Abstract
A novel system that leaks β-galactosidase (β-gal) without a requirement for secretion or export signals was developed in Lactococcus lactis by controlled expression of integrated phage holin and lysin cassettes. The late promoter of the lytic lactococcal bacteriophage φ31 is an 888-bp fragment (P15A10) encoding the transcriptional activator. When a high-copy-number P15A10::lacZ.st fusion was introduced into L. lactis strains C10, ML8, NCK203, and R1/r1t, high levels of the resultant β-gal activity were detected in the supernatant (approximately 85% of the total β-gal activity for C10, ML8, and NCK203 and 45% for R1/r1t). Studies showed that the phenotype resulted from expression of Tac31A from the P15A10 fragment, which activated a homologous late promoter in prophages harbored by the lactococcal strains. Despite the high levels of β-gal obtained in the supernatant, the growth of the strains was not significantly affected, nor was there any evidence of severe membrane damage as determined by using propidium iodide or transmission electron microscopy. Integration of the holin-lysin cassette of phage r1t, under the control of the phage φ31 late promoter, into the host genome of MG1363 yielded a similar “leaky” phenotype, indicating that holin and lysin might play a critical role in the release of β-gal into the medium. In addition to β-gal, tetanus toxin fragment C was successfully delivered into the growth medium by this system. Interestingly, the X-prolyl dipeptidyl aminopeptidase PepXP (a dimer with a molecular mass of 176 kDa) was not delivered at significant levels outside the cell. These findings point toward the development of bacterial strains able to efficiently release relevant proteins and enzymes outside the cell in the absence of known secretion and export signals.
Lactococcus lactis is best known for its role in mesophilic dairy fermentations, including those used in production of cheddar cheese, buttermilk, and sour cream. Its long history of safe use in the food industry and generally recognized as safe (GRAS) status provide new opportunities for using L. lactis in important roles in food biotechnology, particularly in the presentation of vaccines, antimicrobial agents, or intracellular peptidases involved in cheese ripening, outside of the cell. Three major mechanisms have been exploited in these studies. First, signal sequences of the lactococcal secreted protein Usp45 (43), the lactococcal proteinase (46), and the S-layer protein (encoded by slpA) (45) of Lactobacillus brevis have been employed to secrete heterologous proteins from L. lactis via the secretory pathway. ATP-binding cassette–transporter export systems have also been used to export heterologous bacteriocins from L. lactis (for a review, see reference 1). Lastly, induction of cell autolysis can result in efficient release of homologous and heterologous proteins from the cell. Interest in naturally occurring autolytic strains has focused on the release of intracellular enzymes, namely peptidases, into the cheese medium to enhance flavor development and accelerate cheese ripening. Strains of L. lactis differ widely in their ability to undergo autolysis (25). Recent studies have implicated both the major lactococcal autolysin AcmA (5) and “leaky” low-level expression of a prophage-encoded lysin (21, 27) as causes of cell autolysis.
Advances in the molecular techniques available for studying L. lactis have increased efforts to genetically engineer autolytic strains to control cell lysis. For instance, the lactococcal autolysin AcmA was cloned under the control of two regulated promoters, the chloride-inducible promoter (39) and the promoter-operator region of the temperate phage r1t (5, 32), generating strains which, upon induction of the promoter, lyse to release intracellular enzymes into the supernatant. Another major advance in this area has been the cloning and characterization of several lactococcal bacteriophage lysins and holins (reviewed in reference 15). In contrast to the lactococcal autolysin AcmA, the bacteriophage lysins do not possess a signal sequence (4). Externalization occurs via a small, transmembrane holin which oligomerizes to form nonspecific pores in the host membrane (for reviews, see references 3, 15, 52, and 53). The genes encoding several bacteriophage lysins and/or holins have been exploited to design strains which lyse under controlled conditions. For example, the holin and lysin genes of phages r1t and US3 were placed under the control of PnisA (6) and the chloride-inducible promoter (39), generating strains which released intracellular peptidases into the medium upon induction of the promoter. Overproduction of only the holin from phage φUS3, via PnisA, resulted in an immediate inhibition of cell growth and partial lysis of the host cell (6).
The use of bacteriophage lytic cassettes is an exciting advance in the continued development of expression systems allowing the delivery of proteins and enzymes to the outside environment. However, these systems often cause rapid cessation of growth and, eventually, cell lysis. Moreover, since lysin works extracellularly, the growth of other lactococcal strains used in combination would also be affected. In this study, we describe a novel expression system which allows the release of significant levels of a Streptococcus thermophilus β-galactosidase (β-gal) (40) in L. lactis without the use of signal sequences and without causing severe losses of cell viability or integrity. The system utilizes the phage transcriptional activator Tac31A (formerly open reading frame 2 [ORF2] and tac [36, 47]) to activate a late promoter residing on a prophage integrated in the genome of L. lactis. Low-level activation of the late promoter, driving downstream expression of holin and lysin, resulted in a leaky behavior that efficiently externalized an enzyme and a vaccine, with minimal losses of other cytoplasmic enzymes.
MATERIALS AND METHODS
Strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. L. lactis strains were propagated at 30°C in M17 medium (Difco) (42) supplemented with 0.5% glucose (GM17). Where necessary, erythromycin, tetracycline, and/or chloramphenicol was added at 5, 2, and 7.5 μg/ml, respectively. Escherichia coli strains were grown in Luria-Bertani broth at 37°C with shaking or on Luria-Bertani medium supplemented with 1.5% agar. Erythromycin resistance in E. coli was selected by plating on brain heart infusion agar (Difco) supplemented with 120 μg of erythromycin/ml (34).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, phage, plasmid, or promoter fragment | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM110 | E. coli cloning host | 51 |
| XL1-Blue | E. coli cloning host | Gibco-BRL |
| L. lactis | ||
| NCK203 | Subsp. lactis; KP1 derivative | 18 |
| ML8 | Subsp. lactis | U. of Lavalb |
| C10 | Subsp. lactis | 12 |
| R1/r1t | Subsp. cremoris; carries r1t prophage | 22 |
| R1C | Subsp. cremoris; r1t-cured derivative | 22 |
| MG1363 | Subsp. cremoris | 14 |
| MM210 | Subsp. cremoris; industrial strain | NCSUc |
| Bacteriophage φ31.9 | Phage φ31 derivative with F142L mutation in Tac31A | 10 |
| Plasmids | ||
| pTRKH2 | Emr; high-copy-number cloning vector | 34 |
| pNZ18 | Cmr; high-copy-number shuttle vector | 7 |
| pGhost8 | Tetr; lactococcal integration vector | INRAd |
| pTRK390 | Emr; lactococcal promoter screening vector | 36 |
| pTRK391 | Emr; pTRK390 (P15A10::lacZ.st) | 36 |
| pTRK406 | Cmr; pNZ18::abiA, AbiA+ | 8 |
| pTRK483 | Emr; pTRK390 (P566–888::lacZ.st) | 47 |
| pTRK609 | Emr; pTRK390 (P6::lacZ.st) | This study |
| pTRK610 | Emr; pTRK390 (P15A10φ31.9::lacZ.st) | This study |
| pTRK611 | Emr; pTRK390 (P15A102X::lacZ.st) | This study |
| pTRK617 | Emr; P15A102X::TTFC on pTRKH2 | This study |
| pTRK618 | Emr; P6::TTFC on pTRKH2 | This study |
| pTRK619 | Emr; P15A102x::pepXP on pTRKH2 | This study |
| Promoter fragments | ||
| P6 promoter | Strong, constitutive Lactobacillus promoter | 9 |
| P15A10 | 888-bp φ31 late promoter encoding tac31A and two phage-inducible transcription start sites | 36 |
| P15A10φ31.9 | P15A10 derivative encoding mutant tac31A (F142L mutation), resulting in a 50% reduction in promoter activity | 10, this study |
| P15A102x | P15A10 derivative with mutation in inverted repeat downstream of transcription start sites, resulting in a 20 to 3-fold increase in promoter activity | 47, this study |
| P566–888 | Tightly regulated version of φ31 late promoter which does not encode tac31A | 47 |
Abbreviations: Emr, erythromycin resistance; Cmr, chloramphenicol resistance; Tetr, tetracycline resistance.
