Abstract
Several oncogenic mechanisms have been identified for MET, including MET amplification, fusions, mutations in the tyrosine kinase domain and exon 14 skipping alterations. MET exon 14 mutations are found in about 3–5% of non-small-cell lung cancers. Dysregulation of the MET receptor leads to cell proliferation and survival by activation of the PI3K–AKT–TOR and RAS–RAF–MET–ERK canonical pathways. Targeting the MET tyrosine kinase domain in the setting of MET exon 14 mutations using effective MET tyrosine kinase inhibitors is a current targeted therapy option for patients with metastatic lung cancer. In this Review, we focus on the management of patients with MET exon 14 skipping alterations by addressing the biology of the MET receptor and exon 14 skipping mutations, current treatment strategies, and sequential treatment options based on resistance mechanisms to MET inhibitors in patients with non-small-cell lung cancer.
Keywords: acquired resistance, exon 14 skipping mutations, MET, next-generation sequencing, non-small-cell lung cancer, target therapy
Introduction
The MET oncogene encodes for a transmembrane tyrosine kinase receptor (RTK) that regulates physiological processes such as cell scattering during embryogenesis, wound healing, angiogenesis and proliferation.1–3 MET oncogenic alterations have been found in multiple tumours, including lung cancer, gastric cancer, head and neck squamous cell carcinoma, childhood hepatocellular carcinoma, and colorectal cancer.4–8 Overall, MET alterations are found in 2.6% of all human cancers, reaching up to 5% in non-small-cell lung cancer (NSCLC).9 Unlike other RTKs, alterations in MET affect more than one oncogenic mechanism, including point mutations in the tyrosine kinase domain, MET exon 14 skipping, MET amplifications, overexpression and fusions.10 Interestingly, MET alterations have been described not only as the primary driver alteration in tumour cells but also as an acquired resistance mechanism to tyrosine kinase inhibitors in EGFR-mutant or ALK-rearranged lung cancer.11–14 Identification of these mechanisms has led to the development of multiple therapeutic strategies targeting MET, including specific tyrosine kinase inhibitors (TKIs) that are now approved for the treatment of MET-driven NSCLC, such as tepotinib15 and capmatinib,16 and monoclonal antibodies.17–19
Herein, we review the management of patients with tumours that harbour one of the main oncogenic mechanisms found in NSCLC, namely MET exon 14 skipping alterations. We focus on the biology of MET and the oncogenic mechanisms of MET exon 14-driven tumours, the diagnostic methods, clinical characteristics and actual strategies in targeting MET, including resistance mechanisms, in this tumour model.
Review
MET exon 14 skipping alterations
The MET proto-oncogene encodes a tyrosine kinase receptor composed of an extracellular portion that contains the SEMA domain that binds to its ligand hepatocyte growth factor (HGF), a single-pass transmembrane region, and an intracellular tyrosine kinase domain. The receptor tyrosine kinase domain contains a catalytic domain that is activated through the phosphorylation of key residues such as Y1234-Y1235 and Y1349-Y1356. Following this, multiple signalling effectors, including SH2, GRB2 and STAT3, bind to MET docking sites, are activated and, subsequently, modulate intracellular canonical signalling pathways such as PI3K–AKT–TOR and RAS–RAF–MET–ERK.17 Several genomic alterations can lead to ligand-independent MET oncogenic activation, including receptor tyrosine kinase domain mutations, MET amplification, fusions, MET overexpression and exon 14 skipping alterations.10,20
To comprehend the oncogenic role of MET exon 14 skipping alterations, firstly, it is important to overview how the MET juxtamembrane domain regulates MET signalling physiologically. The juxtamembrane domain of the MET receptor includes the tyrosine residue in codon 1003, encoded in the exon 14 of MET. Upon MET activation, the phosphorylated Y1003 serves as a binding site for E3 ubiquitin-protein ligase (c-CBL). c-CBL induces MET ubiquitination, leading to receptor internalization and degradation.21,22 Consequently, MET degradation results in lower levels of total cellular MET levels and serves as a normal negative regulator of MET signalling in normal cells.
There are multiple genomic alterations that hamper c-CBL binding to the Y1003 residue, including mutations or deletions in MET exon 14 splicing regulatory sites leading to MET exon 14 skipping during mRNA splicing, MET exon 14 partial or complete deletions, and point mutations in Y1003. In a comprehensive genome profiling from 38,028 tumour samples, Frampton et al. identified 224 distinct MET exon 14 alterations, including base substitutions and indels at the splice acceptor sites, splice donor sites and in the non-coding intronic sequences immediately adjacent to the splice acceptor site.23 Most recently, in a large genomic database of 1599 tumours with MET exon 14 alterations, deletions were most frequently found to affect the splicing acceptor site (41%) in intron 13 whereas base substitution mutations were the most frequent alteration found at the splicing donor site (48%) of intron 14.23,24 The multitude of genomic events converge in the occurrence of an alternative splicing that excludes exon 14 at the mRNA level and, therefore, in the absence of the juxtamembrane domain protein synthesis during mRNA translation.
