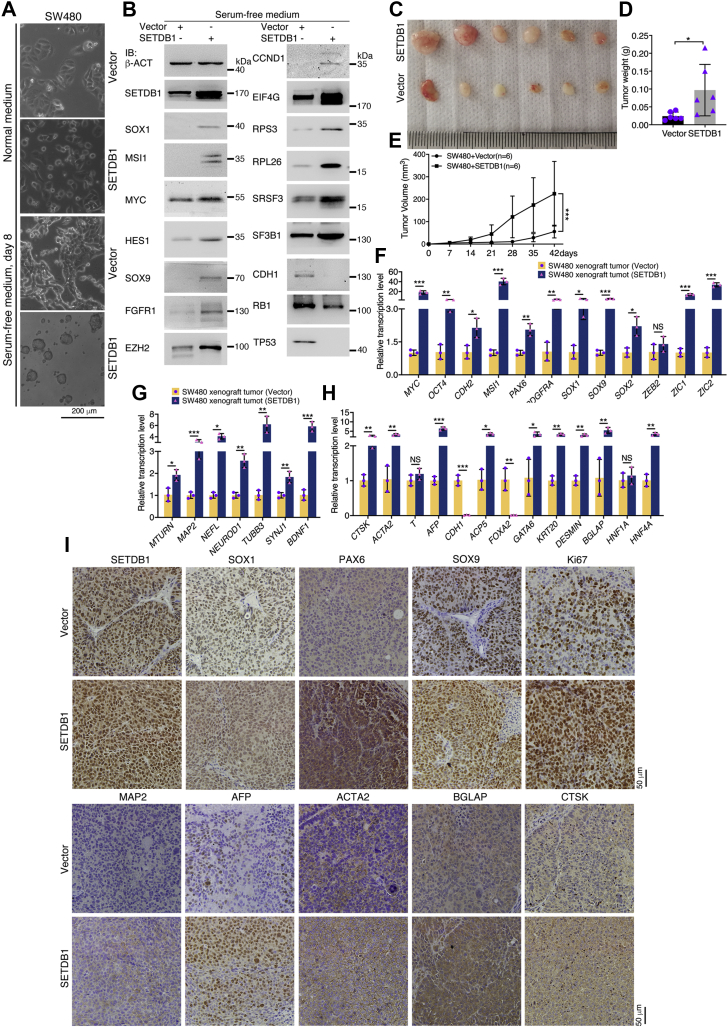
The colorectal cancer cell line SW480 has weaker tumorigenicity than cell lines such as HCT116. Correspondingly, SW480 cells could not form spherical structures in serum-free medium, but HCT116 cells formed neurospheres within the same duration of culture (2) (Fig. S3). We investigated how SETDB1 overexpression can affect the properties of SW480 cells. Overexpression of SETDB1 caused only a small change in cell morphology in normal culture medium. In serum-free medium, control cells formed clusters attached to the bottom of the culture dish. However, cells with SETDB1 overexpression formed free-floating spherical structures (Fig. 4A), similar to NSCs and HCT116 cells. A series of neural stemness markers and proteins involved in maintaining neural stemness—that is, SOX1, MSI1, MYC, HES1, SOX9, FGFR1, and EZH2—were upregulated in SETDB1-overexpressing cells (Fig. 4B). IF also detected enhanced expression of SOX1 and MSI1 in SETDB1-overexpressing cells (Fig. S4A). The increased CCND1 expression indicated that overexpressed SETDB1 promoted cell cycle. In contrast to SETDB1 knockdown, SETDB1 overexpression enhanced the expression of EIF4G, RPS3, RPL26, SRSF3, and SF3B1 (Fig. 4B). This again indicates that basic cellular functional machineries should be concertedly regulated to correspond to the proliferation rate of cells, as reported previously (1, 15). Repressed expression was observed for the epithelial marker CDH1 and for the tumor suppressors RB1 and TP53 (Fig. 4B), which function primarily in cell cycle arrest, maintenance of genomic integrity, and cell differentiation.
Figure 4.
Influence of enhanced neural stemness via SETDB1 overexpression on the property of SW480 cells.A, comparison of phenotypic change between control (Vector) SW480 cells and cells with SETDB1 overexpression (SETDB1), which were cultured in normal and serum-free medium, respectively, in a period as indicated. B, protein expression change between control and overexpression cells, as detected with IB. C–E, tumor formation of control and overexpression cells in each six injected mice (C) and difference in tumor weight (D) and volume (E) between the two groups. Significance of difference in tumor weight (D) was calculated using unpaired Student’s t test. Significance of difference in tumor volume (E) was calculated using two-way ANOVA-Bonferroni/Dunn test. Data are shown as mean ± SD. ∗p < 0.05, ∗∗∗p < 0.001. F–H, difference in transcription of genes representing neural stemness (F), neuronal differentiation (G), and mesodermal and endodermal tissue differentiation (H) between tumors of control and of overexpression cells, as detected with RT-qPCR. Significance in transcription change was calculated based on experiments in triplicate using unpaired Student’s t test. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NS, not significant. I, IHC detection of cell/tissue markers in sections of tumors derived from control and overexpression cells. Objective magnification: 20 × . IB, immunoblotting; IHC, immunohistochemical; RT-qPCR, Reverse transcriptase-quantitative PCR.
Enhancement of neural stemness in SW480 cells due to SETDB1 overexpression would mean that their tumorigenicity and differentiation potential would increase. Indeed, we observed increased capabilities of invasion, migration, and colony formation (Fig. S4, B–D) in cells overexpressing SETDB1. These cells displayed a greater ability to form xenograft tumors than control cells (Fig. 4, C–E and Table S2). Evaluation of gene transcription demonstrated that a series of genes representing neural stemness and/or involved in the maintenance of neural stemness were upregulated in xenograft tumors formed by SETDB1-overexpressing cells (Fig. 4F). Increased transcription of genes representing neuronal differentiation (Fig. 4G) and genes representing mesodermal and endodermal cell differentiation (Fig. 4H) was also detected. Immunohistochemical staining revealed stronger staining for SETDB1 in sections of tumors formed by SETDB1-overexpressing cells than in sections from control tumors (Fig. 4I). Stronger signals were also observed for SOX1, PAX6, SOX9, and KI67 (Fig. 4I), indicating that tumors formed by SETDB1-overexpressing cells contained cells with enhanced neural stemness and proliferation. Moreover, MAP2 was barely detected in tumors formed by control cells but was detected in tumors formed by SETDB1-overexpressing cells. AFP, ACTA2, BGLAP, and CTSK were more strongly detected in sections of tumors formed by SETDB1-overexpressing cells (Fig. 4I). Taken together, these results suggest that cells with enhanced neural stemness have a stronger capability to form tumors, which contain cells with enhanced neural stemness and proliferation and cells characteristic of neuronal, mesodermal, and endodermal differentiation.
SETDB1 sustains a regulatory network maintaining neural stemness
Inhibition of SETDB1 results in a decrease in the expression of proteins involved in the functions of basic cellular functional machineries and developmental programs. This result allowed us to investigate whether SETDB1 might regulate these proteins or their genes. Mass spectrometry identified 222 putative Setdb1 interaction partners in NE-4C cells (Table S4). These interaction partners were mostly involved in ribosome biogenesis and spliceosome assembly, which are responsible for the biological processes of translation, mRNA processing, and RNA splicing. Correspondingly, these proteins were mainly associated with molecular functions and cellular components that are involved in related pathways, biological processes, and cellular components, such as RNA binding, ribosome constituents, intracellular ribonucleoprotein complexes, the cytoplasm, and the nucleus. (Fig. S5 and Table S4). SETDB1 was found to interact with EZH2, SF3B1, RPS3, RPL26, and EIF4G in HCT116 cells. SETDB1 also interacted with the proteasome component proteins PSMD2 and ADRM1, and USP11 (Fig. 5A), a deubiquitinase that is expressed primarily in the embryonic nervous system, regulates neurogenesis and neuronal migration and, accordingly, is upregulated in cancer cells and promotes cancer (31, 32, 33).
Figure 5.
