Abstract
The multilineage differentiation potential of human mesenchymal stem cells (hMSCs) underpins their clinical utility for tissue regeneration. Control of such cell-fate decisions is tightly regulated by different growth factors/cytokines and their cognate receptors. Fibroblast growth factors (FGFs) are among such factors critical for osteogenesis. However, how FGF receptors (FGFRs) help to orchestrate osteogenic progression remains to be fully elucidated. Here, we studied the protein levels of FGFRs during osteogenesis in human adult bone marrow-derived MSCs and discovered a positive correlation between FGFR2 expression and alkaline phosphatase (ALP) activity, an early marker of osteogenesis. Through RNA interference studies, we confirmed the role of FGFR2 in promoting the osteogenic differentiation of hMSCs. Knockdown of FGFR2 resulted in downregulation of pro-osteogenic genes and upregulation of pro-adipogenic genes and adipogenic commitment. Moreover, under osteogenic induction, FGFR2 knockdown resulted in upregulation of Enhancer of Zeste Homolog 2 (EZH2), an epigenetic enzyme that regulates MSC lineage commitment and suppresses osteogenesis. Lastly, we show that serial-passaged hMSCs have reduced FGFR2 expression and impaired osteogenic potential. Our study suggests that FGFR2 is critical for mediating osteogenic fate by regulating the balance of osteo-adipogenic lineage commitment. Therefore, examining FGFR2 levels during serial-passaging of hMSCs may prove useful for monitoring their multipotency.
Keywords: mesenchymal stem cell, osteogenesis, FGFR, EZH2, adipogenesis, lineage commitment, potency
Introduction
Stem cell-based therapies offer significant potential for treating disease or injury through the regeneration of damaged tissues. Among the various stem cells available, mesenchymal stem cells (MSCs) are garnering considerable interest because of their availability, ease of isolation, and multipotency. Importantly, under appropriate culture conditions, MSCs can be selectively induced to differentiate into various lineages, such as osteoblasts, adipocytes, chondrocytes, and myoblasts (Caplan, 1991; Prockop, 1997). This makes them popular for studies of factors that control cell fate. Understanding the mechanism governing commitment and progression through these lineages is crucial for the development of effective strategies that lead to tissue regeneration.
The commitment of MSCs to the osteoblast lineage is characterized by the initial expression of Runt-related transcription factor 2 (RUNX2), followed by type 1 collagen (COL1A, encoded by COL1A1 and COL1A2 genes) and alkaline phosphatase (ALP, encoded by the ALPL gene), and finally mineralization of the extracellular matrix (Lian et al., 2004). Osteogenic lineage commitment of MSCs can be induced by in vitro supplementation of growth media with dexamethasone, ascorbic acid and β-glycerol phosphate (Jaiswal et al., 1997). In addition, growth factors and other signaling proteins, such as bone morphogenetic proteins (BMPs) (Chen et al., 2012; Smith et al., 2018) and WNT proteins (Ling et al., 2009), regulate osteogenic lineage commitment and differentiation.
Fibroblast growth factors (FGFs) are essential for early osteogenesis and bone homeostasis, although FGF signaling itself does not directly induce osteoblast differentiation (Charoenlarp et al., 2017). Nonetheless, FGFs could modulate osteogenesis by regulating the expression of multiple genes involved in bone formation. In particular, FGF is important for the proliferation of osteoprogenitor cells and their subsequent maturation into bone-forming cells (Nakamura et al., 1998; Zellin and Linde, 2000; Lisignoli et al., 2001). Loss of FGF2 expression in mice caused a reduction in bone volume, mineral deposition, and bone formation efficiency (Montero et al., 2000). FGFs function through interaction with FGF receptors (FGFRs), a group of cell surface receptor tyrosine kinases. There are five FGFRs. In the early phase of bone formation, both FGFR1 and FGFR2 are expressed in developing bone tissue, and in the perichondrium and periosteum (Ornitz and Marie, 2002). In contrast, FGFR3 is mainly expressed by chondrocytes (Wang et al., 2001) and has been shown to inhibit bone formation by promoting chondrocyte proliferation (Deng et al., 1996).
Among all the FGFRs, FGFR2 has been shown to positively regulate osteogenesis (Ornitz and Marie, 2002; Wilkie, 2002; Wilkie et al., 2002; Jackson et al., 2006). Frequently, murine cell lines or in vivo models are used to evaluate the role of FGFR2 during osteogenesis. However, conflicting results have been observed. Notably, increased FGFR2 transcripts have been reported during osteogenesis in immortalized murine MSCs (Kahkonen et al., 2018), whereas the opposite result (decreased FGFR2) has been observed in hMSCs (Simann et al., 2017). Such conflicting observations warrant further investigation into the role of FGFR2 during osteogenesis, particularly in hMSCs.
Osteogenesis of MSCs is a complex process involving cross-talk between many epigenetic and transcriptional factors (Perez-Campo and Riancho, 2015). Histones in the promoter region of RUNX2 and PPARγ, which are involved in lineage commitment, undergo modification during MSC differentiation (Meyer et al., 2016). Additional epigenetic switches involving Enhancer of zeste 2 (EZH2), a component of the polycomb repressor complex 2 (PRC2) that enhances the methylation of H3K2, govern MSC differentiation (Hemming et al., 2014; Dudakovic et al., 2015; Samsonraj et al., 2018; Galvan et al., 2021). In this context, EZH2 regulates the methylation status of H3K27 on the promoter of RUNX2, PPARγ and CEBPA. Notably, downregulation of EZH2 expression promotes osteogenesis (Hemming et al., 2014), while overexpression downregulates the expression of RUNX2 and OC (Osteocalcin) (Hemming et al., 2016).
In the current study, we employed siRNA to knockdown FGFRs in hMSCs to determine their functions in mediating osteogenesis. Combined with a focused PCR gene array study, we evaluated how FGFR2 could modulate the osteo-adipogenic lineage commitment of hMSCs. Moreover, we sought to establish a link between FGFR2 and EZH2 in regulating MSC osteogenesis by examining their expression under osteogenic induction. The data show that under osteogenic stimuli, EZH2 levels decrease while FGFR2 and ALP increase resulting in the deposition of a mineralized matrix. Under the same osteogenic conditions, FGFR2 knockdown resulted in a greater reduction of EZH2 levels and decreased ALP and matrix mineralization. These results highlight a possible FGFR2-EZH2 osteogenic axis and that epigenetic changes regulated by EZH2 may contribute to the reduced osteogenic differentiation observed upon FGFR2 knockdown in MSCs. Furthermore, the current study suggests FGFR2 is critical for the control of osteogenic fate , and thus constitutes a potential target for strategies that enhance bone regeneration.
Materials and methods
Human donor information
Bone marrow-derived MSCs from three independent human donors were utilized (Lonza; passage 2) following appropriate ethics approval from NUS-IRB e-Declaration Ref No: N-17-018. Donor information is listed in Supplementary Table 1. Unless otherwise stated, all studies were performed using MSCs from Donor 1. Cells from two additional donors (Donor 2 and 3) were utilized in age-related assessments and multi-lineage assays.
Cell culture
All cells were maintained in growth medium, consisting of low-glucose (1 g/L) Dulbecco’s Modified Eagle Medium (DMEM) (HyClone; Logan, Utah), 10% v/v fetal calf serum (HyClone), and 4 mM L-glutamine. The media was supplemented with 1% penicillin/streptomycin. Cells were seeded at 5000 cells/cm2 in 15 cm culture dishes unless otherwise specified and maintained in a 5% CO2 incubator at 37°C. All media was changed every 3 to 4 days and cells were split at ∼80% confluence. All experiments were performed on cells at passage 5, unless otherwise stated. Cell counts were performed with a nucleocounter (Chemometec, Denmark), unless stated otherwise.
Osteogenic differentiation and staining
hMSCs seeded in triplicate at 3000 cells/cm2 in 6-well plates (for Western blotting) or in 24-well plates (for staining) were cultured overnight. This seeding density was chosen according to previous publications (McBeath et al., 2004; Neuhuber et al., 2008; Rider et al., 2008; Samsonraj et al., 2018) that report high seeding density favours adipogenesis rather than osteogenesis. Growth media was then changed to osteogenic media, consisting of growth media plus 10 nM dexamethasone (Sigma Aldrich, New Jersey, United States), 25 µg/ml L-ascorbate-2-phosphate and 10 mM glycerol-2-phosphate (Sigma Aldrich, St. Louis, Missouri). Cells were then maintained in osteogenic media for the specified times in the different assays. The medium was changed at 3–4 day intervals. Undifferentiated cells served as control and were kept in growth media for the same period of time. After 21 days, cells were washed with PBS and fixed with 100% methanol for 20 min before staining with 0.1% (w/v) Alizarin red for 20 min at room temperature. Washed cells were then imaged with a digital scanner. Alizarin red dye was also quantified by extracting the stain with 10% (v/v) acetic acid, followed by neutralization with 10% w/v ammonium hydroxide and absorbance read at 405nm.
Adipogenic differentiation and staining
hMSC were seeded in triplicate at 18,000 cells/cm2 in 12-well plates (for qPCR) or 24-well plates (for staining with oil red-O) and maintained for 3 days in growth media. When confluent, the media was changed to adipogenic medium (growth media supplemented with 1 µM dexamethasone, 20 µM indomethacin, 115 μg/ml 3-isobutyl-1-methylxanthine, and 10 µM insulin). Cells were grown in adipogenic medium for 14 days for qPCR analysis or 21 days for oil red-O staining, with a medium change every 3–4 days. hMSCs cultured in growth media were used as a negative control. After 21 days, cells were washed with PBS, then fixed with 4% paraformaldehyde for 1 h. The cells were then washed with deionized water and 0.36% (w/v) oil red-O solution was used to stain the cells for 1 h. The excessive stain was washed with 60% 2-propanol followed by a final wash in deionized water. Plates were then imaged using a digital scanner.
