Abstract
We have applied molecular approaches, including PCR-based detection strategies and DNA fingerprinting methods, to study the ecology of Listeria monocytogenes in food processing environments. A total of 531 samples, including raw fish, fish during the cold-smoking process, finished product, and environmental samples, were collected from three smoked fish processing facilities during five visits to each facility. A total of 95 (17.9%) of the samples tested positive for L. monocytogenes using a commercial PCR system (BAX for Screening/Listeria monocytogenes), including 57 (27.7%) environmental samples (n = 206), 8 (7.8%) raw material samples (n = 102), 23 (18.1%) samples from fish in various stages of processing(n = 127), and 7 (7.3%) finished product samples (n = 96). L. monocytogenes was isolated from 85 samples (16.0%) using culture methods. Used in conjunction with a 48-h enrichment in Listeria Enrichment Broth, the PCR system had a sensitivity of 91.8% and a specificity of 96.2%. To track the origin and spread of L. monocytogenes, isolates were fingerprinted by automated ribotyping. Fifteen different ribotypes were identified among 85 isolates tested. Ribotyping data established possible contamination patterns, implicating raw materials and the processing environment as potential sources of finished product contamination. Analysis of the distribution of ribotypes revealed that each processing facility had a unique contamination pattern and that specific ribotypes persisted in the environments of two facilities over time (P ≤ 0.0006). We conclude that application of molecular approaches can provide critical information on the ecology of different L. monocytogenes strains in food processing environments. This information can be used to develop practical recommendations for improved control of this important food-borne pathogen in the food industry.
The advent of molecular methodology has revolutionized our ability to investigate and understand microbial ecology, offering new and unique opportunities to explore the ecology of food-borne pathogens, including Listeria monocytogenes, throughout the food chain and in the food processing environment. Highly discriminatory molecular typing methods, including multilocus enzyme electrophoresis, pulsed-field gel electrophoresis (PFGE), random amplification of polymorphic DNA, ribotyping, and phage typing, have been successfully applied to investigations of contamination patterns in foods and in the food processing environment and are increasingly used for surveillance of human disease cases and for tracking of outbreak sources (2–4, 7, 12, 26, 34, 36, 37, 39). While each method provides discriminatory differentiation of L. monocytogenes subtypes, highly automated and standardized methods provide a simplified approach to molecular subtyping and data analysis. The RiboPrinter Microbial Characterization System (Qualicon, Inc., Wilmington, Del.) is one example of such an approach. This system is based on ribotyping, a subtyping method based upon scoring restriction polymorphisms in the rRNA operons of prokaryotes (8, 9, 25).
L. monocytogenes, an invasive food-borne pathogen capable of causing serious disease in immunocompromised individuals and pregnant women, is common to many natural and man-made environments (16). As this organism is ubiquitous and capable of growth at refrigeration temperatures, the zero-tolerance ruling issued by the U.S. Food and Drug Administration for L. monocytogenes in ready-to-eat foods presents a serious challenge to the food industry. Cold-smoked fish products, which are typically consumed without cooking, are among the ready-to-eat foods of particular concern due to the lack of a heat inactivation step during processing. The prevalence of L. monocytogenes in cold-smoked fish and cooked seafood products has been reported to range from 6 to 36% (6) up to 78% (15, 27). Many studies have also demonstrated a high prevalence of L. monocytogenes in a variety of food processing environments (3, 4, 29, 33, 34, 37) and in ready-to-eat foods that had been subjected to microbial destruction steps sufficient to eliminate L. monocytogenes present in raw materials (for a recent survey, see http://www.fsis.usda.gov/oa/topics/lm_action.htm), strongly suggesting that the processing environment represents a significant source of this organism in finished products.
We hypothesize that molecular studies on the ecology of L. monocytogenes strains present in food processing environments will provide information crucial for the development of better control and prevention strategies for this important food-borne pathogen. The smoked fish industry provides an ideal model for the development and evaluation of molecular detection and tracking systems, as L. monocytogenes is frequently isolated from cold-smoked fish and hazard analysis critical control point (HACCP) programs are mandatory for seafood processors (1). The primary objective of this study was to investigate the prevalence, ecology, and transmission of L. monocytogenes in different seafood processing facilities using molecular detection and subtyping methods. The application of a commercially available PCR-based screening system for L. monocytogenes (BAX for Screening/Listeria monocytogenes; Qualicon, Inc.) allowed us to evaluate its performance using a culture-based detection method as a standard of comparison.
MATERIALS AND METHODS
Samples.
A total of 531 samples were collected from three New York State smoked fish processing facilities over five visits (30 April, 10 August, 26 August, 13 October, and 26 October) to each facility in 1998. Processors C and D were located 2 miles apart. Processor B was located approximately 42 miles from the other two facilities. Samples collected from the processing environment (n = 206) were taken from floor drains, sink drains, cooler floors, and equipment surfaces. Sterile swabs were moistened with 1.0% peptone water containing 0.85% NaCl prior to sampling dry surfaces. Samples of raw fish (n = 102), belonging predominantly to the family Salmonidae, were taken from the collar and belly flap area of fresh, frozen, or thawed whole gutted fish or fish fillets. In-process samples (n = 127) were taken from fish during the wet or dry brine step and from brine solutions. Finished product samples (n = 96) included paper-wrapped and vacuum-packaged cold-smoked fish. Environmental swabs were transported in 2.0 ml of 1.0% peptone water containing 0.85% NaCl. All other samples were transported in sterile Stomacher 400 closure bags (Seward Ltd., London, United Kingdom). Samples were transported to the laboratory and held on ice. Sample analysis was initiated within 24 h of collection.
