Abstract
Cerebral small vessel disease is a leading cause of stroke and a major contributor to cognitive decline and dementia, but our understanding of specific genes underlying the cause of sporadic cerebral small vessel disease is limited. We report a genome-wide association study and a whole-exome association study on a composite extreme phenotype of cerebral small vessel disease derived from its most common MRI features: white matter hyperintensities and lacunes. Seventeen population-based cohorts of older persons with MRI measurements and genome-wide genotyping (n = 41 326), whole-exome sequencing (n = 15 965), or exome chip (n = 5249) data contributed 13 776 and 7079 extreme small vessel disease samples for the genome-wide association study and whole-exome association study, respectively. The genome-wide association study identified significant association of common variants in 11 loci with extreme small vessel disease, of which the chr12q24.11 locus was not previously reported to be associated with any MRI marker of cerebral small vessel disease. The whole-exome association study identified significant associations of extreme small vessel disease with common variants in the 5′ UTR region of EFEMP1 (chr2p16.1) and one probably damaging common missense variant in TRIM47 (chr17q25.1). Mendelian randomization supports the causal association of extensive small vessel disease severity with increased risk of stroke and Alzheimer’s disease. Combined evidence from summary-based Mendelian randomization studies and profiling of human loss-of-function allele carriers showed an inverse relation between TRIM47 expression in the brain and blood vessels and extensive small vessel disease severity. We observed significant enrichment of Trim47 in isolated brain vessel preparations compared to total brain fraction in mice, in line with the literature showing Trim47 enrichment in brain endothelial cells at single cell level. Functional evaluation of TRIM47 by small interfering RNAs-mediated knockdown in human brain endothelial cells showed increased endothelial permeability, an important hallmark of cerebral small vessel disease pathology. Overall, our comprehensive gene-mapping study and preliminary functional evaluation suggests a putative role of TRIM47 in the pathophysiology of cerebral small vessel disease, making it an important candidate for extensive in vivo explorations and future translational work.
Keywords: cerebral small vessel disease, endothelial cells, GWAS, TRIM47, whole-exome association study
In a comprehensive gene-mapping study on a composite extreme phenotype of cerebral small vessel disease, Mishra et al. identify 11 risk loci, including the novel locus chr12q24.11, and show that TRIM47 at the chr17q25.1 locus is a strong candidate for future functional studies.
Introduction
Cerebral small vessel disease encompasses a group of pathological processes affecting small arteries, arterioles, capillaries and small veins in the brain. It is one of the main causes of stroke, representing a quarter to a third of stroke cases, importantly both ischaemic and haemorrhagic stroke.1,2 Cerebral small vessel disease features on brain MRI have been associated with an increased risk of dementia, including of the Alzheimer type, and with accelerated cognitive decline.2 In fact, the vast majority of cognitive impairment and dementia cases in the community are now largely recognized as due to a mix of neurodegenerative processes and vascular brain injury, of which cerebral small vessel disease is by far the most important substrate.3,4 No specific mechanistic treatments are available for cerebral small vessel disease to date.
Cerebral small vessel disease is a common condition driven by a complex mix of environmental and genetic determinants, the two main known risk factors being age and hypertension.2 Deciphering the genetic determinants of cerebral small vessel disease could provide novel insights into the biological pathways underlying cerebral small vessel disease, thus contributing to accelerating the discovery of drug targets. MRI markers of covert cerebral small vessel disease, including burden of white matter hyperintensities (WMH) and lacunes of presumed vascular origin (or covert small subcortical brain infarcts), are commonly used for its diagnosis and assessment of severity.5 They are frequently observed on brain MRI scans of older persons in the general population and were shown to be highly heritable.6,7
To date, genome-wide association studies (GWAS) of WMH burden have reported 33 linkage disequilibrium (LD) independent variants in 30 risk loci that are >1 Mb away from each other.8-13 So far, no genome-wide significant association has been identified with lacunes.14,15 In a recent pilot study we demonstrated the usefulness of a new gene-mapping strategy to identify common and rare cerebral small vessel disease risk variants using a composite extreme cerebral small vessel disease phenotype (extreme-SVD), derived from extreme distributions of WMH burden and presence or absence of lacunes, applied to whole-exome sequencing (WES).16 We hypothesize that applying this strategy on a larger scale, using genotypes derived from both genome-wide genotyping arrays and next-generation sequencing, will enable the discovery of novel genetic risk loci for cerebral small vessel disease and point to loci associated with a composite, possibly more specific cerebral small vessel disease phenotype than individual MRI markers of small vessel disease.
Transitioning from genetic risk locus discovery to the identification of affected gene(s) underlying complex cerebral small vessel disease pathophysiology is another major challenge. A number of strategies were proposed to identify causal associations of molecular traits with complex diseases, such as tissue specific gene expression profiles.17,18 The summary-based Mendelian randomization (SMR) approach for instance integrates GWAS and expression quantitative trait loci (eQTL) data identified from a given tissue to identify potential functionally relevant genes at the genome-wide significant risk loci of a given complex phenotype.17 However, differentiating true causal associations from pleiotropic ones still remains challenging. We hypothesize that combining information from the SMR approach with (i) screening of potential human homozygous or heterozygous knockouts (zero or only one functional copy of a given gene) using WES data; and with (ii) exploring the impact of small interfering RNAs (siRNA)-mediated knockdown of putative candidate gene(s) on cellular phenotypes of relevance for cerebral small vessel disease might help identify the best candidate genes to be prioritized for further functional exploration of extreme-SVD risk loci, with the goal of identifying putative biotargets for this condition.
We aimed, first, to report a multi-cohort GWAS and whole-exome association study (WEAS) on MRI-derived extreme-SVD and to leverage these results to explore the clinical significance of extreme-SVD using Mendelian randomization (MR). Second, we aimed to screen for causal genes underlying observed associations with extreme-SVD, in order to facilitate the identification of putative biotargets by: (i) combining GWAS results with information from brain and blood vessel eQTL and screening of human exomes for loss of function allele carriers; and (ii) conducting in vitro experiments to determine the impact of candidate gene knockdown on endothelial cell permeability.
Materials and methods
Study population
Seventeen population-based cohorts of European and African-American ancestries with brain MRI and genome-wide genotyping (n = 41 326, nEuropean = 39 584), WES (n = 16 251, nEuropean = 15 525), or exome chip (EC; n = 5298, nEuropean = 4265) data from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium19 and the UK Biobank participated in this study contributing 13 776 (nEuropean = 13 196) and 7079 (nEuropean = 6525) extreme-SVD samples for the GWAS and WEAS, respectively. Study design, MRI measurements, genotyping and WES in individual cohorts are described in the Supplementary material. The local research and medical ethics committees approved individual studies. All participants signed an informed consent.
Definition of extremes of cerebral small vessel disease
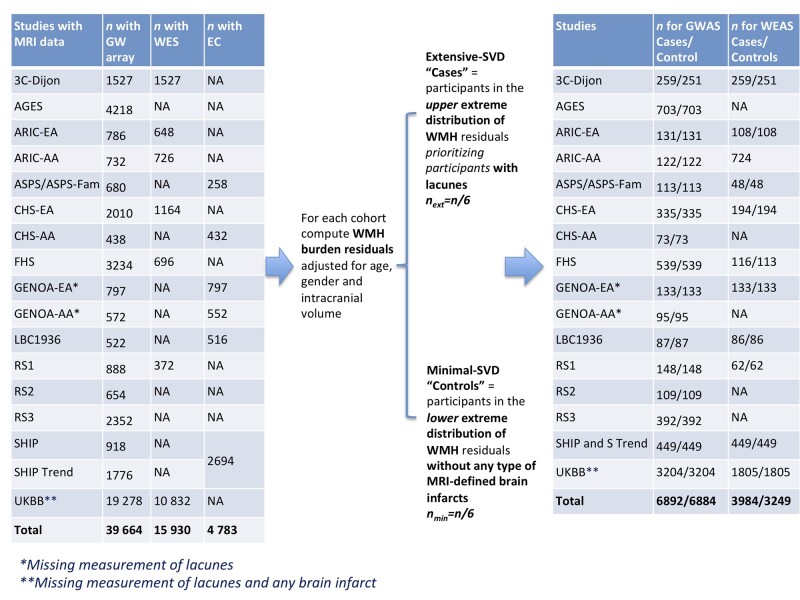
We defined extremes of cerebral small vessel disease using information on the distribution of WMH burden and presence or absence of lacunes in individual cohorts, as described previously.16 Before generating extreme-SVD, individual cohorts removed ancestry outliers, participants with a history of stroke or other pathologies that may influence the measurement of WMH (e.g. brain tumour, head trauma, etc.) at the time of MRI and participants with missing data for age, sex or intracranial volume. Out of the total considered sample with MRI data (n), n/3 participants were defined as extreme-SVD: n/6 as extensive-SVD ‘cases’ and n/6 as minimal-SVD ‘controls’. In each cohort we first computed WMH burden residuals by log-transforming the WMH measurements (natural log of [WMH burden + 1]) and extracted WMH burden residuals adjusting for age, gender and intracranial volume. We defined extensive-SVD participants as those in the top quartile of WMH burden residuals, first selecting participants who also had lacunes and then out of the remaining participants selecting those with the largest WMH burden residuals until reaching n/6. We defined minimal-SVD participants as those in the bottom quartile of WMH burden residuals who did not have any type of MRI-defined brain infarct (including lacunes and other types of covert MRI-defined brain infarcts) and had WMH burden residuals at the bottom tail of the distribution, in order to have a control group with as little vascular brain injury as possible. Definitions of lacunes and MRI-defined brain infarcts in each cohort are provided in the Supplementary material. Due to unavailable measurement of lacunes and other MRI-defined brain infarcts in the UK Biobank we defined extreme-SVD using only WMH burden residuals adjusted for age, gender and intracranial volume in this study sample. Similarly, in the GENOA cohort we defined extreme-SVD using information of only WMH burden residuals and brain infarcts, as lacunes were not assessed in this cohort. The strategy used to define extreme-SVD is presented in Fig. 1.
Figure 1.
Contributing studies and defined number of extreme-SVD cases and controls for GWAS and WEAS.
