Abstract
The eukaryotic cap-binding proteins belonging to the eIF4E family are generally involved in mediating the recruitment of ribosomes to capped mRNA. We described previously a cap-binding protein (now called eIF4E1) in Schizosaccharomyces pombe that appears to have all of the usual structural and functional attributes of an eIF4E. We have now characterised a new type of cap-binding protein (eIF4E2) from this organism, which at the amino acid sequence level, is 52% identical and 59% similar to eIF4E1. eIF4E2 is not essential in S.pombe but has some novel properties that may be related to a special function in the cell. The ratio of eIF4E2:eIF4E1 in the cell shifts in favour of eIF4E2 at higher temperatures. Despite having all of the dorsal face amino acids that have so far been associated with eIF4G binding to eIF4E1, eIF4E2 binds the eIF4E-binding domain of S.pombe eIF4G >102-times weaker than eIF4E1 in vitro. The eIF4E2 cap-binding affinity is in the typical micromolar range. The results suggest that eIF4E2 is not active on the main pathway of translation initiation in fission yeast but might play a role in the adaptation strategy of this organism under specific growth conditions. Moreover, they provide insight into the molecular characteristics required for tight binding to eIF4G.
INTRODUCTION
Recognition of the 5′ m7GpppN cap structure is a critical step in the initiation of translation on most eukaryotic mRNAs. This function is fulfilled by the cap-binding initiation complex eIF4F. Mammalian eIF4F comprises the eukaryotic initiation factors eIF4E, eIF4G and eIF4A (1). The largest eIF4F component, eIF4G (2), has binding sites for eIF4E, eIF3, eIF4A, the poly(A)-binding protein PABP and the MAPK-activated protein kinase Mnk1 (2–7). More recent work indicates that yeast eIF4G also binds to the mRNA decapping protein (Dcp1; 8). The eIF4F complex is tethered to the mRNA cap by the 25 kDa cap-binding protein, eIF4E. eIF3 is believed to form a bridge between the 40S ribosomal subunit and eIF4F, whereas the binding of eIF4G to PABP may play a role in promoting interactions between the 3′ and 5′ ends of mRNA. The significance of the interaction between eIF4G and eIF4A is not clear, but the latter factor (together with eIF4B) catalyses ATP-dependent RNA helicase activity that may promote ribosomal scanning along structured mRNA (9).
Recent work (10–12) has shown that both the yeast and the mammalian eIF4E proteins each interact with eIF4G via a site that maps to the face opposite to the ventral cap-binding slot (13). The binding sites for eIF4G of Saccharomyces cerevisiae and the yeast eIF4E-binding protein p20 overlap within this dorsal region on yeast eIF4E (10). There is also another yeast 4E-binding protein called Eap1 (14) that is suspected to act partially as a functional analogue of the human eIF4E-binding proteins (4E-BPs), and possesses an eIF4E-binding motif similar to that of p20. 4E-BP1 binds to the equivalent region on the dorsal face of human eIF4E (11,12), but this regulatory protein uses a slightly different set of molecular contacts to those involved in eIF4G binding (11,15). The binding of protein ligands at the dorsal site of yeast or human eIF4E is capable of stabilising cap binding at the ventral cap-binding slot (10,11,16,17), although the physiological significance of this effect has yet to be determined.
It seems likely that eIF4E-type proteins are represented in all eukaryotes, which suggests that the function of anchoring eIF4G, and thereby other translation factors, to the 5′ cap is a generally conserved feature (18). However, recent reports have shown that, in at least some organisms, multiple eIF4E-encoding genes exist. There are five eIF4E isoforms in Caenorhabditis elegans (19,20). Three of these (IFE-1, IFE-2, IFE-5) bind both 7-methylguanosine caps and 2,2,7-trimethylguanosine caps at the 5′ end of C.elegans mRNAs, and are partially redundant. The presence of the binding specificity for 2,2,7-trimethylguanosine caps is essential in an organism in which ∼70% of the mRNAs have this modification. A fourth isoform (IFE-3) is specific for the monomethylated cap structure and is essential, whereas a fifth isoform (IFE-4), also 7-methylguanosine-specific, is non-essential. IFE-4 is more closely related than the other C.elegans isoforms to the eIF4E-related proteins found in Arabidopsis thaliana (nCBP; 21) and vertebrates (4EHP; 22). The functions of the eIF4E-related proteins in higher eukaryotes are unknown.
We initially identified only one eIF4E-type protein in the fission yeast Schizosaccharomyces pombe (23). Genetic complementation experiments revealed that this protein is functionally equivalent to its counterpart in S.cerevisiae. Southern blot analyses provided no clear evidence of any further eIF4E-encoding genes in S.pombe. However, the S.pombe genome-sequencing project revealed the existence of a related gene. In this study, we have characterised this second gene and examined the functional characteristics of the protein it encodes. The results of this study identify this new protein as being an unusual type of eIF4E with some surprising features.
MATERIALS AND METHODS
Cloning and expression
General DNA and RNA methods for S.pombe were as described in our previous paper and elsewhere (23,24). The cDNA and chromosomal tif452 sequences were obtained via RT–PCR on S.pombe RNA and PCR on S.pombe chromosomal DNA, respectively, using oligos designed on the basis of sequence data obtained from The Sanger Centre, Cambridge, UK (http://www.sanger.ac.uk). The expression of S.pombe tif452 in S.cerevisiae was achieved using the expression plasmid called SUPEX1 (25). Escherichia coli strains grown in Luria Broth medium containing 100 µg/ml ampicillin used for protein purification were: TG2 supE hsdΔ5 thi Δ(lac-pro AB) Δ(srl-recA)306::Tn10(tetr) F’[traD36 proAB+ lacIq lacZΔM15] for the expression of eIF4G-binding domain of 4E, and BL21 hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) for expression of eIF4E1 and FLAG-eIF4E2. For protein production, eIF4E1 and an N-terminal FLAG-tagged form of eIF4E2 were cloned into the NdeI/EcoRI sites of pET5a (Promega Inc.) and the DNA encoding the C-terminal 6× histidine-tagged eIF4G-binding domain of eIF4E (4G-BD4E6His) was cloned in the NdeI/BamHI sites of the pCYTEXP1 expression vector (10).
