Abstract
The diversity of endosymbiotic bacteria that kill male host offspring during embryogenesis and their frequencies in certain groups of host taxa suggest that the evolution of male killing and the subsequent spread of male-killing symbionts are primarily determined by host life history characteristics. We studied the 10-spot ladybird beetle, Adalia decempunctata L. (Coleoptera: Coccinellidae), in which male killing has not been recorded previously, to test this hypothesis, and we also assessed the evolution of the male killer identified by DNA sequence analysis. Our results show that A. decempunctata harbors male-killing Rickettsia (α-proteobacteria). Male-killing bacteria belonging to the genus Rickettsia have previously been reported only for the congeneric two-spot ladybird beetle, Adalia bipunctata L. Phylogenetic analysis of Rickettsia DNA sequences isolated from different populations of the two host species revealed a single origin of male killing in the genus Rickettsia. The data also indicated possible horizontal transfer of symbionts between host species. In addition, A. bipunctata is known to bear at least four different male-killing symbionts in its geographic range two of which coexist in the two locations from which A. decempunctata specimens were obtained for the present study. Since only a single male-killing taxon was found in A. decempunctata, we assume that the two closely related ladybird beetle species must differ in the number and/or geographic distribution of male killers. We discuss the importance of these findings to our understanding of the evolution and dynamics of symbiotic associations between male-killing bacteria and their insect hosts.
A large variety of organisms are hosts of “heritable” symbionts that are predominantly vertically transmitted through the host lineage. Due to the mode of transmission, symbiont survival and proliferation rely on the survival and proliferation of the host and should therefore be associated with mutualistic interactions. Such symbionts, usually intracellular microorganisms, are common among arthropods, in which they are passed from one host generation to the next through eggs. Intriguingly, some of the maternally inherited symbionts do not appear to provide direct benefits to their arthropod hosts. Instead, symbiont spread and maintenance in host populations have been ascertained via manipulation of host reproduction. These associations have attracted scientific interest because they are associated with distinct phenotypic effects in the host, such as cytoplasmic incompatibility or sex ratio distortion, including feminization of genetic males, induction of parthenogenesis, or killing of male offspring during embryogenesis (early male killing; referred to below as male killing) (9, 13, 32, 37, 43).
Male killing has been found to be associated with a variety of bacteria belonging to four taxonomic groups (Mollicutes, flavobacteria, α-proteobacteria, and γ-proteobacteria). Some of these organisms are extremely distantly related (e.g., members of the Mollicutes and members of the remaining groups). So far, male-killing symbionts have been identified in insect hosts belonging to four orders (Coleoptera, Lepidoptera, Hymenoptera, and Diptera) (6, 12–15, 20, 22, 29). There are also indications that male killers occur in a variety of other organisms, including members of various families of the Coleoptera and Lepidoptera, different taxa of drosophilid Diptera, Hymenoptera, and Hemiptera (all Insecta), as well as possibly some mites (Acari) (13, 16, 21). Interestingly, male-killing symbionts seem to be particularly common in ladybird beetles (Coleoptera: Coccinellidae). One ladybird beetle, the two-spot ladybird beetle, Adalia bipunctata, is the host of at least four different male-killing symbionts (14, 20, 28).
The diversity of male-killing agents contrasts with the diversity of agents which similarly manipulate host reproduction. Cytoplasmic incompatibility, parthenogenesis induction, and feminization are all almost exclusively associated with bacteria belonging to the genus Wolbachia (9, 32, 37). The diversity of male killers thus suggests that the trait not only arose several times independently but may even be a comparatively easily evolved behavior. In this context, it is worth pointing out that (i) close relatives of the male-killing flavobacteria and Wolbachia spp. are found in other insect hosts to be involved in mutualistic interactions and alternative types of host reproductive modifications, respectively; and (ii) certain insect taxa, such as ladybird beetles, appear to be particularly prone to invasion by such symbionts. Thus, it may be postulated that the evolution of male killing, as well as the subsequent spread of male-killing bacteria between host species, is primarily determined by host life history characteristics. Previous studies suggested that the presence of strong antagonistic interactions between siblings is important in this context since such interactions may be reduced by male killing, which in turn provides a fitness advantage to the offspring of male-killer-infected female hosts (12–14).
In the first part of the present study, we tested the hypothesis described above by using the 10-spot ladybird beetle, Adalia decempunctata, in which male killing has not been reported previously. This species is closely related to the two-spot ladybird beetle, A. bipunctata, which is known to bear several male-killing bacteria (14, 20, 28). These two taxa share a large number of life history traits, including strong antagonistic sibling interactions, such as sibling egg cannibalism and resource competition among larval siblings (26, 36). A. decempunctata was therefore expected to be permissive for invasion and spread of male-killing symbionts. Using breeding experiments and molecular genetic techniques, we demonstrated that A. decempunctata is a host of male-killing bacteria belonging to the genus Rickettsia (α-proteobacteria), which previously have been reported to produce male killing only in A. bipunctata (42).
Based on the results obtained, we decided in the second part of this study to assess the origin and evolution of male killing in the genus Rickettsia. To do this, DNA sequences were isolated from the symbionts and subjected to a detailed phylogenetic analysis. Two different gene regions were studied: part of the 17-kDa antigen gene and part of the gltA gene, which encodes citrate synthase. These gene regions have previously been used for reconstruction of the phylogenetic relationships of various Rickettsia spp. They both appeared to be more variable and phylogenetically informative than 16S ribosomal DNA (rDNA), which has been used in the past to infer Rickettsia phylogenies (3, 33, 42). The results obtained provide evidence for a monophyletic origin of male-killing Rickettsia spp. and also suggest that there may be horizontal transfer of symbionts between host species.
MATERIALS AND METHODS
Identification of male-killer-bearing host lineages.
Breeding experiments were used to identify male-killer-bearing host lineages as described by Hurst et al. (17). Four features that indicate the presence of male-killing symbionts were assessed: reduced egg hatch rate (significantly less than 60%), female-biased offspring sex ratio (significantly less than 50% males), maternal inheritance of the trait, and antibiotic sensitivity of the trait. Specimens of A. decempunctata were collected from Bielefeld, Germany, and Berlin, Germany, in May 1997. They were taken to the laboratory in Cambridge, United Kingdom, and their sexes were determined as described by Randall et al. (30). Then random mating pairs were placed individually in petri dishes. Once the females started producing fertilized eggs, they were separated from the males and allowed to continue laying eggs for approximately 2 weeks. Egg clutches were collected daily, and the hatch rate was determined. In all cases in which the egg hatch rates were significantly reduced and in some of the remaining cases, larvae were reared to adulthood and the offspring sex ratios were recorded.
