Abstract
Pseudomonas oleovorans GPo1 and its polyhydroxyalkanoic acid (PHA) depolymerization-minus mutant, GPo500 phaZ, residing in natural water microcosms, were utilized to asses the effect of PHA availability on survival and resistance to stress agents. The wild-type strain showed increased survival compared to the PHA depolymerase-minus strain. The appearance of a round cellular shape, characteristic of bacteria growing under starvation conditions, was delayed in the wild type in comparison to the mutant strain. Percent survival at the end of ethanol and heat challenges was always higher in GPo1 than in GPo500. Based on these results and on early experiments (H. Hippe, Arch. Mikrobiol. 56:248–277, 1967) that suggested an association of PHA utilization with respiration and oxidative phosphorylation, we investigated the association between PHA degradation and nucleotide accumulation. ATP and guanosine tetraphosphate (ppGpp) production was analyzed under culture conditions leading to PHA depolymerization. A rise in the ATP and ppGpp levels appeared concomitant with PHA degradation, while this phenomenon was not observed in the mutant strain unable to degrade the polymer. Complementation of the phaZ mutation restored the wild-type phenotype.
Upon nutrient starvation or entry into the stationary phase, a pleiotropic phenotype comprising cell morphology change, stress tolerance, and starvation survival develops in bacteria (18). In enterobacteria and Pseudomonas this transition is genetically controlled by the rpoS gene (14, 21, 26). This gene encodes a transcription factor that activates the expression of genes involved in cellular shape change and in cross protection against damaging agents, such as ethanol, H2O2, high temperature, or high salt concentration. There is strong evidence that guanosine tetraphosphate (ppGpp) induces expression of the rpoS gene (7).
Many bacterial species, including Pseudomonas oleovorans, synthesize polyhydroxyalkanoic acids (PHAs) under growth conditions characterized by an abundance of carbon sources with respect to other nutrients, such as nitrogen or phosphorus (1). Synthesis and degradation of PHAs are part of a cycle in which the acyl coenzyme A (acyl-CoA) precursors are converted by different metabolic pathways into PHAs. The depolymerization produces acyl-CoAs and acetyl-CoA, which is metabolized in the tricarboxylic acid cycle as a source of carbon and energy (13). PHA accumulation increases bacterial survival in nutrient-depleted cultures (17). Previous studies from our laboratory using wild-type strains and mutants deficient in the synthesis of PHA in natural waters have shown that the capability for synthesis of the polymer endows the bacteria with enhanced survival and competition abilities (15). The growth of bacteria in microcosms resembling natural environments where starvation conditions prevail is an alternative for the study of starvation-related phenomena, like those of growth in limiting conditions or the stationary phase achieved in laboratory cultures. The approach used in the above-mentioned microcosm experiments based on PHA-minus strains excluded the possibility of controlling PHA accumulation. Further experiments performed with a P. oleovorans mutant affected in the ability to degrade the accumulated polymer showed that it was a more appropriate system for studying the role of PHAs in bacterial survival (24). The influence of PHA has not been analyzed in bacterial stress resistance studies.
The mechanisms by which PHA favors bacterial survival are not yet fully understood (17). Early work suggested an association of PHA utilization with respiration and oxidative phosphorylation in Ralstonia eutropha (9).
In this work, we assessed the influence of PHA utilization in survival and tolerance of ethanol and heat challenge in starved cells of P. oleovorans by comparing wild-type and PHA depolymerase-minus (phaZ) strains in river water microcosms. Based on previous results (9, 15, 17), we investigated whether the polymer could be related to the synthesis of nucleotides, including ppGpp as well. This molecule was chosen as a compound involved in the enhanced survival and stress resistance specific to bacteria subjected to starvation conditions.
