Abstract
Schlafen (SLFN) genes belong to a vertebrate gene family encoding proteins with high sequence homology. However, each SLFN is functionally divergent and differentially expressed in various tissues and species, showing a wide range of expression in cancer and normal cells. SLFNs are involved in various cellular and tissue-specific processes, including DNA replication, proliferation, immune and interferon responses, viral infections, and sensitivity to DNA-targeted anticancer agents. The fundamental molecular characteristics of SLFNs and their structures are beginning to be elucidated. Here, we review recent structural insights into the N-terminal, middle and C-terminal domains (N-, M-, and C-domains, respectively) of human SLFNs and discuss the current understanding of their biological roles. We review the distinct molecular activities of SLFN11, SLFN5, and SLFN12 and the relevance of SLFN11 as a predictive biomarker in oncology.
Subject terms: Cancer, Cancer therapy
Schlafen proteins: Understanding structure and function could open therapeutic potential
The diverse roles that Schlafen family proteins play in cell proliferation, immune modulation, and other biological processes make them promising targets for treating and tracking diseases, especially cancer. Ukhyun Jo and Yves Pommier from the National Cancer Institute in Bethesda, USA, review the molecular characteristics and structural features of Schlafen proteins. These proteins take their name from the German word for “sleep”, as the first described Schlafen proteins caused cells to stop dividing, although later reports found that related members of the same protein family serve myriad cellular functions, including in the regulation of DNA replication. A better understanding of Schlafen proteins could open up new avenues in cancer management, for instance, diagnostics that monitor activity levels of one such protein, SLFN11, could help oncologists predict how well patients might respond to anti-cancer therapies.
Introduction
Repeated genes are classified as tandemly arrayed genes and clustered genes. Tandemly arrayed genes consist of duplications arranged in a head-to-tail fashion. This allows the rapid production of multiple copies of gene products, such as ribosomal RNAs, that act in similar biological functions. In contrast, clustered genes generally express physically and functionally divergent gene products. The genes are functionally linked to each other but play specific roles under different biological circumstances, contributing to genetic complexity and durability1,2. Schlafen (SLFN) genes are clustered genes that are evolutionally conserved in a wide range of vertebrate species3 and probably evolved from a common ancestor by multiple unequal recombination events.
The first SLFN genes (Slfn1, 2, 3, and 4) were identified in mice as a gene family expressed during thymocyte development and immune maturation and preferentially upregulated in the lymphoid cell lineage4. Subsequent studies added additional members to the SLFN gene family, the members of which were found to exist in gene clusters on the same chromosome in the mouse and human genomes3,5–8 (Fig. 1A). Individual SLFN genes are differentially involved in multiple cellular processes, including proliferation, differentiation, the immune response, suppression of viral infection, and DNA replication, and are related to chemosensitivity9–12. However, the range of functions of SLFNs remains only partially understood.
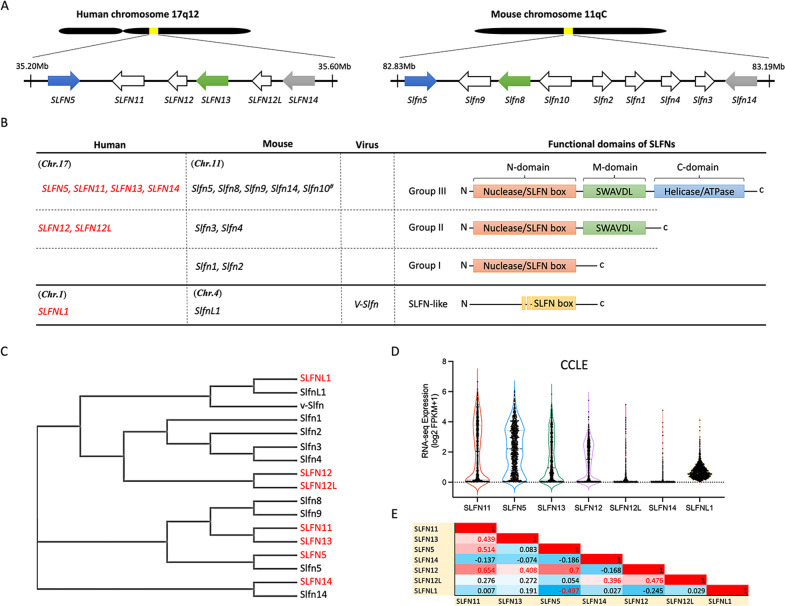
Fig. 1. The conserved SLFN family gene cluster.
A The locations of SLFN genes in human and mouse chromosomes. The colors indicate orthologous relationships between human and mouse SLFNs (blank: unknown). B Classification of SLFN genes and proteins in human, mouse, and virus. C Phylogenetic tree of SLFN proteins generated by ClustalW2. D Violin plot of human SLFN mRNA expression in the CCLE cancer cell database (CellMinerCDB, discover.nci.nih.gov/cellminercdb). E Correlation of the expression of SLFN genes in human.
Here, we describe the main characteristics of the human SLFN gene family and focus on the emerging structural insights related to SLFN11’s potential endoribonuclease and helicase/ATPase activity. We also summarize how SLFNs are increasingly being exploited in oncology.
The SLFN family
Over the past two decades, foremost SLFN investigations have focused on the human and mouse SLFN genes. Human SLFNs (SLFN5, 11, 12, 12L, 13, and 14) are clustered on chromosome 17, and mouse Slfn genes (Slfn1, 2, 3, 4, 5, 8, 9, and 14) and the pseudogene (Slfn10) are clustered on chromosome 11 (Fig. 1A). In addition, a SLFN-like gene containing a partial SLFN box has been identified in the genomes of various species, including human, mouse, and orthopoxvirus (Fig. 1B)13.
SLFN proteins are classified into three groups based on their structures and functional domains (Fig. 1B). Group I SLFNs consist of the common N-domain region containing a nuclease structure and a unique SLFN box conserved in all SLFN proteins2,14. Group II SLFNs contain the N-domain and a linker middle domain region (M-domain), including a SWAVDL motif and a potential protein-interacting region14. Group III SLFNs form the largest subgroup. They include a third functional putative helicase/ATPase C-terminal domain with Walker A/B motifs15. SLFN-like proteins contain partially conserved amino acid sequences with the SLFN box, but their biological activity remains unknown.
SLFN proteins are differentially localized in cells, reflecting their putative cellular functions. Mouse Group 1 and II SLFNs are predominantly found in the cytoplasm, while Group III SLFNs are present in the nucleus7. In contrast to mouse Slfn genes, human SLFN genes only encode polypeptides belonging to Group II (SLFN12) and Group III (SLFN5, 11, 13, and 14). While SLFN12 and SLFN13 are cytoplasmic, SLFN11, SLFN14, and SLFN5 are present in the nucleus because of their nuclear localization signal2,11,14,16,17. Although SLFN11 is primarily detected in the nucleus by immunostaining and immunofluorescence, it also modulates proteotoxic stress control and protein translation in the cytoplasm18,19.
