Abstract
RNA molecules fold into characteristic secondary and tertiary structures that account for their diverse functional activities. Many of these RNA structures are assembled from a collection of RNA structural motifs. These basic building blocks are used repeatedly, and in various combinations, to form different RNA types and define their unique structural and functional properties. Identification of recurring RNA structural motifs will therefore enhance our understanding of RNA structure and help associate elements of RNA structure with functional and regulatory elements. Our goal was to develop a computer program that can describe an RNA structural element of any complexity and then search any nucleotide sequence database, including the complete prokaryotic and eukaryotic genomes, for these structural elements. Here we describe in detail a new computational motif search algorithm, RNAMotif, and demonstrate its utility with some motif search examples. RNAMotif differs from other motif search tools in two important aspects: first, the structure definition language is more flexible and can specify any type of base–base interaction; second, RNAMotif provides a user controlled scoring section that can be used to add capabilities that patterns alone cannot provide.
INTRODUCTION
RNA is characterized by its base sequence and higher order structural constraints. This is particularly true of non-coding RNAs such as rRNAs and tRNAs and other functional RNAs, such as RNase P and the signal recognition particle (SRP). All of these RNAs are characterized by short- and long-range base pair interactions that organize the molecules into domains and provide a framework for functional interactions. Similar mechanisms of secondary and tertiary structure interactions probably also play an important role in regulating mRNA expression. An example of this is the iron response element, a structural regulatory motif occurring in the untranslated regions (UTRs) of various members of the iron metabolism and transport pathway (1,2). Other instances of mRNA secondary structures include stem–loops in the 3′-UTRs of histone and vimentin mRNAs that are important for processing and localization, respectively (3–5). We describe here an algorithm to identify RNA structural motifs in nucleotide sequence databases using elements of both sequence and structure in an integrated fashion.
At a fundamental level, RNA secondary structure consists of nucleotides that are in one of two states, paired or unpaired, where pairing includes all base–base interactions. In general most base pairings are adjacent and antiparallel with other base pairings to form secondary structure helices. The combination of one or more helical elements interspersed with unpaired, single-stranded nucleotides constitutes an RNA structure. Over the last decade, a number of tools such as RNAMOT, Palingol and PatScan, were developed to define and search for such RNA structures (6–9) (PatScan web server: http://www-unix.mcs.anl.gov/compbio/PatScan/HTML/patscan.html). In addition to these general purpose motif searching tools, others such as tRNAscan, FAStRNA and CITRON were designed and optimized to search for specific kinds of structural RNA molecules (10–12). While these efforts were moderately successful in defining simple RNA structures, they were not adequate to capture complex structural domains or various non-canonical pairings that are present in RNA motifs.
This paper describes a new motif search algorithm, RNAMotif. The algorithm is robust, yet flexible, and confers on the user the freedom to search for any definable simple and complex secondary and tertiary structure. These structural motifs include the quintessential base pairs, helices and unpaired nucleotides in hairpins, internal and multi-stem loops, as well as a collection of more complicated motifs that contain specific sequence constraints within a combination of paired and unpaired nucleotides. This includes E-loops, specific tetraloop and closing base pair combinations and other RNA structure motifs that will be discussed below. Further, the RNAMotif program was designed to account for RNA structural elements that may not be currently known or appreciated. This algorithm should be able to define structural motifs that are determined and understood from the new, high resolution RNA structures. These motifs include protein and metal binding sites, and other motifs that could have unusual base pair and backbone conformations. The structural patterns are defined in a ‘descriptor’ with a pattern language that distinguishes, at its lowest level, paired and unpaired positions. The RNAMotif descriptors, like those of its predecessors, can be parameterized as to length, sequence and base pairing, providing a high degree of control over the structures that are identified. However, RNAMotif differs from earlier algorithms with an awk-like (13) scoring section that combines these pattern elements to add capabilities that patterns alone cannot provide. In particular, scoring allows the user to rank imperfect matches to desired sequence/structural elements.
RNAMotif allows for all 16 types of base pairs, including canonical Watson–Crick (G:C and A:U), wobble (G:U) and other non-canonical base pairs (e.g. A:C and U:U) that are defined as part of a helix. These can be defined globally across all helical regions or within a few selected helices or, even more specifically, at specified locations within a given helix. Similar levels of control are also provided for defining sequence mismatches and mispairings. In addition to base pairs, base triples and base quadruples can also be defined. Additional flexibility and fine tuning of the descriptor is with optional sections called parms, site and score. They are all described in detail in Materials and Methods.
MATERIALS AND METHODS
Motif description
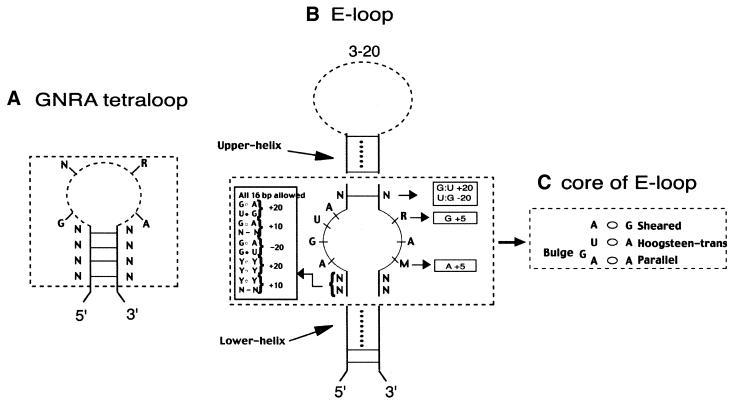
The input to an RNAMotif run is a formal description of the permissible forms of the structure and the sequences contained within it. Figure 1 shows a set of real examples of a few such motifs. The constraint description file consists of four sections called parameters, descriptor, sites and score. Default variables that are used in the rest of the descriptor are defined in the parameters section. The descriptor section defines the criteria required to generate a match. The sites section allows users to specify relations among the elements of the descriptor, while the score section ranks matches to the constraints based upon criteria defined by the user.
Figure 1.