Co-occurring genomic alterations are frequent in cancers with MET exon 14 skipping. High MET amplification can be found in 11% of patients with MET exon 14 skipping mutations,4 which increases the level of expression of MET exon 14 mutant alleles. In addition, TP53 mutations (43%), MDM2 (34%) and CDK4/6 amplification (19%), and CDKN2A/B loss (20–26%) are also common events in MET exon 14 skipping tumours; however, the impact of these co-alterations in clinical outcomes is unknown.25 Moreover, other known driver mutations in genes such as EGFR, KRAS, ERBB2, ALK, ROS1 or RET have been described but are rarely found in patients with MET exon 14-mutant lung cancer.4,23 Additionally, tumours from patients with MET exon 14 skipping alterations have higher levels of PD-L1 and a low tumour mutational burden.25 A study addressing PD-L1 expression by immunohistochemistry in MET exon 14-mutant lung cancers found that 41% of patients had PD-L1 expression of >50% (high), 22% had PD-L1 expression of 1–49% (intermediate) and 37% of cases had an expression of 0% (negative).26
MET exon 14 alterations are found in about 3–5% of tumours from patients with metastatic NSCLC, most commonly in lung adenocarcinomas, but also found in adenosquamous and squamous cell carcinomas.4,23 In addition to lung adenocarcinomas, squamous cell carcinomas and adenosquamous carcinomas, pleomorphic or sarcomatoid lung cancers are particularly enriched in MET exon 14 skipping alterations, with reports ranging from 8% to 22% of cases.27–30 Sarcomatoid lung tumours are a more aggressive lung cancer subtype and have a dismal prognosis compared to epithelial lung cancers;31 hence, targeting MET in this population can provide therapeutic benefits for patients.
Patients with MET exon 14-mutant NSCLC tend to be of older age with a median age of 72 years, and this is significantly higher compared to patients with other driver alterations such as EGFR or KRAS mutations (median 72.5 years versus 61 and 65 years, respectively; p<0.001).4,27 Additionally, there is a higher proportion of women with MET exon 14-mutated cancers (56–68%), and about half of the patients with this molecular subtype of cancer have a history of smoking exposure.4,27,32
Before the development of MET-targeted therapies in this setting, patients with MET exon 14 skipping-mutant lung cancer seemed to have a poorer prognosis compared to historical survival data. The median overall survival of patients with MET exon 14-mutant NSCLC that did not receive MET selected therapy nor immunotherapy in retrospective studies ranged from 7 to 9.5 months.29,32,33 Furthermore, in a small subset analysis that included 20 patients with MET exon 14 skipping mutations and 6 patients with a co-occurring MET amplification, overall survival was inferior in patients with tumours that harbour the co-alteration (median overall survival of 5.2 months (95% CI 2.3 to not reached (NR)) versus 10.5 months (95% CI 5.3 to NR)).32 Therefore, historically, there was a high unmet need to improve clinical outcomes in patients with lung cancer that harbour MET exon 14 alterations. Currently, with the increasing access to next-generation sequencing (NGS) as a molecular diagnostic tool and the development of MET targeted therapies, the treatment landscape of this subgroup of patients has changed.
Molecular diagnosis of MET exon 14 skipping alterations
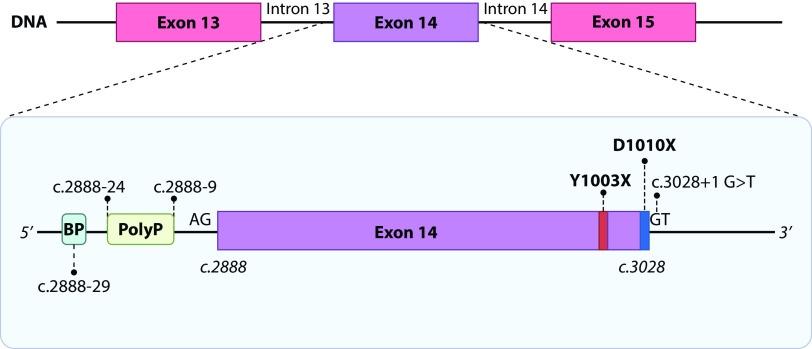
Genomic alterations leading to MET exon 14 skipping as well as whole MET exon 14 deletions and MET Y1003 point mutations can be detected using NGS platforms. DNA-based NGS gene panels can detect genomic alterations that are known to confer MET exon 14 skipping by analysing the MET exon 14 sequence and flanking key intronic regions.34 Some examples of DNA NGS platforms include Foundation Medicine in tissue and plasma samples, Guardant360 and Inivata in plasma,35 MSK-IMPACT NGS panel,36 and the OncoPanel at the Dana-Farber Cancer Institute37 in tissue samples, amongst others.27,38 MET exon 14 skipping at the mRNA level is often a consequence of insertion/deletions (indels) and base substitutions in the polypyrimidine tract in intron 13, the splice acceptor site in intron 13, the D1010 codon in exon 14 and the splice donor site in exon 14 (Figure 1).23 The first codon in exon 14 is c.2888, which codifies for codon D963 taking as a reference the MET transcript variant 2 (NM_000245.2) or D981 if taking as a reference MET transcript variant 1 (NM_001127500.2).39 It is relevant to check what MET variant is used to align and nomenclate MET alterations as it may lead to confusion in clinical practice.
Figure 1.
MET exon 14 alterations, diagram including exon 14 and flanking introns 13 and 14.
BP, branching point; PolyP, polypyrimidine tract.
Most intronic alterations that affect the polypyrimidine tract and the splice acceptor sites at intron 13 are indels. As an example, to illustrate the reporting nomenclature to aid in the interpretation of this report, the ‘MET c.2888-35,2888-17del’ refers to a deletion starting at intronic base 35 counting backwards from c.2888 to the intronic base 17.25 Genomic deletions at the DNA level affecting these regions in intron 13 may also include a portion of exon 14.