University of Laval, Laval, Quebec, Canada.
NCK culture collection at North Carolina State University, Raleigh.
Institut National de la Recherche Agronomique, Paris, France.
Bacteriophage propagation and phage DNA isolation.
When necessary, the resident prophages of the lactococcal strains used in this study were induced by using mitomycin C at a level of 10 μg/ml (for strains NCK203, C10, and ML8) or 2 μg/ml (for R1/r1t). Prophage DNA was isolated and purified as described by Raya et al. (37) as modified by Walker et al. (48). In some cases, L. lactis genomic DNA preparations were prepared 1 h after mitomycin C induction, using the procedure of Hill et al. (17). To determine the number of r1t phages present in the culture supernatant, culture samples were filtered (0.45 μm pore size) and serial dilutions were spotted onto a lawn of R1Cs cells (prophage cured). CaCl2 (10 mM) was added to the medium, and soft agar was used to prepare the lawn of the sensitive host.
DNA isolation.
Small-scale E. coli plasmid preparations were performed by the alkaline-sodium dodecyl sulfate method (38). Large-scale E. coli plasmid preparations were performed using a Qiagen plasmid kit (Qiagen Inc., Chatsworth, Calif.) according to manufacturer's directions. Small-scale isolation of plasmids from L. lactis was performed as previously described (35), except that ethidium bromide was not used prior to phenol-chloroform extraction.
DNA manipulations and transformations.
Standard procedures were used for the DNA manipulations (38). Restriction enzymes and T4 DNA ligase were provided by Boehringer Mannheim Biochemicals (Indianapolis, Ind.) and used according to the manufacturer's instructions. Southern hybridizations were performed at 65°C in a Robbins (Robbins Scientific, Inc., Sunnyvale, Calif.) hybridization oven per the manufacturer's instructions. DNA probes were 32P labeled by using the Multiprime DNA labeling system (Amersham, Piscataway, N.J.). The r1t attP fragment used to probe genomic DNA preparations for the induction of prophages contained the 3′ portion of integrase, attP, ORF50, and the 3′ portion of lysin (nucleotides 31728 to 185 of the published sequence [44]). The fragment was amplified from R1/r1t genomic DNA by PCR using the forward primer 5′-GGCTATCACACAGCAAACCTATATC-3′ and the reverse primer 5′-CGTTCCTACTCGGCACAGGTCAAG-3′. Ligations were transformed into RbCl-competent E. coli strains. RbCl-competent E. coli cells were prepared by the procedure of Hanahan (16). Cell preparations were frozen at −70°C in 100-μl aliquots and transformed by a procedure described for CaCl2-competent cells (38). After being screened to verify their proper insertion in E. coli, plasmids were electroporated into L. lactis by the procedure of Holo and Nes (19), modified as described by Walker and Klaenhammer (47). Electroporations were carried out in a 0.2-cm-path-length cuvette with a Bio-Rad (Richmond, Calif.) Gene Pulser, using 100 μl of cell preparation and the following conditions: 25 μF, 2.45 kV, and 200 Ω. Cell recovery was achieved by incubation in GM17 supplemented with 10 mM MgCl2 and 1 mM CaCl2 for 2 h at 30°C prior to plating on media with appropriate antibiotics.
PCR.
PCR was performed with Taq DNA polymerase (Boehringer Mannheim) according to the manufacturer's instructions. In each case, 40 cycles were used to amplify the regions of interest. Annealing temperatures were 5 to 10°C below the lowest melting temperature of each primer pair. To facilitate cloning of PCR products, restriction enzyme sites (indicated throughout the manuscript by underlining) were designed for the 5′ ends of the primers.
RNA manipulations.
RNA was isolated from L. lactis strains by using TRIzol reagent (Gibco-BRL, Gaithersburg, Md.) according to the procedure of Dinsmore and Klaenhammer (8). Slot blot Northern hybridizations using equivalent amounts of RNA from each sample (approximately 10 μg) were performed on a Bio-Rad slot blot apparatus (Bio-Rad, Richmond) according to the manufacturer's protocol. The r1t lysin probe used to measure the induction of lysin mRNA was amplified from a genomic DNA preparation of R1/r1t, using the forward primer 5′-CGTTCCTACTCGGCACAGGTCAAG-3′ and the reverse primer 5′-CCAAACTCTTTATCGACTTC-3′, and consisted of nucleotides 32065 to 32815 of the published r1t sequence (44). The r1t holin probe used to measure the induction of holin mRNA was amplified using the forward primer 5′-GCACAAGCAATGATTGGCGCTTTGG-3′ and the reverse primer 5′-TTGACTAGGCTTGCTGTATTATCG-3′ and consisted of nucleotides 31866 to 32023 of the published r1t sequence (44). The DNA probe used to measure induction of mRNA from the late region of the uncharacterized prophage of L. lactis strains C10, ML8, and NCK203 was designed using sequence data from the late region of the lytic phage φ31 and contained the 3′ portion of ORF3, cos, ORF4, and the 5′ portion of ORF5 (48). The fragment was amplified from phage φ31 genomic DNA by using the forward primer 5′-CGTGATTGGTCTTCTTATG-3′ and the reverse primer 5′-AGAAATGAGCTTCAAGAACAA-3′.
Enzyme assays.
β-gal determinations were performed using the o-nitrophenyl-β-d galactopyranoside (ONPG) assay described by Miller (30), as modified by O'Sullivan et al. (36). To determine the level of β-gal in the supernatant of samples, 100-μl samples of the whole culture (cells plus growth medium) or just the filter-sterilized supernatant were tested. Both reactions (whole culture and supernatant) were stopped upon development of a yellow color in the reaction containing the whole culture. Absorbances at 420 nm were read after centrifugation to remove any cell debris. Percentages were determined with the formula (A420 supernatant/A420 whole culture) × 100.
Intracellular X-prolyl dipeptidyl aminopeptidase (PepXP) activity present in the whole culture versus that in the supernatant was measured in 100-μl samples of the whole culture or filter-sterilized supernatant as described by Nivens and Mulholland (33). Whole cultures were permeabilized by using chloroform. Both reactions were stopped by addition of 30% acetic acid when color development was detected in the whole-culture sample. Absorbances were read at 410 nm, and percentages were determined as described above.
Determination of the proportion of cells with damaged membranes.
The integrity of Lactococcus cell membranes was measured by a fluorescence procedure described by Niven and Mulholland (33). Basically, L. lactis strains C10, ML8, R1/r1t, and NCK203, with and without P15A10 (containing tac31A), were propagated to an initial optical density at 600 nm (OD600) of 0.65 and centrifuged to remove the medium. Cell pellets were resuspended in phosphate-buffered saline (PBS) to an OD600 of 0.9. Viable-cell counts were performed. The samples were divided into four tubes, each of which received one of the following treatments: no treatment, treatment with propidium iodide (PI) alone (30 μmol/liter), treatment with the permeabilizing agent cetyltrimethylammonium bromide (CTAB) alone (0.2 mmol/liter), or treatment with a combination of PI and CTAB. After a 30-min incubation at room temperature, fluorescence was measured using an excitation wavelength of 500 nm and an emission wavelength of 600 nm. Readings were also taken for PBS containing PI, CTAB, or PI plus CTAB. The ratio of cells with damaged membranes was determined with the formula (cellsPI − cellsalone − PBSPI)/(cellsPI+CTAB − cellsalone − PBSPI+CTAB).
