Abstract
As both natural selection and population history can affect genome-wide patterns of variation, disentangling the contributions of each has remained as a major challenge in population genetics. We here discuss historical and recent progress towards this goal—highlighting theoretical and computational challenges that remain to be addressed, as well as inherent difficulties in dealing with model complexity and model violations—and offer thoughts on potentially fruitful next steps.
Keywords: natural selection, demography, population history, statistical inference, background selection, genetic hitchhiking
Significance.
While natural selection can shape patterns of sequence variation across the genome, changes in population size and structure can result in similar patterns. This has led to the development of sophisticated computational methods to infer parameters of both demography and purifying selection from population genomic data. Such approaches have been applied to many organisms revealing new insights about evolution. However, as the type of genomic data and evolutionary questions become more complex, new challenges have arisen. We here discuss recent progress on simultaneously inferring both selective and demographic parameters using polymorphism data, summarize the challenges involved, and highlight potentially useful future directions.
Glossary.
Approximate Bayesian Computation (ABC)—A class of computational methods based on Bayesian statistics for performing simulation-based inference; often used when the likelihood function does not exist or is too computationally expensive to evaluate.
Background selection (BGS)—The effects of purifying selection on linked sites.
Diffusion equations—Partial differential equations that describe the random movement of particles, and the process of random walks and frequency changes of alleles in finite populations.
Direct selection—Selection acting on variants that directly impact fitness.
Distribution of fitness effects (DFE)—The distribution of selection coefficients of new mutations.
Folded SFS—The distribution of frequencies of the minor allele (i.e. frequency ≤ 0.5) in a population sample.
Linkage disequilibrium (LD)—The non-random association of alleles at different genomic sites in a population.
Poisson random field (PRF) approach—A mathematical framework using diffusion theory to model variant frequencies in a population experiencing genetic drift and selection under the assumption of independence between sites (i.e. no linkage or interference among mutations). Under these assumptions, the number of fixed and polymorphic sites in a population can be modeled by independent Poisson distributions.
Selective sweep—The effects of positive selection on linked sites.
Site frequency spectrum (SFS)—The distribution of allele frequencies in a population sample.
Structured coalescent—A mathematical framework that models the genetic ancestry of samples in a population that is subdivided or compartmentalized (i.e. samples are no longer exchangeable, unlike in a panmictic population).
Summary statistic—A quantitative summary of the observed data.
Supervised machine learning—A subcategory of machine learning in which an example or test data set (where the input and output are known) is used to make predictions or inference.
Transition matrix—A method of calculating the exact distribution of allele frequencies under a specified model.
Two-epoch model—A model of single-population size change from ancestral size (N1) to current size (N2) at time τ.
Unfolded SFS—The distribution of frequencies of derived alleles in a population sample.
Wright–Fisher population—A randomly mating panmictic population consisting of individuals with discrete generations, such that new individuals are created by the random sampling of gametes with replacement from the previous generation.
Introduction
Accurately characterizing the demographic and selective histories of natural populations remains as a key aim of population genetics. Achieving this goal is not only vital for addressing specific evolutionary questions in a given species of interest (e.g. characterizing historical migration patterns in human populations, or identifying drug-resistance mutations in pathogens), but it is also essential for resolving larger-scale questions central to our understanding of evolution itself (e.g. why genetic variation varies relatively little across species with vastly different census sizes, also known as Lewontin's Paradox; see Charlesworth and Jensen 2022). Various summary statistics (terms in bold may be found in the Glossary) and inference approaches have been devised that seek to utilize the patterns of genetic variation observed from a sample of individuals to infer the type and extent of natural selection (reviewed by Nielsen 2005), as well as historical temporal and spatial changes in population size and structure (reviewed by Beichman et al. 2018). However, because selection and demography can lead to similar patterns of variation, distinguishing these selective from neutral processes is difficult, while at the same time fundamental (Jensen et al. 2019). For this reason, multiple approaches have recently been developed to co-estimate these neutral and selective parameters, and we here discuss theoretical and computational progress in using population genomic data to jointly infer population history with the distribution of fitness effects (DFE).
