Abstract
The recent development of experimental methods for measuring chromatin state at single-cell resolution has created a need for computational tools capable of analyzing these datasets. Here we developed Signac, a comprehensive toolkit for the analysis of single-cell chromatin data. Signac enables an end-to-end analysis of single-cell chromatin data, including peak calling, quantification, quality control, dimension reduction, clustering, integration with single-cell gene expression datasets, DNA motif analysis and interactive visualization. Through its seamless compatibility with the Seurat package, Signac facilitates the analysis of diverse multimodal single-cell chromatin data, including datasets that co-assay DNA accessibility with gene expression, protein abundance and mitochondrial genotype. We demonstrate scaling of the Signac framework to analyze datasets containing over 700,000 cells.
Several technologies are now available for measuring aspects of chromatin state at single-cell resolution, particularly single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) and scCUT&Tag1-11. The development of these new technologies has created a need for computational tools to analyze single-cell chromatin data. While the analysis of these datasets presents some unique challenges in comparison to more established single-cell methods such as single-cell RNA sequencing (scRNA-seq), many analysis steps are shared. These include nonlinear dimension reduction, cell clustering, identifying differentially active features between groups of cells and visualizing cells in reduced-dimension space. Alongside these common tasks, the analysis of single-cell chromatin data presents opportunities for several more-specialized analysis tasks. These include identifying DNA sequence features (motifs or variants) that are enriched in different sets of cells, specialized feature weighting and linear dimension reduction methods and genome browser-style data visualization.
Furthermore, new technologies now enable the co-assay of multiple cellular modalities in single cells, including DNA accessibility alongside mRNA abundance12-20, protein abundance21-23, CRISPR guide RNAs24,25 or spatial position26. These datasets present unique opportunities to learn the relationships between cellular modalities27 and will be especially powerful in deciphering the regulatory roles of noncoding DNA sequences. The analysis of these datasets is challenging without software designed to facilitate a multimodal analysis and an ideal computational solution would facilitate an integrative analysis of multimodal single-cell data encompassing gene expression, chromatin state and other modalities, including cell lineage, protein expression or spatial position in a single framework. Many new methods have been developed to address individual steps in the analysis of single-cell chromatin data, including specialized methods for dimensionality reduction28-30, peak co-accessibility analysis31 and DNA motif enrichment32. However, existing toolkits designed to facilitate end-to-end analysis of single-cell chromatin data were designed for analysis of unimodal single-cell datasets33-35 or have limited functionality36-38, limiting the ability of investigators to analyze multimodal single-cell chromatin data in a consistent framework.
Here we developed Signac, a framework for the analysis of single-cell chromatin data. While Signac is a standalone solution for the analysis of single-cell chromatin data, we also designed Signac to interface seamlessly with the Seurat package to enable the analysis of multimodal single-cell datasets39-42. Signac enables end-to-end analysis of chromatin data and includes functionality for diverse analysis tasks, including identifying cells from background noncell-containing barcodes, calling peaks, quantifying counts in genomic regions, quality control filtering of cells, dimension reduction, clustering, integration with single-cell gene expression data, interactive genome browser-style data visualization, finding differentially accessible peaks, finding enriched DNA sequence motifs, transcription factor footprinting and linking peaks to potential regulatory target genes (Fig. 1a and Supplementary Table 1). Furthermore, Signac provides a framework for the identification of mitochondrial genome variants from single-cell DNA accessibility experiments, enabling a joint analysis of clonal relationships and DNA accessibility in single cells15,16,43.
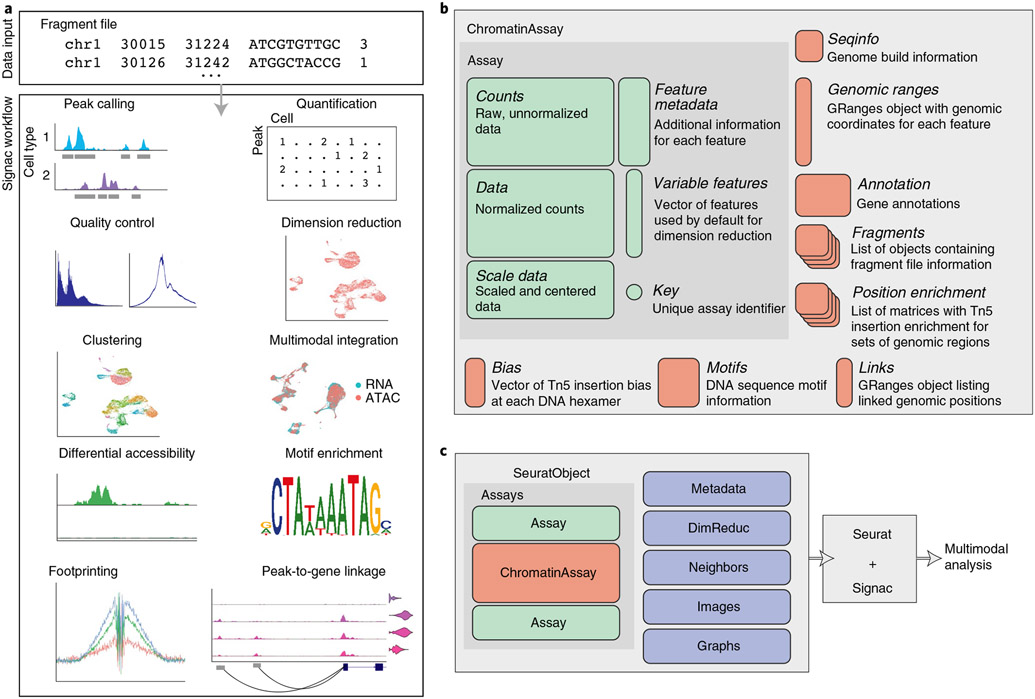
Fig. 1 ∣. Single-cell chromatin analysis workflow with Signac.
a, Overview of key steps comprising analysis of single-cell chromatin data with Signac. All analysis tasks can be completed with one or multiple fragment files as input. b, Design of a custom Assay for single-cell chromatin data. We designed a specialized ChromatinAssay class with the capacity to store data required for analysis of single-cell chromatin datasets. c, ChromatinAssay objects can be stored side by side with standard Assay objects in a Seurat object to enable analysis of multimodal single-cell data.
Results
Package design.
We aimed to create an extensible framework for single-cell chromatin data analysis that builds on existing tools used in the single-cell, genomics and R language communities. We designed an R toolkit for analysis and visualization of single-cell chromatin data in a way that allowed interoperability with the Seurat R package, designed for the analysis of multimodal single-cell data40-42. The Seurat package uses the Seurat object as its central data structure. The Seurat object is composed of any number of Assay objects containing data for single cells. The Assay object was originally designed for analysis of single-cell gene expression data and allows for storage and retrieval of raw and processed single-cell measurements and metadata associated with each measured feature. To facilitate the analysis of single-cell chromatin data within the Seurat framework, we developed a specialized ‘ChromatinAssay’ object class (Fig. 1b). The ChromatinAssay allows for the storage and retrieval of information needed for the analysis of single-cell chromatin data, including genomic ranges associated with each feature in the experiment, gene annotations, genome build information, DNA motif information and on-disk storage of single-cell data as tabix-indexed fragment files44. Crucially, the specialized ChromatinAssay can be stored in a Seurat object side by side with standard Seurat Assay-class objects to facilitate analysis of multimodal single-cell data (Fig. 1c).
Analysis of multimodal human PBMC data.
To demonstrate the core functionality of the Signac package we analyzed a publicly available dataset that jointly profiled messenger RNA abundance and DNA accessibility in single human peripheral blood mononuclear cells (PBMCs), generated by 10x Genomics. We computed per-cell quality control (QC) metrics using the DNA accessibility assay, including the strength of the nucleosome banding pattern (Fig. 2a) and transcriptional start site (TSS) enrichment score (Fig. 2b; Methods) and removed low-quality cells based on these QC metrics resulting in a dataset of 10,466 cells. Next, we processed the gene expression assay by normalizing RNA counts with SCTransform and Seurat41,45. We annotated cell types by mapping cells to an annotated multimodal PBMC reference dataset, using the gene expression assay42. This revealed 20 different cell types present in the dataset, including rare populations such as γδ T cells.
