Abstract
The fatty acid synthase genes FAS1 and FAS2 of the yeast Saccharomyces cerevisiae are transcriptionally co-regulated by general transcription factors (such as Reb1, Rap1 and Abf1) and by the phospholipid-specific heterodimeric activator Ino2/Ino4, acting via their corresponding upstream binding sites. Here we provide evidence for a positive autoregulatory influence of FAS1 on FAS2 expression. Even with a constant FAS2 copy number, a 10-fold increase of FAS2 transcript amount was observed in the presence of FAS1 in multi-copy, compared to a fas1 null mutant. Surprisingly, the first 66 nt of the FAS2 coding region turned out as necessary and sufficient for FAS1-dependent gene expression. FAS2–lacZ fusion constructs deleted for this region showed high reporter gene expression even in the absence of FAS1, arguing for a negatively-acting downstream repression site (DRS) responsible for FAS1-dependent expression of FAS2. Our data suggest that the FAS1 gene product, in addition to its catalytic function, is also required for the coordinate biosynthetic control of the yeast FAS complex. An excess of uncomplexed Fas1 may be responsible for the deactivation of an FAS2-specific repressor, acting via the DRS.
INTRODUCTION
The biosynthesis of multiprotein complexes with a defined stoichiometry of subunits requires coordinate expression of the corresponding structural genes. In eukaryotes, gene activation by related upstream sequences is a general mechanism to ensure balanced production of the respective polypeptides in response to regulatory signals (1). In previous work, we studied the genetic control of structural genes encoding subunits of the fatty acid synthase (FAS) complex in the yeast Saccharomyces cerevisiae. FAS1 and FAS2 are genetically unlinked and encode the multifunctional subunits β and α, respectively (2–5), which finally constitute the α6β6 heteromultimeric complex (6).
Deletion analysis of FAS1 and FAS2 promoters revealed the existence of a common type of upstream activation site (UAS), designated ICRE (inositol/choline responsive element; 7), which could be also identified in the control regions of several structural genes involved in phospholipid metabolism (such as INO1, CHO1, ACC1 and ACS2; 8–11, respectively). Importantly, transcriptional activation of target genes by ICRE motifs (= UASINO; consensus sequence, WYTTCAYRTG; 12) is regulated by phospholipid precursors inositol and choline and requires the positive factors Ino2 and Ino4 (13–16). Ino2 and Ino4 both contain a basic helix–loop–helix structural motif and bind to the ICRE as a heterodimer (16). Negative regulation of ICRE-containing genes by phospholipid precursors is mediated by the Opi1 repressor (17,18), which contacts Ino2 as well as the pleiotropic repressor Sin3 (19). In addition to UAS elements influenced by phospholipid precursors, FAS promoters are also activated by binding sites of the essential transcription factors Rap1, Abf1 and Reb1 (20). These proteins are also required for the expression of glycolytic as well as ribosomal protein genes (21,22). Since FAS genes fulfill a housekeeping function in cellular biochemistry, constitutively activating motifs ensure fatty acid biosynthesis even under conditions of inositol/choline repression. However, neither ICRE motifs nor constitutive elements of transcription appear suitable to ensure a defined stoichiometry of FAS1 and FAS2 gene products within the cell.
The construction and subsequent characterization of fas1 and fas2 null mutants provided evidence for additional mechanisms leading to a balanced ratio of FAS subunits α and β. While the substantially increased sensitivity of individual FAS subunits against vacuolar or proteosomal proteinases (23) may be a trivial consequence of unsuccessful complex formation, the influence of a fas1 deletion on the amount of the FAS2 transcript suggested a biosynthetic cross-talk among both genes (24). In the absence of FAS1 (but with an intact FAS2 gene), a decrease of FAS2 mRNA to ~35% of the wild-type level was found. In contrast, the concentration of the FAS1 transcript was not affected by the allelic status of FAS2. In this work, we investigated the influence of FAS1 on FAS2. To our surprise, the FAS2 upstream region did not respond to a variation of FAS1 gene dosage. Instead, we identified a region within its reading frame that was necessary and sufficient for mediating FAS1-dependent expression of FAS2. The corresponding FAS2 coding sequence turned out to be a negatively-acting element, requiring an excess of individual Fas1 to overcome its inhibitory influence. This finding supports the existence of a new mechanism contributing to a coordinate biosynthesis of proteins within multi-subunit complexes.