Bacteriological analysis.
The method for isolation of L. monocytogenes used in this study was a modification of the protocol recommended by the Food and Drug Administration for the isolation of L. monocytogenes from food (24). Twenty-five-gram portions of raw, in-process, and smoked fish, prepared using sterilized instruments, were homogenized in 225 ml of Listeria Enrichment Broth (LEB) (Difco Laboratories, Detroit, Mich.) using a Stomacher 400 laboratory blender (Seward Ltd.). Brine solutions, in 25-ml aliquots, were inoculated into 225 ml of LEB. Swabs and transport media were transferred aseptically to 8 ml of LEB. Following 24 and 48 h of incubation at 30°C, 0.1 ml of each enrichment culture was plated on Oxford medium containing the Oxford Antimicrobic Supplement (Difco Laboratories) and incubated at 30°C for 48 h. Esculin hydrolysis-positive colonies with morphology typical for Listeria spp. were streaked for isolation on brain heart infusion (Difco Laboratories) agar and incubated at 37°C for 24 h. Colonies were isolated preferentially from the Oxford plate inoculated after 24 h of sample enrichment to facilitate recovery of L. monocytogenes as opposed to other Listeria species (30, 35). Pure culture isolates used for hlyA PCR or BAX system analysis (for confirmation that this system correctly identified isolates from culture-positive, BAX system-negative samples) were grown in brain heart infusion broth at 37°C with shaking for 12 to 15 h.
Identification of L. monocytogenes.
Four Listeria-like isolates obtained from each sample on Oxford plates were tested with a PCR assay targeting the listeriolysin O gene, hlyA, to specifically identify L. monocytogenes isolates. Primers α-1 (CCT AAG ACG CCA ATC GAA AAG AAA) and β-1 (TAG TTC TAC ATC ACC TGA GAC AGA) define an 858-bp fragment of the hlyA gene (10). Assays were performed using GeneAmp PCR core reagents (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Each 25-μl reaction mixture contained 1× PCR buffer II, 1.5 mM MgCl2, 125 μM each dATP, dCTP, dGTP, and dTTP, 0.5 μM each primer, 1 U of Amplitaq DNA polymerase, and 2 μl of a 1:10 dilution of a crude cell lysate prepared as described by Furrer et al. (18). Cycling was performed in the GeneAmp PCR system 2400 (Perkin-Elmer Applied Biosystems). Amplification cycling began with 2 min at 95°C, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. A final extension step of 72°C for 10 min was followed by a hold at 4°C. The PCR products were electrophoresed on 1.5% agarose gels at 120 V, stained with ethidium bromide, and photodocumented (41). A positive result was indicated by the presence of an approximately 858-bp band.
BAX for Screening/Listeria monocytogenes.
Following 48 h of sample enrichment, 250 μl of the enrichment culture was removed and centrifuged at maximum speed in a bench-top microcentrifuge for 2 min. The supernatant was discarded and the pellet was resuspended in 250 μl of sterile deionized water. This prelysis wash step was incorporated to minimize the possibility of PCR inhibition by components of the enrichment culture (personal communication from Technical Assistance, Qualicon, Inc.). The lysate was prepared as outlined in the manufacturer's protocol. Temperature cycling for the DNA amplification step was performed in either the GeneAmp PCR system 2400 or PCR system 9600 (Perkin-Elmer Applied Biosystems). PCR products were loaded onto 1.5% agarose gels as outlined in the BAX for Screening protocol, electrophoresed at 120 V, stained with ethidium bromide, and photodocumented (41). A positive result was indicated by the presence of an approximately 400-bp band in the sample lane and an 800-bp band in the corresponding control lane. Aliquots of the resuspended enrichment culture pellets, stored at −80°C, were used to retest samples for which PCR-based screening and culture methods yielded discrepant results.
Ribotyping.
One L. monocytogenes isolate from each culture-positive sample was subtyped. Automated ribotyping with normalized data was performed using the RiboPrinter (Qualicon, Inc.), as previously described (9, 25), in the Laboratory for Molecular Typing at Cornell University. This automated typing method involves EcoRI digestion of L. monocytogenes chromosomal DNA followed by Southern hybridization with an Escherichia coli rrnB rRNA operon probe. Images are acquired with a charge-coupled device and analyzed using custom software that normalizes fragment pattern data for band intensity and relative band size compared to a molecular size marker.
Statistical analyses.
Culture- and PCR-based detection system results were compared by defining the culture-based system results as the standard of comparison (“gold standard”) and then calculating the sensitivity (true positive rate), specificity (true negative rate), and accuracy (method agreement) of the PCR-based system using standard formulas (11). BAX system-negative, culture-positive results were interpreted as BAX system false-negative results. As characterization of as few as five Listeria-like isolates in a culture-based screen can result in an overall sample-negative result (24), BAX system-positive results were interpreted as false positive if hlyA screening of four Listeria-like isolates from the corresponding sample yielded negative results. If the presence of L. monocytogenes was subsequently confirmed by hlyA PCR screening of up to 16 additional isolates from a sample, the BAX results were interpreted as true positive for calculation of the revised specificity estimate.
