Abstract
Lung diseases carry a significant burden of morbidity and mortality worldwide. The advent of digital pathology (DP) and an increase in computational power have led to the development of artificial intelligence (AI)‐based tools that can assist pathologists and pulmonologists in improving clinical workflow and patient management. While previous works have explored the advances in computational approaches for breast, prostate, and head and neck cancers, there has been a growing interest in applying these technologies to lung diseases as well. The application of AI tools on radiology images for better characterization of indeterminate lung nodules, fibrotic lung disease, and lung cancer risk stratification has been well documented. In this article, we discuss methodologies used to build AI tools in lung DP, describing the various hand‐crafted and deep learning‐based unsupervised feature approaches. Next, we review AI tools across a wide spectrum of lung diseases including cancer, tuberculosis, idiopathic pulmonary fibrosis, and COVID‐19. We discuss the utility of novel imaging biomarkers for different types of clinical problems including quantification of biomarkers like PD‐L1, lung disease diagnosis, risk stratification, and prediction of response to treatments such as immune checkpoint inhibitors. We also look briefly at some emerging applications of AI tools in lung DP such as multimodal data analysis, 3D pathology, and transplant rejection. Lastly, we discuss the future of DP‐based AI tools, describing the challenges with regulatory approval, developing reimbursement models, planning clinical deployment, and addressing AI biases. © 2022 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: lung diseases, digital pathology, computational pathology, machine learning, artificial intelligence
Introduction
Lung diseases are among the leading causes of mortality worldwide, accounting for nearly 1/6th of death [1]. Over the last 2 years, the COVID‐19 pandemic has had a profound impact across the globe, with nearly 5.6 million deaths to date [2]. Tuberculosis is one of the most common causes of death in developing countries and the immunocompromised population [3]. Meanwhile, lung cancer is the leading cause of cancer‐related mortality worldwide, with nearly 1.8 million deaths in 2020 [4]. Non‐small cell lung cancer (NSCLC) is the predominant subtype, accounting for ~85% of the cases, and includes squamous cell and adenocarcinoma, which are the most common histological subtypes [5]. Small cell lung cancer, an aggressive form with high mortality, meanwhile accounts for ~15% of the cases [6].
Histopathologic tissue analysis forms the backbone of lung disease characterization and management [7, 8, 9], and histopathologic confirmation remains the gold standard in clinical workflow for diagnosis [10]. Tissue samples obtained from lung biopsies and resections are analyzed by pathologists to detect and classify cells, identify tumor morphology and subtype, and evaluate features that predict response to treatment and prognosis [7]. Lung pathology is unique, with several distinct biomarkers. For instance, lung adenocarcinoma (LUAD) is driven by accumulated genetic alterations (‘driver mutations’) such as EGFR, KRAS mutations, and ALK rearrangements [11, 12, 13]. In this era of immunotherapy, studies have focused on tumor‐infiltrating lymphocytes (TILs) and the tumor microenvironment to better characterize the immune response to tumors. Similarly, fibroblast foci have been investigated as a morphologic marker in interstitial lung diseases, with prognostic implications [14, 15]. Studies from COVID‐19 lung autopsies have shown a distinct immune architecture with neutrophilic and lymphocytic predominance [16, 17, 18]. The vast number of biomarkers visually discernible on histopathology images creates an opportunity to apply artificial intelligence (AI) tools to assist clinical workflow.
The advent of whole slide scanning along with improvements in AI‐based tools has paved the way for advances in computational pathology (Table 1). Mukhopadhyay et al [29] compared the diagnostic performance of digital pathology (DP) and traditional microscopy‐based methods in a study with specimens from 1,992 patients, evaluated by 16 surgical pathologists from four institutions. The study found that DP was non‐inferior to conventional microscopy for primary diagnosis in surgical pathology. The results paved the way for the Food and Drug Administration (FDA) [30] approval of whole slide imaging through the de novo premarket review pathway [31] for primary diagnosis in surgical pathology in the United States and its widespread implementation. Meanwhile, the FDA granted a 510(k) clearance to the first DP‐based AI tool, Paige Prostate, for detecting prostate cancer on whole slide images (WSIs) [32]. While many applications of AI in pathology have focused on breast [33, 34, 35, 36], prostate [37, 38, 39, 40], and head and neck cancers [41, 42, 43, 44], there has been a steady interest in applying these tools in the context of lung diseases. The introduction of AI‐based tools, with their power to unlock pathological diagnostic, prognostic, and predictive features, could assist pathologists, pulmonologists, and thoracic oncologists to guide patient management [45] (Figure 1). In diagnostics, these tools can assist in detecting and quantifying morphological features, acting as a decision support system for pathologists. Prognosis involves stratifying patients based on their risk or likelihood of disease recurrence or progression and would aid pulmonologists and oncologists to plan the course of treatment. Additionally, AI tools could identify sub‐visual patterns which may predict response to treatments such as radiotherapy and immunotherapy, thereby assisting clinicians in selecting the appropriate treatment.
Table 1.
Description of common computational pathology terms
| Artificial intelligence | Umbrella term to indicate technologies with the ability to simulate intelligent behavior, allowing it to function appropriately and with foresight in its environment [19] |
| Digital pathology | Also known as whole slide imaging, it is the dynamic, image‐based environment that enables the acquisition, management, and interpretation of pathology information generated from a digitized glass slide [20]. Digital pathology has a growing interest in diagnostic medicine and lays the ground for AI tool deployment |
| Pathomics | The high throughput extraction of quantitative features from digitized histopathology images, thereby converting images into data [21] |
| Biomarker | “A biological marker, or biomarker, is a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes or a response to a therapeutic intervention” [22]. In digital pathology, it is an image characteristic that can be quantitatively measured and analyzed, and is typically associated with a disease or therapeutic outcome |
| Computational pathology | The use of artificial intelligence tools to extract information from whole slide images and associated patient metadata for specific clinical indications [23] |
| Machine learning | A subtype of artificial intelligence in which the system uses a provided set of data (training data) to learn and structure it, and subsequently uses a representative function to best describe the data and provide a prediction when presented with previously unseen data [24] |
| Deep learning | A type of machine learning technique with a representation‐learning method. Deep learning attempts to learn by example using neural networks [25] |
| Artificial neural network | An architecture inspired by the neuronal networks found within the brain and capable of constructing non‐linear relationships. Each layer in an artificial neural network has multiple perceptrons, which function like neurons, capable of receiving and sending signals [26] |
| Convolutional neural network (CNN) | A subtype of artificial neural network commonly applied to medical images; its extensive use derives from the architecture upholding the integrity of spatial relationships in image data. CNN is a feedforward network that specializes in filtering spatial data to create feature maps [27] |
| Generative adversarial network (GAN) | An example of generative AI, GAN is a deep learning approach that aims to ‘generate’ artificial data that resemble the real data using a combination of two neural networks – a generator, which creates synthetic data, and a discriminator, which scrutinizes the authenticity of the data [28] |
Figure 1.
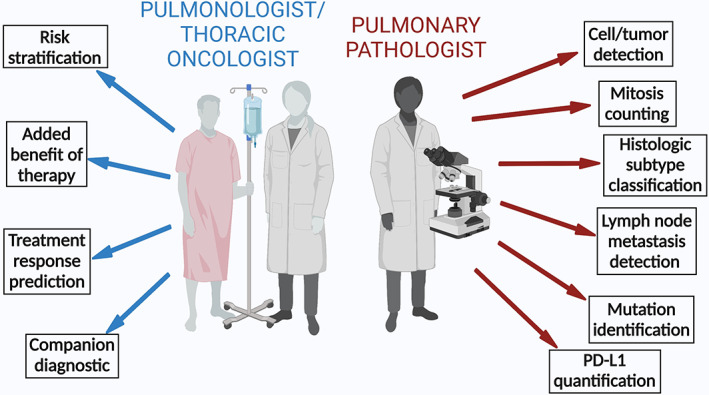
Digital pathology‐based AI tools add value to the workflow of the pulmonologist, oncologist, and pathologist. Created with Biorender.com.
Previous works in lung cancer have focused on the use of radiomics, which is a high throughput analysis of radiologic images [46]. In this article, we look at DP‐based AI tools used in lung diseases. First, we discuss various AI representations applied, elaborating on hand‐crafted and deep learning (DL)‐based methods. Next, we review the applications of AI approaches both in the field of lung cancers [47, 48, 49, 50] and in non‐oncologic diseases such as interstitial lung disease [51, 52, 53], tuberculosis [54, 55, 56], and COVID‐19 [57]. Further, we look at emerging applications, which we anticipate will be explored in the context of lung diseases. Lastly, we discuss the opportunities and challenges while developing AI algorithms regarding biases, regulatory approvals, reimbursements, and the clinical deployment of these tools.
AI approaches in lung pathology
AI is a broad discipline with multiple approaches to construct a model for a particular task (Table 1) [45]. There are two common ways to extract feature representations for building an AI model: DL‐based unsupervised feature learning and the hand‐crafted approach. While the former allows the system to identify features suitable for automatic classification, the latter uses existing domain expertise in lung histopathology to select the features. DL‐based unsupervised feature learning is favored in low‐level tasks such as cell and lung tumor detection and classification. This is useful since visual confirmation of the result is sufficient and does not require interpretation of the selected features. This contrasts with many high‐level tasks such as prognosis or treatment response prediction which require a certain level of interpretability and hence hand‐crafted domain‐inspired features may be favored by the medical community to construct these models [58].
Deep learning (DL)‐based unsupervised feature learning
Most WSIs are unlabeled, and annotating architecturally complex regions is laborious for pathologists, who are already facing increasing clinical workload [59, 60]. This hinders the development of machine learning models, which require well‐annotated datasets. Unsupervised feature learning is an approach in which the system automatically learns and selects appropriate features from the image to maximize class separability.
Convolutional neural network (CNN) [27] is a type of DL approach that has been used extensively in computational pathology models for a variety of tasks, including segmentation, object detection, and image classification [52, 55, 56, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70]. CNNs are composed of multiple layers of networks and are designed to learn spatial hierarchies of features automatically and adaptively through a backpropagation algorithm. They do so by deconvolution of the image content into thousands of salient features, followed by selection and aggregation of the most meaningful features and recognition of these patterns in yet‐unseen images. CNN is well suited for detecting, identifying, and classification of images. For example, upon providing a CNN with a set of images with the corresponding annotations, the network automatically learns the features and generates a probability map on a separate set of images. Inception v3 and Resnet50 are two common types of CNNs, representing supervised DL, that have been used in lung research for various tasks such as classification of histologic subtypes [65, 70, 71] or cell counting [72]. U‐Net is another CNN, where the ‘U architecture’ of the network combines the structural detail with its spatial context [73]. Hence, such models are used for quick and accurate segmentation of WSIs [74, 75, 76, 77] (Figure 2). With a particular interest in TILs as a prognostic biomarker in NSCLC, AlexNet [56, 75] has been employed in classifying TIL shape and density. Trained DL models are often not interpretable and hence are regarded as being ‘black‐box’ in nature [78]. This means that it is often challenging to mechanistically link the individual layers of a neural network function to a corresponding output, or which features from an image helped the DL model attain corresponding results. For some tasks, such as training a DL algorithm to identify nuclei or other structures, a visual confirmation of the output of the model by a domain expert is sufficient to test its accuracy and subsequently build imaging biomarkers that might be interpretable [79]. On the other hand, if the DL model generates a direct prognosis, the lack of interpretability might undermine trust among the physicians [80].
Figure 2.