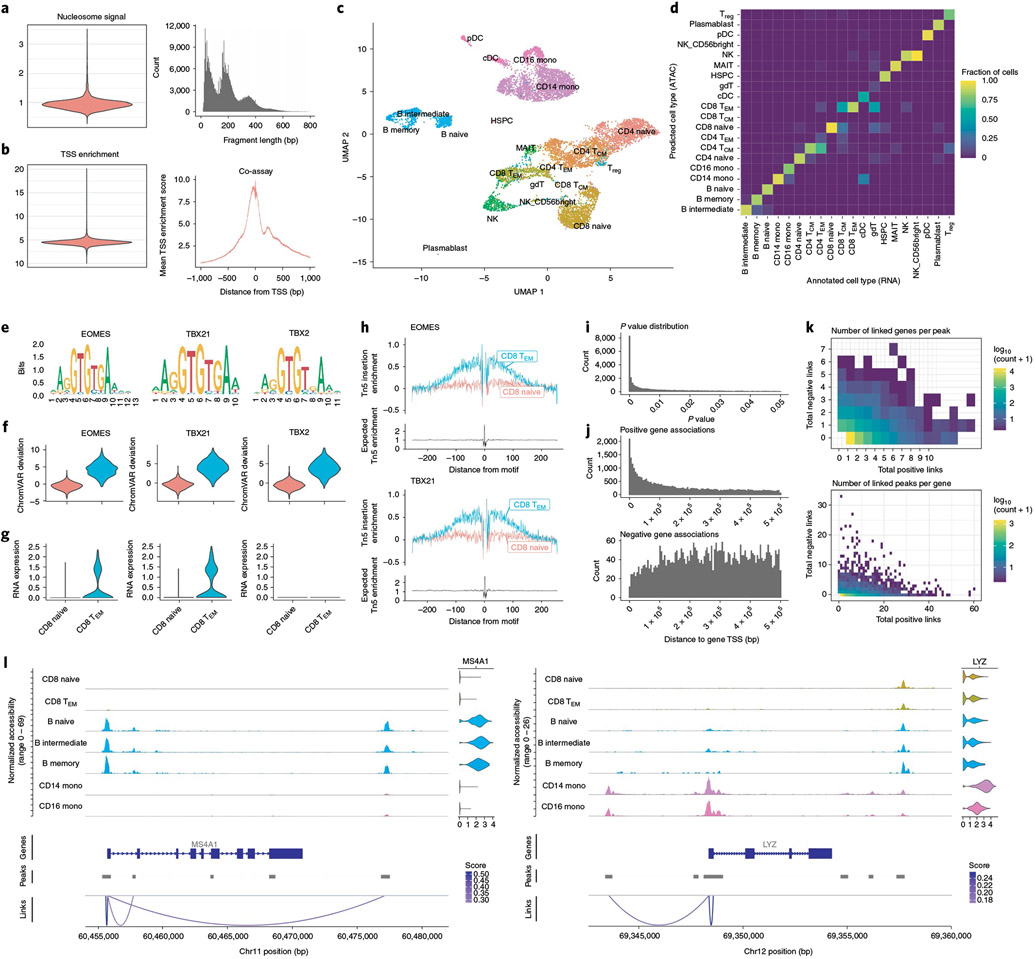
Fig. 2 ∣. Integrative single-cell analysis of gene expression and DNA accessibility in human PBMCs.
a, Nucleosome signal QC metric distribution and DNA fragment length distribution for the DNA accessibility assay. b, TSS enrichment score distribution and Tn5 insertion frequency at TSS sites for the DNA accessibility assay. c, UMAP representation of the multimodal human PBMC dataset, with cells annotated by predicted cell type. UMAP was constructed from the DNA accessibility assay. Treg, regulatory T cell; TEM, effector memory T cell; cDC, conventional dendritic cell; pDC, plasmacytoid dendritic cell; HSPC, hematopoietic stem and progenitor cell; MAIT, mucosal-associated invariant T cell. d, Multimodal label transfer accuracy. Multimodal single-cell data were split into DNA accessibility and gene expression assays and multimodal label transfer performed between pseudo-scRNA-seq and pseudo-scATAC-seq datasets. e, DNA sequence motifs for top overrepresented TF motifs between CD8+ effector and naive T cells. f, chromVAR32 deviations for top enriched DNA sequence motifs (EOMES, TBX21, TBX2) for CD8+ effector (CD8 TEM) and naive CD8+ (CD8 naive) T cells. g, RNA expression for EOMES, TBX21 and TBX2 genes in CD8+ effector and naive T cells. h, TF footprinting analysis for EOMES and TBX21 motifs sites. i, Distribution of peak-to-gene link P values for all reported links. P values were determined by a one-sided z-test without multiple testing correction. j, Distances from peak to linked gene TSSs, for positive- and negative-coefficient peak–gene links. k, Total number of positive-coefficient and negative-coefficient peak–gene links for each linked gene (top) and peak (bottom). l, Representative example peak–gene links for key immune genes.
Analysis of chromatin datasets can be highly dependent on accurate peak calling and this challenge is compounded in single-cell assays where peaks specific to rare populations are sometimes missed when calling peaks on the whole cell population. To address this problem, we identified peaks using MACS2 (ref. 46) for each annotated cell type separately and combined the individual peak calls into a unified peak set using Signac. Indeed, peaks specific to rare cell populations were often missed when calling peaks on the whole dataset using MACS2 (Supplementary Fig. 1a,b). We further compared the MACS2 bulk-cell peak calls with the peak calls produced by 10x Cellranger ATAC v.1, commonly used for the analysis of scATAC-seq data and found 13,751 cases where a Cellranger peak merged distinct MACS2 peaks into a single region, whereas there were only two cases where a MACS2 peak overlapped multiple Cellranger peaks (Supplementary Fig. 1c). This revealed a bias in Cellranger for aberrant merging of multiple distinct peaks into a single region and highlights the importance of accurate cell-type-specific peak calling methods in the analysis of single-cell chromatin datasets. In the absence of multimodal data, an independent clustering of the cells can be performed using the chromatin data and peaks identified per cluster in place of cell-type-specific peak calling. To assess the similarity between cluster-specific and cell-type-specific peak calls, we clustered the cells using the DNA accessibility assay (Supplementary Fig. 2) and called peaks per cluster using MACS2. We found that 92.7% of cell-type-specific peaks overlapped cluster-specific peaks, while only 78.5% of cell-type-specific peaks overlapped a peak identified using the bulk-cell data. Finally, to evaluate how the size of a cell population influenced the ability to detect peaks in that population, we randomly sampled cells from the CD14+ monocyte population, with the total number of sampled cells ranging from 50 to 2,850 cells. For each downsampling, we called peaks using MACS2 and assessed the fraction of peaks identified using the 2,850-cell population that were able to be recovered using each downsampled population (Supplementary Fig. 1d.e). When sampling 950 cells, 75% of peaks detected when using the larger cell population were able to be recovered, while only 18% of peaks were able to be recovered using the 50-cell population, highlighting the need for sufficient sampling of rare cell populations in single-cell chromatin studies.
We next reduced the dimensionality of the DNA accessibility assay by latent semantic indexing (LSI)4,47 and reduced the dimensionality of the gene expression assay by principal component analysis (PCA). We constructed a low-dimensional visualization of the DNA accessibility assay using uniform manifold approximation and projection (UMAP)48 (Fig. 2c). In the absence of paired gene expression measurements, single-cell chromatin data can be independently clustered using Signac and manually annotated, or multimodal integration can be used to annotate the cell types in an unsupervised analysis41. To assess the accuracy of multimodal integration, we treated the gene expression and DNA accessibility assays as separate experiments and performed cell type label transfer from the annotated scRNA-seq assay to the unannotated scATAC-seq assay using the previously developed Seurat v3 data integration methods41. This revealed an overall label transfer accuracy of 87.0% for high-resolution cell annotations (Supplementary Fig. 3a) or 92.5% for lower-resolution cell annotations, with incorrect predictions occurring mostly between highly similar cell types (Fig. 2d). Furthermore, incorrect predictions received lower prediction scores, allowing low-confidence predictions to be identified (Supplementary Fig. 3b).
To explore differences in DNA accessibility landscapes between cell types in the PBMC dataset, we identified ATAC-seq peaks open in CD8+ effector T cells relative to CD8+ naive T cells, revealing many regions of open chromatin that were specific to the CD8+ effector T cells. We assessed the rate of false positive results in our differential accessibility (DA) testing by drawing two populations of 100 cells at random from the CD4+ central memory T (TCM) cell population and comparing peak accessibility between these two cell populations, revealing no DA peaks. To further assess how the fraction of cells in the population with a true difference affected our ability to identify DA peaks, we gradually increased the fraction of cells in the comparison group drawn from a separate natural killer (NK) cell population. When drawing >40% of cells from a truly different population, we were able to detect DA peaks between the groups, with the fraction of true DA peaks able to be recovered increasing as a higher fraction of cells in the comparison group originated from a distinct population (Supplementary Fig. 4).
To identify transcription factors (TFs) that may be implicated in regulating these cells, we searched for overrepresented DNA sequence motifs in the set of CD8+ effector cell-specific peaks (Methods). This revealed a strong overrepresentation of EOMES, TBX21 and TBX2 TF-binding motifs. However, the motifs for each of these TFs are nearly identical (Fig. 2e) and displayed the same patterns of accessibility among the cells (Fig. 2f), making it difficult to correctly identify the TF involved in binding these motifs in effector T cells. To identify putative regulatory TFs, we examined gene expression data in these cells. While EOMES and TBX21 were both expressed in T cells, TBX2 was not detected (Fig. 2g). This indicated that EOMES and TBX21 likely regulate these sites49, rather than TBX2 and highlights the ability of combined gene expression and DNA accessibility data to improve identification of TFs involved in regulating different cell states. We further examined enrichment of Tn5 integration events surrounding EOMES and TBX21 motifs sites by performing TF footprinting50, revealing a strong enrichment of integration events flanking the TF motif in CD8+ effector cells compared to CD8+ naive cells (Fig. 2h).