The rates of recovery of specific ribotypes of L. monocytogenes among the three processing plants and among the sample sources (environment, raw materials, in-process product, and finished product) were compared using Pearson's chi-square test. Exact P values were calculated since expected cell values in some cells were less than five. No analyses were attempted for ribotypes isolated fewer than four times overall due to the small sample size and infrequency of those observations. P values of ≤0.05 were considered significant. All analyses were performed using the statistical software program StatXact-4 for Windows (CYTEL Software Corporation, Cambridge, Mass.).
RESULTS
Detection of L. monocytogenes in cold-smoked fish and in the smoked fish processing environment.
A total of 531 environmental, raw material, in-process, and smoked fish samples were screened for the presence of L. monocytogenes using both the culture-based method and the BAX for Screening/Listeria monocytogenes system. The BAX PCR system detected L. monocytogenes in 17.8% of the 531 samples, compared to 16.0% positive-culture results. Sensitivity, specificity, and overall accuracy estimates for the PCR system are shown in Table 1. Thirteen samples that gave initial BAX system false-negative results were retested using the resuspended frozen enrichment culture pellets. Six samples gave positive results upon retesting, improving the sensitivity of the PCR system (Table 1). Twenty-nine samples gave BAX system false-positive results compared to the initial culture-based screening results. Subsequent hlyA PCR screening of up to 16 additional isolates per sample confirmed the presence of L. monocytogenes in 12 of these samples, thus improving the specificity of the PCR system (Table 1). While there was no apparent correlation between sample type and BAX system false-positive results, four of the seven BAX system false-negative samples were taken from finished products. The corrected BAX system sensitivity and specificity values reported in this study should be regarded as rough estimates, as only the discrepant results were further analyzed. Estimation of true corrected values would have required retesting of all samples.
TABLE 1.
Sensitivity and specificity estimates for the BAX for Screening/Listeria monocytogenes system using culture-based detection results as the standard of comparison
| BAX result | No. of samples with the indicated culture result after:
|
|||
|---|---|---|---|---|
| Initial testinga
|
Reexamination of discrepant resultsb
|
|||
| Positive | Negative | Positive | Negative | |
| Positive | 60 | 29 | 78 | 17 |
| Negative | 13 | 429 | 7 | 429 |
| Total | 73 | 458 | 85 | 446 |
Sensitivity, 82.2% (95% confidence interval, 71 to 91%); specificity, 93.7% (95% confidence interval, 92 to 96%); accuracy, 92.1%.
Corrected sensitivity, 91.8% (95% confidence interval, 84 to 98%); corrected specificity, 96.2% (95% confidence interval, 94 to 98%); corrected accuracy, 95.5%.
The BAX system detected L. monocytogenes in 27.7% of the 206 environmental samples taken throughout the processing areas (Table 2) (the data reflect prevalence following reexamination of discrepant results). Positive samples included those from floors and floor drains in raw material preparation areas, brining rooms, cold smokers, finished product processing areas, and food contact surfaces, including cutting tables and automated finished product slicer blades. L. monocytogenes was isolated from environmental samples with a lower frequency (21.4%) than was detected by the PCR-based system. Nine raw material samples (8.8%), including salmon, whitefish, and chub, were L. monocytogenes culture positive, compared to a slightly lower frequency using the PCR-based system. L. monocytogenes was detected in in-process samples with a slightly higher frequency (18.1%) by the PCR-based system. The prevalence of L. monocytogenes in finished products, including smoked sea bass, sablefish and cold-smoked salmon, was similar to that observed in raw materials. Additional Listeria species (L. monocytogenes hlyA PCR negative) were isolated from 54.1% of the 85 culture-positive samples.
TABLE 2.
Detection of L. monocytogenes in samples from the processing environment, raw materials, fish during the cold-smoking process, and cold-smoked fish
| Sample source | No. (%)
|
Total no. of samples | |
|---|---|---|---|
| BAX system positive | Culture positive | ||
| Environment | 57 (27.7) | 44 (21.4) | 206 |
| Raw materials | 8 (7.8) | 9 (8.8) | 102 |
| In process | 23 (18.1) | 21 (16.5) | 127 |
| Finished product | 7 (7.3) | 11 (11.5) | 96 |
Tracking L. monocytogenes in cold-smoked fish and in the smoked fish processing environment.
