ABSTRACT
Objectives:
To estimate SARS-CoV-2 RNA clearance time among non-severe COVID-19 patients and explore factors associated with delayed negative conversion.
Methods:
A retrospective cohort study was conducted at the COVID-19 unit of a tertiary care center in the Western region of Saudi Arabia. Reverse transcriptase-polymerase chain reaction (RT-PCR) confirmed COVID-19 patients diagnosed between April 1 and June 30, 2020, were considered. The primary outcome was the time (days) from disease onset to first negative RT-PCR, which was analyzed using Kaplan–Meier and Cox regression survival methods. Demographic data, clinical history, baseline clinical, radiological and laboratory findings and management, and outcome data were collected and analyzed as factors associated with the viral RNA clearance time.
Results:
One hundred and forty-four patients were included. The mean (standard deviation [SD]) age was 36.93 (14.41) years, 50.7% were females, and 45.1% were healthcare workers. COVID19 was asymptomatic, mild and moderate in 11.1%, 71.5% and 17.4% of the participants, respectively. Fever (59.4%) and cough (58.0%) were the dominant onset symptoms. The mean viral RNA clearance time was 22.9 days (SD = 8.6; 95% confidence interval [CI] = 21.5–24.3 days). Extended clearance time was associated with older age (mean = 24.0 vs. 22.0 days; log-rank = 0.037), married status (23.2 vs. 22.6 days; log-rank = 0.021), working in health sector (24.2 vs. 21.8 days; log-rank = 0.006), and having a chronic disease (24.8 vs. 21.9 days; log-rank = 0.028), compared to their counterparts, respectively. In the adjusted model, the job sector was the only factor that was independently associated with clearance time. Non-healthcare sector showed hazard ratio 1.8 (95% CI = 1.3–2.7; log-rank = 0.002) with reference to healthcare sector.
Conclusion:
SARS-CoV-2 RNA clearance time is likely to be longer in non-severe COVID-19 patients, representing an additional risk for the virus dissemination among the community and calling for higher caution among the population.
Keywords: Clearance, COVID-19, healthcare, negative, RNA, RT-PCR, SARS-CoV-2
Introduction
Towards the end of 2019 and the start of 2020, the outbreak of a novel airborne viral pandemic, namely COVID-19 (coronavirus disease 2019), was identified from Wuhan, Hubei Province, China.[1] The massive impact of this pandemic on the world and the global economy is unprecedented due to the constant increase of deaths, the continuous panic, adverse effects of lockdown, and social-distancing measures imposed to control the spread of the pandemic.[2,3,4,5]
Saudi Arabia was one of the countries that implemented early preventive measures against the introduction and local spread of SARS-CoV-2 before the first case was declared in the country on March 2, 2020. Since then, control measures were revised regularly to adapt to the changes of the pandemic and mitigate its socioeconomic impact.[6,7]
The critical issue is to have a strategic plan to return to regular life after several months of the economic shutdown and the social activity.[8] As the cases are increasing, it is crucial to have a deeper insight into the infectiousness of convalescent individuals to plan their safe return to workplaces and classrooms. This helps in preventing the secondary spread of the virus along with alleviating staff shortages.
Therefore, understanding the natural history of the viral life cycle in the infected individual is of paramount importance. It is generally reported that the highest RNA titers are reached within 7 to 10 days of symptoms onset, during which the samples should be collected for molecular testing methods for best sensitivity to detect the infection.[9] Additionally, several studies have estimated the time between disease onset and negative conversion of Sars-CoV-2 RNA detection in molecular diagnostic methods, as a reflection of the infectiveness time, and analyzed the factors associated with prolonged viral RNA shedding.[10,11,12,13,14,15] However, due to discrepancies in the findings across these studies, it is necessary to obtain the local figures, notably among the individuals with asymptomatic and non-severe forms of the disease. These are the individuals who are at high risk of being undiagnosed or inadequately isolated representing the major vector of the virus dissemination.
This study aimed to estimate the time of Sars-CoV-2 RNA clearance among biologically confirmed non-severe COVID-19 patients and explore factors associated with delayed negative conversion.
Methods
Design and participants
A retrospective cohort study was conducted in patients who were diagnosed, managed, and followed up for confirmed COVID-19 between April 1, 2020, and June 30, 2020, at the COVID19 Unit of King Abdulaziz University Hospital, Jeddah, Saudi Arabia.