ALP activity
hMSC were seeded in 6 well-plates at 3000 cells/cm2 and cultured in either growth or osteogenic media for 8 days. Next, the cells were lysed with ice-cold RIPA buffer (Thermo Fisher Scientific, Waltham, Massachusetts) and total protein was quantified using the bicinchoninic acid assay (BCA assay) (Thermo Fisher Scientific). Total protein (5 µg) was used for ALP enzymatic activity quantification. The chemiluminescent substrate, p-nitrophenyl phosphate (Thermo Fisher Scientific) was mixed with the protein samples and absorbance measured at 405 nm using the Hidex Sense microplate reader (Turku, Finland). Cells were seeded in triplicate conditions and each sample was analyzed in duplicate for quantification.
FGFR knockdown and cell proliferation
hMSCs were seeded at 10,000 cells/cm2 in a 96 well-plate in growth medium. After 24 hours, transfection was performed with a mixture of 25 nM siRNA and 0.5 μl/ml Dharmafect 1 (Dharmacon, Lafayette, Colorado). After 3 days of siRNA transfection, the number of viable cells was evaluated (WST-1 cell proliferation reagent, Sigma-Aldrich). Briefly, the WST-1 reagent was mixed with fresh growth media at 1:10 ratio and added to the cells and allowed to incubate for 1 hour. Absorbance was measured at 450 nm using the Hidex Sense microplate reader (Turku, Finland).
Western blotting
hMSCs were seeded in 6-well plates 3000 cells/cm2 and cultured in either growth or osteogenic medium. At the specified time points, cells were lysed with ice-cold RIPA buffer containing protease inhibitors (Merck Millipore, Burlington, Massachusetts). Total protein was measured using the BCA assay and 10 μg protein was mixed in Laemmli buffer containing SDS. The protein sample was heated at 95°C for 5 min to denature the sample. The denatured protein samples were separated by 4–12% SDS-PAGE in MOP buffer at 150 volts and then transferred to nitrocellulose membranes at 110 volts. Membranes were blocked with 5% milk or bovine serum albumin and blotted with specific primary antibodies (Abcam, Cambridge, United Kingdom). HRP-conjugated secondary antibodies (Jackson Immunoresearch, West Grove, Pennsylvania) were used to detect primary antibodies. Blots were visualized by adding the Chemiluminescent HRP (substrate SuperSignal West Pico, Thermo Fisher Scientific) and developed on hyperfilm (GE healthcare, Chicago, Illinois). Primary antibodies used for this study were: anti-FGFR1 ( ab137084, Abcam ) , anti-FGFR2 (ab109372, Abcam), anti-FGFR3 (ab133644, Abcam), anti-Ki67 (Ab92742, Abcam), anti-p16 (10883-1-AP, Proteintech), anti-PCNA (proliferation cell nuclear antigen) (sc-56, Santa Cruz Biotechnology), anti-Actin (MAB1501R, Merck-Millipore). Secondary antibodies used for the study were goat-antimouse (115-035-003) and goat-anti-rabbit (111-035-144).
Reverse transcription and quantitative polymerase chain reaction (qPCR)
Total RNA was purified from the cells using a Nucleospin RNA extraction kit (Macherey-Nagel; Bethlehem, PA) following the manufacturer’s recommendations. The RNA concentration and purity were measured by a NanoDrop UV spectrophotometer (Thermo Fisher Scientific). RNA (2 µg) was converted to cDNA using a SuperScript VILO cDNA synthesis kit (Life Technologies) with an ABI Verti thermal cycler (Applied Biosystems, Massachusetts). qPCR was performed on a Quantstudio 6 Flex real-time PCR machine (Applied Biosystems, Foster City, California). Next, cDNA (40 ng per reaction) was analyzed with the TaqMan gene expression probes (Thermo Fisher Scientific) according to the recommended protocol. Gene transcription (mRNA) levels were analysed with the following TaqMan probes: FGFR1 (HS00915142_m1), FGFR2 (HS01552926_m1), FGFR3 (HSHS00179829_m1), CCAAT Enhancer Binding Protein Alpha (CEBPA) (HS00269972_s1); Peroxisome Proliferator Activated Receptor Gamma (PPARG) (HS01115513_m1); EZH2 (HS00544830_m1); ALPL (HS01029144_m1). ACTB (HS01060665_g1) was utilized as a reference gene. Data were analyzed using the comparative ΔΔCt method and mRNA levels were expressed as relative expression units (REU). Data are shown as mean REU or mean fold change ± standard deviation (S.D.) of three independent experiments.
PCR array
hMSC PCR array, the RT2 Profiler (PAHS-082, SABiosciences, Qiagen, Hilden), was employed to assess the relative transcript levels of 84 genes governing the stemness and differentiation of hMSCs. Cells were seeded at 10,000 cells/cm2 in a 10 cm dish and cultured in growth medium. Transfection was performed with a mixture of 25 nM siRNA and 0.5 μl/ml Dharmafect 1 (Dharmacon, Lafayette, Colorado). After 4 days of siRNA transfection, total RNA was extracted and purified as described earlier. The total amount of RNA isolated was measured with a Nanodrop UV spectrometer (Thermo Fisher Scientific). RNA (500 ng) was converted to cDNA using the RT2 First Strand Kit (Qiagen, Hilden, Germany), which includes enzymes to remove genomic DNA contamination. The cDNA was added to 96 wellplate preloaded with primers of genes of interest, reference genes as well as the positive and negative control of the PCR amplification efficiency. A melt curve analysis was performed to verify the PCR product had a single amplicon using the QuantStudio 6 Flex system. Threshold and baseline values were set according to the user guide from the manufacturer of the PCR array. The fold change of relative gene expression (FGFR2 siRNA treated cells versus scramble siRNA treated cells) was calculated using the comparative ΔΔCt method with ACTB as the reference gene (control). A two-fold change was set as a cut-off to select up-regulated and down-regulated genes. A heatmap was generated with Excel (Microsoft) using a three-color scale map, with white set to 1 (no fold change). Blue was used to represent downregulation and red was used to represent upregulation.
FGFR2 expression and ALP activity in early and late passage MSCs
hMSCs from donor 2 and 3 (previously expanded and cryopreserved at p4 and p10) were seeded (5000/cm2) in normal growth medium and allowed to recover for 3 to 4 days. Cells were then passaged (p5 and p11) and seeded for Western blot and ALP assays. For Western blot analysis, cells were seeded (5000/cm2) and cultured for 3 days before protein lysates were collected as describe earlier. For ALP assays, cells were seeded (3000/cm2) and cultured for 8 days in osteogenic medium before protein lysates were collected and ALP activity determined as described earlier.
EZH2 expression study
hMSCs (passage 5) from three donors were seeded (10000 cells/cm2) in 12 well plates for 24 hours and transfected with either scramble siRNA or FGFR2 siRNA as described earlier for FGFR2 knockdown. For either group, 4 hours after siRNA transfection, the cultures were replaced with either fresh growth media or osteogenic media. Three replicate wells were used for each treatment condition. After 5 days, cells were harvested to collect RNA samples for RT-PCR study (primers and assay procedures were described earlier).
Statistical Analysis
All quantitative results are represented in the graph as the mean ± S.D. of several independent experiments (as specified in the data). For statistical significance analysis, one and two-way Analysis of Variance (Jaukovic et al.) followed by Post-hoc testing were performed where appropriate. Alternatively, a Student t-test was used. Normality of data were analysed by D’Agostino’s K-squared test where appropriate. Analyses were performed with Prism 7 software (GraphPad, San Diego, California). The following symbols were used to represent significance levels: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns – not significant (p > 0.05).
Results
Growth arrest enhances osteogenic commitment
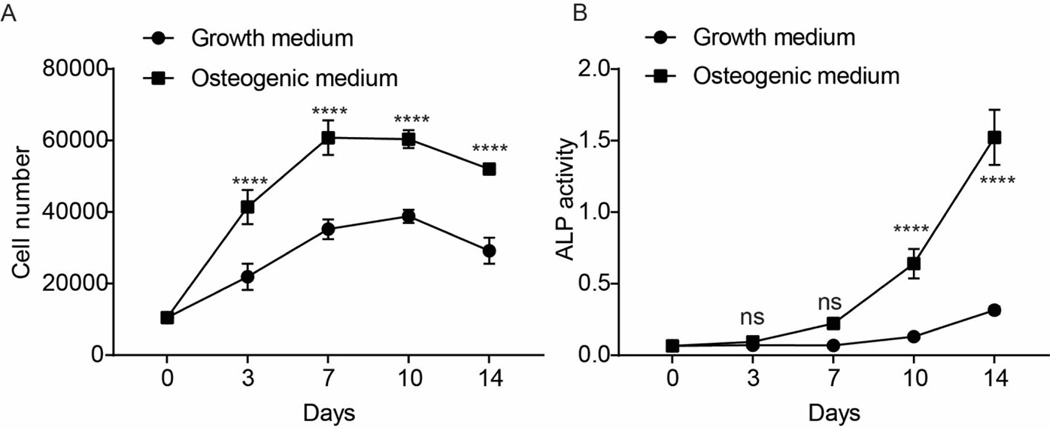
We first monitored cell growth and ALP activity during the osteogenic differentiation of hMSCs. Cell number increased significantly with time (p <0.0001) (Fig. 1A). Across all time points, cells cultured in the osteogenic medium had significantly higher cell numbers compared with control growth medium (p < 0.0001). Notably, irrespective of media conditions, cell number reached a plateau between days 7 and 10. These observations are consistent with other studies which show that osteogenic differentiation is characterized by an initial cell expansion phase. Both L-ascorbic-2-phosphate and dexamethasone enhanced the cell proliferation rate of the hMSCs until they reach confluency (Choi et al., 2008; Xiao et al., 2010).