Construction of knockout strains/tetrad analysis
The S.pombe strains used in this study were HE639 [h+ N leu1-32 ura4-D18 ade6-M210], HE665 [h– S leu1-32 ura4-D18 ade6-M216], CB15 [h+ N leu1-32 ura4-D18 ade6-M210] and CB12 [h– S leu1-32 ura4-D18 ade6-M216]. A linear tif452::kanMX6 disruption fragment was amplified by PCR for use in the transformation of diploid S.pombe CB12. After plating out on agar plates containing 100 µg ml–1 G418 in YES medium (5 g l–1 yeast extract, 30 g l–1 glucose, 225 mg l–1 adenine, histidine, leucine, uracil and lysine hydrochloride), colony PCR was used to test for correct integration of the disruption sequence into the genome. Disruption of tif451 was performed in diploid cells from the cross between S.pombe HE639 and HE665. The impact of tif451 disruption on haploid cell growth was determined after sporulation and tetrad analysis of confirmed heterozygous disruptant colonies. Tetrad analysis to test the ability of tif452 to complement the cdc33::LEU2 allele in S.cerevisiae was performed using the strain GEX1 (26). YCpSupex2 was used for expression of tif452 in S.cerevisiae (25). Saccharomyces cerevisiae was grown in YPD (20 g l–1 peptone, 10 g l–1 yeast extract) containing 20 g l–1 glucose.
Protein methods
Cap analogue-free eIF4E1 and FLAG-eIF4E2 were prepared as described earlier for S.cerevisiae eIF4E (17). The 6× histidine-tagged 4E-binding domain of eIF4G (4G-BD4E6His) was purified as described (10). Anti-eIF4E2 polyclonal antibodies were raised in rabbits and used in western blotting experiments at a dilution of 1:5000 but were blocked previously by 1 µg/ml of purified eIF4E1 in order to prevent cross-reaction. Mouse anti-eIF4E1 serum (23) was used at a dilution of 1:2500.
Cell-free translation
A cell-free translation system was prepared from S.pombe CB12 following the general procedures described in previous work (27–29). Schizosaccharomyces pombe cells were grown to an OD600 of 0.8 in YES growth medium, before being broken using glass beads. Capped luciferase transcripts were prepared in vitro using constructs generated in earlier work as templates (25). Capped mRNAs were further purified from unincorporated nucleotides and m7GpppG by means of phenol:chloroform:isoamylalcohol extraction, gel filtration (NAP-5 column from Pharmacia), precipitations with ammonium acetate, sodium acetate and finally three washes with cold 70% ethanol. Translation was performed for 60 min at 25°C in a final volume of 30 µl.
Surface plasmon resonance (SPR)
For the measurement of the eIF4E-mRNA cap interaction, in vitro transcribed, 3′-biotinylated 19mer RNAs were immobilised on Extravidin-coated sensor chips essentially as described previously (17). 1000 RU Extravidin (Sigma) were coupled to the surface of a Sensorchip CM5 (BIACore). Solutions (5 µg/ml) of capped and uncapped RNA in 100 mM NaPO4 pH 8.0 were injected into adjacent flow-cells for immobilisation to a response of 100 RU. Buffers, flow rates and evaluation procedures were as described. For observation of the interaction between the respective eIF4E proteins and 4G-BD4E6His, the polyHis-tagged protein was immobilised on a Sensorchip NTA (BIACore) and eIF4E injected at different concentrations as described earlier for the corresponding S.cerevisiae proteins (10).
Molecular modelling
Comparative models were made for S.pombe eIF4E1 and eIF4E2 based on mouse eIF4E, as mouse structures are available as complexes with both eIF4GII and eIF4E-BP1. Of the mouse complexes, an eIF4E monomer bound to eIF4GII (coordinate set 1EJH) was chosen, although there is little overall difference between this eIF4E structure and that complexed with the eIF4E-BP1 peptide (12). Sequence alignments, comparative model building and analysis were performed with the program QUANTA (Molecular Simulations Inc.), running on a Silicon Graphics workstation. Two backbone regions, both in loops between elements of regular secondary structure, required small modifications. In the first, a single residue insert (common to both S.pombe proteins) was included, and in the second either a one amino acid (S.pombe eIF4E1) or a two amino acid (S.pombe eIF4E2) deletion was modelled. In addition, a 6-residue segment that is missing near to the C-terminus of the crystal structure was omitted. None of these regions are close to the eIF4G-binding site. Side-chains were copied from the template mouse structure where possible and added or extended.
RESULTS
Cloning and sequencing of a new TIF45-related gene from S.pombe
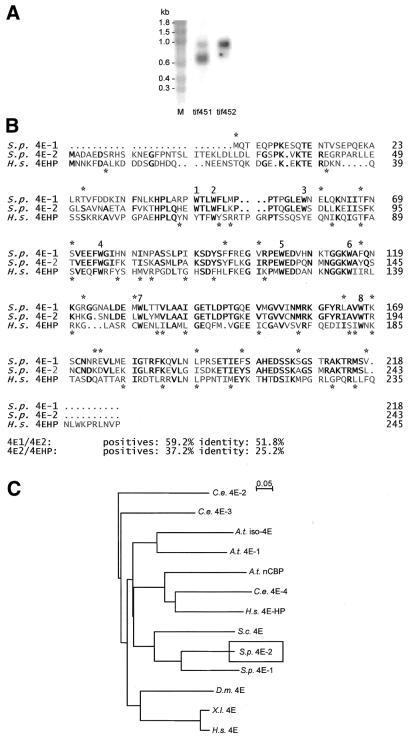
The S.pombe genome sequencing project performed at the Sanger Centre (Cambridge, UK; web site http://www.sanger.ac.uk) revealed the presence of a second gene that is closely related to both S.cerevisiae CDC33 (30) and to the previously characterised S.pombe eIF4E-encoding gene (23). A genomic clone for this latter gene was generated via PCR and sequenced on both strands. Using a probe derived from the genomic DNA we detected the presence of an mRNA species that was not detected in our earlier study (Fig. 1A). The new mRNA species is considerably longer than that of eIF4E1. The corresponding cDNA was prepared using RT–PCR and was also sequenced on both strands. The resulting sequence data revealed that the gene has two introns (data not shown).