Maternal inheritance of the trait was tested by crossing progeny from putative male-killing lineages with specimens derived from lineages with a 1:1 offspring sex ratio. The latter specimens were bred as described above, and the offspring sex ratios were recorded. To assess the antibiotic sensitivity of the trait, female offspring from male-killing lineages were bred for approximately 2 weeks, subsequently fed tetracycline in golden syrup (100 mg of antibiotic in 1 g of syrup) for at least 5 days, and then bred again for another period of about 2 weeks. This part of the experiment also included one control in which a female from a male-killing lineage was fed only golden syrup. Offspring sex ratios were determined before and after treatment.
To ascertain the reliability of subsequent statistical analyses of data from the initial part of the breeding experiments, we used only data from females that produced at least five egg clutches and a total of at least 50 eggs. In cases where offspring sex ratios were recorded, data were included only if at least 20 offspring were reared to adulthood. Ladybird beetles were maintained in the laboratory by using standard procedures (27). All experiments were performed at temperatures below 25°C.
Characterization of male-killing bacteria by PCR-based association tests.
Two PCR-based association tests were employed to identify bacteria associated with the male-killing trait in A. decempunctata. The first of these was based on PCR assays specific for three bacterial groups (the Spiroplasma ixodetis clade, the genus Rickettsia, and the A+B-group Wolbachia strains), all of which contain previously identified male-killing symbionts, including symbionts from A. bipunctata (14, 20, 42). To do this, genomic DNA was isolated from ovaries of females producing male-killing symbionts or from whole female ladybird beetle specimens that did not show the male-killing trait or had been cured with antibiotic. Ovaries were obtained by dissection of specimens in sterile petri dishes and then directly processed as described below. Each specimen whose whole body was used was washed in sterile H2O. Surplus water was removed with sterile tissue paper, and the specimens were ground in digestion buffer with a sterile pipette tip. DNA isolation was performed by a modification of a previously described, cetyltrimethylammonium bromide-based protocol (35, 46). Samples were incubated overnight at 50°C in 250 μl of digestion buffer (2% [wt/vol] cetyltrimethylammonium bromide, 0.1 M Tris-HCl [pH 8.0], 0.02 M EDTA, 1.4 M NaCl, 0.5% [vol/vol] β-mercaptoethanol, 10 mg proteinase K per ml). DNA was extracted with 2 volumes of chloroform-isoamyl alcohol (24:1) and then precipitated by addition of 2/3 volume of isopropanol, incubation for 1 h at −20°C, and subsequent centrifugation at 14,000 × g for 30 min. The resulting DNA pellet was washed with 70% ethanol, air dried, and resuspended in either 20 μl (DNA from ovaries) or 50 μl (DNA from whole specimens) of sterile Millipore H2O.
The DNA isolated was subsequently subjected to bacterium-specific PCR assays performed with primers SP-ITS-J04 and SP-ITS-N55 for the ribosomal spacer region of the S. ixodetis clade (29, 40), primers wsp81F and wsp691R for the wsp gene of A+B-group Wolbachia strains (49), and primers R1 and R2 for the Rickettsia 17-kDa gene (45). All reactions were performed in 25-μl mixtures containing 16 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8), 0.01% Tween 20, each deoxynucleoside triphosphate at a concentration of 0.8 mM, 2.5 mM MgCl2, each primer at a concentration of 1 μM, 2 U of BioTaq polymerase (Bioline UK Ltd.), and 0.5 μl of genomic DNA. Prior to addition of genomic DNA, reaction premixtures were always UV irradiated (150 mJ) to cross-link any contaminant DNA. PCRs were performed with a Progene thermal cycler (Techne Ltd.) by using the following profile: 2 min at 95°C, followed by 30 cycles of 20 s at 95°C, 30 s at 55°C, and 1 min at 72°C and a final extension step of 10 min at 72°C. Amplification products were separated on 1% agarose gels, and this was followed by staining with ethidium bromide and visualization of the results under UV light. PCR assays always included one positive control containing DNA from A. bipunctata specimens that had previously been shown to bear one of the male-killing symbionts and one negative control to which sterile H2O was added instead of genomic DNA. To exclude the possibility that negative results in the PCR assays were due to contaminated DNA isolates, we also attempted to amplify the insect ITS1 region from the DNA samples obtained by using primers BD1 (5′ GTCGTAACAAGGTTTCCGTA) and 4S (5′ TCTAGATGCGTTCGAAATGTCGATG) and the procedures described above.
The PCR assays revealed that Rickettsia spp. are associated with male killing in A. decempunctata (see below). As these assays permitted detection of only three different groups of bacteria, we investigated whether Rickettsia spp. are indeed the only bacteria present in the male-killing host lineages by performing a second association test. For this test, the entire 16S rRNA gene was amplified from ovary genomic DNA of two females (specimen SR12 from Bielefeld and specimen SR18 from Berlin) by using the universal eubacterial PCR primers fD1 and rP2 (41). PCR were performed in 50-μl reaction mixtures with the Expand high-fidelity PCR system (Boehringer Mannheim Ltd.) by using the manufacturer's instructions and the following cycling conditions: 2 min at 95°C, followed by 10 cycles of 20 s at 95°C, 1 min at 50°C, and 1 min at 72°C, followed by 20 cycles of 20 s at 95°C, 1 min at 50°C, and 1 min at 72°C with an additional 15 s for each cycle, and followed by a final extension step of 10 min at 72°C. Amplification products were purified with Microcon-50 microconcentrators (Amicon Ltd.) and cloned via TA cloning with the pGEM-T vector system (Promega Ltd.). For subsequent chemical transformation of Escherichia coli DH5α and selection of recombinants we generally used standard procedures (34). To prevent duplication of clones, transformants were grown for less than 1 h in SOC medium prior to plating on agar plates. In addition, as unligated PCR products and untransformed plasmids may still have been present on plate surfaces after plating and could therefore bias results of subsequent molecular analyses, we randomly selected 60 recombinant clones per host specimen and transferred these clones to new agar plates with sterile toothpicks. Then they were checked for the presence of Rickettsia 16S rDNA inserts via PCR performed with primers RSSUF (5′ CGGCTTTCAAAACTACTAATCTA) and RSSUR (5′ GAAAGCATCTCTGCGATCCG). These primers were designed on the basis of previously published bacterial DNA sequences to specifically amplify about 380 bp of the Rickettsia 16S rRNA gene. PCR were performed in 25-μl mixtures by using the conditions used for the bacterium-specific PCR assays. In this case, however, a tip of a toothpick containing recombinant clones was directly added to the reaction mixture instead of genomic DNA.