Figure 1.
Cell proliferation and ALP activity changes during osteogenesis in hMSCs. A) Cell numbers at different time points for hMSC grown in either growth or osteogenic medium. B) Alkaline phosphatase activity measured at different time points for hMSCs grown in either growth or osteogenic medium. Data from triplicate wells of a single experiment. The data represents absorbance at 405 nm from the chemiluminescent substrate.
In parallel assays, osteogenic status was evaluated by ALP activity. The data show that ALP levels increased significantly with time under osteogenic induction (p <0.0001) (Fig. 1B). ALP activity increased significantly after 7 days in osteogenic medium to reach a value 5-fold higher than in control growth medium by days 10 (p<0.0001) and 14 (p<0.0001) (Fig. 1B). This coincided with the onset of growth arrest (Fig. 1A), suggesting that contact inhibition is important for the onset of osteogenic commitment.
FGFR2 expression is positively correlated with ALP activity
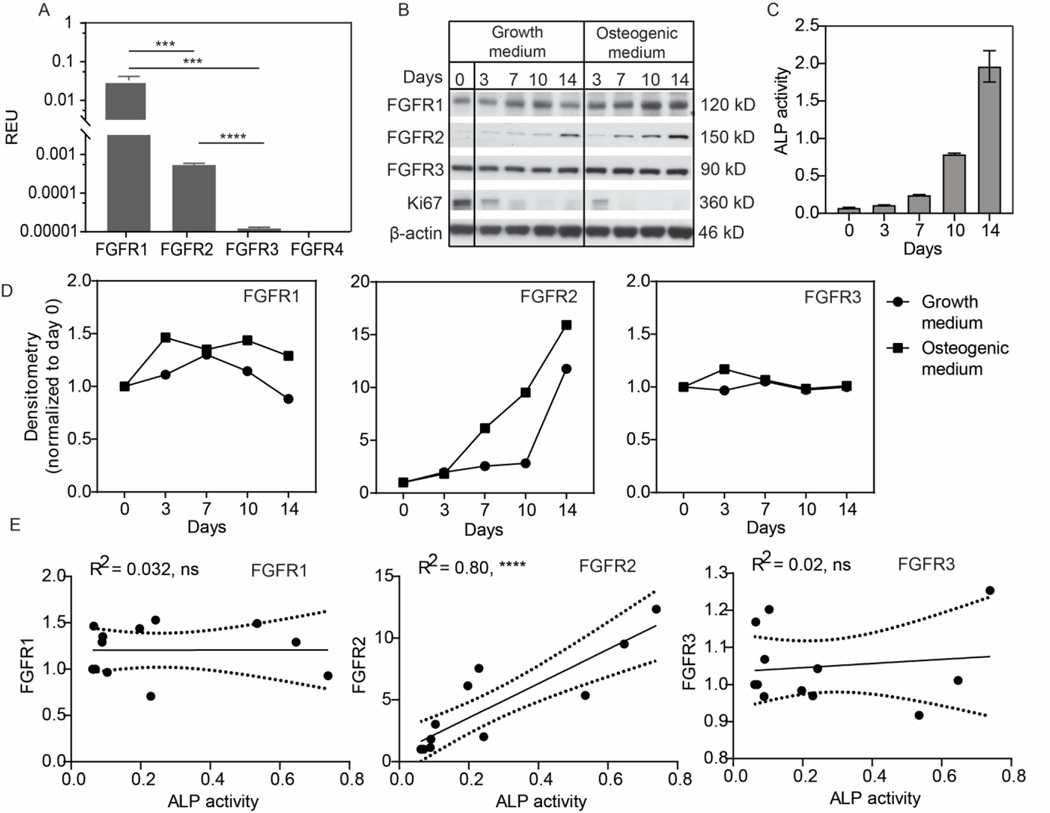
To elucidate the functions of FGFRs during the osteogenic differentiation of hMSCs, we first analyzed the relative mRNA level of individual FGFRs using the TaqMan gene expression assay (Fig. 2A). Notably, the expression levels of the four FGFRs differed significantly when cultured in growth medium (p <0.0001). Among the four receptor subtypes, FGFR1 (0.03 REU) was the most abundantly expressed, followed by FGFR2 (0.0005 REU). FGFR3 displayed a basal level expression, at approximately 0.001% of the reference control (ACTB expression). FGFR4 expression was not detectable.
Figure 2.
Expression of FGFRs before and during osteogenesis in hMSCs. A) Relative mRNA levels of FGFRs measured by qPCR for hMSCs cultured for 2 days in growth medium. Data from triplicate wells of a single experiment. B) Western blot showing changes in FGFR expression in MSCs cultured in growth or osteogenic conditions. Data from a single experiment. C) ALP activity in MSCs under osteogenic induction. Data from triplicate wells of a single experiment. D) Densitometry of the Western blots in B). E) Correlation (day 0 to 10) between ALP activity and FGFR expression from data in C and Supplementary Figs. 1 and 2. Data from three independent experiments.
We next examined the expression of FGFRs during osteogenesis at the protein level by Western blot (Fig. 2B). The expression levels of each protein were first normalized to β-actin, then to the starting point (day 0) using densitometry analysis (Fig. 2D). Across three independent experiments, FGFR1 and FGFR3 levels showed variable responses to time in culture and the presence/absence of osteogenic stimuli (Fig. 2B & D; Suppl. Figs. 1 & 2). In contrast, FGFR2 levels increased markedly (∼15-fold) by day 14 under osteogenic stimulation, compared to a 10-fold increase in control growth media (Fig. 2D). A similar trend was observed in repeat experiments, albeit to varying extents (Supp. Figs. 1 & 2). Notably, irrespective of media formulation, expression of Ki67 (proliferation marker) was initially high and then decreased as the cells approached confluency around days 7–10. This trend was also observed in repeat experiments, albeit to varying extents (Supp. Figs. 1 & 2).
As the upregulation of FGFR2 (∼ day 7–10) coincided with the onset of ALP activity (Fig. 1B), we hypothesized that FGFR2 protein levels might correlate with ALP enzymatic activity. To assess this relationship, we performed linear regression analysis to determine the coefficient of determination (R2) using the Pearson correlation test. Protein lysates used for the Western blot study were measured for ALP activity (Fig. 2C, Suppl. Figs 1B and 2B). The data is taken from three independent experiments (day 0 to day 10). The results show that ALP activity is positively correlated with FGFR2 levels (R2 = 0.0.80) (Fig. 2E). In contrast, ALP activity was not correlated with either FGFR1 or FGFR3 expression in any of the experiments (R2 = 0.032 and 0.02 respectively).
FGFR2 knockdown suppresses hMSC osteogenesis
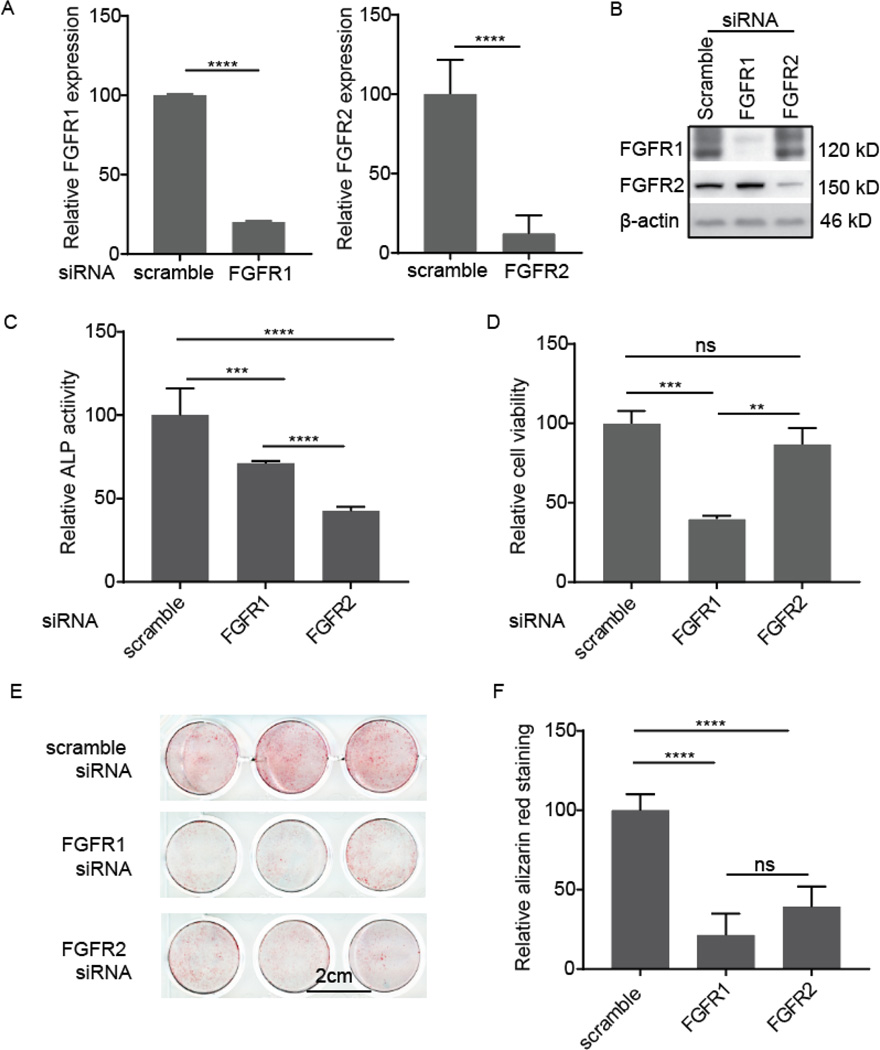
To further determine the potential role of FGFR2 expression in hMSC osteogenesis, we performed gene knockdown using siRNA transfection. Gene downregulation was confirmed by both qPCR and Western blot. FGFR1 siRNA transfection inhibited 80% of FGFR1 expression; similar knockdown efficiency was achieved with FGFR2 siRNA (Fig. 3A). Western blotting confirmed these results (Fig. 3B). FGFR1 siRNA treatment almost completely inhibited FGFR1 protein expression and FGFR2 siRNA also effectively inhibited FGFR2 protein expression.