Case definition and eligibility criteria
Cases were defined as patients diagnosed using molecular methods including real-time reverse transcription-polymerase chain reaction (RT-PCR) analysis for SARS-CoV-2 in nasopharyngeal swabs, which was collected on the day of the patient presentation. Cases were eligible if they had at least two consecutive negative RT-PCR results. Only non-severe cases were included, whereas severe cases that resulted in the intensive care unit (ICU) admission, long-term hospitalization, death, or severe complications were excluded. Further, patients who were swabbed after the day of presentation and those diagnosed using other methods including imaging, serological tests or based on the clinical presentation and epidemiological context only were not included.
Data collection procedure
Eligible participants were identified using the hospital electronic registry for COVID-19, and the relevant study data were collected from their respective electronic medical files using a structured data collection sheet. Collected data included 1) demographic and professional data, including age, gender, marital status, professional status, sector (healthcare vs. non-healthcare), residency (cluster vs. outside cluster); 2) pre-COVID-19 health status, including lifestyle (sleep quality, smoking, physical activity, and dietary habits) and comorbidities (hypertension, diabetes, etc.); 3) parameters on presentation, including the date of first positive RT-PCR, onset symptoms, vital signs at the first consultation, symptoms duration before diagnosis, severity level at the worst clinical status (asymptomatic, mild or moderate), lung involvement, lab findings (leukocytes level, liver function, renal function), thrombosis; 4) management and outcome, including hospital admission, oxygen therapy, continuous positive airway pressure, antibiotic therapy, antiviral therapy, and ICU admission, and date of first negative RT-PCR.
Statistical methods
Data were collected in Microsoft Excel sheets, then coded and transferred to SPSS for statistical analysis, version 21 for Windows (SPSS Inc., Chicago, IL, USA). The primary outcome, viral RNA clearance time, was calculated as the time, in days, from the first positive RT-PCR to the first negative RT-PCR. Descriptive statistical methods were used to present the demographic and clinical parameters of the study population. Kaplan–Meier survival methods were carried out to analyze the factors associated with SARS-CoV-2 RNA clearance time; results were presented as mean (95% confidence interval [CI]) clearance time with the corresponding log-rank level. Multivariate Cox regression was used to analyze the independent factors of clearance time; results are presented as hazard ratio (HR), with 95% CI. A log-rank value <0.05 was considered for statistical significance.
Ethical clearance
Data were entered anonymously and coded in Excel sheets, and the database was only shared with trusted collaborators. The study was ethically approved by the institutional review board of King Abdulaziz University.
Results
Participants’ characteristics
One hundred and forty-four patients were included. The mean (standard deviation [SD]) age was 36.93 (14.41) years, 50.7% were females and 45.1% were healthcare workers. The pre-COVID-19 chronic diseases for these patients included hypertension and type 2 diabetes (14.6% each), and asthma and thyroid disease (4.2% each). Other demographic, lifestyle and medical history data are depicted in Table 1.
Table 1.
Participants’ demographics and baseline clinical characteristics (n=144)
| Parameter | Category | Frequency | Percentage |
|---|---|---|---|
| Age (y) | Mean, SD (range=1, 77) | 36.93 | 14.41 |
| Gender | Male | 71 | 49.3 |
| Female | 73 | 50.7 | |
| Marital status | Single or separated | 49 | 34.0 |
| Married | 95 | 66.0 | |
| Professional status | Employed | 98 | 68.1 |
| Non-employed | 46 | 31.9 | |
| Job sector | Healthcare | 65 | 45.1 |
| Non-healthcare | 79 | 54.9 | |
| Residency | Cluster | 42 | 29.2 |
| Outside cluster | 102 | 70.8 | |
| Sleep quality | Satisfied | 106 | 73.6 |
| Unsatisfied | 38 | 26.4 | |
| Smoking status | Active smoker | 47 | 32.6 |
| Non-smoker | 97 | 67.4 | |
| Physical activity | Regular | 41 | 28.5 |
| Irregular | 103 | 71.5 | |
| Dietary habits | Healthy | 37 | 25.7 |
| Unhealthy | 107 | 74.3 | |
| Chronic diseases | Hypertension | 21 | 14.6 |
| Type 2 diabetes | 21 | 14.6 | |
| Thyroid disease | 6 | 4.2 | |
| Asthma | 6 | 4.2 | |
| Heart disease | 6 | 4.2 | |
| Other | 12 | 8.3 |
COVID-19-related data
In the majority of the participants (71.5%), COVID-19 was mild, and lung involvement was observed in 32.6%, whereas there were no cases of thrombosis diagnosed. Fever (59.4%) and cough (58.0%) were the dominant onset symptoms, followed by the sore throat (34.3%), headache (21.7%), and shortness of breath (19.6%) [Table 2 and Figure 1].
