Abstract
The Brn-3a transcription factor stimulates the expression of the anti-apoptotic Bcl-2 and Bcl-x proteins and protects neuronal cells from apoptosis. Here we show that a minimal Bcl-x promoter is activated by Brn-3a and that this stimulation is prevented by the pro-apoptotic p53 protein. Both these effects are mediated via Bcl-x promoter sequences, which are indistinguishable from those required for minimal basal promoter activity. A newly described upstream Bcl-x promoter is also activated by Brn-3a with this activation being prevented by p53. Hence, Brn-3a-mediated activation of two distinct Bcl-x promoters and of the Bcl-2 promoter is blocked by p53 whereas this is not observed for Brn-3a activated promoters derived from genes not involved in inhibiting apoptosis. p53 therefore appears to specifically target the activation by Brn-3a of promoters derived from genes with an anti-apoptotic effect and this may be involved in the pro-apoptotic activity of p53.
INTRODUCTION
The Brn-3a transcription factor is a member of the POU (Pit-Oct-Unc) family of transcription factors (reviewed in 1,2), which is expressed in specific neurones in the brain and at high levels in sensory neurones in the dorsal root ganglia and trigeminal ganglia (3,4). This factor appears to play a critical role in the development and survival of the neurones that express it, with knock out mice lacking Brn-3a showing extensive losses of somatosensory neurones resulting in death shortly after birth (5,6). In contrast, knock out mice lacking the closely related POU factors Brn-3b or Brn-3c do not show impaired survival, although they show defects in, respectively, the survival of visual and auditory neurones (7).
The critical role of Brn-3a in the developing nervous system is paralleled by its ability to promote neuronal survival both in vitro and in vivo. Thus, we have demonstrated that the overexpression of Brn-3a in cultured dorsal root or trigeminal neurones results in enhanced survival when they are exposed to neurotrophic factor withdrawal (8,9), whilst the reduction of Brn-3a levels with an anti-sense approach results in impaired survival of sensory neurones even in the presence of neurotrophic factors (8,9). Moreover, this survival-promoting effect of Brn-3a can be observed in vivo as well as in vitro. Thus, enhanced expression of Brn-3a produced by a herpes simplex virus vector expressing this protein was able to rescue dorsal root ganglion neurones in the neonatal rat, which would otherwise have died following sciatic nerve lesion (10).
As a transcription factor, Brn-3a is likely to achieve its survival-promoting effects by modulating the expression of specific target genes. Indeed, we have demonstrated that overexpression of Brn-3a in dorsal root or trigeminal neurones results in enhanced expression of the genes encoding the anti-apoptotic proteins Bcl-2 and Bcl-x, with this effect being observed in both cultured neurones and within the intact neonatal animal in vivo (8–10). Conversely, an antisense-mediated reduction of Brn-3a levels in sensory neurones results in reduced levels of Bcl-2 and Bcl-x and a correspondingly reduced survival (8–10). Interestingly, overexpression of Brn-3a does not result in enhanced survival of sympathetic neurones, which do not normally express Brn-3a and correspondingly no upregulation of Bcl-2 or Bcl-x is observed when Brn-3a is overexpressed in these neurones (8–10).
We have previously characterised the mechanisms by which Brn-3a upregulates the Bcl-2 promoter. Thus, we have demonstrated that a sequence between –584 and –594 bp upstream of the Bcl-2 P2 promoter mediates its induction by Brn-3a (8). Moreover, we have shown that the activation of this promoter by Brn-3a is completely abolished by the pro-apoptotic protein p53, which interacts with Brn-3a via a protein–protein interaction (11). This effect is mediated via the ability of p53 to bind to a binding site at –535 to –558 in the Bcl-2 P2 promoter, adjacent to the Brn-3a binding site (11).
In contrast, we have not previously investigated the mechanism by which Brn-3a regulates the Bcl-x promoter or determined whether the activation of this promoter by Brn-3a is inhibited by p53. This is of particular importance as inactivation of the Bcl-2 gene does not lead to enhanced neuronal cell death during the period of naturally occurring cell death in development (12–14), whereas Bcl-x knock out mice show gross abnormalities of the nervous system due to enhanced programmed cell death during the period when such neuronal cell death normally occurs (15) and their phenotype is similar to that observed in the Brn-3a knockout mice (5,6). We have therefore investigated the Bcl-x promoter sequences necessary for activation by Brn-3a and determined whether such activation is inhibited by p53.
MATERIALS AND METHODS
Bcl-x promoter constructs
The initial construct used contains upstream sequences of the human Bcl-x promoter as defined by Grillot et al. (16). This was used to generate three additional constructs containing successive truncations from the 5′ end by PCR. This was performed using a common 3′ oligonucleotide which was complementary to –37 to –19 (relative to the translation start site) of the Bcl-x upstream sequence (5′-CTCAACCAGTCCATTGTCC-3′) together with the following 5′ oligonucleotides: Bcl-x B 5′-ATCCATACCAGCCACCTCCG-3′ (–779 to –759), Bcl-x C 5′-GCCTGCCGGGTCGCATGATC-3′ (–568 to –548) and Bcl-x D 5′-GTGCTTTCGATTTGACTTAAG-3′ (–279 to –258). The PCR profile used was a 3 min hot start at 95°C followed by 30 cycles of 94°C for 1 min, 55°C for 1 min (for D) or 50°C for 1 min (for B and C) and 72°C for 1 min. All PCR products were cloned into the HindIII cloning site of pGL2 basic (Promega, Poole, UK).
The promoter constructs B SmaI and C BglII were generated from the indicated parental construct by digestion with the appropriate restriction enzyme, which cut within the Bcl-x sequence and within the multiple cloning site (MCS) of pGL2 basic. The resulting DNA was religated to generate constructs with 5′ ends at –687 and –451, respectively. D NsiI was generated by digestion of Bcl-x D with NsiI (digests Bcl-x at –156) and MluI (cuts pGL2 basic MCS). DNA was subsequently filled-in using T4 DNA polymerase (Promega) and religated. D XhoII/TATA was generated by digesting the Bcl-x PCR fragment (prior to cloning into pGL2 basic) with XhoII and HindIII and the resulting 76 bp product was cloned into pGL2 basic using the BglII and HindIII restriction sites. The authenticity of all the constructs was confirmed by DNA sequencing prior to use.