SETDB1 maintains protein stability via interaction with USP11.A, CoIP detection of binding of SETDB1 to other proteins. B, the effect of protein degradation in response to inhibition of de novo protein synthesis via CHX treatment in a time series and SETDB1 knockdown in HCT116 cells. C, detection of the dependence of reduced protein expression caused by SETDB1 knockdown on proteasomal activity via treatment of cells with MG132. D, effect of SETDB1 knockdown on ubiquitination of endogenous SF3B1, EIF4G1, EZH2, and overexpressed RPS3, which were precipitated with their respective antibodies or the Myc-tag (MT) antibody. E, CoIP detection of binding of overexpressed HA-USP11 to other proteins. F, effect of USP11 knockdown on proteins expression in HCT116 cells. G, effect of USP11 knockdown on enhanced protein expression caused by overexpressed SETDB1. H, effect of SETDB1 knockdown on enhanced protein expression caused by overexpressed USP11. I, effect of USP11 knockdown on ubiquitination of endogenous EIF4G1, SF3B1, EZH2, and overexpressed RPS3, which were precipitated with their respective antibodies or the MT antibody. J, differential binding of N- and C-terminal region of SETDB1 to proteins. K, differential effect of enforced expression of N- and C-terminal region of SETDB1, respectively, on protein expression. CHX, cycloheximide; CoIP, coimmunoprecipitation.
A time-course assay revealed that EIF4G, RPS3, SF3B1, and EZH2 degraded faster in HCT116 cells with SETDB1 knockdown than in control cells (Fig. 5B). Since knockdown of SETDB1 did not cause a significant change in the transcription of the genes encoding these proteins (Fig. S6), we deduced that SETDB1 might regulate the stability of these proteins. Blocking SETDB1 reduced EIF4G, RPS3, SRSF3, SF3B1, and EZH2 protein levels, while inhibiting proteasome activity with MG132 increased the levels of these proteins. Inhibition of proteasome activity was able to reverse the reductions in these proteins (Fig. 5C), suggesting that SETDB1-mediated expression of these proteins depends on the ubiquitin–proteasome system. Indeed, SETDB1 knockdown caused an increase in the ubiquitination of SF3B1, EIF4G1, and EZH2, which were precipitated with their corresponding antibodies, as well as the overexpressed RPS3 protein, which was precipitated with an anti-Myc-tag (MT) antibody (Fig. 5D). The ubiquitination status might be due to the interaction between SETDB1 and USP11. Coimmunoprecipitation analysis showed that USP11 was able to interact with SETDB1, EIF4G, RPS3, SF3B1, and EZH2 (Fig. 5E), indicating that USP11 and SETDB1 form complexes with other proteins. Similar to the effect of blocking SETDB1, knockdown of USP11 using a validated shRNA (shUSP11) in HCT116 cells led to decreased levels of EIF4G, EIF4A1, RPS3, SRSF3, SF3B1, EZH2, MYC, SOX9, MSI1, and FGFR1 and increased levels of MAP2 and SYN1 (Fig. 5F). Although overexpression of SETDB1 enhanced protein expression, this effect was abolished in the absence of USP11 (Fig. 5G). In contrast, overexpression of USP11 caused enhanced expression of proteins and was able to reverse the reductions in expression caused by SETDB1 knockdown (Fig. 5H). Therefore, the maintenance of protein expression by SETDB1 depends on USP11. Indeed, we found enhanced ubiquitination of EIF4G1, SF3B1, EZH2, and RPS3 when USP11 was blocked (Fig. 5I). We made two truncation mutants that contained either the N-terminal region (aa 1–725) or C-terminal region (aa 726–1291) of SETDB1 and were fused to the HA-tag, designated HA-Nterm and HA-Cterm, respectively. Forced expression of HA-Nterm revealed interactions with EIF4G, USP11, RPS3, EZH2, and SF3B1, whereas HA-Cterm did not exhibit these interactions (Fig. 5J). In agreement with this result, forced expression of HA-Nterm enhanced the expression of EIF4G, EIF4A1, RPS3, SRSF3, SF3B1, and EZH2, but forced expression of HA-Cterm did not exhibit this effect (Fig. 5K). Since the pre-SET, SET, and post-SET domains in the C-terminal region of SETDB1 are required for its methyltransferase activity, the differential effect of the N- and C-terminal regions on protein interaction and expression suggests that methyltransferase activity might not be essential for promoting protein stability.
SETDB1 regulates the expression of EZH2, and both mediate transcriptional silencing. SETDB1 knockdown caused the activation of genes promoting neuronal differentiation and inhibiting cell cycle and proliferation (Fig. 6A). Accordingly, SETDB1 knockdown caused decreased binding of EZH2 to gene promoters and, correspondingly, decreased the levels of H3K9me3 and H3K27me3 in the promoters of genes that were upregulated in NE-4C and/or HCT116 cells, for example, CDKN1A, GADD45B, BCL6, NEUROD1, and TUBB3 (Fig. 6B). Among the genes, CDKN1A and GADD45B encode proteins that are involved in the inhibition of cell cycle and proliferation and regulate neuronal differentiation (34, 35, 36). NEUROD1 and BCL6 encode proneurogenic factors that are essential for neurogenesis (37, 38). TUBB3 is a typical marker gene indicating neuronal differentiation and the encoded protein plays a critical role in axon guidance and maintenance. A decrease in H3K9me3 and H3K27me3 changes the chromatin configuration into a state of transcriptional activation. In summary, these results suggest that SETDB1 functions through two layers of regulation. One function is to bridge USP11 to the interaction partners, thereby protecting them from ubiquitination and degradation, and the other is to establish a transcriptionally silenced state in the promoters of genes promoting differentiation. As a result, SETDB1 maintains high levels of basic cellular functional machineries and factors involved in developmental programs to correspond to the tumorigenicity and differentiation potential of neural stem and cancer cells.
Figure 6.
Influence of SETDB1 knockdown on gene transcription and on the binding of EZH2 to promoters and the change in H3K9me3 and H3K27me3 on promoters.A, RT-qPCR detection of transcription of genes promoting neuronal differentiation, cell cycle, and growth arrest in control and knockdown HCT116 cells. Significance in transcription change was calculated based on experiments in triplicate using unpaired Student’s t test. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NS, not significant. B, ChIP detection of binding of EZH2 to promoters of CDKN1A, GADD45B, BCL6, NEUROD1, TUBB3, and change in H3K9me3 and H3K27me3 in these promoters in response to knockdown of SETDB1 in HCT116 cells. Chromatin fragments were precipitated with antibodies against EZH2, H3K9me3, and H3K27me3, respectively, and detected with qPCR using primer pairs amplifying different regions of promoters (Table S7). Significance in difference in amplified promoter fragments was calculated using unpaired Student’s t test based on experiments in triplicate. Data are shown as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. ChIP, chromatin immunoprecipitation; NS, not significant; RT-qPCR, Reverse transcriptase-quantitative PCR.
Serial transplantation of SW480 cells demonstrates that neural stemness, tumorigenicity, and differentiation potential are coupled
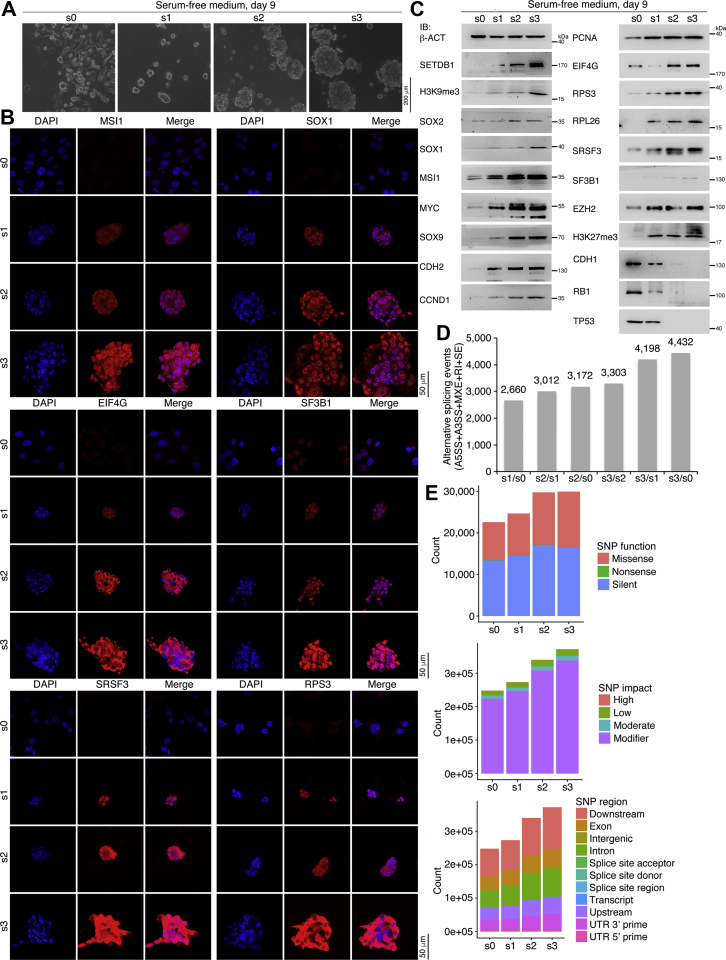
We investigated whether neural stemness, tumorigenicity, and differentiation potential are coupled cell properties using serial transplantation or in vivo passaging via subcutaneous injection in nude mice, as depicted in Fig. S7A. Cells isolated from xenograft tumors were cultured in NSC-specific serum-free medium for 9 days to test their ability to form neurosphere-like structures, an indication of neural stemness. In vitro-cultured SW480 cells were designated s0. Xenograft tumors and cells forming neurosphere-like structures from the first, second, and third in vivo passages were designated s1, s2, and s3, respectively (Fig. S7A). While SW480 cells did not form neurospheres in serum-free medium, cells from the first transplantation (s1) formed small spherical structures, whereas the cells from the second (s2) grew larger spherical structures, and those from the third (s3) grew the largest spherical structures (Fig. 7A). Correspondingly, there was a tendency toward the increasing expression of MSI1 and SOX1 in cells or cell spheres from s0 to s3. Increased expression was also observed for EIF4G, SF3B1, SRSF3, and RPS3 (Fig. 7B). A similar expression trend was observed for some additional proteins and corresponding histone modifications, including SETDB1 and H3K9me3, SOX2, SOX1, MSI1, MYC, SOX9, CDH2, CCND1, PCNA, RPL26, EZH2, and H3K27me3 (Fig. 7C). In contrast, CDH1, RB1, and TP53, which are repressed during tumorigenesis, were repressed during serial transplantation (Fig. 7C). These data demonstrate that serial transplantation causes a gradual enhancement of neural stemness and a gradual increase in the expression of proteins that promote neural stemness in SW480 cells.
