Abstract
Brain low grade gliomas (LGG) often give serious clinical symptoms due to the invasion towards nervous system, affecting the life quality of patients. Collagen type I alpha 1(COL1A1) is the main component of type I collagen. Although there are many reports about abnormal expression of COL1A1 in various tumors, specific role and clinical significance of COL1A1 in LGG have not yet been elucidated. In this work, Tumor Immune Estimation Resource database was used for detecting the expression level of COL1A1 in cancer and normal tissues, and aimed to explore the relationship between COL1A1 and tumor immune infiltration. We applied Kaplan-Meier to analyze the role of COL1A1 in clinical prognosis. Univariate survival rate and multivariate Cox analysis were used to compare clinical characteristics and survival rate. The relativity between the expression of COL1A1 and the tumor microenvironment was evaluated using ESTIMATE algorithm. Finally, the relationship between expression level of COL1A1 and gene marker sets of immune cell infiltration was investigated via TIMER. According to TCGA, COL1A1 overexpression was correlated with overall survival (OS), progression free interval (PFI) and disease specific survival (DSS) of multiple tumors, especially in LGG. Multivariate analysis showed that COL1A1 expression was an independent prognostic factor for LGG. The expression of COL1A1 was positively correlated with the infiltration of CD4 + T and CD8 + T cells, neutrophils, macrophages and dendritic cells in LGG. In addition, there was a strong correlation between expression of COL1A1 and different immune marker sets in LGG. The results suggest that COL1A1 is related with tumor immune infiltration of LGG.
Introduction
Glioma is a primary tumor that originates from neuroglial stem or progenitor cells [1], which is common primary intracranial malignant tumor [2]. According to histological characteristics outlined by the World Health Organization (WHO), gliomas are classified into grades I to IV [3]. Grades I/II is classified as low-grade glioma (LGG), and grades III/IV is classified as high-grade glioma (HGG). Although LGG is less malignant than HGG, LGG inevitably develops into HGG eventually, and the long-term survival rate of LGG is still not satisfactory [4]. Immune mechanism plays a significant role in development of many tumors. Especially, LGGs, HGGs, and brain metastases lie on a spectrum of immunogenicity and T-cell infiltration [5]. Meanwhile, some immune cells, like T follicular helper (TFH) cells and M0 macrophages, had been selected as independent predictors for malignant transformation (MT) from LGG to HGG and formed an immune risk score [6]. In recent years, immunotherapy for the cancer has became as one of the most promising treatments. In these treatments for non-small cell lung cancer (NSCLC), immunotherapy using anti-programmed death-1 (PD-1) and its ligand PD-L1 antibody to target immune checkpoints is an effective method. [7]. This provides a new direction for the treatment of LGG. Therefore, there is an urgent need for new treatments that target the interaction between the tumor microenvironment (TME) and immune response.
Collagen is the major component of extracellular matrix (ECM) [8]. Type I collagen is widely distributed in structural organs such as bones, skin and teeth [9]. Type I collagen protein mainly consists of two collagen type I alpha 1 (COL1A1) and one collagen type I alpha 2 (COL1A2) chains [10, 11]. There is an evidence that the COL1A1 is involved in the carcinogenesis of several tissue types [12]. COL1A1 could promote the tumor progression in multiple types of cancer [13–15]. As an essential component of ECM, fibroblasts-secreted COL1A1 was proved to promote ovarian cancer metastasis via activating ITGB1/AKT signal pathway [16]. Meanwhile, COL1A1 is and involved in a variety of biological behaviors, such as cell proliferation, metastasis, invasion and angiogenesis [17, 18]. Bioinformatics analysis showed that there was differential expression of COL1A1 in gastric cancer [19]. However, it is unclear about the specific mechanism of COL1A1 in LGG and how it is related to tumor immunity.
Therefore, we employed databases of TCGA and Oncomine to comprehensively evaluate the correlation between the level of COL1A1 expression and prognosis of tumors. We also explored relationship between COL1A1 and tumor immune cell infiltration by relevant tumor data from TIMER. Our finding revealed the functional role of COL1A1 in LGG, and provided novel insights for LGG’s immunotherapy regimen.
Materials and method
1. Differential expression of COL1A1 by database analysis
We analyzed the differences in gene expression among the various type cancers from Cancer Genome Atlas (TCGA) project (https://portal.gdc.cancer.gov/repository) through Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) [20]. Through the Diff Exp module on the webpage, we check the differential expression of COL1A1 between the tumor and adjacent normal tissues. TIMER uses the Wilcoxon test by default to assess whether it is statistically significant. Considering the limited number of normal tissues sample in TCGA, we entered the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2223) and downloaded the GSE2223 data set, which contains 4 normal tissue samples and 26 LGG tumor samples. The information corresponding to the annotation samples was obtained through GPL1833, and the Wilcoxon test was applied to evaluate whether the expression of COL1A1 was different. Since the subtypes of LGG in the TCGA database is not comprehensive, we utilized the analysis module that comes with Chinese Glioma Genome Atlas (CGGA; http://www.cgga.org.cn/analyse/RNA-data.jsp) to analyze and visualize the mRNAseq_693 cohort in order to further explore the expression of COL1A1 in the subtypes [1–3].
2. Prognostic values of COL1A1 analysis
Through the University of California Santa Cruz Xena browser (UCSC Xena, https://xenabrowser.net/datapages/) [21], we obtained the expression, clinical and survival data of 33 types of cancer. According to the data in the TCGA, Kaplan–Meier method was used to conduct univariate analysis of survival for overall survival (OS) by survival [22] and survminer [23] package in R software (version 3.6.3). Moreover, we applied the survival R package for Cox proportional hazards model for continuous variables, and the forestplot [24] R package for generating plots. Similarly, limma [25], survival, survminer and forestplot package were used to describe and analyze progression free interval (PFI), disease specific survival (DSS), disease free interval (DFI), and generate corresponding forest plots. We extracted clinical data of LGG on various tumors in the TCGA and CGGA database and deleted missing and incomplete data. Our research pays a special attention to the effect of COL1A1 on the cancer grade of LGG.
3. Tumor mutational burden and tumor microenvironment analysis
We downloaded the mutation data from the TCGA database and calculated the tumor mutational burden (TMB) score for each tumor, which was tested using Spearman’s rank correlation coefficient method. The tumor microenvironment consists of stromal cells, immune cells and cytokines, which has been shown to be closely related to the biological behavior of tumor cells [26]. The estimate [27] and limma packages in R software were also employed in evaluating the immune infiltration (ImmuneScore), overall stromal content (StromalScore) and the combined score (ESTIMATEScore) of the cancer samples in TCGA. Figures were created by R plotting system ggplot2 package [28].
4. Immune infiltration analysis
The Timer Web server is mainly used to systematically analyze the immune infiltration level of different tumors. The timer algorithm was applied to assess the abundance of 6 immune cells (CD4+T cells, CD8+T cells, B cells, neutrophils, macrophages, and dendritic cells). We conducted a series of analyses on the expression of COL1A1 in different tumors and infiltration abundance of immune cells in LGG via the gene module. Next, we collected 47 common immune checkpoint genes and estimated the relevance between COL1A1 expression and the immune checkpoint genes.
5. Statistical analysis
P-values of Kaplan–Meier survival, DSS, DFI and PFI analysis derived from a log-rank test. The cox regression model was performed for estimating the Hazard Ratio (HR) and 95% confidence interval (CI) values. The correlation of the expression levels of COL1A1 was analyzed by Spearman’s rank-order test correlation. P-values <0.05 were regarded as statistically significant.