The accumulation of ppGpp depends on multiple factors, such as energy source availability, amino acid starvation, and growth inhibition. Therefore, the detection of an association between its synthesis and a single phenomenon requires a special experimental approach (4). In the case of the stringent response, the association with ppGpp accumulation is visualized either by amino acid starvation in an auxotroph or by the addition of serine hydroxamate (4). In our case, the study of a relationship between nucleotide synthesis and PHA depolymerization also required a special experimental approach, as the nucleotides could be synthesized from other precursors. The genetic approach chosen in this work in order to provide evidence for an association between the two phenomena relied on the utilization of a phaZ mutation, together with special culture conditions that allowed the observation of spontaneous polymer degradation. Thus, it was possible to observe a relationship between nucleotide accumulation and PHA degradation independently of the availability of other precursors.
MATERIALS AND METHODS
Bacterial strains, plasmids, and genetic manipulations.
The strains used in this study were P. oleovorans GPo1 and its PHA depolymerase-minus mutant, P. oleovorans GPo500 phaZ (10). Plasmid pGEc422 carrying phaC1 and phaZ genes and their promoter region from P. oleovorans GPo1, which restores PHA degradation in the strain GPo500 (10), was digested with EcoRI and ClaI, and the fragment harboring the pha genes was subcloned in the mobilizable vector pBBR1MCS-2 (23), producing pmPoCZ. This plasmid was mobilized from Escherichia coli DH5α into P. oleovorans strains using the helper plasmid pRK24 (28). Transconjugants were isolated on E medium (8) plates containing sodium octanoate (5 g · liter−1) and kanamycin (50 μg · ml−1). Plasmids and DNA fragments were purified with purification kits (Concert; GIBCO BRL). DNA manipulations were performed according to the method of Sambrook et al. (25).
Water samples.
Experiments were performed with surface water samples collected from the top 10 cm of water from the Rio de la Plata (Buenos Aires, Argentina) using a plastic container with a lid, previously rinsed three times with the same water. The water was processed in the laboratory within a few hours of collection. The river water is poor in carbon and nutrient (15).
Microcosms.
Microcosms were constructed as previously described (15). Bacteria were grown with shaking up to the early stationary phase (optical density at 600 nm, 1 to 1.1) in nutrient broth supplemented with 5 g · of sodium octanoate liter−1 at 30°C. The cells were harvested by centrifugation and rinsed twice with saline solution (9 g of NaCl liter−1). The pellets were then resuspended in the microcosms, consisting of 100 ml of filtered river water in 500-ml Erlenmeyer flasks, and incubated with shaking at 30°C. The bacteria were counted immediately after the inocula were added (day zero) and then at appropriate times over 25 days by serial dilutions onto triplicate nutrient agar plates. The variation coefficient for replicate platings was ≤20%. Microcosm samples were microscopically checked for PHA content periodically by staining them with Nile blue (19).
Challenge protocol.
One-milliliter water samples of the microcosms were diluted in saline solution according to cell numbers and then, at time zero, were diluted 10 times in saline solution supplemented with 20% ethanol or preheated at 47°C. The ethanol challenge was performed at 25°C. The challenge conditions (concentration, time, and temperature) were chosen so that a rapid decline in the survival of a growing culture was obtained. At different time intervals, aliquots (0.1 ml) were spread onto nutrient agar plates and incubated overnight at 30°C. Each challenge experiment was performed at least twice. Variations in replicate platings were ≤20%. Aliquots from an exponentially growing culture were challenged by the same protocol to compare exponential-phase cells with cells residing in microcosms. Stress resistance was expressed as the percentage of survival compared to the situation shortly before stress application, which was set at 100%.
Culture conditions.
PHA accumulation was achieved as previously described (11) with slight modifications. Cells were precultured at 30°C for 24 h in 250-ml Erlenmeyer flasks containing 25 ml of E medium supplemented with 5 g of sodium octanoate liter−1. The resulting cultures were used to inoculate 1-liter Erlenmeyer flasks containing 250 ml of 0.5 N E2P medium. This medium contains 0.5 g of K2HPO4 · 3H2O, 0.24 g of KH2PO4, 0.45 g of NH4Cl, 0.5 g of NaCl, and 1 g of yeast extract per liter, supplemented with either 5 or 2.5 g of sodium octanoate liter−1 and 2 mM MgSO4. Phosphate concentrations in 0.5 N E2P medium are appropriate for performing ppGpp accumulation studies. When necessary, kanamycin was added at 10 μg · ml−1.
