Summary
Autoimmune diseases are characterized by dysfunctional immune systems that misrecognize self as non-self and cause tissue destruction. Several cell types have been implicated in triggering and sustaining disease. Due to a strong association of major histocompatibility complex II (MHC-II) proteins with various autoimmune diseases, CD4+ T lymphocytes have been thoroughly investigated for their roles in dictating disease course. CD4+ T cell activation is a coordinated process that requires three distinct signals: Signal 1, which is mediated by antigen recognition on MHC-II molecules; Signal 2, which boosts signal 1 in a costimulatory manner; and Signal 3, which helps to differentiate the activated cells into functionally relevant subsets. These signals are disrupted during autoimmunity and prompt CD4+ T cells to break tolerance. Herein, we review our current understanding of how each of the three signals plays a role in three different autoimmune diseases and highlight the genetic polymorphisms that predispose individuals to autoimmunity. We also discuss the drawbacks of existing therapies and how they can be addressed to achieve lasting tolerance in patients.
Keywords: TCR, MHC, costimulation, coinhibition, cytokines
1. Introduction
A fundamental feature of the immune system is its unique ability to sufficiently distinguish self from non-self to prevent aberrant infection and colonization. The plethora of pathogens that exist can rapidly outmaneuver immune responses, as evidenced the ongoing global pandemic caused by SARS-CoV-2. Consequently, immune cells exist in a poised state to ensure rapid clearance of any infectious agent. However, maintaining this cell state requires exquisite control over immune cell activity and function to avoid unwarranted responses directed towards self. In autoimmune diseases, this tolerance to self is perturbed, leading to inappropriate activation of the immune system and subsequent tissue-specific or system immune cell infiltration and culminating in tissue destruction (1).
Currently, more than 80 different autoimmune diseases have been identified in humans (2). Each of these has its own etiology that involves different genetic predispositions, cell populations, tissues and environmental triggers. Common autoimmune diseases include type 1 diabetes (T1D), in which the pancreas is targeted; multiple sclerosis (MS), in which the central nervous system (CNS) is targeted; and inflammatory bowel diseases (IBD), in which the gastrointestinal (GI) tract is targeted. Frequently, such autoimmune diseases involve a loss in T cell tolerance and inappropriate activation of autoreactive T cells as evidenced by elevated numbers of such cells in tissues. Dampening T cell function is often implemented in patients to treat and manage disease (2).
T cell activation requires three signals – antigen recognition (Signal 1) initiated by the T cell antigen receptor (TCR) recognizing its cognate antigen on a given cell, costimulation (Signal 2), in which costimulatory/coinhibitory molecules are engaged and activated to boost or inhibit Signal 1 and cytokine-mediated differentiation (Signal 3), which directs T cell survival and differentiation into functional subsets (3). In the absence of any of these three signals, T cells are not properly activated, resulting in either unresponsive T cells (4) or reduced survival of T cells (5). In autoimmunity, however, dysregulation of each of these signaling pathways, either in isolation or in combination, has been reported to affect T cell activation (2, 6, 7). Additionally, subsequent T cell infiltration and effector responses resulting in chronic inflammation in tissues is further regulated by these signals in tissue sites themselves. Here, we review how these fundamental T cell activation pathways are perturbed in CD4+ T cells during autoimmune disease progression, with a focus on T1D, MS and IBD.
2. Signal 1 – TCR-Mediated Peptide-MHC Recognition by T cells
Major histocompatibility complex (MHC) molecules non-covalently interacting with linear peptide fragments are tethered to surfaces of antigen presenting cells (APCs) and serve as the primary ligands recognized by TCRs on T cells (8). Cognate TCRs recognizing a specific peptide/MHC (pMHC) complex presented by APCs can transduce activating signals to T cells. The peptides themselves are proteolytic byproducts that bind to the peptide binding groove of the MHC proteins during their assembly (9). In this manner, MHC molecules provide an overview of the proteins that are expressed within a cell, thereby allowing T cells to survey the landscape for any perturbations. Broadly speaking, MHC-I proteins that present intracellular peptides and MHC-II proteins that present peptides originating from extracellular proteins captured via the endocytic pathway (10). Through this division of labor, MHC molecules have evolved means to alert T cells to the presence of both extracellular and intracellular pathogens (Figure 1A).
Figure 1.
MHC-II alleles and peptides contributing to altered Signal 1 in autoimmune diseases. A. Antigen presenting cells (APCs) express MHC-II molecules complexed with linear peptide fragments in antigen-binding pockets. CD4+ T cells bear αβ TCRs with the capacity to recognize the peptide/MHC-II complexes. Upon antigen recognition, the CD3 signaling subunits (γ, δ, ε, ζ) transduce signals that culminate in T cell activation. B. The known MHC-II risk alleles for type 1 diabetes (T1D), multiple sclerosis (MS) and inflammatory bowel disease (IBD) are listed for both humans and mice. C. The known self-antigens reported to be targeted in T1D, MS and IBD are listed. The references for the studies in which these antigens were identified are provided parenthetically. Abbreviations, ChgA – chromogranin A, GAD65 – glutamic acid decarboxylase, GRP78 - glucose-regulated protein 78, HIPs – hybrid insulin peptides, IA-2 – islet tyrosine phosphatase 2, IAPP – islet amyloid polypeptide, IGRP – islet-specific glucose-6-phosphatase catalytic subunit-related protein, ZnT8 – zinc transporter 8, CNPase – 2’,3’-cyclic-nucleotide 3’-phosphodiesterase, MAG – myelin-associated antigen, MBP – myelin basic protein, MOBP – myelin-associated oligodendrocyte basic protein, MOG – myelin oligodendrocyte glycoprotein, PLP – proteolipid protein, S100β – S100 calcium-binding protein B, HH_1713 – a Helicobacter hepaticus-unique protein. This image was created with BioRender.com.
Although structurally and functionally similar, a few key differences exist between MHC-I and MHC-II proteins. While MHC-I molecules are expressed by all nucleated cells, MHC-II proteins are expressed only by certain populations at steady-state, the so-called ‘professional’ APCs (APCs) consisting of B cells, dendritic cells (DCs), and macrophages, at steady-state (8). Additionally, while the ligand-binding pocket in MHC-I molecules is formed by one polypeptide, the same pocket in MHC-II molecules is formed by two distinct polypeptides that form a heterodimer. Lastly, due to the closed ends of their peptide binding grooves, MHC-I molecules chiefly present peptides of 8–10 residues (11, 12). MHC-II molecules, though, possess open-ended grooves that allow presentation of longer peptides, with the peptide termini extending beyond the ends of the groove (13). Peptides bound in this extended conformation to MHC-II interact with the groove by forming conserved hydrogen bonds between the peptide backbone and MHC-II residues (14). As the MHC-II interactions occur with the peptide backbone and not its side chains, the bound peptides can shift in the groove while still maintaining their hydrogen bonds (15). Consequently, the same peptide can be recognized in distinct manners by different TCRs depending on the register in which it is bound to MHC-II (16).
Two α-helices and a β-pleated sheet form the sides and the floor of the MHC antigen binding groove, respectively (8). Residues within this pocket dictate which peptides can bind with sufficient affinity to create a stable complex that can be recognized by TCRs. Interestingly, the genes coding for MHC proteins are among the most polymorphic known (17). These polymorphisms are concentrated in the regions that code for the residues in the antigen binding cleft, which results in different MHC alleles displaying distinct peptide-binding biases that in turn impart greater susceptibility to or protection against pathogenic threats to the host (9). Another notable consequence of these polymorphisms is that certain MHC alleles have been linked to greater incidences of autoimmune diseases (2).
Adding to this complexity is the fact that the bound peptides exert their own potency in dictating the T cell response. In some cases, the immunogenic self-peptides are not always those against which T cells are effectively tolerized. Although, positive and negative selection in the thymus ensure that the T cells entering the periphery do not display strong reactivities to self-peptides, these peptides do not completely sample the antigenic space. Altered self-antigens in peripheral tissues are presented as neoantigens against which the T cells can mount an immune response due to incomplete tolerization (15, 18). In other cases, certain viral or bacterial peptides presented during an infection bear sufficient similarities to host self-peptides that the T cell response is misdirected to target host tissues and initiate the autoimmune cascade (19). Overall, both the peptides and the MHC molecules play critical roles in pushing T cells to break tolerance in various autoimmune diseases.
From the T cell perspective, the TCR is a protein that harbors extraordinary diversity with the capacity to recognize the entire universe of peptide-MHC complexes (20). The TCR heterodimer is composed of an α- and a β-chain and recognizes antigens using three flexible loops called complementarity determining regions (CDRs) on each chain (21). The CDR1 and CDR2 loops are coded within the variable (V) genes for each chain and have been shown to interact with the α-helices that form the peptide binding grooves of MHC molecules. The TCR diversity, though, is largely restricted to the CDR3 of each chain, which is produced via a coordinated series of somatic rearrangements. The CDR3 loops fixate on the MHC-bound peptide to ensure that the most diverse part of the TCR engages the most diverse part of the ligand (22). Antigen recognition results in conformational changes and subsequent intracellular signal transduction.
The TCR heterodimer itself has no signaling capacity. Instead, it is associated with a complex of proteins that cluster with the TCR and initiate the signaling cascade (23). The affinity of the TCR for its cognate peptide-MHC ligand determines the strength of the intracellular signal that is transmitted. High affinity interactions promote strong and lasting T cell activation with improved T cell fitness (24). Alternatively, high concentrations of cognate peptides presented by APCs can overcome the threshold required for T cell activation due to increased TCR avidity (25) (Figure 2). Thus, during autoimmunity, T cells bearing self-reactive TCRs with low to moderate affinities for their target peptides can be atypically activated and mediate disease in environments where cognate peptide concentrations are high (26, 27).
Figure 2.
High antigen concentration in the periphery can overcome poor MHC-II binding. Left, Numerous self-antigens are presented in the thymus by thymic APCs but due to their low availability, any given peptide/MHC complex is not presented at high levels. Self-peptides that bind to MHC molecules with low affinities are further underrepresented, allowing any developing T cell with cognate specificity to mature and not undergo clonal deletion. Right, In peripheral tissues, the low affinity self-peptides could be available at high concentrations, which could promote enhanced presentation of these peptides due to their increased proportional sampling. Low affinity interactions could, therefore, be overcome by higher avidity antigen presentation, which leads to activation of autoreactive T cells. This image was created with BioRender.com.
2.1. Type I Diabetes
T1D is characterized by T cell infiltration into the pancreatic islets culminating in the destruction of insulin-producing β cells (28). A strong disease association with certain MHC-II alleles implicates a more principal role for CD4+ T cells in T1D (29–31). Indeed, CD4+ T cells potentiate the production of insulin-specific autoantibodies by B cells, a key feature of T1D (32). In addition, activated CD4+ T cells promote inflammatory macrophage and neutrophil infiltration into the islets, which cause further depletion of β cells (33–35).
About 90% of patients diagnosed with T1D have been reported to carry the HLA-DQ2 or HLA-DQ8 haplotypes (36, 37) (Figure 1B). One principal element shared by these strong T1D-associated MHC-II alleles is a polymorphism at position 57 in the HLA-DQβ chain (29, 38). While most other MHC-II alleles possess an aspartic acid residue at this position, the disease-associated alleles possess a non-aspartic acid residue instead. Indeed, the widely used non-obese diabetic (NOD) spontaneous diabetes mouse model also possesses a non-aspartic acid at this position in its MHC-II allele (I-Ag7) (39, 40). β57 aspartic acid normally forms a salt bridge with an arginine residue (R76) in the DQα (or I-Aα in mouse) chain (41, 42) and in its absence, the αR76 uniquely shapes the peptide repertoire bound to the MHC-II alleles by preferentially selecting for peptides containing acidic residues at the P9 position (43, 44). As a result, MHC-II alleles with this position 57 polymorphism bind to peptides with significantly lower affinities than those with the canonical aspartic acid (45). Reduced peptide affinity to MHC-II has been proposed as a possible mechanism for how self-reactive T cells can avoid clonal deletion during thymic central tolerance (46). Self-peptides are poorly presented during thymic selection but their abundance in the target tissues allows autoreactive T cells to overcome the weak presentation and become activated.
Such inferior peptide binding to the T1D-associated MHC-II alleles could be linked to poor Class II-associated invariant chain peptide (CLIP) binding to the MHC-II groove as well. CLIP is the peptide derived from invariant chain (CD74) that prevents aberrant peptide binding to MHC-II proteins after their assembly in the ER. It is released in the late endosomes by HLA-DM (H-2DM in mice), thereby allowing peptide exchange to occur for proper antigen presentation. Previous work, however, has demonstrated that CLIP binding affinity to MHC-II varies in an MHC-allele-specific manner based on the polymorphic residues lining the binding pocket (47). For the non-βD57 containing HLA-DQ2, HLA-DQ8 and I-Ag7 alleles, the positively charged pocket lowers the CLIP binding affinity, resulting in low affinity peptides loaded onto MHC-II that were not edited by HLA-DM (48–50).
Recent work has further supported the hypothesis that poor CLIP binding to these T1D-associated MHC-II alleles could contribute to T1D progression. The Wucherpfennig lab generated a knock-in NOD mouse in which a CLIP mutation was introduced to increase its affinity for I-Ag7 (51). Mice carrying this mutation were reported to have a lower incidence of T1D compared to their WT counterparts. Additionally, fewer antigen-specific T cells infiltrating the islets were observed in these knock-in mice. Strikingly, while APCs from mutant mice could process proteins and subsequently present peptides to T cells as efficiently as those from WT mice, they inefficiently presented soluble peptides to T cells compared to WT APCs (51). Therefore, increasing CLIP affinity to MHC-II prevented premature peptide loading and ensured that H2-DM could promote binding of high affinity peptides, which subsequently hampered soluble peptide exchange at cell surfaces. Consequently, diabetogenic self-peptides were bound to I-Ag7 less frequently, resulting in reduced T cell priming and islet infiltration.
Numerous other MHC risk alleles have been identified in humans that have been linked to increased susceptibility of T1D (30). Furthermore, some MHC-I alleles have also been associated with a greater predisposition to develop T1D (52). Indeed, high numbers of islet invading CD8+ T cells have been observed in T1D patients (53, 54). This observation correlates with experiments performed in mice which concluded that CD8+ T cells and MHC-I molecules are critical for T1D development (55–57). Yet, the precise molecular biases that impact disease development for these risk alleles have not been identified and should be thoroughly investigated in the future.
Although all APCs have been shown to contribute to disease development, B cells in particular have been repeatedly demonstrated to be necessary for T1D. Mice lacking B cells are protected from T1D (58, 59), while human T1D patients in whom B cells are selectively depleted display improved β cell function (60). Autoreactive B cells, which make up to 75% of the immature B cell repertoire (61), have the potential to capture protein antigens and process/present peptides to cognate self-reactive T cells (62). In their absence, the CD4+ and CD8+ effector T cell populations are suppressed and the remaining DCs and macrophages instead promote a more regulatory landscape (63), further implicating B cells in promoting T cell-mediated β cell destruction. Overall, this highlights that even though all APCs express the same MHC alleles, the cell type activating T cells can have drastic consequences for the disease course.
The antigens specifically recognized by T cells make up the final piece of the ligand presented by APCs that helps to trigger Signal 1 in T cells. Many self-antigens, such as insulin, chromogranin A, GAD65, IGRP and ZnT8, for example, have been identified as targets in T1D (64–71) (Figure 1C). However, it remained unclear why autoreactive T cells targeted these antigens despite having undergone central tolerance in the thymus. For example, one oft-targeted peptide in T1D in both humans and mice is the insulin β-chain fragment B:9–23 (15, 44). Yet, the specific register bound in the MHC-II groove had been unknown and complicated by the I-Ag7 β57 polymorphism. Extensive work by the Kappler and Unanue groups has revealed that this peptide is likely generated in the β cells themselves, which may employ distinct proteases and generate peptides with termini different from those observed in APCs (72–74). This peptide is subsequently loaded onto I-Ag7 via peptide exchange at the cell surfaces of APCs such as DCs, which then prime T cells. Furthermore, structural and functional data indicate that due to its C-terminal truncation, the bound peptide does not occupy the C-terminal end of the I-Ag7 groove (15, 74, 75), allowing it to perhaps engage the MHC molecule with higher affinity than that of the peptide generated by the proteases in APCs. Altogether, these results hint at how T cells exposed to the same peptide in the thymus escape negative selection while simultaneously getting activated in the pancreas (Figure 2).
In addition, several recent studies have provided strong evidence for a novel mechanism instigating a break in tolerance in T1D. In a process termed transpeptidation, noncontiguous peptides are fused in the lysosomes of β cells (67, 76). As proteins are degraded in these vesicles, the N-terminus of one peptide can form a peptide bond with the C-terminus of an unrelated peptide, resulting in a chimeric neoantigenic peptide. The high concentrations of certain proteins such as insulin and chromogranin A favors the generation of fusion peptides in β cells while their low expression in thymic APCs prevents clonal deletion of T cells expressing TCRs with specificities to these neoantigens (77). Indeed, the fusion peptides identified to date have largely involved either insulin (Hybrid Insulin Peptides) or chromogranin A. Furthermore, neoantigen-specific T cells can be isolated and characterized from pancreatic islets using neopeptide-loaded MHC tetramers in both mouse and human (67, 78–80). These T cells are expanded and have an activated profile, suggesting that they play a pathogenic role in T1D. How the peptides are transferred from β cells to APCs (or to what extent such peptides can also be generated in APCs themselves) to initially activate the T cells remains an area of active investigation.
Recognition of antigens by TCRs expressed by diabetogenic T cells activates the cells but characterizing the antigen-specific TCR repertoire has been challenging in humans because only the circulating T cells can be assessed for their antigen receptor sequences while the majority of the antigen-specific T cell population is localized in the pancreas. Additionally, although a strong association exists between certain MHC alleles and T1D, the penetrance is not complete. Consequently, many individuals express the risk MHC alleles and possess antigen-specific T cells while remaining disease-free, thereby confounding the link between disease and bearing diabetogenic T cells (81–83). Despite these complications, many studies have identified notable features in the TCR repertoires in T1D human patients as well as in diabetic mouse models. Common TCR features across individuals would indicate that both the autoantigens and autoreactive TCRs are conserved during disease and highlight a focused immune response, which could subsequently shape therapy design.
Early work by several groups in the NOD mouse model identified some V genes (TRBV12 and TRBV13) in the TCRβ locus (TRBV genes) that contributed to disease progression (84, 85). Yet, crossing NOD mice to a strain lacking these genes did not impede T1D development, although insufficient backcrossing could have contributed to this phenotype (86). Indeed, further studies observed an enrichment of T cells bearing TRBV13 rearrangements in the islets, suggesting that these TCRs were clonally expanded in the tissue due to a common antigen (87, 88). But many of these initial studies only examined the TCRβ chains from islet-infiltrating T cells, which paints an incomplete picture regarding the antigen-specific repertoire. This limitation was overcome by the Davis group by implementing single cell PCR to amplify both TCR chain sequences from sorted single T cells (89). This study also confirmed that TRBV13 rearrangements were enriched in the islets of NOD mice. In particular, this study identified that many of the clones possessed CDR3β loops with an acidic residue that was further corroborated in a later study (90). Subsequent work to better understand biases in the TCRα repertoire also highlighted the importance of TRAV5D-4 variable gene usage in diabetogenic TCRs (91). Altogether, these studies provided some preliminary evidence for an immune response in T1D in NOD mice involving a “public” TCR repertoire – a repertoire that is shared across individuals due to common antigens instead of unique to each individual.
Preliminary work in humans pointed to a largely “private” TCR repertoire used by the T cells entering islets in T1D patients (92–94). While clonal expansions were observed within an individual, common motifs were not reported across patients. In a recent comprehensive study, however, many novel findings were reported about the T1D TCR repertoire in humans (95). The authors analyzed over 2×108 total TCRβ sequences from different CD4+ T cell subsets isolated from T1D patients. T cells from T1D patients expressed TCR sequences with shorter CDR3β loops and a reduced contribution from random non-templated nucleotide insertions compared to the CDR3β sequences of healthy individuals. Former work in mice had demonstrated that TCRs with reduced nucleotide insertions tended to be more promiscuous and displayed greater self-reactivities (96). Due to decreased nucleotide additions and a subsequent reduction in TCR diversity, public TCRs were more frequently observed in T1D patients than in healthy individuals. Of note, hydrophobic amino acids were enriched in T1D CDR3β sequences in positions that are associated with enhanced self-reactivity (97).
In one study, a large-scale expression quantitative trait loci (eQTL) analysis was performed to determine how MHC haplotypes impact the TCR repertoire (98). Using gene expression data from over 900 human samples, the authors attempted to link MHC allelic differences to TCR variable gene usage. Remarkably, MHC polymorphisms at position 57 in HLA-DQ strongly impacted TRAV gene usage with a high probability. Indeed, individuals with MHC alleles that coded for a non-aspartic acid at this position displayed TRAV gene usage that was often reciprocal to that observed in those individuals with an aspartic acid at position 57. This residue is likely to play a strong role in dictating TRAV gene usage since it is surface exposed and can be directly contacted by the TCR. In particular, since canonical TCR docking onto MHC results in TCRα/MHCβ and TCRβ/MHCα interactions (22), it is not surprising that polymorphisms in the MHCβ chain have a greater influence on TCRα gene usage. Future work can expand upon this study to determine whether certain MHCα polymorphisms affect TCRβ gene usage as well.
2.2. Multiple Sclerosis
MS occurs due to demyelination of neurons in the CNS, which results in progressive loss of neurological function (6). T cells have been shown in both humans and mice to play a central role in exacerbating disease. Genome wide association studies (GWAS) approaches have identified the MHC-II β-chain encoding HLA-DRB1 region as a major risk locus associated with MS (99). Interestingly, unlike most other MHC-II genes where allelic diversity exists for both the α and β chains, HLA-DRα is non-polymorphic (100). Perhaps as a compensatory mechanism, the HLA-DRB1 gene has more polymorphisms than other MHC-II genes, which could provide an underlying rationale for its association with several autoimmune diseases. One of these alleles, HLA-DRB1*15:01, displays a strong association with MS with a reported odds ratio of 3.08 (101, 102) (Figure 1B).
Through the use of fine genetic mapping techniques to explain the effect of this DRB1 allele, DRβ1 position 71 was identified as the most significant residue (103). While most DRB1 alleles code for a charged amino acid at this position (arginine, glutamic acid, or lysine), HLA-DRB1*15:01 instead codes for an alanine at β71. Additionally, β71 helps form the P4 pocket in the peptide binding groove that helps to anchor peptides to the MHC-II molecule. Consequently, larger/hydrophobic peptide anchor residues can be accommodated at this position (104). Unsurprisingly, a recent study in which the HLA-DRB1*15:01 peptidome was investigated using mass spectrometry identified tyrosine and phenylalanine as enriched at the P4 position of bound peptides (105). This corroborates structural work in which the immunodominant peptide from human myelin basic protein (MBP, an antigen targeted by T cells in MS) bound to HLA-DRB1*15:01 also possesses a phenylalanine at P4 (106).
A link with MHC-II has been more indirect in the mouse model for MS called experimental autoimmune encephalomyelitis (EAE). Antigen-specific CD4+ T cells are sufficient to induce disease in both transfer and transgenic mouse models, indicating that that a key disease-causing factor is MHC-II-mediated antigen presentation (107–109). Yet, specific risk-associated MHC-II alleles have not been identified. The MHC-II alleles in EAE-susceptible mouse strains do not confer disease susceptibility when introduced into disease-resistant strains (110). This could be partly explained by the fact that the MHC-II locus responsible for disease in susceptible strains maps to the I-A region, instead of the I-E locus, because the I-E heterodimer is not expressed in many strains (111). Additionally, unlike the I-E locus (the HLA-DR ortholog in mice), the I-A locus displays polymorphisms in both MHC chains (112, 113). Thus, increased diversity in both MHC chains complicates pinpointing the disease-associated polymorphisms. Moreover, unlike the NOD mouse which develops spontaneous T1D, a strain of mouse that develops spontaneous MS has not been identified. Instead, EAE can often be effectively induced in several mouse strains by immunizing mice with myelin-derived antigens (114). As such, while MHC-II-dependent antigen presentation plays a critical role in driving EAE progression, no common MHC-II features across different strains can be readily identified as necessary for disease. Rather, each MHC-II allele presents antigens in a unique manner that distinctly activates the autoreactive T cells.
