Abstract
The American White Ibis (Eudocimus albus) is a nomadic wading bird common to wetland habitats in the southeastern US. In south Florida, US, habitat depletion has driven many ibis to become highly urbanized. Although they forage in neighborhood parks, artificial wetlands, backyards, and golf courses, the majority continue to nest in natural wetlands, often in dense, mixed species colonies. Adults and juveniles commonly disperse thousands of kilometers to other breeding colonies along the Gulf and southeast Atlantic coasts, presenting the potential for close contact with humans, domestic animals, and other wild bird species. Historically, wading birds were not considered to be significant hosts for influenza A virus (IAV), yet as ibis regularly move among various human, domestic animal, and wildlife interfaces, their potential to be exposed to or infected with IAV deserves attention. We experimentally challenged wild-caught, captive-reared White Ibis (n=20) with IAV, tested wild White Ibis for IAV, and serologically tested wild White Ibis for antibodies to IAV. White Ibis were highly susceptible to experimental challenge with H6N1 and H11N9 IAVs, with cloacal shedding lasting an average of 6 d. All 13 infected birds seroconverted by 14 d postinfection as determined by microneutralization. In contrast, no birds challenged with H3N8 were infected. We tested 118 swabs and 578 serum samples from White Ibis captured in southeastern Florida for IAV infection and antibodies to IAV, respectively. Although no IAVs were isolated, 70.4% serum samples were antibody positive by blocking enzyme-linked immunosorbent assay (bELISA). Neutralizing antibodies to H1–H12 were detected in 96.0% of a subset of bELISA positive birds (n=196) and 81.0% tested antibody positive to two or more hemagglutinin subtypes, indicating that exposure to multiple IAVs is common. These results provide evidence that White Ibis are susceptible and naturally infected with IAV and may represent a component of the IAV natural reservoir system.
Keywords: Florida, influenza A virus, reservoir, serology, White Ibis
INTRODUCTION
Since the isolation of influenza A virus (IAV) in free-ranging waterfowl in 1972, extensive research and surveillance efforts have firmly established that wild bird species of two orders, Anseriformes and Charadriiformes, are important for maintaining IAVs (Slemons et al. 1974; Webster et al. 1992; Olsen et al. 2006). Although our understanding of temporal and spatial patterns of infection of birds in these orders has grown considerably, many questions pertaining to IAV maintenance and the potential contribution of other species within avian communities remain. To understand the full scale of the natural IAV reservoir, our research scope may need to be broadened to focus on the role of other species not conventionally recognized as IAV hosts (Caron et al. 2017).
A starting point is to consider species with life histories compatible with established IAV ecology and that have contact with known IAV reservoirs. One such species is the American White Ibis (Eudocimus albus). Like dabbling ducks, gulls, and shorebirds, ibis utilize aquatic habitats and nest and roost in dense colonies that can number as high as 100,000 pairs in a single colony (Frederick et al. 1996; Cook and Baranski 2019). Although not a true migratory bird, White Ibis are nomadic, undertake long-distance, unpredictable movements, and cohabit areas utilized by overwintering Anseriforme and Charadriiforme species (Heath et al. 2009). Although most White Ibis return to natural areas during the breeding season in the spring, a large portion of White Ibis in southern Florida, US have become highly urbanized, resulting in close contact with humans, peridomestic waterfowl, backyard poultry, and other avian species that utilize urban parks, golf courses, backyards, and zoos to forage (Hernandez et al. 2016; Murray et al. 2018). Furthermore, individual White Ibis may disperse as far as 1,600 km to other populations throughout the southeastern US (Frederick et al. 1996). Thus, White Ibis may serve to maintain, amplify, or disseminate IAVs throughout the region, while also presenting a potential threat to human and domestic animal health.
White Ibis are members of the Order Pelicaniformes, which also includes herons and egrets. Birds in this order have rarely been a focus of IAV surveillance, yet viral detections or serologic evidence of infections have been reported from this group of birds on all continents except Antarctica (Pfitzer et al. 2000; Ellis et al. 2004; Epstein et al. 2007). In addition, Grey Herons (Ardea cinerea) are one of the species from which viruses have been commonly isolated during recent highly pathogenic IAV outbreaks in Eurasia, which continue to threaten wild and domestic birds (Lee et al. 2017; Pohlmann et al. 2017; Woo et al. 2017).