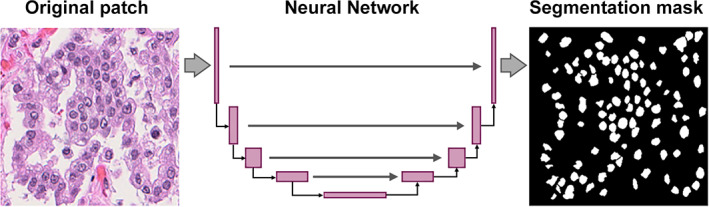
Example of a neural network architecture for nuclei segmentation.
Hand‐crafted approach
In the hand‐crafted approach, relevant features from the data are manually selected. Domain knowledge is employed for feature engineering, which is subsequently used to construct machine learning models. This requires close collaboration between clinicians and machine learning engineers to construct appropriate AI models. In contrast to DL‐based unsupervised feature learning, through incorporating the expertise of subject matter experts, hand‐crafted‐based models lend themselves to some interpretability. Hand‐crafted features can be either domain‐agnostic or domain‐inspired (Table 2). The former may include morphometric features like shape, texture, and gland features of nuclei, which are common to multiple organs and tissue, while domain‐inspired features may have relevance to a particular disease site, like collagen fiber orientation, which was recently implicated in determining the aggressiveness of breast cancer [91]. In lung cancer, TILs [77] and cancer cell nuclei [74] have been explored as biomarkers to predict recurrence and risk of recurrence or death in NSCLC (Figure 3). For instance, the knowledge that hyperchromatic nuclei in malignant cells lead to a folded nuclear membrane [92] can be captured as pixel intensity variation of cancer nuclei using AI tools. Similarly, there has been a growing understanding that tumors behave as a cluster of cells [93], allowing graph‐based measurements of the tumor nuclei to analyze the spatial relationship within the tumor to assess its aggressiveness. Hand‐crafted features also facilitate the construction of AI models rooted in the understanding of cellular architecture and morphology. Studies have demonstrated that the density of TILs has a prognostic value [94]. An automated approach to quantify TIL density can overcome the challenge with inter‐reader variability in manual assessment. Similarly, the relationship of cells in the tumor microenvironment holds relevance, and mapping them could add prognostic value. For example, a low PD‐1 to CD‐8 ratio has been associated with a better prognosis in NSCLC [95]. Hand‐crafted features quantitatively analyzing the spatial architecture of the cells (immune, cancer, stroma) have been performed by constructing Delaunay triangulation (Figure 4). Likewise, textural‐based features have been analyzed using a gray level co‐occurrence matrix. For example, textural features extracted from the peri‐nuclear region in lung cancer may represent the consistency of cytoplasmic staining, with heterogeneous cytoplasm indicative of aggressive tumors. Lu et al [75] demonstrated a novel method of constructing feature‐driven local cell graphs (FLocK) that take into account nuclear intensity in addition to nuclear proximity, thereby adding granularity to the algorithm. They demonstrated that FLocK features extracted from WSIs of early‐stage NSCLC patients were associated with overall survival. While such hand‐crafted approaches alleviate some of the apprehension associated with the black‐box nature of DL models and inspire confidence among clinicians, they require extensive involvement of domain experts in defining regions of interest and interpreting results.
Table 2.
Description of hand‐crafted features used in lung pathology applications
| Feature class | Description | Mathematical expression | Lung pathology studies |
|---|---|---|---|
| Spatial architecture | Describes the spatial organization of primitives (e.g. nuclei or glands) in the tissue. Facilitates identification of areas with a high disorder such as cancerous regions | Voronoi diagram, Delaunay triangulation, minimum spanning tree, local graphs | Yao et al [81], Wang et al [82], Vaidya et al [83] |
| Spatial interplay | Extracts measures from the points where two or more structures in the tissue come together and affect each other | Intersection, neighborhood diversity | Lu et al [75], Saltz et al [84], Barrera et al [85], Corredor et al [57], Corredor et al [77] |
| Texture | Quantifies the spatial arrangement of color or intensities in an image region. It is useful to differentiate among different structures in the tissue, for example, lymphocytes or cancerous cells | Haralick (gray level co‐occurrence matrix), Gabor, Laplacian, Laws features | Yao et al [81], Wang et al [82], Vaidya et al [83], Sandino et al [86], Corredor et al [87], Wang et al [88], Alvarez‐Jimenez et al [89] |
| Shape | Provides information on the physical appearance (e.g. size or silhouette) of some structures in the tissue such as cells, cartilages, vessels, nodules, among others | Area, length of axes, eccentricity, equivalent diameter, Zernike moments | Wang et al [82], Vaidya et al [83], Corredor et al [87], Lu et al [75] |
| Color | Extracts measures associated with the perception derived from the spectrum of light interaction with the human eye | Intensity, RGB channels, YUV channels, CMYK channels, HSV channels | Wang et al [82], Lu et al [90], Corredor et al [86] |
| Orientation | Provides metrics of the position of an element (e.g. nucleus) in relation to its surroundings | Angle between axes | Wang et al [82], Vaidya et al [83] |
Figure 3.
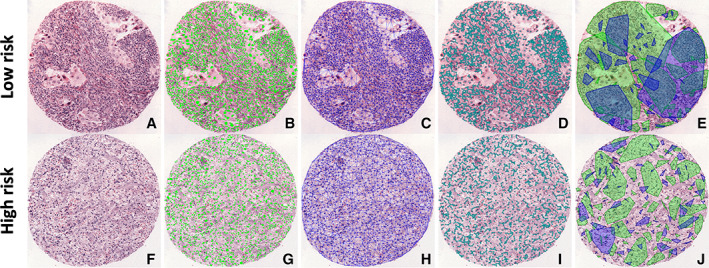
Illustration of the use of hand‐crafted features for risk stratification of lung cancer. Tissue microarray from low‐risk and high‐risk lung cancer patients showing: (A, F) digitized H&E images; (B, G) features with automated nuclei detection; (C, H) features with Delaunay triangulation – a type of global graph; (D, I) local graph based on the distance between closest nuclei; and (E, J) feature showing the spatial architecture of tumor‐infiltrating lymphocytes with lymphocytes (blue) and non‐lymphocytes (green). Clusters are built based on distance thresholds. If cells of the same type are closer than a threshold, they form a cluster.
Figure 4.
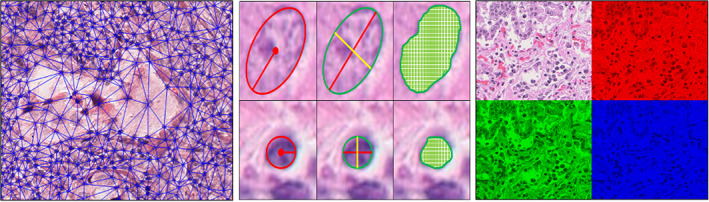
Illustrative examples of hand‐crafted features used in lung pathology applications. Left to right: Delaunay triangulations capturing the spatial architecture; shape features: equivalent diameter, length of axes, and area; and color features demonstrating the RGB channels.
AI applications in lung pathology
Biomarker quantification and histogenomics
In recent years, the need for an increased number of ancillary studies such as immunohistochemistry, driver mutations, and programmed death‐ligand 1 (PD‐L1) has placed growing demands on pathologists, a scenario where they have to ‘do more with less’ [96, 97, 98]. Computational approaches have been developed to automate the estimation of biomarkers, as well as for the discovery of newer ones by integrating complementary modalities of data. For instance, computational pathology approaches which provide spatial and morphological information of the disease from WSIs have been combined with genomics, which provides the molecular profile of disease to aid in discovery of newer biomarkers [99, 100].
Immunohistochemical determination of tumor PD‐L1 status has become routine in identifying patients who may benefit from programmed death‐1 (PD‐1)/PD‐L1 inhibitors in NSCLC [101]. Currently, pathologists visually estimate the fraction of tumor cells expressing PD‐L1 via a tumor proportion score (TPS) system, which is constrained by ill‐defined cutoffs, differences in positivity estimation, and inter‐observer variability, making it a tedious and error‐prone task. It is also challenged by inadequate tissue samples or a lack of resources in peripheral settings [102]. Sha et al [103] developed a ResNet‐based DL model on WSIs to predict PD‐L1 status, thereby developing a ‘virtual stain’ using H&E WSIs alone. They used H&E and immunohistochemistry WSIs from 130 patients with pathologist‐annotated PD‐L1 positive and negative tumor regions on training samples as the ground truth. The model remained effective over a range of PD‐L1 cutoff thresholds [area under the receiver operating characteristics curve (AUC) = 0.67–0.81, p ≤ 0.01], even when different proportions of the labels were randomly shuffled to simulate inter‐pathologist disagreement (AUC = 0.63–0.77, p ≤ 0.03). Generative adversarial networks (GAN) (Table 1) have been explored as an approach for virtual H&E staining [104, 105]. Bayramoglu et al [106] used a conditional GAN model for virtual H&E staining of unstained lung tissue, which can allow automating DP workflow. Meanwhile, Kapil et al [107] developed a semi‐supervised GAN‐based approach to automate an objective PD‐L1 expression scoring method for patients with late‐stage NSCLC, reducing the need for manual annotations. Wu et al [108] developed a U‐Net‐based end‐to‐end DL model to automatically detect tumors, segment cells, and calculate the TPS of PD‐L1 by highlighting PD‐L1‐positive tumor cells. In a study with 437 NSCLC patients, Xia et al [109] demonstrated the utility of computational pathology approaches in the discovery of newer biomarkers. Using tissue microarrays, they discovered a new histologic feature, the stroma inflammation score, which was highly correlated with patient overall survival in NSCLC. This biomarker was subsequently validated by two blinded observers for prognostic value.
The World Health Organization classification of lung tumors has been recently modified [110] to emphasize the role of genetic mutations, given our increasing understanding of molecular profiles [110, 111]. Identifying the driver mutation is crucial for NSCLC management [112]. For example, LUADs with EGFR, ALK, ROS1, or RET mutations are associated with specific targeted therapies, and the KRAS G12C mutation is targeted by KRAS inhibitors [113, 114]. Coudray et al [71] trained a CNN model for a multi‐task classification, predicting the ten most common genetic mutations from H&E‐stained WSIs. The six commonly found mutations including STK11, EGFR, FAT1, SETBP1, KRAS, and TP53 were predicted by the model with AUCs between 0.733 and 0.856.
Lung adenocarcinoma is unique for having driver mutations which are potential targets for therapy. The recent approval of sotorasib for KRAS G12C‐mutated NSCLC [115] demonstrates the growing interest in developing novel therapies that target driver mutations. This often requires a companion diagnostic test that checks for the presence of specific mutations. These genomic assays often have longer turnaround times [116], and the costs could be prohibitively expensive, especially in lower‐middle‐income countries [117]. However, AI tools could be developed to detect the presence of point mutations from H&E slides alone, along with being non‐disruptive [118].
The diagnosis of idiopathic pulmonary fibrosis (IPF) requires a pathologic or radiologic diagnosis of usual interstitial pneumonia (UIP) [119]. In more than half of the patients, the diagnosis of UIP cannot be made by imaging alone, requiring a surgical biopsy, associated with a risk of morbidity and mortality [120]. Although transbronchial lung biopsy is a low‐yield substitute, it poses challenges for pathologists to confirm a diagnosis. Raghu et al [51] trained a machine learning model using histopathology and RNA sequence data from 90 patients to identify UIP. With the addition of molecular data, obtained along with biopsy samples, they demonstrated a significant improvement in the performance of the model. In addition, fibroblast foci are a known biomarker in IPF and increasing numbers of fibroblast foci have been associated with a poorer prognosis in some studies [15]. Mäkelä et al [52] trained a CNN to quantify fibroblast foci, interstitial mononuclear inflammation, and intra‐alveolar macrophages, and analyzed the association of these parameters with survival. Interstitial mononuclear inflammation and intra‐alveolar macrophages were associated with prolonged survival, suggesting their role as novel prognostic histologic biomarkers in IPF. In addition, the CNN‐based DL model also confirmed the association between high numbers of fibroblast foci and poor prognosis for patients with IPF.