MATERIALS AND METHODS
Strains and media
The strains of S.cerevisiae used for this study are isogenic to the regulatory wild-type JS91.15-23 (MATα ura3 leu2 his3 trp1 can1) and were obtained by introduction of the mutant alleles indicated (IKY1, pra1::HIS3; IKY2, Δfas1::LEU2; IKY3, pra1::HIS3 Δfas1::LEU2; IKY4, Δfas2::LEU2; PWY12, Δfas1::HIS3 Δfas2::LEU2). Disruption plasmids that were used for strain construction have been described [pBF3, Δfas1::HIS3; pBF4, Δfas2::LEU2; pBF5, Δfas1::LEU2 (24); pra1::HIS3 (23)]. Strain WCG4-11/22A (MATa ura3 leu2 his3 pre1 pre2) (25) containing two defective subunits of the proteasome represents a different strain background. Transformants were grown in synthetic complete media selecting for the introduced genetic markers (e.g. SCDFA-Ura-Trp). To allow growth of fas mutants, all media were supplemented with 1% Tween-40 and 0.03% hydrolyzed butter.
Plasmid construction
Reporter plasmid pSAK1 was derived from YIplac211 (integrating LEU2 vector) (26) by transfer of a FAS2–lacZ fusion from pJS203 (7), containing an ~1 kb upstream region together with 921 bp of the coding sequence. Similarly, pBF16 was obtained by insertion of the same fusion gene into YCplac22 (ARS CEN TRP1 vector). FAS gene dosage variation was achieved with effector plasmids pJS222 and pJS225 (6.9 kb BamHI/SalI fragment with FAS2, inserted into 2 µm LEU2 vector YEp351 and 2 µm URA3 vector YEp352, respectively), and pJS229 (9.9 kb SacI/SphI fragment with FAS1, inserted into YEp352) (27).
To obtain FAS2–lacZ reporter plasmids with a varying portion of the coding sequence, fragments were amplified by PCR, using a constant upstream primer in combination with downstream primers scanning the FAS2 reading frame. The resulting single-copy plasmids [ARS CEN TRP1 FAS2(1/x)–lacZ; otherwise identical to pBF16] contain 1008 bp of the FAS2 control region together with 3, 45, 66, 75, 84, 93, 99, 150 or 228 bp of the coding sequence. To further define the FAS2 regulatory element necessary and sufficient for FAS1-mediated autoregulation, a lacZ variant lacking its start codon was inserted into expression plasmid p414-MET25 (28). Finally, MET25–FAS2(x/y)–lacZ reporter plasmids were constructed by insertion of PCR fragments containing varying sequences of the FAS2 reading frame. To avoid a different translational efficiency, all constructs contain the natural (–6/+6) sequence of FAS2.
Miscellaneous procedures
For northern blot hybridization, 20 µg of total RNA from yeast transformants was separated by gel electrophoresis under denaturing conditions (1% agarose with 2.2 M formaldehyde). Following membrane transfer, hybridization was done under standard conditions, using 32P-labeled FAS2 and ACT1 (internal loading control) probes. Phosphoimager quantification of signal intensities was performed with Fujifilm Bio-Imaging Analyser BAS-1500. Yeast transformation was done by a simplified lithium acetate procedure (29). Activity of FAS was determined by assaying β-ketoacyl reductase-dependent oxidation of NADPH in the presence of acetyl-CoA and malonyl-CoA (30). The β-galactosidase assay has been described (7).