Figure 1.
A new isoform of eIF4E in S.pombe. (A) Northern blot results indicate the presence of two types of eIF4E-encoding mRNA in S.pombe. These were detected in separate hybridisations using DNA probes derived from the gene isolated previously (tif451; 23) and from the cDNA described in the present work (tif452), and the two lanes are compared here side-by-side. One strong major band is visible in each of the lanes; the tif451 mRNA is shorter than the tif452 mRNA. There is some cross-reaction with the tif452 mRNA evident in the tif451 lane. (B) Comparison of the protein sequences of S.pombe eIF4EI and eIF4E2, and of 4EHP (the human 4E homologous protein; 22). Identities are indicated by bold letters, similarities by asterisks. Asterisks above the sequences relate to the eIF4E1 and eIF4E2 sequences, whereas those below relate to eIF4E2 and 4EHP. The eight conserved tryptophan residues are numbered. (C) Gonnet series analysis using ClustalX (35). Schizosaccharomyces pombe eIF4E1 and eIF4E2 are more closely related to each other and to S.cerevisiae eIF4E than to the higher eukaryotic eIF4Es or to the various isoforms and 4E-related proteins in other organisms.
The new coding sequence is closely related to that of the first eIF4E-encoding gene discovered in S.pombe (Fig. 1B), showing 52% identity and 59% similarity. There is an N-terminal extension in the new protein, which comprises 243 amino acids and has a predicted molecular weight of 27.7 kDa. The close similarity to other eIF4E proteins contrasts with the relatively poor similarity to the human 4EHP sequence (Fig. 1B). Indeed, the relationship to other eIF4E and eIF4E-related sequences is consistent with this view (Fig. 1C). For this reason, and on the basis of the results presented in the remainder of this paper, we have decided to refer to the new gene product as eIF4E2.
It has recently been proposed that the genes encoding initiation factors in S.pombe be given a systematic nomenclature that reflects in a logical way the protein nomenclature generally applied in the translation field (31). We originally named the first eIF4E-encoding gene tif1 (23). However, in line with the systematic nomenclature proposal (31), the eIF4E1-encoding gene (previously tif1) should now be referred to as tif451, whereas the eIF4E2-encoding gene described in this paper becomes tif452. Given the relative lengths of the tif451 and tif452 coding sequences, it is likely that the tif452 mRNA is longer in the 5′ and/or 3′ untranslated regions than the tif451 mRNA.
eIF4E2 has a distinct function
A haploid tif452::kanMX6 disruption strain was found to be viable (Fig. 2A–D). This contrasted with the effect of the tif451::kanMX6 disruption (Fig. 2E). Tetrad analysis after sporulation of a heterozygous diploid showed that the haploid tif451::kanMX6 products were non-viable. Closer examination revealed that the tif451::kanMX6 haploid spores were able to divide a small number of times before growth ceased. This initial phase of viability was presumably attributable to a remainder of eIF4E1 that progressively became diluted by virtue of cell division and degradation to the point where it became insufficient to support further growth. We also observed that tif452 was unable to suppress the lethality of a cdc33 disruption in S.cerevisiae (Fig. 2F).
Figure 2.
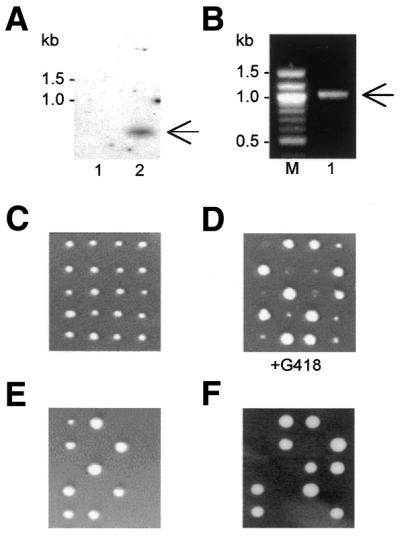
The viability of knockout strains. (A) Southern blot of genomic DNA from wild type S.pombe (lane 1) and from a haploid product of sporulation (C) bearing the tif452::kanMX6 disruption (lane 2). The radioactive probe used was derived from the kanMX6 gene. (B) Detection of the correct insertion of kanMX6 into the tif452 gene via PCR on genomic DNA using primers complementary to a sequence within kanMX6 and a sequence downstream of the insertion site, respectively. The expected bands in (A and B) are labelled with arrows. (C) Tetrad analyses revealed that all haploid products generated by sporulation of a diploid strain heterozygous for Δtif452::kanMX6 were viable. (D) Selection on G418 confirmed the presence of the tif452 disruption in half the haploid products. (E) Haploid cells carrying Δtif451::kanMX6 are non-viable, generating a 2:0 ratio. (F) Schizosaccharomyces pombe tif452 does not complement the disruption cdc33::LEU2 in haploid cells of S.cerevisiae generated by sporulation of the transformed strain GEX1 (26).
