Abstract
Pantoea agglomerans (synonym: Erwinia herbicola) strain Eh318 produces through antibiosis a complex zone of inhibited growth in an overlay seeded with Erwinia amylovora, the causal agent of fire blight. This zone is caused by two antibiotics, named pantocin A and B. Using a genomic library of Eh318, two cosmids, pCPP702 and pCPP704, were identified that conferred on Escherichia coli the ability to inhibit growth of E. amylovora. The two cosmids conferred different antibiotic activities on E. coli DH5α and had distinct restriction enzyme profiles. A smaller, antibiotic-conferring DNA segment from each cosmid was cloned. Each subclone was characterized and mutagenized with transposons to generate clones that were deficient in conferring pantocin A and B production, respectively. Mutated subclones were introduced into Eh318 to create three antibiotic-defective marker exchange mutants: strain Eh421 (pantocin A deficient); strain Eh439 (pantocin B deficient), and Eh440 (deficient in both pantocins). Cross-hybridization results, restriction maps, and spectrum-of-activity data using the subclones and marker exchange mutants, supported the presence of two distinct antibiotics, pantocin A and pantocin B, whose biosynthetic genes were present in pCPP702 and pCPP704, respectively. The structure of pantocin A is unknown, whereas that of pantocin B has been determined as (R)-N-[((S)-2-amino-propanoylamino)-methyl]-2-methanesulfonyl-succinamic acid. The two pantocins mainly affect other enteric bacteria, based on limited testing.
Pantoea agglomerans or Pantoea dispersa (20), also known as Erwinia herbicola (Löhnis) Dye are members of the Enterobacteriaceae and are ubiquitous in nature, inhabiting plants, soil, and water (16, 20, 21) and animals and humans (16, 35). Strains belonging to E. herbicola are members of the E. herbicola-Enterobacter agglomerans cluster; some have been redesignated P. agglomerans and P. dispersa, while others did not fall into either of the two species (20). P. agglomerans and P. dispersa are frequent companions of Erwinia amylovora (Burr.) Winslow et al. the causal agent of the disease fire blight of apple and pear trees (36, 38). There is current interest in P. agglomerans and P. dispersa as biological control agents for fire blight because they are harmless to apple and pear trees and are able to protect them against invasion of the pathogen (4, 29). P. agglomerans strain Eh318, isolated from a symptomless apple stem in New York State, protected immature pear fruits in the laboratory (53) and apple blossoms in controlled environment and orchard tests (5, 23, 43).
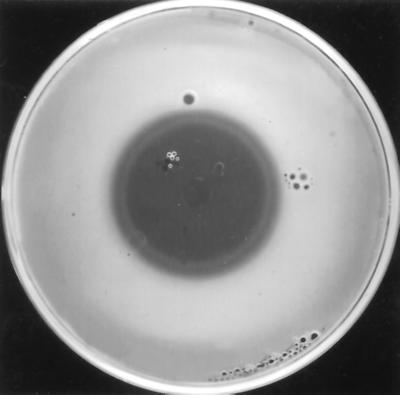
Production of antibiotics inhibitory to E. amylovora by several strains of Pantoea spp. seems important for inhibition of E. amylovora in planta (30, 45, 53). In vitro inhibition of E. amylovora by antibiotics of Pantoea spp. is well documented (24, 28, 45, 47, 48). Different strains of P. agglomerans and P. dispersa have different spectra of antimicrobial activity (15, 25) and produce different types of inhibition zones against the same indicator organism (3); both observations presumably reflect the fact that different antibiotics are produced by different strains. One strain of P. agglomerans, strain C9-1, produces three different antibiotics, which were purified and characterized preliminarily (27). The presence of an inner and an outer zone of inhibition in an E. amylovora 110-seeded agar overlay led Ishimaru and coworkers to suggest that P. agglomerans C9-1 produced more than one antibiotic (28). P. agglomerans Eh318 also forms a double halo in an overlay seeded with E. amylovora strain Ea273 (Fig. 1), which we hypothesized was due to the action of two antibiotics (S. Wright-Dobrzeniecka and S. V. Beer, Abstr. 93rd Gen. Meet. Am. Soc. Microbiol. 1993, abstr. Q420, 1993). One of these, pantocin B has been characterized chemically as (R)-N-[((S)-2-amino-propanoylamino)-methyl]-2-methanesulfonyl-succinamic acid (10); it inhibits N-acetylornithine transaminase through competitive binding with N-acetylornithine, thus interfering with the last step in the arginine biosynthetic pathway (10, 49). Antibiotics of Pantoea species frequently are grouped on the basis of the type of amino acid that, when added to the overlay, renders E. amylovora insensitive to them. Most strains of P. agglomerans and P. dispersa produce histidine-reversible or histidine- and/or leucine-reversible antibiotics (14, 50). The antibiosis of E. amylovora by P. agglomerans Eh318 is abolished in the presence of a combination of histidine and arginine, but not by either amino acid alone (48, 49).
FIG. 1.
P. agglomerans Eh318 produces a double zone of inhibition against E. amylovora Ea273 in a chloroform assay.
We have demonstrated that two distinct cosmids, pCPP702 and pCPP704, containing inserts of Eh318 DNA bestow on E. coli the ability to produce two distinct antibiotics inhibitory to E. amylovora. The observed requirement for two amino acids to abolish antibiosis by Eh318 is a consequence of the two antibiotics. They were named pantocins after the genus name of the producing organism. Histidine reversed the activity of pantocin A, and arginine reversed that of pantocin B. The distinctive antibiotic phenotypes of defined marker-exchange mutants of Eh318 that are defective in production of pantocin A and/or B provide clear genetic evidence for the production of these two antibiotics by P. agglomerans.
(Brief reports on these findings were made previously at scientific conferences [56; Wright-Dobrzeniecka and Beer, Abstr. 93rd Gen. Meet. Am. Soc. Microbiol 1993].)
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were cultured routinely in Luria-Bertani (LB) medium (39). The reaction of strains Eh318 and Eh252 (Table 1) in API 20E (bioMérieux Vitek, Inc., Hazelwood, Mo.) was consistent with their identification as species of Pantoea. The results of a GN2 Microlog test (Biolog, Inc., Hayward, Calif.) identified them with highest probability as belonging to P. agglomerans. E. amylovora and Pantoea spp. were cultured at 28°C, and Escherichia coli was cultured at 37°C. The following antibiotics were used at the concentrations indicated: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 20 μg/ml; rifampin, 25 μg/ml; spectinomycin, 50 μg/ml; tetracycline, 10 μg/ml. Antibiotic production assays were done on minimal media, either glucose-asparagine (GA) medium (52, 53) or E. coli minimal medium (EcMM), which contained per liter: 0.25 g of yeast extract (Difco Laboratories, Detroit, Mich.), 20 ml of glycerol, 4.0 g of K2HPO4, 1.72 g of KH2PO4, 0.5 g of NaCl, 2.0 g of (NH4)2SO4, 0.2 g of sodium citrate, and 0.02 g of MgSO4 · 7H2O. Thiamine was added to GA medium and EcMM at 0.1 μg/ml for the growth of E. coli strains DH5α and JM109.