RESULTS
Regulation of FAS2 mRNA level by the FAS1 gene dosage
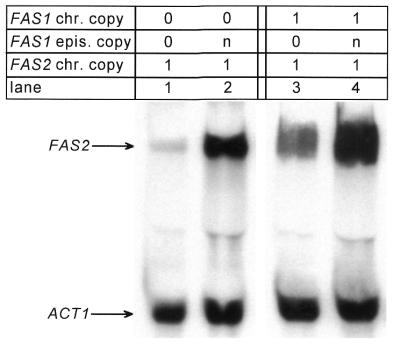
Our previous characterization of fas1 and fas2 deletion mutants by northern blot hybridization provided evidence for influence of FAS1 on FAS2 gene expression. Even with an intact FAS2 gene, the amount of FAS2 mRNA decreased to ~35% of the wild-type level in the presence of a fas1 mutation (24). We now ask whether an increase of FAS1 gene dosage also affects the steady-state concentration of FAS2 mRNA. Isogenic strains carrying a fas1 null mutation (IKY3) or the chromosomal FAS1 wild-type allele (IKY1) were transformed with an episomal multi-copy plasmid containing FAS1 under natural promoter control (pJS229). The transformants obtained contain a single chromosomal FAS2 gene but differ with respect to the amount of FAS1 encoded β-subunit (verified by immunoblot analysis, using β-specific antibodies; not shown). As depicted in Figure 1 (lanes 1 and 2), elevating the FAS1 copy number from 0 to n (copy number of a plasmid containing the 2 µm origin) led to a strong increase of FAS2 mRNA (~10-fold, according to phosphoimager quantification). With a FAS1 wild-type strain, a further 2.5-fold increase of the FAS2 transcript amount was detected in the presence of additional FAS1 copies (lanes 3 and 4). Thus, in addition to its catalytic function, FAS1 must be considered as a regulatory factor of FAS2 gene expression.
Figure 1.
FAS1-dependent steady-state concentration of the FAS2 mRNA. For the northern blot hybridization shown, total RNA was isolated from strains with identical FAS2 copy number (single chromosomal copy, chr. copy) but varying FAS1 gene dosage. Strains IKY1 (ura3 pra1 FAS1 FAS2, lanes 3 and 4) and IKY3 (ura3 pra1 Δfas1 FAS2, lanes 1 and 2) were transformed with plasmids YEp352 (vector control, 2 µm URA3, lanes 1 and 3) or pJS229 (copy number n; 2 µm URA3 FAS1, lanes 2 and 4) and subsequently grown in SCD-Ura, supplemented with fatty acids. The filter was simultaneously hybridized against FAS2 and ACT1 (internal control) DNA probes. Quantification of signal intensities was done by phosphoimager analysis. After background subtraction, the ratio of PSL values (photo stimulated luminescence) for FAS2 and ACT1 signals was calculated for each lane (PSLFAS2/PSLACT1; lane 1, 0.15; lane 2, 1.39; lane 3, 0.76; lane 4, 1.9).
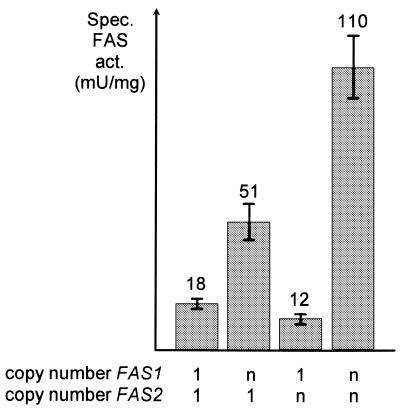
We also investigated whether FAS1 dosage variation similarly affects FAS enzyme activity. For a functional FAS, both α and β subunits are required (31). Individual FAS subunits were sensitive against proteolytic attack, with vacuolar proteinases yscA and yscB being required for degradation of β (24) while the proteasome (yscE) is involved in cleavage of α (23). Thus, overexpression of a single FAS gene should not lead to an increase in FAS activity. A FAS wild-type strain lacking the vacuolar proteinase yscA was transformed with FAS1 or FAS2 containing multi-copy plasmids, and transformants were subsequently assayed for specific FAS activity. As is apparent from Figure 2, multiple copies of FAS2 only did not lead to an increased FAS activity in transformants. In contrast, a 2.8-fold increase was assayed as a result of overexpressing only FAS1. This finding agrees with the idea of a dual role for FAS1: overexpression not only results in increased production of β but also allows stimulation of α biosynthesis. Due to its positive regulatory function, FAS1 overexpression is sufficient for a substantially enhanced FAS activity. As expected, a further increase of FAS activity could be assayed when both FAS genes were introduced in multi-copy. With a pre1 pre2 double mutant allowing stable overproduction of α, almost identical results were obtained (not shown).