Brief Overview of Two-Step Inference Approaches and Caveats
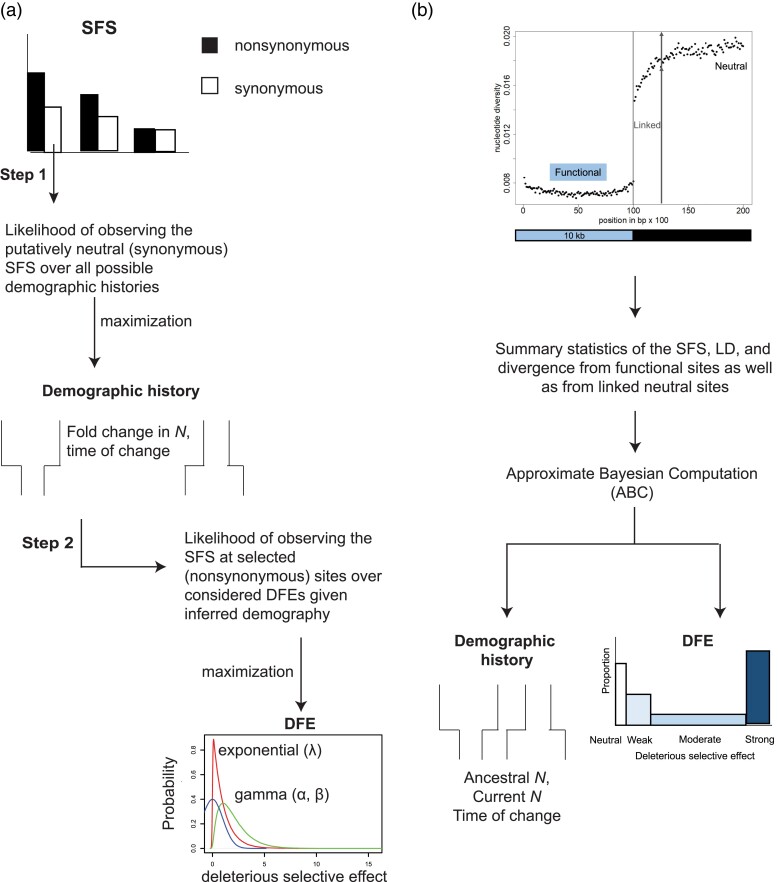
One common solution has been to infer these two sets of underlying parameters from different classes of sites — inferring population history from sites that are most likely to be neutral, and inferring selection from sites that are most likely to be functional (fig. 1a). We refer to this as the two-step approach. One of the first important breakthroughs in this regard was made by likelihood-based methods (Williamson et al. 2005; Keightley and Eyre-Walker 2007) that used the site frequency spectrum (SFS) at putatively neutral synonymous/noncoding sites to infer parameters of the underlying demographic history (Table 1). Conditional on the inferred demography, the SFS at putatively functional non-synonymous sites was used to estimate a DFE.
Fig. 1.
General workflow of methods that infer parameters of demography and selection employing different approaches: (a) a two-step approach, assuming independence between sites and (b) a simultaneous inference approach that accounts for linkage effects.
Table 1.
Details of current methods for inferring parameters of demography and selection from population genomic data
| Approach | Two-step approach using the W–F matrix | Two-step approach using diffusion approximations | SFS reweighting | Single-step joint inference using a Bayesian approach |
|---|---|---|---|---|
| Implementation/software | DFE-α | dadi and fitdadi | polyDFE; GRAPES; DoFE | Approximate Bayesian Computation (ABC) |
| Inference framework | Maximum likelihood | Maximum likelihood | Maximum likelihood | Approximate Bayesian |
| Data required | Single-population SFS of interdigitated neutral and selected sites | Single- or multi-population SFS of interdigitated neutral and selected sites | Single-population SFS of interdigitated neutral and selected sites; DoFE uses only folded SFS; GRAPES requires divergence as well | SFS-based and LD-based statistics from functional regions, and their flanking intergenic regions |
| Key differentiating assumptions |
|
|
|
|
| Parameters estimated |
|
|
|
|
| Computational time/complexity | Can be used for coding sites belonging to a few or all genes in the genome | Can be used for a large number of individuals (e.g. 1000) and sites | Can be used for coding sites belonging to a few or all genes in the genome | Can be used for hundreds of functional elements; whole genome inference would be computationally intensive |
| Relevant citations | Keightley and Eyre-Walker 2007; Schneider et al. 2011 | Williamson et al. 2005; Boyko et al. 2008; Kim et al. 2017; Huang et al. 2021 | Eyre-Walker et al. 2006; Galtier 2016; Tataru et al. 2017; Tataru and Bataillon 2019 | Johri et al. 2020; Johri et al. 2021 |
Such approaches yielded the first computational estimates of the DFE of new mutations in a number of organisms (Eyre-Walker and Keightley 2007), including both that of beneficial and deleterious mutations (Boyko et al. 2008; Eyre-Walker and Keightley 2009; Schneider et al. 2011), while better accounting for the confounding effects of demography. However, this first class of approaches suffered from two main limitations. Firstly, the demographic modeling involved a single panmictic population with a relatively simple population history that was approximated by a two-epoch model (Williamson et al. 2005; Keightley and Eyre-Walker 2007; Kousathanas and Keightley 2013). Secondly, these methods assume that all sites are independent and unlinked. However, substantial evidence has now accumulated suggesting that the effects of selection at linked sites may be widespread in genomes (reviewed by Cutter and Payseur 2013; Charlesworth and Jensen 2021). Specifically, the ever-present action of purifying selection results in background selection (BGS) effects on linked sites (Charlesworth et al. 1993), and the episodic action of positive selection may similarly result in selective sweep effects (Maynard Smith and Haigh 1974).