Given their life history, investigating the potential for White Ibis to be exposed to and infected with IAVs was warranted. In this study, we hypothesized that White Ibis are susceptible and naturally infected with IAV, and that related wading birds may be an important component of the IAV natural reservoir system. We assessed this by 1) experimentally challenging wild-caught, captive-reared White Ibis with three IAVs; 2) testing wild-caught White Ibis for IAVs; and 3) testing sera from wild-caught White Ibis for antibodies to IAV.
MATERIALS AND METHODS
Experimental infection
All husbandry, procedures, and methods used in this study were approved by the University of Georgia (UGA) Institutional Animal Care and Use Committee (UGIACUC; Animal Use permit A201609-012-A1). White Ibis were acquired under Florida scientific collection permit LSSC-11-00119G and federal collection permit MB779238-2 through the Southeastern Cooperative Wildlife Disease Study, UGA. Twenty, approximately 2-wk-old nestlings were hand-caught in Broward County, Florida, US. They were transported to UGA under Georgia Department of Natural Resources Import permit 1000529276, where they were raised in confinement at UGA to approximately 7 mo old, then transferred to a biosafety level 2 facility, where they were assigned to one of three groups housed in separate rooms. The H3N8 and H6N1 groups each included five challenge birds with two control birds that had direct contact with challenge birds. The H11N9 group included four challenge birds and two control birds. Birds in each room were housed in large dog runs and had direct contact with each other. Water was provided in four, approximately 40-cm-diameter, round rubber tubs in each room, filled to a depth of approximately 7.5 cm. These were cleaned and refilled with fresh water a minimum of twice daily. Birds were fed a mixture of a commercially available pelleted diet (Mazuri Flamingo Breeder, PMI Nutrition International LLC, St. Louis, Missouri, USA), seafood (smelt, shrimps), bread, eggs, and supplements in platters twice daily. Platters were cleaned and sanitized twice daily. Cages were cleaned and sanitized once daily.
The three Mallard (Anas platyrhynchos)-origin, low pathogenic avian influenza viruses used in this study were A/mallard/MN/AI07-4724/2007 (H3N8), A/mallard/MN/AI09-4345/2009 (H6N1), and A/mallard/MN/AI08-3267/2008 (H11N9). Viruses were propagated by second passage in 9–11-d-old specific-pathogen-free, embryonated chicken eggs (ECEs). Viruses were titrated in ECEs and the 50% embryo infectious dose (EID50) was calculated (Reed and Muench 1938). On day 0, inocula were diluted in brain–heart infusion (BHI) media to the desired EID50, and back-titrations were performed in ECEs to confirm the titer. The calculated titers were 105.6 EID50/0.1 mL of H3N8, 106.2 EID50/0.1 mL of H6N1, and 106.2 EID50/0.1 mL of H11N9.
Birds were allowed to acclimate in the biosafety level 2 rooms for 10 d prior to inoculation. Three days prior to inoculation, blood was collected from each bird by jugular venipuncture at a total volume less than or equal to 1% of the individual’s body mass. All birds tested negative for antibodies to the IAV nucleoprotein (NP) by blocking enzyme-linked immunosorbent assay (bELISA; IDEXX Laboratories, Westbrooke, Maine, USA) performed according to the manufacturer’s instructions and using a serum neutralization cutoff value of 0.5. On day 0, challenge birds were inoculated via the choanal cleft with 0.1 mL of inoculum containing one of the three viruses listed. Control birds were inoculated via the choanal cleft with 0.1 mL of BHI media.
White Ibis were monitored at least twice daily for evidence of clinical signs. Cloacal and oropharyngeal swabs were collected immediately prior to inoculation and at 2, 4, 6, 8, 10, and 14 d postinoculation (DPI). All swabs were placed in separate tubes containing 2 mL of BHI media supplemented with antimicrobials and were kept on ice packs until long-term storage at −80 C. At 4 DPI, water from each tub (four per room) was sampled by saturating a sterile cotton-tipped applicator (Puritan Medical Products Company LLC, Guilford, Maine, USA) prior to the tub being cleaned. An additional serum sample was collected at 14 DPI, at which time all birds were humanely euthanized via carbon dioxide inhalation, followed by cervical dislocation.