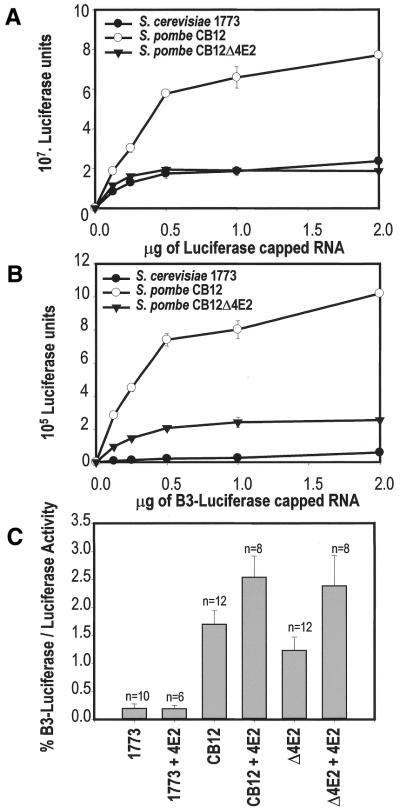
We considered the possibility that eIF4E2 participates in an alternative type of cap-binding complex that promotes translation initiation via a route that is subject to a distinct set of regulatory influences. For example, it has been proposed that eIF4F may interact with the helicase activity of eIF4A/eIF4B to promote the unwinding of structure in the 5′ UTR that can potentially inhibit ribosomal scanning (see for example 1). We therefore examined whether eIF4E2 can influence the way in which translation in an S.pombe cell-free extract responds to the presence of a stable stem–loop structure in the leader of an mRNA. Put another way, does the presence of two eIF4E isoforms confer an advantage in terms of dealing with structured mRNAs? In vitro transcribed luciferase mRNAs were added to the S.pombe and S.cerevisiae cell-free extracts, generating readily measurable enzyme activity (Fig. 3). There was some variation in the absolute activities supported by different lysates from the respective strains. For example, the Δ4E2 extract used here was less active than the wild type CB12 extract (Fig. 3A). However, this was related to variations in the exact point in the growth curve at which harvesting was performed and to small changes in the preparatory procedure rather than to intrinsic properties of these strains. These variations have no bearing on the ratios that we have calculated (Fig. 3C).
Figure 3.
The effect of structure in the mRNA 5′ UTR on translation. (A) Different amounts of a capped luciferase mRNA with an unstructured 5′ UTR of length 75 nt were added to cell-free extracts from S.pombe and S.cerevisiae to titrate the activity of the translation apparatus. Incubations were for 60 min. The S.pombe extract was either from a wild type CB12 strain or from an isogenic strain bearing the kanMX6::tif452 disruption. The S.cerevisiae extract was from the dsRNA-deficient strain 1773. (B) The same titrations as in (A) were performed, except that we used a derivative of the luciferase mRNA into which the B3 stem–loop structure (24) had been inserted. (C) The luciferase activity obtained with 250 ng of the B3-containing mRNA expressed as a percentage of the activity of the unstructured mRNA at identical mRNA concentration. The x-axis indicates the type of extract, and whether recombinant eIF4E2 (200 ng) has been added to the incubation mixture. Each n-value indicates the number of times each experiment was performed.
We observed that the absence of eIF4E2 from the extract resulted in a small reduction in the ability of the translation apparatus to overcome the inhibitory influence of secondary structure in the 5′ UTR. The B3 stem–loop that was originally described in the work of Oliveira et al. (25; predicted stability of –17.2 kcal mol–1) reduced the generation of luciferase in the wild type S.pombe extract by at least 98% (Fig. 3B). The ratio of translation with and without this structure increased by a small amount in the presence of added recombinant eIF4E2 (Fig. 3C). Although the effect of adding eIF4E2 was equivalent to a factor of only 1.5–2.0, this was a highly reproducible result. A comparative experiment performed using a cell-free extract derived from the S.cerevisiae strain 1773 revealed that translation in S.pombe is generally less sensitive to secondary structure in the 5′ UTR than it is in the budding yeast. Moreover, S.pombe eIF4E2 does not affect the translation of a structured mRNA in this system, a result that implies that the functional role of this factor is specific to S.pombe.
In relation to the above, we wanted to obtain information about the relative amounts of eIF4E1 and eIF4E2 in S.pombe. Extracts from cells grown at different temperatures were subjected to western blot analysis using polyclonal antibodies raised against the respective proteins (Fig. 4). Equal amounts of the respective lysates were compared. There was a marked increase in the detected amount of eIF4E2 relative to eIF4E1 at higher temperatures. This was quantitated relative to calibrations of the western response against different amounts of recombinant eIF4E1 and eIF4E2.
Figure 4.
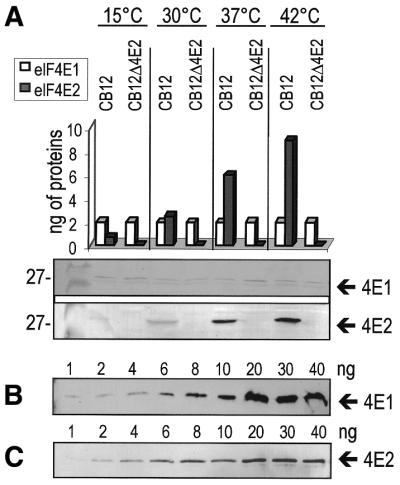
Abundance of eIF4E1 and eIF4E2 at different temperatures. (A) Western blot analysis of extracts from wild type CB12 and a tif452::kanMX6 disruption derivative using antibodies raised in mice against eIF4E1 and in rabbits against eIF4E2. The strains were grown in YES medium at four different temperatures. Equal amounts of proteins from full cell extracts were loaded on 12.5% SDS–PAGE gels and subjected to transfer. The amounts of eIF4E1 and eIF4E2 in the respective extracts were estimated on the basis of calibrations using recombinant eIF4E1 (B) and eIF4E2 (C).
eIF4E1 and eIF4E2 show almost identical cap-binding characteristics
We observed previously that the interactions of the human and yeast eIF4E proteins with the mRNA cap are characterised by rapid rate constants (11,17). We therefore investigated whether the two S. pombe eIF4E proteins would show distinct or similar cap-binding properties. In order to perform these experiments, we produced eIF4E1 and eIF4E2 in E.coli and purified the recombinant proteins to high levels of purity. As both proteins were purified solely from soluble fractions derived from E.coli on the basis of cap-affinity chromatography, we were confident that they were properly folded and active.