While recent studies have encouragingly found that the inference of selection on deleterious mutations might remain unbiased without accounting for linkage effects (Kim et al. 2017; Huang et al. 2021), the inference of demographic history can be severely biased by both BGS and selective sweeps (Messer and Petrov 2013; Nicolaisen and Desai 2013; Ewing and Jensen 2016; Schrider et al. 2016; Johri et al. 2021). Importantly for the two-step inference approaches, the demographic history of the population may thus be strongly mis-inferred in the first step by fitting a model that unknowingly encompasses the effects of selection on linked sites, which could in some situations result in the mis-inference of selection at directly selected sites in the second-step given that the demographic model is employed as a null expectation (e.g. to estimate the proportion and strength of beneficial mutations; Schneider et al. 2011).
In the two-step approach, the expected SFS is estimated by either explicitly calculating the probability density of mutations under a Wright–Fisher model using a transition matrix, or by approximating the change in allele frequencies using a diffusion equation. Namely, by approximating evolution with a continuous stochastic process, the partial differential equation describing the change in allele frequency forward in time (also known as the Kolmogorov forward equation) can be used to obtain the probability of observing a given SFS. As such, likelihoods can be obtained by numerically solving this equation for a transient (time-dependent) distribution of allele frequencies in a population (Williamson et al. 2005) to calculate expected allele frequencies with and without selection together with changing population size. Initially these methods could only accommodate a simple demographic model in which the population underwent a single instantaneous change in population size. However, the diffusion equation approach, assuming independently segregating sites [i.e. the Poisson random field (PRF) approach], has been extended to infer multiple changes in size for a single population, while also inferring the DFE (Boyko et al. 2008). Advances in likelihood-based approaches in which single-locus diffusion equations for multiple populations were solved numerically (Gutenkunst et al. 2009) allowed for further improvements in the inference of complex demographic histories with the DFE (Ma et al. 2013; Kim et al. 2017). Yet, solving the partial differential equations for single loci still assumes independence between sites, thereby neglecting the effects of linkage with selected sites (Table 1). The extent to which this is problematic will differ by organism, where genomes with high functional densities, stronger selection effects, and/or little or no recombination would be expected to exacerbate mis-inference. More generally however, two-step/two-class approaches require the existence of both a well-annotated genome such that functional regions are known, as well as a genome that is sufficiently recombining and functionally sparse such that neutral, unlinked sites exist at all. While these conditions may be met for certain large vertebrate and land-plant genomes, they currently exclude the great majority of species.
Brief Overview of Simultaneous Inference Approaches and Caveats
This variety of complicating issues has brought attention to the important need of developing inference approaches capable of jointly estimating parameters of selection and demography—that is, approaches that incorporate the direct and linked effects of selection that are applicable to genomic sites that may be shaped by both neutral and selective processes, and that perform simultaneous rather than step-wise inference. To account for the effects of selection on linked sites within an analytical framework, the diffusion equations approximating the two-locus Wright–Fisher model require a solution—a non-trivial challenge. Helpfully, Cvijovic et al. (2018) obtained analytical expressions for the SFS at linked neutral sites experiencing BGS for a non-recombining locus under demographic equilibrium. Furthermore, while general analytical solutions for even single-locus, single-population scenarios have not yet been obtained, Friedlander and Steinrücken (2022) recently described a numerical framework in which a system of ordinary differential equations can be solved to obtain the expected SFS and linkage disequilibrium (LD) around a selected region, for a Wright–Fisher two-locus model with mutation, recombination, selection, and changing population size. The development of future likelihood methods leveraging such a numerical approach may be utilized to jointly infer parameters of complex demographic histories with selection. Furthermore, while most advances in modeling the joint effects of selection and demography have been made using diffusion theory, the structured coalescent has also been employed and appears equally promising for small sample sizes (Zeng and Charlesworth 2011; Zeng 2013); however, the derivations of specific SFS statistics and likelihoods remain in need of further investigation.
On the computational front, recent progress has also been made using approximate Bayesian computation (ABC) approaches combined with forward simulations (fig. 1b). Johri et al. (2020) constructed an ABC method to jointly estimate parameters of arbitrary size change with the DFE of new deleterious mutations, while Sheehan and Song (2016) inferred the parameters of a population bottleneck jointly together with the comparatively rare processes of positive and balancing selection. Such simulation-based approaches appear promising, particularly as the effects of selection at linked sites can be directly modeled as modulated by the number of directly selected sites and locus-specific recombination rates of the region(s) under investigation (Johri et al. 2020). In addition, this class of methods can utilize multiple aspects of the data when performing estimation (e.g. the SFS, LD, and divergence) and avoids the assumption of neutrality on any class of sites (Table 1). However, forward simulations involving entire chromosomes remain extremely computationally expensive, particularly considering the large demographic and selective parameter spaces that must be investigated, and demographic modeling to date with direct and linked selection effects remains limited to relatively simple single-population size-change histories.