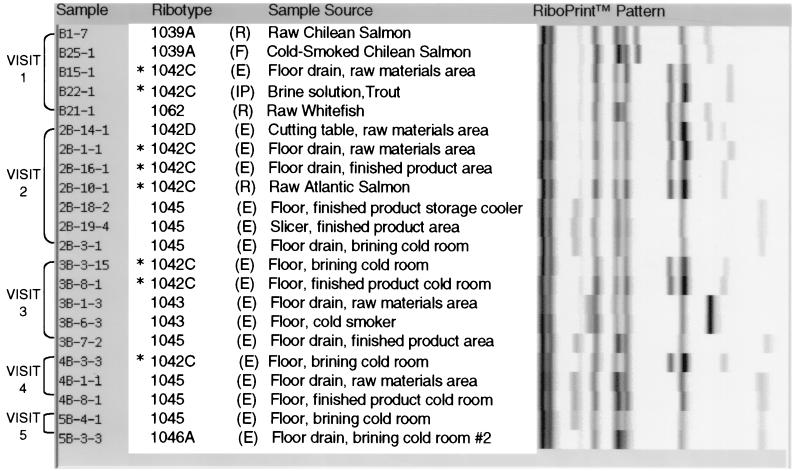
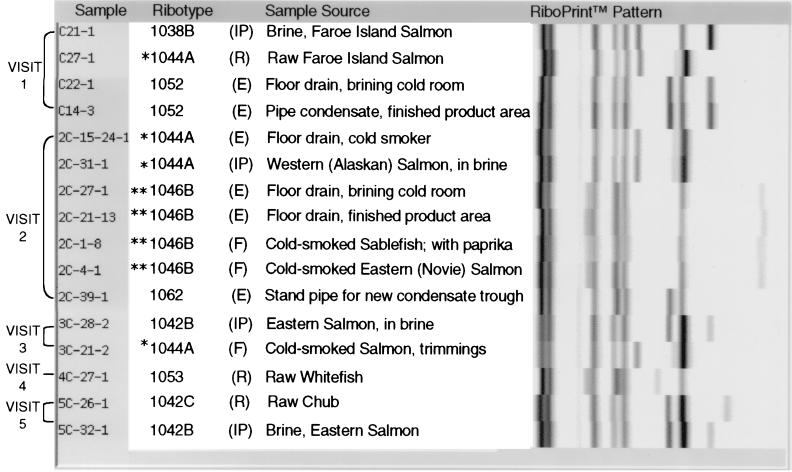
L. monocytogenes strain characterization by ribotyping identified 15 ribotypes among 85 isolates analyzed. The distribution of ribotypes among processing facilities, grouped by sampling visit, is shown in Fig. 1 to 3.
FIG. 1.
Ribotypes of L. monocytogenes isolates from processor B environmental, raw material, in-process, and finished product samples grouped by sampling visit. (E), environmental sample; (R), raw materials; (IP), in-process sample; (F), finished product. ∗, ribotype isolated most frequently from processor B samples.
FIG. 3.
Ribotypes of L. monocytogenes isolates from processor D environmental, raw material, in-process, and finished product samples grouped by sampling visit. (E), environmental sample; (R), raw materials; (IP), in-process sample; (F), finished product. ∗, ribotype isolated most frequently from processor D samples.
Seven different ribotypes were isolated from samples collected at processor B (Fig. 1). L. monocytogenes DUP-1042C, isolated from 36.4% of positive samples, predominated in the samples collected from this facility. This strain persisted in environmental samples collected over a period of 5 months and was also isolated from raw materials and in-process samples. L. monocytogenes DUP-1045 was isolated from 31.8% of positive processor B samples and persisted in environmental samples collected over 2.5 months. This ribotype was isolated only from the environment of processor B, was not isolated from processor C samples, and was isolated from only two in-process samples collected from processor D (Fig. 3). As shown in Table 3, the distribution of ribotypes DUP-1042C and DUP-1045 varied significantly by processing facility (P = 0.0003 and P = 0.0006, respectively).
TABLE 3.
Prevalence of L. monocytogenes ribotypes in each processing facility over five sampling visits
| Ribotype | % Prevalence
|
P valuea | ||
|---|---|---|---|---|
| Processor B (n = 129b) | Processor C (n = 173) | Processor D (n = 229) | ||
| 1039C | 0.0 | 0.0 | 10.0 | 0.0000 |
| 1042B | 0.8 | 1.2 | 0.4 | 0.8221 |
| 1042C | 6.2 | 0.6 | 0.4 | 0.0003 |
| 1044A | 0.0 | 2.3 | 3.1 | 0.1494 |
| 1045 | 5.4 | 0.0 | 0.9 | 0.0006 |
| 1046B | 0.0 | 2.3 | 0.0 | 0.0144 |
| 1053 | 0.0 | 0.6 | 1.7 | 0.2686 |
| 1062 | 0.8 | 0.6 | 2.6 | 0.1822 |
Two-sided exact P value computed for Pearson's chi-square test.
Total number of samples.
Eight different L. monocytogenes ribotypes were isolated from 16 of the samples collected at processor C (Fig. 2). L. monocytogenes DUP-1044A and DUP-1046B were isolated with the highest frequency (25.0% each) from positive samples. L. monocytogenes DUP-1044A persisted in samples collected from this facility over 4 months and was not isolated from processor B samples. The distribution of ribotype DUP-1044A, however, was not shown to vary significantly by processor (Table 3). The distribution of ribotype DUP-1046B, isolated only from samples collected at processor C, was shown to vary significantly among processing facilities (P = 0.0144) (Table 3).
FIG. 2.
Ribotypes of L. monocytogenes isolates from processor C environmental, raw material, in-process, and finished product samples grouped by sampling visit. (E), environmental sample; (R), raw materials, (IP), in-process sample, (F), finished product. ∗ and ∗∗, ribotypes isolated most frequently from processor C samples.
Nine L. monocytogenes ribotypes were isolated from 47 of the samples collected at processor D (Fig. 3). L. monocytogenes DUP-1039C, isolated from 48.9% of positive processor D samples, persisted in the processing environment over all sampling visits (6 months). This strain was not isolated from raw material samples collected at processor D or from any samples collected from processor B or C. The distribution of L. monocytogenes DUP-1039C was shown to vary significantly by processing facility (P = 0.0000) (Table 3). While L. monocytogenes ribotypes DUP-1044A and DUP-1062 also persisted in samples collected from processor D over a 3-month period, the distribution of these ribotypes did not vary significantly by processing facility (Table 3).
As preliminary analyses indicated that the type of sample collected varied significantly (P = 0.0001) according to processing facility, we suspected that the apparent difference in ribotype distribution among processors could be attributed to the prevalence of a given ribotype in a specific sample type. The distribution of each ribotype, however, was not shown to vary significantly by sample type (P > 0.05).
