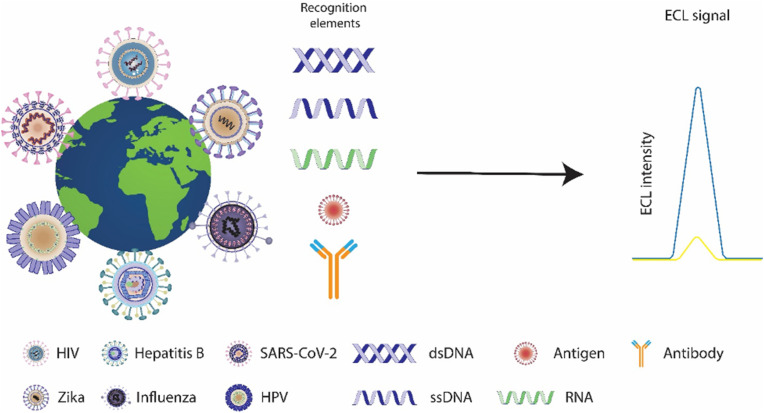
Abstract
Researchers are constantly looking to find new techniques of virus detection that are sensitive, cost-effective, and accurate. Additionally, they can be used as a point-of-care (POC) tool due to the fact that the populace is growing at a quick tempo, and epidemics are materializing greater often than ever. Electrochemiluminescence-based (ECL) biosensors for the detection of viruses have become one of the most quickly developing sensors in this field. Thus, we here focus on recent trends and developments of these sensors with regard to virus detection. Also, quantitative analysis of various viruses (e.g., Influenza virus, SARS-CoV-2, HIV, HPV, Hepatitis virus, and Zika virus) with a specific interest in Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) was introduced from the perspective of the biomarker and the biological receptor immobilized on the ECL-based sensors, such as nucleic acids-based, immunosensors, and other affinity ECL biosensors.
Keywords: Virus detection, Electrochemiluminescence, Respiratory virus, COVID-19, SARS-CoV-2, HPV, Zika virus, HIV, Influenza, Hepatits B, Immunosensors, Genosensors
1. Introduction
The rate of infectious diseases occurrence has been increasing despite all the developments in the healthcare systems and has not only affected humans but animals and plants as well. Among the pathogens responsible for these diseases, viruses have been found to play an essential role [1]. Throughout history, the evolution of viruses has co-occurred with that of humans, resulting in the death of millions of people due to several viral prevalence and epidemics [2]. The current pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), promoting coronavirus disease (COVID-19), is to blame for more than 4.5 million deaths worldwide. Like the new coronavirus, various undiscovered viruses could constitute future health threats and bring disaster to the global economy [3]. Therefore, the development of novel tools and techniques that provide sensitive, inexpensive, rapid, and accurate results seems inevitable for the future confrontation of these outbreaks and prevention of their catastrophic outcomes.
Biosensors are highly sensitive, selective, and accurate measurement systems to determine ultralow analyte concentrations in biological samples. In general, biosensors are considered as analytical devices, consisting of bio-recognition elements, signal transducers, and a detector with a digital output. The bio-recognition element interacts with the target, recognizes the analyte through a reaction, and then the transducer translates the changes to a comprehensible signal measured by the digital detector. Due to the various types of transducers, biosensors can be classified as optical, thermometric, piezoelectric, electrochemical, and electrochemiluminescence (ECL) biosensors. Among them, ECL-based biosensors is a developing alternative tool for quantitative analysis of viruses thanks to the perfectly combined electrochemical and spectroscopic techniques [[4], [5], [6], [7], [8], [9], [10], [11]].
Hence, this article reviews recent advances in ECL-based biosensors for virus detection, with a specific focus on respiratory viruses (Scheme 1 ). Previous reviews have covered essential advancements in the electrochemical-based biosensors for the detection of respiratory viruses [12], the contribution of biosensors in the detection of respiratory viruses [13], and multiple techniques and applications of biosensors in virus detection [14]; however, no review articles cover ECL-based biosensors with regards to virus detection. Therefore, we aim to fill the blank with essential background information about ECL-based biosensors in the field of virus detection.
Scheme 1.
Electrochemiluminescence biosensors for human virus detection.
2. Electrochemiluminescence-based biosensors
Electrochemiluminescence, also called electrogenerated chemiluminescence, is one of the most widely used techniques for the emission of light from electrochemically excited ECL emitters via an efficient electron transfer. ECL has received enormous attention as a powerful tool in the biosensing field due to intrinsic strengths: rapid response, high sensitivity, simplified setup, a wide range of detection, flexibility, low cost, and miniaturized instrumentation [15,16]. In comparison with other optical-based analytical methods, the ECL process has some advantages: The light emission of ECL does not require an external source of light. Therefore, there is no background noise from the sample auto fluorescence or scattered light. Secondly, the potential applied at an electrode can control the ECL emission of light. Also, the ECL technique allows greater control over the emission position since the ECL emission is close to the electrode surface. This better control over the emission is advantageous for selectivity, sensitivity, imaging analysis, and multi-analytes detection. Thirdly, some ECL reactants can be electrochemically regenerated at the electrode. This advantage increases the sensitivity of the technique, saves reactants significantly, and simplifies the instrument [17,18].
There are two main mechanisms through which ECL can be generated: annihilation and co-reactant mechanisms. Moreover, ECL can also be produced through hot electron-induced mechanisms and electrostatic chemiluminescence. In each mechanism, two species are produced electrochemically and undergo an electron-transfer reaction to create both a ground state and an electronically excited state, emitting light and relaxing to the ground state. In the annihilation ECL mechanism, radical species are produced from a single luminophore, while the co-reactant ECL mechanism requires electrochemical interaction between the emitter and a suitable coreactant [[19], [20], [21]]. In both mechanisms, the ECL-luminophore plays a significant part in changing electrical energy into radiative energy. Therefore, the sensitivity of the ECL biosensor specifically depends on ECL luminophores. Numerous ECL luminophores were synthesized in the past years, and several organic, inorganic, and nanomaterial systems can produce ECL [22]. Nevertheless, among the different emitting species commonly utilized in ECL biosensors, ruthenium complexes [[23], [24], [25], [26], [27]] and luminol [[28], [29], [30], [31], [32], [33]] derivatives are the most popular ones employed due to their high solubility in an aqueous medium and the ability to use at a physiological pH in biological samples and also viral detection.
3. Virus detection
Many methods have been developed for detecting viruses. Biosensors have found enormous applications in virus detection, and it extends more sensitive, specific, fast, and reproducible results than conventional techniques like biochemical assays and immunoassays. Generally, biosensors are considered analytical devices that transform bio-related reactions into detectable signals and can be categorized according to their analytes or the reactions they measure. According to the analytes or reactions measured, biosensors for virus detection are categorized into 1, immunosensors that are based on antibody-antigen interaction; 2, genosensors that are dependent on gene sequences derived from viruses; and 3, whole virus biosensors.
3.1. Immunosensors
Immunosensors are a class of biosensors based on antibody-antigen-specific interactions. B-lymphocytes and plasma cells produce antibodies upon antigen contact. It is possible to detect viruses and infections relying on the subclass of antibodies due to their excellent specificity, extreme sensitivity, and high affinity [34,35]. For instance, in the case of SARS-CoV-2 infection, the immunoglobulin G (IgG) subclass of antibodies against N protein is detectable no later than four days after infection [36].
Immunosensors can be classified as both label-free and labeled in their sensing formats. Label-free immunosensors determine the antigen-antibody complex by detecting the physical changes induced by the emergence of other complexes. However, in the case of labeled immunosensors, the immunocomplex is determined by the measurement of the label. Among various types of virus antibody diagnostic technologies, enzyme-linked immunosorbent assay (ELISA) is the most popular one, providing recognition and quantification of various antigens. ELISA, like other types of immunoassays, is based on specific antibody-antigen interactions. However, in the past decade, various immunosensors have been developed to detect antibodies against viruses based on electrochemical and ECL techniques. For instance, a multichannel electrochemical immunoassay platform for quantitative detection of influenza A (H1N1) and SARS-CoV-2 viruses was developed [37]. Zhou and colleagues reported an ECL-based immunosensor employing a sandwich assay to detect human immunodeficiency virus type 1 antibody (anti-HIV-1) employing molecularly imprinted magnetic polymers as receptors [38].
3.2. Whole-virus biosensors
Whole viruses and their structural proteins can be applied as recognition elements. Different surface antigens in viruses’ structures (e.g., proteins) can target various viruses. Upon cell lysis, proteins encoded via viral genomes emerge in blood circulation and can be detected upon the active replication of viruses [35]. In early diagnosis of COVID-19, the whole SARS-CoV-2, and its four structural proteins: spike (S) protein, envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein, could be employed as targets. It should be noted that S and N proteins remain the most notable biomarkers in the COVID-19 early diagnosis [39].
3.3. Genosensors
Genosensors or deoxyribonucleic acid (DNA) biosensors detect the viral genome or specific gene sequences obtained from viruses. DNA, the genetic information carrier, is unique within every. Organism; therefore, this sensor is one of the most sensitive diagnostic tests routinely used to detect and identify viral diseases. The principle of genosensors depends on the stable immobilization of the nucleic acid (ssDNA or ssRNA) on the surface of the sensor to detect the target virus nucleic acid, based on nucleic acid hybridization [40]. The amount of probe immobilized on the ECL genosensors surface is straightly dependent on the availability of analyte binding sites. Consequently, the methods used for the immobilization of the probes are essential to verify the performance of genosensors. Overall nucleic acid-based ECL biosensors feature great stability, specificity, and sensitivity that through miniaturization can be used in the fabrication of POC devices for virus detection.