Computer‐aided diagnosis (CAD)
Traditional pathology involves the use of a microscope by pathologists to analyze tissue samples for architecture, cellular patterns, and structures, and interpretation of these findings in the context of the clinical picture. This process is inherently subjective and prone to inter‐observer variability [121, 122, 123]. In some instances, pathologic analysis requires repetitive tasks such as counting mitoses in malignant cells or manual enumeration of cells positive for certain immunohistochemical markers (e.g. Ki‐67). Since computational approaches automate these processes, they have the potential to improve diagnostic accuracy and workflow efficiency, cutting down the time that pathologists spend on time‐consuming tasks, allowing more time for intellectually demanding aspects of interpretation [10, 124].
An issue that has become a focus of interest over the last 12 years due to advances in targeted therapies has been accurate histologic subtyping of non‐small cell lung carcinomas (NSCLCs). Despite substantial advances in immunohistochemical subtyping of NSCLC that have facilitated tumor subtyping [125], the accurate interpretation of subtypes of NSCLC can occasionally be challenging, particularly in poorly differentiated examples with overlapping immunophenotypes [126]. In one of the first studies to demonstrate the utility of CNNs in distinguishing between lung cancer subtypes, Coudray et al [71] trained a de novo Inception v3 network using transfer learning to automate the histologic type classification of each slide. Notably, in contrast to real‐life practice, where pathologists need to pick between dozens of choices when faced with a lung biopsy, the task in this study was limited to the separation of LUAD and squamous cell carcinoma of the lung. Another noteworthy point is that pathologists were allowed to use H&E morphology only, in contrast to real‐life practice, where immunohistochemistry is used for subtyping of cases that cannot be subtyped on H&E. Having acknowledged these limitations, the tumor subtype agreement between the DL model and the classification provided by the gold standard for the study (The Cancer Genome Atlas; TCGA) was higher than the agreement between each pathologist and the TCGA. In analyzing the slides that posed challenges to pathologists but were correctly classified by the model, it performed well even with poorly differentiated tumors, demonstrating the advantage of CNN approaches in ambiguous cases.
In LUAD, in addition to the stage at the time of diagnosis, which has been the most important prognostic parameter for decades, the predominant tumor growth pattern also impacts the outcome. The lepidic growth pattern is the least aggressive; papillary and acinar growth patterns are considered intermediate; and micropapillary and solid growth patterns are categorized as the most aggressive, associated with an increased likelihood of recurrence and poor survival [127]. Recently, the cribriform pattern has also been identified as a marker of unfavorable prognosis in early‐stage LUAD [128].
In another application, Gertych et al [64] constructed a lightweight CNN using an AlexNet model. The goal was to assess the accuracy of the CNN in identifying four LUAD growth patterns (acinar, micropapillary, solid, and cribriform) in surgically resected cases. Compared with the gold standard (pathologist interpretation of tumor versus non‐tumor and LUAD growth patterns), they also observed that de novo trained models performed significantly better than pre‐trained models and that the model performed better in some validation sets than in others. Wei et al [129] used a ResNet model for a multi‐label classification problem. They trained the model on 422 WSIs of LUAD and compared the output of the model in predicting the probability of six histological subtypes against three pathologists. The model achieved a Kappa score of 0.525 and showed an agreement of 66.6% with three pathologists, which was higher than the inter‐pathologist Kappa score (0.485). Yu et al [130] recently constructed a fully automated framework to identify tumor regions and their histologic subtype, and correlate them with individual transcriptomic profiles, allowing the tool to function as an end‐to‐end decision support system for thoracic pathologists in lung cancer evaluation.
AI tools have also been applied to cytopathology samples obtained via fine needle aspiration, typically in the form of individual cells and smaller tissue fragments. They are less invasive than a biopsy and are increasingly performed for the evaluation and staging of lung cancer [131]. The individual cells seen in these samples can be computationally analyzed for cellular and nuclear morphology. In 2020, Gonzalez et al [132] studied cytology and biopsy specimens from 40 patients and trained a CNN‐based DL algorithm to distinguish small cell lung carcinoma (SCLC) and large cell neuroendocrine carcinoma based on morphologic features. The number of cases used was very small, although in this limited sample both the cytology algorithm and the biopsy algorithm were able to correctly identify most cases of both tumors, demonstrating the feasibility of using cytology specimens for CAD systems.
One challenge for pathologists is to identify metastatic tumor deposits in lymph node biopsies and excisions, which can be difficult in a background of lymphocytes, histiocytes, endothelial cells, and other distractors. To resolve this, Pham et al [61] developed a two‐step DL model with a CNN approach, in which the first algorithm eliminated all lymphoid follicles from the WSI and subsequently detected ‘true positive’ nodal metastasis in the second step. Using this DL method, they reduced errors by 36.4% on average and up to 89% in slides with reactive lymphoid follicles. Although developed on lung cancer patients, the detection of metastatic tumor cells in a background of reactive immune cells can be expanded to other disease indications as well.
Corredor et al [57] performed one of the first studies applying computational pathology approaches in COVID‐19 to quantitatively characterize the spatial architecture of lymphocytes and non‐lymphocytic cells on digitized autopsy images from patients who died of COVID‐19 and compared it with those who died of H1N1, a comparable viral infection. While visual assessment of lymphocytes in COVID‐19 samples has been performed [133, 134], it was qualitative with inter‐reader variability. In contrast, in this study, the authors used a watershed approach to segment the nuclei, a machine learning classifier to identify lymphocytes, and subsequently constructed clusters of cells to analyze spatial patterns.
Tuberculosis screening is a laborious task that involves the detection of acid‐fast bacilli on a pathology slide stained with acid‐fast stains such as the Ziehl–Neelsen stain [135, 136]. Experts have described best practices for the detection of acid‐fast bacteria in pathology specimens, but these remain labor‐intensive and time‐consuming [137]. In a complicated study that involved errors both in the pathologist‐based ground truth and in the AI model, Xiong et al [54] developed a CNN model, named tuberculosis AI (TB‐AI) by feeding patches from positive and negative regions of the WSI to train the model. Although the TB‐AI system initially misdiagnosed several cases of dye residue and contaminants as acid‐fast bacteria, the algorithm was improved in a subsequent run, achieving 97.94% sensitivity and 83.65% specificity. Lo et al [56] developed a CNN classification model using 1,815 image blocks to identify acid‐fast bacilli with an accuracy of 95.3%. In addition, they repeated the same experiment without color and saw a drop of accuracy to 73.8%, thereby demonstrating the role of color information in detections (Figure 5).
Figure 5.
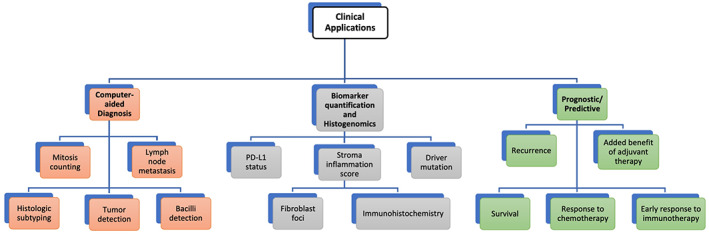
Applications of computational pathology in lung diseases.
Prognostic and predictive applications
Pleural mesothelioma is a rare malignancy of the pleura, with a poor prognosis [138]. Courtiol et al [67] developed MesoNet, a fully unsupervised deep CNN using WSIs to predict the overall survival of mesothelioma patients without any pathologist‐provided locally annotated regions. They validated the model on an internal validation cohort and an independent cohort from the TCGA. Subsequently, they demonstrated the model to be more accurate in predicting patient survival than current pathology practices. However, unlike classical black‐box DL methods, MesoNet identified regions contributing to patient outcome prediction, primarily in the stroma.
The classification of LUAD developed by the International Association for the Study of Lung Cancer (IASLC), the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was an effort to develop an integrated approach to classify NSCLC and identify prognostic and predictive biomarkers [139]. While it demonstrated that growth patterns in LUAD are prognostic, they are not currently used for clinical decision making, and there remains a lack of robust and objective biomarkers to determine the risk of recurrence and metastasis in patients with early‐stage LUAD. In early‐stage NSCLC treated with surgery, the added benefit of adjuvant chemotherapy is unknown, and administration depends on each patient's risk of recurrence. However, there are no validated biomarkers to predict the risk of recurrence accurately and consistently.
Yu et al [140] developed a prognostic model for early‐stage NSCLC using quantitative histomorphometric analysis by combining an automated feature extraction method with a machine learning classifier. They analyzed local anatomical structures (nuclei and cytoplasmic shape) as well as global features (nuclear and cellular texture) and found them to be predictive of patient survival. In a similar study, Luo et al [141] demonstrated that nuclear and cell texture features were independently prognostic of survival across all stages of NSCLC. In a study with 305 patients, Wang et al [82] focused on graph features extracted from tumor nuclei features of tumor cells on tissue microarray core images to develop a supervised classification model for predicting recurrence in early‐stage NSCLC. Using a multivariable Cox proportional hazard model, including additional markers such as gender and disease stage, the computationally derived risk score was found to be independently prognostic. A unique prospect of computational pathology techniques is the ability to perform multiple analyses stepwise. In a subsequent study with over 1,000 patients, Wang et al [74] developed an image risk score from nuclear and perinuclear histomorphometric features which was prognostic in early‐stage NSCLC. In addition, it was one of first models in lung cancers to additionally predict the added benefit of adjuvant chemotherapy by demonstrating a subset of high‐risk patients treated with only surgery who might have had a better survival if treated with adjuvant chemotherapy. Saltz et al [84] used WSIs from the TCGA to study the association of TIL patterns with molecular features and overall survival. Corredor et al [77] analyzed the spatial architecture of TILs and non‐lymphocytes as well as the interplay and co‐localization of these cells, and demonstrated features associated with recurrence in early‐stage NSCLC patients.
Lu et al [76] developed a feature classifier named CellDiv, which analyzes the local cellular morphological diversity displayed by lung cancer owing to its heterogeneity. CellDiv features showed a strong correlation with overall survival in early‐stage NSCLC in addition to being able to distinguish KRAS mutation status and being associated with biologic pathways of cellular differentiation, apoptosis, and signaling. Vaidya et al [83] took an approach combining the top predictive features from lung CT scans and H&E WSIs to construct an integrated ‘RaPtomic’ classifier to predict 5‐year recurrence free‐survival in NSCLC. The integrated model had a better performance (AUC 0.78; p < 0.005) than radiomic (AUC 0.74; p < 0.01) and pathomic (AUC 0.67; p < 0.05) features alone.
In SCLC, Jain et al [142] studied the association of quantitative features of tumor nuclei extracted from platinum‐based chemotherapy‐treated patients with prognosis and observed that the top features capturing nuclei shape, size, and texture were able to stratify patients based on their overall survival.
In the last decade, several novel therapeutic agents have been discovered to treat previously incurable lung diseases [142, 143, 144, 145, 146, 147, 148]. While targeted agents such as immunotherapy have shown high efficacy, they are expensive and have low response rates [149, 150, 151]. Hence, there is a need to study the therapeutic response and identify patients who are likely to benefit from treatment or are at a higher risk for adverse effects. With the approval of the first immune checkpoint inhibitor (ICI) pembrolizumab as a first‐line drug for metastatic NSCLC [152], there has been much interest in exploring the tumor microenvironment and tumor‐infiltrating lymphocytes in the hope of finding reliable biomarkers to predict immunotherapy response [94, 153, 154].