TABLE 1.
Bacterial strains, phages, cosmids, and plasmids used in this study
| Strain, phage, cosmid or plasmid | Relevant characteristicsa | Reference or sourceb |
|---|---|---|
| Escherichia coli | ||
| JM109 | recA1 thiΔ (lac-proAB) lacZΔM15 | 39 |
| HB101 | SmrrecA13 | 39 |
| DH5β | F−recA1 lacZΔM15 thi-1 Nalr | 39 |
| LE392 | hsdR514 supE44 supF58 lacY1 galK2 galT22 metB1 trpR55 λ− | 39 |
| CC118 | Δ(ara-leu)7697 ΔlacX74 ΔphoA20 galE galK thi rpsE rpoB argE recA1 Spr | 32 |
| SM10(λpir) | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmrλpir | 34 |
| CGSC6151 | LamB204; resistant to lambda | M. Schwartz, E. coli Genetic Stock Center |
| Erwinia amylovora Ea273 | Infected apple, ATCC 49946 | |
| Pantoea agglomerans | ||
| Eh318 | Rpr | Rundle, CUCPB 2140, from apple leaves |
| Eh421 (PanA−) | Marker exchange mutant of Eh318, deficient in pantocin A; Kmr | This work, CUCPB 4189 |
| Eh439 (PanB−) | Marker exchange mutant of Eh318, deficient in pantocin B; Cmr | This work, CUCPB 4433 |
| Eh440 (PanAB−) | Marker exchange mutant of Eh318, deficient in pantocin A and B; Kmr Cmr | This work, CUCPB 4434 |
| Eh252 | Rundle, CUCPB 2050, from apple leaves | |
| Pantoea sp. Eh112Y | Billing, CUCPB 0119, from apple | |
| P. stewartii 2 | Woods (54), CUCPB 0176 | |
| Erwinia carotovora subsp. carotovora B12 | Received from A. K. Chatterjee, CUCPB 2260 | |
| Erwinia carotovora subsp. betavasculorum 101 | CUCPB 0306 | |
| Erwinia chrysanthemi 3665 | Received from A. Toussaint, CUCPB 2256 | |
| Enterobacter aerogenes 1422/77 | CUCPB 0294 | |
| Serratia marcescens Ser 101 | CUCPB 0155 | |
| Klebsiella pneumoniae | ATCC 15574, CUCPB 0309 |
|
| Pseudomonas putida W713 | S. E. Lindow, CUCPB 2002 | |
| Pseudomonas syringae pv. tomato DC3000 | D. Cuppels, CUCPB 4103 | |
| Xanthomonas campestris pv. pelargonii Xpel-1 | Geranium isolate | M. Daughtery, CUCPB 4672 |
| Agrobacterium tumefaciens C58 | CUCPB 0312 | |
| Phage λ467 | λb221 rex::Tn5 cI857 Oam29 Pam80 Kmr | 6 |
| Cosmids | ||
| pCPP702 | Spr Smr; contains genomic fragments cloned in pCPP9, responsible for synthesis of pantocin A | This work |
| pCPP704 | Spr Smr; contains genomic fragments cloned in pCPP9, responsible for synthesis of pantocin B | This work |
| Plasmids | ||
| pCPP9 | pGB2-mob λcos Spr | 2 |
| pRK2013 | helper plasmid; Kmr | 18 |
| pBluescript KS(+) | Apr; cloning vehicle | Stratagene Corp. |
| pCPP1051 | 8.4 and 11.8-kb EcoRI fragments of pCPP702 in pBluescript KS(+) | This work |
| pCPP719 | 32.7-kb BamHI fragment of pCPP704, religated on itself; Spr Smr | |
| pUT/mini-Tn5Cm | Apr Cmrmob (RP4) ori (R6K) | 13 |
| pCPP723 | 16-kb partial ClaI fragment of pCPP719 carrying mini-Tn5Cm insertion A14, cloned in pBR322; Apr Tcr Kmr | This work |
| pCPP726 | 17.6-kb EcoRI fragment of pCPP1051 carrying Tn5 insertion 122, cloned in pBR325; Apr Tcr Cmr Kmr | This work |
| pBR322 | Apr Tcr; cloning vehicle | 9 |
| pBR325 | Apr Tcr Cmr; cloning vehicle | 8 |
| pCPP745 | pCPP1051 with Tn5 insertion 122; Apr Spr Kmr | This work |
| pCPP810 | pCPP719 with mini-Tn5Cm insertion A14; Cmr Spr | This work, CUCPB 5070 |
Apr, Cmr, Kmr, Nalr, Rpr, Smr, Spr, Tcr, indicate resistance to ampicillin, chloramphenicol, kanamycin, nalidixic acid, rifampin, streptomycin, spectinomycin, and tetracycline, respectively.
ATCC, American Type Culture Collection; CUCPB; Cornell University Collection of Phytopathogenic Bacteria. Last name denotes isolator or distributor.
Antibiotic production assays.
Antibiotic production was assayed by two methods, the live assay and the chloroform assay. In both assays, a basal layer of either GA medium or EcMM was covered with a soft-agar overlay which consisted of 0.8 ml 5× GA salts stock solution, 3.2 ml of 0.7% agar (cooled to 48 to 50°C), and 0.3 ml of bacterial indicator cells, grown to an optical density at 620 nm of 0.4, spun down, and resuspended in an equal volume of 5 mM potassium phosphate buffer, pH 6.5. Overlays seeded with E. coli DH5α were amended with thiamine to a final concentration of 0.1 μg/ml.
In the live assay, the strains to be tested for antibiotic production were spotted directly onto the gelled surface of the overlay. In the chloroform assay, the producer was allowed to grow in a 6-mm-diameter spot, after which all growth was removed and the plate was treated with chloroform and then overlaid with the indicator strain (15). Plates of E. coli DH5α harboring cosmid clones routinely were incubated for 2 days to allow for antibiotic production. When determining whether the presence of amino acids affected the sensitivity of the indicator strain to antibiosis, arginine was added to the overlay to a final concentration of 0.76 g/ml and histidine was added at 1 g/ml, when the amino acids were added alone. These concentrations were halved when the amino acids were added in combination. The amount of amino acid added corresponded to a final concentration of 0.2 g of nitrogen/ml in the overlay. The plates were incubated at 28°C for 16 h before zones of inhibition could be seen. The chloroform assay generally resulted in larger zones of inhibition. It was therefore employed to confirm the absence of antibiotic production by the marker-exchange mutants of Eh318.
Gene transfer methods.
The first spot-agar conjugation technique described by Steinberger and Beer (41) was employed, with the modification of 12 h of incubation of the spot and subsequent resuspension in 0.5 ml of sterile water before spreading on selective medium. The cosmids pCPP702 and pCPP704 were thus transferred from E. coli JM109 to E. coli DH5α, and the pUT::mini-Tn5Cm plasmid was transferred from E. coli SM10λpir to E. coli CGSC6151(pCPP719). Mobilization of the two cosmids between JM109 and DH5α required the presence of the helper E. coli HB101(pRK2013). The nalidixic acid resistance of DH5α was employed to select for the transconjugants after the matings. Routine transformations of E. coli strains with plasmids followed the procedure of V. Simanis as described by Hanahan et al. (22).