Examples of RNA structure motifs and descriptor constraints with important conserved nucleotides and scoring values. (A) A helical stem closed by a tetraloop. (B) An E-loop motif. (C) The core of the E-loop depicted with the observed non-canonical base pairing interactions. The most significant structural elements within the motif are shown within the dotted box. The structural components (stems or single-stranded regions) that flank this internal loop can vary in length and composition. (D) E-loop descriptor derived from the constraints shown in (B). Additional constraints for the stems flanking the core motif not shown were used in the overall score calculations reported in Table 2.
Search algorithm
RNAMotif uses a two-stage algorithm to perform motif searches. The first stage is a compilation stage, which analyzes the specified motif, called a descriptor, and converts it into a search tree based on the helical nesting of the motif. This stage also checks that the descriptor is syntactically valid and performs a number of consistency checks on the specification of the motif to ensure that a solution exists. Two examples of consistency checking are: (i) testing that all length specifiers of the individual strands of a single helix are the same; (ii) testing that if a motif element contains both explicit length specifications and implicit length constraints derived from sequence specifications, the two are compatible. If the descriptor is syntactically valid and passes all consistency checks, the compilation stage then analyzes the lengths of the motif elements and computes limits as to where each element must begin and end.
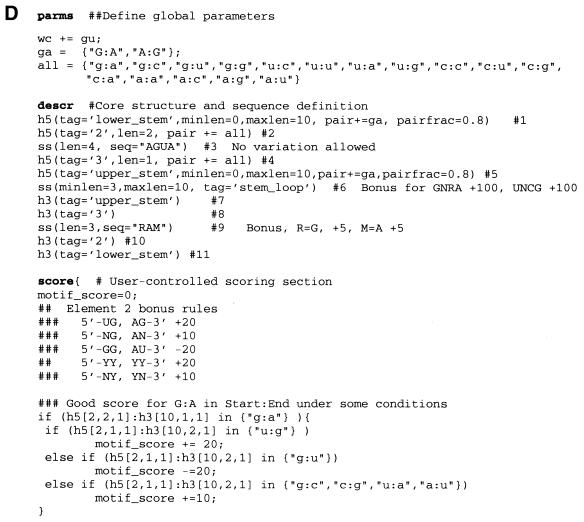
The second stage is a depth first search of the tree that was created by the compilation stage. This tree is constructed by noting that every duplex can be represented as a binary tree where the root of the tree is the helix itself, the left sub-tree is the motif that is contained in the interior of the helix and the right sub-tree is the motif that follows the helix. Table 1 shows some examples of trees generated in this manner. Triples and quadruples are represented as three- and four-way trees with the first n – 1 sub-trees corresponding to the interior motifs contained by the helical strands, with the nth or right-most sub-tree again representing the motif that follows the helix. Pseudoknots are accommodated by collecting all the duplexes involved in the pseudoknot into a single structure whose root then becomes the 5′-most helix, and whose n – 1 interior nodes (n ≥ 4) represent the motifs that are contained inside all the helices that form the pseudoknot. Once again, the right-most sub-tree is the motif that follows the pseudoknot. Finally, single-stranded regions are treated as binary trees that have a null left (interior) sub-tree with, at most, one non-null right sub-tree describing the motif that follows the single-stranded region.
Table 1. Conversion of the descriptor structure motifs into an RNAMotif search list.
Every motif is a tree. This tree has one sub-tree for the sub-structure(s) contained in each of its interior regions (shown by ↓) and one sub-tree for the substructure(s) that follows it (shown by →). By definition, single-stranded regions have no interior elements and have, at most, one element following them.
The actual search algorithm follows immediately from the tree representation of the secondary structure motif. It begins by testing the first or left-most position of the target sequence to see if it contains any instances of the left-most sub-motif of the descriptor. In general, a sub-motif will have several possible solutions and RNAMotif examines all candidates in shortest to longest order. The search algorithm is then recursively called to find all solutions of the sub-motif of the left-most interior region or, if all interior regions have been searched, the algorithm is applied to the region following the left-most motif. Each time a complete solution to the original descriptor is found, the candidate is passed to an optional score section for scoring and ranking. If the score section is absent, then the candidate is automatically accepted. When all candidates at a particular recursion level have been examined, the search algorithm backs up and continues the search on any unexamined candidates from the previous level. When all candidates have been examined at the original level, the top level search is moved one position to the right on the target sequence and the search is continued, until the entire target sequence has been searched.
Scoring
The score section is an optional set of tests that are applied to each candidate found by the search. This section serves two related purposes. First, it provides a way to add constraints to a motif that are difficult to implement in a pure pattern language. Some examples would be specifying the GC content of a sub-motif, requiring that the solution has no consecutive mismatches, or setting an upper bound on the total number of mispairs in a solution. The second purpose of the score section is to evaluate and rank the candidates, possibly rejecting those below some threshold.
The score section was modeled after awk and consists of a list of rules, where a rule is a pattern/action pair. Each time the search finds a candidate, the rule list is evaluated in top to bottom order until either a rule explicitly accepts or rejects a candidate or the rule list is exhausted, in which case the candidate is accepted. In the absence of any rules, all candidates returned by the searches are accepted.
The score section provides the usual set of small language features: for and while statements for looping, if and if/else statements for testing, variables and operators for expressions and assignments and a small number of built-in functions. Most of these functions are used to access an attribute (length, mispairs, mismatches, etc.) of the string that matches a specified sub-motif. The actual sequences that match each sub-motif of the descriptor are available as a set of read-only string variables. These variables are indexed, providing access to any set of sub-strings of the current match.
Many RNAMotif descriptors generate large numbers of candidates. This is especially true for descriptors that contain only helical constraints without sequence constraints. In many cases there is no obvious scoring system by which these candidates can be ranked and pruned. To help in this case, RNAMotif provides two general scoring functions by which a candidate can be evaluated.
The first function evaluates the thermodynamic stability (ΔG) of the candidate, or any part of it, and allows candidate solutions to be ranked based on their calculated free energies. We have implemented the latest thermodynamic parameters and calculations as described in Matthews et al. (14). Free energy (ΔG, kcal/mol) is calculated by calling the built-in function efn(), which takes two arguments, the beginning and the ending positions of the structure whose energy is to be evaluated. This can be invoked anywhere within the score section and provides a measure of the thermodynamic stability of the structural element. The function uses the descriptor to convert the selected sequence into a base pair and connection table and then applies the energy function to that table. Energy values have been calculated for a number of structural element combinations, such as stacking energies of consecutive base pairs, stacking energies for hairpin loops and symmetrical and asymmetrical interior loops, single base stacking energies, loop destabilizing energies and a few other conformations. Data files for these can be downloaded from a number of sites, including http://bioinfo.math.rpi.ed u/~zukerm/rna/energy/.
