Abstract
An RNA degrading, high molecular weight complex was purified from Rhodobacter capsulatus. N-terminal sequencing, glycerol-gradient centrifugation, and immunoaffinity purification as well as functional assays were used to determine the physical and biochemical characteristics of the complex. The complex comprises RNase E and two DEAD-box RNA helicases of 74 and 65 kDa, respectively. Most surprisingly, the transcription termination factor Rho is a major, firmly associated component of the degradosome.
INTRODUCTION
Bacterial mRNAs have average half-lives of a few minutes only. In this way bacteria quickly adapt their mRNA pattern to rapidly changing environmental parameters. Cellular RNases degrade mRNAs with varying speed, depending on structural features of the RNA. mRNA degradation itself is possibly controlled by environmental parameters (for recent reviews see 1,2). The puf operon, coding for proteins of the Rhodobacter capsulatus photosynthetic complex, is one of the few model systems for degradation of polycistronic prokaryotic mRNAs (3,4). We have identified a variety of mRNA stabilizing and destabilizing structural elements within the primary transcript that are critical in the nucleolytic formation of mRNA fragments with quite different half-lives. This in turn leads to different molar amounts of translated proteins, required to form a functional photosynthetic complex in this organism.
The mRNA degradation machinery requires not only endo- and exoribonucleases, but also proteins other than nucleases. The identification of such additional components is currently developing into a model of highly ordered mRNA degradation in the bacterial cell (1,5).
The model of prokaryotic mRNA decay postulates a combined action of endo- and 3′→5′ exoribonucleases (6). The key enzyme for the initiation of mRNA degradation is endoribonuclease E (RNase E) (EC 3.1.26.-) (7). In vivo, RNase E recognizes specific cleavage sites, preferentially in single-stranded A/U-rich regions, and makes rate-determining endonucleolytic cuts (8,9). Fragments are then rapidly degraded by 3′→5′ exoribonucleases with mechanistic details varying depending on the stability of structures at the RNA 3′-end (10).
Escherichia coli RNase E is very sensitive to proteases, and purification of full-length RNase E requires optimal protective conditions. A major percentage of E.coli RNase E is part of a high molecular weight complex, the degradosome (11). In this complex, RNase E is associated with polynucleotide phosphorylase (PNPase) (EC 2.7.7.8), which together with RNase II is the most important 3′→5′ exonuclease in E.coli (12). Enolase and the ATP-dependent DEAD-box helicase RhlB were also identified as part of the degradosome (13), as was polyphosphate kinase (PPK) (EC 2.7.4.1), which catalyzes the conversion of poly-Pi and ADP, both inhibitors of RNA degradation, to ATP (14).
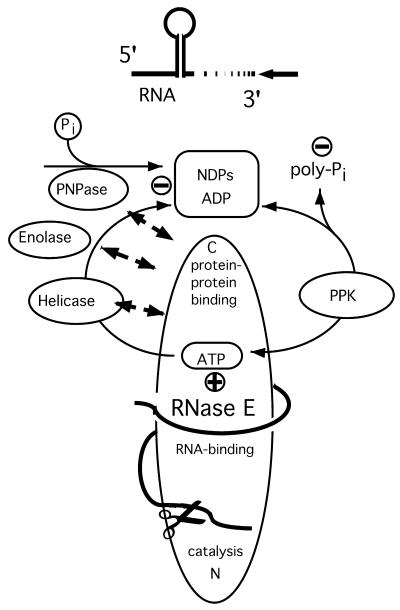
The C-terminal half of RNase E contains distinct binding sites for the degradosome components RhlB, enolase and PNPase (15). The degradosome is thus assembled on the C-terminal half of RNase E by direct RNase E–ligand contacts. Degradosome-like complexes have also been described in chloroplasts and yeast mitochondria (16–19). Figure 1 depicts the current model of the E.coli degradosome acting on RNA 3′-ends. In this model, RNase E is the assembly platform for a degradative complex directed towards the 3′-end of RNA.
Figure 1.
A model of the bacterial degradosome. This scheme presents current knowledge of the structural organization of the degradosome and its mode of action. NDPs inhibit PNPase, poly-phosphate probably inhibits the helicase. The model also depicts the current ideas about the interaction of known degradosome components. The ATP-dependent helicase dissolves RNA secondary structure and makes the RNA accessible for PNPase. PPK recycles ATP from NDPs; the role of enolase is still elusive. Ortho-phosphate Pi, poly-phosphate (Pi)n, dinucleotides NDP. + or – indicate stimulatory or inhibitory influence on mRNA degradation. (PPK, poly-phosphate-kinase; PNPase, polynucleotide-phosphorylase). The figure is adapted from Rauhut and Klug (1).
Although the organization of the degradative apparatus in a complex appears to be a recurring theme, there is no evidence for this from bacteria other than E.coli. Our previous studies in R.capsulatus made it clear that the degradation of the puf operon depends on rate-limiting cleavage by an RNase E-like activity (20,21). For our further analysis of mRNA degradation in R.capsulatus it is essential to understand whether this bacterium uses a degradosome complex. We could indeed purify a high molecular weight complex with degradative activity. Here we describe the characteristics of this complex and compare the identified components with those purified from other sources. The complex contains an RNase E of the apparent ‘180 kDa’ type and the Rho factor. Most interestingly, we find two DEAD-box RNA helicases of 65 and 74 kDa, respectively. Enolase and PNPase apparently are not major components of the R.capsulatus complex. Rhodobacter capsulatus is an α purple bacterium and thus only distantly related to E.coli. This provides some insight into the evolution of degradation complexes.