At the other end of exon 14, the last base is in position c.3028; most mutations affecting the splicing donor site of intron 14 are point mutations with base substitutions but indels can also be found (Figure 1). In this case, one of the most common mutations is a base substitution from a guanine in position c.3028+1 to another base.4 This affects the splicing donor site identified with the bases GT in intron 14. Point mutations affecting the last codon of exon 14 (D1010 referencing MET transcript 2 or D1028 in MET transcript 1) is also a common alteration that confers MET exon skipping. Some of these mutations include c.3028G>A (D1010N (V2) D1028N (V1)), c.3028G>C (D1010H (V2) or D1028H (V1)), and c.3028G>T (D1010Y (V2) or D1028Y (V1)). Aside from point mutations in D1010 or base substitutions in the intron 14 splice donor site c.3028+1, deletions involving one or both loci also cause MET exon 14 skipping. DNA-based NGS can also detect point mutations in Y1003 (V2), also referred as Y1021 (V1), that change the tyrosine residue for another amino acid, impeding c-CBL binding.40,41 This alteration can be found using DNA-based and RNA-based NGS testing with exon 14 coverage but does not confer MET exon 14 skipping. In summary, DNA-based NGS panels are very informative of a diverse range of genomic alterations that confer MET exon 14 skipping or an equivalent biological effect; however, interpretation of these results can be challenging. Nevertheless, in most cases, NGS reports will clarify that the genomic alteration at the DNA level confers MET exon 14 skipping. The FDA has recently approved the FoundationOne CDx and FoundationOne Liquid CDx assays in tissue and plasma samples, respectively, as companion diagnostics for the MET inhibitor capmatinib.16
RNA-based NGS assays have a higher accuracy to detect MET exon 14 skipping than DNA-based NGS platforms.42,43 RNA NGS panels are designed to interrogate the loss of exon 14 at the mRNA level, often reported as an ‘intragenic fusion’ between MET exon 13 and exon 15 (M13:M15), reflecting MET exon 14 skipping. Independently from the underlying genomic alteration, RNA-based NGS will detect MET exon 14 skipping using RNA extracted from the tumour sample. RNA quality is an important limitation to perform this type of testing, which mandates the guarantee of optimal preanalytical conditions to process the sample.44 The Archer MET RNA-based NGS platform has been approved in Japan to detect MET exon 14 skipping in tissue and plasma samples as a companion diagnostic for the MET TKI tepotinib. Other commercial NGS panel tests interrogate both at the DNA and RNA level, like Caris.45
Some NGS platforms combine DNA-based and RNA-based NGS panels in the same tissue sample, like the Oncomine Focus Assay15 (ThermoFisher) as tested in the tepotinib VISION trial. Real-time PCR assays using RNA from tissue samples are currently in development; however, they have not yet been accepted or qualified to become companion diagnostic assays.46
Because of exon 14 skipping mutations, MET receptor internalization and degradation are impaired; hence, hypothetically, MET protein overexpression could be observed in MET exon 14-mutant cells. Guo et al.47 addressed this issue by analysing MET protein expression by immunohistochemistry in samples from MET exon 14-mutant lung cancers. MET-high expression (defined as MET H-score ≥200) was observed in 13 of 22 samples (59%) and was associated with a higher overall response rate (ORR) when compared to lower H-scores (ORR 62% for H-score ≥200, 25% for H-score 150–199 and 33% for H-score 1–149) as well as with a longer progression-free survival (PFS) for MET-high tumours as compared to MET-low tumours. The median PFS was longer in patients with tumours that have an H-score ≥200 than in those with tumours with an H-score <200 (10.4 versus 5.5 months, respectively; HR 3.87; p=0.02).47 Nevertheless, MET immunohistochemistry is not a reliable surrogate for MET exon 14 skipping.48
Targeting MET as a primary oncogenic driver
A variety of molecules targeting MET are currently approved for the treatment of patients in the clinical setting and in development in clinical trials. There are two MET TKIs approved in this setting, capmatinib and tepotinib. MET TKIs are classified according to their binding properties to the tyrosine kinase domain as type I inhibitors, which bind the ATP-binding pocket in its active conformation (crizotinib, capmatinib, tepotinib and savolitinib), and type II inhibitors, which bind the ATP-binding pocket in its inactive state (cabozantinib, merestinib, glesatinib).10,39 Type I inhibitors are subclassified in type Ia inhibitors, which interact with the G1163 solvent front residue (crizotinib), or type Ib inhibitors, which do not interact with the G1163 residue (capmatinib, tepotinib, savolitinib).
Crizotinib is an ALK, ROS1 and MET inhibitor currently approved as a standard treatment for patients with ROS1-rearranged NSCLC and is the first ALK inhibitor approved to treat patients with ALK-rearranged lung cancer. Crizotinib does not have regulatory approval for the treatment of patients with MET exon 14 lung cancers. However, crizotinib was the first drug to show promising activity against MET exon 14-mutant NSCLC and is currently being used off-label in settings where tepotinib and capmatinib are not approved nor commercially available. In the phase I PROFILE 1001 study, 69 patients with MET exon 14 alterations, of whom 38% had not received prior treatment for advanced disease, were included. The ORR with crizotinib was 32% (95% CI 21–45), the median duration of response (DOR) was 9.1 months (95% CI 6.4–12.7) and median PFS was 7.3 months (95% CI 5.4–9.1). The median overall survival with crizotinib was 20.5 months (95% CI 14.3–21.8)49 (Table 1).
Table 1.
Clinical trials including MET exon 14 skipping mutant NSCLC treated with specific MET tyrosine kinase inhibitors.
| Trial and drug | Trial design (phase, endpoints, number of patients) | Outcomes (ORR, median PFS and median OS) | All-grade toxicities | Refs. |
|---|---|---|---|---|
| GEOMETRY mono-1, capmatinib |
|
|
Pre-treated/Naive
|
16,50 |
| VISION, tepotinib |
|
|
|
15,51 |
| NCT02897479, savolitinib |
|
|
|
52 |
| PROFILE 1001, crizotinib |
|
|
|
49 |
| METROS, crizotinib |
|
|
|
66 |
DOR, duration of response; mo, months; ORR, overall response rate; OS, overall survival; PFS, progression-free survival; TTR, time to response.