Analytical determinations.
PHA was measured chromatographically (3). The PHA content was expressed as the percentage of cellular dry weight. Proteins were determined by the method of Lowry et al. (16). Intracellular levels of ATP and ppGpp were measured chromatographically as described previously (5) in cultures uniformly labeled with [32P]Na2HPO4. One-milliliter aliquots of the parent culture were taken at the indicated time intervals and incubated further in the presence of 0.1 mCi of [32P]Na2HPO4/ml for 2 h. Then extraction was performed in accordance with the method of Bochner and Ames (2). Cultures of Salmonella enterica serovar Typhimurium argC 95 grown with and without arginine were used as controls. In the absence of arginine, ppGpp synthesis is strongly induced in this strain. Extracts were analyzed by thin-layer chromatography in polyethyleneimine-cellulose plaques using 1.5 M KH2PO4 (pH 3.5) as the mobile phase. The nucleotide spots were identified as previously described (2). Those spots corresponding to ATP and ppGpp were scraped, and radioactivity was quantified in a liquid scintillation counter.
RESULTS
Different cellular traits were used in order to determine the sampling periods for the challenge experiments. As bacterial cells grown under starvation conditions acquire a round shape (14), this easily detectable trait, along with survival and PHA content, was used to analyze starved P. oleovorans GPo1 and GPo500 cells in river water microcosms. Challenge experiments were performed on bacteria residing for 3, 10, and 21 days in river water microcosms. These days were selected on the basis of changes in cell shape and survival.
Effect of starvation on survival, cell shape, and PHA content.
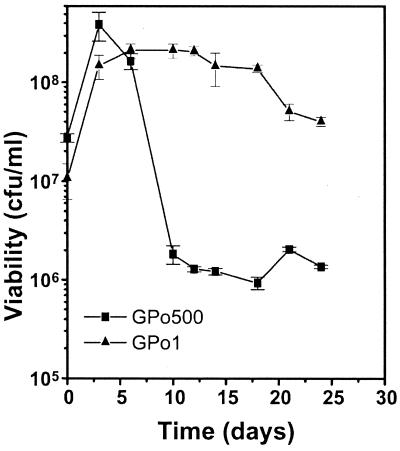
Survival of the wild-type strain was higher than that of the PHA depolymerase-minus strain. Cell numbers increased during the first days of microcosm residence for both strains. This increase has also been observed in other species (8, 15, 20) and reflects residual cell division. At day 10, the mutant strain showed a large decrease. At day 21, survival differences between the strains were at 1 order of magnitude (Fig. 1). This experiment was repeated twice, and the same survival pattern was displayed under these conditions (data not shown).
FIG. 1.
Survival of P. oleovorans GPo1 and GPo500 in sterile river water microcosms. Bacterial counts were expressed as CFU per milliliter. The values represent means ± 1 standard deviation of three replicated plate counts.
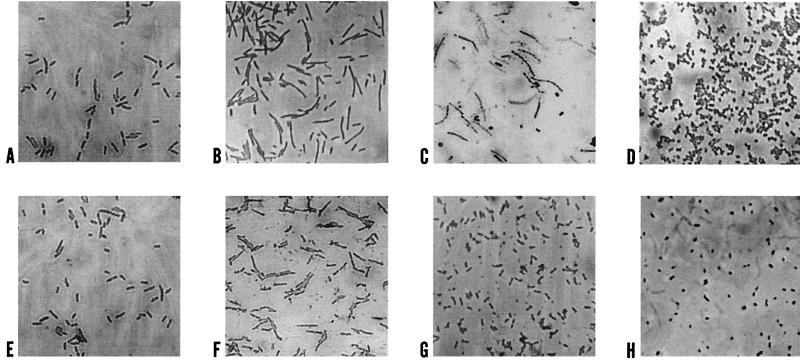
Growing cells of P. oleovorans are rod shaped. Microscopic observations of the microcosm aliquots revealed gradual changes in the cell shape. At day 3, cells of both strains were longer than normally growing cells and were cylindrical. By day 10, strain GPo500 cells had become round, while the wild-type cells acquired a filamentous pattern, probably due to the lack of division. Small round cells were observed after 21 days of microcosm residence for both strains (Fig. 2). The changes in the cell shape correlated with the survival decrease observed in the mutant strain after 10 days of microcosm residence. Granules of PHA were not observed in the wild-type strain after 1 day of residence in the microcosm, while in the mutant strain, PHA granules were observed throughout the experiment (data not shown).