DNA isolation and manipulations.
For the construction of a genomic library, the DNA from plasmid pCPP9 (2) was isolated by the large-scale alkaline lysis procedure (39) and purified further by cesium chloride-ethidium bromide equilibrium centrifugation (39). Isolation of total genomic DNA of Eh318 and its mutants, whether for library construction or to run on gels for Southern blotting, followed the procedure of Silhavy and coworkers (40). Cosmid DNA was isolated by a medium-scale alkaline plasmid preparation procedure that combined the methods described by Marko et al. (33) and Zasloff et al. (57). Plasmid DNA was routinely isolated on a small scale by an alkaline miniprep extraction procedure (7).
Restriction endonucleases were purchased from Promega Corp. (Madison, Wis.), and digestion of DNA was carried out as recommended by the manufacturer. Calf intestinal alkaline phosphatase was obtained from Boehringer Mannheim (Indianapolis, Ind.) and DNA T4 ligase was obtained from Bethesda Research Laboratories (GIBCO BRL, Gaithersburg, Md.). Dephosphorylations and ligations followed standard procedures (39).
DNA in agarose gels for Southern transfer was depurinated, denatured, neutralized, and transferred to Gene Screen Plus nylon membranes (Dupont, NEN Research Products, Boston, Mass.) according to the capillary blotting procedure suggested by the manufacturer (1). Prehybridization, hybridization and washes of membranes were done at 65°C and followed the protocol of Sambrook et al. (39), with the addition of 2.5 mM EDTA, 50 mM Tris-HCl (pH 8.0), and 10% polyethylene glycol to the prehybridization solution. Membranes were washed for 15 min in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.2% sodium dodecyl sulfate and twice in 0.4× SSC with 0.2% sodium dodecyl sulfate. The DNA fragment to be used as a probe was purified from agarose with the GeneClean Kit (Bio 101, Inc., La Jolla, Calif.) and labeled with 50 μCi of [α-32P]dGTP (Dupont, NEN) by the random primer labeling method (17). The probe was purified with a Sephadex G-50 spin column (Boehringer Mannheim). The membrane was exposed to Kodak X-Omat AR film (Eastman Kodak Co., Rochester, N.Y.) at −80°C with a Cronex Lightning Plus intensifying screen (E. I. Du Pont de Nemours & Co., Wilmington, Del.).
Construction of a genomic library.
Cloning of genomic DNA fragments of Eh318 of approximately 40 kb into the cosmid vector pCPP9 (2) followed the procedure of Ish-Horowicz and Burke (26), with the inclusion of a DNA sizing step on a 10 to 40% sucrose gradient (39). Fractions containing fragments of 32 to 47 kb from a partial Sau3AI digest were dephosphorylated with calf intestinal alkaline phosphatase. The vector pCPP9 was digested separately with either EcoRI or SalI, further digested with BamHI, and ligated with the genomic Sau3AI fragments. Recombinant cosmids were packaged in vitro with the Gigapack packaging kit (Stratagene, La Jolla, Calif.), and transduced into E. coli JM109. Transductants were selected on the basis of their spectinomycin resistance.
Screening the genomic library for antibiotic-producing transductants.
Transductants were screened for antibiotic production in the live assay using an overlay seeded with Ea273. The cosmid DNA of colonies that produced antibiotics was extracted, digested with several restriction enzymes, and electrophoresed to visualize restriction enzyme profiles. Antibiosis toward Ea273 in the presence and absence of arginine and histidine was evaluated by a modified chloroform assay in which the producer was allowed to grow for 2 days on GA medium before it was removed and the plate was exposed to chloroform vapors. DH5α carrying two distinct cosmids that conferred antibiosis toward Ea273 was assayed for activity against a number of different bacteria (see Table 2). The marker exchange mutants of Eh318 that were deficient in pantocin A and/or B synthesis (see below) were included to confirm that the activities of pantocin A and B were consistent in different genetic backgrounds. The results were recorded qualitatively (absence or presence of zones of inhibition) rather than quantitatively, since both the live and the chloroform tests were used. The sensitivities of Eh252 and Eh318 (control) to pantocin A and pantocin B produced by DH5α(pCPP1051) and DH5α(pCPP719) were tested separately. A colony of Ea273 that appeared in a zone of inhibition produced by Eh421 (deficient in pantocin A synthesis [PanA−]) was propagated several times on fresh plates where Eh421 had grown, in order to select for maintenance of the antibiotic resistance phenotype. The strain Ea273R421 was subsequently seeded in an overlay that was poured over plates containing antibiotics produced by Eh421 (PanA−) and Eh318.
TABLE 2.
Inhibition of bacteria by Eh318 and derivativesa producing one or two pantocins that inhibit E. amylovora.
| Inhibition byb
|
||||||
|---|---|---|---|---|---|---|
| Indicator | Eh318 (Pan AB+) | DH5α (pCPP702) (PanA+) | Eh439 (PanA+) | DH5α (pCPP704) (PanB+) | Eh421 (PanB+) | Eh440 (PanAB−) |
| E. amylovora Ea273 | + | + | + | + | + | − |
| Pantoea sp. Eh112Y | + | + | NTc | (+) | NT | NT |
| P. agglomerans Eh252 | + | − | NT | + | NT | NT |
| P. stewartii 2 | + | + | + | + | + | − |
| E. carotovora subsp. carotovora B12 | + | + | + | (+) | (+) | − |
| E. carotovora subsp. betavasculorum 101 | − | − | NT | − | NT | NT |
| E. chrysanthemi 3665 | + | + | + | + | + | − |
| E. aerogenes 1422/77 | (+) | (+) | (+) | Vd | (+) | (+) |
| E. coli DH5α | + | − | + | (+) | + | + |
| S. marcescens Ser 101 | (+) | (+) | (+) | (+) | (+) | (+) |
| K. pneumoniae | (+) | − | (+) | − | (+) | (+) |
| P. putida W713 | − | − | − | − | − | − |
| P. syringae pv. tomato DC 3000 | + | − | + | − | + | + |
| X. campestris pv. pelargonii Xpel-1 | + | − | + | + | + | + |
| A. tumefaciens C58 | − | − | − | − | − | − |
Combined results of a chloroform assay for E. coli strains carrying cosmids and Eh318 and a live assay for marker exchange mutants and Eh318, using GA base medium and GA overlay.
Symbols: +, clear inhibition zone; (+), diffuse inhibition zone; −, no inhibition zone.
NT, not tested.
V, variable.
Construction of smaller, antibiotic-encoding clones.
DNA from cosmid pCPP702 and pBluescript KS(+) was digested with EcoRI, ligated, and added to competent DH5α cells. White colonies on LB agar amended with ampicillin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were screened for antibiotic production in a live assay with GA medium or EcMM as the basal medium. The cloned DNA present in colonies that produced zones of inhibition against Ea273 was characterized. A smaller, antibiotic-conferring clone was designated pCPP1051. Similarly, a subclone of pCPP704 was constructed by digesting the cosmid DNA with BamHI and religating. A 32.7-kb BamHI fragment that also included the cosmid vector pCPP9 was designated pCPP719.