Capmatinib is a standard treatment for patients with MET exon 14-mutant lung cancers recently approved by the FDA and Japan based on the results of the GEOMETRY mono-1 study. This multi-arm trial design included previously treated and treatment-naive patients with MET amplifications and MET exon 14 skipping mutations. Globally, the study enrolled 160 patients with MET exon 14 mutant NSCLC who received capmatinib at a dose of 400 mg BID. The updated overall response in previously treated patients with MET exon 14 skipping mutations was 44% (95% CI 34–54) (cohorts 4 and 6) and in treatment-naive patients it was 66.7% (95% CI 53–78) (cohorts 5b and 7). The median DOR in both cohorts was 9.7 and 12.6 months and median PFS was 5.5 months (95% CI 4.2–8.1) and 12.3 months (95% CI 8.2–21.6), respectively. Median overall survival was 13.6 months (95% CI 8.2–22.2) and 20.8 (95% CI 12.4 to not estimable) for previously treated and treatment-naive patients, respectively.16,50
Tepotinib is also approved for the treatment of patients with advanced MET exon 14-mutant lung cancer based on the results of the phase II VISION trial, which also included previously treated and treatment-naive patients. Amongst 152 patients with MET exon 14 skipping mutations enrolled and included in the efficacy analysis, the objective response rate was 44.7% (95% CI 36.7–53), with a median DOR of 11.1 months (95% CI 8.4–18.5) and a median PFS of 8.9 months (95% CI 8.3–11.2). There were no statistical differences in the outcomes of patients regarding the line of treatment, with an ORR of 44.9% (95% CI 32.9–57.4) and median PFS of 8.5 months (95% CI 6.8–11.3) in treatment-naive patients and an ORR of 44.6% (95% CI 33.7–55.9) and median PFS of 10.9 (95% CI 8.2–12.7) in previously treated patients.15,51 A summary of MET TKI trials is provided in Table 1.
Both capmatinib and tepotinib show high response rates and PFS benefit for patients in this molecular subtype; however, the impact of the line of therapy in which MET TKIs are prescribed remains to be further explored based on the discordant results from these trials. Considering the proven benefits of immunotherapy and chemoimmunotherapy options in the front-line treatment setting for patients with metastatic NSCLC, the optimal treatment sequencing strategy is still debated.
The selective and potent MET inhibitor savolitinib has shown activity in patients with exon 14-mutant lung cancer (n=70), including a large proportion of patients with pulmonary sarcomatoid tumours (36%). Globally, the ORR was 49.2% (95% CI 36.1–62.3), with a median DOR of 8.3 months (95% CI 5.3–16.6) and a median PFS of 6.8 months (95% CI 4.2–9.6). In patients with sarcomatoid tumours, the objective response rate was 40.0% (95% CI 21.1–61.3), with a median DOR of 17.9 months (95% CI 4.1–not estimable) and a median PFS of 5.5 months (95% CI 2.8–6.9).52 Savolitinib is approved in China for the treatment of patients with MET exon 14-mutant lung cancer.
Capmatinib and tepotinib showed similar safety profiles, with peripheral oedemas and nausea being the most frequent adverse effects in both trials. Grade 3 or 4 adverse effects occurred in 67% of patients from all cohorts in the capmatinib trial and in 28% of patients in the tepotinib trial. Treatment-related adverse events led to dose reduction in 33% of patients and to permanent discontinuation in 11% of patients in the tepotinib trial, with the values being 23% and 11%, respectively, in the capmatinib trial. Adverse effects were effectively reverted after drug discontinuation or dose reduction.15,16 A summary of related adverse effects is shown in Table 1.
Cabozantinib is a multi-kinase inhibitor active against VEGFR2, RET, c-KIT and a MET type II inhibitor and has shown preclinical activity against MET both in vitro and in vivo, including activity against point mutations Y1248C/H, D1246N and K1262R in the kinase domain.53 However, there is scarce data supporting the use of type II inhibitors, such as cabozantinib, in patients that experience disease progression with type I MET TKIs given case reports of an acquired MET D1228V mutation.54,55 The CABinMET clinical trial (NCT03911193) is currently recruiting patients to evaluate the efficacy in patients with MET-deregulated NSCLC. Other molecules, such as glesatinib, have shown promising preclinical activity56 but clinical development was discontinued (NCT02954991). Results from a phase II trial examining the efficacy of the multitarget inhibitor merestinib are still pending (NCT02920996).
Currently, there is a number of novel therapeutic agents, including monoclonal antibodies directed against the SEMA domain and antibody–drug conjugates designed to target MET, under development. Sym015 is a combination of two monoclonal antibodies against non-overlapping epitopes of MET that was tested both in patients with MET exon 14-mutant (n=12) and MET amplified (n=8) NSCLC, including treatment-naive patients and patients previously treated with MET TKIs. ORR for the MET exon 14 treatment-naive cohort was 100% with a median DOR of 6.5 months and a median PFS of 9.2 months. A lesser response rate was seen in the previously treated population.57 Amivantamab is a bispecific antibody targeting EGFR-MET approved by the FDA for the treatment of patients with EGFR exon 20-mutant NSCLC. The Chrysalis trial included a cohort of 19 patients with MET exon 14-mutant NSCLC, both pre-treated (n=17) and untreated. A partial response was seen in 64% of patients and median DOR was not reached at data cut-off.58 The Chrysalis trial is currently recruiting patients (NCT02609776).
In January 2022, the antibody–drug conjugate telisotuzumab vedotin was granted ‘Breakthrough Therapy Designation’ for the treatment of patients with previously treated NSCLC with high MET overexpression defined as H-score of ≥150 based on still unpublished preliminary data from the phase II LUMINOSITY trial (NCT03539536). Data from the phase I trial, which included 52 patients, showed an objective response of 23% with median DOR of 8.7 months and a median PFS of 5.2 months.17 Given the different mechanism of action of these agents compared to MET TKIs, in the future, combination or sequential treatment strategies will be needed to overcome acquired resistance to first-line-specific treatment.
Consistently across all trials evaluating MET TKIs in patients with MET exon 14 skipping-mutant NSCLC, peripheral oedema (51–75% of patients) and nausea (26–53%) were the most common adverse effects. Grade 3 or 4 adverse effects occurred in 27–67% of patients, with oedema being the most common across all trials. Serious adverse events occurred in 15–24% of patients, including pleural effusion and pneumonia15,16,52 (Table 1).