Despite these difficulties, some anomalous antigen binding and recognition features have been identified that have been useful in formulating hypotheses for how T cells break tolerance in EAE/MS. In one mouse model of EAE, mice bearing the I-Au MHC-II allele (such as mice belonging to the PL/J background) are immunized with the MBP peptide (115) (Figure 1B). This results in a relapsing/remitting chronic disease, which is reminiscent of the disease course in humans. Interestingly, the immunodominant epitope in this disease binds poorly to I-Au (116–118). The reason for this weak binding was evident once the trimeric structure involving a cognate TCR/MBP peptide/I-Au was solved (119). The bound MBP peptide has a charged lysine in the p6 position, which is incompatible with the hydrophobic p6 pocket created by the I-Au groove. In addition, the first three positions of the peptide binding groove remain unoccupied by the peptide, further destabilizing the peptide/MHC interactions. Such poor binding likely impacts peptide presentation in the thymus and has been experimentally demonstrated to result in improper negative selection of autoreactive thymocytes (120). Thus, distinct antigen presentation modes between thymic and peripheral APCs could provide an explanation for why central thymic tolerance does not effectively eliminate autoreactive clones (Figure 2). Whether other self-antigens in MS/EAE display similar poor MHC binding remains to be determined (121–126) (Figure 1C).
One further consequence of poor peptide binding is that TCRs also dock onto the peptide/MHC complex in unusual manners. In the MBP/I-Au example described above, as the first three positions of the groove are empty, the cognate TCR focuses on the carboxy-terminal region of the peptide instead of the center (119). This promotes distinct interactions between the TCR and the ligand that are usually not observed with other TCRs. Such unconventional TCR binding was also observed in a structure involving human HLA-DRB1*15:01 presenting MBP peptide and a cognate TCR (127). Unlike the mouse MBP peptide, human MBP peptide binds MHC-II with high affinity and fills the entire groove. Surprisingly, the cognate TCR binds to the complex with a focus on the N-terminal peptide fragment and the MHC α-helices. Uncommon TCR binding modalities have been previously observed to result in poor TCR signal transduction in the responding T cells (128). While it is unclear whether the abnormal docking of the MBP-specific TCRs triggers weak TCR signaling, such a possibility could explain how these T cells escape negative selection in the thymus. In the periphery, weak signaling could be overcome due to the high concentrations of the antigenic peptides that are sufficient to activate the T cells.
Since the aforementioned studies underscore the extent to which improper antigen presentation and recognition impact T cell activation, it is important to have an in-depth understanding of whether all APCs process and present antigens similarly or if instead different APC subsets have distinct capacities in activating T cells. Yet, the precise APC population that primes autoreactive T cells in the lymph nodes has remained elusive. Much attention has been paid to the contribution of DCs in initiating the immune response in EAE due to their superior antigen presentation competency. For instance, when in vitro bone marrow derived DCs were pulsed with MBP peptide and transferred into mice possessing antigen-specific T cells, the recipient mice rapidly developed EAE, suggesting that DCs play an important role in triggering disease (129). Indeed, a previous study by our group also observed that upon activation, DCs could in turn stimulate T cells to break tolerance and cause EAE (130). However, this has also been challenged more recently. Using genetic ablation strategies to deplete DCs, one study observed that mice lacking DCs experienced exacerbated disease, implicating a more protective role for DCs in EAE (131). This was further corroborated in a model of spontaneous EAE in which conditional deletion of DCs resulted in accelerated disease, indicating that other pAPCs are both capable and potent in priming T cells (132).
Several efforts have also been placed to determine which APCs are critical for restimulating the T cells in the CNS to license them to attack the tissue. Antigen presentation has most often been attributed to microglia, which are the CNS-resident leukocyte population (133). Other APC populations such as astrocytes, B cells, macrophages and monocytes have also been linked to restimulating the invading T cells (134–136). Recently, a publication by the Becher group has shed light on the APC population most critical in the CNS (137). Using various genetic targeting techniques to specifically delete MHC-II in different APC populations, the authors identified that a subset of DCs called cDCs are necessary for infiltrating T cells to become locally activated and invade the tissue. Mice in which cDCs lacked high MHC-II expression were protected from severe disease and displayed reduced EAE symptoms. Whether CNS cDCs are as critical in reactivating T cells in humans remains to be investigated.
While antigen presentation is critical for Signal 1, antigen recognition by the cognate TCRs is just as crucial for transducing the signal. Our current understanding of the characteristics of the TCR repertoire of pathogenic CD4+ T cells in MS and EAE has primarily resulted from studies of TRBV gene usage and CDR3β sequences. In human MS patients, much of the work has determined that a shared repertoire does not exist across all individuals. Instead, each individual possesses distinct clonally expanded populations in the CNS that attack tissue and mediate disease (138–141). This is in stark contrast to the repertoire analyses performed using T cells isolated from EAE mice, in which a more public TCR repertoire is observed involving dominant TRBV gene usage (142, 143). Indeed, saturation sequencing of the TCRβ repertoire in myelin oligodendrocyte glycoprotein (MOG)-peptide immunized mice revealed that public TCRs were preferentially enriched in the CNS (144). Furthermore, when public TCRβ chains were overexpressed in developing thymocytes, the resultant T cells displayed a greater predisposition to becoming autoreactive and causing disease, independent of the TCRα chains with which they were paired (145). These differences between humans and mice could be explained by the fact that disease in mice is often initiated due to autoreactivity to a defined peptide such as MBP or MOG. However, the antigens in humans could be varied even in HLA-matched individuals, which could engender diverse T cell responses. Moreover, epitope spreading is a common characteristic in human MS, in which T cell responses against epitopes distinct from those that initiate disease can arise during chronic inflammation (146, 147). As such, it is difficult to draw conclusions regarding shared TCR repertoire features when the TCRs being assessed may have distinct specificities. When this issue was avoided by evaluating the TCR repertoire to a fixed antigen from multiple MS patients, a preferential TRBV gene usage and CDR3β motif was readily identified (148). Thus, by not focusing on the TCR repertoire directed against a specific epitope, the conclusions in human studies are obfuscated and underscore how human TCR repertoire analyses from MS patients need to be revisited with a greater focus on antigen-specificities.
Recent work by our group, though, has argued against the notion of a public TCR repertoire even in EAE (149). In this study, we isolated CD4+ T cells from mice that had been immunized with the MOG peptide to induce EAE. By implementing single cell RNA-sequencing (scRNA-seq), we were able to identify both the TCRα and TCRβ chain sequences of each of the isolated cells, which could then be used to assign cells to a clone. Interestingly, although within an individual mouse there was substantial clonal sharing across different tissues, such TCR sharing was absent when TCR sequences were compared across mice. Thus, prior work focusing exclusively on the TCRβ repertoire to define public clones does not take into consideration how TCRα chains also affect antigen recognition. As such, the MS-predisposing polymorphism at position 71 in HLA-DRB1*15:01 has a large impact of TRAV gene usage in humans (98), implying that the TCRα repertoire is not innocuous in disease initiation. These results point to a great need to properly utilize existing techniques to readdress many of the questions regarding the nature of the TCR repertoire in EAE and MS.
2.3. Inflammatory Bowel Diseases
Disorders characterized by chronic intestinal inflammation are called inflammatory bowel diseases (IBD). IBD can be further categorized into two main subgroups – ulcerative colitis (UC), which manifests largely in the colon, and Crohn’s disease (CD), which can impact any region of the small or large intestine (150). Although immunosuppressive therapies have provided relief for many patients suffering from IBD, a large proportion of individuals either do not respond to therapy or become resistant within a year, with others progressing to the point of requiring surgical intervention (151).
Several studies identified a higher concordance rate for IBD among monozygotic twins, suggesting a genetic component to this disease (152). Additionally, familial aggregation of IBD has bolstered the notion that genetic predispositions to disease exist. However, IBD genetic linkage studies have not been able to strongly associate genetic variants with disease susceptibility. As the gastrointestinal (GI) tract is one of the largest tissues exposed to the environment, this weak association likely underscores the importance of environmental factors that affect disease development (153). Consequently, even though immune cells drive the chronic inflammatory state in IBD, the underlying cause of the disease can vary between individuals. For example, microbial composition and variation in the gut due to a variety of factors including age, diet, and geography can dictate the nature of the antigens presented to the immune system and subsequently influence the course of the immune response (154). Overall, the low penetrance of risk variants indicates that IBD likely arises in patients due to genetic susceptibility coupled with environmental factors.
Nevertheless, a few HLA locus polymorphisms have been linked to disease susceptibility. Much as in MS, several HLA-DRB1 alleles are associated with IBD. In particular, HLA-DRB1*07:01 is associated with CD patients often present with ileal inflammation (155, 156) (Figure 1B). Indeed, the odds ratios for ileal disease in CD patients in three independent cohorts were at least 1.5, signifying that there is a 1.5-fold greater risk of ileal inflammation in patients expressing the HLA-DRB1*07:01 allele (155, 157, 158). The HLA-DRB1*01:03 allele, on the other hand, is linked with increased susceptibility to both CD and UC with strong colonic inflammation (159). Multiple HLA haplotypes containing this allele have been observed in IBD patients, further connecting this allele to disease progression. Moreover, the odds ratios for colonic disease in IBD patients were observed to be >5 in multiple studies, indicating a strong association (155, 157, 158). However, why the polymorphisms in these alleles are linked to disease has not been determined. Unlike in T1D and MS where structural and functional studies have provided a greater mechanistic understanding of how HLA risk alleles predispose individuals to disease, similar work to gain insight into the molecular underpinnings of IBD risk variants has been lacking.
MHC-II also appears to play an important role in mouse models of IBD. In one widely used model of colitis, naïve CD4+ T cells (as determined by high expression of CD45RB) transferred into lymphopenic mice (such as Rag1−/−, for example) leads to the recipient mice beginning to lose weight within a few weeks after transfer (160, 161). These mice experience severe colitis mediated by CD4+ T cell infiltration. MHC-II allelic variants do not seem to impact susceptibility in this transfer model as multiple mouse strains with distinct MHC haplotypes have been used to successfully initiate disease (160, 162). Since the recipient mice do not possess any T cells of their own, the disease is a direct consequence of the transferred cells. Indeed, transferring regulatory T cells along with the naïve T cells significantly dampens disease severity (163). The gut microbiota appear to be critical in this transfer model because germ-free mice are resistant to disease (164). Thus, it is likely that certain bacterial peptides presented by MHC-II activate the transferred T cells to initiate colitis.
At steady-state, these microbial antigens function as self-ligands to promote peripheral tolerance in the hosts. Indeed, recent work has highlighted that there exists a cross-talk between the gut and the thymus to ensure that developing T cells bearing TCRs specific for commensal peptides are tolerant to these antigens, blurring the line between what is “self” and “non-self” (165). However, bacterial communities in the GI tract are profoundly sensitive to changes in the host, such as diet and health status (166). Upon perturbation, microbial community shifts have been observed, which could trigger a breakdown in T cell tolerance by presenting antigens to which the host has not been tolerized. Another non-mutually exclusive phenomenon could cause genetically predisposed individuals to gradually develop disease in which antigen-specific T cells that are resistant to peripheral tolerance mechanisms are continually activated. Indeed, this serves as the likely explanation for how disease is initiated in the CD4+ T cell transfer model (160). In the absence of any immunoregulatory influences, the transferred cells are activated by bacterial antigens and proceed to attack the intestine. Conversely, IBD can also manifest when T cells are super-activated, even though regulatory mechanisms are intact. This is most evident as a side-effect of anti-tumor immune checkpoint blockade (ICB) therapy (referred to as immune-related adverse events) in which the “brakes” on T cells are hindered resulting in colitic infiltration and IBD development in many treated patients (167).
Altogether, IBD paints a complicated picture with respect to antigen-driven T cell-mediated autoimmunity, in that the antigenic specificity is by itself insufficient to initiate/exacerbate disease. The context under which the T cells are activated is just as vital in determining their effector potential, which is a concept that could extend to other autoimmune diseases as well. Indeed, TCR transgenic cells with fixed specificities that recognize commensal antigens develop into regulatory T cells at steady-state, but into effector cells (primary Th17 cells) during IBD (168). In a separate study, the authors demonstrated that the same T cells can be polarized into distinct lineages depending on the microbe that expresses its cognate antigen, adding another layer of complexity to T cell activation (169). Confounding matters further, a greater appreciation is being paid to pathobionts – commensal microbes that can cause harm during certain perturbations. One such pathobiont in mice is Helicobacter hepaticus, which does not cause colitis upon colonization unless the mice are deficient in interleukin-10 (IL-10) signaling (170). Helicobacter hepaticus-specific T cells normally adopt a regulatory phenotype in the gut when exposed to H. hepaticus, but become inflammatory in IL-10-deficient hosts (168) (Figure 1C). This inflammatory phenotype is also conferred upon bystander T cells with specificities for other commensals, which then persist as long-lived memory cells in the intestine and could contribute to disease. Simply determining disease-causing antigen specificities and epitopes, thus, is insufficient to deeply understand the causes behind IBD development. Collectively, although IBD shares many features with other autoimmune diseases, it might be better characterized as an inappropriate immune response mounted against bacterial antigens in genetically susceptible individuals (168, 171).
Taking these varied antigens into consideration, it is also difficult to gain insight into any shared TCR features that promote IBD. Because the composition of the gut microbiota is host-dependent, the antigens presented are also frequently private. Consequently, the TCRs that recognize these antigens could be distinct from individual to individual. This is likely true even in the CD4+ T cell transfer model of colitis in mice. Since a polyclonal repertoire is transferred into recipient mice in this model without confirming any antigen-specificity, the targeted antigens are likely to be distinct in each recipient. Indeed, one study reached similar conclusions when comparing the repertoires of the transferred cells across multiple recipients (172). While oligoclonal T cell expansions were observed within an individual mouse, the TCR gene usage varied across mice, indicating that there was little antigenic overlap. Private clonal expansions were observed in individual human IBD patients as well, highlighting that although antigen-driven T cell dysfunction is common in patients, the antigens themselves are not common (173–175).
3. Signal 2 – Costimulatory and Coinhibitory Pathways Activated in T cells
To properly initiate the TCR signaling cascade, T cells need to engage both their cognate antigens and receive an additional signal, which is commonly referred to as ‘Signal 2.’ This second signal serves to amplify (or dampen) the signal stemming from antigen recognition by the TCR (176). Without this second signal, T cells enter a state of anergy, which is characterized by their unresponsiveness (177). This prevents aberrant T cell activation since the costimulatory receptors are not engaged in non-inflammatory settings. To accommodate the various tissues/cell states/inflammatory states that a given T cell may experience, numerous costimulatory and coinhibitory receptors exist on T cells that can skew the T cell fate in myriad directions and provide exquisite control over the extent of the T cell response (176). These receptors engage their ligands on the APCs bearing antigen and colocalize with TCRs within the immunological synapse, where the surfaces of the two cells are juxtaposed and TCR signaling is initiated (178).
Broadly speaking, the costimulatory and coinhibitory receptors belong to two families – the immunoglobulin superfamily (IgSF) and the tumor necrosis factor receptor superfamily (TNFRSF) (176). The classic family member belonging to the IgSF is CD28, expressed by T cells, which interacts with CD80 and CD86 on APCs. CD28 is constitutively expressed at high levels by naïve T cells and helps potentiate the production of a key survival cytokine (IL-2) after activation (179). Interestingly, a coinhibitory molecule called cytotoxic T lymphocyte antigen 4 (CTLA-4) competes with CD28 to bind to CD80/CD86 and hinder T cell activation. In fact, CTLA-4 interacts with CD80/CD86 with approximately 10-fold higher affinity than does CD28 but CD28 can still be engaged due to its higher expression levels compared to CTLA-4 (180). CTLA-4 can also act in a cell-extrinsic manner, when, for example, immunosuppressive Tregs (discussed in greater detail below) expressing high levels of CTLA-4 outcompete CD80/CD86 binding on APCs by CD28 expressed by the T cell being primed/activated (181). Thus, whether a T cell is activated and the degree of its activation are often dictated by which of these two proteins can more potently mediate its actions.
Notably, CD28 serves as more of an exception with respect to its expression on naïve CD4+ T cells. Expression of many costimulatory molecules is instead often induced de novo or upregulated upon T cell activation (182). In addition, many coinhibitory molecules are also expressed after activation, which help to maintain tolerance and prevent unchecked CD4+ T cell activation. Indeed, high expression of coinhibitory molecules is frequently a feature of activated cells, with the expression levels directly correlating with the extent of activation (183). The kinetics of costimulatory/coinhibitory molecule expression on T cells has been dubbed the tidal model of co-signaling (176, 182). Initially, T cell activation through the TCR results in increased expression of costimulatory receptors, which continue to favor and promote T cell activation. However, at the peak of the T cell response, coinhibitory receptors are also expressed at high levels, which then function to resolve the immune response by suppressing T cell activation and function. Therefore, T cell costimulation is not merely an on/off switch to amplify/dampen TCR signals but instead is a dynamic process that provides cues to CD4+ T cells in spatiotemporally regulated manners.
Stimulatory signals from receptors belonging to either superfamily promote cell survival, cell growth and activation of effector function. Signaling through these receptors primarily converge on the activation of canonical immune transcription factors such as NF-kB, NFAT and AP-1 (184, 185). Since these factors are also downstream of TCR signaling, pathways activated by costimulatory receptor engagement integrate and synergize with those of the TCR to enhance cell activation. Coinhibitory receptors act at various nodes along the pathways to inactivate or dampen the signals to serve as ‘brakes’ on the system. Importantly, though, each surface molecule can also activate independent pathways that together contribute to eventual CD4+ T cell fate (176). This is best evident when coinhibitory receptor blockade is employed in anti-tumor and chronic viral infection therapies to activate T cells (186). Merely blocking one receptor is often not as effective as blocking multiple receptors. Furthermore, blocking distinct coinhibitory receptors can differentially rescue specific functionality such as proliferation, cytotoxicity or cytokine secretion, indicating that each coinhibitory receptor plays a non-redundant role in dampening T cell activation and response. Similarly, while CD28 signaling results in high IL-2 production, signaling through inducible T cell costimulatory (ICOS), another IgSF member, instead results in high IL-4 and IL-21 production (187). Altogether, Signal 2 pathways both overlap with those of Signal 1 and can also provide unique signals depending on the molecule(s) engaged. Such molecular checks and balances are necessary to guide the T cells to mount appropriate immune responses when faced with dynamic immunogenic conditions and are often impacted in various autoimmune diseases, thereby contributing to improper T cell activation (Figure 3).
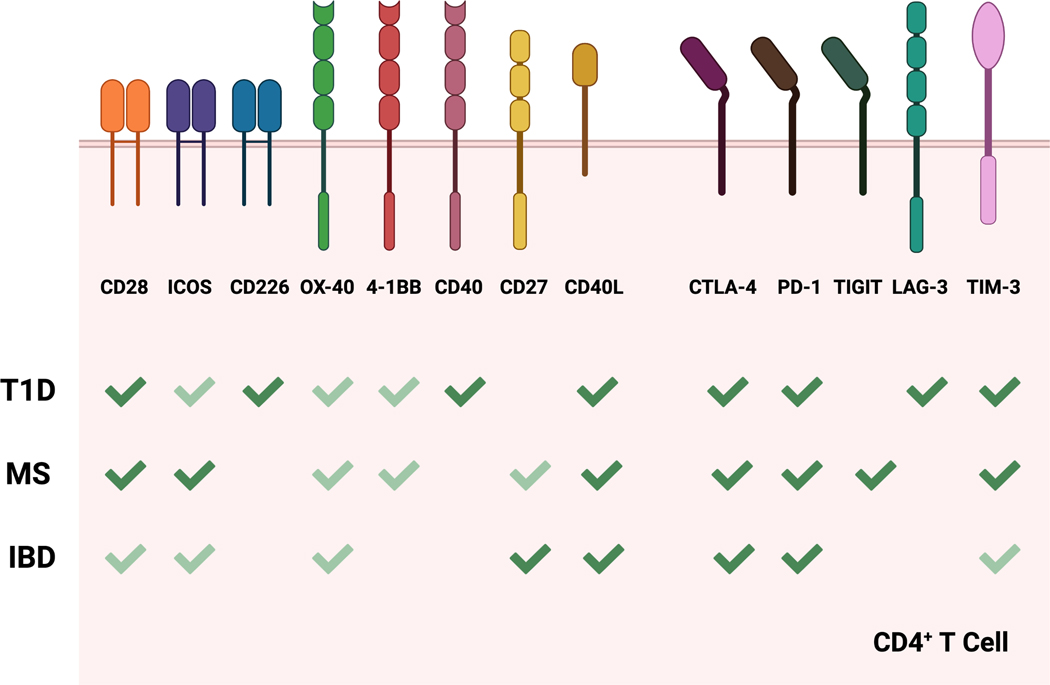
Figure 3.
Costimulatory and coinhibitory receptors involved in autoimmune diseases. The different costimulatory and coinhibitory receptors expressed by CD4+ T cells known to play a role in T1D, MS and IBD are depicted. The significance of any given receptor in contributing to disease progression is highlighted using dark green check marks (strong reported contribution) vs light green check marks (weak reported contribution). This image was created with BioRender.com.
3.1. Type I Diabetes
Due to its important role in T cell priming, it would be expected that CD28 plays an important role in activating diabetogenic T cell clones. In the absence of proper CD28 signaling, T cells were thought to become anergic and individuals would be protected from T1D. Instead, CD80/CD86 double-deficiency in NOD mice exacerbated T1D incidence (188). This finding was further corroborated in CD28-deficient NOD mice and was attributed to the paucity of CD25+ CD4+ Tregs that require CD28 signaling to properly develop. Indeed, absence of Tregs has long been known to increase incidence of multiple autoimmune disorders (189). Consistent with these results, treatment of NOD mice with antibodies that block CD28 interactions with CD80/CD86 also resulted in increased disease due to a reduction in Treg numbers (190). Interestingly, humans expressing the T1D-associated SNP (CT60G) in the CTLA4 locus expressed lower levels of the CTLA4 isoform lacking the transmembrane domain, referred to as soluble CTLA-4 (sCTLA-4) (191). Reduced expression of this sCTLA-4 was thought to allow for increased CD28-mediated activation and subsequent autoimmunity. NOD mice in which sCTLA-4 expression is reduced by RNA interference displayed accelerated T1D, further corroborating the human data (192). Our lab has previously reported that NOD mice also express an alternatively spliced CTLA-4 variant called ligand independent CTLA-4 (liCTLA-4) that lacks the extracellular domain required for CD80/CD86 binding (193). This isoform is expressed constitutively in primary CD4+ T cells and functions by binding to and dephosphorylating the TCR signaling proteins to inhibit signal cascade. Notably, disease-resistant mice possessed CD4+ T cells with higher expression of liCTLA-4 compared to those in disease-susceptible mice. Thus, various isoforms derived from the same genes can all impact autoimmunity but via distinct mechanisms.
One recent study measured the role of CD226 in impacting T1D incidence (194). CD226 is another costimulatory molecule expressed by T cells that interacts with CD155 (and also CD112) expressed by APCs and has been shown to promote differentiation of CD4+ T cells into the inflammatory Th1 subset (195). Of note, a SNP in CD226 in humans is associated with T1D and has been shown to enhance T cell activation (196, 197). Additionally, a risk locus in NOD mice also contains the Cd226 gene, indicating that this gene might be important even in the disease observed in mice (198). Indeed, NOD mice lacking expression of Cd226 displayed decreased T1D incidence and islet infiltration (194). Intriguingly, the T cells in this mouse also displayed reduced affinities for an immunodominant antigen, suggesting that CD226 signaling impacts T cell development in the thymus and in its absence, strongly self-reactive cells are not matured.
Unlike IgSF receptors such as CD28 and ICOS, TNFRSF members such as CD40-ligand (CD40L), OX-40 and 4–1BB appear to contribute more to effector T cell generation in T1D without impacting Treg function. This is rather surprising given the fact that TNFRSF members are vital for thymic Treg development (199). Since the importance of TNFRSF members in Treg development was assessed in C57BL/6 mice, while their absence in influencing disease was addressed in NOD mice, strain-dependent features could explain these differences. Nevertheless, germline deficiency of CD40L was sufficient to inhibit T1D in NOD mice, suggesting that CD40/CD40L signaling was required to initiate disease (200–202). CD40-expressing DCs promote Th1 differentiation and induce production of potent pro-inflammatory cytokines by the activated T cells (203). Interestingly, while CD40 is more often associated with expression in APCs, an aggressive autoreactive CD4+ T cell population was observed to express CD40 in T1D. Transferring these CD40+ T cells into non-diabetic mice was sufficient to induce progressive disease while transferring CD40− T cells could not transfer disease, highlighting the importance of CD40-expressing T cells in disease initiation (204, 205). In total, many costimulatory receptors cooperate with one another to maintain a delicate balance between Treg function and effector T cell activation that progressively disrupts homeostasis as T1D develops.
Prominent coinhibitory receptors such as PD-1 and LAG-3 also dramatically impact T1D progression, with SNPs in the human PDCD1 locus (the gene coding for PD-1) associated with disease susceptibility (206). This finding is recapitulated in mice as Pdcd1−/− mice develop aggressive T1D with complete penetrance (207). Similarly, Lag3 deficient NOD mice displayed highly aggressive disease progression (208). Extracellularly, LAG-3 competes with CD4 to bind to MHC-II while intracellularly, it inhibits TCR signal transduction (209). Due to its dual role, it is possible that LAG-3 can also function in a cell-extrinsic manner since Tregs express high levels of this protein and their function could be impacted in its absence. Lastly, T cell immunoglobulin domain and mucin domain-containing protein 3 (TIM-3) is another coinhibitory receptor that could play a role in exacerbating T1D. TIM-3 blockade in NOD mice accelerated T1D progression compared to that in control mice (210). Curiously, though, SNP analysis within the TIM-3 gene revealed no positive correlation with T1D, suggesting that there could be functional redundancy with other proteins in humans (211).