Real-time monitoring via SPR was used to observe the interaction between recombinant purified eIF4E proteins and in vitro transcribed, capped RNAs that had been immobilised via a biotin residue incorporated near their 3′ end. Both proteins showed specific association with capped, but not uncapped transcripts, and this association could be prevented by the addition of 0.1 mM m7GTP, but not 0.1 mM GTP, to the protein (data not shown). This highlights that both S. pombe eIF4E isoforms are fully equivalent to eIF4E proteins from most other organisms in their requirement for cap structures containing a single N7 methylation for efficient recognition of RNA 5′ ends.
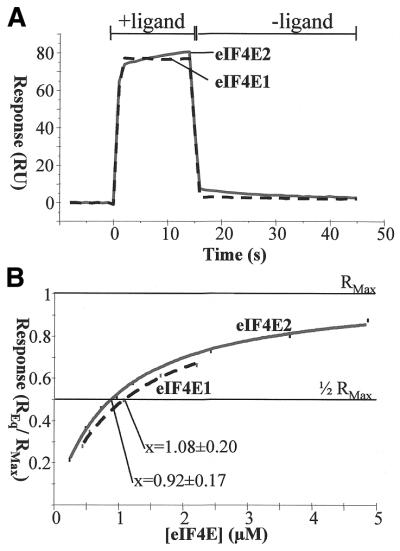
The basic sensorgram obtained from an injection of the S.pombe eIF4Es over capped mRNAs (Fig. 5A) closely resembles curves resulting from injection of the S.cerevisiae or human proteins. The fact that the response at the end of the injection phase returns very rapidly to the baseline shows that the dissociation rates for both isoforms are very quick. They may also be distinct for the two proteins, but we have not been able to quantify this accurately. The fast dissociation also explains why an equilibrium state is reached within seconds after the start of the injection, even though the concentration of eIF4E used was well below the saturation point. Overall, the on- and off-rates are so fast that we have not attempted to obtain accurate values for the respective proteins, but we can conclude that eIF4E2 is capable of at least as fast a binding/release cycle as eIF4E1.
Figure 5.
Interaction between S.pombe eIF4E proteins and the mRNA cap in the BIAcore system. (A) The basic sensorgrams obtained from the injection of 10 µM S.pombe eIF4E1 and eIF4E2, respectively, over capped, immobilised mRNAs. Responses obtained with uncapped RNAs were subtracted from the represented curves (compare 17). Two phases are shown: with (+) and without (–) protein ligand. (B) Equilibrium responses plotted against the protein concentration. The activity loss of the RNA layer per injection (∼1%) was determined in independent experiments and the curves were corrected accordingly. The point of half-maximal binding, which corresponds to the kD value, is identical within the standard deviation for both proteins.
A detailed analysis of the equilibrium responses obtained with different protein concentrations (Fig. 5B) reveals that the kD values are virtually identical for both proteins (1.1 ± 0.2 µM for eIF4E1, compared with 0.9 ± 0.2 µM for eIF4E2). Thus, the strength of cap binding is approximately equal for both isoforms and in the range described for homologous proteins from other organisms.
The eIF4E2/eIF4G interaction is comparatively weak
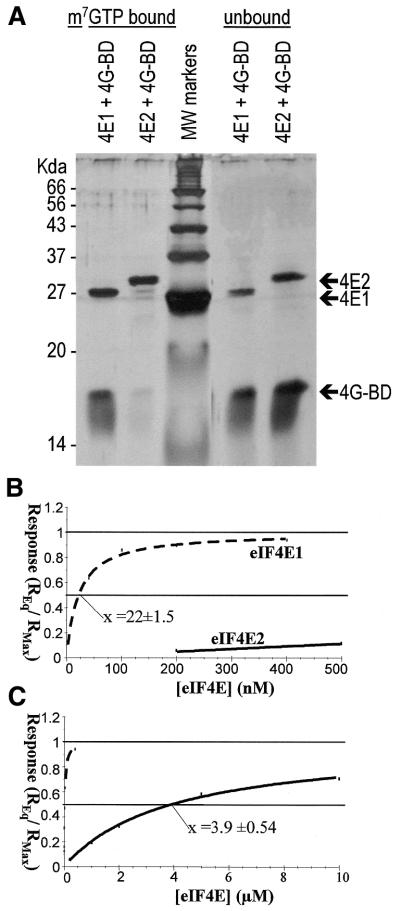
eIF4E proteins generally interact with the large adapter protein eIF4G via a cluster of amino acids on the face of eIF4E distal to the cap-binding site, and the major amino acid motif is clearly present in both S.pombe eIF4E isoforms described here (Fig. 1A, eIF4E1 amino acids 71–77). The binding between eIF4E and eIF4G can be studied in vitro using a small fragment of eIF4G comprising the eIF4E-binding site (termed here 4G-BD4E), and cap-analogue immobilised on Sepharose. The S.pombe 4G-BD4E domain sequence (corresponding to amino acids 348–513 of S.pombe eIF4G) was cloned and expressed in E.coli specifically for this purpose. If eIF4E1 is mixed with an equimolar amount of 4G-BD4E, the latter can be co-eluted from the m7GTP resin as was shown before for the corresponding S.cerevisiae proteins (10). 4G-BD4E alone does not bind to the resin. To our surprise, we found that in the case of eIF4E2, the amount of 4G-BD4E that is co-eluted from the column is greatly reduced (Fig. 6A), indicating a much weaker interaction between these two proteins. It was also observed that, in contrast to eIF4E1, 4G-BD4E did not stabilise the association of eIF4E2 with m7GTP–Sepharose.
Figure 6.
