Abstract
The purpose of this study is to examine the association between G protein-coupled receptor 87 (GPR87) and lung adenocarcinoma (LUAD) metastasis and immune infiltration. The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets extract clinical data. According to the TCGA database, increased GPR87 expression predicts poor overall survival, progression-free interval, and disease-specific survival in LUAD patients. The meta-analysis also reveals a significant association between high GPR87 expression and poor overall survival. Moreover, functional experiments demonstrate that GPR87 silencing reduces LUAD cell invasion and migration. Immunoblotting shows that GPR87 knockdown decreased Vimentin and N-cadherin expression and increased E-cadherin expression in LUAD cells. GPR87 expression in LUAD is positively correlated with immune infiltration. In addition, GPR87 expression is associated with immune and chemotherapy resistance in LUAD patients. Our findings indicate that GPR87 promotes tumor progression and is correlated with immune infiltration, suggesting GPR87 as a possible biomarker for prognosis prediction in LUAD.
Subject terms: Cancer, Immunology
GPR87 is reported as a central player in lung adenocarcinoma and in resistance to immunotherapy, by promoting tumor cell invasion and mediating the immunogenomic landscape.
Introduction
With an estimated 1.6 million new cases and 1.38 million deaths each year, lung cancer is the leading cause of cancer death globally1. Lung adenocarcinoma (LUAD) as the most prevalent subtype, accounts for 40% of non-small cell lung cancer (NSCLC) at diagnosis2. Although treatments such as chemotherapy, radiotherapy, and immunotherapy have been improved over the previous decades, the overall survival (OS) of LUAD patients remains low3. Thus, it is critical to identify prognostic indicators and treatment targets.
The epithelial-mesenchymal transition (EMT) is a critical cellular process controlled by a series of EMT-inducing transcription factors. It is characterized by the loss of epithelial characteristics and the acquisition of mesenchymal markers4. Not only is EMT related to tumor growth, but it is also associated with poor prognosis and medication resistance in LUAD5. Analysis of RNA expression profiles to identify therapeutic target genes has become a hot issue in recent years to improve prognosis6. Therefore, finding prognostic genes associated with EMT by sequencing may help improve LUAD patients’ prognoses.
G protein-coupled receptors comprise about 800 proteins, as the most prominent family of eukaryotic membrane signaling proteins7. G protein-coupled receptor (GPR) 87 locates at chromosome 3q24 and encodes a protein with an extracellular N-terminal, seven helixes, and three intracellularly loops8. It is overexpressed on the cell surface in different malignancies and is crucial to the survival of tumor cells. Zhang et al. found that GPR87 has an important role in the response of p53-dependent cells to DNA damage. By enhancing the stabilization and activation of p53, inhibition of GPR87 expression could sensitize cancer cells to growth inhibition induced by DNA damage9. GPR87 is highly expressed in NSCLC and associated with cell proliferation10–12. However, its EMT and immune function is to be investigated.
We examined the differential expression of GPR87 in LUAD and normal tissues and the relationship between its gene expression and DNA methylation in TCGA. Additionally, we assessed the predictive values of GPR87 expression and methylation. The predictive effects of GPR87 were then verified using 4 independent GEO datasets. A complete meta-analysis was conducted using data from five public databases to determine the overall prognostic importance of GPR87. Finally, in vitro experiments confirmed that GPR87 downregulation hindered the EMT process of LUAD cells. In conclusion, our findings revealed the regulatory effects and the underlying mechanisms of GPR87 on tumor metastasis in LUAD, implying that GPR87 might be an important prognostic and therapeutic target for LUAD patients.
Results
GPR87 is highly expressed in LUAD and associated with lymph node metastasis
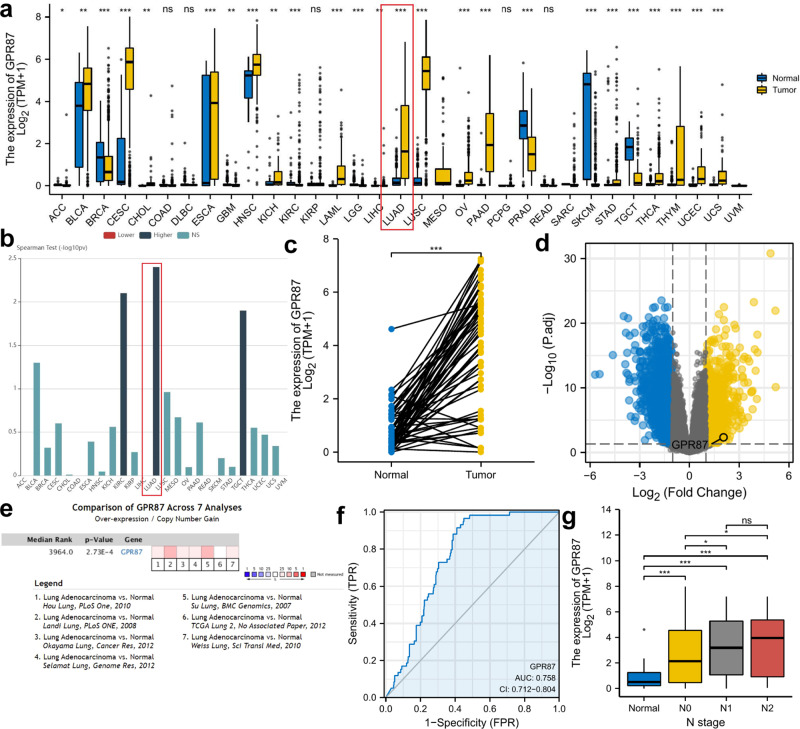
First, we assessed the distribution of GPR87 expression in all tumor tissues and found that GPR87 expression was elevated in most cancers (Fig. 1a). Next, we examined the relationship between GPR87 and the clinical stages of all TCGA cancers using TISIDB (Fig. 1b). GPR87 mRNA levels were most significantly correlated with the clinical stages of LUAD patients. Therefore, we focused on the role of GPR87 in LUAD progression. To validate the high expression of GPR87 in LUAD, we examined GPR87 expression in 57 paired LUAD patients from the GEO dataset GSE31210, Oncomine, and TCGA (Fig. 1c–e). We confirmed that the expression of GPR87 was significantly higher in LUAD than in normal tissues. Meanwhile, GPR87 was moderately accurate in predicting tumor and normal outcomes (Fig. 1f, AUC = 0.758, CI = 0.712–0.804). To investigate the relationship between GPR87 expression and lymph node metastasis, we investigated the breakdown of GPR87 expression by N stage in LUAD. High GPR87 levels in LUAD were also associated with a high grade of lymph node metastasis (Fig. 1g).
Fig. 1. GPR87 expression in LUAD and adjacent normal tissues.
a Expression of the GPR87 gene in pan-cancer using TCGA data. b The relationship between GPR87 and the clinical stages of all TCGA cancers using TISIDB. c GPR87 expression levels in 57 paired normal and tumor and tissues. d Volcano plot of differential analysis in GSE31210. e Obtaining differential expression of GPR87 in lung cancer using Oncomine database. f ROC curve to verify the accuracy of GPR87 to distinguish tumor and normal tissues. g Detection of GPR87 expression levels concerning tumor lymph node metastasis.