3.2. Multiple Sclerosis
Remarkably, unlike what is observed for T1D, CD28 signaling is necessary for induction of EAE (212). This is further corroborated by the fact that CD80/CD86 deficient mice are also protected from MOG peptide induced EAE (213). These puzzling discrepancies between disease models could be explained by the fact that the experiments were performed on mice from distinct genetic backgrounds (NOD vs C57BL/6). Indeed, even in EAE experiments, CD80/CD86 blockade accelerated EAE in mice from the SJL/J background, suggesting that other CD28 signaling deficiencies do not manifest themselves in the same manners in different strains (214, 215). Of note, while CD80 and CD86 are often considered to be interchangeable, their expression profiles are not always similar in different APC populations during EAE, with CD80 contributing more in the presentation of myelin-derived antigens (216–218). The tissue and cell type-dependent expression of CD80 and CD86 during EAE remain poorly understood and should be a future area of investigation.
ICOS deficiency surprisingly results in enhanced EAE induction with severe CNS infiltration by lymphocytes (219, 220). This has been attributed to reduced immunoregulatory cytokines such as IL-4 and IL-10, produced by Th2 and Tr1 cells, respectively, that require ICOS signaling for their proper differentiation. These results are also in line with an independent study in which ICOS engagement with its ligand was blocked via treatment with a monoclonal antibody (221). Such blockade was discovered to have dynamic consequences depending on when the antibody was administered. When administered during the T cell priming phase, antibody blockade augmented disease induction in mice, which appears to phenocopy what is observed in ICOS germline deficiency. Conversely, when administered at later time points during the efferent phase, antibody blockade largely abrogated disease. Thus, the temporal context under which ICOS signaling is impacted can dramatically influence disease course.
For some of the TNFRSF members whose roles have been investigated in MS/EAE, a recurring theme is that while their absence does impact disease, they mostly appear to function in conjunction with the CD28/CD80/CD86 axis. For example, genetic deletion of either OX40 or OX40L reduces disease severity but does not intrinsically protect against EAE (222–224). Overexpression of OX40L, on the other hand, exacerbated disease but only when the CD28 signaling axis was intact (225). Interestingly, CD27 deficient mice experience worse disease while CD70 (the ligand for CD27) overexpressing mice experience ameliorated disease (226). This observation could be due to the fact that intact CD27 signaling is critical for proper thymic Treg development, as mentioned previously (199). Without proper CD27 signaling, dysfunctional Tregs might not be able to successfully impede effector T cell activity.
In addition to costimulatory molecules, several coinhibitory molecules have also been implicated in regulating disease induction. The previously described CTLA4 SNP associated with T1D (CT60G) is also a risk variant associated with MS (191, 227). Additionally, CTLA-4 has been shown to affect MBP-specific T cell function in MS patients (228). This extends to mice because blocking CTLA-4 results in more severe EAE (229–231). PD-1 signaling can also affect disease progression in both mice and humans (206, 232). Former work by our group identified the T cell Ig and ITIM domain (TIGIT) protein as regulatory during EAE (233). The CD226/TIGIT axis is analogous to the CD28/CTLA-4 pathway in which both CD226 and TIGIT bind to the same ligands but TIGIT provides an inhibitory signal to T cells. Mice lacking TIGIT developed more severe EAE upon immunization with MOG peptide, supporting its role as a strong coinhibitory receptor. In an independent study also by our group, we identified that TIM-3 also serves to inhibit T cell responses during EAE (234). Tolerance induction was inhibited when TIM-3-mediated signaling was abrogated and T cells became polarized into inflammatory effectors. Moreover, TIM-3 has also been linked to MS in humans where TIM-3 expression was observed to be reduced in T cells from MS patients (235, 236). Altogether, different coinhibitory pathways coordinate to prevent autoreactive T cells from causing CNS tissue damage in MS.
3.3. Inflammatory Bowel Diseases
How the CD28 pathway influences IBD initiation is not entirely straightforward. In one initial study, when T cells were transferred into CD80 or CD86 single deficient mice to induce colitis, mice developed accelerated disease in either of the two single deficient recipient mice compared to WT recipient mice (237). However, CD80−/− CD86−/− recipient mice did not exhibit colitis-like symptoms, suggesting that single deficiency of either of the two CD28 ligands impacts T cell activation differently than absence of both. This observation was attributed to the fact that CTLA-4 expression on the transferred T cells was reduced in single deficient recipient mice compared to CD80/CD86-sufficient mice, suggesting that high cell-intrinsic CTLA-4 expression is necessary to inhibit rampant T cell activation (237). These results were revisited in a separate study by the same group in which colitis was induced spontaneously in a transgenic mouse that constitutively secreted soluble CD86-Ig fusion protein (238). In this system, the authors argued that increased disease in the presence of the fusion protein was due to its binding to CTLA-4 on T cells (because of its higher affinity for CTLA-4 over CD28) that inhibited the cell-intrinsic role CTLA-4 plays to curb immune activation.
Interestingly, CD28/CD80/CD86 may not even be entirely required during intestinal inflammation. One study identified that when T cells are deprived of CD80/CD86 signaling, they rely instead on ICOS and OX40 signaling to break tolerance. An earlier study had also demonstrated that ICOS blockade is effective in ameliorating colitis only in the absence of CD28 (239). Furthermore, OX40L expressing activated DCs were enriched in the draining lymph nodes during intestinal inflammation (240). Blocking OX40L-mediated signals was sufficient to abrogate colitis development in the treated mice. A later study determined that part of the effects of OX40 signaling also involve Tregs because Treg numbers were depleted due to increased apoptosis in the absence of OX40 (241). Blocking other costimulatory molecules such as CD27 and CD40L have also proven to be effective therapies in mouse models of colitis (242, 243). Mice overexpressing CD40L acquired a lethal inflammatory bowel disease marked by infiltration of CD40L+ T cells (244). This is particularly noteworthy since CD40L expression has been observed to be elevated in human IBD patients, especially within the inflamed mucosa (245, 246). Altogether, several costimulatory molecules can each initiate IBD.
Coinhibitory receptors in IBD have received renewed interest of late due to the increased use of ICB therapies in patients with tumors, which have now been successfully implemented in the clinic to treat patients suffering from various cancers such as melanoma, non-small cell lung cancer, and renal cell carcinoma, to name a few (247). In these therapies, antibodies targeting CTLA-4, PD-1 or PD-L1 (the ligand for PD-1) are used to reinvigorate dysfunctional T cells that express high levels of CTLA-4 or PD-1 due to chronic antigenic stimulation within the tumors (248). In doing so, the antigen-specific T cells are now poised to attack and kill the malignant cells. Unfortunately, because of the broad nature of these therapies, T cells are indiscriminately activated, even those that bear specificities for non-tumor self-antigens in other tissues. Consequently, many autoimmune diseases have been reported in patients receiving αCTLA-4 or αPD-1 treatments (167). One major site in which autoimmunity is initiated is the GI tract resulting in IBD. Severe T cell infiltration and substantial morbidity often results in patients choosing to discontinue therapy. αCTLA-4 therapy induces more frequent and severe inflammatory events, which is congruent with the role of CTLA-4 in maintaining mucosal tolerance in mice during colitis (249). More recently, antibodies targeting TIM-3 have also been demonstrated to reduce tumor burden when combined with αPD-1 therapy (250, 251). Intriguingly, patients with UC had reduced expression of TIM-3 on the surfaces of T cells, indicating that decreased TIM-3 levels could contribute to enhanced T cell responses and exacerbated disease (252). Thus, it remains to be seen whether treatment with αTIM-3 antibodies also results in IBD in some patients.
4. Signal 3 – Cytokine Signaling and T Helper Cell Subset Differentiation
Cognate antigen recognition coupled with costimulation is sufficient to activate T cells. However, this activated cell-state does not intrinsically predispose T cells to be functionally more (or less) adept at combating intracellular pathogens such as viruses or extracellular pathogens such as parasites. Clearing the host of these pathogens requires further specialization on the part of the T cells to ensure that the downstream responses are ideally suited to target the pathogen. The specialized activated T cells can then mediate their effector functions by recruiting specific immune cell populations to tissues and promoting inflammatory/regulatory environments by secreting distinct soluble molecules (253). This specialization is often established during the initial T cell priming stage through cytokines secreted by APCs, which act on the responding T cells (Signal 3) and differentiate them into functionally distinct subsets (Figure 4).
Figure 4.
Therapies targeting different Th subsets implemented in humans. Top, In addition to Signal 1 and Signal 2, different cytokine cocktails are required to effectively differentiate naïve CD4+ T cells into the Th1, npTh17, pTh17, Tfh, pTreg and Tr1 subsets. Subsequently, these functional subsets display distinct cytokine secretion profiles that can differentially impact autoimmune disease progression. Bottom, The current therapies employed to inhibit effector T cell differentiation and/or function as well as expand regulatory T cell differentiation and/or function are listed for each subset. This image was created with BioRender.com.
Some initial work has indicated that strength of TCR signaling can affect effector T cell polarization into functional subsets, suggesting that distinct antigen specificities can instruct T cell fate (254–257). However, in most cases, the strength of TCR/costimulatory signaling does not fundamentally dictate T cell fate. Instead, the cytokines present during activation play critical roles in shaping the functionality of the T cells (253). Due to this, a substantial body of reductionist in vitro work exists in which naïve CD4+ T cells are polarized using non-specific TCR stimulation in the presence or absence of various other cytokines. These studies have imparted tremendous knowledge to the community and identified both the core cytokines required to polarize T cells and the cytokines secreted by the polarized cells.
The canonical T helper subsets are the type 1 (Th1) and type 2 (Th2) cells (258). Th1 cells are important for host defense against intracellular pathogens such as viruses and secrete large amounts of the antiviral cytokine interferon gamma (IFNγ), which promotes enhanced antigen presentation on APCs and activates many innate (NK cells) and adaptive (CD8+ T cells) lymphocyte populations (259). Th2 cells play an important role in clearing extracellular pathogens such as parasites and helminths by producing the prototypical Th2 cytokine interleukin 4 (IL-4) that helps to recruit innate immune cells such as eosinophils and mast cells to the site of infection (259). Initial activation sensitizes T cells to IL-12 and IL-4 produced by APCs to differentiate cells into the Th1 and Th2 subsets, respectively (260–262). Importantly, these signals elicit expression of the master transcription factors associated with these subsets – T-bet and GATA-3, respectively – that promote transcriptional and chromatin remodeling to establish stable fates upon differentiation (263, 264).
Three other Th subsets were more recently identified. Th17 cells, named because of their ability to secrete large amounts of the potent cytokine IL-17A, were identified for their role in autoimmunity (265–267). Different cytokine cocktails have been identified to promote Th17 differentiation both in vitro and in vivo. While combined exposure to TGFβ1 and IL-6 was shown to effectively polarize Th17 cells initially, it was later discovered that these Th17 cells possessed a regulatory profile imparted by signals received from the immunosuppressive cytokine TGFβ1 (268–270). Concurrently, a more inflammatory Th17 subset was discovered to be polarized by IL-23 (271). Importantly, both the regulatory and inflammatory Th17 subsets express high levels of the master transcription factor RORγt, which enhances and stabilizes their phenotype (272). A second T cell subset was identified due to its importance in curtailing autoimmunity in both humans and mice. Regulatory T cells (Tregs) suppress effector immune responses through myriad mechanisms and express the transcription factor FoxP3 (273). Interestingly, Tregs can also develop in multiple ways in vivo. In a process termed agonist selection, some strongly self-reactive T cells are diverted into the Treg lineage in the thymus instead of undergoing negative selection (274). Thus, unlike the overwhelming majority of T cells that exit the thymus in a naïve unpolarized state, thymic-derived natural Tregs (nTregs) commit to their lineage as a consequence of their thymic development. Tregs can also arise in the periphery (pTregs) from naïve CD4+ T cells that are activated in the presence of TGFβ (275). Lastly, type 1 regulatory (Tr1) cells are another CD4+ T cell subset characterized by strong production of the immunosuppressive cytokines IL-10 and TGFβ1 (276). Strikingly, although they share functional features with Tregs with respect to their ability to suppress effector T cell responses, these cells do not express the Treg transcription factor FoxP3. Tr1 cells can be differentiated in the presence of IL-10, IL-27 and/or TGFβ1 (277–279). Overall, the proportion and composition of the various Th subsets can drastically impact how tissues are surveyed by T cells and whether they break tolerance.
4.1. Type I Diabetes
Historically, the major T cell subset thought to be driving T1D was the Th1 population. An initial study explored the roles of differentiated T cell subsets in initiating disease course in NOD mice (280). The investigators polarized naïve CD4+ T cells into either Th1 or Th2 subsets in vitro by exposing the cells to different cytokine environments. Subsequently, these polarized cells were independently transferred into neonatal NOD mice to determine whether polarized cells of a given lineage were sufficient to cause disease. The in vitro-derived Th1 cells caused hyperglycemia in 50% of the recipient mice within 2 weeks after transfer and caused diabetes in 90% of the mice by 5 weeks (280). By contrast, the mice receiving Th2 cells largely were resistant to developing disease, with only 1 out 21 mice becoming hyperglycemic. This dramatic difference in ability to cause disease could not be attributed to differential tissue-trafficking abilities between the two Th subsets but rather a differential ability to create a pro-inflammatory environment upon arriving at the islets in the pancreas.
Further supporting a key role of Th1 cells in disease progression are the studies that have identified IFNγ, the signature Th1-produced cytokine, as a major contributing factor in T1D. Blocking IFNγ was sufficient to prevent T1D in NOD mice (281, 282) and overexpression of the gene suppressor of cytokine signaling 1 (SOCS1), which negatively regulates IFNγ signaling, resulted in protection from disease (283). T1D patients also possessed autoantigen-specific IFNγ-producing cells, highlighting the importance of Th1 cells in humans (284). Patients with gain of function mutations in STAT1, the transcription factor downstream of IFNγ receptor, also presented with concomitant T1D (285). Unsurprisingly, NOD mice lacking STAT1 were resistant to T1D (286). The same study also confirmed these results using an inhibitor for Jak2, the kinase upstream of STAT1, which delayed T1D progression in mice.
More recently, Th17 cells have also been proposed as a possible disease-causing T cell subset in T1D but conflicting results have made a definitive verdict about the role of this population challenging. NOD mice colonized with the Th17-inducing segmented filamentous bacteria (SFB) displayed delayed diabetes kinetics (287). However, the Th17 cells induced by SFB are known to possess a more regulatory phenotype and serve to play a protective role in mucosal tissues (288, 289). Instead, when antigen-specific Th17 cells were polarized in the presence of the pro-inflammatory cytokine IL-23, the mice developed rapid insulitis and subsequent diabetes (290, 291). Interestingly, disease progression was dependent on the transferred Th17 cells converting to Th1 cells and producing IFNγ. This observation is consistent with work indicating that Th17 cells exposed to IL-23 are a potent inflammatory population that adopts a pathogenic phenotype characterized by coinciding Th1 and Th17 functionalities (292–294).
APCs isolated from humans with T1D expressed elevated levels of IL-1β and IL-6, two cytokines that are important in Th17 differentiation, suggesting that Th17 cells could participate in human disease (295). Furthermore, gain of function mutations in STAT3, which is downstream of IL-6, have also been linked to early-onset T1D in humans (296–298). These patients present with higher numbers of Th17 cells, likely due to the hyperactive STAT3. Mapping some STAT3 missense mutations to functions has helped determine that one possible mechanism by which STAT3 is hyperactive is through enhanced DNA binding affinity, which would result in increased chromatin accessibility in STAT3-dependent gene expression and a more stable Th17 phenotype (299). In a separate study, STAT3 gain of function was observed to result in increased expression of SOCS3, which negatively regulates IL-2 signaling and Treg numbers and stability (297).
Another cytokine with a curious role in T1D is IL-21, which belongs to the common γ-chain receptor family of cytokines that includes IL-2, IL-4, and IL-7. It is one of the candidate genes located in a disease-susceptibility locus and is elevated during disease in mice (300–304). The importance of IL-21 signaling was highlighted in two studies in which mice lacking the IL-21 receptor (IL-21R) failed to develop T1D (304, 305). While IL-21 can stabilize and enhance Th17 differentiation (306, 307), IL-21 signaling deficient mice did not exhibit any differences in Th17 numbers or functions (304). Yet, a separate study identified that IL-21 signaling in CD4+ T cells was necessary in T1D progression, suggesting that a different IL-21-sensitive Th subset is likely responsible for causing disease (308).
Indeed, one distinct T cell population that appears to be critical in T1D is the T follicular helper (Tfh) subset. These T cells are important in germinal center formation and function, where they provide crucial cues for proper B cell maturation during an immune response (309). They express high levels of the transcription factor Bcl6 and secrete IL-21, which also signals through STAT3. Remarkably, IL-21 itself can lead to increased expression of Bcl6 (310). In germinal centers, antigen-specific B cells mutate their antigen receptors and undergo a selection process, which relies on Tfh cells, to retain only those B cells with high affinities for the target antigens. Thus, the observation that Tfh cells are enriched in pancreatic lymph nodes in mice implicates their role in the generation of islet-specific autoantibodies, which are a hallmark of T1D (311). Mice in which Tfh generation is augmented exhibit accelerated T1D progression, further linking this T helper subset to disease (312).
Dysfunctional Tregs are another feature in individuals with T1D (313). As previously described, Tregs can arise during thymic selection (nTregs) or be generated in peripheral tissues when exposed to TGFβ1 during antigenic stimulation (pTregs) (274, 275). Thymic Tregs are reliant on IL-2 stimulation, which serves as a surrogate signal 3 even in sterile conditions, for their complete maturation. Strong TCR signaling in the thymus generates CD25hi FoxP3− Treg progenitors that require IL-2 signaling to mature into nTregs (314). Importantly, several studies have been able to link SNPs in humans impacting IL-2 signaling with Treg dysfunction (301, 315, 316). These results were also corroborated in NOD mice, which display a defect in IL-2 signaling in Tregs (317). Current therapies in T1D involve delivering IL-2 specifically to Tregs to ensure Treg activation without activating effector T cell populations and may serve as effective therapies in the future (318).
Although both express FoxP3 and display strong immunosuppressive capacities, nTregs and pTregs are not entirely interchangeable populations, especially with respect to their stabilities. Unfortunately, it is challenging to distinguish between the two Treg populations to unambiguously determine whether one subset plays a more crucial role in T1D. One clever approach implemented by the Mathis lab took advantage of studying the TCR repertoire to discriminate between the populations (319). Since pTregs are derived from conventional naïve CD4+ T cells, their TCR repertoire would more likely overlap with that of the effector populations than the distinct repertoire employed by nTregs. Upon comparing the repertoire between Tregs and effector T cells isolated from the islets of NOD mice, the authors identified minimal overlap, suggesting that nTregs are more involved in disease progression (319). However, in one recent study, the conserved noncoding sequence 1 (CNS1) in the Foxp3 promoter was deleted in NOD mice to more clearly assess the role of pTregs (320). Downstream of TGFβ signaling, the transcription factor Smad3 binds to CNS1 and drives upregulation of FoxP3 to produce pTregs (321, 322). CNS1 deficiency specifically impacts pTreg generation while leaving nTreg generation intact, due to a lack of dependence of nTregs on TGFβ signaling (323). Deleting CNS1 in NOD mice resulted in increased incidence of T1D, indicating that fully grasping the role of pTregs in T1D may require deeper analyses (320).
Lastly, the role of Tr1 cells in T1D is understudied. Although islet antigen-specific IL-10 producing CD4+ T cells have been identified in T1D patients, these cells also secrete IFNγ, suggesting that during disease the cells become dysfunctional (284). Moreover, genetically predisposed healthy relatives of patients with T1D also possess islet-specific CD4+ T cells but these cells are more prone to producing IL-10, unlike their T1D relatives whose islet-specific cells produce more IFNγ (324). Consequently, the ratio of Tr1 cells to Th1 cells is important in determining whether disease is initiated. Interestingly, non-specifically activating T cells via CD3 engagement resulted in elevated IL-10 in T1D patients with a concomitant reduction in IFNγ and TNFα secretion, suggesting that activating Tr1 cells using this approach could arrest disease development (325). Experiments in NOD mice have supported these findings as well, with Tr1 functional activity inversely correlating with disease progression (326, 327). Curiously, autoantigenic stimulation in NOD mice also expanded a Tr1 population that was sufficient to prevent effector T cell migration to the islets, indicating that antigen-specific or non-specific stimulation in T1D preferentially activates the Tr1 subset of cells (328).
4.2. Multiple Sclerosis
Not unlike the pathogenic effectors observed in T1D, the major CD4+ T cell effectors in MS/EAE are also Th1 and Th17 differentiated cells. Initially, the major T cell population implicated in driving disease was the Th1 subset. IFNγ levels were elevated in CNS lesions in human patients and treating MS patients with IFNγ exacerbated disease progression (329, 330). In mice, deletion of the IL-12 subunit p40 was sufficient to protect mice from disease, indicating that differentiated Th1 cells were involved in disease (331, 332). Furthermore, mice lacking STAT4, a transcription factor important in Th1 differentiation, were resistant to EAE (333). However, these results were confounded by the finding that disrupting IFNγ signaling or deleting IFNγ itself accelerated EAE progression (334–336). The discovery of IL-23 and its shared use of the p40 subunit began to unravel some of this mystery. Mice lacking the IL-12-specific subunit p35 remained susceptible to EAE while mice lacking the IL-23-specific subunit p19 phenocopied the p40 deficient mice in their resistance to EAE (267, 332). These data resulted in a clear paradigm shift that Th1 cells were not solely responsible for disease since IL-23 signaling was dispensable for Th1 differentiation.
These initial findings coincided with the discovery of Th17 cells. Although Th17 cells were originally revealed to require TGFβ1 for their differentiation, subsequent experiments identified that TGFβ1 promotes differentiation of a homeostatic/non-pathogenic Th17 population while exposure to IL-23 instead results in a pro-inflammatory/pathogenic Th17 subset with a license to initiate disease (268–271). Yet, the complete IL-23 heterodimeric receptor (composed of IL-23R and IL-12RB1) is not appreciably expressed by naïve CD4+ T cells (337). Instead, the gene coding for IL-23R (Il23r) is upregulated upon activation of STAT3, which is downstream of IL-6 and IL-21, two cytokines strongly involved in Th17 differentiation and maintenance (338). Thus, activation of STAT3 serves as an important initial signaling hub that helps to promote the transcriptional landscape necessary for Th17 pathogenicity. Indeed, in support of this, STAT3 has been identified as a susceptibility risk gene in MS and a SNP in the STAT3 locus was independently validated to display a positive association with MS susceptibility (339–341). Furthermore, elevated levels of activated STAT3 were observed in MS patients during relapse, indicating that enhanced signaling through this pathway could be contributing to disease recurrence (342). The importance of STAT3 was also confirmed in mice in whom targeted deletion of STAT3 protected mice from developing EAE (343, 344). This was attributed to an inability of the STAT3-deficient T cells to differentiate effectively into Th17 cells and subsequently cause disease. Additionally, the STAT3 knockout T cells also displayed defective trafficking to the CNS. Conversely, deletion of SOCS3, which inhibits STAT3 signaling, resulted in hyperactivated macrophages and severe disease (345, 346). Therefore, STAT3 signaling impacts pathogenic T cells in multiple manners.
Interestingly, IL-21 signaling was observed to be unnecessary in EAE induced via peptide immunization, implying that Il23r expression could be regulated in a STAT3-independent manner as well (347, 348). Instead, one manner in which Il23r can be upregulated is through the coordinated efforts of the transcription factor RORγt, which is induced in differentiated Th17 cells, and RBPJ, a regulator downstream of Notch signaling, which together further entrench these cells into the Th17 lineage (349). Mice with a T cell-specific deletion of RBPJ were protected from EAE induction, largely due to an unstable Th17 effector population that had dramatically reduced expression of Il23r. Our lab also identified TGFβ3 as a potent cytokine that drives high levels of IL-23R in differentiating Th17 cells in an autocrine manner and endows them with strong pathogenic potential in EAE (350). Collectively, these results argue for critical roles of both Th17 cells and IL-23 in disease progression. However, conflicting data in the functional relevance of Th17 cells in EAE have tempered the notions that inhibiting Th17 activity could impede disease.