Figure 3.
Effect of FGFR1 and FGFR2 knockdown on the osteogenesis of hMSCs. A) siRNA knockdown efficiency by qPCR analysis from either three (FGFR1) and six (FGFR2) separate wells from a single experiment. B) siRNA knockdown efficiency as assessed by Western blot from a single experiment. C) ALP activity after siRNA transfection. hMSCs were cultured in osteogenic medium for 8 days. Absorbance at 405 nm was normalized against that of the scrambled siRNA-treated cells. Data were from six wells from a single experiment. D) Cell viability in maintenance media measured 4 days after siRNA transfection from triplicate wells from a single experiment. Absorbance data at 450 nm were normalized against that of the control scrambled siRNA-treated cells. E) Alizarin red staining in siRNA-treated cells for mineral deposition after 21 days in osteogenic media. Data from triplicate wells from a single experiment. F) Quantitative analysis of Alizarin red staining in (E).
ALP activity was assessed after growing the cells in osteogenic medium for 8 days. FGFR1 siRNA treatment resulted in a ∼30% reduction in ALP activity, whereas FGFR2 siRNA treatment reduced ALP activity by ∼60%, as compared to the scrambled-siRNA treated cells (Fig. 3C). Moreover, loss of FGFR2 expression resulted in ALP activity levels that were ∼41% lower than those resulting from a loss of FGFR1 expression. As FGFR1 is crucial for hMSC proliferation and survival, we postulated that the reduction in ALP activity following FGFR1 downregulation might be linked to cell survival. This was assessed by a cell viability assay. Four days post siRNA treatment, loss of FGFR1 expression resulted in ∼60% cell death. In comparison, loss of FGFR2 did not affect cell viability (Fig. 3D).
The mineral deposition was evaluated by staining cells with Alizarin Red after 21 days in osteogenic medium. Similar to the ALP assay, we observed a reduction in osteogenic commitment (Fig. 3E) in both the FGFR1 and FGFR2 siRNA knockdown conditions. In the case of FGFR1 knockdown, loss of cell viability was observed (Fig. 3D) that may be linked to the ∼ 70% reduction in mineralization. In comparison, FGFR2 loss did not adversely affect cell viability (Fig. 3D), yet there was ∼ 60% reduction in matrix mineralization.
FGFR2 knockdown modulates the expression of several osteogenic genes
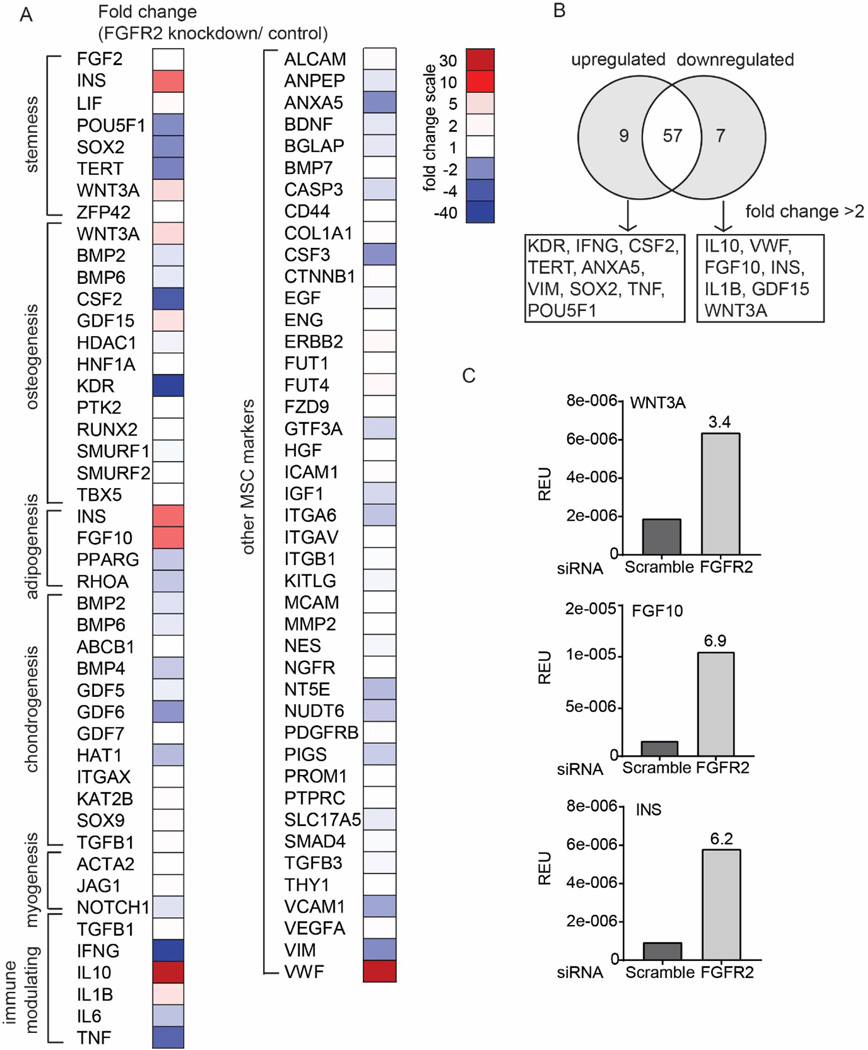
To help determine a link between FGFR2 and osteogenic potential, an 84 primer-pair qPCR array was utilized to identify mRNA transcripts expressed in hMSCs with siRNA-knockdown FGFR2 expression (Fig. 4). Out of a possible 84 genes, 73 transcripts were expressed on the array and the fold-change (FGFR2 knockdown/control) presented as a heatmap (Fig. 4A). A 2-fold cut-off was also applied to highlight differential expression of these transcripts (Fig. 4B). Amongst the 73 genes, 7 were upregulated following FGFR2 knockdown and 9 downregulated (>2-fold change) (Fig. 4B). Of the 7 upregulated genes, most were growth factors and cytokines known to mediate cell differentiation (FGF10, INS, GDF15, WNT3A) or to modulate immune responses (IL10 and IL1B). In comparison, genes downregulated were involved in stem cell maintenance (SOX2, POU5F1, TERT), cell fate (ANXA5, VIM), or immunomodulation (TNF, IFNG). Notably, the osteogenic factor (WNT3A) and the adipogenic factors (FGF10 and INS) were amongst the most strongly 16 mRNA transcripts differentially regulated by FGFR2 knockdown, WNT3A (osteogenic factor) and FGF10 and INS (adipogenic factors) were upregulated more than 3-fold (Fig. 4C).
Figure 4.
Gene expression profile of FGFR2 siRNA-transfected cells cultured in maintenance media in comparison to control scrambled siRNA-transfected cells from a single experiment. A) Heatmap summarizing the relative fold-change in gene expression (FGFR2 siRNA/scramble control siRNA). Gene function clustering was according to manufacturer’s guide with modification based on established studies from literature. B) Venn diagram showing genes affected by FGFR2 knockdown (fold change >2). C) Bar graph depicts relative gene expression (normalized to actin) for transcripts related to osteogenesis and adipogenesis. The number on the bar chart showed fold change (FGFR2 siRNA/scramble control siRNA).
FGFR2 knockdown enhances hMSC adipogenesis
To further determine the effect of FGFR2 on adipogenesis, hMSCs were transfected with FGFR2 siRNA or a scrambled control siRNA and maintained in adipogenic medium for either 14 (qPCR), or 21 days (Oil Red O staining). PPARG and CEBPA mRNA level were analysed. Both are transcription factors activated at early stages of adipogenesis and are essential for induction and maintain of the adipocyte phenotype (Wu et al., 1999; Imai et al., 2004). Knockdown of FGFR2 (knockdown efficiency is shown in Supp. Fig. 3) resulted in ∼ 2-fold increase in CEBPA expression and a 4-fold increase in PPARG expression (Fig. 5A). These data were supported by an increase in lipid deposits in the FGFR2 siRNA treated group (Fig. 5B).
Figure 5.