4. ECL biosensors for viruses detection
4.1. Respiratory virus
Two of the most well-known respiratory viruses, influenza virus, and coronavirus, are responsible for a large portion of morbidities worldwide. Determination of the viral genome sequence, protein structures, and their response to antibodies helps with the realization of the virus pathogenicity through which the development of appropriate and effective vaccines and therapies can be reached. Detection of these types of viruses helps with the decision-making in clinical trials, thus, here we discuss recent developments in their ECL-based biosensors.
4.1.1. Influenza
Influenza is a viral infectious disease that causes numerous medical issues and an enormous financial burden. The single-stranded antisense RNA virus encodes the proteins associated with the virus structure and functions. These enveloped viruses of the Orthomyxoviridae family, are classified into four genera, including influenza virus A–D (IAV, IBV, ICV, and IDV). IAVs and IBVs are of primary concern. The ICVs are endemic and can cause mild disease in humans, and IDVs primarily account for infections in cattle [41]. Hemagglutinin (H.A.), a surface glycoprotein, plays a significant part in the case of animal cell infections with influenza. Neuraminidase (N.A.), another viral surface protein, functions mostly as a virion releaser through the cleavage of virus-bound carbohydrates (sialic acids) from the cell surface [42]. Various ECL biosensors have been reported for the diagnosis of influenza viruses. Naoyoshi Egashira et al. [43] developed an ECL-based biosensor that employs a Ru complex encapsulated in an immunoliposome system. The sensor detects HA through hemagglutinin or antigen peptide immobilization on the surface of a working electrode made of gold. Then, the immunoliposome binds with hemagglutinin onto the working electrode through a competitive antigen-antibody assay. After destroying immunoliposomes, the Ru complex is adsorbed on the working electrode by heating, and ECL measurement is done. Hemagglutinin molecules of the influenza virus were determined in a concentration range of 3 × 10−14 to 2 × 10−12 g mL−1. This level of sensitivity suggests that a limit of detection as low as 6 × 10−19 mol 50 μL−1 could be reached. The biosensor enables fast detection of hemagglutinin proteins in attomolar concentrations.
Also, in another work, Yumi Katayama et al. [44] reported the detection of influenza virus A (H1N1) using an ECL-based immunosensor that benefits from a tris (2, 2′bipyridyl) -ruthenium (II) encapsulated in an immunoliposome system. A competitive assay between the virus and HA is the principle behind the detection system that is immobilized on self-assembled monolayers (SAMs) through the liposome surface-bound antibodies immunoreaction. The first modified DTPA/HT binary SAMs on Au electrode, after fixing hemagglutinin peptide on activated DTPA, competitive reaction of immunoliposome and influenza virus done. They demonstrated significant improvement in sensitivity and accuracy by introducing binary SAMs instead of mono SAMs. The background signal decreased by 50%, and the sensitivity was higher than mono SAMs of dithiodipropionic acid (DTPA). This method features a double amplification of the ECL signal by liposome and adsorption of Ru complex onto Au electrode. The proposed sensor can determine the virus in the range of 2.7 × 102 to 2.7 × 103 PFU mL−1. Recently, Luo et al. [45] proposed an immunomagnetic sensor for the detection of H9N2 avian influenza virus (H9N2 AIV) employing functional silica nanospheres as signal carriers and magnetic nanobeads (M.B.s). The virus detection is based on the formation of a sandwich assay. In this work, [Ru (bpy) 3] 2+ as a luminophore and silica nanoparticles (SNPs) have been used for the amplification of the ECL signal. The functional silica nanospheres were prepared by embedding multiple [Ru (bpy) 3] 2+ in SNPs and further modifying with pAb, improving detection sensitivity as signal probes (Fig. 1 ). Monoclonal antibody (mAb) modified magnetic nanobeads (MBs) achieved the aim of specific capture and separation of H9N2 AIV in the system. Therefore, the combination of immunomagnetic separation and RuSi NPs made the method possible for practical application. The ECL-based immunosensor achieved a limit of detection as low as 14 fg mL−1 H9N2 AIV with a dynamic range of 25 fg mL−1 to 25 ng mL−1, and the ECL signals by about 10−3 fold in comparison with the same concentration of [Ru (bpy)3] 2+.
Fig. 1.
Illustration of the immunomagnetic based ECL immunosensor for H9N2 AIV detection [45].
4.1.2. SARS-CoV-2
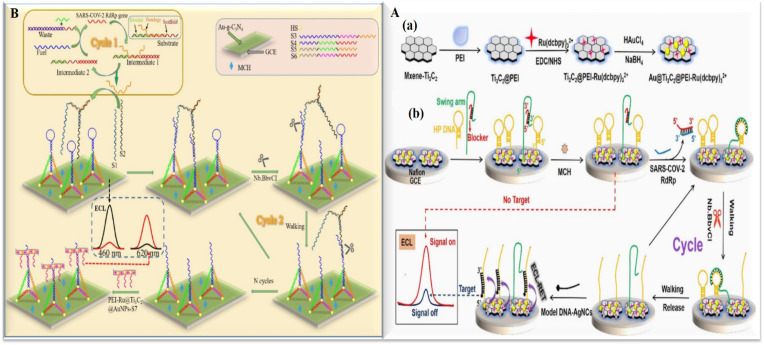
Since January 2020, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), has been spreading worldwide, bringing about the first documented coronavirus pandemic in history. The novel virus is known to cause coronavirus disease (COVID-19) associated with severe health problems, e.g., respiratory distress [46]. SARS-CoV-2 is a beta coronavirus capable of infecting both mammals and birds [47]. The virus belongs to the ssRNA virus group with a positive sense [46]. According to the high incidence and morbidity worldwide, rapid and sensitive detection of COVID-19 is of utmost importance. Three conserved viral sequences are found in the SARS-related genes: (1) RNA-dependent RNA polymerase gene (RdRp gene), (2) envelope protein gene (E gene), and (3) nucleocapsid protein gene (N gene) [48]. The gold standard detection method of COVID-19 is based on quantitative reverse transcription PCR (qRT-PCR), however, it is time-consuming and requires costly equipment [49]. Previously reported papers have discussed advancements in the development of the affinity sensors and immunosensors for the detection of the virus [[50], [51], [52], [53]]. Most of the ECL-based techniques for this virus detection are based on Genosensor methods. Zhenqiang Fan and colleagues [54] reported the fabrication of an ECL-based biosensor that amplifies the signal by employing a DNA walker strategy for the detection of the SARS-CoV-2 RdRp gene. RdRp gene was used to trigger the entropy-driven reaction, the bandage was output, which in combination with the two single-stranded S1 and S2 formed a bipedal DNA walker. The modified Au-g-C3N4/GCE was applied to the ECL donor to combine PEI-Ru@Ti3C2@AuNPs-S7 probes as the ECL acceptor, and TDNAs got modified by hairpin structures to change the signal by ECL resonance energy transfer (ECL-RET) (Fig. 2 A). The proposed biosensor achieved a high sensitivity through the combination of a double amplification strategy, entropy-driven, and bipedal DNA walker. It can quantify the SARS-CoV-2 gene from 10 aM to 10 pM, and the detection limit was reported at 7.8 aM. Yao et al. [55] designed an ECL-based biosensor based on Au@Ti3C2@PEI-Ru (dcbpy) 3 2+ nanocomposites for the detection of the RdRp gene of SARS-CoV-2. As shown in Fig. 2B, the Au NPs were linked to the DNA and Ru (dcbpy) 3 2+ as the ECL emitter, was fixed on the Ti3C2 surface to improve the sensitivity of the ECL biosensor. Polyethylenimine (PEI) can bound to Ru (dcbpy) 3 2+ through an amide bond and as a co-reactant enhances the emission efficiency of Ru (dcbpy) 3 2+. Subsequently, the HP DNAs and swing arm-blocker were anchored on the Au@Ti3C2@PEI- Ru (dcbpy) 3 2+ nanocomposites surface via the Au–S bond possessed a strong ECL signal called “signal on” state. In the presence of the target DNA, the ECL biosensor could realize the transition from “signal-on” to “signal-off” condition. The intensity of the ECL signal decreased with the increasing concentration of the target. Based on this model, the ECL intensity change of the ECL biosensor could reflect different target DNA concentrations. The team of Fan [55] used DNA tetrahedron (TET) as a platform for the biosensor's construction to also detect the RdRp gene of SARS-CoV-2. They indicated an entropy-driven amplified ECL strategy for detection. The tetrahedral structure reduces non-specific adsorptions on the electrode surface, which significantly reduced the process of sensor preparation and made it more user-friendly. The enzyme-free entropy-driven reaction cuts on the expenses of expensive enzyme reagents and facilitates the realization of high-throughput screening of SARS-CoV-2 patients.
Fig. 2.
A: Schematic of the double amplification strategy based on DNA tetrahedral ratiometric ECL-based biosensor for the assay SARS-CoV-2 RdRp gene [53]. B: Schematic Illustration of (a) Preparation of Au@Ti3C2@PEI-Ru(dcbpy)32+ Nanocomposites; (b) based on Au@Ti3C2@PEI-Ru(dcbpy)32+ Nanocomposites ECL Biosensor Detection of the SARS-CoV-2 RdRp Gene Combined Unipedal DNA Walker Amplification Strategy [55].