Phylogenic analyses have shown homologous relationships between human, mouse, and virus SLFN protein sequences (Fig. 1C). Human SLFN5 and SLFN14 have direct mouse orthologs (Slfn5 and Slfn14, respectively). Mouse Slfn8 and SLFN13 are also functional homologs2. Human SLFN12/12L and SLFN11/13 show sequence similarity with mouse Slfn3/4 and Slfn8/9, respectively. Whether mouse Slfn9 is the ortholog of SLFN11 remains to be determined. Humans do not have gene orthologs or homologs for mouse Slfn1 and Slfn2.
Although SLFNs are clustered on the same chromosome, transcription profiling data in the Cancer Cell Line Encyclopedia (CCLE) cancer cell line database20 show that each SLFN is expressed independently of the others (Fig. 1D, E). SLFN11, SLFN5, SLFN13, and SLFN12 exhibit a wide range of transcription levels in cancer cells, whereas SLFN12L, SLFN14 and SLFNL1 are barely expressed in cancer cell lines.
Structure of SLFN proteins
Structural and biochemical studies have begun to reveal the molecular characteristics of SLFN proteins. The N- and C-domains appear to function as a nuclease and a helicases/ATPase, respectively, while the M-domain may be a linker connecting the two N- and C-terminal enzymatic domains and may potentially interact with other proteins.
The N-domain of SLFNs: an endoribonuclease domain
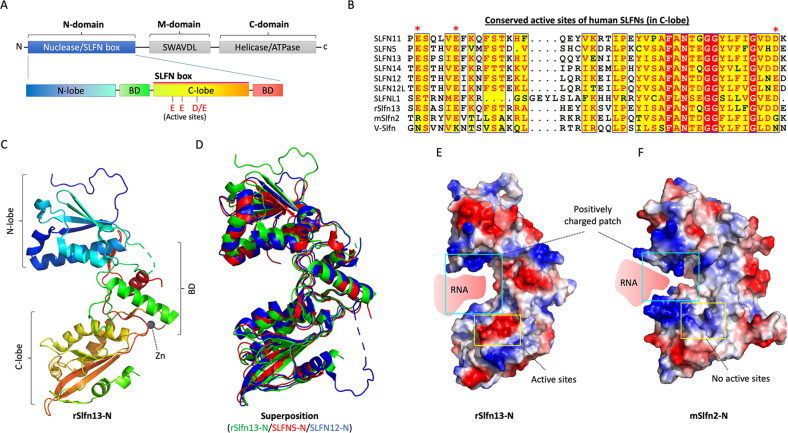
SLFN14 purified from rabbit reticulocytes shows novel endoribonuclease activity against rRNA and ribosome-associated mRNAs 21. The N-domain of SLFNs has been revealed as a key domain related to tRNA/rRNA endoribonuclease activity2,21. Structural analyses of rat Slfn13 (14~353 residues), which is the homolog of human SLFN13 and mouse Slfn8 (Fig. 2A, C), have shown that the N-domain consists of two lobes (N-lobe and C-lobe) between two bridging domains (BDs) (Fig. 2A). Structural conservation of the SLFN-N domain is also observed in SLFN12 and SLFN5, which shows only slightly different conformations (Fig. 2D)14,22,23. Notably, the C-lobe region (also referred to as the SLFN box) with high sequence identity between SLFN proteins includes the conserved active residues (EED: Glu-Glu-Asp) for ribonuclease activity (Fig. 2A, B). The SLFN-N domain has a U-shaped architecture and binds nucleotides via a positively charged patch in the valley (Fig. 2E). In contrast, the electrostatic surface of the ribonuclease active site is negatively charged, and its enzymatic activity relies on Mn2+/Mg2+ 2 (Fig. 2E). Although the active site is conserved among SLFN family members, the enzymatic activity of SLFNs varies. SLFN11 selectively suppresses the cellular type II tRNAs that are utilized to synthesize DNA repair response proteins such as ATR and ATM and HIV proteins21. SLFN13 cleaves the acceptor stem of tRNAs, implying that SLFNs recognize the secondary or tertiary structure of substrate RNAs2. In in vitro experiments, SLFN12 cleaves rRNA as an active RNase complex with phosphodiesterase 3A (PDE3A), enhancing DNMDP (6-(4-(diethylamino)-3-nitrophenyl)-5-methyl-4,5-dihydropyridazin-3(2H)-one)-induced cancer cell death14. Rabbit SLFN14 cleaves rRNA and ribosome-associated mRNA in a manner dependent on Mg2+ and Mn2+ in reticulocytes24. However, the substrate specificities of SLFNs have not yet been fully defined. In contrast to the other SLFNs, SLFN5 does not have endoribonuclease activity against tRNAs2, although its active site is conserved, implying that it might target single-stranded or double-stranded DNA. Conversely, the active site of mouse Slfn2 is positively charged (Fig. 2F). Yue et al. discovered that mouse Slfn2 shields tRNAs from cleavage by ribonucleases activated by oxidative stress in T cells, thereby counteracting translation inhibitory effects25. These observations suggest that the functions of SLFNs have evolutionally adapted according to their environments.
Fig. 2. The N-domain of SLFNs.
A Schematic diagram of an SLFN protein. The expanded diagram shows the N-domain of SLFN proteins with the conserved active site. B Sequence alignment of the conserved active site in human, mouse, and virus SLFNs. The sequence alignment was generated using ESPript. C Ribbon diagram of the crystal structure of rSlfn13-N (PDB: 5YD0). BD: bridging domain. D Superimposition of rSlfn13-N, SLFN5-N (PDB: 6RR9), and SLFN12-N (PDB: 7LRE). E The electrostatic surface potentials of rSlfn13-N. The colors indicate electric charge (red: negative and blue: positive). F The electrostatic surface potentials of mSlfn2-N (1-378) (modeled by AlphaFold: AF-Q9Z0I6-F1).
In addition, a putative zinc finger motif has been identified on the backside valley in SLFN5, SLFN12, and rSlfn13 (Fig. 2C)2,14,23, implying that the putative zinc finger might help in the recognition of nucleotide targets or assist protein folding and nuclease/ATPase activity in the M and C domains. The zinc finger motif is also conserved in all human and mouse SLFNs, indicating that nucleotide-binding capacity is one of the molecular features of SLFNs.