Results
1. COL1A1 expression in different cancer
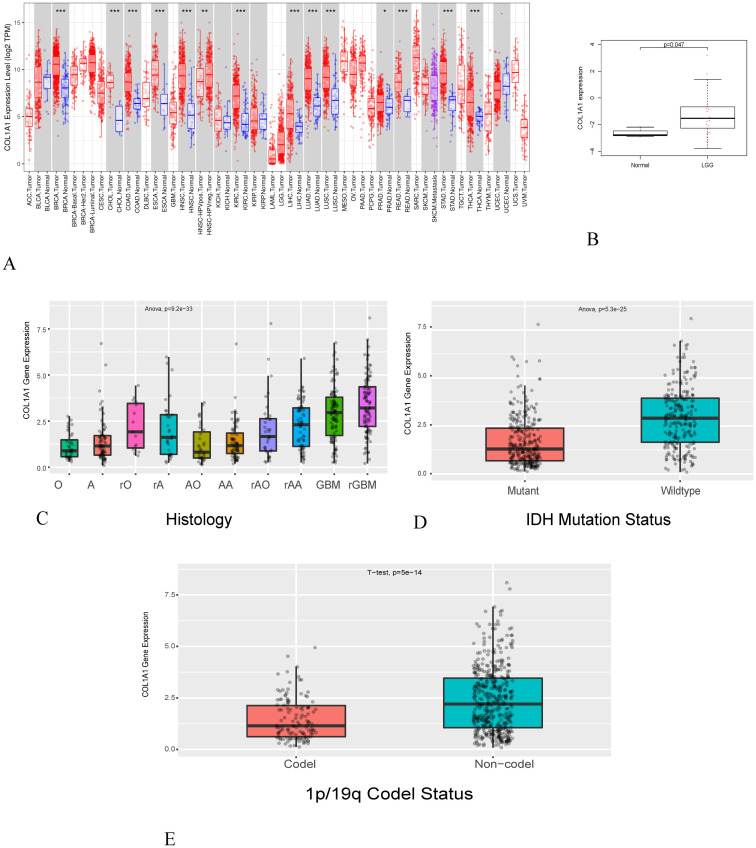
To estimate the expression level of genes in normal and tumor samples, we first conducted an analysis of multiple tumors using the TIMER database. The results demonstrated that the expression of COL1A1 was significantly increased in breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma(HNSC), kidney renal clear cell carcinoma(KIRC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma(READ), stomach adenocarcinoma (STAD) and thyroid carcinoma (THCA) in Fig 1A as compared with normal tissues. We also applied GEO databases to estimate expression of gene in LGG. The results displayed that expression level of COL1A1 increased in LGG in contrast to normal tissues in Fig 1B. Considering that the number of normal glioma in TCGA is limited, we also referred to with CGGA database. We found that the expression of COL1A1 increased in LGG, including different histological subtypes and molecular classifications (Fig 1C–1E). It was worth noting that COL1A1 expressed highly in wildtype and Non-code, while Mutant and Code showed opposite trends (Fig 1D and 1E). In other words, the expression of COL1A1 was decreased in IDH-mutant and 1p/19q codeleted LGGs. Previous researchers found that IDH-mutant and 1p/19q codeleted LGGs have better prognosis [29], which further conformed our results. In summary, highly expressed COL1A1 played an important role on the patient’s prognosis.
Fig 1. The expression level of COL1A1 in normal and tumor tissues.
(A) In the TIMER database, the expression level of COL1A1 in different cancer tissues compared with normal tissues. (B) the expression level of COL1A1 in GEO. (C-E) The expression level of COL1A1 in different subtypes of glioma (A: Astrocytoma, O: Oligodendro, rO: recurrent Oligodendro, rA: recurrent Astrocytoma, AA: Anaplastic Astrocytoma, AO: Anaplastic Oligodendro, rAA: recurrent Anaplastic Astrocytoma, rAO: recurrent Anaplastic Oligodendro, GBM:Glioblastoma, rGBM: recurrent Glioblastoma.** P < 0.05,** P < 0.005, *** P < 0.001).
2. Prognostic evaluation of COL1A1 in tumors
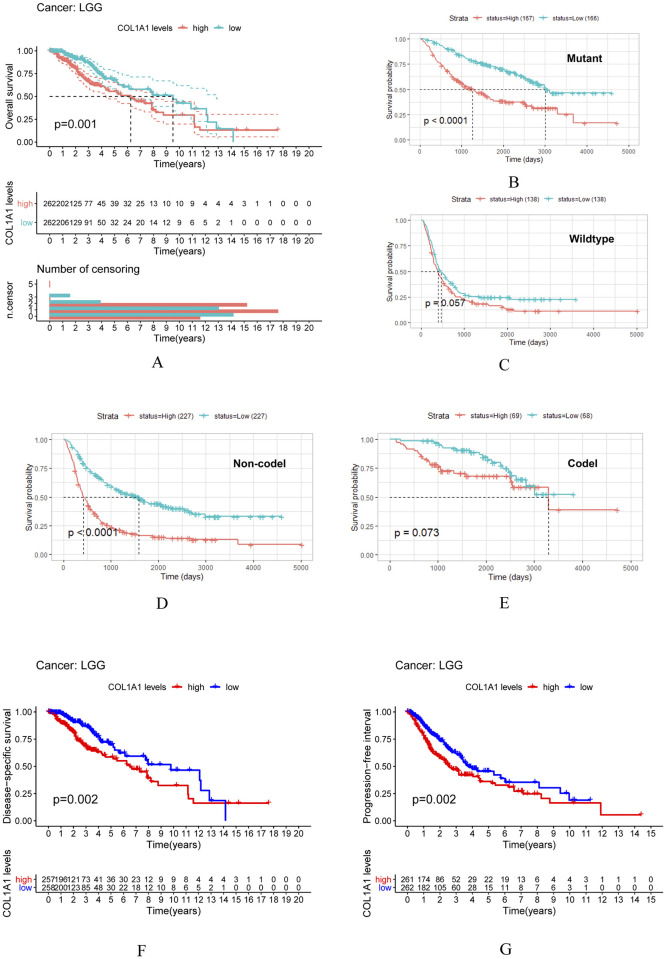
To investigate whether COL1A1 expression is correlated to the prognosis of tumor, we downloaded the clinical data of tumors from TCGA and CGGA. Figs 2 and 3 show the relationship between COL1A1 expression and the prognosis of different cancers. We found that the following four cancers showed an essential correlation between the prognosis of cancers and the expression of COL1A1, including LGG, KIRP, cutaneous melanoma (SKCM) and mesothelioma (MESO). The outcome indicates that highly expressed COL1A1 is significantly related to the poor prognosis of these cancers.
Fig 2. Overall survival curve of high and low expression of COL1A1 in glioma and it’s subclass.
(A) Survival curves of OS by Kaplan-Meier respectively in LGG. (B-E) Survival curves of Survival probability in Mutant, Wildtype, Non-codel and Codel. (F-G) Survival curves of DSS and PFI in LGG. DSS, disease specific survival; PFI, progression free interval.
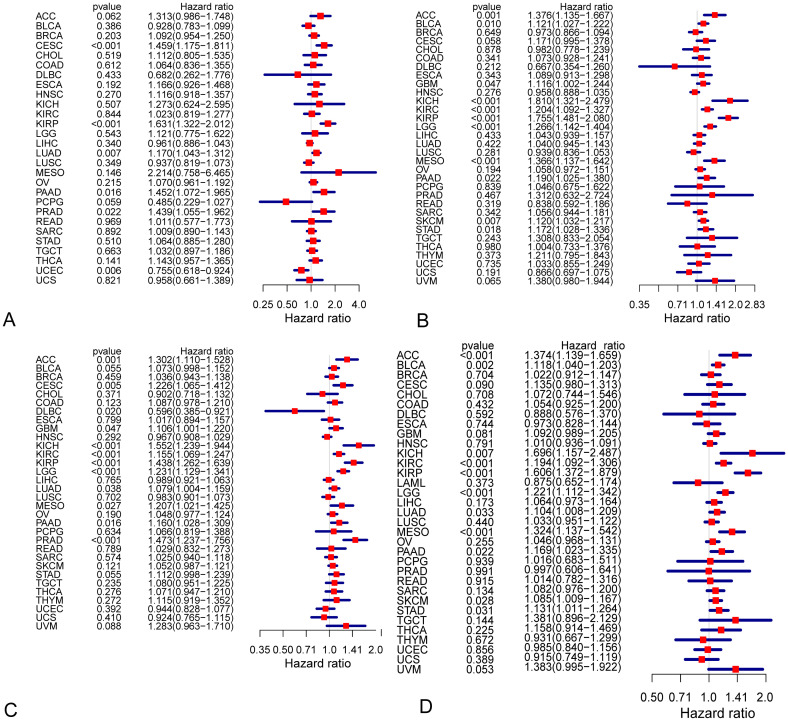
Fig 3. Survival curves and cox proportional hazards regression model of DFI, DSS, and PFI in different types of cancers.