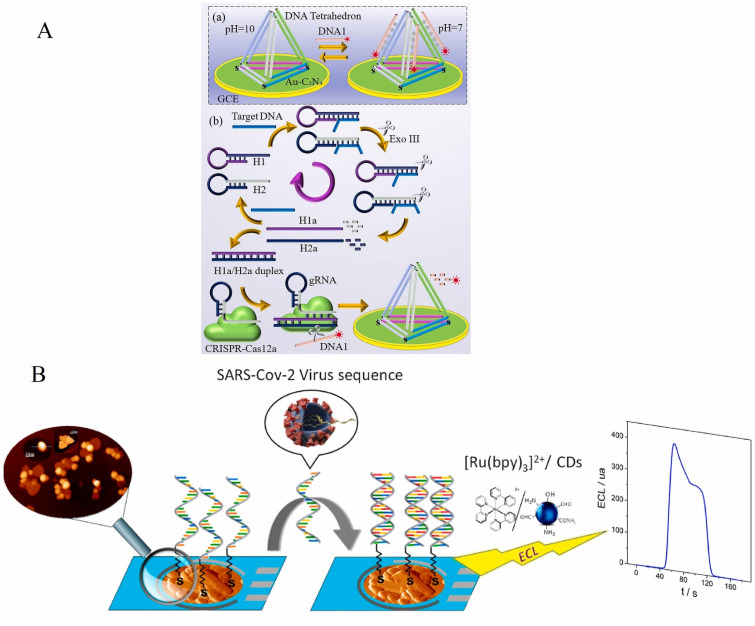
In another work done by Kai Zhang et al. [58] an ECL-based biosensor was designed for the detection of SARS-CoV-2. In this technique, the RNA-dependent RNA polymerase (RdRp) gene was detected using CRISPR-Cas12a and 3D-DNA walker as gene amplification methods, and a ruthenium complex was used as the anodic emitter of the ECL system. The detection of the gene is done in a 2-step process. First, DNA2/DNA6 duplex forms on the surface of the gold nanoparticle, and the second part of the system is an ECL sensor and CRISPR/Cas12a. The activated CRISPR/Cas12a cuts the single-stranded DNA that increases the ECL signal. They reported a linear range of 10 aM–500 aM and a detection limit as low as 12.8 aM. Zhang et al. [59] have reported the identification of the same gene (SARS-CoV-2 RNA-dependent RNA polymerase (RdRp)) using a pH engineered regenerative DNA tetrahedron and CRISPR-Cas12a as the amplification method (Fig. 3 A). In this work, GCE was modified with Au-g-C3N4 as the ECL emitter and donor and the DNA-Ru as the acceptor, and pH was optimized at 10.0 because the biosensor can be regenerated at this pH. They showed that with the increasing concentration of the target from 0 to 1500 aM, the intensity of the ECL peak at 460 nm, which is the characteristic peak of Au-g-C3N4 decreases, and the peak at 620 nm increases drastically. The linear range was reported from 10 aM to 10 pM and the detection limit was found to be as low as 43.70 aM.
Fig. 3.
A: Schematic of the pH-induced biosensor for the detection of SARS-CoV-2 RdRp gene [59]. B: Scheme of the ECL DNA biosensor development [60].
Laura Guti'errez-G'alvez et al. [60] recently developed an electrochemiluminescent nanostructured DNA biosensor for SARS-CoV-2 detection based on two distinct nanomaterials, gold nanomaterials (AuNMs) and carbon nanodots (CDs), for the development of an enhanced ECL DNA biosensor. CDs synthesized by green chemistry are used as coreactants agents in the [Ru(bpy)3]2+ anodic ECL, and changes in the ECL signal of [Ru(bpy)3]2+/CDs in combination with AuNMs nanostructures detect hybridization. In this work, SARS-CoV-2 was detected using the open reading frame 1 ab (ORF1ab) sequences as a model target. Fig. 3B shows the development of this ECL DNA biosensor. The first step is to use AuNMs to modify the surface of the AuSPE. Then, by immobilizing the thiolated capture probe on the AuNMs, the probe-target hybridization was performed to identify a specific DNA sequence of SARS-CoV-2. Finally, an ECL system was employed to identify and quantify SARS-CoV-2 specific DNA sequences by adding a mixture of [Ru (bpy) 3]2+/CDs to the solution.
In a recent work by our team, we developed an ECL-based immunosensor for the detection of the virus [61]. Gold deposited GCEs were used as the working electrode. The surface of the electrode was further modified with MUA/MPA for the covalent immobilization of antibodies. The sensor benefits from a signal-on sensing strategy in which luminol is covalently attached to an Au-based nanocomposite working as the ECL reporter. The secondary SARS-CoV-2 specific antibodies attached to the nanocomposite, ensures the high specificity of the assay and lessens the possibility of false positive signals. A linear range of 10 ng mL−1 to 10 μg mL−1 with a LOD of 1.93 ng mL−1 was reached.
Moreover, Yingying Chen et al. [62] improved the sensitivity of the ECL biosensor through-reactant enrichment and used it to detect SARS-CoV2 Nucleocapsid Proteins. The combination of carboxyl-functionalized poly [2,5-dioctyl-1,4 phenylene] polymer nanoparticles (PDP PNPs) as an excellent ECL luminophore and β-cyclodextrin (β-CD)-Pt nanocomposites as ideal carriers of co-reactants (TDBA), build an attractive ECL platform and shed light on the detection of the target. The GCE surface was modified with PDP PNPs to trap the first antibody (Ab1) and capture the target and the secondary antibody complex (TDBA-β-CD-Pt@Ab2). The obtained biosensor with a sandwich structure was able to detect SARS-CoV-2 Nucleocapsid Proteins with high sensitivity. Table 1 summarizes the various reported ECL biosensors for respiratory virus.
Table 1.
Summary of various biosensors for respiratory virus detection.
| Platform | ECL Luminophor | Type of Virus detection | target | Linear ranges | ECL pathway | LOD | matrix | Ref | |
|---|---|---|---|---|---|---|---|---|---|
| Gold electrode | Ru complex | Immunosensor | Hemagglutinin (virus) |
3 × 10−14 to 2 × 10−12 g mL−1 | Coreactant | 10−14 | [43] | ||
| Au electrode/DTPA/HT binary SAMs | Ru complex | Immunosensor | influenza virus A (H1N1) | 2.7 × 102 to 2.7 × 103 PFU mL−1 | Coreactant | Not determined | Commercial H1N1 |
[44] | |
| Au nanostructures (AGN)/ITO | Ru complex | Immunosensor | H9N2 avian influenza virus | 25 fg mL−1 to 25 ng mL−1 | Coreactant | 14 fg mL−1 | chicken liver, serum, and lung | [45] | |
| PEI-Ru@Ti3C2@AuNPs | g-C3N4 | Genosensor | SARS-CoV-2 | 10 aM to 10 pM | Coreactant | 7.8 aM | human serum | [54] | |
| Au@Ti3C2@PEI-Ru(dcbpy)32+ | Ru complex | Genosensor | SARS-CoV-2 | 1 fM to 100 pM | Coreactant | 0.21 fM | human serum | [57] | |
| DNA tetrahedron | Ru complex | Genosensor | SARS-CoV-2 | 1 fM to 100 pM | Coreactant | 2.67 fM | human serum | [56] | |
| PEI-Ru@Ti3C2@AuNPs | Ru complex | Genosensor | SARS-CoV-2 | 0 aM to 1000 aM. |
Coreactant | 12.8 aM | pharyngeal swabs samples | [58] | |
| Au-g-C3N4/DNA tetrahedron | Ru complex | Genosensor | SARS-CoV-2 | 10 aM to 10 pM | Coreactant | 43.70 aM | pharyngeal swabs samples | [59] | |
| AuNMs and CDs | Ru complex | Genosensor | SARS-CoV-2 | 50.0 fM to 100.0 nM | Coreactant | 514 aM | human serum | [60] | |
| Gold electrode | Ru complex | Genosensor | SARS-CoV-2 ORF1 gene | 0.1 fM to10 μM | Coreactant | 0.1 fM | simulation samples(saliva and urine) | [63] | |
| modified GCE | PDP polymer nanoparticles | Immunosensor | SARS-CoV-2 | 50 fg mL−1 -1.0 ng mL−1 | Coreactant | 22 fg mL−1 | Serum Sample | [62] | |
4.2. Human immunodeficiency virus (HIV) and human Papilloma virus (HPV)
Although human papillomavirus (HPV) and human immunodeficiency virus (HIV) can both be transmitted sexually, there's no medical link between the two conditions. However, the behaviors that put someone at risk of getting HIV can also raise the risk of getting HPV.
To date, more than 79 million people worldwide have been infected with HIV, and there are now approximately 37.7 million globally living with HIV. An accurate diagnosis of HIV is the primary way to control its spread, particularly in patients with the acute infection before the occurrence of seroconversion. ECL-based detection techniques can enable fast monitoring of disease analytes without the need for complex equipment.
The p24 antigen which is the HIV-1 capsid protein appears earlier than the antibody in the case of HIV infection due to an explosive replication of the virus following acute infection and is correlated with highly infectious viremia. Thus, the p24 antigen can be used as a biomarker for early detection of HIV in its “window period.”
Zhou et al. [64] developed an immunosensor for HIV-1 p24 analysis using Ru–SiO2 NPs and gold-nanoparticle-decorated graphene. The as-prepared composite works as an ECL emitter and a carrier to immobilize the target antibody to build a sandwich-type ECL immunosensor through the antibody-antigen interaction. A high ECL signal was obtained due to the large amounts of Ru(bpy)3 2+ molecules per Ru–SiO2 nanoparticle. The composite with good conductivity and high surface area not only accelerated the electron transfer rate but also improved the loading of both ECL molecules and capture antibodies, which could further increase the ECL signal and result in high sensitivity.
HIV-1 Tat protein is considered another biomarker used in the early detection of HIV. Wang et al. [65] introduced an ECL-based aptasensor that uses Ti3C2Tx MXenes modified with Imidazole Zinc -organic frameworks as an ECL emitter. To quench the ECL signal conductive carbon black combined with magnetic nanoparticles was used. The aptamers and their complementary sequence bonded covalently to the surface of the modified electrode.