Electron microscopy.
The cells were prepared for transmission electron microscopy as described by Dykstra (11).
Integration of the r1t holin-lysin cassette in MG1363.
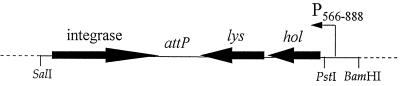
An integration cassette containing the r1t holin and lysin genes under the control of the tightly regulated phage φ31 late promoter P566–888 was cloned into pGhost8, a temperature-sensitive vector (29). PCR primers (P1 [5′-GATCGTCGACTGTCTGACGGCTGGGTAATGT-3′] and P2 [5′-GTTTCTGCAGTCGGTTCAGCCAGTGATTGTTC-3′]) were used to amplify a single fragment (SalI-PstI) (see Fig. 1) containing the integrase, the attP region, and the holin-lysin cassette from a genomic DNA preparation of R1/r1t (nucleotides 31738 to 1391 of the published r1t sequence [44]). This fragment also contained the putative ORF50 coding region, which is located just downstream of the lysin gene (44). PCR primers P3 (5′-ATAGGATCCGTGTCACATAACTGAGCGCC-3′) and P4 (5′-GATGCTGCAGTATTGGCTTGCCACATATTC-3′) were used to amplify P566–888 (BamHI-PstI) (Fig. 1) (47). The two PCR products were restricted, gel purified, and ligated into pGhost8 which had been restricted with BamHI and XhoI. Ligation reaction products were transformed directly into MG1363 competent cells. After outgrowth at 30°C, the transformants were selected on GM17 with tetracycline (2 μg/ml) at the nonpermissive temperature of 37°C.
FIG. 1.
Organization of construct used for integration of the P566–888::holin-lysin cassette into L. lactis MG1363.
Construction of P15A102X::TTFC and P15A102X::PepXP fusions.
The coding region for tetanus toxin fragment C (TTFC) was amplified from pLET1-TTFC (50) using the forward primer 5′-GATCCTGCAGTTGTTTAACTTTAAGAAGGAG-3′ (PstI site underlined) and the reverse primer 5′-ATATCTCGAGTAGTTCCTCCTTTCAGCA-3′ (XhoI site underlined). The fragment, which contained the translation initiation region utilized on pLET1-TTFC, was cloned into pTRKH2 which had been restricted with PstI and XhoI. To achieve adequate expression levels, a stronger, mutated version of the phage φ31 late promoter, P15A102X, containing tac31A and the phage-inducible transcription start sites (36, 47) was cloned upstream of the TTFC coding region to yield pTRK617. In the mutated P15A102x, a small inverted repeat downstream of the transcription start sites was eliminated by site-directed mutagenesis, resulting in a two- to threefold increase in expression levels (47). The forward primer 5′-ATATGGATCCGCAGAGCATTTGTAAGGTTGG-3′ (BamHI site underlined) and the reverse primer 5′-GATGCTGCAGGATTGGCTTGCCACATATTC-3′ (PstI site underlined) were used to amplify the 888-bp P15A102X, which was cloned into the BamHI-PstI sites upstream of the TTFC coding region. Since unintentional promoter activity from the vector allowed low-level expression of the transcriptional activator (unpublished data), this cloning strategy, and all others to follow, retained the phage promoter in the same orientation as that in pTRK391. As another control, the strong Lactobacillus P6 promoter (9) was cloned upstream of the TTFC coding region to yield pTRK618.
The PepXP coding region was amplified with its own translation initiation region from L. lactis, using forward primer 5′-TAAGCTAAAAGTATTATCATGTTTATTACGGAGG-3′ and reverse primer 5′-GTCGAGCAACTGTGTTGTAAGGAG-3′ (SalI site underlined). Primers were designed using sequence information from Nardi et al. (31). The pepXP fragment was initially cloned into pT7Blue (Novagen, Madison, Wis.) to introduce a BamHI site at the 5′ end of the coding region. The pepXP fragment was removed by using BamHI and SalI and cloned into pTRKH2 restricted with BamHI and SalI. P15A102x was cloned upstream as a BamHI fragment.
Western blotting to detect TTFC.
Cell extracts were prepared from strains propagated to mid-log phase (OD600 = 0.5) in GM17 supplemented with erythromycin. The cells were centrifuged, washed in 100 mM Tris-HCl (pH 8.0), and resuspended at a 40× concentration in 100 mM Tris-HCl. The cells were broken by bead beating two times for 1 min each, with a 30-s intermission, on ice. The cell extract was collected by removing the supernatant after centrifugation at 4°C. Concentrated (50-fold) supernatant samples were prepared by dialyzing filter-sterilized supernatants of the cultures against water (overnight), freeze drying, and resuspending in 100 mM Tris-HCl. Dialysis was necessary to remove concentrated solutes which interfered with electrophoresis. Denatured cell extracts and concentrated supernatants were electrophoresed on a 10% polyacrylamide gel, using a Bio-Rad mini-PROTEAN II electrophoresis unit, by standard procedures (38). The gels were transferred overnight to polyvinylidene difluoride membranes (0.2 μm pore size) by using a Bio-Rad Mini Trans-blot cell at 25 V. TTFC was detected with a rabbit anti-TTFC antibody (1:1,000 dilution; Calbiochem, La Jolla, Calif.) followed by an alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (Boehringer Mannheim). Color development was accomplished by using the colorimetric substrates 4-nitroblue tetrazolium chloride (Boehringer Mannheim) and 5-bromo-4-chloro-3-indolylphosphate (Boehringer Mannheim) according to the manufacturer's instructions.
RESULTS
Leakage of β-gal into Lactococcus culture supernatants.
The 888-bp late-promoter fragment, P15A10, from the lytic bacteriophage φ31 (36) encodes its own transcriptional activator, Tac31A (formerly ORF2 and tac [47]). It was observed that when L. lactis NCK203 carried a P15A10::lacZ.st fusion (pTRK391), β-gal activity was detected largely in the culture supernatant and not in the cells. A total of five L. lactis strains were electroporated with pTRK391(P15A10::lacZ.st), and the level of β-gal activity in each supernatant was measured (Table 2). Three of the strains (C10, ML8, and NCK203) contained prophages harboring a late-promoter region homologous to P15A10 in φ31 (48), while two strains (MM210 and MG1363) did not. In the three prophage-bearing strains C10, ML8, and NCK203, 84 to 88% of the β-gal activity was detected in the culture supernatant. Less than 10% of the β-gal activity was detected in the supernatants of L. lactis strains that did not harbor prophages. Introduction of pTRK609(p6::lacZ.st), in which β-gal expression is driven by the strong, constitutive P6 promoter (9), into strains C10, ML8, and NCK203 failed to yield comparable levels of β-gal in the supernatants. The results showed that significant levels of β-gal activity were located in the supernatants of the lysogenic strains when the promoter P15A10 was used to drive lacZ expression from the high-copy-number replicon.
TABLE 2.