Figure 7.
Characterization of property of SW480 cells and cells derived from tumors by serial transplantation.A, different ability of cell sphere formation in serum-free medium by SW480 cells (s0) and cells derived from tumors by serial transplantation (s1, s2, and s3). B and C, detection of protein expression with IF (B) and IB (C) in cells and cell spheres in (A). D, alternative splicing events compared between cells or cell spheres in (A) that were identified by RNA-sequencing. E, numbers of SNPs in cells or cell spheres in (A) that were identified by RNA-sequencing, according to SNP function, impact, or region of occurrence. A3SS, Alternative 3′ splice site; A5SS, Alternative 5' splice site; IB, immunoblotting; IF, immunofluorescence; MXE, Mutually exclusive exon; RI, Retained intron; SE, Skipped exon.
Overall transcription in cell spheres changed progressively from s0 to s3. The transcriptome of s3 cell spheres displayed the largest difference from that of s0 cells, with s2 cell spheres showing a less pronounced difference and s1 spheres showing the least difference (Fig. S7B). Interestingly, downregulated genes between s3 and s0 and between s2 and s0 were strongly enriched in immune response and immune system process, suggesting that successive in vivo passaging leads to a gradually altered immune response in cells (Fig. S7C). This might reflect the increasing immune evasion ability of cancer cells during cancer progression (39). Upregulated genes were weakly associated with cell adhesion (Fig. S7D). The numbers of alternative splicing events constantly increased in cells at high passage numbers compared with cells at low passage numbers (Fig. 7D and Table S5). This corresponds with the increased expression of proteins involved in alternative splicing in cells at later passages. In addition, the numbers of SNPs increased during passaging (Fig. 7E and Table S6), an effect associated with decreased expression of TP53. Increases in SNPs and genomic abnormalities have been observed during cancer progression (40, 41).
The results above suggest that SW480 cells might exhibit enhanced malignant features and tumorigenicity after in vivo passaging. Indeed, cells at passages 0, 1, 2, and 3 displayed constant increases in the abilities of invasion, migration (Fig. S8A), and colony formation in soft agar (Fig. S8, B and C). During serial transplantation, cells formed small xenograft tumors in nude mice during the first passage. Cells derived from the first transplantation formed larger tumors, and tumors formed by cells from the second transplantation were even larger (Fig. 8, A–C and Table S2). Genes representing neural stemness and/or promoting cancers—that is, SETDB1, SOX1, MSI1, PAX6, SOX2, MYC, and ZIC2—showed a tendency toward enhanced transcription during serial transplantation (Fig. 8D). Analysis of tumor sections revealed a similar tendency toward increased expression of SETDB1, SOX1, SOX9, and KI67, and the staining signals for these proteins were denser in sections of tumors at higher passage numbers than those at lower passage numbers (Fig. 8E). Additionally, MAP2, BGLAP, CTSK, and AFP were more strongly stained in tumors at later passages (Fig. 8E), suggesting that these tumors showed more efficient differentiation of tissue or cell types. In summary, serial transplantation leads to progressive enhancement of neural stemness, tumorigenicity, and differentiation potential in tumorigenic cells.
Figure 8.
Characterization of xenograft tumors formed by SW480 cells using serial transplantation and a general model unifying embryogenesis and tumorigenesis.A–C, tumor formation via serial transplantation in each six injected mice (A) and difference in tumor volume (B) and weight (C) between different groups. Significance of difference in tumor volume (B) was calculated using two-way ANOVA-Bonferroni/Dunn test. Significance of difference in tumor weight (C) was calculated using unpaired Student’s t test. Data are shown as mean ± SD. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. D, comparison of expression of genes for SETDB1 and neural stemness protein in SW480 cells and xenograft tumors in (A), as revealed by RT-qPCR. Significance in transcription change was calculated based on experiments in triplicate using unpaired Student’s t test. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. NS, not significant. E, IHC analysis of SETDB1 and other marker protein expression in sections of tumors in (A). Objective magnification: 20 × . F, a model depicting the correlation between embryonic development and tumorigenesis. Normal embryogenesis needs the inhibition of epidermal fate in gastrula ectoderm by extracellular signals secreted by Spemann organizer or node, leading to restoration of the neural ground state or neural stemness in ectoderm, a process called neural induction. Formation of the neural precursor tissues is required for differentiation of the nervous system and many nonneural tissues, such that the body axis of an embryo can form. This neural induction can occur ectopically during embryogenesis, caused by either an ectopic organizer or node activity or ectopic expression of embryonic neural genes, leading to formation of secondary embryonic structures or a conjoined twin. This process might occur in any cell and at any time of an animal life. Somatic cells could suffer various extracellular (e.g., microenvironmental change) and/or intracellular (e.g., gene mutations) insults. If occasionally the insults cause activation of neural stemness regulatory network and/or downregulation/silencing of tissue-specific or differentiation genes/factors, then somatic cells progressively lose their original cell identity and restore the neural ground state or neural stemness, similar to the neural induction process in gastrula ectodermal cells. The resulting cells can self-renew and differentiate, resembling a defected process of embryonic development, that is, tumorigenesis. For detailed information, see text and also refer to refs. 1 and 13 (1, 13) and references therein. IHC, immunohistochemical; RT-qPCR, Reverse transcriptase-quantitative PCR.
Discussion
Tumorigenicity and pluripotent differentiation potential are the basic cell properties for understanding tumorigenesis and embryogenesis, respectively. Although ideas such as “cancer as a dynamic developmental disorder” have been proposed (42), the most prominent connection between cancer and embryonic development might be exemplified by teratocarcinoma. Since 2017, our studies have come to the general conclusion that cancer cells exhibit neural stemness and pluripotent differentiation potential. In combination with other studies, our studies suggest that neural stemness might be the ultimate determinant of these basic cell properties (1, 2, 11, 13). This idea is further supported by the present study. When the network regulating neural stemness was disrupted, for example, via inhibition of Setdb1/SETDB1, NSCs and cancer cells exhibited neuronal or neuronal-like differentiation, accompanied by reduced malignant features, tumorigenicity, and differentiation potential. Conversely, overexpression simultaneously enhanced neural stemness, tumorigenicity, and differentiation potential in weakly tumorigenic cells. Importantly, these cell properties were enhanced during the process of serial transplantation, which is believed to mimic cancer progression (24). These lines of evidence demonstrate that tumorigenicity and pluripotent differentiation potential are integral properties of neural stemness.