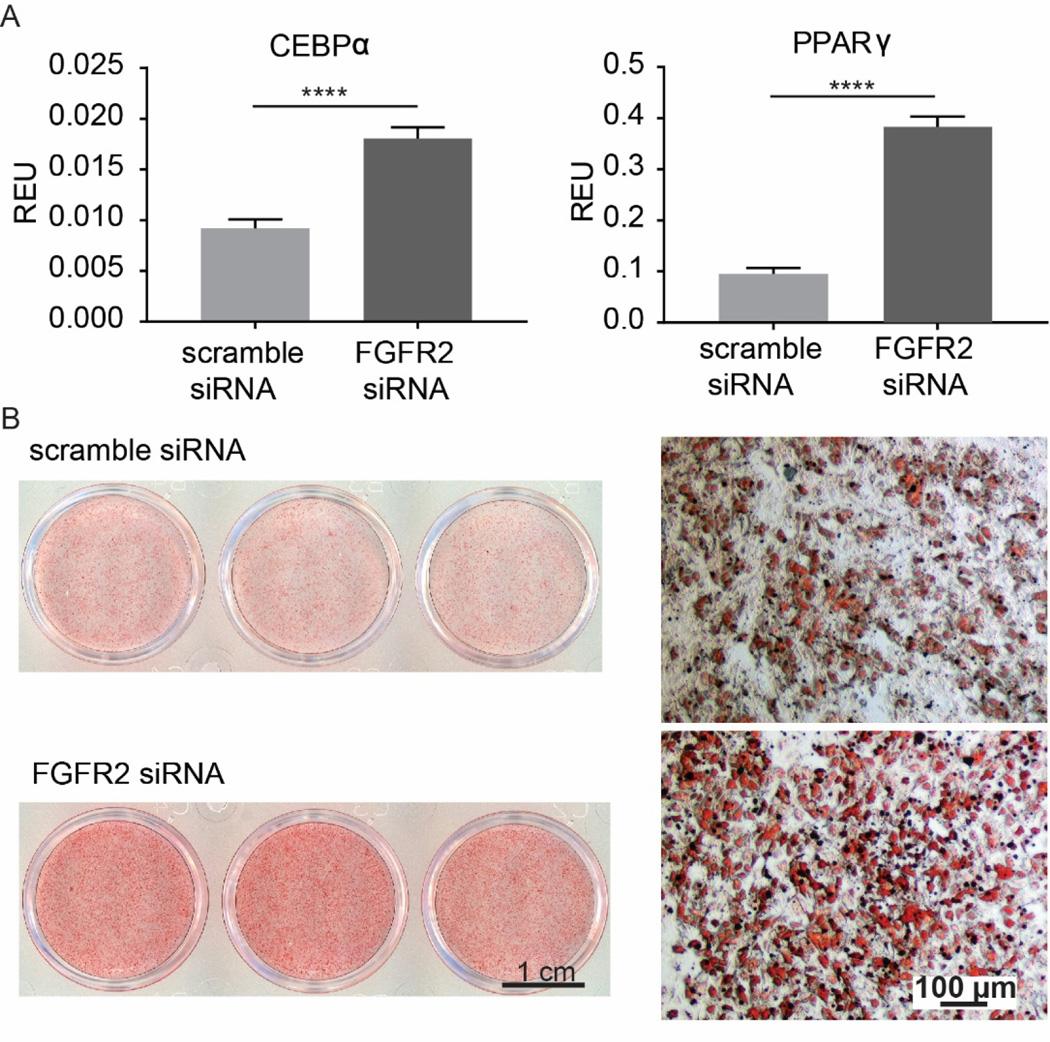
Effect of FGFR2 knockdown on adipogenic differentiation of hMSCs. Data from triplicate wells from a single experiment. A) Expression of the adipogenic markers (CEBPα and PPARγ) in FGFR2 siRNA- and control scrambled siRNA-treated cells after 14 days in adipogenic media. B) Lipid formation (Oil red O staining) in FGFR2 siRNA- and control scrambled siRNA-treated cells after 21 days in adipogenic media. ****, p < 0.0001.
Moreover, we have confirmed the effect of FGFR2 knockdown on osteogenic and adiogenic lineage fate decisions in hMSCs from two additional donors. Downregulation of FGFR2 in MSCs from these additional donors resulted in decreased osteogenic and increased adipogenic progression (Suppl.Fig. 4). The results were consistent with the earlier observations from donor 1.
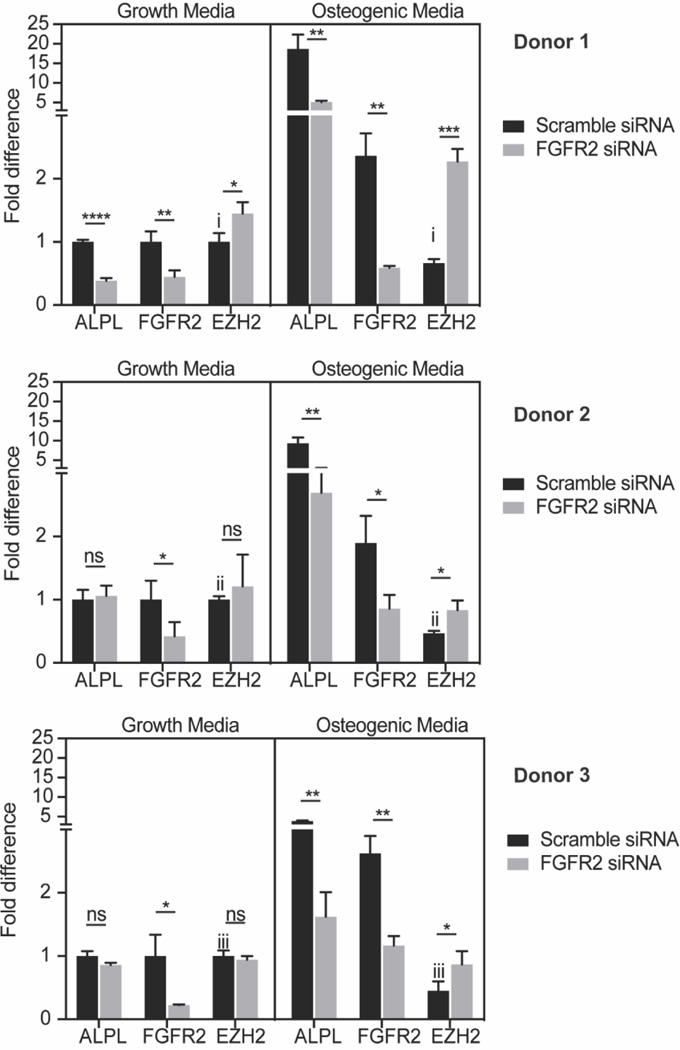
FGFR2 expression negatively correlates with EZH2 expression
The histone 3 lysine 27 (H3K27) methyl transferase EZH2 has been shown to control lineage specification in MSCs (Hemming et al., 2014; Dudakovic et al., 2015; Hemming et al., 2016; Sen et al., 2020) and suppresses osteogenesis in vivo (Dudakovic et al., 2016) (Hemming et al., 2017). Because EZH2 levels are downregulated during osteogenesis and FGFR2 loss of function decreases osteogenic differentiation, we hypothesized that FGFR2 may regulate EZH2 levels. Therefore, we examined whether siRNA depletion of FGFR2 expression would modulate EZH2 levels. Consistent with our hypothesis, MSCs cultured in osteogenic media showed reduced EZH2 expression as compared to MSCs cultured in growth media in three different donor cells (scramble siRNA treated MSCs in Fig. 6). When MSCs were cultured in normal growth media, FGFR2 knockdown did not change EZH2 levels in cell from donor 2 and donor 3 (Fig. 6). While in MSCs from donor 1, EZH2 was upregulated (∼20%). Notably, when MSCs were stimulated under osteogenic conditions, FGFR2 knockdown resulted in a significant increase in EZH2 levels in the MSCs from all three donors. These results suggest that FGFR2 levels control EZH2 expression and that epigenetic changes regulated by EZH2 may contribute to the reduced osteogenic differentiation observed upon FGFR2 knockdown in MSCs.
Figure 6.
FGFR2 knockdown and EZH2 expression in MSCs from three donors cultured in growth and osteogenic media. For each donor, data were from triplicate wells. Relative expression of genes were normalized to the data from cells transfected with scramble siRNA and cultured in growth media. (i: *; ii: ***; iii: ** )
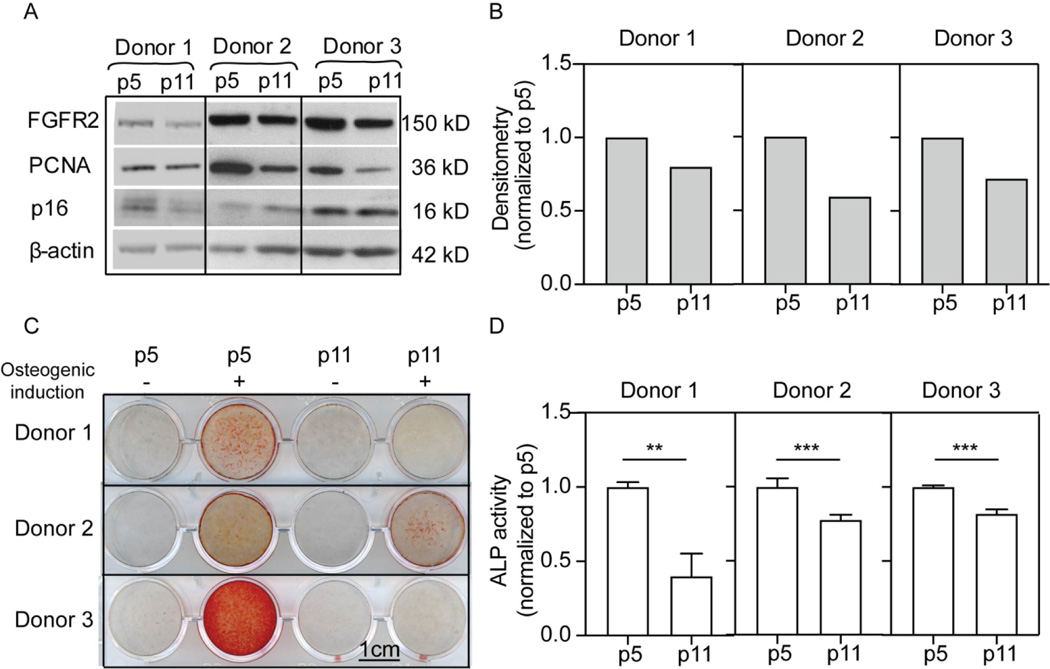
FGFR2 expression is reduced in serially-passaged MSCs
Reduced osteogenic potential and enhanced adipogenesis is also a characteristic of MSC aging. Several studies report that MSCs from aged mice have drastically lowered osteogenic potential (Baht et al., 2015; Lin et al., 2017; Infante and Rodriguez, 2018). For human MSCs, serially passaging (passage 8) has been shown to reduce both ALP activity and matrix mineralization (Yang et al., 2018). However, it remains to be determined whether FGFR2 is involved in this in vitro ageing phenotype. To evaluate this further, MSCs from three donors were serial passaged and changes in FGFR2 levels evaluated by Western blot. Levels of ALP activity and matrix mineralization (Alizarin red staining) were also determined to compare osteogenic potential between early (p5) and late (p11) passage MSCs (Fig. 7). ALP activity and Alizarin red staining confirmed that late passage MSCs showed reduced osteogenic potential (Figure 7C, D), as previously reported (Yang et al., 2018). Western blot data showed that late-passage hMSCs expressed reduced levels of PCNA, increased p16, and reduced FGFR2 expression (Fig. 7A,B). These results suggest a possible relationship between cellular age, FGFR2 expression and osteogenic potential.