Common variant association testing
Each cohort performed the extreme-SVD ‘cases versus controls’ GWAS using logistic regression under an additive model adjusting for age, sex (RS-I, RS-II, RS-III, SHIP, SHIP-Trend), when relevant principal components of population stratification (3C-Dijon, AGES, ASPS, LBC1936 and UK Biobank), family structure (FHS, GENOA-AA and GENOA-EA) and/or study site (ARIC-AA, ARIC-EA, CHS-AA and CHS-EA). Fig. 1 reports the number of extreme-SVD cases and controls defined in each cohort that underwent association testing. The quality control (QC) of genotypes and imputation methods of individual cohorts are presented in the Supplementary material. Briefly, sample-specific quality control filters on heterozygosity, call rate, gender mismatch, cryptic relatedness and analysis of principal components for population stratification and ancestral outliers as well as single nucleotide polymorphism (SNP)-level QC on genotyping call rate and Hardy–Weinberg equilibrium were applied prior to the imputation. The high-quality samples and SNPs underwent imputation using Haplotype Reference Consortium or a combined UK10K and Haplotype Reference Consortium or 1000 genomes phase 1 version 3 (1000Gp1v3) reference panels.
The R package EasyQC along with in-house custom harmonization scripts were used to perform the QC of individual GWAS summary results.20 We removed variants with minor allele frequency (MAF) less than 1%, imputation quality score less than 0.50 and effective allele count (= 2 × number of cases × MAF × imputation quality score) less than 10.
We conducted inverse-variance weighted fixed-effects meta-analyses using the METAL software21 for ancestry-specific meta-analyses (European and African-American) followed by combined meta-analysis of ancestry-specific results. We applied the ‘genomic inflation’ correction option to all input files. We removed variants with a heterogeneity P < 0.001 using Cochran’s Q-test from ancestry-specific meta-analyses. Moreover, we kept variants that were present in more than or equal to 50% of participating studies (>7 European studies and >2 African-American studies). Overall, 6 075 887 and 6 197 308 genetic variants were kept from the European only and trans-ethnic GWAS meta-analyses, respectively.
Follow-up post-GWAS analyses, namely conditional and joint analysis, gene-based tests of common variant associations, SNP-heritability and partition SNP-heritability analyses are described in the Supplementary material.
Whole-exome sequencing and exome-chip studies
The WES and EC data processing of individual cohorts is described in the Supplementary material. We performed pooled meta-analyses of single variant tests and SKAT-O gene-based tests using the R package SeqMeta (https://cran.r-project.org/web/packages/seqMeta/index.html, accessed 10 December 2019). Individual population-based cohorts (3C-Dijon, ASPS_Fam, ARIC-AA, ARIC-EA, CHS-AA, CHS-EA, LBC1936, RS-I, RS-II, RS-III, SHIP, SHIP-Trend and UK Biobank) generated ‘prepScores’ of individual variants using a binomial regression adjusting for age, sex and if relevant principal components of population stratification and study site using the SeqMeta software, whereas family-based cohorts (FHS, GENOA-AA, GENOA-EA) generated ‘prepScores’ of individual variants using a binomial regression adjusting for age, sex, family structure and principal components of population stratification, using the RVfam software.22 The number of extreme-SVD cases and controls defined in each cohort that underwent WEAS are reported in Fig. 1. For single variant association testing, we removed variants with minor allele count less than six across all participants. Overall, 1 687 160 variants underwent single variant association testing. We used the SKAT-O approach,23 for gene-based analyses of protein-modifying (splice acceptor, splice donor, start lost, stop lost, stop gained, frameshift, inframe insertion, inframe deletion and missense) rare and low-frequency variants. We considered genes with a cumulative MAF of rare or low frequency protein-modifying variants higher than 0.1. We separately performed meta-analyses of WES and EC studies, and combined WES and EC association P-values using Stouffer’s method for sample size weighted combination of P-values.
We used the polyphen-2 web-server24 to query the protein-damaging effect of extreme-SVD associated missense variant (http://genetics.bwh.harvard.edu/pph2/; accessed 25 February 2022).
We adopted the resampling strategy implemented in the VEGAS2Pathway software25 to perform pathway analyses of SKAT-O gene-based P-values of protein-modifying rare and low-frequency variants. We analysed 9981 pathways from the NCBI BioSystems database26 comprising a minimum of 10 and a maximum of 1000 genes.
Mendelian randomization analysis
Using the sentinel variants at genome-wide or gene-wide significant common variant risk loci of extreme-SVD we performed two-sample MR tests to identify potential causal association of extreme-SVD with late-onset Alzheimer’s disease (n = 71 880/383 378)27 and stroke including any stroke (n = 40 585/406 111),28 any ischaemic stroke (n = 34 217/404 630),28 ischaemic stroke subtypes comprising small vessel stroke (n = 5386),28 large artery stroke (n = 4373)28 and cardioembolic stroke (n = 7193)28 and intracerebral haemorrhage (n = 1545/1481).29 We considered the P-value threshold of 7.14 × 10−3 of significant association testing correcting for seven traits analysed. The MR analysis was performed considering effect estimates in European ancestry samples only (refer to the Supplementary material for details).
SMR/HEIDI association testing
We performed an SMR test17 using the brain and blood vessel eQTL data to identify the most likely causal genes at extreme-SVD risk loci (Supplementary material). SMR tests whether the transcription level of a gene in the brain or blood vessel is associated with risk of extreme-SVD using top brain or blood vessel eQTL of a given gene as an instrument. The SMR software extracted 7440 and 7812 probes with at least one brain and blood vessel eQTL with P-value < 5 × 10−8; hence, we used a P-value < 3.28 × 10−6 [0.05/(7440 + 7812)] as a multiple testing corrected threshold for SMR associations. Utilizing the LD structure at individual SMR associations, the heterogeneity in dependent instruments (HEIDI) test distinguish linkage-based associations from pleiotropic or causal ones by testing whether the SMR effect estimates differ between top-eQTL for a given probe and set of variants in LD with it (Supplementary material).17 The significance threshold of HEIDI P-value < 0.05 was considered to identify linkage-based SMR associations. For the SMR and HEIDI analyses we used European ancestry-specific extreme-SVD GWAS meta-analysis summary statistics and the Haplotype Reference Consortium imputed data of 6489 participants of European ancestry from the 3C-study30 (a French population cohort, Supplementary material) to compute LD between variants.
Screening of loss-of-function variant carrier
To identify human gene knockouts we screened participants carrying loss-of-function allele genotypes in recently released UK Biobank WES data (Supplementary material) processed using the Functionally Equivalent protocol,31 who also underwent brain MRI and had available information on age, gender, hypertension status at the time of MRI (defined as SBP > 140 mmHg and DBP >90 mmHg or antihypertensive treatment), intracranial volume, WMH volume (n = 10 832). We used Loss-Of-Function Transcript Effect Estimator (LOFTEE) plugin in variant effect predictor software to identify high-confidence loss-of-function stop gain, frame shift and splicing variants.32 Participants carrying a loss-of-function allele in candidate genes were also screened for ClinVar annotated pathogenic or likely pathogenic mutations for familial cerebral small vessel disease (Supplementary Table 15). We also searched for NOTCH3 EGFr domain cysteine-modifying missense variants, the typical type of mutation causing cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy, (CADASIL, MIM: 125310).33
Expression profile of mice homologues of extreme-SVD putative causal genes
Male C57BL/6J mice aged 2–16 months were used in accordance with both the University of Bordeaux institutional committee (committee CEEA50) and the European Community guidelines (L358-86/609/EEC) for experimental animal use. We compared the expression profile of mice homologues of identified candidate gene(s) in isolated brain vessel preparations (n = 9 mice) versus total brain fraction (n = 10 mice). Details on brain microvascular fragments purification, immunofluorescence staining, immunoblot analysis and RNA preparation and quantitative PCR are described in the Supplementary material.
Impact of RNA silencing of extreme-SVD putative causal genes on endothelial cell permeability
Cell culture and transfection with siRNA
HBMEC (human brain microvascular endothelial cells; Alphabioregen-Clinisciences, #ALHE02) were grown in endothelial basal medium-2 (EBM-2) supplemented with EGM-2 BulletKits (Lonza #CC-3162) with 0.1% gelatin coating. An immortalized human cerebral microvascular endothelial cell line (hCMEC/D3 cells; Sigma-Aldrich #SCC066) was grown in EBM-2 medium (Lonza CC-3156) supplemented with 5% foetal bovine serum (FBS), 10 mM HEPES, 1 ng/ml basic fibroblast growth factor (bFGF), 1.4 µM hydrocortisone, 5 µg/ml ascorbic acid, penicillin–streptomycin with 0.1% gelatin coating as well. Cells from passage 3 to passage 8 were used.
The siRNAs were transfected using Interferin (Polyplus #409) at a final concentration of 30 nM. Two siRNA couples, siTRIM47 #1 and #2 targeting human TRIM47 were used34 and a non-specific siRNA as negative control (Eurogentec®). SiRNA-depletion was then resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis using TRIM47 antibody (Thermofisher Scientific, #PA5-50892). Protein loading quantity was controlled using mouse monoclonal anti α-tubulin antibodies (Sigma, Cat no. T5168) and Ve cadherin (Invitrogen, 35-2500).
Permeability assay
After 24 h of siRNA treatment, hCMEC/D3 and HBMECs were seeded on the top of 24-well filter inserts at a density of 3 × 105 cells/cm2 (membrane pore size 8.0 μm, Falcon). The day after, fluorescently labelled tracers 3 kDa Texas Red® Dextran (TXR 3 kDa, #D3328, Thermofisher), 20 kDa tetramethylrhodamine isothiocyanate dextran (TMR 20 kDa, #73766, Sigma-Aldrich) and 70 kDa fluorescein isothiocyanate dextran (70 kDa, 5 μM, #FD70S, Sigma-Aldrich) in a final concentration of 10 μM were added to the top chamber of the filter inserts. Fluorescently labelled tracer molecules were chosen as biomarkers for paracellular transport. At least three inserts per condition were used for each experimental set. Samples were taken from the bottom chamber after 1 h of incubation at 37°C. Fluorescence was measured on Tecan’s infinite M200 PRO, using the following order of excitation/emission (nm): TXR 560/620, TMR 540/590, FITC 485/535. Permeability flux was calculated as a ratio to the control condition fluorescence measurement. For each condition, three inserts were used for each experimental set and samples were taken in duplicates each time. Significance was calculated with GraphPad Prism 8.0 software using a two-way ANOVA with Dunnett’s multiple comparison test.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
URLs
(i)https://github.com/bulik/ldsc/wiki/Partitioned-Heritability (accessed 25 February 2022).