In early experiments examining the effects of IL-17A (the signature cytokine produced by Th17 cells) during EAE, IL-17A was overexpressed in T cells to trigger T cells to produce IL-17A at steady-state (351). Upon immunization with MOG peptide, no difference in EAE induction was observed when compared to control mice, suggesting that IL-17A secretion is not critical for disease in rodents. This was further confirmed using IL-17A deficient mice that also did not display altered disease course or severity (351). The authors also deleted IL-17F, a cytokine related to and sharing expression patterns with IL-17A but observed no differences in disease progression. This contrasted with observations in human MS patients who exhibited reduced lesion formation upon administration of an anti-IL-17A compound (352). Nevertheless, a recent study identified that while production of IL-17A/F was critical for EAE, the role of T cell-derived IL-17A/F is less certain (353). Thus, other cell populations producing these cytokines can determine disease course.
Since the true impact of IL-17 in EAE/MS remains unresolved, several groups have tried instead to determine if other soluble proteins produced by encephalitogenic Th17 cells were more critical in disease. In particular, the goal was to elucidate the specific role of IL-23 in licensing Th17 cells to infiltrate the CNS and facilitate destruction. Through the use of a novel reporter mouse, the Becher group identified that a key cytokine produced by IL-23-treated cells was granulocyte-macrophage colony stimulating factor (GM-CSF) (354). This cytokine is sensed by tissue-damaging myeloid cells, resulting in their activation and migration to the CNS, where they continue to preserve an inflammatory environment. IL-23 signaling was shown to be critical in driving expression of GM-CSF in antigen-specific T cells both in vitro and in vivo and GM-CSF-deficient T cells were incapable of causing severe disease.
More recently, our lab identified two distinct Th17 populations in mice during EAE – a stem-like, homeostatic population that expresses SLAMF6 and a pathogenic population that expresses the chemokine receptor CXCR6 (149). Interestingly, the homeostatic population served as a reservoir from which pathogenic effector Th17 cells could be generated. This conversion was catalyzed by exposure of the stem-like population to IL-23, which resulted in an augmented IL-23-driven gene signature, including the expression of CXCR6. Subsequently, Th17 cells expressing CXCR6 produced IFNγ and GM-CSF and displayed increased migratory capacity to the CNS during EAE due to high levels of the cognate chemokine ligand CXCL16 (149). Therefore, IL-23 induces conversion of Th17 cells into pathogenic effectors that are then endowed with the ability to produce potent pro-inflammatory cytokines and migrate to the tissue sites of inflammation. Overall, these two studies highlight that the necessity for IL-23 in driving pathogenic responses by Th17 cells.
Tregs also play critical roles in modulating EAE/MS due to their immunosuppressive capabilities. Indeed, co-transfer of Tregs along with pathogenic effector T cells alleviates EAE progression (355–358). But the inflammatory environment created during EAE can also alter Treg functionality and reduce their ability to suppress immune responses. Such dysfunctionality of Tregs was also observed in MS patients (359). Additionally, a SNP in the IL2RA locus, which codes for the high affinity IL-2 receptor CD25, resulted in impaired IL-2-mediated signal transduction in T cells and was associated with greater MS susceptibility (360). An independent study also determined that IL-2 signaling in CD4+ T cells isolated from MS patients bearing this SNP resulted in increased GM-CSF production, linking this SNP to a disease-relevant functional outcome (361). Since IL-2 is key for proper Treg function, compromised IL-2 signaling could be detrimental for their suppression. A recent study also identified a novel mechanism that impacted pTreg differentiation in MS patients (362). Humans with MS were enriched for exosomes (extracellular vesicles secreted by many different cell populations) containing RNA-silencing miRNAs that specifically target TGFβR1, the receptor required for TGFβ1 signal transduction during pTreg differentiation. A separate study also identified miRNAs that target different mRNA molecules coding for various proteins along the TGFβ1 signaling pathway (363). These miRNAs were preferentially enriched in the CD4+ T cells of MS patients. Altogether, both nTreg and pTreg differentiation defects in EAE/MS can accelerate a break in tolerance.
Although Tr1 cells have not been investigated to the same extent in MS/EAE as Tregs, they do appear to be dysfunctional in disease. CD4+ T cells from MS patients stimulated to produce IL-10 secreted less of this cytokine compared to stimulated cells from healthy individuals (364). This perhaps highlights a cell-intrinsic/genetic component that drives Tr1 dysfunction in patients. Promoting Tr1 differentiation and subsequent IL-10 production in mice also had a favorable outcome in the progression of EAE (365–367). Indeed, sensitivity to IL-27 was shown to be important in increasing the proportion of IL-10-producing Tr1 cells during EAE (278, 279). Furthermore, IL-27 also promotes Tr1-like differentiation of human CD4+ T cells, with its production by DCs being potentiated by interferon beta (IFNβ), an FDA-approved therapy in MS (368–371).
4.3. Inflammatory Bowel Diseases
The contribution of Th1 cells to IBD has been controversial in mouse models of disease. In the CD4+ T cell transfer model of colitis, the transferred T cells were observed to produce high levels of IFNγ (161, 372). By administering anti-IFNγ antibodies into recipient mice, wasting disease and colitis was prevented, indicating a key role for Th1 cells in driving disease. IFNγ deficient mice were also protected from colitis in a separate mouse model of disease, there providing indirect evidence for a pathogenic role for Th1 cells (373). However, transferring IFNγ-deficient CD4+ T cells into recipient mice was insufficient to halt colitis (374). Instead, mice receiving STAT4-deficient cells or treated with anti-IL-12 antibodies did not develop colitis, implying that IFNγ secretion by Th1 cells plays either a redundant or non-essential role in the T cell transfer model of IBD (374). Similar contradictory results were also observed when the signaling cascade of another Th1-secreted cytokine TNFα was perturbed in mice with IBD (375–378). These results are inconsistent with GWAS analyses in humans which revealed that SNPs near the IFNG, IFNGR2 and the TNF loci were strongly associated with IBD (379–385). Another SNP in IBD patients was associated with increased IFNγ and more aggressive disease (386), suggesting that the underlying mechanisms driving disease progression in mice and humans may not completely overlap.
STAT4 plays a curious role in colitis in both humans and mice. While T cell-specific complete STAT4 deficiency results in disease resistance in the CD4+ T cell transfer model (374), different STAT4 isoforms display distinct capacities to induce disease. STAT4α codes for the full-length protein while STAT4β lacks the C-terminal transactivation domain, signifying perhaps that the latter isoform is a hypomorphic variant (387). Instead, STAT4β expressing CD4+ T cells produced more TNFα than STAT4α expressing cells during colitis, resulting in more severe disease (388). Elevated STAT4β/STAT4α ratios were also observed in IBD patients (389). Yet, the precise mechanisms for how STAT4β produces more pro-inflammatory Th1 cells are currently unclear.
Th17 cells represent an interesting population during intestinal inflammation. This T cell subset is abundant in the lamina propria of the intestine during homeostatic conditions to maintain barrier integrity at a critical mucosal surface with numerous microbes (166). These Th17 cells secrete IL-17A, IL-17F and IL-22 at steady-state to stimulate production of antimicrobial proteins by intestinal epithelial cells. Th17 differentiation in the intestines require microbiota because in germ-free mice, Th17 cells are absent in the lamina propria (390, 391). Overall, Th17 differentiation at steady-state as a consequence of gut microbes is largely protective.
However, these cells acquire a pathogenic phenotype when exposed to IL-23 whereupon they acquire a more Th1-like program and express higher levels of IFNγ (293, 392). Indeed, the IL-23/IL-23R signaling axis is a critical component in IBD initiation in humans as well, with several disease associated SNPs being identified in the IL23R locus (393, 394). Transferred T cells deficient in IL-23R displayed reduced intestinal inflammation and infiltration, highlighting the role of appropriate IL-23 signaling in disease (395). Importantly, IL-23R signaling impaired Treg differentiation without impacting Th1 differentiation (162). Antibody neutralization of IL-23 is also sufficient to ameliorate disease in the T cell transfer model of disease (396).
As mentioned previously, STAT3 is the transcription factor that is activated downstream of both IL-6 and IL-21 (397, 398). Interestingly, while IL-23R shares a subunit with IL-12 (IL-12RB1) and therefore also signals through STAT4, the IL-23R unique subunit can signal through STAT3 as well (399). Unsurprisingly, a STAT3 SNP has been associated with increased disease severity in human IBD patients (400, 401). In mice, CD4+ T cells lacking STAT3 fail to induce colitis upon transfer into recipient mice (402). In particular, these CD4+ T cells inadequately differentiated into Th17 cells while Th1 differentiation was unaffected. Furthermore, through the use of epigenetic analyses, the authors identified that STAT3 binds to regions that stabilize the Th17 lineage and also those that promote survival and, in its absence, the T cells displayed reduced survival in lymphopenic settings (402).
The intestinal CD4+ T cell populations also contain Tregs at steady-state, which are a mixture of nTregs and pTregs. pTregs can be readily identified in colonic tissues by their co-expression of both FoxP3 and RORγt and are conspicuously absent in germ-free mice, suggesting that their role is to provide tolerance against microbial antigens (403–405). The RORγt-expressing Tregs are particularly potent at suppressing Th17 responses during colitis. Indeed, functional Tregs are also observed in human IBD patients but are likely outnumbered by their effector T cell counterparts and are unable to counteract disease progression. One key suppressive feature of Tregs is their IL-10 production to inhibit pathogenic T cell responses during disease (406–408). Recombinant IL-10 was sufficient to reduce disease burden in mice (161). Humans with polymorphisms in the IL10 or IL10R loci have also been reported to develop severe Crohn’s disease (409–412). Mechanistically, IL-10 production by Tregs is thought to function in a positive feedback loop whereby Treg-produced IL-10 promotes increased IL-10 production through IL-10 receptor (IL-10R) (413). Consequently, mice with targeted deletions specifically in Tregs for either IL-10 or IL-10R develop severe colitis. Although Tregs are still present in these mice, they are incapable of suppressing the inflammatory Th17 responses (413). Of note, IL-10 signal transduction also requires STAT3 (414), indicating that STAT3 SNPs impact both effector and regulatory T cell responses.
Tregs can also suppress colitogenic T cells in several other manners. TGFβ1 is a noteworthy cytokine because it both drives differentiation of pTregs and is produced by Tregs to suppress effector T cell responses (275, 415). In one study, Treg-derived TGFβ1 was required to mitigate Th1 responses in the transfer model of colitis (270). In line with this, when TGFβ1-deficient Tregs were co-transferred with naïve CD4+ T cells into lymphopenic mice, the Tregs were not capable of preventing colitis (416). These findings were supported in human Crohn’s disease patients as well who possessed colonic T cells with elevated levels of Smad7, an inhibitor of the TGFβ1 signaling pathway (417). Altogether, Tregs are a necessary T cell subset in thwarting intestinal inflammation and their dysregulation can rapidly escalate IBD.
Tr1 cells were initially identified because of their dominant role in suppressing colitis in the T cell transfer model of disease in mice (277). While this suppression required IL-10, it was not mediated by CD4+ CD25+ T cells, thereby excluding canonical Tregs. Future work also corroborated these findings with Tr1-derived IL-10 determined to be sufficient in inhibiting Th1 and Th17 responses (408). Upon blocking IL-10 signaling in the Th1/Th17 cells, co-transfer of Tr1 cells into recipient lymphopenic mice did not prevent disease, highlighting how critical a factor Tr1-secreted IL-10 is in modulating colitis. Importantly, Tr1 cells and Tregs are located in anatomically distinct regions in the GI tract. Tr1 cells are more abundant in the small intestine while Tregs are more prevalent in the colonic lamina propria (418, 419). This suggests that different regulatory T cells subsets are differentially localized, which could impact how therapies targeting these cells are discretely designed to specifically treat Crohn’s disease vs. ulcerative colitis, as these disease subtypes themselves display distinct affected patterns.
5. Perspectives
Therapies directed at modulating effector T cell functions have been moderately effective in patients with autoimmune diseases. Since T cell specificities in patients are either not known or diverse, current therapeutic approaches are often based upon inhibition of pathways that are conserved across individuals, such as costimulatory and cytokine signaling networks. The underlying assumption in these approaches is that the same pathways are operative among enough patients with a given disease that perturbing them would show significant efficacy. Furthermore, there is a growing interest in classifying autoimmune diseases by the molecular therapies that are successful instead of the tissues that are targeted. For example, blocking costimulation via CD28 with abtacept, a CTLA-4-fusion protein, is approved for use in rheumatoid arthritis, juvenile rheumatoid arthritis and psoriatic arthritis while blockade of CD40/CD40L interactions has shown promise in a study involving patients with systemic lupus erythematosus (420–424). Thus, despite their varied etiologies, targeting the same molecular pathways has demonstrated success in the clinic for different autoimmune diseases. Similarly, since the different Jak enzymes are important in initiating cytokine signaling and T cell differentiation, inhibiting their activity precludes effector T cell function and Jak inhibitors are being developed as drugs to treat autoimmune diseases and have been approved for use in rheumatoid arthritis, psoriatic arthritis and UC (425) (Figure 4). Blocking the potent inflammatory cytokine TNFα has also demonstrated efficacy in various autoimmune diseases and has been approved for use in arthritis, psoriasis, ankylosing spondylitis and IBD (426). Mechanistically, inhibiting TNFα activity during arthritis led to downregulation of other pro-inflammatory cytokines and also impacted lymphocyte/leukocyte trafficking through the expression of their chemokine receptors and integrins (427, 428). More recently, antibodies targeting Th17-associated cytokines such as IL-17A and IL-23 have also shown promise in MS and efficacy in IBD, respectively (352, 429). Indeed, in agreement with animal studies, ustekinumab, a human monoclonal antibody targeting the p40 subunit of IL-12 and IL-23, is approved for treatment of IBD (430) (Figure 4).
Despite the observed success in many of these therapies, side effects are associated with each of these approaches. For example, patients treated with Jak inhibitors are more susceptible to bacterial and viral infections due to poor T cell differentiation and responses (425). In patients with IBD, despite high levels of IL-17A in the colon, anti-IL-17A therapy has also resulted in worse disease, possibly due to the homeostatic role of IL-17A in maintaining barrier integrity (431, 432). Serious side effects in anti-TNFα therapy are limited, although patients can be more susceptible to infections by pathogens such as mycobacterium tuberculosis (433). Additionally, patients can fail to respond to this therapy or lose sensitivity over time (2). The mechanisms underlying anti-TNFα therapy resistance are only now beginning to be unraveled.
Importantly, different diseases require different therapies since the cell types and molecules involved vary across the autoimmune disease spectrum. As previously mentioned, anti-IL-17A therapy has opposing effects in different diseases in that it appears to be promising in MS patients yet is ineffective or detrimental in IBD patients (352, 431). Similarly, anti-TNFα therapy, while helpful in many IBD patients, appears to exacerbate disease activity and resulted in increased relapses in MS (434, 435). Additionally, blocking IL-23 is efficacious in IBD but displays no clinical significance in human MS patients (2, 6). Altogether, these conflicting data indicate that no clear “magic bullet” therapy exists to effectively treat all autoimmune diseases. In particular, many of the aforementioned therapies concentrate on dampening effector responses instead of engendering long-lasting tolerance to self-antigens. With such therapies, individuals are likely to relapse when therapy is halted because the autoreactive cells are not being eliminated or made unresponsive. Overall, there is a desperate need to implement safe and well-tolerated broad-spectrum therapeutics that can ameliorate multiple autoimmune diseases by restoring a state of immune tolerance to autoantigens.
One strategy that has gained traction over the years has been to stimulate generation of cells that are regulatory and will sustain their immunosuppressive function even after any extrinsic therapy is interrupted (2). Numerous studies have shown that co-transfer of suppressive immune cells along with effector cells can decelerate or entirely arrest disease progression (163, 366). Yet, the source of the Tregs in humans in adoptive cell therapy (ACT) proved to be challenging. Purifying Tregs from peripheral blood mononuclear cells (PBMCs) is complicated because the bona fide Treg marker (FoxP3) cannot be used to determine identity of the cell since staining for this protein will render the cells nonviable. Surface markers expressed by Tregs are often times not uniquely expressed by this population and are shared by activated cells, suggesting that pathogenic effector cells could possibly be included by this strategy (436). Since the conditions to polarize Tregs in vitro had been well established, many investigators instead attempted to transfer in vitro-derived Tregs into mice. However, these Tregs were determined to be less stable than nTregs due to gradual loss of FoxP3 and eventual transition into Th1 or Th17 effector subsets, thereby possibly exacerbating disease (437–439).
Different approaches have been undertaken to specifically target one of the three signals to specifically enrich and expand regulatory T cells in vivo. To successfully manipulate Signal 1, antigen-specific T cells are a crucial component. Indeed, antigen-specific Tr1 cells are capable of inhibiting colitis development in mice (277). Additionally, some signs of efficacy employing antigen-specific Tregs were observed in a small open-label Phase 1/2a single injection study in which Tregs specifically activated with peptides were administered to patients with refractory CD (440). In an effort to specifically differentiate cells into the Tr1 or Treg lineages, autoantigenic peptides have also been administered to individuals in a tolerogenic manner. This can be achieved either via the route of antigen delivery (oral or intramuscular, for example) or through the use of tolerogenic APCs that can promote Treg differentiation (441, 442). Transfer of tolerogenic DCs as a therapy for autoimmune diseases is now beginning to be explored as an option (443). One interesting approach that was successfully implemented in mice was administering nanoparticles coated with autoantigenic peptide/MHC complexes that triggered the generation of antigen-specific Tr1 and Treg cells in multiple autoimmune disease models, perhaps via an anergy-inducing mechanism (444). These cells effectively suppressed autoimmune disease development. Yet, a lack of full understanding of the nature of the self-antigens that promote a break in tolerance has hindered complete translation of these peptide-driven therapeutic approaches into humans.
Numerous strides made in synthetic biology have also been crucial in generating antigen-specific regulatory T cells. As a result, investigators have successfully introduced autoantigen-specific TCRs into Tregs that endogenously express TCRs of different specificities. Initial work using TCR transgenic mice or enriched antigen-specific Tregs demonstrated that substantially fewer antigen-specific Tregs are required to inhibit or reverse T1D compared to polyclonal Tregs (445, 446). Thus, by ectopically expressing autoreactive TCRs in nTregs or Tr1 cells, fewer cells might be needed in ACT for different autoimmune diseases due to their enhanced antigen-dependent suppressive potency. One major challenge in this approach is the proper identification of the antigenic epitopes and the full TCRαβ sequences of an autoreactive TCR (447). Current scRNA-seq techniques can help to isolate and identify antigen-specific TCR sequences and future work will begin to address whether these TCRs can be implemented therapeutically.
The importance of proper Signal 2 in boosting Treg responses is evident in the use of chimeric antigen receptor (CAR)-T cells to treat autoimmune diseases. CAR-T cells are engineered to express a fusion protein containing an antibody for its ectodomain and intracellular T cell signaling components (448). The major advantage of this approach is bypassing any MHC restriction in patient therapy and consequently allowing the engineered T cells to be offered to individuals with disparate MHC haplotypes. This technique has seen moderate success in the clinic as an anti-tumor therapy. While the antibody specificities important to successfully apply this therapy to the treatment of autoimmune diseases are still being identified (449), many lessons can be learned regarding the costimulatory pathways required for appropriate Treg differentiation and function.
The fusion protein in CAR-T cells has both TCR and costimulatory signaling intracellular domains to properly activate T cells (448). To determine which costimulatory domains promote enhanced Treg functions in CAR-Tregs, different groups have replaced the intracellular costimulatory domains with those from various IgSF and TNFRSF receptors. Indeed, through this approach, CD28-domain-possesssing CAR-Tregs were superior at preventing graft-vs-host disease (GvHD) and skin allograft rejection (450, 451). Interestingly, CAR-Tregs with 4–1BB endodomains were not effective suppressors in a skin transplant model (452). Thus, the nature of the costimulatory domain in CARs can dictate Treg function. Future work should focus on how CAR-Tregs and CAR-Tr1s can be successfully used in autoimmune diseases.
Although antigen-specific Treg/Tr1 responses might be superior in inhibiting autoimmunity, polyclonal Treg responses can also provide substantial suppression, especially in situations where the specificities are less clear or well-known. By instead relying on polyclonal Tregs, a broader state of tolerance can be achieved (453, 454). Signal 3-dependent therapies can be developed to expand regulatory T cells with diverse specificities. Indeed, a low-dose IL-2 therapy has resulted in increased circulating Tregs and reduced disease scores (455–457) (Figure 4). This is likely due to the fact that the high affinity IL-2 receptor CD25 is expressed at elevated levels on Tregs in comparison to its expression on conventional T cells, thereby preferentially expanding them. To prevent aberrant activation of activated T cells, a few different strategies have been developed. In one study, the authors generated an anti-IL-2 antibody that stabilized IL-2 in a conformation that preferentially activated and expanded Tregs in vivo (458). In a separate study, synthetic IL-2 and IL-2 receptor analogs were introduced into cells that did not display cross-reactivity with endogenous IL-2/IL-2R (318). Consequently, T cells expressing this orthogonal IL-2R could be preferentially activated while limiting bystander activation. While these proteins have now been tested in tumor and transplantation settings (318, 459), it would be interesting to confirm the results in autoimmune diseases as well. Lastly, generating pTregs by modulating the gut microbiome has also been shown to impact colitis development (460). Gut colonization by specific bacteria such as Clostridium resulted in enhanced colonic Treg numbers and resistance to colitis development. This was dependent on increased TGFβ produced by intestinal epithelial cells that promoted differentiation of T cells into Tregs. Since the composition of the gut microbiome can impact numerous autoimmune diseases, enriching for Treg-promoting gut-resident bacteria through drugs or diet could provide wide reaching and lasting protection.
6. Conclusions
T cells are by nature a versatile population. They have the potential to express an unimaginably large antigen receptor repertoire, which helps them respond to known and unknown antigenic threats. Additionally, due to their plasticity, T cells are rapidly able to differentiate into various effector subsets to effectively mount immune responses in distinct tissue microenvironments. Multiple checks and balances are incorporated into T cell activation as well to ensure that the differentiation cues they receive are appropriately amplified or inhibited. Because of all of these attractive features, harnessing the power of T cells to treat immune disorders has been effective in some autoimmune and cancerous diseases. However, many of these therapies broadly target only one of the three main signaling pathways required for complete T cell activation and/or differentiation, which inevitably results in unintended side effects. In the future, more effective and safer therapies could be developed by targeting pathogenic T cells to revert them to a non-pathogenic state and concurrently promote activity/function of antigen-specific regulatory T cells. Precise targeting of selected T cells at nodes within the activation and differentiation pathways elicited by Signals 1, 2 and 3 may allow for synergistic therapeutic opportunities for patients with autoimmune diseases.
7. Acknowledgements
We would like to thank Mary Collins and Yoon-Chul Kye for critical reading of and feedback for this manuscript. We apologize to the authors whose publications could not be cited in the scope of this review. This work was supported by National Institutes of Health grants 5T32AR007530–37 (to S.H.K.) and R01NS045937, R01NS30843, R01AI144166, P01AI073748, P01AI039671, and P01AI056299 (to V.K.K.).
Footnotes
Conflicts of Interest
All authors declare they have no conflicts of interest to disclose.