The second general fitness function involves sequence complexity. Lower complexity sequence regions, which refer to regions that have highly biased and/or repetitive sequence compositions, may match a generic descriptor with very high scores that owe virtually nothing to residue order but are due instead to segment composition. These also often result in ‘slippage’, where multiple matches to the descriptor may be found by simply sliding the descriptor along the sequence. Examples of these include the highly AU-rich regions of the 3′-UTRs of eukaryotic mRNAs. RNAMotif allows the user to measure the complexity of a given string using the built-in function bits(). This measures the compositional complexity, which is a simplification of base composition where the original bases of the string are reduced to bases of the different types (first base, second base, and so on) (15). Compositional complexity (K) is defined as
![]()
where N is the number of symbols in the alphabet, which for nucleic acids is 4, L the length of the string and n[i] is the number of instances of the ith base. For instance, the two strings AAAATTTT and CGCGCGCG, while having completely different base compositions, have the same compositional complexity (1.000) because both contain four instances of the first base (A or C) and four instances of the second base (T or G). Since the relationship between sequence complexity and occurrence of RNA secondary structure is not known, RNAMotif does not automatically filter out matches in regions of low complexity. Instead, the score evaluated by the function bits() may be used to evaluate the relative information content of the current candidate (or any sub-string of it) and used in conjunction with other calculated score parameters.
RESULTS
Motif search examples
The examples discussed below highlight various aspects of the new RNAMotif search tool. They are arranged in order of increasing complexity of the descriptor file, starting with a simple tetraloop descriptor without any scoring routine, to a logical scoring scheme with more complex descriptors.
Tetraloop motifs. Consider a simple structural element such as a stem whose 5′- and 3′-sides are separated by a hairpin loop with 4 nt (Fig. 1A). This is a common, recurring motif found in many structured RNA molecules, often with specific sequence constraints in the hairpin loop (GNRA, UNCG and CUUG) (16). Sometimes this motif has a preferred closing pair at the base of the loop that is associated with the sequence in the hairpin loop. For instance, UUCG loops show a bias for a C:G closing pair, while CUUG tetraloops prefer a G:C closing pair (16).
A typical descriptor for the UNCG family based on the above constraint on the closing pair is shown below:
h5(tag=‘5p_helix’,minlen=2, maxlen=4, seq=“C$“) ### 5′ helix ends in C
ss(len=4, seq=“UNCG”)
h3(tag=‘3p_helix’,seq=“^G”) ###Helix starts with G
The helical elements have an optional ‘tag’ identifying them unambiguously as being the 5′- and 3′-sides of the same helix. For short motifs such as these the tag is not necessary, but in longer, more complex constructs they are helpful and sometimes necessary. Run against the Escherichia coli 16S rRNA, this descriptor produced five hits, all of which correspond to previously identified UNCG tetraloop motifs in 16S rRNA (16).
A similar construct for the motif that has the loop sequence constraint ‘GNRA’, where R represents the purine A or G, was run against E.coli 16S rRNA. This produced 10 hits, only five of which corresponded to true GNRA tetraloop motifs (16). Further, the descriptor did not find four other documented GNRA motifs at E.coli 16S rRNA positions 157, 295, 1011 and 1075, respectively. Upon closer examination, these regions were found to have G:U pairing in the helix or had A:U/U:A as the closing base pair, neither of which was allowed in the original descriptor. The original descriptor was modified to accommodate both of these:
parms
wc +=gu; ###This permits GU pairing globally
descr
h5 (tag=‘5p_helix’, len=3) ### sequence constraints for this helix have been removed
ss(len=4, seq=“GNRA”)
h3(tag=‘3p_helix’)
Running this against the 16S rRNA sequence produced 16 hits, including all nine known GNRA tetraloops (16). The above motif represents a very generic ‘GNRA’ descriptor, with very little constraint on the helical region. Hence, it is not surprising that there are a number of false positives. Other examples described below will show additional constraints derived from an alignment, as well as the use of the scoring scheme to rank/reject results.
E-loop motif. Another motif that appears several times in many structural RNA molecules is the E-loop motif (Fig. 1B) (17–20). The majority of E-loops identified to date are characterized by an asymmetrical internal loop, with the nucleotides AGUA on one side and RAM, where R is either G or A and M is either C or A, on the other. The elements of the above internal loop are involved in cross-strand pairing with a sheared A:G, trans-Hoogsteen U:A, a bulged G and parallel A:A (18) as shown in Figure 1C. The internal loop is flanked on the 5′-side by at least 2 bp, both of which can be non-canonical pairings. The 3′-side of the motif typically has at least 1 bp. Figure 1B shows additional constraints, as well as an empirical scoring scheme that captures the essential features of this motif. The RNAMotif descriptor is shown in Figure 1D. This descriptor defines an E-loop that is flanked by helical stems with very little sequence constraint. We used two different metrics to rank the results: first, the score value calculated based on user-defined criteria (motif score) and, second, the free energy of the structural unit (ΔG) as described above. The final score reported in Table 2 is a combination of the motif score calculated in Figure 1D with the scores for the flanking helices (details not shown). We ran two versions of the descriptor, one exactly as shown in Figure 1B and another that was a circular permutation of the above, where the 5′- and 3′-sides of the motif were flipped (not shown).