(A-C) Cox proportional hazards regression model of DFI, DSS, and PFI. (D) Cox proportional hazards model of OS in LGG for expression of COL1A1. DFI, disease free interval.
In order to evaluate the prognostic effect of the expression level of COL1A1 in different tumors, we show the overall survival curve of COL1A1 in LGG (Fig 2A) and in its subclass (Fig 2B–2E). It should be noted that overall survival curve of COL1A1 in Wildtype and Codel have a significant statistical significance (Fig 2B and 2D). However, we did not observe this phenomenon in Mutant and Codel (Fig 2C and 2E). Meanwhile the data source of Fig 2A is from TCGA dataset, Fig 2B and 2E are CGGA. Next, we used the other three outcome metrics provided by TCGA: DSS, PFI and DFI. These results clearly indicate that the expression of COL1A1 has an effect on the prognosis of LGG (Fig 2F and 2G). Although, we did not ascertain any significant relationship between the expression of COL1A1 and DFI in LGG patients (Fig 3A), it was worth noting that high COL1A1 expression relates to DSS and PFI in multiple cancers including LGG (Fig 3B and 3C). Meanwhile, R software was used to detect the prognostic value of COL1A1 by cox proportional hazards model (Fig 3D). Unanimously, the higher expression level of COL1A1 was shown to be related to its poor prognosis in LGG (OS HR = 1.221, 95% CI = 1.112 to 1.342, P = 3.00E-05).
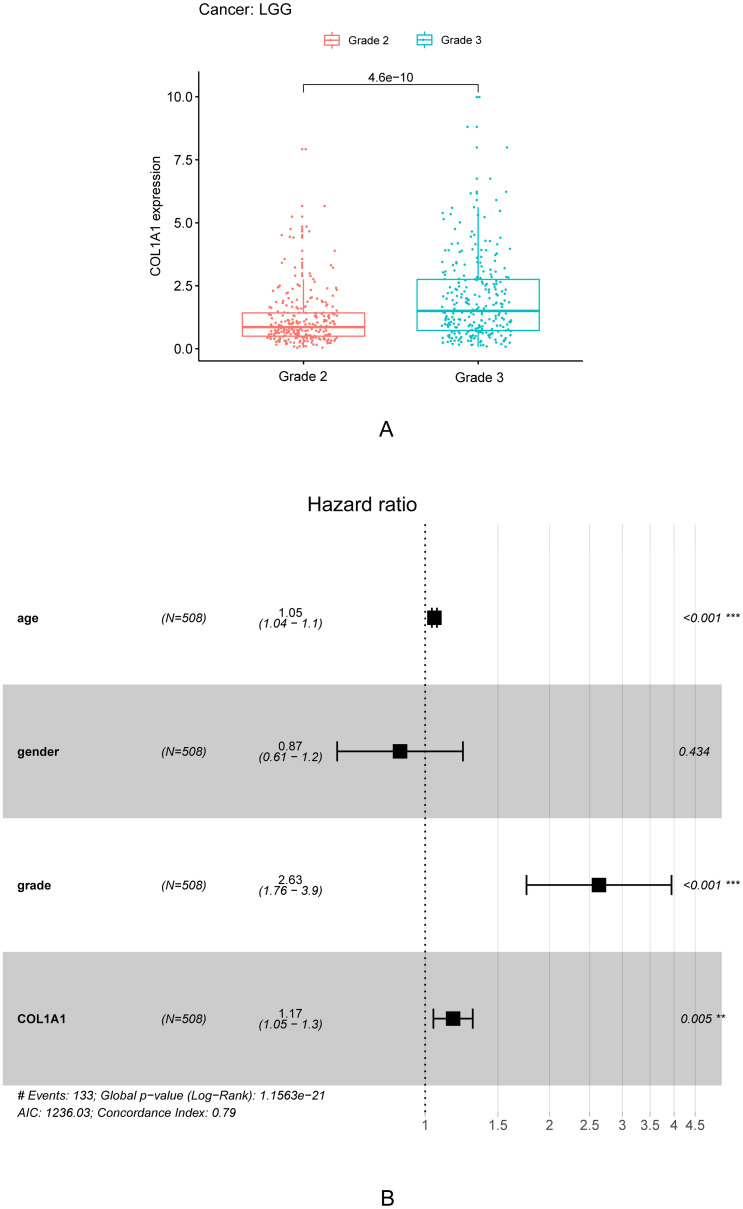
In addition, we also collected and compiled clinical data. As shown in Table 1, univariate correlation analysis using Cox regression showed that the grade was significantly correlated with overall survival. Age, tumor grade, and COL1A1 expression were independent prognostic factors in multivariate analysis (Fig 4B). The expression of COL1A1 showed a significant correlation with tumor grade through Table 1, Fig 4. With the increase of tumor grade, the expression of COL1A1 increased accordingly (Fig 4A).
Table 1. Univariate (a) and multivariate analysis (b) of the correlation of COL1A1 expression with OS among LGG patients.
| Hazard Ratio(HR) | 95%CI | pvalue | |
|---|---|---|---|
| a | |||
| age | 1.056 | 1.042–1.070 | 0.000 |
| gender | 0.857 | 0.607–1.211 | 0.381 |
| grade | 3.496 | 2.389–5.115 | 0.000 |
| COL1A1 | 1.264 | 1.144–1.398 | 0.000 |
| b | |||
| grade | 2.633 | 1.758–3.944 | 0.000 |
| COL1A1 | 1.168 | 1.047–1.304 | 0.005 |
Fig 4. The expression of COL1A1 in LGG grade and the forest plot of LGG.
A. Differential expression of COL1A1 in different cancer grade of LGG. Based on data from 258 grade2(G2) patients and 270 grade3(G3) patients in TCGA. B. Multivariate Cox analysis of COL1A1 expression and clinical pathological factors. As age, tumor grade and COL1A1 expression are independent prognostic factors (* P <0.05; ** P < 0.005;*** P < 0.001).
3. TMB and TME analysis
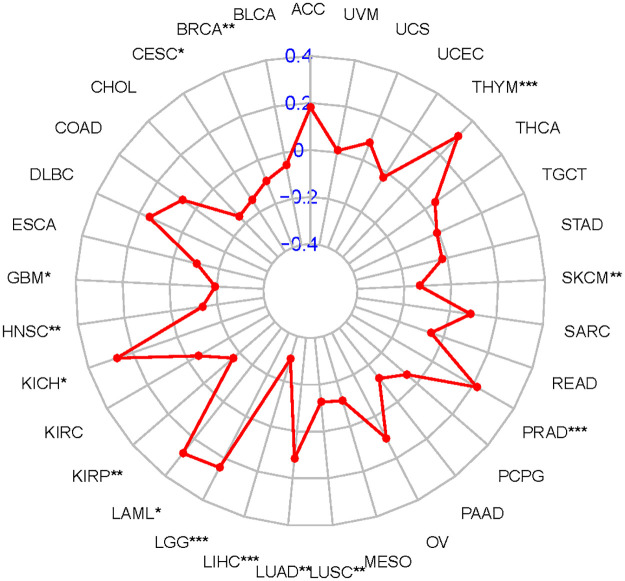
TMB is an independent biomarker that can be used to predict the efficacy of immunotherapy found in various tumor in recent years. Those with high TMB expression have greater clinical benefit from immune checkpoint inhibitor treatment [30]. Here, we separately counted the TMB of each tumor sample, and analyzed the relationship between gene expression and TMB as follows, using Spearman rank correlation coefficient (Fig 5). We can see that there are six tumors that are positively correlated with TMB from the analysis, including LGG, thymoma (THYM), PRAD, LUAD, acute myeloid leukemia (LAML) and kidney chromophobe (KICH). These results indicate that the treatment plan of immune checkpoint inhibitor may play a significant role in these tumors.
Fig 5. Radar graph for relationship between the expression of COL1A1 and TMB of each tumor sample.
Correlation between Tumor mutational burden and LGG, THYM, PRAD, LUAD, LAML and KICH(* P <0.05; ** P < 0.005;*** P < 0.001).