The measurement of both gene expression and DNA accessibility in the same cell creates an opportunity to link noncoding DNA elements to their potential regulatory targets through the correlation between DNA accessibility and the expression of a nearby gene12,17,20. We implemented a peak-to-gene linkage method in Signac based on recently described methods20. Briefly, we computed the Pearson correlation between the expression of a gene and the accessibility of each peak within 500 kb of the gene TSS and compared this value with the expected value given the GC content, overall accessibility and length of the peak (Methods). Applying this linkage method to all expressed genes in the PBMC dataset revealed a set of 37,424 peak–gene links with P < 0.05 across the genome (Fig. 2i). The majority (89%) of these links displayed a positive relationship between accessibility of the peak and expression of the linked gene. Although links were enriched in close proximity to the gene TSS, we also observed a substantial number of long-range putative regulatory relationships, with 58% of links spanning a distance of >100 kb from the gene TSS (Fig. 2j). Linked genes were on average linked to ~six peaks (mean = 6.37, s.d. = 7.09), whereas linked peaks were linked to ~one gene on average (mean = 1.57, s.d. = 1.26) (Fig. 2k).
Cell-type-specific immune genes seemed to form accurate links with nearby peaks accessible in the same cell types expressing the genes (Fig. 2l). We sought to systematically assess the accuracy of peak–gene links identified by examining a set of expression quantitative trait loci (eQTL) fine-mapped variants for whole blood, produced by the GTEx Consortium51. Of the 154,504 peaks in the PBMC dataset, only 3,598 contained a fine-mapped eQTL variant identified for whole blood by the GTEx Consortium and 2,054 of these peaks also had one or more peak–gene links identified. For the set of 2,054 peaks that overlapped a fine-mapped eQTL and were linked to a gene, the eQTL variant was associated with the same gene as the peak in 52.6% of cases, whereas 13.4% was expected by random chance. The systematic identification of putative regulatory targets for any open chromatin region in the genome using multimodal single-cell datasets has the potential to enable a more accurate assignment of trait- or disease-associated noncoding variants to a gene likely to be impacted by the variant.
Evaluation of scATAC-seq dimension reduction methods.
LSI was originally developed for natural language processing47 and uses a term frequency-inverse document frequency (TF-IDF) weighting scheme to weight features according to their frequency in a document and their frequency across all documents in a text corpus. LSI has since been applied for the analysis of single-cell chromatin data, where a cell is analogous to a document and a term is analogous to a genomic region4. The most popular TF-IDF method applied to single-cell chromatin data computes the term frequency as TF = Cij/Fj where Cij is the total number of counts for peak i in cell j and Fj is the total number of counts for cell j. The inverse document frequency is typically computed as IDF = log(1 + N/ni) where N is the total number of cells in the dataset and ni is the total number counts for peak i across all cells. The TF-IDF matrix is then computed as TF × IDF. We found that, when applied to scATAC-seq data, this implementation often results in nonzero values in the TF-IDF matrix having low variance and a mean very close to zero and a poor ability to discriminate between cell types. We developed a simple modification to the TF-IDF weighting scheme that improves the results of LSI when applied to single-cell chromatin data. In our modified method we compute the inverse document frequency as IDF = N/ni and TF-IDF as log(1 + (TF × IDF) × 104).
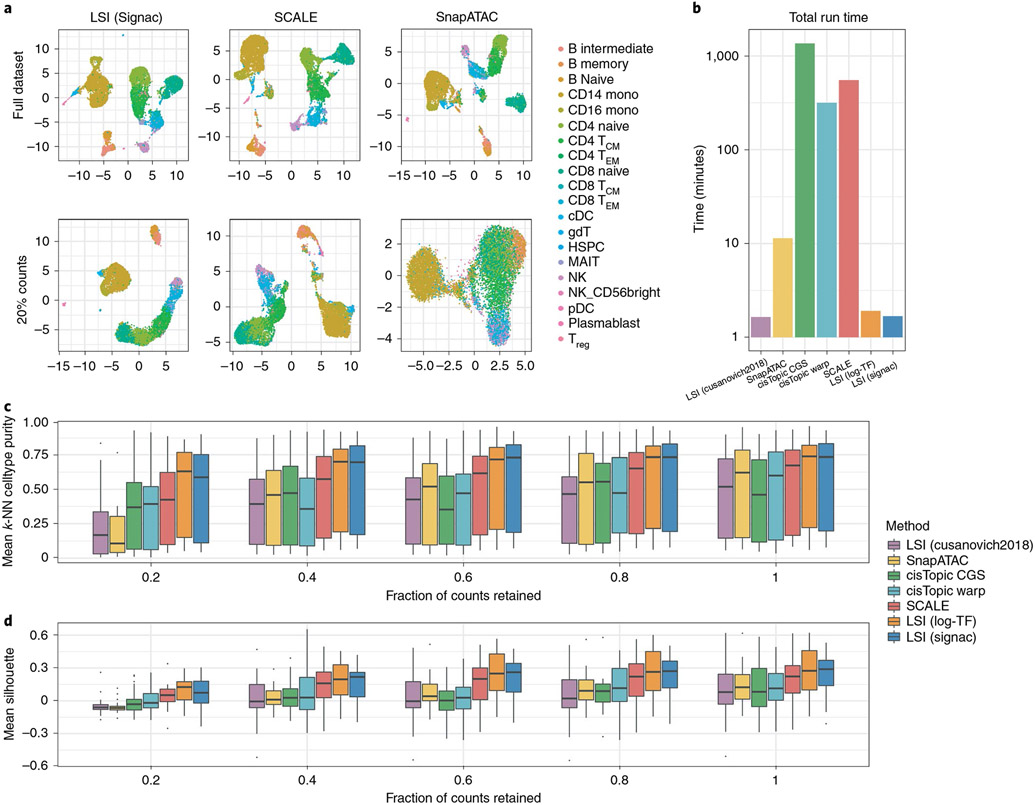
To assess the performance of our modified TF-IDF method in comparison to alternative methods for computing LSI and the currently top-performing methods for scATAC-seq dimension reduction, we downsampled the total counts for the multimodal PBMC dataset ran a variety of scATAC-seq dimension reduction methods. We performed LSI using our modified method, the original LSI method4 and a recently proposed ‘log-TF’ method (Methods). Furthermore, we included a comparison with cisTopic28 and SnapATAC34 as these methods were highlighted as top-performing in a recent benchmarking study52, as well as the recently developed SCALE method30. Overall, we found that our modified LSI method (LSI (Signac)), the log-TF method (LSI (log-TF)) and SCALE were the top-performing methods, whereas SnapATAC and cisTopic struggled to separate cell types in the downsampled datasets (Fig. 3a). Furthermore, we observed substantial differences in the runtimes for different methods, with cisTopic and SCALE having the longest runtimes (Fig. 3b), although the runtime for SCALE can be improved by using a graphics processing unit with sufficient memory30.
Fig. 3 ∣. Evaluation of dimension reduction methods for single-cell chromatin data.
a, UMAP representations of reduced-dimension single-cell DNA accessibility data for LSI (Signac), SCALE and SnapATAC for the full dataset and with the total number of counts per cell downsampled to 20% of the total counts. b, Runtimes for each of the dimension reduction methods profiled. CisTopic CGS, cisTopic collapsed Gibbs sampling; cisTopic Warp: cisTopic Warp-LDA. c, Mean k-NN cell type purity (k = 100) and (d) mean Silhouette score for each cell type in the dataset, for each dimension reduction method and downsampling level. For each box-plot, n = 20 points (cell types). For each box-plot, n = 6 points (cell types). Box-plot lower and upper hinges represent first and third quartiles. Upper/lower whiskers extend to the largest/smallest value no further than 1.5× the interquartile range. Data beyond the whiskers are plotted as single points.
We further assessed the preservation of local cell neighborhoods in each downsampled dataset by computing the average fraction of k-nearest neighbors (k-NN) (k = 100, additional values of k are shown in Supplementary Fig. 5) for each cell belonging to the same cell type as the query cell (mean k-NN purity per cell type), as well as the average Silhouette score for each cell type, with cell types annotated using the independent gene expression assay. This revealed a gradual decline in local structure preservation as fewer counts were retained from the original dataset, with a greater decline seen when using the original LSI method, SnapATAC and cisTopic (Fig. 3c,d). To test how these results generalize to other datasets, we repeated a similar analysis using a series of synthetic scATAC-seq human bone marrow cell datasets generated in a recent benchmarking study52, with similar results (Supplementary Fig. 6). These results indicate that LSI, when applied with the right TF-IDF method, can be a powerful dimension reduction technique for single-cell DNA accessibility data.
Joint analysis of DNA accessibility and mitochondrial genotype.
New technologies capable of measuring chromatin state alongside other data modalities at single-cell resolution are now being rapidly developed. These include the development of assays that measure DNA accessibility data alongside mitochondrial genome sequence15,16,43. As the mitochondrial genome mutates at a much higher rate than the nuclear genome and mitochondrial mutations are inherited over cell divisions, measuring mitochondrial genome sequences in single cells can be informative in reconstructing clonal cell relationships15,16,43. These experiments therefore provide an opportunity to study DNA accessibility differences between or within clonal groups of cells. To facilitate joint analysis of these datasets, we developed computational methods to enable identification of informative mitochondrial variants, calculation of mitochondrial variant allele frequencies and clonal cell clustering within the Signac framework.