Levels of β-gal detected in supernatants of Tac31A+ (encoded by P15A10 or P15A10φ31.9) and Tac31A− cultures
| Strain | Prophage with homologous Pφ31a | % of β-gal detected for construct:
|
||
|---|---|---|---|---|
| P6::lacZ.st | P15A10::lacZ.st | P15A10φ31.9::lacZ.st | ||
| MM210 | No | —b | <10 | — |
| MG1363 | No | — | <10 | — |
| NCK203 | Yes | 10 | 88.3 | — |
| C10 | Yes | 4.3 | 84.0 | — |
| ML8 | Yes | 6.1 | 88.0 | — |
| R1/r1t | Yes | <10 | Unstable | 45.3 |
| R1C | No | — | — | <10 |
Pφ31 refers to the phage φ31 late promoter, designated P15A10 in this paper.
—, not tested.
These results indicated that the leaky behavior resulted from expression of Tac31A, in trans, in combination with prophages encoding a homologous Pφ31 promoter. Prophage-cured derivatives of C10, ML8, and NCK203 were not available for use as negative controls to confirm this result. However, phage r1t, of L. lactis R1, also harbors a Pφ31-homologous promoter (48), and a prophage-cured derivative, designated R1C, is available (22). Therefore, L. lactis R1/r1t and R1C (prophage cured) were transformed with plasmid constructs encoding P15A10::lacZ.st or P6::lacZ.st. Several transformants of each were propagated in GM17 plus erythromycin overnight, and the percentage of β-gal activity in each of the supernatants was determined. Very little β-gal was detected in the supernatants when P6::lacZ.st was present in R1/r1t or R1C (Table 2) or when P15A10::lacZ.st was present in prophage-cured R1C. When P15A10::lacZ.st was present in R1/r1t, various levels of β-gal were detected in the supernatants of the transformants, ranging from very little (no more than that obtained with P6::lacZ.st) to about 40 to 50% of the total β-gal activity. After successive passage of the R1/r1t(P15A10::lacZ.st) transformants, none exhibited the leaky β-gal phenotype. Further investigation revealed a loss of the r1t prophage from the transformants. It appeared that the presence of tac31A on P15A10 was sufficiently lethal to select for r1t-cured derivatives in the population, explaining the variability and instability of the leaky behavior in this phage r1t lysogen background. This instability was resolved by reconstructing the lacZ.st fusion with the promoter P15A10φ31.9, which encodes a mutated version of tac31A and yields a 50% reduction in promoter activity compared to P15A10 (10). P15A10φ31.9 was directionally cloned into BamHI- and PstI-restricted pTRK390 upstream of lacZ.st to generate pTRK610 and transformed into R1/r1t and R1C. Table 2 shows that β-gal levels were low (10% or less) in the supernatants of R1C transformants, whereas an average of 45% of the total β-gal activity was detected in the supernatants of the R1/r1t transformants. This leaky phenotype was evident after multiple passages. These results clearly indicated that the presence of the r1t prophage was responsible for the increased levels of β-gal detected in the supernatants when Tac31A was expressed in trans.
Leaky behavior is independent of prophage induction.
It was important to determine whether Tac31A induced replication of a prophage or simply activated expression over a late region. This question could not be answered by using L. lactis strains NCK203, ML8, and C10, since no sensitive lactococcal hosts were available to determine the titers of phages that might appear from Tac31A+ derivatives of these cultures. Two alternative approaches were therefore used to address this question. The first approach utilized the AbiA abortive phage defense mechanism, which targets phage replication (17, 18) and can reduce the burst size of phage r1t 10-fold after induction with mitomycin C (unpublished observations). Since C10, ML8, and NCK203 contain prophages with homology to r1t, there existed the possibility that AbiA would severely inhibit their replication as well. To test this, pTRK483 (P566–888::lacZ.st) was combined with pTRK406 (pNZ18::abiA [8]), or the control plasmid pNZ18, in L. lactis NCK203. The tightly regulated P566–888 does not encode Tac31A (47); therefore, no β-gal will be detected from the P566–888::lacZ.st fusion unless Tac31A is provided in trans via induction of the resident prophage with mitomycin C (47, 48). When pTRK483 (P566–888::lacZ.st) was combined with pNZ18, addition of mitomycin C to the lysogen led to efficient induction of β-gal from the phage promoter, as described previously by Walker et al. (48). However, when AbiA was introduced on pTRK406, β-gal expression was eliminated after mitomycin C induction, indicating that replication of the prophage was inhibited (data not shown). The final experiment was conducted by combining pTRK406 (AbiA+) with pTRK391 (P15A10::lacZ.st) in L. lactis NCK203. Efficiency of plaquing (EOP) assays using phage φ31 confirmed that AbiA was functional in the transformants. The levels of β-gal in the supernatants of chosen colonies were comparable to those obtained with P15A10::lacZ.st alone (Table 2) or with P15A10::lacZ.st plus pNZ18, demonstrating that prophage induction and replication were not responsible for the leaky behavior.
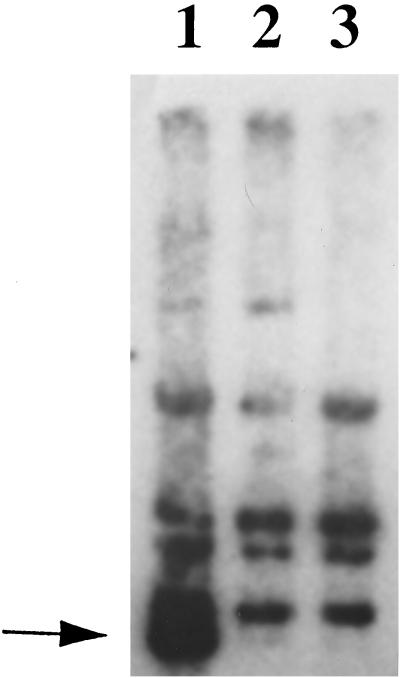
A second approach to evaluate whether prophage induction was occurring used Southern hybridization to assess any changes in the conformation of the attP site in leaky cells. PvuII-digested genomic DNAs isolated from L. lactis NCK203, NCK203 treated with mitomycin C (positive control for prophage induction), and NCK203(pTRK391; P15A10::lacZ.st) were electrophoresed on an agarose gel and transferred to a Magnacharge nylon membrane (38). The membrane was probed with a 32P-labeled fragment containing the r1t attP region, since this region is conserved among several other temperate members of the P335 lactococcal phage species (e.g., TUC2009 and LC3). Figure 2 shows that hybridization of the attP region to restriction digests of the NCK203 genomic DNA, after induction of the resident prophage, yielded an extra band representing the annealing of attP after prophage excision from the host chromosome. This extra band was not detected when P15A10 was present (Fig. 2, lane 2). These results, confirmed by using ML8 and C10 (data not shown), provided further evidence that the resident prophages of NCK203, C10, and ML8 were not induced to detectable levels by Tac31A. The results are speculative, however, since linkage of the homologous r1t attP region in NCK203 to the resident prophage encoding the homologous P15A10 region has not been established.
FIG. 2.
Southern hybridization of genomic DNA restricted with PvuII and probed with a 32P-labeled fragment containing the r1t attP region. Lane 1, NCK203 genomic DNA isolated after induction with mitomycin C; lane 2, NCK203(P15A10::lacZ.st) genomic DNA; lane 3, NCK203 (control) genomic DNA. The arrow indicates the extra band appearing after excision of the prophage from the host genome.
Tac31A causes a slight activation of the late region of the resident prophages.