A cell property is controlled by a regulatory network rather than a single molecule or molecular event. Cancer cells and NSCs share a regulatory network, which primarily comprises factors with specific or enriched expression in NSCs or embryonic neural cells (1, 2, 11, 13). The enriched expression of SETDB1 in embryonic neural cells and its functions during neuronal differentiation and neural development mean that it is a component of the neural regulatory network and a component of the regulatory network promoting cancers. By recruiting the deubiquitinase USP11, SETDB1 interacts with and maintains the expression of components in the regulatory network, including neural stemness proteins and proteins involved in translation initiation, ribosome biogenesis, and spliceosome assembly in cancer cells and NSCs. Other potential interaction partners of SETDB1 include proteins participating in DNA replication and transcription (MCM3, TOP1, TOP2A, and DDX3X) (Table S4), nucleocytoplasmic transport of proteins and RNAs (RAN), and proteasome assembly (PSMD2 and ADRM1), suggesting that SETDB1 might regulate a broader range of proteins that are involved in basic cellular functional machineries, which are usually enriched in embryonic neural cells (1, 2). Similar to PRMT1, which coordinates ribosomes and proteasomes to maintain neural stemness in cancer cells and NSCs (15), SETDB1 plays a role in the regulation of basic cellular functional machineries, thereby maintaining neural stemness, tumorigenicity, and pluripotent differentiation potential in tumorigenic cells.
Compared with nonneural genes, both genes regulating cancers and neural genes are enriched in long genes consisting of more exons/introns (1, 2, 19). Consistent with this feature is the enriched expression of components involved in spliceosome assembly in embryonic neural cells and cancer cells (1), suggesting high activity of alternative splicing in these cells. Our data on serial transplantation of SW480 cells provide convincing evidence for the association between spliceosome protein expression and alternative splicing. Cells at later passages, which exhibited stronger differentiation potential, also exhibited higher levels of spliceosome protein expression. Accordingly, there were more alternative splicing events. It is logical that cells with enriched expression of long genes and high activity of alternative splicing are more plastic in terms of differentiation potential. Generation of more splice variants means the need for increased protein translation to perform their functions and the need for more efficient protein turnover. In general, all machineries should operate at a higher efficiency to correspond to the status of rapid cell cycle, high proliferation, and pluripotent differentiation potential (1, 15, present study). Thus, neural stemness should be the general stemness or the ground state for cell differentiation (1, 3). The basic cell functional machineries such as the cell cycle, ribosomes, proteasomes, spliceosomes, and epigenetic modifications have been proven to play active roles during tumorigenesis. Inhibition of the activity of these machineries either has been applied to cancer therapy or is under evaluation in preclinical trials (43, 44, 45, 46, 47, 48, 49, 50, 51). Considering the contribution of neural stemness to cell tumorigenicity and the similarity in the regulatory network between cancer cells and NSCs (1, 11, 13), inhibition of cancer via targeted therapy is per se achieved by disruption of the neural regulatory network.
NSCs are obvious precursors of the nervous system. The contribution of neural stemness to nonneural differentiation is not obvious. We and others have demonstrated the pluripotency of NSCs in chimeric embryos and xenograft tumors (2, 3, 4). In fact, nonneural differentiation of embryonic neural cells can be observed during and is critical for normal embryogenesis. Located between the neural plate (the precursor tissue of the central nervous system) and the epidermal ectoderm, neural crest cells are induced by interactions between the neural plate and adjacent tissues and exhibit pluripotency (52, 53, 54, 55, 56). Thus, the pluripotent differentiation potential of neural crest cells is ultimately derived from neural plate cells. In the most posterior region of elongating embryos, neuromesodermal progenitors generate both the spinal cord and paraxial mesoderm. These progenitors are thought to be derivatives of the anterior neural plate (57, 58).
During normal embryogenesis, neural induction in the ectoderm results from inhibition of epidermalizing (or antineuralizing) signaling, that is, the BMP signaling, by the secreted signals from the Spemann organizer in amphibian gastrulae (6, 59, 60). In other words, neural induction is a process of restoration of the neural ground state in ectodermal cells during gastrulation. In mammalian embryos, the functionally homologous structure of amphibian organizer is the node. Neural induction ensures the formation of the nervous system and the differentiation of nonneural cells. Failure of neural induction due to loss of organizer activity or the activity of embryonic neural genes causes failure of body axis formation and, hence, embryogenesis (61, 62, 63). Conversely, neural induction via ectopic organizer or node activity results in the formation of secondary body axis or a conjoined twin. A similar effect can also be achieved by ectopic expression of genes with enriched or localized transcription in embryonic neural cells, that is, eed, yy1, ski, egfr, erbb2, and erbb4 (64, 65, 66), all of which are upregulated in cancer cells and promote cancers. In a general sense, neural induction-like processes might occur at any stage and in any cell in an animal. Cells in adult tissues can suffer various extracellular/intracellular insults, including gene mutation and dysregulation, genomic instability, or microenvironmental changes. These insults might occasionally cause upregulation of neural stemness genes or genes with localized/enriched expression in embryonic neural cells, downregulation of tissue-specific genes or both; the cells will consequently gain the property of neural stemness, that is, the neural ground state. Consequently, the cells acquire tumorigenicity and differentiation potential (1, 2). The resulting cells proliferate quickly and differentiate into different cell types, a process resembling embryonic growth and tissue differentiation. During normal development, neural crest cells and NSCs, both derived from the neuroectoderm, migrate extensively along guided routes to differentiate further. Likewise, cancer cells migrate robustly but in an uncontrolled manner. A recent single-cell RNA-sequencing analysis revealed that genes in cancer cells with prometastatic memory are predominantly related to a neural signature (67). In summary, we suggest that neural stemness is a cell property unifying embryonic development and tumorigenesis (Fig. 8F). Tumorigenesis represents a process of progressive loss of original cell identity and gain of neural stemness in somatic cells, resembling a distorted replay of neural induction and subsequent tissue/cell differentiation during normal embryogenesis.
Experimental procedures
Cell culture
HEK293T and HCT116 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Thermo Fisher Scientific, #11965-092); SW480 cells was cultured in Leibovitz's L-15 medium (L-15, Thermo Fisher Scientific, #41300039); NE-4C cells were in MEM (Gibco, #11090073) added with 1% MEM non-essential amino acids (Thermo Fisher Scientific, #11140050), and 1% Glutamax (Gibco, #35050061). All media were supplemented with 10% fetal bovine serum (FBS, Gibco, #10099141). Mouse embryonic stem cells (mESCs) were cultured in DMEM, added with 100 μM β-mercaptoethanol, 1 ng/ml human LIF (Cell Signaling Technology, #8911), 2 mM L-glutamine (Thermo Fisher Scientific, #25030164), 15% FBS, and 1 × MEM non-essential amino acids. All media were added with 50 U/ml penicillin/50 μg/ml streptomycin. For culture of NE-4C cells, petri dishes were coated with 10 μg/ml poly-D-lysine (Sigma-Aldrich, #P0899); for mESCs, dishes were coated with 0.1% gelatin. All cells were cultured at 37 °C with 5% CO2, except SW480, which was cultured at 37 °C in 100% air. Cancer cell lines were authenticated with short tandem repeat profiling, and cells were detected free of mycoplasma contamination with PCR.
Cells were also cultured in a defined serum-free medium Ndiff227 (CellArtis, #Y40002) used for derivation of primNSCs from mESCs (25) and for the test of neurosphere or neurosphere-like structure formation by NSCs or cancer cells. Control or treated cells were cultured at a density of 1 × 105/cm2.
Plasmid construction, virus packaging, cell infection, or transfection
Validated MISSION shRNAs (Sigma-Aldrich) were used for knockdown of mouse Setdb1, human SETDB1, and human USP11. The shRNAs were TRCN0000092973 (mouse Setdb1), TRCN0000276105 (human SETDB1), and TRCN0000011090 (human USP11), which were subcloned to the lentiviral vector pLKO.1 and designated as shSetdb1, shSETDB1, and shUSP11, respectively. For stable overexpression of SETDB1, the coding region corresponding aa 110 to 1291 was subcloned to pLVX-IRES-Puro or pLVX-IRES-ZsGreen vector, because removal of aa 1 to 109 that contains nuclear export signals facilitates nuclear entry of overexpressed SETDB1 (68). The N-terminal region aa 1 to 725 and C-terminal region aa 726 to 1291 of SETDB1 were subcloned to pCS2+4 × HAmcs that contains four repeats of HA-tags and designated as HA-Nterm and HA-Cterm, respectively, and used for transient overexpression. The complete coding region for human USP11 was subcloned to pCS2+4 × HAmcs vector and RPS3 was subcloned to pCS2+6 × MTmcs vector that contains six repeats of MTs for transient overexpression and designated as HA-USP11 and RPS3-MT, respectively.