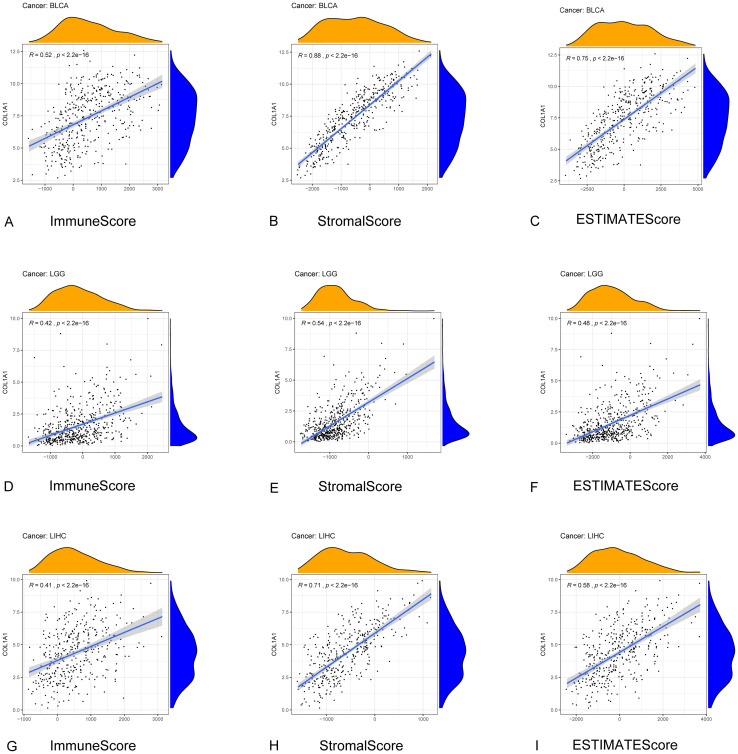
TME is the cellular environment of tumor. It’s composed of immune cells, inflammatory mediators, endothelial cells, extracellular matrix (ECM) and mesenchymal cells [31]. In TME, stromal and immune cells are two major non-tumor components, which are considered to be of great significance for the diagnosis and prognostic evaluation of cancers. We calculated the correlations between gene expression levels and ImmuneScore, StromalScore respectively to predict the infiltration of stromal and immune cells by the ESTIMATE algorithm among 33 cancers (Table 2). We selected several representative results for visualization (Fig 6). The results reveal that the expression of COL1A1 has a strong correlation with the TME of various cancers.
Table 2. Correlation analysis of COL1A1 expression level and TME score.
| CancerType | StromalScore P value | cor | ImmuneScore P value | cor | ESTIMATEScore P value | cor |
|---|---|---|---|---|---|---|
| ACC | ** | 0.389 | 0.768 | 0.034 | 0.077 | 0.200 |
| BLCA | *** | 0.883 | *** | 0.521 | *** | 0.748 |
| BRCA | *** | 0.770 | *** | 0.214 | *** | 0.495 |
| CESC | *** | 0.695 | 0.886 | -0.008 | *** | 0.328 |
| CHOL | * | 0.478 | 0.460 | 0.127 | 0.084 | 0.292 |
| COAD | *** | 0.796 | *** | 0.475 | *** | 0.685 |
| DLBC | *** | 0.674 | 0.088 | -0.249 | 0.068 | 0.266 |
| ESCA | *** | 0.742 | * | 0.205 | *** | 0.521 |
| GBM | *** | 0.636 | *** | 0.353 | *** | 0.485 |
| HNSC | *** | 0.822 | * | 0.137 | *** | 0.500 |
| KICH | *** | 0.560 | * | 0.344 | ** | 0.454 |
| KIRC | *** | 0.711 | *** | 0.214 | *** | 0.467 |
| KIRP | *** | 0.562 | *** | 0.243 | *** | 0.388 |
| LAML | * | 0.247 | 0.973 | -0.003 | 0.197 | 0.106 |
| LGG | *** | 0.544 | *** | 0.421 | *** | 0.480 |
| LIHC | *** | 0.712 | *** | 0.406 | *** | 0.578 |
| LUAD | *** | 0.553 | ** | 0.169 | *** | 0.380 |
| LUSC | *** | 0.764 | *** | 0.355 | *** | 0.568 |
| MESO | *** | 0.635 | 0.982 | -0.002 | * | 0.302 |
| OV | *** | 0.835 | *** | 0.286 | *** | 0.592 |
| PAAD | *** | 0.659 | *** | 0.354 | *** | 0.521 |
| PCPG | *** | 0.631 | *** | 0.441 | *** | 0.561 |
| PRAD | *** | 0.752 | *** | 0.390 | *** | 0.604 |
| READ | *** | 0.836 | *** | 0.404 | *** | 0.687 |
| SARC | *** | 0.495 | 0.056 | 0.118 | *** | 0.275 |
| SKCM | *** | 0.689 | *** | 0.203 | *** | 0.415 |
| STAD | *** | 0.630 | *** | 0.215 | *** | 0.451 |
| TGCT | *** | 0.738 | *** | -0.359 | 0.440 | 0.062 |
| THCA | *** | 0.759 | *** | 0.587 | *** | 0.697 |
| THYM | *** | 0.775 | 0.551 | -0.055 | *** | 0.416 |
| UCEC | *** | 0.624 | * | 0.123 | *** | 0.357 |
| UCS | *** | 0.788 | 0.030 | 0.292 | *** | 0.557 |
| UVM | *** | 0.666 | ** | 0.402 | *** | 0.506 |
* P <0.05;
** P < 0.005;
*** P < 0.001.
Fig 6. Correlation between COL1A1 expression and TME in diffrent tumer.
(A-C). Relationship between COL1A1 expression and Stromal Score, Immune Score and ESTIMATE Score in BLCA. (D-F). Correlation between COL1A1 expression and Stromal Score, Immune Score and ESTIMATE Score in LGG. (G-I). Relationship between COL1A1 expression and TME in LIHC.
4. COL1A1 expression correlated with immune cell infiltration and immune cell markers in LGG
In cancer patients, tumor-infiltrating lymphocytes are major indicators that independently predict the overall survival rate and sentinel lymph node status [32, 33]. Thus, we further explored if the COL1A1 expression is correlated with the degree of immune cell infiltration in different types of tumor by TIMER database.
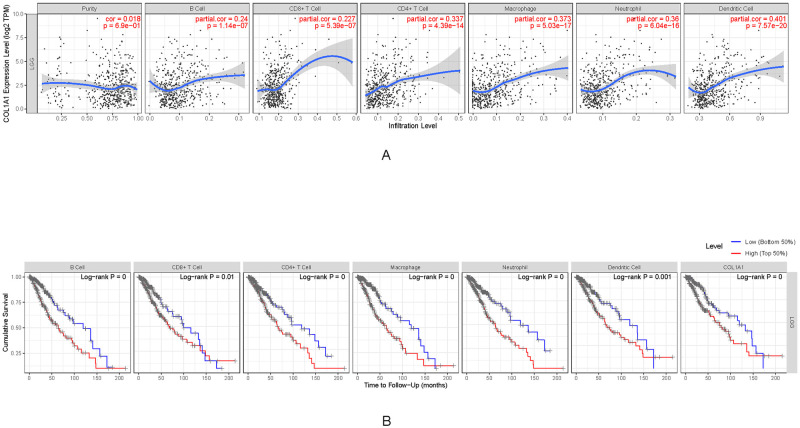
In the LGG, the expression of COL1A1 was notably associated with the infiltration levels of B cell (R = 0.24, P = 1.14e−07), CD4+T cells (R = 0.337, P = 4.39e−14), CD8+ T cells (R = 0.227, P = 5.39e−07), macrophages (R = 0.373, P = 5.03e−17), dendritic cells (R = 0.401, P = 7.57e−20) and neutrophils (R = 0.36,P = 6.04e−16) (Fig 7A). To further clarify the correlation between COL1A1 and immune cell infiltration in LGG, we generated the Kaplan-Meier diagram through the TIMER database. We found that B cell (P = 4.25E-05), CD8+ T cells(P = 0.0095), CD4+T cells(P = 0.0005), macrophages(P = 9.20E-06), neutrophils(P = 5.83E-06), dendritic cells infiltration (P = 0.0006), and COL1A1 expression (P = 0.0004) were significantly correlated with the prognosis of LGG(Fig 7B). It indicates that COL1A1 clearly plays an important role in modulating immune cell infiltration in LGG, especially in macrophages and DCs infiltration.