Virus isolation from all swabs collected at 4, 10, and 14 DPI was attempted in 9–11-d-old ECEs (Webster et al. 2002). Extraction of RNA and molecular detection via matrix real-time reverse transcriptase PCR (RRT-PCR) was attempted on all swabs (Latorre-Margalef et al. 2017). Cycle threshold (Ct) values below 40 were considered positive. All sera were tested for antibodies to the NP by bELISA, and sera collected at 14 DPI were tested for neutralizing antibodies to H1–H12 by microneutralization (MN; Wong et al. 2016). Viruses used as antigens included A/mallard/New Jersey/AI12-4823/2012 (H1N1), A/mallard/Minnesota/AI08-2755/2008 (H2N3), A/mallard/Minnesota/AI10-2593/2010 (H3N8), A/mallard/Minnesota/AI10-3208/2010 (H4N6), A/mallard/Minnesota/AI11-3933/2011 (H5N1), A/mallard/Minnesota/Sg-00796/2008 (H6N1), A/mallard/Minnesota/AI08-3770/2009 (H7N9), A/mallard/Minnesota/SG-01048/2008 (H8N4), A/ruddy turnstone/Delaware/AI11-809/2011 (H9N2), A/mallard/Minnesota/SG-00999/2008 (H10N7), A/mallard/Minnesota/SG-00930/2008 (H11N9), and A/mallard/Minnesota/AI07-3285/2007 (H12N5). Viral subtypes H13 and H16 were not included in testing because appropriate viral titers cannot be achieved in Madin-Darby canine kidney cells (American Type Culture Collection, Manassas, Virginia, USA) with conventional techniques.
Virus isolation and serologic testing of wild White Ibis
White Ibis were live captured in Palm Beach and Martin counties in southern Florida in March 2010 (n=12); December 2012 (n=36); and July, August, and December 2013 (n=72), as part of a previous study (Hernandez et al. 2016); in March–April 2014 (n=120) under Florida scientific collection permit LSSC-11-00119C, federal collection permit MB779238-2, and UGIACUC Animal Use permit A2013 01-005-R3; and in October 2015–August 2017 (n=338) under Florida scientific collection permit LSSC1-11-100119G, federal collection permit MB779238-2, and UGIACUC Animal Use permit A2016 11-019-Y2-A1.
Birds were captured using nylon slip-knot leg lassos, modified manually operated flip traps, and mist nets with decoys (Murray et al. 2018). An oropharyngeal swab and either cloacal or fecal swab were collected and placed in a single vial containing 2 mL of BHI media supplemented with antimicrobials. These were kept on ice packs until being placed in long-term storage at −80 C. A subset of samples from March 2010 (n=60), August 2013 (n=21), and March 2014 (n=37) were later thawed and virus isolation was attempted in 9–11-d-old ECEs (Webster et al. 2002).
Blood samples were collected by jugular venipuncture at a total volume less than or equal to 1% of the individual’s body mass. Samples were kept on ice until centrifugation, within 8 h of collection. The plasma fraction was aliquoted and frozen, and later thawed and tested by bELISA, then refrozen. Samples from 2015–17 that were positive by bELISA were rethawed and tested for neutralizing antibodies to H1–12 by MN.
Statistical analyses
Birds were considered infected if positive virus isolation or RRT-PCR results were obtained from either cloacal or oropharyngeal swabs. The average duration of cloacal and oropharyngeal shedding for each viral challenge group was calculated by taking the mean number of days positive RRT-PCR results were obtained from challenged birds. The mean Ct value was calculated for oropharyngeal and cloacal samples from challenged birds in each group each day. For the purpose of determining the mean value, RRT-PCR results greater than 40 were assigned a value of 45.
Wild White Ibis plasma samples collected in February and March were designated spring; samples collected in June, July, and August were designated summer; and samples collected in October, November, and December were designated fall. For serum samples collected between 2015 and 2017, the proportion of urban land cover within a 650-m radius of each capture site was calculated using the 2016 Cooperative Land Cover (CLC version 3.2, Florida Fish and Wildlife Conservation Commission, Tallahassee, Florida, USA) map for the state of Florida in ArcGIS (Murray et al. 2018).
A Pearson’s chi-squared test was used to compare the number of bELISA positive samples between seasons. It was also performed to compare the prevalence of neutralizing antibodies for each subtype between seasons. The median proportion of urban land cover at capture sites was compared between bELISA-positive and bELISA-negative birds within each season using a nonparametric test (Wilcoxon rank-sum test). All calculations were performed in STATA 15.1 (StataCorp LP, College Station, Texas, USA).
RESULTS
Experimental challenge
No evidence of clinical disease was observed in any bird. All birds inoculated with H6N1 and H11N9 shed virus as detected by RRT-PCR and virus isolation, and no virus was detected from the H3N8 group (Table 1). Cloacal shedding lasted longer than oropharyngeal shedding for both infected groups, averaging 6 d for the H6N1 group and 5 d for the H11N9 (Fig. 1). For both groups, the inverse mean Ct value of oropharyngeal shedding peaked at 4 DPI, and the value for cloacal shedding peaked at 6 DPI.