Table 2. Results of an RNAMotif search for E-loop motifs in E.coli 16S and 23S rRNA sequences.
| Molecule | Score | ΔG (kcal/mol) | Start | Length | Presence in crystal structure |
|---|---|---|---|---|---|
| The score value calculated based on user defined criteria (shown in Fig. 1C) is shown in the second column, while the free energy of the structural unit (ΔG) is shown in the third column. The columns marked start and length define the 5′-end of the match to our descriptor (E.coli coordinates) and the total length of the match, respectively. All of the top scoring hits from RNAMotif (shown in bold) are present in the 16S and 23S crystal structures (21,22). | |||||
| E.coli 16S rRNA | 82 | –13.4 | 1342 | 38 | Yes |
| 82 | –3.7 | 884 | 31 | Yes | |
| 40 | 2.1 | 246 | 23 | No | |
| 40 | 7.5 | 1329 | 23 | No | |
| 37 | 15.5 | 1422 | 39 | No | |
| 36 | 1.4 | 430 | 25 | No | |
| 36 | 7.6 | 108 | 33 | No | |
| 17 | 13.8 | 875 | 20 | No | |
| E.coli 23S rRNA | 136 | –21.8 | 364 | 46 | Yes |
| 115 | –8.2 | 2642 | 37 | Yes | |
| 84 | –6.1 | 230 | 38 | Yes | |
| 70 | –5.2 | 184 | 29 | Yes | |
| 50 | 3.4 | 455 | 19 | Yes | |
| 42 | 3.1 | 2264 | 31 | No | |
| 35 | 8.7 | 1247 | 27 | No | |
| 30 | 3.1 | 200 | 22 | No | |
| 20 | 4.9 | 1263 | 25 | No | |
Table 2 shows the results of an RNAMotif search for E-loop motifs against E.coli 16S and 23S rRNA sequences. These results were compared to previously documented occurrences of E-loops (17,18) that have been confirmed by published crystal structures (21,22). We were able to identify two of three E-loops in the 16S rRNA and five of eight E-loops in the 23S rRNA. A cut-off motif score of 50, and/or a thermodynamically stable free energy score (ΔG < 0), was used to threshold the hits. Both of these scores were clearly able to differentiate true hits (seen in the published crystal structures for these molecules) from false hits. With the exception of one E-loop motif at position 455 in 23S rRNA, which does not have a flanking lower stem and therefore has a poor ΔG value (and a marginal motif score), all of the other ‘true’ E-loops in rRNA had significantly better scores than the false positives.
Of the E-loop motifs that were not identified (Table 3), the motif at position 1265:2014 (E.coli coordinates) spans >700 nt between domains III and IV in 23S rRNA. Our descriptor was not long enough to accommodate this. The other three missed motifs had less common sequences in the AGUA internal loop: (i) GUUA in the 16S rRNA motif at position 449:487; (ii) GGUA in the two 23S rRNA motifs (at 858:918 and 818:1189). Our current descriptor did not allow for any variations in this loop. When we modified our search to include all motifs of the type GUA:GA, which represents the core of the E-loop motif without the parallel A:A base pair at the end, we found many more matches to our descriptor. This included those described above and other ‘E-like’ loops described by Gutell and co-workers (17). This search, however, was much less restrictive and our false positive hits went up significantly.
Table 3. Other rRNA E-loop motifs that were not identified by the RNAMotif descriptor.
| Molecule | 5′-Side motif | 3′-Side motif | Core motif sequence | Exception to RNAMotif descriptor |
|---|---|---|---|---|
| The coordinates and the sequences of these E-loop motifs are shown above. Each of these had an exception to the descriptor (shown in the last column). | ||||
| E.coli 16S rRNA | 447–9 | 484–7 | GAG:GUUA | Non-canonical motif sequence |
| E.coli 23S rRNA | 818–20 | 1186–9 | GAA:GGUA | 5′→3′ distance too long to match current descriptor, non-canonical motif sequence |
| 858–61 | 916–8 | GGUA:GAA | Non-canonical motif sequence | |
| 1265–8 | 2012–4 | AGUA:GAA | 5′→3′ distance too long to match current descriptor | |
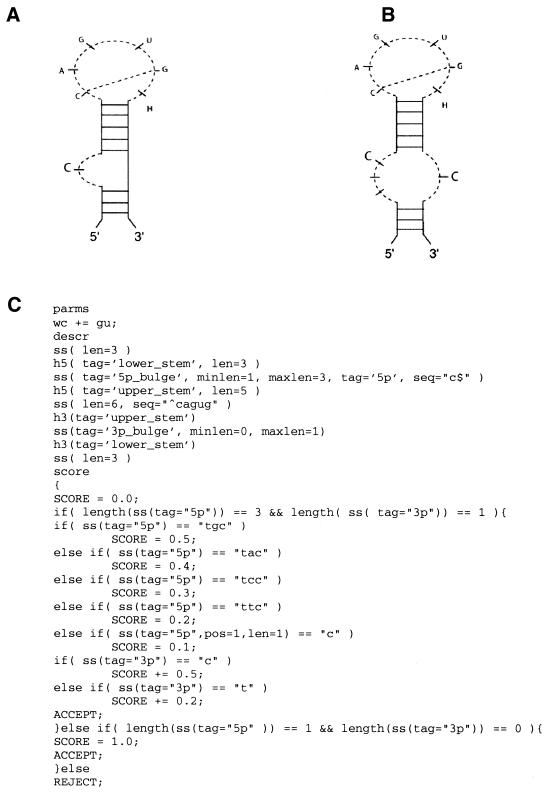
Iron-responsive element (IRE). The eukaryotic IRE is an example of a stem–loop with an internal loop. IREs have been shown to occur in the 5′- and 3′-UTRs of several mRNAs (1,23–26). They bind cellular iron-regulatory proteins (IRPs) and regulate iron homeostasis. The secondary structure of IREs has been extensively studied and two forms of the structure have been proposed (27). Most IREs are shown with a single C-bulge (see Fig. 2A) inserted on the 5′-side of a compound helix. The alternative form, shown in Figure 2B, appears predominantly in ferritin mRNA and has an asymmetrical internal loop at this position, with a three-base bulge on one side ending in a C and a single C-bulge opposite it.
Figure 2.
Models for the IRE structure and generic descriptor. (A) IRE bulge model. (B) IRE internal loop model. (C) Descriptor that captures both types of IRE.