MATERIALS AND METHODS
Cell material
The wild-type R.capsulatus 37b4 strain (Deutsche Sammlung von Mikroorganismen, DSM 938) was used during this purification. Bacteria were grown under vigorous aeration in minimal malate medium (22) to an OD660 of ∼1.5.
Purification
All purification steps were performed between 0 and 8°C. Buffers contained 2 µg/ml aprotinin, 0.8 µg/ml leupeptin and 0.8 µg/ml pepstatin A (Fluka). A suspension of 100 g R.capsulatus cells in 100 ml of room temperature lysozyme–EDTA buffer containing 50 mM Tris–HCl pH 7.5, 100 mM NaCl, 5% glycerol, 3 mM EDTA, 1 mM dithiothreitol (DTT), 1.5 mg/ml lysozyme, and 1 mM phenylmethylsulfonyl fluoride (PMSF) (Promega) was prepared. After 40 min on ice, 50 ml of room temperature DNase–Triton buffer containing 50 mM Tris–HCl pH 7.5, 100 mM NaCl, 5% glycerol, 1 mM DTT, 3% Triton X-100, 30 mM magnesium acetate, 1 mM PMSF and 20 µg/ml DNase I (Promega) were added, followed by a 1 min low-speed blending. The lysate was kept on ice for 30 min and 37.5 ml of 5 M NH4Cl were slowly added. The lysate was stirred for an additional 30 min and clarified for 1 h at 27 000 g. A high-speed supernatant was then prepared at 100 000 g for 3.5 h. Proteins of this supernatant were precipitated with 40% ammonium sulphate, dissolved in 112.5 ml of buffer A containing 10 mM Tris–HCl pH 7.5, 5% glycerol, 0.5% Genapol X-080, 1 mM EDTA, 0.1 mM DTT, 0.1 mM PMSF, 50 mM NaCl and loaded on a sulphopropyl (SP)–Sepharose cation-exchanger column (Pharmacia) (9.5 × 1.6 cm), equilibrated with buffer A containing 50 mM NaCl. After washing with buffer A containing first 50 and then 300 mM NaCl, RNase E was eluted with 1 M NaCl and 0.5% Genapol X-080 in buffer A. Fractions were analyzed with denaturing PAGE and the silver staining of proteins. Peak fractions were pooled and stored at –80°C without apparent loss of activity even after several cycles of freezing and thawing. A similar protocol was successfully applied in the seminal paper of Carpousis et al. describing the first purification of the degradosome from E.coli (11).
Glycerol gradients
Peak fractions from the SP-column were diluted 2-fold with buffer A and layered on a 10–30% (w/v) glycerol gradient containing buffer A with 300 mM NaCl. Centrifugation was performed in a Beckman SW60 rotor at 4°C for 15 h at 37 000 r.p.m. Fractions were collected from the top of the gradient tube. For calibration purposes β-galactosidase (116 kDa subunit) and catalase (240 kDa) were subjected to centrifugation under identical conditions. The distribution of proteins was visualized by running aliquots of each fraction on denaturing polyacrylamide (PAA) gels with subsequent silver staining.
Protein sequencing
For protein sequencing, glycerol-gradient fractions were used. Proteins were separated during gel electrophoresis and blotted to Immobilon-PVDF membranes (Millipore). Protein bands were excised after staining with Ponceau S (Sigma) and sequenced by Edman degradation in a pulsed-liquid sequencer (Applied Biosystems, Inc., model 477A/120A), following the supplier’s guidelines.
Immunological characterization
Whole-cell extracts or column fractions were run on SDS–PAGE gels. Proteins were transferred onto Immobilon-PVDF membranes using semi-dry blotting. The efficient transfer of RNase E requires 1.6 mA/cm2 for 2 h. Incubation with primary antibodies, second antibody—horseradish peroxidase conjugate—and detection with NBT staining followed standard protocols. The antibody against R.capsulatus RNase E was raised in rabbits against electroeluted and freeze-dried protein of the 180 kDa band at Eurogentec (Belgium). The antibody against the E.coli RNase E was a gift from C. Higgins (Oxford). Antibodies against E.coli DnaK and GroEL proteins were provided by B. Bukau (Freiburg) and E.coli PNPase antibody by C. Portier (Paris).
Immunoprecipitation
Antibodies raised against R.capsulatus RNase E proteins were directly coupled to protein A–Sepharose beads as described previously (23). One hundred microliters of this resin in buffer A containing 300 mM NaCl were incubated with fractions from the SP–Sepharose column at 4°C for 2 h. The fractions were pre-treated with 1.5 µg/µl RNase A. After washing with the same buffer, proteins were eluted with 20 µl 0.1 M glycine pH 2.0 and analyzed with SDS–PAGE.
PNPase activity
The PNPase activity assay uses the unique ability of PNPase to form poly(A) from ADP (13). Protein fractions from glycerol gradients were separated on a native PAA gel. The gel was then incubated in 0.1 M Tris–HCl buffer pH 8.0, 20 mM MgCl2 and 20 mM ADP. After 5 h at 37°C, synthesized poly(A) was stained in situ with 1% acridine orange, thus visualizing the presence of PNPase in certain fractions (24).