Isolation of Rickettsia DNA sequences.
For phylogenetic analysis, DNA sequences of part of the Rickettsia 17-kDa gene and part of the gltA gene were isolated from two A. decempunctata specimens (specimen SR12 from Bielefeld and specimen SR18 from Berlin). We also obtained data from five male-killing Rickettsia strains from A. bipunctata that were previously identified from different populations in the following locations in Europe: Cambridge, United Kingdom; Bielefeld, Germany; Berlin, Germany; Ribe, Denmark; and Moscow, Russia (20, 28). The gene regions were PCR amplified from genomic DNA isolated from ovaries by using primers R1 and R2 for the 17-kDa gene (45) and primers RCIT133F and RCIT1197R for the gltA gene (3). For PCR, we employed the Expand high-fidelity PCR system (Boehringer Mannheim Ltd.) and the conditions described as above. For PCR product purification and cloning we also used the procedures described above, although recombinant clones were directly isolated from the first agar plate and not transferred to a second plate. Then, plasmids from recombinant clones were purified with the Wizard Minipreps DNA purification system (Promega Ltd.). Both strands of the Rickettsia gene inserts were subsequently sequenced for three clones per host specimen with pUC/M13 primers and, for the gltA gene only, with two additional internal primers (primer sequences are available from J. H. G. von der Schulenburg). DNA sequencing was performed with a ABI Prism BigDye terminator cycle sequencing kit, and results were visualized with an ABI Prism 377 DNA sequencer (Perkin-Elmer Ltd.). To guard against PCR errors, majority rule consensus sequences were generated for the three clones isolated from each host specimen.
DNA sequence analysis.
Similarities between Rickettsia sequences were assessed with the BLAST algorithm (1). For phylogenetic analysis, the isolated sequences were manually aligned with previously published data by considering the serial triplet structure of the genes and using the program XESEE (2). We included only taxa for which DNA sequences were available for both gene regions. Data sets were characterized by calculating the numbers and percentages of nucleotide differences between pairs of sequences. Subsequent phylogenetic analysis was performed with the program PAUP*, version 4.0b4a (38), by using combined data sets to increase data information content. Character partition homogeneity between the two gene regions was assessed with the incongruence-length difference test (4). Tree estimation was then based on the maximum-likelihood (ML) criterion by using four different substitution models: the HKY85 model (8) either without or with rate heterogeneity across sites (HKY85 and HKY85+Γ, respectively; rate heterogeneity was approximated with four discrete gamma-distributed rate categories); and the general time reversible substitution model (47), taking into account rate heterogeneity across sites and, in addition, a fraction of invariable positions (GTR+Γ and GTR+Γ+I, respectively). Phylogenetic trees were reconstructed with an iterative search strategy as described by von der Schulenburg et al. (39). An initial tree topology, obtained by using unweighted maximum parsimony and a heuristic tree search via branch swapping by tree bisection and reconnection, was used to obtain ML estimates required for the different substitution models. These parameters and the initial tree topology were then employed for ML tree estimation by using each of the substitution models and a heuristic search by tree bisection and reconnection. The likelihood scores obtained in this way were compared by the likelihood ratio test to identify the substitution model that provided the most realistic representation of the pattern of sequence evolution in the data (48). The best-fit model was then used for subsequent ML-based analyses. The robustness of the inferred tree topology was assessed by nonparametric bootstrapping (5) based on 100 replicates, and specific hypotheses about the evolution of male-killing Rickettsia spp. were tested with the method of Kishino and Hasegawa (25) (KH test).
Nucleotide sequence and alignment accession numbers.
DNA sequences obtained in the present study and the alignment used for phylogenetic analyses have been submitted to the EMBL database under accession numbers AJ269516 to AJ269522 (nucleotide data) (see Table 5) and DS42855 (combined Rickettsia gene sequence alignment).
TABLE 5.
Taxa used for phylogenetic analysis of the Rickettsia genes
| Abbreviation | Taxon(a) | GenBank accession no.
|
|
|---|---|---|---|
| 17-kDa gene | gltA gene | ||
| ADEC | Rickettsia, A. decempunctata | AJ269516 | AJ269522 |
| ABCB | Rickettsia, A. bipunctata from Cambridge and Bielefeld | AJ269517a | AJ269520 |
| ABMB | Rickettsia, A. bipunctata from Moscow and Berlin | AJ269517a | AJ269521 |
| ABRI | Rickettsia, A. bipunctata from Ribe | AJ269518 | AJ269519 |
| RCON | R. conorii | M28480 | U20243 |
| RJAP | R. japonica | D16515 | U59724 |
| RMON | R. montana | U11017 | U74756 |
| RPAR | R. parkeri | U17008 | U59732 |
| RPRO | R. prowazekii | M28482 | M17149 |
| RRHI | R. rhipicephali | U11020 | U59721 |
| RRIC | R. rickettsii | M16486 | U59729 |
| RTTT | Rickettsia sp. (Thai tick typhus) | AF027124 | U59726 |
| RTYP | R. typhi | M28481 | U59714 |
The 17-kDa gene sequences from the ABCB and ABMB taxa were identical.
RESULTS
Male killing in A. decempunctata.
A total of 49 female ladybird beetles from Bielefeld and 29 beetles from Berlin were assessed for the presence of male killing. Four of these, two each from Bielefeld and Berlin (4.08% and 6.9%, respectively), produced significantly reduced egg hatch rates and female-biased offspring sex ratios (Table 1). These females were thus suspected to be hosts of male-killing bacteria. All other specimens surveyed either showed high egg hatch rates or, if the hatch rates were significantly reduced, an offspring sex ratio that was not significantly different from 1:1 (results not shown). Offspring from the four putatively male-killer-bearing females were subsequently studied to test for maternal inheritance and antibiotic sensitivity of the trait. All female offspring produced significantly biased offspring sex ratios (Tables 2 and 3). In contrast, male ladybird beetles derived from these lineages always showed offspring sex ratios of approximately 1:1 (Table 2). In addition, females with biased offspring sex ratios always generated approximately equal proportions of male and female progeny after being fed antibiotics (Table 3).
TABLE 1.