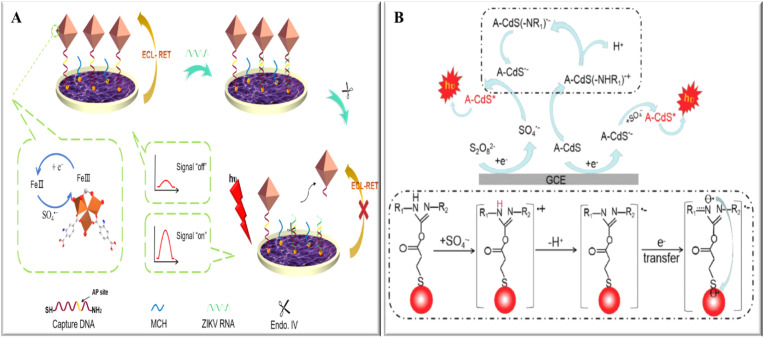
A dual strategy technique for HIV detection was reported by Cai et al. [66]. They modified the surface of the ITO electrode with SnO2 nanoflowers, and then the Au nanoparticles covalently bonded to which the HS-DNA was then linked. The target DNA then forms a three-chain structure in hybridization with the two other hairpins. A bipedal DNA walker with (abga fragments) was generated then through the cleavage of each clip by Exo III. The walker triggered cycle II and resulted in amplification of the target DNA, then target DNA was used to link the 3D CdSe QDs-DNA signal probes with numerous QDs to the electrode (Fig. 4 ), thus greatly enhancing the ECL signals and enabling an ultra-sensitive detection of HIV.
Fig. 4.
Scheme of the ECL biosensing based on 3D CdSe QDs-DNA nanonetwork- SnO2 nanoflower coupled with DNA walker multiple amplification for HIV detection [66].
A non-enzymatic multiple amplified ECL-based biosensor was proposed by Wu and coworkers for the detection of HIV [67] CdSe/ZnS quantum dots were employed as the ECL luminophore and DNA functionalized magnetic beads were used to separate the target HIV DN. In the following, by adding another hairpin DNA modified nanospheres, a strand displacement amplification reaction resulted from the increasing binding sites between two hairpin DNAs. Based on strand displacement amplification, more nanospheres can be captured on the surface of magnetic beads through cycling reaction, which was beneficial for low-abundant biomarker detection. Finally, the magnetic particle was captured on a magnetic glassy carbon electrode surface and ECL signals were obtained in the presence of coreactant.
EuS nanocrystals have been used as ECL fluorophores by Babamiri and colleagues. Through polymerization of functional monomers, they managed to fabricate plastic antibodies around template molecules. The HIV DNA was used as the template molecule and phenylenediamine was employed as the functional monomer on the surface of an ITO electrode [68]. Cervical cancer ranks 4th in the most common cancers seen in women worldwide. Infections with human high-risk papillomavirus (HR-HPV) and HPV16 are amongst the most common causes of cervical cancers reported. Hence, early diagnosis of cervical cancer through monitoring HR-HPV genes has been adopted ubiquitously. E6 and E7, the two main oncogenic genes, are vital for viral replication with the latter having high genetic conservation making it an excellent biomarker for therapeutic interventions. Since HPV cannot be detected through cell culture techniques, its detection and identification mainly relies on molecular techniques. Among all detection methods, ECL-based detection techniques have been recognized as powerful and promising analytical technique due to their distinctive advantages, such as rapidness, low background noise, and astonishing versatility. Hong et al. [69] introduced a split-type ECL technique that produced ascorbic acid through hydrolyzation of the conversion of alkaline phosphate to l-ascorbic acid 2-phosphate for the DNA hybridization reaction. For this, an amino-functionalized capture probes which is linked to the surface of carboxylic MBs1 , formed a sandwich structure via hybridization of HPV16 E7 and the biotin-labeled reporter probe.
After attraction between biotin-avidin in streptavidin-alkaline phosphatase and the biotinylated DNA complex, ascorbic acid was produced. Finally, through magnetic separation, a purified solution containing ascorbic acid was obtained. Based on ECL resonant transfer between AuNCs and MnO2 NMs, the ECL signal of MnO2/AuNCs/GCE was decreased. The ECL signal recovered significantly after immersing the MnO2/AuNC/GCE into the above solution due to the MnO2 etching.
Most of the DNA-based ECL biosensors are fabricated by self-assembly of thiolated single-stranded DNA probes on the Au electrode surface. Due to this random assembly process, a significant discrepancy exists in the distribution of a modified DNA film on different electrodes, which directly affects the reproducibility of a biosensor. He et al. [70] worked on porous bovine serum albumin modified electrode to improve the self-assembly of the ssDNA probe's position distribution and spatial orientation. With the help of DNA amplification techniques in the presence of the target, the surface of the electrode accumulates abundant amplified DNA through reaction, which contains ds-DNA followed by bountiful Ru II complex as fluorophore insertion into grooves of ds-DNA fragments, and an ECL signal can be detected.
CRISPR/Cas systems, components of bacterial immune systems, have gained enormous popularity in nucleic acid detections. Through the programmability of these biomolecular components, an enhanced and amplified ECL signal can be reached in the fields of biosensing. Leu et al. used Cas12a which improved specificity and signal amplification through part recognition mechanisms and trans-cleavage capability respectively [71]. In this work, l-methionine stabilized gold nanoclusters are used as ECL luminophores for the detection of HPV-16. Met-AuNCs modified electrode was used to achieve an original ECL signal and ferrocene-tagged thiolated single-strand DNA as non-specific ssDNA tethered on the surface of the modified electrode to quench the luminophore emission. In the presence of the target molecule, Cas12a activated and possessed trans-cleavage ability on ferrocene-tagged thiolated single-strand DNA with the help of the two-part recognition system, leading to the indiscriminate cleavage into short fragments. The recovered ECL signal under different concentrations of target HPV-16 DNA was used for the quantitative detection of HPV-16 in blood samples. Table 2 summarizes the variously reported ECL biosensors for HIV and HPV.
Table 2.
Summary of various biosensors for HIV and HPV detection.
| Platform | ECL luminophor | Type of Virus detection | ECL pathway | Linear ranges | LOD | Matrix | Ref |
|---|---|---|---|---|---|---|---|
| ECL | RGO@Au composite | HIV Immunosensor | coreactant | 10 ng mL−1 to 1 pg mL−1 |
1 ng mL−1 | serum samples | [64] |
| ECL | Ti3C2Tx MXenes modified ZIF-8 | HIV aptasensor | coreactant | 1 nM 1 fM to |
0.3 fM | Serum sample | [65] |
| ECL | CdSe QDs-DNA nano-network | HIV Genosensor | coreactant | 0.5 μM to 5 fM, | 2.4 fM | serum samples | [66] |
| ECL | CdSe/ZnS quantum dots @poly(styrene/acrylamide) nanospheres | HIV Genosensor | coreactant | 50 nM to 0.5 pM | 39.81 fM | [67] | |
| ECL | EuS NCs | HIV Genosensor | coreactant | 0.3 nM to 0.3 fM | 0.3 fM | Serum sample | [68] |
| ECL | AuNCs | HPV Genosensor | coreactant | 0.1 fM to 10 nM | 0.06 fM | Clinical samples | [69] |
| ECL | Ru(phen)32+ | HPV Genosensor | coreactant | 1 fM to 15 pM | 7.6 fM | Clinical samples | [70] |
| ECL | AuNCs | HPV Genosensor | coreactant | 10 nM to 1pM | 0.48 pM | Blood sample | [71] |
4.3. Hepatitis
Hepatitis viruses can be classified into 5 types including types A, B, C, D, and E. Hepatitis C virus (HCV) is responsible for several chronic liver diseases namely hepatocellular carcinoma, cirrhosis, and end-stage liver disease. HCV is a small single-stranded positive-sense RNA virus (+ssRNA virus) of the Flaviviridae family with a relatively small size (40–80) nm. It has six known genotypes and multiple subtypes amongst which genotype 1 is the most common worldwide. It is believed that there are about 71 million people worldwide infected with HCV. HCV can be diagnosed with various methods and among them, enzyme-linked immunosorbent assay (ELISA) and reverse-transcriptase polymerase chain reaction (RT-PCR) are the two most common detection methods. Hepatitis B virus (HBV) is responsible for a global chronic viral disease. The double-stranded DNA virus (dsDNA) belongs to the family of Hepadnaviridae with 257 million people worldwide being infected by it [72]. Immunoassays and polymerase chain reaction (PCR) are two of the reported assays mainly used to detect HBV. The comparison of the detection strategies has shown that electrochemical and electrogenereatedchemiluminescence provides a higher sensitivity in the analyte detection and a lessen dependency on nonspecific adsorptions [73]. The hepatitis B virus (HBV) is a DNA virus that replicates its genome via an RNA intermediate using reverse transcription 1 [74]. Hepatitis virus A (HAV) is a small, single-stranded positive RNA virus (+ssRNA virus) that can survive on hands and non-porous environments and various kinds of foods [75]. The hepatitis D (delta) viral agent, which is an infectious agent that needs hepadnavirus for propagation, contains a covalently closed circular single-stranded RNA genome of 1167 nucleotides. This genome encodes the proteins p24 and p27 that bind specifically to antisera from patients with chronic hepatitis D infections [76]. Various ECL-based biosensors have also been adopted to detect different types of hepatitis viruses. Yang Liu et al. developed a distance-dependent Plasmon-enhanced ECL genosensors to detect the HCV gene, based on the amplification with hybridization chain reaction (HCR) [77]. The nonmetallic plasmonic MoS2 nanosheets were employed to enhance the ECL signal of sulfur-doped boron nitrogen QDs (S-BN QDs). The distance-dependent plasmon-enhanced ECL were discussed with different length DNA chains. With the increased distance between MoS2 nanosheets and S-BN QDs, the energy transfer effect was limited, and the surface plasma coupling effect was strengthened. As illustrated in Fig. 5 A, each initiator can propagate a cascade hybridization event between alternate hairpins to form a long-nicked dsDNA with repeating units. To amplify the ECL signal of QDs, QDs were tagged on H2; MoS2 nanosheets were connected to H1 on electrodes. In another research, a multiplex ECL DNA sensor was developed based on multicolor CdTe QDs and Au nanoparticles to determine hepatitis B virus (HBV) and hepatitis C virus (HCV) [78]. QDs were utilized as ECL luminophores and electrochemically reduced graphene nanosheets were employed to connect the luminophore onto GCE and enhance ECL intensity. After the introduction of target DNA, the capture DNAs on the surface of CdTe QDs hybridized with complementary target, and only the unreacted capture probes could hybridize with the complementary Au NPs-probe DNA. Au NPs could quench the ECL intensity of CdTe QDs due to the inner filter effect. Hence, target DNAHBV and target DNAHCV could be determined through monitoring the ECL DNA sensor based on Au NPs-probe DNA/target DNA/CdTe QDs-capture DNA/GNs/GCE composite film. The ECL signals of dual-color CdTe QDs increased with the increased concentration of target DNAHBV and target DNAHCV added with the limit of detection of target DNAHBV and target DNAHCV being 0.082 pM and 0.34 pM, respectively. In another effort to detect hepatitis C virus, an ECL genosensor was proposed to combine GQDs as a label and ECL signal source with site-specific recognition of BamHI endonuclease and bidentate chelation of dithiocarbamate ligands for enhanced the robustness of DNA immobilization on the surface of the gold electrode (Fig. 5B) [79]. BamHI endonuclease recognized the symmetrical duplex sequence and catalyzed the dsDNA cleavage, making the dsDNA fragments and the GQDs break off from the electrode surface. This resulted in a decreased ECL signal intensity. This signal-off ECL DNA biosensor employs hepatitis C virus-1b genotype complementary DNA (HCV-1b cDNA) as a model and displayed good analytical performance and a linear range from 5 fM to 100 pM with a detection limit as low as 0.45 fm. Recently, a novel multiple amplification strategies were reported for ultrasensitive near-infrared ECL immunoassay in K2S2O8 solution for the detection of target procalcitonin (PCT) (Fig. 5C) [80]. The realization of this strategy is based on the antenna effect of Eu-MOF (EuBTC), and the high-efficiency catalysis of CoS2 hollow triple shelled nano boxes (TSNBs). In comparison with the traditional ECL-emitters such as luminol, [Ru (bpy)3]2+, and noble metal catalysts, the Eu-MOF and CoS2 make the ECL biosensor possess a low-cost advantage. This sandwich-type ECL biosensor has a near-infrared luminescence in 800–900 nm that does not damage the sample in the meantime. The strategy provides a feasible method for fearful bacterial infection, hepatitis B, and peritonitis. The limit of detection (LOD) for target procalcitonin (PCT) was calculated to be 3.65 fg mL−1 and a linear range of 10 fg mL−1 to 100 ng mL−1 was reported for the detection.