Fig 7. Immune infiltration level and survival time in LGG.
A. Correlation of COL1A1 expression with infiltrating levels of B cell, CD4+ T cells, CD8+ T cells, Macrophages, Neutrophils and DCs in LGG. B. Cumulative survival of COL1A1 expression related to B cell, T cells, Macrophages, Neutrophils and DCs in LGG.
Next, in order to explore the relationship between a variety of immune infiltrating cells and COL1A1 in TIMER, we pay attention to the correlation between COL1A1 and the gene sets of immune cell marker in LGG and MESO (control group). We analyzed the correlation between different immune cell marker genes and COL1A1, such as CD8+ T cells, T cells (general), B cells, monocytes, TAMs, M1 and M2 macrophages, neutrophils, NK cells, DCs, Th1 cells, Th2 cells, Tfh cells, Th17 cells, Tregs and exhausted T cells. According to the results adjusted for purity, the expression level of COL1A1 in LGG was significantly correlated with most of the immune cell marker sets (Table 3). However, expression of COL1A1 was significantly correlated with only 9 immune cell marker sets in MESO. Specifically, the results show that COL1A1 is significant correlated with CD8A, CD8B of CD8+ T cell markers, CD3D, CD3E, CD2 of T cell markers, CD19, CD79A of B cell markers, CD86, CD115 of monocyte markers, CCL2, CD68, IL10 of TAM markers, IRF5, COX2 of M1 macrophages markers, CD163, VSIG4, MS4A4A of M2 macrophage markers, CD11b, CCR7 of neutrophils markers, KIR2DL1, KIR2DL3, KIR2DL4, KIR3DL1, KIR3DL2, KIR2DS4 of natural killer cell markers, HLA-DPB1, HLA-DQB1, HLA-DRA, HLA-DPA1, BDCA-1, BDCA-4, CD11c of DCs markers, T-bet, STAT1, IFN-γ of Th1 markers, GATA3, STAT6, STAT5A of Th2 markers, STAT3 of Th17 markers, CCR8, TGFβ of Treg markers and PD-1, CTLA4, LAG3, TIM-3, GZMB of T cell exhaustion markers in LGG.
Table 3. Correlation analysis of COL1A1 expression level and immune cell markers in TIMER.
| Description | Gene markers | LGG | MESO | ||||||
|---|---|---|---|---|---|---|---|---|---|
| None | Purity | None | Purity | ||||||
| Cor | P | Cor | P | Cor | P | Cor | P | ||
| CD8+ T cell | CD8A | 0.230 | *** | 0.253 | *** | 0.024 | 0.825 | -0.101 | 0.357 |
| CD8B | 0.194 | *** | 0.205 | *** | 0.117 | 0.281 | 0.020 | 0.855 | |
| T cell (general) | CD3D | 0.491 | *** | 0.511 | *** | 0.061 | 0.573 | -0.064 | 0.561 |
| CD3E | 0.518 | *** | 0.531 | *** | 0.081 | 0.457 | -0.047 | 0.671 | |
| CD2 | 0.544 | *** | 0.554 | *** | 0.031 | 0.775 | -0.090 | 0.412 | |
| B cell | CD19 | 0.372 | *** | 0.352 | *** | 0.015 | 0.893 | -0.112 | 0.309 |
| CD79A | 0.218 | *** | 0.221 | *** | -0.122 | 0.259 | -0.239 | 0.027 | |
| Monocyte | CD86 | 0.285 | *** | 0.301 | *** | 0.061 | 0.574 | -0.020 | 0.856 |
| CD115 (CSF1R) | 0.172 | *** | 0.184 | *** | 0.190 | 0.078 | 0.130 | 0.237 | |
| TAM | CCL2 | 0.358 | *** | 0.353 | *** | -0.147 | 0.175 | -0.187 | 0.086 |
| CD68 | 0.401 | *** | 0.417 | *** | 0.043 | 0.696 | -0.012 | 0.916 | |
| IL10 | 0.358 | *** | 0.361 | *** | 0.139 | 0.199 | 0.078 | 0.477 | |
| M1 Macrophage | INOS (NOS2) | -0.042 | 0.345 | -0.029 | 0.533 | 0.459 | *** | 0.453 | *** |
| IRF5 | 0.287 | *** | 0.324 | *** | -0.189 | 0.080 | -0.234 | 0.031 | |
| COX2(PTGS2) | 0.148 | ** | 0.143 | * | 0.225 | 0.037 | 0.253 | 0.019 | |
| M2 Macrophage | CD163 | 0.480 | *** | 0.463 | *** | 0.138 | 0.203 | 0.058 | 0.600 |
| VSIG4 | 0.213 | *** | 0.212 | *** | 0.142 | 0.190 | 0.100 | 0.364 | |
| MS4A4A | 0.422 | *** | 0.424 | *** | 0.174 | 0.107 | 0.090 | 0.415 | |
| Neutrophils | CD66b (CEACAM8) | -0.025 | 0.571 | -0.038 | 0.407 | 0.099 | 0.360 | 0.127 | 0.246 |
| CD11b (ITGAM) | 0.256 | *** | 0.281 | *** | 0.193 | 0.074 | 0.154 | 0.160 | |
| CCR7 | 0.391 | *** | 0.391 | *** | 0.045 | 0.681 | -0.092 | 0.403 | |
| Natural killer cell | KIR2DL1 | 0.164 | ** | 0.168 | ** | -0.213 | 0.048 | -0.286 | * |
| KIR2DL3 | 0.266 | *** | 0.266 | *** | -0.206 | 0.056 | -0.272 | 0.012 | |
| KIR2DL4 | 0.278 | *** | 0.272 | *** | -0.289 | ** | -0.385 | ** | |
| KIR3DL1 | 0.146 | ** | 0.148 | * | -0.184 | 0.088 | -0.284 | * | |
| KIR3DL2 | 0.190 | *** | 0.209 | *** | -0.080 | 0.462 | -0.181 | 0.098 | |
| KIR3DL3 | 0.032 | 0.470 | 0.033 | 0.476 | -0.329 | ** | -0.400 | ** | |
| KIR2DS4 | 0.209 | *** | 0.226 | *** | -0.142 | 0.190 | -0.190 | 0.082 | |
| Dendritic cell | HLA-DPB1 | 0.499 | *** | 0.506 | *** | 0.164 | 0.130 | 0.087 | 0.426 |
| HLA-DQB1 | 0.428 | *** | 0.425 | *** | 0.129 | 0.233 | 0.078 | 0.477 | |
| HLA-DRA | 0.498 | *** | 0.503 | *** | 0.149 | 0.169 | 0.070 | 0.526 | |
| HLA-DPA1 | 0.506 | *** | 0.513 | *** | 0.148 | 0.171 | 0.063 | 0.565 | |
| BDCA-1(CD1C) | 0.379 | *** | 0.357 | *** | 0.181 | 0.093 | 0.130 | 0.236 | |
| BDCA-4(NRP1) | 0.521 | *** | 0.519 | *** | 0.567 | *** | 0.546 | *** | |
| CD11c (ITGAX) | 0.232 | *** | 0.256 | *** | 0.062 | 0.566 | -0.005 | 0.967 | |
| Th1 | T-bet (TBX21) | 0.448 | *** | 0.438 | *** | -0.103 | 0.342 | -0.269 | 0.013 |
| STAT4 | -0.066 | 0.134 | -0.057 | 0.214 | 0.036 | 0.739 | -0.112 | 0.307 | |
| STAT1 | 0.429 | *** | 0.427 | *** | -0.138 | 0.203 | -0.218 | 0.046 | |
| IFN-γ (IFNG) | 0.214 | *** | 0.226 | *** | 0.076 | 0.484 | -0.023 | 0.837 | |
| TNF-α (TNF) | 0.061 | 0.169 | 0.052 | 0.252 | 0.026 | 0.815 | -0.105 | 0.338 | |
| Th2 | GATA3 | 0.407 | *** | 0.421 | *** | 0.243 | 0.023 | 0.208 | 0.056 |
| STAT6 | 0.241 | *** | 0.304 | *** | -0.134 | 0.214 | -0.113 | 0.302 | |
| STAT5A | 0.299 | *** | 0.311 | *** | -0.168 | 0.121 | -0.209 | 0.054 | |
| IL13 | -0.066 | 0.133 | -0.051 | 0.266 | 0.130 | 0.230 | 0.137 | 0.210 | |
| Tfh | BCL6 | -0.042 | 0.344 | -0.065 | 0.155 | 0.177 | 0.101 | 0.209 | 0.055 |
| IL21 | 0.091 | 0.039 | 0.080 | 0.080 | 0.108 | 0.317 | 0.113 | 0.304 | |
| Th17 | STAT3 | 0.469 | *** | 0.456 | *** | 0.107 | 0.325 | 0.072 | 0.515 |
| IL17A | 0.045 | 0.304 | 0.029 | 0.523 | 0.084 | 0.441 | 0.123 | 0.262 | |
| Treg | FOXP3 | 0.045 | 0.307 | 0.069 | 0.130 | 0.224 | 0.037 | 0.167 | 0.127 |
| CCR8 | 0.162 | ** | 0.173 | ** | 0.227 | 0.034 | 0.163 | 0.135 | |
| STAT5B | 0.019 | 0.669 | 0.012 | 0.797 | 0.210 | 0.050 | 0.205 | 0.060 | |
| TGFβ (TGFB1) | 0.340 | *** | 0.353 | *** | 0.488 | *** | 0.474 | *** | |
| T cell exhaustion | PD-1 (PDCD1) | 0.430 | *** | 0.428 | *** | 0.031 | 0.777 | -0.036 | 0.741 |
| CTLA4 | 0.322 | *** | 0.309 | *** | 0.117 | 0.280 | 0.022 | 0.844 | |
| LAG3 | 0.300 | *** | 0.294 | *** | -0.285 | ** | -0.377 | ** | |
| TIM-3 (HAVCR2) | 0.312 | *** | 0.336 | *** | 0.058 | 0.593 | -0.025 | 0.818 | |
| GZMB | 0.425 | *** | 0.425 | *** | -0.225 | ** | -0.357 | ** | |
* P <0.05;