Table 2.
COVID-19 related data: presentation, management and outcomes (n=144)
| Parameter | Category | Frequency | Percentage |
|---|---|---|---|
| Clinical parameters | |||
| Close contact | No | 40 | 27.8 |
| Yes | 104 | 72.2 | |
| Collection site | Nasopharynx | 144 | 100.0 |
| Severity | Asymptomatic | 16 | 11.1 |
| Mild | 103 | 71.5 | |
| Moderate | 25 | 17.4 | |
| Temperature (°C) | Mean, SD (range=36.0, 39.7) | 37.2 | 0.9 |
| Oxygen saturation (%) | Mean, SD (range=90, 100) | 98.4 | 2.0 |
| Pulse (beat/min) | Mean, SD (range=60, 146) | 91.8 | 16.6 |
| Respiratory rate (m/min) | Mean, SD (range=18, 32) | 20.8 | 2.1 |
| Systolic BP (mmHg) | Mean, SD (range=91, 180) | 131.0 | 16.8 |
| Diastolic BP (mmHg) | Mean, SD (range=52, 118) | 78.2 | 12.0 |
| Investigations | |||
| Lung involvement (Radiology) | None | 97 | 67.4 |
| Yes | 47 | 32.6 | |
| Anemia | No | 104 | 72.2 |
| Yes | 40 | 27.8 | |
| Leukocyte | Normal | 98 | 68.1 |
| Leukocytosis | 2 | 1.4 | |
| Leucopenia | 44 | 30.6 | |
| Liver function | Normal | 128 | 88.9 |
| Abnormal | 16 | 11.1 | |
| Renal function | Normal | 136 | 94.4 |
| Abnormal | 8 | 5.6 | |
| Thrombopenia | No | 141 | 97.9 |
| Yes | 3 | 2.1 | |
| Thrombosis | No | 143 | 99.3 |
| Yes | 1 | 0.7 | |
| Management | |||
| Hospital admission | Yes | 77 | 53.5 |
| Oxygen therapy | Yes | 7 | 4.9 |
| CPAP | Yes | 1 | 0.7 |
| Antibiotic therapy | Yes | 42 | 29.2 |
| Antiviral | Yes | 29 | 20.1 |
| Corticosteroids | Yes | 0 | 0.0 |
| ICU admissions | Yes | 0 | 0.0 |
Figure 1.
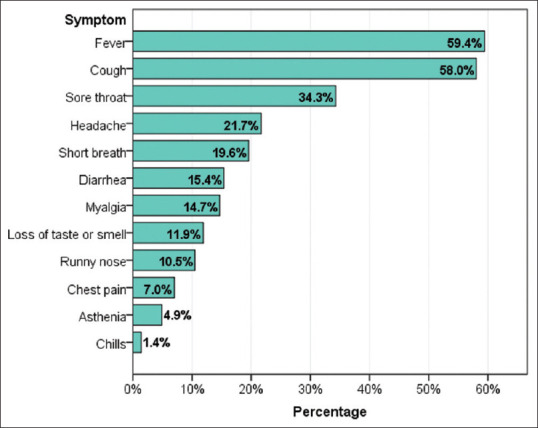
Onset symptoms among COVID-19 patients
Laboratory investigations showed anemia in 27.8% of cases, abnormal liver function in 11.1% of cases, and abnormal renal function in 5.6% of cases. More than half of the participants were hospitalized (53.5%), seven (4.9%) required oxygen therapy (average 5 L/min) and one (0.7%) required continuous positive airway pressure. Antibiotic therapy was required for 29.2% of the patients, whereas 20.1% required antiviral therapy [Table 2].
Viral RNA clearance time
The mean viral RNA clearance time was 22.9 days (SD = 8.6; 95% CI = 21.5–24.3 days). Normality testing showed Kolmogorov–Smirnov (statistics = 0.094, P = 0.004) and Shapiro–Wilk (0.976, P = 0.013), indicating the normal distribution of the variable [Figures 2 and 3].
Figure 2.

Histogram of COVID-19 PCR clearance time in days among 144 confirmed non-severe cases
Figure 3.