Interaction between S.pombe eIF4E proteins and the S.pombe eIF4E-binding fragment of eIF4G. (A) The respective eIF4E proteins and 4G-BD4E were mixed in a 1:2 ratio, incubated with cap-analogue resin, washed, and eluted using soluble cap-analogue. Portions of the eluate and the non-bound fraction were separated on 12.5% SDS–PAGE gels and visualised via silver staining. The amount of S.pombe 4G-BD4E that co-eluted with eIF4E2 was markedly reduced compared with eIF4E1; correspondingly, the amount of 4G-BD4E remaining unbound was increased. (B) PolyHis-tagged 4G-BD4E was immobilised on nickel-coated sensor-chips, and the eIF4E proteins were injected at varying concentrations to monitor the association with the chip surface in the BIAcore system. An association can be observed if eIF4E1 is used at low nanomolar concentrations, with half-maximal binding occurring at a concentration of 22 nM. eIF4E2 association can only be detected at much higher concentrations. (C) The same data as in (B) but represented with the x-axis stretched to accommodate the point of half-maximal binding of eIF4E2 at 3.9 µM.
A more detailed analysis using SPR with 4G-BD4E immobilised via a 6× histidine tag on a nickel-coated Sensorchip, confirmed the difference in binding affinities between the two isoforms. We determined the eIF4E concentrations at which half-maximal binding to the immobilised 4G-BD4E occurred, and obtained equilibrium binding constants of 22 nM and 4 µM for eIF4E1 and eIF4E2, respectively (Fig. 6B and C). Whereas the value for eIF4E1 is comparable with values obtained with the S.cerevisiae proteins (10) the value for eIF4E2 is >100-fold lower.
Rationalising the low affinity of eIF4E2 for eIF4G
We have made comparative models of S.pombe eIF4E1 and eIF4E2 based on the mouse crystal structure (13). With high sequence identity, these models give a good estimate of the overall fold and provide a firm basis for prediction of eIF4G-binding site changes. Mouse eIF4E is the most appropriate framework as co-crystal structures are available with peptides derived from eIF4G and 4E-BP1 (12). These structures offer themselves as a useful starting point for considering the possible structural basis for the different binding behaviours of eIF4E1 and eIF4E2. Our modelling analysis of the binding of these peptides, which show similar affinity for eIF4E when 4E-BP1 is not phosphorylated, indicates that whilst 4E-BP1 possesses more N-terminal order, eIF4G extends further at the peptide C-terminus. With a largely common central binding motif, dominated by non-polar interactions, we suggest that these end effects may balance to give the comparable affinities, and that alterations to binding can be effected through changes in this balance. Indeed, strongly reduced binding of phosphorylated 4E-BP1 is mediated by phosphorylation of a single serine at the C-terminus of this site (32).
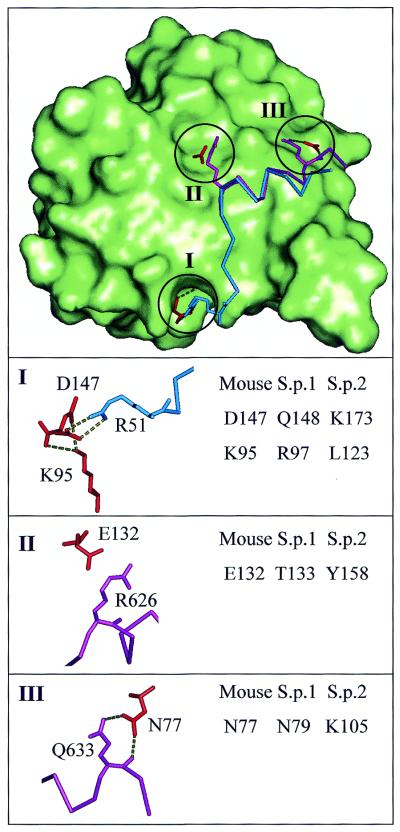
These considerations are relevant to the observed difference in eIF4G binding between S.pombe eIF4E1 and eIF4E2 (Fig. 6). We see very little difference in the central region that could account for the ∼100-fold reduction in binding seen with eIF4E2. The largest such change substitutes a threonine (eIF4E1) or tyrosine (eIF4E2) for E132 of mouse eIF4E (Fig. 7, region II). Whilst this residue forms a salt bridge with an arginine side chain of both eIF4G and 4E-BP1 in the mouse crystal structures, it is unlikely that the equivalent S.pombe residues contribute significantly to the altered binding as this arginine site in the binding region is occupied by a smaller valine in S.pombe eIF4G.
Figure 7.
Modelling the interactions between eIF4E2 and eIF4G. Differences between modelled S.pombe eIF4E-1 and eIF4E-2 adjacent to the eIF4G-binding site. The top panel shows in outline the binding modes of eIF4GII (purple) and 4E-BP1 (blue) in the context of the molecular surface (green) and selected residues (red) of mouse eIF4E (12). Three regions (labelled I, II, III) where differences between S.pombe eIF4E proteins could potentially alter affinity for eIF4G are indicated and magnified in separate views. Each of these views shows residue numbers for highlighted eIF4E residues (shown in red on the right hand side), those for S.pombe deriving from sequence alignment and comparative modelling. The potential effects of substitutions are described in the text. Residue numbers are also given for interacting 4E-BP1 (I) and eIF4G2 (II, III) residues (shown in blue and purple, respectively, on the left hand side).
Considering the binding site termini that figure in our model for affinity modification in the mouse system, we note that mouse D147 becomes glutamine in S.pombe eIF4E1 and lysine in eIF4E2 (Fig. 7, region I). This eIF4E site (along with E140 of mouse eIF4E) forms a charge network with R51 of 4E-BP1 in the mammalian system, which aligns to a lysine in S.pombe eIF4G. Therefore, alteration at this site could alter binding affinity, with a greater reduction for the charge-clash lysine substitution of S.pombe eIF4E2.