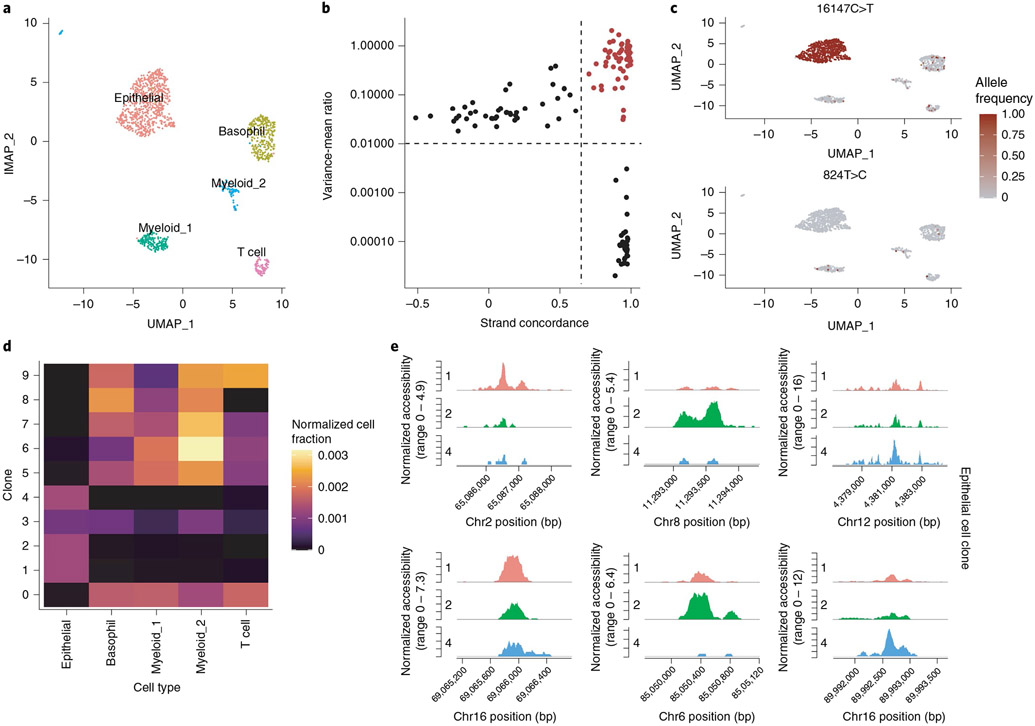
We analyzed a recently published single-cell DNA accessibility and mitochondrial genome sequence co-assay dataset from a patient with a colorectal cancer (CRC) tumor16. We first performed QC, dimension reduction and clustering on the DNA accessibility assay and annotated the major cell types present in the dataset based on the DNA accessibility at key marker genes (Fig. 4a). This revealed five major clusters present in the dataset encompassing tumor-derived epithelial cells, basophils, myeloid cells and T cells, as previously reported16. To identify clonal relationships between cells in the CRC dataset, we identified highly variable mitochondrial genome positions among the cells by computing the variance-to-mean ratio and the Pearson correlation between strand coverage (Fig. 4b). Visualization of per-cell allele frequencies (fraction heteroplasmy) for these variants in the two-dimensional UMAP space computed using the DNA accessibility assay revealed the variant 16147C>T present at nearly 100% frequency in tumor-derived epithelial cells, whereas other variants were shared across different immune cell types (Fig. 4c). We further identified cell clones by clustering allele frequency data, revealing ten distinct clones (Fig. 4d). Clones 1, 2 and 4 were highly specific to epithelial cells, whereas other clones were dispersed more evenly across different immune cell types, indicating that those immune cells likely originated from a common hematopoietic progenitor. We further identified differential DNA accessibility peaks between the three different epithelial cell clones, highlighting the ability of the additional clonotype data to aid in identifying additional sources of chromatin state heterogeneity within a cell type (Fig. 4e).
Fig. 4 ∣. Joint analysis of mitochondrial genotypes and DNA accessibility in single cells.
a, UMAP plot for cells from a tumor from a patient with CRC profiled by scATAC-seq, with the major cell types annotated. b, Variance-to-mean ratio versus strand concordance (Pearson correlation between strand coverage) for mitochondrial genome variants. High confidence, highly variable mitochondrial genome sites are shown in red. c, Per-cell allele frequencies (fraction heteroplasmy) for two representative mitochondrial genome variants used to identify cell clones. d, Fraction of cells belonging to each clone that were assigned to each cell type, normalized for the total number of cells belonging to each cell type. e, Differentially accessible regions of the nuclear genome between epithelial cell clones.
Scalable analysis of single-cell chromatin data.
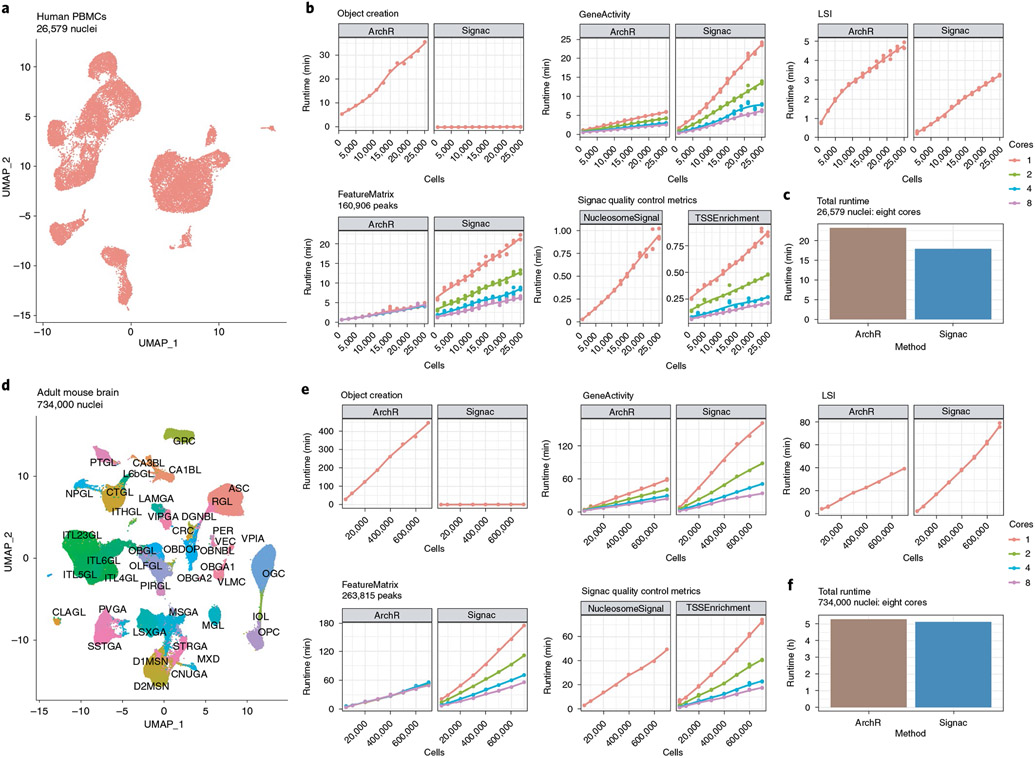
Methods are now available that enable generation of very large scATAC-seq datasets8. This presents opportunities to deeply characterize the chromatin state of tissues at single-cell resolution, but also raises the need for computational tools that similarly scale to large cell numbers. To explore how the runtime of key analysis steps in the Signac framework scales to large numbers of cells, we analyzed two separate scATAC-seq datasets of differing sizes: a human PBMC scATAC-seq dataset of 26,579 cells from 10X Genomics and an adult mouse brain dataset of 734,000 cells from the Brain Initiative Cell Census Network (BICCN)53. We downsampled the full dataset down to 1,000 cells (for the PBMC dataset) or 50,000 cells (for the BICCN dataset) and ran each step in a full analysis workflow, including creating the initial object required, quantifying counts in peaks, quantifying DNA accessibility at each gene, computing QC metrics and performing dimensionality reduction. We also compared the runtime for Signac to the recently published ArchR package35 for equivalent analysis steps. For steps that are able to be run in parallel, we tested with 1, 2, 4 or 8 cores. For the PBMC dataset (Fig. 5a), we found a generally linear increase in runtime with the addition of more cells for most steps (Fig. 5b). Notably, ArchR requires a large amount of time to create the files needed to run an analysis, whereas Signac requires substantially less time for the object creation step. Overall, we found that Signac performs an end-to-end analysis of the PBMC dataset in slightly less time than ArchR (Fig. 5c). Repeating a similar analysis on the BICCN dataset (Fig. 5d), we found similar results, with both ArchR and Signac able to process consortium-scale datasets (Fig. 5e) and Signac again performing an end-to-end analysis of the full 734,000-cell dataset slightly faster than ArchR (Fig. 5f). These results provide a valuable benchmark resource for those planning experiments and estimate the time required to analyze single-cell chromatin datasets of different sizes with the Signac package.