DISCUSSION
L. monocytogenes is a food-borne bacterial pathogen that is ubiquitous in nature and shows the ability to persist in the food processing environment for prolonged time periods. Control of this organism is thus extremely challenging for the food industry. Development of a better understanding of the ecology of L. monocytogenes in food processing plants in combination with rapid, standardized detection and typing systems will provide critical tools and knowledge for the development and verification of improved control strategies.
In-plant L. monocytogenes contamination patterns.
L. monocytogenes was detected in a variety of environmental, raw material, in-process, and cold-smoked fish samples (Table 2) collected from three smoked fish processing plants. We isolated L. monocytogenes from 11.5% of finished product samples, consistent with the prevalence range reported by previous surveys (6, 21, 27). As L. monocytogenes is a common contaminant of the food processing environment (6, 16), we were not surprised that this organism was isolated from over 20% of our environmental samples.
To specifically investigate L. monocytogenes contamination patterns and strain progressions in these plants, all isolates were characterized by molecular subtyping. Our subtyping data, in combination with statistical analysis, revealed a unique L. monocytogenes contamination pattern for each processing plant. Different ribotypes predominated in the samples collected from each processor, and specific ribotypes persisted in samples collected over time (Fig. 1 to 3). In support of a unique contamination pattern for each processor, the distributions of L. monocytogenes ribotypes DUP-1039C, DUP-1042C, DUP-1045, and DUP-1046B were shown to vary significantly by processing facility (Table 3) but not according to sample type. The results for processor D provide the most striking illustration, as L. monocytogenes DUP-1039C persisted in the processing environment over the 6-month sampling period and was isolated from nearly half of the culture-positive samples collected at this facility (Fig. 3). This ribotype was not isolated from any samples collected from processors B and C. In contrast, a different subtype (DUP-1042C) persisted in the environment of processor B over a period of 5 months (Fig. 1).
The persistence of L. monocytogenes DUP-1042C and DUP-1039C in the environments of processors B and D, respectively, for at least 5 months suggests that these strains are a part of the resident microflora and are not eliminated by current cleaning and sanitation programs. Previous reports have shown that L. monocytogenes has the ability to adhere to surfaces and colonize the food processing environment (22, 28, 29, 31). Once these populations are established, they appear to become more resistant to cleaning and sanitation measures (17, 28, 32). The affected processing areas, therefore, can serve as primary sources of finished product contamination. Our results show that ribotyping provides an effective means for monitoring the persistence of resident populations of L. monocytogenes in the processing environment and facilitates a better understanding of the ecology of this organism in food processing facilities. With this information available, the processor can develop and implement specific programs to eliminate resident populations and minimize the potential for L. monocytogenes colonization.
The predominance of specific L. monocytogenes subtypes over time is consistent with findings of previous investigations of contamination patterns in a variety of food processing environments, including those for smoked fish, poultry, meat, and dairy foods (3, 29, 33, 37). The persistence of specific strains in a variety of processing environments, along with the isolation of genetically diverse sporadic contaminants, suggests that some strains might have unique genetic characteristics conferring an enhanced ability to survive environmental stresses and colonize the food processing environment. In support of this hypothesis, a recent study indicated that L. monocytogenes strains differ in their ability to form biofilms (M. Chae and M. Schraft, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. P-104, p. 531, 1999). In a previous study evaluating the relationship between the resistance of specific strains to commercial sanitizers and their ability to persist in the processing environment, Earnshaw and Lawrence observed that while the sensitivity to sanitizers varied significantly among strains in suspension, there was not a significant difference between strains which persisted in the poultry processing environment over a 6-month period and sporadic isolates (14). Further studies will likely elucidate characteristics that contribute to the apparent enhanced ability of specific subtypes to colonize processing environments and resist cleaning and sanitation measures.
Potential sources of finished product contamination.
Molecular subtyping data also provided useful information regarding potential sources of specific L. monocytogenes strains isolated from finished product samples. Our subtyping data, in conjunction with our prevalence results, indicate that both raw materials and the processing environment serve as potential sources of finished product contamination. For example, DUP-1039A was isolated from processor B raw and cold-smoked Chilean salmon but not from the processing environment (Fig. 1, visit 1), indicating that raw materials likely served as the source of finished product contamination at the time of sampling. While results from a study by Eklund et al. also implicated raw fish as an important source of L. monocytogenes in the smoked fish processing environment and in finished products (15), other studies employing molecular typing methods concluded that raw materials may not serve as a significant source of finished product contamination (4, 37). Additional comprehensive studies monitoring contamination of raw fish and finished products using molecular subtyping methods are needed to clarify the importance of raw materials as a source of L. monocytogenes in finished products.
Analysis of ribotyping data also provided strong evidence in support of the processing environment as a significant source of finished product contamination. L. monocytogenes DUP-1046B, for example, was isolated from cold-smoked Nova salmon, cold-smoked sablefish, and two floor drain samples collected during the second visit to processor C (Fig. 2). As DUP-1046B was not isolated from any raw material samples collected from this facility over the 6-month sampling period, the processing environment most likely served as the source of this subtype in finished product samples. L. monocytogenes DUP-1039C, which persisted in the processing environment of processor D over the 6-month sampling period, was also isolated from in-process samples and from five of seven positive cold-smoked fish samples. DUP-1039C was not isolated from raw material samples (Fig. 3), strongly suggesting that the environment serves as the primary source of finished product contamination in this processing facility. Consistent with our results, previous studies employing discriminatory subtyping methods have identified the processing environment as a significant source of L. monocytogenes isolated from samples during processing and from finished products (4, 37). Rørvik et al. observed the persistence of a single electrophoretic type in the processing environment of a smoked salmon processing plant and in finished products, strongly suggesting that the environment served as a primary source of contamination (37). Similarly, analysis of PFGE subtyping data led Autio et al. to conclude that the contamination of cold-smoked rainbow trout was most closely linked to sites associated with brining and slicing (4).