Egg hatch rates and offspring sex ratios for four wild-caught female A. decempunctata beetles
| Female | Origin | Egg hatch ratea | Sex ratiob |
|---|---|---|---|
| DBI19 | Bielefeld | 86/289 (0.298) | 3/59 (0.051) |
| DBI43 | Bielefeld | 163/342 (0.477) | 0/81 (0) |
| DBE15 | Berlin | 117/278 (0.421) | 0/69 (0) |
| DBE37 | Berlin | 73/211 (0.356) | 0/50 (0) |
Number of eggs hatched/ total number of eggs laid. The numbers in parentheses are the proportions of eggs hatched. The hatch rates shown are all significantly less than 60% (5% level, binomial distribution).
Number of male offspring/ total number of offspring reared. The numbers in parentheses are the proportions of male offspring. The sex ratios obtained are significantly different from 0.5 (5% level according to a χ2 test)
TABLE 2.
Maternal inheritance of the male-killing trait
| DBI19a
|
DBI43a
|
DBE15a
|
DBE37a
|
||||
|---|---|---|---|---|---|---|---|
| Cross | Sex ratiob | Cross | Sex ratiob | Cross | Sex ratiob | Cross | Sex ratiob |
| Females derived from male-killing lineages | |||||||
| SR29 | 2/27 (0.074) | SR12 | 2/23 (0.087) | SR7 | 4/33 (0.121) | SR41 | 1/28 (0.036) |
| SR29B | 0/23 (0) | SR12B1 | 0/21 (0) | SR9 | 1/60 (0.017) | SR41B | 0/27 (0) |
| SR31 | 0/35 (0) | SR12B2 | 0/36 (0) | SR9B | 0/49 (0) | SR41C | 0/37 (0) |
| SR12B4 | 0/20 (0) | SR9B2 | 0/25 (0) | ||||
| SR12B5 | 1/26 (0.038) | SR9B3 | 0/33 (0) | ||||
| SR12B7 | 5/20 (0.250) | SR9B4 | 0/26 (0) | ||||
| SR18 | 0/28 (0) | ||||||
| Males derived from male-killing lineages | |||||||
| MSR291 | 12/26 (0.462) | SR12A | 24/52 (0.462) | MSR71 | 14/26 (0.538) | MSR41 | 12/26 (0.462) |
| MSR292 | 11/21 (0.524) | MSR72 | 14/28 (0.500) | ||||
DBI19, DBI43, DBE15, and DBE37 are original male-killing lineages (Table 1) from which female or male offspring used in breeding experiments were derived.
Number of male offspring/total number of offspring reared. The numbers in parentheses are the proportions of male offspring. Boldface type indicates sex ratios that are significantly different from 0.5 (5% level according to a χ2 test).
TABLE 3.
Antibiotic sensitivity of the male-killing traita
| Cross | Sex ratiob
|
|
|---|---|---|
| Before treatment | After treatment | |
| Tetracycline in golden syrup | ||
| SR12B10 | 0/20 (0) | 17/29 (0.586) |
| SR9B5 | 0/28 (0) | 28/59 (0.475) |
| SR9B6 | 0/21 (0) | 13/30 (0.433) |
| Golden syrup only | ||
| SR12B | 0/46 (0) | 0/46 (0) |
The females used for crosses SR12B10 and SR12B were-derived from the male-killing lineage DBI43, whereas the females used for crosses SR9B5 and SR9B6 came from lineage DBE15 (Table 1).
Number of male offspring/total number of offspring reared. The numbers in parentheses are the proportions of male offspring. Sex ratios were determined before and after treatment with tetracycline in golden syrup or with golden syrup alone. Boldface type indicates sex ratios that are significantly different from 0.5 (5% level, χ2 test).
In the bacterium-specific PCR assays (first association tests), we tested eight females with the male-killing trait, three females which originally showed male killing but had been cured with antibiotics, and 13 wild-caught females with unbiased offspring sex ratios. All of these animals produced positive results in the control PCR in the insect ITS1 region. They all were negative in the PCR assays specific for the S. ixodetis clade or the A+B-group Wolbachia strains. Rickettsia-specific primers yielded amplification products of the expected size for the eight females showing male killing but no amplification products of the expected size for the other animals (Table 4). An exclusive relationship between Rickettsia bacteria and the male-killing trait was subsequently assessed in the second association test. All 120 eubacterial 16S rDNA clones surveyed (60 clones each from two different females with the male-killing trait) tested positive with the Rickettsia-specific 16S rDNA primers. From these results, we concluded that male killing in A. decempunctata is associated with maternally inherited bacteria belonging to the genus Rickettsia.
TABLE 4.
Results of the bacterium-specific PCR assays
| Characteristica | No. of femalesb | No. positive forc:
|
||
|---|---|---|---|---|
| Rickettsia | Spiroplasma | Wolbachia | ||
| Male killing | 8 | 8 | 0 | 0 |
| No male killing | 13 | 0 | 0 | 0 |
| Antibiotic cure | 3 | 0 | 0 | 0 |
The females screened showed male killing, did not show this trait, or were cured with antibiotics after showing this trait.
Total number of females included (all of these females produced positive results in the control PCR for the insect ITS1 region).
PCR assays specific for the genus Rickettsia, the S. ixodetis clade, and the A+B-group Wolbachia strains.
Analysis of Rickettsia DNA sequence data.
DNA sequences were obtained for 394 bp of the 17-kDa gene and 1,020 bp of the gltA gene. Both genes were identical for the Rickettsia isolates from two A. decempunctata specimens from different populations (Bielefeld and Berlin). The male-killing Rickettsia isolates from A. bipunctata specimens from Cambridge, Bielefeld, Berlin, and Moscow all produced identical 17-kDa sequences. However, they had a single nucleotide difference compared to the A. bipunctata Rickettsia from Ribe, Denmark (0.25% sequence dissimilarity). For the gltA gene, the DNA sequences were identical for the A. bipunctata symbionts from either Cambridge and Bielefeld or Berlin and Moscow. Nucleotide variation between these symbionts or the symbionts from Ribe was found at no more than seven positions (0.2 to 0.69%). The 17-kDa and gltA gene sequences from the different host species exhibited up to two and five nucleotide differences, respectively (0.25 to 0.51 and 0.2 to 0.49% sequence dissimilarity respectively).
BLAST analysis showed that the sequences from the male killers were always more similar to each other than to the sequences published previously for other taxa of the genus Rickettsia. Exclusive monophyly of the male-killing Rickettsia isolates was consistently confirmed by phylogenetic tree estimation. However, the initial analyses, which were performed for the two different gene regions separately, in each case provided only poor resolution with respect to the relationships between male killers (results not shown). For detailed phylogenetic analysis, we therefore decided to combine gene regions to increase the information content of the data. The data set used contained each of the unique combinations of 17-kDa and gltA gene sequences that were isolated from the same coccinellid host specimens in the course of the present study (one combination for the male-killing Rickettsia isolates from A. decempunctata and three combinations for the male-killing Rickettsia isolates from A. bipunctata). We also included nine additional taxa for which DNA sequences of both genes were available (Table 5). The sequences could be aligned without ambiguities and consisted of 1,414 alignment positions (394 positions-for the 17-kDa gene and 1,020 positions for the gltA gene). A total of 230 (16.27%) of these, sites were variable (18.27 and 15.49% of the sites for the 17-kDa and gltA genes, respectively).