The key features of the IRE motif are: (i) the conserved sequence CAGUG in the hairpin loop; (ii) an internal loop with one or three bases ending with a C on the 5′-side; (iii) a 5 bp upper helix. A single descriptor for both IRE motifs is shown in Figure 2C. Since an element in a descriptor can have zero length, the optional right bulge, labeled ‘3p_bulge’, can be specified as having a length range of 0–1. The left bulge, labeled ‘5p_bulge’, has a range of 1–3. However, there are a large number of sequences that satisfy this loose constraint. To limit the number of possible solutions to the biologically meaningful sequences, we have used the RNAMotif scoring function to identify and rank the matches to the IRE descriptor.
When GenBank (February 2001 release) was searched for IRE elements with this descriptor we obtained 914 total hits. Of these, 165 were on mRNAs (positive strand), and out of these 105 were in the UTR regions of mRNAs where IREs are known to occur. Over 90% of the UTR hits had the maximum score of 1.0. This included all previously identified and documented IREs in UTRs of mRNAs such as ferritin, transferrin, aconitase and ALAS (totaling ∼75 hits). Table 4 shows a list of new UTR hits that scored well. Prominent amongst these are the solute carrier family 11 (SLC11) family of proteins that are annotated in the literature as iron-regulated transporters (IREG1), ferroportin 1 (FPN1) or SLC11 member 3 (SLC11A3). Known orthologs of this family in human, mouse, rat and zebrafish all had an IRE-like element in their 5′-UTR.
Table 4. Perfect matches to IRE motif in human UTR sequences.
| 5′-UTR | 3′-UTR |
|---|---|
| Ferritin (light and heavy chain) | Transferrin receptor (five copies) |
| ALAS | Fukutin/FCMD |
| Apoferritin | Caveolin (Cav3) |
| Ferroportin 1 (FPN1) | |
| Iron-regulated transporter (IREG1) | |
| Solute carrier family 11 | |
| FLI_cDNA | |
| Mitochondrial aconitase (overlaps start site) | |
| Nuclear matrix protein (NRPB) |
In addition to the IRE elements that were previously known and identified here, we have identified a few new IRE-like elements (score = 1.0) that are located in the 5′-UTRs of nuclear matrix protein (NRPb/ENC-1) and a liver cDNA clone (FLI) identified as PRO0149 protein and in the 3′-UTRs of caveolin (Cav3) and fukutin. ENC-1 has a mouse ortholog, but there was no evidence for an IRE-like structure in its 5′-UTR. Based on the annotation in GenBank, FL1 has no known function and we could not find any FL1 orthologs. The genes caveolin and fukutin are both associated with muscular dystrophy and have long 3′-UTR sequences (28–31). We could not find any orthologous sequences in GenBank for the human version of fukutin, while the mouse and rat caveolin sequences do not contain the IRE element. There were a few (<5%) hits that could not be associated with an IRE element with strong confidence and may be considered false positives. We also identified a few hits with perfect scores in the coding regions of human mRNAs. However, as with some of the UTR hits discussed above, none of these were present in orthologous genes in other organisms, suggesting that these matches to the IRE motif descriptor are not biologically functional.
SRP-RNA domain IV stem–loop descriptor. The RNA component of the SRP, SRP-RNA (4.5S RNA in prokaryotes and 7S RNA in eukaryotes), is an essential small RNA molecule involved in targeting signal peptide-containing proteins to endoplasmic reticulum (eukaryotes) or the plasma membrane (prokaryotes) (32,33). While all known SRP-RNA sequences contain a terminal stem–loop structure (also called the domain IV stem–loop), other sections of the SRP-RNA are variable in length, composition and secondary structure. Domain IV is conserved across evolution, from bacteria to mammals, and has been shown to be the binding site for the protein component of the particle (34).
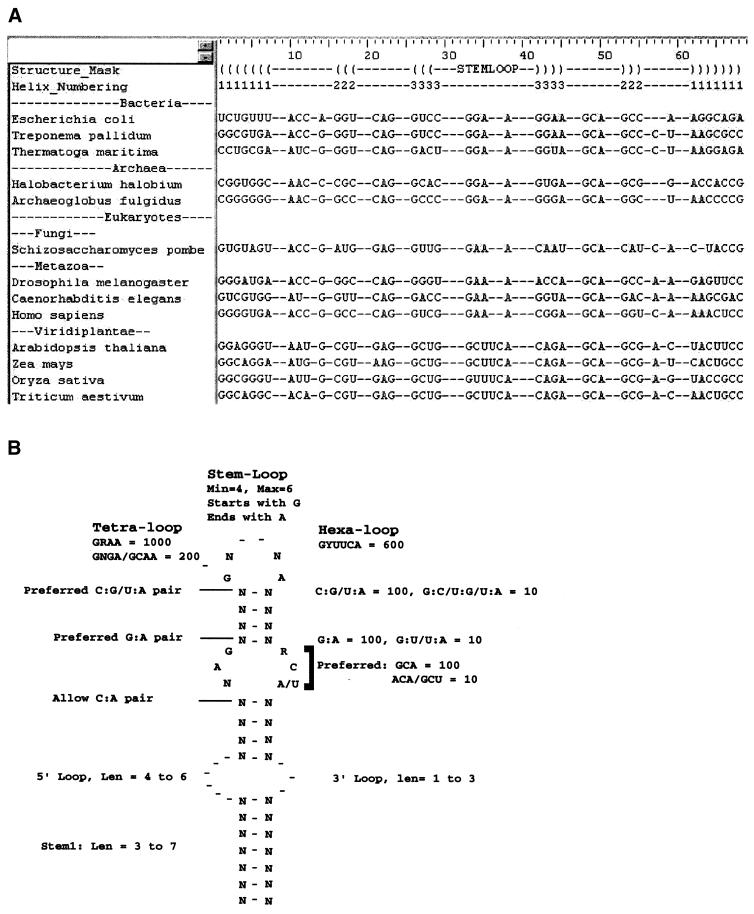
The alignment of sequences in this region of SRP-RNA for a few representative organisms from the major phylogenetic domains (Bacteria, Archaea and Eukarya) is shown in Figure 3A. The key features of this structure are the two internal loops, a symmetrical loop closer to the top of the stem and a variable asymmetrical loop closer to the base of the stem. Figure 3B shows the consensus structure for this region based on the complete alignment from over 100 organisms obtained from the SRP website (http://psyche.uthct.edu/dbs/SRPDB/SRPDB.html). The helices are of varying lengths and the stem–loop has two predominant types, a stable tetraloop in most organisms is replaced by a hexaloop in plant SRPs. An RNAMotif descriptor with an empirical scoring scheme was created based on the observed biases in nucleotide and base pair frequencies and the range of size variations in loops and helices seen in the alignment, shown in Figure 3B.