Evaluation of a commercial PCR-based system for detection of L. monocytogenes.
Conventional culture methods for the isolation and identification of L. monocytogenes from food and environmental samples require a minimum of 4 to 5 days to complete (6, 13, 16). These methods may lead to false-negative results if mixed populations of Listeria species are present in a given sample, as most selective plating media do not allow visual differentiation of L. monocytogenes from other Listeria spp. (6). Rapid sample screening methods, including immunochemical, nucleic acid probe-based, and PCR-based assays, may overcome some of these limitations (5, 13, 23, 40). An increasing number of commercial PCR-based assays are available for the specific detection of L. monocytogenes, including the BAX for Screening system and the Probelia PCR system (Sanofi Diagnostics Pasteur). These assays have the potential to provide rapid and reliable molecular methods for monitoring of L. monocytogenes in the food industry.
We used the BAX for Screening/Listeria monocytogenes system for monitoring the presence of this organism in raw materials, in-process fish, cold-smoked fish, and environmental samples. Our results indicate a sensitivity of 91.8% for the BAX system (Table 1). It appears that our PCR system false-negative results were likely due to low levels of L. monocytogenes after sample enrichment or due to a high level of background microflora. Fewer than 3 × 102 Listeria-like colonies (<3 × 103 CFU/ml), below the system detection limit of 1 × 105 CFU/ml (40), were observed on the Oxford plates for six of the seven BAX system false-negative samples. The other sample appeared to contain Listeria in a high background of a mixed bacterial population. Further, pure-culture L. monocytogenes isolates from all false-negative samples tested BAX system positive, suggesting that the false-negative results were not due to the presence of nonreacting isolates. The employment of a two-step sample enrichment, as recommended by the manufacturer of the BAX for Screening/Listeria monocytogenes system, may increase the sensitivity of this system by facilitating recovery and growth of this organism. We used a single-step enrichment in LEB for this study based upon reports of comparable or enhanced recovery of L. monocytogenes from naturally contaminated seafood as compared to other enrichment protocols (6).
Our results indicated a specificity of 96.2% for the BAX system (Table 1). We believe that the BAX system false-positive results may be due to the specific detection of L. monocytogenes in a high background of other Listeria species. As additional Listeria spp. were observed in over half of our L. monocytogenes-positive samples, it is possible that this organism was overgrown by competing species during sample enrichment (resulting in culture-negative screening results). In agreement with our results, several researchers have observed that Listeria innocua can outcompete L. monocytogenes if the two species are grown together in commonly used enrichment media, including LEB (30, 35). It is unlikely that nonspecific priming contributed to the BAX system false-positive results, as this system has demonstrated 100% exclusivity for 60 Listeria spp. strains (non-L. monocytogenes) and 44 non-Listeria strains (BAX for Screening/Listeria monocytogenes package insert, Qualicon, Inc., Wilmington, Del.). Further, a recent report indicated that the presence of other Listeria species did not interfere with the detection of L. monocytogenes by the BAX system (40).
Conclusions.
Our results indicate that molecular detection and subtyping methods facilitate a better understanding of the ecology of food-borne pathogens in the food processing environment. This work further demonstrates the utility of molecular subtyping, specifically ribotyping, for tracking the sources and spread of food-borne pathogens. Ribotyping was applied in our study because this subtyping method is highly discriminatory, automated, and reproducible from one laboratory to another, thus facilitating rapid identification and data sharing among research groups. Although a single typing method can often elucidate contamination patterns, the use of multiple typing methods (20), multiple enzymes for ribotyping (19), or other typing methods (e.g., PFGE) (20) may increase discriminatory power and provide additional information on contamination patterns. It is also important to remember that molecular typing methods rely on culture-based isolation of L. monocytogenes. As it has been shown that different enrichment procedures may favor the growth of different subtypes and that multiple subtypes may be present in a given sample (38), the employment of more than one enrichment method and subtyping of more than one isolate per sample may facilitate further elucidation of contamination patterns.
A thorough understanding of strain persistence and progression will be crucial for improved control strategies for L. monocytogenes. Based on our results, we propose that sanitation protocols specifically targeting possible reservoirs of persistent strains may efficiently reduce environmental contamination. Sanitation control procedures are of particular importance for products that do not receive a heat treatment sufficient to kill food-borne pathogens, including L. monocytogenes. Routine environmental monitoring programs featuring molecular detection and subtyping methods will help to provide the necessary information regarding the origins and spread of contaminants to facilitate targeted control and sanitation strategies.
ACKNOWLEDGMENTS
We are indebted to the smoked fish processors who participated in this study. We thank Qualicon, Inc., Cornell Research Support Specialist Mary Bodis, and the members of the Food Safety Laboratory for their expert technical advice and assistance.
This paper is a result of research funded by the National Oceanic and Atmospheric Administration (award NA86RG0056 to the Research Foundation of State University of New York for New York Sea Grant).