In a study of 221 patients treated with nivolumab, Gataa et al [155] analyzed the association of TILs with treatment response to ICI. They employed a semi‐quantitative method by pathologists using H&E‐stained sections from archival pretreatment tumor tissue sample to assess the TIL density and observed that a high TIL density (>10%) was associated with response to ICI therapy in advanced NSCLC, thereby raising the possibility of TIL density as a potential biomarker for predicting response to ICI.
Wang et al [156] quantitatively extracted features related to the spatial arrangement of TILs and nuclei from 82 NSCLC patients treated with the PD‐1 inhibitor nivolumab to construct a random forest classifier, which yielded an AUC of 0.68 on the validation set. Ofek et al [157] used a DL approach to analyze the spatial arrangement of TILs in the tumor microenvironment. This imaging biomarker was predictive of response to pembrolizumab in patients with advanced NSCLC. Barrera et al [85] took a hand‐crafted approach by having two expert pathologists manually delineate tumor regions which were used to identify the TILs. Subsequently, TIL arrangement features were computed to create a machine learning classifier that accurately distinguished patients who responded to nivolumab versus those who did not. The detection and subsequent analysis of TILs also pave the way for studying other biomarkers such as tumor‐associated macrophages in lung cancer [158].
Jain et al [142], in a study of 106 SCLC patients, trained a naïve Bayes classifier analyzing morphological and functional features (shape, size, intensity, cellular texture) of the cancer nuclei to predict response to platinum‐based chemotherapy. Barrera et al [159] looked at a combination of textural and shape features from WSIs with features from CT scans to construct a machine learning classifier for predicting response to chemotherapy in SCLC.
Emerging applications
Multi‐omics
Intratumoral heterogeneity in NSCLC has been well studied [160]. Recent studies have mapped spatial transcriptomics onto histopathology feature maps to analyze the role of different cell populations in cancers [161, 162]. The underlying molecular mechanisms for several morphologic features in NSCLC have largely remained unexplained [163]. Meanwhile, there are more imaging features currently not readily evaluable by clinicians, yet associated with patient outcomes [164]. For example, lung cancer can present either as ground‐glass opacities or as solid nodules on chest CT scans [165, 166]. Despite being the same clinical entity, their appearance, and the clinical course, can differ, indicating distinct tumor biology. With 538 LUAD cases from the TCGA, Yu et al [167] used WSIs, RNA sequencing, and proteomics data, and constructed a machine learning model to identify molecular pathways associated with morphologic features. Interestingly, the authors chose a random forest [168], a machine learning algorithm which works well with high‐dimensional data such as multi‐omics, to correlate with outcomes. AbdulJabbar et al [169] developed a DL tool integrating spatial histology with multi‐region exome sequencing data to map regions of immune ‘hot spots’ and their association with intratumoral heterogeneity. The tool, validated on squamous cell carcinoma and LUAD patients from the TraceRx study and LATTICe‐A study, demonstrated that the overall number of immune cold regions within a tumor determined the prognosis of LUAD.
A radiologic examination is often the first modality of investigation in lung disease due to its non‐invasive nature. However, one of the major drawbacks of radiographic imaging is the inability to accurately define the disease extent, which still requires histopathologic analysis. Recent computational approaches have allowed co‐registering or ‘mapping’ the histologic volume from biopsies onto the CT scan volume to allow facilitating early characterization of lung nodules' extent on non‐invasive imaging modalities by avoiding biopsy [170]. Rusu et al [170] performed one of the first known studies in co‐registering lung ex vivo pathology specimens with CT scans. They demonstrated a method of first constructing a 3D histological volume using group‐wise registration and subsequently co‐registering the CT scan with the reconstructed histology. Their approach showed a Kappa value of 0.68, indicating substantial agreement.
Microbiome
The lung microbiota has been shown to play a role in lung cancer development as well as in prognosis and response to treatments [171]. It is also associated with the histological subtypes of lung cancer [172]. Given its role in disease progression, there have been large‐scale efforts to collect microbiome and metabolome data [173]. With the demand to process and analyze this ‘big data’, machine learning tools in combination with image‐based biomarkers captured from WSIs offer a bright prospect.
3D pathology
Novel imaging techniques such as 3D microscopy [174] have recently been utilized in DP workflows. The non‐destructive nature of the technology along with its ability to analyze larger samples of tissue could help to overcome challenges faced by spatial and temporal heterogeneity. Currently, it has been applied in the context of prostate cancer in enhancing risk stratification [175]. With its ability to interrogate spatial relationships of various cell types by reconstructing entire tissue blocks, techniques such as 3D microscopy combined with DP would be an invaluable tool in a pathologist's armamentarium.
Transplant rejection prediction
A major cause of mortality in lung transplant recipients is graft failure, and predicting the risk of rejection poses a challenge for clinicians as clinical criteria alone are inaccurate [176]. Studies developing machine learning models using WSIs of tissues from heart [177, 178] and kidney [179, 180] transplant patients have demonstrated unique morphological patterns related to lymphocytes and stroma to predict the risk of transplant rejection. Similar techniques could potentially be applied to lung transplant recipients in the future.
Drug development and discovery
Recently, there has been renewed interest in drug development for pulmonary diseases such as IPF and pulmonary artery hypertension to address unmet medical needs [181, 182, 183]. A crucial step is the estimation of response to novel agents using scoring systems that may involve manual assessment of histopathology slides [184, 185]. Computational approaches can accelerate the process by automated analysis of digitized image data [186, 187, 188, 189].
Challenges and opportunities
Challenges pertaining to both the technical development of AI tools and their clinical translation need to be addressed before adoption by the clinical community. Past works have explored the various technical hurdles such as batch effects, slide artifacts, and staining variability, and their solutions [190, 191, 192]. One of the issues with the clinical translation of DP‐based AI tools is the appropriate regulatory approval pathways from agencies such as the FDA, in the USA and the Conformité Européenne (CE) mark in Europe [193]. Currently, no specific regulatory pathways exist for the approval of AI‐based tools in the United States. The recent FDA approval of Paige Prostate [32] for prostate cancer detection was classified as a Class II device, indicating a moderate to high risk to patients [45]. It underwent the de novo pathway [194] and was categorized as a ‘software algorithm device to assist users in digital pathology’. This creation of a new product code is expected to allow more DP‐based solutions to follow suit in obtaining approvals for clinical translation [195]. While the tools approved so far aid in diagnostics, regulation for predictive tools is expected to be rigorous, with careful evaluation of their performance owing to the downstream impact they may have in patient management. Even after approvals, diagnostic decision support systems will have to be monitored via prospective trials to avoid misuse as primary diagnostic tools [196]. While there are no DP‐based AI tools for lung cancer at the time of writing, Optellum [197] received a 510(k) approval for ‘Virtual Nodule Clinic’ as the first radiomics‐based AI tool for lung cancer detection, which demonstrates the interest in these tools in pulmonary diseases.
Additionally, AI tools need to address issues with reimbursement, which may be complicated since the existing fee‐for‐service model cannot be applied [198, 199]. The American Medical Association (AMA) current procedural terminology (CPT) codes and Centers for Medicare & Medicaid Services (CMS) currently do not have a payment pathway for DP‐based AI tools. The recent decision by the CMS to grant New Technology Add‐on Payment reimbursement to a stroke detection model by Viz.AI [200] performed under a unique ICD‐10 Procedure Coding System (ICD‐10‐PCS) procedure code [201] is a step forward. One approach would be to create a new CPT code for CAD tools [202], whereas another might be to analyze and modify existing ones to incorporate DP services [203].
The clinical deployment of DP‐based AI tools requires collaboration between stakeholders, including machine learning scientists, software developers, and clinicians who are the end‐users. While diagnostic tools such as Paige Prostate have been deployed as decision support tools integrated with image viewers, such a process is yet to be developed for prognostic and predictive tools [204]. DP‐based AI tools can be deployed either as Storage as a Service (SaaS) cloud business model or as on‐premise solutions [205]. While cloud‐based programs may facilitate remote access of these tools, the larger size of high‐resolution pathology images might favor an on‐premise tool. The mode of deployment may vary according to institutions, and financial implications need to be taken into consideration as well [206].
Given that results from AI tools may have a far‐reaching impact on the life of a patient, there are important legal challenges to be considered for their safer translation into clinical practice [207]. An equitable liability scheme needs to be designed to define the roles of AI tool developers, physicians, and healthcare institutions. This discussion would revolve around what is considered as the gold standard adopted by the clinical community, and whether AI would continue to remain as assistive technology, or become the gold standard against which the performance of a physician is evaluated [208].
Finally, an important consideration before the integration of AI tools into clinical practice is to make sure that these tools have been ethically trained and are not subject to bias [199]. While human errors in diagnostics have been largely attributed to cognitive biases [209], AI errors tend to be systematic and often introduced during the training phase [210]. Efforts need to be taken to ensure that future AI tools are trained and validated on a plurality of populations. There have been attempts to incorporate molecular and phenotype differences among populations which may manifest as tissue morphology differences into AI models. Studies in prostate [211], breast [212], and endometrial [213] cancers suggest that population‐specific AI models could be more accurate in stratifying patients based on their risk.
Conclusion
The use of AI tools in lung DP is expanding rapidly [214]. While CNN‐based DL models performing detection and pattern recognition are intuitively suited for low‐level tasks such as automated lung tissue segmentation, disease diagnosis, and quantifying biomarkers such as PD‐L1, hand‐crafted models are being integrated with clinical data and outcomes for risk stratification and prediction of treatment response, which can be further made interpretable by using domain‐inspired hand‐crafted features [215]. Integration of DP in the clinical workflow across major institutions remains a challenge, due to organizational structures, the cost of initial set‐up, requirements of advanced security systems in hospitals, and demands for storage of ‘big data’ [214, 215, 216, 217]. Currently, most existing AI tools have been validated on retrospective data [196], which does not represent real‐world scenarios. Validating the algorithms in randomized controlled trials and prospective studies will be a crucial step towards clinical adoption [199]. There is still hesitancy within the clinical community regarding the black‐box nature of some AI‐based tools, which may determine regulatory approvals as well as their acceptance for clinical deployment. Hence, there is a growing interest in interpretable AI [218]. DP‐based AI tools hold immense potential for improving workflow issues for pathologists by reducing labor‐intensive, time‐consuming tasks, and for pulmonologists and thoracic oncologists by providing risk stratification and companion diagnostics tools for lung diseases. While there are significant challenges in translating these technologies into clinical practice, recent regulatory approvals and deployment of these tools [219] signal a changing trend.
Author contributions statement
VSV and AM researched data for the manuscript. GC and VSV created the illustrations for the manuscript. All the authors were responsible for reviewing and editing the manuscript.
Acknowledgements
We acknowledge the following grant support: National Cancer Institute: U24CA199374‐01, R01CA202752‐01A1, R01CA208236‐01A1, R01CA216579‐01A1, R01CA220581‐01A1; National Center for Research Resources under award number 1 C06 RR12463‐01; The DOD Prostate Cancer Synergistic Idea Development Award (PC120857), The DOD Lung Cancer Idea Development, New Investigator Award (LC130463), The DOD Prostate Cancer Idea Development Award; The DOD Peer Reviewed Cancer Research Program W81XWH‐16‐1‐0329; The Ohio Third Frontier Technology Validation Fund; The Wallace H. Coulter Foundation Program; and the Clinical and Translational Science Award Program (CTSA) at Case Western Reserve University.
Conflict of interest statement: AM is an equity holder in Elucid Bioimaging and in Inspirata Inc. In addition, he has served as a scientific advisory board member for Inspirata Inc, AstraZeneca, Bristol Meyers‐Squibb, and Merck. Currently, he serves on the advisory board of Aiforia Inc and currently consults for Caris, Roche, Biohme, Castle Biosciences, and Aiforia. He also has sponsored research agreements with Philips, AstraZeneca, Boehringer‐Ingelheim, Eli‐Lilly, and Bristol Meyers‐Squibb. His technology has been licensed to Elucid Bioimaging. He is also involved in three different R01 grants with Inspirata Inc. The other authors declared no conflicts of interest.