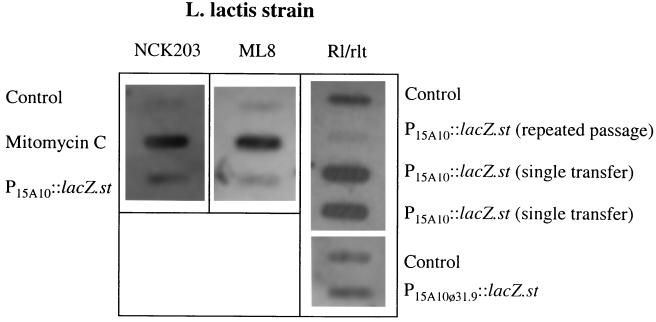
The results suggested that the leaky phenotype was directed by Tac31A activation of the late region of resident prophages, which typically include holin-lysin cassettes. The genes encoding the holin and lysin of the C10, ML8, and NCK203 resident prophages are uncharacterized and different from r1t (see above). Therefore, to assess holin-lysin expression, a fragment consisting of the late region of phage φ31 (48) just downstream of the late promoter and the right cos site was used to probe RNAs isolated from mitomycin C-treated NCK203, NCK203(P6::lacZ.st), and NCK203(P15A10::lacZ.st). RNAs isolated from mitomycin C-treated ML8, ML8(P6::lacZ.st), and ML8(P15A10::lacZ.st) were probed in the same manner. The results (Fig. 3) showed that the presence of P15A10 encoding Tac31A resulted in approximately a twofold increase in the level of RNA from the late region over that of the controls. This level of expression was significantly lower than that obtained after induction of the resident prophage with mitomycin C. The same results were obtained with RNA slot blots of C10 RNA (data not shown).
FIG. 3.
RNA slot blots measuring induction of the late regions of strains NCK203, ML8, and R1/r1t, with and without Tac31A (encoded on P15A10::lacZ.st) or mutant Tac31A (encoded on P15A10φ31.9::lacZ.st). RNA isolated from strains NCK203 and ML8 was probed with the beginning of the late region of phage φ31 (see Materials and Methods). RNA isolated from strain R1/r1t was probed with both an r1t lysin probe and an r1t holin probe. The results were identical, and thus only data for the r1t lysin probe are shown.
This experiment was repeated for the R1/r1t and R1C transformants. The RNA was probed with either a 32P-labeled r1t lysin probe or a 32P-labeled r1t holin probe, and identical results were achieved (Fig. 3). RNA isolated from R1/r1t(P15A10::lacZ.st) transformants propagated once showed a marked induction of both holin and lysin compared to RNA from R1/r1t. If R1/r1t(P15A10::lacZ.st) transformants were subjected to repeated passages, the levels of lysin and holin mRNAs began to decrease until very little could be detected, indicating that the r1t prophage was being cured from the population. As expected, lysin and holin mRNAs were not detected in any R1C transformants (data not shown). Therefore, the presence of low levels of Tac31A in R1/r1t resulted in significant expression of holin and lysin mRNAs, much higher levels than the expression of the late region achieved with the C10, ML8, and NCK203 prophages (see above). This high level of expression may explain why the leaky phenotype is so unstable in R1/r1t. RNA isolated from the R1/r1t(P15A10φ31.9::lacZ.st) transformants and probed with a 32P-labeled r1t lysin fragment (Fig. 3) showed little or no induction over that seen with the control R1 cells, which is more consistent with results obtained with the C10, ML8, and NCK203 prophages.
Effect of leaky expression on the growth, viability, and appearance of L. lactis.
Growth curves were constructed for L. lactis NCK203 and R1 parents and their counterparts harboring plasmids encoding P6::lacZ.st or P15A10::lacZ.st. The presence of Tac31A on P15A10 had no significant effect on the growth of these strains, as measured by changes in both OD600 and CFU per milliliter (Fig. 4; data not shown for CFU per milliliter or R1). The fluorescent dye PI was used to determine the proportion of dead or membrane-compromised cells in Tac31A+ populations (33). In this experiment, the cells were washed before being tested, thereby eliminating any contribution made by lysed cells. With L. lactis C10 and NCK203, the presence of Tac31A on P15A10 did not significantly increase the proportion of dead or compromised cells in the population. In all cases, the percentage of dead or damaged cells was less than 10% of the total (see Table 3 for NCK203 data). The same was true for R1/r1t harboring the P15A10φ31.9::lacZ.st construct (Table 3). These results were confirmed by viable-cell counts. Similar CFU-per-milliliter levels were obtained at the same OD regardless of whether P15A10 was present (data not shown). In addition, transmission electron microscopy showed that the presence of Tac31A did not significantly alter the appearance of NCK203(pTRK391) compared to the control strain NCK203(pTRKH2) (data not shown). No ghost cells or cellular debris were observed, providing further evidence that cell lysis was not occurring.
FIG. 4.
Growth curve for L. lactis NCK203 (circles), compared to its derivatives harboring pTRK609 (P6::lacZ.st) (triangles) and pTRK391 (P15A10::lacZ.st) (diamonds).
TABLE 3.
Direct comparison of levels of β-gal and PepXP detected in supernatants and proportion of membrane-damaged cells in strains carrying various constructs
| Strain | Construct carried | % of enzyme in supernatant
|
% of cells damageda | |
|---|---|---|---|---|
| β-gal | PepXP | |||
| NCK203 | P6::lacZ.st | 10 | 5.9 | 5.5 |
| P15A10::lacZ.st | 88.3 | 9.2 | 4.8 | |
| P15A102X::TTFC | —b | — | — | |
| P15A102X::PepXP | — | 6.4 | — | |
| R1/r1t | P6::lacZ.st | <10 | 10.5 | — |
| P15A10φ31.9::lacZ.st | 45.3 | 5.0 | 4.3 | |
| R1C | P15A10φ31.9::lacZ.st | <10 | — | 3.0 |
| MG1363/hol-lys | None | — | — | 8.1 |
| P6::lacZ.st | 6.0 | — | 4.3 | |
| P15A10::lacZ.st | 66 | 40–90c | 40.0 | |
| P15A10φ31.9::lacZ.st | 46 | 5.0 | 15.0 | |
Determined with PI.
—, not tested.
Due to the large deviation in these readings, this value is given as a range rather than an average.
Integration of the r1t holin-lysin cassette in MG1363 results in leaky expression of β-gal.
To establish that expression of holin and lysin was responsible for the leaky phenotype, the r1t holin-lysin cassette was integrated into the prophage-free strain MG1363 by using the r1t integrase and attP region. One stable integrant was obtained, and PCR analysis (data not shown) confirmed that the holin and lysin genes were in the proper orientation with respect to the phage φ31 late promoter (illustrated in Fig. 1). The integrant was then transformed with the P15A10::lacZ.st, P15A10φ31.9::lacZ.st, and P6::lacZ.st constructs independently, and the resulting derivatives were compared. Both β-gal and PepXP levels were measured in the whole cultures and in the supernatants (Table 3). Although the presence of wild-type Tac31A in MG1363/hol-lys resulted in 66% of the β-gal activity being found in the supernatant, the phenotype was very different from that obtained with C10, ML8, and NCK203 and was more similar to results obtained with R1/r1t, in which it was lethal. Analysis of cell damage with PI showed that 40% of the MG1363/hol-lys(P15A10::lacZ.st) cells took up the dye compared to the control. In addition, these cells grew extremely slowly and exhibited considerable decreases in OD600 after overnight storage at 4°C (data not shown). When the mutant Tac31A was present on P15A10φ31.9, an average of 46% of the β-gal activity was detected in the supernatant and only a slight increase in the percentage of damaged cells could be detected with the PI test. These results strongly suggest that activation of the integrated cassette including the holin and lysin genes by Tac31A was largely responsible for the leaky phenotype. Moreover, relatively undamaged (as determined via PI analysis) and highly leaky cells did not release comparable amounts of an intracellular peptidase (PepXP) into the supernatant, providing further evidence for an externalization process independent of cell lysis.