The M-domain of SLFNs: a linker and protein-interacting domain
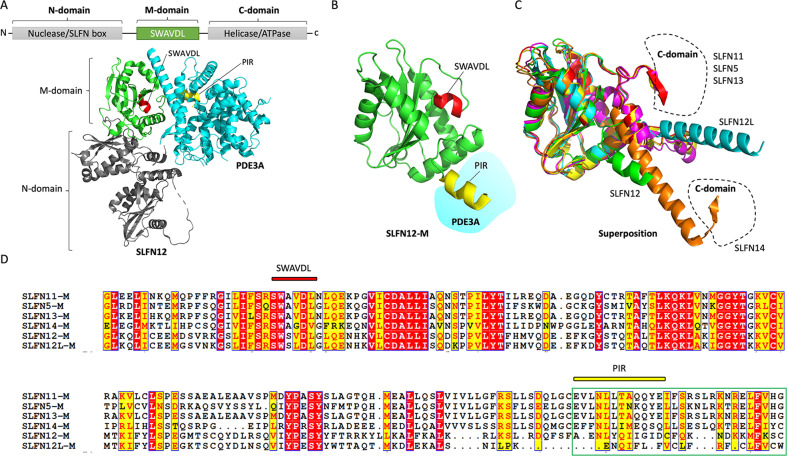
Recently, the M-domain of human SLFN12, which binds PDE3A through its PDE3A interacting region (PIR), has been defined using cryo-electron microscopy14 (Fig. 3A, B). The M-domain of human SLFN12 also includes the conserved SWAVDL sequence common to all SLFN family members (Fig. 3B, D). The C-terminal region of the M-domain of human Group II SLFNs (SLFN12 and SLFN12L) is structurally distinct from that of the Group III SLFNs (SLFN11, SLFN5, SLFN13, and SLFN14). Group II SLFNs show a stretched-out helix end (Fig. 3C), whereas the M-domains of the Group III SLFNs are kinked where they connect to their putative helicase/ATPase in the C-domain23. Among the Group III SLFNs, SLFN14 exhibits a longer helix, with a different connection between its linker region and its C-domain (Fig. 3C).
Fig. 3. The M-domain of SLFNs.
A Schematic diagram of the M-domain of SLFN proteins. The ribbon diagram shows the cryo-EM structure of SLFN12-PDE3A (PDB: 7LRD) including the M-domain (green), SWAVDL motif (red), and PIR (PDE3A interacting region; yellow). B The expanded M-domain of SLFN12. C Superimposition of SLFN12-M (PDB: 7LRE), SLFN11-M (AlphaFold: AF-Q7Z7L1-F1), SLFN5-M (AlphaFold: AF-Q08AF3-F1), SLFN12L-M (AlphaFold: AF-Q6IEE8-F1), SLFN13-M (AF-Q68D06-F1), and SLFN14-M (AlphaFold: AF-P0C7P3-F1). D Sequence alignment of the M-domains in human SLFNs. The sequence alignment was generated using ESPript.
SLFN14 utilizes its M-domain to bind to ribosomes, and alteration of the M-domain reduces endonucleolytic RNA cleavage activity24, indicating that the M-domain of SLFNs might be a docking site for nucleic acids or for functional cofactors, as seen in the SLFN12-PDE3A complex. Molecular interactions between SLFN12 and ribosomal proteins (RPS27A, RPS6, and RPL7A) have also been detected after treatment with 17-β-estradiol (E2), thereby inhibiting the translation of ER-mediated antiapoptotic proteins (Bcl-2 and Mcl-1)26. Similarly, SLFN13 might negatively modulate translation by cleaving ribosomal RNAs2. Furthermore, SLFN11 suppresses the proliferation of hepatocellular cancer cells by interacting with the ribosomal protein S4 X-linked (RPS4X), resulting in attenuation of S6 and eIF4E phosphorylation in the ribosome complex and inhibition of the mTOR signaling pathway27. SLFN11 also functionally associates with protein folding and translation initiation complexes to protect cells from proteotoxic stress18. Further studies are warranted to clarify how the M-domain of SLFNs affects ribosome interactions.
The C-domain of group III SLFNs: a putative helicase/ATPase domain
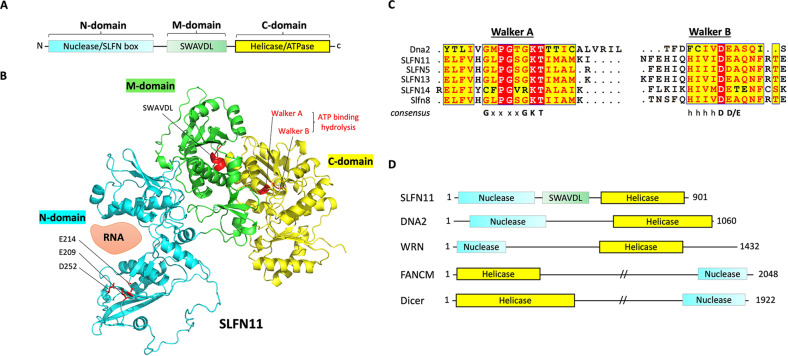
The C-domain of Group III SLFNs bears homology to superfamily I RNA/DNA helicases5 (Fig. 4A). Protein modeling shows that the C-domain region of SLFN11 structurally resembles the structure of Dna2, a nuclease-helicase that controls genomic integrity28,29 (Fig. 4B). The C-domain of SLFN11 contains conserved Walker A and B motifs, suggesting ATPase activity. This ATPase motif characterizes all Group III SLFNs (Fig. 4C), implying that the C-domain of SLFNs might function in chromatin remodeling via RNA/DNA helicase activity, as seen in other helicases, including DNA2, WRN, FANCM, and Dicer (Fig. 4D). The ATPase activity of the SLFN helicase motif has been established by using SLFN11 variants (K605M/D668A and E669Q) in the Walker A and B motifs17,30. The helicase activity is required for SLFN11-mediated chemosensitivity to DNA-damaging agents17 and replication fork degradation. It abolishes the recruitment of RAD51 at stalled forks in Fanconi anemia cells, thereby exposing the stalled forks to the nucleases MRE11 and DNA231.
Fig. 4. The C-domain of SLFNs.
A Schematic diagram of the C-domain of SLFN proteins. B The ribbon diagram shows the modeled structure of SLFN11 (AlphaFold: AF-Q7Z7L1-F1). The annotations indicate the localization of key elements in SLFN11. C Sequence alignment of the Walker A/B motifs in human SLFNs, mouse Slfn8, and mouse Dna2. The sequence alignment was generated using ESPript. D Schematic diagram of selected proteins targeted by DNA/RNA helicases.
The ATPase activity of SLFN11 is essential for the killing of cancer cells in response to replicative DNA-damaging agents and chromatin opening, which leads to lethal replication arrest and activation of cellular stress response genes in the FOS-JUN pathways17. In addition, the D668A/E669A mutant of SLFN11 fails to attenuate prototype foamy virus (PFV) replication32, suggesting that the ATPase activity of the C-domain plays a role in the antiviral properties of Group III SLFNs19,33,34.
SLFN5 binds to HSV-1 DNA to interrupt its accessibility to RNA polymerase II, thereby blocking the transcription of viral promoters in host cells35. SLFN5 also negatively controls STAT1-mediated transcriptional activation of IFN-stimulated genes and ZEB1 transcription, suppressing the antitumor immune response in glioblastoma cells and the mesenchymal-epithelial transition36,37. The inability of SLFN5 C-domain mutant to inhibit transcription implies the importance of its helicase/ATPase activity. Further studies are warranted to determine how the putative ATPase activity regulates DNA replication and transcription and how the C-domain of Group III SLFNs makes them functionally distinct from the Group I and II SLFNs, as helicases commonly participate in various cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair, and ribosome biogenesis, by remodeling RNA/DNA strands using ATP hydrolysis38–41.