The pGL2-TK plasmid was generated by digestion of pBLCAT2 (17) with BglII and BamHI and the released TK promoter fragment was cloned into the BglII site of pGL2 basic. All Bcl-x-TK constructs were generated by PCR using the following oligonucleotides: A/B 5′ oligonucleotide (–1000 to –980): 5′-ACAAACCACGCATTTGTTGG-3′ with 3′-oligonucleotide (–780 to –800): 5′-TTAGTTGGTTTTTGTTTTTC-3′; NsiI/XhoII 5′ oligonucleotide (–171 to –152): 5′-GGTAAATGGCATGCATATT-3′ with 3′ oligonucleotide (–79 to –98): 5′-GTGATTCTCTTCTAAGATC-3′; XhoII/TATA 5′ oligonucleotide (–94 to –74): 5′-GATCTTAGAAGAGAATCAC-3′ with 3′ oligonucleotide (–19 to –38): 5′-CTCAACCAGTCCATTGTCC-3′. All PCRs were performed as described above for the initial constructs using a 55°C annealing temperature. Resulting PCR products were cloned into pGL2-TK using KpnI and NheI restriction sites. The reverse construct TATA/XhoII-TK was generated using the oligonucleotides described above for the original XhoII/TATA-TK but was cloned between the KpnI and NheI in the reverse orientation. Again the authenticity of all constructs was confirmed by DNA sequencing prior to use.
Subsequently, we also tested constructs containing the recently identified upstream Bcl-x promoter (18).
Transfections
The ND7 neuronal cell line (19) was transfected by the calcium phosphate method (20). Routinely, 0.5 × 105 cells were transfected in 24-well plates with 1 µg of the reporter plasmid and 1 µg of expression vector together with 0.2 µg of the TK-Renilla plasmid. This contains the Renilla luciferase gene under the control of the minimal thymidine kinase promoter (Promega). All expression vectors have been described previously (8,11). In transfections where more than one expression vector was needed, 1 µg of each was used and total DNA was kept constant at the maximum amount needed using sonicated salmon sperm DNA (Pharmacia Biotech., St Albans, UK). Cells were analysed for reporter gene activity after 48 h.
Luciferase assay of reporter gene activity
Transfected cells were lysed in 1× passive lysis buffer (Promega) and assayed for both firefly and Renilla luciferase activities. This was performed on 20 µl lysate using the dual luciferase assay system as per instructions (Promega). The firefly luciferase values obtained were divided by their respective Renilla luciferase values to give relative light units for each transfection condition. Each transfection was performed in triplicate and relative light unit values obtained from each one were converted into per cent vector control. Statistical significance of observed changes in promoter function was determined using the Mann–Whitney U-test.
RESULTS AND DISCUSSION
To identify the region of the Bcl-x promoter that was responsive to Brn-3a, we constructed a series of plasmids in which different regions of the previously defined Bcl-x promoter (16) were linked to the luciferase reporter gene. Each of these constructs began progressively further upstream and each ended at –19 bp relative to the ATG start codon for translation of the Bcl-x protein (Fig. 1). These constructs contain previously defined transcriptional start sites at –655, –400, –96 and –83 relative to the ATG codon (16). In each case, the constructs were transfected with a Brn-3a expression vector and their activity compared with that observed when the same construct was transfected with expression vector lacking any insert.
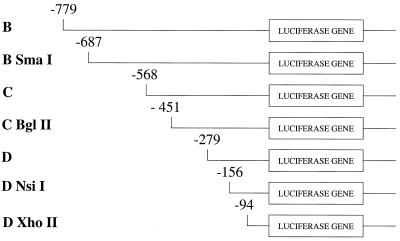
Figure 1.
Bcl-x promoter constructs. In each case the region of the Bcl-x promoter linked to luciferase is shown. The 5′ end of each fragment is given in base pairs upstream (–) of the translational start site. All the constructs have a common 3′ end at –19 upstream of the translational start site.
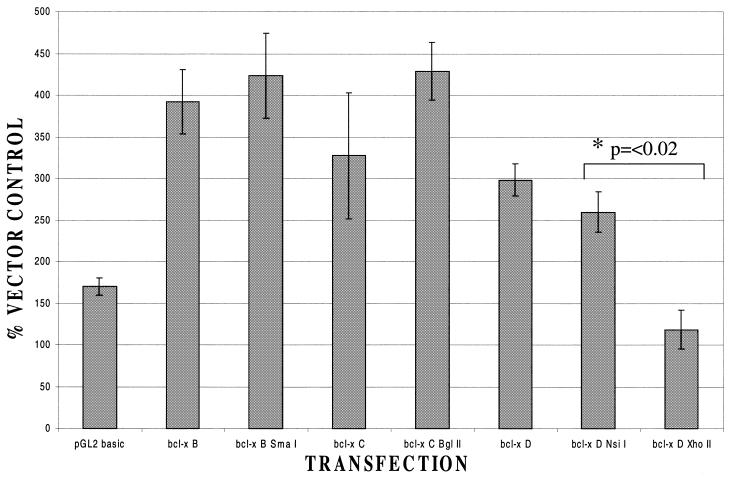
In these experiments (Fig. 2), clear activation of the Bcl-x promoter in the longest construct (B) was observed in accordance with our previous results (10). As the region of the Bcl-x promoter being tested was progressively truncated, from –779 to –451, no decrease in the extent of transactivation by Brn-3a was noted. Hence, the transcriptional start site at –655 can be eliminated without affecting the response to Brn-3a. This is not surprising as this start site was shown to be predominantly used in the thymus rather than in neuronal cells (16). Some diminution in the degree of transactivation was noted when the promoter was deleted from –451 to –279 removing the start site at –400 but this decrease was not significant and was not investigated further. In contrast, transactivation by Brn-3a was abolished when the upstream region was truncated from –156 to –94 (Fig. 2) with significant transactivation being observed in the Bcl-x D NsiI construct but not in the shorter Bcl-x D XhoII. Hence, the elimination of sequences from –156 to –95 relative to the translational start site abolishes transactivation by Brn-3a.