Figure 3.
SRP-RNA alignment and structure. (A) Structure-based alignment of the domain IV stem–loop region of SRP-RNA based on the full-length alignment from the SRP-RNA website (http://psyche.uthct.edu/dbs/SRPDB/SRPDB.html). Sequences of a few representative organisms from the three major phylogenetic kingdoms (Bacteria, Archaea and Eukarya) are shown here. The top two lines of the alignment show the base pairing schema. ( and ) are used to denote the 5′ and 3′ sides of a helix, respectively, and the numbers indicate the paired segments. (B) A consensus diagram of the SRP-RNA structure derived from analyzing the alignment of over 100 organisms. The biases in nucleotides and range of size variations in loops and helices are shown. An RNAMotif descriptor (not shown) was derived based on the constraints shown here.
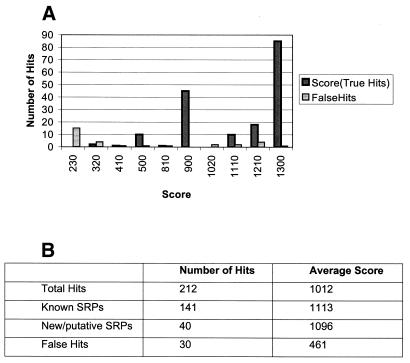
We searched the GenBank sequence database (February 2001 release) with this descriptor for occurrences of the conserved components of SRP-RNA. These results are shown in Figure 4A. All of the previously annotated SRPs were readily identified by our descriptor and had a score distribution that was distinct from that for the false hits. In addition to the identification of SRP-RNAs that were already known, we identified more than 40 new SRPs that were not annotated in GenBank (Fig. 4B). Several of these could be verified at the SRP website (http://psyche.uthct.edu/dbs/SRPDB/SRPDB.html).
Figure 4.
Summary of results of a RNAMotif search for SRP-RNA. (A) Score distribution of true and false hits based on RNAMotif scores in a search of the GenBank nucleotide database with an SRP-RNA descriptor. (B) Summary of scores for true and false hits.
We also tested our SRP-RNA descriptor against complete prokaryotic genomes. The results are shown in Table 5. As of February 2001, only 21 of the 38 completed genomes had SRP locations annotated in the NCBI website (http://www.ncbi.nlm.nih.gov/). The SRP website (updated February 2001) lists many additional SRPs, but a few of the recently completed genomes were missing. We identified the SRP-RNA in 37 of the 38 genomes. The only exception was Buchnera sp., which did not match the SRP domain IV motif. This organism was also not listed in the SRP website.
Table 5. Identification of SRP-RNA in prokaryotic genome sequences.
| RNAMotif matches present in SRP DB | RNAMotif matches missing in SRP DB |
|---|---|
| Using the RNAMotif descriptor-based search described in the text we were able to identify SRP-RNA in 37 of the 38 completely sequenced bacterial genomes (as of February 2001). The column on the left shows organisms that were found by our descriptor that was also present in the SRP-RNA database. The right column shows organisms that were identified by our search but were not present in the SRP-RNA database. The last entry in the right column shows the only organism, Buchnera sp., for which the complete genome sequence is known but where we could not detect an SRP domain IV-like motif. | |
| Aeropyrum pernix | Bacillus halodurans C125 |
| Aquifex aeolicus | Chlamydia muridarum |
| Archaeoglobus fulgidus | Chlamydophila pneumonia AR39 |
| Bacillus subtilis | Chlamydophila pneumonia J138 |
| Borrelia burgdorferi | Deinococcus radiodurans |
| Campylobacter jejuni | Escherichia coli O157 |
| Chlamydia pneumoniae | Thermoplasma acidophilum |
| Chlamydia trachomatis | |
| Escherichia coli | Completed genomes with no known SRP RNA |
| Haemophilus influenzae | Buchnera sp. |
| Halobacterium halobium | |
| Helicobacter pylori J99 | |
| Helicobacter pylori 26695 | |
| Methanococcus jannaschii | |
| Methanobacterium thermoautotrophicum | |
| Mycoplasma genitalium | |
| Mycoplasma pneumoniae | |
| Mycobacterium tuberculosis | |
| Neisseria meningitidis MC58 | |
| Neisseria meningitidis Z2491 | |
| Pyrococcus abyssi | |
| Pseudomonas aeruginosa | |
| Pyrococcus horikoshii | |
| Rickettsia prowazekii | |
| Synechocystis sp. | |
| Thermotoga maritima | |
| Treponema pallidum | |
| Ureaplasma urealyticum | |
| Vibrio cholera | |
| Xylella fastidosa | |
DISCUSSION
We have created a new RNA structure motif description and discovery tool, RNAMotif, which can be used to search nucleic acid databases for sequences that can adopt a specified secondary structure. RNAMotif represents a significant improvement over the previous generations of motif search tools, which cannot adequately describe the complexity of constraints required to accurately define RNA structure. RNAMotif has a scoring section that allows almost any arbitrary calculation to be performed on the sequences that match the user defined constraints, both within a structure element and globally across various elements, and users can define criteria for accepting or rejecting any given solution.
RNAMotif is capable of finding sequences that can contain any secondary structure element: single strands, duplexes (both antiparallel and parallel), pseudoknots, triplexes and quadruplexes. The specification of the secondary structure is given with a descriptor that contains both the sequence and the structure pattern. Structural motifs can be defined with varying degrees of complexity. Each structure element has a set of search parameters that include length variation, sequence variation and some form of approximation. This last parameter allows RNAMotif to search for sequences that are similar to the descriptor, although not identical.
One of the major shortcomings of pattern-based descriptors is a general inability to incorporate context information. RNAMotif provides two mechanisms to resolve these problems, a rather simple sites list, as well as the much more powerful score section. Sites also provide a general way to specify long-range co-variation. The biggest limitation to using sites, however, is that it is ‘all or nothing’. The candidate sequence either passes all the site specifiers or it is rejected. This is often too severe and the score section can be used to create more flexible acceptance criteria.