Figure 2.
Specific FAS activity in transformants with varying FAS1 and FAS2 gene dosage. Strain IKY1 (ura3 leu2 pra1 FAS1 FAS2) was transformed in pairs with combinations of empty vectors (YEp352, 2 µm URA3; YEp351, 2 µm LEU2; total FAS copy numbers 1), or multi-copy plasmids (pJS229, 2 µm URA3 FAS1; pJS225, 2 µm LEU2 FAS2; total FAS copy numbers n). Transformants were grown in selective medium (SCD-Ura-Leu). Specific FAS activity is given in nanomoles NADPH oxidized per minute per milligram of protein (mU/mg). Standard deviation of the mean value is indicated by bars.
Influence of FAS1 and FAS2 on FAS2–lacZ reporter gene expression
For the identification of the cis-acting element mediating the positive influence of FAS1, we first tested whether the autoregulation among FAS genes can be also detected with a FAS2–lacZ reporter gene. An FAS2–lacZ fusion containing 1008 bp of the upstream region together with 921 bp of the coding sequence was integrated at the LEU2 locus of a regulatory wild-type strain. Subsequently, we introduced either a Δfas1::HIS3 mutant allele or additional FAS1 copies by transformation with pJS229, allowing a comparison of the influence of FAS1 copy numbers 0, 1 and n on FAS2–lacZ expression. As is apparent from Table 1, deletion of FAS1 reduced FAS2–lacZ expression to ~24% of the wild-type level. In contrast, a 2.6-fold increase was assayed in the presence of additional FAS1 copies. Similar results were obtained with an identical FAS2–lacZ fusion transferred to a single-copy ARS CEN plasmid. Importantly, with a strain lacking the vacuolar proteinase yscA (pra1 mutant), elevating the FAS1 copy number had an even greater effect on FAS2–lacZ expression. A 4.9-fold increase of specific β-galactosidase activity was assayed with multiple copies of FAS1 in the pra1 mutant (compared with the chromosomal FAS1 single-copy situation) while a 2.9-fold increase could be detected in the isogenic PRA1 background. Since the FAS1 gene product is more stable in the absence of yscA, FAS2–lacZ expression may indeed be stimulated by the FAS β-subunit.
Table 1. Influence of FAS1 gene dosage variation on FAS2(1/921)–lacZ reporter gene expression.
| Genetic background | Specific β-galactosidase activity (U/mg) with |
||
|---|---|---|---|
| Δfas1 | FAS1 | (FAS1)n | |
| Reporter plasmids pSAK1 (FAS2–lacZ LEU2, integrative) and pBF16 (FAS2–lacZ ARS CEN TRP1) were transformed into isogenic strains JS91.15-23 (PRA1) and IKY1 (pra1), respectively. FAS1 dosage variation was achieved by co-transformation with empty vector YEp352 (2 µm URA3) or pJS229 (2 µm URA3 FAS1). Transformants were grown in selective medium (SCD-Ura-Leu or SCD-Ura-Trp), supplemented with fatty acids. Specific β-galactosidase activities are given in nanomoles ONPG hydrolyzed per minute per milligram of protein. Standard deviation was ≤25% of the mean value. | |||
| FAS2–lacZ (integrated) in PRA1 strain | 25 | 105 | 280 |
| FAS2–lacZ (ARS CEN) in PRA1 strain | 20 | 85 | 250 |
| FAS2–lacZ (ARS CEN) in pra1 strain | 40 | 150 | 735 |
These experiments were extended by a systematic comparison of FAS2–lacZ expression in strains containing FAS1 FAS2, fas1 FAS2, FAS1 fas2 and fas1 fas2 alleles, transformed with empty vector or either FAS1 or FAS2 in multi-copy (data shown in Table 2). Interestingly, introduction of a fas2 deletion led to an increase of FAS2–lacZ expression by a factor of 2.4. The FAS2 gene product may act as a repressor of its own expression. Consequently, the stimulating effect of FAS1 might be the indirect result of removal of individual α-subunits, becoming assembled into the FAS complex. Assuming such a mechanism, deletion of both FAS effector genes should allow high expression of FAS2–lacZ. However, the idea of FAS2 encoding its own repressor appears unlikely, since a fas1 fas2 double deletion mutant shows low expression of FAS2–lacZ, similarly to the fas1 single mutant. Nevertheless, FAS2 indirectly influences its own expression by controlling the level of individual β-subunits. Overexpression of FAS2 in the fas2 single mutant led to a 2.9-fold decrease of FAS2–lacZ expression, compared with the vector control. In contrast, with all genetic situations tested, FAS1 overexpression stimulated FAS2–lacZ expression. We conclude that FAS1 does not simply counteract repression by the FAS2 encoded α-subunit.