Figure 2.
Response of Bcl-x promoter constructs to Brn-3a. Result of co-transfecting each of the Bcl-x constructs with a Brn-3a expression vector or with empty vector control. In each case the result is presented as a ratio between the luciferase activity observed with Brn-3a and that observed with the same construct co-transfected with empty expression vector. The result for the luciferase construct lacking any promoter (pGL2 basic) is also shown for comparison. Values are the average of three independent determinations whose standard error is shown by the bars.
These findings suggest the possibility that the response to Brn-3a requires the region of the Bcl-x promoter between –156 and –95 relative to the translational start site. However, the shortest Bcl-x promoter construct containing only bases downstream of –94 did not produce detectable promoter activity compared with the pGL2 basic vector containing luciferase alone (data not shown). Hence, it is also possible that this construct is incapable of producing any transcription and, therefore, cannot respond to Brn-3a even though it is necessary for the Brn-3a response of the intact promoter.
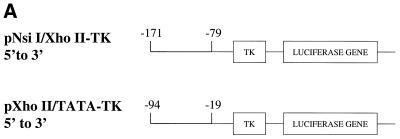
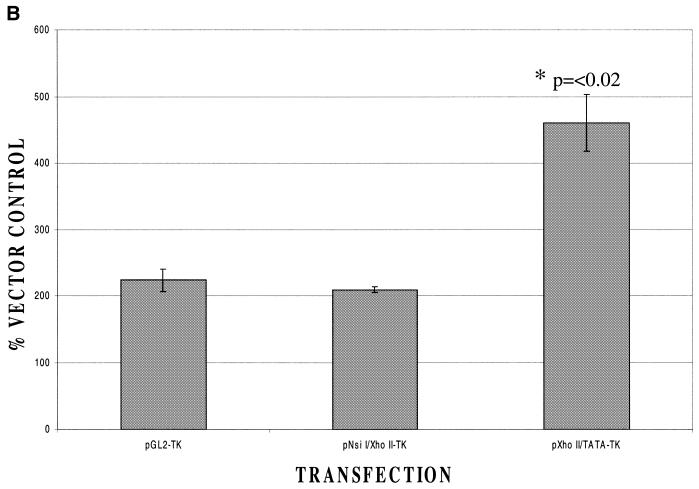
To distinguish these possibilities, fragments from –171 to –79 or from –94 to –19 were cloned into the vector pBL2 (17), which contains the thymidine kinase promoter upstream of a luciferase reporter gene (Fig. 3A). This therefore allows us to assess the effects of each of these regions on inducibility by Brn-3a when basal promoter activity was provided by the TK promoter. In this experiment (Fig. 3B) the –171 to –79 region was not able to confer enhanced inducibility by Brn-3a on the TK promoter whereas a clear increase in inducibility was seen in the case of the construct containing the –94 to –19 region. These findings suggest that the minimal Bcl-x promoter contained within the region –94 to –19 relative to the translational start site (and which contains a transcriptional start site at –83) (16) can confer inducibility by Brn-3a on a heterologous promoter even though it does not have sufficient promoter activity to drive detectable luciferase expression.
Figure 3.
Response to Brn-3a of Bcl-x promoter constructs linked to a heterologous promoter. (A) Structure of the constructs containing the Bcl-x promoter either from –171 to –79 or –94 to –19 relative to the translational start site upstream of the TK promoter and the luciferase gene. (B) Response of these constructs to Brn-3a compared with the response of the parental construct pGL2-TK lacking any extra insert. In each case, the response is presented as a ratio between the luciferase activity observed with Brn-3a and that observed with the same luciferase construct co-transfected with expression vector lacking any insert. Values are the average of three independent determinations whose standard error is shown by the bars.
Interestingly, we have previously identified upstream response elements for Brn-3a in promoters such as that of the genes encoding Bcl-2 (8), SNAP-25 (21) or NGFIA (22). However, in other cases such as that of the genes encoding the various neurofilament subunits or that encoding α-internexin it is impossible to dissect away the Brn-3a response from minimal promoter activity, with the response being lost concomitantly with the loss of promoter activity (23,24). This suggests that two classes of Brn-3a-inducible promoters may exist with some promoters, such as that of the Bcl-2 gene, being induced by the binding of Brn-3a to upstream sequences; whereas in other cases Brn-3a may participate in a minimal basal transcriptional complex required for gene transcription. It is clear from the experiments presented here that Bcl-x, unlike Bcl-2, belongs to this second class of promoters.
In previous experiments, we have also demonstrated that Brn-3a-responsive promoters differ in the region of Brn-3a that is required for their induction. Thus, some promoters, such as those of the genes encoding the neurofilaments, can be activated by the isolated POU domain of Brn-3a alone (24,25). They are thus equally well activated by the naturally occurring long and short forms of Brn-3a, which both contain the POU domain but differ in the presence or absence of 84 amino acids at the N-terminus (26,27). In contrast, other Brn-3a activated promoters such as those of the genes encoding Bcl-2 (8) and α-internexin (23) require, in addition to the POU domain, the N-terminal 84 amino acids found only in the long form of Brn-3a and are, therefore, not activated by the short form.