Virus production and cell infection were performed as described (27). Virus packaging plasmids, shRNA, or overexpression constructs were cotransfected into HEK293T cells with PEI. Forty-eight hours after transfection, polybrene was added at a final concentration of 10 μg/ml to the lentiviral supernatant. The supernatant was filtered through 0.45 μm filters and centrifuged at 4 °C to concentrate lentiviral particles, which were used for infecting cells. Cells after infection for 48 h were selected with puromycin at 1 μg/ml in culture for 3 days if a puromycin selection vector was used. Cells were cultured further for a desired period when a significant phenotypic change was observed or were harvested for additional assays. In parallel, virus production with the empty vector and cell infection were performed, which was used as a control for knockdown or overexpression assay, respectively.
For transient overexpression assays, HEK293T cells or HCT116 cells were transfected with an overexpression plasmid or a vector plasmid using PEI when cells grew to 70 to 80% confluency. Forty-eight hours later, the cells were collected for further assays.
Immunoblotting
Whole cell lysates were prepared for detection of protein expression using conventional SDS-PAGE and immunoblotting. Protein bands were revealed with a Western blotting substrate (Tanon, #180-501). Primary antibodies were as follows: β-ACT (Cell Signaling Technology, #4970, 1:10,000), CCND1 (Cell Signaling Technology, #2978, 1:1000), CDH1 (Cell Signaling Technology, #3195, 1:1000), CDH2 (Cell Signaling Technology, #13116, 1:1000), EIF4A1 (Abclonal, #A5294, 1:1000), EIF4G (Cell Signaling Technology, #2469), EIF4G1 (Abclonal, #A0881, 1:1000), EZH2 (Cell Signaling Technology, #5246, 1:2000), FGFR1 (Cell Signaling Technology, #9740, 1:2000), H3K27me3 (Cell Signaling Technology, #9733, 1:1000), H3K9me3 (Abcam, #ab8898, 1:1000), HA-tag (Cell Signaling Technology, #3724, 1:2000), HES1 (Cell Signaling Technology, #11988, 1:2000), LSD1 (Cell Signaling Technology, #2139, 1:2000), MAP2 (Cell Signaling Technology, #8707, 1:1000), MSI1 (Cell Signaling Technology, #5663, 1:1000), MYC (Cell Signaling Technology, #13987, 1:1000), MT (Abclonal, #AE010, 1:1000), NEUN (Cell Signaling Technology, #12943, 1:1000), OCT4 (Cell Signaling Technology, #83932, 1:1000), PCNA (Cell Signaling Technology, #13110, 1:2000), PRMT1 (Cell Signaling Technology, #2449, 1:2000), RB1 (Cell Signaling Technology, #9309, 1:1000), RPL24 (Abclonal, #A14255, 1:1000), RPL26 (Abclonal, #A16680, 1:1000), RPS3 (Abclonal, #A2533, 1:1000), SETDB1 (Cell Signaling Technology, #2196, 1:1000), SF3B1 (Abcam, #ab172634; Bethyl, #A300-997A-M), SOX1 (Abcam, #ab87775, 1:1000), SOX2 (Cell Signaling Technology, #3579, 1:1000), SOX9 (Cell Signaling Technology, #82630, 1:1000), SRSF3 (Abcam, #ab198291, 1:1000), SYN1 (Cell Signaling Technology, #5297, 1:1000), TP53 (Cell Signaling Technology, #2527, 1:1000), TUBB3 (Cell Signaling Technology, #5568, 1:1000), USP11 (Abclonal, #A19562, 1:1000), VIM (Cell Signaling Technology, #5741, 1:1000), ZIC1 (Abcam, #ab134951, 1:1000).
Immunofluorescence
Immunofluorescence was performed as described (27). Briefly, control and treated cells were cultured in either normal medium or serum-free medium on coverslips in six-well plates for a desired culture period. Cells were then washed with PBS for three times, followed by fixation with 4% paraformaldehyde for 15 min and inactivation with 50 mM ammonium chloride in PBS for 10 min. Afterward, cells were permeabilized with 0.1% Triton X-100 for 10 min and blocked with 0.2% fish skin gelatin (Sigma-Aldrich, #G7041) for 30 min at room temperature. Subsequently, primary antibodies were added to cells and incubated at 4 °C overnight. Primary antibodies were as follows: EIF4G (Cell Signaling Technology, #2469, 1:200), MAP2 (Abcam, #ab183830, 1:200), MSI1 (Cell Signaling Technology, #5663, 1:200), NEFL (Cell Signaling Technology, #2837, 1:200), RPS3 (Abclonal, #A2533, 1:200), SETDB1 (Cell Signaling Technology, #2196, 1:200), SF3B1 (Abcam, #ab172634, 1:200), SOX1 (Abcam, #ab109290, 1:200), SRSF3 (Abcam, #ab198291, 1:200), SYN1(Cell Signaling Technology, #5297, 1:100), and TUBB3 (Cell Signaling Technology, #5568 and #4466, 1:200). Secondary antibody, Alexa Flour 594 (Thermo Fisher Scientific, #A21207, #A21203, 1:500), or anti-mouse IgG (FITC-conjugated) (Sigma-Aldrich, #F9137. 1:1000) was added to cells after washing. Cell nuclei were counterstained with DAPI. Slides were rinsed, and coverslips were mounted with an antifade mounting medium (Invitrogen, #S36936). Cells were viewed and photographed with a fluorescence microscope (Zeiss LSM 880).
Reverse transcriptase-quantitative PCR
Total RNA preparation and reverse transcriptase-quantitative PCR was performed as described (69). Total RNA was prepared from cells or tumors using TRIzol according to the manufacturer’s protocol. HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, #R212-01/02), which contains reagent for removing genomic DNA, was used for reverse transcription of complementary DNA (cDNA). qPCR was performed on a LightCycler96 System (Roche) using following parameters: one cycle of predenaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing and extension at 60 °C for 30 s, and an additional cycle for melting curve. In each experiment, transcription of β-Act/β-ACT was detected as a loading control. Significance in difference of transcription level was calculated based on experiments in triplicate using unpaired Student’s t test. Results are presented as histograms with relative units of transcription level. Primers for qPCR are listed in Table S7.
Cell migration/invasion assay
24-well transwell plates with inserts of 8-μm pore size (Corning, #3422) were used for cell migration assays. Each 1 × 105 control or treated cells were suspended in 200 μl of culture medium without FBS and added to the upper compartment. The lower compartment contained 500 μl of culture medium supplemented with 10% FBS. After incubation of the plates at 37 °C for desired period as indicated in the text, cells were washed with PBS, then fixed with 37% formaldehyde, and stained with 0.5% crystal violet for 5 min. After removal of cells without migration, migrated cells were washed with PBS and observed under a microscope and photographed.
For assessment of cell invasion, each 5 × 105 control or treated cells were added to 80 μl of Matrigel (Corning, #354234) that was diluted in PBS at a ratio of 1:8 and evenly distributed onto a 24-well transwell insert. After incubation at 37 °C for a desired period as indicated in the text, cells were processed and documented in the same way as in the migration assay.
Soft agar colony formation assay
Soft agar was made of two layers of low melting agarose (Sangon Biotech, # A600015), with top layer being 0.35% of agarose in complete culture medium and bottom layer being 0.7% of agarose. In each well of a six-well culture plate, 2000 control or treated cells were plated on the top layer of agar and cultured at 37 °C for a desired period as indicated in the text. Experiments were repeated thrice. Colonies larger than 50 μm in diameter were counted for significance analysis on using unpaired Student’s t test.
Xenograft tumor assay and serial transplantation of SW480 cells
Animal use in the study was approved by and in accordance with the guidelines of the Institutional Animal Care and Use Committee at the Model Animal Research Center of Medical School, Nanjing University. Immunodeficient nude Foxn1nu male mice of five to sixweek old were purchased from the National Resource Center for Mutant Mice and maintained in a specific pathogen-free facility. Control or treated cells were suspended in 100 μl of PBS and injected subcutaneously into the dorsal flank of a mouse. Cell types and injected cell numbers are listed in Table S2. Tumor size was measured periodically before mice were sacrificed. After sacrifice of mice, tumors were excised and weighed. Tumor volume was calculated with the formula: length × width2/2. Significance of difference in tumor volume between control and treated groups was calculated with two-way ANOVA-Bonferroni/Dunn test. Significance of difference in tumor weight was calculated with unpaired Student’s t test.