Activity test for RNase E
For nuclease activity assays we used the pZBP RNA construct. Derived from the R.capsulatus puf operon, it comprises two closely spaced RNase E cleavage sites, which are both processed during in vitro degradation assays. Within the puf operon, one of the sites is critical for the rate-determining endonucleolytic cleavage of puf transcripts in vivo (Fig. 2B) (21). A 217 nt RNA was transcribed in vitro from a HindIII linearized pGEM3Zf(+)plasmid in the presence of [α-32P]UTP (25). Approximately 5000 c.p.m. of labeled RNA were dissolved in 8 µl of a buffer containing 10 mM Tris–HCl pH 7.5, 5 mM MgCl2, 0.2 mg/ml carrier yeast tRNA and incubated with 2 µl of RNase E containing fractions at 30°C. When necessary, protein fractions were diluted with buffer A containing 300 mM NaCl. The reaction was terminated for 10 min at 50°C with 10 µl of 10 mM Tris–HCl pH 7.5, 10 mM EDTA, 0.2% SDS, and 1 mg/ml proteinase K. Generated RNA fragments were then visualized autoradiographically after separation on 7 M urea, 9% PAA gels.
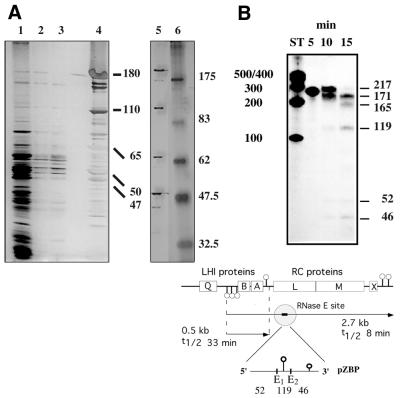
Figure 2.
(A) Purification of the R.capsulatus degradosome. The figure shows the protein pattern observed during different steps of the purification (left gel 3.5–14.5% PAA, right gel 8% PAA). Lane 1, flow-through from SP column; lanes 2 and 3, salt washes of same column; lane 4, eluted proteins from this column (1 M salt); lane 5, degradosome proteins as observed in fraction 8 of the glycerol gradient; lane 6, molecular weight standard (kDa). (B) Degradation assay. The diagram shows the pZBP substrate in the puf context. RNase E cleavage at site E1 initiates in vivo degradation of the 2.7 kb pufBALMX mRNA. In vitro, both RNase E sites, E1 and E2, are processed. Pins indicate RNA secondary structure. (LHI, light harvesting complex I proteins; RC, reaction center proteins; t1/2, observed half-lives for the 2.7 and the pufBA 0.5 kb messenger.) For a typical activity test, shown in the gel picture, 5000 c.p.m. of [α-32P]UTP labeled pZBP RNA were incubated with 2 µl of diluted gradient fraction 9 at 30°C for the time indicated. Fragments were separated on a 7 M urea PAA-gel. ST, RNA calibration standard, numbers indicating nucleotides.
Helicase assay
To screen for helicase and concomitant 3′→5′ exonuclease activity, we used the helicase assay of Py et al. (13) with a substrate similar to the one described by McLaren et al. (26). The malE–malF intergenic region of the E.coli genome (nt 424 2645–424 2958) was cloned under T7 control and mRNA was transcribed in vitro in the presence of [α-32P]UTP. The folding of this 318 nt RNA is predicted to form an RNA with two stem–loops impeding 3′→5′ degradation. The substrate was incubated at 32°C with degradosome fractions and helicase buffer with or without 5 mM ATP (13). Reactions were then phenolized, ethanol precipitated and run on a 6% PAA–7 M urea gel. RNA was visualized with autoradiography.
To degrade the substrate with PNPase only, RNA was incubated with 0.1 µg of E.coli PNPase in 20 µl helicase buffer and analyzed as above. Escherichia coli PNPase was kindly provided by C. Portier (Paris).
RESULTS
Purification
After ammonium-sulphate fractionation, SP–Sepharose chromatography, and glycerol-gradient centrifugation we obtained fractions with a protein pattern very similar (also stoichiometrically) to that obtained from immunoprecipitated degradosomes (Figs 2A, 3 and 4).During immunoprecipitation, only proteins forming a stable complex with RNase E are precipitated (see below). Glycerol-gradient fractions obtained, therefore represent highly purified degradosomes. Most prominently, there is a ‘180 kDa’ RNase E in R.capsulatus. This presumably resembles the situation observed for the E.coli RNase E, a 118 kDa protein migrating with an apparent molecular mass of 180 kDa due to a prolin-rich C-terminus. As for E.coli, the presence of protease inhibitors during purification was absolutely essential for obtaining full-length RNase E and an intact complex in R.capsulatus. We found that, once purified, the complex from R.capsulatus, especially the RNase E, even after repeated cycles of freezing and thawing is much more stable than the complex we obtained from a parallel purification using E.coli cells. The amount of degradosome complex per gram of starting material appeared to be similar in both organisms. Apart from the 180 kDa band, two additional protein bands of 175 and 110 kDa in our preparation of the R.capsulatus degradosome cross-reacted with RNase E antibody. They are most likely degradation products of full-length RNase E (see below).
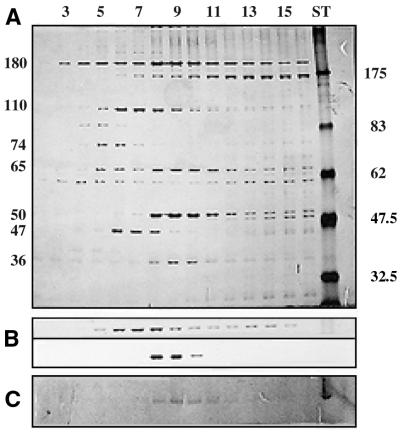
Figure 3.
Glycerol-gradient centrifugation of the R.capsulatus degradosome. (A) The collected fractions from fraction 2 (top) to 16 (bottom). Numbers indicating molecular masses in kDa. ST, protein standards. (B) The sedimentation of gradient calibration proteins β-galactosidase (116 kDa subunit; top) and catalase (240 kDa; bottom). (C) The PNPase assay. In all three panels identical fractions are aligned vertically. Protein size indicated in kDa.