Fig. 5 ∣. Scalable analysis of single-cell chromatin data.
a, UMAP representation of the full human PBMC scATAC-seq dataset of 26,579 nuclei. b, Runtimes for key analysis steps for ArchR and Signac, for each downsampled PBMC scATAC-seq dataset. c, Total runtime for an end-to-end analysis of the full PBMC scATAC-seq dataset for ArchR and Signac using eight cores. d, UMAP representation of the full BICCN adult mouse brain dataset of 734,000 nuclei. Cells are colored by their cell type label given by the original study authors53. e, Runtimes for key analysis steps for ArchR and Signac, for each downsampled BICCN scATAC-seq dataset. f, Total runtime for an end-to-end analysis of the full BICCN scATAC-seq dataset for ArchR and Signac using eight cores.
Discussion
As experimental methods for measuring aspects of chromatin state at single-cell resolution continue to be developed and improved, the parallel development of computational tools designed to analyze these datasets becomes increasingly important. Here, we developed Signac for the analysis of single-cell chromatin data and demonstrated running key analysis steps using Signac for the analysis of both unimodal and multimodal single-cell chromatin datasets. These analysis steps can be scaled to datasets containing >700,000 cells and the scalability of these methods will become particularly important as large-scale cell atlas projects are completed53. We further developed a simple modification to the popular LSI dimension reduction method that improved the performance of LSI when applied to single-cell chromatin data, particularly for datasets with low sensitivity. Furthermore, Signac enables running other tools developed by the community for the analysis of single-cell chromatin data, including chromVAR for estimating DNA motif variability between cells32, Monocle for building pseudotime trajectories54, Cicero for finding co-accessible networks of peaks31 and Harmony for performing dataset integration55. While we have focused here on the analysis of DNA accessibility data alone, our suite of tools is also fully compatible with recently developed methods to construct a joint neighbor graph encompassing multiple data modalities42, enabling chromatin data to be combined with additional modalities to jointly define cell states. As additional experimental methods for measuring multiple aspects of cell state are developed, a major challenge is to analyze these diverse datasets together in a consistent framework to learn how different modalities influence one another. The Seurat framework, via the extensible Assay class, is an appealing solution for the analysis of multimodal single-cell data and we envision future computation methods will further build on the Seurat and Signac frameworks to jointly analyze multimodal single-cell datasets. Future work will aim to further improve the scalability of the Signac package and to expand the suite of computational methods implemented. A major challenge currently facing biology is understanding how the genome encodes the organism56. Developing a deep understanding of how genes are regulated by noncoding DNA elements would greatly improve our ability to predict the effect of mutations and to predict the target genes for trait-associated noncoding loci. A joint analysis of multimodal single-cell chromatin and gene expression data hold greats promise in furthering these goals and the analytical framework presented here will be a valuable component in deciphering these gene regulatory relationships.
Methods
Signac 1.2.0 was used for all analyses and is available on CRAN (https://cloud.r-project.org/package=Signac) and GitHub (https://github.com/timoast/signac/). R v.4.0.3 was used for all analyses, with standard BLAS and LAPACK libraries linked, running on Ubuntu v.18.04.4 LTS with Intel Xeon W-2135 central processing units at 3.70 GHz.
Data structures.
We extended the Seurat Assay class via the R language class inheritance framework to create the ChromatinAssay class for single-cell chromatin data analysis. We extended the Assay class to add slots for the storage of genomic ranges, DNA motifs, genome build information, gene annotations, Tn5 insertion bias, positional enrichment information, genomic links and linked on-disk data storage as fragment files.
The fragment file is a data format introduced by 10x Genomics for the storage of scATAC-seq data. Fragment files are defined as coordinate-sorted, block gzip-compressed (bgzip) and indexed browser-extensible data files with the following five columns: chromosome, start, end, cell barcode, PCR duplicate count. The start and end fields of the fragment file correspond to positions of the two Tn5 integration events that generated the sequenced DNA fragment. As the fragment file contains a deduplicated and near-complete representation of a single-cell chromatin experiment and existing tools are established to efficiently retrieve subsets of a fragment file that overlap a given set of genomic regions44, we utilized the fragment file format as the central disk-based data structure in the Signac framework and is the only requirement for running a single-cell data analysis using Signac.
To facilitate construction of a fragment file outside of running the 10x Genomics Cellranger software, we developed a Python package (Sinto) capable of generating the fragment file from a BAM file. This software is available on the Python Package Index (https://pypi.org/project/sinto/), Bioconda (https://anaconda.org/bioconda/sinto), and GitHub (https://github.com/timoast/sinto).
Quality control metrics.
Nucleosome signal.
The length of DNA wrapped around a single nucleosome has been experimentally determined as 147 bp57. As Tn5 has a strong preference to integrate into nucleosome-free DNA58, successful ATAC-seq experiments typically exhibit a depletion of DNA fragments with lengths that are multiples of 147 bp. We defined the nucleosome signal QC metric in Signac as the ratio of mononucleosomal (147–294 bp) to nucleosome-free (<147 bp) fragments sequenced for the cell, as a way of quantifying the expected depletion of nucleosome-length DNA fragments. To compute the nucleosome signal per cell, we sampled the first n fragments from the fragment file, where n was the total number of cells in the dataset multiplied by 5,000. We then divide the number of mononucleosomal fragments per cell by the number of nucleosome-free fragments. This was implemented in the NucleosomeSignal function in Signac.
TSS enrichment.
The TSS enrichment score was originally defined by the ENCODE consortium59 as a signal-to-noise metric for ATAC-seq experiments. As the TSS for most genes are typically open, these regions represent a ‘positive control’ set when taken together and enables computing a metric that reflects the sensitivity of the ATAC-seq experiment. The TSS score was defined as the mean number of Tn5 insertion events centered on the TSS sites (±500 bp of the TSS) divided by the mean Tn5 insertion count at TSS-flanking regions, defined as +900 to +1,000 and −900 to −1,000 bp from the TSS. Calculation of the TSS enrichment score per-cell was implemented in the TSSEnrichment function in Signac.
Dimension reduction.
Latent semantic indexing.
LSI involves two steps. First, we compute the TF-IDF matrix from the count matrix. Term frequency was defined as TF = Cij/Fj where Cij was the total number of counts for peak i in cell j and Fj was the total number of counts for cell j. IDF was defined as IDF = N/ni, where N was the total number of cells in the dataset and ni was the total number of counts for peak i across all cells. The TF-IDF matrix was then computed as TF-IDF = log(1 + (TF × IDF) × 104). For comparison with alternative LSI methods4, we also computed IDF as IDF = log(1 + N/ni) and subsequently TF-IDF as TF × IDF (for ‘Cusanovich2018’) and TF-IDF as log(TF) × IDF (for ‘log-TF’; http://andrewjohnhill.com/blog/2019/05/06/dimensionality-reduction-for-scatac-data/). This was implemented in the RunTFIDF function in Signac, with the ‘method’ argument used to choose the TF-IDF method used. We decomposed the resulting TF-IDF matrix via truncated singular value decomposition using the irlba R package (https://cran.r-project.org/package=irlba)60, implemented in the RunSVD function in Signac.
UMAP.
We performed UMAP using the RunUMAP function in the Seurat package (v.3.2.0) using LSI components 2 to 40 for the PBMC multiome dataset, components 2 to 50 for the CRC tumor dataset, 2 to 30 for the PBMC scATAC-seq dataset and 2 to 100 for the BICCN mouse brain dataset. The first LSI component was excluded from each analysis as it typically captures sequencing depth (technical variation) and was highly correlated with the total number of counts for the cell. The RunUMAP function uses the uwot R package to compute two-dimensional UMAP coordinates48 (https://CRAN.R-project.org/package=uwot).
Genome browser visualization.
A common analysis task for single-cell chromatin data is genome browser-style data visualization for different groups of cells. Signac enables such visualizations with cells dynamically grouped into different pseudo-bulk tracks by reading Tn5 integration data from a position-indexed fragment file44. To visualize pseudo-bulk accessibility tracks for different groups of cells, we constructed a sparse matrix of base-resolution Tn5 integration events, where each row was a cell and each column a DNA base in the requested region. We then grouped cells and computed the total accessibility at each site within each group and scaled the total accessibility within each group by the total number of cells in the group and the average total counts for cells in each group to account for differences in overall chromatin signal and cell number between different groups of cells. We then smoothed the chromatin signal across small regions by computing a rolling window sum across the genomic region for each group of cells (using a window size of 100 bp by default). This was implemented in the CoveragePlot function in Signac. We also implemented an interactive version of the CoveragePlot function using the Shiny Gadgets framework in R (https://cran.r-project.org/package=shiny) as the CoverageBrowser function in Signac. The interactive CoverageBrowser provides the same functionality as CoveragePlot, but additionally allows interactive navigation to different genomic regions and dynamic regrouping of cells.
One major advantage of genome browser-style visualizations is the ability to stack different data visualizations conveying different information as different browser tracks. We further built this concept into the CoveragePlot and CoverageBrowser functions in Signac by including the ability to plot additional tracks displaying gene expression information, gene annotations, peak coordinates, genomic links, genomic ranges, the presence/absence of Tn5 integration events in individual cells as genomic ‘tile’ plots, as well as data from an existing bigwig file.