References
- 1. Forum of International Respiratory Societies . The Global Impact of Respiratory Disease (2nd edn). European Respiratory Society: Sheffield, 2017. [Google Scholar]
- 2. WHO Coronavirus (COVID‐19) Dashboard | WHO Coronavirus (COVID‐19) Dashboard with Vaccination Data. [Accessed 22 February 2022]. Available from: https://covid19.who.int/table
- 3. MacNeil A, Glaziou P, Sismanidis C, et al. Global epidemiology of tuberculosis and progress toward achieving global targets – 2017. MMWR Morb Mortal Wkly Rep 2019; 68: 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71: 209–249. [DOI] [PubMed] [Google Scholar]
- 5. Cancer Stat Facts – Lung and Bronchus Cancer. SEER. [Accessed 11 February 2022]. Available from: https://seer.cancer.gov/statfacts/html/lungb.html
- 6. Rudin CM, Brambilla E, Faivre‐Finn C, et al. Small‐cell lung cancer. Nat Rev Dis Primers 2021; 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davidson MR, Gazdar AF, Clarke BE. The pivotal role of pathology in the management of lung cancer. J Thorac Dis 2013; 5: S463–S478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cavazza A, Rossi G, Carbonelli C, et al. The role of histology in idiopathic pulmonary fibrosis: an update. Respir Med 2010; 104: S11–S22. [DOI] [PubMed] [Google Scholar]
- 9. King TE Jr. Clinical advances in the diagnosis and therapy of the interstitial lung diseases. Am J Respir Crit Care Med 2005; 172: 268–279. [DOI] [PubMed] [Google Scholar]
- 10. Gurcan MN, Boucheron LE, Can A, et al. Histopathological image analysis: a review. IEEE Rev Biomed Eng 2009; 2: 147–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol Mech Dis 2011; 6: 49–69. [DOI] [PubMed] [Google Scholar]
- 12. Karachaliou N, Mayo C, Costa C, et al. KRAS mutations in lung cancer. Clin Lung Cancer 2013; 14: 205–214. [DOI] [PubMed] [Google Scholar]
- 13. Won JK, Keam B, Koh J, et al. Concomitant ALK translocation and EGFR mutation in lung cancer: a comparison of direct sequencing and sensitive assays and the impact on responsiveness to tyrosine kinase inhibitor. Ann Oncol 2015; 26: 348–354. [DOI] [PubMed] [Google Scholar]
- 14. Flaherty KR, Colby TV, Travis WD, et al. Fibroblastic foci in usual interstitial pneumonia: idiopathic versus collagen vascular disease. Am J Respir Crit Care Med 2003; 167: 1410–1415. [DOI] [PubMed] [Google Scholar]
- 15. Harada T, Watanabe K, Nabeshima K, et al. Prognostic significance of fibroblastic foci in usual interstitial pneumonia and non‐specific interstitial pneumonia. Respirology 2013; 18: 278–283. [DOI] [PubMed] [Google Scholar]
- 16. Elsoukkary SS, Mostyka M, Dillard A, et al. Autopsy findings in 32 patients with COVID‐19: a single‐institution experience. Pathobiology 2021; 88: 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tian S, Xiong Y, Liu H, et al. Pathological study of the 2019 novel coronavirus disease (COVID‐19) through postmortem core biopsies. Mod Pathol 2020; 33: 1007–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barton LM, Duval EJ, Stroberg E, et al. COVID‐19 autopsies, Oklahoma, USA. Am J Clin Pathol 2020; 153: 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. WMA – The World Medical Association. WMA Statement on Augmented Intelligence in Medical Care. [Accessed 6 January 2022]. Available from: https://www.wma.net/policies-post/wma-statement-on-augmented-intelligence-in-medical-care/
- 20. Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol 2019; 20: e253–e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gupta R, Kurc T, Sharma A, et al. The emergence of pathomics. Curr Pathobiol Rep 2019; 7: 73–84. [Google Scholar]
- 22. Biomarkers Definitions Working Group . Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 2001; 69: 89–95. [DOI] [PubMed] [Google Scholar]
- 23. Abels E, Pantanowitz L, Aeffner F, et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J Pathol 2019; 249: 286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deo RC. Machine learning in medicine. Circulation 2015; 132: 1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–444. [DOI] [PubMed] [Google Scholar]
- 26. Bertolaccini L, Solli P, Pardolesi A, et al. An overview of the use of artificial neural networks in lung cancer research. J Thorac Dis 2017; 9: 924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cui M, Zhang DY. Artificial intelligence and computational pathology. Lab Invest 2021; 101: 412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goodfellow IJ, Pouget‐Abadie J, Mirza M, et al. Generative adversarial networks. Adv Neural Inf Process Syst. 2014; 27.
- 29. Mukhopadhyay S, Feldman MD, Abels E, et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: A multicenter blinded randomized noninferiority study of 1992 cases (pivotal study). Am J Surg Pathol 2018; 42: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Commissioner of the U.S. Food and Drug Administration. FDA; 2022. [Accessed 11 February 2022]. Available from: https://www.fda.gov/home
- 31. Commissioner of the U.S. Food and Drug Administration. FDA Allows Marketing of First Whole Slide Imaging System for Digital Pathology. FDA; 2020. [Accessed 11 February 2022]. Available from: https://www.fda.gov/news-events/press-announcements/fda-allows-marketing-first-whole-slide-imaging-system-digital-pathology
- 32. FDA Authorizes Software that Can Help Identify Prostate Cancer. FDA; 2021 [Accessed 26 January 2022]. Available from: https://www.fda.gov/news-events/press-announcements/fda-authorizes-software-can-help-identify-prostate-cancer
- 33. Sechopoulos I, Teuwen J, Mann R. Artificial intelligence for breast cancer detection in mammography and digital breast tomosynthesis: state of the art. Semin Cancer Biol 2021; 72: 214–225. [DOI] [PubMed] [Google Scholar]
- 34. Ibrahim A, Gamble P, Jaroensri R, et al. Artificial intelligence in digital breast pathology: techniques and applications. Breast 2020; 49: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duggento A, Conti A, Mauriello A, et al. Deep computational pathology in breast cancer. Semin Cancer Biol 2021; 72: 226–237. [DOI] [PubMed] [Google Scholar]
- 36. Yousif M, van Diest PJ, Laurinavicius A, et al. Artificial intelligence applied to breast pathology. Virchows Arch 2022; 480: 191–209. [DOI] [PubMed] [Google Scholar]
- 37. Goldenberg SL, Nir G, Salcudean SE. A new era: artificial intelligence and machine learning in prostate cancer. Nat Rev Urol 2019; 16: 391–403. [DOI] [PubMed] [Google Scholar]
- 38. Tolkach Y, Dohmgörgen T, Toma M, et al. High‐accuracy prostate cancer pathology using deep learning. Nat Mach Intell 2020; 2: 411–418. [Google Scholar]
- 39. Tătaru OS, Vartolomei MD, Rassweiler JJ, et al. Artificial intelligence and machine learning in prostate cancer patient management – current trends and future perspectives. Diagnostics (Basel) 2021; 11: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Van Booven DJ, Kuchakulla M, Pai R, et al. A systematic review of artificial intelligence in prostate cancer. Res Rep Urol 2021; 13: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Classe M, Lerousseau M, Scoazec JY, et al. Perspectives in pathomics in head and neck cancer. Curr Opin Oncol 2021; 33: 175–183. [DOI] [PubMed] [Google Scholar]
- 42. Lollie TK, Krane JF. Applications of computational pathology in head and neck cytopathology. Acta Cytol 2021; 65: 330–334. [DOI] [PubMed] [Google Scholar]
- 43. Patil S, Habib Awan K, Arakeri G, et al. Machine learning and its potential applications to the genomic study of head and neck cancer – a systematic review. J Oral Pathol Med 2019; 48: 773–779. [DOI] [PubMed] [Google Scholar]
- 44. Crowson MG, Ranisau J, Eskander A, et al. A contemporary review of machine learning in otolaryngology – head and neck surgery. Laryngoscope 2020; 130: 45–51. [DOI] [PubMed] [Google Scholar]
- 45. Bera K, Schalper KA, Rimm DL, et al. Artificial intelligence in digital pathology – new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 2019; 16: 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thawani R, McLane M, Beig N, et al. Radiomics and radiogenomics in lung cancer: a review for the clinician. Lung Cancer 2018; 115: 34–41. [DOI] [PubMed] [Google Scholar]
- 47. Cong L, Feng W, Yao Z, et al. Deep learning model as a new trend in computer‐aided diagnosis of tumor pathology for lung cancer. J Cancer 2020; 11: 3615–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Acs B, Rantalainen M, Hartman J. Artificial intelligence as the next step towards precision pathology. J Intern Med 2020; 288: 62–81. [DOI] [PubMed] [Google Scholar]
- 49. Sakamoto T, Furukawa T, Lami K, et al. A narrative review of digital pathology and artificial intelligence: focusing on lung cancer. Transl Lung Cancer Res 2020; 9: 2255–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang S, Yang DM, Rong R, et al. Artificial intelligence in lung cancer pathology image analysis. Cancers (Basel) 2019; 11: 1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raghu G, Flaherty KR, Lederer DJ, et al. Use of a molecular classifier to identify usual interstitial pneumonia in conventional transbronchial lung biopsy samples: a prospective validation study. Lancet Respir Med 2019; 7: 487–496. [DOI] [PubMed] [Google Scholar]
- 52. Mäkelä K, Mäyränpää MI, Sihvo HK, et al. Artificial intelligence identifies inflammation and confirms fibroblast foci as prognostic tissue biomarkers in idiopathic pulmonary fibrosis. Hum Pathol 2021; 107: 58–68. [DOI] [PubMed] [Google Scholar]
- 53. Pankratz DG, Choi Y, Imtiaz U, et al. Usual interstitial pneumonia can be detected in transbronchial biopsies using machine learning. Ann Am Thorac Soc 2017; 14: 1646–1654. [DOI] [PubMed] [Google Scholar]
- 54. Xiong Y, Ba X, Hou A, et al. Automatic detection of mycobacterium tuberculosis using artificial intelligence. J Thorac Dis 2018; 10: 1936–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang M, Nurzynska K, Walts AE, et al. A CNN‐based active learning framework to identify mycobacteria in digitized Ziehl–Neelsen stained human tissues. Comput Med Imaging Graph 2020; 84: 101752. [DOI] [PubMed] [Google Scholar]
- 56. Lo C‐M, Wu Y‐H, Li Y‐C, et al. Computer‐aided bacillus detection in whole‐slide pathological images using a deep convolutional neural network. Appl Sci 2020; 10: 4059. [Google Scholar]
- 57. Corredor G, Toro P, Bera K, et al. Computational pathology reveals unique spatial patterns of immune response in H&E images from COVID‐19 autopsies: preliminary findings. J Med Imaging (Bellingham) 2021; 8(Suppl 1): 017501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tonekaboni S, Joshi S, McCradden MD, et al. What clinicians want: contextualizing explainable machine learning for clinical end use. ArXiv Preprint 2019; ArXiv:1905.05134 Cs Stat.