Alternative Methods to Infer Only the DFE and Caveats
An alternative approach to estimating the DFE under complex population histories is to assume that demography affects neutral and selected polymorphisms to the same extent, obviating the need to estimate demographic parameters (Eyre-Walker et al. 2006; Galtier 2016; Tataru et al. 2017; Tataru and Bataillon 2019). This is achieved by either reweighting the SFS (Eyre-Walker et al. 2006; James et al. 2016; Galtier 2016; Tataru et al. 2017; Tataru and Bataillon 2019) or by simply fitting DFE models to the ratio of the SFS at selected and neutral sites (James et al. 2016). Although, demography indeed affects neutral and selected polymorphisms to different extents (Otto and Whitlock 1997), assuming that it does not seems to yield reasonable estimates of the DFE under simple demographic models, even when linkage is strong (Eyre-Walker et al. 2006; Eyre-Walker and Keightley 2009). However, these methods are biased when there is a dramatic increase in population size (Eyre-Walker et al. 2006) and are not useful if the population history is itself also of interest.
With this brief overview of existing methodologies, we next discuss three of the most challenging issues that remain to be solved, together with our views on possibly fruitful paths forward.
Comparing the Relative Merits of Semi-Analytical and Forward Simulation-Based Approaches
Computational and statistical challenges remain for both inference approaches based on diffusion approximations and on forward simulations. Under a diffusion approximation, Friedlander and Steinrücken (2022) and Ragsdale (2021) demonstrated efficient numerical approaches for calculating the statistics of pairs of sites as noted above. Building the corresponding statistical inference framework, however, remains challenging. With no selection, inferring demography using a composite likelihood based on pairs of sites has been demonstrated (Ragsdale and Gutenkunst 2017). But modeling selection will require incorporating distinct types of pairs with different DFEs on each element of the pair, which may prove computationally prohibitive. More fundamentally, pairwise approaches cannot directly model the effects of multiple linked selected loci, which is most likely important, particularly in genomes with dense functional sites as discussed above. Hence, while analytical/numerical solutions are the ultimate goal, and when achieved are also the most efficient, such solutions remain elusive for many complex and biologically realistic scenarios of interest. For such models, simulation-based approaches remain necessary.
That being said, forward simulations are computationally intensive when population sizes are large. Guided by theory, parameter rescaling can be used to mimic the effects of selection in smaller simulated populations (Hoggart et al. 2007). But rescaling may introduce biases in certain scenarios (Uricchio and Hernandez 2014; Adrion et al. 2020), and the tradeoffs are not well understood. In addition, efficiently using the simulated data for inference is also a challenge. For instance, in ABC approaches, many simulation results are discarded because they do not match the observed data sufficiently. Supervised machine learning algorithms can potentially capture the information contained within a set of simulations under diverse parameter values more efficiently (reviewed in Schrider and Kern 2018). Such algorithms can be trained on summary statistics like ABC (Beaumont et al. 2002), but neural networks can also be trained directly on representations of sequence alignments (Flagel et al. 2019). They may thus capture more information, but it is unclear how to represent data with multiple classes of sites (such as synonymous and non-synonymous), and the resulting models may be difficult to interpret. Furthermore, machine learning classification approaches which neglect underlying uncertainty as well as constantly operating evolutionary processes (e.g. purifying selection) can be prone to serious mis-inference (e.g. as shown by Harris et al. 2018). However, adversarial methods, in which an established population genetic simulator is paired with a neural network trained to distinguish simulated from real data may offer easier interpretation (Wang et al. 2021). Moreover, recent advances in the inference of the full ancestral recombination graph from sampled sequences might help capture more information from sequence data (Kelleher et al. 2019; Speidel et al. 2019), potentially improving the ability to disentangle signatures of demography and selection.
Dealing with Model Complexity and Uncertainty
The joint inference of selection and demography comes with the statistical challenge of both accurately inferring multiple parameters within the context of parameter-rich models, and more generally of identifying potentially explanatory models in the first place (i.e. which models are worth investigating/fitting for comparison). Hence, the two-step approach of first inferring demography from putatively neutral sites, followed by inferring selection from selected sites (fig. 1a), has certain advantages in this regard. Firstly, conditioning on the demographic model when inferring the DFE reduces the total number of parameters to be inferred simultaneously, simplifying the inference problem. Secondly, the use of interdigitated putatively neutral sites for this purpose may provide a control for the effects of selection on linked sites, at least to some extent. For example, while the PRF approach assumes independence among sites (Sawyer and Hartl 1992)—neglecting interference among variants—the deleterious variants and interdigitated putatively neutral variants are on the same underlying genealogy. Simulation studies have shown that correct DFE inference can be obtained when these BGS effects are neglected, even when the true demography is more complex than the model used for inference (as will always be the case in natural populations). Specifically, Kim et al. (2017) simulated data with cryptic population structure and limited recombination, where the true demography was that of a population that expanded and split into eight sub-populations. Individuals from the eight sub-populations were then pooled together as a single population for inference, mimicking what might be done in practice. By fitting a population size change model to the synonymous SFS and using that incorrect demographic model for inference of the DFE, they found that unbiased estimates of the DFE parameters were still obtained. However, if the demographic history is itself of interest, this inference may be strongly biased by these neglected BGS effects, as noted previously (Ewing and Jensen 2016; Johri et al. 2021).