Resistance to MET inhibitors
As seen with other target therapies, tumours treated with MET inhibitors eventually develop resistance through a variety of on-target and off-target molecular mechanisms. In a series of 20 patients with MET exon 14-mutated NSCLC who were treated with MET TKIs, samples from tissue and/or plasma were analysed using NGS (Dana Farber Cancer Institute OncoPanel assay) and Guardant360 panels, respectively, at the time of progression with MET TKIs. Resistance mechanisms were identified in 75% of cases. On-target resistance mechanisms included secondary point mutations in the MET kinase domain (G1163R, D1228H/N, Y1230C/H/S, L1195V, D1228N, H1094Y, L195V) as well as amplification of the MET exon 14-mutant allele. On the other hand, off-target mechanisms included EGFR, HER3, KRAS and BRAF amplifications and activating mutations in KRAS.59
In another study, on-target resistance mechanisms were identified in 20% (3/15) of tissue samples from patients with acquired resistance to MET inhibitors. These mechanisms included MET D1228N point mutation (two cases) and a case with HGF amplification. Off-target acquired resistance mechanisms were found in 33% of cases, including EGFR and KRAS amplifications and KRAS G12S and RASA1 S742* mutations. KRAS G13V mutation was identified in a plasma sample from 1 patient (1/11).47
MET resistance mechanisms were also described when targeting MET amplification as a resistance mechanism to erlotinib in a patient with EGFR-mutant lung cancer upon treatment with osimertinib and savolitinib combined. Plasma NGS at the time of disease progression showed several acquired MET mutations (D1228H, D1228N, D1228Y, Y1230C) in the MET-amplified allele, suggesting multiclonal MET-driven resistance.60 Given the differential binding properties of type I and type II MET TKIs, resistance by acquired point mutations in the tyrosine kinase domain can be potentially overcome by switching to a different type of MET TKI. This strategy has been reported in several case reports, with patients achieving both stable disease and partial responses when switching from type Ib to type II inhibitors, such as cabozantinib or merestinib, in the setting of acquired MET mutations59–61 and a partial response in a patient switching from a type II to a type Ia inhibitor in the setting of an acquired MET mutant-exon 14 allele amplification.59 Given the biological rationale for this approach, there is an unmet need to conduct clinical trials that can shed a light on the optimal therapeutic sequencing strategy tailored according to resistance mechanisms.
The role of immunotherapy in MET exon 14-driven tumours
There is a high grade of heterogeneity in cancer immunological responses to anti-PD-1/PD-L1 inhibitors according to distinct molecular subtypes in patients with NSCLC. Patients with EGFR-mutant, ALK-, ROS1- and RET-rearranged lung cancers do not seem to benefit from treatment with immunotherapy. On the contrary, patients with KRAS or BRAF mutant lung cancers may achieve high response rates and prolonged benefits with immune-checkpoint inhibitors.62,63 As previously mentioned, MET exon 14-mutant lung cancers express higher levels of PD-L1 and have low tumour mutational burden compared to other mutant-driven lung cancers; however, the role of immunotherapy treatment in patients with MET exon 14-mutant lung cancer is yet to be well defined. In a series of 147 patients with MET exon 14-mutant NSCLC, 24 patients received immune checkpoint inhibitors, 11 as first-line treatment, 6 patients in the second-line and 7 patients in the third-line setting. The overall response rate was 17% (95% CI 6–36), with a median PFS of 1.9 months (95% CI 1.7–2.7) and a median overall survival of 18.2 months (95% CI 12.9 to NR).26 In another series of 46 patients treated with immunotherapy, there were no significant differences in outcomes of patients with MET exon 14-mutant NSCLC treated with immunotherapy according to previous treatment status with MET TKIs.32
On the other hand, in the Immunotarget International Registry, which included 23 patients with MET exon 14 skipping-mutant NSCLC treated with immunotherapy, median overall survival for this subgroup (25 months; 95% CI 18.4 to NR) was significantly longer than other molecular subgroups.63 Moreover, data from a case series of 13 patients with MET exon 14-mutant lung cancer treated with immunotherapy showed that 6 (46%) patients experienced prolonged PFS ranging from 18 to 49 months.64
The current treatment landscape of patients with MET exon 14-mutant metastatic lung cancer includes selective MET inhibitors, chemotherapy and immunotherapy. The efficacy and safety of the combination of capmatinib and nivolumab have been addressed in a phase II clinical trial including previously treated patients, showing an ORR of 25% for MET high patients with a median PFS of 13.8 (95% CI 3.5–19.2) months. Serious adverse effects were reported in 24% of patients.65
Given the current data, starting with a selective MET inhibitor is, most likely, the preferred option when available; however, at the moment, immunotherapy could also be considered at some point in the treatment of this patient population.
Conclusion
MET exon 14 skipping alterations are a novel therapeutic target in patients with NSCLC. MET exon 14 skipping alterations can be identified using NGS both in tissue or blood samples or real-time PCR assays. As comprehensive molecular testing becomes more available, there is a need to improve the interpretation of NGS testing results to correctly identify patients that can benefit from MET-targeted therapies. There are several drugs in development to continue to build on the current standard therapies, aiming to effectively target MET in lung and other cancer types.
Acknowledgements
Figure 1 was created with biorender.com.
Footnotes
Contributions: JBB and GR both contributed equally to the drafting, writing and correction of the manuscript. All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole and have given their approval for this version to be published.