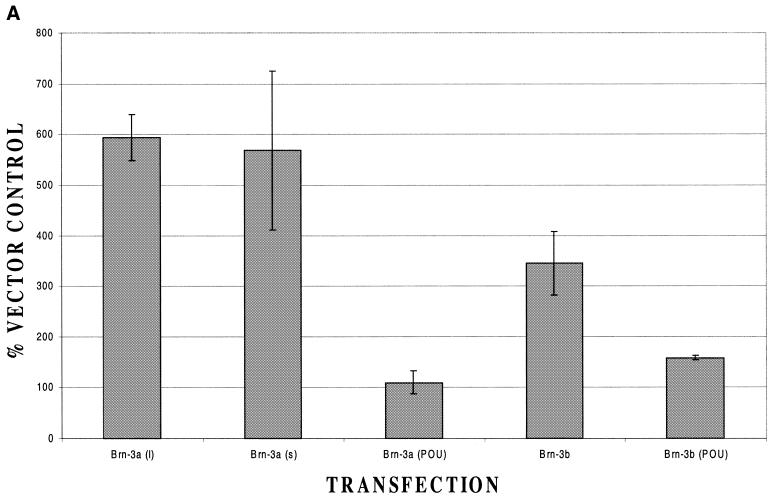
To determine which of these classes Bcl-x belongs to, we co-transfected the Bcl-x promoter with constructs encoding the long or short form of Brn-3a or the isolated POU domain. For comparison, we also included Brn-3b and its isolated POU domain. In these experiments (Fig. 4A), an equally strong activation was observed in the case of both the long and the short forms of Brn-3a. However, no significant activation was observed with the POU domain. Similarly, a weak activation was observed with full-length Brn-3b but not with its isolated POU domain. This response renders Bcl-x unique in the Brn-3a-responsive promoters investigated so far in that its activation does not appear to require the 84 amino acids found only in the long form of Brn-3a but does require other elements of the protein, which are not present in the isolated C-terminal POU domain.
Figure 4.
Response of the Bcl-x promoter to different forms of Brn-3a and Brn-3b. (A) Response of the Bcl-x promoter construct to the long form of Brn-3a, the naturally occurring short form, which lacks the 84 N-terminal amino acids, or to the isolated POU domain. The response to the related Brn-3b factor and its isolated POU domain is shown for comparison. (B) Response of the Bcl-x promoter to chimeric constructs containing different regions derived from Brn-3a or Brn-3b. In each case, the Brn-3 factor has been divided into four regions with the most N-terminal region being the sequence unique to the long form of Brn-3a whilst the most C-terminal region is the POU domain (22,23). In each case the region derived from Brn-3a or Brn-3b is indicated by the appropriate letter. Thus, construct AABB contains the N-terminal domain and domain 2 of Brn-3a whilst having domain 3 and the C-terminal POU domain derived from Brn-3b. Similarly, the construct –BBA lacks the N-terminal domain and has domains 2 and 3 derived from Brn-3b with the C-terminal POU domain derived from Brn-3a. In both panels, values are the average of three independent determinations whose standard error is shown by the bars. Domain boundaries are: Brn-3a: domain 1, amino acids 1–40; domain 2, amino acids 41–108; domain 3, amino acids 109–267; domain 4, amino acids 268-end. Brn-3b: domain 2, amino acids 1–92; domain 3, amino acids 93–169; domain 4, amino acids 170-end.
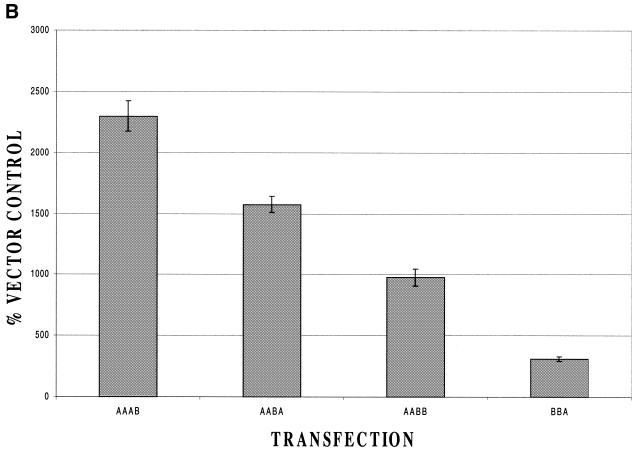
To investigate this further, we made use of the much stronger response to Brn-3a compared with Brn-3b and investigated the response of the Bcl-x promoter to plasmids encoding chimeric forms of Brn-3 which were partly derived from Brn-3a and partly from Brn-3b. Each of these chimeras is denoted by a four-letter code of the form A/B, which indicates which of the four domains of Brn-3 are derived from A and B. The first letter indicates the presence or absence of the N-terminal domain unique to the long form of Brn-3a whilst the last letter indicates the source of the POU domain. We have previously used these constructs to investigate the responses of a number of different promoters to Brn-3a/Brn-3b (for example, 25).
In these experiments (Fig. 4B) the strongest response was obtained with the chimera AAAB confirming that the POU domain need not be derived from Brn-3a for activation to occur. In contrast, very weak activation was observed with the chimera –BBA, indicating that sequences upstream of the POU domain must be derived from Brn-3a for full activation to occur. In general, this experiment supports the idea that sequences in domains 2 and 3 downstream of the N-terminal domain but upstream of the C-terminal domain are important for promoter activation by Brn-3a as the strongest activation was observed with the chimera which had both these regions derived from Brn-3a. Similarly, significant activation was observed with the two chimeras where only one of these regions was derived from Brn-3a and no significant activation with the chimera in which both these regions were derived from Brn-3b. Hence, experiments with the chimeras and with the long and short forms of Brn-3a, as well as its isolated POU domain, clearly indicate that Bcl-x is a unique promoter in that its activation by Brn-3a appears to be independent of the N-terminal domain and the source of the C-terminal POU domain and to depend critically on the intervening region.