9. Bibliography
- 1.Bluestone JA, Bour-Jordan H, Cheng M, Anderson M. T cells in the control of organ-specific autoimmunity. J. Clin. Invest. 2015; 125: 2250–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fugger L, Jensen LT, Rossjohn J. Challenges, progress, and prospects of developing therapies to treat autoimmune diseases. Cell 2020; 181: 63–80. [DOI] [PubMed] [Google Scholar]
- 3.Sckisel GD, Bouchlaka MN, Monjazeb AM, Crittenden M, Curti BD, Wilkins DEC, Alderson KA, et al. Out-of-Sequence Signal 3 Paralyzes Primary CD4(+) T-Cell-Dependent Immunity. Immunity 2015; 43: 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenkins MK, Schwartz RH. Antigen presentation by chemically modified splenocytes induces antigen-specific T cell unresponsiveness in vitro and in vivo. J. Exp. Med. 1987; 165: 302–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park AY, Hondowicz BD, Scott P. IL-12 is required to maintain a Th1 response during Leishmania major infection. J. Immunol. 2000; 165: 896–902. [DOI] [PubMed] [Google Scholar]
- 6.Baecher-Allan C, Kaskow BJ, Weiner HL. Multiple sclerosis: mechanisms and immunotherapy. Neuron 2018; 97: 742–768. [DOI] [PubMed] [Google Scholar]
- 7.Walker LSK, Herrath M von. CD4 T cell differentiation in type 1 diabetes. Clin. Exp. Immunol. 2016; 183: 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neefjes J, Jongsma MLM, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011; 11: 823–836. [DOI] [PubMed] [Google Scholar]
- 9.Wieczorek M, Abualrous ET, Sticht J, Álvaro-Benito M, Stolzenberg S, Noé F, Freund C. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 2017; 8: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 2008; 8: 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumura M, Fremont DH, Peterson PA, Wilson IA. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science 1992; 257: 927–934. [DOI] [PubMed] [Google Scholar]
- 12.Bouvier M, Wiley DC. Importance of peptide amino and carboxyl termini to the stability of MHC class I molecules. Science 1994; 265: 398–402. [DOI] [PubMed] [Google Scholar]
- 13.Chicz RM, Urban RG, Lane WS, Gorga JC, Stern LJ, Vignali DA, Strominger JL. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature 1992; 358: 764–768. [DOI] [PubMed] [Google Scholar]
- 14.McFarland BJ, Beeson C. Binding interactions between peptides and proteins of the class II major histocompatibility complex. Med Res Rev 2002; 22: 168–203. [DOI] [PubMed] [Google Scholar]
- 15.Marrack P, Kappler JW. Do MHCII-presented neoantigens drive type 1 diabetes and other autoimmune diseases? Cold Spring Harb. Perspect. Med. 2012; 2: a007765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levisetti MG, Suri A, Petzold SJ, Unanue ER. The insulin-specific T cells of nonobese diabetic mice recognize a weak MHC-binding segment in more than one form. J. Immunol. 2007; 178: 6051–6057. [DOI] [PubMed] [Google Scholar]
- 17.Borghans JAM, Keşmir C, & De Boer RJ (2007). MHC diversity in Individuals and Populations. In Silico Immunology, 177–195. [Google Scholar]
- 18.Roep BO, Kracht MJ, Lummel M van, Zaldumbide, A. A roadmap of the generation of neoantigens as targets of the immune system in type 1 diabetes. Curr. Opin. Immunol. 2016; 43: 67–73. [DOI] [PubMed] [Google Scholar]
- 19.Rojas M, Restrepo-Jiménez P, Monsalve DM, Pacheco Y, Acosta-Ampudia Y, Ramírez-Santana C, Leung PSC, et al. Molecular mimicry and autoimmunity. J. Autoimmun. 2018; 95: 100–123. [DOI] [PubMed] [Google Scholar]
- 20.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature 1988; 334: 395–402. [DOI] [PubMed] [Google Scholar]
- 21.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu. Rev. Immunol. 2008; 26: 171–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006; 24: 419–466. [DOI] [PubMed] [Google Scholar]
- 23.Gaud G, Lesourne R, Love PE. Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 2018; 18: 485–497. [DOI] [PubMed] [Google Scholar]
- 24.Gett AV, Sallusto F, Lanzavecchia A, Geginat J. T cell fitness determined by signal strength. Nat. Immunol. 2003; 4: 355–360. [DOI] [PubMed] [Google Scholar]
- 25.Campillo-Davo D, Flumens D, Lion E. The Quest for the Best: How TCR Affinity, Avidity, and Functional Avidity Affect TCR-Engineered T-Cell Antitumor Responses. Cells 2020; 9: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zehn D, Bevan MJ. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity 2006; 25: 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korem Kohanim Y, Tendler A, Mayo A, Friedman N, Alon U. Endocrine Autoimmune Disease as a Fragility of Immune Surveillance against Hypersecreting Mutants. Immunity 2020; 52: 872–884.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma S, Pettus J, Gottschalk M, Abe B, Gottlieb P, Teyton L. Single-Cell Analysis of CD4 T Cells in Type 1 Diabetes: From Mouse to Man, How to Perform Mechanistic Studies. Diabetes 2019; 68: 1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 1987; 329: 599–604. [DOI] [PubMed] [Google Scholar]
- 30.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009; 41: 703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu X, Deutsch AJ, Lenz TL, Onengut-Gumuscu S, Han B, Chen W-M, Howson JMM, et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 2015; 47: 898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soeldner JS, Tuttleman M, Srikanta S, Ganda OP, Eisenbarth GS. Insulin-dependent diabetes mellitus and autoimmunity: islet-cell autoantibodies, insulin autoantibodies, and beta-cell failure. N. Engl. J. Med. 1985; 313: 893–894. [DOI] [PubMed] [Google Scholar]
- 33.Thayer TC, Delano M, Liu C, Chen J, Padgett LE, Tse HM, Annamali M, et al. Superoxide production by macrophages and T cells is critical for the induction of autoreactivity and type 1 diabetes. Diabetes 2011; 60: 2144–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Padgett LE, Anderson B, Liu C, Ganini D, Mason RP, Piganelli JD, Mathews CE, et al. Loss of NOX-Derived Superoxide Exacerbates Diabetogenic CD4 T-Cell Effector Responses in Type 1 Diabetes. Diabetes 2015; 64: 4171–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J, Xiao Y, Xu A, Zhou Z. Neutrophils in type 1 diabetes. J. Diabetes Investig. 2016; 7: 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fowler MJ. Diabetes: magnitude and mechanisms. Clinical Diabetes 2010; 28: 42–46. [Google Scholar]
- 37.Pociot F, Lernmark Å Genetic risk factors for type 1 diabetes. Lancet 2016; 387: 2331–2339. [DOI] [PubMed] [Google Scholar]
- 38.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat. Immunol. 2001; 2: 501–507. [DOI] [PubMed] [Google Scholar]
- 39.Corper AL, Stratmann T, Apostolopoulos V, Scott CA, Garcia KC, Kang AS, Wilson IA, et al. A structural framework for deciphering the link between I-Ag7 and autoimmune diabetes. Science 2000; 288: 505–511. [DOI] [PubMed] [Google Scholar]
- 40.Latek RR, Suri A, Petzold SJ, Nelson CA, Kanagawa O, Unanue ER, Fremont DH. Structural basis of peptide binding and presentation by the type I diabetes-associated MHC class II molecule of NOD mice. Immunity 2000; 12: 699–710. [DOI] [PubMed] [Google Scholar]
- 41.Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 1993; 364: 33–39. [DOI] [PubMed] [Google Scholar]
- 42.Scott CA, Peterson PA, Teyton L, Wilson IA. Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues. Immunity 1998; 8: 319–329. [DOI] [PubMed] [Google Scholar]
- 43.Suri A, Walters JJ, Gross ML, Unanue ER. Natural peptides selected by diabetogenic DQ8 and murine I-A(g7) molecules show common sequence specificity. J. Clin. Invest. 2005; 115: 2268–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan X, Unanue ER. Antigen recognition in autoimmune diabetes: a novel pathway underlying disease initiation. Precis. Clin. Med. 2018; 1: 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hovhannisyan Z, Weiss A, Martin A, Wiesner M, Tollefsen S, Yoshida K, Ciszewski C, et al. The role of HLA-DQ8 beta57 polymorphism in the anti-gluten T-cell response in coeliac disease. Nature 2008; 456: 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng L, Mariuzza RA. Recognition of self-peptide-MHC complexes by autoimmune T-cell receptors. Trends Biochem. Sci. 2007; 32: 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sette A, Southwood S, Miller J, Appella E. Binding of major histocompatibility complex class II to the invariant chain-derived peptide, CLIP, is regulated by allelic polymorphism in class II. J. Exp. Med. 1995; 181: 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hausmann DH, Yu B, Hausmann S, Wucherpfennig KW. pH-dependent peptide binding properties of the type I diabetes-associated I-Ag7 molecule: rapid release of CLIP at an endosomal pH. J. Exp. Med. 1999; 189: 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiesner M, Stepniak D, Ru AH de, Moustakis AK, Drijfhout JW, Papadopoulos GK, Veelen PA van, et al. Dominance of an alternative CLIP sequence in the celiac disease associated HLA-DQ2 molecule. Immunogenetics 2008; 60: 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fallang L-E, Roh S, Holm A, Bergseng E, Yoon T, Fleckenstein B, Bandyopadhyay A, et al. Complexes of two cohorts of CLIP peptides and HLA-DQ2 of the autoimmune DR3-DQ2 haplotype are poor substrates for HLA-DM. J. Immunol. 2008; 181: 5451–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito Y, Ashenberg O, Pyrdol J, Luoma AM, Rozenblatt-Rosen O, Hofree M, Christian E, et al. Rapid CLIP dissociation from MHC II promotes an unusual antigen presentation pathway in autoimmunity. J. Exp. Med. 2018; 215: 2617–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nejentsev S, Howson JMM, Walker NM, Szeszko J, Field SF, Stevens HE, Reynolds P, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 2007; 450: 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coppieters KT, Dotta F, Amirian N, Campbell PD, Kay TWH, Atkinson MA, Roep BO, et al. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J. Exp. Med. 2012; 209: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babon JAB, DeNicola ME, Blodgett DM, Crèvecoeur I, Buttrick TS, Maehr R, Bottino R, et al. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat. Med. 2016; 22: 1482–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serreze DV, Leiter EH, Christianson GJ, Greiner D, Roopenian DC. Major histocompatibility complex class I-deficient NOD-B2mnull mice are diabetes and insulitis resistant. Diabetes 1994; 43: 505–509. [DOI] [PubMed] [Google Scholar]
- 56.DiLorenzo TP, Graser RT, Ono T, Christianson GJ, Chapman HD, Roopenian DC, Nathenson SG, et al. Major histocompatibility complex class I-restricted T cells are required for all but the end stages of diabetes development in nonobese diabetic mice and use a prevalent T cell receptor alpha chain gene rearrangement. Proc. Natl. Acad. Sci. USA 1998; 95: 12538–12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 2005; 435: 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serreze DV, Chapman HD, Varnum DS, Hanson MS, Reifsnyder PC, Richard SD, Fleming SA, et al. B lymphocytes are essential for the initiation of T cell-mediated autoimmune diabetes: analysis of a new “speed congenic” stock of NOD.Ig mu null mice. J. Exp. Med. 1996; 184: 2049–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noorchashm H, Noorchashm N, Kern J, Rostami SY, Barker CF, Naji A. B-cells are required for the initiation of insulitis and sialitis in nonobese diabetic mice. Diabetes 1997; 46: 941–946. [DOI] [PubMed] [Google Scholar]
- 60.Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, Becker DJ, Gitelman SE, Goland R, Gottlieb PA, et al. Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N. Engl. J. Med. 2009; 361: 2143–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science 2003; 301: 1374–1377. [DOI] [PubMed] [Google Scholar]
- 62.Mariño E, Silveira PA, Stolp J, Grey ST. B cell-directed therapies in type 1 diabetes. Trends Immunol. 2011; 32: 287–294. [DOI] [PubMed] [Google Scholar]
- 63.Mariño E, Villanueva J, Walters S, Liuwantara D, Mackay F, Grey ST. CD4(+)CD25(+) T-cells control autoimmunity in the absence of B-cells. Diabetes 2009; 58: 1568–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stadinski BD, Delong T, Reisdorph N, Reisdorph R, Powell RL, Armstrong M, Piganelli JD, et al. Chromogranin A is an autoantigen in type 1 diabetes. Nat. Immunol. 2010; 11: 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harfouch-Hammoud E, Walk T, Otto H, Jung G, Bach JF, Endert PM van, Caillat-Zucman S. Identification of peptides from autoantigens GAD65 and IA-2 that bind to HLA class II molecules predisposing to or protecting from type 1 diabetes. Diabetes 1999; 48: 1937–1947. [DOI] [PubMed] [Google Scholar]
- 66.Buitinga M, Callebaut A, Marques Câmara Sodré F, Crèvecoeur I, Blahnik-Fagan G, Yang M-L, Bugliani M, et al. Inflammation-Induced Citrullinated Glucose-Regulated Protein 78 Elicits Immune Responses in Human Type 1 Diabetes. Diabetes 2018; 67: 2337–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delong T, Wiles TA, Baker RL, Bradley B, Barbour G, Reisdorph R, Armstrong M, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 2016; 351: 711–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baker RL, Delong T, Barbour G, Bradley B, Nakayama M, Haskins K. Cutting edge: CD4 T cells reactive to an islet amyloid polypeptide peptide accumulate in the pancreas and contribute to disease pathogenesis in nonobese diabetic mice. J. Immunol. 2013; 191: 3990–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mukherjee R, Wagar D, Stephens TA, Lee-Chan E, Singh B. Identification of CD4+ T cell-specific epitopes of islet-specific glucose-6-phosphatase catalytic subunit-related protein: a novel beta cell autoantigen in type 1 diabetes. J. Immunol. 2005; 174: 5306–5315. [DOI] [PubMed] [Google Scholar]
- 70.Nakayama M. Insulin as a key autoantigen in the development of type 1 diabetes. Diabetes Metab Res Rev 2011; 27: 773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chujo D, Foucat E, Nguyen T-S, Chaussabel D, Banchereau J, Ueno H. ZnT8-Specific CD4+ T cells display distinct cytokine expression profiles between type 1 diabetes patients and healthy adults. PLoS One 2013; 8: e55595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohan JF, Levisetti MG, Calderon B, Herzog JW, Petzold SJ, Unanue ER. Unique autoreactive T cells recognize insulin peptides generated within the islets of Langerhans in autoimmune diabetes. Nat. Immunol. 2010; 11: 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Levisetti MG, Lewis DM, Suri A, Unanue ER. Weak proinsulin peptide-major histocompatibility complexes are targeted in autoimmune diabetes in mice. Diabetes 2008; 57: 1852–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stadinski BD, Zhang L, Crawford F, Marrack P, Eisenbarth GS, Kappler JW. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc. Natl. Acad. Sci. USA 2010; 107: 10978–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crawford F, Stadinski B, Jin N, Michels A, Nakayama M, Pratt P, Marrack P, et al. Specificity and detection of insulin-reactive CD4+ T cells in type 1 diabetes in the nonobese diabetic (NOD) mouse. Proc. Natl. Acad. Sci. USA 2011; 108: 16729–16734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reed B, Crawford F, Hill RC, Jin N, White J, Krovi SH, Marrack P, et al. Lysosomal cathepsin creates chimeric epitopes for diabetogenic CD4 T cells via transpeptidation. J. Exp. Med. 2021; 218: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reed BK, Kappler JW. Hidden in Plain View: Discovery of Chimeric Diabetogenic CD4 T Cell Neo-Epitopes. Front. Immunol. 2021; 12: 669986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wiles TA, Delong T, Baker RL, Bradley B, Barbour G, Powell RL, Reisdorph N, et al. An insulin-IAPP hybrid peptide is an endogenous antigen for CD4 T cells in the non-obese diabetic mouse. J. Autoimmun. 2017; 78: 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baker RL, Rihanek M, Hohenstein AC, Nakayama M, Michels A, Gottlieb PA, Haskins K, et al. Hybrid insulin peptides are autoantigens in type 1 diabetes. Diabetes 2019; 68: 1830–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiles TA, Hohenstein A, Landry LG, Dang M, Powell R, Guyer P, James EA, et al. Characterization of Human CD4 T Cells Specific for a C-Peptide/C-Peptide Hybrid Insulin Peptide. Front. Immunol. 2021; 12: 668680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tree TIM, Lawson J, Edwards H, Skowera A, Arif S, Roep BO, Peakman M. Naturally arising human CD4 T-cells that recognize islet autoantigens and secrete interleukin-10 regulate proinflammatory T-cell responses via linked suppression. Diabetes 2010; 59: 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakayama M, McDaniel K, Fitzgerald-Miller L, Kiekhaefer C, Snell-Bergeon JK, Davidson HW, Rewers M, et al. Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects. Proc. Natl. Acad. Sci. USA 2015; 112: 4429–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Culina S, Lalanne AI, Afonso G, Cerosaletti K, Pinto S, Sebastiani G, Kuranda K, et al. Islet-reactive CD8+ T cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci. Immunol. 2018; 3: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reich EP, Sherwin RS, Kanagawa O, Janeway CA. An explanation for the protective effect of the MHC class II I-E molecule in murine diabetes. Nature 1989; 341: 326–328. [DOI] [PubMed] [Google Scholar]
- 85.Bacelj A, Charlton B, Mandel TE. Prevention of cyclophosphamide-induced diabetes by anti-V beta 8 T-lymphocyte-receptor monoclonal antibody therapy in NOD/Wehi mice. Diabetes 1989; 38: 1492–1495. [DOI] [PubMed] [Google Scholar]
- 86.Shizuru JA, Taylor-Edwards C, Livingstone A, Fathman CG. Genetic dissection of T cell receptor V beta gene requirements for spontaneous murine diabetes. J. Exp. Med. 1991; 174: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sarukhan A, Bedossa P, Garchon HJ, Bach JF, Carnaud C. Molecular analysis of TCR junctional variability in individual infiltrated islets of non-obese diabetic mice: evidence for the constitution of largely autonomous T cell foci within the same pancreas. Int. Immunol. 1995; 7: 139–146. [DOI] [PubMed] [Google Scholar]
- 88.Yang Y, Charlton B, Shimada A, Dal Canto R, Fathman CG. Monoclonal T cells identified in early NOD islet infiltrates. Immunity 1996; 4: 189–194. [DOI] [PubMed] [Google Scholar]
- 89.Baker FJ, Lee M, Chien Y, Davis MM. Restricted islet-cell reactive T cell repertoire of early pancreatic islet infiltrates in NOD mice. Proc. Natl. Acad. Sci. USA 2002; 99: 9374–9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Quinn A, McInerney M, Huffman D, McInerney B, Mayo S, Haskins K, Sercarz E. T cells to a dominant epitope of GAD65 express a public CDR3 motif. Int. Immunol. 2006; 18: 967–979. [DOI] [PubMed] [Google Scholar]
- 91.Nakayama M, Castoe T, Sosinowski T, He X, Johnson K, Haskins K, Vignali DAA, et al. Germline TRAV5D-4 T-cell receptor sequence targets a primary insulin peptide of NOD mice. Diabetes 2012; 61: 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Codina-Busqueta E, Scholz E, Muñoz-Torres PM, Roura-Mir C, Costa M, Xufré C, Planas R, et al. TCR bias of in vivo expanded T cells in pancreatic islets and spleen at the onset in human type 1 diabetes. J. Immunol. 2011; 186: 3787–3797. [DOI] [PubMed] [Google Scholar]
- 93.Eugster A, Lindner A, Catani M, Heninger A-K, Dahl A, Klemroth S, Kühn D, et al. High diversity in the TCR repertoire of GAD65 autoantigen-specific human CD4+ T cells. J. Immunol. 2015; 194: 2531–2538. [DOI] [PubMed] [Google Scholar]
- 94.Cerosaletti K, Barahmand-Pour-Whitman F, Yang J, DeBerg HA, Dufort MJ, Murray SA, Israelsson E, et al. Single-Cell RNA Sequencing Reveals Expanded Clones of Islet Antigen-Reactive CD4+ T Cells in Peripheral Blood of Subjects with Type 1 Diabetes. J. Immunol. 2017; 199: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gomez-Tourino I, Kamra Y, Baptista R, Lorenc A, Peakman M. T cell receptor β-chains display abnormal shortening and repertoire sharing in type 1 diabetes. Nat. Commun. 2017; 8: 1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gavin MA, Bevan MJ. Increased peptide promiscuity provides a rationale for the lack of N regions in the neonatal T cell repertoire. Immunity 1995; 3: 793–800. [DOI] [PubMed] [Google Scholar]
- 97.Stadinski BD, Shekhar K, Gómez-Touriño I, Jung J, Sasaki K, Sewell AK, Peakman M, et al. Hydrophobic CDR3 residues promote the development of self-reactive T cells. Nat. Immunol. 2016; 17: 946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sharon E, Sibener LV, Battle A, Fraser HB, Garcia KC, Pritchard JK. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat. Genet. 2016; 48: 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.International Multiple Sclerosis Genetics Consortium, Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007; 357: 851–862. [DOI] [PubMed] [Google Scholar]
- 100.Noble JA. Immunogenetics of type 1 diabetes: A comprehensive review. J. Autoimmun. 2015; 64: 101–112. [DOI] [PubMed] [Google Scholar]
- 101.Hollenbach JA, Oksenberg JR. The immunogenetics of multiple sclerosis: A comprehensive review. J. Autoimmun. 2015; 64: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Caillier SJ, Briggs F, Cree BAC, Baranzini SE, Fernandez-Viña M, Ramsay PP, Khan O, et al. Uncoupling the roles of HLA-DRB1 and HLA-DRB5 genes in multiple sclerosis. J. Immunol. 2008; 181: 5473–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Patsopoulos NA, Barcellos LF, Hintzen RQ, Schaefer C, Duijn CM van, Noble JA, Raj T, et al. Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS Genet. 2013; 9: e1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sollid LM, Pos W, Wucherpfennig KW. Molecular mechanisms for contribution of MHC molecules to autoimmune diseases. Curr. Opin. Immunol. 2014; 31: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Scholz EM, Marcilla M, Daura X, Arribas-Layton D, James EA, Alvarez I. Human Leukocyte Antigen (HLA)-DRB1*15:01 and HLA-DRB5*01:01 Present Complementary Peptide Repertoires. Front. Immunol. 2017; 8: 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith KJ, Pyrdol J, Gauthier L, Wiley DC, Wucherpfennig KW. Crystal structure of HLA-DR2 (DRA*0101, DRB1*1501) complexed with a peptide from human myelin basic protein. J. Exp. Med. 1998; 188: 1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Waldner H, Whitters MJ, Sobel RA, Collins M, Kuchroo VK. Fulminant spontaneous autoimmunity of the central nervous system in mice transgenic for the myelin proteolipid protein-specific T cell receptor. Proc. Natl. Acad. Sci. USA 2000; 97: 3412–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pöllinger B, Krishnamoorthy G, Berer K, Lassmann H, Bösl MR, Dunn R, Domingues HS, et al. Spontaneous relapsing-remitting EAE in the SJL/J mouse: MOG-reactive transgenic T cells recruit endogenous MOG-specific B cells. J. Exp. Med. 2009; 206: 1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jäger A, Dardalhon V, Sobel RA, Bettelli E, Kuchroo VK. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J. Immunol. 2009; 183: 7169–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Encinas JA, Lees MB, Sobel RA, Symonowicz C, Greer JM, Shovlin CL, Weiner HL, et al. Genetic analysis of susceptibility to experimental autoimmune encephalomyelitis in a cross between SJL/J and B10.S mice. J. Immunol. 1996; 157: 2186–2192. [PubMed] [Google Scholar]
- 111.Mathis DJ, Benoist C, Williams VE, Kanter M, McDevitt HO. Several mechanisms can account for defective E alpha gene expression in different mouse haplotypes. Proc. Natl. Acad. Sci. USA 1983; 80: 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Klein J. Natural history of the major histocompatibility complex. 99thed. Wiley, 1986. [Google Scholar]
- 113.Lawrance SK, Karlsson L, Price J, Quaranta V, Ron Y, Sprent J, Peterson PA. Transgenic HLA-DRα faithfully reconstitutes IE-controlled immune functions and induces cross-tolerance to Eα in Eα0 mutant mice. Cell 1989; 58: 583–594. [DOI] [PubMed] [Google Scholar]
- 114.Rangachari M, Kuchroo VK. Using EAE to better understand principles of immune function and autoimmune pathology. J. Autoimmun. 2013; 45: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zamvil SS, Mitchell DJ, Moore AC, Schwarz AJ, Stiefel W, Nelson PA, Rothbard JB, et al. T cell specificity for class II (I-A) and the encephalitogenic N-terminal epitope of the autoantigen myelin basic protein. J. Immunol. 1987; 139: 1075–1079. [PubMed] [Google Scholar]