SW480 cells were passaged in vivo by serial transplantation. SW480 cells were injected subcutaneously into six nude mice at a dose of 3 × 106 cells per mouse. Forty nine days later, tumors were dissected and tumor cells were dissociated and cultured in NSC-specific Ndiff227 serum-free medium for 9 days. Cell spheres formed in the serum-free medium were collected and dissociated by trituration. Each 3 × 106 cells of the tumors at the first transplantation were injected subcutaneously into nude mice, and tumors were dissected again after 49 days after transplantation. The tumors of the second transplantation were the cell source for the third transplantation, which was done exactly as the first and second time. SW480 cells cultured in serum-free medium for 9 days (s0), sphere-forming cells from the first (s1), second (s2), and the third (s3) transplantation were subjected to assays such as RNA sequencing. Tumor volume and weight were measured, and significance in difference of tumor size and weight between different passages was analyzed in the same way as in xenograft assays above. The strategy of serial transplantation was illustrated in Fig. S7A.
Immunohistochemistry
Immunohistochemistry was used to detect expression of proteins marking different cell/tissue types in tumors using paraffin sections according to conventional method. In brief, sections were deparaffinized with a wash in xylene for 10 min first and then a wash for 5 min and rehydrated with serial washes in 100%, 95%, 85%, 70%, 50% ethanol, and dH2O. Endogenous peroxidase activity was blocked with 3% H2O2 solution in methanol at room temperature for 15 min. After washing slides with PBS thrice, antigen retrieval was performed to unmask the antigenic epitope by treating slides with 0.01 M sodium citrate solution at 95 to 100 °C for 20 min, followed by rinsing with PBS after cooling to room temperature. Sections were blocked with 5% bovine serum albumin in PBS for 1 h at room temperature, then incubated with primary antibody at 4 °C overnight, and washed with PBS. Afterward, biotin-conjugated goat anti-rabbit secondary antibody (Sangon Biotech, #D110066, 1:500) was added and sections were incubated at room temperature for 1 h, and a DAB substrate (Sangon Biotech, # E670033) was added and sections were incubated at room temperature for a desired period (3–15 min) for signal visualization. Cell nuclei were counterstained with hematoxylin. Primary antibodies were as follows: ACTA2 (Abclonal, #A11111, 1:250), AFP (Cell Signaling Technology, #4448, 1:250), BGLAP (Abclonal, #A6205, 1:500), CTSK (ABclonal, #A5871, 1:500), HES1 (Cell Signaling Technology, #11988, 1:200), KI67 (Cell Signaling Technology, #9129, 1:200), MAP2 (Abcam, #ab183830, 1:200), PAX6 (Abcam, #ab195045, 1:200), SETDB1 (Cell Signaling Technology, #2196, 1:200), SOX1 (Abcam, #ab109290, 1:200), SOX9 (Cell Signaling Technology, #82630, 1:200).
Coimmunoprecipitation, mass spectrometric identification of Setdb1 interaction proteins, and functional annotation
Coimmunoprecipitation was performed using conventional method as described (15). Cells were collected and washed twice with ice-cold PBS. Cells were lysed by resuspension in lysis buffer (300 mM NaCl, 1% NP-40, 2 mM EDTA, 50 mM Tris–Cl pH7.5, and protease inhibitor cocktail) on ice for 20 min. Cell lysate was centrifuged for 20 min at 12,000 rpm. Supernatant was collected and immunoprecipitation was carried out using an antibody against an endogenous protein or an HA- or MT-tag that was linked to protein G sepharose beads. In parallel, immunoprecipitation with the antibody against IgG was performed as a negative control. After incubation at 4 °C overnight, beads were collected by centrifugation and washed with TBST buffer (25 mM Tris–Cl PH7.2, 150 mM NaCl, 0.5% Tween-20). Immunocomplexes were eluted by incubating the beads in 1 × loading buffer at 95 °C for 8 min and then subjected to SDS-PAGE.
Identification of Setdb1 interaction proteins with mass spectrometry was performed exactly as described (15). Using the same method above, Setdb1 interaction proteins in NE-4C cells were precipitated with Setdb1 antibody, while a background control was performed in parallel with an IgG antibody. Immunoprecipitates were concentrated by SDS-PAGE, followed by Coomassie blue staining. A single gel band that contained the majority of precipitated proteins was excised and subjected to in-gel digestion. Firstly, cysteine residues were reduced by addition of DTT at a final concentration of 25 mM for 60 min at 70 °C and alkylated by addition of iodoacetamide at a final concentration of 90 mM for 30 min at room temperature in darkness. Proteins were digested with 0.2 μg of modified sequencing grade trypsin (Promega) in 50 mM ammonium bicarbonate at 37 °C overnight. Peptides were extracted, dried, and resuspended in 10 μl of 3% acetonitrile and 2% FA, and subjected to LC-MS/MS. Peptides were analyzed by a NanoLC-2D (Eksigent Technologies) coupled with a TripleTOF 5600+ System (AB SCIEX) as previously described (70).
Data analysis was made by submitting the original files to ProteinPilot Software (version 4.5, AB Sciex). LC-MS/MS data were searched against UniProt database for Mus musculus (April 9, 2016, containing 50,943 sequences, http://www.uniprot.org/proteomes/UP000000589). Proteins of interests were identified by exporting mgf files from ProteinPilot, which were then subjected to Search Compare program in Protein Prospector (version 5.19.1, UCSF) for summarization, validation, and comparison of results using parameters described previously (70). Briefly, trypsin was set as the enzyme with a maximum of two missed cleavage sites. Mass tolerance for parent ion was set at ±20 ppm, and tolerance for fragment ions was set at ±0.6 Da. The expectation value cutoff corresponding to a percent false positive (% FP) rate was determined by searching against a normal database concatenated with the reversed form of the database. Expectation values versus % FP rate were plotted by an algorithm in Search Compare. Based on the results, an expectation value cutoff corresponding to ≤0.01% FP for all peptides was chosen. At this false positive rate, false protein hits from the decoy database were not observed.
According to the result, a protein is considered to be a putative Setdb1 interaction protein when at least two peptides of a protein are identified in proteins precipitated with Setdb1 antibody but not in the precipitate with IgG antibody or the fold change between the number of a protein peptide(s) in the sample precipitated by Setdb1 antibody and by IgG antibody is ≥ 2 (Table S4). The proteomics data was deposited to the ProteomeXchange Consortium via the PRIDE partner repository with identifier PXD030453.
Enrichment analysis for the genes of Setdb1-binding proteins was performed using the DAVID annotation tools (71) with default settings.
Chemical treatment of cells
PrimNSCs were derived from mESCs by culturing mESCs in serum-free Ndiff227 medium at 37 °C with 5% CO2, which formed free-floating neurospheres in the medium. To induce neuronal differentiation in primNSCs, RA (Sigma-Aldrich, #R2625) was added to the medium to a final concentration of 1 μM for 6 days. For detection of protein ubiquitination and the effect of proteasomal activity on protein expression, HCT116 cells were treated with MG132 (Selleckchem, #S2619) at 25 μM for 18 h. For detection of protein half-life, HCT116 cells are infected with lentivirus carrying empty vector or SETDB1 knockdown construct, selected with puromycin, and treated with cycloheximide (Selleckchem, #S7418) at a dose of 50 μg/ml for 0, 2, 4, 6, and 8 h, respectively. Cells were collected and subjected to additional analysis.
Chromatin immunoprecipitation
HCT116 cells were infected with lentivirus derived from empty vector or SETDB1 knockdown construct. Chromatin immunoprecipitation (ChIP) and subsequent qPCR were performed exactly as described (27). Antibodies used for ChIP were EZH2 (Cell Signaling Technology, #5246), H3K9me3 (Abcam, #ab8898), and H3K27me3 (Cell Signaling Technology, #9733). Different regions of a gene promoter were amplified from precipitated DNA using quantitative PCR. Significance in change of precipitated chromatin fragments by an antibody was calculated using unpaired Student’s t test based on experiments in triplicate. Results are shown as histograms with relative units. Primers for ChIP-qPCR are listed in Table S7.