Putative posttranslational modifications of SLFNs
Posttranslational modifications such as phosphorylation, acetylation, and ubiquitination fine-tune the activity and functional localization of helicases according to different steps of the cell cycle and biological processes41,42. SLFN11 has been shown to be phosphorylated in both its N-domain (S219 and T230) and its C-domain (S753)43. Upon DNA damage, protein phosphatase 1 catalytic subunit γ (PPP1CC) dephosphorylates SLFN11 to increase its activity, thereby sensitizing cancer cells to the topoisomerase I inhibitor, camptothecin. Similarly, SLFN12 dephosphorylation (S368 and S573) is induced by cytotoxic PDE3A modulators, promoting its RNase activity and leading to cell death44. Ubiquitination has been observed for SLFN14 when it is misfolded due to missense mutations45. Hence, it is likely that posttranslational modifications regulate the cellular function of SLFN proteins.
v-Slfn and SLFNL1
As mentioned above in the review of the classification of the SLFN family (Figs. 1B and 2B), a partially conserved SLFN sequence has been detected in the C-terminal region of SLFN-like proteins in human, mouse, and virus. The expression of virus Schlafen (v-Slfn) was first observed during infection by the camelpox virus, in which v-Slfn modulated virulence and showed predominantly cytoplasmic localization in the host cells13. v-Slfn is conserved across orthopoxviruses. Notably, v-Slfn encodes a 57 kDa protein consisting of poxin fused with the SLFN N- and C-terminal domains46. The poxin domain, which includes a viral cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) nuclease, inactivates the cGAS-STING pathway, consistent with the importance of the cGAS-STING signaling pathway for antiviral responses against orthopoxviruses. However, details of the functional roles of the SLFN domain in v-Slfn remain to be elucidated.
Human and mouse also express the SLFN-like protein SLFNL1, which resembles v-Slfn (Figs. 1B and 2B). SLFNL1 contains a partial SLFN domain in the C-terminus and an unknown N-terminal domain, thus being evolutionally divergent from v-Slfn. Further studies are warranted to understand the underlying molecular mechanism of SLFNL1 in cells.
Exploitation of human SLFNs for cancer therapy
The diverse functions of SLFNs in key cellular processes, such as DNA replication, cell proliferation, transcription, protein folding, and cell motility, highlight the potential of SLFNs as therapeutic targets and biomarkers for diagnosis, therapeutic decision, and prognosis.
SLFN11
Replication checkpoint activity of SLFN11
During the last decade, SLFN11 has been extensively studied due to its relevance in cancer research. SLFN11 acts as a negative replication checkpoint in response to replication stress in parallel to the ATR pathway47,48. When replication stress is induced by endogenous or exogenous factors, SLFN11 proteins are recruited and accumulate in the proximity of DNA lesions in coordination with the single-stranded binding protein replication protein A (RPA), forming foci30,49. Chromatin-bound SLFN11 destabilizes replication by interacting with RPA, the replicative helicase minichromosome maintenance complex component 3 (MCM3), and chromatin licensing and DNA replication factor 1 (CDT1), leading to irreversible replication arrest17,50, whereas ATR activates the downstream kinase CHK1 to transiently arrest the cell cycle and enable repair. SLFN11 simultaneously targets chromatin to modify structural accessibility and activate transcription of immediate early genes (IEGs), including JUN, FOS, EGR1, NFKB2, ATF3, CDKN1A (p21WAF1), and the growth arrest and DNA damage-inducible gene GADD4517,30,50,51 (Fig. 5B). In addition, a recent study in Fanconi anemia cells showed that SLFN11 hinders the binding of the single-strand binding recombination protein RAD51 at stalled forks and destabilizes nascent DNA tracts, leading to degradation of the stalled forks by the nucleases MRE11 and DNA231.
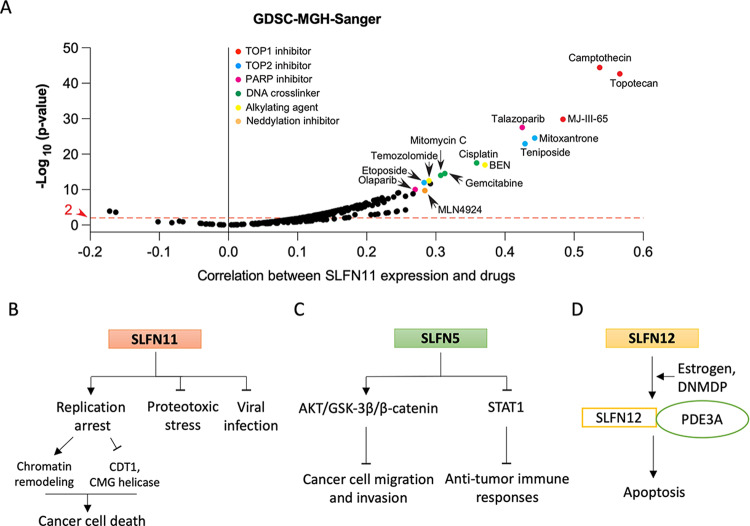
Fig. 5. SLFNs in cancers.
A Correlation of SLFN11 expression with sensitivity to anticancer drugs in the GDSC-MGH-Sanger database (each point is a drug; n = 297) analyzed using CellMinerCDB (https://discover.nci.nih.gov/cellminercdb). B Proposed signaling model for SLFN11 as a replication checkpoint. C Proposed signaling model for SLFN5 as a tumor suppressor and oncogene. D Proposed functional model for SLFN12 with PDE3A activated by estrogen and DNMDP.
SLFN11 as a biomarker predicting sensitivity to DNA-damaging agents (DDAs)
Given that SLFN11 is a key responder to replication stress, its expression status is being actively investigated as a biomarker for drug selection and prognosis in cancer therapy with broadly used DNA-damaging agents (DDAs), including topoisomerase I (TOP1) inhibitors (topotecan, irinotecan, and indotecan), TOP2 inhibitors (etoposide, doxorubicin, and epirubicin), alkylating and crosslinking agents (cyclophosphamide, temozolomide, cisplatin, carboplatin, and oxaliplatin), and DNA synthesis inhibitors (5-fluorouracil, gemcitabine, cytarabine, and hydroxyurea)12,52–54 (Fig. 5A). SLFN11 expression is also correlated with vulnerability to poly-(ADP)-ribose polymerase (PARP) inhibitors (olaparib, veliparib, talazoparib, and niraparib)47,54,55. Conversely, a lack of SLFN11 expression can cause resistance to DDAs12,52. SLFN11 mRNA expression data in the National Cancer Institute Antitumor Cell Line Panel (NCI-60), Cancer Cell Line Encyclopedia (CCLE), and Genomics of Drug Sensitivity in Cancer (GDSC) datasets show that SLFN11 is not expressed in ~50% of cancer cells that exhibit poor response to clinically used DDAs. The relationship between SLFN11 expression and sensitivity to DDAs has been assessed in colorectal, ovarian, lung, breast, head and neck, gastric, esophageal, and prostate cancers, as well as sarcomas56–66.