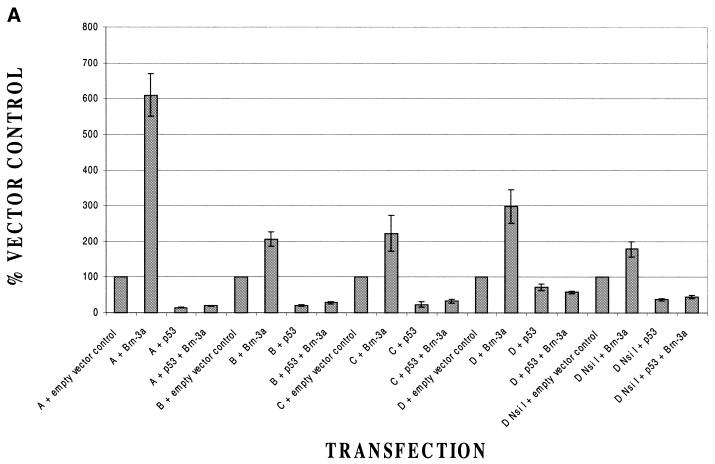
We have reported previously that p53 is able to abolish the activation of the Bcl-2 promoter by Brn-3a (11). In view of the similarities and differences between Bcl-2 and Bcl-x modulation by Brn-3a, we therefore wished to determine whether activation of the Bcl-x promoter by Brn-3a was also inhibited by p53. We therefore transfected each of our Bcl-x constructs with Brn-3a or with an empty expression vector in the presence or absence of p53. The results of this experiment (Fig. 5A) revealed that in the case of all constructs tested, activation by Brn-3a was abolished by p53. This applied even to the shortest construct able to be activated, Bcl-x D NsiI, which contains only promoter sequences from –156 to –19 from the translational start site. Evidently, the shortest construct Bcl-x D XhoII (Fig. 1) was not tested in this experiment, as it is not activated by Brn-3a (Fig. 2). Therefore, these experiments indicate that the minimal Bcl-x promoter that is inducible by Brn-3a is also inhibited by p53 and is in contrast to the result on the Bcl-2 promoter where an upstream promoter region mediates activation by Brn-3a and inhibition by p53 (11). The inhibitory effect of p53 on the Bcl-2 and Bcl-x promoters was specific as p53 did not inhibit either the basal activity of the α-internexin promoter or its activation by Brn-3a (Fig. 5B).
Figure 5.
Effect of p53 on the activation of various Bcl-x promoter constructs by Brn-3a. Activity of Bcl-x (A) or control α-internexin (B) promoter constructs with Brn-3a or with empty vector control in the presence or absence of co-transfected p53. All values have been equalised to that observed with each construct in the absence of Brn-3a or p53. Note the inhibition of Brn-3a activation by p53 in all the Bcl-x promoter constructs but not with the α-internexin construct. Values are the average of three independent determinations whose standard error is shown by the bars.
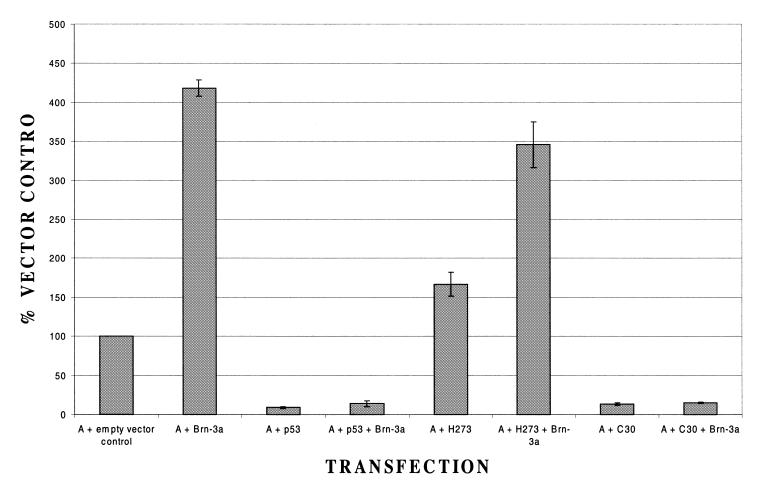
In our previous experiments with the Bcl-2 promoter (11), we demonstrated that different mutants of p53 had distinct effects on activation by Brn-3a. In particular, a mutant lacking the last 30 C-terminal amino acids of p53 was able to block activation by Brn-3a in the same manner as wild-type p53. In contrast, a mutant in which the arginine at position 273 had been changed to histidine and which cannot bind to DNA (28,29) was completely unable to prevent activation by Brn-3a.
When these mutants were tested for their effect on the activation of the Bcl-x promoter by Brn-3a, we observed exactly the same result as with the Bcl-2 promoter (Fig. 6). Thus, the deletion of the last 30 amino acids of p53 had no effect on its ability to block activation by Brn-3a whereas the arginine to histidine change at position 273 completely abolished the ability to block activation by Brn-3a. Hence, at least in the case of these two mutants, the Bcl-2 and Bcl-x promoters are indistinguishable with repression of Brn-3a activation being possible without the C-terminal regulatory domain of p53 but being abolished by a mutation which prevents DNA binding of p53 (Fig. 6).
Figure 6.
Response of the Bcl-x promoter construct to different mutants of p53. In each case the Bcl-x promoter construct has been co-transfected either with Brn-3a or with empty expression vector, in the presence or absence of either wild-type p53 or the two p53 mutants in which arginine 273 has been changed to histidine (H273) or the C-terminal 30 amino acids have been deleted (C30). Values are the average of three independent determinations whose standard error is shown by the bars.