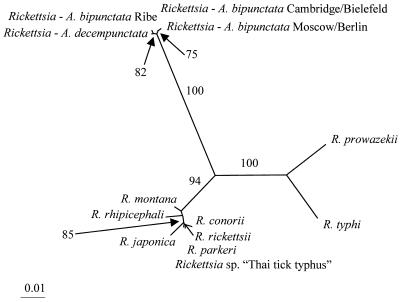
The two genes produced similar patterns of sequence divergence values, so that pairwise comparisons of 17-kDa gene sequences showed proportions of nucleotide differences similar to those of corresponding pairwise comparisons of gltA gene sequences (Table 6). Signal homogeneity between gene partitions was also supported by the incongruence-length difference test (P = 1). Hence, both genes provided consistent phylogenetic information, thus justifying their combined use for phylogenetic tree reconstruction. ML estimations with different substitution models yielded the same single tree topology. The likelihood score inferred with the GTR+Γ model was significantly higher than those obtained with the simpler models (HKY85, HKY85+Γ) but not significantly lower than that generated by the GTR+Γ+I model (Table 7). The GTR+Γ model was therefore used for subsequent analyses. The ML tree clearly supports a single origin of the male-killing symbionts. Within these symbionts, the male killers from A. bipunctata were shown to be paraphyletic. Monophyly was indicated for the Rickettsia strains from A. bipunctata from Cambridge, Bielefeld, Moscow, and Berlin and for the Rickettsia strains from A. bipunctata from Ribe and from A. decempunctata. These groups were all supported by bootstrap values greater than 75 (Fig. 1). However, a comparison of all possible relationships between the taxa by the KH test showed that exclusive monophyly of the Rickettsia strains from A. bipunctata cannot be eliminated. Although the likelihood was found to be highest for the topology illustrated in Fig. 1, four of the alternative topologies did not show significant differences at the 5% level; these topologies included one with a most common ancestry only for the Rickettsia strains from A. bipunctata (Table 8).
TABLE 6.
Percentages of nucleotide differences for 17-kDa and gltA gene sequences
| Taxon(a)a | Nucleotide differences
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADEC | ABCB | ABMB | ABRI | RCON | RJAP | RMON | RPAR | RPRO | RRHI | RRIC | RTTT | RTYP | |
| ADEC | 0.25b | 0.51 | 0.51 | 7.61 | 7.87 | 7.63 | 7.36 | 11.00 | 7.61 | 8.12 | 6.85 | 10.66 | |
| ABCB | 0.20 | 0.25 | 0.25 | 7.36 | 7.61 | 7.38 | 7.11 | 11.00 | 7.36 | 7.87 | 6.60 | 10.66 | |
| ABMB | 0.29 | 0.49 | 0 | 7.11 | 7.36 | 7.12 | 6.85 | 11.25 | 7.11 | 7.61 | 6.35 | 10.91 | |
| ABRI | 0.49 | 0.69 | 0.20 | 7.11 | 7.36 | 7.12 | 6.85 | 11.25 | 7.11 | 7.61 | 6.35 | 10.91 | |
| RCON | 7.75 | 7.94 | 7.65 | 7.84 | 1.52 | 1.53 | 0.25 | 12.53 | 1.78 | 1.02 | 0.76 | 11.93 | |
| RJAP | 8.14 | 8.33 | 8.24 | 8.43 | 1.08 | 2.04 | 1.27 | 12.53 | 2.03 | 2.03 | 1.27 | 12.18 | |
| RMON | 7.45 | 7.65 | 7.55 | 7.75 | 1.08 | 1.37 | 1.27 | 11.54 | 1.53 | 2.04 | 1.27 | 11.20 | |
| RPAR | 7.84 | 8.04 | 7.94 | 8.14 | 0.49 | 0.98 | 0.98 | 12.28 | 1.52 | 0.76 | 0.51 | 11.68 | |
| RPRO | 10.39 | 10.59 | 10.29 | 10.49 | 7.16 | 7.45 | 7.06 | 7.16 | 11.51 | 13.04 | 12.28 | 5.37 | |
| RRHI | 8.04 | 8.24 | 8.14 | 8.33 | 1.27 | 1.57 | 1.18 | 1.18 | 7.35 | 2.28 | 1.52 | 11.42 | |
| RRIC | 7.77 | 7.96 | 7.87 | 8.06 | 0.79 | 1.28 | 1.08 | 0.69 | 7.47 | 1.47 | 1.27 | 12.44 | |
| RTTT | 7.75 | 7.94 | 7.65 | 7.84 | 0.39 | 0.88 | 0.88 | 0.29 | 7.16 | 1.08 | 0.59 | 11.68 | |
| RTYP | 10.49 | 10.69 | 10.39 | 10.59 | 7.16 | 7.45 | 7.16 | 7.35 | 3.14 | 7.55 | 7.67 | 7.35 | |
For an explanation of the taxon abbreviations see Table 5.
The values above the diagonal are the values for the 17-kDa gene sequences and the values below the diagonal are the values for the gltA gene sequences.
TABLE 7.
Comparison of different substitution models by the likelihood ratio test
| Modela | lb | 2ΔlHKY85c | 2ΔlHKY85+Γc | 2ΔlGTR+Γc |
|---|---|---|---|---|
| HKY85 | −3,479.534 | |||
| HKY85+Γ | −3,451.437 | 28.097 | ||
| (Pν=4 < 0.0001) | ||||
| GTR+Γ | −3,427.483 | 52.051 | 23.954 | |
| (Pν=8 < 0.0001) | (Pν=4 < 0.0001) | |||
| GTR+Γ+I | −3,427.314 | 52.220 | 24.123 | 0.169 |
| (Pν=9 < 0.0001) | (Pν=5 = 0.0002) | (Pν=1 = 0.6810) |
Different substitution models which were used for ML estimation.
l, In likelihood obtained for the models.
Twice the likelihood difference between substitution models (rows versus columns). The probability (in parentheses) of obtaining 2Δl under the hypothesis of no difference between models was calculated by assuming a χ2 distribution. Values less than 0.05 are indicated by boldface type; ν indicates the degrees of freedom.