Table 1.
Summary of infection results obtained by challenging White Ibis (Eudocimus albus) with influenza A viruses (IAV). White Ibis were challenged with H3N8, H6N1, or H11N9 IAV, or mock-inoculated and housed with challenge birds. Oropharyngeal and cloacal swabs were collected at 0, 2, 4, 6, 8, 10, or 14 d postinoculation and tested for evidence of viral shedding. Blood was collected at 14 d postinoculation and tested for the presence of antibodies. (Data shown as number positive/number in group.)
| Viral sheddinga |
Serologyb |
||||
|---|---|---|---|---|---|
| Virus | Challenge method | RRT-PCR | VI | bELISA | MN |
| H3N8 | Contact-exposure | 0/2 | 0/2 | 0/2 | 0/2 |
| Inoculated | 0/5 | 0/5 | 0/5 | 0/5 | |
| H6N1 | Contact-exposure | 2/2 | 2/2 | 1/2 | 2/2 |
| Inoculated | 5/5 | 5/5 | 3/5 | 5/5 | |
| H11N9 | Contact-exposure | 2/2 | 2/2 | 1/2 | 2/2 |
| Inoculated | 4/4 | 3/4 | 4/4 | 4/4 | |
Viral shedding as determined by matrix real-time reverse transcriptase PCR (RRT-PCR) and virus isolation (VI). A bird was considered positive by RRT-PCR if a cycle threshold value less than 40 was detected from oropharyngeal or cloacal swabs collected at any time point. Virus isolation was attempted on all swabs collected at 4, 10, and 14 d postinoculation.
Serologic results from sera collected at 14 d postinoculation as determined by blocking enzyme linked immunosorbent assay (bELISA) and microneutralization (MN).
Figure 1.
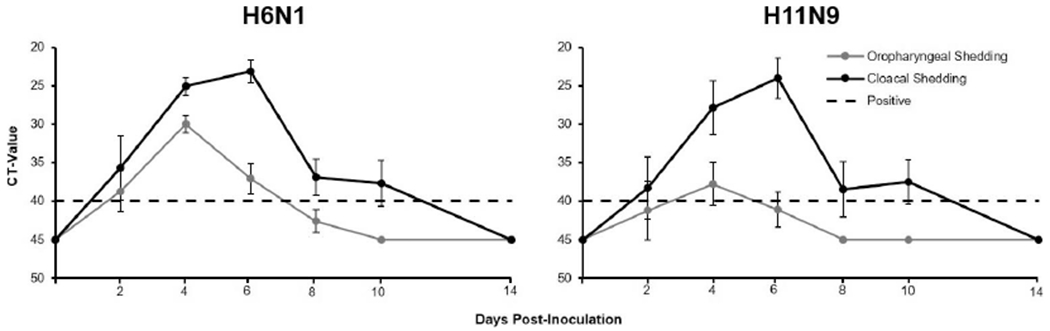
Mean (±SE) real-time reverse transcriptase PCR cycle threshold (CT) values of oropharyngeal and cloacal swabs collected from American White Ibis (Eudocimus albus). White Ibis were experimentally challenged with H3N8 (n=5), H6N1 (n=5), or H11N9 (n=4) influenza A virus and sampled at 2, 4, 6, 8, 10, and 14 d postinfection. Cycle threshold values less than 40 are considered positive. All values above 40 were assigned a value of 45. No evidence of viral shedding was detected from the group challenged with H3N8.
Viral shedding was also detected in control birds in the H6N1 and H11N9 groups beginning on day 4. Virus was detected by RRT-PCR in all water samples collected from the H6N1 and H11N9 challenge rooms. It was isolated from three of four water samples collected in the H6N1 room and two of four water samples from the H11N9 room. Virus was not detected or isolated from the four water samples collected in the H3N8 challenge room.
By 14 DPI, three of five inoculated birds and one of two control birds in the H6N1 group seroconverted, as determined by bELISA (Table 1). Neutralizing antibodies to H6 were detected in all birds in this group. Two birds also had neutralizing antibodies to the H1 antigen. All inoculated birds and one of two control birds in the H11N9 group seroconverted, as determined by bELISA. Neutralizing antibodies to H11 were detected in all birds in this group. Antibodies to other hemagglutinin subtypes were not detected. None of the birds in the H3N8 group seroconverted as detected by bELISA or MN.