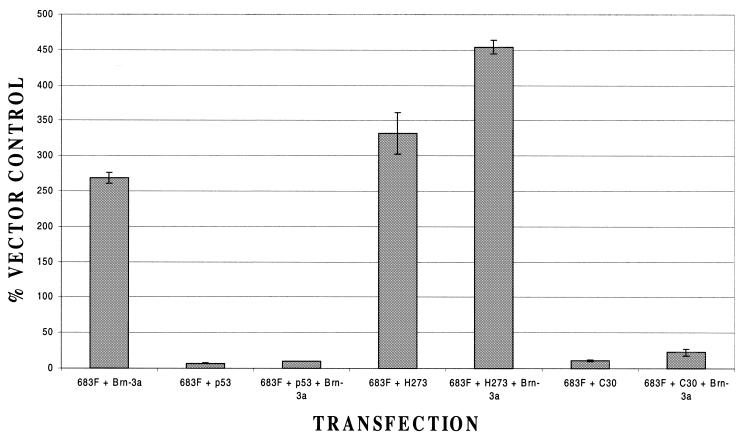
These experiments thus establish the promoter region immediately upstream of the translational start site for Bcl-x as a target for activation by Brn-3a and for repression of such activation by p53. Interestingly, it has been reported recently that a further Bcl-x promoter exists >1500 bases upstream of the translational start site and that transcription from this promoter results in the inclusion of an alternate first exon (exon 1B) in the resulting RNA (18). To determine whether this upstream promoter was also responsive to Brn-3a, we used a construct in which a 683 bp fragment containing only this promoter has been linked to a luciferase reporter gene (18). This construct was co-transfected with the Brn-3a expression vector. As indicated in Figure 7, this promoter was activated by Brn-3a, although not to such a significant extent as the downstream promoter. Moreover, as in the case of the downstream promoter, activation was abolished by p53 (Fig. 7). Similarly, as in the case of the downstream promoter and the Bcl-2 promoter, a mutant lacking the 30 C-terminal amino acids of p53 continued to abolish Brn-3a-mediated transactivation. In contrast, this effect appeared to be lost in the histidine 273 mutant with Brn-3a still activating the promoter, although this effect was complicated by the ability of this mutant to itself transactivate the promoter (Fig. 7).
Figure 7.
Response of the upstream Bcl-x promoter to Brn-3a and p53. Luciferase activity is shown for the upstream promoter construct (683F) transfected either with empty expression vector (set at 100%) or the Brn-3a expression vector. Results for this co-transfection in the absence of any p53 are compared with the effect of adding either wild-type p53, the H273 mutant or the mutant lacking the C-terminal 30 amino acids (C30). In all cases values are the average of three independent determinations whose standard error is shown by the bars.
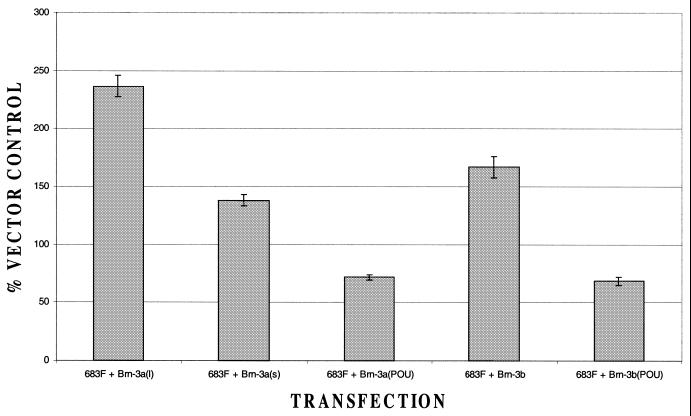
Interestingly, when we tested the effects of the long and short forms of Brn-3a on the upstream promoter, the short form produced only very weak activation and was considerably less effective in activating the promoter than the long form (Fig. 8) This indicates that, in contrast to the downstream promoter (see Fig. 4A), activation of the upstream Bcl-x promoter requires the N-terminal domain of Brn-3a, which is also required for activation of the Bcl-2 promoter (8,30). As in the case of the downstream Bcl-x promoter (Fig. 4A) and the Bcl-2 promoter (8,30), the isolated POU domain of Brn-3a had no effect on the upstream Bcl-x promoter (Fig. 8).
Figure 8.
Response of the upstream Bcl-x promoter to different forms of Brn-3a. Luciferase activity is shown for the upstream promoter construct transfected either with empty vector (set at 100%) or with expression vectors encoding different forms of Brn-3a or Brn-3b. Values are the average of three independent determinations whose standard error is shown by the bars.
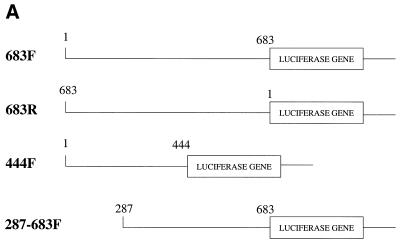
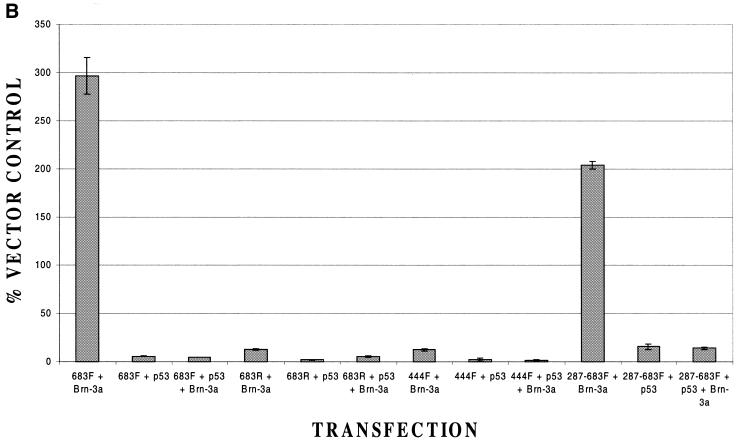
To determine whether, as in the case of the downstream promoter, a minimal upstream Bcl-x promoter was inducible by Brn-3a, we used constructs containing shorter fragments from the region containing this promoter (Fig. 9A). In these experiments, constructs containing this region in the inverse orientation to that originally tested (683R) or containing the first 444 bases of this region (444F) were inactive, as reported previously (18), and did not respond to Brn-3a or p53 (Fig. 9B). In contrast, a construct containing the downstream 400 bases of this region (287–683F) was active, as reported previously (18), and was able to be activated by Brn-3a in a p53-inhibitable manner (Fig. 9B) in the same way as the larger fragment. Hence, as in the case of the downstream promoter, the smallest tested fragment of the upstream promoter that had activity was regulated by Brn-3a and p53.