The key to making the RNAMotif score section work was the realization that the symbols that specify the structure elements in the search pattern can have a second related meaning. In the constraint section they describe the motif, but in the score section they represent the actual sequences that matched these pattern elements. For example, in the pattern section, the symbols h5(tag=‘hlx-A’) represent the 5′-end of a standard duplex labeled hlx-A, but the same symbols in the score section would represent the actual nucleotides that matched that part of the pattern. Allowing these symbols to act as read-only string variables with length and sub-string operations and combining them with standard programming constructs (such as assignments, expression, testing and looping) allows implementation of arbitrary context relations that make RNAMotif more powerful than earlier motif search tools. It also permits most types of standard calculations and evaluations to be performed on the matched elements and can report multiple scores for the same region. This provides the user complete control and flexibility to fully evaluate any given match based on one or more criteria simultaneously.
We have here discussed a few well-characterized RNA structures as examples of the capabilities of the new descriptor language in RNAMotif. We used a combination of sequence/structure constraints in the descriptor section with a user defined score section to separate the true signals from the background noise. In the case of IRE, we combined alternative forms of the structure into a single motif and searched for them simultaneously. This represents a significant advantage during analysis and minimized redundant hits. Further, by appropriately tuning the scoring functions, most false positives were eliminated. Using this method we were able to correctly identify all previously described IRE-containing messages (suggesting that there were no false negatives). A few potentially new IRE-containing UTRs may have been identified, most notably in the fukutin and caveolin (Cav3) 3′-UTRs. Both of these are implicated in muscular dystrophy. Iron potentiates the generation of the highly reactive and toxic hydroxyl radical, resulting in oxidative damage, and thus may play an important role in muscular dystrophy (35). However, it remains to be determined if fukutin or caveolin is directly regulated by iron levels in the cell.
Stem–loop IV in SRP-RNA is a conserved structural motif that is present in almost all known organisms. This region in fact serves as a good signature for SRP-RNA identification and can be used to annotate SRP in new genomes. The RNAMotif scoring scheme and descriptor shown in Figure 3B was used to search the 38 complete prokaryotic genomes present in GenBank as of February 2001. The SRP-RNAs annotated in 21 of these genomes were correctly identified (Table 5). In addition, we were also able to predict the locations of SRP-RNAs in 16 of the remaining 17 genomes. The only exception was Buchnera sp., which is an endocellular bacterial symbiont of aphids with a very small genome and no documented SRP. These results correlate very well with the data maintained in the SRP database maintained by C. Zwieb. The descriptor was also used to analyze the entire GenBank nucleotide sequence database. As shown in Figure 4B, we identified all previously known instances of SRP-RNA. Our score discriminates well between true and false hits and, based on this, we predict the existence of several additional SRP-RNAs.
Another significant improvement in RNAMotif over previous pattern discovery tools is that it allows new pattern definitions to be built up from existing ones. A pre-processor that allows file inclusions achieves this. This allows us to break RNA structures down to their minimal required units and search for combinations of the various motifs in random order. We are currently building a database of all known RNA motifs and representing these using the RNAMotif descriptor language.
PROGRAM AVAILABILITY
RNAMotif is written in ANSI C. It is a small program of about 15 000 lines. The descriptor language is specified via lex and yacc (36). RNAMotif is run from the command line and works on most Unix/Linux systems. The descriptor is read from a file and the sequences to search, in fasta format, are read either from stdin or one or more files specified on the command line. The program as source code, with a complete user manual, is available via ‘anonymous ftp’ at ftp.scripps.edu/pub/macke/rnamotif-version.tar.gz.
Acknowledgments
ACKNOWLEDGEMENTS
The ideas for a new RNA description language were formalized in the fall of 1998 at the RNA Summit held at Ibis Therapeutics in Carlsbad, CA. The authors thank Dr Christian Massire, Dr Elena Lesnik, Dr Harold Levene, Tim Henderson and Nan Lin for valuable input at several stages of RNAMotif development. This work was supported in part by the NIH (grant GM48207 awarded to R.R.G.) and the Welch foundation (R.R.G.).
REFERENCES
- 1.Theil E.C. (1998) The iron responsive element (IRE) family of mRNA regulators. Regulation of iron transport and uptake compared in animals, plants and microorganisms. Met. Ions Biol. Syst., 35, 403–434. [PubMed] [Google Scholar]
- 2.Kim H.Y., LaVaute,T., Iwai,K., Klausner,R.D. and Rouault,T.A. (1996) Identification of a conserved and functional iron-responsive element in the 5′-untranslated region of mammalian mitochondrial aconitase. J. Biol. Chem., 271, 24226–24230. [DOI] [PubMed] [Google Scholar]
- 3.Son S.Y. (1993) The structure and regulation of histone genes. Saenghwahak Nyusu, 13, 64–70. [Google Scholar]
- 4.Shepherd R.K., Gabryszuk,J., Al-Ali,M., Allen,C.A., Joyce,I., Holmes,W.M. and Zehner,Z.E. (1997) A dual stem–loop structure in the 3′ untranslated region of vimentin mRNA binds specific protein(s). Nucleic Acids Symp. Ser., 36, 142–145. [Google Scholar]
- 5.Zehner Z.E., Shepherd,R.K., Gabryszuk,J., Fu,T.-F., Al-Ali,M. and Holmes,W.M. (1997) RNA–protein interactions within the 3′ untranslated region of vimentin mRNA. Nucleic Acids Res., 25, 3362–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gautheret D., Major,F. and Cedergren,R. (1990) Pattern searching/alignment with RNA primary and secondary structures: an effective descriptor for tRNA. Comput. Appl. Biosci., 6, 325–331. [DOI] [PubMed] [Google Scholar]
- 7.Laferriere A., Gautheret,D. and Cedergren,R. (1994) An RNA pattern matching program with enhanced performance and portability. Comput. Appl. Biosci., 10, 211–212. [DOI] [PubMed] [Google Scholar]
- 8.Billoud B., Kontic,M. and Viari,A. (1996) Palingol: a declarative programming language to describe nucleic acids’ secondary structures and to scan sequence database. Nucleic Acids Res., 24, 1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pesole G., Liuni,S. and D’Souza,M. (2000) PatSearch: a pattern matcher software that finds functional elements in nucleotide and protein sequences and assesses their statistical significance. Bioinformatics, 16, 439–450. [DOI] [PubMed] [Google Scholar]
- 10.Fichant G.A. and Burks,C. (1991) Identifying potential tRNA genes in genomic DNA sequences. J. Mol. Biol., 220, 659–671. [DOI] [PubMed] [Google Scholar]
- 11.el-Mabrouk N. and Lisacek,F. (1996) Very fast identification of RNA motifs in genomic DNA. Application to tRNA search in the yeast genome. J. Mol. Biol., 264, 46–55. [DOI] [PubMed] [Google Scholar]
- 12.Lisacek F., Diaz,Y. and Michel,F. (1994) Automatic identification of group I intron cores in genomic DNA sequences. J. Mol. Biol., 235, 1206–1217. [DOI] [PubMed] [Google Scholar]
- 13.Aho A.V., Kernighan,B.W. and Weinberger,P.J. (1987) The AWK Programming Language. Addison Wesley, Reading, MA.