** P < 0.005;
*** P < 0.001.
The expression of COL1A1 is particularly related to markers of the most TAM, monocyte, M1 and M2 macrophage in LGG. It further suggests that COL1A1 may be involved in the polarization process of macrophages. On the basis of these results, we find that the marker gene sets of DCs are significantly associated with COL1A1, suggesting that high expression level of COL1A1 is correlated to the increase of DCs infiltration in LGG, but this requires further research to support this conclusion. We also observed that COL1A1 is significantly related to the T cell exhaustion marker gene sets, indicating that COL1A1 may play a crucial part in immune escape of LGG.
5. COL1A1 expression correlated with Immune checkpoint gene in LGG
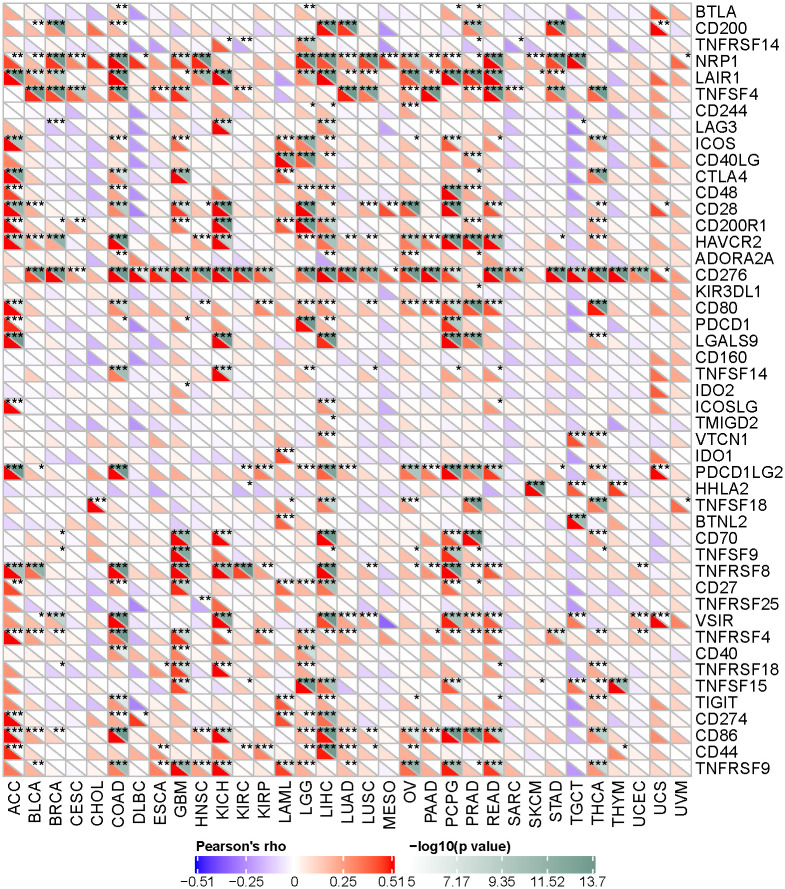
Immune checkpoint pathway used by cancers inhibit tumor-reactive T cells considering as a mechanism of immune resistance, and it may induce immune suppression and immune escape [34]. Immune checkpoint inhibitors (ICIs) are one of the most promising methods for treating cancer [35]. To identify potential therapeutic targets of those cancers and immune escape, we have collected more than forty representative immune checkpoint genes, including BTLA, CD200, TNFRSF14, NRP1, LAIR1 and so on, and we evaluated the relationship between expression of COL1A1 and immune checkpoint. We extracted these immune checkpoint genes separately and calculated the coefficient correlation with expression of our target gene. The most of the immune checkpoint genes were observed to be positively co-expressed with COL1A1 of 33 tumors. We found that the immunological target showing remarkably significant correlation is CD276 among 27 tumors. In LGG, COL1A1 has a significant correlation and statistical significance with most selected immune checkpoint genes (Fig 8). Among them, CD28(R = 0.82,P = 6.37E-128),NRP1(R = 0.49, P = 5.92E-31), PDCD1(R = 0.51, P = 8.33E-34) and TNFSF15(R = 0.75, P = 1.55E-92) have a strong correlation with COL1A1 expression. Accordingly, the results further confirmed the specific correlation between COL1A1 and immune infiltrating in LGG, indicating that COL1A1 has a profound effect on immune escape of TMB.
Fig 8. Correlation of the expression between COL1A1 and immune checkpoint genes in different tumors.
Based on Pearson correlation coefficients, COL1A1 has a significant correlation with most selected immune checkpoint genes in LGG(* P <0.05; ** P < 0.005;*** P < 0.001).
Discussion
COL1A1, located in chromosome 17, region 17q21–22, and presenting 51 exons, encodes the pro-α1 chain of type I collagen, which is consisting of a triple helix composed of one α2 chain and two α1 chains [36]. A fibrillar collagen C‑terminal domain (COLF) a collagen triple‑helix repeat and A von Willebrand factor type C (vWFC) domain are three conservative domains of COL1A1 [37]. Previous studies have shown that the expression of COL1A1 is correlated to a variety of tumor types, and COL1A1 is highly expressed in tumor tissues and cells [38–43]. Based on the integrated transcriptome analysis, six crosstalk collagen genes (including COL1A1) may be related to the progress and prognosis of LGG (4). Functional enrichment analysis showed that over-expressed COL1A1 may participate in the progression and poor prognosis of LGG through the ECM-receptor interaction pathway. COL1A1 regulates the expression of three key genes of Rac1-GTP, p-JNK and RhoA-GTP in WNT/PCP signaling pathway. The Wnt/PCP signaling pathway mainly activates JNK through small G protein to regulate cytoskeletal rearrangement, and transmits signals from Frizzled and ROR2/RYK co-receptors on the cell surface to the nucleus through Rho GTPases and JNK, which is essential for cell migration indispensable process [44]. Among these key proteins, JNK is central to promoting tumor invasion and metastasis [45]. Therefore, COL1A1 may affect tumor development through WNT/PCP signaling pathway. However, there are few reports on the function and mechanism of COL1A1 in LGG. Recent studies have shown that collagen density is related to the infiltration of T cells, thereby affecting tumor development [7]. In this research, we are aimed at exploring the expression level of COL1A1 and prognosis in LGG, and analyze the related mechanism of COL1A1 in tumor immune infiltration.