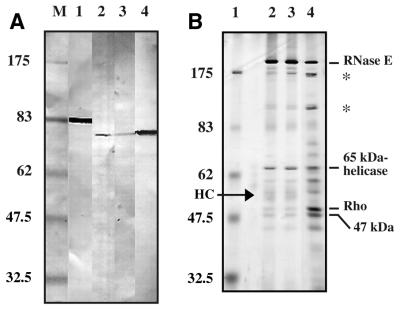
Figure 4.
(A) Immunodetection of PNPase with E.coli PNPase antibody. Lane 1, E.coli extract; lanes 2 and 3, R.capsulatus glycerol gradient, fractions 8 and 10, respectively; lane 4, R.capsulatus SP fraction; M, protein marker (kDa). (B) The gel shows the proteins eluted from an anti-RNase E immunoaffinity resin. Lane 1, molecular weight standard (kDa); lane 2, precipitation with RNase added; lane 3, without RNase; lane 4, fractions loaded. Band of antibody heavy chain (HC).
To estimate the increase in specific activity during purification, we used a degradation assay with the pZBP substrate (Fig. 2B). Specific activity, measured as the decrease of full-length substrate per (min × mg protein), increased ∼550-fold from first supernatant to gradient peak fractions.
Glycerol-gradient centrifugation
During centrifugation, free RNase E sediments according to its true molecular mass of 118 kDa (for the E.coli enzyme). Using carefully standardized gradient runs with material from individual preparations, we obtained a highly reproducible protein pattern in the collected fractions (Fig. 3). Aberrant sedimentation of several proteins, not matching molecular masses, clearly indicated the presence of protein complexes. Significant was the heterogeneity of the observed complex. In fractions 5–11 we observed that RNase E associated with various other proteins. These fractions reach from the sedimentation position of β-galactosidase (116 kDa subunit) to that of catalase (240 kDa) during calibration runs (see Fig. 3B). In fraction 9 we observed the exact co-sedimentation of four protein bands of highly different molecular weights. Using protein sequencing, we have identified three of these bands as RNase E (180 kDa), a DEAD-box RNA helicase (65 kDa) and the transcription termination factor Rho (50 kDa). The nature of the fourth band (36 kDa), instead, remains unresolved. The 74 kDa band in fractions 5 and 6 was identified as a second DEAD-box RNA helicase. A prominent 47 kDa protein could also not be identified. Other distinct forms of the complex with much higher molecular weights, indicating differing stoichiometry of the constituent protein components were not observed.
Protein sequencing
N-terminal sequencing identified several components of the complex visible in the gradient fractions. For database searches we used the sequence entries of the R.capsulatus genome project available through the IGwit home page (www.IntegratedGenomics.com). The band migrating at a 180 kDa position during electrophoresis was clearly identified as RNase E. The first 17 amino acids MAKKMLIDATHAEETRV exhibit a 64% identity of the R.capsulatus enzyme with the known RNase E sequence of E.coli. The R.capsulatus enzyme is currently not in the database. The 65 kDa band, which peaked in fraction 9, revealed an N-terminus MTKFIDLNLDPAVL. This matches the entry for R.capsulatus ORF 1970, an ATP-dependent DEAD-box RNA helicase with a calculated molecular mass of 60.5 kDa. The N-terminus of the 74 kDa band provided the sequence XDFPXLPAALAE matching R.capsulatus ORF 4133, another DEAD-box RNA helicase with a predicted mass of 73.2 kDa. The 50 kDa band has an N-terminus MTERLNLSDLKAXSPXDLLAM, which matches the R.capsulatus ORF 3312 coding for transcription termination factor Rho. The database entry for the predicted protein of this ORF has an N-terminal extension of 37 amino acids due to an incorrectly assigned start codon. The corrected molecular mass is 47.2 kDa. Our 47 kDa band, sedimenting slightly slower than the 50 kDa band provided the N-terminus VKLIGIA, which resembles the N-terminus of E.coli enolase. As this sequence does not match the entry for enolase, already in the R.capsulatus database, the nature of this protein remains unresolved. The 36 kDa band provided an N-terminus MXSSKSXS, which did not match with any database entry.
Immunological characterization
The antibody against the R.capsulatus RNase E was used on a western blot of the glycerol gradient fractions. Except for the full-length 180 kDa RNase E band, the antibody reacts with the 175 and 110 kDa bands (not shown). These bands apparently originate from RNase E. Whereas antibodies raised against E.coli RNase E cross reacted with the R.capsulatus enzyme, the R.capsulatus antibody reacted strongly with the R.capsulatus RNase E, but failed to recognize the E.coli protein (21). The antibody against E.coli PNPase strongly reacts with a 75 kDa protein in R.capsulatus SP fractions and recognizes a protein of the same size in glycerol-gradient fractions 8–10 (Fig. 4A). This signal is very weak though, compared with the signal in the SP fractions, indicating that the Rhodobacter degradosome preparation contains only a minute fraction of the cellular PNPase activity. To screen for the presence of DnaK and GroEL protein, both associated with the E.coli degradosome in non-stoichiometric amounts (27), we tested crude cell fractions and glycerol-gradient degradosome fractions with antibodies against E.coli DnaK and GroEL proteins. Both proteins are clearly present in R.capsulatus crude extracts, but were not detected as part of the degradosome (not shown).
