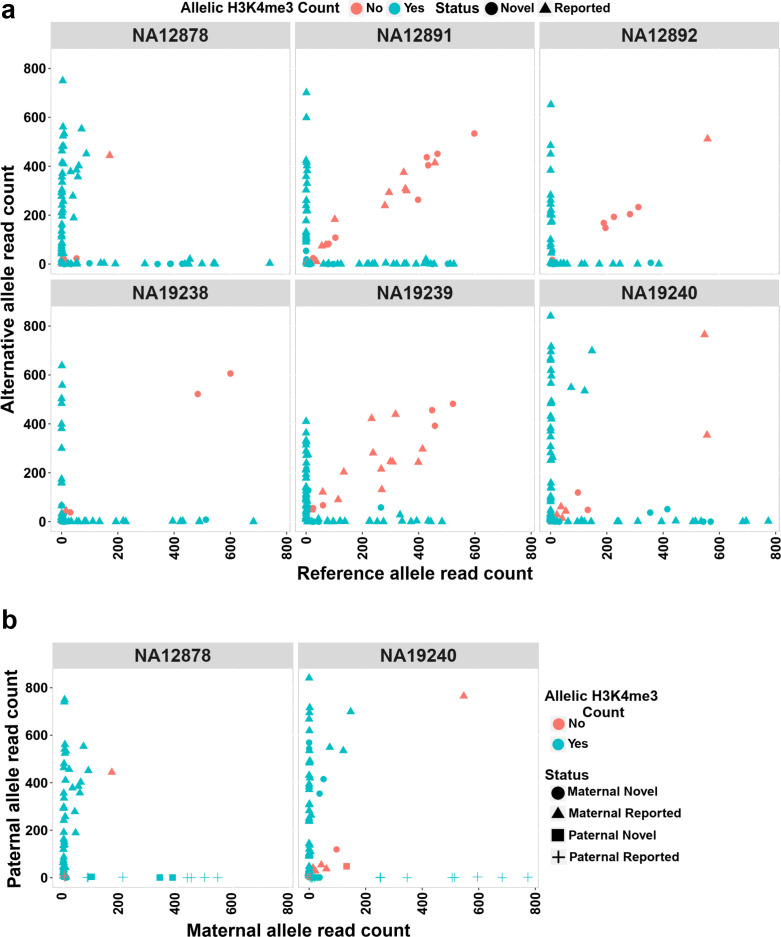
Figure 4. Allelic H3K4me3 histone mark at detected imprinted differentially methylated regions (DMRs).
(a) The plots representing reference and alternative alleles H3K4me3 read counts for the heterozygous single-nucleotide variants (SNVs) mapped to the detected DMRs for the six examined samples. Each point represents an SNV. Blue color displays SNVs with Fisher’s combined p-value binomial <0.05 and at least 80% of the reads on one allele and red color represent those SNVs that did not satisfy either of these thresholds. (b) The plots representing paternal and maternal H3K4me3 read counts for the heterozygous SNVs at DMRs in NA12878 and NA19240. Each point represents an SNV. The ‘Status’ indicates the methylation origin of the DMR and if the DMR is novel or reported.