There are several additional limitations of note, which suggest opportunities for future work. First, the choice of the putatively neutral class is not always obvious. Synonymous mutations could themselves be under selection (Hershberg and Petrov 2008; Plotkin and Kudla 2011; Ragsdale et al. 2018; Machado et al. 2020), and under this scenario mis-inference may be severe (Johri et al. 2021), although the topic remains in need of additional investigation. When trying to infer the DFE for noncoding mutations in putatively functional regions of the genome, the choice of neutral sites is even more elusive. This highlights the value of joint-inference approaches which do not require an a priori definition of neutral sites (Johri et al. 2020)—however, these approaches need to be extended to more complex demographic scenarios as discussed above, insofar as they are currently restricted to single size-change models and assume panmictic populations (Table 1). It remains to be explored how population structure with migration may bias DFE inference and what assumptions might be appropriate to model the differences in DFEs between populations (Huang et al. 2021). Moreover, when modeling the effects of selection genome-wide, current studies generally group the fitness effects of new mutations at non-synonymous, synonymous and regulatory sites into a single distribution, which may not be a reasonable assumption. Existing models of selection based on a DFE also assume that selection coefficients are constant over time; and while limited inference approaches have been explored for characterizing temporally changing selective effects, they have been on the scale of individual mutations rather than full DFEs and rely on time-sampled data (e.g. Shim et al. 2016).
Finally, the impact of even subtle mis-specifications of the underlying models are likely to be exacerbated as the size of the datasets used for inference increase. For example, not accurately modeling subtle recent population structure and human population growth is likely to matter more when considering samples in the thousands as opposed to a smaller sample size (e.g. < 500); neglecting to model multiple mutations at the same site additionally becomes more problematic with increasing sample size (Harpak et al. 2016). However, larger samples may also provide an opportunity to better address linkage effects. Indeed, the recent TopMed (Trans-Omics for Precision Medicine) study found that as the sample sizes grew to >3000 individuals, estimates of recent population growth from synonymous variants approached estimates for putatively neutral (i.e. identified after removing sites that were phylogenetically conserved, potentially linked to regions experiencing selective sweeps, and so on; Torres et al. 2018) and unlinked variants (Taliun et al. 2021). For populations for which this is feasible, future sampling studies may allow for more accurate inference of population history from synonymous sites, which in turn might allow for more accurate inference of parameters of the DFE.
The Importance of Fine-Scale Mutation and Recombination Rate Heterogeneity
The above discussed methods attempting to jointly infer the DFE with demography typically assume that the mutation and recombination rates are uniform across the genomic region being considered. However, in reality both vary in ways that are potentially challenging. The mutation rate is known to vary at a variety of spatial scales from the single nucleotide to the chromosomal level (reviewed in Hodgkinson and Eyre-Walker 2011; Pfeifer 2020). For example, it has long been known that the mutation rate of a site depends on the adjacent nucleotides (Gojobori et al. 1982); this is best exemplified by the dinucleotide CG in mammals, which has an elevated mutation rate because of the epigenetic methylation of the cytosine, which tends to spontaneously deaminate to thymine (Coulondre et al. 1978). Variation in the mutation rate between sites is challenging for methods that employ two or more categories of sites to make inferences about the DFE and demography, because the mutation rate is expected to vary systematically between categories of sites for two reasons. First, natural selection tends to preserve hypermutable sites that otherwise dissipate if the mutation is neutral, and this leads to higher mutation rates in selected regions (Schmidt et al. 2008; Michaelson et al. 2012). Second, the mutation rate appears to depend on rates of recombination (Pratto et al. 2014; Arbeithuber et al. 2015) and epigenetic marks (Francioli et al. 2015; Smith et al. 2018) that may differ systematically between selected and neutral sites that are spatially separated. This may cause problems for methods that employ the increase in diversity away from a selected region.
Variation in the mutation rate can also potentially have consequences for methods that use the unfolded SFS, because identifying derived alleles depends on estimating their ancestral state. There are three approaches to this problem; in the first, methods have been developed to estimate the ancestral state (Hernandez et al. 2007) or the SFS (Williamson et al. 2007; Keightley et al. 2016; Keightley and Jackson 2018) taking into account some form of variation in the mutation rate. However, these methods still do not control for all the influences of adjacent nucleotides on the underlying rate (Hwang and Green 2004; Aggarwala and Voight 2016), or variation that is independent of context (Hodgkinson et al. 2009; Johnson and Hellmann 2011; Harpak et al. 2016). Uncorrected mutation rate variation may lead to the mis-inference of the SFS, and particularly impact the inference of high frequency polymorphisms. This pattern of mis-inference is likely to affect selected sites more than neutral sites, because highly mutable nucleotides are more likely to be preserved at the former. An alternative to the problem of mis-inference is to estimate the rate as a part of the method; this is potentially feasible given that the signature of mutation rate variation is different than that of selection when population sizes are stationary (Glémin et al. 2015). However, it remains possible that some of this signal may be mis-inferred as a part of the demographic model. Finally, if the demographic history is not of interest, it has been argued that the weighted SFS method will control much of this variation, provided that it is similar for neutral and selected sites (Galtier 2016).