Figure 9.
Response of Bcl-x upstream promoter constructs to Brn-3a and p53. (A) Structure of the upstream Bcl-x promoter constructs. (B) Luciferase activity of the indicated upstream Bcl-x promoter constructs when co-transfected with Brn-3a or with empty expression vector in the presence or absence of p53. Values are the average of three independent determinations whose standard error is shown by the bars.
These experiments thus indicate that the minimal upstream and downstream Bcl-x promoters are targets for transactivation by Brn-3a and that such transactivation is inhibited by p53. In these respects, the Bcl-x promoters thus resemble the Bcl-2 promoter which we showed previously to be activated by Brn-3a (8) and repressed by p53 with the same response to the different mutants as occurs for the Bcl-2 promoter (11). Nonetheless, the findings presented here reveal some differences between the Bcl-2 and Bcl-x promoters. Thus, whilst activation of the Bcl-2 promoter by Brn-3a and its repression by p53 are mediated by distinct upstream binding sites (8,11) the response of the downstream Bcl-x promoter to Brn-3a and p53 appears to be mediated by the minimal promoter. In addition, whilst activation of Bcl-2 and the upstream Bcl-x promoter depends upon the presence of the N-terminal domain of Brn-3a (8,30), this is not the case for the downstream Bcl-x promoter although further sequences in addition to the C-terminal POU domain are required for activation of this promoter.
The similar response of the downstream Bcl-x promoter to the long and short forms of Brn-3a, only one of which contains the N-terminal domain required to activate the Bcl-2 promoter, may be of significance as these forms are generated by alternative splicing and their relative proportion is regulated in different neuronal cell types and in response to specific stimuli (27). Thus, under some conditions, Bcl-x expression may be induced by Brn-3a where induction of Bcl-2 expression does not occur. This may be of particular significance in view of the key role played by Bcl-x in regulating the extent of programmed cell death during the development of the nervous system (12–14).
Despite these differences, both Bcl-2 and Bcl-x are similar in that activation of their promoters by Brn-3a is abolished by p53. This is not the case for a variety of other Brn-3a-activated promoters. Thus, for example, the α-internexin promoter whose activation is dependent upon the N-terminal domain of Brn-3a and the neurofilament promoters whose activation is dependent upon the POU domain of Brn-3a are all unaffected by p53 (11; Fig. 5B). This suggests that p53 specifically targets the activation of Brn-3a-responsive promoters whose corresponding proteins have an anti-apoptotic effect regardless of the other characteristics of these promoters. To fully test this possibility, it will be necessary to study the effect of Brn-3a and p53 on the endogenous Bcl-2 and Bcl-x genes in cells with endogenous wild-type or mutant p53. Similarly, it will evidently be of interest to characterise the effect on apoptosis of over-expressing the anti-apoptotic Brn-3a protein together with the pro-apoptotic p53 protein as well as to examine the effect of Brn-3a on promoters that are activated by p53 such as that encoding the pro-apoptotic Bax member of the Bcl-2 family.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the Association for International Cancer Research, the Cancer Research Campaign, the Leukaemia Research Fund, the Medical Research Council and the Sir Jules Thorn Charitable Trust.
REFERENCES
- 1.Ryan A.K. and Rosenfeld,M.G. (1997) POU domain family values:- flexibility, partnerships and developmental codes. Genes Dev., 11, 1207–1225. [DOI] [PubMed] [Google Scholar]
- 2.Verrijzer C.P. and Van der Vliet,P.C. (1993) POU domain transcription factors. Biochim. Biophys. Acta, 1173, 1–21. [DOI] [PubMed] [Google Scholar]
- 3.He X., Treacy,M.N., Simmons,D.M., Ingraham,H.A., Swanson,L.S. and Rosenfeld,M.G. (1989) Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature, 340, 35–42. [DOI] [PubMed] [Google Scholar]
- 4.Gerrero M.R., McEvilly,R.J., Turner,E., Lin,C.R., O’Connell,S., Jenne,K.J., Hobbs,M.V. and Rosenfeld,M.G. (1993) Brn-3.0 A POU domain protein expressed in the sensory, immune and endocrine systems that functions on elements distinct from known octamer motifs. Proc. Natl Acad. Sci. USA, 90, 10841–10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McEvilly R.J., Erkman,L., Luo,L., Sawchenko,P.E., Ryan,A.F. and Rosenfeld,M.G. (1996) Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature, 384, 574–577. [DOI] [PubMed] [Google Scholar]
- 6.Xiang M., Gan,L., Zhou,L., Klein,W.H. and Nathans,J. (1996) Targeted deletion of the mouse POU domain gene Brn-3a causes a selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement and impaired suckling. Proc. Natl Acad. Sci. USA, 93, 11950–11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erkman L., McEvilly,J., Luo,L., Ryan,A.K., Hooshmand,F., O’Connel,S.M., Keithley,E.M., Rapaport,D.H., Ryan,A.F. and Rosenfeld,M.G. (1996) Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature, 381, 603–606. [DOI] [PubMed] [Google Scholar]
- 8.Smith M.D., Ensor,E.A., Coffin,R.S., Boxer,L.M. and Latchman,D.S. (1998) Bcl-2 transcription from the proximal P2 promoter is activated in neuronal cells by the Brn-3a POU transcription factor. J. Biol. Chem., 273, 16715–16722. [DOI] [PubMed] [Google Scholar]