Figure 7.
FGFR2 expression and osteogenesis in early and late passage MSCs. A) Western blot for protein levels in early and late passage cells from three separate donors. B) Densitometry values for FGFR2 levels in A. C) Alizarin red staining of matrix from early and late passage MSCs from three donors (triplicates were performed for each condition and the data are presented in Supplementary Figure 5). D) ALP activity of early and late passage MSCs from three separate donors.
Discussion
Understanding the conditions which drive lineage commitment is pivotal for the therapeutic development of hMSCs for the purposes of tissue regeneration. The regulation of bone homeostasis and regeneration is necessary to improve outcomes not only for patients suffering severe trauma but for the demographically increasing numbers of the elderly. Although numerous physical, chemical and biological signals have been identified that influence lineagespecific differentiation, the exact mechanisms and molecular events underlying the differentiation process remain to be elucidated. FGFs and their cognate receptors are essential for both the proliferation and lineage-commitment of hMSCs (Coutu and Galipeau, 2011). In this study, we analyzed the expression of FGFRs in order to elucidate their respective functions during the process of osteogenesis.
Here, we show in hMSCs that osteogenic differentiation is initiated following a phase of cell proliferation that plateaus when the cells form a monolayer and contact inhibit. In parallel, FGFR1 levels also increase during hMSC proliferation and decline as the cells became confluent. Moreover, knockdown of FGFR1 resulted in decreased cell viability, a finding that supports our earlier study demonstrating that FGFR1 regulates hMSC proliferation (Dombrowski et al., 2013). Unlike FGFR1 inhibition, FGFR2 knockdown did not affect cell viability; instead, we observed decreased expression of ALP and a loss of mineral deposition. This suggests that FGFR2 is important for osteogenic differentiation. Future studies that normalize mineralization levels with cell numbers would provide more direct support for this conclusion. Nonetheless, our observation is consistent with a previous study wherein overexpressing FGFR2 in murine MSCs enhanced osteogenic activity via ERK1/2 expression (Miraoui et al., 2009). This study also showed that FGFR2 overexpression increased MSC proliferation without adversely affecting cell survival. Also, evidence suggests that FGFR2 gain of function mutations (S252W) observed in Apert syndrome reduce growth potential and increase osteogenic potential of MSCs (Yeh et al., 2012). Collectively, these studies highlight the importance of FGFR2 in controlling osteogenesis, compared to FGFR1 that may be more prominent in regulating MSC proliferation and survival.
Since FGFR1 or FGFR2 knockout is embryonic lethal (Su et al., 2014), conditional knockout strategies have been employed to examine the roles of these receptors during in vivo osteogenesis. These data show that loss of FGFR1 decreases the proliferation of mesenchymal progenitors, which results in reduced cartilage condensation sizes within the bone (Verheyden et al., 2005). Moreover, loss of FGFR1 in osteo-chondro-progenitors delays their osteogenic maturation, yet lineage commitment is unaffected, as evidenced by normal Runx2 expression (Jacob et al., 2006). These results further support a role for FGFR1 in mediating self-renewal of MSCs and proliferation of early osteoblast progenitors. In contrast, conditional knockdown of FGFR2 causes reduced bone mineral density and shortened stature, as well as the levels of the osteogenic markers RUNX2, COL1A and osteocalcin (Yu et al., 2003). In addition to such osteogenic effects, suppression of FGFR2 activity (by miR-223) has also been shown to enhance adipogenesis (Guan et al., 2015). These results suggest that FGFR2 is important in regulating osteogenic and adipogenic fates necessary for bone homeostasis.
Dysregulation of the osteo-adipogenic balance is associated with several pathophysiologic conditions, including aging (Moerman et al., 2004), obesity (Misra and Klibanski, 2013), osteopetrosis (Schwartz et al., 2013) and osteoporosis (Cohen et al., 2012). For example, the most common bone remodeling disorder, osteoporosis, is associated with increased deposition of adipose tissue within the bone marrow and reduced osteogenic potential (Meunier et al., 1971). Our results showed that knockdown of FGFR2 upregulates the expression of pro-adipogenic factors such as FGF10 and insulin in hMSCs. FGF10 signaling is essential for preadipocyte proliferation and differentiation, as the development of adipose tissue in FGF10 knockout mice is severely impaired (Sakaue et al., 2002). Insulin is widely used as a medium supplement to induce adipogenesis in vitro (Klemm et al., 2001). Our results also show that FGFR2 knockdown upregulates factors that suppress osteogenic differentiation, such as WNT3A. Notably, exogenous application of WNT3A has been shown to suppress matrix mineralization and ALP activity (Boland et al., 2004; de Boer et al., 2004; Ling et al., 2009). Furthermore, we observed an increase in GDF15 expression following FGFR2 knockdown, a factor which negatively regulates osteoblast differentiation (Westhrin et al., 2015). Together, these data support the role of FGFR2 in controlling the balance of osteo-adipogenic cell fate.
We also showed that under osteogenic induction, FGFR2 and EZH2 levels are inversely related. EZH2 levels are reduced and FGFR2 expression increased when cells underwent osteogenic stimulation. These data suggest that FGFR2 may regulate lineage commitment of MSCs at the epigenetic level by modulating EZH2 expression. Notably, overexpression of EZH2 in MSCs suppresses their in vitro osteogenic potential and in vivo bone forming capacity (Hemming et al., 2014). While increased EZH2 expression has been observed in MSCs under adipogenic induction (Hemming et al., 2014) knockdown of EZH2 was able to recover osteogenic commitment in the MSCs and restore the balance between adipogenesis and osteogenesis. In addition, pharmacological inhibitors that decrease either EZH2 activity (Dudakovic et al., 2015; Galvan et al., 2021) or protein levels (Samsonraj et al., 2018) promote osteogenic differentiation. Collectively, these data highlight the need for continuing studies aimed at understanding the reciprocal functional linkage between FGFR2 and EZH2 levels in regulating the osteogenic potential of MSCs and the epigenetic changes associated with altered EZH2. Furthermore, agonists that activate FGFR2 signalling could be combined with established pharmacological strategies for blocking EZH2 activity. The latter would leverage both epigenetic priming of chromatin in the nucleus (Dudakovic et al., 2020) and (Velletri et al.)agonist induced FGFR2 signalling at the cell surface to accelerate osteogenic lineage commitment and progression.
During aging, impaired MSC function leads to dysregulated balance between osteo-adipogenic commitment. As a result, bone formation is affected and there is an increased risk for osteoporosis (Chen et al., 2016; Infante and Rodriguez, 2018). The expression of the adipogenic PPARG was highly upregulated in aged murine MSCs (Moerman et al., 2004). Forkhead box P1 (FOXP1), which negatively regulates adipogenesis by inhibiting CEBPB were found to decline significantly in old mice (Li et al., 2017). In contrast, the Core-binding factor subunit beta (CBFβ), which is a co-factor for RUNX2, was reduced in aged MSCs in mice(Wu et al., 2017). While changes transcription factors that regulate osteo-adipogenesis are well studied, there are few studies on the upstream signaling events that govern these changes. Our study suggests FGFR2 may function as an essential upstream signal to control the balance of osteo-adipogenic differentiation. We show that late passage hMSCs have reduced FGFR2 expression and concomitantly reduced ALP activity when cultured in osteogenic conditions. Supporting our finding, a study of hMSCs from 61 donors, aged 17 to 84, revealed that FGFR2 is one of the top 3 genes correlated with the age of donors(Wu et al., 2017) . Future studies manipulating the expression of FGFR2 in MSCs from donors of different age would be beneficial to establishing a cause-effect relationship between FGFR2 decline and reduced osteogenic capacity of aged MSCs.