Functional enrichment analysis shows that GPR87 is associated with EMT
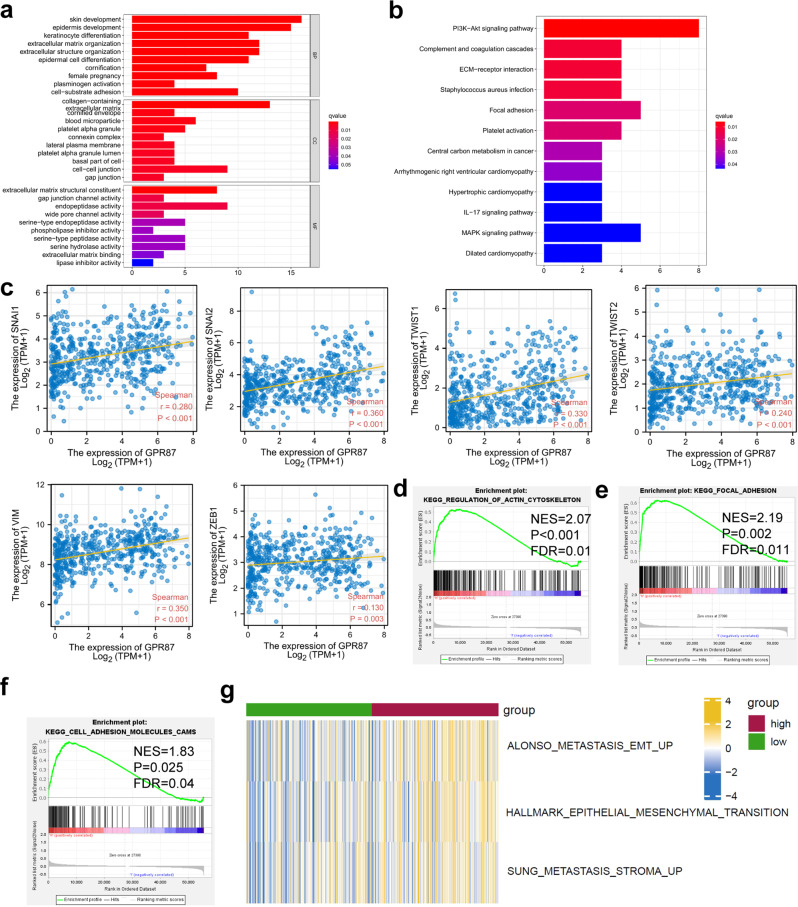
To verify whether the GPR87 was correlated with the EMT process, we first separated the patients into high- and low-GPR87 expressing groups based on the median GPR87 mRNA level. Differential expression analysis was performed in these two groups using limma. Differentially expressed genes (DEGs) were then integrated into the subsequent GO and KEGG analysis, and the results showed that GPR87 was associated with a variety of EMT-related functions, including cell-cell junction, gap junction, and Focal adhesion (Fig. 2a, b). The results of the KEGG enrichment analysis showed that GPR87 was positively correlated with the transcription of SNAI1, SNAI2, TWIST1, TWIST2, VIM, and ZEB1, which were EMT-associated transcription factors (Fig. 2c). We further performed enrichment analysis using GSEA and found that GPR87 was also associated with cell adhesion molecules (CAMs), focal adhesion, and regulation of actin cytoskeleton, further confirming the involvement of GPR87 in the regulation of EMT (Fig. 2d–f). In addition, we also scored the EMT of each LUAD patient in TCGA using the Single-sample gene set enrichment analysis (ssGSEA) algorithm. The results showed that the metastasis-related scores were significantly higher in the high GPR87 expressing group (Fig. 2g).
Fig. 2. Functional enrichment analysis to verify the association of GPR87 with EMT.
a GO enrichment analysis. b KEGG enrichment analysis. c GPR87 is positively correlated with EMT-related gene transcription. Regulation of actin cytoskeleton (d), focal adhesion (e), and CAMs (f). g RNA-seq data from LUAD patients in the TCGA database were analyzed using the ssGSEA algorithm to evaluate the association of GPR87 with tumor metastasis.
GPR87 plays an oncogene role in LUAD
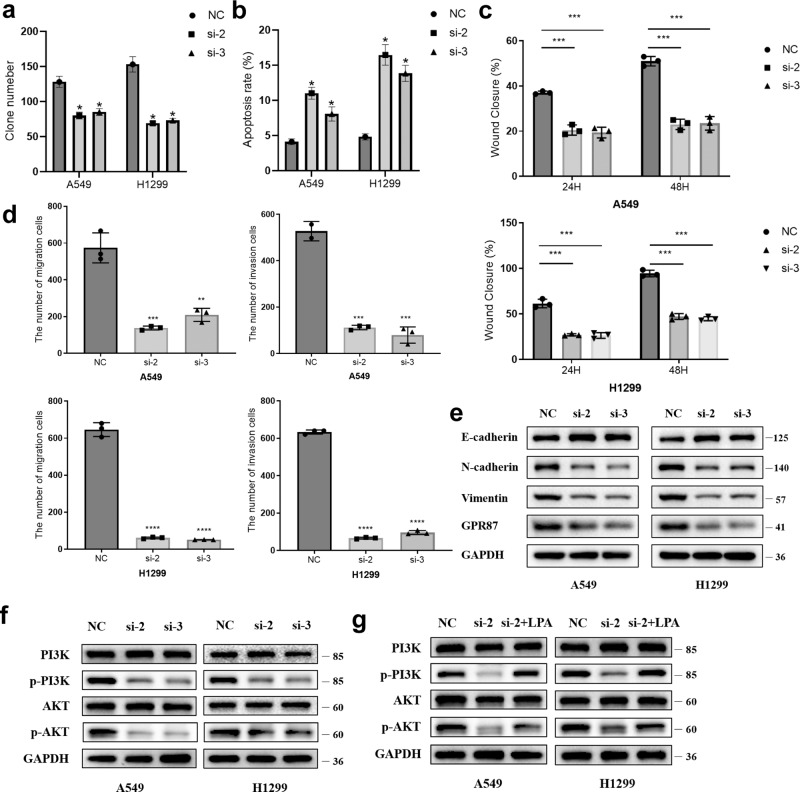
To investigate the effects of GPR87 knockdown on EMT in LUAD cells, GPR87 siRNA was transfected into A549 and H1299 cells. The results of qRT-PCR showed that GPR87 mRNA expression was significantly reduced after siRNA transfection (Fig. S1). Immunoblotting confirmed that GPR87 protein was downregulated in both A549 and H1299 cells (Fig. 3e). The results of the clone formation assay showed that knockdown of GPR87 significantly inhibited the proliferation of A549 and H1299 cells (Fig. 3a and Fig. S2a). Flow cytometry assay of apoptosis showed that knockdown of GPR87 significantly promoted the apoptosis rate of A549 and H1299 cells (Fig. 3b and Fig. S2b). Wound healing and modified Boyden chamber assay were used to examine the effects of GPR87 knockdown on the migration and invasion of LUAD cells. The migration and invasion rates of A549-KD and H1299-KD cells were significantly lower than NC ones (Fig. 3c, d and Fig. S2c, d). Immunoblotting confirmed the elevated levels of the epithelial marker E-cadherin. GPR87 knockdown decreased the levels of mesenchymal markers, such as Vimentin and N-cadherin (Fig. 3e). According to the results of the KEGG enrichment analysis, GPR87 is highly correlated with the PI3K-Akt signaling pathway, so we examined the activation of the PI3K-Akt pathway after the knockdown of GPR87 by immunoblotting, and the results showed that inhibition of GPR87 inhibited the activation of PI3K-Akt pathway (Fig. 3f). LPA, as a ligand of GPR87, could partially reverse the inhibition of the PI3K-Akt pathway caused by the knockdown of GPR87 (Fig. 3g). These results suggested that GPR87 knockdown induced the transition to epithelial phenotype in A549 and H1299 cells.