Virus isolation and IAV antibodies in wild White Ibis
No viruses were isolated from the 118 samples collected from wild White Ibis. Antibodies to the NP were detected by bELISA in 70.4% (95% confidence interval [CI] 66.7–74.1) of serum samples collected from 578 wild birds. The seasonal NP antibody prevalence for summer, fall, and spring were 56.3% (95% CI 48.1–64.6), 68.5% (95% CI 61.2–75.7), and 78.8% (95% CI 74.0–83.7), respectively. The spring NP antibody prevalence was higher than the prevalence in summer (P<0.001) and fall (P=0.016) and the prevalence in the fall was higher than the summer (P=0.028). In bELISA-positive samples from 2015 to 2017 (n=198), neutralizing antibodies to one or more hemagglutinin subtypes were detected in 95.9% (95% CI 93.1–98.7; Fig. 2). Neutralizing antibodies to two or more subtypes were detected in 80.8% (95% CI 75.2–86.4) of these birds. Antibodies to H6, H12, H9, H5, and H1 were most commonly detected with prevalences ranging from 39.4% to 61.6%. Antibodies to H3, H4, and H8 were rarely or never detected (≤2.0%).
Figure 2.
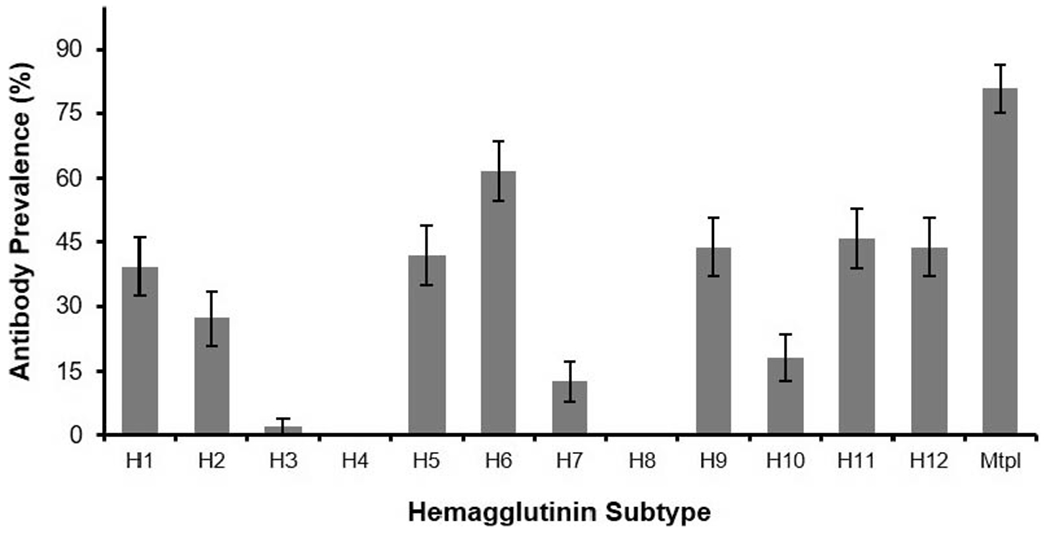
Prevalence of influenza A virus (IAV) neutralizing antibodies detected in serum samples from American White Ibis (Eudocimus albus) that tested positive for antibodies by blocking enzyme-linked immunosorbent assay (bELISA). Error bars represent 95% confidence interval. White Ibis were live captured in Palm Beach and Martin counties in southern Florida, USA in 2015–17. A total of 196 bELISA-positive samples were tested by microneutralization. The bar labeled Mtpl represents the percentage of samples that neutralized more than one IAV subtype. These samples are also represented in individual subtype columns.
The prevalence of H5 neutralizing antibodies in the summer and spring was 54.8% (95% CI 39.1–70.5) and 30.8% (95% CI 21.1–40.4) and was significantly different (P=0.008). The prevalence of H5 neutralizing antibodies in the fall was 49.2% (95% CI 36.7–61.7) and was significantly different from the fall (P=0.019) but not the summer (P=0.576). There was no significant difference in the prevalence of neutralizing antibodies by season for any other subtype. The median urban land cover proportion of capture sites for birds positive by bELISA in 2015–17 was higher than negative birds in the fall and spring (P=0.036 and P=0.002, respectively) but not the summer (P=0.188).
DISCUSSION
A pathogen reservoir can consist of a system of interconnected maintenance and non-maintenance populations (Haydon et al. 2002). Species in the Anseriformes and Charadriiformes are well-recognized components of the IAV reservoir system, yet we have a nascent understanding of how other species contribute to IAV maintenance dynamics. The unique life history of White Ibis, particularly those that exploit urban environments during their nonbreeding period, puts them in close contact with peridomestic and native waterfowl, gulls, and several other aquatic and semiaquatic avian species, some of which they would never contact in natural settings. Our results indicate that White Ibis can be experimentally infected with IAV and that a high proportion have serologic evidence of previous natural infections. Given their shift from natural foraging to urban living, White Ibis may become more significant to IAV epidemiology.