REFERENCES
- 1.Anonymous. Code of Federal regulations. Vol. 60. Washington, D.C.: Office of the Federal Register; 1995. [Google Scholar]
- 2.Anonymous. Update: multistate outbreak of listeriosis—United States, 1998–1999. Morb Mortal Wkly Rep. 1999;47:1117–1118. [PubMed] [Google Scholar]
- 3.Arimi S M, Ryser E T, Pritchard T J, Donnelly C W. Diversity of Listeria ribotypes recovered from dairy cattle, silage, and dairy processing environments. J Food Prot. 1997;60:811–816. doi: 10.4315/0362-028X-60.7.811. [DOI] [PubMed] [Google Scholar]
- 4.Autio T, Hielm S, Miettinen M, Sjöberg A, Aarnisalo K, Björkroth T, Mattila-Sandholm T, Korkeala H. Sources of Listeria monocytogenes contamination in a cold-smoked rainbow trout processing plant detected by pulsed-field gel electrophoresis typing. Appl Environ Microbiol. 1999;65:150–155. doi: 10.1128/aem.65.1.150-155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassler H A, Flood S J A, Livak K J, Marmaro J, Knorr R, Batt C A. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben Embarek P K. Presence, detection and growth of Listeria monocytogenes in seafoods: a review. Int J Food Microbiol. 1994;23:17–34. doi: 10.1016/0168-1605(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 7.Boerlin P, Boerlin-Petzold F, Bannerman E, Bille J, Jemmi T. Typing Listeria monocytogenes isolates from fish products and human listeriosis cases. Appl Environ Microbiol. 1997;63:1338–1343. doi: 10.1128/aem.63.4.1338-1343.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruce J. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 1996;50:77–81. [Google Scholar]
- 9.Bruce J L, Hubner R J, Cole E M, McDowell C I, Webster J A. Sets of EcoRI fragments containing ribosomal RNA sequences are conserved among different strains of Listeria monocytogenes. Proc Natl Acad Sci USA. 1995;92:5229–5233. doi: 10.1073/pnas.92.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bsat N, Batt C A. A combined modified reverse dot-blot and nested PCR assay for the specific non-radioactive detection of Listeria monocytogenes. Mol Cell Probes. 1993;7:199–207. doi: 10.1006/mcpr.1993.1029. [DOI] [PubMed] [Google Scholar]
- 11.Dawson-Saunders B, Trapp R G. Basic and clinical biostatistics. 2nd ed. Norwalk, Conn: Appleton & Lange; 1994. Evaluating diagnostic procedures. [Google Scholar]
- 12.Destro M T, Leitao M F F, Farber J M. Use of molecular typing methods to trace the dissemination of Listeria monocytogenes in a shrimp processing plant. Appl Environ Microbiol. 1996;62:705–711. doi: 10.1128/aem.62.2.705-711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dever F P, Schaffner D W, Slade P J. Methods for the detection of foodborne Listeria monocytogenes in the U.S. J Food Safety. 1993;13:263–292. [Google Scholar]
- 14.Earnshaw A M, Lawrence L M. Sensitivity to commercial disinfectants, and the occurrence of plasmids within various Listeria monocytogenes genotypes isolated from poultry products and the poultry processing environment. J Appl Microbiol. 1998;84:642–648. doi: 10.1046/j.1365-2672.1998.00395.x. [DOI] [PubMed] [Google Scholar]
- 15.Eklund M W, Poysky F T, Paranjpye R N, Lashbrook L C, Peterson M E, Pelroy G A. Incidence and sources of Listeria monocytogenes in cold-smoked fishery products and processing plants. J Food Prot. 1995;58:502–508. doi: 10.4315/0362-028X-58.5.502. [DOI] [PubMed] [Google Scholar]
- 16.Farber J M, Peterkin P I. Listeria monocytogenes, a foodborne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank J F, Koffi R A. Surface-adherent growth of Listeria monocytogenes is associated with increased resistance to surfactant sanitizers and heat. J Food Prot. 1990;53:550–554. doi: 10.4315/0362-028X-53.7.550. [DOI] [PubMed] [Google Scholar]
- 18.Furrer B, Candrian U, Hoefelein C, Luethy J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J Appl Bacteriol. 1991;70:372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- 19.Gendel S M, Ulaszek J. Ribotype analysis of strain distribution of Listeria monocytogenes. J Food Prot. 2000;63:179–185. doi: 10.4315/0362-028x-63.2.179. [DOI] [PubMed] [Google Scholar]
- 20.Graves L M, Swaminathan B, Hunter S B. Subtyping Listeria monocytogenes. In: Ryser E T, Marth E H, editors. Listeria, listeriosis and food safety. 2nd ed. New York, N.Y: Marcel Dekker, Inc.; 1999. pp. 279–297. [Google Scholar]
- 21.Heinitz M L, Johnson J M. The incidence of Listeria spp., Salmonella spp., and Clostridium botulinum in smoked fish and shellfish. J Food Prot. 1998;61:318–323. doi: 10.4315/0362-028x-61.3.318. [DOI] [PubMed] [Google Scholar]
- 22.Herald P J, Zottola E A. Attachment of Listeria monocytogenes to stainless steel surfaces at various temperatures and pH values. J Food Sci. 1988;53:1549–1552. [Google Scholar]