FIG. 2.
Microscopic observations of the change in cell shape of P. oleovorans GPo1 (A to D) and GPo500 (E to H). Growing cells before microcosm inoculation (A and E) and cells after 3 (B and F), 10 (C and G), and 21 (D and H) days of microcosm residence are shown. Magnification, ×1,000.
Ethanol challenge.
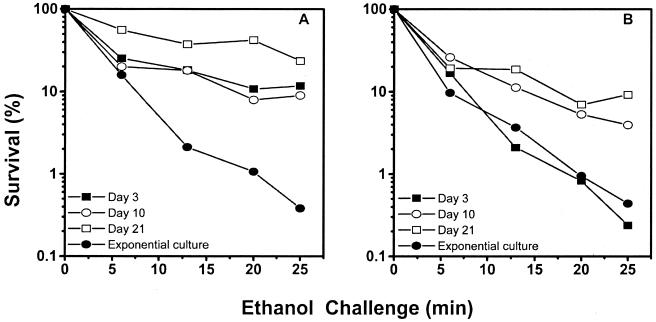
Exponentially growing cells and cells residing in the microcosms were challenged with 20% ethanol (Fig. 3). The wild-type strain developed enhanced resistance with increasing residence time in microcosms (Fig. 3A). Cells of the mutant strain were sensitive to ethanol on day 3, and resistance was observed on day 10 (Fig. 3B). On day 21, both strains showed resistance; however, the percentages of survival after 25 min of exposure were 23 and 9% for the wild-type and mutant strains, respectively.
FIG. 3.
Challenge of P. oleovorans GPo1 (A) and P. oleovorans GPo500 (B) residing in microcosms with 20% ethanol. An exponential culture and cells residing in the microcosms for 3, 10, and 21 days were challenged with 20% ethanol at 25°C. Percent survival was determined as the number of viable cells at each time divided by the number of viable cells before exposure to ethanol. The data are means of two independent experiments.
Heat challenge.
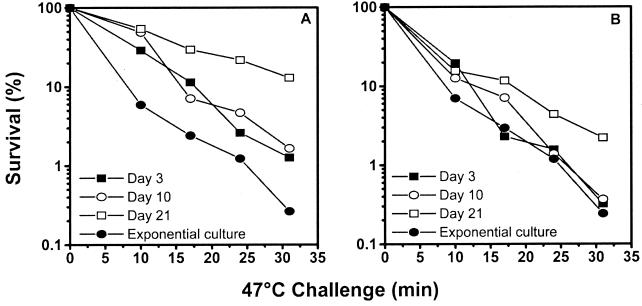
Exponentially growing cells and cells residing in microcosms were challenged with heat shock at 47°C (Fig. 4). The wild-type strain showed a gradual increase in heat shock resistance with increasing residence time in microcosms in agreement with the ethanol challenge results (Fig. 4A). Resistance was at a maximum on day 21 (13% survival at the end of the exposure). The mutant strain did not show stress resistance on days 3 and 10 of the experiment. By day 21, the mutant strain had slight resistance, as demonstrated by the low percentage of survival (2%) after 30 min of exposure to 47°C (Fig. 4B).
FIG. 4.