The lack of SLFN11 expression is primarily due to epigenetic changes in DNA methylation in the SLFN11 gene promoter67. The promoter CpG islands of SLFN11 are frequently hypermethylated in colorectal cancers, leading to poor prognosis and chemoresistance68,69. The gene body of SLFN11 is also targeted by the histone modifier EZH2 (enhancer of zeste homology 2) during acquired chemoresistance in small-cell lung cancer (SCLC) cells, which increases H3K27me3 and local chromatin condensation70. Since epigenetic changes can be easily detected in patient samples, DNA methylation can be utilized as a surrogate marker to determine susceptibility to DDAs. Therapeutic strategies using inhibitors of EZH2, histone deacetylases (HDACs), and DNA methyltransferases (DNMTs) have been shown to reactivate SLFN11 expression and overcome resistance to DDAs68,70,71.
Given the therapeutic relevance of SLFN11 in oncology, the way to determine SLFN11 DNA, RNA, and protein expression in clinical samples are being actively investigated. Advances in microarray and sequencing technologies have made genome-wide profiling to detect SLFN11 mRNA and gene methylation. Immunohistochemistry (IHC) has also been successfully evaluated as a biomarker in two clinical trials of PARP inhibitors (NCT03880019 and NCT04334941) with various tumor types72,73. The reliability of determining SLFN11 expression status by IHC has been confirmed in a large number of patient tumors with various histologies48,56,62,72,73. Thus, IHC could readily be applied to evaluate the expression of SLFN11 in the clinical setting.
Therapeutic strategies including SLFN11 and ATR inhibitors
The chemoresistance of SLFN11-negative cancer cells can be overcome by combining DDAs with ATR inhibitors47,48,50,74,75. Because SLFN11-deficient cancer cells rely on the ATR pathway to modulate their DNA replication and damage repair processes in response to replication stress, the combination of ATR inhibitors with DDAs is selectively active in SLFN11-negative cancer cells47,76. These observations suggest that the expression status of SLFN11 can be used alongside cancer therapy with ATR inhibitors, which are in late-phase clinical development.
Overexpression of SLFN11
Overexpression of SLFN11 is observed in leukemia and sarcoma cells, implying that it might be involved in the development of certain cancer types48,53,77,78. The oncogenic EWS-FLI fusion transcriptionally activates SLFN11 expression in Ewing’s sarcoma78. In addition, gain-of-function mutations in the JAK signaling pathway in acute leukemia cells have recently been shown to cause high expression of SLFN11 due to abnormal activation of the upstream ETS transcription factor77. How overexpression of SLFN11 is related to tumorigenesis and the cytoplasmic roles of SLFN11 need to be studied in further research.
Cytoplasmic roles of SLFN11
In addition to its replication stress checkpoint functions, SLFN11 interacts with ribosomal protein S4 X-linked (RPS4X) and suppresses the mTOR signaling pathway in hepatocellular carcinoma (HCC), inhibiting HCC growth and metastasis27. SLFN11 also protects cells from proteotoxic stress caused by the accumulation of unfolded proteins, while its deficiency increases the cellular levels of ubiquitin conjugates due to uncontrolled endoplasmic reticulum stress and protein quality control18. A difference in the activity of the proteotoxic stress response pathway was recently found to explain why the clinically developed and first-in-class inhibitor of the ubiquitin-activating enzyme UBA1, TAK-243, selectively targets SLFN11-deficient cells18. Further studies are warranted to identify anticancer drugs that function specifically in SLFN11-negative cancers that could be used alone or in combination with TAK-243.
SLFN5
As a member of SLFN Group III, SLFN5 shares most of the structural domains of SLFN11, SLFN13, and SLFN14. However, the endoribonuclease activity of SLFN5 appears defective, suggesting that SLFN5 is functionally unique in the SLFN family2,23. Nevertheless, SLFN5 expression is still induced by interferon (IFN), and IFN-activated SLFN5 localizes mainly in the nucleus and suppresses the anchorage-independent growth of melanoma cancer cells79. Negative regulation of cell motility and invasiveness has also been reported for SLFN5 in renal cell carcinoma (RCC), in which SLFN5 expression is positively correlated with survival benefit80.
Mechanistically, SLFN5 also interacts with the NOTCH/TGF-β signaling pathway and suppresses matrix metalloproteinases-1 (MMP-1) and MMP-13, which are required for the degradation and rearrangement of extracellular matrix (ECM) proteins, thereby blocking morphological changes. SLFN5 also downregulates MMP14 expression through inhibition of the β-catenin pathway81 (Fig. 5C). As a transcriptional repressor, SLFN5 prevents epithelial-mesenchymal transition (EMT) in breast cancer and targets the ZEB1 promoter to abrogate ZEB1 transcription and the downstream PTEN/AKT/cyclin D1 signaling cascade, ultimately prompting cancer cell death37,82. These observations suggest that SLFN5 could be exploited as a biomarker for cancer therapy with IFN stimulation.
SLFN5 expression has been correlated with cancer progression. In gastric cancer (GC) cells, SLFN5 has been associated with the aggressive transition from intestinal metaplasia to GC83. In glioblastoma, SLFN5 promotes tumor formation, growth, and invasion, suppressing STAT1-driven gene transcription36 (Fig. 5C). Oncogenic SLFN5 expression has also been observed in castration-resistant prostate cancer patients with poor outcomes. A direct interaction between SLFN5 and ATF4 has been proposed to regulate the L-type AA transporter LAT1, which activates the mTOR signaling pathway84. The apparently divergent roles of SLFN5 as a tumor suppressor and tumor promoter need to be investigated in further studies.
SLFN13 and SLFN14
Yang et al. provided the first structural insights demonstrating that the conserved N-domain of SLFN cleaves tRNA and rRNA by endoribonuclease activity2. In glioblastoma, high expression of SLFN13 mRNA is detected along with poor overall survival36. In thrombocytopenia patients, SLFN14 mutations (K218E, K219N, and V220D) localized near the active sites of the SLFN box have been identified16. These variants of SLFN14 are associated with platelet secretion defects, suggesting that SLFN14 mutations might be related to preneoplastic changes.
SLFN12 and SLFN12L
SLFN12 is a human Group II SLFN lacking the C-terminal helicase domain (Fig. 1A). SLFN12 might play similar roles to the Group III SLFNs SLFN11, 5, 13, and 14 by associating with a functional partner molecule that compensates for the missing C-terminal domain85. SLFN12 interacts with phosphodiesterase 3A (PDE3A), which is a regulator of the development of interstitial cells of Cajal and gastrointestinal stromal tumors (GISTs)85,86. SLFN12 has been proposed as a therapeutic target and predictive biomarker for PDE3 inhibitors such as zardaverine and quazinone87. The drug activity of PDE3 inhibitors has also been shown to be enhanced by coexpression of PDE3A and SLFN1285,87,88. Diverse chemical modulators, including 17-β-estradiol (E2), anagrelide, nauclefine, and DNMDP (6-(4-(diethylamino)-3-nitrophenyl)-5-methyl-4,5-dihydropyridazin-3(2H)-one), lead to apoptotic cell death by enhancing the molecular interaction between PDE3A and SLFN12 and increasing SLFN12 RNase activity independent of any inhibition of PDE3A enzymatic activity14,22,26,85,89,90 (Fig. 5D). Structural studies revealed that the M-domain of SLFN12 is required for binding to PDE3A14,22. SLFN12 expression is also correlated with favorable therapeutic outcomes in lung, prostate, and breast cancers91–93.