Transposon mutagenesis of pCPP1051 and pCPP719.
The two subclones, pCPP1051 and pCPP719, were mutagenized with transposons that carried distinct antibiotic resistances (kanamycin and chloramphenicol, respectively) to allow for the construction and selection of a double marker exchange mutant of Eh318 that lacked the ability to produce either pantocin. Tn5 insertion mutagenesis of pCPP1051 was performed essentially as described by de Bruijn and Lupski (12). E. coli CC118 was the host for pCPP1051 and the λ::Tn5 phage used was λb221 rex::Tn5 cI857 (6). Phage stocks were propagated in strain LE392 as described (12). E. coli DH5α was transformed with plasmid DNA that had been extracted from CC118(pCPP1051) infected with λ::Tn5 and plated. Resulting colonies were screened in a live assay, and insertions were mapped.
E. coli CGSC6151(pCPP719) was mated with E. coli SM10λpir(pUT/mini- Tn5Cm)(13). The plasmid carrying the mini-Tn5Cm element bears oriR6K and was unable to replicate in strain CGSC6151. Transconjugant colonies of CGSC6151(pCPP719::miniTn5Cm) were selected on medium amended with spectinomycin and chloramphenicol and the plasmid DNA was isolated en masse and used to transform DH5α. Single colonies of DH5α(pCPP719::mini-Tn5Cm) were grown and tested for antibiotic production in a live test. The insertion site of an antibiotic-deficient colony was mapped.
Marker exchange mutagenesis, screening, and phenotypic characterization.
DNA fragments that carried transposon insertion A14 or 122 were cloned into pBR322 or pBR325, respectively, because these cloning vectors maintained themselves stably in Eh318. The new constructs were electroporated into Eh318, and cured, which allowed for marker exchange mutagenesis to occur. Successive subculturing in phosphate-limited medium, while maintaining selection for Kmr (for the Tn5 insertion) or Cmr (for the mini-Tn5Cm insertion) (37), resulted in mutant selection. Tetracycline-sensitive colonies were screened for antibiotic production in a live assay, and colonies with reduced antibiotic production were selected for further analysis. Plasmid preparations of these confirmed the absence of the pBR325 or pBR322 constructs. The points of the insertions in putative marker exchange mutants were confirmed based on Southern blot analyses (data not shown) (55).
RESULTS
Identification and characterization of antibiotic-encoding cosmids.
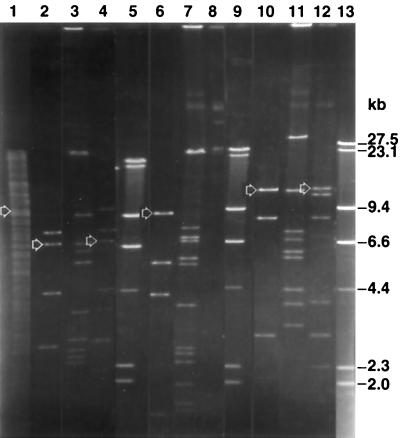
Five of 1,500 members of the genomic library of Eh318 in E. coli JM109 exhibited antibiosis against Ea273 on GA medium. Examination of the EcoRI and HindIII digest patterns of the five cosmids (data not shown) (55) indicated that two of the cosmids, designated pCPP702 and pCPP704, had no bands in common. The restriction enzyme patterns of two of the other cosmids (pCPP701 and pCPP703) appeared similar to those of pCPP702, whereas the pattern of the fifth cosmid (pCPP705) was distinct from all others. Figure 2 depicts the absence of common bands in pCPP702 and pCPP704 after digesting with EcoRI and a combination of EcoRI and XbaI. This initial genetic difference between these two cosmid clones was supported by detailed genetic mapping of clones, mapping of transposon insertions, and cross-hybridization tests.
FIG. 2.
Restriction enzyme patterns of cosmids pCPP704 (lanes 3, 7, and 11) and pCPP702 (lanes 4, 8, and 12) and a subclone of pCPP702, pCPP1051 (lanes 2, 6, and 10). Lane 1 has Eh318 genomic DNA; lanes 5, 9, and 13 have λ digested with HindIII as molecular weight markers. The DNA was digested to completion with EcoRI and XbaI (lanes 1 through 4), EcoRV (lanes 6 through 8), and EcoRI (lanes 10 through 12). The arrows indicate the fragments that hybridized with a 3.9-kb XbaI-HindIII fragment of pCPP1051 (see Fig. 3) that covered a region considered important for synthesis of pantocin A. The composite photograph was created with Adobe Photoshop 5.5.
Construction of pCPP1051 and pCPP719.
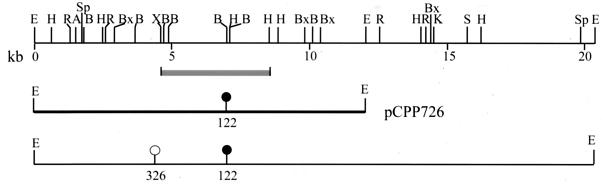
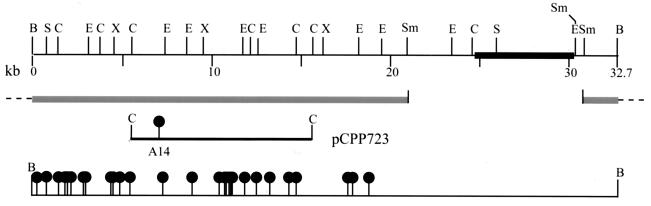
Digestion of pCPP702 with EcoRI resulted in seven fragments, whose sizes—11.8, 10.8, 8.4, 4, 3, 2.3, and 1.8 kb (Fig. 2)—total 42.1 kb, a number which includes the cosmid vector pCPP9 (5.3 kb). Recombinant plasmid pCPP1051 consisted of an 11.8- and an 8.4-kb EcoRI fragment from pCPP702 cloned into pBluescript. DH5α(pCPP1051) inhibited Ea273 when grown on EcMM. A restriction enzyme map of the 20.2-kb insert of pCPP1051 was generated (Fig. 3). The genetic region involved in the biosynthesis of pantocin B (10) was cloned from pCPP704 into pCPP719. This clone conferred upon DH5α the ability to inhibit the growth of Ea273. A detailed restriction enzyme map of pCPP719 was generated (Fig. 4).
FIG. 3.
Physical map of the 20.2-kb insert (top) from pCPP702 in pCPP1051. The 11.8-kb EcoRI fragment containing the Tn5 insertion 122 (middle) that was cloned into pBR325 to create pCPP726. This clone was used for marker exchange of insertion 122 into Eh318 to generate Eh421 (PanAl). The positions of Tn5 insertion 122, which abolishes pantocin A production (filled lollipop), and insertion 326, which does not affect production (open lollipop), are shown at the bottom. The XbaI-HindIII fragment (solid gray bar, below map) was radioactively labeled for use as a probe in Southern blots. Abbreviations: A, ApaI; B, BamHI; C, ClaI; E, EcoRI; R, EcoRV; S, SaII; Sm, SmaI; X, XbaI. The figure was generated from Innovative Data Design MacDraft, converted to MacroMedia FreeHand 8, and printed from Adobe Photoshop 5.5.