The recombination rate is also known to vary between sites. As with the mutation rate, variation seems to occur over a variety of scales (reviewed in Peñalba and Wolf 2020). Most pertinent to the inference of demography and the DFE is fine-scale variation (see Dapper and Payseur 2017, 2018). In many organisms, recombination is concentrated in hotspots, whilst in others it appears to be more uniformly distributed (reviewed in Stapley et al. 2017). Recombination frequency also seems to depend on epigenetic marks in the genome (reviewed in Brachet et al. 2012), and these are likely to differ between genomic locations; as a consequence, regions subject to selection, such as a protein-coding sequence, may have a different recombination rate than those outside the sequence. The uneven distribution of recombination events will have two consequences. First, for all methods inferring parameters of selection, unaccounted for recombination rate variation will downwardly bias error estimates, and second, for methods that consider diversity as a function of the distance from a selected region, it will lead to biased parameter estimates, unless correctly modeled. Importantly however, the impact of rate heterogeneity and uncertainty on downstream inference can be quantified in any given empirical application using simulated data, though future method development will ultimately need to tackle the incorporation of realistic mutation and recombination rate maps directly into inference procedures.
Concluding Thoughts
While significant progress has been made in developing statistical approaches to jointly infer demography and selection—both in the development of analytical frameworks, as well as simulation-based inference procedures—much work still remains to extend existing approaches to account for various simultaneously acting evolutionary processes at a genome-wide level and to include more complex/realistic evolutionary scenarios. For species with gene-sparse and sufficiently recombining genomes, where unlinked neutral sites can be accurately identified, current two-step approaches can be employed confidently with careful analysis and an evaluation of possible model violations. However, for organisms with complex life history traits (e.g. non-standard mating systems), limited recombination, and/or compact genomes, the utilization of simultaneous inference methods will be essential (Johri et al. 2022b). Indeed, disentangling the effects of selection and demography is more complicated in organisms characterized by genomes with high functional densities, stronger selective effects, and/or little or no recombination, such that genetic hitchhiking effects may be pervasive genome-wide (i.e. as with the effects of population history). Complicating the matter, non-model organisms with poorly annotated genomes and uncharacterized recombination rate maps add another layer of uncertainty in even quantifying these potential effects.
Yet, the statistical identifiability of models, and parameters within those models, remains a fundamentally important consideration in all scenarios. As the model space to be explored is so much larger than the limited information in sequence variation within individuals, it is inherently difficult to distinguish between competing models. While utilizing the variety of newly developed approaches here discussed may help distinguishing between certain models, comprehensively evaluating competing models and following a probabilistic approach of assigning posterior probabilities to each should prove useful (Gelman and Shalizi 2013). Importantly, instead of exploring the parameter space by selecting models based on the preference of the authors’ narrative, a more principled alternative involves beginning with a baseline model and comparing that against a series of nested alternative models. Specifically, by constructing a baseline evolutionary model for each species and population under study, one may quantify the extent to which hypothesized evolutionary processes (e.g. selective sweeps) may be distinguished and quantified on top of commonly acting processes (e.g. purifying and background selection) (see Johri et al. 2022a, 2022b). Helpfully, with the advent of highly efficient forward simulation software (Thornton 2014; Haller and Messer 2019), this type of model evaluation and comparison is now feasible. By quantifying the variety of scenarios that are capable of explaining the observed data, one creates a useful template for future experimentation, sampling, and statistical method development that may better differentiate and dissect potential evolutionary explanations.
Acknowledgments
This work was funded by National Institutes of Health grants R35GM139383 to J.D.J., R01GM127348 to R.N.G., and R35GM119856 to K.E.L.
Contributor Information
Parul Johri, School of Life Sciences, Arizona State University, Tempe, AZ, USA.
Adam Eyre-Walker, School of Life Sciences, University of Sussex, Brighton, UK.
Ryan N Gutenkunst, Department of Molecular and Cellular Biology, University of Arizona, Tucson, AZ, USA.
Kirk E Lohmueller, Department of Ecology and Evolutionary Biology, University of California, Los Angeles, CA, USA; Department of Human Genetics, University of California, Los Angeles, CA, USA.
Jeffrey D Jensen, School of Life Sciences, Arizona State University, Tempe, AZ, USA.