Immunoprecipitation of the degradosome
To provide additional evidence for the physical interaction of proteins in a complex, fractions from the SP column were bound to an anti-RNase E antibody resin. After careful washing, only proteins forming a stable complex with RNase E are retained on this resin. These were eluted and the protein pattern was analyzed. Almost all of the proteins observed in the SP fractions were retained by the antibody resin; in particular, RNase E (180, 175, 110 kDa), the Rho factor (50 kDa), the 65 kDa DEAD-box RNA helicase and the unknown 47 kDa protein (Fig. 4B). The 74 kDa helicase and the unknown 36 kDa protein do not tightly associate with RNase E, at least not under these conditions.
Enzymatic assays
To provide experimental evidence for identified or putative enzymes various specific tests were performed. PNPase was described as a component of the E.coli degradosome (11). We have been able to identify a PNPase-like protein by immunological techniques, but not with any of the sequenced N-termini. Also, we could clearly identify a weak PNPase activity using a highly specific test for PNPase activity, during which poly(A) is formed from ADP (13). The PNPase activity co-sedimented with the two helicases and RNase E, and its peak activity was observed again in fraction 9 of the gradient run (see Fig. 3C). As a visible protein band cannot be assigned to PNPase in this fraction, PNPase apparently is not a major component of the R.capsulatus degradosome.
To screen for RNase E activity we used the R.capsulatus puf operon-derived substrate pZBP, specific for RNase E. It was accurately processed upon incubation with degradosome containing fractions throughout the purification. Figure 2B shows a time course of degradation. The 3′ proximal cleavage site is processed first, as it is the 171 nt 2/3-intermediate which appears before the 165 nt intermediate. Activity correlated with the protein pattern after glycerol-gradient centrifugation and was highest around fraction 9 (not shown).
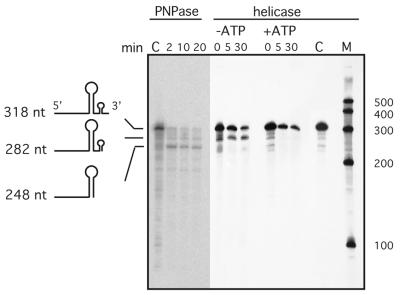
Using a substrate with two internal stem–loops (13,26) we could demonstrate the presence of helicase and exonuclease activity in our degradosome preparation (Fig. 5). In the absence of ATP, 3′→5′ exonuclease degradation of the RNA substrate already stalls at the base of the minor 3′ stem–loop. In the presence of ATP, the unwinding of RNA duplexes through helicases resulted in 3′→5′ degradation without intermediates.
Figure 5.
Helicase assay. Degradation of the 318 nt substrate is monitored during a 30 min incubation in the presence or absence of ATP. Intermediates are shown to the left. Zero minute samples were prepared in the presence and control C in the absence of added protein. M, RNA length standard (nt). The gel on the left shows the degradation pattern when substrate is incubated with PNPase only.
When a substrate was incubated with pure PNPase only (Fig. 5), the same pattern of two intermediates like in the helicase assay is observed, but exonucleolytic degradation cannot proceed through the major stem–loop. This again demonstrates that an exonucleolytic activity must also be present in our preparation of the R.capsulatus degradosome.
DISCUSSION
Applying appropriate anti-proteolytic measures, we have purified full-length RNase E as part of a degradosome-like complex from R.capsulatus. The enzyme, when incubated with the specific RNase E substrate pZBP from R.capsulatus, efficiently cleaves at correct positions. Escherichia coli RNase E is 1061 amino acids long with a calculated molecular mass of 118 kDa, but runs at a 180 kDa position probably due to a prolin-rich C-terminal region (28). The N-terminal half of the protein contains the catalytic center (29–31). Rhodobacter capsulatus RNase E migrates with an apparent molecular mass of 180 kDa, thus presumably following the aberrant behavior of the E.coli 118 kDa RNase E. Like almost all bacterial RNases E, the R.capsulatus enzyme is a ‘large size’ RNase E, whereas RNases E from chloroplasts are lacking the entire C-terminal half (32). The bacterial catalytic N-terminal half is highly conserved, quite contrary to the C-terminal half. It is this C-terminus that provides specific protein–protein binding sites and assembles the degradosome in E.coli (15,32). An RNase E with a missing C-terminus should not facilitate degradosome formation. For this reason proteolysis of RNase E disrupts complex formation (11,24). On the other hand, degradative complexes of a similar size observed in bacteria have also been described in chloroplasts, despite the missing bacterial C-terminus (32,33). The possibly ancient evolutionary design of joining degradative activities in a complex may have led to several independent solutions. The R.capsulatus RNase E during immunopurification and glycerol-gradient centrifugation clearly behaves like a complex. Immunoprecipitation with or without RNase A added does not result in different protein patterns. RNA-mediated co-precipitation of proteins can, therefore, be excluded. Without protease inhibitors, RNase E in R.capsulatus is rapidly destroyed with a main fragment of ∼60 kDa (not shown). A complex is not observed in this case. Our preparation of the R.capsulatus degradosome shows RNase E associated with two DEAD-box RNA helicases. The 75 kDa enzyme, though, does not appear to be tightly attached to RNase E, as it does not bind to the immunoaffinity resin. Evidence for helicases not only stems from direct N-terminal sequencing, but also from the functional assay. The 3′→5′ degradation through the stem–loop of the substrate requires ATP-dependent helicases when R.capsulatus degradosome fractions are used (Fig. 5). Due to the heterogeneity of the sedimenting complex, the quaternary structure of the E.coli degradosome is unknown (11,15,32). The molecular mass was estimated at 160–500 kDa (11,24). The protein pattern observed after glycerol gradients, during our preparation, tends to favor the lower end of this estimate as the 240 kDa marker occupies the same position, like the proteins observed in fraction 9.