Disclosure and potential conflicts of interest: JBB reports educational grants from Amgen and Pfizer, GR reports grants from Amgen and Janssen, personal fees/advisory boards from Amgen, Bayer, BMS, Merck Serono, MSD, Pfizer, Roche and Takeda. GR is also a Partner at PxMedica, and reports clinical trials from Amgen, AstraZeneca, Bayer, Janssen and Roche. The International Committee of Medical Journal Editors (ICMJE) Potential Conflicts of Interests form for the authors is available for download at: https://www.drugsincontext.com/wp-content/uploads/2022/05/dic.2022-2-2-COI.pdf
Funding declaration: There was no funding associated with the preparation of this article.
Correct attribution: Copyright © 2022 Blaquier JB, Recondo G. https://doi.org/10.7573/dic.2022-2-2. Published by Drugs in Context under Creative Commons License Deed CC BY NC ND 4.0.
Article URL: https://www.drugsincontext.com/non-small-cell-lung-cancer-how-to-manage-met-exon-14-skipping-mutant-disease
Provenance: Invited; externally peer reviewed.
Drugs in Context is published by BioExcel Publishing Ltd. Registered office: Plaza Building, Lee High Road, London, England, SE13 5PT.
BioExcel Publishing Limited is registered in England Number 10038393. VAT GB 252 7720 07.
For all manuscript and submissions enquiries, contact the Editorial office editorial@drugsincontext.com
For all permissions, rights and reprints, contact David Hughes david.hughes@bioexcelpublishing.com
References
- 1.Zhu H, Naujokas MA, Park M. Receptor chimeras indicate that the met tyrosine kinase mediates the motility and morphogenic responses of hepatocyte growth/scatter factor. Cell Growth Differ. 1994;5(4):359–366. [PubMed] [Google Scholar]
- 2.Bussolino F, Di Renzo MF, Ziche M, et al. Hepatocyte growth factor is a potent angiogenic factor which stimulates endothelial cell motility and growth. J Cell Biol. 1992;119(3):629–641. doi: 10.1083/jcb.119.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You WK, McDonald DM. The hepatocyte growth factor/c-Met signaling pathway as a therapeutic target to inhibit angiogenesis. BMB Rep. 2008;41(12):833–839. doi: 10.5483/bmbrep.2008.41.12.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awad MM, Oxnard GR, Jackman DM, et al. MET Exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-met overexpression. J Clin Oncol. 2016;34(7):721–730. doi: 10.1200/JCO.2015.63.4600. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima M, Sawada H, Yamada Y, et al. The prognostic significance of amplification and overexpression of c-met and c-erb B-2 in human gastric carcinomas. Cancer. 1999;85(9):1894–1902. doi: 10.1002/(sici)1097-0142(19990501)85:9<1894::aid-cncr3>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 6.Di Renzo MF, Olivero M, Martone T, et al. Somatic mutations of the MET oncogene are selected during metastatic spread of human HNSC carcinomas. Oncogene. 2000;19(12):1547–1555. doi: 10.1038/sj.onc.1203455. [DOI] [PubMed] [Google Scholar]
- 7.Park WS, Dong SM, Kim SY, et al. Somatic mutations in the kinase domain of the Met/hepatocyte growth factor receptor gene in childhood hepatocellular carcinomas. Cancer Res. 1999;59(2):307–310. [PubMed] [Google Scholar]
- 8.Umeki K, Shiota G, Kawasaki H. Clinical significance of c-met oncogene alterations in human colorectal cancer. Oncology. 1999;56(4):314–321. doi: 10.1159/000011985. [DOI] [PubMed] [Google Scholar]
- 9.AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo R, Luo J, Chang J, Rekhtman N, Arcila M, Drilon A. MET-dependent solid tumours - molecular diagnosis and targeted therapy. Nat Rev Clin Oncol. 2020;17(9):569–587. doi: 10.1038/s41571-020-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci. 2007;104(52):20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura T, Nakashima C, Komiya K, et al. Mechanisms of acquired resistance to afatinib clarified with liquid biopsy. PLoS ONE. 2018;13(12):e0209384. doi: 10.1371/journal.pone.0209384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramalingam SS, Cheng Y, Zhou C, et al. Mechanisms of acquired resistance to first-line osimertinib: Preliminary data from the phase III FLAURA study. Ann Oncol. 2018;29:viii740. doi: 10.1093/annonc/mdy424.063. [DOI] [Google Scholar]
- 14.Dagogo-Jack I, Yoda S, Lennerz JK, et al. MET Alterations are a recurring and actionable resistance mechanism in ALK-positive lung cancer. Clin Cancer Res. 2020;26(11):2535–2545. doi: 10.1158/1078-0432.CCR-19-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paik PK, Felip E, Veillon R, et al. Tepotinib in non-small-cell lung cancer with MET exon 14 skipping mutations. N Engl J Med. 2020;383(10):931–943. doi: 10.1056/NEJMoa2004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf J, Seto T, Han JY, et al. Capmatinib in MET exon 14-mutated or MET-amplified non-small-cell lung cancer. N Engl J Med. 2020;383(10):944–957. doi: 10.1056/NEJMoa2002787. [DOI] [PubMed] [Google Scholar]
- 17.Camidge DR, Morgensztern D, Heist RS, et al. Phase I study of 2- or 3-week dosing of telisotuzumab vedotin, an antibody–drug conjugate targeting c-Met, monotherapy in patients with advanced non–small cell lung carcinoma. Clin Cancer Res. 2021;27(21):5781–5792. doi: 10.1158/1078-0432.CCR-21-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poulsen TT, Grandal MM, Skartved NJØ, et al. Sym015: A highly efficacious antibody mixture against MET-amplified tumors. Clin Cancer Res. 2017;23(19):5923–5935. doi: 10.1158/1078-0432.CCR-17-0782. [DOI] [PubMed] [Google Scholar]