- 59. Kelly M, Soles R, Garcia E, et al. Job stress, burnout, work‐life balance, well‐being, and job satisfaction among pathology residents and fellows. Am J Clin Pathol 2020; 153: 449–469. [DOI] [PubMed] [Google Scholar]
- 60. Lindman K, Rose JF, Lindvall M, et al. Annotations, ontologies, and whole slide images – development of an annotated ontology‐driven whole slide image library of normal and abnormal human tissue. J Pathol Inform 2019; 10: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pham HHN, Futakuchi M, Bychkov A, et al. Detection of lung cancer lymph node metastases from whole‐slide histopathologic images using a two‐step deep learning approach. Am J Pathol 2019; 189: 2428–2439. [DOI] [PubMed] [Google Scholar]
- 62. Šarić M, Russo M, Stella M, et al. CNN‐based method for lung cancer detection in whole slide histopathology images. In 2019 4th International Conference on Smart and Sustainable Technologies (SpliTech), 2019; 1–4. 10.23919/SpliTech.2019.8783041. [DOI] [Google Scholar]
- 63. Wang S, Wang T, Yang L, et al. ConvPath: a software tool for lung adenocarcinoma digital pathological image analysis aided by a convolutional neural network. EBioMedicine 2019; 50: 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gertych A, Swiderska‐Chadaj Z, Ma Z, et al. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci Rep 2019; 9: 1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kriegsmann M, Haag C, Weis C‐A, et al. Deep learning for the classification of small‐cell and non‐small‐cell lung cancer. Cancers (Basel) 2020; 12: 1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang S, Chen A, Yang L, et al. Comprehensive analysis of lung cancer pathology images to discover tumor shape and boundary features that predict survival outcome. Sci Rep 2018; 8: 10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Courtiol P, Maussion C, Moarii M, et al. Deep learning‐based classification of mesothelioma improves prediction of patient outcome. Nat Med 2019; 25: 1519–1525. [DOI] [PubMed] [Google Scholar]
- 68. Naso JR, Levine AB, Farahani H, et al. Deep‐learning based classification distinguishes sarcomatoid malignant mesotheliomas from benign spindle cell mesothelial proliferations. Mod Pathol 2021; 34: 2028–2035. [DOI] [PubMed] [Google Scholar]
- 69. Heinemann F, Birk G, Schoenberger T, et al. Deep neural network based histological scoring of lung fibrosis and inflammation in the mouse model system. PLoS One 2018; 13: e0202708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Le Page AL, Ballot E, Truntzer C, et al. Using a convolutional neural network for classification of squamous and non‐squamous non‐small cell lung cancer based on diagnostic histopathology HES images. Sci Rep 2021; 11: 23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non‐small cell lung cancer histopathology images using deep learning. Nat Med 2018; 24: 1559–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xue Y, Ray N, Hugh J, et al. Cell counting by regression using convolutional neural network. In Computer Vision – ECCV 2016 Workshops, Hua G, Jégou H (eds). Springer International Publishing: Cham, 2016; 274–290. [Google Scholar]
- 73. Oskal KRJ, Risdal M, Janssen EAM, et al. A U‐net based approach to epidermal tissue segmentation in whole slide histopathological images. SN Appl Sci 2019; 1: 672. [Google Scholar]
- 74. Wang X, Bera K, Barrera C, et al. A prognostic and predictive computational pathology image signature for added benefit of adjuvant chemotherapy in early stage non‐small‐cell lung cancer. EBioMedicine 2021; 69: 103481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lu C, Koyuncu C, Corredor G, et al. Feature‐driven local cell graph (FLocK): new computational pathology‐based descriptors for prognosis of lung cancer and HPV status of oropharyngeal cancers. Med Image Anal 2021; 68: 101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lu C, Bera K, Wang X, et al. A prognostic model for overall survival of patients with early‐stage non‐small cell lung cancer: a multicentre, retrospective study. Lancet Digit Health 2020; 2: e594–e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Corredor G, Wang X, Zhou Y, et al. Spatial architecture and arrangement of tumor‐infiltrating lymphocytes for predicting likelihood of recurrence in early‐stage non‐small cell lung cancer. Clin Cancer Res 2019; 25: 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tizhoosh HR, Pantanowitz L. Artificial intelligence and digital pathology: challenges and opportunities. J Pathol Inform 2018; 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Koyuncu CF, Lu C, Bera K, et al. Computerized tumor multinucleation index (MuNI) is prognostic in p16+ oropharyngeal carcinoma. J Clin Invest 2021; 131: e145488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Petch J, Di S, Nelson W. Opening the black box: the promise and limitations of explainable machine learning in cardiology. Can J Cardiol 2022; 38: 204–213. [DOI] [PubMed] [Google Scholar]
- 81. Yao J, Ganti D, Luo X, et al. Computer‐assisted diagnosis of lung cancer using quantitative topology features. In Machine Learning for Medical Imaging, Zhou L, Wang L, Wang Q, et al. (eds). Springer International Publishing: Cham, 2015; 288–295. [Google Scholar]
- 82. Wang X, Janowczyk A, Zhou Y, et al. Prediction of recurrence in early stage non‐small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci Rep 2017; 7: 13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Vaidya P, Wang X, Bera K, et al. RaPtomics: integrating radiomic and pathomic features for predicting recurrence in early stage lung cancer. In Medical Imaging 2018: Digital Pathology. SPIE: Houston, 2018; 21. [Accessed 18 February 2022]. Available from: https://www.spiedigitallibrary.org/conference‐proceedings‐of‐spie/10581/2296646/RaPtomics‐‐integrating‐radiomic‐and‐pathomic‐features‐for‐predicting‐recurrence/10.1117/12.2296646.full
- 84. Saltz J, Gupta R, Hou L, et al. Spatial organization and molecular correlation of tumor‐infiltrating lymphocytes using deep learning on pathology images. Cell Rep 2018; 23: 181–193.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Barrera C, Velu P, Bera K, et al. Computer‐extracted features relating to spatial arrangement of tumor infiltrating lymphocytes to predict response to nivolumab in non‐small cell lung cancer (NSCLC). J Clin Oncol 2018; 36: 12115–12115. [Google Scholar]
- 86. Sandino AA, Alvarez‐Jimenez C, Mosquera‐Zamudio A, et al. Cell density features from histopathological images to differentiate non‐small cell lung cancer subtypes. In 15th International Symposium on Medical Information Processing and Analysis. SPIE, 2020; 41–46. [Accessed 12 February 2022]. Available from: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/11330/1133007/Cell-density-features-from-histopathological-images-to-differentiate-non-small/10.1117/12.2542360.full. [Google Scholar]
- 87. Corredor G, Wang X, Lu C, et al. A watershed and feature‐based approach for automated detection of lymphocytes on lung cancer images. In Medical Imaging 2018: Digital Pathology. SPIE: Houston, 2018; 213–218. [Accessed 22 October 2021]. Available from: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10581/105810R/A-watershed-and-feature-based-approach-for-automated-detection-of/10.1117/12.2293147.full. [Google Scholar]
- 88. Wang C‐W, Yu C‐P. Automated morphological classification of lung cancer subtypes using H&E tissue images. Mach Vis Appl 2013; 24: 1383–1391. [Google Scholar]
- 89. Alvarez‐Jimenez C, Sandino AA, Prasanna P, et al. Identifying cross‐scale associations between radiomic and pathomic signatures of non‐small cell lung cancer subtypes: preliminary results. Cancers (Basel) 2020; 12: 3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lu C, Wang X, Prasanna P, et al. Feature driven local cell graph (FeDeG): predicting overall survival in early stage lung cancer. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, Frangi AF, Schnabel JA, Davatzikos C, et al. (eds). Springer International Publishing: Cham, 2018; 407–416. [Google Scholar]
- 91. Li H, Bera K, Toro P, et al. Collagen fiber orientation disorder from H&E images is prognostic for early stage breast cancer: clinical trial validation. NPJ Breast Cancer 2021; 7: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Idowu MO, Powers CN. Lung cancer cytology: potential pitfalls and mimics – a review. Int J Clin Exp Pathol 2010; 3: 367–385. [PMC free article] [PubMed] [Google Scholar]
- 93. Wrenn ED, Yamamoto A, Moore BM, et al. Regulation of collective metastasis by nanolumenal signaling. Cell 2020; 183: 395–410.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jang N, Kwon HJ, Park MH, et al. Prognostic value of tumor‐infiltrating lymphocyte density assessed using a standardized method based on molecular subtypes and adjuvant chemotherapy in invasive breast cancer. Ann Surg Oncol 2018; 25: 937–946. [DOI] [PubMed] [Google Scholar]
- 95. Bocchialini G, Lagrasta C, Madeddu D, et al. Spatial architecture of tumour‐infiltrating lymphocytes as a prognostic parameter in resected non‐small‐cell lung cancer. Eur J Cardiothorac Surg 2020; 58: 619–628. [DOI] [PubMed] [Google Scholar]
- 96. Mukhopadhyay S. Utility of small biopsies for diagnosis of lung nodules: doing more with less. Mod Pathol 2012; 25: S43–S57. [DOI] [PubMed] [Google Scholar]
- 97. Roh MH. The utilization of cytologic and small biopsy samples for ancillary molecular testing. Mod Pathol 2019; 32: 77–85. [DOI] [PubMed] [Google Scholar]
- 98. Ofiara LM, Navasakulpong A, Ezer N, et al. The importance of a satisfactory biopsy for the diagnosis of lung cancer in the era of personalized treatment. Curr Oncol 2012; 19(Suppl 1): S16–S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Barsoum I, Tawedrous E, Faragalla H, et al. Histo‐genomics: digital pathology at the forefront of precision medicine. Diagnosis (Berl) 2019; 6: 203–212. [DOI] [PubMed] [Google Scholar]
- 100. Perco P, Kainz A, Wilflingseder J, et al. Histogenomics: association of gene expression patterns with histological parameters in kidney biopsies. Transplantation 2009; 87: 290–295. [DOI] [PubMed] [Google Scholar]
- 101. Lantuejoul S, Sound‐Tsao M, Cooper WA, et al. PD‐L1 testing for lung cancer in 2019: perspective from the IASLC Pathology Committee. J Thorac Oncol 2020; 15: 499–519. [DOI] [PubMed] [Google Scholar]
- 102. Rizzardi AE, Johnson AT, Vogel RI, et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn Pathol 2012; 7: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sha L, Osinski BL, Ho IY, et al. Multi‐field‐of‐view deep learning model predicts nonsmall cell lung cancer programmed death‐ligand 1 status from whole‐slide hematoxylin and eosin images. J Pathol Inform 2019; 10: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jose L, Liu S, Russo C, et al. Generative adversarial networks in digital pathology and histopathological image processing: a review. J Pathol Inform 2021; 12: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tschuchnig ME, Oostingh GJ, Gadermayr M. Generative adversarial networks in digital pathology: a survey on trends and future potential. Patterns (N Y) 2020; 1: 100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bayramoglu N, Kaakinen M, Eklund L, et al. Towards virtual H&E staining of hyperspectral lung histology images using conditional generative adversarial networks. In 2017 IEEE International Conference on Computer Vision Workshop (ICCVW), 2017; 64–71. 10.1109/ICCVW.2017.15. [DOI] [Google Scholar]
- 107. Kapil A, Meier A, Zuraw A, et al. Deep semi supervised generative learning for automated PD‐L1 tumor cell scoring on NSCLC tissue needle biopsies. Sci Rep 2018; 8: 17343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wu J, Liu C, Liu X, et al. Artificial intelligence‐assisted system for precision diagnosis of PD‐L1 expression in non‐small cell lung cancer. Mod Pathol 2022; 35: 403–411. [DOI] [PubMed] [Google Scholar]