We could not unambiguously identify enolase, known from E.coli to be a degradosome component. PNPase activity, as followed by specific synthesis of poly(A), is observed around fraction 9 (Fig. 3C). A protein band could not be assigned to PNPase in this fraction, although PNPase protein is present in fractions 8–10 as shown with the E.coli PNPase antibody, which recognizes a 75 kDa R.capsulatus protein in these fractions. The important exonuclease PNPase is not always found in bacterial genomes (34). The R.capsulatus database, though, contains an entry for a predicted 78.3 kDa PNPase protein (ORF 3090), which does not match any of our N-termini. PNPase is possibly only a very minor non-stoichiometric component of the complex. Consistent with this interpretation, the E.coli PNPase antibody does not recognize a protein band in the immunoprecipitated degradosome from R.capsulatus. PNPase uses ADP for poly(A) synthesis. As poly(A) synthesis by poly(A)-polymerase requires ATP, the observed poly(A) stain cannot be attributed to a possible presence of this enzyme. In E.coli, PNPase is a homotrimer with 78 kDa subunits and a major component of the degradosome (11). The homotrimer would, therefore, have a sedimenting molecular mass of ∼240 kDa, corresponding to fraction 9. Therefore, we cannot exclude that PNPase is simply a residual activity present in our preparation, sedimenting according to its molecular mass, but not related to the complex.
Distinct complexes with a much higher molecular mass than observed in fraction 9 are not present in our preparation. Forms of RNase E trailing to the bottom of the gradient may be due to oligomerization of RNase E, with other degradosome proteins remaining attached (15,35). DnaK and GroEL, which have been demonstrated to be non-stoichiometric components of the E.coli degradosome, are not found (15,27,36,37).
The transcription termination factor Rho, a protein acting on 3′-ends, is a major component of the R.capsulatus degradosome; it firmly connects to RNase E during immunopurification of the complex. It is tempting to speculate about the fact that proteins acting on 3′-ends join in one complex. The Rho protein is itself a hexameric helicase acting on RNA–DNA duplexes (38).
Although not visible in the immunoprecipitation experiment, the 74 kDa helicase co-purifies with the complex during protein purification. DEAD-box RNA helicases unwind RNA duplexes (39–41). Their general importance as a driving force in changes of protein–RNA interactions is increasingly recognized (42). The presence of a second helicase in the degradosome would follow the pattern observed in other degradation complexes. The yeast mitochondrial complex contains three 3′→5′ exonucleases and an RNA helicase (18,19). The yeast exosome instead, a nuclear complex, harbors exonucleases and interacts with ATP-dependent RNA helicase cofactors (43,44). These helicases probably unwind RNA prior to degradation (45,46). Many prokaryotic mRNAs display 3′-UTRs with intrinsic rho-independent transcription termination stem–loops. They can also serve as a protective measure against 3′→5′ exonucleases, which stall at the base of the stem (12,26,47–50). This led to the model of the degradosome in which helicases unwind impeding stem–loops (Fig. 1) (51,52). It was demonstrated that RhlB, the helicase described in the E.coli degradosome, unwinds RNA hairpins in an ATP-dependent reaction during degradosome-mediated RNA degradation (13). We could show that helicases are acting similarly in R.capsulatus. The helicase assay at the same time made clear that exonucleolytic activity must be present, although we could not identify the corresponding protein. A recent computer analysis of completely sequenced bacterial and archaeal genomes had the surprising result that the majority of the organisms do not form stem–loops in their 3′-UTRs (34). This not only has implications for the mechanism of transcription termination, but also for mRNA degradation. In fact, these genomes often lack detectable genes for PNPase and RNase II (34). Data for R.capsulatus are not available yet. Escherichia coli, the only other bacterium in which a degradosome has been described, clearly uses stem–loops in 3′-UTRs.
Coburn and Mackie recently showed, that in E.coli, possibly only the removal of very stable stem–loops requires helicases and a degradosome (10). Rhodobacter capsulatus, as a GC-rich organism, could be a case where the presence of a second helicase is required to overcome particularly stable stem–loops. A second helicase may also be necessitated by impeding factors, found in E.coli to bind to 3′ stem–loops (53). Escherichia coli RhlB helicase shares extensive family-specific homology with the two R.capsulatus helicases.
Can the degradosome model be generalized for all bacteria? With >40 microbial genomes published, GenBank currently contains only 11 bacterial sequences with RNase E annotation, 10 are ‘long type’ RNases E. The situation is somewhat complicated by various sequences for RNase G, the recently renamed CafA protein (54). It closely resembles the N-terminal half of RNase E, which can lead to incorrect annotations. The activities of RNases G and E are distinct, but overlapping, and it has already been pointed out that this may also lead to wrong experimental conclusions (54,55).
It has been shown that repeated cycles of polyadenylation and PNPase action can degrade stem–loops, without the need for helicase action (10). Only now that a degradosome has been purified from an organism other than E.coli can one assume that a degradosome complex is the road taken during bacterial RNA degradation. But already this R.capsulatus degradosome deviates significantly from the complex observed in E.coli. Notably, an exonuclease activity appears not to be present as a major component. This may reflect the fact that the α purple bacterium R.capsulatus is evolutionarily quite far away from γ purple bacteria like E.coli. Degradosomes from other bacteria need to be purified in order to develop a more comprehensive picture of the varieties of prokaryotic RNA degradation.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank C. Higgins and B. Bukau for providing antibodies and C. Portier for PNPase protein and antibody. Early sequencing work in this project was done by Dr D. Linder, Giessen Sequencing Laboratory.