FIG. 4.
Physical map of pCPP719 (top), with the DNA of the pCPP9 vector indicated as a thick bar. The ClaI fragment resulting from a partial digest of pCPP810, containing the mini-Tn5Cm insertion A14 (bottom) was cloned into pBR322 to create pCPP723. This clone was used for marker exchange of insertion A14 into Eh318 and Eh421 (PanA−) to generate Eh439 (PanB−) and Eh440 (PanAB−), respectively. At the bottom is a map of the locations of mini-Tn5Cm insertions (filled lollipops), all of which resulted in abolished pantocin B production. The 23-kb SmaI fragment that was radioactively labeled for use in Southern blot experiments is indicated as two solid gray bars (below map). The dashed lines indicate the points of continuation in the circular pCPP719, and tick marks indicate the points of SmaI cleavage. Abbreviations: B, BamHI; C, ClaI; E, EcoRI; S, SalII; Sm, SmaI; X, XbaI. The figure was generated from Innovative Data Design MacDraft, converted to MacroMedia FreeHand 8, and printed from Adobe Photoshop 5.5.
Transposon mutagenesis and construction of clones for mutagenesis of Eh318.
The two subclones, pCPP1051 and pCPP719, were subsequently mutagenized with Tn5 and mini-Tn5Cm, respectively, to generate constructs potentially useful for marker exchange mutagenesis of Eh318 and to determine the approximate locations and sizes of pantocin-encoding regions in the subclones. Colonies of DH5α(pCPP1051::Tn5) and DH5α(pCPP719::miniTn5Cm) that had lost the ability to inhibit growth of E. amylovora were selected, and the transposon insertions were mapped. The Tn5 insertion 122 abolished pantocin A production by DH5α(pCPP1051) while insertion 326 did not (Fig. 3). The 29 mini-Tn5Cm-insertions all abolished pantocin B production by DH5α(pCPP719) (Fig. 4). Insertion A14 was chosen for marker exchange mutagenesis. The 11.8-kb EcoRI fragment of pCPP1051 containing insertion 122 was cloned from pCPP745 (a pantocin A-deficient Tn5 mutant of pCPP1051) into pBR325 to generate pCPP726 (Fig. 3). Similarly, a 10.3-kb ClaI fragment of pCPP719 containing insertion A14 was cloned from pCPP810 (the mini-Tn5Cm mutant of pCPP719) into pBR322 to generate pCPP723 (Fig. 4).
Marker exchange mutagenesis.
Plasmids pCPP726 and pCPP723 were separately introduced into Eh318 and subsequently cured, allowing for homologous recombination of the transposon insertions into the genome of Eh318. A marker exchange mutant originating from Eh318(pCPP726) was designated Eh421 (PanA−), and a mutant originating from Eh318(pCPP723) was designated Eh439 (PanB−). Eh421 (PanA−) was mutagenized using pCPP723 to generate a mutant of Eh318 that carried both transposons, designated Eh440 (PanAB−). Eh440 (PanAB−) was identified by its complete lack of antibiosis toward Ea273 in a live test. The genomic DNA of Eh421 (PanA−) was hybridized to a radioactive probe of a 3.9-kb XbaI-HindIII fragment of pCPP1051 (Fig. 3) and that of Eh439 (PanB−) and Eh440 (PanAB−) to a probe of the 23-kb SmaI fragment of pCPP719 (Fig. 4), respectively. Since Eh440 (PanAB−) was derived from Eh421 (PanA−), it was not necessary to confirm the location of the Tn5 insertion. In all cases, the mutants were true marker exchange mutants, based on analysis of the Southern blots (see details of analysis below) (55). DNAs of Eh421 (PanA−) and Eh318 were digested with NotI and XbaI, HindIII, BglII, and EcoRI and probed with the 3.9-kb insert DNA of pCPP717. The Tn5 insertion was detected in the expected position based on the analysis of several enzyme digests (data not shown) (55). The 12-kb NotI-XbaI fragment of Eh318 was replaced by a 10- and a 3.2-kb fragment in Eh421 (PanA−). The 1.45-kb HindIII fragment of Eh318, which had been mutated by the inserted transposon, was absent in Eh421 (PanA−) as expected.
The analysis of the insertion sites in Eh439 (PanB−) and Eh440 (PanAB−) using a radioactive probe of the 23-kb SmaI fragment of pCPP719 indicated successful marker exchange also of the mini-Tn5Cm insertion. The combined digest with XbaI and SalI showed that the 5-kb XbaI fragment, which contained mini-Tn5Cm, was absent in Eh439 (PanB−) and Eh440 (PanAB−) (data not shown) (55). That fragment had been replaced by two new genomic hybridizing fragments, 2.5 and 6.7 kb in size, through the presence of a SalI site in one end of the transposon. Moreover, in the BamHI and ClaI double digest of Eh439 (PanB−) and Eh440 (PanAB−) DNA, the native 6.6-kb ClaI fragment was absent from the blot as expected (55).
The mutants of Eh318 that were defective in synthesis of one of the antibiotics, i.e., Eh421 (PanA−) and Eh439 (PanB−), produced inhibition zones against Ea273 in the chloroform assay that at first glance looked similar in size to or slightly smaller than those produced by Eh318 (data not shown). However, the zones were single in nature, whereas Eh318 produced a double halo (Fig. 1). In the test in which overlays were seeded with Ea273R421 (the variant of Ea273 that was resistant to the antibiotic[s] produced by Eh421 [Pan A−]) and E318 and Eh421 (Pan A−) were the producers, only the plates in which Eh318 had grown had a zone of inhibition (data not shown).
Cross-hybridization data.
EcoRI-XbaI-, EcoRV-, and EcoRI- digested DNA of Eh318, pCPP702, pCPP704, and pCPP1051 was hybridized to a 3.9-kb XbaI-HindIII fragment of pCPP1051 that encompassed the DNA region of the Tn5 insertion site 122 in the pantocin A-deficient clone pCPP726 (Fig. 3). The only hybridizing fragments were those of Eh318, pCPP702 or pCPP1051 origin, as indicated in Fig. 2: a 9.9-kb EcoRI-XbaI (lane 1), a 7.2-kb EcoRI-XbaI (lanes 2 and 4), a 9.9-kb EcoRV (lane 9), and a 12-kb EcoRI (lane 16 and 18) band (data not shown).
Effect of amino acid supplementation on activity of pantocins.
The antibiotic activity of DH5α(pCPP702) to E. amylovora was inhibited by the presence of histidine but not arginine; however, arginine but not histidine inhibited the activity of DH5α(pCPP704). However, the zone of inhibition produced by Eh318 in an overlay seeded with E. amylovora was not affected by the presence of either amino acid, when added separately or together.
Spectrum of activity.