The expression of SLFN12L, another human Group II SLFN, is associated with the transition of preneoplastic cells to gastric cancer cells during Helicobacter infection94,95. However, the details of how SLFN12L mechanistically regulates cell transformation remain unclear.
Conclusions
SLFNs have emerged as biomarkers and therapeutic targets and have been linked with immune responses and suppression of viral infections. Structural studies have provided fundamental molecular clues for how the N- and M-domains of SLFNs can modulate cellular processes through RNA/DNA and functional cofactors to inhibit abnormal cellular replication and viral replication and promote cell death. However, further studies focusing on the C-domain of SLFNs are warranted to further understand the biological roles of SLFNs, and such studies will uncover how SLFNs are structurally and functionally involved. The cellular interactors and posttranslational modifications of SLFNs also remain to be fully established.
Acknowledgements
This project was supported by the Intramural Program, Center for Cancer Research of the NCI, NIH (Z01-BC006150).
Author contributions
U.J. designed and wrote the manuscript. Y.P. revised and supervised manuscript preparation.
Funding
Open Access funding provided by the National Institutes of Health (NIH).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ukhyun Jo, Email: jou2@nih.gov.
Yves Pommier, Email: pommier@nih.gov.
References
- 1.Graham GJ. Tandem genes and clustered genes. J. Theor. Biol. 1995;175:71–87. doi: 10.1006/jtbi.1995.0122. [DOI] [PubMed] [Google Scholar]
- 2.Yang JY, et al. Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun. 2018;9:1165. doi: 10.1038/s41467-018-03544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustos O, et al. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene. 2009;447:1–11. doi: 10.1016/j.gene.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwarz DA, Katayama CD, Hedrick SM. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity. 1998;9:657–668. doi: 10.1016/S1074-7613(00)80663-9. [DOI] [PubMed] [Google Scholar]
- 5.Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int. Immunol. 2004;16:1535–1548. doi: 10.1093/intimm/dxh155. [DOI] [PubMed] [Google Scholar]
- 6.Bell TA, et al. The paternal gene of the DDK syndrome maps to the Schlafen gene cluster on mouse chromosome 11. Genetics. 2006;172:411–423. doi: 10.1534/genetics.105.047118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumann B, Zhao L, Murphy K, Gonda TJ. Subcellular localization of the Schlafen protein family. Biochem. Biophys. Res. Commun. 2008;370:62–66. doi: 10.1016/j.bbrc.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 8.van Zuylen WJ, et al. Macrophage activation and differentiation signals regulate schlafen-4 gene expression: evidence for Schlafen-4 as a modulator of myelopoiesis. PLoS ONE. 2011;6:e15723. doi: 10.1371/journal.pone.0015723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu F, Zhou P, Wang Q, Zhang M, Li D. The Schlafen family: complex roles in different cell types and virus replication. Cell Biol. Int. 2018;42:2–8. doi: 10.1002/cbin.10778. [DOI] [PubMed] [Google Scholar]
- 10.de la Casa-Esperon E. From mammals to viruses: the Schlafen genes in developmental, proliferative and immune processes. Biomol. Concepts. 2011;2:159–169. doi: 10.1515/bmc.2011.018. [DOI] [PubMed] [Google Scholar]
- 11.Mavrommatis E, Fish EN, Platanias LC. The schlafen family of proteins and their regulation by interferons. J. Interferon Cytokine Res. 2013;33:206–210. doi: 10.1089/jir.2012.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murai J, Thomas A, Miettinen M, Pommier Y. Schlafen 11 (SLFN11), a restriction factor for replicative stress induced by DNA-targeting anti-cancer therapies. Pharmacol. Ther. 2019;201:94–102. doi: 10.1016/j.pharmthera.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gubser C, et al. Camelpox virus encodes a schlafen-like protein that affects orthopoxvirus virulence. J. Gen. Virol. 2007;88:1667–1676. doi: 10.1099/vir.0.82748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garvie CW, et al. Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase. Nat. Commun. 2021;12:4375. doi: 10.1038/s41467-021-24495-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiese C, et al. Disparate requirements for the Walker A and B ATPase motifs of human RAD51D in homologous recombination. Nucleic Acids Res. 2006;34:2833–2843. doi: 10.1093/nar/gkl366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher SJ, et al. SLFN14 mutations underlie thrombocytopenia with excessive bleeding and platelet secretion defects. J. Clin. Invest. 2015;125:3600–3605. doi: 10.1172/JCI80347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murai J, et al. SLFN11 blocks stressed replication forks independently of ATR. Mol. Cell. 2018;69:371–384.e376. doi: 10.1016/j.molcel.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murai Y, et al. SLFN11 inactivation induces proteotoxic stress and sensitizes cancer cells to ubiquitin activating enzyme inhibitor TAK-243. Cancer Res. 2021;81:3067–3078. doi: 10.1158/0008-5472.CAN-20-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491:125–128. doi: 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luna A, et al. CellMiner cross-database (CellMinerCDB) version 1.2: exploration of patient-derived cancer cell line pharmacogenomics. Nucleic Acids Res. 2021;49:D1083–D1093. doi: 10.1093/nar/gkaa968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, et al. DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 2018;25:1047–1058. doi: 10.1038/s41594-018-0142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, et al. Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells. Nat. Commun. 2021;12:6204. doi: 10.1038/s41467-021-26546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzner FJ, Huber E, Hopfner KP, Lammens K. Structural and biochemical characterization of human Schlafen 5. Nucleic Acids Res. 2022;50:1147–1161. doi: 10.1093/nar/gkab1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pisareva VP, Muslimov IA, Tcherepanov A, Pisarev AV. Characterization of novel ribosome-associated endoribonuclease SLFN14 from rabbit reticulocytes. Biochemistry. 2015;54:3286–3301. doi: 10.1021/acs.biochem.5b00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue, T. et al. SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell-mediated immunity. Science10.1126/science.aba4220 (2021). [DOI] [PMC free article] [PubMed]
- 26.Li D, et al. Estrogen-related hormones induce apoptosis by stabilizing Schlafen-12 protein turnover. Mol. Cell. 2019;75:1103–1116. doi: 10.1016/j.molcel.2019.06.040. [DOI] [PubMed] [Google Scholar]
- 27.Zhou C, et al. SLFN11 inhibits hepatocellular carcinoma tumorigenesis and metastasis by targeting RPS4X via mTOR pathway. Theranostics. 2020;10:4627–4643. doi: 10.7150/thno.42869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou, C., Pourmal, S. & Pavletich, N. P. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. Elife10.7554/eLife.09832 (2015). [DOI] [PMC free article] [PubMed]