(ii)https://data.broadinstitute.org/alkesgroup/LDSCORE/ (accessed 25 February 2022)
(iii)https://cran.r-project.org/web/packages/seqMeta/index.html (accessed 10 December 2019)
(iv)http://genetics.bwh.harvard.edu/pph2/ (accessed 25 February 2022)
(v)https://www.ukbiobank.ac.uk/wp-content/uploads/2019/12/Description-of-the-alt-aware-issue-with-UKB-50k-WES-FE-data.pdf (accessed 2 February 2020)
(vi)http://betsholtzlab.org/VascularSingleCells/database.html (accessed 25 February 2022)
Results
Seventeen population-based cohorts of European and African American ancestry with brain MRI and genome-wide genotyping (n = 41 326, nEuropean = 39 584), WES (n = 15 965, nEuropean = 15 239), or EC (n = 5298, nEuropean = 4265) data from the CHARGE consortium19 and the UK Biobank participated in this study. We used a composite MRI-defined phenotype combining WMH burden and lacunes in CHARGE cohorts and only WMH burden in the UK Biobank to define extreme-SVD, including ‘cases’ with extensive-SVD and ‘controls’ with minimal-SVD severity (Fig. 1). We observed a high genetic correlation of 0.72 (SE = 0.14, P-value = 1.13 × 10−7) between the composite extreme-SVD defined in CHARGE cohorts and extreme-SVD defined using WMH burden only in the UK Biobank. Overall we derived 13 776 (nEuropean = 13 196) extreme-SVD participants for the GWAS (mean age: 65.14 ± 10.28 years, 52.31% female) and 7079 (nEuropean = 6525) extreme-SVD participants for the WEAS (mean age: 64.44 ± 9.83 years, 53.53% female). Baseline characteristics of extensive- and minimal-SVD participants are reported in Supplementary Table 1 (GWAS) and Supplementary Table 2 (WEAS).
Common variant association study
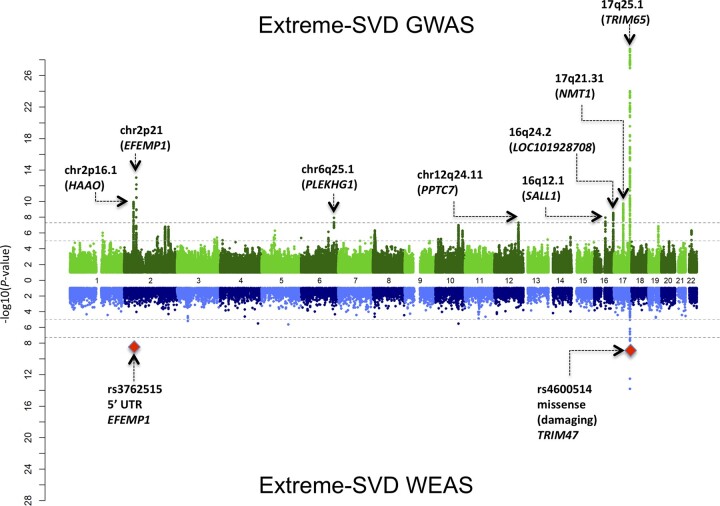
We performed inverse-variance weighted meta-analysis of GWAS on extreme-SVD adjusting for age, sex and when relevant principal component of population stratification, study site and familial relationships. The intercept of LD Score regression in the European ancestry GWAS meta-analysis of 0.98 and the genomic inflation factor, λ, of 1.04 in the transethnic GWAS meta-analysis show no systematic inflation of summary statistics (Supplementary Fig. 1). We identified genome-wide significant associations with extreme-SVD at eight chromosomal locations (Fig. 2): chr2p21 (nearest gene to lead variant, HAAO), chr2p16.1 (EFEMP1), chr6q25.1 (PLEKHG1), chr12q24.11 (PPTC7), chr16q12.1 (SALL1), chr16q24.1 (LOC101928708), chr17q21.31 (NMT1) and chr17q25.1 (TRIM65), of which chr12q24.11 was not previously reported to be associated with MRI markers of small vessel disease, including in two recent publications on WMH burden with n > 40 000 (Table 1, Fig. 2).8,12 Notably the risk allele at the lead extreme-SVD chr12q24.11 risk variant, rs73191849-C, showed suggestive association with increasing WMH burden,8 [β(SE) = 0.05 (0.01), P-value = 6.92 × 10−5, n = 48 236] and nominal association with MRI-defined brain infarcts14 [OR (CI 95%) = 1.18 (1.02–1.36), P-value = 0.02, n = 13 659; Supplementary Table 3). The conditional and joint analysis35 did not identify more than one independent genome-wide significant signal at the eight genome-wide significant risk loci for extreme-SVD (Supplementary Table 4). After additional adjustment for hypertension (n = 10 258), associations at chr2p16.1, chr6q25.1, chr12q24.11, chr16q12.1 and chr17q21.31 were no longer genome-wide significant, whereas associations at chr2p21, chr16q24.2 and chr17q25.1 remained genome-wide significant, suggesting that these three loci are not primarily mediated by hypertension (Supplementary Table 5). We explored associations with extreme-SVD of previously reported 33 LD independent risk variants for WMH burden,8–13 of which four variants reported by Persyn et al.12 were available only in the UK Biobank and hence filtered out from our meta-analysis, two suggestive risk loci for lacunes,14 five risk loci for clinical lacunar stroke and seven risk loci reported for multi-trait analyses of clinical lacunar stroke and WMH burden.36 All available 29 WMH burden risk variants showed nominally significant association with extreme-SVD (P-value < 0.05), with 26 associations remaining significant after multiple testing correction (P-value < 1.79 × 10−3; Supplementary Table 6). We saw evidence for association of one suggestive lacune risk locus rs9371194 at chr6q25.1 (P-value = 9.21 × 10−3) with extreme-SVD, while no association was observed for other lacune risk locus rs75889566 at chr6q27 (P-value = 0.46). Notably, rs9371194 is in moderate LD (r2 = 0.45, in 1000 genomes European ancestry sample) with a recently reported WMH volume risk variant rs27535010 and extreme-SVD risk variant rs275350 at chr6q25.1. Four of five lacunar stroke risk loci and all seven loci from multi-trait analysis of lacunar stroke and WMH burden showed nominally significant association with extreme-SVD (P-value < 0.05). After multiple testing correction three lacunar stroke risk loci (P-value < 0.01) and six loci from multi-trait analysis of lacunar stroke and WMH burden (P-value < 7.14 × 10−3) remain significantly associated with extreme-SVD (Supplementary Table 6).
Figure 2.
Miami plot of GWAS and WEAS of extreme-SVD. Top: The GWAS plot reports chromosomal location and nearest gene to the top genome-wide significant variant. Bottom: The WES plot reports whole exome-wide significant associations with the most likely functional variants and affected gene.
Table 1.
Genome-wide significant associations of extreme-SVD
| Locus | rs ID | Chromosome:base (hg19)a | Nearest gene | RA/OA | European (n = 13196) | African American (n = 580) | Combined (n = 13776) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAF | OR (CI 95%)b | P-value* | RAF | OR (CI 95%) | P-value* | OR (CI 95%) | P-value* | |||||
| 2p21 | rs13403122 | 2:43078758 | HAAO | C/T | 0.73 | 1.23 (1.15–1.31) | 1.31 × 10−10 | 0.88 | 1.14 (0.70–1.83) | 0.61 | 1.23 (1.15–1.30) | 1.21 × 10−10 |
| 2p16.1 | rs78857879 | 2:56135099 | EFEMP1 | A/G | 0.10 | 1.43 (1.30–1.56) | 9.70 × 10−14 | NA | NA | NA | 1.43 (1.30–1.56) | 9.70 × 10−14 |
| 6q25.1 | rs275350 | 6:151016058 | PLEKHG1 | C/G | 0.41 | 1.18 (1.11–1.24) | 1.35 × 10 − 8 | 0.59 | 1.09 (0.81–1.49) | 0.56 | 1.17 (1.11–1.24) | 1.31 × 10−8 |
| 12q24.11 | rs73191849 | 12:111017205 | PPTC7 | C/T | 0.94 | 1.42 (1.25–1.61) | 4.97 × 10−8 | NA | NA | NA | 1.42 (1.25–1.61) | 4.97 × 10−8 |
| 16q12.1 | rs1948948 | 16:51442679 | SALL1 | C/T | 0.56 | 1.17 (1.11–1.24) | 1.14 × 10−8 | NA | NA | NA | 1.17 (1.11–1.24) | 1.13 × 10−8 |
| 16q24.2 | rs12149643 | 16:87231499 | LOC101928708 | T/C | 0.58 | 1.19 (1.12–1.26) | 2.92 × 10−9 | NA | NA | NA | 1.19 (1.12–1.26) | 3.01 × 10−9 |
| 17q21.31 | rs6503417 | 17:43144218 | NMT1 | C/T | 0.63 | 1.20 (1.13–1.27) | 3.47 × 10−10 | 0.77 | 1.23 (0.86–1.75) | 0.26 | 1.20 (1.14–1.27) | 1.84 × 10−10 |
| 17q25.1 | rs3744027 | 17:73888743 | TRIM65 | A/G | 0.19 | 1.51 (1.40–1.62) | 4.24 × 10−30 | NA | NA | NA | 1.51 (1.40–1.62) | 4.24 × 10−30 |
OA = other allele; RA = risk allele; RAF = frequency of risk allele.
Sorted by chromosome and base pair position of sentinel variant.
OR represents effect size of risk allele.
P-value < 5 × 10−8 suggests a genome-wide significant association.
The joint SNP gene-based analysis using the VEGAS2 software37 identified significantly associated genes at three additional loci: chr2q32.1 (CALCRL), chr2q33.2 (ICA1L and WDR12) and chr10q24.33 (SH3PXD2A; Supplementary Table 7), which were recently reported to be associated with WMH burden.8 Overall, using an extreme-phenotype strategy we identified genome-wide and gene-wide significant associations with extreme-SVD at 11 common variant risk loci, of which one locus is novel, despite a much smaller total sample size compared to the published analyses on continuous WMH burden alone.8–13
The SNP-based heritability estimate of extreme-SVD computed using European only GWAS summary statistics was 0.36 (SE = 0.06). We performed partitioned heritability analyses of extreme-SVD by functional categories,38 and genes expressed specifically in human brain and blood vessel tissues,39 using the LD SCore regression software.40 We identified high and significant enrichment of extreme-SVD SNP-heritability in multiple functional categories (Supplementary Table 8). The highest enrichment was observed for transcription start sites, with 2% of SNP coverage explaining 29% of extreme-SVD SNP-based heritability. The partitioned heritability analysis of genes expressed specifically in human brain and blood vessel tissues did not identify any significant association after multiple testing correction, with coronary artery and brain stem reaching nominal significance (Supplementary Table 9).