Changes in eIF4E that abut, but do not directly contact, the N-terminus of the crystallographically defined peptide-binding site could influence affinity through weakening the underlying protein framework. The side chain of mouse eIF4E K95 (Fig. 7, region I) makes hydrogen bonds with main chain carbonyls of residues 146 and 147. This ‘pinning’ interaction could be maintained in S.pombe eIF4E1 (arginine) but would be lost in eIF4E2 (leucine).
Finally, at the C-terminus of the binding region, the higher order observed for eIF4G relative to 4E-BP1 binding could well be related to an interaction that comprises hydrogen bonds between N77 of eIF4E and a glutamine side chain as well as the main chain of eIF4G (Fig. 7, region III). The equivalent residue is arginine in 4E-BP1, these interactions are not made and the binding peptide is less ordered at the site C-terminus. In the S.pombe system, the eIF4G protein possesses the glutamine residue and differences are observed between the eIF4E proteins. Whilst eIF4E1 can make these stabilising interactions with an asparagine equivalent to N77 of mouse eIF4E, a lysine substitution in S.pombe eIF4E2 gives a significantly longer side chain that is unlikely to fit into this scheme, and is therefore consistent with a lower affinity.
CONCLUSIONS
eIF4E2 constitutes a previously unknown type of fungal eIF4E, with unexpected properties. Under at least some conditions, eIF4E2 is more abundant than eIF4E1. The significance of this observation alone is not easy to interpret because a previous study has reported that, at least in S.cerevisiae, variations in eIF4E abundance over quite a large range have little consequence for cell viability (33). However, it would be expected that an eIF4E isoform that interacts relatively poorly with eIF4G is likely to support attenuated rates of translation initiation. Further studies, designed to explore the possible relationship between the expected resulting shift in translational capacity and cell physiology, will therefore be needed to provide an understanding of the function of this new factor in vivo. The reproducible influence of eIF4E2 on the ability of S.pombe ribosomes to translate mRNA with a structured leader is potentially of functional significance, but this is a relatively minor effect.
Despite being closely homologous to eIF4E1 and other typical eIF4E proteins, eIF4E2 shows a significantly reduced affinity for eIF4G. Indeed, the affinity between eIF4E2 and eIF4G is lower than that for an interaction between any type of eIF4E homologue and a binding partner containing the Tyr-X-X-X-X-Leu-Φ motif (34) reported so far (10,11). Interestingly, unlike eIF4E1, eIF4E2 did not manifest a stabilised cap-binding interaction in the presence of 4G-BD4E. This observation is consistent with earlier reports that protein ligands that bind tightly to the dorsal site of eIF4E are capable of stabilising the eIF4E:cap interaction (10,11,17).
Why does eIF4E2 bind less tightly to eIF4G? We have concluded that sequence differences between S.pombe eIF4E1 and eIF4E2 are consistent with a model for affinity modification of eIF4G domain binding to eIF4E in which such effects are largely associated with relatively subtle changes at the binding site termini, superimposed on an intervening and largely constant non-polar binding surface. This model was originally derived in the context of human eIF4E binding to 4E-BP1 that was phosphorylated or unphosphorylated at a serine residue located towards the C-terminus of the α-helical binding domain (32). The discovery of S.pombe isoforms of eIF4E with clearly different affinity for eIF4G, combined with our suggestions of how these effects may be mediated at the molecular level, will direct additional testing of the model.
We also observed that eIF4E2 manifests binding and release kinetics in its cap interactions that are as least as fast as those of eIF4E1. This means that, like eIF4E1, eIF4E2 should be able to exchange rapidly on and off mRNA. Such behaviour could be relevant to the rate of turnover of bound eIF4F complexes in vivo, and further investigation of this will follow.
In the present paper, we have been concerned with characterising the basic properties of this new protein, rather than with determining its cellular function. However, the fact that eIF4E2 varies in abundance as a function of temperature and has a small effect on the ability of the S.pombe translation apparatus to translate structured mRNAs suggests that this factor may play a regulatory role in cellular adaptation to stress conditions, perhaps by modifying the relative levels of translation of different subsets of mRNAs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Simon Morley (Sussex, UK) and Dr David Hughes (UMIST, UK) for helpful discussions, and Prof. Norbert Kaeufer (Braunschweig, Germany) for additional supervision and support for L.G.. We are grateful to the Biotechnology and Biological Sciences Research Council (BBSRC) and the Wellcome Trust for supporting T.vdH., M.P. and K.B.
REFERENCES
- 1.Raught B., Gingras,A.-C. and Sonenberg,N. (2000) In Sonenberg,N., Hershey,J.W.B. and Mathews.M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 245–293.
- 2.Imataka H., Gradi,A. and Sonenberg,N. (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamphear B.J., Kirchweger,R., Skern,T. and Rhoads,R.E. (1995) Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J. Biol. Chem., 270, 21975–21983. [DOI] [PubMed] [Google Scholar]
- 4.Mader S., Lee,H., Pause,A. and Sonenberg,N. (1995) The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol., 15, 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morley S.J., Curtis,P.S. and Pain,V.M. (1997) eIF4G: translation’s mystery factor begins to yield its secrets. RNA, 3, 1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 6.Tarun S.Z. and Sachs,A.B. (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 7.Pyronnet S., Imataka,H., Gingras,A.-C., Fukunaga,R., Hunter,T. and Sonenberg,N. (1999) Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J., 18, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilela C., Velasco,C., Ptushkina,M. and McCarthy,J.E.G. (2000) The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J., 19, 4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonenberg N. and Gingras,A.-C. (1998) The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol., 10, 268–275. [DOI] [PubMed] [Google Scholar]
- 10.Ptushkina M., von der Haar,T., Vasilescu,S., Frank,R., Birkenhäger,R. and McCarthy,J.E.G. (1998) Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J., 17, 4798–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ptushkina M., von der Haar,T., Karim,M.M., Hughes,J.M.X. and McCarthy,J.E.G. (1999) Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state. EMBO J., 18, 4068–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcotrigiano J., Gingras,A.-C., Sonenberg,N. and Burley,S.K. (1999) Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell, 3, 707–716. [DOI] [PubMed] [Google Scholar]
- 13.Marcotrigiano J., Gingras,A.-C., Sonenberg,N. and Burley,S.K. (1997) Co-crystal structure of the messenger RNA 5′ cap-binding protein (elF4E) bound to 7-methyl-GDP. Cell, 89, 951–961. [DOI] [PubMed] [Google Scholar]
- 14.Cosentino G.P., Schmelzle,T., Haghighat,A., Helliwell,S.B., Hall,M.N. and Sonenberg,N. (2000) Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes J.M.X., Ptushkina,M., Karim,M.M., Koloteva,N., von der Haar,T. and McCarthy,J.E.G. (1999) Translational repression by human 4E-BP1 in yeast specifically requires human eIF4E as target. J. Biol. Chem., 274, 3261–3264. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy J.E.G. (1998). Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev., 62, 1492–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von der Haar T., Ball,P. and McCarthy,J.E.G. (2000) Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-cap by domains of eIF4G. J. Biol. Chem., 275, 30551–30555. [DOI] [PubMed] [Google Scholar]
- 18.Hershey J.W.B. and Merrick,W.C. (2000) In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 33–88.