- 14.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 15.Wootton J.C. and Federhen,S. (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol., 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 16.Woese C.R., Winker,S. and Gutell,R.R. (1990) Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc. Natl Acad. Sci. USA, 87, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutell R.R., Cannone,J.J., Shang,Z., Du,Y. and Serra,M.J. (2000) A story: unpaired adenosine bases in ribosomal RNAs. J. Mol. Biol., 304, 335–354. [DOI] [PubMed] [Google Scholar]
- 18.Leontis N.B. and Westhof,E. (1998) The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA, 4, 1134–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varani G., Wimberly,B. and Tinoco,I.,Jr (1989) Conformation and dynamics of an RNA internal loop. Biochemistry, 28, 7760–7772. [DOI] [PubMed] [Google Scholar]
- 20.Wimberly B., Varani,G. and Tinoco,I.,Jr (1993) The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry, 32, 1078–1087. [DOI] [PubMed] [Google Scholar]
- 21.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 22.Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R., Carter,A.P., Vonrheins,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 23.Ke Y., Sierzputowska-Gracz,H., Gdaniec,Z. and Theil,E.C. (2000) Internal loop/bulge and hairpin loop of the iron-responsive element of ferritin mRNA contribute to maximal iron regulatory protein 2 binding and translational regulation in the iso-iron-responsive element/iso-iron regulatory protein family. Biochemistry, 39, 6235–6242. [DOI] [PubMed] [Google Scholar]
- 24.McKie A.T., Marciani,P., Rolfs,A., Brennan,K., Wehr,K., Barrow,D., Miret,S., Bomford,A., Peters,T.J., Farzaneh,F., Hediger,M.A., Hentze,M.W. and Simpson,R.J. (2000) A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol. Cell, 5, 299–309. [DOI] [PubMed] [Google Scholar]
- 25.Thomson A.M., Rogers,J.T. and Leedman,P.J. (1999) Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int. J. Biochem. Cell Biol., 31, 1139–1152. [DOI] [PubMed] [Google Scholar]
- 26.Schlegl J., Gegout,V., Schlager,B., Hentze,M.W., Westhof,E., Ehresmann,C., Ehresmann,B. and Romby,P. (1997) Probing the structure of the regulatory region of human transferrin receptor messenger RNA and its interaction with iron regulatory protein-1. RNA, 3, 1157–1172. [PMC free article] [PubMed] [Google Scholar]
- 27.Gdaniec Z., Sierzputowska-Gracz,H. and Theil,E.C. (1998) Iron regulatory element and internal loop/bulge structure for ferritin mRNA studied by cobalt(III) hexammine binding, molecular modeling and NMR spectroscopy. Biochemistry, 37, 1505–1512. [DOI] [PubMed] [Google Scholar]
- 28.Minetti C., Sotgia,F., Bruno,C., Scartezzini,P., Broda,P., Bado,M., Masetti,E., Mazzocco,M., Egeo,A., Donati,M.A., Volonte,D., Galbiati,F., Cordone,G., Bricarelli,F.D., Lisanti,M.P. and Zara,F. (1998) Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nature Genet., 18, 365–368. [DOI] [PubMed] [Google Scholar]
- 29.McNally E.M., de Sa Moreira,E., Duggan,D.J., Bonnemann,C.G., Lisanti,M.P., Lidov,H.G., Vainzof,M., Passos-Bueno,M.R., Hoffman,E.P., Zatz,M. and Kunkel,L.M. (1998) Caveolin-3 in muscular dystrophy. Hum. Mol. Genet., 7, 871–877. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi K., Sasaki,J., Kondo-Iida,E., Fukuda,Y., Kinoshita,M., Sunada,Y., Nakamura,Y. and Toda,T. (2001) Structural organization, complete genomic sequences and mutational analyses of the Fukuyama-type congenital muscular dystrophy gene, fukutin(1). FEBS Lett., 489, 192–196. [DOI] [PubMed] [Google Scholar]
- 31.Toda T., Kobayashi,K., Kondo-Iida,E., Sasaki,J. and Nakamura,Y. (2000) The Fukuyama congenital muscular dystrophy story. Neuromuscul. Disord., 10, 153–159. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz U., Behrens,S., Freymann,D.M., Keenan,R.J., Lukavsky,P., Walter,P. and James,T.L. (1999) Structure of the phylogenetically most conserved domain of SRP RNA. RNA, 5, 1419–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitz U., James,T.L., Lukavsky,P. and Walter,P. (1999) Structure of the most conserved internal loop in SRP RNA. Nature Struct. Biol., 6, 634–638. [DOI] [PubMed] [Google Scholar]
- 34.Batey R.T., Rambo,R.P., Lucast,L., Rha,B. and Doudna,J.A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science, 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- 35.Polla B.S. (1999) Therapy by taking away: the case of iron. Biochem. Pharmacol., 57, 1345–1349. [DOI] [PubMed] [Google Scholar]
- 36.Levine J., Mason,T. and Brown,D. (1992) Lex & Yacc, 2nd Edn. O’Reilly & Associates, Sebastapool, CA.