- 116.Fairchild PJ, Wildgoose R, Atherton E, Webb S, Wraith DC. An autoantigenic T cell epitope forms unstable complexes with class II MHC: a novel route for escape from tolerance induction. Int. Immunol. 1993; 5: 1151–1158. [DOI] [PubMed] [Google Scholar]
- 117.Mason K, Denney DW, McConnell HM. Kinetics of the reaction of a myelin basic protein peptide with soluble IAu. Biochemistry 1995; 34: 14874–14878. [DOI] [PubMed] [Google Scholar]
- 118.Fugger L, Liang J, Gautam A, Rothbard JB, McDevitt HO. Quantitative analysis of peptides from myelin basic protein binding to the MHC class II protein, I-Au, which confers susceptibility to experimental allergic encephalomyelitis. Mol Med 1996; 2: 181–188. [PMC free article] [PubMed] [Google Scholar]
- 119.He X, Radu C, Sidney J, Sette A, Ward ES, Garcia KC. Structural snapshot of aberrant antigen presentation linked to autoimmunity: the immunodominant epitope of MBP complexed with I-Au. Immunity 2002; 17: 83–94. [DOI] [PubMed] [Google Scholar]
- 120.Liu GY, Fairchild PJ, Smith RM, Prowle JR, Kioussis D, Wraith DC. Low avidity recognition of self-antigen by T cells permits escape from central tolerance. Immunity 1995; 3: 407–415. [DOI] [PubMed] [Google Scholar]
- 121.Muraro PA, Kalbus M, Afshar G, McFarland HF, Martin R. T cell response to 2’,3’-cyclic nucleotide 3’-phosphodiesterase (CNPase) in multiple sclerosis patients. J. Neuroimmunol. 2002; 130: 233–242. [DOI] [PubMed] [Google Scholar]
- 122.Andersson M, Yu M, Söderström M, Weerth S, Baig S, Solders G, Link H. Multiple MAG peptides are recognized by circulating T and B lymphocytes in polyneuropathy and multiple sclerosis. Eur. J. Neurol. 2002; 9: 243–251. [DOI] [PubMed] [Google Scholar]
- 123.Rosbo NK de, Kaye JF, Eisenstein M, Mendel I, Hoeftberger R, Lassmann H, Milo R, et al. The myelin-associated oligodendrocytic basic protein region MOBP15–36 encompasses the immunodominant major encephalitogenic epitope(s) for SJL/J mice and predicted epitope(s) for multiple sclerosis-associated HLA-DRB1*1501. J. Immunol. 2004; 173: 1426–1435. [DOI] [PubMed] [Google Scholar]
- 124.Sun JB, Olsson T, Wang WZ, Xiao BG, Kostulas V, Fredrikson S, Ekre HP, et al. Autoreactive T and B cells responding to myelin proteolipid protein in multiple sclerosis and controls. Eur. J. Immunol. 1991; 21: 1461–1468. [DOI] [PubMed] [Google Scholar]
- 125.Schmidt S, Linington C, Zipp F, Sotgiu S, Waal Malefyt R de, Wekerle H, Hohlfeld R. Multiple sclerosis: comparison of the human T-cell response to S100 beta and myelin basic protein reveals parallels to rat experimental autoimmune panencephalitis. Brain 1997; 120 ( Pt 8): 1437–1445. [DOI] [PubMed] [Google Scholar]
- 126.Banki K, Colombo E, Sia F, Halladay D, Mattson DH, Tatum AH, Massa PT, et al. Oligodendrocyte-specific expression and autoantigenicity of transaldolase in multiple sclerosis. J. Exp. Med. 1994; 180: 1649–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat. Immunol. 2005; 6: 490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Beringer DX, Kleijwegt FS, Wiede F, Slik AR van der, Loh KL, Petersen J, Dudek NL, et al. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat. Immunol. 2015; 16: 1153–1161. [DOI] [PubMed] [Google Scholar]
- 129.Dittel BN, Visintin I, Merchant RM, Janeway CA. Presentation of the self antigen myelin basic protein by dendritic cells leads to experimental autoimmune encephalomyelitis. J. Immunol. 1999; 163: 32–39. [PubMed] [Google Scholar]
- 130.Waldner H, Collins M, Kuchroo VK. Activation of antigen-presenting cells by microbial products breaks self tolerance and induces autoimmune disease. J. Clin. Invest. 2004; 113: 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yogev N, Frommer F, Lukas D, Kautz-Neu K, Karram K, Ielo D, Stebut E von, et al. Dendritic cells ameliorate autoimmunity in the CNS by controlling the homeostasis of PD-1 receptor(+) regulatory T cells. Immunity 2012; 37: 264–275. [DOI] [PubMed] [Google Scholar]
- 132.Luu T, Cheung JF, Baccon J, Waldner H. Priming of myelin-specific T cells in the absence of dendritic cells results in accelerated development of Experimental Autoimmune Encephalomyelitis. PLoS One 2021; 16: e0250340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010; 330: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Becher B, Bechmann I, Greter M. Antigen presentation in autoimmunity and CNS inflammation: how T lymphocytes recognize the brain. J. Mol. Med. 2006; 84: 532–543. [DOI] [PubMed] [Google Scholar]
- 135.Lehmann-Horn K, Kronsbein HC, Weber MS. Targeting B cells in the treatment of multiple sclerosis: recent advances and remaining challenges. Ther Adv Neurol Disord 2013; 6: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Waisman A, Johann L. Antigen-presenting cell diversity for T cell reactivation in central nervous system autoimmunity. J. Mol. Med. 2018; 96: 1279–1292. [DOI] [PubMed] [Google Scholar]
- 137.Mundt S, Mrdjen D, Utz SG, Greter M, Schreiner B, Becher B. Conventional DCs sample and present myelin antigens in the healthy CNS and allow parenchymal T cell entry to initiate neuroinflammation. Sci. Immunol. 2019; 4: [DOI] [PubMed] [Google Scholar]
- 138.Ben-Nun A, Liblau RS, Cohen L, Lehmann D, Tournier-Lasserve E, Rosenzweig A, Zhang JW, et al. Restricted T-cell receptor V beta gene usage by myelin basic protein-specific T-cell clones in multiple sclerosis: predominant genes vary in individuals. Proc. Natl. Acad. Sci. USA 1991; 88: 2466–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vandevyver C, Mertens N, Elsen P van den, Medaer R, Raus J, Zhang J. Clonal expansion of myelin basic protein-reactive T cells in patients with multiple sclerosis: restricted T cell receptor V gene rearrangements and CDR3 sequence. Eur. J. Immunol. 1995; 25: 958–968. [DOI] [PubMed] [Google Scholar]
- 140.Hafler DA, Saadeh MG, Kuchroo VK, Milford E, Steinman L. TCR usage in human and experimental demyelinating disease. Immunol. Today 1996; 17: 152–159. [DOI] [PubMed] [Google Scholar]
- 141.Junker A, Ivanidze J, Malotka J, Eiglmeier I, Lassmann H, Wekerle H, Meinl E, et al. Multiple sclerosis: T-cell receptor expression in distinct brain regions. Brain 2007; 130: 2789–2799. [DOI] [PubMed] [Google Scholar]
- 142.Mendel I, Kerlero de Rosbo N, Ben-Nun A. A myelin oligodendrocyte glycoprotein peptide induces typical chronic experimental autoimmune encephalomyelitis in H-2b mice: fine specificity and T cell receptor V beta expression of encephalitogenic T cells. Eur. J. Immunol. 1995; 25: 1951–1959. [DOI] [PubMed] [Google Scholar]
- 143.Menezes JS, Elzen P van den, Thornes J, Huffman D, Droin NM, Maverakis E, Sercarz EE. A public T cell clonotype within a heterogeneous autoreactive repertoire is dominant in driving EAE. J. Clin. Invest. 2007; 117: 2176–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhao Y, Nguyen P, Ma J, Wu T, Jones LL, Pei D, Cheng C, et al. Preferential Use of Public TCR during Autoimmune Encephalomyelitis. J. Immunol. 2016; 196: 4905–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao Y, Nguyen P, Vogel P, Li B, Jones LL, Geiger TL. Autoimmune susceptibility imposed by public TCRβ chains. Sci. Rep. 2016; 6: 37543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tuohy VK, Yu M, Yin L, Kawczak JA, Kinkel RP. Spontaneous regression of primary autoreactivity during chronic progression of experimental autoimmune encephalomyelitis and multiple sclerosis. J. Exp. Med. 1999; 189: 1033–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Powell AM, Black MM. Epitope spreading: protection from pathogens, but propagation of autoimmunity? Clin. Exp. Dermatol. 2001; 26: 427–433. [DOI] [PubMed] [Google Scholar]
- 148.Hong J, Zang YC, Tejada-Simon MV, Kozovska M, Li S, Singh RA, Yang D, et al. A common TCR V-D-J sequence in V beta 13.1 T cells recognizing an immunodominant peptide of myelin basic protein in multiple sclerosis. J. Immunol. 1999; 163: 3530–3538. [PubMed] [Google Scholar]
- 149.Schnell A, Huang L, Singer M, Singaraju A, Barilla RM, Regan BML, Bollhagen A, et al. Stem-like intestinal Th17 cells give rise to pathogenic effector T cells during autoimmunity. Cell 2021; 184: 6281–6298.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Plichta DR, Graham DB, Subramanian S, Xavier RJ. Therapeutic Opportunities in Inflammatory Bowel Disease: Mechanistic Dissection of Host-Microbiome Relationships. Cell 2019; 178: 1041–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roda G, Jharap B, Neeraj N, Colombel J-F. Loss of Response to Anti-TNFs: Definition, Epidemiology, and Management. Clin Transl Gastroenterol 2016; 7: e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Halme L, Paavola-Sakki P, Turunen U, Lappalainen M, Farkkila M, Kontula K. Family and twin studies in inflammatory bowel disease. World J. Gastroenterol. 2006; 12: 3668–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Thia KT, Loftus EV, Sandborn WJ, Yang S-K. An update on the epidemiology of inflammatory bowel disease in Asia. Am. J. Gastroenterol. 2008; 103: 3167–3182. [DOI] [PubMed] [Google Scholar]
- 154.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu. Rev. Immunol. 2010; 28: 573–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ahmad T, Armuzzi A, Bunce M, Mulcahy-Hawes K, Marshall SE, Orchard TR, Crawshaw J, et al. The molecular classification of the clinical manifestations of Crohn’s disease. Gastroenterology 2002; 122: 854–866. [DOI] [PubMed] [Google Scholar]
- 156.Ahmad T, Marshall S-E, Jewell D. Genetics of inflammatory bowel disease: the role of the HLA complex. World J. Gastroenterol. 2006; 12: 3628–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fernandez L, Mendoza JL, Martinez A, Urcelay E, Fernandez-Arquero M, Garcia-Paredes J, Peña AS, et al. IBD1 and IBD3 determine location of Crohn’s disease in the Spanish population. Inflamm. Bowel Dis. 2004; 10: 715–722. [DOI] [PubMed] [Google Scholar]
- 158.Newman B, Silverberg MS, Gu X, Zhang Q, Lazaro A, Steinhart AH, Greenberg GR, et al. CARD15 and HLA DRB1 alleles influence susceptibility and disease localization in Crohn’s disease. Am. J. Gastroenterol. 2004; 99: 306–315. [DOI] [PubMed] [Google Scholar]
- 159.Goyette P, Boucher G, Mallon D, Ellinghaus E, Jostins L, Huang H, Ripke S, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet. 2015; 47: 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int. Immunol. 1993; 5: 1461–1471. [DOI] [PubMed] [Google Scholar]
- 161.Powrie F, Leach MW, Mauze S, Menon S, Barcomb Caddle L, Coffman RL. Inhibition of Thl responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity 1994; 1: 553–562. [DOI] [PubMed] [Google Scholar]
- 162.Izcue A, Hue S, Buonocore S, Arancibia-Cárcamo CV, Ahern PP, Iwakura Y, Maloy KJ, et al. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity 2008; 28: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mottet C, Uhlig HH, Powrie F. Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. J. Immunol. 2003; 170: 3939–3943. [DOI] [PubMed] [Google Scholar]
- 164.Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, Uhlig H, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm. Bowel Dis. 2007; 13: 1202–1211. [DOI] [PubMed] [Google Scholar]
- 165.Zegarra-Ruiz DF, Kim DV, Norwood K, Kim M, Wu W-JH, Saldana-Morales FB, Hill AA, et al. Thymic development of gut-microbiota-specific T cells. Nature 2021; 594: 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature 2016; 535: 75–84. [DOI] [PubMed] [Google Scholar]
- 167.Dougan M, Luoma AM, Dougan SK, Wucherpfennig KW. Understanding and treating the inflammatory adverse events of cancer immunotherapy. Cell 2021; 184: 1575–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 2018; 554: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 2014; 510: 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kullberg MC, Andersen JF, Gorelick PL, Caspar P, Suerbaum S, Fox JG, Cheever AW, et al. Induction of colitis by a CD4+ T cell clone specific for a bacterial epitope. Proc. Natl. Acad. Sci. USA 2003; 100: 15830–15835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, Fort M, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J. Clin. Invest. 2004; 113: 1296–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Matsuda JL, Gapin L, Sydora BC, Byrne F, Binder S, Kronenberg M, Aranda R. Systemic activation and antigen-driven oligoclonal expansion of T cells in a mouse model of colitis. J. Immunol. 2000; 164: 2797–2806. [DOI] [PubMed] [Google Scholar]
- 173.Doorenspleet ME, Westera L, Peters CP, Hakvoort TBM, Esveldt RE, Vogels E, Kampen AHC van, et al. Profoundly Expanded T-cell Clones in the Inflamed and Uninflamed Intestine of Patients With Crohn’s Disease. J. Crohns Colitis 2017; 11: 831–839. [DOI] [PubMed] [Google Scholar]
- 174.Werner L, Lee YN, Rechavi E, Lev A, Yerushalmi B, Ling G, Shah N, et al. Alterations in T and B Cell Receptor Repertoires Patterns in Patients With IL10 Signaling Defects and History of Infantile-Onset IBD. Front. Immunol. 2020; 11: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rosati E, Pogorelyy MV, Dowds CM, Moller FT, Sorensen SB, Lebedev YB, Frey N, et al. Identification of Disease-associated Traits and Clonotypes in the T Cell Receptor Repertoire of Monozygotic Twins Affected by Inflammatory Bowel Diseases. J. Crohns Colitis 2020; 14: 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 2013; 13: 227–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schwartz RH, Mueller DL, Jenkins MK, Quill H. T-cell clonal anergy. Cold Spring Harb. Symp. Quant. Biol. 1989; 54 Pt 2: 605–610. [DOI] [PubMed] [Google Scholar]
- 178.Saito T, Yokosuka T, Hashimoto-Tane A. Dynamic regulation of T cell activation and co-stimulation through TCR-microclusters. FEBS Lett. 2010; 584: 4865–4871. [DOI] [PubMed] [Google Scholar]
- 179.Rudd CE, Taylor A, Schneider H. CD28 and CTLA-4 coreceptor expression and signal transduction. Immunol. Rev. 2009; 229: 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Collins AV, Brodie DW, Gilbert RJC, Iaboni A, Manso-Sancho R, Walse B, Stuart DI, et al. The interaction properties of costimulatory molecules revisited. Immunity 2002; 17: 201–210. [DOI] [PubMed] [Google Scholar]
- 181.Walker LSK. Treg and CTLA-4: two intertwining pathways to immune tolerance. J. Autoimmun. 2013; 45: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhu Y, Yao S, Chen L. Cell surface signaling molecules in the control of immune responses: a tide model. Immunity 2011; 34: 466–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 2009; 10: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Croft M. The role of TNF superfamily members in T-cell function and diseases. Nat. Rev. Immunol. 2009; 9: 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Boomer JS, Green JM. An enigmatic tail of CD28 signaling. Cold Spring Harb. Perspect. Biol. 2010; 2: a002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Attanasio J, Wherry EJ. Costimulatory and coinhibitory receptor pathways in infectious disease. Immunity 2016; 44: 1052–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Simpson TR, Quezada SA, Allison JP. Regulation of CD4 T cell activation and effector function by inducible costimulator (ICOS). Curr. Opin. Immunol. 2010; 22: 326–332. [DOI] [PubMed] [Google Scholar]
- 188.Salomon B, Lenschow DJ, Rhee L, Ashourian N, Singh B, Sharpe A, Bluestone JA. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity 2000; 12: 431–440. [DOI] [PubMed] [Google Scholar]
- 189.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell 2010; 140: 845–858. [DOI] [PubMed] [Google Scholar]
- 190.Lenschow DJ, Ho SC, Sattar H, Rhee L, Gray G, Nabavi N, Herold KC, et al. Differential effects of anti-B7–1 and anti-B7–2 monoclonal antibody treatment on the development of diabetes in the nonobese diabetic mouse. J. Exp. Med. 1995; 181: 1145–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ueda H, Howson JMM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003; 423: 506–511. [DOI] [PubMed] [Google Scholar]
- 192.Gerold KD, Zheng P, Rainbow DB, Zernecke A, Wicker LS, Kissler S. The soluble CTLA-4 splice variant protects from type 1 diabetes and potentiates regulatory T-cell function. Diabetes 2011; 60: 1955–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vijayakrishnan L, Slavik JM, Illés Z, Greenwald RJ, Rainbow D, Greve B, Peterson LB, et al. An autoimmune disease-associated CTLA-4 splice variant lacking the B7 binding domain signals negatively in T cells. Immunity 2004; 20: 563–575. [DOI] [PubMed] [Google Scholar]
- 194.Shapiro MR, Yeh W-I, Longfield JR, Gallagher J, Infante CM, Wellford S, Posgai AL, et al. CD226 deletion reduces type 1 diabetes in the NOD mouse by impairing thymocyte development and peripheral T cell activation. Front. Immunol. 2020; 11: 2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lozano E, Joller N, Cao Y, Kuchroo VK, Hafler DA. The CD226/CD155 interaction regulates the proinflammatory (Th1/Th17)/anti-inflammatory (Th2) balance in humans. J. Immunol. 2013; 191: 3673–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hafler JP, Maier LM, Cooper JD, Plagnol V, Hinks A, Simmonds MJ, Stevens HE, et al. CD226 Gly307Ser association with multiple autoimmune diseases. Genes Immun. 2009; 10: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gaud G, Roncagalli R, Chaoui K, Bernard I, Familiades J, Colacios C, Kassem S, et al. The costimulatory molecule CD226 signals through VAV1 to amplify TCR signals and promote IL-17 production by CD4+ T cells. Sci. Signal. 2018; 11: [DOI] [PubMed] [Google Scholar]
- 198.Hollis-Moffatt JE, Hook SM, Merriman TR. Colocalization of mouse autoimmune diabetes loci Idd21.1 and Idd21.2 with IDDM6 (human) and Iddm3 (rat). Diabetes 2005; 54: 2820–2825. [DOI] [PubMed] [Google Scholar]
- 199.Mahmud SA, Manlove LS, Schmitz HM, Xing Y, Wang Y, Owen DL, Schenkel JM, et al. Costimulation via the tumor-necrosis factor receptor superfamily couples TCR signal strength to the thymic differentiation of regulatory T cells. Nat. Immunol. 2014; 15: 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Balasa B, Krahl T, Patstone G, Lee J, Tisch R, McDevitt HO, Sarvetnick N. CD40 ligand-CD40 interactions are necessary for the initiation of insulitis and diabetes in nonobese diabetic mice. J. Immunol. 1997; 159: 4620–4627. [PubMed] [Google Scholar]
- 201.Bour-Jordan H, Salomon BL, Thompson HL, Szot GL, Bernhard MR, Bluestone JA. Costimulation controls diabetes by altering the balance of pathogenic and regulatory T cells. J. Clin. Invest. 2004; 114: 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Baker RL, Wagner DH, Haskins K. CD40 on NOD CD4 T cells contributes to their activation and pathogenicity. J. Autoimmun. 2008; 31: 385–392. [DOI] [PubMed] [Google Scholar]
- 203.Martin S, Agarwal R, Murugaiyan G, Saha B. CD40 expression levels modulate regulatory T cells in Leishmania donovani infection. J. Immunol. 2010; 185: 551–559. [DOI] [PubMed] [Google Scholar]
- 204.Wagner DH, Vaitaitis G, Sanderson R, Poulin M, Dobbs C, Haskins K. Expression of CD40 identifies a unique pathogenic T cell population in type 1 diabetes. Proc. Natl. Acad. Sci. USA 2002; 99: 3782–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Waid DM, Vaitaitis GM, Wagner DH. Peripheral CD4loCD40+ auto-aggressive T cell expansion during insulin-dependent diabetes mellitus. Eur. J. Immunol. 2004; 34: 1488–1497. [DOI] [PubMed] [Google Scholar]
- 206.Okazaki T, Honjo T. PD-1 and PD-1 ligands: from discovery to clinical application. Int. Immunol. 2007; 19: 813–824. [DOI] [PubMed] [Google Scholar]
- 207.Wang J, Yoshida T, Nakaki F, Hiai H, Okazaki T, Honjo T. Establishment of NOD-Pdcd1−/− mice as an efficient animal model of type I diabetes. Proc. Natl. Acad. Sci. USA 2005; 102: 11823–11828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bettini M, Szymczak-Workman AL, Forbes K, Castellaw AH, Selby M, Pan X, Drake CG, et al. Cutting edge: accelerated autoimmune diabetes in the absence of LAG-3. J. Immunol. 2011; 187: 3493–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Graydon CG, Mohideen S, Fowke KR. Lag3’s enigmatic mechanism of action. Front. Immunol. 2020; 11: 615317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sánchez-Fueyo A, Tian J, Picarella D, Domenig C, Zheng XX, Sabatos CA, Manlongat N, et al. Tim-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nat. Immunol. 2003; 4: 1093–1101. [DOI] [PubMed] [Google Scholar]
- 211.Brück P, Ramos-Lopez E, Bartsch W, Böhme A, Badenhoop K. TIM-3 polymorphisms in type 1 diabetes families. J. Hum. Genet. 2008; 53: 559–564. [DOI] [PubMed] [Google Scholar]
- 212.Oliveira-dos-Santos AJ, Ho A, Tada Y, Lafaille JJ, Tonegawa S, Mak TW, Penninger JM. CD28 costimulation is crucial for the development of spontaneous autoimmune encephalomyelitis. J. Immunol. 1999; 162: 4490–4495. [PubMed] [Google Scholar]
- 213.Chang TT, Jabs C, Sobel RA, Kuchroo VK, Sharpe AH. Studies in B7-deficient mice reveal a critical role for B7 costimulation in both induction and effector phases of experimental autoimmune encephalomyelitis. J. Exp. Med. 1999; 190: 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Racke MK, Scott DE, Quigley L, Gray GS, Abe R, June CH, Perrin PJ. Distinct roles for B7–1 (CD-80) and B7–2 (CD-86) in the initiation of experimental allergic encephalomyelitis. J. Clin. Invest. 1995; 96: 2195–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jabs C, Greve B, Chang TT, Sobel RA, Sharpe AH, Kuchroo VK. Genetic background determines the requirement for B7 costimulation in induction of autoimmunity. Eur. J. Immunol. 2002; 32: 2687–2697. [DOI] [PubMed] [Google Scholar]
- 216.Miller SD, Vanderlugt CL, Lenschow DJ, Pope JG, Karandikar NJ, Dal Canto MC, Bluestone JA. Blockade of CD28/B7–1 interaction prevents epitope spreading and clinical relapses of murine EAE. Immunity 1995; 3: 739–745. [DOI] [PubMed] [Google Scholar]
- 217.Karandikar NJ, Vanderlugt CL, Eagar T, Tan L, Bluestone JA, Miller SD. Tissue-specific up-regulation of B7–1 expression and function during the course of murine relapsing experimental autoimmune encephalomyelitis. J. Immunol. 1998; 161: 192–199. [PubMed] [Google Scholar]
- 218.Cross AH, Lyons JA, San M, Keeling RM, Ku G, Racke MK. T cells are the main cell type expressing B7–1 and B7–2 in the central nervous system during acute, relapsing and chronic experimental autoimmune encephalomyelitis. Eur. J. Immunol. 1999; 29: 3140–3147. [DOI] [PubMed] [Google Scholar]
- 219.Dong C, Juedes AE, Temann UA, Shresta S, Allison JP, Ruddle NH, Flavell RA. ICOS co-stimulatory receptor is essential for T-cell activation and function. Nature 2001; 409: 97–101. [DOI] [PubMed] [Google Scholar]
- 220.Galicia G, Kasran A, Uyttenhove C, De Swert K, Van Snick J, Ceuppens JL. ICOS deficiency results in exacerbated IL-17 mediated experimental autoimmune encephalomyelitis. J. Clin. Immunol. 2009; 29: 426–433. [DOI] [PubMed] [Google Scholar]
- 221.Sporici RA, Beswick RL, Allmen C von, Rumbley CA, Hayden-Ledbetter M, Ledbetter JA, Perrin PJ. ICOS ligand costimulation is required for T-cell encephalitogenicity. Clin. Immunol. 2001; 100: 277–288. [DOI] [PubMed] [Google Scholar]
- 222.Weinberg AD, Wegmann KW, Funatake C, Whitham RH. Blocking OX-40/OX-40 ligand interaction in vitro and in vivo leads to decreased T cell function and amelioration of experimental allergic encephalomyelitis. J. Immunol. 1999; 162: 1818–1826. [PubMed] [Google Scholar]
- 223.Nohara C, Akiba H, Nakajima A, Inoue A, Koh CS, Ohshima H, Yagita H, et al. Amelioration of experimental autoimmune encephalomyelitis with anti-OX40 ligand monoclonal antibody: a critical role for OX40 ligand in migration, but not development, of pathogenic T cells. J. Immunol. 2001; 166: 2108–2115. [DOI] [PubMed] [Google Scholar]
- 224.Carboni S, Aboul-Enein F, Waltzinger C, Killeen N, Lassmann H, Peña-Rossi C. CD134 plays a crucial role in the pathogenesis of EAE and is upregulated in the CNS of patients with multiple sclerosis. J. Neuroimmunol. 2003; 145: 1–11. [DOI] [PubMed] [Google Scholar]
- 225.Ndhlovu LC, Ishii N, Murata K, Sato T, Sugamura K. Critical involvement of OX40 ligand signals in the T cell priming events during experimental autoimmune encephalomyelitis. J. Immunol. 2001; 167: 2991–2999. [DOI] [PubMed] [Google Scholar]
- 226.Coquet JM, Middendorp S, Horst G van der, Kind J, Veraar EAM, Xiao Y, Jacobs H, et al. The CD27 and CD70 costimulatory pathway inhibits effector function of T helper 17 cells and attenuates associated autoimmunity. Immunity 2013; 38: 53–65. [DOI] [PubMed] [Google Scholar]