Transcriptome profiling
Alteration of transcriptome of NE-4C cells and HCT116 cells in response to Setdb1/SETDB1 knockdown, and transcriptome of SW480 cells and cells derived from tumors from serial transplantation of SW480 cells were analyzed with RNA sequencing. Total RNA was prepared with TRIzol. Total amounts and integrity of RNA were assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies). mRNA was purified from total RNA by using poly(T) oligo-attached magnetic beads. After fragmentation of mRNA, first strand cDNA was synthesized using random hexamer primer and M-MuLV reverse transcriptase. Subsequently, RNA was degraded using RNaseH, and second strand cDNA synthesis was performed using DNA Polymerase I and dNTP. cDNA overhangs were blunted, and 3′ ends of DNA fragments were adenylated. Adapters with hairpin loop structure were ligated to prepare for hybridization. The library fragments were purified with AMPure XP system (Beckman Coulter) to select cDNA fragments of preferentially 370 to 420 bp in length. After qualification, cDNA library was subjected to sequencing by Illumina NovaSeq 6000. The end reading of 150 bp pairing was generated. The image data measured by the sequencer were converted into sequence data (reads) by CASAVA base recognition. Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts and mapped against mouse or human reference genomes using Hisat2 (v2.0.5) software. The mapped reads of each sample were assembled by StringTie (v1.3.3 b) in a reference-based approach. The featureCounts v1.5.0-p3 was used to count the reads numbers mapped to each gene. FPKM of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis of two conditions was performed using the edgeR package (3.24.3). p values were adjusted using the Benjamini-Hochberg method. Padj ≤ 0.05 and |log2(fold change)| ≥ 1 were set as the threshold for significantly differential expression.
Gene Ontology and KEGG pathway enrichment analysis of differentially expressed genes was performed with the clusterProfiler R package (3.8.1). rMATS(4.0.2) software was used to analyze alternative splicing events. GATK2 (v3.8) software was used to perform SNP calling, and SnpEff (4.3q) software was used to annotate SNP. Sequencing, signal processing, and data analyses were performed by Novogene Co, Ltd. RNA-seq data were deposited to the Gene Expression Omnibus under accession numbers GSE192372, GSE192373, and GSE192374.
Data availability
Proteomics data are available via ProteomeXchange with identifier PXD030453 (http://www.ebi.ac.uk/pride). Transcriptomics data are deposited in GEO under accession numbers GSE192372 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192372), GSE192373 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192373), and GSE192374 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192374).
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare no potential competing interests.
Acknowledgments
Author contributions
Y. C. conceptualization; M. Z., Y. L., L. S., L. F., and L. X. investigation; M. Z., L. F., and Y. C. methodology; M. Z. validation; M. Z. data curation; Y. C. writing–original draft; Y. C. project administration; Y. C. funding acquisition; Y. C. supervision.
Funding and additional information
This work was supported by Shenzhen Science and Technology Program (Grant No.: JCYJ20210324120205015) and National Science Foundation of China (Grant No.: 31671499) to Y. C.
Edited by Brian Strahl
Supporting information
References
- 1.Cao Y. Neural is fundamental: neural stemness as the ground state of cell tumorigenicity and differentiation potential. Stem Cell Rev. Rep. 2022;18:37–55. doi: 10.1007/s12015-021-10275-y. [DOI] [PubMed] [Google Scholar]
- 2.Xu L., Zhang M., Shi L., Yang X., Chen L., Cao N., et al. Neural stemness contributes to cell tumorigenicity. Cell Biosci. 2021;11:21. doi: 10.1186/s13578-021-00531-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke D.L., Johansson C.B., Wilbertz J., Veress B., Nilsson E., Karlström H., et al. Generalized potential of adult neural stem cells. Science. 2000;288:1660–1663. doi: 10.1126/science.288.5471.1660. [DOI] [PubMed] [Google Scholar]
- 4.Tropepe V., Hitoshi S., Sirard C., Mak T.W., Rossant J., van der Kooy D. Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert S.F., Barresi M.J. Developmental Biology. 11th Edition. Sinauer Associates, Inc; MA: 2016. Early amphibian development; pp. 333–364. [Google Scholar]
- 6.Muñoz-Sanjuán I., Brivanlou A.H. Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 2002;3:271–280. doi: 10.1038/nrn786. [DOI] [PubMed] [Google Scholar]
- 7.Smukler S.R., Runciman S.B., Xu S., van der Kooy D. Embryonic stem cells assume a primitive neural stem cell fate in the absence of extrinsic influences. J. Cell Biol. 2006;172:79–90. doi: 10.1083/jcb.200508085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng L., Hu W., Qiu B., Zhao J., Yu Y., Guan W., et al. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014;24:665–679. doi: 10.1038/cr.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Guo X., Huang H., Wang C., Yang F., Zhang Y., et al. A switch in tissue stem cell identity causes neuroendocrine tumors in Drosophila Gut. Cell Rep. 2020;30:1724–1734.e4. doi: 10.1016/j.celrep.2020.01.041. [DOI] [PubMed] [Google Scholar]
- 10.Southall T.D., Davidson C.M., Miller C., Carr A., Brand A.H. Dedifferentiation of neurons precedes tumor formation in Lola mutants. Dev. Cell. 2014;28:685–696. doi: 10.1016/j.devcel.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Lei A., Xu L., Chen L., Chen Y., Zhang X., et al. Similarity in gene-regulatory networks suggests that cancer cells share characteristics of embryonic neural cells. J. Biol. Chem. 2017;292:12842–12859. doi: 10.1074/jbc.M117.785865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domazet-Lošo T., Brajkovićm J., Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007;23:533–539. doi: 10.1016/j.tig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Cao Y. Tumorigenesis as a process of gradual loss of original cell identity and gain of properties of neural precursor/progenitor cells. Cell Biosci. 2017;7:61. doi: 10.1186/s13578-017-0188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalton S. Linking the cell cycle to cell fate decisions. Trends Cell Biol. 2015;25:592–600. doi: 10.1016/j.tcb.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L., Zhang M., Fang L., Yang X., Cao N., Xu L., et al. Coordinated regulation of the ribosome and proteasome by PRMT1 in the maintenance of neural stemness in cancer cells and neural stem cells. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agosto L.M., Lynch K.W. Alternative pre-mRNA splicing switch controls hESC pluripotency and differentiation. Genes Dev. 2018;32:1103–1104. doi: 10.1101/gad.318451.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baralle F.E., Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017;18:437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bush S.J., Chen L., Tovar-Corona J.M., Urrutia A.O. Alternative splicing and the evolution of phenotypic novelty. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372 doi: 10.1098/rstb.2015.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahakyan A.B., Balasubramanian S. Long genes and genes with multiple splice variants are enriched in pathways linked to cancer and other multigenic diseases. BMC Genomics. 2016;17:225. doi: 10.1186/s12864-016-2582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dodge J.E., Kang Y.K., Beppu H., Lei H., Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan S.L., Nishi M., Ohtsuka T., Matsui T., Takemoto K., Kamio-Miura A., et al. Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development. 2012;139:3806–3816. doi: 10.1242/dev.082198. [DOI] [PubMed] [Google Scholar]
- 22.Strepkos D., Markouli M., Klonou A., Papavassiliou A.G., Piperi C. Histone methyltransferase SETDB1: a common denominator of tumorigenesis with therapeutic potential. Cancer Res. 2021;81:525–534. doi: 10.1158/0008-5472.CAN-20-2906. [DOI] [PubMed] [Google Scholar]
- 23.Lazaro-Camp V.J., Salari K., Meng X., Yang S. SETDB1 in cancer: Overexpression and its therapeutic implications. Am. J. Cancer Res. 2021;11:1803–1827. [PMC free article] [PubMed] [Google Scholar]
- 24.Chan M.M., Lu X., Merchant F.M., Iglehart J.D., Miron P.L. Serial transplantation of NMU-induced rat mammary tumors: a model of human breast cancer progression. Int. J. Cancer. 2007;121:474–485. doi: 10.1002/ijc.22684. [DOI] [PubMed] [Google Scholar]
- 25.Ying Q.L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 26.Buschendorf M., Terranova R., Boutsma E., Mao X., Isono K., Brykczynska U., et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- 27.Lei A., Chen L., Zhang M., Yang X., Xu L., Cao N., et al. EZH2 regulates protein stability via recruiting USP7 to mediate neuronal gene expression in cancer cells. Front. Genet. 2019;10:422. doi: 10.3389/fgene.2019.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sher F., Boddeke E., Copray S. Ezh2 expression in astrocytes induces their dedifferentiation toward neural stem cells. Cell Reprogram. 2011;13:1–6. doi: 10.1089/cell.2010.0052. [DOI] [PubMed] [Google Scholar]