Leaky expression of enzymes and other proteins.
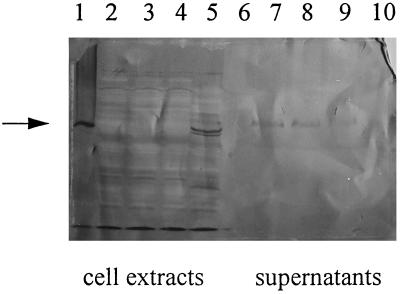
It was of considerable interest to determine if other heterologous proteins or enzymes could leak into the culture supernatant by this mechanism. The first protein evaluated was TTFC, a 47-kDa model antigen in vaccine delivery systems designed for L. lactis (49, 50). P15A102X::TTFC constructs were transformed separately into NCK203, C10, and MG1363 (prophage-cured control). Western blot analysis was used to detect TTFC in the supernatants and cell extracts of each strain (Fig. 5). High levels of TTFC were detected in the cell extracts of MG1363(P15A102x::TTFC), whereas no TTFC was detected in the supernatant. In contrast, in the leaky constructs of NCK203 and C10 (harboring P15A102x::TTFC), TTFC was detected mostly in the supernatants, and little was found associated with the cell extracts.
FIG. 5.
Western blot of the cell extracts and supernatants of lactococcal strains carrying P15A102X::TTFC constructs. The arrow marks the position of the TTFC standard. Cell extracts (lanes 2 to 5) and supernatants (lanes 7 to 10) of lactococcal strains carrying P15A102x::TTFC constructs were examined for expression of TTFC. Lane 1, TTFC standard; lanes 2 and 7, C10; lanes 3 and 8, NCK203a; lanes 4 and 9, NCK203b; lanes 5 and 10, MG1363 (prophage-negative control); lane 6, Rainbow high-molecular-weight markers (Amersham). NCK203a and NCK203b are two clones carrying the same construct.
Since the leaky behavior described herein could be useful for the externalization of flavor enzymes from Lactococcus cultures during cheese manufacture, secretion of the intracellular enzyme PepXP was also studied. The proportions of PepXP activity present in the supernatants of log-phase cultures of NCK203, with and without Tac31A, and R1/r1t, with and without the mutant Tac31A, were measured (Table 3). Interestingly, while PepXP activity was detected in the whole-culture samples, it was not detected at significant levels (<10%) in the supernatants of NCK203 or R1/r1t, even when Tac31A or its mutant was present. Attempts were made to increase the background cytoplasmic levels of PepXP levels by creating a P15A102x::pepXP fusion in pTRKH2 to yield pTRK619. This fusion construct was transformed into NCK203, and levels of PepXP activity in the supernatant were measured. Again, significant levels of PepXP activity were not detected in the supernatant (Table 3). Therefore, Tac31A does not promote leakage of PepXP to the supernatant. Moreover, these data provide additional evidence that leaky L. lactis cells are not lysing to release β-gal or TTFC into the supernatant.
DISCUSSION
This paper describes a novel expression system that allows release of certain proteins and enzymes into the growth medium without the use of export or secretion pathways and without significant effects on cell viability or cell membrane integrity. The leaky behavior, first observed for a heterologous β-gal enzyme expressed from the P15A10 phage φ31 late promoter (P15A10::lacZ.st), depends on two features: the Tac31A transcriptional activator of the phage φ31 promoter, and a resident prophage containing a promoter homologous to P15A10 which directs low-level expression of a downstream holin-lysin cassette.
Several lines of evidence confirm the importance of Tac31A to the leaky phenotype. Tac31A expression from pTRK391 resulted from low-level promoter activity associated with vector sequences. Earlier studies had suggested that higher Tac31A expression levels were lethal in L. lactis NCK203 (47), possibly due to induction of lethal gene products from an uncharacterized prophage harboring sequences identical to the φ31 late promoter (48). The lethality of Tac31A was confirmed in L. lactis R1/r1t, in which even low-level expression from pTRK391 resulted in an unstable leaky phenotype and, ultimately, selection of an r1t-cured derivative of the strain. The use of a mutated version of Tac31A established a stable leaky phenotype in R1/r1t. Expression of equivalent or higher levels of β-gal from a strong Lactobacillus promoter (P6::lacZ.st) in these strains did not result in high levels of β-gal in the growth medium, proving that the strain by itself did not allow the release of significant levels of β-gal.
One interesting question arising from this study was why L. lactis R1/r1t was more sensitive to the wild-type Tac31A than were C10, ML8, and NCK203. There are several possible explanations for this difference. First, tac31A mRNA levels were not measured, and the possibility that Tac31A was expressed more efficiently in R1/r1t cannot be ruled out. Second, R1/r1t undergoes spontaneous prophage induction (20), which can lead to approximately 103 to 104 phages/ml in the culture supernatant. This level of spontaneous induction may lead to an increased number of phage genomes replicating in many of the cells and, therefore, to an increased induction of lysin and holin by Tac31A. Third, although the holin- and lysin-encoding regions of the responsible resident prophages of C10, ML8, and NCK203 have not been identified, we have obtained evidence that they are different from those encoded by r1t. Different activity levels of the lysin and/or holin, or even different transcription and/or translation efficiencies of these gene products, may explain the differences. For example, on the phage r1t genome, the holin-lysin cassette lies at the end of a very long, late transcript. Lastly, the difference in sensitivity simply may be due to strain differences.
Initially, it was considered highly probable that the leaky phenotype resulted from Tac31A induction of a prophage from a small proportion of the population. The phage infection cycle would increase β-gal levels in that proportion of cells by activation of the P15A10::lacZ.st cassette, and lysis would result in higher levels of β-gal in the supernatant. However, none of the experiments described in this paper support this theory. First, no differences were observed in culture growth, OD, or CFU per milliliter even though a clear majority of the β-gal activity was found in the supernatant. Second, Southern hybridization with an r1t attP probe failed to show any significant induction of the resident prophage in C10, ML8, or NCK203 when Tac31A was present. Third, the presence of AbiA, an abortive-infection protein which inhibits phage replication, did not affect the leaky phenotype in NCK203; hence, replication of the prophage does not appear to be a requirement for release of β-gal activity. Fourth, the presence of the mutant Tac31A in R1/r1t did not significantly increase the number of spontaneously induced phages present in the culture supernatant (data not shown). Taken together, these data suggest that the leaky phenotype is due not to induction and replication of the resident prophage but rather to induction of the late region of the integrated prophage. RNA slot blot analyses measuring induction of the late region of the resident prophages of C10, ML8, and NCK203 support this conclusion. The RNA data showed that only a low-level activation of the late region is required for the release of β-gal (Fig. 3).
An MG1363 derivative containing an integrated r1t holin-lysin cassette under the control of the tightly regulated P566–888 late promoter confirmed the importance of holin and lysin to the leaky phenotype. The phenotype of MG1363/hol-lys was very similar to that observed in R1/r1t containing P15A10::lacZ.st or P15A10φ31.9::lacZ.st, strongly suggesting that activation of lysin and holin at a very low level is largely responsible for the release of β-gal into the growth medium. In almost all reported circumstances, induction of holin and lysin results in cell death as well as lysis (6; for reviews, see references 15 and 39). Also, induction of holin alone usually results in cessation of growth due to increased cell membrane permeability and collapse of the membrane potential (6; reviewed in reference 53), sometimes followed by partial loss of turbidity. In most of the studies, however, induction was achieved by utilizing expression vectors which would efficiently express the gene(s), thus allowing for fairly high-level production of the holin and lysin products (6; reviewed in references 15 and 39). In contrast, a recent report by Husson-Kao et al. (21) suggested that leaky low-level expression of a prophage lytic cassette due to incomplete prophage repression was not lethal to the cell until environmental forces (lactose depletion or solvent addition) intervened.