- 19.Neijssen J, Cardoso RMF, Chevalier KM, et al. Discovery of amivantamab (JNJ-61186372), a bispecific antibody targeting EGFR and MET. J Biol Chem. 2021;296:100641. doi: 10.1016/j.jbc.2021.100641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16(1):68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 21.Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011;3(1 Suppl):S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peschard P, Ishiyama N, Lin T, Lipkowitz S, Park M. A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain binding site required for suppression of oncogenic activation. J Biol Chem. 2004;279(28):29565–29571. doi: 10.1074/jbc.M403954200. [DOI] [PubMed] [Google Scholar]
- 23.Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5(8):850–859. doi: 10.1158/2159-8290.CD-15-0285. [DOI] [PubMed] [Google Scholar]
- 24.Cortot AB, Kherrouche Z, Descarpentries C, et al. Exon 14 deleted MET receptor as a new biomarker and target in cancers. J Natl Cancer Inst. 2017;109(5):djw262. doi: 10.1093/jnci/djw262. [DOI] [PubMed] [Google Scholar]
- 25.Lee JK, Madison R, Classon A, et al. Characterization of non–small-cell lung cancers with MET exon 14 skipping alterations detected in tissue or liquid: clinicogenomics and real-world treatment patterns. JCO Precis Oncol. 2021;5:1354–1376. doi: 10.1200/PO.21.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabari JK, Leonardi GC, Shu CA, et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Ann Oncol. 2018;29(10):2085–2091. doi: 10.1093/annonc/mdy334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrock AB, Frampton GM, Suh J, et al. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J Thorac Oncol. 2016;11(9):1493–1502. doi: 10.1016/j.jtho.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Jia Y, Stoopler MB, et al. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J Clin Oncol. 2016;34(8):794–802. doi: 10.1200/JCO.2015.62.0674. [DOI] [PubMed] [Google Scholar]
- 29.Tong JH, Yeung SF, Chan AWH, et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of non–small cell lung carcinoma with poor prognosis. Clin Cancer Res. 2016;22(12):3048–3056. doi: 10.1158/1078-0432.CCR-15-2061. [DOI] [PubMed] [Google Scholar]
- 30.Yu Y, Zhang Q, Zhang J, Lu S. Prevalence of MET exon 14 skipping mutation in pulmonary sarcomatoid carcinoma patients without common targetable mutations: a single-institute study. J Cancer Res Ther. 2019;15(4):909. doi: 10.4103/jcrt.JCRT_591_18. [DOI] [PubMed] [Google Scholar]
- 31.Sharma A, Contreras E, Sandoval K, Nicholson L, Waalen J, Bhangoo M. 223P Sarcomatoid lung carcinoma: an uncommon and deadly entity. J Thorac Oncol. 2018;13(4):S134. doi: 10.1016/S1556-0864(18)30495-7. [DOI] [Google Scholar]
- 32.Awad MM, Leonardi GC, Kravets S, et al. Impact of MET inhibitors on survival among patients with non-small cell lung cancer harboring MET exon 14 mutations: a retrospective analysis. Lung Cancer. 2019;133:96–102. doi: 10.1016/j.lungcan.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hur JY, Ku BM, Shim JH, et al. Characteristics and clinical outcomes of non-small cell lung cancer patients in Korea with MET exon 14 skipping. In Vivo. 2020;34(3):1399–1406. doi: 10.21873/invivo.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pruis MA, Geurts-Giele WRR, von der TJH, et al. Highly accurate DNA-based detection and treatment results of MET exon 14 skipping mutations in lung cancer. Lung Cancer. 2020;140:46–54. doi: 10.1016/j.lungcan.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Gale D, Lawson ARJ, Howarth K, et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS ONE. 2018;13(3):e0194630. doi: 10.1371/journal.pone.0194630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1(19):e87062. doi: 10.1172/jci.insight.87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le X, Hong L, Hensel C, et al. Landscape and clonal dominance of co-occurring genomic alterations in non–small-cell lung cancer harboring MET exon 14 skipping. JCO Precis Oncol. 2021;5:1802–1812. doi: 10.1200/PO.21.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Recondo G, Che J, Jänne PA, Awad MM. Targeting MET dysregulation in cancer. Cancer Discov. 2020;10(7):922–934. doi: 10.1158/2159-8290.CD-19-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao YL, Xu QQ. MET Y1003S point mutation shows sensitivity to crizotinib in a patient with lung adenocarcinoma. Lung Cancer. 2019;130:84–86. doi: 10.1016/j.lungcan.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66(1):283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 42.Jurkiewicz M, Saqi A, Mansukhani MM, et al. Efficacy of DNA versus RNA NGS-based Methods in MET Exon 14 skipping mutation detection. J Clin Oncol. 2020;38(Suppl 15):9036–9036. doi: 10.1200/JCO.2020.38.15_suppl.9036. [DOI] [Google Scholar]