Fig. 5.
A: The HCR-based sensing process and distance-dependent plasmon-enhanced ECL [77] BB. Schematic of the ECL-based biosensor based on GQDs combining with endonuclease cleavage and bidentate chelation [79], C: The mechanism illustration of the increased ECL by antenna effect and dual enhancement effect of CoS2 TSNBs [80].
Moreover, Nikolaou and colleagues [81] recently suggested a new molecular biosensor for detecting Hepatitis B whole genome. The DNA sensor is based on a surface cooperative hybridization at a miniaturized gold electrode in conjunction with an ECL detection approach utilizing the [Ru (phen)2dppz]2+ complex. This PCR-free biosensor could detect both synthetic HBV genomes (SG ds-HBV) and samples collected from real samples (EG ds-HBV), with a detection limit of 0.05 μL−1 for extracted samples. In Table 3 , various biosensors for hepatitis virus detection are summarized (see Table 4).
Table 3.
Summary of various biosensors for hepatitis virus detection.
| Platform | ECL Luminophor | Type of Virus detection | target | Linear ranges | ECL pathway | LOD | matrix | Ref |
|---|---|---|---|---|---|---|---|---|
| Eu-MOF (EuBTC) | immunosensor | procalcitonin (PCT) ((hepatitis B)) |
(10 fg mL−1-100 ng mL−1) | Coreactant | 3.65 fg mL−1 | [80] | ||
| rGO-TEPA | Au@Pt Nanoparticles | immunosensor | HBsAg hepatitis B |
0.25 pg mL−1 to 400 ng mL−1 | Coreactant | 0.08 pg mL−1 |
human serum | [82] |
| MoS2 nanosheets | sulfur doped boron nitrogen QDs (S-BN QDs) | Genosensor | hepatitis C virus (HCV) gene | 0.5 pM to 1 nM | Coreactant | 0.17 pM | human serum | [77] |
| polymer material | [Ru(bpy)3]2+ -Polymer | Genosensor | HBsAg gene of the Hepatitis B virus | 102 aM–106 aM | Coreactant | 100 aM | serum | [83] |
| Graphene nano sheets | CdTe QDs | Genosensor | DNA HBV and DNA HCV | 0.0005–0.5 nM And 0.001–1.0 nM |
Coreactant | 0.082 pM and 0.34 pM | human serum | [78] |
| DTC-DNA/MCH/ethylenedianmine/GQDs | graphene quantum dots (GQDs) | Genosensor | hepatitis C virus- | 5 fM to 100 pM | Coreatant | 0.45 fM | Not mentioned | [79] |
| paper-based BPE | [Ru(bpy)3]2+ | Genosensor | hepatitis B virus gene (HBV) | 0.2 to 100 fM | Coreactant | 0.2 fM | Serum | [84] |
| GCE/ABEI-PEI/ternary NCs/CS | N-(aminobutyl)-N-(ethylisoluminol) (ABEI) | Genosensor | HBV-related DNA | 0.01–100 fM | Coreactant | down to 65 aM | human serum | [85] |
| poly(aniline-luminol)-MWCNTs | Luminol | Genosensor | hepatitis B virus | 10−16–10−7 M | Coreactant | 2.4 × 10−17 M | Serum | [86] |
| Au electrode (CHI101) | (phen)2dppz]2+ complex | Genosensor | hepatitis B virus gene (HBV) | 0–0.75–0.017 cps μL−1 | Coreactant | 0.05 cps μL−1 | genome extracted from real samples | [81] |
Table 4.
Summary of various biosensors for zika virus detection.
| Platform | Type of Virus detection | ECL luminophor | ECL pathway | Linear ranges | LOD | Matrix | Ref |
|---|---|---|---|---|---|---|---|
| polystyrene beads (PSB) | immunosensor | Ru(bpy)3+2 | Coreactant | 0–104 PFU | 1 PFU in 100 μl | human plasma, human urine. | [92] |
| Fe-MIL-88 MOFs | genosensor | AuNPs&g-C3N4@Zr-MOG | Coreactant | 0.3 nM to 3 μM | 0.1 nM | Human serum | [93] |
| SiO2 microspheres | immunosensor | CdS QDs | Coreactant | 1.0 fg mL−1 to 1.0 ng mL−1 | 0.3 fg mL−1 | Human serum | [94] |
4.4. Zika virus (ZIKV)
Zika virus, is a single-stranded RNA virus that belongs to the family of Flaviviridae with strong neurotropic toxicity and teratogenicity [83]. The virus was first identified in 1947, and the name came from a forest in Uganda [84]. This virus can produce devastating consequences for the function of fetal development. Zika Virus spreads by infected mosquito bites and mother to fetus transmission, sexual contact, or blood transfusion [[87], [89]]. Zika's structure is tiled with a tightly packed coat of envelope proteins. The cryo-EM structure also revealed that an asparagine amino acid on the surface of this protein is glycosylated [90]. Recent techniques for Zika virus detection and diagnosis of its infections contain nuclear acid-based assays, for example, quantitative polymerase chain reaction (QPCR), and immunoassays, such as neutralization tests or enzyme-linked immunosorbent assay (ELISA) [91] and ECL methods [92].
Acharya et al. worked on an immunoassay method to detect the Zika virus in human biological fluids [92]. They used polystyrene beads (PSB) with several ECL luminophores conjugated with antiZIKV monoclonal antibodies to form anti-ZIKV-PSBs. The rubrene/benzoyl peroxide (RUB/BPO) system was also chosen for the detection of viruses which is measured after further magnetic beads separation [71]. Their results indicated a linear range between the ECL intensity and the logarithm of RUB concentration is from 0 to 104 PFU and the detection limit of anti-ZIKV-PSBs is 1 plaque-forming unit (PFU) in 100 μl of the sample. A platform for ultrasensitive detection of the zika virus based on a switchable ECL RNA method was developed by Yi-Wen Zhang and coworkers [93]. This platform surface was made from the metal-organic gel (AuNPs&gC3N4@Zr-MOG) and used a metal-organic framework (Fe-MIL-88 MOFs) as electrode surface and biofunctionalized the platform with DNA (Fig. 6 A). Because of this, the ECL signal got enhanced. There was a direct relationship between the increase of ZIKV RNA concentration and the ECL signal. The linear range was reported from 0.3 nM to 3 μM, and the detection limit was 100 pM. Recently, a platform based on the sandwich immunoassay method was reported to detect the Zika virus (Fig. 6B), which was designed based on CdS quantum dots capped with 3-mercaptopropionic acid (MPA@CdS QDs) as ECL labels and silica microspheres as the carrier. One of the advantages of this method is that the generated ECL signals are robust enough that their photos may be captured by smartphone usage. The proposed immunosensor could quantify ZIKV from 1.0 fg mL−1 to 1.0 ng mL−1, and the LOD reported was 0.3 fg mL−1 [94].
Fig. 6.
A. Schematic illustration of the construction process of the proposed ECL sensing platform [88]. B. Preparation of the activation of MPA@CdS QDs (A), the fabrication process of the proposed ECL immunosensor (B) and ECL mechanism of A-CdS QDs (C) [89].