- 109. Xia D, Casanova R, Machiraju D, et al. Computationally‐guided development of a stromal inflammation histologic biomarker in lung squamous cell carcinoma. Sci Rep 2018; 8: 3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Nicholson AG, Tsao MS, Beasley MB, et al. The 2021 WHO Classification of lung tumors: impact of advances since 2015. J Thorac Oncol 2022; 17: 362–387. [DOI] [PubMed] [Google Scholar]
- 111. Travis WD, Brambilla E, Burke AP, et al. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J Thorac Oncol 2015; 10: 1240–1242. [DOI] [PubMed] [Google Scholar]
- 112. Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015; 10: 1243–1260. [DOI] [PubMed] [Google Scholar]
- 113. Stenzinger A, Alber M, Allgäuer M, et al. Artificial intelligence and pathology: from principles to practice and future applications in histomorphology and molecular profiling. Semin Cancer Biol 2021. 10.1016/j.semcancer.2021.02.011. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 114. Román M, Baraibar I, López I, et al. KRAS oncogene in non‐small cell lung cancer: clinical perspectives on the treatment of an old target. Mol Cancer 2018; 17: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Center for Drug Evaluation and Research . FDA Grants Accelerated Approval to Sotorasib for KRAS G12C Mutated NSCLC. FDA, 2021. [Accessed 21 January 2022]. Available from: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-sotorasib-kras-g12c-mutated-nsclc. [Google Scholar]
- 116. DiStasio M, Chen Y, Rangachari D, et al. Molecular testing turnaround time for non‐small cell lung cancer in routine clinical practice confirms feasibility of CAP/IASLC/AMP guideline recommendations: a single‐center analysis. Clin Lung Cancer 2017; 18: e349–e356. [DOI] [PubMed] [Google Scholar]
- 117. Schluckebier L, Caetano R, Garay OU, et al. Cost‐effectiveness analysis comparing companion diagnostic tests for EGFR, ALK, and ROS1 versus next‐generation sequencing (NGS) in advanced adenocarcinoma lung cancer patients. BMC Cancer 2020; 20: 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chen M, Zhang B, Topatana W, et al. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis Oncol 2020; 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Katzenstein A‐LA, Mukhopadhyay S, Myers JL. Erratum to “Diagnosis of usual interstitial pneumonia and distinction from other fibrosing interstitial lung diseases” [Hum Pathol 39 (2008) 1275‐1294]. Hum Pathol 2008; 39: 1562–1581. [DOI] [PubMed] [Google Scholar]
- 120. Temes RT, Joste NE, Qualls CR, et al. Lung biopsy: is it necessary? J Thorac Cardiovasc Surg 1999; 118: 1097–1100. [DOI] [PubMed] [Google Scholar]
- 121. Rosenthal SA, Curran WJ Jr. The significance of histology in non‐small cell lung cancer. Cancer Treat Rev 1990; 17: 409–425. [DOI] [PubMed] [Google Scholar]
- 122. Van Bockstal MR, Berlière M, Duhoux FP, et al. Interobserver variability in ductal carcinoma in situ of the breast. Am J Clin Pathol 2020; 154: 596–609. [DOI] [PubMed] [Google Scholar]
- 123. Shanes JG, Ghali J, Billingham ME, et al. Interobserver variability in the pathologic interpretation of endomyocardial biopsy results. Circulation 1987; 75: 401–405. [DOI] [PubMed] [Google Scholar]
- 124. Pantanowitz L, Hartman D, Qi Y, et al. Accuracy and efficiency of an artificial intelligence tool when counting breast mitoses. Diagn Pathol 2020; 15: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Mukhopadhyay S, Katzenstein ALA. Subclassification of non‐small cell lung carcinomas lacking morphologic differentiation on biopsy specimens: utility of an immunohistochemical panel containing TTF‐1, napsin A, p63, and CK5/6. Am J Surg Pathol 2011; 35: 15–25. [DOI] [PubMed] [Google Scholar]
- 126. Zachara‐Szczakowski S, Verdun T, Churg A. Accuracy of classifying poorly differentiated non‐small cell lung carcinoma biopsies with commonly used lung carcinoma markers. Hum Pathol 2015; 46: 776–782. [DOI] [PubMed] [Google Scholar]
- 127. Russell PA, Wainer Z, Wright GM, et al. Does lung adenocarcinoma subtype predict patient survival?: a clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Lung Adenocarcinoma Classification. J Thorac Oncol 2011; 6: 1496–1504. [DOI] [PubMed] [Google Scholar]
- 128. Kadota K, Yeh YC, Sima CS, et al. The cribriform pattern identifies a subset of acinar predominant tumors with poor prognosis in patients with stage I lung adenocarcinoma: a conceptual proposal to classify cribriform predominant tumors as a distinct histologic subtype. Mod Pathol 2014; 27: 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wei JW, Tafe LJ, Linnik YA, et al. Pathologist‐level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci Rep 2019; 9: 3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yu KH, Wang F, Berry GJ, et al. Classifying non‐small cell lung cancer types and transcriptomic subtypes using convolutional neural networks. J Am Med Inform Assoc 2020; 27: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Jain D, Roy‐Chowdhuri S. Molecular pathology of lung cancer cytology specimens: a concise review. Arch Pathol Lab Med 2018; 142: 1127–1133. [DOI] [PubMed] [Google Scholar]
- 132. Gonzalez D, Dietz RL, Pantanowitz L. Feasibility of a deep learning algorithm to distinguish large cell neuroendocrine from small cell lung carcinoma in cytology specimens. Cytopathology 2020; 31: 426–431. [DOI] [PubMed] [Google Scholar]
- 133. Nakajima N, Sato Y, Katano H, et al. Histopathological and immunohistochemical findings of 20 autopsy cases with 2009 H1N1 virus infection. Mod Pathol 2012; 25: 1–13. [DOI] [PubMed] [Google Scholar]
- 134. Polak SB, Van Gool IC, Cohen D, et al. A systematic review of pathological findings in COVID‐19: a pathophysiological timeline and possible mechanisms of disease progression. Mod Pathol 2020; 33: 2128–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Law YN, Jian H, Lo NWS, et al. Low cost automated whole smear microscopy screening system for detection of acid fast bacilli. PLoS One 2018; 13: e0190988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Nahid P, Pai M, Hopewell PC. Advances in the diagnosis and treatment of tuberculosis. Proc Am Thorac Soc 2006; 3: 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Jain D, Ghosh S, Teixeira L, et al. Pathology of pulmonary tuberculosis and non‐tuberculous mycobacterial lung disease: facts, misconceptions, and practical tips for pathologists. Semin Diagn Pathol 2017; 34: 518–529. [DOI] [PubMed] [Google Scholar]
- 138. Guzmán‐Casta J, Carrasco‐CaraChards S, Guzmán‐Huesca J, et al. Prognostic factors for progression‐free and overall survival in malignant pleural mesothelioma. Thorac Cancer 2021; 12: 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011; 6: 244–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Yu KH, Zhang C, Berry GJ, et al. Predicting non‐small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 2016; 7: 12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Luo X, Zang X, Yang L, et al. Comprehensive computational pathological image analysis predicts lung cancer prognosis. J Thorac Oncol 2017; 12: 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Jain P, Barrera C, Osme A, et al. P68.02 computer extracted morphology features of tumor nuclei predict response to chemotherapy and prognostic of OS in small cell lung cancer. J Thorac Oncol 2021; 16: S560. [Google Scholar]
- 143. Chu QS. Targeting non‐small cell lung cancer: driver mutation beyond epidermal growth factor mutation and anaplastic lymphoma kinase fusion. Ther Adv Med Oncol 2020; 12: 1758835919895756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Sehgal K, Patell R, Rangachari D, et al. Targeting ROS1 rearrangements in non‐small cell lung cancer with crizotinib and other kinase inhibitors. Transl Cancer Res 2018; 7: S779–S786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. O'Leary CG, Andelkovic V, Ladwa R, et al. Targeting BRAF mutations in non‐small cell lung cancer. Transl Lung Cancer Res 2019; 8: 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Mase S, Chorba T, Parks S, et al. Bedaquiline for the treatment of multidrug‐resistant tuberculosis in the United States. Clin Infect Dis 2020; 71: 1010–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Gandhi L, Rodríguez‐Abreu D, Gadgeel S, et al. Pembrolizumab plus chemotherapy in metastatic non‐small‐cell lung cancer. N Engl J Med 2018; 378: 2078–2092. [DOI] [PubMed] [Google Scholar]
- 148. Richeldi L, du Bois RM, Raghu G, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med 2014; 370: 2071–2082. [DOI] [PubMed] [Google Scholar]
- 149. Ai X, Guo X, Wang J, et al. Targeted therapies for advanced non‐small cell lung cancer. Oncotarget 2018; 9: 37589–37607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Majeed U, Manochakian R, Zhao Y, et al. Targeted therapy in advanced non‐small cell lung cancer: current advances and future trends. J Hematol Oncol 2021; 14: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bestvina CM, Zullig LL, Yousuf Zafar S. The implications of out‐of‐pocket cost of cancer treatment in the usa: a critical appraisal of the literature. Future Oncol 2014; 10: 2189–2199. [DOI] [PubMed] [Google Scholar]
- 152. FDA Approves Pembrolizumab as First‐Line Treatment for PD‐L1‐Positive Non‐Small Cell Lung Cancer – The ASCO Post. [Accessed 25 January 2022]. Available from: https://ascopost.com/issues/november-10-2016/fda-approves-pembrolizumab-as-first-line-treatment-for-pd-l1-positive-non-small-cell-lung-cancer/
- 153. Gu Y, Sheng SY, Tang YY, et al. PD‐1 expression and function of T‐cell subsets in TILs from human lung cancer. J Immunother 2019; 42: 297–308. [DOI] [PubMed] [Google Scholar]
- 154. Murciano‐Goroff YR, Warner AB, Wolchok JD. The future of cancer immunotherapy: microenvironment‐targeting combinations. Cell Res 2020; 30: 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Gataa I, Mezquita L, Rossoni C, et al. Tumour‐infiltrating lymphocyte density is associated with favourable outcome in patients with advanced non‐small cell lung cancer treated with immunotherapy. Eur J Cancer 2021; 145: 221–229. [DOI] [PubMed] [Google Scholar]
- 156. Wang X, Barrera C, Lu C, et al. MA11.03 Interaction of tumor infiltrating lymphocytes and cancer nuclei predicts response to nivolumab in non‐small cell lung cancer (NSCLC). J Thorac Oncol 2018; 13: S393. [Google Scholar]
- 157. Ofek E, Bar J, Zer A, et al. Predicting response to pembrolizumab in non‐small cell lung cancer, by analyzing the spatial arrangement of tumor infiltrating lymphocytes using deep learning. J Clin Oncol 2021; 39: 9045–9045. [Google Scholar]
- 158. Conway EM, Pikor LA, Kung SHY, et al. Macrophages, inflammation, and lung cancer. Am J Respir Crit Care Med 2016; 193: 116–130. [DOI] [PubMed] [Google Scholar]
- 159. Barrera C, Khorrami M, Jain P, et al. Combination of quantitative features from H&E biopsies and CT scans predicts response to chemotherapy and overall survival in small cell lung cancer (SCLC). J Clin Oncol 2021; 39: 8572. [Google Scholar]
- 160. Laurinavicius A, Plancoulaine B, Rasmusson A, et al. Bimodality of intratumor Ki67 expression is an independent prognostic factor of overall survival in patients with invasive breast carcinoma. Virchows Arch 2016; 468: 493–502. [DOI] [PubMed] [Google Scholar]
- 161. Rao A, Barkley D, França GS, et al. Exploring tissue architecture using spatial transcriptomics. Nature 2021; 596: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Li Y, Stanojevic S, He B, et al. Benchmarking computational integration methods for spatial transcriptomics data. BioRxiv 2021; 2021.08.27.457741. [Not peer reviewed].