Whole-exome association study
We performed a WEAS study on 7079 extreme-SVD participants, to identify exonic variants associated with extreme-SVD. Overall, 324 714 variants with a minor allele count of more than five across all participants qualified for single variant association testing. We used the genome-wide significance threshold P < 5 × 10−8 for testing association of non-coding variants, and P-value < 4.3 × 10−7 for coding variants.41 Eleven variants at chr2p16.1 and chr17q25.1 were associated with extreme-SVD (Fig. 2 and Table 2), including one variant (rs3762515) in the 5′ UTR region of EFEMP1 (chr2p16.1) and two missense variants in TRIM47 (rs4600514) and FBF1 (rs1135889) (chr17q25.1). These variants are common and in LD with sentinel variants at the same loci identified in the GWAS (Table 2). The polyphen-2 software24 predicted rs4600514 to be probably damaging to TRIM47 stability and function using both HumDiv and HumVar training data sets, and rs1135889 to be possibly damaging (HumDiv) or benign (HumVar) to FBF1 stability and function (Table 2).
Table 2.
Whole exome-wide significant associations of extreme-SVD
| Locus | Sentinel GWAS variant | LD (r2) Euro | rsID | Position hg19a | Consequence | PolyPhen-2 score | Gene | RA/OA | RAF | OR (CI 95%)b | P-value* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2p16.1 | rs78857879 | 0.95 | rs3762515 | 2:56150864 | 5′ UTR variant | EFEMP1 | C/T | 0.10 | 1.59 (1.36–1.86) | 4.41 × 10−9 | |
| 17q25.1 | rs3744027 | 0.56 | rs74410877 | 17:73832384 | Intron variant | UNC13D | C/T | 0.13 | 1.44 (1.26–1.63) | 2.69 × 10−8 | |
| 0.47 | rs9903200 | 17:73839498 | Intron variant | UNC13D | G/A | 0.30 | 1.30 (1.19–1.43) | 1.96 × 10−8 | |||
| 0.94 | rs3744017 | 17:73871467 | Intron variant | TRIM47 | G/A | 0.19 | 1.53 (1.37–1.70) | 1.50 × 10−14 | |||
| 0.73 | rs3903010 | 17:73874012 | Synonymous variant | TRIM47 | T/G | 0.15 | 1.47 (1.30–1.67) | 3.09 × 10−9 | |||
| 0.74 | rs4600514 | 17:73874071 | Missense variant (NP_258411.2: p.R187W) | 0.99 probably damaging (HumDiv); 0.96 probably damaging (HumVar) |
TRIM47 | A/G | 0.15 | 1.47 (1.29–1.67) | 4.36 × 10−9 | ||
| 0.73 | rs4072479 | 17:73874138 | Synonymous variant | TRIM47 | G/C | 0.15 | 1.47 (1.30–1.67) | 2.89 × 10−9 | |||
| 0.74 | rs116044941 | 17:73874684 | Upstream gene variant | TRIM47 | G/C | 0.15 | 1.49 (1.31–1.70) | 1.04 × 10−9 | |||
| 0.99 | rs34974290 | 17:73888354 | Synonymous variant | TRIM65 | G/A | 0.19 | 1.50 (1.34–1.67) | 2.99 × 10−13 | |||
| 0.54 | rs9902371 | 17:73897046 | Intron variant | MRPL38 | A/G | 0.29 | 1.35 (1.23–1.48) | 3.76 × 10−10 | |||
| 0.27 | rs1135889 | 17:73926121 | Missense variant (NP_001306122.1: p.G79V) | 0.65 possibly damaging (HumDiv); 0.37 benign (HumVar) |
FBF1 | C/A | 0.22 | 1.26 (1.16–1.36) | 4.39 × 10−8 |
OA = other allele; RA = risk allele; RAF = risk allele frequency.
Sorted by hg19 variant position.
OR represents effect size of risk allele.
Significance threshold for testing association of non-coding is P-value < 5 × 10−8, and for coding variants is P-value < 4.3 × 10−7.
Overall, 14 149 genes qualified for the SKAT-O gene-based analysis of protein-modifying rare and low-frequency variants. We did not observe any gene-wide significant association at P-value < 3.53 × 10−6. The top five SKAT-O gene-based associations with extreme-SVD were PEMT (17p11.2, P-value = 1.01 × 10−4), LTBP4 (19q13.2, P-value = 3.17 × 10−4), PIK3C2G (12p12.3, P-value = 5.58 × 10−4), SLC11A2 (12q13.12, P-value = 6.09 × 10−4), and KLHL38 (8q24.13, P-value = 6.21 × 10−4; Supplementary Table 10). These genes are not located within extreme-SVD GWAS risk loci.
We performed Biosystems gene-set enrichment tests based on SKAT-O gene-based P-values, using the resampling strategy and accounting for correlated P-values of overlapping and neighbouring genes and the number of genes per gene-set.25 We did not observe any gene-set wide significant association after Bonferroni correction for multiple testing of 9981 Biosystems gene-sets (P-value < 5.01 × 10−6). Interestingly, however, the top gene-set associations included the ‘regulation of Notch signalling pathway, GO: 0008593, Biosystems ID: 492852’ (P-value = 1.20 × 10−4) and the ‘complement and coagulation cascades gene-set, WikiPathways ID: WP558 Biosystems ID: 198880’ (P-value = 4.20 × 10−4; Supplementary Table 11).
Clinical significance of extreme-SVD using Mendelian randomization
In our pilot study, we reported that European ancestry participants with extensive-SVD severity were at significantly higher risk of incident dementia and incident stroke than participants with minimal SVD.16 Using the effect size estimates at extreme-SVD risk loci in Europeans (Supplementary Table 12), we now performed MR tests to explore the causal nature of the association between extensive-SVD severity and risk of Alzheimer’s disease and stroke, as this approach is less exposed to unmeasured confounding and reverse causation than observational studies.42 We observed that doubling of the genetically predicted risk of extensive-SVD severity was associated with significantly increased risk of Alzheimer’s disease [OR (95% CI) = 1.01 (1.01–1.02), P-value = 2.01 × 10−6], any ischaemic stroke [OR (95% CI) = 1.03 (1.01–1.06), P-value = 3.54 × 10−3] and small vessel stroke [OR (95% CI) = 1.10 (1.04–1.15), P-value =5.21 × 10−4] at P < 7.14 × 10−3 (after correcting for seven traits, Supplementary Table 13). Although some indication of horizontal pleiotropy for the association with small vessel stroke was evident, after removing pleiotropic outliers, the inverse-variance weighted association remained significant for small vessel stroke while the MR-Egger intercept (average pleiotropic effects) did not significantly differ from zero. Goodness of fit for the inverse-variance weighted model was additionally confirmed with QR values close to one. Using MR-Steiger we provided additional evidence for the causal direction of the association between extreme-SVD and Alzheimer’s disease, any ischaemic stroke and small vessel stroke (Supplementary Table 13).
Prioritization of putative biotarget of extreme-SVD
We used the SMR/HEIDI approach17 to seek evidence supporting potential causal associations of changes in transcription level of specific genes in brain or blood vessels with extreme-SVD risk. At the chr17q25.1 locus, which comprises a total of 40 genes within ±500 kb of the lead variant, the SMR test was significant for six genes in the brain (TRIM47, TRIM65, MRPL38, FBF1, RP11-552F3.9 and WBP2) and three genes in blood vessels (TRIM47, TRIM65 and FBF1; Fig. 3A and Table 3). The HEIDI test rejected linkage-based association (supporting causal or pleiotropic relationship) of gene expression profiles of TRIM47 with extreme-SVD in both brain and blood vessels (Fig. 3B and C), and of RP11-552F3.9 and WBP2 in the brain (Supplementary Fig. 2A and B). At the chr2q32.1 locus (two genes within ±500 kb of the lead variant), the SMR test was significant for CALCRL in blood vessels (Supplementary Fig. 3A), and the HEIDI test rejected linkage-based association for this gene (Supplementary Fig. 3B and Table 3). At the chr17q21.31 locus (29 genes within ±500 kb of the lead variant), the SMR test was significant for DCAKD in both brain (Supplementary Fig. 4A) and blood vessels (Supplementary Fig. 5A), and the HEIDI test rejected linkage-based association in the brain (Supplementary Fig. 4B) but not in blood vessels (Supplementary Fig. 5B) for this gene (Table 3). The sign of the effect estimate for SMR associations was suggestive of genetically determined higher expression of TRIM47, RP11-552F3.9 and WBP2 in the brain, as well as TRIM47 and CALCRL in blood vessels, being associated with lower risk of extensive-SVD; and genetically determined higher expression of DCAKD in the brain being associated with higher risk of extensive-SVD (Table 3).
Figure 3.
SMR associations at chr17q25.1 (A) SMR associations in brain and blood vessels; (B) SMR effect size plot of TRIM47 eQTLs in brain; and (C) SMR effect size plot of TRIM47 eQTLs in blood vessels.
Table 3.