The relatively high antibody prevalence detected in wild White Ibis is comparable with those detected in a number Anseriformes and Charadriiformes species (e.g., Maxted et al. 2012; Hall et al. 2014; Johnson et al. 2014). Interpreting serology in field investigations is often confounded by incomplete information about antibody dynamics in a given species. Knowing the length of antibody persistence following exposure would be helpful in determining if the antibody profile we detected in the White Ibis population represents a continuous cycle of frequent exposures or if it reflects single, sporadic exposures that occur infrequently but remain detectable for many years, an important consideration, given White Ibis’ relatively long lifespan. Regardless, our experimental challenge data suggested that every White Ibis from which neutralizing antibodies were detected underwent a productive infection that, in an experimental setting, was sufficient to infect other individuals in close proximity.
We suspect that mock-inoculated birds in the H6N1 and H11N9 group became infected through IAV-contaminated water, as we isolated IAV in water samples at day 4, and fecal-contaminated water is considered a major route by which IAVs are transmitted in Anseriformes (Stallknecht et al. 2010). Shedding patterns in White Ibis would have been best characterized by performing viral titrations on fecal samples. Although we did not attempt this, the RRT-PCR Ct-values obtained from cloacal swabs were comparable and often much lower than values that were sufficient to infect Mallards (Brown et al. 2013). Furthermore, we observed no detectable change in behavior during infection, suggesting that IAV-infected wild White Ibis maintain the same movement, roosting, and foraging patterns, regardless of infection status.
Our failure to isolate IAV from free-ranging White Ibis may seem to undermine our hypothesis. However, our sampling effort was far from exhaustive, assuming infection dynamics within White Ibis have a spatial and temporal aspect, as is well documented in Anseriformes and Charadriiformes (Krauss et al. 2004). For example, Stallknecht et al. (1990) isolated only one IAV from 272 ducks sampled in December and January in coastal Louisiana, and there are numerous examples of failed efforts to detect virus in Ruddy Turnstones (Arenaria interpres) outside of the Delaware Bay hot spot, where most viral isolations are reported in this species (Krauss et al. 2010). Therefore, additional sampling from White Ibis and related species throughout the year in the southeastern US is warranted.
A continued area of interest in IAV ecology is the role of avian communities on wintering grounds in maintaining IAVs and driving their evolution. In North American wintering grounds, this may be a function of interactions between migrating birds and resident avian species such as Mottled Ducks (Anas fulvigula) in coastal Louisiana or egrets (Egretta sp.) and herons (Family Ardeidae) in California (Stallknecht et al. 1990; Siembieda et al. 2010). Although the overall prevalence of infections at these areas is much lower than in northern latitudes, a higher diversity of subtypes is typically isolated, suggesting that wintering grounds are an important place for IAV reassortment and evolution (Hill et al. 2012). Our findings that White Ibis are susceptible and have antibodies against various subtypes are consistent with this framework and provide a number of mechanisms by which White Ibis and related wading birds may contribute to IAV dynamics on wintering grounds. At the very least, they may serve to maintain IAVs in the short term while also amplifying the viral load in aquatic environments. In addition, the diversity of neutralizing antibodies in wild White Ibis indicates that coinfections are plausible, which is the well-recognized mechanism for IAV reassortment. Finally, there was a marked paucity of neutralizing antibodies to H3 and H4, which are the most common subtypes isolated from ducks in northern latitudes (Wilcox et al. 2011). Although this could be a function of White Ibis’ resistance to these subtypes, as demonstrated in our experimental challenge, it may also reveal a temporal mismatch. The vast majority of migrating waterfowl may clear infections with these subtypes by the time they reach wintering grounds, resulting in a void in which less common IAV subtypes can circulate in avian communities.