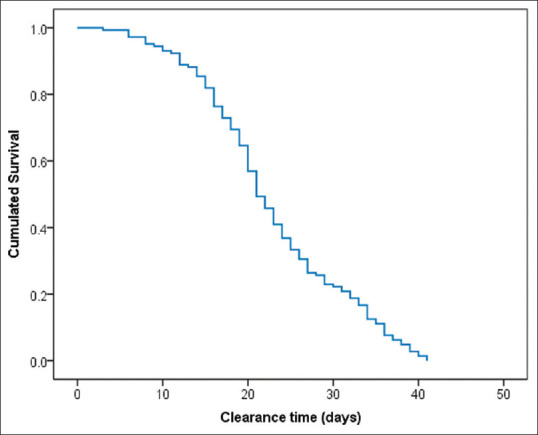
Survival function of COVID-19 PCR clearance among 144 non-severe PCR-confirmed patients
Factors associated with extended clearance time
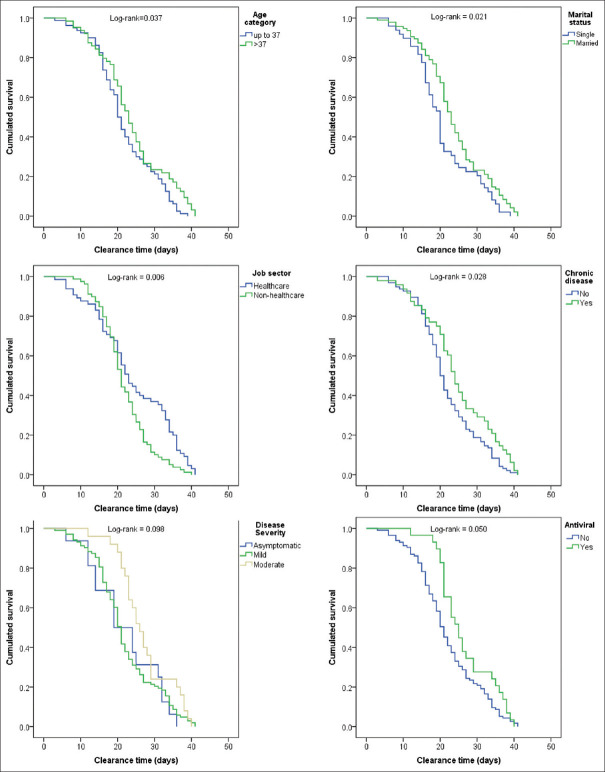
Extended clearance time was significantly associated with age >37 years (mean = 24.0 vs. 22.0 days; log-rank = 0.037), married status (23.9 vs. 20.8 days; log-rank = 0.021), working in health sector (24.2 vs. 21.8 days; log-rank = 0.006), and having a chronic disease (24.8 vs. 21.9 days; log-rank = 0.028), compared to their counterparts, respectively. Additionally, clearance time was longer in patients who received antiviral treatment (mean = 26.6 vs. 22.0 days; log-rank = 0.050) compared to those who did not receive. There are other factors worth noting although they did not reach statistical significance. For instance, regular exercise was associated with shorter clearance time (mean = 21.6 vs. 23.6 days; log-rank = 0.076) and moderate severity (27.2 days) versus asymptomatic (22.1 days) and mild (22.1 days) and the comparison approached statistical significance (log-rank = 0.098) [Table 3 and Figure 4].
Table 3.
Factors associated with COVID-19 clearance time (Kaplan-Meier analysis)
| Factor | Category | Clearance time (days) | Log-rank | ||
|---|---|---|---|---|---|
|
| |||||
| Mean | 95% CI | ||||
| Age (y) | ≤37 | 22.0 | 20.2 | 23.8 | |
| >37 | 24.0 | 21.7 | 26.2 | 0.037* | |
| Gender | Male | 22.6 | 20.8 | 24.3 | |
| Female | 23.2 | 21.0 | 25.4 | 0.229 | |
| Marital status | Single | 20.8 | 18.5 | 23.2 | |
| Married | 23.9 | 22.2 | 25.7 | 0.021* | |
| Profession | Employed | 23.3 | 21.5 | 25.1 | |
| Non-employed | 22.0 | 19.8 | 24.1 | 0.172 | |
| Job sector | Healthcare | 24.2 | 21.7 | 26.7 | |
| Non-healthcare | 21.8 | 20.3 | 23.3 | 0.006* | |
| Residency | Cluster | 20.6 | 17.9 | 23.4 | |
| Outside cluster | 23.8 | 22.2 | 25.4 | 0.134 | |
| Sleep quality | Satisfactory | 23.1 | 21.4 | 24.7 | |
| Unsatisfactory | 22.4 | 19.6 | 25.2 | 0.944 | |
| Smoking status | Active smoker | 22.6 | 20.5 | 24.6 | |
| Non-smoker | 23.0 | 21.2 | 24.9 | 0.472 | |
| Physical exercise | Regular | 21.2 | 18.8 | 23.6 | |
| Irregular | 23.6 | 21.8 | 25.3 | 0.076 | |
| Dietary habits | Healthy | 21.6 | 18.4 | 24.8 | |
| Unhealthy | 23.3 | 21.8 | 24.9 | 0.631 | |
| Any chronic disease | No | 21.9 | 20.3 | 23.6 | |
| Yes | 24.8 | 22.1 | 27.5 | 0.028* | |
| No. symptoms | 0 | 21.9 | 18.3 | 25.5 | |
| 1-2 | 23.3 | 20.7 | 25.8 | ||
| 3 | 23.0 | 20.4 | 25.5 | ||
| 4+ | 23.0 | 20.0 | 26.0 | 0.808 | |
| Severity level | Asymptomatic | 22.1 | 17.6 | 26.5 | |
| Mild | 22.0 | 20.3 | 23.6 | ||
| Moderate | 27.2 | 24.4 | 30.0 | 0.098 | |
| Lung involvement | No | 22.6 | 20.7 | 24.4 | |