5. Conclusion and future perspective
Currently, the world faces a health crisis caused by the pandemic of SARS-CoV-2. Besides the challenges of diagnosing and treatment of COVID-19, lots of efforts have been put into the prevention of future outbreaks. Despite significant developments in biosensor-based disease diagnosis through previous years, there is still much room for improvement. In recent years, ECL-based biosensors as sensitive, cost-effective, and easily adaptable techniques have been increasingly employed for clinical diagnosis of viruses and other infectious diseases. ECL biosensors are ideal platforms with many advantages, such as high detection capability, simplicity, stability, reliability, and they can be developed without compromising the sensitivity and reproducibility of standards in clinical analysis.
Although some attempts to design different ECL biosensors for virus detection and especially COVID-19 diagnosis have been made, not many portable ECL biosensors for virus detection were fabricated due to many challenges in their use in point-of-care tests. For example, a large number of interferes (antibodies, proteins, cells, DNA, etc.) in several complex samples, a small volume of the viruses in the whole sample volume, and how to isolate the viruses from the real samples are some of the main challenges in the sample preparation step. Also, the immobilization of the biorecognition elements, uniform distribution of the receptor on the electrode surface, and the affinity of bio-recognition elements remain unresolved challenges in ECL biosensors. Moreover, the development of the miniaturized, reproducible, stable, environment-friendly, and cost-effective ECL-based biosensors is another challenge for the fabrication of ECL-based point-of-care biosensors. Therefore, more effort shall be put to overhaul the above challenges in the design and fabrication of the portable ECL biosensors. Despite the critical emerging advances that have considerable capability to design ECL devices for virus diagnosis, the effort to develop an integrated smart ECL sensing system, which is also user-friendly, employed biocompatible substances that provide wearable devices, and the use of minimally invasive samples, such as tears, saliva, urine, or breath is needed.
We have generally introduced the different ECL-based biosensors for each virus and reported various bio-recognition elements. Some examples and recent trends in ECL biosensors are described and gathered in tables with their detection limits for each section. This review primarily prepared valuable references for designing platforms for viral diagnosis based on the ECL methods.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ebtesam Sobhani, Foad Salehnia, Guobao Xu, Yalda Hamidipanah, Shayesteh Arshian, Ali Firoozbakhtian, Morteza Hosseini, Mohammad Reza Ganjali, Saima Hanif, declare no conflict of interest.
Acknowledgements
The authors of this paper would like to express their gratitude to the Iran National Science Foundation and Chinese Academy of Sciences (INSF99008701, CAS-VPST Silk Road Science, GJHZ202125) and the Agence Universitaire de la Francophonie (AUF), Canada for the financial support. We also gratefully acknowledge the support from the University of Tehran.
Footnotes
Magnetic beads
References
- 1.Malik A.A., Nantasenamat C., Piacham T. Molecularly imprinted polymer for human viral pathogen detection. Mater. Sci. Eng. C. 2017;77:1341–1348. doi: 10.1016/j.msec.2017.03.209. [DOI] [PubMed] [Google Scholar]
- 2.Lu L., Su S., Yang H., Jiang S. Antivirals with common targets against highly pathogenic viruses. Cell. 2021;184(6):1604–1620. doi: 10.1016/j.cell.2021.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Hassan M.M., Sium F.S., Islam F., Choudhury S.M. A review on plasmonic and metamaterial based biosensing platforms for virus detection. Sensing and Bio-Sensing Research. 2021;33 doi: 10.1016/j.sbsr.2021.100429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saylan Y., Erdem Ö., Ünal S., Denizli A. An alternative medical diagnosis method: biosensors for virus detection. Biosensors. 2019;9(2):65. doi: 10.3390/bios9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L., Chen Y., Zhu J.-J. Recent advances in electrochemiluminescence analysis. Anal. Chem. 2017;89(1):358–371. doi: 10.1021/acs.analchem.6b04675. [DOI] [PubMed] [Google Scholar]
- 6.Hesari M., Ding Z. Spooling electrochemiluminescence spectroscopy: development, applications and beyond. Nat. Protoc. 2021;16(4):2109–2130. doi: 10.1038/s41596-020-00486-x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., Arbault S., Sojic N., Jiang D. Electrochemiluminescence imaging for bioanalysis. Annu. Rev. Anal. Chem. 2019;12:275–295. doi: 10.1146/annurev-anchem-061318-115226. [DOI] [PubMed] [Google Scholar]
- 8.Sojic N. vol. 15. Royal Society of Chemistry; 2019. (Analytical Electrogenerated Chemiluminescence: from Fundamentals to Bioassays). [Google Scholar]
- 9.Gross E.M., Maddipati S.S., Snyder S.M. A review of electrogenerated chemiluminescent biosensors for assays in biological matrices. Bioanalysis. 2016;8(19):2071–2089. doi: 10.4155/bio-2016-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delaney J.L., Hogan C.F., Tian J., Shen W. Electrogenerated chemiluminescence detection in paper-based microfluidic sensors. Anal. Chem. 2011;83(4):1300–1306. doi: 10.1021/ac102392t. [DOI] [PubMed] [Google Scholar]
- 11.Firoozbakhtian A., Sojic N., Xu G., Hosseini M. Ref. Modul. Biomed. Sci. Elsevier; 2022. Electrochemiluminescence sensors in bioanalysis. [DOI] [Google Scholar]
- 12.Zhao Z., Huang C., Huang Z., Lin F., He Q., Tao D., Jaffrezic-Renault N., Guo Z. Advancements in electrochemical biosensing for respiratory virus detection: a review. TrAC, Trends Anal. Chem. 2021 doi: 10.1016/j.trac.2021.116253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribeiro B.V., Cordeiro T.A.R., Oliveira e Freitas G.R., Ferreira L.F., Franco D.L. Biosensors for the detection of respiratory viruses: a review. Talanta Open. 2020;2 doi: 10.1016/j.talo.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maddali H., Miles C.E., Kohn J., O'Carroll D.M. Optical biosensors for virus detection: prospects for SARS-CoV-2/COVID-19. Chembiochem. 2021;22(7):1176. doi: 10.1002/cbic.202000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du F., Chen Y., Meng C., Lou B., Zhang W., Xu G. Recent advances in electrochemiluminescence immunoassay based on multiple-signal strategy. Current Opinion in Electrochemistry. 2021 [Google Scholar]
- 16.Muzyka K., Saqib M., Liu Z., Zhang W., Xu G. Progress and challenges in electrochemiluminescent aptasensors. Biosens. Bioelectron. 2017;92:241–258. doi: 10.1016/j.bios.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Miao W. Electrogenerated chemiluminescence and its biorelated applications. Chem. Rev. 2008;108(7):2506–2553. doi: 10.1021/cr068083a. [DOI] [PubMed] [Google Scholar]
- 18.Richter M.M. Electrochemiluminescence (ecl) Chem. Rev. 2004;104(6):3003–3036. doi: 10.1021/cr020373d. [DOI] [PubMed] [Google Scholar]
- 19.Nikolaou P., Valenti G., Paolucci F. Nano-structured materials for the electrochemiluminescence signal enhancement. Electrochim. Acta. 2021 [Google Scholar]
- 20.Ma C., Cao Y., Gou X., Zhu J.-J. Recent progress in electrochemiluminescence sensing and imaging. Anal. Chem. 2019;92(1):431–454. doi: 10.1021/acs.analchem.9b04947. [DOI] [PubMed] [Google Scholar]
- 21.Hu L., Xu G. Applications and trends in electrochemiluminescence. Chem. Soc. Rev. 2010;39(8):3275–3304. doi: 10.1039/b923679c. [DOI] [PubMed] [Google Scholar]
- 22.Abdussalam A., Xu G. Recent advances in electrochemiluminescence luminophores. Anal. Bioanal. Chem. 2021:1–16. doi: 10.1007/s00216-021-03329-0. [DOI] [PubMed] [Google Scholar]
- 23.Mesgari F., Salehnia F., Beigi S.M., Hosseini M., Ganjali M.R. Electroanalysis; 2021. Enzyme Free Electrochemiluminescence Sensor of Histamine Based on Graphite-carbon Nitride Nanosheets. [Google Scholar]
- 24.Mesgari F., Beigi S.M., Salehnia F., Hosseini M., Ganjali M.R. Enhanced electrochemiluminescence of Ru (bpy) 32+ by Sm2O3 nanoparticles decorated graphitic carbon nitride nano-sheets for pyridoxine analysis. Inorg. Chem. Commun. 2019;106:240–247. [Google Scholar]
- 25.Pur M.R.K., Hosseini M., Faridbod F., Ganjali M.R., Hosseinkhani S. Early detection of cell apoptosis by a cytochrome C label-Free electrochemiluminescence aptasensor. Sensor. Actuator. B Chem. 2018;257:87–95. [Google Scholar]
- 26.Pur M.R.K., Hosseini M., Faridbod F., Dezfuli A.S., Ganjali M.R. A novel solid-state electrochemiluminescence sensor for detection of cytochrome c based on ceria nanoparticles decorated with reduced graphene oxide nanocomposite. Anal. Bioanal. Chem. 2016;408(25):7193–7202. doi: 10.1007/s00216-016-9856-6. [DOI] [PubMed] [Google Scholar]
- 27.Hosseini M., Karimi Pur M.R., Norouzi P., Moghaddam M.R., Faridbod F., Ganjali M.R., Shamsi J. Enhanced solid-state electrochemiluminescence of Ru (bpy) ₃2+ with nano-CeO₂ modified carbon paste electrode and its application in tramadol determination. Anal. Methods. 2015;7(5):1936–1942. doi: 10.1039/C4AY02772H. [DOI] [Google Scholar]