In the US, agricultural and urban development in the last century has been disproportionately intense in wintering areas, which may be resulting in a higher density of mixed species of avian communities in remaining aquatic habitats (Dahl 2011; Hill et al. 2012). Wetlands in Florida have been especially affected, yet White Ibis have been able to respond by utilizing urban areas to forage (Dahl 2005; Hernandez et al. 2016). A large portion of urbanized White Ibis shed Salmonella, which may have important health consequences to humans and the numerous gull and duck species with which they have close contact (Hernandez et al. 2016). Our findings indicated that White Ibis with previous exposure to IAV may use more urban areas during some portions of the year compared to White Ibis with no evidence of previous exposure. The underlying causes for this are unknown, but could reflect differences related to behavior or age structure within these subpopulations. Nonetheless, this study demonstrated that the humans, domestic animals, and peridomestic species in direct contact with White Ibis are epidemiologically linked to IAV ecology in altered wintering areas. It is important to acknowledge the potential disease risk this presents.
ACKNOWLEDGMENTS
We thank the numerous technicians, students, and volunteers that have helped with the ongoing fieldwork in Florida. We thank Laura Hollander and Alinde Fojtik for assistance with animal sampling. This project was partially funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract HHSN272201400006C and the National Science Foundation Ecology and Evolution of Infectious Diseases (DEB-1518611). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
LITERATURE CITED
- Brown JD, Poulson R, Carter DL, Lebarbenchon C, Stallknecht DE. 2013. Infectivity of avian influenza virus-positive field samples for mallards: What do our diagnostic results mean? J Wildl Dis 49:180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron A, Cappelle J, Gaidet N. 2017. Challenging the conceptual framework of maintenance hosts for influenza A viruses in wild birds. J Appl Ecol 54:681–690. [Google Scholar]
- Cook MI, Baranski M. 2019. South Florida wading bird report. Vol. 20. South Florida Water Management District, West Palm Beach, Florida, 59 pp. [Google Scholar]
- Dahl TE. 2005. Florida’s wetlands: an update on status and trends 1985 to 1996. US Department of Interior, Fish and Wildlife Service, Washington, DC, 80 pp. [Google Scholar]
- Dahl TE. 2011. Status and trends of wetlands in the conterminous United States 2004 to 2009. US Department of Interior, Fish and Wildlife Service, Washington, DC, 108 pp. [Google Scholar]
- Ellis TM, Bousfield RB, Bissett LA, Dyrting KC, Luk GSM, Tsim ST, Sturm-Ramirez K, Webster RG, Guan Y, Peiris JSM. 2004. Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Pathol 33:492–505. [DOI] [PubMed] [Google Scholar]
- Epstein JH, McKee J, Shaw P, Hicks V, Micalizzi G, Daszak P, Kilpatrick AM, Kaufman G. 2007. The Australian white ibis (Threskiornis molucca) as a reservoir of zoonotic and livestock pathogens. Eco-Health 3:290–298. [Google Scholar]
- Frederick PC, Bildstein KL, Fleury B, Ogden J. 1996. Conservation of large, nomadic populations of white ibis (Eudocimus albus) in the United States. Conserv Biol 10:203–216. [Google Scholar]
- Hall JS, Hallgrimsson GT, Suwannanarn K, Sreevatsen S, Ip HS, Magnusdottir E, TeSlaa JL, Nashold SW, Dusek RJ. 2014. Avian influenza virus ecology in Iceland shorebirds: Intercontinental reassortment and movement. Infect Genet Evol 28:130–136. [DOI] [PubMed] [Google Scholar]
- Haydon DT, Cleaveland S, Taylor LH, Laurenson MK. 2002. Identifying reservoirs of infection: A conceptual and practical challenge. Emerg Infect Dis 12:1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath JA, Frederick PC, Kushlan JA, Bildstein KL. 2009. White Ibis (Eudocimus albus). In: Birds of North America online, Version 2.0, Poole AF, editor. Cornell Lab of Ornithology, Ithaca, New York. 10.2173/bna.9. Accessed October 2018. [DOI] [Google Scholar]
- Hernandez SM, Welch CN, Peters VE, Lipp EK, Curry S, Yabsley MJ, Sanchez S, Presotto A, Gerner-Smidt P, Hise KB, et al. 2016. Urbanized White Ibis (Eudocimus albus) as carriers of Salmonella enterica of significance to public health and wildlife. PLoS One 11:e0164402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill NJ, Takekawa JY, Cardona CJ, Meixell BW, Ackerman JT, Runstadler JA, Boyce WM. 2012. Cross-seasonal patterns of avian influenza virus in breeding and wintering migratory birds: A flyway perspective. Vector Borne Zoonotic Dis 12:243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, DeCicco LH, Ruthrauff DR, Krauss S, Hall JS. 2014. Avian influenza virus antibodies in Pacific coast red knots (Calidris canutus roselaari). J Wildl Dis 50:671–675. [DOI] [PubMed] [Google Scholar]