| Yes | 23.6 | 21.5 | 25.6 | 0.887 | |
| Anemia | No | 23.1 | 21.4 | 24.8 | |
| Yes | 22.4 | 19.9 | 24.9 | 0.441 | |
| Leukocytes | Normal | 22.4 | 20.6 | 24.1 | |
| Leukocytosis | 30.5 | 11.9 | 49.1 | ||
| Leukopenia | 23.7 | 21.3 | 26.1 | 0.293 | |
| Antibiotic | No | 22.1 | 20.4 | 23.9 | |
| Yes | 24.7 | 22.5 | 27.0 | 0.290 | |
| Antiviral | No | 22.0 | 20.4 | 23.5 | |
| Yes | 26.6 | 23.9 | 29.3 | 0.050* | |
*Statistically significant result (Log rank <0.05)
Figure 4.
Factors associated with PCR clearance time among non-severe COVID-19-confirmed patients
In the adjusted model, the job sector was the only factor that was independently associated with clearance time as indicated in the non-healthcare sector (hazard ratio [HR] = 1.8, 95% CI = 1.3–2.7; log-rank = 0.002) with reference to healthcare sector [Table 4].
Table 4.
Predictors of COVID-19 clearance time (Cox regression)
| Factor | Category | Clearance time | Log-rank | ||
|---|---|---|---|---|---|
|
| |||||
| HR | 95% | CI | |||
| Age (y) | ≤37 | Ref | |||
| >37 | 0.9 | 0.6 | 1.5 | 0.708 | |
| Marital status | Single | Ref | |||
| Married | 0.9 | 0.6 | 1.5 | 0.812 | |
| Job sector | Healthcare | Ref | |||
| Non-healthcare | 1.8 | 1.3 | 2.7 | 0.002* | |
| Any chronic disease | No | Ref | |||
| Yes | 0.8 | 0.5 | 1.2 | 0.242 | |
| Antiviral | No | Ref | |||
| Yes | 0.6 | 0.4 | 1.0 | 0.065 | |
HR: hazard ratio; *statistically significant result (Log-rank <0.05)
Discussion
Summary of findings
The appraisal of viral RNA clearance time and the associated factors represent a crucial epidemiological parameter to understand and control the spread of SARS-CoV-2 infection. This retrospective cohort study used Kaplan–Meier survival analysis to estimate the viral RNA clearance time and characterize its variance among 144 non-severe RT-PCR confirmed COVID-19 patients. Findings suggest that, on average, SARS-CoV-2 RNA remains detectable in the nasopharynx of the patient for 21 to 24 days. Older age and the existence of comorbidities were among the factors associated with prolonged clearance time, whereas non-healthcare professions were independently associated (HR = 1.8).
Clearance time
Estimated clearance time in this study was relatively high compared with those reported in the literature. A study by Chen et al.[16] that included 267 hospitalized COVID-19 patients found the median viral RNA clearance time was 12 days. The viral RNA remained detectable until 21 days among approximately 11% of the patients. Another study by Li et al.[10] showed comparable results with a median viral RNA clearance time of 11 days. More than half of the patients achieved viral RNA clearance after 11 days.