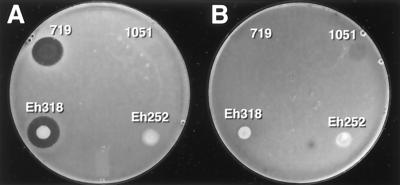
The antibacterial spectra of activity of Eh318, DH5α(pCPP702), Eh439 (PanB−), DH5α(pCPP704), Eh421 (PanA−), and Eh440 (PanAB−) are summarized in Table 2. The antibacterial spectra of the two antibiotics produced by Eh318 that are inhibitory to E. amylovora differ somewhat but overall are highly similar. Typically, the antibiotics inhibit close relatives of Eh318, such as species of Pantoea, Erwinia, Enterobacter, and Serratia. Eh252 was resistant to pantocin A but not pantocin B (Fig. 5; Table 2). In addition, we can conclude from the data that a third antibiotic of Eh318 inhibits some nonenterics, judging by the spectrum of activity of Eh440 (PanAB−), and this antibiotic does not inhibit E. amylovora.
FIG. 5.
Sensitivity of Eh252 (A) and Eh318 (B), to antibiotics produced by Eh318, Eh252, DH5α(pCPP1051) (denoted 1051), and DH5α(pCPP719) (denoted 719), in a live assay.
DISCUSSION
P. agglomerans strain Eh318 produces two antibiotics that are active against E. amylovora Ea273 based on genetic, biological, and chemical evidence. We have proposed to name them pantocin A, whose biosynthetic genes are present in pCPP702 and pCPP1051, and pantocin B (10), whose biosynthetic genes are present in pCPP704 and pCPP719. The DNA regions responsible for the synthesis of pantocin A and B are distinct based on size, restriction maps (Fig. 3 and 4), and lack of hybridization of DNA for biosynthesis of pantocin A to that for biosynthesis of pantocin B (Fig. 2). The activities of the two antibiotics also are clearly distinct. The activity of DH5α(pCPP702) is lost in the presence of histidine, while that of DH5α(pCPP704) is lost in the presence of arginine. The spectra of activity for the antibiotics produced by strains that synthesize one or both antibiotics are distinct (Table 2). The double zone of inhibition produced by Eh318 in an overlay seeded with Ea273 (Fig. 1) likely is due to the presence of two antibiotics, of which one diffuses further than the other. Mutants that produce only one of the pantocins, i.e., Eh421 (PanA−) and Eh439 (PanB−), produce single, discrete zones in overlays of Ea273. A variant of Ea273 with spontaneous resistance to the antibiotic produced by Eh421 (PanA−), i.e., to pantocin B is sensitive to Eh318.
Chemical data also suggest that the two antibiotics are distinct. Pantocin B was recently identified as (R)-N-[((S)-2-amino-propanoylamino )-methyl ] - 2 - methanesulfonyl-succinamic acid, a peptide of 296 Da (10). It is sufficiently stable to allow for its isolation and characterization from culture supernatants of DH5α(pCPP719). In contrast, pantocin A is labile to extremes of pH, and therefore it has been recalcitrant to isolation and structural characterization using similar procedures as employed for pantocin B (M. Jin, personal communication).
Subcloning and transposon mutagenesis data suggest that the genetic region involved in the biosynthesis of pantocin A is at most 7.5 kb, while that of pantocin B is at least 18.5 kb. The Tn5 insertion 326 at map position 4.3 (Fig. 3) does not abolish the antibiotic activity of DH5α(pCPP1051). A subclone of pCPP1051 that carries only the 11.8-kb EcoRI fragment confers pantocin A production on DH5α (55). Hence, the genes for pantocin A biosynthesis lie within a 7.5-kb region. Based on the mapping of 29 mini-Tn5 insertions in pCPP719 (Fig. 4), the biosynthetic region for pantocin B is at least 18.5 kb. The genetic regions involved in the synthesis of antibiotics of other strains of P. agglomerans-P. dispersa are under investigation elsewhere. For Eh1087, a New Zealand strain, a 2.2-kb region was found to be essential (31). In Eh252, another New York strain, deletion and complementation analysis of transposon-bearing clones delimited the mccEh252 biosynthetic genes to a 2.4-kb region (44). In C9-1, a Michigan strain, a cosmid clone, AA818, was identified from a genomic DNA library that confers on DH5α the ability to synthesize herbicolin O (11).
Pantocin A and pantocin B have similar but distinct spectra of activity. However, only pantocin B inhibited P. agglomerans Eh252, Xanthomonas campestris pv. pelargonii and E. coli DH5α (Table 2). The two pantocins produced by DH5α carrying the cosmids mainly inhibited enteric strains of bacteria. They are together solely responsible for the inhibition of Erwinia stewartii, Erwinia chrysanthemi, Erwinia carotovora subsp. carotovora, and E. amylovora by Eh318, judging by the absence of inhibition of these strains by Eh440 (PanAB−). This result is consistent with those of El-Goorani and coworkers, who found that the antimicrobial activity of Eh318 primarily affects enterics, with the exception of Rhodococcus fascians (15), and the same was true for several other strains of Pantoea spp. (15, 25, 28). The antibiotics of several Pantoea strains were initially designated bacteriocins due to their inhibition primarily of closely related species (3). Based on the inhibition by Eh440 (PanAB−) of Streptococcus faecalis, X. campestris pv. pelargonii, Pseudomonas syringae pv. tomato, and Klebsiella pneumoniae, Eh318 likely produces a third antibiotic compound that is ineffective on erwinias.
Earlier studies have found that the zone(s) of inhibition produced by Eh318 in an overlay seeded with E. amylovora is abolished or reduced in diameter to 50% or less in the presence of a combination of histidine and arginine (48–50). However, in our experiments, using as much as 10 mg of the two amino acids per ml in the overlay did not prevent the formation of zones. Perhaps numbers of Eh318 cells used or the time they were allowed to produce antibiotic overcame the arginine-histidine supplementation effect. The arginine effect on pantocin B activity likely is due to the redundancy of the arginine biosynthetic pathway, the target of pantocin B (10), when arginine is supplied exogenously.
Pantocin A is inactive in the presence of histidine, which is a characteristic of most antibiotics produced by Pantoea species that have been tested (14, 50, 51). MccEh252, the antibiotic produced by P. agglomerans strain Eh252 (J. L. Vanneste, J. Yu, D. C. Cornish, and M. D. Voyle, 7th Int. Congr. Plant Pathol, paper 3.5.4, 1998 [www.bspp.org.uk/icpp98/abstracts /3.5/4.html]), also is antagonized by histidine. Interestingly, Eh252 was unaffected by pantocin A, but it was antagonized by pantocin B (Fig. 5; Table 2). Strains Eh252 and Eh318 both were isolated from apple tissue in the same fruit growing area in New York state. The two histidine-type antibiotics, pantocin A and mccEh252, have low molecular masses (<3,000 Da) (Jin, personal communication; Vanneste et al., 7th Int. Congr. Plant Pathol.). Although their structures are not known, mccEh252 has been proposed to be a peptide and a microcin (46; Vanneste et al., 7th Int. Congr. Plant Pathol.) based on its protease sensitivity (45). Pantocin A is water soluble (Jin, personal communication) and also is probably a small peptide (55). A DNA fragment of Eh252 hybridized to a similar sized fragment (2.5 kb, XbaI-HindIII) in Eh318 when probed with radioactively labeled DNA of pCPP1051 (S. A. I. Wright and S. V. Beer, unpublished data). This suggests that pantocin A and mccEh252 have one or more homologous genes that are involved in biosynthesis. It is unlikely, however, that pantocin A and mccEh252 are identical, since Eh252, but not Eh318, is active against A. tumefaciens (15), and pantocin A but not mccEh252 is active against Pantoea stewartii and Serratia marcescens (Table 2) (45; Wright and Beer, unpublished data).