PBMC multiome analysis.
We downloaded data for human PBMCs processed using the 10X Genomics Multiome (ATAC + RNA) method from the 10X Genomics website (https://support.10xgenomics.com/single-cell-multiome-atac-gex/datasets/1.0.0/pbmc_granulocyte_sorted_10k).
Quality control and cell filtering.
We computed the nucleosome signal score and TSS enrichment score for each cell as described above. We retained cells with a TSS enrichment score > 1, a nucleosome signal score <2, between 5,000 and 70,000 total ATAC-seq counts (based on the 10x Cellranger ATAC-seq count matrix) and between 1,000 and 25,000 total RNA counts.
Gene expression data preprocessing and cell annotation.
We normalized gene expression UMI count data using SCTransform45 and performed PCA on the SCTransform Pearson residual matrix using the RunPCA function in Seurat. We found the 20 nearest neighbors for each cell using the FindNeighbors function, with dims = 1:50 to use the first 50 principal components and annotated cell types in the PBMC dataset by label transfer from a publicly available multimodal PBMC reference dataset42. We identified anchor cells41 between the query and reference datasets using the FindTransferAnchors function in Seurat v4, with reference.reduction = ’spca’ to use a precomputed reference dimension reduction object. We then computed cell type predictions for each cell in the query using the TransferData function in Seurat. As erythrocytes are not nucleated and the query PBMC dataset was derived from cell nuclei, we assigned a small number of cells that were incorrectly predicted as erythrocytes to the most common predicted class of those cells’ 20 nearest neighbors.
DNA accessibility data processing.
ATAC-seq peaks in the PBMC dataset were identified using MACS2 (ref. 46) with the following arguments: -g 2.7e9 -f BED – nomodel –extsize 200 –shift −100 –max-gap 50. We used the fragment file as input to the peak calling algorithm, as this contained the deduplicated Tn5 insertion sites for each cell. Peak calling was performed for each cell type using the CallPeaks function in Signac, with group.by = ‘celltype’ to call peaks on each predicted cell type separately and combine the resulting peak calls across all cell types. We removed any peaks overlapping annotated genomic blacklist regions for the hg38 genome61. We quantified counts for the resulting peak set for each cell using the FeatureMatrix function in Signac.
Dimension reduction was performed on the DNA accessibility assay dataset using LSI and UMAP as described above. We performed graph-based clustering on the LSI components 2 to 40 by first computing a shared nearest-neighbor graph using the LSI low-dimensional space (with k = 20 neighbors) and then applying the Smart Local Moving algorithm for community detection62. This was performed using the function FindNeighbors with dimensions = 2:40 and reduction = ‘lsi’ followed by FindClusters with algorithm = 3 in Seurat v3.2.0 (ref. 41).
To assess how parameters chosen in the clustering algorithm affected final cluster membership, we re-ran the clustering workflow with a range of values for k-NN (5–50) and a range of values for the number of LSI components used (10–50). For each clustering, we calculated the adjusted Rand index between the clustering results obtained and the cell type labels derived from the gene expression assay using the mclust R package63.
Peak-calling sensitivity analysis.
To evaluate the ability of MACS2 to detect peaks with different numbers of input cells, we performed peak calling using the CD14+ monocyte cell population in the PBMC multiome dataset. We compared peak calls obtained using 2,850 CD14+ monocytes to random downsamplings of the cell population in steps of 50 cells down to 50 cells in total. For each set of peaks identified using the downsampled set of cells, we counted the total number of peaks identified that overlapped a peak in the peak set obtained using 2,850 cells.
Multimodal label transfer.
To assess multimodal label transfer accuracy, we treated the DNA accessibility and gene expression assays of the multiome dataset as though they were separate scATAC-seq and scRNA-seq experiments. We computed a gene activity assay for the scATAC-seq dataset by counting fragments overlapping the gene body and a 2-kb upstream region for each gene in each cell, using the GeneActivity function in Signac. We log-normalized the gene activity counts for the DNA accessibility assay using the NormalizeData function in Seurat41,42. We then identified anchor cells between the scATAC-seq and scRNA-seq datasets using canonical correlation analysis, with the function FindTransferAnchors in Seurat with the parameters, reduction = ‘cca’. Cell type labels were transferred from the scRNA-seq to scATAC-seq dataset using the TransferData function, with weight.reduction = query[[‘lsi’]] and dims = 2:30 to weight anchors based on nearest-neighbor distances in the LSI space. To evaluate the accuracy of multimodal label transfer, we counted the number of cells obtaining the correct predicted cell type label.
Differential accessibility.
DA between groups of cells was computed by constructing a logistic regression model predicting group membership based on the accessibility of a given peak in the set of cells being compared, with the total number of counts in each cell included as a latent variable in the model and comparing this with a null model using a likelihood ratio test. This was performed using the FindMarkers function in Seurat v3.2.0, with test.use = ‘LR’ and latent.vars = ‘nCount_ATAC’. We classified peaks with an adjusted P value (Bonferroni-corrected) <0.01 and absolute log2 fold change >0.4 as being DA between the cell groups.
To assess the false positive rate for DA testing, we randomly sampled without replacement two groups of 100 cells from the CD4+ TCM population and repeated DA testing between the two groups. To assess how the composition of the comparison group affected DA results, we also sampled n = 10 to n = 100 cells from the NK population, mixed with 100 – n cells from the CD4+ TCM population. We computed a set of ground-truth DA peaks by performing DA testing between the whole CD4+ and NK population and classified peaks with an adjusted P value <0.01 and absolute log2 fold change >0.4 as DA. For each sampling, we computed the receiver operator characteristic and area under the curve using the ROCR R package, by lowering the fold-change cutoff for significance64.
Motif enrichment.
A hypergeometric test was used to test for overrepresentation of each DNA motif in the set of differentially accessible peaks compared to a background set of peaks. We tested motifs present in the JASPAR database65 for human (species code 9606) by first identifying which peaks contained each motif using the motifmatchr R package (https://bioconductor.org/packages/motifmatchr). We computed the GC content (percentage of G and C nucleotides) for each differentially accessible peak and sampled a background set of 40,000 peaks such that the background set was matched for overall GC content, accessibility and peak width. This was performed using the FindMotifs function in Signac, with features.match = c(‘GC.percent’, ‘count’, ‘sequence.length’).
Motif footprinting.
We performed transcription factor motif footprinting following previously described methods50. To account for Tn5 sequence insertion bias, we first computed the observed Tn5 insertion frequency at each DNA hexamer using all Tn5 insertions on chromosome 1. This was performed by extracting the base-resolution Tn5 insertion positions for each fragment mapped to chromosome 1 and extending the insertion coordinate 3 bp upstream and 2 bp downstream. We then extracted the DNA sequence corresponding to these coordinates using the getSeq function from the Biostrings R package (https://bioconductor.org/packages/Biostrings) and counted the frequency of each hexamer using the table function in R. We next computed the expected Tn5 hexamer insertion frequencies based on the frequency of each hexamer on chromosome 1. We counted the frequency of each hexamer using the oligonucleotideFrequency function in the Biostrings package with width = 6 and names = ‘chr1’, using the hg38 genome via the BSgenome R package (https://bioconductor.org/packages/BSgenome). Finally, we computed the Tn5 insertion bias as the observed Tn5 insertions divided by the expected insertions at each hexamer. This was performed using the InsertionBias function in Signac.
To perform motif footprinting, we first identified the coordinates of each instance of the motif to be footprinted using the matchMotifs function from the motifmatchr package with out = ‘positions’ to return the genomic coordinates of each motif instance (https://bioconductor.org/packages/motifmatchr). Motif coordinates were then resized to include the ±250-bp sequence. The Tn5 insertion frequency was counted at each position in the region for each motif instance to produce a matrix containing the total observed Tn5 insertion events at each position relative to the motif center for each cell. We then found the expected Tn5 insertion frequency matrix by computing the hexamer frequency matrix, M. The hexamer frequency matrix M was defined as a matrix with i rows corresponding to i different DNA hexamers and j columns corresponding to j positions centered on the motif and each entry Mij corresponded to the hexamer count for hexamer i at position j. To find the expected Tn5 insertion frequency at each position relative to the motif given the Tn5 insertion bias (see above), we computed the matrix cross product between the hexamer frequency matrix M and the Tn5 insertion bias vector. Finally, the expected Tn5 insertion frequencies were normalized by dividing by the mean expected frequency in the 50-bp flanking regions (the regions 200 to 250 bp from the motif). To correct for Tn5 insertion bias we subtracted the expected Tn5 insertion frequencies from the observed Tn5 insertion frequencies at each position. This was performed using the Footprint function in Signac.
Peak-to-gene linkage.