Fig. 3. GPR87 knockdown inhibits LUAD cell clonogenesis, apoptosis, and EMT.
a Quantitative results of clone formation experiments. b Quantitative results of cell apoptosis experiments. c Cell migration was evaluated by wound healing assay and reduced by GPR87 knockdown. Quantification of relative wound closure. d Migration and invasion of A549 and H1299 cells transfected with GPR87 siRNA were evaluated by a modified Boyden chamber assay. Quantification of the migration and invasion cells. e Representative immunoblotting of EMT-related proteins. f Representative immunoblotting of PI3K signaling pathway-related proteins. g After the knockdown of GPR87, the LPA receptor (5 µM) was added to the culture medium and WB assayed PI3K signaling pathway activation.
GPR87 is associated with a poor prognosis
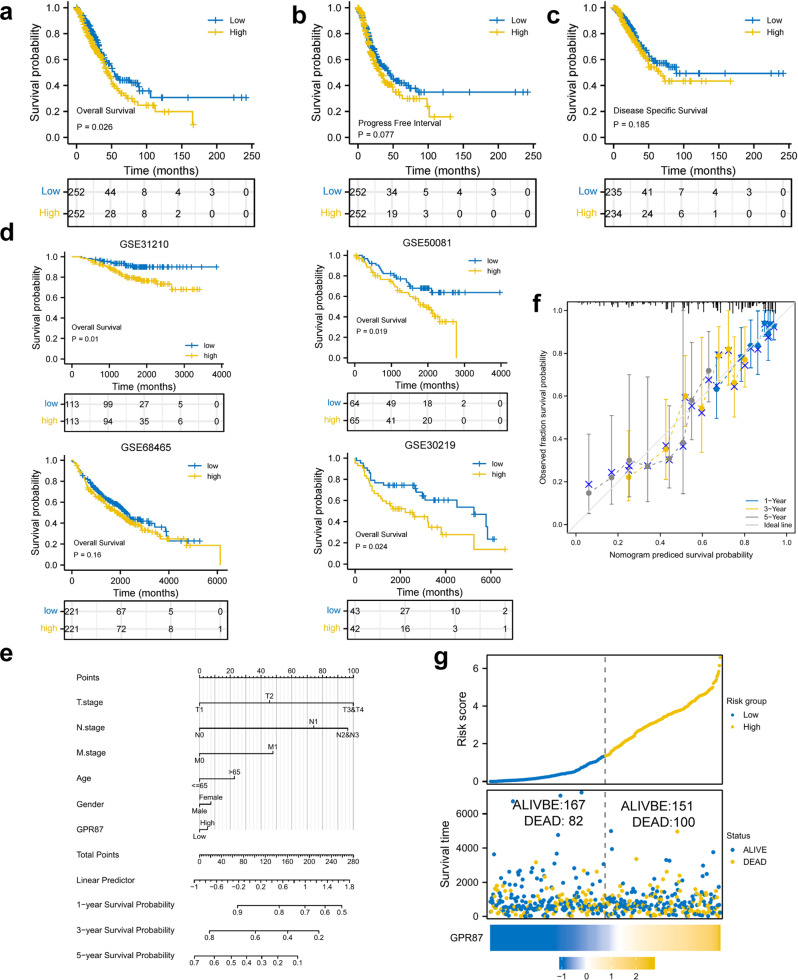
We further analyzed the association between GPR87 and different clinical outcomes in LUAD patients. Survival analysis showed that the high-expression group was associated with shorter OS (HR = 1.39, 95% CI [1.04−1.86], P < 0.05), disease-specific survival (HR = 1.29, 95% CI [0.89−1.87], P = 0.185), and progress-free interval (HR = 1.28, 95% CI [0.97−1.69], P = 0.077, Fig. 4a–c). In addition to the TCGA data, four independent datasets were selected as the validation sets to confirm the effects of GPR87 on the prognosis of LUAD patients. The results of these independent datasets were consistent with the TCGA results, and GPR87 expression was associated with poor prognosis (GSE30219: HR = 2.01, 95% CI [1.10−3.68], P < 0.05; GSE31210: HR = 2.63, 95% CI [1.26−5.49], P < 0.05; GSE50081: HR = 1.95, 95% CI [1.12−3.42], P < 0.05, GSE68465: HR = 1.2, 95% CI [0.93−1.55]) (Fig. 4d). Meanwhile, we drew a nomogram using GPR87 expression together with clinical factors and plotted calibration curves to validate the accuracy of the prediction model (Fig. 4e, f). The predicted values matched well with the actual values, indicating that our model could be applied to predict the prognosis of LUAD patients. The survival status, survival time, and GPR87 expression levels of the LUAD patients were shown in Fig. 4g. With the increase in the risk scores, the number of deaths also increased.
Fig. 4. Kaplan–Meier curves of GPR87 in LUAD patients.
OS (a), disease-specific survival (c), and progress-free interval (b) in the TCGA-LUAD dataset. d OS in the GSE31210, GSE50081, GSE68465, and GSE30219. e GPR87 was combined with other clinical factors to plot the nomogram and predict the prognosis of LUAD patients. f The calibration curve was drawn to verify the accuracy of the prediction model in predicting 1-, 3-, and 5-year survival rates. g Survival conditions of LUAD patients.
Meta-analysis also shows the prognostic value of GPR87 in LUAD
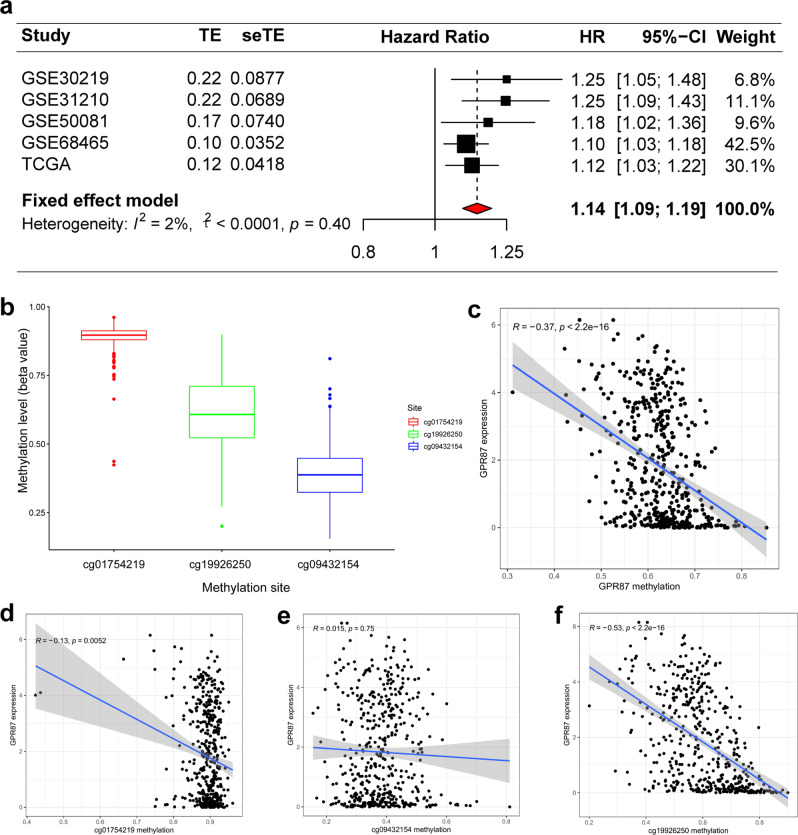
Due to few reports on the association between GPR87 expression and OS in LUAD patients, we integrated the prognosis from the five different datasets into the meta-analysis. The combined HR and 95% CI association between GPR87 expression and OS was 1.31 (1.22–1.40) in 1390 LUAD patients, with no significant heterogeneity between the 4 datasets (I2 = 5%, P = 0.37, Fig. 5a). Therefore, we concluded that high expression of GPR87 was a robust predictor of poor prognosis in LUAD patients.