The only Pantoea histidine-type antibiotic for which there exists some structural information is herbicolin O, a β-lactam antibiotic (27). Herbicolin O is similar to pantocin A in that its molecular weight is less than 3,500 and its activity is labile to acid (pH 3.5) and base (pH 10) (28). Although their antimicrobial spectra have not been compared in the same assay at the same time, they are both active against Eh112Y, E. amylovora, Enterobacter aerogenes, S. marcescens, E. carotovora subsp. carotovora, and inactive against several pseudomonads, Bacillus megaterium, and K. pneumoniae (Table 2), (15, 28). Pantocin A, mccEh252, and herbicolin O may fall into a family of structurally related compounds. Interestingly, the genes responsible for their synthesis do not reside on native plasmids (11, 45, 55), whereas the biosynthetic genes for several other antibiotics of Pantoea sp. are plasmid borne (19, 30, 42).
We have demonstrated the value of using a genomic library to identify and isolate clones corresponding to distinct biosynthetic regions of two antibiotics. The differences in their inhibitory activities and sensitivities to extremes of pH, in addition to the genetic difference, clearly indicate that pantocin A and pantocin B are two distinct compounds, which are produced by one strain of P. agglomerans.
ACKNOWLEDGMENTS
We are grateful to Susanna Sanchez de Viala, Sung-Hwan Yung, and Raymond Fernalld for technical assistance. We thank David Bauer and Barbara Sneath for discussing the experiments and techniques, Richard Wodzinski for suggestions on improving the assay for antibiotic production, and Mi Jin for sharing unpublished data. We also thank Genevieve Louise Mark for assistance with API and BIOLOG tests, Kent Loeffler for preparing the photographs, and Ken Sandlan for computer advice.
This work was supported, in part, by the Cornell Biotechnology Program, a Center for Advanced Technology, supported by a consortium of industries and New York State.
REFERENCES
- 1.Anonymous. Gene Screen Plus hybridization transfer membrane. Protocols for electrophoretic and capillary transfer of DNA and RNA, DNA and RNA hybridization, and DNA and RNA rehybridization. Boston, Mass: DuPont NEN Research Products; 1987. [Google Scholar]
- 2.Bauer D W. Ph.D. thesis. Molecular genetics of pathogenicity of Erwinia amylovora: techniques, tools and their application. Ithaca, N.Y: Cornell University; 1990. [Google Scholar]
- 3.Beer S V, Rundle J R. Inhibition of Erwinia amylovora by bacteriocin-like substances. Phytopathology. 1980;70:459. . (Abstract.) [Google Scholar]
- 4.Beer S V, Rundle J R, Norelli J L. Recent progress in the development of biological control for fire blight—a review. Acta Hortic. 1984;151:195–201. [Google Scholar]
- 5.Beer S V, Wright-Dobrzeniecka S, Wodzinski R S, Zumoff C H. Antibiotic production by Erwinia herbicola strain Eh318 and biological control of fire blight. Phytopathology. 1993;83:1342. . (Abstract.) [Google Scholar]
- 6.Berg D E, Davies J, Allet B, Rochaix J-D. Transposition of R factor genes to bacteriophage λ. Proc Natl Acad Sci USA. 1975;72:3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birnboim H C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–254. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- 8.Bolivar F. Construction and characterization of new cloning vehicles III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant molecules. Gene (Amsterdam) 1978;4:121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- 9.Bolivar F, Rodriquez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene (Amsterdam) 1977;2:95–113. [PubMed] [Google Scholar]
- 10.Brady S F, Wright S A, Lee J C, Sutton A E, Zumoff C H, Wodzinski R S, Beer S V, Clardy J. Pantocin B, an antibiotic from Erwinia herbicola discovered by heterologous expression of cloned genes. J Am Chem Soc. 1999;121:11912–11913. [Google Scholar]
- 11.Davis L A, Ishimaru C A. Cloning and expression of herbicolin O biosynthesis genes in Escherichia coli. Phytopathology. 1993;83:1339. . (Abstract.) [Google Scholar]
- 12.de Bruijn F J, Lupski J R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids—a review. Gene (Amsterdam) 1984;27:131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]
- 13.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Goorani M A, Beer S V. Antibiotic production by strains of Erwinia herbicola and their interactions with Erwinia amylovora in immature pear fruits. Phytopathology. 1991;81:121. . (Abstract.) [Google Scholar]
- 15.El-Goorani M A, Hassanein F M, Shoeib A A. Antibacterial and antifungal spectra of antibiotics produced by different strains of Erwinia herbicola (=Pantoea agglomerans) J Phytopathol (Berlin) 1992;136:335–339. [Google Scholar]
- 16.Ewing W H, Fife M A. Enterobacter agglomerans (Beijerinck) comb. nov. (the Herbicola-Lathyri bacteria) Int J Syst Bacteriol. 1972;22:4–11. [Google Scholar]
- 17.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 18.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gantotti B V, Kindle K L, Beer S V. Transfer of the drug-resistance transposon Tn5 to Erwinia herbicola and the induction of insertion mutants. Curr Microbiol. 1981;6:377–381. [Google Scholar]
- 20.Gavini F, Mergaert J, Beji A, Mielcarek C, Izard D, Kersters K, de Ley J. Transfer of Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersa sp. nov. Int J Syst Bacteriol. 1989;39:337–345. [Google Scholar]
- 21.Graham D C, Hodgkiss W. Identity of Gram negative, yellow pigmented, fermentative bacteria isolated from plants and animals. J Appl Bacteriol. 1967;30:175–189. doi: 10.1111/j.1365-2672.1967.tb00287.x. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan D, Jessee J, Bloom F R. Techniques for transformation of E. coli. In: Glover D M, Hames B D, editors. DNA cloning. 2nd ed. Vol. 1. New York, N.Y: Oxford University Press; 1995. pp. 1–36. [Google Scholar]
- 23.Hickey K D, van der Zwet T. Efficacy of antagonistic bacteria for control of fire blight on apple. Acta Hortic. 1996;411:299–302. [Google Scholar]
- 24.Hodges S S, Beer S V, Rundle J R. Effects of a bacteriocin produced by Erwinia herbicola on Erwinia amylovora. Phytopathology. 1980;70:463. [Google Scholar]
- 25.Howitt D J, Epton H A S, Sigee D C, Cook K. In vitro production of antibiotics by strains of Erwinia herbicola selected for biocontrol of Erwinia amylovora. In: Lemattre M, Freigoun S, Rudolph K, Swings J G, editors. Proceedings of the 8th International Conference on Plant Pathogenic Bacteria, les colloques de l'INRA, no. 66. Versailles, France: INRA; 1994. pp. 917–922. [Google Scholar]