- 28.Sobhanie E., Faridbod F., Hosseini M., Ganjali M. An ultrasensitive ECL sensor based on conducting polymer/electrochemically reduced graphene oxide for non-enzymatic detection in biological samples. ChemistrySelect. 2020;5(17):5330–5336. [Google Scholar]
- 29.Hamtak M., Fotouhi L., Hosseini M., Reza Ganjali M. Sensitive nonenzymatic electrochemiluminescence determination of hydrogen peroxide in dental products using a polypyrrole/polyluminol/titanium dioxide nanocomposite. Anal. Lett. 2019;52(4):633–648. [Google Scholar]
- 30.Salehnia F., Hosseini M., Ganjali M.R. Enhanced electrochemiluminescence of luminol by an in situ silver nanoparticle-decorated graphene dot for glucose analysis. Anal. Methods. 2018;10(5):508–514. [Google Scholar]
- 31.Karimi A., Husain S., Hosseini M., Azar P.A., Ganjali M. A sensitive signal-on electrochemiluminescence sensor based on a nanocomposite of polypyrrole-Gd 2 O 3 for the determination of L-cysteine in biological fluids. Microchim. Acta. 2020;187(7):1–9. doi: 10.1007/s00604-020-04372-x. [DOI] [PubMed] [Google Scholar]
- 32.Karimi A., Husain S., Hosseini M., Azar P.A., Ganjali M. Rapid and sensitive detection of hydrogen peroxide in milk by enzyme-free electrochemiluminescence sensor based on a polypyrrole-cerium oxide nanocomposite. Sensor. Actuator. B Chem. 2018;271:90–96. [Google Scholar]
- 33.Hamtak M., Hosseini M., Fotouhi L., Aghazadeh M. A new electrochemiluminescence biosensor for the detection of glucose based on polypyrrole/polyluminol/Ni (OH) 2–C 3 N 4/glucose oxidase-modified graphite electrode. Anal. Methods. 2018;10(47):5723–5730. [Google Scholar]
- 34.Ribeiro B.V., Cordeiro T.A.R., e Freitas G.R.O., Ferreira L.F., Franco D.L. Talanta Open; 2020. Biosensors for the Detection of Respiratory Viruses: A Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ménard-Moyon C.c., Bianco A., Kalantar-Zadeh K. Two-dimensional material-based biosensors for virus detection. ACS Sens. 2020;5(12):3739–3769. doi: 10.1021/acssensors.0c01961. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Z., Huang C., Huang Z., Lin F., He Q., Tao D., Jaffrezic-Renault N., Guo Z. Advancements in electrochemical biosensing for respiratory virus detection: a review. TrAC, Trends Anal. Chem. 2021;139 doi: 10.1016/j.trac.2021.116253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J., Lin R., Yang Y., Zhao R., Song S., Zhou Y., Shi J., Wang L., Song H., Hao R. ACS applied materials & interfaces; 2021. Multichannel Immunosensor Platform for the Rapid Detection of SARS-CoV-2 and Influenza A (H1N1) Virus. [DOI] [PubMed] [Google Scholar]
- 38.Zhou J., Gan N., Li T., Hu F., Li X., Wang L., Zheng L. A cost-effective sandwich electrochemiluminescence immunosensor for ultrasensitive detection of HIV-1 antibody using magnetic molecularly imprinted polymers as capture probes. Biosens. Bioelectron. 2014;54:199–206. doi: 10.1016/j.bios.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 39.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 40.Brazaca L.C., Dos Santos P.L., de Oliveira P.R., Rocha D.P., Stefano J.S., Kalinke C., Muñoz R.A.A., Bonacin J.A., Janegitz B.C., Carrilho E. Biosensing strategies for the electrochemical detection of viruses and viral diseases–A review. Anal. Chim. Acta. 2021 doi: 10.1016/j.aca.2021.338384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flerlage T., Boyd D.F., Meliopoulos V., Thomas P.G., Schultz-Cherry S. Influenza virus and SARS-CoV-2: pathogenesis and host responses in the respiratory tract. Nat. Rev. Microbiol. 2021;19(7):425–441. doi: 10.1038/s41579-021-00542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Salihi S.A.A., Alberti F. Naturally occurring terpenes: a promising class of organic molecules to address influenza pandemics. Natural Products and Bioprospecting. 2021;11(4):405–419. doi: 10.1007/s13659-021-00306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egashira N., Morita S.I., Hifumi E., Mitoma Y., Uda T. Attomole detection of hemagglutinin molecule of influenza virus by combining an electrochemiluminescence sensor with an immunoliposome that encapsulates a Ru complex. Anal. Chem. 2008;80(11):4020–4025. doi: 10.1021/ac702625d. [DOI] [PubMed] [Google Scholar]
- 44.Katayama Y., Ohgi T., Mitoma Y., Hifumi E., Egashira N. Detection of influenza virus by a biosensor based on the method combining electrochemiluminescence on binary SAMs modified Au electrode with an immunoliposome encapsulating Ru (II) complex. Anal. Bioanal. Chem. 2016;408(22):5963–5971. doi: 10.1007/s00216-016-9587-8. [DOI] [PubMed] [Google Scholar]
- 45.Luo F., Long C., Wu Z., Xiong H., Chen M., Zhang X., Wen W., Wang S. Functional silica nanospheres for sensitive detection of H9N2 avian influenza virus based on immunomagnetic separation. Sensor. Actuator. B Chem. 2020;310 [Google Scholar]
- 46.Vermisoglou E., Panáček D., Jayaramulu K., Pykal M., Frébort I., Kolář M., Hajdúch M., Zbořil R., Otyepka M. Human virus detection with graphene-based materials. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 48.Mahshid S.S., Flynn S.E., Mahshid S. The potential application of electrochemical biosensors in the COVID-19 pandemic: a perspective on the rapid diagnostics of SARS-CoV-2. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021;12(1):1–10. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drobysh M., Ramanaviciene A., Viter R., Chen C.F., Samukaite-Bubniene U., Ratautaite V., Ramanavicius A. Biosensors for the determination of SARS-CoV-2 virus and diagnosis of COVID-19 infection. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23020666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drobysh M., Ramanaviciene A., Viter R., Ramanavicius A. Affinity sensors for the diagnosis of covid-19. Micromachines. 2021;12 doi: 10.3390/mi12040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramanavicius A., Ryskevic N., Oztekin Y., Kausaite-Minkstimiene A., Jursenas S., Baniukevic J., Kirlyte J., Bubniene U., Ramanaviciene A. Immunosensor based on fluorescence quenching matrix of the conducting polymer polypyrrole. Anal. Bioanal. Chem. 2010;398 doi: 10.1007/s00216-010-4265-8. [DOI] [PubMed] [Google Scholar]
- 53.Dronina J., Samukaite-Bubniene U., Ramanavicius A. Advances and insights in the diagnosis of viral infections. J. Nanobiotechnol. 2021;19 doi: 10.1186/s12951-021-01081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan Z., Yao B., Ding Y., Xu D., Zhao J., Zhang K. Rational engineering the DNA tetrahedrons of dual wavelength ratiometric electrochemiluminescence biosensor for high efficient detection of SARS-CoV-2 RdRp gene by using entropy-driven and bipedal DNA walker amplification strategy. Chem. Eng. J. 2022;427 doi: 10.1016/j.cej.2021.131686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao B., Zhang J., Fan Z., Ding Y., Zhou B., Yang R., Zhao J., Zhang K. Rational engineering of the DNA walker amplification strategy by using a Au@ Ti3C2@ PEI-Ru (dcbpy) 32+ nanocomposite biosensor for detection of the SARS-CoV-2 RdRp gene. ACS Appl. Mater. Interfaces. 2021;13(17):19816–19824. doi: 10.1021/acsami.1c04453. [DOI] [PubMed] [Google Scholar]
- 56.Fan Z., Yao B., Ding Y., Zhao J., Xie M., Zhang K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens. Bioelectron. 2021:178. doi: 10.1016/j.bios.2021.113015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao B., Zhang J., Fan Z., Ding Y., Zhou B., Yang R., Zhao J., Zhang K. Rational engineering of the DNA walker amplification strategy by using a Au@Ti3C2@PEI-Ru(dcbpy)32+nanocomposite biosensor for detection of the SARS-CoV-2 RdRp gene. ACS Appl. Mater. Interfaces. 2021;13(17):19816–19824. doi: 10.1021/acsami.1c04453. [DOI] [PubMed] [Google Scholar]
- 58.Zhang K., Fan Z., Huang Y., Ding Y., Xie M. A strategy combining 3D-DNA Walker and CRISPR-Cas12a trans-cleavage activity applied to MXene based electrochemiluminescent sensor for SARS-CoV-2 RdRp gene detection. Talanta. 2022:236. doi: 10.1016/j.talanta.2021.122868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang K., Fan Z., Ding Y., Xie M. A pH-engineering regenerative DNA tetrahedron ECL biosensor for the assay of SARS-CoV-2 RdRp gene based on CRISPR/Cas12a trans-activity. Chem. Eng. J. 2022:429. doi: 10.1016/j.cej.2021.132472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gutiérrez-Gálvez L., del Caño R., Menéndez-Luque I., García-Nieto D., Rodríguez-Peña M., Luna M., Pineda T., Pariente F., García-Mendiola T., Lorenzo E. Electrochemiluminescent nanostructured DNA biosensor for SARS-CoV-2 detection. Talanta. 2022;240 doi: 10.1016/j.talanta.2021.123203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hosseini M., Sobhanie E., Salehnia F., Xu G., Rabbani H., Naghavi Sheikholeslami M., Firoozbakhtian A., Sadeghi N., Hossein Farajollah M., Reza Ganjali M., Vosough H. Development of sandwich electrochemiluminescence immunosensor for COVID-19 diagnosis by SARS-CoV-2 spike protein detection based on Au@BSA-luminol nanocomposites. Bioelectrochemistry. 2022;147 doi: 10.1016/j.bioelechem.2022.108161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y., He Y., Zhao J., Zhang J., Yuan R., Chen S. Hydrophobic localized enrichment of Co-reactants to enhance electrochemiluminescence of conjugated polymers for detecting SARS-CoV-2 nucleocapsid proteins. Anal. Chem. 2022;94(10):4446–4454. doi: 10.1021/acs.analchem.1c05407. [DOI] [PubMed] [Google Scholar]