- 227.Karabon L, Kosmaczewska A, Bilinska M, Pawlak E, Ciszak L, Jedynak A, Jonkisz A, et al. The CTLA-4 gene polymorphisms are associated with CTLA-4 protein expression levels in multiple sclerosis patients and with susceptibility to disease. Immunology 2009; 128: e787–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Oliveira EML, Bar-Or A, Waliszewska AI, Cai G, Anderson DE, Krieger JI, Hafler DA. CTLA-4 dysregulation in the activation of myelin basic protein reactive T cells may distinguish patients with multiple sclerosis from healthy controls. J. Autoimmun. 2003; 20: 71–81. [DOI] [PubMed] [Google Scholar]
- 229.Karandikar NJ, Vanderlugt CL, Walunas TL, Miller SD, Bluestone JA. CTLA-4: a negative regulator of autoimmune disease. J. Exp. Med. 1996; 184: 783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Perrin PJ, Maldonado JH, Davis TA, June CH, Racke MK. CTLA-4 blockade enhances clinical disease and cytokine production during experimental allergic encephalomyelitis. J. Immunol. 1996; 157: 1333–1336. [PubMed] [Google Scholar]
- 231.Hurwitz AA, Sullivan TJ, Krummel MF, Sobel RA, Allison JP. Specific blockade of CTLA-4/B7 interactions results in exacerbated clinical and histologic disease in an actively-induced model of experimental allergic encephalomyelitis. J. Neuroimmunol. 1997; 73: 57–62. [DOI] [PubMed] [Google Scholar]
- 232.Carter LL, Leach MW, Azoitei ML, Cui J, Pelker JW, Jussif J, Benoit S, et al. PD-1/PD-L1, but not PD-1/PD-L2, interactions regulate the severity of experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2007; 182: 124–134. [DOI] [PubMed] [Google Scholar]
- 233.Joller N, Hafler JP, Brynedal B, Kassam N, Spoerl S, Levin SD, Sharpe AH, et al. Cutting edge: TIGIT has T cell-intrinsic inhibitory functions. J. Immunol. 2011; 186: 1338–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sabatos CA, Chakravarti S, Cha E, Schubart A, Sánchez-Fueyo A, Zheng XX, Coyle AJ, et al. Interaction of Tim-3 and Tim-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nat. Immunol. 2003; 4: 1102–1110. [DOI] [PubMed] [Google Scholar]
- 235.Koguchi K, Anderson DE, Yang L, O’Connor KC, Kuchroo VK, Hafler DA. Dysregulated T cell expression of TIM3 in multiple sclerosis. J. Exp. Med. 2006; 203: 1413–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Yang L, Anderson DE, Kuchroo J, Hafler DA. Lack of TIM-3 immunoregulation in multiple sclerosis. J. Immunol. 2008; 180: 4409–4414. [DOI] [PubMed] [Google Scholar]
- 237.Kim G, Levin M, Schoenberger SP, Sharpe A, Kronenberg M. Paradoxical effect of reduced costimulation in T cell-mediated colitis. J. Immunol. 2007; 178: 5563–5570. [DOI] [PubMed] [Google Scholar]
- 238.Kim G, Turovskaya O, Levin M, Byrne FR, Whoriskey JS, McCabe JG, Kronenberg M. Spontaneous colitis occurrence in transgenic mice with altered B7-mediated costimulation. J. Immunol. 2008; 181: 5278–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jong YP de, Rietdijk ST, Faubion WA, Abadia-Molina AC, Clarke K, Mizoguchi E, Tian J, et al. Blocking inducible co-stimulator in the absence of CD28 impairs Th1 and CD25+ regulatory T cells in murine colitis. Int. Immunol. 2004; 16: 205–213. [DOI] [PubMed] [Google Scholar]
- 240.Malmström V, Shipton D, Singh B, Al-Shamkhani A, Puklavec MJ, Barclay AN, Powrie F. CD134L expression on dendritic cells in the mesenteric lymph nodes drives colitis in T cell-restored SCID mice. J. Immunol. 2001; 166: 6972–6981. [DOI] [PubMed] [Google Scholar]
- 241.Griseri T, Asquith M, Thompson C, Powrie F. OX40 is required for regulatory T cell-mediated control of colitis. J. Exp. Med. 2010; 207: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Manocha M, Rietdijk S, Laouar A, Liao G, Bhan A, Borst J, Terhorst C, et al. Blocking CD27-CD70 costimulatory pathway suppresses experimental colitis. J. Immunol. 2009; 183: 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Stuber E, Strober W, Neurath M. Blocking the CD40L-CD40 interaction in vivo specifically prevents the priming of T helper 1 cells through the inhibition of interleukin 12 secretion. J. Exp. Med. 1996; 183: 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Clegg CH, Rulffes JT, Haugen HS, Hoggatt IH, Aruffo A, Durham SK, Farr AG, et al. Thymus dysfunction and chronic inflammatory disease in gp39 transgenic mice. Int. Immunol. 1997; 9: 1111–1122. [DOI] [PubMed] [Google Scholar]
- 245.Liu Z, Colpaert S, D’Haens GR, Kasran A, Boer M de, Rutgeerts P, Geboes K, et al. Hyperexpression of CD40 ligand (CD154) in inflammatory bowel disease and its contribution to pathogenic cytokine production. J. Immunol. 1999; 163: 4049–4057. [PubMed] [Google Scholar]
- 246.Battaglia E, Biancone L, Resegotti A, Emanuelli G, Fronda GR, Camussi G. Expression of CD40 and its ligand, CD40L, in intestinal lesions of Crohn’s disease. Am. J. Gastroenterol. 1999; 94: 3279–3284. [DOI] [PubMed] [Google Scholar]
- 247.Atkins MB, Clark JI, Quinn DI. Immune checkpoint inhibitors in advanced renal cell carcinoma: experience to date and future directions. Ann. Oncol. 2017; 28: 1484–1494. [DOI] [PubMed] [Google Scholar]
- 248.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018; 8: 1069–1086. [DOI] [PubMed] [Google Scholar]
- 249.Pauken KE, Dougan M, Rose NR, Lichtman AH, Sharpe AH. Adverse events following cancer immunotherapy: obstacles and opportunities. Trends Immunol. 2019; 40: 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kurtulus S, Madi A, Escobar G, Klapholz M, Nyman J, Christian E, Pawlak M, et al. Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1-CD8+ Tumor-Infiltrating T Cells. Immunity 2019; 50: 181–194.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016; 44: 989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shi F, Guo X, Jiang X, Zhou P, Xiao Y, Zhou T, Chen G, et al. Dysregulated Tim-3 expression and its correlation with imbalanced CD4 helper T cell function in ulcerative colitis. Clin. Immunol. 2012; 145: 230–240. [DOI] [PubMed] [Google Scholar]
- 253.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 2010; 28: 445–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kumar V, Bhardwaj V, Soares L, Alexander J, Sette A, Sercarz E. Major histocompatibility complex binding affinity of an antigenic determinant is crucial for the differential secretion of interleukin 4/5 or interferon gamma by T cells. Proc. Natl. Acad. Sci. USA 1995; 92: 9510–9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Pfeiffer C, Stein J, Southwood S, Ketelaar H, Sette A, Bottomly K. Altered peptide ligands can control CD4 T lymphocyte differentiation in vivo. J. Exp. Med. 1995; 181: 1569–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Tao X, Grant C, Constant S, Bottomly K. Induction of IL-4-producing CD4+ T cells by antigenic peptides altered for TCR binding. J. Immunol. 1997; 158: 4237–4244. [PubMed] [Google Scholar]
- 257.Brogdon JL, Leitenberg D, Bottomly K. The potency of TCR signaling differentially regulates NFATc/p activity and early IL-4 transcription in naive CD4+ T cells. J. Immunol. 2002; 168: 3825–3832. [DOI] [PubMed] [Google Scholar]
- 258.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986; 136: 2348–2357. [PubMed] [Google Scholar]
- 259.Ruterbusch M, Pruner KB, Shehata L, Pepper M. In vivo CD4+ T cell differentiation and function: revisiting the th1/th2 paradigm. Annu. Rev. Immunol. 2020; 38: 705–725. [DOI] [PubMed] [Google Scholar]
- 260.Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O’Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science 1993; 260: 547–549. [DOI] [PubMed] [Google Scholar]
- 261.Swain SL, Weinberg AD, English M, Huston G. IL-4 directs the development of Th2-like helper effectors. J. Immunol. 1990; 145: 3796–3806. [PubMed] [Google Scholar]
- 262.Le Gros G, Ben-Sasson SZ, Seder R, Finkelman FD, Paul WE. Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4-producing cells. J. Exp. Med. 1990; 172: 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 2000; 100: 655–669. [DOI] [PubMed] [Google Scholar]
- 264.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 1997; 89: 587–596. [DOI] [PubMed] [Google Scholar]
- 265.Guglani L, Khader SA. Th17 cytokines in mucosal immunity and inflammation. Curr Opin HIV AIDS 2010; 5: 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Aggarwal S, Ghilardi N, Xie M-H, Sauvage FJ de, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 2003; 278: 1910–1914. [DOI] [PubMed] [Google Scholar]
- 267.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 2003; 421: 744–748. [DOI] [PubMed] [Google Scholar]
- 268.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 2006; 441: 235–238. [DOI] [PubMed] [Google Scholar]
- 269.Veldhoen M, Hocking RJ, Flavell RA, Stockinger B. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat. Immunol. 2006; 7: 1151–1156. [DOI] [PubMed] [Google Scholar]
- 270.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity 2007; 26: 579–591. [DOI] [PubMed] [Google Scholar]
- 271.Ghoreschi K, Laurence A, Yang X-P, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature 2010; 467: 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Peters A, Lee Y, Kuchroo VK. The many faces of Th17 cells. Curr. Opin. Immunol. 2011; 23: 702–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Josefowicz SZ, Lu L-F, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 2012; 30: 531–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Hsieh C-S, Lee H-M, Lio C-WJ. Selection of regulatory T cells in the thymus. Nat. Rev. Immunol. 2012; 12: 157–167. [DOI] [PubMed] [Google Scholar]
- 275.Wan YY, Flavell RA. “Yin-Yang” functions of transforming growth factor-beta and T regulatory cells in immune regulation. Immunol. Rev. 2007; 220: 199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zeng H, Zhang R, Jin B, Chen L. Type 1 regulatory T cells: a new mechanism of peripheral immune tolerance. Cell Mol Immunol 2015; 12: 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Groux H, O’Garra A, Bigler M, Rouleau M, Antonenko S, Vries JE de, Roncarolo MG. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature 1997; 389: 737–742. [DOI] [PubMed] [Google Scholar]
- 278.Awasthi A, Carrier Y, Peron JPS, Bettelli E, Kamanaka M, Flavell RA, Kuchroo VK, et al. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat. Immunol. 2007; 8: 1380–1389. [DOI] [PubMed] [Google Scholar]
- 279.Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, Burns EJ, et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 2010; 11: 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Katz JD, Benoist C, Mathis D. T helper cell subsets in insulin-dependent diabetes. Science 1995; 268: 1185–1188. [DOI] [PubMed] [Google Scholar]
- 281.Debray-Sachs M, Carnaud C, Boitard C, Cohen H, Gresser I, Bedossa P, Bach JF. Prevention of diabetes in NOD mice treated with antibody to murine IFN gamma. J. Autoimmun. 1991; 4: 237–248. [DOI] [PubMed] [Google Scholar]
- 282.Campbell IL, Kay TW, Oxbrow L, Harrison LC. Essential role for interferon-gamma and interleukin-6 in autoimmune insulin-dependent diabetes in NOD/Wehi mice. J. Clin. Invest. 1991; 87: 739–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Chong MM, Thomas HE, Kay TW. gamma-Interferon signaling in pancreatic beta-cells is persistent but can be terminated by overexpression of suppressor of cytokine signaling-1. Diabetes 2001; 50: 2744–2751. [DOI] [PubMed] [Google Scholar]
- 284.Arif S, Tree TI, Astill TP, Tremble JM, Bishop AJ, Dayan CM, Roep BO, et al. Autoreactive T cell responses show proinflammatory polarization in diabetes but a regulatory phenotype in health. J. Clin. Invest. 2004; 113: 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Uzel G, Sampaio EP, Lawrence MG, Hsu AP, Hackett M, Dorsey MJ, Noel RJ, et al. Dominant gain-of-function STAT1 mutations in FOXP3 wild-type immune dysregulation-polyendocrinopathy-enteropathy-X-linked-like syndrome. J. Allergy Clin. Immunol. 2013; 131: 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Kim S, Kim HS, Chung KW, Oh SH, Yun JW, Im S-H, Lee M-K, et al. Essential role for signal transducer and activator of transcription-1 in pancreatic beta-cell death and autoimmune type 1 diabetes of nonobese diabetic mice. Diabetes 2007; 56: 2561–2568. [DOI] [PubMed] [Google Scholar]
- 287.Kriegel MA, Sefik E, Hill JA, Wu H-J, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. USA 2011; 108: 11548–11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 2012; 149: 1578–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Martin-Orozco N, Chung Y, Chang SH, Wang Y-H, Dong C. Th17 cells promote pancreatic inflammation but only induce diabetes efficiently in lymphopenic hosts after conversion into Th1 cells. Eur. J. Immunol. 2009; 39: 216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Bending D, De la Peña H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, Cooke A. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J. Clin. Invest. 2009; 119: 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. Late developmental plasticity in the T helper 17 lineage. Immunity 2009; 30: 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 2011; 12: 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Duhen R, Glatigny S, Arbelaez CA, Blair TC, Oukka M, Bettelli E. Cutting edge: the pathogenicity of IFN-γ-producing Th17 cells is independent of T-bet. J. Immunol. 2013; 190: 4478–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Bradshaw EM, Raddassi K, Elyaman W, Orban T, Gottlieb PA, Kent SC, Hafler DA. Monocytes from patients with type 1 diabetes spontaneously secrete proinflammatory cytokines inducing Th17 cells. J. Immunol. 2009; 183: 4432–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Flanagan SE, Haapaniemi E, Russell MA, Caswell R, Allen HL, De Franco E, McDonald TJ, et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat. Genet. 2014; 46: 812–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Milner JD, Vogel TP, Forbes L, Ma CA, Stray-Pedersen A, Niemela JE, Lyons JJ, et al. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood 2015; 125: 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Haapaniemi EM, Kaustio M, Rajala HLM, Adrichem AJ van, Kainulainen L, Glumoff V, Doffinger R, et al. Autoimmunity, hypogammaglobulinemia, lymphoproliferation, and mycobacterial disease in patients with activating mutations in STAT3. Blood 2015; 125: 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Fabbri M, Frixou M, Degano M, Fousteri G. Type 1 diabetes in STAT protein family mutations: regulating the th17/treg equilibrium and beyond. Diabetes 2019; 68: 258–265. [DOI] [PubMed] [Google Scholar]
- 300.Zhernakova A, Alizadeh BZ, Bevova M, Leeuwen MA van, Coenen MJH, Franke B, Franke L, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am. J. Hum. Genet. 2007; 81: 1284–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007; 39: 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.King C, Ilic A, Koelsch K, Sarvetnick N. Homeostatic expansion of T cells during immune insufficiency generates autoimmunity. Cell 2004; 117: 265–277. [DOI] [PubMed] [Google Scholar]
- 303.Clough LE, Wang CJ, Schmidt EM, Booth G, Hou TZ, Ryan GA, Walker LSK. Release from regulatory T cell-mediated suppression during the onset of tissue-specific autoimmunity is associated with elevated IL-21. J. Immunol. 2008; 180: 5393–5401. [DOI] [PubMed] [Google Scholar]
- 304.Sutherland APR, Van Belle T, Wurster AL, Suto A, Michaud M, Zhang D, Grusby MJ, et al. Interleukin-21 is required for the development of type 1 diabetes in NOD mice. Diabetes 2009; 58: 1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Spolski R, Kashyap M, Robinson C, Yu Z, Leonard WJ. IL-21 signaling is critical for the development of type I diabetes in the NOD mouse. Proc. Natl. Acad. Sci. USA 2008; 105: 14028–14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Korn T, Bettelli E, Gao W, Awasthi A, Jäger A, Strom TB, Oukka M, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature 2007; 448: 484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wei L, Laurence A, Elias KM, O’Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J. Biol. Chem. 2007; 282: 34605–34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Van Belle TL, Nierkens S, Arens R, Herrath MG von. Interleukin-21 receptor-mediated signals control autoreactive T cell infiltration in pancreatic islets. Immunity 2012; 36: 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Crotty S. T follicular helper cell biology: A decade of discovery and diseases. Immunity 2019; 50: 1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Ozaki K, Spolski R, Ettinger R, Kim H-P, Wang G, Qi C-F, Hwu P, et al. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J. Immunol. 2004; 173: 5361–5371. [DOI] [PubMed] [Google Scholar]
- 311.Kenefeck R, Wang CJ, Kapadi T, Wardzinski L, Attridge K, Clough LE, Heuts F, et al. Follicular helper T cell signature in type 1 diabetes. J. Clin. Invest. 2015; 125: 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Silva DG, Daley SR, Hogan J, Lee SK, Teh CE, Hu DY, Lam K-P, et al. Anti-islet autoantibodies trigger autoimmune diabetes in the presence of an increased frequency of islet-reactive CD4 T cells. Diabetes 2011; 60: 2102–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Bettini M, Bettini ML. Function, failure, and the future potential of tregs in type 1 diabetes. Diabetes 2021; 70: 1211–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Lio C-WJ, Hsieh C-S. A two-step process for thymic regulatory T cell development. Immunity 2008; 28: 100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Long SA, Cerosaletti K, Wan JY, Ho JC, Tatum M, Wei S, Shilling HG, et al. An autoimmune-associated variant in PTPN2 reveals an impairment of IL-2R signaling in CD4(+) T cells. Genes Immun. 2011; 12: 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Garg G, Tyler JR, Yang JHM, Cutler AJ, Downes K, Pekalski M, Bell GL, et al. Type 1 diabetes-associated IL2RA variation lowers IL-2 signaling and contributes to diminished CD4+CD25+ regulatory T cell function. J. Immunol. 2012; 188: 4644–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Godoy GJ, Olivera C, Paira DA, Salazar FC, Ana Y, Stempin CC, Motrich RD, et al. T Regulatory Cells From Non-obese Diabetic Mice Show Low Responsiveness to IL-2 Stimulation and Exhibit Differential Expression of Anergy-Related and Ubiquitination Factors. Front. Immunol. 2019; 10: 2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Sockolosky JT, Trotta E, Parisi G, Picton L, Su LL, Le AC, Chhabra A, et al. Selective targeting of engineered T cells using orthogonal IL-2 cytokine-receptor complexes. Science 2018; 359: 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Wong J, Mathis D, Benoist C. TCR-based lineage tracing: no evidence for conversion of conventional into regulatory T cells in response to a natural self-antigen in pancreatic islets. J. Exp. Med. 2007; 204: 2039–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Schuster C, Jonas F, Zhao F, Kissler S. Peripherally induced regulatory T cells contribute to the control of autoimmune diabetes in the NOD mouse model. Eur. J. Immunol. 2018; 48: 1211–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 2008; 9: 194–202. [DOI] [PubMed] [Google Scholar]
- 322.Xu L, Kitani A, Stuelten C, McGrady G, Fuss I, Strober W. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity 2010; 33: 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 2012; 482: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Petrich de Marquesini LG, Fu J, Connor KJ, Bishop AJ, McLintock NE, Pope C, Wong FS, et al. IFN-gamma and IL-10 islet-antigen-specific T cell responses in autoantibody-negative first-degree relatives of patients with type 1 diabetes. Diabetologia 2010; 53: 1451–1460. [DOI] [PubMed] [Google Scholar]
- 325.Herold KC, Burton JB, Francois F, Poumian-Ruiz E, Glandt M, Bluestone JA. Activation of human T cells by FcR nonbinding anti-CD3 mAb, hOKT3gamma1(Ala-Ala). J. Clin. Invest. 2003; 111: 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Battaglia M, Stabilini A, Draghici E, Migliavacca B, Gregori S, Bonifacio E, Roncarolo M-G. Induction of tolerance in type 1 diabetes via both CD4+CD25+ T regulatory cells and T regulatory type 1 cells. Diabetes 2006; 55: 1571–1580. [DOI] [PubMed] [Google Scholar]
- 327.Yu H, Gagliani N, Ishigame H, Huber S, Zhu S, Esplugues E, Herold KC, et al. Intestinal type 1 regulatory T cells migrate to periphery to suppress diabetogenic T cells and prevent diabetes development. Proc. Natl. Acad. Sci. USA 2017; 114: 10443–10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Chen C, Lee W-H, Yun P, Snow P, Liu C-P. Induction of autoantigen-specific Th2 and Tr1 regulatory T cells and modulation of autoimmune diabetes. J. Immunol. 2003; 171: 733–744. [DOI] [PubMed] [Google Scholar]
- 329.Panitch HS, Hirsch RL, Haley AS, Johnson KP. Exacerbations of multiple sclerosis in patients treated with gamma interferon. Lancet 1987; 1: 893–895. [DOI] [PubMed] [Google Scholar]
- 330.Olsson T. Cytokines in neuroinflammatory disease: role of myelin autoreactive T cell production of interferon-gamma. J. Neuroimmunol. 1992; 40: 211–218. [DOI] [PubMed] [Google Scholar]
- 331.Segal BM, Dwyer BK, Shevach EM. An interleukin (IL)-10/IL-12 immunoregulatory circuit controls susceptibility to autoimmune disease. J. Exp. Med. 1998; 187: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Becher B, Durell BG, Noelle RJ. Experimental autoimmune encephalitis and inflammation in the absence of interleukin-12. J. Clin. Invest. 2002; 110: 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Chitnis T, Najafian N, Benou C, Salama AD, Grusby MJ, Sayegh MH, Khoury SJ. Effect of targeted disruption of STAT4 and STAT6 on the induction of experimental autoimmune encephalomyelitis. J. Clin. Invest. 2001; 108: 739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Ferber IA, Brocke S, Taylor-Edwards C, Ridgway W, Dinisco C, Steinman L, Dalton D, et al. Mice with a disrupted IFN-gamma gene are susceptible to the induction of experimental autoimmune encephalomyelitis (EAE). J. Immunol. 1996; 156: 5–7. [PubMed] [Google Scholar]
- 335.Krakowski M, Owens T. Interferon-gamma confers resistance to experimental allergic encephalomyelitis. Eur. J. Immunol. 1996; 26: 1641–1646. [DOI] [PubMed] [Google Scholar]
- 336.Willenborg DO, Fordham S, Bernard CC, Cowden WB, Ramshaw IA. IFN-gamma plays a critical down-regulatory role in the induction and effector phase of myelin oligodendrocyte glycoprotein-induced autoimmune encephalomyelitis. J. Immunol. 1996; 157: 3223–3227. [PubMed] [Google Scholar]
- 337.Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, Pflanz S, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 2002; 168: 5699–5708. [DOI] [PubMed] [Google Scholar]
- 338.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 2007; 8: 967–974. [DOI] [PubMed] [Google Scholar]
- 339.Jakkula E, Leppä V, Sulonen A-M, Varilo T, Kallio S, Kemppinen A, Purcell S, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am. J. Hum. Genet. 2010; 86: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D, Wu W, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum. Mol. Genet. 2009; 18: 2078–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Oksenberg JR, Baranzini SE. Multiple sclerosis genetics--is the glass half full, or half empty? Nat. Rev. Neurol. 2010; 6: 429–437. [DOI] [PubMed] [Google Scholar]
- 342.Frisullo G, Angelucci F, Caggiula M, Nociti V, Iorio R, Patanella AK, Sancricca C, et al. pSTAT1, pSTAT3, and T-bet expression in peripheral blood mononuclear cells from relapsing-remitting multiple sclerosis patients correlates with disease activity. J. Neurosci. Res. 2006; 84: 1027–1036. [DOI] [PubMed] [Google Scholar]
- 343.Liu X, Lee YS, Yu C-R, Egwuagu CE. Loss of STAT3 in CD4+ T cells prevents development of experimental autoimmune diseases. J. Immunol. 2008; 180: 6070–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Harris TJ, Grosso JF, Yen H-R, Xin H, Kortylewski M, Albesiano E, Hipkiss EL, et al. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J. Immunol. 2007; 179: 4313–4317. [DOI] [PubMed] [Google Scholar]