SMR associations of genetically predicted gene expression level in brain and blood vessel with extreme-SVD
| Locus | Gene | probeID | Probe location (hg19)a | Top eQTL (location hg19) | RA/OA | RAF | Beta (SE) eQTL | P-value eQTL | Beta (SE) extreme-SVD | P-value extreme-SVD | Beta (SE) SMRb | P-value SMR | P-value HEIDI* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain | |||||||||||||
| 17q21.31 | DCAKD | ENSG00000172992 | 17:43100708–43138477 | rs4793173 (17:43127715) | A/C | 0.62 | 0.52 (0.04) | 6.53 × 10−38 | 0.18 (0.03) | 6.50 × 10−10 | 0.34 (0.06) | 2.61 × 10−8 | 0.7 |
| 17q25.1 | WBP2 | ENSG00000132471 | 17:73841780–73852588 | rs58587945 (17:73920219) | T/C | 0.14 | 0.45 (0.08) | 1.39 × 10−8 | −0.42 (0.04) | 3.23 × 10−23 | −0.94 (0.19) | 8.40 × 10−7 | 0.3 |
| TRIM47 | ENSG00000132481 | 17:73870242–73874656 | rs4072479 (17:73874138) | C/G | 0.18 | 0.58 (0.05) | 2.06 × 10−27 | −0.43 (0.04) | 1.94 × 10−28 | −0.74 (0.10) | 9.79 × 10−15 | 0.59 | |
| RP11-552F3.9 | ENSG00000267801 | 17:73872453–73875627 | rs76154832 (17:73876473) | G/A | 0.18 | 0.50 (0.09) | 6.42 × 10−9 | −0.44 (0.04) | 5.47 × 10−29 | −0.88 (0.17) | 2.59 × 10−7 | 0.63 | |
| TRIM65 | ENSG00000141569 | 17:73876416–73893084 | rs55872768 (17:73892414) | T/G | 0.36 | 0.42 (0.04) | 1.35 × 10−25 | 0.25 (0.03) | 2.31 × 10−17 | 0.60 (0.09) | 4.70 × 10−11 | 2.08 × 10−3 | |
| MRPL38 | ENSG00000204316 | 17:73894724–73905899 | rs4788913 (17:73950216) | G/A | 0.37 | 0.50 (0.05) | 5.25 × 10−27 | −0.22 (0.03) | 8.27 × 10−14 | −0.44 (0.07) | 8.81 × 10−10 | 5.96 × 10−5 | |
| FBF1 | ENSG00000188878 | 17:73905655–73937221 | rs59867239 (17:73931787) | G/A | 0.25 | 0.55 (0.06) | 2.05 × 10−21 | 0.26 (0.03) | 9.22 × 10−15 | 0.47 (0.08) | 1.90 × 10−9 | 1.58 × 10−4 | |
| Blood vessel | |||||||||||||
| 2q32.1 | CALCRL | ENSG00000064989.8 | 2:188207856–188313187 | rs17705966 (2:188165166) | G/A | 0.22 | 0.72 (0.05) | 6.19 × 10−50 | −0.18 (0.03) | 4.02 × 10−7 | −0.24 (0.05) | 1.58 × 10−6 | 0.29 |
| 17q21.31 | DCAKD | ENSG00000172992.7 | 17:43100708–43138477 | rs8071429 (17:43128906) | A/T | 0.62 | 0.36 (0.03) | 8.77 × 10−30 | 0.18 (0.029) | 9.15 × 10−10 | 0.49 (0.09) | 7.41 × 10−8 | 6.38 × 10−3 |
| 17q25.1 | TRIM47 | ENSG00000132481.2 | 17:73870242–73874656 | rs3903010 (17:73874012) | T/G | 0.18 | 0.32 (0.04) | 7.43 × 10−14 | −0.43 (0.04) | 2.47 × 10−28 | −1.37 (0.22) | 5.95 × 10−10 | 0.08 |
| TRIM65 | ENSG00000141569.6 | 17:73876416–73893084 | rs2608881 (17:73936653) | C/G | 0.30 | 0.25 (0.03) | 1.30 × 10−14 | 0.20 (0.03) | 1.77 × 10−10 | 0.81 (0.16) | 8.98 × 10−7 | 2.40 × 10−4 | |
| FBF1 | ENSG00000188878.12 | 17:73905655–73937221 | rs1135889 (17:73926121) | A/C | 0.25 | 0.71 (0.05) | 3.48 × 10−56 | 0.26 (0.03) | 1.36 × 10−14 | 0.36 (0.05) | 4.37 × 10−12 | 6.38 × 10−6 | |
OA = other allele; RA = risk allele; RAF = risk allele frequency.
Sorted by probe location.
Sign of SMR beta suggest potential relationship between transcription level of given gene in brain or blood vessel with risk of extreme-SVD, where ‘+’ SMR beta means high transcription level of given gene is associated with extensive SVD and ‘–’ means minimal SVD.
HEIDI P-value < 0.05 suggests potential linkage based SMR association between transcription level of given gene and risk of extreme-SVD.
We hypothesized that, if a linear relationship exists between expression of SMR-associated genes in the brain or blood vessels and risk of extensive-SVD severity, then participants born with zero or only one functional copy (homozygous or heterozygous knockout) of a true causal gene are likely to show extensive-SVD if the expression level of this gene is negatively associated with extensive-SVD risk. To identify human knockouts of putative causal genes for extensive-SVD severity, we screened the WES data of 10 832 UK Biobank participants of European ancestry with available information on WMH burden for loss of function allele carriers in SMR/HEIDI-associated protein coding genes (TRIM47, WBP2, CALCRL and DCAKD), and MRPL38 as RP11-552F3.9 encodes its antisense non-coding RNA. Using LOFTEE, we identified two participants carrying a loss-of-function allele in TRIM47 and WBP2, and one participant carrying a loss-of-function allele in MRPL38 (all heterozygous carriers). These participants did not carry any pathogenic variant (Supplementary Table 14) in genes known to cause familial cerebral small vessel disease (NOTCH3, HTRA1, COL4A1, COL4A2 and TREX1). Both participants carrying loss-of-function alleles in TRIM47 displayed extensive-SVD on their brain MRI (Supplementary Table 15): one with a scan at age 56 and a stop gain variant (TRIM47: stop gained: NM_033452.3:c.1595G>A: exon6: c.G1595A: NP_258411.2: p.Trp532Ter) and the other with a scan at age 65 and a frameshift variant (TRIM47: frameshift variant: NM_033452.3:c.1541del: NP_258411.2: p.Gly514fs). In WBP2, both participants carried the same loss-of-function splice acceptor variant (WBP2: splice acceptor variant: NM_012478.4: INTRON = 4/7: INTRON_SIZE = 639), but only one participant developed extensive-SVD at age 60, while the other did not develop extensive-SVD by age 76 (Supplementary Table 15). The participant carrying a loss-of-function splice donor variant in MRPL38 (MRPL38: splice donor variant: NM_032478.4: INTRON = 4/8: INTRON_SIZE = 438) did not develop extensive-SVD by age 56 (Supplementary Table 15).
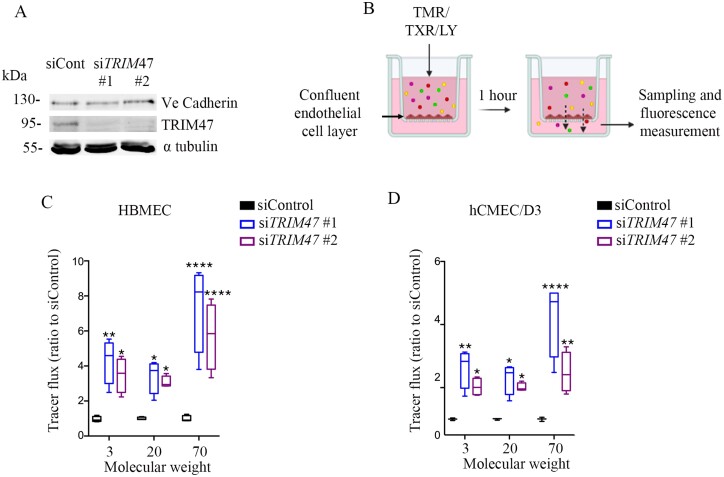
To seek prima facie evidence of potential functional involvement of TRIM47 in cerebral small vessel disease pathology, we performed two follow-up in vivo and in vitro experiments. First, we compared the expression profile of the mice homologue of TRIM47 in isolated brain vessel preparations versus total brain fraction. For this experiment, we prepared intact brain vessel fragments from the mouse brain parenchyma to preserve structural integrity and molecular properties of the blood–brain barrier (Supplementary Fig. 6A).43 The purity of the microvessel preparation was assessed by analysing the expression of markers for vascular (endothelial, pericyte, smooth muscle) and non-vascular (astrocyte, neuron) cells using qRT-PCR, and comparing it to the expression of the same markers in non-vascular brain tissue (Supplementary Fig. 6B and C). This experiment identified significant enrichment of Trim47 expression in isolated brain vessel preparations compared to total brain fraction (Supplementary Fig. 6D), consistent with the molecular atlas of cell types and zonation in the brain vasculature (see ‘URLs' section), where Trim47 was shown to be highly expressed in venous, capillary and arterial endothelial cells.44 Endothelial cells form the inner lining of the brain’s blood vessels and are responsible for maintaining the integrity of the blood–brain barrier and regulating transport across it. Blood–brain barrier leakage due to endothelial dysfunction is one of the main characteristic features of cerebral small vessel disease.45 We examined whether experimental knockdown of TRIM47 has any effect on human brain endothelial cell permeability. In this experiment, two distinct human brain endothelial cells, HBMEC and hCMEC/D3, were challenged for TRIM47 depletion using two different single siRNA oligos (siTRIM47-#1 and siTRIM47-#2) (Fig. 4A). We measured the passage of 3, 20 and 70 kDa FITC-dextran across confluent TRIM47 depleted endothelial cell monolayers grown on transwell inserts to assess change in permeability (Fig. 4B). Compared to siControl transfected endothelial cells, significantly more dextran molecules of different sizes passed across siTRIM47 transfected endothelial cell monolayers after 1 h (Fig. 4C and D), suggesting that siRNA silencing of TRIM47 increases endothelial cell permeability in vitro.
Figure 4.
TRIM47 depletion in human brain endothelial cells increases endothelial permeability. (A) Efficient reduction of TRIM47 expression in endothelial cells was achieved by siTRIM47 #1 and 2 versus siControl. Ve-cadherin expression, a marker of endothelial cell junction was followed and α tubulin was used as a loading control. (B) Endothelial transwell permeability assay. Quantification of dextran leakage expressed as fold change compared to HBMEC and (C) hCMEC/D3 (D) cells treated with control siRNA TRIM47 #1 and #2 versus siControl. Fluorescent intensity was normalized to siControl knock down. Two-way ANOVA with a Dunnett’s multiple comparisons test was performed. Data are presented as mean ± SD. **P-value < 0.01, ***P < 0.0001, ***P > 0.00001.
Discussion
In summary, we report a first GWAS and WEAS meta-analysis of a composite, MRI-defined extreme-SVD phenotype in population-based cohorts of middle-aged and older persons. The GWAS and follow-up gene-based analysis identified 11 extreme-SVD risk loci, of which the chr12q24.11 locus was not previously reported to be associated with any MRI marker of SVD. The WEAS identified 11 significant associations in two GWAS loci, including one probably damaging missense variant in TRIM47 at the chr17q25.1 risk locus, and one 5′UTR variant in EFEMP1 at chr2p16.1. Utilizing the GWAS summary data of European ancestry we validated and quantified the causal associations of extensive-SVD severity with increased risk of ischaemic stroke, small vessel stroke and Alzheimer’s disease. Using both in silico and in vitro studies we identified TRIM47 as a putative causal gene underlying the extremely gene-rich and most significant risk locus of extreme-SVD: chr17q25.1. Notably our in silico analyses showed genetically determined reduced expression of TRIM47 is associated with increased risk of extensive-SVD, while our in vitro analyses showed that experimental knockdown of TRIM47 was responsible for a significant increase of human brain endothelial cell permeability.