Certain bacterial strains have been found to utilize phage-encoded holins and/or lysins to release cellular products. Expression and release of Serratia marcescens extracellular nuclease (2, 23) and bacteriocin 28b (13) were found to be due to putative prophage-encoded operons containing genes for a transcriptional activator, holin, and lysin. In both cases, however, evidence suggests that release may be mediated by cell lysis. In other studies, low-level expression of holin led to the leakage of intracellular enzymes or compounds without substantially affecting cell growth. Kyogoku and Sekiguchi (24) found that expression of a Bacillus licheniformis holin in E. coli resulted in β-gal leakage into the supernatant without loss of cell viability. However, the leakage of β-gal was considerably less than that observed in this study and was possibly due to low levels of cell lysis (24). In addition, low-level expression of a Streptococcus thermophilus holin gene, lyt50, in E. coli (before induction of the expression vector) resulted in a high background of isocitrate dehydrogenase activity in the culture medium, with no effects on cell viability (41). Lastly, low-level expression of small B. licheniformis and Bacillus subtilis proteins possessing the characteristics of a holin was found to complement certain alkaline phosphatase-deficient mutants of E. coli, presumably by altering the cell membrane permeability so that the XP substrate entered more readily and was hydrolyzed by cytoplasmic phosphatases (26, 28). The proteins were not lethal unless induced in E. coli, but no measurement of the degree of leakiness was given. These studies are interesting because they suggest that low-level expression of certain holins may allow for release of intracellular enzymes into the growth medium while allowing continued growth of the culture.
Work with other proteins and enzymes suggested that this system could be utilized to release other relevant products into the growth medium. TTFC was detected at higher levels in the supernatants than in the cell extracts. Interestingly, another enzyme was not released efficiently, if at all; PepXP activity was not detected to any significant extent in the supernatant, regardless of whether Tac31A was present, even when PepXP was expressed from the P15A10 promoter. The only exception was with MG1363/hol-lys, for which expression of the wild-type Tac31A resulted in high levels of PepXP in the medium and concomitant cell lysis. Work by Wells et al. (49) has suggested that the cell wall can act as a barrier to the diffusion of some proteins or enzymes into the medium. Perhaps PepXP is similarly affected, but this seems unlikely since one would expect at least a slight increase of PepXP in the supernatant even if diffusion was somewhat limited. At this point, we do not understand why some proteins and enzymes (e.g., β-gal and TTFC) are externalized while others (PepXP) are retained. All previous complementation studies have indicated that holins are nonspecific, allowing for the release of heterologous lysin products outside the cell (reviewed in reference 53). Data from this study suggest that holins allow export of other proteins while restricting others. Further investigation is needed to elucidate the mechanism through which leaky cells externalize select proteins and enzymes.
To our knowledge, this is the first report describing the release of significant levels of β-gal and other, heterologous proteins into the growth medium without the use of export signals and without seriously compromising the viability of the cells by inducing autolysis or prophage induction. Leaky lactic acid bacteria are expected to find valuable applications as delivery vehicles in bioprocessing, food, and the gastrointestinal tract.
ACKNOWLEDGMENTS
This work was supported in part by Southeast Dairy Foods Research Center; Dairy Management, Inc.; Rhodia, Inc.; and the USDA (NRICGP project no. 97-35503-4368).
We thank Jerry Wells for kindly providing the genetic constructs encoding TTFC. We also thank Mark Conkling, Eric Miller, Evelyn Durmaz, W. Mike Russell, Mick O'Callahan, and Soren Madsen for helpful discussions and critical reading of the manuscript. Photographs were kindly taken by Brendyln Bradley-Kerr at the Laboratory for Advanced Light and Electron Optical Methods, College of Veterinary Medicine, North Carolina State University, Raleigh.
Footnotes
Paper no. FSR00-20 of the Department of Food Science, Southeast Dairy Foods Research Center, North Carolina State University, Raleigh, N.C.
REFERENCES
- 1.Allison G E, Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria and their use in novel industrial applications. In: Demain A L, Davies J E, Atlas R M, Cohen G, Hershberger C L, Hu W-S, Sherman D H, Willson R C, Wu J H D, editors. Manual of industrial microbiology and biotechnology. Washington, D.C.: ASM Press; 1999. pp. 789–808. [Google Scholar]
- 2.Berkmen M, Benedik M J, Bläsi U. The Serratia marcescens NucE protein functions as a holin in Escherichia coli. J Bacteriol. 1997;179:6522–6524. doi: 10.1128/jb.179.20.6522-6524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasi U, Young Y. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 4.Buist G, Kok J, Leenhouts K J, Dabrowska M, Venema G, Haandrikman A J. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buist G, Karsens H, Nauta A, van Sinderen D, Venema G, Kok J. Autolysis of Lactococcus lactis caused by induced overproduction of its major autolysin, AcmA. Appl Environ Microbiol. 1997;63:2722–2728. doi: 10.1128/aem.63.7.2722-2728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Ruyter P G G A, Kuipers O P, Meijer W C, de Vos W M. Food-grade controlled lysis of Lactococcus lactis for accelerated cheese ripening. Nat Biotechnol. 1997;15:976–979. doi: 10.1038/nbt1097-976. [DOI] [PubMed] [Google Scholar]
- 7.de Vos W M. Gene cloning and expression in lactic streptococci. FEMS Microbiol Rev. 1987;46:281–295. [Google Scholar]
- 8.Dinsmore P K, Klaenhammer T R. Molecular characterization of a genomic region in a Lactococcus bacteriophage that is involved in its sensitivity to the phage defense mechanism AbiA. J Bacteriol. 1997;179:2949–2957. doi: 10.1128/jb.179.9.2949-2957.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djordjevic G M, Bojovic B, Miladinov N, Topisorovic L. Cloning and molecular analysis of promoter-like sequences isolated from the chromosomal DNA of Lactobacillus acidophilus ATCC 4356. Can J Microbiol. 1997;43:61–69. doi: 10.1139/m97-009. [DOI] [PubMed] [Google Scholar]
- 10.Djordjevic G M, Klaenhammer T R. Bacteriophage-triggered defense systems: phage adaptation and design improvements. Appl Environ Microbiol. 1997;63:4370–4376. doi: 10.1128/aem.63.11.4370-4376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dykstra M J. A manual of applied techniques for biological electron microscopy. New York, N.Y: Plenum Press; 1993. p. 257. [Google Scholar]