- 345.Qin H, Yeh W-I, De Sarno P, Holdbrooks AT, Liu Y, Muldowney MT, Reynolds SL, et al. Signal transducer and activator of transcription-3/suppressor of cytokine signaling-3 (STAT3/SOCS3) axis in myeloid cells regulates neuroinflammation. Proc. Natl. Acad. Sci. USA 2012; 109: 5004–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Qin H, Holdbrooks AT, Liu Y, Reynolds SL, Yanagisawa LL, Benveniste EN. SOCS3 deficiency promotes M1 macrophage polarization and inflammation. J. Immunol. 2012; 189: 3439–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Sonderegger I, Kisielow J, Meier R, King C, Kopf M. IL-21 and IL-21R are not required for development of Th17 cells and autoimmunity in vivo. Eur. J. Immunol. 2008; 38: 1833–1838. [DOI] [PubMed] [Google Scholar]
- 348.Coquet JM, Chakravarti S, Smyth MJ, Godfrey DI. Cutting edge: IL-21 is not essential for Th17 differentiation or experimental autoimmune encephalomyelitis. J. Immunol. 2008; 180: 7097–7101. [DOI] [PubMed] [Google Scholar]
- 349.Meyer Zu Horste G, Wu C, Wang C, Cong L, Pawlak M, Lee Y, Elyaman W, et al. RBPJ Controls Development of Pathogenic Th17 Cells by Regulating IL-23 Receptor Expression. Cell Rep. 2016; 16: 392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 2012; 13: 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Haak S, Croxford AL, Kreymborg K, Heppner FL, Pouly S, Becher B, Waisman A. IL-17A and IL-17F do not contribute vitally to autoimmune neuro-inflammation in mice. J. Clin. Invest. 2009; 119: 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Havrdová E, Belova A, Goloborodko A, Tisserant A, Wright A, Wallstroem E, Garren H, et al. Activity of secukinumab, an anti-IL-17A antibody, on brain lesions in RRMS: results from a randomized, proof-of-concept study. J. Neurol. 2016; 263: 1287–1295. [DOI] [PubMed] [Google Scholar]
- 353.Regen T, Isaac S, Amorim A, Núñez NG, Hauptmann J, Shanmugavadivu A, Klein M, et al. IL-17 controls central nervous system autoimmunity through the intestinal microbiome. Sci. Immunol. 2021; 6: [DOI] [PubMed] [Google Scholar]
- 354.Komuczki J, Tuzlak S, Friebel E, Hartwig T, Spath S, Rosenstiel P, Waisman A, et al. Fate-Mapping of GM-CSF Expression Identifies a Discrete Subset of Inflammation-Driving T Helper Cells Regulated by Cytokines IL-23 and IL-1β. Immunity 2019; 50: 1289–1304.e6. [DOI] [PubMed] [Google Scholar]
- 355.Kohm AP, Carpentier PA, Anger HA, Miller SD. Cutting edge: CD4+CD25+ regulatory T cells suppress antigen-specific autoreactive immune responses and central nervous system inflammation during active experimental autoimmune encephalomyelitis. J. Immunol. 2002; 169: 4712–4716. [DOI] [PubMed] [Google Scholar]
- 356.McGeachy MJ, Stephens LA, Anderton SM. Natural recovery and protection from autoimmune encephalomyelitis: contribution of CD4+CD25+ regulatory cells within the central nervous system. J. Immunol. 2005; 175: 3025–3032. [DOI] [PubMed] [Google Scholar]
- 357.Mekala DJ, Alli RS, Geiger TL. IL-10-dependent infectious tolerance after the treatment of experimental allergic encephalomyelitis with redirected CD4+CD25+ T lymphocytes. Proc. Natl. Acad. Sci. USA 2005; 102: 11817–11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Stephens LA, Malpass KH, Anderton SM. Curing CNS autoimmune disease with myelin-reactive Foxp3+ Treg. Eur. J. Immunol. 2009; 39: 1108–1117. [DOI] [PubMed] [Google Scholar]
- 359.Kimura K. Regulatory T cells in multiple sclerosis. Clin. Exp. Neuroimmunol. 2020; 11: 148–155. [Google Scholar]
- 360.Cerosaletti K, Schneider A, Schwedhelm K, Frank I, Tatum M, Wei S, Whalen E, et al. Multiple autoimmune-associated variants confer decreased IL-2R signaling in CD4+ CD25(hi) T cells of type 1 diabetic and multiple sclerosis patients. PLoS One 2013; 8: e83811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Hartmann FJ, Khademi M, Aram J, Ammann S, Kockum I, Constantinescu C, Gran B, et al. Multiple sclerosis-associated IL2RA polymorphism controls GM-CSF production in human TH cells. Nat. Commun. 2014; 5: 5056. [DOI] [PubMed] [Google Scholar]
- 362.Kimura K, Hohjoh H, Fukuoka M, Sato W, Oki S, Tomi C, Yamaguchi H, et al. Circulating exosomes suppress the induction of regulatory T cells via let-7i in multiple sclerosis. Nat. Commun. 2018; 9: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Severin ME, Lee PW, Liu Y, Selhorst AJ, Gormley MG, Pei W, Yang Y, et al. MicroRNAs targeting TGFβ signalling underlie the regulatory T cell defect in multiple sclerosis. Brain 2016; 139: 1747–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Astier AL, Meiffren G, Freeman S, Hafler DA. Alterations in CD46-mediated Tr1 regulatory T cells in patients with multiple sclerosis. J. Clin. Invest. 2006; 116: 3252–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Moore KW, Waal Malefyt R de, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001; 19: 683–765. [DOI] [PubMed] [Google Scholar]
- 366.Barrat FJ, Cua DJ, Boonstra A, Richards DF, Crain C, Savelkoul HF, Waal-Malefyt R de, et al. In vitro generation of interleukin 10-producing regulatory CD4(+) T cells is induced by immunosuppressive drugs and inhibited by T helper type 1 (Th1)- and Th2-inducing cytokines. J. Exp. Med. 2002; 195: 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Astier AL, Hafler DA. Abnormal Tr1 differentiation in multiple sclerosis. J. Neuroimmunol. 2007; 191: 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Murugaiyan G, Mittal A, Lopez-Diego R, Maier LM, Anderson DE, Weiner HL. IL-27 is a key regulator of IL-10 and IL-17 production by human CD4+ T cells. J. Immunol. 2009; 183: 2435–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Sweeney CM, Lonergan R, Basdeo SA, Kinsella K, Dungan LS, Higgins SC, Kelly PJ, et al. IL-27 mediates the response to IFN-β therapy in multiple sclerosis patients by inhibiting Th17 cells. Brain Behav. Immun. 2011; 25: 1170–1181. [DOI] [PubMed] [Google Scholar]
- 370.Bermel RA, You X, Foulds P, Hyde R, Simon JH, Fisher E, Rudick RA. Predictors of long-term outcome in multiple sclerosis patients treated with interferon β. Ann. Neurol. 2013; 73: 95–103. [DOI] [PubMed] [Google Scholar]
- 371.Schubert RD, Hu Y, Kumar G, Szeto S, Abraham P, Winderl J, Guthridge JM, et al. IFN-β treatment requires B cells for efficacy in neuroautoimmunity. J. Immunol. 2015; 194: 2110–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, Jankovic D, et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect. Immun. 1998; 66: 5157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Ito R, Shin-Ya M, Kishida T, Urano A, Takada R, Sakagami J, Imanishi J, et al. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin. Exp. Immunol. 2006; 146: 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Simpson SJ, Shah S, Comiskey M, Jong YP de, Wang B, Mizoguchi E, Bhan AK, et al. T cell-mediated pathology in two models of experimental colitis depends predominantly on the interleukin 12/Signal transducer and activator of transcription (Stat)-4 pathway, but is not conditional on interferon gamma expression by T cells. J. Exp. Med. 1998; 187: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Shen C, Hertogh G de, Bullens DMA, Van Assche G, Geboes K, Rutgeerts P, Ceuppens JL. Remission-inducing effect of anti-TNF monoclonal antibody in TNBS colitis: mechanisms beyond neutralization? Inflamm. Bowel Dis. 2007; 13: 308–316. [DOI] [PubMed] [Google Scholar]
- 376.Yang Y, Wang H, Dou Y, Wang Y, Han G, Wang R, Wang L, et al. Colitogenic role of tumour necrosis factor (TNF) receptors in trinitrobenzene sulphonic acid colitis: TNF-R1 ablation does not affect systemic inflammatory response. Clin. Exp. Immunol. 2011; 165: 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Ebach DR, Newberry R, Stenson WF. Differential role of tumor necrosis factor receptors in TNBS colitis. Inflamm. Bowel Dis. 2005; 11: 533–540. [DOI] [PubMed] [Google Scholar]
- 378.Stillie R, Stadnyk AW. Role of TNF receptors, TNFR1 and TNFR2, in dextran sodium sulfate-induced colitis. Inflamm. Bowel Dis. 2009; 15: 1515–1525. [DOI] [PubMed] [Google Scholar]
- 379.Silverberg MS, Cho JH, Rioux JD, McGovern DPB, Wu J, Annese V, Achkar J-P, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat. Genet. 2009; 41: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012; 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Andreou N-P, Legaki E, Gazouli M. Inflammatory bowel disease pathobiology: the role of the interferon signature. Ann Gastroenterol 2020; 33: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Ferreira AC, Almeida S, Tavares M, Canedo P, Pereira F, Regalo G, Figueiredo C, et al. NOD2/CARD15 and TNFA, but not IL1B and IL1RN, are associated with Crohn’s disease. Inflamm. Bowel Dis. 2005; 11: 331–339. [DOI] [PubMed] [Google Scholar]
- 383.López-Hernández R, Valdés M, Campillo JA, Martínez-Garcia P, Salama H, Salgado G, Boix F, et al. Genetic polymorphisms of tumour necrosis factor alpha (TNF-α) promoter gene and response to TNF-α inhibitors in Spanish patients with inflammatory bowel disease. Int J Immunogenet 2014; 41: 63–68. [DOI] [PubMed] [Google Scholar]
- 384.Sanchez R, Levy E, Costea F, Sinnett D. IL-10 and TNF-alpha promoter haplotypes are associated with childhood Crohn’s disease location. World J. Gastroenterol. 2009; 15: 3776–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Naderi N, Farnood A, Dadaei T, Habibi M, Balaii H, Firouzi F, Mahban A, et al. Association of Tumor Necrosis Factor Alpha Gene Polymorphisms with Inflammatory Bowel Disease in Iran. Iran. J. Public Health 2014; 43: 630–636. [PMC free article] [PubMed] [Google Scholar]
- 386.Gonsky R, Deem RL, Landers CJ, Haritunians T, Yang S, Targan SR. IFNG rs1861494 polymorphism is associated with IBD disease severity and functional changes in both IFNG methylation and protein secretion. Inflamm. Bowel Dis. 2014; 20: 1794–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Hoey T, Zhang S, Schmidt N, Yu Q, Ramchandani S, Xu X, Naeger LK, et al. Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. EMBO J. 2003; 22: 4237–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.O’Malley JT, Eri RD, Stritesky GL, Mathur AN, Chang H-C, Hogenesch H, Srinivasan M, et al. STAT4 isoforms differentially regulate Th1 cytokine production and the severity of inflammatory bowel disease. J. Immunol. 2008; 181: 5062–5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Jabeen R, Miller L, Yao W, Gupta S, Steiner S, Kaplan MH. Altered STAT4 Isoform Expression in Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015; 21: 2383–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Ivanov II, Frutos R de L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008; 4: 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature 2008; 455: 808–812. [DOI] [PubMed] [Google Scholar]
- 392.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Invest. 2006; 116: 1310–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006; 314: 1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Xu W-D, Xie Q-B, Zhao Y, Liu Y. Association of Interleukin-23 receptor gene polymorphisms with susceptibility to Crohn’s disease: A meta-analysis. Sci. Rep. 2015; 5: 18584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 2010; 33: 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Elson CO, Cong Y, Weaver CT, Schoeb TR, McClanahan TK, Fick RB, Kastelein RA. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology 2007; 132: 2359–2370. [DOI] [PubMed] [Google Scholar]
- 397.Leonard WJ, Wan C-K. IL-21 Signaling in Immunity. [version 1; peer review: 3 approved]. F1000Res. 2016; 5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Pullamsetti SS, Seeger W, Savai R. Classical IL-6 signaling: a promising therapeutic target for pulmonary arterial hypertension. J. Clin. Invest. 2018; 128: 1720–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Pastor-Fernández G, Mariblanca IR, Navarro MN. Decoding IL-23 Signaling Cascade for New Therapeutic Opportunities. Cells 2020; 9: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Cénit MC, Alcina A, Márquez A, Mendoza JL, Díaz-Rubio M, Heras V de las, Izquierdo G, et al. STAT3 locus in inflammatory bowel disease and multiple sclerosis susceptibility. Genes Immun. 2010; 11: 264–268. [DOI] [PubMed] [Google Scholar]
- 401.Zhang J, Wu J, Peng X, Song J, Wang J, Dong W. Associations between STAT3 rs744166 polymorphisms and susceptibility to ulcerative colitis and Crohn’s disease: a meta-analysis. PLoS One 2014; 9: e109625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 2010; 32: 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Ohnmacht C, Park J-H, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 2015; 349: 989–993. [DOI] [PubMed] [Google Scholar]
- 404.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 2015; 349: 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, Föhse L, et al. Foxp3(+) T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2016; 9: 444–457. [DOI] [PubMed] [Google Scholar]
- 406.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 2008; 28: 546–558. [DOI] [PubMed] [Google Scholar]
- 407.Park S-G, Mathur R, Long M, Hosh N, Hao L, Hayden MS, Ghosh S. T regulatory cells maintain intestinal homeostasis by suppressing γδ T cells. Immunity 2010; 33: 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Huber S, Gagliani N, Esplugues E, O’Connor W, Huber FJ, Chaudhry A, Kamanaka M, et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3− and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity 2011; 34: 554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Glocker E-O, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 2009; 361: 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Glocker E-O, Frede N, Perro M, Sebire N, Elawad M, Shah N, Grimbacher B. Infant colitis--it’s in the genes. Lancet 2010; 376: 1272. [DOI] [PubMed] [Google Scholar]
- 411.Begue B, Verdier J, Rieux-Laucat F, Goulet O, Morali A, Canioni D, Hugot J-P, et al. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am. J. Gastroenterol. 2011; 106: 1544–1555. [DOI] [PubMed] [Google Scholar]
- 412.Moran CJ, Walters TD, Guo C-H, Kugathasan S, Klein C, Turner D, Wolters VM, et al. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm. Bowel Dis. 2013; 19: 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich J-M, Jack RS, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity 2011; 34: 566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Hutchins AP, Diez D, Miranda-Saavedra D. The IL-10/STAT3-mediated anti-inflammatory response: recent developments and future challenges. Brief. Funct. Genomics 2013; 12: 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Sun C-M, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 2007; 204: 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Nakamura K, Kitani A, Fuss I, Pedersen A, Harada N, Nawata H, Strober W. TGF-beta 1 plays an important role in the mechanism of CD4+CD25+ regulatory T cell activity in both humans and mice. J. Immunol. 2004; 172: 834–842. [DOI] [PubMed] [Google Scholar]
- 417.Monteleone G, Kumberova A, Croft NM, McKenzie C, Steer HW, MacDonald TT. Blocking Smad7 restores TGF-beta1 signaling in chronic inflammatory bowel disease. J. Clin. Invest. 2001; 108: 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Kamanaka M, Kim ST, Wan YY, Sutterwala FS, Lara-Tejero M, Galán JE, Harhaj E, et al. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity 2006; 25: 941–952. [DOI] [PubMed] [Google Scholar]
- 419.Uhlig HH, Coombes J, Mottet C, Izcue A, Thompson C, Fanger A, Tannapfel A, et al. Characterization of Foxp3+CD4+CD25+ and IL-10-secreting CD4+CD25+ T cells during cure of colitis. J. Immunol. 2006; 177: 5852–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Maxwell LJ, Singh JA. Abatacept for rheumatoid arthritis: a Cochrane systematic review. J. Rheumatol. 2010; 37: 234–245. [DOI] [PubMed] [Google Scholar]
- 421.Mease P, Genovese MC, Gladstein G, Kivitz AJ, Ritchlin C, Tak PP, Wollenhaupt J, et al. Abatacept in the treatment of patients with psoriatic arthritis: results of a six-month, multicenter, randomized, double-blind, placebo-controlled, phase II trial. Arthritis Rheum. 2011; 63: 939–948. [DOI] [PubMed] [Google Scholar]
- 422.Orban T, Bundy B, Becker DJ, DiMeglio LA, Gitelman SE, Goland R, Gottlieb PA, et al. Co-stimulation modulation with abatacept in patients with recent-onset type 1 diabetes: a randomised, double-blind, placebo-controlled trial. Lancet 2011; 378: 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Boumpas DT, Furie R, Manzi S, Illei GG, Wallace DJ, Balow JE, Vaishnaw A, et al. A short course of BG9588 (anti-CD40 ligand antibody) improves serologic activity and decreases hematuria in patients with proliferative lupus glomerulonephritis. Arthritis Rheum. 2003; 48: 719–727. [DOI] [PubMed] [Google Scholar]
- 424.Vazquez-Lombardi R, Phan TG, Zimmermann C, Lowe D, Jermutus L, Christ D. Challenges and opportunities for non-antibody scaffold drugs. Drug Discov. Today 2015; 20: 1271–1283. [DOI] [PubMed] [Google Scholar]
- 425.Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O’Shea JJ. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat. Rev. Drug Discov. 2017; 16: 843–862. [DOI] [PubMed] [Google Scholar]
- 426.Monaco C, Nanchahal J, Taylor P, Feldmann M. Anti-TNF therapy: past, present and future. Int. Immunol. 2015; 27: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Feldmann M, Brennan FM, Maini RN. Role of cytokines in rheumatoid arthritis. Annu. Rev. Immunol. 1996; 14: 397–440. [DOI] [PubMed] [Google Scholar]
- 428.Taylor PC, Peters AM, Paleolog E, Chapman PT, Elliott MJ, McCloskey R, Feldmann M, et al. Reduction of chemokine levels and leukocyte traffic to joints by tumor necrosis factor alpha blockade in patients with rheumatoid arthritis. Arthritis Rheum. 2000; 43: 38–47. [DOI] [PubMed] [Google Scholar]
- 429.Feagan BG, Sandborn WJ, D’Haens G, Panés J, Kaser A, Ferrante M, Louis E, et al. Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe Crohn’s disease: a randomised, double-blind, placebo-controlled phase 2 study. Lancet 2017; 389: 1699–1709. [DOI] [PubMed] [Google Scholar]
- 430.Tamilarasan AG, Cunningham G, Irving PM, Samaan MA. Recent advances in monoclonal antibody therapy in IBD: practical issues. Frontline Gastroenterol. 2019; 10: 409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Neurath MF. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat. Immunol. 2019; 20: 970–979. [DOI] [PubMed] [Google Scholar]
- 432.Eftychi C, Schwarzer R, Vlantis K, Wachsmuth L, Basic M, Wagle P, Neurath MF, et al. Temporally Distinct Functions of the Cytokines IL-12 and IL-23 Drive Chronic Colon Inflammation in Response to Intestinal Barrier Impairment. Immunity 2019; 51: 367–380.e4. [DOI] [PubMed] [Google Scholar]
- 433.Xie X, Li F, Chen J-W, Wang J. Risk of tuberculosis infection in anti-TNF-α biological therapy: from bench to bedside. J Microbiol Immunol Infect 2014; 47: 268–274. [DOI] [PubMed] [Google Scholar]
- 434.TNF neutralization in MS: results of a randomized, placebo-controlled multicenter study. The Lenercept Multiple Sclerosis Study Group and The University of British Columbia MS/MRI Analysis Group. Neurology 1999; 53: 457–465. [PubMed] [Google Scholar]
- 435.Oosten BW van, Barkhof F, Truyen L, Boringa JB, Bertelsmann FW, Blomberg BM von, Woody JN, et al. Increased MRI activity and immune activation in two multiple sclerosis patients treated with the monoclonal anti-tumor necrosis factor antibody cA2. Neurology 1996; 47: 1531–1534. [DOI] [PubMed] [Google Scholar]
- 436.Rosenblum MD, Gratz IK, Paw JS, Abbas AK. Treating human autoimmunity: current practice and future prospects. Sci. Transl. Med. 2012; 4: 125sr1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martínez-Llordella M, Ashby M, Nakayama M, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 2009; 10: 1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Chen Q, Kim YC, Laurence A, Punkosdy GA, Shevach EM. IL-2 controls the stability of Foxp3 expression in TGF-beta-induced Foxp3+ T cells in vivo. J. Immunol. 2011; 186: 6329–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-hora M, Kodama T, Tanaka S, et al. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat. Med. 2014; 20: 62–68. [DOI] [PubMed] [Google Scholar]
- 440.Desreumaux P, Foussat A, Allez M, Beaugerie L, Hébuterne X, Bouhnik Y, Nachury M, et al. Safety and efficacy of antigen-specific regulatory T-cell therapy for patients with refractory Crohn’s disease. Gastroenterology 2012; 143: 1207–1217.e2. [DOI] [PubMed] [Google Scholar]
- 441.Domogalla MP, Rostan PV, Raker VK, Steinbrink K. Tolerance through Education: How Tolerogenic Dendritic Cells Shape Immunity. Front. Immunol. 2017; 8: 1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Serra P, Santamaria P. Antigen-specific therapeutic approaches for autoimmunity. Nat. Biotechnol. 2019; 37: 238–251. [DOI] [PubMed] [Google Scholar]
- 443.Phillips BE, Garciafigueroa Y, Trucco M, Giannoukakis N. Clinical tolerogenic dendritic cells: exploring therapeutic impact on human autoimmune disease. Front. Immunol. 2017; 8: 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Clemente-Casares X, Blanco J, Ambalavanan P, Yamanouchi J, Singha S, Fandos C, Tsai S, et al. Expanding antigen-specific regulatory networks to treat autoimmunity. Nature 2016; 530: 434–440. [DOI] [PubMed] [Google Scholar]
- 445.Green EA, Choi Y, Flavell RA. Pancreatic lymph node-derived CD4(+)CD25(+) Treg cells: highly potent regulators of diabetes that require TRANCE-RANK signals. Immunity 2002; 16: 183–191. [DOI] [PubMed] [Google Scholar]
- 446.Tang Q, Henriksen KJ, Bi M, Finger EB, Szot G, Ye J, Masteller EL, et al. In vitro-expanded antigen-specific regulatory T cells suppress autoimmune diabetes. J. Exp. Med. 2004; 199: 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.De Simone M, Rossetti G, Pagani M. Single cell T cell receptor sequencing: techniques and future challenges. Front. Immunol. 2018; 9: 1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Newick K, O’Brien S, Moon E, Albelda SM. CAR T cell therapy for solid tumors. Annu. Rev. Med. 2017; 68: 139–152. [DOI] [PubMed] [Google Scholar]
- 449.Mohseni YR, Tung SL, Dudreuilh C, Lechler RI, Fruhwirth GO, Lombardi G. The future of regulatory T cell therapy: promises and challenges of implementing CAR technology. Front. Immunol. 2020; 11: 1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.MacDonald KG, Hoeppli RE, Huang Q, Gillies J, Luciani DS, Orban PC, Broady R, et al. Alloantigen-specific regulatory T cells generated with a chimeric antigen receptor. J. Clin. Invest. 2016; 126: 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Noyan F, Zimmermann K, Hardtke-Wolenski M, Knoefel A, Schulde E, Geffers R, Hust M, et al. Prevention of Allograft Rejection by Use of Regulatory T Cells With an MHC-Specific Chimeric Antigen Receptor. Am. J. Transplant. 2017; 17: 917–930. [DOI] [PubMed] [Google Scholar]
- 452.Boroughs AC, Larson RC, Choi BD, Bouffard AA, Riley LS, Schiferle E, Kulkarni AS, et al. Chimeric antigen receptor costimulation domains modulate human regulatory T cell function. JCI Insight 2019; 5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Eggenhuizen PJ, Ng BH, Ooi JD. Treg enhancing therapies to treat autoimmune diseases. Int. J. Mol. Sci. 2020; 21: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Clough JN, Omer OS, Tasker S, Lord GM, Irving PM. Regulatory T-cell therapy in Crohn’s disease: challenges and advances. Gut 2020; 69: 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Rosenzwajg M, Churlaud G, Mallone R, Six A, Dérian N, Chaara W, Lorenzon R, et al. Low-dose interleukin-2 fosters a dose-dependent regulatory T cell tuned milieu in T1D patients. J. Autoimmun. 2015; 58: 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Todd JA, Evangelou M, Cutler AJ, Pekalski ML, Walker NM, Stevens HE, Porter L, et al. Regulatory T Cell Responses in Participants with Type 1 Diabetes after a Single Dose of Interleukin-2: A Non-Randomised, Open Label, Adaptive Dose-Finding Trial. PLoS Med. 2016; 13: e1002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.He J, Zhang X, Wei Y, Sun X, Chen Y, Deng J, Jin Y, et al. Low-dose interleukin-2 treatment selectively modulates CD4(+) T cell subsets in patients with systemic lupus erythematosus. Nat. Med. 2016; 22: 991–993. [DOI] [PubMed] [Google Scholar]
- 458.Trotta E, Bessette PH, Silveria SL, Ely LK, Jude KM, Le DT, Holst CR, et al. A human anti-IL-2 antibody that potentiates regulatory T cells by a structure-based mechanism. Nat. Med. 2018; 24: 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Hirai T, Ramos TL, Lin P-Y, Simonetta F, Su LL, Picton LK, Baker J, et al. Selective expansion of regulatory T cells using an orthogonal IL-2/IL-2 receptor system facilitates transplantation tolerance. J. Clin. Invest. 2021; 131: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]