The identification of 11 cerebral small vessel disease risk loci, of which one novel finding in chr12q24.11 locus near PPTC7 gene, which encodes a mitochondrial phosphatase involved in the biosynthesis of coenzyme Q10 by targeting CoQ7,46 in 13 776 participants with extreme-SVD. Moreover, we observed significant association after multiple testing correction with extreme-SVD of 26 of 29 WMH burden risk loci (∼86%), 3 of 5 lacunar stroke risk loci (60%) and 6 of 7 multi-trait lacunar stroke and WMH burden risk loci (∼90%), highlighting that our study design could capture a broader spectrum of SVD manifestations. Our GWAS on 13 776 participants yielded more significant hits than previous GWAS conducted in over 20 000 participants from the general population that reported only five genome-wide significant risk loci for WMH burden11 and none for lacunes.14 We acknowledge that the total sample of participants with brain MRI and genomic information used for deriving 13 776 extreme-SVD participants was 41 326, and that a recent GWAS on continuous WMH volume using genotype–phenotype data of 42 310 participants has yielded 19 genome-wide significant loci,12 which is substantially more than the 11 loci identified in our study. However, our study provides evidence that for applying cutting-edge, expensive next-generation sequencing platforms (WES and whole-genome sequencing) to understand the genetic architecture of covert SVD, costs of sequencing in a population-based setting could be substantially reduced by adopting our study design with limited loss in power. We previously showed that extreme-SVD is associated with two and a half times increased risk of incident stroke and doubling the risk of incident dementia,16 for which we now provide the evidence for causality using the Mendelian randomization approach, highlighting the clinical relevance of the extreme-SVD phenotype.
We report the largest WEAS (7079 participants) on any MRI-marker of cerebral small vessel disease to date. We were underpowered to identify novel rare variants associated with extreme-SVD, in line with previous publications suggesting that hundreds of thousands of sequencing samples will be required to understand the rare variant risk architecture of complex diseases.47 Nevertheless, this WEAS analysis provided interesting complementary insight. At chr2p16.1 we identified whole exome-wide significant association of extreme-SVD with a common variant in the 5′UTR of EFEMP1, encoding extracellular matrix glycoprotein fibulin-3. Mutations in EFEMP1 are known to cause Doyne honeycomb degeneration of the retina, a rare autosomal dominant disease (MIM: 126600) and Efemp1 knockout mice were reported to age faster and die earlier compared to wild-type ones.48 A recent article reported two loss of function variants in EFEMP1 causing an uncharacterized connective tissue disorder,49 the phenotype of which has similarities with cutis laxa (MIM: 613177), an inherited connected tissue disorder caused by variants in LTBP4. Interestingly LTBP4 was one of the top five genes identified in our SKAT-O gene-based analysis of rare and low-frequency protein-modifying variants. At chr17q25.1 we identified whole exome-wide significant association of extreme-SVD with a common exonic variant predicted to be probably damaging to the stability and function of the TRIM47 protein, adding to the bioinformatics and experimental evidences supporting an involvement of TRIM47 in extreme-SVD pathophysiology, as detailed below.
Indeed, using multiple complementary analyses to detect gene–trait relationships we report TRIM47 as the most plausible causal gene at the gene-rich and most significant extreme-SVD risk locus, chr17q25.1, also by far the most significant risk locus for WMH burden, replicated in multiple populations of different ancestry backgrounds.9–12 First, we made the following in silico observations: (i) an SMR/HEIDI test showed causal or pleiotropic association of lower expressions of TRIM47 in the brain and blood vessels with increased risk of extensive-SVD; and (ii) building on the SMR/HEIDI findings our targeted human knockout screening in the UK Biobank revealed two middle-aged participants carrying heterozygous loss-of-function alleles in TRIM47 and both participants developed extensive-SVD, both analyses suggest a genetically determined inverse relationship between TRIM47 activity and risk of extensive-SVD. Second, our preliminary functional follow-up showed (i) an enrichment of Trim47 in isolated brain vessel preparations compared to total brain fraction in mice, in line with single cell sequencing resources showing high enrichment of TRIM47 in venous, capillary and arterial human endothelial cells44; and (ii) a significant increase in permeability of human brain endothelial cells after experimental knockdown of TRIM47 using two distinct TRIM47 siRNAs, in agreement with the hypothesis that enhanced blood–brain barrier permeability is one of the key mechanisms of SVD.45 Overall, both in silico and in vitro experiments support the hypothesis that genetically determined or experimentally attenuated activity of TRIM47 could be involved in cerebral small vessel disease pathophysiology. Detailed functional in vivo studies are thus warranted in TRIM47 knockout cellular and animal models to explore the mechanisms underlying its association with extreme-SVD. The TRIM47 gene encodes an E3 ubiquitin-protein ligase,50 which is responsible for ubiquitination and subsequent proteosomal degradation of SMAD4 and CYLD.51 SMAD4 is the key mediator of canonical TGF-β signalling.52 CYLD is known to inhibit IKK and NF-kB signalling through deubiquitination of TRAF2, TRAF6 and NEMO53,54 and is involved in neuronal death and its ablation is protective for trauma-induced brain damage.55
Overall, our comprehensive study highlights the added value of a composite extreme phenotype design for the discovery of novel genetic risk variants for MRI features of cerebral small vessel disease, demonstrates a likely causal relation of MRI-defined extreme-SVD with risk of stroke and dementia, and provides compelling evidence for a role of TRIM47 in the pathophysiology of cerebral small vessel disease. Nonetheless, we acknowledge several limitations. First, our GWAS and WEAS were underpowered to understand the full genetic risk architecture of extreme-SVD. Moreover ∼50% of our extreme-SVD sample in both GWAS and WEAS were defined using only WMH burden as information on lacunes was missing in the UK Biobank; however, both phenotypes showed high genetic correlation. Second, we explored the role of extreme-SVD risk variants in adult brain and blood vessel tissues using available eQTL resources, while some cerebral small vessel disease risk variants might exert their effect through specific cell types, other tissues or at different developmental stages. Third, human knockout screening was performed on a relatively limited sample size of ∼10 000 UK Biobank participants, additional loss-of-function variants may be detected in larger, well-characterized cohorts with WES data. Fourth, the presented experimental work is preliminary and had a specific objective to assess only prima facie evidence for a functional role of prioritized candidate genes from SMR and human knockout screening analyses in the context of cerebral small vessel disease pathology. In vivo characterization of brain vessel structure and function and brain features in Trim47 knockout animal models are warranted to validate our findings and explore the underlying mechanisms.
In conclusion, we report a comprehensive gene-mapping study identifying 11 risk loci of extreme-SVD, of which chr12q24.11 is a novel finding. We also provide converging bioinformatics and experimental evidence to prioritize TRIM47 at the chr17q25.1 extreme-SVD risk locus for further experimental explorations to decipher the mechanisms underlying this most significant risk locus for cerebral small vessel disease. If confirmed in future studies, TRIM47 could represent an important putative biotarget for SVD.
Funding
This project is an EU Joint Programme–Neurodegenerative Disease Research (JPND) project. The project is supported through the following funding organisations under the aegis of JPND www.jpnd.eu: Australia, National Health and Medical Research Council, Austria, Federal Ministry of Science, Research and Economy; Canada, Canadian Institutes of Health Research; France, French National Research Agency; Germany, Federal Ministry of Education and Research; Netherlands, The Netherlands Organization for Health Research and Development; United Kingdom, Medical Research Council. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 643417. This project has also received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 640643 and from the European Union’s Horizon 2020 research and innovation programme under grant agreements Nos. 667375 and 754517. This work was also supported by a grant overseen by the French National Research Agency (ANR) as part of ANR-14-CE12-60016 and the ‘Investment for the Future Programme’ ANR-18-RHUS-0002. Part of the computations were performed at the Bordeaux Bioinformatics Centre (CBiB), University of Bordeaux and at the CREDIM (Centre de Ressource et Développement en Informatique Médicale) at University of Bordeaux, on a server infrastructure supported by the Fondation Claude Pompidou.
The neurology Working Group in the CHARGE Consortium is partly funded by the CHARGE infrastructure grant R01HL105756 and grants from the National Institute on Aging, AG033193, AG049505, AG052409 and AG059421.
P.M.M. acknowledges personal support from the Edmond J Safra Foundation and Lily Safra and an NIHR Senior Investigator Award and research support from the UK Dementia Research Institute and NIHR Imperial College Healthcare Trust Biomedical Research Centre.
Study-specific funding information is provided in the Supplementary material.
Competing interests
B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. P.M.M. receives an honorarium as Chair of the UKRI Medical Research Council Neuroscience and Mental Health Board. He acknowledges consultancy fees from Adelphi Communications, MedScape, Neurodiem, Nodthera, Biogen, Celgene and Roche. He has received speakers’ honoraria from Celgene, Biogen, Novartis and Roche and has received research or educational funds from Biogen, GlaxoSmithKline and Novartis. He is paid as a member of the Scientific Advisory Board for Ipsen Pharmaceuticals. Professor Hans Grabe has received travel grants and speakers’ honoraria from Fresenius Medical Care, Neuraxpharm, Servier and Janssen Cilag as well as research funding from Fresenius Medical Care. P.M.M. has received consultancy fees from Roche, Adelphi Communications, Celgene, Neurodiem and Medscape, honoraria or speakers’ fees from Novartis and Biogen and received research or educational funds from Biogen, Novartis and GlaxoSmithKline. This article was partially prepared while R.G. was employed at the Johns Hopkins University School of Medicine. The opinions expressed in this article are the authors’ own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services or the United States Government.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Glossary
- CHARGE
Cohorts for Heart and Aging Research in Genomic Epidemiology
- EC
exome chip
- extensive/extreme/minimal-SVD
MRI-derived composite extreme phenotype of cerebral small vessel disease with extensive/extreme/minimal severity
- eQTL
expression quantitative trait loci
- GWAS
genome-wide association study
- HEIDI
heterogeneity in dependent instruments
- LD
linkage disequilibrium
- MR
Mendelian randomization
- siRNA
small interfering RNA
- SMR
summary-based Mendelian randomization
- SNP
single nucleotide polymorphism
- WEAS
whole-exome association study
- WES
whole-exome sequencing
- WMH
white matter hyperintensities
Contributor Information
Aniket Mishra, University of Bordeaux, INSERM, Bordeaux Population Health Research Centre, Team ELEANOR, UMR 1219, F-33000 Bordeaux, France.
Cécile Duplaà, University of Bordeaux, INSERM, Biologie des Maladies Cardiovasculaires, U1034, F-33600 Pessac, France.