Challenge of P. oleovorans GPo1 (A) and P. oleovorans GPo500 (B) residing in microcosms at 47°C. An exponential culture and cells residing in the microcosms for 3, 10, and 21 days were challenged at 47°C. Percent survival was determined as the number of viable cells at each time divided by the number of viable cells before exposure to heat. The data are means of two independent experiments.
Nucleotide accumulation in P. oleovorans wild type and depolymerase-minus mutant.
These results and those previously reported (15) clearly show that PHA enhances survival and stress resistance in bacteria in natural environments. As this phenomenon is also observed in nutrient-limited cultures (17), and as PHA is a reservoir of carbon and energy, it can be hypothesized that the products of polymer degradation are used for the synthesis of nucleotides and other compounds.
As different precursors could be utilized for the synthesis of nucleotides, the study of a relationship between their synthesis and a single factor, such as PHA, requires a special experimental approach. The strategy chosen in this work was to obtain genetic evidence for the phenomena by the analysis of the behavior of a depolymerase-minus mutant in cultures where PHA depolymerization occurs.
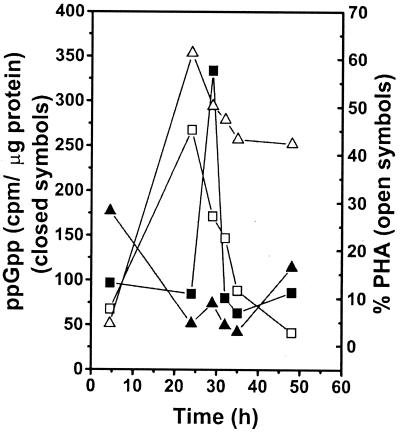
The parameters for PHA accumulation (mainly polyhydroxyoctanoate and polyhydroxyhexanoate) and depolymerization in P. oleovorans GPo1 and GPo500 under laboratory conditions have been clearly established by Huisman et al. (11). Polymer degradation starts after approximately 30 h of cultivation for GPo1, while this phenomenon is not detected in strain GPo500 except for a slight degradation due to the leakiness of the phaZ mutant. Based on these results, we chose this system in order to observe an association between nucleotide accumulation and PHA depolymerization. ppGpp, which is involved in general stress resistance (7), and ATP were measured. Figure 5 shows the kinetics of ppGpp production in the PHA-accumulating cultures. A peak of ppGpp synthesis was detected coinciding with PHA degradation in P. oleovorans GPo1. However, levels of ppGpp remained constant for P. oleovorans GPo500, except for a small peak of synthesis associated with slight biopolymer degradation. This experiment was repeated once, and the same accumulation pattern was obtained.
FIG. 5.
Intracellular levels of ppGpp and PHA content of P. oleovorans GPo1 (squares) and GPo500 (triangles). Cells were grown in 0.5 N E2P supplemented with 5 g of sodium octanoate per liter. Culture samples were analyzed for ppGpp and PHA content at the start of PHA degradation. Each value is the result of two determinations.
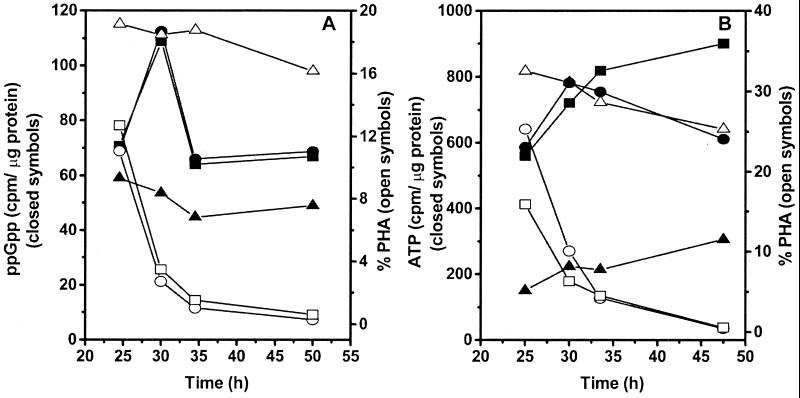
Plasmid pmPoCZ was introduced by conjugation into P. oleovorans GPo1 and GPo500 for complementation analysis. The intracellular levels of ATP and ppGpp were measured in the transconjugants grown under conditions that allow the observation of PHA depolymerization.