FIG. 1.
Unrooted tree inferred from combined Rickettsia genes. Tree estimation was based on ML as implemented in the program PAUP* (38), assuming the GTR+Γ substitution model. Branch lengths are proportional to the estimated number of substitutions per site (bar = 0.01 substitution). The numbers next to branches are the results of ML bootstrapping, as determined with 100 replicates.
TABLE 8.
Results of the KH testa
| Topological constraintb | In likelihood | Δlc | SD | Test statistic | Pd |
|---|---|---|---|---|---|
| [(ADEC,ABRI),(ABCB,ABMB)] | −3,427.483 | 0 | NAe | NA | NA |
| [(ADEC,ABCB),(ABRI,ABMB)] | −3,448.979 | 21.496 | 10.913 | 1.970 | 0.0491 |
| [(ADEC,ABMB),(ABRI,ABCB)] | −3,448.979 | 21.496 | 10.913 | 1.970 | 0.0491 |
| {[(ADEC,ABRI),ABCB],ABMB} | −3,429.504 | 2.021 | 2.549 | 0.793 | 0.4281 |
| {[(ADEC,ABRI),ABMB],ABCB} | −3,429.504 | 2.021 | 2.549 | 0.793 | 0.4281 |
| {[(ADEC,ABCB),ABRI],ABMB} | −3,448.979 | 21.496 | 10.913 | 1.970 | 0.0491 |
| {[(ADEC,ABCB),ABMB],ABRI} | −3,448.177 | 20.694 | 10.175 | 2.034 | 0.0421 |
| {[(ADEC,ABMB),ABRI],ABCB} | −3,448.979 | 21.496 | 10.913 | 1.970 | 0.0491 |
| {[(ADEC,ABMB),ABCB],ABRI} | −3,448.177 | 20.694 | 10.175 | 2.034 | 0.0421 |
| {[(ABCB,ABRI),ADEC],ABMB} | −3,448.979 | 21.496 | 10.913 | 1.970 | 0.0491 |
| {[(ABCB,ABRI),ABMB],ADEC} | −3,447.920 | 20.437 | 10.082 | 2.027 | 0.0428 |
| {[(ABMB,ABRI),ABCB],ADEC} | −3,447.920 | 20.437 | 10.082 | 2.027 | 0.0428 |
| {[(ABMB,ABRI),ADEC],ABCB} | −3,448.979 | 21.496 | 10.913 | 1.970 | 0.0491 |
| {[(ABCB,ABMB),ADEC],ABRI} | −3,428.629 | 1.146 | 1.705 | 0.672 | 0.5016 |
| {[(ABCB,ABMB),ABRI],ADEC} | −3,428.629 | 1.146 | 1.705 | 0.672 | 0.5016 |
ML tree estimation was performed with the combined data set by using the topological constraints indicated and the GTR+Γ substitution model.
For an explanation of the taxon abbreviations see Table 5.
Likelihood difference compared to the optimal ML tree.
Probability of getting a more extreme test statistic under the assumption of no difference between the null hypothesis and the optimal ML tree according to the KH test (25) as implemented in PAUP* (38). Values less than 0.05 are indicated by boldface type.
NA, not applicable.
DISCUSSION
In agreement with our a priori hypothesis that A. decempunctata should be permissive for invasion by male-killing symbionts, we found that this ladybird beetle species is a host of male-killing Rickettsia strains. Our finding adds weight to the opinion that male-killing bacteria are common among insects with suitable life histories (12–14). This is of general importance as such symbionts have the potential to shape the evolution of certain aspects of host genetics and biology. In particular, they have been shown to alter host mating behavior (23). Theoretical work suggests that they affect the evolution of egg-laying behavior or reduce mitochondrial DNA diversity due to linkage disequilibrium between symbionts and mitochondrial DNA in response to combined maternal inheritance (13, 19, 24). A more general assessment of the distribution of male-killing symbionts among insect hosts should thus be pursued in the future to ascertain the correct interpretation of particular features of host biology or the evolutionary history of host organisms as inferred from the diversity of mitochondrial DNA.
Male-killing Rickettsia strains have previously been reported only from the closely related two-spot ladybird beetle, A. bipunctata (42). The two host taxa show similarly low levels of infection with male-killing Rickettsia spp. (18, 20, 28). Such low levels seem to be required for long-term persistence of associations between male killers and hosts (13). Phylogenetic analysis of bacterial DNA sequences, including data from different populations of both host species, consistently revealed a single origin of male killing in the genus Rickettsia. The close relationship of the ladybird beetle species and the male-killing bacteria thus suggests that cospeciation of symbionts and hosts occurred. Interestingly, ML tree inference and bootstrapping strongly support paraphyly of the Rickettsia spp. from A. bipunctata. This indicates that the presence of male killers in either A. decempunctata or the A. bipunctata population from Ribe is due to horizontal transfer. Only the latter of these alternatives is consistent with host-symbiont cospeciation. Moreover, so far horizontal transfer of male-killing symbionts is known only from a single apparently exceptional case. This case involves male killing in the hymenopteran Nasonia vitripennis, where the bacterium is extracellular and transmitted by larval feeding rather than transovarially, in contrast to all other incidences of male killing (6, 7, 10, 12–15, 20, 22, 29, 44). A horizontal mode of transmission may therefore be more common in male-killing microorganisms, thus paralleling findings in other primarily transovarially transmitted symbionts (11, 39, 43).
However, in this context it should be noted that the inferred relationships of the male-killing Rickettsia spp. may not be entirely reliable. The results of the KH test indicate that four alternative topologies are not significantly worse than the optimal tree. One of these topologies shows exclusive monophyly of the Rickettsia symbionts from A. bipunctata and is therefore consistent with cospeciation of symbionts and hosts without horizontal transfer. Consequently, the data obtained do not seem to contain sufficient information to resolve questions concerning the evolution of male-killing Rickettsia symbionts with absolute certainty. Assessment of symbiont-host coevolution and/or the occurrence of horizontal transfer thus requires further investigation (e.g., via analysis of faster-evolving rickettsial DNA regions). The sample size of male-killing Rickettsia symbionts should also be increased to ensure that the whole extent of symbiont diversity is taken into account. This particularly applies to the Rickettsia symbionts from A. decempunctata, which so far have been identified in only two different locations but may also be present in other host populations. Similarly, information on phylogenetic relationships of host lineages, as deducible from host DNA sequence data, should be compared with symbiont phylogenies to aid unambiguous identification of horizontal transfer events.