Table 2. Expression of the FAS2–lacZ reporter gene with varying FAS gene dosage.
| Relevant genotype | Specific β-galactosidase activity with effector plasmids | ||
|---|---|---|---|
| YEp352 (vector control) | pJS229 (FAS1)n | pJS222 (FAS2)n | |
| Reporter plasmid pBF16 (FAS2–lacZ ARS CEN TRP1) was transformed into isogenic strains JS91.15-23 (FAS1 FAS2), IKY2 (Δfas1 FAS2), IKY4 (FAS1 Δfas2) and PWY12 (Δfas1 Δfas2), respectively. FAS1 or FAS2 dosage variation was achieved by co-transformation with empty vector YEp352 (2 µm URA3), pJS229 (2 µm URA3 FAS1) or pJS222 (2 µm URA3 FAS2). Transformants were grown in selective medium (SCD-Ura-Trp), supplemented with fatty acids. Specific β-galactosidase activities are given in nanomoles ONPG hydrolyzed per minute per milligram of protein. Standard deviation was ≤25% of the mean value. | |||
| FAS1 FAS2 | 85 | 310 | 60 |
| Δfas1 FAS2 | 20 | 250 | 30 |
| FAS1 Δfas2 | 200 | 370 | 70 |
| Δfas1 Δfas2 | 25 | 480 | 40 |
In contrast with what was found for the FAS2–lacZ fusion, no influence of either FAS1 or FAS2 on the expression of an FAS1–lacZ reporter gene could be detected (not shown). These findings agree with northern blot hybridizations described previously (24). To complete these experiments on the mutual influence of genes involved in fatty acid biosynthesis, we also considered the essential acetyl-CoA carboxylase gene ACC1/FAS3. However, gene dosage variation of either FAS1 or FAS2 did not affect expression of an ACC1–lacZ fusion. Similarly, FAS1–lacZ and FAS2–lacZ fusions were not influenced by overexpression of the ACC1 structural gene (not shown). Thus, stimulation of FAS2 expression by FAS1 is the sole autoregulatory influence among structural genes ACC1, FAS1 and FAS2, which are specifically required for fatty acid synthesis in yeast.
Negative regulation of FAS2 by a downstream element
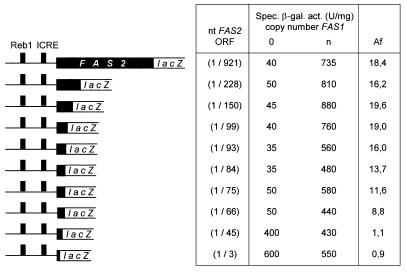
Initially, we expected to map the cis-acting element(s) responsible for FAS1-mediated autoregulation of FAS2 in the upstream region of the gene. However, FAS2–lacZ fusions devoid of FAS2 coding sequences did not respond to variations of FAS1 copy number. With the FAS2(1/3)–lacZ fusion containing only the natural ATG codon, a high β-galactosidase activity was measured even in the absence of FAS1 (Fig. 3). This finding argues for the existence of a negatively-acting element leading to low reporter gene expression when fusion constructs contain a considerable portion of the FAS2 coding region. Thus, the corresponding cis-acting element within the FAS2 reading frame must be considered as a downstream repression site (DRS). For a precise localization of the DRS, we shortened the coding region of FAS2 which, together with an upstream region of constant length, was then inserted 5′ to the lacZ reporter gene. As is apparent from Figure 3, a FAS2(1/66)–lacZ fusion still showed FAS1-dependent expression while FAS2(1/45)–lacZ was constantly activated at a high level. We conclude that autoregulation among FAS genes requires at least 66 nt of the FAS2 coding region. It remained undetermined whether FAS2-specific upstream promoter elements or sequences of the 5′-untranslated part of the mRNA are also necessary.