- 43.Davies KD, Lomboy A, Lawrence CA, et al. DNA-based versus RNA-based detection of MET exon 14 skipping events in lung cancer. J Thorac Oncol. 2019;14(4):737–741. doi: 10.1016/j.jtho.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 44.Heydt C, Wölwer CB, Velazquez Camacho O, et al. Detection of gene fusions using targeted next-generation sequencing: a comparative evaluation. BMC Med Genomics. 2021;14(1):62. doi: 10.1186/s12920-021-00909-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spizzo G, Siebert U, Gastl G, et al. Cost-comparison analysis of a multiplatform tumour profiling service to guide advanced cancer treatment. Cost Eff Resour Alloc. 2019;17(1):23. doi: 10.1186/s12962-019-0191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Y, Li G, Ju K, et al. Mesenchymal-epithelial transition exon 14 skipping mutation and amplification in 5,008 patients with lung cancer. Front Oncol. 2021;11:755031. doi: 10.3389/fonc.2021.755031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo R, Offin M, Brannon AR, et al. MET exon 14–altered lung cancers and MET inhibitor resistance. Clin Cancer Res. 2021;27(3):799–806. doi: 10.1158/1078-0432.CCR-20-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo R, Berry LD, Aisner DL, et al. MET IHC is a poor screen for MET amplification or MET exon 14 mutations in lung adenocarcinomas: data from a tri-institutional cohort of the lung cancer mutation consortium. J Thorac Oncol. 2019;14(9):1666–1671. doi: 10.1016/j.jtho.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drilon A, Clark JW, Weiss J, et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat Med. 2020;26(1):47–51. doi: 10.1038/s41591-019-0716-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf J, Garon EB, Groen HJM, et al. Capmatinib in MET exon 14-mutated, advanced NSCLC: updated results from the GEOMETRY mono-1 study. J Clin Oncol. 2021;39(Suppl 15):9020. doi: 10.1200/JCO.2021.39.15_suppl.9020. [DOI] [Google Scholar]
- 51.Paik P, Sakai H, Felip E, et al. MA11.05 tepotinib in patients with MET exon 14 (METex14) skipping advanced NSCLC: updated efficacy results from VISION cohort A. J Thorac Oncol. 2021;16(3):S174. doi: 10.1016/j.jtho.2021.01.250. [DOI] [Google Scholar]
- 52.Lu S, Fang J, Li X, et al. Once-daily savolitinib in Chinese patients with pulmonary sarcomatoid carcinomas and other non-small-cell lung cancers harbouring MET exon 14 skipping alterations: a multicentre, single-arm, open-label, phase 2 study. Lancet Respir Med. 2021;9(10):1154–1164. doi: 10.1016/S2213-2600(21)00084-9. [DOI] [PubMed] [Google Scholar]
- 53.Yakes FM, Chen J, Tan J, et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol Cancer Ther. 2011;10(12):2298–2308. doi: 10.1158/1535-7163.MCT-11-0264. [DOI] [PubMed] [Google Scholar]
- 54.Wang SXY, Zhang BM, Wakelee HA, et al. Case series of MET exon 14 skipping mutation-positive non-small-cell lung cancers with response to crizotinib and cabozantinib. Anti-Cancer Drugs. 2019;30(5):537–541. doi: 10.1097/CAD.0000000000000765. [DOI] [PubMed] [Google Scholar]
- 55.Bahcall M, Sim T, Paweletz CP, et al. Acquired MET D1228V mutation and resistance to MET inhibition in lung cancer. Cancer Discov. 2016;6(12):1334–1341. doi: 10.1158/2159-8290.CD-16-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engstrom LD, Aranda R, Lee M, et al. Glesatinib exhibits antitumor activity in lung cancer models and patients harboring MET exon 14 mutations and overcomes mutation-mediated resistance to type I MET inhibitors in nonclinical models. Clin Cancer Res. 2017;23(21):6661–6672. doi: 10.1158/1078-0432.CCR-17-1192. [DOI] [PubMed] [Google Scholar]
- 57.Camidge DR, Janku F, Martinez-Bueno A, et al. Safety and preliminary clinical activity of the MET antibody mixture, Sym015 in advanced non-small cell lung cancer (NSCLC) patients with MET amplification/exon 14 deletion (METAmp/Ex14) J Clin Oncol. 2020;38(Suppl 15):9510. doi: 10.1200/JCO.2020.38.15_suppl.9510. [DOI] [Google Scholar]
- 58.Spira A, Krebs M, Cho BC, et al. OA15.03 Amivantamab in non-small cell lung cancer (NSCLC) with MET exon 14 skipping (METex14) mutation: initial results from CHRYSALIS. J Thorac Oncol. 2021;16(10):S874–S875. doi: 10.1016/j.jtho.2021.08.084. [DOI] [Google Scholar]
- 59.Recondo G, Bahcall M, Spurr LF, et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14–mutant NSCLC. Clin Cancer Res. 2020;26(11):2615–2625. doi: 10.1158/1078-0432.CCR-19-3608. [DOI] [PubMed] [Google Scholar]
- 60.Piper-Vallillo AJ, Halbert BT, Rangachari D, Kobayashi SS, Costa DB. Acquired resistance to osimertinib plus savolitinib is mediated by MET-D1228 and MET-Y1230 mutations in EGFR-mutated MET-amplified lung cancer. JTO Clin Res Rep. 2020;1(4):100071. doi: 10.1016/j.jtocrr.2020.100071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pruis MA, Paats MS, Geurts WRR, Dubbink HJ, Dingemans AMC. Overcoming acquired resistance mutation MET D1228N to crizotinib with cabozantinib in NSCLC with MET exon 14 skipping mutation. JCO Precis Oncol. 2021;5:849–853. doi: 10.1200/PO.21.00076. [DOI] [PubMed] [Google Scholar]
- 62.Lee CK, Man J, Lord S, et al. Clinical and molecular characteristics associated with survival among patients treated with checkpoint inhibitors for advanced non–small cell lung carcinoma. JAMA Oncol. 2018;4(2):210. doi: 10.1001/jamaoncol.2017.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol. 2019;30(8):1321–1328. doi: 10.1093/annonc/mdz167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mayenga M, Assié JB, Monnet I, et al. Durable responses to immunotherapy of non-small cell lung cancers harboring MET exon-14–skipping mutation: A series of 6 cases. Lung Cancer. 2020;150:21–25. doi: 10.1016/j.lungcan.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 65.Felip E, Minotti V, Tan D, et al. P76.03 Efficacy and safety of capmatinib plus nivolumab in pretreated patients with EGFR wild-type non–small cell lung cancer. J Thorac Oncol. 2021;16(3):S585–S586. doi: 10.1016/j.jtho.2021.01.1060. [DOI] [Google Scholar]
- 66.Landi L, Chiari R, Tiseo M, et al. Crizotinib in MET -deregulated or ROS1 -rearranged pretreated non–small cell lung cancer (METROS): a phase II, prospective, multicenter, two-arms trial. Clin Cancer Res. 2019;25(24):7312–7319. doi: 10.1158/1078-0432.CCR-19-0994. [DOI] [PubMed] [Google Scholar]