- 23.Hill W E. The polymerase chain reaction: applications for the detection of foodborne pathogens. Crit Rev Food Sci Nutr. 1996;36:123–173. doi: 10.1080/10408399609527721. [DOI] [PubMed] [Google Scholar]
- 24.Hitchins A D. Bacteriological analytical manual. 8th ed. Gaithersburg, Md: AOAC International; 1995. pp. 10.01–10.13. [Google Scholar]
- 25.Hubner R J, Cole E M, Bruce J L, McDowell C I, Webster J A. Types of Listeria monocytogenes predicted by the positions of EcoRI cleavage sites relative to ribosomal RNA sequences. Proc Natl Acad Sci USA. 1995;92:5234–5238. doi: 10.1073/pnas.92.11.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacquet C, Catimel B, Brosch R, Buchrieser C, Dehaumont P, Goulet V, Lepoutre A, Veit P, Rocourt J. Investigations related to the epidemic strain involved in the French listeriosis outbreak in 1992. Appl Environ Microbiol. 1995;61:2242–2246. doi: 10.1128/aem.61.6.2242-2246.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jørgensen L V, Huss H H. Prevalence and growth of Listeria monocytogenes in naturally contaminated seafood. Int J Food Microbiol. 1998;42:127–132. doi: 10.1016/s0168-1605(98)00071-3. [DOI] [PubMed] [Google Scholar]
- 28.Krysinski E P, Brown L J, Marchisello T J. Effect of cleaners and sanitizers on Listeria monocytogenes attached to product contact surfaces. J Food Prot. 1992;55:246–251. doi: 10.4315/0362-028X-55.4.246. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence L M, Gilmour A. Characterization of Listeria monocytogenes isolated from poultry products and from the poultry-processing environment by random amplification of polymorphic DNA and multilocus enzyme electrophoresis. Appl Environ Microbiol. 1995;61:2139–2144. doi: 10.1128/aem.61.6.2139-2144.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDonald F, Sutherland A D. Important differences between the generation times of Listeria monocytogenes and Listeria innocua in two Listeria enrichment broths. J Dairy Res. 1994;61:433–436. doi: 10.1017/s0022029900030879. [DOI] [PubMed] [Google Scholar]
- 31.Mafu A A, Roy D, Goulet J, Magny P. Attachment of Listeria monocytogenes to stainless steel, glass, polypropylene, and rubber surfaces after short contact times. J Food Prot. 1990;53:742–746. doi: 10.4315/0362-028X-53.9.742. [DOI] [PubMed] [Google Scholar]
- 32.Mafu A A, Roy D, Goulet J, Savoie L, Roy R. Efficacy of sanitizing agents for destroying Listeria monocytogenes on contaminated surfaces. J Dairy Sci. 1990;73:3428–3432. doi: 10.3168/jds.S0022-0302(90)79040-6. [DOI] [PubMed] [Google Scholar]
- 33.Nesbakken T, Kapperud G, Caugant D A. Pathways of Listeria monocytogenes contamination in the meat processing industry. Int J Food Microbiol. 1996;31:161–171. doi: 10.1016/0168-1605(96)00978-6. [DOI] [PubMed] [Google Scholar]
- 34.Ojeniyi B, Wegener H C, Jensen N E, Bisgaard M. Listeria monocytogenes in poultry and poultry products: epidemiological investigations in seven Danish abattoirs. J Appl Bacteriol. 1996;80:395–401. doi: 10.1111/j.1365-2672.1996.tb03234.x. [DOI] [PubMed] [Google Scholar]
- 35.Petran R L, Swanson K M J. Simultaneous growth of Listeria monocytogenes and L. innocua. J Food Prot. 1993;56:616–618. doi: 10.4315/0362-028X-56.7.616. [DOI] [PubMed] [Google Scholar]
- 36.Ralyea R D, Wiedmann M, Boor K J. Bacterial tracking in a dairy production system using phenotypic and ribotyping methods. J Food Prot. 1998;61:1336–1340. doi: 10.4315/0362-028x-61.10.1336. [DOI] [PubMed] [Google Scholar]
- 37.Rørvik L M, Caugant D A, Yndestad M. Contamination pattern of Listeria monocytogenes and other Listeria spp. in a salmon slaughter-house and smoked salmon processing plant. Int J Food Microbiol. 1995;25:19–27. doi: 10.1016/0168-1605(94)00080-p. [DOI] [PubMed] [Google Scholar]
- 38.Ryser E T, Arimi S M, Marie-Claire Bunduki M, Donnelly C W. Recovery of different Listeria ribotypes from naturally contaminated, raw refrigerated meat and poultry products with two primary enrichment media. Appl Environ Microbiol. 1996;62:1781–1787. doi: 10.1128/aem.62.5.1781-1787.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salamina G, Dalle Donne E, Niccolini A, Poda G, Cesaroni D, Bucci M, Fini R, Maldini M, Schuchat A, Swaminathan B, Bibb W, Rocourt J, Binkin N, Salmaso S. A foodborne outbreak of gastroenteritis involving Listeria monocytogenes. Epidemiol Infect. 1996;117:429–436. doi: 10.1017/s0950268800059082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart D, Gendel S M. Specificity of the BAX polymerase chain reaction system for detection of the foodborne pathogen Listeria monocytogenes. J AOAC Int. 1998;81:817–822. [PubMed] [Google Scholar]
- 41.Winans S C, Rooks M J. Sensitive, economical laboratory photodocumentation using a standard video camera and thermal printer. BioTechniques. 1993;14:902. [PubMed] [Google Scholar]