REFERENCES
- 1.Rauhut R. and Klug,G. (1999) mRNA degradation in bacteria. FEMS Microbiol. Rev., 23, 353–370. [DOI] [PubMed] [Google Scholar]
- 2.Steege D.A. (2000) Emerging features of mRNA decay in bacteria. RNA, 6, 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belasco J.G., Beatty,J.T., Adams,C.W., von Gabain,A. and Cohen,S.N. (1985) Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell, 40, 171–181. [DOI] [PubMed] [Google Scholar]
- 4.Klug G. (1993) The role of mRNA degradation in the regulated expression of bacterial photosynthesis genes. Mol. Microbiol., 9, 1–7. [DOI] [PubMed] [Google Scholar]
- 5.Coburn G.A. and Mackie,G.A. (1999) Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol., 62, 55–108. [DOI] [PubMed] [Google Scholar]
- 6.Nierlich D.P. and Murakawa,G.J. (1996) The decay of bacterial messenger RNA. Prog. Nucleic Acid Res. Mol. Biol., 52, 153–216. [DOI] [PubMed] [Google Scholar]
- 7.Cohen S.N. and McDowall,K.J. (1997) RNase E: still a wonderfully mysterious enzyme. Mol. Microbiol., 23, 1099–1106. [DOI] [PubMed] [Google Scholar]
- 8.Lin-Chao S., Wong,T.T., McDowall,K.J. and Cohen,S.N. (1994) Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNA I encoded by the pBR322 plasmid. J. Biol. Chem., 269, 10797–10803. [PubMed] [Google Scholar]
- 9.Arraiano C.M., Cruz,A.A. and Kushner,S.R. (1997) Analysis of the in vivo decay of the Escherichia coli dicistronic pyrF- orfF transcript: evidence for multiple degradation pathways. J. Mol. Biol., 268, 261–272. [DOI] [PubMed] [Google Scholar]
- 10.Coburn G.A. and Mackie,G.A. (1998) Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J. Mol. Biol., 279, 1061–1074. [DOI] [PubMed] [Google Scholar]
- 11.Carpousis A.J., Van Houwe,G., Ehretsmann,C. and Krisch,H.M. (1994) Copurification of E.coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell, 76, 889–900. [DOI] [PubMed] [Google Scholar]
- 12.Higgins C.F., Causton,H.C., Dance,G.S.C. and Mudd,E.A. (1993) The role of the 3′ end in mRNA stability and decay. In Belasco,J.G. and Brawerman,G. (eds.), RNA. Academic Press, San Diego, CA, pp. 13–52.
- 13.Py B., Higgins,C.F., Krisch,H.M. and Carpousis,A.J. (1996) A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature, 381, 169–172. [DOI] [PubMed] [Google Scholar]
- 14.Kornberg A. (1995) Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J. Bacteriol., 177, 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanzo N.F., Li,Y.S., Py,B., Blum,E., Higgins,C.F., Raynal,L.C., Krisch,H.M. and Carpousis,A.J. (1998) Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev., 12, 2770–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lisitsky I., Kotler,A. and Schuster,G. (1997) The mechanism of preferential degradation of polyadenylated RNA in the chloroplast. The exoribonuclease 100RNP/polynucleotide phosphorylase displays high binding affinity for poly(A) sequence. J. Biol. Chem., 272, 17648–17653. [DOI] [PubMed] [Google Scholar]
- 17.Hayes R., Kudla,J., Schuster,G., Gabay,L., Maliga,P. and Gruissem,W. (1996) Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J., 15, 1132–1141. [PMC free article] [PubMed] [Google Scholar]
- 18.Min J., Heuertz,R.M. and Zassenhaus,H.P. (1993) Isolation and characterization of an NTP-dependent 3′-exoribonuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem., 268, 7350–7357. [PubMed] [Google Scholar]
- 19.Margossian S.P., Li,H., Zassenhaus,H.P. and Butow,R.A. (1996) The DExH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell, 84, 199–209. [DOI] [PubMed] [Google Scholar]
- 20.Klug G., Jock,S. and Rothfuchs,R. (1992) The rate of decay of Rhodobacter capsulatus-specific puf mRNA segments is differentially affected by RNase E activity in Escherichia coli. Gene, 121, 95–102. [DOI] [PubMed] [Google Scholar]
- 21.Fritsch J., Rothfuchs,R., Rauhut,R. and Klug,G. (1995) Identification of an mRNA element promoting rate-limiting cleavage of the polycistronic puf mRNA in Rhodobacter capsulatus by an enzyme similar to RNase E. Mol. Microbiol., 15, 1017–1029. [DOI] [PubMed] [Google Scholar]
- 22.Drews G. (1976) Mikrobiologisches Praktikum, 3rd edn. Springer-Verlag, Berlin.
- 23.Harlow E. and Lane,D. (1988) Antibodies. A Laboratory Manual. CSH Press, Cold Spring Harbor, NY, pp. 522–523.