- 29.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mu Y, et al. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 2016;17:94–109. doi: 10.15252/embr.201540964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto Y, et al. SLFN11 promotes stalled fork degradation that underlies the phenotype in Fanconi anemia cells. Blood. 2021;137:336–348. doi: 10.1182/blood.2019003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo G, et al. Human Schlafen 11 exploits codon preference discrimination to attenuate viral protein synthesis of prototype foamy virus (PFV) Virology. 2021;555:78–88. doi: 10.1016/j.virol.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Valdez, F. et al. Schlafen 11 restricts flavivirus replication. J. Virol. 10.1128/JVI.00104-19 (2019). [DOI] [PMC free article] [PubMed]
- 34.Seong RK, et al. Schlafen 14 (SLFN14) is a novel antiviral factor involved in the control of viral replication. Immunobiology. 2017;222:979–988. doi: 10.1016/j.imbio.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim ET, et al. Comparative proteomics identifies Schlafen 5 (SLFN5) as a herpes simplex virus restriction factor that suppresses viral transcription. Nat. Microbiol. 2021;6:234–245. doi: 10.1038/s41564-020-00826-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arslan AD, et al. Human SLFN5 is a transcriptional co-repressor of STAT1-mediated interferon responses and promotes the malignant phenotype in glioblastoma. Oncogene. 2017;36:6006–6019. doi: 10.1038/onc.2017.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan G, et al. Human Schlafen 5 regulates reversible epithelial and mesenchymal transitions in breast cancer by suppression of ZEB1 transcription. Br. J. Cancer. 2020;123:633–643. doi: 10.1038/s41416-020-0873-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 39.Bourgeois CF, Mortreux F, Auboeuf D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 2016;17:426–438. doi: 10.1038/nrm.2016.50. [DOI] [PubMed] [Google Scholar]
- 40.O’Donnell ME, Li H. The ring-shaped hexameric helicases that function at DNA replication forks. Nat. Struct. Mol. Biol. 2018;25:122–130. doi: 10.1038/s41594-018-0024-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bohm S, Bernstein KA. The role of post-translational modifications in fine-tuning BLM helicase function during DNA repair. DNA Repair. 2014;22:123–132. doi: 10.1016/j.dnarep.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Xu X. Post-translational modifications of the mini-chromosome maintenance proteins in DNA replication. Genes. 2019;10:331. doi: 10.3390/genes10050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malone D, Lardelli RM, Li M, David M. Dephosphorylation activates the interferon-stimulated Schlafen family member 11 in the DNA damage response. J. Biol. Chem. 2019;294:14674–14685. doi: 10.1074/jbc.RA118.006588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan, B. et al. Multiple PDE3A modulators act as molecular glues promoting PDE3A-SLFN12 interaction and induce SLFN12 dephosphorylation and cell death. Cell Chem. Biol. 10.1016/j.chembiol.2022.01.006 (2022). [DOI] [PubMed]
- 45.Fletcher SJ, et al. Role of the novel endoribonuclease SLFN14 and its disease-causing mutations in ribosomal degradation. RNA. 2018;24:939–949. doi: 10.1261/rna.066415.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernaez, B. et al. Viral cGAMP nuclease reveals the essential role of DNA sensing in protection against acute lethal virus infection. Sci. Adv. 10.1126/sciadv.abb4565 (2020). [DOI] [PMC free article] [PubMed]
- 47.Murai J, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget. 2016;7:76534–76550. doi: 10.18632/oncotarget.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jo U, Murai Y, Takebe N, Thomas A, Pommier Y. Precision oncology with drugs targeting the replication stress, ATR, and Schlafen 11. Cancers. 2021;13:4601. doi: 10.3390/cancers13184601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redon CE, et al. gamma-H2AX and other histone post-translational modifications in the clinic. Biochim. Biophys. Acta. 2012;1819:743–756. doi: 10.1016/j.bbagrm.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jo U, et al. SLFN11 promotes CDT1 degradation by CUL4 in response to replicative DNA damage, while its absence leads to synthetic lethality with ATR/CHK1 inhibitors. Proc. Natl Acad. Sci. USA. 2021 doi: 10.1073/pnas.2015654118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murai J, et al. Chromatin remodeling and immediate early gene activation by SLFN11 in response to replication stress. Cell Rep. 2020;30:4137–4151.e4136. doi: 10.1016/j.celrep.2020.02.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zoppoli G, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc. Natl Acad. Sci. USA. 2012;109:15030–15035. doi: 10.1073/pnas.1205943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B, et al. A wake-up call for cancer DNA damage: the role of Schlafen 11 (SLFN11) across multiple cancers. Br. J. Cancer. 2021;125:1333–1340. doi: 10.1038/s41416-021-01476-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lok BH, et al. PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin. Cancer Res. 2017;23:523–535. doi: 10.1158/1078-0432.CCR-16-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kagami T, et al. The first evidence for SLFN11 expression as an independent prognostic factor for patients with esophageal cancer after chemoradiotherapy. BMC Cancer. 2020;20:1123. doi: 10.1186/s12885-020-07574-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang MH, et al. Activity of MM-398, nanoliposomal irinotecan (nal-IRI), in Ewing’s family tumor xenografts is associated with high exposure of tumor to drug and high SLFN11 expression. Clin. Cancer Res. 2015;21:1139–1150. doi: 10.1158/1078-0432.CCR-14-1882. [DOI] [PubMed] [Google Scholar]
- 58.Deng Y, et al. High SLFN11 expression predicts better survival for patients with KRAS exon 2 wild type colorectal cancer after treated with adjuvant oxaliplatin-based treatment. BMC Cancer. 2015;15:833. doi: 10.1186/s12885-015-1840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pietanza MC, et al. Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J. Clin. Oncol. 2018;36:2386–2394. doi: 10.1200/JCO.2018.77.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isnaldi E, et al. Schlafen-11 expression is associated with immune signatures and basal-like phenotype in breast cancer. Breast Cancer Res. Treat. 2019;177:335–343. doi: 10.1007/s10549-019-05313-w. [DOI] [PubMed] [Google Scholar]
- 61.Lee TW, et al. Radiosensitization of head and neck squamous cell carcinoma lines by DNA-PK inhibitors is more effective than PARP-1 inhibition and is enhanced by SLFN11 and hypoxia. Int. J. Radiat. Biol. 2019;95:1597–1612. doi: 10.1080/09553002.2019.1664787. [DOI] [PubMed] [Google Scholar]
- 62.Conteduca V, et al. SLFN11 expression in advanced prostate cancer and response to platinum-based chemotherapy. Mol. Cancer Ther. 2020;19:1157–1164. doi: 10.1158/1535-7163.MCT-19-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lheureux S, et al. EVOLVE: a multicenter open-label single-arm clinical and translational phase II Trial of cediranib plus olaparib for ovarian cancer after PARP inhibition progression. Clin. Cancer Res. 2020;26:4206–4215. doi: 10.1158/1078-0432.CCR-19-4121. [DOI] [PubMed] [Google Scholar]