Figure 3.
Deletion analysis of the FAS2 coding region responsible for FAS1-dependent gene expression. Deletion constructs contain identical upstream sequences (–1008) but differ with respect to the length of the FAS2 reading frame, fused to lacZ. Reporter plasmids [ARS CEN TRP1 FAS2(1/x)–lacZ; data are given in nucleotide positions] were transformed into fas1 mutant strain IKY3, containing either the empty vector YEp352 (FAS1 copy number 0) or FAS1 multi-copy plasmid pJS229 (copy number n). Transformants were grown in selective medium (SCD-Ura-Trp), supplemented with fatty acids. Specific β-galactosidase activities are given in nanomoles ONPG hydrolyzed per minute per milligram of protein. Standard deviation was ≤25% of the mean value. Af, activation factor of specific enzyme activity in the presence of multiple FAS1 copies, compared with the null mutant.
Thus, FAS2 reading frame fragments of varying length and position were inserted between the heterologous MET25 promoter (containing its natural transcription initiation site) and lacZ as a reporter gene. The MET25–FAS2(1/66)–lacZ construct clearly showed FAS1-dependent expression (cf. Fig. 4), supporting the view that 66 nt of the FAS2 reading frame are necessary and sufficient for autoregulation. A weakened but still significant influence of FAS1 copy number was also observed with the MET25–FAS2(22/150)–lacZ construct. However, a fusion gene lacking the first 39 nt of FAS2 ORF was constantly expressed at an intermediate level. We conclude that nt 22–66 of the FAS2 coding region may represent the essential core of the DRS, which is able to negatively regulate gene expression even when separated from its natural context.
Figure 4.
FAS1-dependent expression of FAS2–lacZ reporter constructs under heterologous promoter control. Various sequences of the FAS2 coding region (indicated by nucleotide positions) were inserted between the MET25 promoter and the lacZ reporter gene. To ensure similar translational efficiency, reading frame fragments with an artificial start codon contain the natural –6/–1 sequence of FAS2. Reporter plasmids [ARS CEN TRP1 MET25–FAS2(x/y)–lacZ] were transformed into fas1 mutant strain IKY3, containing either the empty vector YEp352 (FAS1 copy number 0) or FAS1 multi-copy plasmid pJS229 (copy number n). For a maximal promoter strength, transformants were grown in selective medium without methionine (SCD-Ura-Trp-Met), supplemented with fatty acids. Specific β-galactosidase activities are given in nanomoles ONPG hydrolyzed per minute per milligram of protein. Standard deviation was ≤25% of the mean value. Af, activation factor of specific enzyme activity in the presence of multiple FAS1 copies, compared to the null mutant.
DISCUSSION
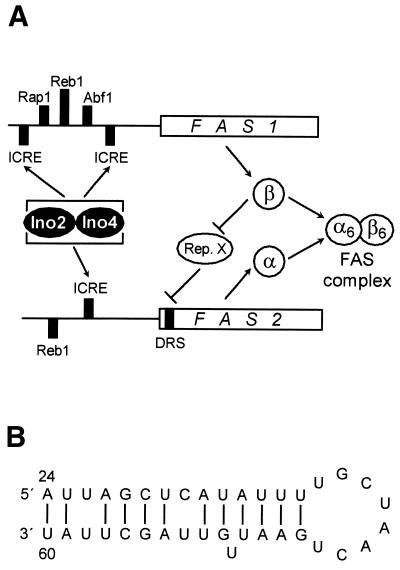
Coordinate expression of genes that encode functionally associated proteins is usually achieved by similar upstream promoter elements such as UAS or URS motifs. In this work, we present evidence for the existence of an autoregulatory mechanism depending on a sequence within the coding region of the yeast FAS gene FAS2. FAS autoregulation comprises genes and gene products of the FAS complex as a whole, involving FAS1 as a positive factor of FAS2 expression. In the absence of Fas1 (β subunit), FAS2 expression is down regulated while, conversely, overexpression of FAS1 and the subsequent accumulation of individual β subunits may counteract negative regulation. Thus, a temporary excess of Fas1 should derepress FAS2 expression and subsequently allow synthesis of Fas2 (α subunit), finally ensuring a balanced ratio of both subunits of the FAS complex. In contrast with the positive influence of the trans-acting factor Fas1, the autoregulatory cis-acting sequence (DRS) was identified as a negative element. Thus, Fas1 may function as an indirect positive factor, mediating deactivation of an unknown repressor that is responsible for a low level of FAS2 mRNA. Together with previous work, these results