Finally, it is interesting to note that A. decempunctata has been found to contain a single male-killing symbiont, whereas A. bipunctata is known to be the host of at least four different male killers, two of which coexist in the two German locations, Bielefeld and Berlin, from which A. decempunctata specimens were obtained for the present study (14, 20, 28). These two species thus appear to differ in the number and/or distribution of male killers. Such differences may be due to different invasion times or locations of the various symbionts, to local adaptation between hosts and particular symbionts, or to the presence of host and/or environmental factors that limit the distribution of specific symbionts (13, 20, 28, 31). Future investigation of male killing in these ladybird beetles, including a larger variety of A. decempunctata host populations, should permit assessment of the relevance of such factors.
In conclusion, our study provides evidence that there is a previously unknown association between male-killing Rickettsia symbionts and the 10-spot ladybird beetle, A. decempunctata, and thus supports the notion that such symbionts are common in host species with suitable life history characteristics. DNA sequence data clearly indicate that there was a single transition to male killing in the genus Rickettsia. However, the relationships between male killers isolated from two Adalia species remain ambiguous. Our results nevertheless highlight the conclusion that these two closely related host taxa provide a promising system for comparative analysis of the evolution and dynamics of male killing that may be shaped by horizontal transfer events and factors leading to differences in the number and/or distribution of male-killing symbionts among host species. Such an analysis is expected to provide valuable insights into the nature of these peculiar symbiotic associations.
ACKNOWLEDGMENTS
J. H. G. von der Schulenburg was funded by a TMR fellowship from the European Union and, for the collection of ladybird beetle specimens in Germany, by travel grants from both Magdalene College (Cambridge, United Kingdom) and the Cambridge Philosophical Society (Cambridge, United Kingdom). G. D. D. Hurst was supported by a BBSRC D. Phillips fellowship (United Kingdom). Part of the work was carried out in a laboratory funded by the Wolfson Foundation (United Kingdom).
We thank Chris Maddren, Dennis Farrington, and Roger Day for technical assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Cabot E. XESEE. The Eyeball Sequence Editor DOS protected mode version of ESEE, version 3.1. Rochester, N.Y: University of Rochester; 1997. [Google Scholar]
- 3.Davis M J, Ying Z, Brunner B R, Pantoja A, Ferwerda F H. Rickettsial relative associated with papaya bunchy top disease. Curr Microbiol. 1998;36:80–84. doi: 10.1007/s002849900283. [DOI] [PubMed] [Google Scholar]
- 4.Farris J S, Kallersjo M, Kluge A G, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–319. [Google Scholar]
- 5.Felsenstein J. Confidence limits on phylogenies—an approach using the bootstrap. Evolution. 1985;40:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 6.Fialho R F, Stevens L. Male-killing Wolbachia in a flour beetle. Proc R Soc Lond B Biol Sci. 2000;267:1469–1473. doi: 10.1098/rspb.2000.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gherna R L, Werren J H, Weisburg W G, Cote R, Woese C R, Mandelco L, Brenner D J. Arsenophonus nasoniae gen. nov., sp. nov., the causative agent of the son killer trait in the parasitic wasp Nasonia vitripennis. Int J Syst Bacteriol. 1991;41:563–565. [Google Scholar]
- 8.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;21:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann A A, Turelli M. Cytoplasmic incompatibility in insects. In: O'Neill S L, Hoffmann A A, Werren J H, editors. Influential passengers. Oxford, United Kingdom: Oxford University Press; 1997. pp. 42–80. [Google Scholar]
- 10.Huger A M, Skinner S W, Werren J H. Bacterial infections associated with the son-killer trait in the parasitoid wasp Nasonia (= Mormoniella) vitripennis (Hymenoptera: Pteronalidae) J Invertebr Pathol. 1985;46:272–280. doi: 10.1016/0022-2011(85)90069-2. [DOI] [PubMed] [Google Scholar]
- 11.Huigens M E, Luck R F, Klaassen R H G, Maas M F P M, Timmermans M J T N, Stouthamer R. Infectious parthenogenesis. Nature. 2000;405:178–179. doi: 10.1038/35012066. [DOI] [PubMed] [Google Scholar]
- 12.Hurst G D D, Bandi C, Sacchi L, Cochrane A G, Bertrand D, Karaca I, Majerus M E N. Adonia variegata (Coleoptera: Coccinellidae) bears maternally inherited flavobacteria that kill males only. Parasitology. 1999;118:125–134. doi: 10.1017/s0031182098003655. [DOI] [PubMed] [Google Scholar]
- 13.Hurst G D D, Hurst L D, Majerus M E N. Cytoplasmic sex-ratio distorters. In: O'Neill S L, Hoffmann A A, Werren J H, editors. Influential passengers. Oxford, United Kingdom: Oxford University Press; 1997. pp. 125–154. [Google Scholar]
- 14.Hurst G D D, Jiggins F M, von der: Schulenburg J H G, Bertrand D, West S A, Goriacheva II, Zakharov I A, Werren J H, Stouthamer R, Majerus M E N. Male-killing Wolbachia in two species of insect. Proc R Soc Lond B Biol Sci. 1999;266:735–740. [Google Scholar]
- 15.Hurst G D D, Johnson A P, von der Schulenburg J H G, Fuyama Y. Male-killing Wolbachia in Drosophila: a temperature sensitive trait with a threshold bacterial density. Genetics. 2000;156:699–709. doi: 10.1093/genetics/156.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurst G D D, Majerus M E N. Why do maternally inherited microorganisms kill males? Heredity. 1993;71:81–95. [Google Scholar]
- 17.Hurst G D D, Majerus M E N, Walker L E. Cytoplasmic male-killing elements in Adalia bipunctata (Linnaeus) (Coleoptera: Coccinellidae) Heredity. 1992;69:84–91. [Google Scholar]
- 18.Hurst G D D, Majerus M E N, Walker L E. The importance of cytoplasmic male killing elements in natural populations of the two spot ladybird, Adalia bipunctata (Linnaeus) (Coleoptera: Coccinellidae) Biol J Linn Soc. 1993;49:195–202. [Google Scholar]
- 19.Hurst G D D, McVean G A T. Parasitic male-killing bacteria and the evolution of clutch size. Ecol Entomol. 1998;23:350–353. [Google Scholar]