- 64.Ramkumar K, et al. AXL inhibition induces DNA damage and replication stress in non-small cell lung cancer cells and promotes sensitivity to ATR inhibitors. Mol. Cancer Res. 2021;19:485–497. doi: 10.1158/1541-7786.MCR-20-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takashima T, et al. Schlafen 11 predicts response to platinum-based chemotherapy in gastric cancers. Br. J. Cancer. 2021;125:65–77. doi: 10.1038/s41416-021-01364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gartrell J, et al. SLFN11 is widely expressed in pediatric sarcoma and induces variable sensitization to replicative stress caused by DNA-damaging agents. Mol. Cancer Ther. 2021;20:2151–2165. doi: 10.1158/1535-7163.MCT-21-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nogales V, et al. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget. 2016;7:3084–3097. doi: 10.18632/oncotarget.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He T, et al. Methylation of SLFN11 is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics. 2017;9:849–862. doi: 10.2217/epi-2017-0019. [DOI] [PubMed] [Google Scholar]
- 70.Gardner EE, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell. 2017;31:286–299. doi: 10.1016/j.ccell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang SW, et al. Overcoming resistance to DNA-targeted agents by epigenetic activation of Schlafen 11 (SLFN11) expression with class I histone deacetylase inhibitors. Clin. Cancer Res. 2018;24:1944–1953. doi: 10.1158/1078-0432.CCR-17-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takashima T, et al. Immunohistochemical analysis of SLFN11 expression uncovers potential non-responders to DNA-damaging agents overlooked by tissue RNA-seq. Virchows Arch. 2021;478:569–579. doi: 10.1007/s00428-020-02840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qu S, et al. Molecular subtypes of primary SCLC tumors and their associations with neuroendocrine and therapeutic markers. J. Thorac. Oncol. 2022;17:141–153. doi: 10.1016/j.jtho.2021.08.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mao S, et al. Resistance to pyrrolobenzodiazepine dimers is associated with SLFN11 downregulation and can be reversed through inhibition of ATR. Mol. Cancer Ther. 2021;20:541–552. doi: 10.1158/1535-7163.MCT-20-0351. [DOI] [PubMed] [Google Scholar]
- 75.Coussy, F. et al. BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Sci. Transl. Med. 10.1126/scitranslmed.aax2625 (2020). [DOI] [PMC free article] [PubMed]
- 76.Jo U, et al. Novel and highly potent ATR inhibitor M4344 kills cancer cells with replication stress, and enhances the chemotherapeutic activity of widely used DNA damaging agents. Mol. Cancer Ther. 2021;20:1431–1441. doi: 10.1158/1535-7163.MCT-20-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murai Y, et al. Schlafen 11 expression in human acute leukemia cells with gain-of-function mutations in the interferon-JAK signaling pathway. iScience. 2021;24:103173. doi: 10.1016/j.isci.2021.103173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang SW, et al. SLFN11 is a transcriptional target of EWS-FLI1 and a determinant of drug response in Ewing Sarcoma. Clin. Cancer Res. 2015;21:4184–4193. doi: 10.1158/1078-0432.CCR-14-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Katsoulidis E, et al. Role of interferon {alpha} (IFN{alpha})-inducible Schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J. Biol. Chem. 2010;285:40333–40341. doi: 10.1074/jbc.M110.151076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sassano A, et al. Human Schlafen 5 (SLFN5) is a regulator of motility and invasiveness of renal cell carcinoma cells. Mol. Cell Biol. 2015;35:2684–2698. doi: 10.1128/MCB.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wan G, et al. SLFN5 suppresses cancer cell migration and invasion by inhibiting MT1-MMP expression via AKT/GSK-3beta/beta-catenin pathway. Cell. Signal. 2019;59:1–12. doi: 10.1016/j.cellsig.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 82.Gu X, et al. SLFN5 influences proliferation and apoptosis by upregulating PTEN transcription via ZEB1 and inhibits the purine metabolic pathway in breast cancer. Am. J. Cancer Res. 2020;10:2832–2850. [PMC free article] [PubMed] [Google Scholar]
- 83.Companioni Napoles O, et al. SCHLAFEN 5 expression correlates with intestinal metaplasia that progresses to gastric cancer. J. Gastroenterol. 2017;52:39–49. doi: 10.1007/s00535-016-1202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinez RS, et al. SLFN5 regulates LAT1-mediated mTOR activation in castration-resistant prostate cancer. Cancer Res. 2021;81:3664–3678. doi: 10.1158/0008-5472.CAN-20-3694. [DOI] [PubMed] [Google Scholar]
- 85.de Waal L, et al. Identification of cancer-cytotoxic modulators of PDE3A by predictive chemogenomics. Nat. Chem. Biol. 2016;12:102–108. doi: 10.1038/nchembio.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vandenberghe P, et al. Phosphodiesterase 3A: a new player in development of interstitial cells of Cajal and a prospective target in gastrointestinal stromal tumors (GIST) Oncotarget. 2017;8:41026–41043. doi: 10.18632/oncotarget.17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nazir M, et al. Targeting tumor cells based on phosphodiesterase 3A expression. Exp. Cell Res. 2017;361:308–315. doi: 10.1016/j.yexcr.2017.10.032. [DOI] [PubMed] [Google Scholar]
- 88.An R, et al. PDE3A inhibitor anagrelide activates death signaling pathway genes and synergizes with cell death-inducing cytokines to selectively inhibit cancer cell growth. Am. J. Cancer Res. 2019;9:1905–1921. [PMC free article] [PubMed] [Google Scholar]
- 89.Lewis TA, et al. Optimization of PDE3A modulators for SLFN12-dependent cancer cell killing. ACS Med. Chem. Lett. 2019;10:1537–1542. doi: 10.1021/acsmedchemlett.9b00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ai Y, et al. An alkaloid initiates phosphodiesterase 3A-schlafen 12 dependent apoptosis without affecting the phosphodiesterase activity. Nat. Commun. 2020;11:3236. doi: 10.1038/s41467-020-17052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Al-Marsoummi, S. et al. Schlafen 12 is prognostically favorable and reduces C-Myc and proliferation in lung adenocarcinoma but not in lung squamous cell carcinoma. Cancers10.3390/cancers12102738 (2020). [DOI] [PMC free article] [PubMed]
- 92.Kovalenko PL, Basson MD. Schlafen 12 expression modulates prostate cancer cell differentiation. J. Surg. Res. 2014;190:177–184. doi: 10.1016/j.jss.2014.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Al-Marsoummi S, Vomhof-DeKrey E, Basson MD. Schlafen12 reduces the aggressiveness of triple negative breast cancer through post-transcriptional regulation of ZEB1 that drives stem cell differentiation. Cell Physiol. Biochem. 2019;53:999–1014. doi: 10.33594/000000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ding L, et al. MiR130b from Schlafen4(+) MDSCs stimulates epithelial proliferation and correlates with preneoplastic changes prior to gastric cancer. Gut. 2020;69:1750–1761. doi: 10.1136/gutjnl-2019-318817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Merchant JL, Ding L. Hedgehog signaling links chronic inflammation to gastric cancer precursor lesions. Cell. Mol. Gastroenterol. Hepatol. 2017;3:201–210. doi: 10.1016/j.jcmgh.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]