Fig. 5. GPR87 methylation and its effects on prognosis in LUAD tissues were revealed by bioinformatics analysis.
a Forest plots of high GPR87 expression from the four datasets with worse OS in LUAD patients. b Distribution of GPR87 promoter CpG sites. c Correlation analysis between GPR87 methylation and expression levels. d–f Correlation analysis between methylation levels of CpG sites and GPR87 expression.
DNA methylation of GPR87 inhibits its expression
To investigate the relationship between GPR87 DNA methylation and gene expression, we analyzed the GPR87 methylation sites, which are mainly distributed in three GPR87 CpG sites (Fig. 5b). Spearman correlation analysis was used to identify the sites that correlated with GPR87 expression (Fig. 5c). Except cg09432154, the methylation of the other CpG sites was correlated with GPR87 expression, and the most correlated site was cg19926250 (Fig. 5d–f). Based on the GPR87 methylation levels, the patients were divided into the GPR87 hypo- and hypermethylated groups. We used chi-square tests to investigate GPR87 expression and gene methylation correlations with a range of clinical features. GPR87 expression was significantly correlated with N stage (P < 0.05), and GPR87 methylation (P < 0.0001, Table 1). Similarly, GPR87 expression (P < 0.0001) also correlated with GPR87 methylation levels.
Table 1.
Correlation of GPR87 mRNA expression and methylation with clinicopathological features in the TCGA database.
| GPR87 expression | P | GPR87 methylation | P | |||||
|---|---|---|---|---|---|---|---|---|
| Covariates | Type | Total | High | Low | High | Low | ||
| Age | ≤65 | 225(48.39%) | 114(49.14%) | 111(47.64%) | 0.7726 | 110(47.41%) | 115(49.36%) | 0.4894 |
| >65 | 221(47.53%) | 110(47.41%) | 111(47.64%) | 110(47.41%) | 111(47.64%) | |||
| unknown | 19(4.09%) | 8(3.45%) | 11(4.72%) | 12(5.17%) | 7(3%) | |||
| M | M0 | 299(64.3%) | 149(64.22%) | 150(64.38%) | 0.9701 | 148(63.79%) | 151(64.81%) | 0.7755 |
| M1 | 19(4.09%) | 10(4.31%) | 9(3.86%) | 11(4.74%) | 8(3.43%) | |||
| unknown | 147(31.61%) | 73(31.47%) | 74(31.76%) | 73(31.47%) | 74(31.76%) | |||
| N | N0 | 306(65.81%) | 140(60.34%) | 166(71.24%) | 0.039 | 155(66.81%) | 151(64.81%) | 0.4934 |
| N1 | 81(17.42%) | 47(20.26%) | 34(14.59%) | 36(15.52%) | 45(19.31%) | |||
| N2 | 65(13.98%) | 40(17.24%) | 25(10.73%) | 32(13.79%) | 33(14.16%) | |||
| N3 | 1(0.22%) | 1(0.43%) | 0(0%) | 1(0.43%) | 0(0%) | |||
| unknown | 12(2.58%) | 4(1.72%) | 8(3.43%) | 8(3.45%) | 4(1.72%) | |||
| T | T1 | 157(33.76%) | 76(32.76%) | 81(34.76%) | 0.885 | 83(35.78%) | 74(31.76%) | 0.3076 |
| T2 | 247(53.12%) | 122(52.59%) | 125(53.65%) | 120(51.72%) | 127(54.51%) | |||
| T3 | 42(9.03%) | 23(9.91%) | 19(8.15%) | 20(8.62%) | 22(9.44%) | |||
| T4 | 16(3.44%) | 9(3.88%) | 7(3%) | 6(2.59%) | 10(4.29%) | |||
| unknown | 3(0.65%) | 2(0.86%) | 1(0.43%) | 3(1.29%) | 0(0%) | |||
| Gender | female | 249(53.55%) | 134(57.76%) | 115(49.36%) | 0.0848 | 128(55.17%) | 121(51.93%) | 0.5434 |
| male | 216(46.45%) | 98(42.24%) | 118(50.64%) | 104(44.83%) | 112(48.07%) | |||
| Stage | stage 1 | 256(55.05%) | 115(49.57%) | 141(60.52%) | 0.0837 | 130(56.03%) | 126(54.08%) | 0.1395 |
| stage 2 | 111(23.87%) | 59(25.43%) | 52(22.32%) | 51(21.98%) | 60(25.75%) | |||
| stage 3 | 73(15.7%) | 46(19.83%) | 27(11.59%) | 34(14.66%) | 39(16.74%) | |||
| stage 4 | 20(4.3%) | 10(4.31%) | 10(4.29%) | 12(5.17%) | 8(3.43%) | |||
| unknown | 5(1.08%) | 2(0.86%) | 3(1.29%) | 5(2.16%) | 0(0%) | |||
| Expression | high | 232(49.89%) | - | - | - | 90(38.79%) | 142(60.94%) | <0.0001 |
| low | 233(50.11%) | - | - | 142(61.21%) | 91(39.06%) | |||
| Methylation | high | 232(49.89%) | 90(38.79%) | 142(60.94%) | <0.0001 | - | - | - |
| low | 233(50.11%) | 142(61.21%) | 91(39.06%) | - | - | |||
GPR87 expression positively correlates with immune cell infiltration and immune checkpoint expression
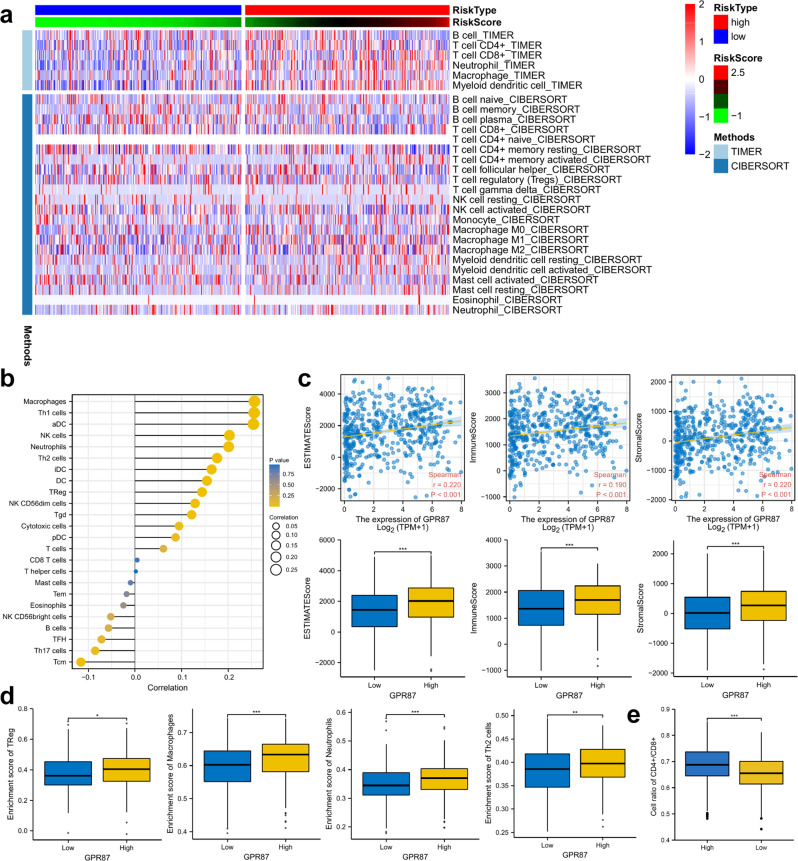
We used CIBERSORT and TIMER algorithm to evaluate the levels of immune cell infiltration in LUAD patients. The results showed that the levels of immune cell infiltration were higher in the GPR87 high-expression group (Fig. 6a). GPR87 was positively correlated with macrophages, Th1, aDC cells, etc., and negatively correlated with Tcm, Th17 cells, etc. (Fig. 6b). In addition, the immune and stromal scores of LUAD patients were calculated using the ESTIMATE algorithm, which further confirmed that patients with high expression of GPR87 had significantly higher levels of immune infiltration than those with low expression (Fig. 6c). The result of ssGSEA showed that infiltration of regulatory T cells, macrophages, neutrophils, T helper 2 cells, and natural killer cells was significantly higher in patients with high GPR87 expression than in patients with low expression (Fig. 6d). The proportion of CD4+/CD8+ cells was higher in patients with high GPR87 expression (Fig. 6e).