- 63.Jiang C., Mu X., Liu S., Liu Z., Du B., Wang J., Xu J. A study of the detection of SARS-CoV-2 ORF1ab gene by the use of electrochemiluminescent biosensor based on dual-probe hybridization. Sensors. 2022;22(6):2402. doi: 10.3390/s22062402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou L., Huang J., Yu B., Liu Y., You T. A novel electrochemiluminescence immunosensor for the analysis of HIV-1 p24 antigen based on P-RGO@ Au@ Ru-SiO2 composite. ACS Appl. Mater. Interfaces. 2015;7(44):24438–24445. doi: 10.1021/acsami.5b08154. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y., Sun W., Li Y., Zhuang X., Tian C., Luan F., Fu X. Imidazole metal-organic frameworks embedded in layered Ti3C2Tx Mxene as a high-performance electrochemiluminescence biosensor for sensitive detection of HIV-1 protein. Microchem. J. 2021:167. doi: 10.1016/j.microc.2021.106332. [DOI] [Google Scholar]
- 66.Cai Q., Wu D., Li H., Jie G., Zhou H. Versatile photoelectrochemical and electrochemiluminescence biosensor based on 3D CdSe QDs-DNA nanonetwork-SnO2 nanoflower coupled with DNA walker amplification for HIV detection. Biosens. Bioelectron. 2021 doi: 10.1016/j.bios.2021.113455. [DOI] [PubMed] [Google Scholar]
- 67.Wu Z., Luo F., Wen W., Zhang X., Wang S. Enrichment-stowage-cycle strategy for ultrasensitive electrochemiluminescent detection of HIV-DNA with wide dynamic range. Anal. Chem. 2019;91(19):12238–12245. doi: 10.1021/acs.analchem.9b01969. [DOI] [PubMed] [Google Scholar]
- 68.Babamiri B., Salimi A., Hallaj R. A molecularly imprinted electrochemiluminescence sensor for ultrasensitive HIV-1 gene detection using EuS nanocrystals as luminophore. Biosens. Bioelectron. 2018;117:332–339. doi: 10.1016/j.bios.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 69.Hong G., Zou Z., Huang Z., Deng H., Chen W., Peng H. Split-type electrochemiluminescent gene assay platform based on gold nanocluster probe for human papillomavirus diagnosis. Biosens. Bioelectron. 2021:178. doi: 10.1016/j.bios.2021.113044. [DOI] [PubMed] [Google Scholar]
- 70.He Y., Liu Y., Cheng L., Yang Y., Qiu B., Guo L., Wang Y., Lin Z., Hong G. Highly reproducible and sensitive electrochemiluminescence biosensors for HPV detection based on bovine serum albumin carrier platforms and hyperbranched rolling circle amplification. ACS Appl. Mater. Interfaces. 2021;13(1):298–305. doi: 10.1021/acsami.0c20742. [DOI] [PubMed] [Google Scholar]
- 71.Liu P.F., Zhao K.R., Liu Z.J., Wang L., Ye S.Y., Liang G.X. Cas12a-based electrochemiluminescence biosensor for target amplification-free DNA detection. Biosens. Bioelectron. 2021:176. doi: 10.1016/j.bios.2020.112954. [DOI] [PubMed] [Google Scholar]
- 72.Sher M., Faheem A., Asghar W., Cinti S. Nano-engineered screen-printed electrodes: a dynamic tool for detection of viruses. TrAC, Trends Anal. Chem. 2021:143. doi: 10.1016/j.trac.2021.116374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramanaviciene A., German N., Kausaite-Minkstimiene A., Voronovic J., Kirlyte J., Ramanavicius A. Comparative study of surface plasmon resonance, electrochemical and electroassisted chemiluminescence methods based immunosensor for the determination of antibodies against human growth hormone. Biosens. Bioelectron. 2012;36 doi: 10.1016/j.bios.2012.03.036. [DOI] [PubMed] [Google Scholar]
- 74.Zoulim F., Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137(5):1593–1608. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- 75.Nagashima S., Ko K., Yamamoto C., Bunthen E., Ouoba S., Chuon C., Ohisa M., Sugiyama A., Akita T., Hossain M.S., Ork V., Mao B., Tanaka J. Prevalence of total hepatitis A antibody among 5 to 7 years old children and their mothers in Cambodia. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-83710-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valenzuela P., Hepatitis A B.C.D., viruses E. Structure of their genomes and general properties. Gastroenterol. Jpn. 1990;25(2 Supplement):62–71. doi: 10.1007/BF02779931. [DOI] [PubMed] [Google Scholar]
- 77.Liu Y., Nie Y., Wang M., Zhang Q., Ma Q. Distance-dependent plasmon-enhanced electrochemiluminescence biosensor based on MoS2 nanosheets. Biosens. Bioelectron. 2020:148. doi: 10.1016/j.bios.2019.111823. [DOI] [PubMed] [Google Scholar]
- 78.Liu L., Wang X., Ma Q., Lin Z., Chen S., Li Y., Lu L., Qu H., Su X. Multiplex electrochemiluminescence DNA sensor for determination of hepatitis B virus and hepatitis C virus based on multicolor quantum dots and Au nanoparticles. Anal. Chim. Acta. 2016;916:92–101. doi: 10.1016/j.aca.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 79.Lou J., Liu S., Tu W., Dai Z. Graphene quantums dots combined with endonuclease cleavage and bidentate chelation for highly sensitive electrochemiluminescent DNA biosensing. Anal. Chem. 2015;87(2):1145–1151. doi: 10.1021/ac5037318. [DOI] [PubMed] [Google Scholar]
- 80.Zhao L., Song X., Ren X., Wang H., Fan D., Wu D., Wei Q. Biosensors and Bioelectronics; 2021. Ultrasensitive Near-Infrared Electrochemiluminescence Biosensor Derived from Eu-MOF with Antenna Effect and High Efficiency Catalysis of Specific CoS2 Hollow Triple Shelled Nanoboxes for Procalcitonin. [DOI] [PubMed] [Google Scholar]
- 81.Nikolaou P., Sciuto E.L., Zanut A., Petralia S., Valenti G., Paolucci F., Prodi L., Conoci S. Ultrasensitive PCR-Free detection of whole virus genome by electrochemiluminescence. Biosens. Bioelectron. 2022 doi: 10.1016/j.bios.2022.114165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei S., Xiao H., Gu M., Chen Z., Cao L. Ultrasensitive label-free electrochemical immunosensor based on core-shell Au@PtNPs functionalized rGO-TEPA/PB nanocomposite for HBsAg detection. J. Electroanal. Chem. 2021:890. doi: 10.1016/j.jelechem.2021.115216. [DOI] [Google Scholar]
- 83.Liao Y., Zhou X., Fu Y., Xing D. Linear Ru(bpy)32+-Polymer as a universal probe for sensitive detection of biomarkers with controllable electrochemiluminescence signal-amplifying ratio. Anal. Chem. 2017;89(23):13016–13023. doi: 10.1021/acs.analchem.7b04142. [DOI] [PubMed] [Google Scholar]
- 84.Feng Q., Chen H., Xu J. Disposable paper-based bipolar electrode array for multiplexed electrochemiluminescence detection of pathogenic DNAs. Sci. China Chem. 2015;58(5):810–818. doi: 10.1007/s11426-014-5295-4. [DOI] [Google Scholar]
- 85.Gou L., Sheng Y., Peng Q., Ling J., Yue H., Chen F., Tang H. Ternary nanocube-based “off-on” blinking-type electrochemiluminescence towards enzyme-free detection of hepatitis B virus (HBV)-related DNA. Sens. Actuators, B. 2020:312. doi: 10.1016/j.snb.2020.127987. [DOI] [Google Scholar]
- 86.Dong, R., Y. Zhang, J. Huang, M. Habibul, and G. Li, Electrochemiluminescence DNA Biosensor for HBV Based on Coralloid Poly (Aniline-Luminol)-MWCNTs and Catalysis of Ferrocene. Electroanalysis.
- 87.Adegoke O., Morita M., Kato T., Ito M., Suzuki T., Park E.Y. Localized surface plasmon resonance-mediated fluorescence signals in plasmonic nanoparticle-quantum dot hybrids for ultrasensitive Zika virus RNA detection via hairpin hybridization assays. Biosens. Bioelectron. 2017;94:513–522. doi: 10.1016/j.bios.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 88.Dick G.W., Kitchen S.F., Haddow A.J. Zika virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952;46(5):509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 89.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543(7644):248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sirohi D., Chen Z., Sun L., Klose T., Pierson T.C., Rossmann M.G., Kuhn R.J. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016;352(6284):467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kumar A., Hou S., Airo A.M., Limonta D., Mancinelli V., Branton W., Power C., Hobman T.C. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016;17(12):1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Acharya D., Bastola P., Le L., Paul A.M., Fernandez E., Diamond M.S., Miao W., Bai F. An ultrasensitive electrogenerated chemiluminescence-based immunoassay for specific detection of Zika virus. Sci. Rep. 2016;6(1):1–11. doi: 10.1038/srep32227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y.-W., Liu W.-S., Chen J.-S., Niu H.-L., Mao C.-J., Jin B.-K. Metal-organic gel and metal-organic framework based switchable electrochemiluminescence RNA sensing platform for Zika virus. Sensor. Actuator. B Chem. 2020;321 doi: 10.1016/j.snb.2020.128456. [DOI] [Google Scholar]
- 94.Zhang H.-J., Zhu J., Bao N., Ding S.-N. Enhanced electrochemiluminescence of CdS quantum dots capped with mercaptopropionic acid activated by EDC for Zika virus detection. Analyst. 2021;146(9):2928–2935. doi: 10.1039/d0an02437f. [DOI] [PubMed] [Google Scholar]