- 29.Han X., Gui B., Xiong C., Zhao L., Liang J., Sun L., et al. Destabilizing LSD1 by Jade-2 promotes neurogenesis: an antibraking system in neural development. Mol. Cell. 2014;55:482–494. doi: 10.1016/j.molcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Hirano K., Namihira M. LSD1 mediates neuronal differentiation of human fetal neural stem cells by controlling the expression of a novel target gene, HEYL. Stem Cells. 2016;34:1872–1882. doi: 10.1002/stem.2362. [DOI] [PubMed] [Google Scholar]
- 31.Chiang S.Y., Wu H.C., Lin S.Y., Chen H.Y., Wang C.F., Yeh N.H., et al. Usp11 controls cortical neurogenesis and neuronal migration through Sox11 stabilization. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abc6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun H., Ou B., Zhao S., Liu X., Song L., Liu X., et al. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine. 2019;48:236–247. doi: 10.1016/j.ebiom.2019.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dwane L., O'Connor A.E., Das S., Moran B., Mulrane L., Pinto-Fernandez A., et al. A functional genomic screen identifies the deubiquitinase USP11 as a novel transcriptional regulator of ERα in breast cancer. Cancer Res. 2020;80:5076–5088. doi: 10.1158/0008-5472.CAN-20-0214. [DOI] [PubMed] [Google Scholar]
- 34.Ma D.K., Jang M.H., Guo J.U., Kitabatake Y., Chang M.L., Pow-Anpongkul N., et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sultan F.A., Sweatt J.D. The role of the Gadd45 family in the nervous system: A focus on neurodevelopment, neuronal injury, and cognitive neuroepigenetics. Adv. Exp. Med. Biol. 2013;793:81–119. doi: 10.1007/978-1-4614-8289-5_6. [DOI] [PubMed] [Google Scholar]
- 36.Wainwright L.J., Lasorella A., Iavarone A. Distinct mechanisms of cell cycle arrest control the decision between differentiation and senescence in human neuroblastoma cells. Proc. Natl. Acad. Sci. U S A. 2001;98:9396–9400. doi: 10.1073/pnas.161288698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho J.H., Tsai M.J. The role of BETA2/NeuroD1 in the development of the nervous system. Mol. Neurobiol. 2004;30:35–47. doi: 10.1385/MN:30:1:035. [DOI] [PubMed] [Google Scholar]
- 38.Tiberi L., van den Ameele J., Dimidschstein J., Piccirilli J., Gall D., Herpoel A., et al. BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nat. Neurosci. 2012;15:1627–1635. doi: 10.1038/nn.3264. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez H., Hagerling C., Werb Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., Galipeau P.C., Sanchez C.A., Blount P.L., Maley C.C., Arnaudo J., et al. Single nucleotide polymorphism-based genome-wide chromosome copy change, loss of heterozygosity, and aneuploidy in Barrett's esophagus neoplastic progression. Cancer Prev. Res. (Phila) 2008;1:413–423. doi: 10.1158/1940-6207.CAPR-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wistuba I.I., Meyerson M. Chromosomal deletions and progression of premalignant lesions: less is more. Cancer Prev. Res. (Phila) 2008;1:404–408. doi: 10.1158/1940-6207.CAPR-08-0177. [DOI] [PubMed] [Google Scholar]
- 42.Rubin H. Cancer as a dynamic developmental disorder. Cancer Res. 1985;45:2935–2942. [PubMed] [Google Scholar]
- 43.Bennett R.L., Licht J.D. Targeting epigenetics in cancer. Annu. Rev. Pharmacol. Toxicol. 2018;58:187–207. doi: 10.1146/annurev-pharmtox-010716-105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., Topisirovic I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 45.Bustelo X.R., Dosil M. Ribosome biogenesis and cancer: basic and translational challenges. Curr. Opin. Genet. Dev. 2018;48:22–29. doi: 10.1016/j.gde.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y., Zhang Y., Guo X. Proteasome dysregulation in human cancer: implications for clinical therapies. Cancer Metastasis Rev. 2017;36:703–716. doi: 10.1007/s10555-017-9704-y. [DOI] [PubMed] [Google Scholar]
- 47.Dvinge H., Kim E., Abdel-Wahab O., Bradley R.K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohammad H.P., Barbash O., Creasy C.L. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat. Med. 2019;25:403–418. doi: 10.1038/s41591-019-0376-8. [DOI] [PubMed] [Google Scholar]
- 49.Pelletier J., Thomas G., Volarević S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat. Rev. Cancer. 2018;18:51–63. doi: 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- 50.Soave C.L., Guerin T., Liu J., Dou Q.P. Targeting the ubiquitin-proteasome system for cancer treatment: discovering novel inhibitors from nature and drug repurposing. Cancer Metastasis Rev. 2017;36:717–736. doi: 10.1007/s10555-017-9705-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang B.D., Lee N.H. Aberrant RNA splicing in cancer and drug resistance. Cancers (Basel) 2018;10:458. doi: 10.3390/cancers10110458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buitrago-Delgado E., Nordin K., Rao A., Geary L., LaBonne C. NEURODEVELOPMENT. Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells. Science. 2015;348:1332–1335. doi: 10.1126/science.aaa3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knecht A.K., Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat. Rev. Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- 54.Le Douarin N.M., Dupin E. The pluripotency of neural crest cells and their role in brain development. Curr. Top. Dev. Biol. 2016;116:659–678. doi: 10.1016/bs.ctdb.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 55.Pla P., Monsoro-Burq A.H. The neural border: induction, specification and maturation of the territory that generates neural crest cells. Dev. Biol. 2018;444:S36–S46. doi: 10.1016/j.ydbio.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 56.Selleck M.A., Bronner-Fraser M. Origins of the avian neural crest: the role of neural plate-epidermal interactions. Development. 1995;121:525–538. doi: 10.1242/dev.121.2.525. [DOI] [PubMed] [Google Scholar]
- 57.Henrique D., Abranches E., Verrier L., Storey K.G. Neuromesodermal progenitors and the making of the spinal cord. Development. 2015;142:2864–2875. doi: 10.1242/dev.119768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambasivan R., Steventon B. Neuromesodermal progenitors: a basis for robust axial patterning in development and evolution. Front. Cell Dev. Biol. 2021;8 doi: 10.3389/fcell.2020.607516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson C., Stern C.D. Organizers in development. Curr. Top. Dev. Biol. 2016;117:435–454. doi: 10.1016/bs.ctdb.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 60.De Robertis E.M., Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell Dev. Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Robertis E.M., Larraín J., Oelgeschläger M., Wessely O. The establishment of Spemann's organizer and patterning of the vertebrate embryo. Nat. Rev. Genet. 2000;1:171–181. doi: 10.1038/35042039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao L., Zhu X., Chen G., Ma X., Zhang Y., Khand A.A., et al. A novel role for Ascl1 in the regulation of mesendoderm formation via HDAC-dependent antagonism of VegT. Development. 2016;143:492–503. doi: 10.1242/dev.126292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donohoe M.E., Zhang X., McGinnis L., Biggers J., Li E., Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amaravadi L.S., Neff A.W., Sleeman J.P., Smith R.C. Autonomous neural axis formation by ectopic expression of the protooncogene c-ski. Dev. Biol. 1997;192:392–404. doi: 10.1006/dbio.1997.8780. [DOI] [PubMed] [Google Scholar]
- 65.Nie S., Chang C. Regulation of early Xenopus development by ErbB signaling. Dev. Dyn. 2006;235:301–314. doi: 10.1002/dvdy.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Satijn D.P., Hamer K.M., den Blaauwen J., Otte A.P. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pascual G., Domínguez D., Elosúa-Bayes M., Beckedorff F., Laudanna C., Bigas C., et al. Dietary palmitic acid promotes a prometastatic memory via Schwann cells. Nature. 2021;599:485–490. doi: 10.1038/s41586-021-04075-0. [DOI] [PubMed] [Google Scholar]
- 68.Cho S., Park J.S., Kang Y.K. Regulated nuclear entry of over-expressed Setdb1. Genes Cells. 2013;18:694–703. doi: 10.1111/gtc.12068. [DOI] [PubMed] [Google Scholar]
- 69.Yang X., Cao N., Chen L., Liu L., Zhang M., Cao Y. Suppression of cell tumorigenicity by non-neural pro-differentiation factors via inhibition of neural property in tumorigenic cells. Front. Cell Dev. Biol. 2021;9:7. doi: 10.3389/fcell.2021.714383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang J., Zhao R., Yu C., Bryant C.L.N., Wu K., Liu Z., et al. IKK-mediated regulation of the COP9 signalosome via phosphorylation of CSN5. J. Proteome. Res. 2020;19:1119–1130. doi: 10.1021/acs.jproteome.9b00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucl. Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data are available via ProteomeXchange with identifier PXD030453 (http://www.ebi.ac.uk/pride). Transcriptomics data are deposited in GEO under accession numbers GSE192372 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192372), GSE192373 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192373), and GSE192374 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192374).