- Krauss S, Stallknecht DE, Negovetich NJ, Niles LJ, Webby RJ, Webster RG. 2010. Coincident ruddy turnstone migration and horseshoe crab spawning creates an ecological “hot spot” for influenza viruses. Proc Biol Sci 277:3373–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Walker D, Pryor SP, Niles L, Chenghong L, Hinshaw VS, Webster RG. 2004. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis 4:177–189. [DOI] [PubMed] [Google Scholar]
- Latorre-Margalef N, Brown JD, Fojtik A, Poulson RL, Carter D, Franca M, Stallknecht DE. 2017. Competition between influenza A virus subtypes through heterosubtypic immunity modulates re-infection and antibody dynamics in the mallard duck. PLoS Pathog 13:e1006419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Sharshov K, Swayne DE, Kurskaya O, Sobolev I, Kabilov M, Alekseev A, Irza V, Shestopalov A. 2017. Novel reassortment clade 2.3.4.4 avian influenza A (H5N8) virus in wild aquatic birds, Russia, 2016. Emerg Infect Dis 23:359–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxted AG, Luttrell MP, Goekjian VH, Brown JD, Niles LJ, Dey AD, Kalasz KS, Swayne DE, Stallknecht DE. 2012. Avian influenza virus infection dynamics in shorebird hosts. J Wildl Dis 48:322–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MH, Kidd AD, Curry SE, Hepinstall-Cymerman J, Yabsley MJ, Adams HC, Ellison T, Welch CN, Hernandez SM. 2018. From wetland specialists to hand-fed generalist: shifts in diet and condition with provisioning for a recently urbanized wading bird. Philos Trans R Soc Lond B Biol Sci 373:20170100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus ADME, Fouchier RAM. 2006. Global patterns of influenza A virus in wild birds. Science 312:384–388. [DOI] [PubMed] [Google Scholar]
- Pfitzer S, Verwoerd DJ, Gerdes GH, Labuschagne AE, Erasmus A, Manvell RJ, Grund C. 2000. Newcastle disease and avian influenza a virus in wild waterfowl in South Africa. Avian Dis 44:655–660. [PubMed] [Google Scholar]
- Pohlmann A, Starick E, Harder T, Grund C, Höper D, Globig A, Staubach C, Dietze K, Strebelow G, Ulrich RG, et al. 2017. Outbreaks among wild birds and domestic poultry caused by reasserted influenza A(H5N8) clade 2.3.4.4 viruses, Germany, 2016. Emerg Infect Dis 23:633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. [Google Scholar]
- Siembieda JL, Johnson CK, Cardona C, Anchell N, Dao N, Reisen W, Boyce W. 2010. Influenza A viruses in wild birds of the Pacific flyway, 2005–2008. Vector Borne Zoonotic Dis 10:793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slemons RD, Johnson DC, Osborn JS, Hayes F. 1974. Type-A influenza viruses isolated from wild free-flying ducks in California. Avian Dis 18:119–124. [PubMed] [Google Scholar]
- Stallknecht DE, Goekjian VH, Wilcox BR, Poulson RL, Brown JD. 2010. Avian influenza virus in aquatic habitats: what do we need to learn? Avian Dis 54:461–465. [DOI] [PubMed] [Google Scholar]
- Stallknecht DE, Shane SM, Zwank PJ, Senne DA, Kearney MT. 1990. Avian influenza viruses from migratory and resident ducks of coastal Louisiana. Avian Dis 34:398–405. [PubMed] [Google Scholar]
- Webster R, Cox N, Storh K. 2002. WHO manual on animal influenza diagnosis and surveillance. World Health Organization, WHO Global Influenza Programme. Geneva, Switzerland, 105 pp. [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox BR, Knutsen GA, Berdeen J, Goekjian V, Poulson R, Goyal S, Sreevatsan S, Cardona C, Berghaus RD, Swayne DE, et al. 2011. Influenza A viruses in ducks in northwestern Minnesota: Fine scale spatial and temporal variation in prevalence and subtype diversity. PLoS One 6:e24010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JK, Wilcox BR, Fojtik A, Poulson RL, Stallknecht DE. 2016. Antibodies to influenza A viruses in wintering snow geese (Chen caerulescens) in Texas. Avian Dis 60:337–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C, Kwon JH, Lee DH, Kim Y, Lee K, Jo SD, Son KD, Oem JK, Wang SJ, Kim Y, et al. 2017. Novel reassortant clade 2.3.4.4 avian influenza A (H5N8) virus in a grey heron in South Korea in 2017. Arch Virol 162:3887–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]