- 26.Ish-Horowicz D, Burke J F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981;9:2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishimaru C A. Ph.D. thesis. Herbicolins O, I & 2C: novel β-lactam antibiotics produced by Erwinia herbicola strain C9–1. East Lansing: Michigan State University; 1985. [Google Scholar]
- 28.Ishimaru C A, Klos E J, Brubaker R R. Multiple antibiotic production by Erwinia herbicola. Phytopathology. 1988;78:746–750. [Google Scholar]
- 29.Johnson K B, Stockwell V O, McLaughlin R J, Sugar D, Loper J E, Roberts R G. Effect of antagonistic bacteria on establishment of honey bee-dispersed Erwinia amylovora in pear blossoms and on fire blight control. Phytopathology. 1993;83:995–1002. [Google Scholar]
- 30.Kearns L P, Mahanty H K. Identification and cloning of Erwinia herbicola DNA responsible for suppression of Erwinia amylovora. Acta Hortic. 1993;338:249–253. [Google Scholar]
- 31.Kearns L P, Mahanty H K. Antibiotic production by Erwinia herbicola Eh1087: its role in inhibition of Erwinia amylovora and partial characterization of antibiotic biosynthesis genes. Appl Environ Microbiol. 1998;64:1837–1844. doi: 10.1128/aem.64.5.1837-1844.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manoil C. Analysis of protein localization by use of gene fusions with complementary properties. J Bacteriol. 1990;172:1035–1042. doi: 10.1128/jb.172.2.1035-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marko M A, Chipperfield R, Birnboim H C. A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal Biochem. 1982;121:382–387. doi: 10.1016/0003-2697(82)90497-3. [DOI] [PubMed] [Google Scholar]
- 34.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murashi T F, Friend M, Bolles D. Erwinia-like microorganisms isolated from animal and human hosts. Appl Microbiol. 1965;13:128–131. doi: 10.1128/am.13.2.128-131.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riggle J H, Klos E J. Relationship of Erwinia herbicola to Erwinia amylovora. Can J Bot. 1972;50:1077–1083. [Google Scholar]
- 37.Roeder D L, Collmer A. Marker-exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi. J Bacteriol. 1985;164:51–56. doi: 10.1128/jb.164.1.51-56.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosen H R. Variations within a bacterial species–I. Morphologic variations. Mycologia. 1928;20:251–275. [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 41.Steinberger E M, Beer S V. Creation and complementation of pathogenicity mutants of Erwinia amylovora. Mol Plant-Microbe Interact. 1988;1:135–144. [Google Scholar]
- 42.Tenning P, Van Rijsbergen R, Zhao Y, Joos H. Cloning and transfer of genes for antifungal compounds from Erwinia herbicola to Escherichia coli. Mol Plant-Microbe Interact. 1993;6:474–480. doi: 10.1094/mpmi-6-474. [DOI] [PubMed] [Google Scholar]
- 43.Thomson S V, Gouk S C. Interactions between an antagonist, Erwinia herbicola, and E. amylovora and potential for biological control of fire blight. In: Popay A J, editor. Proceedings of the 45th New Zealand Plant Protection Conference. Wellington, New Zealand: The New Zealand Plant Protection Society, Inc.; 1992. pp. 295–300. [Google Scholar]
- 44.Vanneste J L, Yu J. Cloning and sequencing of a 2.5 kb DNA fragment from Erwinia herbicola Eh252 necessary for production of an antibiotic involved in biological control of fire blight. Phytopathology. 1996;86(11)Suppl.:S84. [Google Scholar]
- 45.Vanneste J L, Yu J, Beer S V. Role of antibiotic production by Erwinia herbicola Eh252 in biological control of Erwinia amylovora. J Bacteriol. 1992;174:2785–2796. doi: 10.1128/jb.174.9.2785-2796.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanneste J L, Yu J, Cornish D A, Voyle M D, Melbourne M. Designing a biological control of fire blight: expression of a new peptide antibiotic gene in E. amylovora and of the harpin gene in E. herbicola. Acta Hortic. 1999;489:669–670. [Google Scholar]
- 47.Wilson M, Epton H A S, Sigee D C. Biological control of fire blight of hawthorn (Crataegus monogyna) with Erwinia herbicola under protected conditions. Plant Pathol (Oxford) 1990;39:301–308. [Google Scholar]
- 48.Wodzinski R S, Beer S V, Zumoff C H, Clardy J C, Coval S J. Antibiotics produced by strains of Erwinia herbicola that are highly effective in suppressing fire blight. Acta Hortic. 1990;273:411–412. [Google Scholar]
- 49.Wodzinski R S, Mudgett M B, Beer S V. Mechanism by which the antibiotic of Erwinia herbicola Eh318 inhibits Erwinia amylovora Ea273. In: Klement Z, editor. Proceedings of the 7th International Conference on Plant Pathogenic Bacteria, Budapest, Hungary. Budapest, Hungary: Akademiai Kiadóés Nyomda Vállalat; 1990. pp. 265–266. [Google Scholar]
- 50.Wodzinski R S, Paulin J-P. Frequency and diversity of antibiotic production by putative Erwinia herbicola strains. J Appl Bacteriol. 1994;76:603–607. [Google Scholar]
- 51.Wodzinski R S, Sobiczewski P, Beer S V. Factors affecting production of herbicolacin 112Y by Erwinia herbicola 112Y. In: Civerolo E L, Collmer A, Davis R E, Gillaspie A G, editors. Proceedings of the Sixth International Conference on Plant Pathogenic Bacteria. College Park, Md: Martinus Nijhoff Publishers; 1987. pp. 551–555. [Google Scholar]
- 52.Wodzinski R S, Umholtz T E, Garrett K, Beer S V. Attempts to find the mechanism by which Erwinia herbicola inhibits Erwinia amylovora. Acta Hortic. 1987;217:223–227. [Google Scholar]
- 53.Wodzinski R S, Umholtz T E, Rundle J R, Beer S V. Mechanisms of inhibition of Erwinia amylovora by Erwinia herbicola in vitro and in vivo. J Appl Bacteriol. 1994;76:22–29. [Google Scholar]
- 54.Woods T L. Ph.D. thesis. Factors affecting the incidence and severity of Stewart's disease of Zea mays L. Ithaca, N. Y: Cornell University; 1978. [Google Scholar]
- 55.Wright S A I. Ph.D. thesis. The genetics of antibiotic production and the role of antibiotics in biological control of Erwinia amylovora by Erwinia herbicola. Ithaca, N.Y: Cornell University; 1997. [Google Scholar]
- 56.Wright S A I, Beer S V. Genetics of pantocin A and pantocin B production and their role in biocontrol of the fireblight pathogen, Erwinia amylovora. In: Duffy B, Rosenberger U, Défago G, editors. Molecular approaches in biological control. 21 (9) Délémont, Switzerland: International Organization for Biological and Integrated Control of Noxious Animals and Plants; 1998. pp. 13–17. [Google Scholar]
- 57.Zasloff M, Ginder G D, Felsenfield G. A new method for the purification and identification of covalently closed circular DNA molecules. Nucleic Acids Res. 1978;5:1139–1152. doi: 10.1093/nar/5.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]