- 163. Chen L, Zeng H, Zhang M, et al. Histopathological image and gene expression pattern analysis for predicting molecular features and prognosis of head and neck squamous cell carcinoma. Cancer Med 2021; 10: 4615–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 2011; 3: 108ra113. [DOI] [PubMed] [Google Scholar]
- 165. Chen K, Bai J, Reuben A, et al. Multiomics analysis reveals distinct immunogenomic features of lung cancer with ground‐glass opacity. Am J Respir Crit Care Med 2021; 204: 1180–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Park CM, Goo JM, Lee HJ, et al. Nodular ground‐glass opacity at thin‐section CT: histologic correlation and evaluation of change at follow‐up. Radiographics 2007; 27: 391–408. [DOI] [PubMed] [Google Scholar]
- 167. Yu KH, Berry GJ, Rubin DL, et al. Association of omics features with histopathology patterns in lung adenocarcinoma. Cell Syst 2017; 5: 620–627.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Hornung R, Wright MN. Block Forests: random forests for blocks of clinical and omics covariate data. BMC Bioinformatics 2019; 20: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. AbdulJabbar K, Raza SEA, Rosenthal R, et al. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat Med 2020; 26: 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Rusu M, Rajiah P, Gilkeson R, et al. Co‐registration of pre‐operative CT with ex vivo surgically excised ground glass nodules to define spatial extent of invasive adenocarcinoma on in vivo imaging: a proof‐of‐concept study. Eur Radiol 2017; 27: 4209–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Liu NN, Ma Q, Ge Y, et al. Microbiome dysbiosis in lung cancer: from composition to therapy. NPJ Precis Oncol 2020; 4: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Huang D, Su X, Yuan M, et al. The characterization of lung microbiome in lung cancer patients with different clinicopathology. Am J Cancer Res 2019; 9: 2047–2063. [PMC free article] [PubMed] [Google Scholar]
- 173. Cammarota G, Ianiro G, Ahern A, et al. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat Rev Gastroenterol Hepatol 2020; 17: 635–648. [DOI] [PubMed] [Google Scholar]
- 174. Liu JTC, Glaser AK, Bera K, et al. Harnessing non‐destructive 3D pathology. Nat Biomed Eng 2021; 5: 203–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Xie W, Reder NP, Koyuncu C, et al. Prostate cancer risk stratification via nondestructive 3D pathology with deep learning‐assisted gland analysis. Cancer Res 2022; 82: 334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Agarwal R. A novel method for the diagnosis of lung transplant rejection. Thorax 2004; 59: 601. [Google Scholar]
- 177. Arabyarmohammadi S, Yuan C, Peyster EG, et al. Abstract 13830: Prediction of cardiac rejection trajectories via machine learning derived features from digital endomyocardial biopsy images. Circulation 2021; 144(Suppl_1): A13830. [Google Scholar]
- 178. Peyster EG, Arabyarmohammadi S, Janowczyk A, et al. An automated computational image analysis pipeline for histological grading of cardiac allograft rejection. Eur Heart J 2021; 42: 2356–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Kers J, Bülow RD, Klinkhammer BM, et al. Deep learning‐based classification of kidney transplant pathology: a retrospective, multicentre, proof‐of‐concept study. Lancet Digit Health 2022; 4: e18–e26. [DOI] [PubMed] [Google Scholar]
- 180. Seyahi N, Ozcan SG. Artificial intelligence and kidney transplantation. World J Transplant 2021; 11: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Yanagihara T, Chong SG, Vierhout M, et al. Current models of pulmonary fibrosis for future drug discovery efforts. Expert Opin Drug Discov 2020; 15: 931–941. [DOI] [PubMed] [Google Scholar]
- 182. Satoh K. Drug discovery focused on novel pathogenic proteins for pulmonary arterial hypertension. J Cardiol 2021; 78: 1–11. [DOI] [PubMed] [Google Scholar]
- 183. Barnes PJ, Bonini S, Seeger W, et al. Barriers to new drug development in respiratory disease. Eur Respir J 2015; 45: 1197–1207. [DOI] [PubMed] [Google Scholar]
- 184. Hübner RH, Gitter W, El Mokhtari NE, et al. Standardized quantification of pulmonary fibrosis in histological samples. Biotechniques 2008; 44: 507–517. [DOI] [PubMed] [Google Scholar]
- 185. Buchsbaum DJ, Bonner JA, Grizzle WE, et al. Treatment of pancreatic cancer xenografts with Erbitux (IMC‐C225) anti‐EGFR antibody, gemcitabine, and radiation. Int J Radiat Oncol Biol Phys 2002; 54: 1180–1193. [DOI] [PubMed] [Google Scholar]
- 186. Arnal‐Estapé A, Foggetti G, Starrett JH, et al. Preclinical models for the study of lung cancer pathogenesis and therapy development. Cold Spring Harb Perspect Med 2021; 11: a037820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Bug D, Feuerhake F, Oswald E, et al. Semi‐automated analysis of digital whole slides from humanized lung‐cancer xenograft models for checkpoint inhibitor response prediction. Oncotarget 2019; 10: 4587–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Lee HN, Seo HD, Kim EM, et al. Classification of mouse lung metastatic tumor with deep learning. Biomol Ther (Seoul) 2022; 30: 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Vamathevan J, Clark D, Czodrowski P, et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov 2019; 18: 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Schömig‐Markiefka B, Pryalukhin A, Hulla W, et al. Quality control stress test for deep learning‐based diagnostic model in digital pathology. Mod Pathol 2021; 34: 2098–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Janowczyk A, Zuo R, Gilmore H, et al. HistoQC: an open‐source quality control tool for digital pathology slides. JCO Clin Cancer Inform 2019; 3: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Kothari S, Phan JH, Stokes TH, et al. Removing batch effects from histopathological images for enhanced cancer diagnosis. IEEE J Biomed Health Inform 2014; 18: 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. PathAI and Roche Team up to Push Adoption of Digital Pathology Tech. MobiHealthNews; 2021. [Accessed 25 January 2022]. Available from: https://www.mobihealthnews.com/news/pathai-and-roche-team-push-adoption-digital-pathology-tech
- 194. De Novo Classification Request. FDA. [Accessed 26 January 2022]. Available from: https://www.fda.gov/medical-devices/premarket-submissions-selecting-and-preparing-correct-submission/de-novo-classification-request
- 195. Will FDA Authorization of Paige Software Accelerate AI Adoption in Pathology? 360Dx; 2021. [Accessed 25 January 2022]. Available from: https://www.360dx.com/clinical-lab-management/will-fda-authorization-paige-software-accelerate-ai-adoption-pathology
- 196. Wu E, Wu K, Daneshjou R, et al. How medical AI devices are evaluated: limitations and recommendations from an analysis of FDA approvals. Nat Med 2021; 27: 582–584. [DOI] [PubMed] [Google Scholar]
- 197. Lung Cancer Prediction AI. Optellum. [Accessed 25 January 2022]. Available from: https://optellum.com/lung-cancer-prediction-ai/
- 198. Baxi V, Edwards R, Montalto M, et al. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol 2022; 35: 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Rajpurkar P, Chen E, Banerjee O, et al. AI in health and medicine. Nat Med 2022; 28: 31–38. [DOI] [PubMed] [Google Scholar]
- 200. Viz.ai Receives New Technology Add‐on Payment Renewal for Stroke AI Software from CMS. [Accessed 21 February 2022]. Available from: https://www.viz.ai/press‐release/viz‐ai‐receives‐new‐technology‐add‐on‐payment‐renewal‐for‐stroke‐ai‐software‐from‐cms
- 201. Hassan AE. New technology add‐on payment (NTAP) for Viz LVO: a win for stroke care. J NeuroInterv Surg 2021; 13: 406–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Lasiter L, Samboy J, Leibowitz C, et al. Aligning reimbursement for digital pathology with its value. J Precision Med 2020. Available from: https://www.thejournalofprecisionmedicine.com/the‐journal‐of‐precision‐medicine/aligning‐reimbursement‐for‐digital‐pathology‐with‐its‐value/. [Google Scholar]
- 203. Reimbursement for Digital Pathology in the Clinic – How Does That Work? w/ Esther Abels, Visiopharm. Digital Pathology Place. [Accessed 12 February 2022]. Available from: https://digitalpathologyplace.com/portfolio-item/reimbursement-for-digital-pathology-in-the-clinic-how-does-that-work-w-esther-abels-visiopharm/
- 204. Paige Prostate for Prostate Cancer. Overview. Advice. NICE. [Accessed 12 February 2022]. Available from: https://www.nice.org.uk/advice/mib280
- 205. Hanna MG, Ardon O, Reuter VE, et al. Integrating digital pathology into clinical practice. Mod Pathol 2022; 35: 152–164. [DOI] [PubMed] [Google Scholar]
- 206. SaaS/Cloud vs. On‐Premise Solutions for Healthcare IT – A Step‐by‐Step Guide for a Financial Evaluation. Sectra Medical. [Accessed 12 February 2022]. Available from: https://medical.sectra.com/resources/saas‐cloud‐vs‐on‐premise‐solutions‐for‐healthcare‐it/
- 207. Naik N, Hameed BMZ, Shetty DK, et al. Legal and ethical consideration in artificial intelligence in healthcare: who takes responsibility? Front Surg 2022; 9: 862322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Mezrich JL. Is artificial intelligence (AI) a pipe dream? Why legal issues present significant hurdles to AI autonomy. AJR Am J Roentgenol 2022; 219: 152–156. [DOI] [PubMed] [Google Scholar]
- 209. Saposnik G, Redelmeier D, Ruff CC, et al. Cognitive biases associated with medical decisions: a systematic review. BMC Med Inform Decis Mak 2016; 16: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Gianfrancesco MA, Tamang S, Yazdany J, et al. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med 2018; 178: 1544–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Bhargava HK, Leo P, Elliott R, et al. Computationally derived image signature of stromal morphology is prognostic of prostate cancer recurrence following prostatectomy in African American patients. Clin Cancer Res 2020; 26: 1915–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Li H, Bera K, Toro P, et al. Abstract PS4‐45: Computerized image analysis of nuclear morphological features reveals differences in phenotype and prognosis of disease free survival of early stage ER+ breast cancers for South Asian and North American women. Cancer Res 2021; 81(4_Supplement): PS4–PS45. [Google Scholar]
- 213. Azarianpour Esfahani S, Fu P, Mahdi H, et al. Computational features of TIL architecture are differentially prognostic of uterine cancer between African and Caucasian American women. J Clin Oncol 2021; 39(15 Suppl): 5585. [Google Scholar]
- 214. Ahmad Z, Rahim S, Zubair M, et al. Artificial intelligence (AI) in medicine, current applications and future role with special emphasis on its potential and promise in pathology: present and future impact, obstacles including costs and acceptance among pathologists, practical and philosophical considerations. A comprehensive review. Diagn Pathol 2021; 16: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: challenges and opportunities. Med Image Anal 2016; 33: 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Higgins C. Applications and challenges of digital pathology and whole slide imaging. Biotech Histochem 2015; 90: 341–347. [DOI] [PubMed] [Google Scholar]
- 217. Fontelo P, Faustorilla J, Gavino A, et al. Digital pathology – implementation challenges in low‐resource countries. Anal Cell Pathol (Amst) 2012; 35: 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Amann J, Blasimme A, Vayena E, et al. Explainability for artificial intelligence in healthcare: a multidisciplinary perspective. BMC Med Inform Decis Mak 2020; 20: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. University of Louisville Health Adopts Paige AI‐enabled Cancer Detection Software for Enhanced Cancer Detection; 2021. [Accessed 12 February 2022]. Available from: https://www.businesswire.com/news/home/20211215005218/en/University‐of‐Louisville‐Health‐Adopts‐Paige‐AI‐enabled‐Cancer‐Detection‐Software‐for‐Enhanced‐Cancer‐Detection