Fig. 6. The relationship between GPR87 expression and immune cell infiltration.
a Heat map demonstrating the level of immune cell infiltration in patients with high and low GPR87 expression, with immune cell infiltration evaluated by TIMER and CIBERSORT algorithms. b Lollipop graph shows the correlation between GPR87 expression and immune cells. c GPR87 was analyzed in correlation with immune scores and stromal scores, and differences in immune scores and stromal scores were analyzed in patients with high and low expression of GPR87. The immune scores and stromal scores were calculated by the ESTIMATE algorithm. d Differential analysis of regulatory T cells, macrophages, neutrophils, and T helper 2 cells in patients with high and low expression of GPR87. e Analysis of differences in CD4+/CD8+ cell ratios between patients with high and low expression of GPR87.
High GPR87 expression promotes immunotherapy resistance and chemoresistance in LUAD
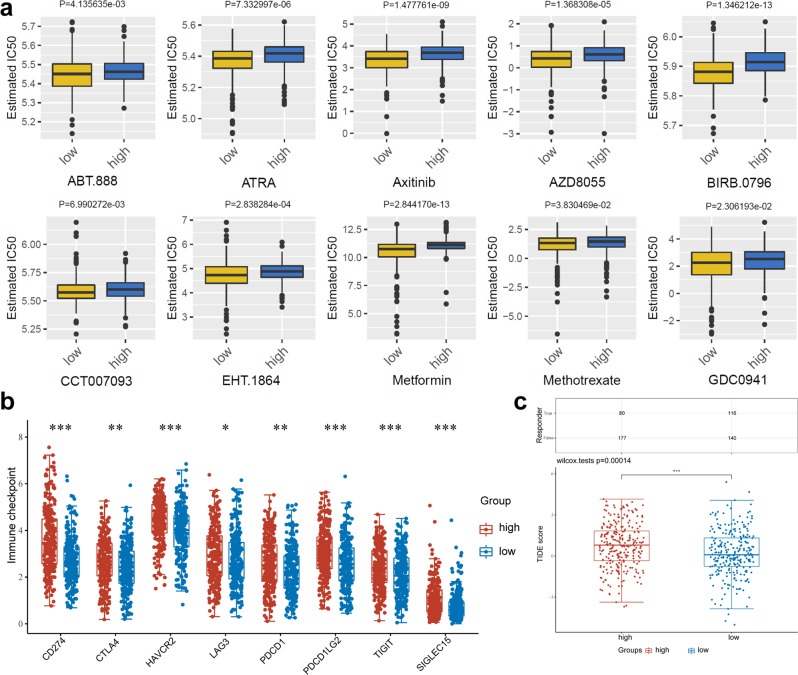
Based on the pRRophetic package, we evaluated the effects of GPR87 on the sensitivity of common chemotherapeutic agents. All the ten drugs (ABT.888, ATRA, axitinib, BIRB.0796, CCT007093, EHT.1864, metformin, methotrexate, GDC0941, and AZD8055) were found to have a higher estimated half-maximal inhibitory concentration (IC50) in high-risk patients (Wilcoxon test, all P < 0.05, Fig. 7a). Considering the vital role of immune checkpoint inhibitors (ICIs) in immunotherapy, we further investigated the differences in immune checkpoint expression between GPR87 high- and low-expression groups. We found that CD274, HAVCR2, LAG3, PDCD1, PDCD1LG2, TIGIT, and SIGLEC15 were significantly higher in patients with high GPR87 expression, which was consistent with the results of immune infiltration and suggested immunosuppression (Fig. 7b). This implies that LUAD patients with high GPR87 expression may be resistant to treatment with these ten drugs, but this conclusion needs to be verified in subsequent clinical trials. We used the tumor immune dysfunction and exclusion (TIDE) algorithm to assess the effects of GPR87 on the response rates to immune checkpoint (PD-1 and CTLA-4) inhibitors. Patients with higher TIDE scores were more likely to have tumor immune escape. Meanwhile, the TIDE score was more accurate than the PD-L1 expression levels and tumor mutation burden (TMB) to predict the survival of cancer patients treated with ICIs. TIDE scores were significantly lower in the high-GPR87 patients than in low-GPR87 ones (Wilcoxon test, P < 0.001, Fig. 7c), suggesting that high-GPR87 patients might respond poorly to immune checkpoint inhibitors and have bad outcomes.
Fig. 7. Immune infiltration and response of high- and low-GPR87 patients to chemotherapy and immunotherapy.
a The box plots of the estimated IC50 for ten common chemotherapeutic agents for high- and low-GPR87 expression. b Distribution of immune checkpoint expression in patients with high- and low-GPR87 expression. c The box plots of the TIDE scores for immunotherapy response for high- and low-GPR87 expression.
Discussion
In the last decades, the importance of DEGs in tumor progression has been recognized. However, genome-wide analysis remains to be investigated to explore its molecular mechanisms and clinical significance. NSCLC is one of the most common malignancies globally, and early diagnosis and treatment of NSCLC can improve its prognosis. Although the molecular mechanisms of NSCLC development and pathogenesis have been investigated using various histological techniques, the global mortality rate of NSCLC has remained high in recent decades. Therefore, the studies on more effective diagnostic and prognostic markers are ongoing challenges for biomedical research. Thanks to advances in bioinformatics technology, researchers have been able to identify promising biomarkers for NSCLC, and several relevant studies have been published13,14.