A study by Ling et al.[11] that included 66 COVID-19 convalescent patients showed a mean (95% CI) clearance time of 11.0 (9.0–16.0) days, which is shorter compared to the one observed in the present study. Another study that included 684 adult patients of different severity levels reported a clearance time of <14 days in approximately 30% of the patients. The clearance time was between 14 and 28 days in the other 30% and >28 days among 40% (approximately) of the patients, which is relatively consistent with our findings.[17] Another study by Hu et al.[12] studied the negative conversion rate among 59 cases with different severity levels. They found that only 10.2% achieved negative conversion 7 days after the infection, 62.7% at 14 days and 91.2% at 21 days. Similar to our findings, Fu et al.[13] reported a median viral RNA clearance time of 19 days in throat-swab specimens of 410 confirmed COVID-19 patients.
Factors associated with clearance time
Although healthcare workers showed longer clearance time in univariate analysis, the adjusted model revealed an inverse trend where non-healthcare professionals were associated with 1.8 HR for clearance time compared to healthcare professionals. This might be explained due to better awareness of healthcare workers with respect to diagnosis, symptoms, accessibility to testing, etc., This is supported by findings from other studies that showed that longer interval from disease onset to hospital admission was significantly associated with an extended viral RNA clearance.[14,16] Further, the present study showed that clearance time was significantly prolonged among older patients and those having comorbidities.
Consistent with our findings, other studies reported age to be significantly associated with RNA viral clearance time in COVID-19. A study by Hu et al.[12] showed that the conversion rate was lower among patients aged ≥45 years at 14 days (45.2% vs. 82.2%) and 21 days (87.1% vs. 96.5%) compared with their counterparts (P = 0.009). In the study by Chen et al.[16] the median viral RNA clearance time was increased by 2 days in 16 to 49 age category and by 4 days in older categories (>49 years) with reference to <16 years categories where the difference was statistically significant (P = 0.018). However, this was not supported by findings from the studies by Ji et al. and Xu et al.[14,17]
The association of SARS-CoV-2 clearance time with baseline clinical and biological parameters explored in other studies showed conflicting findings. For instance, the clearance time was longer in patients with obesity, coronary heart disease, hypertension, diabetes, or hypoalbuminemia, whereas it was shorter in patients with asthma or chronic obstructive pulmonary disease.[13,18,19] The association of cardiovascular diseases with delayed viral RNA clearance was suspected to be associated with a higher expression of angiotensin-converting enzyme 2 (ACE2) in such diseases.[18] Another study showed that higher hemoglobin levels were associated with delayed viral RNA clearance. However, this was not reflected in the multivariate analysis.[10]
Other factors associated with clearance time
In addition to the severity level and presentation symptoms, several other factors associated have been explored by different authors and showed significant association with SARS-CoV-2 clearance time. Although the present study included only non-severe cases, we observed a relatively longer mean clearance time in patients with moderate severity compared with mild or asymptomatic COVID-19 patients. Additionally, symptomatic forms were associated with extended viral RNA clearance time, irrespective of the number or type of symptoms. This is consistent with findings by Hu et al.,[12] who reported lower negative conversion rates among severe cases at 2 weeks (44.5% vs. 66.7%) and 3 weeks (81.9% vs. 93.8%) compared to non-severe respectively (P = 0.02). Similarly, the study by Ji et al.[17] found significant positive association of RNA viral clearance time with disease severity (P = 0.009). Interestingly, the study by Hu et al.[12] explored several symptoms separately and found that fatigue and chest tightness were associated with significantly lower negative conversion rates. Other data showed that the presence of diarrhea in the clinical picture was significantly associated with prolonged viral RNA clearance time (P = 0.042), and so was muscular pain with a near statistical significance (P = 0.085).[16] In contrast, other authors observed that patients with a mild form of COVID-19 had prolonged RNA clearance time compared to those with a severe form of COVID-19. This might be due to the robust immune response in severe forms of the disease, which prompts earlier clearance of the viral RNA.[15] This hypothesis might explain the relatively longer clearance time in the present study, where all patients had a mild form of COVID-19. In line with this observation, a study by Antar et al.[19] demonstrated that the absence of early fever in COVID-19 presentation was associated with delayed viral RNA clearance.
Another factor is the leukocyte level, which showed no significant association with clearance time in the present study. However, Ji et al.[17] showed a decrease of 40% and 50% in CD4 cells among the patients who had clearance time of 14 to 28 days and >28 days, respectively, compared to those who had shorter clearance time (<14 days). This difference was statistically significant.