Dina Vojinovic, Department of Epidemiology, Erasmus MC, University Medical Centre, 3015 GD Rotterdam, The Netherlands; Department of Biomedical Data Sciences, Leiden University Medical Centre, 2333 ZA Leiden, The Netherlands.
Hideaki Suzuki, Department of Cardiovascular Medicine, Tohoku University Hospital, 1-1, Seiryo, Aoba, Sendai 980-8574, Japan; Tohoku Medical Megabank Organization, Tohoku University, 2-1, Seiryo, Aoba, Sendai 980-8573, Japan; Department of Brain Sciences and UK Dementia Research Institute Centre, Imperial College, London, W12 0NN, UK.
Muralidharan Sargurupremraj, University of Bordeaux, INSERM, Bordeaux Population Health Research Centre, Team ELEANOR, UMR 1219, F-33000 Bordeaux, France.
Nuno R Zilhão, Icelandic Heart Association, 200 Kopavogur, Iceland.
Shuo Li, Department of Biostatistics, Boston University School of Public Health, Boston, MA 02115, USA.
Traci M Bartz, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 98195, USA; Department of Biostatistics, University of Washington, Seattle, WA 98195, USA.
Xueqiu Jian, Brown Foundation Institute of Molecular Medicine, McGovern Medical School, University of Texas Health Science Center at Houston, Houston, TX 77030, USA; Glenn Biggs Institute for Alzheimer's and Neurodegenerative Diseases, University of Texas Health Sciences Center, San Antonio, TX, USA.
Wei Zhao, Department of Epidemiology, School of Public Health, University of Michigan, Michigan, MI 48104, USA.
Edith Hofer, Clinical Division of Neurogeriatrics, Department of Neurology, Medical University of Graz, 8036 Graz, Austria; Institute for Medical Informatics, Statistics and Documentation, Medical University of Graz, 8036 Graz, Austria.
Katharina Wittfeld, German Centre for Neurodegenerative Diseases (DZNE), Rostock/Greifswald, 17489 Greifswald, Germany; Department of Psychiatry and Psychotherapy, University Medicine Greifswald, 17489 Greifswald, Germany.
Sarah E Harris, Department of Psychology, University of Edinburgh, EH8 9JZ Edinburgh, UK.
Sandra van der Auwera-Palitschka, German Centre for Neurodegenerative Diseases (DZNE), Rostock/Greifswald, 17489 Greifswald, Germany; Department of Psychiatry and Psychotherapy, University Medicine Greifswald, 17489 Greifswald, Germany.
Michelle Luciano, Department of Psychology, University of Edinburgh, EH8 9JZ Edinburgh, UK.
Joshua C Bis, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 98195, USA.
Hieab H H Adams, Department of Epidemiology, Erasmus MC, University Medical Centre, 3015 GD Rotterdam, The Netherlands; Department of Radiology and Nuclear Medicine, Erasmus MC, University Medical Centre, 3015 GD Rotterdam, The Netherlands; Department of Clinical Genetics, Erasmus University Medical Centre Rotterdam, 3015 GD Rotterdam, The Netherlands.
Claudia L Satizabal, Glenn Biggs Institute for Alzheimer's and Neurodegenerative Diseases, University of Texas Health Sciences Center, San Antonio, TX, USA; The Framingham Heart Study, Framingham, MA 01701, USA; Department of Neurology, Boston University School of Medicine, Boston, MA 2115, USA.
Rebecca F Gottesman, National Institute of Neurological Disorders and Stroke Intramural Research Program, Bethesda, MD 20814, USA.
Piyush G Gampawar, Institute of Molecular Biology & Biochemistry, Gottfried Schatz Research Centre (for Cell Signalling, Metabolism and Aging), Medical University of Graz, 8036 Graz, Austria.
Robin Bülow, Department of Diagnostic Radiology and Neuroradiology, University Medicine Greifswald, 17489 Greifswald, Germany.
Stefan Weiss, Interfaculty Institute for Genetics and Functional Genomics, University Medicine Greifswald, 17489 Greifswald, Germany.
Miao Yu, Department of Epidemiology, School of Public Health, University of Michigan, Michigan, MI 48104, USA.
Mark E Bastin, Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh EH8 9AB, UK; Division of Neuroimaging Sciences, Brain Research Imaging Centre, University of Edinburgh, Western General Hospital, Edinburgh EH4 2XU, UK.
Oscar L Lopez, Department of Neurology, University of Pittsburgh, Pittsburgh, PA 15213, USA; Department of Psychiatry, University of Pittsburgh, Pittsburgh, PA 15213, USA.
Meike W Vernooij, Department of Epidemiology, Erasmus MC, University Medical Centre, 3015 GD Rotterdam, The Netherlands; Department of Radiology and Nuclear Medicine, Erasmus MC, University Medical Centre, 3015 GD Rotterdam, The Netherlands.
Alexa S Beiser, Department of Biostatistics, Boston University School of Public Health, Boston, MA 02115, USA; The Framingham Heart Study, Framingham, MA 01701, USA; Department of Neurology, Boston University School of Medicine, Boston, MA 2115, USA.
Uwe Völker, Interfaculty Institute for Genetics and Functional Genomics, University Medicine Greifswald, 17489 Greifswald, Germany.
Tim Kacprowski, Interfaculty Institute for Genetics and Functional Genomics, University Medicine Greifswald, 17489 Greifswald, Germany; TUM School of Life Sciences Weihenstephan (WZW), Technical University of Munich (TUM), 85354 Freising-Weihenstephan, Germany.
Aicha Soumare, University of Bordeaux, INSERM, Bordeaux Population Health Research Centre, Team ELEANOR, UMR 1219, F-33000 Bordeaux, France.
Jennifer A Smith, Department of Epidemiology, School of Public Health, University of Michigan, Michigan, MI 48104, USA.
David S Knopman, Mayo Clinic, Rochester, MN 55905, USA.
Zoe Morris, Neuroradiology Department, Department of Clinical Neurosciences, Western General Hospital, Edinburgh EH4 2XU, UK.
Yicheng Zhu, Department of Neurology, Peking Union Medical College Hospital, Beijing 100730, China.
Jerome I Rotter, The Institute for Translational Genomics and Population Sciences, Department of Pediatrics, The Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, CA 90502, USA.
Carole Dufouil, University of Bordeaux, INSERM, Bordeaux Population Health Research Centre, Team ELEANOR, UMR 1219, F-33000 Bordeaux, France.
Maria Valdés Hernández, Division of Neuroimaging Sciences, Brain Research Imaging Centre, University of Edinburgh, Western General Hospital, Edinburgh EH4 2XU, UK.
Susana Muñoz Maniega, Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh EH8 9AB, UK.
Mark Lathrop, University of McGill Genome Center, Montreal, Quebec H3A 0G1, Canada.
Erik Boerwinkle, Human Genetics Center, School of Public Health, University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
Reinhold Schmidt, Clinical Division of Neurogeriatrics, Department of Neurology, Medical University of Graz, 8036 Graz, Austria.
Masafumi Ihara, Department of Neurology, National Cerebral and Cardiovascular Center, Suita, Osaka 564-8565, Japan.
Bernard Mazoyer, University of Bordeaux, Institut des Maladies Neurodégénératives, CNRS-CEA UMR 5293, 33000 Bordeaux, France.
Qiong Yang, Department of Biostatistics, Boston University School of Public Health, Boston, MA 02115, USA; The Framingham Heart Study, Framingham, MA 01701, USA.
Anne Joutel, Institute of Psychiatry and Neurosciences of Paris, INSERM UMR1266, Université de Paris, France.
Elizabeth Tournier-Lasserve, Université de Paris, INSERM UMR-1141 Neurodiderot, Paris F-75019, France.
Lenore J Launer, Intramural Research Program, National Institute on Aging, National Institutes of Health, Bethesda, MD 20892, USA.
Ian J Deary, Department of Psychology, University of Edinburgh, EH8 9JZ Edinburgh, UK.
Thomas H Mosley, Memory Impairment and Neurodegenerative Dementia (MIND) Center, University of Mississippi Medical Center, Jackson, MS 39216, USA.
Philippe Amouyel, University of Lille, INSERM, Institut Pasteur de Lille, UMR1167-RID-AGE—Risk Factors and Molecular Determinants of Aging-Related Diseases, F-59000 Lille, France; LabEx DISTALZ, Institut Pasteur de Lille, 59000 Lille, France; Department of Epidemiology and Public Health, Centre Hospital University of Lille, F-59000 Lille, France.
Charles S DeCarli, Department of Neurology and Center for Neuroscience, University of California at Davis, Sacramento, CA 95816, USA.
Bruce M Psaty, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 98195, USA; Department of Epidemiology, University of Washington, Seattle, WA, USA; Department of Health Systems and Population Health, University of Washington, Seattle, WA.
Christophe Tzourio, University of Bordeaux, INSERM, Bordeaux Population Health Research Centre, Team ELEANOR, UMR 1219, F-33000 Bordeaux, France; CHU de Bordeaux, Pole de santé publique, Service d’information médicale, F-33000 Bordeaux, France.
Sharon L R Kardia, Department of Epidemiology, School of Public Health, University of Michigan, Michigan, MI 48104, USA.
Hans J Grabe, German Centre for Neurodegenerative Diseases (DZNE), Rostock/Greifswald, 17489 Greifswald, Germany; Department of Psychiatry and Psychotherapy, University Medicine Greifswald, 17489 Greifswald, Germany.
Alexander Teumer, Institute for Community Medicine, University Medicine Greifswald, D-17475 Greifswald, Germany.
Cornelia M van Duijn, Department of Biomedical Data Sciences, Leiden University Medical Centre, 2333 ZA Leiden, The Netherlands; Nuffield Department of Population Health, University of Oxford, Oxford OX3 7LF, UK.
Helena Schmidt, Institute of Molecular Biology & Biochemistry, Gottfried Schatz Research Centre (for Cell Signalling, Metabolism and Aging), Medical University of Graz, 8036 Graz, Austria.
Joanna M Wardlaw, Division of Neuroimaging Sciences, Brain Research Imaging Centre, University of Edinburgh, Western General Hospital, Edinburgh EH4 2XU, UK.
M Arfan Ikram, Department of Biomedical Data Sciences, Leiden University Medical Centre, 2333 ZA Leiden, The Netherlands; Department of Radiology and Nuclear Medicine, Erasmus MC, University Medical Centre, 3015 GD Rotterdam, The Netherlands.
Myriam Fornage, Brown Foundation Institute of Molecular Medicine, McGovern Medical School, University of Texas Health Science Center at Houston, Houston, TX 77030, USA; Human Genetics Center, School of Public Health, University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
Vilmundur Gudnason, Icelandic Heart Association, 200 Kopavogur, Iceland; Faculty of Medicine, University of Iceland, 101 Reykjavik, Iceland.