- 20.Hurst G D D, von der Schulenburg J H G, Majerus T M O, Bertrand D, Zakharov I A, Baungaard J, Volkl W, Stouthamer R, Majerus M E N. Invasion of one insect species, Adalia bipunctata, by two different male-killing bacteria. Insect Mol Biol. 1999;8:133–139. doi: 10.1046/j.1365-2583.1999.810133.x. [DOI] [PubMed] [Google Scholar]
- 21.Hurst L D. The incidences, mechanisms and evolution of cytoplasmic sex ratio distorters in animals. Biol Rev. 1993;68:121–193. [Google Scholar]
- 22.Jiggins F M, Hurst G D D, Jiggins C D, von der Schulenburg J H G, Majerus M E N. The butterfly Danaus chrysippus is infected by a male-killing Spiroplasma bacterium. Parasitology. 2000;120:439–446. doi: 10.1017/s0031182099005867. [DOI] [PubMed] [Google Scholar]
- 23.Jiggins F M, Hurst G D D, Majerus M E N. Sex-ratio-distorting Wolbachia causes sex-role reversal in its butterfly host. Proc R Soc Lond B Biol Sci. 2000;267:69–73. doi: 10.1098/rspb.2000.0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnstone R A, Hurst G D D. Maternally inherited male-killing microorganisms may confound interpretation of mitochondrial DNA variability. Biol J Linn Soc. 1996;58:453–470. [Google Scholar]
- 25.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 26.Majerus M E N. Ladybirds. London, United Kingdom: HarperCollins Publisher; 1994. [Google Scholar]
- 27.Majerus M E N, Kearns P W E, Ireland H, Forge H. Ladybirds as teaching aids. 1. Collecting and culturing. J Biol Educ. 1989;23:85–95. [Google Scholar]
- 28.Majerus M E N, von der Schulenburg J H G, Zakharov I A. Multiple causes of male-killing in a single sample of the 2-spot ladybird, Adalia bipunctata (Coleoptera: Coccinellidae), from Moscow. Heredity. 2000;84:605–609. doi: 10.1046/j.1365-2540.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 29.Majerus T M O, von der Schulenburg J H G, Majerus M E N, Hurst G D D. Molecular identification of a male-killing agent in the ladybird Harmonia axyridis (Pallas) (Coleoptera: Coccinellidae) Insect Mol Biol. 1999;8:551–556. doi: 10.1046/j.1365-2583.1999.00151.x. [DOI] [PubMed] [Google Scholar]
- 30.Randall K, Majerus M E N, Forge H. Characteristics for sex determination in British ladybirds (Coleoptera: Coccinellidae) Entomologist (London) 1992;111:109–122. [Google Scholar]
- 31.Randerson J P, Smith N G C, Hurst L D. The evolutionary dynamics of male-killers and their hosts. Heredity. 2000;84:152–160. doi: 10.1046/j.1365-2540.2000.00645.x. [DOI] [PubMed] [Google Scholar]
- 32.Rigaud T. Inherited microorganisms and sex determination of arthropod hosts. In: O'Neill S L, Hoffmann A A, Werren J H, editors. Influential passengers. Oxford, United Kingdom: Oxford University Press; 1997. pp. 81–101. [Google Scholar]
- 33.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–261. doi: 10.1099/00207713-47-2-252. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Shahjahan R M, Hughes K J, Leopold R A, DeVault J D. Lower incubation temperature increases yield of insect genomic DNA isolated by the CTAB method. BioTechniques. 1995;19:333–334. [PubMed] [Google Scholar]
- 36.Sloggett J J, Majerus M E N. The evolution of dietary and habitat preferences in the Coccinellidae (Coleoptera) Biol J Linn Soc. 2000;70:63–68. [Google Scholar]
- 37.Stouthamer R. Wolbachia-induced parthenogenesis. In: O'Neill S L, Hoffmann A A, Werren J H, editors. Influential passengers. Oxford, United Kingdom: Oxford University Press; 1997. pp. 102–124. [Google Scholar]
- 38.Swofford D L. PAUP*—phylogenetic analysis using parsimony (* and other methods), version 4. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 39.Von der Schulenburg J H G, Hurst G D D, Huigens T M E, Van Meer M M M, Jiggins F M, Majerus M E N. Molecular evolution and phylogenetic utility of Wolbachia ftsZ and wsp gene sequences with special reference to the origin of male-killing. Mol Biol Evol. 2000;17:584–600. doi: 10.1093/oxfordjournals.molbev.a026338. [DOI] [PubMed] [Google Scholar]
- 40.Von der Schulenburg J H G, Majerus T M O, Dorzhu C M, Zakharov I A, Hurst G D D, Majerus M E N. Evolution of male-killing Spiroplasma (Procaryota: Mollicutes) inferred from ribosomal spacer sequences. J Gen Appl Microbiol. 2000;46:95–98. doi: 10.2323/jgam.46.95. [DOI] [PubMed] [Google Scholar]
- 41.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werren J H, Hurst G D D, Zhang W, Breeuwer J A J, Stouthamer R, Majerus M E N. Rickettsial relative associated with male killing in the ladybird beetle (Adalia bipunctata) J Bacteriol. 1994;176:388–394. doi: 10.1128/jb.176.2.388-394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werren J H, O'Neill S L. The evolution of heritable symbionts. In: O'Neill S L, Hoffmann A A, Werren J H, editors. Influential passengers. Oxford, United Kingdom: Oxford University Press; 1997. pp. 1–41. [Google Scholar]
- 44.Werren J H, Skinner S W, Huger A M. Male-killing bacteria in a parasitic wasp. Science. 1986;231:990–992. doi: 10.1126/science.3945814. [DOI] [PubMed] [Google Scholar]
- 45.Williams S G, Sacci J B J, Schriefer M E, Andersen E N, Fujioka K K, Sorvillo F J, Barr A R, Azad A F. Typhus and typhus-like rickettsiae associated with opossums and their fleas in Los Angeles. J Clin Microbiol. 1992;30:1758–1762. doi: 10.1128/jcm.30.7.1758-1762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winnepenninckx B, Backeljau T, De Wachter R. Extraction of high molecular weight DNA from molluscs. Trends Genet. 1993;9:407. doi: 10.1016/0168-9525(93)90102-n. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z, Goldman N, Friday A. Comparison of models for nucleotide substitution used in maximum-likelihood phylogenetic estimation. Mol Biol Evol. 1994;11:316–324. doi: 10.1093/oxfordjournals.molbev.a040112. [DOI] [PubMed] [Google Scholar]
- 48.Yang Z, Goldman N, Friday A. Maximum likelihood trees from DNA sequences: a peculiar statistical problem. Syst Biol. 1995;44:384–399. [Google Scholar]
- 49.Zhou W G, Rousset F, Oneill S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc R Soc Lond B Biol Sci. 1998;265:509–515. doi: 10.1098/rspb.1998.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]