The results are shown in Fig. 6. As expected, the ATP intracellular content correlated with PHA degradation in the time interval analyzed (Fig. 6B). Both strains, GPo1/pmPoCZ and GPo500/pmPoCZ, showed an increase in ppGpp accumulation as PHA depolymerization proceeded (Fig. 6A). The nucleotide levels were not associated with PHA depolymerization in P. oleovorans GPo500 harboring the vector plasmid. These experiments were repeated once, and the same accumulation pattern was observed (data not shown). The values for protein content did not significantly differ among the strains in the nucleotide accumulation experiments.
FIG. 6.
Intracellular levels of ppGpp (A) or ATP (B) and PHA content of P. oleovorans GPo1/pmPoCZ (squares), GPo500/pmPoCZ (circles), and GPo500/pBBR1MCS-2 (triangles). Cells were grown in 0.5 N E2P supplemented with 2.5 g of sodium octanoate per liter and 50 μg of kanamycin per ml. Culture samples were analyzed for ppGpp (A) or ATP (B) and PHA content at the start of PHA degradation. Each value is the result of two determinations.
These experiments clearly show that the degradation of PHA correlates with an increase in the intracellular levels of ATP and ppGpp. The nucleotide levels might have risen in the mutant strain without being detected in the time intervals analyzed. If that were the case, the precursors for the synthesis would not come from PHA, as PHA is not degraded in the GPo500 strain, except for the minor degradation due to the leakiness of the GPo500 mutant. Concurrently, the complementation of the phaZ mutation gives the same accumulation pattern as the wild-type strain.
DISCUSSION
The experiments described in this work show that the phaZ mutation is a reliable system for studying the role of PHA in tolerance for starvation conditions and damaging agents in natural water microcosms and for suggesting a mechanism to explain this tolerance.
The wild-type strain showed increased survival compared to the PHA depolymerase-minus strain. This result had been previously observed in nonsterile microcosms (24). Survival was tested in association with changes in cell shape and PHA content. The change to a round cell shape favors the absorption of nutrients from the environment. Our results showed that morphological changes are delayed until 21 days of microcosm residence in the wild-type strain, while in the mutant strain these changes were observed on day 10. The wild-type strain could use PHA, a reservoir of carbon and energy, and hence this transformation was delayed. PHA granules were not detected in the wild-type strain the day after the start of the microcosm experiment; however, that does not mean that this carbon and energy source was exhausted at this stage of the experiment. Routinely, P. oleovorans cultures prepared as indicated in Materials and Methods contain 30% PHA relative to cellular dry weight. If this quantity of carbon is available for the cell, a delay in entrance into a resistant state is quite possible. Cells of both strains could have monitored transient C abundance that allowed them to accumulate the polymer and, in the case of GPo1, to degrade it in a dynamic way. The Nile blue staining method detects mainly extremely compact granules. The absence of granules does not mean that PHA is not available as a carbon source, as PHA synthesis is presumed to start with several polymer chains that coalesce in a hydrophilic cytoplasm (6) and polymer chains can also be originated by depolymerization (12).
P. oleovorans GPo1 showed enhanced resistance to ethanol and heat challenge compared to its corresponding PHA depolymerase-minus mutant, GPo500, in river water microcosms. For both strains of P. oleovorans tolerance to stress conditions was enhanced with the time of residence in the microcosms, as evidenced by comparing the resistance of bacteria residing for 3 days in river water with that of bacteria residing for 21 days (Fig. 3 and 4). However, the percent survival at the end of the challenges was always higher in the wild type than in the mutant strain (Fig. 3 and 4). The differences were more evident for heat challenge.