EMT is the phenomenon in which epithelial cells undergo a phenotypic transformation of fibroblasts or mesenchymal cells, loss of cell polarity, rearrangement of cytoskeleton, and the increased ability of migration and movement15. It involves a variety of biological and pathological processes, such as embryonic development, wound healing, cancer cell metastasis, and drug resistance16. EMT also plays vital role in the progression of lung cancer. EMT-related markers such as E-cadherin and vimentin are associated with prognosis, tumor lymph node metastasis, and tumor stem marker CD133 expression in lung cancer patients17,18. EMT is also associated with EGFR-TKI resistance in lung cancer. In EGFR-mutant lung cancer, EMT inhibits BIM through EMT-inducing transcription factors, ZEB1, and TWIST1, and becomes resistant to EGFR-TKIs19–21. In addition, the close relationship between the activation of EMT and inflammatory responses of the tumor microenvironment has also been confirmed, indicating that EMT is a candidate biomarker for NSCLC immunotherapy22,23.
In this study, we found a gene associated with tumor metastasis by analyzing the TCGA-LUAD database. The prognostic significance of GPR87 was then validated in four independent validation cohorts. Functional analysis showed that GPR87 enhanced migration and invasion of LUAD cells. These results suggest that GPR87 plays an important role in the metastasis of LUAD.
To investigate the potential clinical roles of GPR87 expression and methylation in LUAD, we analyzed GPR87 mRNA expression and methylation data in LUAD patients. We originally found that GPR87 mRNA was highly expressed in LUAD and that GPR87 mRNA expression was significantly and negatively correlated with GPR87 methylation. In addition, we validated the prognostic roles of GPR87 expression in four independent validation datasets, and these results suggest the potential prognostic values of GPR87 expression in LUAD patients. Finally, we conducted a meta-analysis of 1639 LUAD patients from five different databases to further prove that GPR87 expression was an independent prognostic variable for OS in LUAD patients.
The high-risk group will have more Treg cell infiltration. It has been reported in the literature that Treg cells promote tumor expression of more immunosuppressive molecules by suppressing CD8 + T cells, causing immune escape of tumors24,25. Tumor-associated macrophages originate from peripheral monocytes, and their tumor-promoting function supports tumor-associated angiogenesis and promotes cancer cell invasion, migration, and vascular metastasis. Tumor-associated macrophages are mainly divided into M1 and M2 subtypes. M1 macrophages located on tumor cell islets are usually associated with a better prognosis, whereas M2 macrophages, which are more abundant in the tumor stroma, are associated with a poorer prognosis26. CD4 + T cell subsets, such as Th1, Th2, Th17, and regulatory T (Treg) cells, play a crucial role in cancer immunity. The Th2 subset of CD4 + T cells secretes IL-4, IL-5, and IL-13 and activates B cells to become antibody-secreting plasma cells. Notably, the balance between Th1 and Th2 differentiation is crucial for immune homeostasis, and the shift of Th1/Th2 balance to Th2 cells is associated with immunosuppression and cancer progression27. In addition, ALEXANDRA et al. showed that the balance of CD4+ and CD8+ lymphocytes infiltrating the tumor mesenchyme is a crucial factor in determining antitumor immune surveillance and has a solid prognostic value as a predictive marker for immunotherapy in treatable NSCLC. High CD4+/CD8+ ratio defined a worst prognosis28. The CD4+/CD8+ ratio was higher in the GPR87 high-expression group than the low GPR87 expression group, suggesting that the high-expression group patients were correspondingly poorer for non-small cell lung cancer immunotherapy, consistent with the results of the TIDE algorithm analysis. Our study suggests that the expression level of GPR87 can be a predictor of immune cell infiltration and immunotherapy in non-small cell lung cancer.
A distinct mode of the immune conditions was further found between the low and high-GPR87 groups. Our studies suggested that GPR87 could be used as prognostic markers and indexes of immune status. In addition, The TIDE score of the high-GPR87 group was higher than that of the low-GPR87 one. The TIDE score reflects ICI response in LUAD patients. The lower the TIDE score and the more sensitive the patient was to ICIs. Therefore, the low-risk patients might benefit more from immunotherapy, which may be related to the relief of immunosuppression. Current bottlenecks in lung cancer treatment also forced such patients to return to conventional chemotherapy. Therefore, mRNA expression data were utilized to investigate the sensitivity of high- and low-GPR87 patients to conventional chemotherapeutic agents (ABT.888, ATRA, axitinib, BIRB.0796, CCT007093, EHT.1864, metformin, methotrexate, GDC0941, and AZD8055). These studies suggest that high-GPR87 patients performed better than low-GPR87 ones with the same drugs. Our studies demonstrated the sensitivity of patients with high- and low-GPR87 expression to ten chemotherapeutic agents, providing a therapeutic target for investigators to overcome drug resistance.
However, the current study has several shortcomings that should be considered when interpreting our results. First, transcriptome analysis can only reflect changes in mRNA levels, not overall changes. Second, the mechanism of GPR87-induced EMT was to be investigated. Finally, our results need to be validated with in vivo experiments as well as clinical samples.
Methods
Data collection and processing
The RNA-seq TPM data of LUAD, containing corresponding clinical data, were acquired from the TCGA29, including 497 LUAD and 54 normal tissues. Preprocessed methylation data were downloaded from UCSC XENA databases30. The datasets (GSE31210, GSE50081, GSE68465, and GSE30219) of NSCLC with survival data were downloaded from the GEO database as the validation sets31–33. GSE31210, GSE50081, and GSE30219 data were obtained from the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array) with 226, 181, and 293 NSCLC tissue samples, respectively. GSE68465 data were obtained from the GPL96 platform (Affymetrix Human Genome U133A Array) with 442 LUAD samples. The relationship between GPR87 expression and patient STAGE was analyzed using TISIDB34.
Differential expression analysis
DEGs were analyzed using the R software limma package (version 3.46.0) in LUAD and its adjacent normal tissues35. The Oncomine database (https://www.oncomine.org/resource/main.html) was used to analyze the expression of GPR87 between tumor and normal tissues36.
Receiver operating characteristic curve
We used the receiver operating characteristic curve (ROC) to analyze the predictive efficacy of molecular expression and predicted outcomes, the larger the area under the curve, the higher the diagnostic accuracy. We selected tumor and normal as predicted outcomes.
Single-sample gene set enrichment analysis
The patients were divided into two groups according to the expression of GPR87. ssGSEA was used to calculate a metastatic signature score for each sample through the R package GSVA (version 1.38.2). Default parameters in the GSVA package were used37. The metastatic gene signatures were obtained from the MSigDB38.
Functional enrichment analysis
The org.Hs.eg.db package was sued to convert the gene symbol into entrezID. GO and KEGG enrichment analyses were then performed using the clusterProfiler package (version 3.18.1)39. P < 0.05 was considered a statistical significance. The ggplot package was used to draw the diagram of GO and KEGG. The GSEA algorithm was used to identify enriched pathways between the GPR87 low- and high-expression groups. A ranked gene list was generated by comparing the GPR87 low- and high-expression groups. GSEA was then used to assess the enrichment of different pathway genomes in this ranked gene list.
Survival analysis
The relationship between GPR87 expression and prognosis was analyzed by Kaplan–Meier curves using the survminer package (version 0.4.9) in R software. In addition, we drew a nomogram including the clinical factors and the expression of GPR87. The calibration curve was painted to illustrate the accurateness of this model in predicting the survival of LUAD patients.