We estimated a linkage score for each peak–gene pair using linear regression models, based on recent work described in the SHARE-seq method20. For each gene, we computed the Pearson correlation coefficient r between the gene expression and the accessibility of each peak within 500 kb of the gene TSS. For each peak, we then computed a background set of expected correlation coefficients given properties of the peak by randomly sampling 200 peaks located on a different chromosome to the gene, matched for GC content, accessibility and sequence length (MatchRegionStats function in Signac). We then computed the Pearson correlation between the expression of the gene and the set of background peaks. A z score was computed for each peak as z = (r − μ)/σ, where μ was the background mean correlation coefficient and σ was the s.d. of the background correlation coefficients for the peak. We computed a P value for each peak using a one-sided z-test and retained peak–gene links with a P value <0.05 and a Pearson correlation coefficient >0.05 or <−0.05. This was performed using the LinkPeaks function in Signac.
Fine-mapped eQTL analysis.
eQTL variants for whole blood that were fine-mapped using CAVIAR66 were downloaded from the GTEx v8 website (https://storage.googleapis.com/gtex_analysis_v8/single_tissue_qtl_data/GTEx_v8_finemapping_CAVIAR.tar)51. For each fine-mapped eQTL overlapping a peak that was linked to a gene in our analysis we counted the number of times the eQTL-associated gene was the same as the linked gene. Cases where multiple fine-mapped eQTLs associated with the same gene overlapped the same peak were treated as a single variant. To find the expected overlap based on random chance, we selected a set of peaks for each gene at random from the peaks within 500 kb of the gene TSS, with the number of peaks selected equal to the number of linked peaks for that gene. We then repeated the same eQTL overlap analysis using the randomized link set, as described above.
Count downsampling analysis.
PBMC multiome dataset.
To test the impact of sequencing depth and assay sensitivity on the performance of different scATAC-seq dimension reduction methods, we downsampled the total number of counts per cell from 100% (full dataset) down to 80%, 60%, 40% and 20% using the downsampleMatrix function from the DropletUtils R package67,68. For each downsampling, we re-ran each dimension reduction method (LSI (Signac), LSI (Cusanovich2018), LSI (log-TF), cisTopic (Warp-LDA), cisTopic (CGS), SnapATAC, SCALE), using default parameters for each method. For each downsampling, we estimated how well the data structure was preserved compared to the full dataset by computing the mean k-NN purity for each cell type in the dataset. This was defined as the fraction of k neighbors in the reduced-dimension space (LSI, cisTopic, SCALE or SnapATAC) for each cell i that belonged to the same annotated cell type as cell i, where cell types were predicted as described above using the gene expression assay. We computed nearest neighbors in each reduced-dimension space using the RANN R package (https://CRAN.R-project.org/package=RANN). For each dimension reduction method, we used the first 20 components removing any dimensions with a correlation >0.9 with the total counts in each cell (dimension 1 for LSI (Signac) and dimension 2 for SnapATAC). For SCALE, an autoencoder-based method, we used the entire latent space (ten dimensions). As an additional performance metric, we computed the mean Silhouette score for each cell type, for each downsampling level, using the Silhouette function in the cluster package in R. For both the k-NN purity metric and the Silhouette score, we computed the mean score for each cell type. This prevented the metric being biased toward the performance of each dimension reduction method on the most abundant cell types in the dataset.
Simulated bone marrow cell dataset.
We downloaded a previously generated simulated scATAC-seq dataset for human bone marrow cells, generated with 250–5,000 average counts per cell and a noise rate of 0.2 (ref. 52). Data were downloaded from GitHub (https://github.com/pinellolab/scATAC-benchmarking). For each simulated dataset, we ran each dimension reduction method as described above for the PBMC dataset, except that we used the first five dimensions for each method rather than 20. For SCALE, we used the full ten-dimension latent space.
Colorectal cancer analysis.
scATAC-seq data processing.
We downloaded processed scATAC-seq counts from Zenodo (https://zenodo.org/record/3977808) and the fragment file from NCBI Gene Expression Omnibus (GSE148509) and computed the nucleosome signal and TSS enrichment score per cell as described above and retained cells with >1,000 counts and <50,000 counts, <5% of counts in genomic blacklist regions, TSS enrichment score >3 and a nucleosome signal score <4 and a mitochondrial genome sequencing depth of ≥10. We performed dimension reduction using LSI and UMAP as described above and identified clusters using the Smart Local Moving algorithm using the FindClusters function in Seurat with resolution = 0.5 and algorithm = 3 (ref. 62).
Mitochondrial variant detection.
Single-cell mitochondrial variant data processed using mgatk16 was downloaded from Zenodo (https://zenodo.org/record/3977808), read into R using the Signac function ReadMGATK and used to create a Seurat assay. Informative mitochondrial variants were identified using the IdentifyVariants function, which computes the strand concordance in variant counts (Pearson correlation) and the variance-mean ratio (VMR) for each variant, as previously described16. Informative mitochondrial variants were selected with a VMR > 0.01 and strand concordance ≥0.65, provided the variant was confidently detected in ≥5 cells. We then computed per-cell mitochondrial allele frequencies for informative variants using the AlleleFreq function in Signac.
Clonal clustering.
We identified cell clones by performing graph-based clustering on the square-root-transformed allele frequency matrix by first creating a neighbor graph using the FindNeighbors function in Seurat with annoy.metric = ‘cosine’ to use the cosine distance to define nearest neighbors and with k = 10. We then performed community detection using the Smart Local Moving algorithm62 using the shared nearest-neighbor graph computed using Seurat. This was implemented in the FindClonotypes function in Signac.
Scalability analysis and benchmarking.
PBMC dataset processing.
We downloaded fragment files for four human PBMC scATAC-seq datasets from the 10x Genomics website and combined the four files into a single fragment file, adding a prefix to the cell barcodes to mark which cell originated from which dataset. We called peaks using the combined dataset with MACS2, using the CallPeaks function in Signac. Peaks overlapping genomic blacklist regions for hg19 were then removed61, resulting in a set of 160,906 peak regions.
We quantified counts in peaks using the FeatureMatrix function in Signac and removed cells with <1,000 total counts. This function was parallelized via the future R package, allowing the user to determine the parallelization strategy used (https://cran.r-project.org/package=future). Signac also includes a convenience function (GenomeBinMatrix) to quantify signal in genomic bins tiling the entire genome or given chromosomes. Finally, we reduced the dimensionality by applying LSI and UMAP as described above, using LSI components 2 to 30.
BICCN dataset processing.
We downloaded FASTQ files for the BICCN dataset from NeMO (https://nemoarchive.org/) and mapped the reads to the mm10 genome using BWA-MEM69. We created a fragment file from the aligned BAM file using sinto (https://github.com/timoast/sinto) and tabix44. We then identified peaks for each brain region using MACS2 (ref. 46) using the CallPeaks function in Signac, with the parameters effective.genome.size = 1.87 × 109. We filtered out peaks with a score <150 to remove low-confidence peaks, resulting in a total of 263,815 peaks. Code to produce the BICCN fragment file and unified peak set is available at https://github.com/timoast/BICCN.
We then quantified the number of fragments overlapping each peak for each cell using the Signac FeatureMatrix function. We retained all cells that were retained in the analysis performed by the original authors of the BICCN dataset53. We reduced dimensionality using LSI and UMAP as described above using LSI components 2 to 100.
Cell downsampling analysis.
Signac.
To test the scalability of key steps in the Signac workflow, we downsampled the total number of cells in the BICCN dataset and the PBMC scATAC-seq dataset. We downsampled the PBMC dataset from 25,000 cells down to 1,000 cells and the BICCN dataset from 700,000 cells down to 50,000 cells. We randomly sampled different cell numbers from the full dataset and used the FilterCells function in Signac to create downsampled fragment files containing only the cells sampled. We then ran CreateFragmentObject to construct the object required for an analysis in Signac, followed by FeatureMatrix, GeneActivity, NucleosomeSignal, TSSEnrichment, RunTFIDF and RunSVD on each downsampled dataset and recorded the total runtime for each step. For steps able to be run in parallel, we tested these with 1, 2, 4 and 8 cores.
To profile the time required for an end-to-end analysis of each dataset starting from the raw data (the fragment file) and a set of peaks, we ran CreateFragmentObject, CreateChromatinAssay and CreateSeuratObject to create the objects in R required for an analysis, followed by FeatureMatrix for quantification, NucleosomeSignal and TSSEnrichment for QC, FindTopFeatures, RunTFIDF, RunSVD and RunUMAP for dimension reduction and FindNeighbors and FindClusters for cell clustering. This was performed using eight cores by setting plan(‘multiprocess’, workers = 8) using the future R package (https://cran.r-project.org/package=future).
ArchR
We ran a similar runtime benchmarking analysis to that described above for Signac, using ArchR v.1.0.1 (ref. 35). To create the Arrow files needed for an analysis in ArchR, we ran the createArrowFiles function providing each downsampled fragment file as input, with addGeneScoreMat = FALSE and addTileMat = FALSE to avoid running additional steps. For comparison with the FeatureMatrix function in Siganc (peak region quantification), we ran the addPeakMatrix function in ArchR. For comparison with the GeneActivity function in Signac, we ran the addGeneScoreMatrix function in ArchR. For comparison with Signac LSI (RunTFIDF and RunSVD functions), we ran the addIterativeLSI function in ArchR with sampleCellsPre = NULL.