Our work presents clear genetic evidence, relying on phaZ mutation and complementation studies, that correlates PHA degradation with nucleotide accumulation. Depolymerization of a carbon and energy source like PHA could provide for their synthesis. The detection of ppGpp accumulation was a particular challenge, as it depends on multiple factors. The choice of approach in this work, to observe an association between ppGpp accumulation and PHA depolymerization, was based on previously well-established culture conditions that allow the observation of spontaneous polymer degradation. This degradation proceeds, as has been previously elucidated (27), under conditions of deficiency of reducing power (NADH2) and acetyl-CoA and an excess of free CoA.
The cross protection from stress agents associated with the resistant state is less efficient in P. oleovorans GPo500 than in GPo1. We can speculate that as ppGpp is an activator of RpoS synthesis, low ppGpp levels would contribute to the reduced survival and stress resistance characteristic of the GPo500 strain. The phaZ genotype could be associated with alterations in the metabolism or uptake of phosphate or any step leading to nucleotide synthesis. Hippe studies (9), performed without the help of mutants in PHA-rich and-depleted cells, showed that the utilization of the polymer was not only associated with respiration and oxidative phosphorylation but also inhibited the degradation of nitrogenous cellular constituents in R. eutropha. Nevertheless, complementation of the phaZ mutation restores the wild-type phenotype and supports the hypothesis of the association between the phenomena.
Bacterial survival and stress resistance studies are important for many aspects of bioremediation, biocontrol, and plant growth promotion. Inorganic polyphosphate, another reserve polymer, has been shown to support stationary-phase survival and stress resistance in E. coli (22). These results, together with those presented in this work, indicate that an energy and carbon reservoir like polyhydroxyalkanoate should be taken into account while performing such studies.
ACKNOWLEDGEMENTS
We thank Bernard Witholt for kindly providing P. oleovorans strains and pGEc422 and Alexander Steinbüchel for the gift of plasmid pBBR1MCS-2. We are grateful to R. Guerrero and A. Steinbüchel for helpful discussions.
This work was supported by grants from Universidad de Buenos Aires and Agencia Nacional de Promoción Cientifica y Tecnológica. B.S.M. and N.I.L. are career investigators from CONICET. J.A.R. has a graduate student fellowship from the Universidad de Buenos Aires.
REFERENCES
- 1.Anderson A J, Dawes E A. Occurrence, metabolism, metabolic role, and industrial use of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochner B R, Ames B N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 3.Brandl H, Gross R A, Lenz R W, Fuller C. Pseudomonas oleovorans as a source of poly-β-hydroxyalkanoate for potential applications as biodegradable polyesters. Appl Environ Microbiol. 1988;54:1977–1982. doi: 10.1128/aem.54.8.1977-1982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cashel M, Rudd K E. The stringent response. In: Ingraham J, Low K B, Magasanik B, Neidhardt F C, Schaecter N, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1987. pp. 1410–1438. [Google Scholar]
- 5.Cashel M. The control of ribonucleic acid synthesis in Escherichia coli. J Biol Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- 6.Gengross T U, Reilly P, Stubbe J, Sinskey A J, Peoples O P. Inmunocytochemical analysis of poly-β-hydroxybutyrate PHB synthase in Alcaligenes eutrophus H16: localization of the synthase enzyme at the surface of PHB granules. J Bacteriol. 1993;175:5289–5293. doi: 10.1128/jb.175.16.5289-5293.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentry D R, Hernández V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of the stationary-phase sigma factor ςs is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Givskov M, Eberl L, Møller S, Kongbak Poulsen L, Molin S. Responses to nutrient starvation in Pseudomonas putida KT2442: analysis of general cross-protection, cell shape, and macromolecular content. J Bacteriol. 1994;176:7–14. doi: 10.1128/jb.176.1.7-14.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hippe H. Abbau und Wiederverwertung von poly-β-hydroxybuttersaure durch Hydrogenomonas H16. Arch Mikrobiol. 1967;56:248–277. [PubMed] [Google Scholar]