- 24.Py B., Causton,H., Mudd,E.A. and Higgins,C.F. (1994) A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol., 14, 717–729. [DOI] [PubMed] [Google Scholar]
- 25.Rauhut R., Jäger,A., Conrad,C. and Klug,G. (1996) Identification and analysis of the rnc gene for RNase III in Rhodobacter capsulatus. Nucleic Acids Res., 24, 1246–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLaren R.S., Newbury,S.F., Dance,G.S., Causton,H.C. and Higgins,C.F. (1991) mRNA degradation by processive 3′–5′ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J. Mol. Biol., 221, 81–95. [PubMed] [Google Scholar]
- 27.Miczak A., Kaberdin,V.R., Wei,C.L. and Lin-Chao,S. (1996) Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl Acad. Sci. USA, 93, 3865–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casaregola S., Jacq,A., Laoudj,D., McGurk,G., Margarson,S., Tempete,M., Norris,V. and Holland,I.B. (1992) Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J. Mol. Biol., 228, 30–40. [DOI] [PubMed] [Google Scholar]
- 29.Kido M., Yamanaka,K., Mitani,T., Niki,H., Ogura,T. and Hiraga,S. (1996) RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J. Bacteriol., 178, 3917–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDowall K.J. and Cohen,S.N. (1996) The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding site. J. Mol. Biol., 255, 349–355. [DOI] [PubMed] [Google Scholar]
- 31.Huang H., Liao,J. and Cohen,S.N. (1998) Poly(A)- and poly(U)-specific RNA 3′ tail shortening by E.coli ribonuclease E. Nature, 391, 99–102. [DOI] [PubMed] [Google Scholar]
- 32.Kaberdin V.R., Miczak,A., Jakobsen,J.S., Lin-Chao,S., McDowall,K.J. and von Gabain,A. (1998) The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl Acad. Sci. USA, 95, 11637–11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayes R., Kudla,J. and Gruissem,W. (1999) Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem. Sci., 24, 199–202. [DOI] [PubMed] [Google Scholar]
- 34.Washio T., Sasayama,J. and Tomita,M. (1998) Analysis of complete genomes suggests that many prokaryotes do not rely on hairpin formation in transcription termination. Nucleic Acids Res., 26, 5456–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackie G.A., Genereaux,J.L. and Masterman,S.K. (1997) Modulation of the activity of RNase E in vitro by RNA sequences and secondary structures 5′ to cleavage sites. J. Biol. Chem., 272, 609–616. [PubMed] [Google Scholar]
- 36.Blum E., Py,B., Carpousis,A.J. and Higgins,C.F. (1997) Polyphosphate kinase is a component of the Escherichia coli RNA degradosome. Mol. Microbiol., 26, 387–398. [DOI] [PubMed] [Google Scholar]
- 37.Ybarra J. and Horowitz,P.M. (1996) Nucleotides reveal polynucleotide phosphorylase activity from conventionally purified GroEL. J. Biol. Chem., 271, 25063–25066. [DOI] [PubMed] [Google Scholar]
- 38.Bogden C.E., Fass,D., Bergman,N., Nichols,M.D. and Berger,J.M. (1999) The structural basis for terminator recognition by the Rho transcription termination factor. Mol. Cell., 3, 487–493. [DOI] [PubMed] [Google Scholar]
- 39.Luking A., Stahl,U. and Schmidt,U. (1998) The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol., 33, 259–296. [DOI] [PubMed] [Google Scholar]
- 40.Bird L.E., Subramanya,H.S. and Wigley,D.B. (1998) Helicases: a unifying structural theme? Curr. Opin. Struct. Biol., 8, 14–18. [DOI] [PubMed] [Google Scholar]
- 41.Schmid S.R. and Linder,P. (1992) D-E-A-D protein family of putative RNA helicases. Mol. Microbiol., 6, 283–291. [DOI] [PubMed] [Google Scholar]
- 42.Linder P., Tanner,N.K. and Banroques,J. (2001) From RNA helicases to RNPases. Trends Biochem. Sci., 26, 339–341. [DOI] [PubMed] [Google Scholar]
- 43.de la Cruz J., Kressler,D., Tollervey,D. and Linder,P. (1998) Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J., 17, 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson J.S.J. and Parker,R.P. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell P. and Tollervey,D. (2000) mRNA stability in eukaryotes. Curr. Opin. Genet. Dev., 10, 193–198. [DOI] [PubMed] [Google Scholar]
- 46.Mitchell P. and Tollervey,D. (2000) Musing on the structural organization of the exosome complex. Nature Struct. Biol., 7, 843–846. [DOI] [PubMed] [Google Scholar]
- 47.Chen C.Y., Beatty,J.T., Cohen,S.N. and Belasco,J.G. (1988) An intercistronic stem–loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell, 52, 609–619. [DOI] [PubMed] [Google Scholar]
- 48.Klug G., Adams,C.W., Belasco,J., Doerge,B. and Cohen,S.N. (1987) Biological consequences of segmental alterations in mRNA stability: effects of deletion of the intercistronic hairpin loop region of the Rhodobacter capsulatus puf operon. EMBO J., 6, 3515–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newbury S.F., Smith,N.H. and Higgins,C.F. (1987) Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell, 51, 1131–1143. [DOI] [PubMed] [Google Scholar]
- 50.Mackie G.A. (1987) Posttranscriptional regulation of ribosomal protein S20 and stability of the S20 mRNA species. J. Bacteriol., 169, 2697–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson J.S. and Parker,R. (1996) RNA turnover: the helicase story unwinds. Curr. Biol., 6, 780–782. [DOI] [PubMed] [Google Scholar]
- 52.Carpousis A.J., Vanzo,N.F. and Raynal,L.C. (1999) mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet., 15, 24–28. [DOI] [PubMed] [Google Scholar]
- 53.Causton H., Py,B., McLaren,R.S. and Higgins,C.F. (1994) mRNA degradation in Escherichia coli: a novel factor which impedes the exoribonucleolytic activity of PNPase at stem-loop structures. Mol. Microbiol., 14, 731–741. [DOI] [PubMed] [Google Scholar]
- 54.Li Z., Pandit,S. and Deutscher,M.P. (1999) RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J., 18, 2878–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tock M.R., Walsh,A.P., Carroll,G. and McDowall,K.J. (2000) The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem., 275, 8726–8732. [DOI] [PubMed] [Google Scholar]