In this report, differences in expression levels of COL1A1 are related to the prognosis of different tumors, which is consistent with previous literature results. We find that COL1A1 has higher expression levels in multiple cancer tissues and the highly expressed COL1A1 is closely correlated to the poor prognosis of LGG. In addition, our results indicate that COL1A1 expression is interrelated with numerous immune marker sets and immune infiltration levels of LGG. Thus, our research suggests that COL1A1 may have a potential role in tumor immunology and provide a theoretical basis for being a biomarker of LGG. To further investigate the potential mechanism of COL1A1 expression in LGG and its clinical correlation, we got the dataset from TCGA database. The expression of COL1A1 was associated with the grade of this tumor by the relative R package analysis, and the study indicated that expression of COL1A1 was a critical independent prognostic factor through multivariate analysis in patients of low-grade glioma.
In this research, we used TCGA independent data sets and GEO database to assess the expression of COL1A1 and the prognosis in kinds of tumor, showing that the expression of COL1A1 of multiple cancers is significantly different between tumors and normal tissues. Then, we analyzed 33 cancers in the TCGA database and observed that expression of COL1A1 was higher in CHOL, COAD, LIHC, STAD, THCA, ESCA, LUSC, HNSC, BRCA, KIRC, LUAD, PRAD and READ compared with adjacent normal tissues. Due to the differences in collection methods of different database and potential biological characteristics, there are certain differences in the expression of COL1A1. However, we observed a correlation between increased expression of COL1A1 and poor prognosis of LGG. In mutant group and non codel group, the high expression level of COL1A1 showed significant low survival rate (P < 0.05), but there was no significant difference in wildtype group and codel group(P>0.05). Through the clinical prognosis data sets in the TCGA database, we found that the HR value of COL1A1 in OS, DSS, PFI revealed that the high expression level of COL1A1 is a high risk factor of the prognosis in LGG. Therefore, these results suggest that COL1A1 could be a valuable factor for the prognosis of LGG.
Another aspect of this study was to estimate the relevance between the COL1A1 and tumor immune infiltration, especially in LGG. We noticed that the degree of macrophage, CD8+, CD4+, DCs and neutrophil infiltration was positively correlated with expression level of COL1A1 via TIMER. Further research confirmed that these immune cells are closely related to the prognosis of LGG. Moreover, we can conclude from the relationship between the expression level of COL1A1 and certain markers of immune cell that COL1A1 plays a critical role in regulating tumor immunology of LGG. In the study of gene markers of macrophages, we found that the expression of COL1A1 is weakly correlated with M1 macrophage gene markers (IRF5, COX2), but it is more moderately correlated with M2 macrophage gene markers(CD163, MS4A4A), which suggests that COL1A1 may be potential regulatory role in polarization of tumor-associated macrophages(TAM). Additionally, our further results show that the increased expression of COL1A1 is positively correlated with the T cell exhaustion markers (PD-1, LAG3, CTLA4, TIM-3 and GZMB) in LGG, thus the T cell exhaustion is associated with the potential of COL1A1 expression. The PD-1L/PD-1 immune checkpoint is regarded as a target for immunotherapy, and the relationship between the immune checkpoint PD-1/PD-L1 and the prognosis of various cancers is currently a hot topic in immunotherapy. High expression of PD-1 is more reflective of T cell exhaustion than intermediate or low expression [46]. Some studies have found that PD-1 is expressed on Tregs, and blocking PD-1/PD-L1 signaling can increase Treg proliferation [47]. Furthermore, these results showed that the expression of COL1A1 was significantly correlated with the regulation of multiple markers of T helper cells (Tfh, Th1, Th2 and Th17) in LGG. In brief, these results reveal the potential of COL1A1 to recruit and activate immune cells in brain lower grade glioma.
In summary, the high expression of COL1A1 is associated with poor prognosis of various cancers and higher levels of immune infiltration, especially in LGG. COL1A1 is likely to be a significant regulator of the immune infiltration in LGG patients and serves as a prognostic factor for LGG. Our research suggests that COL1A1 is a promising target for LGG research. Therefore, we strongly recommend further research on the biological impact of COL1A1 in this field.
Acknowledgments
We would like to thank The Second Affiliated Hospital of Harbin Medical University for their strong support and the constructive comments of the team members.
Data Availability
All data can be obtained from the following sources: 1. TCGA, The LGG cohort (https://portal.gdc.cancer.gov/repository) 2. CGGA, The mRNAseq_693 cohort (http://www.cgga.org.cn/analyse/RNA-data.jsp) 3. GEO, The GSE2223 cohort (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2223) 4. TIMER, Diff Exp module (https://cistrome.shinyapps.io/timer) 5. UCSC Xena, the data sets module, and the expression, clinical, and survival data of 33 ‘GDC TCGA’ cohorts (https://xenabrowser.net).
Funding Statement
This work was funded by Natural Science Foundation of China 51972087(to NH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhang L, Liu Z, Li J, Huang T, Wang Y, Chang L, et al. Genomic analysis of primary and recurrent gliomas reveals clinical outcome related molecular features. Sci Rep. 2019;9(1):16058. doi: 10.1038/s41598-019-52515-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu H, Wang Z, Li M, Zeng F, Wang K, Huang R, et al. Gene Expression and Methylation Analyses Suggest DCTD as a Prognostic Factor in Malignant Glioma. Sci Rep. 2017;7(1):11568. doi: 10.1038/s41598-017-11962-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathologica. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan K, Wang X, Zhang J, Ramos RI, Zhang H, Li C, et al. Hypomethylation of CNTFRα is associated with proliferation and poor prognosis in lower grade gliomas. Scientific reports. 2017;7(1):7079-. doi: 10.1038/s41598-017-07124-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haddad AF, Young JS, Oh JY, Okada H, Aghi MK. The immunology of low-grade gliomas. Neurosurg Focus, 2022,52(2):E2. doi: 10.3171/2021.11.FOCUS21587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J, Li H, Chen Z, Chen Z, Fan L, Feng S, et al.Identification of 3 subpopulations of tumor-infiltrating immune cells for malignant transformation of low-grade glioma. Cancer Cell Int, 2019,19:265. doi: 10.1186/s12935-019-0972-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsao MS, Le Teuff G, Shepherd FA, Landais C, Hainaut P, Filipits M, et al. PD-L1 protein expression assessed by immunohistochemistry is neither prognostic nor predictive of benefit from adjuvant chemotherapy in resected non-small cell lung cancer. Ann Oncol. 2017;28(4):882–9. doi: 10.1093/annonc/mdx003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nissen NI, Karsdal M, Willumsen N. Collagens and Cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J Exp Clin Cancer Res. 2019;38(1):115. doi: 10.1186/s13046-019-1110-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leme-Kraus AA, Aydin B, Vidal CMP, Phansalkar RM, Nam JW, McAlpine J, et al. Biostability of the Proanthocyanidins-Dentin Complex and Adhesion Studies. J Dent Res. 2017;96(4):406–12. doi: 10.1177/0022034516680586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panaroni C, Gioia R, Lupi A, Besio R, Goldstein SA, Kreider J, et al. In utero transplantation of adult bone marrow decreases perinatal lethality and rescues the bone phenotype in the knockin murine model for classical, dominant osteogenesis imperfecta. Blood. 2009;114(2):459–68. doi: 10.1182/blood-2008-12-195859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swinnen FKR, Coucke PJ, De Paepe AM, Symoens S, Malfait F, Gentile FV, et al. Osteogenesis Imperfecta: the audiological phenotype lacks correlation with the genotype. Orphanet J Rare Dis. 2011;6:88-. doi: 10.1186/1750-1172-6-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nature genetics. 2003;33(1):49–54. doi: 10.1038/ng1060 [DOI] [PubMed] [Google Scholar]
- 13.Mustafa DA, Sieuwerts AM, Zheng PP, Kros JM. Overexpression of Colligin 2 in Glioma Vasculature is Associated with Overexpression of Heat Shock Factor 2. Gene regulation and systems biology. 2010;4:103–7. doi: 10.4137/GRSB.S4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu YH, Chang TH, Huang YF, Huang HD, Chou CY. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene. 2014;33(26):3432–40. doi: 10.1038/onc.2013.307 [DOI] [PubMed] [Google Scholar]
- 15.Ibanez de Caceres I, Dulaimi E, Hoffman AM, Al-Saleem T, Uzzo RG, Cairns P. Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer research. 2006;66(10):5021–8. doi: 10.1158/0008-5472.CAN-05-3365 [DOI] [PubMed] [Google Scholar]
- 16.Li M, Wang J, Wang C, Xia L, Xu J, Xie X, et al. Microenvironment remodeled by tumor and stromal cells elevates fibroblast-derived COL1A1 and facilitates ovarian cancer metastasis. Exp Cell Res, 2020,394(1):112153. doi: 10.1016/j.yexcr.2020.112153 [DOI] [PubMed] [Google Scholar]
- 17.Ilhan-Mutlu A, Siehs C, Berghoff AS, Ricken G, Widhalm G, Wagner L, et al. Expression profiling of angiogenesis-related genes in brain metastases of lung cancer and melanoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(1):1173–82. doi: 10.1007/s13277-015-3790-7 [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Yu J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2018;96(1):19–25. doi: 10.1139/bcb-2016-0254 [DOI] [PubMed] [Google Scholar]
- 19.Sun C, Yuan Q, Wu D, Meng X, Wang B. Identification of core genes and outcome in gastric cancer using bioinformatics analysis. Oncotarget. 2017;8(41):70271–80. doi: 10.18632/oncotarget.20082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer research. 2017;77(21):e108–e10. doi: 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–8. doi: 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Therneau TM, Lumley T. Package ‘survival’. Survival analysis Published on CRAN. 2014;2:3. [Google Scholar]
- 23.Kassambara A, Kosinski M, Biecek P, Fabian S. survminer: Drawing Survival Curves using’ggplot2’. R package version 03. 2017;1.