Meta-analysis
We assessed the prognostic significance of GPR87 in LUAD patients using a meta-analysis of five datasets. The combined HR and 95% CI were computed to assess the relationship between GPR87 expression and the prognosis of LUAD patients. The heterogeneity of the five datasets was evaluated by Q-test (I2 statistic). A fixed-effects model was chosen for the combination if there was no significant heterogeneity (I2 < 50%). Meta-analysis was performed using the Meta-Package (version 4.18-0) of R software40.
Methylation analysis
The GPR87 hypo- and hypermethylated groups were classified based on the median GPR87 DNA methylation levels in the TCGA-LUAD dataset. The correlations between GPR87 gene expression or DNA methylation and a range of categorical variables were analyzed using the chi-square test or Fisher’s exact test. The spearman analysis was used to investigate the relationship between GPR87 gene expression and DNA methylation.
Immunity analysis and gene expression
The differences in immune cell infiltration or immune responses between the GPR87 high- and low-expression groups were evaluated using the CIBERSORT41, ESTIMATE42, ssGSEA, and TIMER algorithms43. Heatmaps revealed the differences in immune cell infiltration using different algorithms. In addition, we analyzed the differences in the expression levels of immune checkpoints between the 2 groups.
Evaluation of immunotherapeutic strategies with GPR87 expression
We predicted potential ICI responses with the TIDE algorithm, which integrated the characteristics of T cell dysfunction and exclusion into the tumor immune escape model to predict the ICI responses44.
Evaluation of the sensitivity of chemotherapeutic agents
The pRRophetic algorithm was applied to predict the IC50 of chemotherapeutic drugs via constructing a ridge regression model based on the expression profiles of cancer drug sensitivity genomics and TCGA gene expression profiles45. To explore the effects of GPR87 on chemotherapy sensitivity in LUAD patients, we used the pRRophetic package (version 0.5) to predict the IC50.
Cells
LUAD cell lines, A549 and H1299, were purchased from the Type Culture Center of the Chinese Academy of Sciences (Shanghai, China), and cultured in RPMI-1640 medium (HyClone, USA) containing 10% fetal bovine serum. All cells were cultured in a standard tissue culture incubator maintained at 37 °C with 95% humidity and 5% CO2. We transfected GPR87-specific or non-specific siRNAs synthesized by Beijing TsingKe Company (Beijing, China) using the jetPRIME transfection reagent (Polyplus-transfection® SA, France). siRNA sequences are listed in Table S2.
RNA extraction and qRT-PCR
Total RNA was isolated from cells using TRIzol reagent (Vazyme, China). We used HiScript® Q RT SuperMix (Vazyme, China) to transcribe RNA and ChamQTM SYBR® qPCR Master Mix (Vazyme, China) for qRT-PCR. The primer sequences are listed in Table S1.
Flow cytometry
The negative control cells and siRNA-treated cells were collected, added with binding buffer and Annexin V-FITC staining solution, and kept at 4 °C for 15 min in the dark. After 5 min incubation with a propidium iodide solution, cells were analyzed with the CytoFLEX instrument.
Colony-forming assay
After transfection, 1000 cells per well were seeded in a six-well plate. After 2 weeks, colonies were fixed with 4% paraformaldehyde, stained with 0.5% crystal violet, and counted.
Wound healing assay
We seeded transfected cells into six-well plates. A pipette tip was used to make a straight scratch line in the cell monolayer. The following formula was used to calculate the migration rate: wound closure rate (%) = (area of initial scratch − cell-free area of final imaging)/distance of initial scratch.
Modified Boyden chamber assay
A549 or H1299 cells (3 × 105 cells in 200 μL serum-free medium) were seeded in the upper chambers pre-coated with Matrigel (BD). A culture medium (500 μL) with 10% fetal bovine serum was added to the lower chamber. Migrating cells were fixed with formaldehyde 48 h later and stained with 0.5% crystal violet (Sigma-Aldrich; Merck KGaA). Five fields per chamber were randomly observed by microscopy (Olympus, Japan; 200×), and the number of cells in each field was quantified.
Immunoblotting
The whole-cell proteins were extracted using RIPA lysis buffer (Beyotime, China). SDS-PAGE (10%) gel (EpiZyme, China) was used to separate the proteins, and proteins were transferred to PVDF membranes. We probed GPR87, E-cadherin, N-cadherin, Vimentin, and GAPDH with the corresponding antibodies at 4 °C for 12 h and detected them with chemiluminescence 2 h after incubation with HRP-labeled secondary antibodies (Bio-Rad, USA). Antibodies are listed in Table S3.
Statistics and reproducibility
All experimental data were expressed as mean ± standard deviation (SD). Statistical analysis of variance was conducted by GraphPad Prism 7 and one-way analysis of variance (ANOVA) to identify significant differences among groups. The student t-test was used to test for the significance between two groups, and statistical significance was achieved at P ≤ 0.05 (ns, P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001). All experiments were taken from distinct samples and the number of biological replicates (3).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81972852), Key Research & Development Project of Hubei Province (2020BCA069), Health Commission of Hubei Province Medical Leading Talent Project, Translational Medicine and Interdisciplinary Research Joint Fund of Zhongnan Hospital of Wuhan University (ZNJC201922), and Chinese Society of Clinical Oncology TopAlliance Tumor Immune Research Fund (Y-JS2019-036).
Author contributions
R.B., J.Z., Y.G., and C.X. contributed to the conceptualization of the study. R.B., J.Z., F.H., and Y.L. were responsible for the methodology design. R.B., J.Z., P.D., and Z.H. conducted to verify the experimental results. R.B., J.Z., L.H., and Z.W. were involved in the investigation. R.B. and J.Z. wrote the original draft. Y.G. and C.X. were involved in writing—review and editing and provided supervision and funding acquisition. All authors read and approved the final manuscript.
Peer review
Peer review information
Communications Biology thanks Divijendra Natha Reddy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Eve Rogers and Karli Montague-Cardoso.
Data availability
All data used in this work can be acquired from the Gene Expression Omnibus (GSE31210, GSE50081, and GSE68465;GSE30219) and the UCSC Xena (https://xenabrowser.net/datapages/?cohort=GDC%20TCGA%20Lung%20Adenocarcinoma%20(LUAD)&removeHub=https%3 A%2 F%2Fxena.treehouse.gi.ucsc.edu%3A443). The data of differentially expressed genes in patients with high and low expression of CARM1 and data for meta-analysis were provided as Supplementary Data 1 and 2. The uncropped gel images were provided in Supplementary Fig. S4. Any remaining information can be obtained from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rui Bai, Jianguo Zhan.
These authors jointly supervised this work: Yan Gong, Conghua Xie.
Contributor Information
Yan Gong, Email: yan.gong@whu.edu.cn.
Conghua Xie, Email: chxie_65@whu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03506-6.