- 9.Ensor E., Smith,M.D. and Latchman,D.S. (2001) The Brn-3a transcription factor protects sensory but not sympathetic neurones from programmed cell death/apoptosis. J. Biol. Chem., 276, 5204–5212. [DOI] [PubMed] [Google Scholar]
- 10.Smith M.D., Melton,L.A., Ensor,E.A., Packham,G., Anderson,P., Kinloch,R.A. and Latchman,D.S. (2001) Brn-3a activates the expression of Bcl-XL and promotes neuronal survival in vivo as well as in vitro. Mol. Cell. Neurosci., 17, 460–470. [DOI] [PubMed] [Google Scholar]
- 11.Budhram-Mahadeo V.S., Morris,P.J., Smith,M.D., Midgeley,C.A., Boxer,L.M. and Latchman,D.S. (1999) p53 suppresses the activation of the Bcl-2 promoter by the Brn-3a POU family transcription factor. J. Biol. Chem., 274, 15237–15244. [DOI] [PubMed] [Google Scholar]
- 12.Veis D.J., Sorenson,C.M., Shutter,J.R. and Korsemeyer,S.J. (1993) Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys and hypopigmented hair. Cell, 75, 229–240. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama K., Nakayama,K.I., Negishi,I., Kuida,K., Sawa,H. and Loh,D.Y. (1994) Targeted disruption of Bcl-2 in mice: occurrence of gray hair, polycystic kidney disease and lymphocytopenia. Proc. Natl Acad. Sci. USA, 92, 3700–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenlund L.J. S., Korsemeyer,S.J. and Johnson,E.M.,Jr (1995) Role of Bcl-2 in the survival and function of developing and mature sympathetic neurons. Neuron, 15, 649–661. [DOI] [PubMed] [Google Scholar]
- 15.Motoyama N., Wang,F., Roth,K.A., Sawa,H., Nakayama,K., Nakayama,K., Negishi,I., Senju,S., Zhang,Q., Fujii,S. et al. (1995) Massive cell death of immature haematopoietic cells and neurons in Bcl-x deficient mice. Science, 267, 1506–1510. [DOI] [PubMed] [Google Scholar]
- 16.Grillot D.A.M., González-García,M., Ekhterae,D., Duan,L., Inohara,N., Ohta,S., Seldin,M.F. and Núñez,G. (1997) Genomic organisation, promoter region analysis and chromosome localisation of the mouse bcl-x gene. J. Immunol., 158, 4750–4757. [PubMed] [Google Scholar]
- 17.Luckow B. and Schutz,G. (1987) Cat constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res., 15, 5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacCarthy-Morrogh L., Wood,L., Brimmell,M., Johnson,P.W.M. and Packham,G. (2000) Identification of a novel human BCL-X promoter and exon. Oncogene, 19, 5534–5538. [DOI] [PubMed] [Google Scholar]
- 19.Wood J.N., Bevan,S.J., Coote,P., Darn,P.M., Hogan,P., Latchman,D.S., Morrison,C., Rougon,G., Theveniau,M. and Wheatley,S.C. (1990) Novel cell lines display the properties of nociceptive sensory neurons. Proc. Royal Soc. Ser. B, 241, 187–194. [DOI] [PubMed] [Google Scholar]
- 20.Gorman C.M. (1985) High efficiency gene transfer into mammalian cells. In Glover,D.M. (ed.), DNA Cloning, A Practical Approach. IRL Press, Oxford, Vol. 2, pp. 143–190.
- 21.Lakin N.D., Morris,P.J., Theil,T., Sato,T.N., Moroy,T., Wilson,M.C. and Latchman,D.S. (1995) Regulation of neurite outgrowth and SNAP-25 gene expression by the Brn-3a transcription factor. J. Biol. Chem., 270, 15858–15863. [DOI] [PubMed] [Google Scholar]
- 22.Smith M.D., Ensor,E., Wagner,J. and Latchman,D.S. (1999) Regulation of NGFI-A (egr-1) gene expression by the POU domain transcription factor Brn-3a. Mol. Brain Res., 74, 117–125. [DOI] [PubMed] [Google Scholar]
- 23.Budhram-Mahadeo V.S., Morris,P.J., Lakin,N.D., Theil,T., Ching,G.Y., Lillycrop,K.A., Moroy,T., Liem,R.K.H. and Latchman,D.S. (1995) Activation of the α-internexin promoter by the Brn-3a transcription factor is dependent on the N-terminal region of the protein. J. Biol. Chem., 270, 2853–2858. [DOI] [PubMed] [Google Scholar]
- 24.Smith M.D., Morris,P.J., Dawson,S.J., Schwartz,M.L., Schlaepfer,W.W. and Latchman,D.S. (1997) Co-ordinate induction of the three neurofilament genes by the Brn-3a transcription factor. J. Biol. Chem., 272, 21325–21333. [DOI] [PubMed] [Google Scholar]
- 25.Morris P.J., Theil,T., Ring,C.J.A., Lillycrop,K.A., Möröy,T. and Latchman,D.S. (1994) The opposite and antagonistic effects of the closely related POU family transcription factors on the activity of a target promoter are dependent upon differences in the POU domain. Mol. Cell. Biol., 14, 6907–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theil T., McLean-Hunter,S., Zornig,M. and Möröy,T. (1993) Mouse Brn-3 family of POU transcription factors: a new amino terminal domain is crucial for the oncogenic activity of Brn-3A. Nucleic Acids Res., 21, 5921–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y.Z., Dawson,S.J. and Latchman,D.S. (1996) Alternative splicing of the Brn-3a and Brn-3b transcription factor mRNAs is regulated in neuronal cells. J. Mol. Neurosci., 7, 77–85. [DOI] [PubMed] [Google Scholar]
- 28.Crook T., Marston,N.J., Sara,E.A. and Vousden,K.H. (1994) Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell, 79, 817–827. [DOI] [PubMed] [Google Scholar]
- 29.Park D.J., Nakamura,H., Chumakov,A.M., Said,J.W., Miller,C.W., Chen,D.L. and Koeffler,P. (1994) Transactivational and DNA binding properties of endogenous p53 in p53 mutant cell lines. Oncogene, 9, 1899–1906. [PubMed] [Google Scholar]
- 30.Smith M.D., Dawson,S.J., Boxer,L.M. and Latchman,D.S. (1998) The N-terminal domain unique to the long form of the Brn-3a transcription factor is essential to protect neuronal cells from apoptosis and for the activation of Bcl-2 gene expression. Nucleic Acids Res., 26, 4100–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]