Literature Cited
- Adrion JR, et al. 2020. A community-maintained standard library of population genetic models. eLife 9:e54967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwala V, Voight BF. 2016. An expanded sequence context model broadly explains variability in polymorphism levels across the human genome. Nat Genet. 48:349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeithuber B, Betancourt AJ, Ebner T, Tiemann-Boege I. 2015. Crossovers are associated with mutation and biased gene conversion at recombination hotspots. Proc Natl Acad Sci USA. 112:2109–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. 2002. Approximate Bayesian Computation in population genetics. Genetics 162:2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beichman AC, Huerta-Sanchez E, Lohmueller KE. 2018. Using genomic data to infer historic population dynamics of nonmodel organisms. Annu Rev Ecol Evol Syst. 49:433–456. [Google Scholar]
- Boyko AR, et al. 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4:e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachet E, Sommermeyer V, Borde V. 2012. Interplay between modifications of chromatin and meiotic recombination hotspots. Biol Cell 104:51–69. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Jensen JD. 2021. The effects of selection at linked sites on patterns of genetic variability. Annu Rev Ecol Evol Syst 52:177–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Jensen JD. Forthcoming 2022. How can we resolve Lewontin's Paradox? Genome Biol Evol. (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134:1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274:775–780. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA. 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat Rev Genet. 14:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvijovic I, Good BH, Desai MM. 2018. The effect of strong purifying selection on genetic diversity. Genetics 209:1235–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapper AL, Payseur BA. 2017. Connecting theory and data in recombination rate evolution. Phil Trans R Soc B 372:20160469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapper AL, Payseur BA. 2018. Effects of demographic history on the detection of recombination hotspots from linkage disequilibrium. Mol Biol Evol. 35:335–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing GB, Jensen JD. 2016. The consequences of not accounting for background selection in demographic inference. Mol Ecol. 25:135–141. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. 2007. The distribution of fitness effects of new mutations. Nat Rev Genet. 8:610–618. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. 2009. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 26:2097–2108. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Woolfit M, Phelps T. 2006. The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics 173:891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L, Brandvain Y, Schrider DR. 2019. The unreasonable effectiveness of convolutional neural networks in population genetic inference. Mol Biol Evol. 36:220–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francioli LC, et al. 2015. Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 47:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander E, Steinrücken M. 2022. A numerical framework for genetic hitchhiking in populations of variable size. Genetics 220:iyac012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N. 2016. Adaptive protein evolution in animals and the effective population size hypothesis. PLoS Genet. 12:e1005774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Shalizi CR. 2013. Philosophy and the practice of Bayesian statistics: philosophy and the practice of Bayesian statistics. Br J Math Stat Psychol. 66:8–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S, et al. 2015. Quantification of GC-biased gene conversion in the human genome. Genome Res. 25:1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojobori T, Li W-H, Graur D. 1982. Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol. 18:360–369. [DOI] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5:e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller BC, Messer PW. 2019. SLiM 3: forward genetic simulations beyond the Wright–Fisher model. Mol Biol Evol 36:632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpak A, Bhaskar A, Pritchard JK. 2016. Mutation rate variation is a primary determinant of the distribution of allele frequencies in humans. PLoS Genet. 12:e1006489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RB, Sackman A, Jensen JD. 2018. On the unfounded enthusiasm for soft selective sweeps II: examining recent evidence from humans, flies, and viruses. PLoS Genet. 14:e1007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Williamson SH, Bustamante CD. 2007. Context dependence, ancestral misidentification, and spurious signatures of natural selection. Mol Biol Evol. 24:1792–1800. [DOI] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. 2008. Selection on codon bias. Annu Rev Genet. 42:287–299. [DOI] [PubMed] [Google Scholar]
- Hodgkinson A, Eyre-Walker A. 2011. Variation in the mutation rate across mammalian genomes. Nat Rev Genet. 12:756–766. [DOI] [PubMed] [Google Scholar]
- Hodgkinson A, Ladoukakis E, Eyre-Walker A. 2009. Cryptic variation in the human mutation rate. PLOS Biol. 7:e1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart CJ, et al. 2007. Sequence-level population simulations over large genomic regions. Genetics 177(3):1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, et al. 2021. Inferring genome-wide correlations of mutation fitness effects between populations. Mol Biol Evol. 38:4588–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DG, Green P. 2004. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci USA. 101:13994–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JE, Piganeau G, Eyre-Walker A. 2016. The rate of adaptive evolution in animal mitochondria. Mol Ecol. 25:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JD, et al. 2019. The importance of the Neutral Theory in 1968 and 50 years on: a response to Kern and Hahn 2018. Evolution 73:111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PLF, Hellmann I. 2011. Mutation rate distribution inferred from coincident SNPs and coincident substitutions. Genome Biol Evol. 3:842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri P, et al. 2021. The impact of purifying and background selection on the inference of population history: problems and prospects. Mol Biol Evol. 38:2986–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri P et al. 2022b. Recommendations for improving statistical inference in population genomics. PLoS Biol 20(5):e3001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri P, Charlesworth B, Jensen JD. 2020. Toward an evolutionarily appropriate null model: jointly inferring demography and purifying selection. Genetics 215:173–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri P, Stephan W, Jensen JD. 2022a. Soft selective sweeps: addressing new definitions, evaluating competing models, and interpreting empirical outliers. PLoS Genet. 