In summary, this study provides new evidence suggesting that FGFR2 is required for the onset of osteogenic differentiation. We show that FGFR2 plays a pivotal role in mediating the osteo-adipogenic balance by upregulating osteogenic gene expression and downregulating adipogenic gene expression in concert with changes in EZH2 levels. These findings support the important function of FGF receptors in the control of osteogenesis and lay the foundation for strategies that seek to improve hMSC fate decisions.
Supplementary Material
Research Highlight.
Fibroblast growth factor (FGF) signaling is essential in regulating bone homeostasis. FGF receptor 2 (FGFR2) level correlates with in vitro osteogenesis of human mesenchymal stem cells (hMSCs). Using siRNA knockdown and PCR array profiling, we demonstrate that FGFR2 regulates bone homeostasis by upregulating osteogenic factors and suppressing adipogenic factors.
Acknowledgments
We would like to thank Dr. Padmapriya Sathiyanathan from the Genome Institute of Singapore (GIS) for her comments and feedback on the manuscript. This work was funded by the National Medical Research Council (NMRC), Singapore under the Bedside and Bench Grant Call (NMRC Project No. NMRC/BnB/0003b/2013) and continued under Industry Alignment Fund Pre-Positioning (IAF-PP) funding (H18/01/a0/021 and H18/AH/a0/001) from the Agency for Science, Technology and Research (A*STAR), Singapore. We would also like to thank the Institute of Molecular and Cell Biology (IMCB) and the Biomedical Research Council (BMRC) from the Agency for Science, Technology and Research (A*STAR) for their general support. Additional funding was provided by National Institutes of Health grant R01 AR049069 (to AJvW).
Abbreviation list
- hMSCs
human mesenchymal stem cells
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- ALP
alkaline phosphatase
- EZH2
enhancer of Zeste Homolog 2
- RUNX2
Runt-related transcription factor 2
- COL1A
type 1 collagen
- BMP
bone morphogenetic protein
- DMEM
Dulbecco’s Modified Eagle Medium
- BCA
bicinchoninic acid assay
- ANOVA
analysis of variance
- CEBPA
CCAAT Enhancer Binding Protein Alpha
- PPARG
peroxisome Proliferator Activated Receptor Gamma
- PCNA
proliferation cell nuclear antigen
Footnotes
Credit Author Statement
Ying Zhang: Conceptualization, Investigation, Data curation, Visualization, Writing-original draft.
Ling Ling: Project administration, Writing-Review & Editing
Arya Ajay D/O Ajayakumar: Investigation for part of Figure 7
Yating Michelle Eio: Investigation for part of Figure 7
Andre J. van Wijnen, Writing-Review & Editing
Victor Nurcombe: Writing-Review & Editing, Funding acquisition
Simon M. Cool: Conceptualization, Validation, Resources, Writing - Review & Editing, Supervision, Funding acquisition
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors have no conflict of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Uncategorized References
- Baht GS, Silkstone D, Vi L, Nadesan P, Amani Y, Whetstone H, Wei Q and Alman BA, 2015. Exposure to a youthful circulaton rejuvenates bone repair through modulation of beta-catenin. Nat Commun 6, 7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland GM, Perkins G, Hall DJ and Tuan RS, 2004. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J Cell Biochem 93, 1210–30. [DOI] [PubMed] [Google Scholar]
- Caplan AI, 1991. Mesenchymal stem cells. J Orthop Res 9, 641–50. [DOI] [PubMed] [Google Scholar]
- Charoenlarp P, Rajendran AK and Iseki S, 2017. Role of fibroblast growth factors in bone regeneration. Inflamm Regen 37, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Deng C and Li YP, 2012. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci 8, 272–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q, Cao J, Xie N, Velletri T, Zhang X, Xu C, Zhang L, Yang H, Hou J, Wang Y and Shi Y, 2016. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ 23, 1128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KM, Seo YK, Yoon HH, Song KY, Kwon SY, Lee HS and Park JK, 2008. Effect of ascorbic acid on bone marrow-derived mesenchymal stem cell proliferation and differentiation. J Biosci Bioeng 105, 586–94. [DOI] [PubMed] [Google Scholar]
- Cohen A, Dempster DW, Stein EM, Nickolas TL, Zhou H, McMahon DJ, Muller R, Kohler T, Zwahlen A, Lappe JM, Young P, Recker RR and Shane E, 2012. Increased marrow adiposity in premenopausal women with idiopathic osteoporosis. J Clin Endocrinol Metab 97, 2782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutu DL and Galipeau J, 2011. Roles of FGF signaling in stem cell self-renewal, senescence and aging. Aging (Albany NY) 3, 920–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, Siddappa R, Gaspar C, van Apeldoorn A, Fodde R and van Blitterswijk C, 2004. Wnt signaling inhibits osteogenic differentiation of human mesenchymal stem cells. Bone 34, 818–26. [DOI] [PubMed] [Google Scholar]
- Deng C, Wynshaw-Boris A, Zhou F, Kuo A and Leder P, 1996. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84, 911–21. [DOI] [PubMed] [Google Scholar]
- Dombrowski C, Helledie T, Ling L, Grunert M, Canning CA, Jones CM, Hui JH, Nurcombe V, van Wijnen AJ and Cool SM, 2013. FGFR1 signaling stimulates proliferation of human mesenchymal stem cells by inhibiting the cyclin-dependent kinase inhibitors p21(Waf1) and p27(Kip1). Stem Cells 31, 2724–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri ET, Riester SM, Paradise CR, Gluscevic M, O’Toole TM, Thaler R, Evans JM, Yan H, Subramaniam M, Hawse JR, Stein GS, Montecino MA, McGee-Lawrence ME, Westendorf JJ and van Wijnen AJ, 2016. Enhancer of Zeste Homolog 2 Inhibition Stimulates Bone Formation and Mitigates Bone Loss Caused by Ovariectomy in Skeletally Mature Mice. J Biol Chem 291, 24594–24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler R, Deyle DR, Larson AN, Lewallen DG, Dietz AB, Stein GS, Montecino MA, Westendorf JJ and van Wijnen AJ, 2015. Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2. J Biol Chem 290, 27604–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Samsonraj RM, Paradise CR, Galeano-Garces C, Mol MO, Galeano-Garces D, Zan P, Galvan ML, Hevesi M, Pichurin O, Thaler R, Begun DL, Kloen P, Karperien M, Larson AN, Westendorf JJ, Cool SM and van Wijnen AJ, 2020. Inhibition of the epigenetic suppressor EZH2 primes osteogenic differentiation mediated by BMP2. J Biol Chem 295, 7877–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan ML, Paradise CR, Kubrova E, Jerez S, Khani F, Thaler R, Dudakovic A and van Wijnen AJ, 2021. Multiple pharmacological inhibitors targeting the epigenetic suppressor enhancer of zeste homolog 2 (Ezh2) accelerate osteoblast differentiation. Bone 150, 115993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Gao Y, Zhou J, Wang J, Zheng F, Guo F, Chang A, Li X and Wang B, 2015. miR-223 Regulates Adipogenic and Osteogenic Differentiation of Mesenchymal Stem Cells Through a C/EBPs/miR-223/FGFR2 Regulatory Feedback Loop. Stem Cells 33, 1589–600. [DOI] [PubMed] [Google Scholar]
- Hemming S, Cakouros D, Codrington J, Vandyke K, Arthur A, Zannettino A and Gronthos S, 2017. EZH2 deletion in early mesenchyme compromises postnatal bone microarchitecture and structural integrity and accelerates remodeling. FASEB J 31, 1011–1027. [DOI] [PubMed] [Google Scholar]
- Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A and Gronthos S, 2014. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 32, 802–15. [DOI] [PubMed] [Google Scholar]
- Hemming S, Cakouros D, Vandyke K, Davis MJ, Zannettino AC and Gronthos S, 2016. Identification of Novel EZH2 Targets Regulating Osteogenic Differentiation in Mesenchymal Stem Cells. Stem Cells Dev 25, 909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Takakuwa R, Marchand S, Dentz E, Bornert JM, Messaddeq N, Wendling O, Mark M, Desvergne B, Wahli W, Chambon P and Metzger D, 2004. Peroxisome proliferator-activated receptor gamma is required in mature white and brown adipocytes for their survival in the mouse. Proc Natl Acad Sci U S A 101, 4543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante A and Rodriguez CI, 2018. Osteogenesis and aging: lessons from mesenchymal stem cells. Stem Cell Res Ther 9, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RA, Kumarasuriyar A, Nurcombe V and Cool SM, 2006. Long-term loading inhibits ERK1/2 phosphorylation and increases FGFR3 expression in MC3T3-E1 osteoblast cells. J Cell Physiol 209, 894–904. [DOI] [PubMed] [Google Scholar]
- Jacob AL, Smith C, Partanen J and Ornitz DM, 2006. Fibroblast growth factor receptor 1 signaling in the osteo-chondrogenic cell lineage regulates sequential steps of osteoblast maturation. Dev Biol 296, 315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal N, Haynesworth SE, Caplan AI and Bruder SP, 1997. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J Cell Biochem 64, 295312. [PubMed] [Google Scholar]
- Jaukovic A, Abadjieva D, Trivanovic D, Stoyanova E, Kostadinova M, Pashova S, Kestendjieva S, Kukolj T, Jeseta M, Kistanova E and Mourdjeva M, 2020. Specificity of 3D MSC Spheroids Microenvironment: Impact on MSC Behavior and Properties. Stem Cell Rev Rep 16, 853–875. [DOI] [PubMed] [Google Scholar]