- 12.Efstathiou J D, McKay L L. Plasmids in Streptococcus lactis: evidence that lactose metabolism and proteinase activity are plasmid linked. Appl Environ Microbiol. 1976;32:38–44. doi: 10.1128/aem.32.1.38-44.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrer S, Viejo M B, Guasch J F, Enfedaque J, Regué M. Genetic evidence for an activator required for induction of colicin-like bacteriocin 28b production in Serratia marcescens by DNA-damaging agents. J Bacteriol. 1996;178:951–960. doi: 10.1128/jb.178.4.951-960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasson M J. Lytic systems in lactic acid bacteria and their bacteriophages. Antonie Leeuwenhoek. 1996;70:99–110. doi: 10.1007/BF00395931. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Hill C, Massey I J, Klaenhammer T R. Rapid method to characterize lactococcal bacteriophage genomes. Appl Environ Microbiol. 1991;57:283–288. doi: 10.1128/aem.57.1.283-288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill C, Pierce K, Klaenhammer T R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction modification (R+/M+) and abortive infection (Hsp+) Appl Environ Microbiol. 1989;55:2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huggins A R, Sandine W E. Incidence and properties of temperate bacteriophages induced from lactic streptococci. Appl Environ Microbiol. 1977;33:184–191. doi: 10.1128/aem.33.1.184-191.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Husson-Kao C, Mengaud J, Cesselin B, van Sinderen D, Benbadis L, Chapot-Chartier M-P. The Streptococcus thermophilus autolytic phenotype results from a leaky prophage. Appl Environ Microbiol. 2000;66:558–565. doi: 10.1128/aem.66.2.558-565.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvis A W, Klaenhammer T R. Bacteriophage resistance plasmid pTR2030 inhibits lytic infection of r1t temperate bacteriophage but not induction of r1t prophage in Streptococcus cremoris R1. Appl Environ Microbiol. 1987;53:385–389. doi: 10.1128/aem.53.2.385-389.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin S, Chen Y, Christie G E, Benedik M J. Regulation of the Serratia marcescens extracellular nuclease: positive control by a homolog of P2 Ogr encoded by a cryptic prophage. J Mol Biol. 1996;256:264–278. doi: 10.1006/jmbi.1996.0084. [DOI] [PubMed] [Google Scholar]
- 24.Kyogoku K, Sekiguchi J. Cloning and sequencing of a new holin-encoding gene of Bacillus licheniformis. Gene. 1999;168:61–65. doi: 10.1016/0378-1119(95)00690-7. [DOI] [PubMed] [Google Scholar]
- 25.Langsrud T, Landaas A, Castberg H B. Autolytic properties of different strains of group N streptococci. Milchwissenschaft. 1987;42:556–560. [Google Scholar]
- 26.Lee J W K, Edwards C W, Hulett F M. Identification of four unique clones encoding 10 kDa proteins from Bacillus that cause phenotypic complementation of a phoA mutant strain of Escherichia coli. J Gen Microbiol. 1991;137:667–677. doi: 10.1099/00221287-137-3-667. [DOI] [PubMed] [Google Scholar]
- 27.Lepeuple A-S, Van Gemert E, Chapot-Chartier M-P. Analysis of the bacteriolytic enzymes of the autolytic Lactococcus lactis subsp. cremoris strain AM2 by renaturing polyacrylamide gel electrophoresis: identification of a prophage-encoded enzyme. Appl Environ Microbiol. 1998;64:4142–4148. doi: 10.1128/aem.64.11.4142-4148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longchamp P F, Mauel C, Karamata D. Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-l-alanine amidase gene of prophage PBSX. Microbiology. 1994;140:1855–1867. doi: 10.1099/13500872-140-8-1855. [DOI] [PubMed] [Google Scholar]
- 29.Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol. 1992;174:5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 31.Nardi M, Chopin M-C, Chopin A, Cals M-M, Gripon J-C. Cloning and DNA sequence analysis of an X-prolyl dipeptidyl aminopeptidase gene from Lactococcus lactis subsp. lactis NCDO 763. Appl Environ Microbiol. 1991;57:45–50. doi: 10.1128/aem.57.1.45-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nauta A, van Sinderen D, Karsens H, Smit E, Venema G, Kok J. Inducible gene expression mediated by a repressor-operator system isolated from Lactococcus lactis bacteriophage r1t. Mol Microbiol. 1996;19:1331–1341. doi: 10.1111/j.1365-2958.1996.tb02477.x. [DOI] [PubMed] [Google Scholar]
- 33.Niven G W, Mulholland F. Cell membrane integrity and lysis in Lactococcus lactis: the detection of a population of permeable cells in post-logarithmic phase cultures. J Appl Microbiol. 1998;84:90–96. doi: 10.1046/j.1365-2672.1997.00316.x. [DOI] [PubMed] [Google Scholar]
- 34.O'Sullivan D J, Klaenhammer T R. High and low copy number Lactococcus shuttle cloning vectors with features for clone screening. Gene. 1993;137:227–231. doi: 10.1016/0378-1119(93)90011-q. [DOI] [PubMed] [Google Scholar]
- 35.O'Sullivan D J, Klaenhammer T R. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl Environ Microbiol. 1993;59:2730–2733. doi: 10.1128/aem.59.8.2730-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Sullivan D J, Walker S A, West S G, Klaenhammer T R. Development of an expression strategy using a lytic phage to trigger explosive plasmid amplification and gene expression. Bio/Technology. 1996;14:82–87. doi: 10.1038/nbt0196-82. [DOI] [PubMed] [Google Scholar]
- 37.Raya R R, Kleeman E G, Luchansky J B, Klaenhammer T R. Characterization of the temperate bacteriophage φadh and plasmid transduction in Lactobacillus acidophilus ADH. Appl Environ Microbiol. 1989;55:2206–2213. doi: 10.1128/aem.55.9.2206-2213.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Sanders J W, Venema G, Kok J. A chloride-inducible gene expression cassette and its use in induced lysis of Lactococcus lactis. Appl Environ Microbiol. 1997;63:4877–4882. doi: 10.1128/aem.63.12.4877-4882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroeder C J, Robert C, Lenzen G, McKay L L, Mercenier A. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricus β-galactosidase sequences. J Gen Microbiol. 1991;137:369–380. doi: 10.1099/00221287-137-2-369. [DOI] [PubMed] [Google Scholar]
- 41.Sheehan M M, Stanley E, Fitzgerald G F, van Sinderen D. Identification and characterization of a lysis module present in a large proportion of bacteriophages infecting Streptococcus thermophilus. Appl Environ Microbiol. 1999;65:569–577. doi: 10.1128/aem.65.2.569-577.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Environ Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Asseldonk M, Rutten G, Oteman M, Siezen R J, de Vos W M, Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene. 1990;95:155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 44.van Sinderen D, Karsens H, Kok J, Terpstra P, Ruiters M H J, Venema G, Nauta A. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol Microbiol. 1996;19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- 45.Vidgrén G, Palva I, Pakkanen R, Lounatmaa K, Palva A. S-layer protein gene of Lactobacillus brevis: cloning by polymerase chain reaction and determination of the nucleotide sequence. J Bacteriol. 1992;174:7419–7427. doi: 10.1128/jb.174.22.7419-7427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vos P, Simons G, Siezen R J, de Vos W M. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J Biol Chem. 1989;264:13579–13585. [PubMed] [Google Scholar]
- 47.Walker S A, Klaenhammer T R. Molecular characterization of a phage-inducible middle promoter and its transcriptional activator from the lactococcal bacteriophage φ31. J Bacteriol. 1998;180:921–931. doi: 10.1128/jb.180.4.921-931.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker S A, Dombroski C S, Klaenhammer T R. Common elements regulating gene expression in temperate and lytic bacteriophages of Lactococcus species. Appl Environ Microbiol. 1998;64:1147–1152. doi: 10.1128/aem.64.3.1147-1152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wells J M, Wilson P W, Norton P M, Le Page R W F. A model system for the investigation of heterologous protein secretion pathways in Lactococcus lactis. Appl Environ Microbiol. 1993;59:3954–3959. doi: 10.1128/aem.59.11.3954-3959.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wells J M, Wilson P W, Norton P M, Gasson M J, Le Page R W F. Lactococcus lactis: high-level expression of tetanus toxin fragment C and protection against lethal challenge. Mol Microbiol. 1993;8:1155–1162. doi: 10.1111/j.1365-2958.1993.tb01660.x. [DOI] [PubMed] [Google Scholar]
- 51.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 52.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young R, Blasi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]