18(2):e1010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Campos JL, Booker TR, Charlesworth B. 2016. Inferring the frequency spectrum of derived variants to quantify adaptive molecular evolution in protein-coding genes of Drosophila melanogaster. Genetics 203:975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177:2251–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Jackson BC. 2018. Inferring the probability of the derived vs. the ancestral allelic state at a polymorphic site. Genetics 209:897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher J, et al. 2019. Inferring whole-genome histories in large population datasets. Nat Genet 51:1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Huber CD, Lohmueller KE. 2017. Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. Genetics 206:345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A, Keightley PD. 2013. A comparison of models to infer the distribution of fitness effects of new mutations. Genetics 193:1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, et al. 2013. Population genomic analysis of ten genomes reveals a rich speciation and demographic history of orang-utans (Pongo pygmaeus and Pongo abelii). PLoS One 8:e77175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado HE, Lawrie DS, Petrov DA. 2020. Pervasive strong selection at the level of codon usage bias in Drosophila melanogaster. Genetics 214:511–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. 1974. The hitch-hiking effect of a favourable gene. Genet Res. 23:23–35. [PubMed] [Google Scholar]
- Messer PW, Petrov DA. 2013. Frequent adaptation and the McDonald–Kreitman test. Proc Natl Acad Sci U S A. 110:8615–8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JJ, et al. 2012. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151:1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaisen LE, Desai MM. 2013. Distortions in genealogies due to purifying selection and recombination. Genetics 195:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. 2005. Molecular signatures of natural selection. Annu Rev Genet. 39:197–218. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitlock MC. 1997. The probability of fixation in populations of changing size. Genetics 146:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalba JV, Wolf JBW. 2020. From molecules to populations: appreciating and estimating recombination rate variation. Nat Rev Genet. 21:476–492. [DOI] [PubMed] [Google Scholar]
- Pfeifer SP. 2020. Spontaneous mutation rates. In The Molecular Evolutionary Clock. Theory and Practice. Springer Nature. [Google Scholar]
- Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 12:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratto F, et al. 2014. DNA recombination. Recombination initiation maps of individual human genomes. Science 346:1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale AP. 2021. Can we distinguish modes of selective interactions using linkage disequilibrium?. bioRxiv. [Google Scholar]
- Ragsdale AP, Gutenkunst RN. 2017. Inferring demographic history using two-locus statistics. Genetics 206(2):1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale AP, Moreau C, Gravel S. 2018. Genomic inference using diffusion models and the allele frequency spectrum. Curr Opin Genet Dev. 53:140–147. [DOI] [PubMed] [Google Scholar]
- Sawyer SA, Hartl DL. 1992. Population genetics of polymorphism and divergence. Genetics 132:1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, et al. 2008. Hypermutable non-synonymous sites are under stronger negative selection. PLoS Genet 4(11):e1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Charlesworth B, Eyre-Walker A, Keightley PD. 2011. A method for inferring the rate of occurrence and fitness effects of advantageous mutations. Genetics 189:1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Kern AD. 2018. Supervised machine learning for population genetics: a new paradigm. Trends Genet 34(4):301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Shanku AG, Kern AD. 2016. Effects of linked selective sweeps on demographic inference and model selection. Genetics 204:1207–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan S, Song YS. 2016. Deep learning for population genetic inference. PLoS Comput Biol. 12:e1004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Laurent S, Matuszewski S, Foll M, Jensen JD. 2016. Detecting and quantifying changing selection intensities from time-sampled polymorphism data. G3 6:893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TCA, Arndt PF, Eyre-Walker A. 2018. Large scale variation in the rate of germ-line de novo mutation, base composition, divergence and diversity in humans. PLoS Genet. 14:e1007254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speidel L, Forest M, Shi S, Myers SR. 2019. A method for genome-wide genealogy estimation for thousands of samples. Nat Genet. 51:1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Feulner PGD, Johnston SE, Santure AW, Smadja CM. 2017. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Phil Trans R Soc B 372(1736):20160455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliun D, et al. 2021. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataru P, Bataillon T. 2019. polyDFEv2.0: testing for invariance of the distribution of fitness effects within and across species. Bioinformatics 35:2868–2869. [DOI] [PubMed] [Google Scholar]
- Tataru P, Mollion M, Glémin S, Bataillon T. 2017. Inference of distribution of fitness effects and proportion of adaptive substitutions from polymorphism data. Genetics 207:1103–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton KR. 2014. A C++ template library for efficient forward-time population genetic simulation of large populations. Genetics 198:157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R, Szpiech ZA, Hernandez RD. 2018. Human demographic history has amplified the effects of background selection across the genome. PLoS Genet. 14:e1007387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uricchio LH, Hernandez RD. 2014. Robust forward simulations of recurrent hitchhiking. Genetics 197:221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. 2021. Automatic inference of demographic parameters using generative adversarial networks. Mol Ecol Res 21:2689–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SH, et al. 2005. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc Natl Acad Sci U S A. 102:7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SH, et al. 2007. Localizing recent adaptive evolution in the human genome. PLoS Genet. 3(6):e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K. 2013. A coalescent model of background selection with recombination, demography and variation in selection coefficients. Heredity 110:363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Charlesworth B. 2011. The joint effects of background selection and genetic recombination on local gene genealogies. Genetics 189:251–266. [DOI] [PMC free article] [PubMed] [Google Scholar]