- 19.Jankowska-Anyszka M., Lamphear,B.J., Aamodt,E.J., Harrington,T., Darzynkiewicz,E., Stolarski,R. and Rhoads,R.E. (1998) Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in Caenorhabditis elegans can distinguish between mono- and trimethylated mRNA cap structures. J. Biol. Chem., 273, 10538–10542. [DOI] [PubMed] [Google Scholar]
- 20.Keiper B.D., Lamphear,B.J., Deshpande,A.M., Jankowska-Anyszka,M., Aamodt,E.J., Blumenthal,T. and Rhoads,R.E. (2000) Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem., 275, 10590–10596. [DOI] [PubMed] [Google Scholar]
- 21.Ruud K.A., Kuhlow,C., Goss,D.J. and Browning,K.S. (1998) Identification and characterization of a novel cap-binding protein from Arabidopsis thaliana. J. Biol. Chem., 273, 10325–10330. [DOI] [PubMed] [Google Scholar]
- 22.Rom E., Kim,H.C., Gingras,A.-C., Marcotrigiano,J., Favre,D., Olsen,H., Burley,S. and Sonenberg,N. (1998) Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J. Biol. Chem., 273, 13104–13109. [DOI] [PubMed] [Google Scholar]
- 23.Ptushkina M., Fierro-Monti,I., van den Heuvel,J., Vasilescu,S., Birkenhäger,R., Mita,K. and McCarthy,J.E.G. (1996) Schizosaccharomyces pombe has a novel eukaryotic initiation factor 4F complex containing a cap-binding protein with the human eIF4E C-terminal motif KSGST. J. Biol. Chem., 271, 32818–32824. [DOI] [PubMed] [Google Scholar]
- 24.Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira C.C., von der Heuvel,J.J. and McCarthy,J.E.G. (1993) Inhibition of translational initiation in Saccharomyces cerevisiae by secondary structure: the roles of the stability and position of stem-loops in the mRNA leader. Mol. Microbiol., 9, 521–532. [DOI] [PubMed] [Google Scholar]
- 26.Vasilescu S., Ptushkina,M., Linz,B., Müller,P.P. and McCarthy,J.E.G. (1996) Mutants of eukaryotic initiation factor eIF-4E with altered mRNA cap binding specificity reprogram mRNA selection by ribosomes in Saccharomyces cerevisiae. J. Biol. Chem., 271, 7030–7037. [DOI] [PubMed] [Google Scholar]
- 27.Gasior E., Herrera,F., Sadnik,I., MacLaughlin,C.S. and Modave,K. (1979) The preparation and characterization of a cell-free system from Saccharomyces cerevisiae that translates natural messenger ribonucleic acid. J. Biol. Chem., 254, 3965–3969. [PubMed] [Google Scholar]
- 28.Gerstel B., Tuite,M.F. and McCarthy,J.E.G. (1992) The effects of 5′-capping, 3′-polyadenylation and leader composition upon the translation and stability of mRNA in a cell-free extract derived from the yeast Saccharomyces cerevisiae. Mol. Microbiol., 6, 2339–2348. [DOI] [PubMed] [Google Scholar]
- 29.Iizuka N. and Sarnow,P. (1997) Translation-competent extracts from Saccharomyces cerevisiae: effects of L-A RNA, 5′ cap, and 3′ poly(A) tail on translational efficiency of mRNAs. Methods, 11, 353–360. [DOI] [PubMed]
- 30.Altmann M., Handschin,C. and Trachsel,H. (1987) mRNA cap-binding protein: cloning of the gene encoding protein synthesis initiation factor eIF-4E from Saccharomyces cerevisiae. Mol. Cell. Biol., 7, 998–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linder P., Vornlocher,H.-P., Hershey,J. and McCarthy,J.E.G. (1999) A systematic nomenclature for new translation initiation factor genes from S. pombe and other fungi. Yeast, 15, 865–872. [DOI] [PubMed] [Google Scholar]
- 32.Karim M.M., Hughes,J.M.X., Warwicker,J., Scheper,G., Proud,C. and McCarthy,J.E.G. (2001) A quantitative molecular model for modulation of mammalian translation by the eIF4E-binding protein 1. J. Biol. Chem., 276, 20750–20757. [DOI] [PubMed] [Google Scholar]
- 33.Lang V., Zanchin,N., Lünsdorf,H., Tuite,M.F. and McCarthy,J.E.G. (1994) Initiation factor eIF-4E of Saccharomyces cerevisiae. Distribution within the cell, binding to mRNA and consequences of its overproduction. J. Biol. Chem., 269, 6117–6123. [PubMed] [Google Scholar]
- 34.Altmann M., Schmitz,N., Berset,C. and Trachsel,H. (1997) A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J., 16, 1114–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]