- 24.Gordon M, Lumley T. forestplot: Advanced forest plot using “grid” graphics. R package version. 2017;1(2):70. [Google Scholar]
- 25.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43(7):e47–e. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Justus CR, Sanderlin EJ, Yang LV. Molecular connections between cancer cell metabolism and the tumor microenvironment. International journal of molecular sciences. 2015;16(5):11055–86. doi: 10.3390/ijms160511055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4(1):1–11. doi: 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickham H. ggplot2: elegant graphics for data analysis: springer; 2016. [Google Scholar]
- 29.Yang X, Lin Y, Xing Z, She D, Su Y, Cao D. Predicting 1p/19q codeletion status using diffusion-, susceptibility-, perfusion-weighted, and conventional MRI in IDH-mutant lower-grade gliomas. Acta Radiol, 2021,62(12):1657–1665. doi: 10.1177/0284185120973624 [DOI] [PubMed] [Google Scholar]
- 30.Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. The New England journal of medicine. 2017;377(25):2500–1. doi: 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer cell. 2012;21(3):309–22. doi: 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- 32.Ravelli A, Roviello G, Cretella D, Cavazzoni A, Biondi A, Cappelletti MR, et al. Tumor-infiltrating lymphocytes and breast cancer: Beyond the prognostic and predictive utility. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2017;39(4):1010428317695023. doi: 10.1177/1010428317695023 [DOI] [PubMed] [Google Scholar]
- 33.Azimi F, Scolyer RA, Rumcheva P, Moncrieff M, Murali R, McCarthy SW, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30(21):2678–83. doi: 10.1200/JCO.2011.37.8539 [DOI] [PubMed] [Google Scholar]
- 34.Chandramohan V, Bao X, Yu X, Parker S, McDowall C, Yu Y-R, et al. Improved efficacy against malignant brain tumors with EGFRwt/EGFRvIII targeting immunotoxin and checkpoint inhibitor combinations. J Immunother Cancer. 2019;7(1):142-. doi: 10.1186/s40425-019-0614-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esfahani K, Al-Aubodah T-A, Thebault P, Lapointe R, Hudson M, Johnson NA, et al. Targeting the mTOR pathway uncouples the efficacy and toxicity of PD-1 blockade in renal transplantation. Nat Commun. 2019;10(1):4712-. doi: 10.1038/s41467-019-12628-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villegas-Martínez I, de-Miguel-Elízaga I, Carrasco-Torres R, Marras C, Canteras-Jordana M, Yedra-Guzmán MJ, et al. The COL1A1 SP1 polymorphism is associated with lower bone mineral density in patients treated with valproic acid. Pharmacogenetics and genomics. 2016;26(3):126–32. doi: 10.1097/FPC.0000000000000199 [DOI] [PubMed] [Google Scholar]
- 37.Simon MP, Maire G, Pedeutour F. COL1A1 (collagen, type I, alpha 1). Atlas of Genetics and Cytogenetics in Oncology and Haematology. 2001(2). [Google Scholar]
- 38.Hayashi M, Nomoto S, Hishida M, Inokawa Y, Kanda M, Okamura Y, et al. Identification of the collagen type 1 α 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma. BMC cancer. 2014;14:108. doi: 10.1186/1471-2407-14-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Ding Y, Li A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J Surg Oncol. 2016;14(1):297. doi: 10.1186/s12957-016-1056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oleksiewicz U, Liloglou T, Tasopoulou KM, Daskoulidou N, Gosney JR, Field JK, et al. COL1A1, PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2017;143(7):1133–41. doi: 10.1007/s00432-017-2381-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun S, Wang Y, Wu Y, Gao Y, Li Q, Abdulrahman AA, et al. Identification of COL1A1 as an invasionrelated gene in malignant astrocytoma. Int J Oncol. 2018;53(6):2542–54. doi: 10.3892/ijo.2018.4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Fang C, Wang Y, Zhang J, Yu J, Zhang Y, et al. COL1A1: A potential therapeutic target for colorectal cancer expressing wild-type or mutant KRAS. Int J Oncol. 2018;53(5):1869–80. doi: 10.3892/ijo.2018.4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Wang Y, Zhang J, Zhong J, Yang R. COL1A1 promotes metastasis in colorectal cancer by regulating the WNT/PCP pathway. Mol Med Rep. 2018;17(4):5037–42. doi: 10.3892/mmr.2018.8533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan Y, Guo X, Yang Z, Chen S, Lei Y, Lin M, et al. AEG-1 activates Wnt/PCP signaling to promote metastasis in tongue squamous cell carcinoma. Oncotarget. 2016;7(2):2093–104. doi: 10.18632/oncotarget.6573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeke A, Misheva M, Reményi A, Bogoyevitch MA. JNK Signaling: Regulation and Functions Based on Complex Protein-Protein Partnerships. Microbiology and molecular biology reviews: MMBR. 2016;80(3):793–835. doi: 10.1128/MMBR.00043-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim HR, Ha S-J, Hong MH, Heo SJ, Koh YW, Choi EC, et al. PD-L1 expression on immune cells, but not on tumor cells, is a favorable prognostic factor for head and neck cancer patients. Scientific reports. 2016;6:36956-. doi: 10.1038/srep36956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Q, Munger ME, Highfill SL, Tolar J, Weigel BJ, Riddle M, et al. Program death-1 signaling and regulatory T cells collaborate to resist the function of adoptively transferred cytotoxic T lymphocytes in advanced acute myeloid leukemia. Blood. 2010;116(14):2484–93. doi: 10.1182/blood-2010-03-275446 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data can be obtained from the following sources: 1. TCGA, The LGG cohort (https://portal.gdc.cancer.gov/repository) 2. CGGA, The mRNAseq_693 cohort (http://www.cgga.org.cn/analyse/RNA-data.jsp) 3. GEO, The GSE2223 cohort (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2223) 4. TIMER, Diff Exp module (https://cistrome.shinyapps.io/timer) 5. UCSC Xena, the data sets module, and the expression, clinical, and survival data of 33 ‘GDC TCGA’ cohorts (https://xenabrowser.net).