- 10.Huisman G W, Wonink E, Meima R, Kazemier B, Terpstra P, Witholt B. Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. Identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J Biol Chem. 1991;266:2191–2198. [PubMed] [Google Scholar]
- 11.Huisman G W, Wonink E, de Koning G, Preusting H, Witholt B. Synthesis of poly(3-hydroxyalkanoates by mutant and recombinant Pseudomonas strains. Appl Microbiol Biotechnol. 1992;38:1–5. [Google Scholar]
- 12.Jendrossek D, Schirmer A, Schlegel H G. Biodegradation of polyhydroxyalkanoic acids. Appl Microbiol Biotechnol. 1996;46:451–463. doi: 10.1007/s002530050844. [DOI] [PubMed] [Google Scholar]
- 13.Kessler B, Kraak M N, Ren Q, Klinke S, Prieto M, Witholt B. Enzymology and molecular genetics of PHAmcl biosynthesis. In: Steinbüchel A, editor. Biochemical principles and mechanisms of biosynthesis and degradation of polymers. Weinheim, Germany: Wiley-VCH; 1998. pp. 48–56. [Google Scholar]
- 14.Lange R, Henger-Aronis R. Identification of a central regulator of stationary phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 15.López N I, Floccari M E, García A F, Steinbüchel A, Méndez B S. Effect of poly3-hydroxybutyrate content on the starvation survival of bacteria in natural waters. FEMS Microbiol Ecol. 1995;16:95–102. [Google Scholar]
- 16.Lowry O H, Rosebrough N J, Far A L, Randall R J. Protein measure with the Pholin-phenol reagent. J Biol Chem. 1951;193:265–267. [PubMed] [Google Scholar]
- 17.Matin A, Veldhuis C, Stegeman V, Veenhuis M. Selective advantage of a Spirillum sp. in a carbon-limited environment. Accumulation of poly-β-hydroxybutyric acid and its role in starvation. J Gen Microbiol. 1979;112:349–355. doi: 10.1099/00221287-112-2-349. [DOI] [PubMed] [Google Scholar]
- 18.Nystrom T. The trials and tribulations of growth arrest. Trends Microbiol. 1995;3:131–136. doi: 10.1016/s0966-842x(00)88901-5. [DOI] [PubMed] [Google Scholar]
- 19.Ostle A, Holt J G. Nile Blue A as a fluorescent stain for poly-β-hydroxybutyrate. Appl Environ Microbiol. 1982;44:238–241. doi: 10.1128/aem.44.1.238-241.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Postma J, Hok-A-Hin C H, Shotman J M, Wijffelman C A, van Veen J A. Population dynamics of Rhizobium leguminosarum Tn5 mutants with altered surface properties introduced in sterile and nonsterile soils. Appl Environ Microbiol. 1991;57:649–654. doi: 10.1128/aem.57.3.649-654.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos-González M I, Molin S. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J Bacteriol. 1998;180:3421–3431. doi: 10.1128/jb.180.13.3421-3431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao N N, Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary phase Escherichia coli. J Bacteriol. 1996;178:1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rehm B H A, Krüger N, Steinbüchel A. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. J Biol Chem. 1998;273:24044–24051. doi: 10.1074/jbc.273.37.24044. [DOI] [PubMed] [Google Scholar]
- 24.Ruiz J A, López N I, Méndez B S. Polyhydroxyalkanoates degradation affects survival of Pseudomonas oleovorans in river water microcosms Rev. Argent Microbiol. 1999;31:201–204. [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Samiguet A, Kraus J, Henkels M D, Muehlchen A M, Loper J E. The sigma factor sigma S affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc Natl Acad Sci USA. 1995;92:12255–12259. doi: 10.1073/pnas.92.26.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinbüchel A, Schlegel H G. Physiology and molecular genetics of polyβ-hydroxyalkanoic acid synthesis in Alcaligenes eutrophus. Mol Microbiol. 1991;5:535–542. doi: 10.1111/j.1365-2958.1991.tb00725.x. [DOI] [PubMed] [Google Scholar]
- 28.Thomas C M, Smith C A. Incompatibility group P plasmids: genetics, evolution and use in genetic manipulations. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]