Further, a statistically significant decline (50%) in B cells count was observed among patients with prolonged clearance time (>28 days) compared to those with ≤14 days. Similarly, a study by Ling et al.[11] found that the CD4+ cells count was a significant predictor of viral RNA clearance time, explaining 12% of its variance. Other inflammation markers included in the multivariate model, namely erythrocyte sedimentation rate, C-reactive protein and procalcitonin were not significant predictors. Other authors observed that the clearance of the SARS-CoV-2 among patients with leukopenia was achieved after the restoration of the leukocyte count, notably CD3+, CD4+, and CD8+ T cells and B cells.[20]
Gender was one of these factors identified in several studies. For instance, a study by Shastri et al.,[21] which included 68 patients from the isolation ward found statistically significant prolonged clearance time in males (median = 6 days, range = 1–15 days) compared to females (4 days, 1–10 days). The authors completed these observations by evaluating the expression of mRNA and tissue protein level of ACE2, the plasma membrane-bound receptor that interacts with the SARS-CoV-2 spike protein enabling the virus entry to the host cells. These analyses confirmed the highest ACE2 expression in the testes and testicular proteins, whereas very little expression was observed in the ovarian tissue. Such findings support the prolonged clearance time of the viral RNA in males compared with females and suggest that testicular tissue may serve as a reservoir for the virus. This mechanism may also explain the cases of male gonadal dysfunctions that were observed during the pandemic.[22] Another study by Xu et al.[14] found that male gender was an independent factor associated with the duration of viral RNA detection with a 2.9 odds ratio. However, gender differences observed in the literature are not consistent (e.g., the study by Ji et al.[17]).
Treatments were also investigated among the factors associated with viral RNA clearance time in COVID-19. The study by Ji et al.[17] did not show any difference in viral RNA clearance time in patients who received glucocorticoids compared to those who did not receive in all forms of COVID-19 severity (mild and moderate, P = 0.737; severe and critical forms, P = 0.471). Conversely, Chen et al.[16] observed that the viral RNA clearance time was prolonged by 6 days in patients treated with corticosteroids and by 2 days in patients treated with antiviral drugs. Both the treatments remained significant in the multivariate model; however, antibiotic therapy showed no association with RNA clearance time. Another study showed that invasive mechanical ventilation was associated with prolonged duration of viral RNA detection (log-rank < 0.001); however, this was not confirmed in adjusted multivariate analysis (P = 0.076).[14]
Limitations
The major limitations of the present study are 1) small sample size impacting the statistical power of the study and 2) retrospective design that might impact the validity of the clearance time measurement, as the monitoring of nasopharynx swabs for RT-PCR was not carried out according to the pre-defined protocol. Additionally, the time between disease onset and first positive RT-PCR may vary between the patients that might have impacted the estimation of the clearance time.
Conclusion
The SARS-CoV-2 RNA clearance time is likely to be longer in non-severe COVID-19 patients. This represents an additional risk for the virus dissemination among the community alerting for caution. Older age and the presence of comorbidities are likely to prolong the virus clearance time, especially in non-severe forms, symptomatic, and relatively severe forms of COVID-19. With the advent of accurate molecular methods and advances in the understanding of SARS-CoV-2, further studies are warranted to provide updated data for the duration of the infectiveness period and the associated factors, awaiting the achievement of immunization goals.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Ashour HM, Elkhatib WF, Rahman MM, Elshabrawy HA. Insights into the recent 2019 novel Coronavirus (SARS-CoV-2) in light of past human Coronavirus outbreaks. Pathogens. 2020;9:186. doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandes N. Economic effects of Coronavirus outbreak (COVID-19) on the world economy (March 22, 2020) IESE Business School Working Paper No. WP-1240-E. [Last accessed on 2021 Jul 16]. Available from: https://ssrn.com/abstract=3557504 or http://dx.doi.org/10.2139/ssrn. 3557504 .
- 3.Baldwin R, Mauro BW di. Economics in the time of COVID-19. A VoxEU.org Book. CEPR Press. [Last accessed on 2021 Jul 16]. Available from: https://cepr.org/sites/default/files/news/COVID-19.pdf .