Sudha Seshadri, Department of Epidemiology, School of Public Health, University of Michigan, Michigan, MI 48104, USA; The Framingham Heart Study, Framingham, MA 01701, USA; Department of Neurology, Boston University School of Medicine, Boston, MA 2115, USA.
Paul M Matthews, Department of Brain Sciences and UK Dementia Research Institute Centre, Imperial College, London, W12 0NN, UK.
William T Longstreth, Jr, Department of Epidemiology, University of Washington, Seattle, WA, USA; Department of Neurology, University of Washington, Seattle, WA 98104-2420, USA.
Thierry Couffinhal, University of Bordeaux, INSERM, Biologie des Maladies Cardiovasculaires, U1034, F-33600 Pessac, France.
Stephanie Debette, University of Bordeaux, INSERM, Bordeaux Population Health Research Centre, Team ELEANOR, UMR 1219, F-33000 Bordeaux, France; Department of Neurology, Boston University School of Medicine, Boston, MA 2115, USA; CHU de Bordeaux, Department of Neurology, F-33000 Bordeaux, France.
References
- 1. Greenberg SM. Small vessels, big problems. N Engl J Med. 2006;354(14):1451–1453. [DOI] [PubMed] [Google Scholar]
- 2. Wardlaw JM, Smith C, Dichgans M.. Small vessel disease: Mechanisms and clinical implications. Lancet Neurol. 2019;18(7):684–696. [DOI] [PubMed] [Google Scholar]
- 3. Viswanathan A, Rocca WA, Tzourio C.. Vascular risk factors and dementia: How to move forward? Neurology. 2009;72(4):368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alber J, Alladi S, Bae HJ, et al. White matter hyperintensities in vascular contributions to cognitive impairment and dementia (VCID): Knowledge gaps and opportunities. Alzheimers Dement (N Y). 2019;5:107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wardlaw JM, Smith EE, Biessels GJ, et al. ; STandards for ReportIng Vascular changes on nEuroimaging (STRIVE v1) . Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12(8):822–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Atwood LD, Wolf PA, Heard-Costa NL, et al. Genetic variation in white matter hyperintensity volume in the Framingham Study. Stroke. 2004;35(7):1609–1613. [DOI] [PubMed] [Google Scholar]
- 7. Duperron MG, Tzourio C, Sargurupremraj M, et al. Burden of dilated perivascular spaces, an emerging marker of cerebral small vessel disease, is highly heritable. Stroke. 2018;49(2):282–287. [DOI] [PubMed] [Google Scholar]
- 8. Sargurupremraj M, Suzuki H, Jian X, et al. ; International Headache Genomics Consortium (IHGC) . Cerebral small vessel disease genomics and its implications across the lifespan. Nat Commun. 2020;11(1):6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Traylor M, Zhang CR, Adib-Samii P, et al. Genome-wide meta-analysis of cerebral white matter hyperintensities in patients with stroke. Neurology. 2016;86(2):146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Traylor M, Tozer DJ, Croall ID, et al. ; International Stroke Genetics Consortium . Genetic variation in PLEKHG1 is associated with white matter hyperintensities (n = 11,226). Neurology. 2019;92(8):e749–e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Verhaaren BF, Debette S, Bis JC, et al. Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ Cardiovasc Genet. 2015;8(2):398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Persyn E, Hanscombe KB, Howson JMM, Lewis CM, Traylor M, Markus HS.. Genome-wide association study of MRI markers of cerebral small vessel disease in 42,310 participants. Nat Commun. 2020;11(1):2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fornage M, Debette S, Bis JC, et al. Genome-wide association studies of cerebral white matter lesion burden: The CHARGE consortium. Ann Neurol. 2011;69(6):928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chauhan G, Adams HHH, Satizabal CL, et al. Genetic and lifestyle risk factors for MRI-defined brain infarcts in a population-based setting. Neurology. 2019;92(5):e486–e503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Debette S, Bis JC, Fornage M, et al. Genome-wide association studies of MRI-defined brain infarcts: Meta-analysis from the CHARGE Consortium. Stroke. 2010;41(2):210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mishra A, Chauhan G, Violleau MH, et al. Association of variants in HTRA1 and NOTCH3 with MRI-defined extremes of cerebral small vessel disease in older subjects. Brain. 2019;142(4):1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. [DOI] [PubMed] [Google Scholar]
- 18. Gusev A, Ko A, Shi H, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Psaty BM, O'Donnell CJ, Gudnason V, et al. ; CHARGE Consortium . Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winkler TW, Day FR, Croteau-Chonka DC, et al. ; Genetic Investigation of Anthropometric Traits (GIANT) Consortium . Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9(5):1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Willer CJ, Li Y, Abecasis GR.. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ.. RVTESTS: An efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32(9):1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee S, Wu MC, Lin X.. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13(4):762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mishra A, MacGregor S.. A novel approach for pathway analysis of GWAS data highlights role of BMP signaling and muscle cell differentiation in colorectal cancer susceptibility. Twin Res Hum Genet. 2017;20(1):1–9. [DOI] [PubMed] [Google Scholar]
- 26. Geer LY, Marchler-Bauer A, Geer RC, et al. The NCBI BioSystems database. Nucleic Acids Res. 2010;38(Database issue):D492–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51(3):404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malik R, Chauhan G, Traylor M, et al. ; MEGASTROKE Consortium . Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Woo D, Falcone GJ, Devan WJ, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94(4):511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Study Group 3C. Vascular factors and risk of dementia: Design of the Three-City Study and baseline characteristics of the study population. Neuroepidemiology. 2003;22(6):316–325. [DOI] [PubMed] [Google Scholar]
- 31. Regier AA, Farjoun Y, Larson DE, et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat Commun. 2018;9(1):4038- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rutten JW, Dauwerse HG, Gravesteijn G, et al. Archetypal NOTCH3 mutations frequent in public exome: Implications for CADASIL. Ann Clin Transl Neurol. 2016;3(11):844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang C, Chang J, Jiang Y, Liu J, Mao L, Wang M.. Selective RNA interference and gene silencing using reactive oxygen species-responsive lipid nanoparticles. Chem Commun (Camb). 2019;55(56):8170–8173. [DOI] [PubMed] [Google Scholar]
- 35. Yang J, Ferreira T, Morris AP, et al. ; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44(4):369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Traylor M, Persyn E, Tomppo L, et al. Genetic basis of lacunar stroke: a pooled analysis of individual patient data and genome-wide association studies. Lancet Neurol. 2021;20(5):351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mishra A, Macgregor S.. VEGAS2: Software for more flexible gene-based testing. Twin Res Hum Genet. 2015;18(1):86–91. [DOI] [PubMed] [Google Scholar]
- 38. Finucane HK, Bulik-Sullivan B, Gusev A, et al. ; RACI Consortium . Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47(11):1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Finucane HK, Reshef YA, Anttila V, et al. ; Brainstorm Consortium . Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50(4):621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bulik-Sullivan BK, Loh PR, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sveinbjornsson G, Albrechtsen A, Zink F, et al. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat Genet. 2016;48(3):314–317. [DOI] [PubMed] [Google Scholar]
- 42. Smith GD, Ebrahim S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. [DOI] [PubMed] [Google Scholar]
- 43. Boulay AC, Saubamea B, Decleves X, Cohen-Salmon M.. Purification of mouse brain vessels. J Vis Exp. 2015;(105):e53208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vanlandewijck M, He L, Mae MA, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554(7693):475–480. [DOI] [PubMed] [Google Scholar]
- 45. Thrippleton MJ, Backes WH, Sourbron S, et al. Quantifying blood–brain barrier leakage in small vessel disease: Review and consensus recommendations. Alzheimers Dement. 2019;15(6):840–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gonzalez-Mariscal I, Martin-Montalvo A, Vazquez-Fonseca L, et al. The mitochondrial phosphatase PPTC7 orchestrates mitochondrial metabolism regulating coenzyme Q10 biosynthesis. Biochim Biophys Acta Bioenerg. 2018;1859(11):1235–1248. [DOI] [PubMed] [Google Scholar]
- 47. Flannick J, Mercader JM, Fuchsberger C, et al. ; AMP-T2D-GENES . Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019;570(7759):71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McLaughlin PJ, Bakall B, Choi J, et al. Lack of fibulin-3 causes early aging and herniation, but not macular degeneration in mice. Hum Mol Genet. 2007;16(24):3059–3070. [DOI] [PubMed] [Google Scholar]
- 49. Driver SGW, Jackson MR, Richter K, et al. Biallelic variants in EFEMP1 in a man with a pronounced connective tissue phenotype. Eur J Hum Genet. 2020;28(4):445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vandeputte DA, Meije CB, van Dartel M, et al. GOA, a novel gene encoding a ring finger B-box coiled-coil protein, is overexpressed in astrocytoma. Biochem Biophys Res Commun. 2001;286(3):574–579. [DOI] [PubMed] [Google Scholar]
- 51. Ji YX, Huang Z, Yang X, et al. The deubiquitinating enzyme cylindromatosis mitigates nonalcoholic steatohepatitis. Nat Med. 2018;24(2):213–223. [DOI] [PubMed] [Google Scholar]
- 52. Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K.. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science. 1999;286(5440):771–774. [DOI] [PubMed] [Google Scholar]
- 53. Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G.. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424(6950):793–796. [DOI] [PubMed] [Google Scholar]
- 54. Kovalenko A, Chable-Bessia C, Cantarella G, Israël A, Wallach D, Courtois G.. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424(6950):801–805. [DOI] [PubMed] [Google Scholar]
- 55. Ganjam GK, Terpolilli NA, Diemert S, et al. Cylindromatosis mediates neuronal cell death in vitro and in vivo. Cell Death Differ. 2018;25(8):1394–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
URLs
(i)https://github.com/bulik/ldsc/wiki/Partitioned-Heritability (accessed 25 February 2022).
(ii)https://data.broadinstitute.org/alkesgroup/LDSCORE/ (accessed 25 February 2022)
(iii)https://cran.r-project.org/web/packages/seqMeta/index.html (accessed 10 December 2019)
(iv)http://genetics.bwh.harvard.edu/pph2/ (accessed 25 February 2022)
(v)https://www.ukbiobank.ac.uk/wp-content/uploads/2019/12/Description-of-the-alt-aware-issue-with-UKB-50k-WES-FE-data.pdf (accessed 2 February 2020)
(vi)http://betsholtzlab.org/VascularSingleCells/database.html (accessed 25 February 2022)