References
- 1.Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: current status and perspectives. Crit. Rev. Oncol. Hematol. 2021;157:103194. doi: 10.1016/j.critrevonc.2020.103194. [DOI] [PubMed] [Google Scholar]
- 2.Wan X, et al. Drug combination synergy in worm-like polymeric micelles improves treatment outcome for small cell and non-small cell lung cancer. ACS Nano. 2018;12:2426–2439. doi: 10.1021/acsnano.7b07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535–554. doi: 10.1016/S0140-6736(21)00312-3. [DOI] [PubMed] [Google Scholar]
- 4.Noman MZ, et al. CD47 is a direct target of SNAI1 and ZEB1 and its blockade activates the phagocytosis of breast cancer cells undergoing EMT. Oncoimmunology. 2018;7:e1345415. doi: 10.1080/2162402X.2017.1345415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeon YK, et al. Pellino-1 promotes lung carcinogenesis via the stabilization of Slug and Snail through K63-mediated polyubiquitination. Cell Death Differ. 2017;24:469–480. doi: 10.1038/cdd.2016.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsen JE, et al. Gene expression signature predicts recurrence in lung adenocarcinoma. Clin. Cancer Res. 2007;13:2946–2954. doi: 10.1158/1078-0432.CCR-06-2525. [DOI] [PubMed] [Google Scholar]
- 7.Jiang J, et al. G protein-coupled receptor GPR87 promotes the expansion of PDA stem cells through activating JAK2/STAT3. Mol. Ther. Oncolytics. 2020;17:384–393. doi: 10.1016/j.omto.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, et al. Overexpression of G protein-coupled receptor GPR87 promotes pancreatic cancer aggressiveness and activates NF-kappaB signaling pathway. Mol. Cancer. 2017;16:61. doi: 10.1186/s12943-017-0627-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Qian Y, Lu W, Chen X. The G protein-coupled receptor 87 is necessary for p53-dependent cell survival in response to genotoxic stress. Cancer Res. 2009;69:6049–6056. doi: 10.1158/0008-5472.CAN-09-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gugger M, et al. GPR87 is an overexpressed G-protein coupled receptor in squamous cell carcinoma of the lung. Dis. Markers. 2008;24:41–50. doi: 10.1155/2008/857474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kita Y, et al. Inhibition of cell-surface molecular GPR87 With GPR87-suppressing adenoviral vector disturb tumor proliferation in lung cancer cells. Anticancer Res. 2020;40:733–741. doi: 10.21873/anticanres.14004. [DOI] [PubMed] [Google Scholar]
- 12.Nii K, et al. Overexpression of G protein-coupled receptor 87 correlates with poorer tumor differentiation and higher tumor proliferation in non-small-cell lung cancer. Mol. Clin. Oncol. 2014;2:539–544. doi: 10.3892/mco.2014.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, et al. Establishment of the prognostic index of lung squamous cell carcinoma based on immunogenomic landscape analysis. Cancer Cell Int. 2020;20:330. doi: 10.1186/s12935-020-01429-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, et al. Establishment of the prognostic index reflecting Tumor immune microenvironment of lung adenocarcinoma based on metabolism-related genes. J. Cancer. 2020;11:7101–7115. doi: 10.7150/jca.49266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manshouri R, et al. ZEB1/NuRD complex suppresses TBC1D2b to stimulate E-cadherin internalization and promote metastasis in lung cancer. Nat. Commun. 2019;10:5125. doi: 10.1038/s41467-019-12832-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menju T, Date H. Lung cancer and epithelial-mesenchymal transition. Gen. Thorac. Cardiovasc. Surg. 2021;69:781–789. doi: 10.1007/s11748-021-01595-4. [DOI] [PubMed] [Google Scholar]
- 17.Richardson AM, et al. Vimentin is required for lung adenocarcinoma metastasis via heterotypic tumor cell-cancer-associated fibroblast interactions during collective invasion. Clin. Cancer Res. 2018;24:420–432. doi: 10.1158/1078-0432.CCR-17-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sowa T, et al. Association between epithelial-mesenchymal transition and cancer stemness and their effect on the prognosis of lung adenocarcinoma. Cancer Med. 2015;4:1853–1862. doi: 10.1002/cam4.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat. Rev. Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart EL, Tan SZ, Liu G, Tsao MS. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl. Lung Cancer Res. 2015;4:67–81. doi: 10.3978/j.issn.2218-6751.2014.11.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erin N, Grahovac J, Brozovic A, Efferth T. Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug Resist. Updat. 2020;53:100715. doi: 10.1016/j.drup.2020.100715. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y, Zhan H. Communication between EMT and PD-L1 signaling: New insights into tumor immune evasion. Cancer Lett. 2020;468:72–81. doi: 10.1016/j.canlet.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Raimondi C, et al. PD-L1 and epithelial-mesenchymal transition in circulating tumor cells from non-small cell lung cancer patients: a molecular shield to evade immune system? Oncoimmunology. 2017;6:e1315488. doi: 10.1080/2162402X.2017.1315488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han L, et al. Agrin promotes non-small cell lung cancer progression and stimulates regulatory T cells via increasing IL-6 secretion through PI3K/AKT pathway. Front. Oncol. 2021;11:804418. doi: 10.3389/fonc.2021.804418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, et al. DNA repair and replication-related gene signature based on tumor mutation burden reveals prognostic and immunotherapy response in gastric cancer. J. Oncol. 2022;2022:6469523. doi: 10.1155/2022/6469523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conway EM, et al. Macrophages, inflammation, and lung cancer. Am. J. Respir. Crit. Care Med. 2016;193:116–130. doi: 10.1164/rccm.201508-1545CI. [DOI] [PubMed] [Google Scholar]
- 27.Zhu ZY, et al. Comprehensive pan-cancer genomic analysis reveals PHF19 as a carcinogenic indicator related to immune infiltration and prognosis of hepatocellular carcinoma. Front. Immunol. 2021;12:781087. doi: 10.3389/fimmu.2021.781087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giatromanolaki A, et al. Prognostic relevance of the relative presence of CD4, CD8 and CD20 expressing tumor infiltrating lymphocytes in operable non-small cell lung cancer patients. Anticancer Res. 2021;41:3989–3995. doi: 10.21873/anticanres.15196. [DOI] [PubMed] [Google Scholar]
- 29.Pernemalm M, et al. Quantitative proteomics profiling of primary lung adenocarcinoma tumors reveals functional perturbations in tumor metabolism. J. Proteome Res. 2013;12:3934–3943. doi: 10.1021/pr4002096. [DOI] [PubMed] [Google Scholar]
- 30.Bai Z, et al. Proteomics-based identification of a group of apoptosis-related proteins and biomarkers in gastric cancer. Int. J. Oncol. 2011;38:375–383. doi: 10.3892/ijo.2010.873. [DOI] [PubMed] [Google Scholar]
- 31.Okayama H, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 32.Director’s Challenge Consortium for the Molecular Classification of Lung, A. et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat. Med.14, 822–827 (2008). [DOI] [PMC free article] [PubMed]
- 33.Botling J, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin. Cancer Res. 2013;19:194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 34.Ru B, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 35.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng, Z. & Shao, X. ENO1 acts as a prognostic biomarker candidate and promotes tumor growth and migration ability through the regulation of Rab1A in colorectal cancer. Cancer Manag. Res. 11, 9969–9978 (2019). [DOI] [PMC free article] [PubMed]
- 39.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balduzzi S, Rucker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid. Based Ment. Health. 2019;22:153–160. doi: 10.1136/ebmental-2019-300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang P, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9:e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data used in this work can be acquired from the Gene Expression Omnibus (GSE31210, GSE50081, and GSE68465;GSE30219) and the UCSC Xena (https://xenabrowser.net/datapages/?cohort=GDC%20TCGA%20Lung%20Adenocarcinoma%20(LUAD)&removeHub=https%3 A%2 F%2Fxena.treehouse.gi.ucsc.edu%3A443). The data of differentially expressed genes in patients with high and low expression of CARM1 and data for meta-analysis were provided as Supplementary Data 1 and 2. The uncropped gel images were provided in Supplementary Fig. S4. Any remaining information can be obtained from the corresponding author upon reasonable request.