- 4.Yuan J, Li M, Lv G, Lu ZK. Monitoring transmissibility and mortality of COVID-19 in Europe. Int J Infect Dis. 2020;95:311–5. doi: 10.1016/j.ijid.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Block P, Hoffman M, Raabe IJ, Dowd JB, Rahal C, Kashyap R, et al. Social network-based distancing strategies to flatten the COVID-19 curve in a post-lockdown world. Nat Hum Behav. 2020;4:588–96. doi: 10.1038/s41562-020-0898-6. [DOI] [PubMed] [Google Scholar]
- 6.Algaissi AA, Alharbi NK, Hassanain M, Hashem AM. Preparedness and response to COVID-19 in Saudi Arabia:Building on MERS experience. J Infect Public Health. 2020;13:834–8. doi: 10.1016/j.jiph.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alshahrani NZ, Alshahrani SM, Alshahrani AM, Leggat PA, Rashid H. Compliance of the Gulf Cooperation Council airlines with COVID-19 mitigation measures. J Travel Med. 2021;28:taaa205. doi: 10.1093/jtm/taaa205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walensky RP, del Rio C. From mitigation to containment of the COVID-19 pandemic:Putting the SARS-CoV-2 genie back in the bottle. JAMA. 2020;323:1889–90. doi: 10.1001/jama.2020.6572. [DOI] [PubMed] [Google Scholar]
- 9.Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human Coronavirus infections –the state of the art. Emerg Microbes Infect. 2020;9:747–56. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li TZ, Cao ZH, Chen Y, Cai MT, Zhang LY, Xu H, et al. Duration of SARS-CoV-2 RNA shedding and factors associated with prolonged viral shedding in patients with COVID-19. J Med Virol. 2021;93:506–12. doi: 10.1002/jmv.26280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling Y, Xu SB, Lin YX, Tian D, Zhu ZQ, Dai FH, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J. 2020;133:1039–43. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu X, Xing Y, Jia J, Ni W, Liang J, Zhao D, et al. Factors associated with negative conversion of viral RNA in patients hospitalized with COVID-19. Sci Total Environ. 2020;728:138812. doi: 10.1016/j.scitotenv.2020.138812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y, Han P, Zhu R, Bai T, Yi J, Zhao X, et al. Risk factors for viral RNA shedding in COVID-19 patients. Eur Respir J. 2020;56:2001190. doi: 10.1183/13993003.01190-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu K, Chen Y, Yuan J, Yi P, Ding C, Wu W, et al. Factors associated with prolonged viral RNA shedding in patients with Coronavirus disease 2019 (COVID-19) Clin Infect Dis. 2020;71:799–806. doi: 10.1093/cid/ciaa351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carmo A, Pereira-Vaz J, Mota V, Mendes A, Morais C, da Silva AC, et al. Clearance and persistence of SARS-CoV-2 RNA in patients with COVID-19. J Med Virol. 2020;92:2227–31. doi: 10.1002/jmv.26103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Zhu B, Hong W, Zeng J, He X, Chen J, et al. Associations of clinical characteristics and treatment regimens with the duration of viral RNA shedding in patients with COVID-19. Int J Infect Dis. 2020;98:252–60. doi: 10.1016/j.ijid.2020.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji J, Zhang J, Shao Z, Xie Q, Zhong L, Liu Z. Glucocorticoid therapy does not delay viral clearance in COVID-19 patients. Crit Care. 2020;24:565. doi: 10.1186/s13054-020-03287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Hu W, Ling J, Mo P, Zhang Y, Jiang Q, et al. Hypertension and diabetes delay the viral clearance in COVID-19 patients. MedRxiv. Published online 2020. doi:10.1101/2020.03.22.20040774. [Google Scholar]
- 19.Antar AAR, Yu T, Pisanic N, Azamfirei R, Tornheim JA, Brown DM, et al. Delayed rise of oral fluid antibodies, elevated BMI, and absence of early fever correlate with longer time to SARS-CoV-2 RNA clearance in an longitudinally sampled cohort of COVID-19 outpatients. Open Forum Infect Dis. 2021;8:ofab195. doi: 10.1093/ofid/ofab195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Ling J, Mo P, Zhang Y, Jiang Q, Ma Z, et al. Restoration of leukomonocyte counts is associated with viral clearance in COVID-19 hospitalized patients. MedRxiv. 2020 Published online. doi:10.1101/2020.03.03.20030437. [Google Scholar]
- 21.Shastri A, Wheat J, Agrawal S, Chaterjee N, Pradhan K, Goldfinger M, et al. Delayed clearance of SARS-CoV-2 in male compared to female patients:High ACE2 expression in testes suggests possible existence of gender-specific viral reservoirs. MedRxiv. 2020 Published online. doi:10.1101/2020.04.16.20060566. [Google Scholar]
- 22.Ma L, Xie W, Li D, Shi L, Mao Y, Xiong Y, et al. Effect of SARS-CoV-2 infection upon male gonadal function:A single center-based study. MedRxiv. 2020 Published online. doi:10.1101/2020.03.21.20037267. [Google Scholar]