To profile the time required for an end-to-end analysis of each dataset starting from the raw data (the fragment file) and a set of peaks, we ran the createArrowFiles and ArchRProject functions to create the object in R required for analysis, followed by addPeakSet and addPeakMatrix for quantification, addIterativeLSI and addUMAP for dimension reduction and addClusters for cell clustering. This was performed using eight cores by setting addArchRThreads(threads = 8).
Supplementary Material
Acknowledgements
This work was supported by the Chan Zuckerberg Initiative (EOSS-0000000082 and HCA-A-1704-01895 to R.S.) and the National Institutes of Health (DP2HG009623-01, RM1HG011014-01 and OT2OD026673-01 to R.S.; K99HG011489-01 to T.S.). C.A.L. was supported by a Stanford Science Fellowship. We are grateful to L. Ludwig (MDC Berlin) for insightful conversations about mtDNA lineage tracing. We thank B. Ren (UCSD) for assistance in accessing the BICCN mouse brain dataset. We thank the CRAN maintainers for their assistance in distributing the Signac R package and members of the Satija laboratory for feedback on the manuscript.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41592-021-01282-5.
Competing interests
In the past 3 years, R.S. has worked as a consultant for Bristol-Myers Squibb, Regeneron and Kallyope and served as an SAB member for ImmunAI, Resolve Biosciences, Nanostring, and the NYC Pandemic Response Lab. The remaining authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41592-021-01282-5.
Code availability
Signac is available on CRAN (https://cloud.r-project.org/package=Signac) and on GitHub (https://github.com/timoast/signac), with documentation and tutorials available at https://satijalab.org/signac/. All code used in this paper is available on GitHub at https://github.com/timoast/signac-paper.
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data used in the paper are publicly available. The PBMC multiomic dataset is available from 10X Genomics at https://support.10xgenomics.com/single-cell-multiome-atac-gex/datasets/1.0.0/pbmc_granulocyte_sorted_10k. The PBMC scATAC-seq datasets are available from 10X Genomics at https://support.10xgenomics.com/single-cell-atac/datasets. The synthetic scATAC-seq datasets are available from GitHub at https://github.com/pinellolab/scATAC-benchmarking. Data from the BICCN are available from the Neuroscience Multiomic Archive at https://nemoarchive.org/. Data for the CRC patient sample are available on NCBI Gene Expression Omnibus (GSE148509) and Zenodo (https://zenodo.org/record/3977808).
References
- 1.Ai S et al. Profiling chromatin states using single-cell itChIP-seq. Nat. Cell Biol 21, 1164–1172 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter B et al. Mapping histone modifications in low cell number and single cells using antibody-guided chromatin tagmentation (ACT-seq). Nat. Commun 10, 3747 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cusanovich DA et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaya-Okur HS et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun 10, 1930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q et al. CoBATCH for high-throughput single-cell epigenomic profiling. Mol. Cell 10.1016/j.molcel.2019.07.015 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Ku WL et al. Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone modification. Nat. Methods 16, 323–325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lareau CA et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol 10.1038/s41587-019-0147-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo C et al. Robust single-cell DNA methylome profiling with snmc-seq2. Nat. Commun 9, 3824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satpathy AT et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol 37, 925–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smallwood SA et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao J et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 10.1126/science.aau0730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Lake BB & Zhang K High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol 10.1038/s41587-019-0290-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark SJ et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun 9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig LS et al. Lineage tracing in humans enabled by mitochondrial mutations and Single-Cell genomics. Cell 10.1016/j.cell.2019.01.022 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lareau CA et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat. Biotechnol 10.1038/s41587-020-0645-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu C et al. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat. Struct. Mol. Biol 26, 1063–1070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing QR et al. Parallel bimodal single-cell sequencing of transcriptome and chromatin accessibility. Genome Res. 30, 1027–1039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L et al. Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity. Nat. Commun 10, 470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma S et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 10.1016/j.cell.2020.09.056 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mimitou EP et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol 10.1038/s41587-021-00927-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiskin E, Lareau CA, Eraslan G, Ludwig LS & Regev A Single-cell multimodal profiling of proteins and chromatin accessibility using PHAGEATAC. Preprint at BioRxiv 10.1101/2020.10.01.322420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson E et al. Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq. eLife 10, e63632 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubin AJ et al. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Cell 176, 361–376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierce SE, Granja JM & Greenleaf WJ High-throughput single-cell chromatin accessibility CRISPR screens enable unbiased identification of regulatory networks in cancer. Nat. Commun 12, 2969 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thornton CA et al. Spatially mapped single-cell chromatin accessibility. Nat. Commun 12, 1274 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuart T & Satija R Integrative single-cell analysis. Nat. Rev. Genet 10.1038/s41576-019-0093-7 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Bravo González-Blas C et al. cistopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods 10.1038/s41592-019-0367-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cusanovich DA et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong L et al. SCALE method for single-cell ATAC-seq analysis via latent feature extraction. Nat. Commun 10, 4576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pliner HA et al. Cicero predicts cis-regulatory DNA interactions from Single-Cell chromatin accessibility data. Mol. Cell 71, 858–871.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schep AN, Wu B, Buenrostro JD & Greenleaf WJ chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danese A et al. EpiScanpy: integrated single-cell epigenomic analysis. Nat. Commun 10.1038/s41467-021-25131-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang R et al. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun 12, 1337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granja JM et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet 10.1038/s41588-021-00790-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji Z, Zhou W & Ji H Single-cell regulome data analysis by SCRAT. Bioinformatics 33, 2930–2932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker SM, Rogerson C, Hayes A, Sharrocks AD & Rattray M Classifying cells with scasat, a single-cell ATAC-seq analysis tool. Nucleic Acids Res. 47, e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao C, Hu S, Huo X & Zhang Y Dr.seq2: a quality control and analysis pipeline for parallel single cell transcriptome and epigenome data. PLoS ONE 12, e0180583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 10.1038/nbt.4096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stuart T et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao Y et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J et al. Single-cell lineage tracing by endogenous mutations enriched in transposase accessible mitochondrial DNA. eLife 10.7554/eLife.45105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics 27, 718–719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hafemeister C & Satija R Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deerwester S, Dumais ST, Furnas GW, Landauer TK & Harshman R Indexing by latent semantic analysis. J. Am. Soc. Inf. Sci 41, 391–407 (1990). [Google Scholar]
- 48.McInnes L & Healy J UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at arXiv https://arXiv.org/abs/1802.03426 (2018). [Google Scholar]
- 49.Pearce EL et al. Control of effector CD8+ T cell function by the transcription factor eomesodermin. Science 302, 1041–1043 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Corces MR et al. The chromatin accessibility landscape of primary human cancers. Science 10.1126/science.aav1898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.GTEx Consortium. The GTEx Consortium Atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H et al. Assessment of computational methods for the analysis of single-cell ATAC-seq data. Genome Biol. 20, 241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y et al. An atlas of gene regulatory elements in adult mouse cerebrum. Preprint at bioRxiv 10.1101/2020.05.10.087585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao J et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 10.1038/s41586-019-0969-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korsunsky I et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat. Methods 10.1038/s41592-019-0619-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brenner S Sequences and consequences. Philos. Trans. R. Soc. Lond. B Biol. Sci 365, 207–212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richmond TJ & Davey CA The structure of DNA in the nucleosome core. Nature 423, 145–150 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baglama J & Reichel L Augmented implicitly restarted Lanczos bidiagonalization methods. SIAM J. Sci. Comput 27, 19–42 (2005). [Google Scholar]
- 61.Amemiya HM, Kundaje A & Boyle AP The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep 9, 9354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waltman L & van Eck NJ A smart local moving algorithm for large-scale modularity-based community detection. Eur. Phys. J. B 86, 471 (2013). [Google Scholar]
- 63.Scrucca L, Fop M, Murphy TB & Raftery AE mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R. J 8, 289–317 (2016). [PMC free article] [PubMed] [Google Scholar]
- 64.Sing T, Sander O, Beerenwinkel N & Lengauer T ROCR: visualizing classifier performance in R. Bioinformatics 21, 3940–3941 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Fornes O et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hormozdiari F, Kostem E, Kang EY, Pasaniuc B & Eskin E Identifying causal variants at loci with multiple signals of association. Genetics 198, 497–508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griffiths JA, Richard AC, Bach K, Lun ATL & Marioni JC Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun 9, 2667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lun ATL et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv https://arxiv.org/abs/1303.3997 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in the paper are publicly available. The PBMC multiomic dataset is available from 10X Genomics at https://support.10xgenomics.com/single-cell-multiome-atac-gex/datasets/1.0.0/pbmc_granulocyte_sorted_10k. The PBMC scATAC-seq datasets are available from 10X Genomics at https://support.10xgenomics.com/single-cell-atac/datasets. The synthetic scATAC-seq datasets are available from GitHub at https://github.com/pinellolab/scATAC-benchmarking. Data from the BICCN are available from the Neuroscience Multiomic Archive at https://nemoarchive.org/. Data for the CRC patient sample are available on NCBI Gene Expression Omnibus (GSE148509) and Zenodo (https://zenodo.org/record/3977808).