- Kahkonen TE, Ivaska KK, Jiang M, Buki KG, Vaananen HK and Harkonen PL, 2018. Role of fibroblast growth factor receptors (FGFR) and FGFR like-1 (FGFRL1) in mesenchymal stromal cell differentiation to osteoblasts and adipocytes. Mol Cell Endocrinol 461, 194–204. [DOI] [PubMed] [Google Scholar]
- Klemm DJ, Leitner JW, Watson P, Nesterova A, Reusch JE, Goalstone ML and Draznin B, 2001. Insulin-induced adipocyte differentiation. Activation of CREB rescues adipogenesis from the arrest caused by inhibition of prenylation. J Biol Chem 276, 28430–5. [DOI] [PubMed] [Google Scholar]
- Li H, Liu P, Xu S, Li Y, Dekker JD, Li B, Fan Y, Zhang Z, Hong Y, Yang G, Tang T, Ren Y, Tucker HO, Yao Z and Guo X, 2017. FOXP1 controls mesenchymal stem cell commitment and senescence during skeletal aging. J Clin Invest 127, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL and Stein GS, 2004. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr 14, 1–41. [PubMed] [Google Scholar]
- Lin TH, Gibon E, Loi F, Pajarinen J, Cordova LA, Nabeshima A, Lu L, Yao Z and Goodman SB, 2017. Decreased osteogenesis in mesenchymal stem cells derived from the aged mouse is associated with enhanced NF-kappaB activity. J Orthop Res 35, 281–288. [DOI] [PubMed] [Google Scholar]
- Ling L, Nurcombe V and Cool SM, 2009. Wnt signaling controls the fate of mesenchymal stem cells. Gene 433, 1–7. [DOI] [PubMed] [Google Scholar]
- Lisignoli G, Zini N, Remiddi G, Piacentini A, Puggioli A, Trimarchi C, Fini M, Maraldi NM and Facchini A, 2001. Basic fibroblast growth factor enhances in vitro mineralization of rat bone marrow stromal cells grown on non-woven hyaluronic acid based polymer scaffold. Biomaterials 22, 2095–105. [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K and Chen CS, 2004. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell 6, 483–95. [DOI] [PubMed] [Google Scholar]
- Meunier P, Aaron J, Edouard C and Vignon G, 1971. Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. A quantitative study of 84 iliac bone biopsies. Clin Orthop Relat Res 80, 147–54. [DOI] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Sen B, Rubin J and Pike JW, 2016. Epigenetic Plasticity Drives Adipogenic and Osteogenic Differentiation of Marrow-derived Mesenchymal Stem Cells. J Biol Chem 291, 17829–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miraoui H, Oudina K, Petite H, Tanimoto Y, Moriyama K and Marie PJ, 2009. Fibroblast growth factor receptor 2 promotes osteogenic differentiation in mesenchymal cells via ERK1/2 and protein kinase C signaling. J Biol Chem 284, 4897–904. [DOI] [PubMed] [Google Scholar]
- Misra M and Klibanski A, 2013. Anorexia nervosa, obesity and bone metabolism. Pediatr Endocrinol Rev 11, 21–33. [PMC free article] [PubMed] [Google Scholar]
- Moerman EJ, Teng K, Lipschitz DA and Lecka-Czernik B, 2004. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell 3, 379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD and Hurley MM, 2000. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest 105, 1085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Hara Y, Tagawa M, Tamura M, Yuge T, Fukuda H and Nigi H, 1998. Recombinant human basic fibroblast growth factor accelerates fracture healing by enhancing callus remodeling in experimental dog tibial fracture. J Bone Miner Res 13, 942–9. [DOI] [PubMed] [Google Scholar]
- Neuhuber B, Swanger SA, Howard L, Mackay A and Fischer I, 2008. Effects of plating density and culture time on bone marrow stromal cell characteristics. Exp Hematol 36, 1176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM and Marie PJ, 2002. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev 16, 1446–65. [DOI] [PubMed] [Google Scholar]
- Perez-Campo FM and Riancho JA, 2015. Epigenetic Mechanisms Regulating Mesenchymal Stem Cell Differentiation. Curr Genomics 16, 368–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prockop DJ, 1997. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 276, 71–4. [DOI] [PubMed] [Google Scholar]
- Rider DA, Dombrowski C, Sawyer AA, Ng GH, Leong D, Hutmacher DW, Nurcombe V and Cool SM, 2008. Autocrine fibroblast growth factor 2 increases the multipotentiality of human adipose-derived mesenchymal stem cells. Stem Cells 26, 1598–608. [DOI] [PubMed] [Google Scholar]
- Sakaue H, Konishi M, Ogawa W, Asaki T, Mori T, Yamasaki M, Takata M, Ueno H, Kato S, Kasuga M and Itoh N, 2002. Requirement of fibroblast growth factor 10 in development of white adipose tissue. Genes Dev 16, 908–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonraj RM, Dudakovic A, Manzar B, Sen B, Dietz AB, Cool SM, Rubin J and van Wijnen AJ, 2018. Osteogenic Stimulation of Human Adipose-Derived Mesenchymal Stem Cells Using a Fungal Metabolite That Suppresses the Polycomb Group Protein EZH2. Stem Cells Transl Med 7, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz AV, Sigurdsson S, Hue TF, Lang TF, Harris TB, Rosen CJ, Vittinghoff E, Siggeirsdottir K, Sigurdsson G, Oskarsdottir D, Shet K, Palermo L, Gudnason V and Li X, 2013. Vertebral bone marrow fat associated with lower trabecular BMD and prevalent vertebral fracture in older adults. J Clin Endocrinol Metab 98, 2294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen B, Paradise CR, Xie Z, Sankaran J, Uzer G, Styner M, Meyer M, Dudakovic A, van Wijnen AJ and Rubin J, 2020. beta-Catenin Preserves the Stem State of Murine Bone Marrow Stromal Cells Through Activation of EZH2. J Bone Miner Res 35, 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simann M, Le Blanc S, Schneider V, Zehe V, Ludemann M, Schutze N, Jakob F and Schilling T, 2017. Canonical FGFs Prevent Osteogenic Lineage Commitment and Differentiation of Human Bone Marrow Stromal Cells Via ERK1/2 Signaling. J Cell Biochem 118, 263–275. [DOI] [PubMed] [Google Scholar]
- Smith RAA, Murali S, Rai B, Lu X, Lim ZXH, Lee JJL, Nurcombe V and Cool SM, 2018. Minimum structural requirements for BMP-2-binding of heparin oligosaccharides. Biomaterials 184, 41–55. [DOI] [PubMed] [Google Scholar]
- Su N, Jin M and Chen L, 2014. Role of FGF/FGFR signaling in skeletal development and homeostasis: learning from mouse models. Bone Res 2, 14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velletri T, Huang Y, Wang Y, Li Q, Hu M, Xie N, Yang Q, Chen X, Chen Q, Shou P, Gan Y, Candi E, Annicchiarico-Petruzzelli M, Agostini M, Yang H, Melino G, Shi Y and Wang Y, 2020. Loss of p53 in mesenchymal stem cells promotes alteration of bone remodeling through negative regulation of osteoprotegerin. Cell Death Differ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyden JM, Lewandoski M, Deng C, Harfe BD and Sun X, 2005. Conditional inactivation of Fgfr1 in mouse defines its role in limb bud establishment, outgrowth and digit patterning. Development 132, 4235–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Green RP, Zhao G and Ornitz DM, 2001. Differential regulation of endochondral bone growth and joint development by FGFR1 and FGFR3 tyrosine kinase domains. Development 128, 3867–76. [DOI] [PubMed] [Google Scholar]
- Westhrin M, Moen SH, Holien T, Mylin AK, Heickendorff L, Olsen OE, Sundan A, Turesson I, Gimsing P, Waage A and Standal T, 2015. Growth differentiation factor 15 (GDF15) promotes osteoclast differentiation and inhibits osteoblast differentiation and high serum GDF15 levels are associated with multiple myeloma bone disease. Haematologica 100, e511–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AO, 2002. Abnormal spliceform expression associated with splice acceptor mutations in exon IIIc of FGFR2. Am J Med Genet 111, 105. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Patey SJ, Kan SH, van den Ouweland AM and Hamel BC, 2002. FGFs, their receptors, and human limb malformations: clinical and molecular correlations. Am J Med Genet 112, 266–78. [DOI] [PubMed] [Google Scholar]
- Wu M, Wang Y, Shao JZ, Wang J, Chen W and Li YP, 2017. Cbfbeta governs osteoblast-adipocyte lineage commitment through enhancing beta-catenin signaling and suppressing adipogenesis gene expression. Proc Natl Acad Sci U S A 114, 10119–10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ and Spiegelman BM, 1999. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 3, 151–8. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Peperzak V, van Rijn L, Borst J and de Bruijn JD, 2010. Dexamethasone treatment during the expansion phase maintains stemness of bone marrow mesenchymal stem cells. J Tissue Eng Regen Med 4, 374–86. [DOI] [PubMed] [Google Scholar]
- Yang YK, Ogando CR, Wang See C, Chang TY and Barabino GA, 2018. Changes in phenotype and differentiation potential of human mesenchymal stem cells aging in vitro. Stem Cell Res Ther 9, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Atique R, Ishiy FA, Fanganiello RD, Alonso N, Matushita H, da Rocha KM and Passos-Bueno MR, 2012. FGFR2 mutation confers a less drastic gain of function in mesenchymal stem cells than in fibroblasts. Stem Cell Rev Rep 8, 685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA and Ornitz DM, 2003. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063–74. [DOI] [PubMed] [Google Scholar]
- Zellin G and Linde A, 2000. Effects of recombinant human fibroblast growth factor-2 on osteogenic cell populations during orthopic osteogenesis in vivo. Bone 26, 161–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.