argue for the existence of a temporal order of events (summarized in Fig. 5A) leading to a defined stoichiometry of FAS subunits: (i) ICRE-dependent control requiring the heterodimeric activator Ino2/Ino4 coordinates transcription of structural genes involved in phospholipid biosynthesis (7,14,16); (ii) pleiotropic transcription factors such as Rap1, Abf1 and Reb1 are needed to fulfill the house-keeping function of fatty acids even under conditions of inositol/choline repression (20); (iii) autoregulation of FAS genes at the transcriptional level as a mechanism of fine-tuning leads to a balanced biosynthesis of α and β subunits (this work); and (iv) degradation of individual subunits by different proteolytic systems may support biosynthetic autoregulation under certain conditions (23,24; not depicted in Fig. 5A).
Figure 5.
(A) Hypothesis on coordinate control of FAS genes by Fas1-dependent anti-repression of FAS2 gene expression. In the absence of non-complexed Fas1 (β-subunit), the FAS2 control region (including downstream sequences) is substantially weaker than the FAS1 promoter. An excess of free β-subunit may directly or indirectly deactivate the repressor (Rep. X; acts via the DRS), leading to maximal FAS2 expression and subsequent synthesis of a balanced amount of α-subunit (FAS2 gene product), which may then associate with β to form a functional FAS complex (α6β6). Thereby, withdrawal of β again reduces FAS2 expression. The hypothesis considers FAS1 expression as the leading and FAS2 expression as the lagging step of FAS complex formation. It is unknown whether anti-repression acts at the level of transcriptional initiation or elongation. (B) Hypothetical stem–loop structure within FAS2 mRNA. Nucleotide positions refer to the start of the FAS2 reading frame.
In contrast with what has been reported for higher eukaryotes, only few examples of intragenic regulatory sites were identified in yeast genes. Downstream activation sites (DASs) have been described for the glycolytic genes PGK1 (32) and PYK1 (33) as well as for the glucose-inducible SRP1 gene (= TIR1; 34), encoding a cell wall protein. While trans-acting factors affecting DAS elements of PGK1 and PYK1 were not identified, the pleiotropic repressor/activator protein Rap1 binds to a 33 bp intragenic sequence of SRP1 (34). Transcription of yeast retrotransposons Ty1 and Ty2 is controlled by an activating (35,36) as well as by a repressing site (37), both of which map to the coding region. Similarly, a combination of DAS and DRS elements influences transcription of the lipoamide dehydrogenase gene LPD1 (38). Two DRSs (DRS1 and DRS2) involved in negative regulation with ethanol as a carbon source were precisely mapped in the coding region of the hexokinase PII gene HXK2 (39). Interestingly, both DRS elements contain the core sequence (A/C)(A/G)GAAAT, which has been also identified as a UAS of the invertase gene SUC2 (40). A subunit of the RNA polymerase II mediator complex (Med8, containing a putative leucine zipper motif) could be identified as the corresponding binding factor (41,42). It has been also reported that transcription factor binding sites occurring fortuitously within reading frames may negatively affect expression of the respective gene (shown for a RPL16–lacZ fusion containing Leu3 binding sites and ACC1 containing Gal4 binding sites; 43,44). However, the regulatory significance of such interactions is unknown. While some downstream regulatory sites may also affect gene expression from an upstream position (cf. ref. 38), no such evidence could be obtained for the FAS2 DRS (data not shown).
Our present results do not yet precisely define the regulatory mechanism responsible for FAS autoregulation. In an attempt to map a domain specifically required for its regulatory function and to separate catalytic and regulatory properties of Fas1, we constructed truncation variants of the FAS1 reading frame. Using specific antibodies, Fas1 variants lacking 216 amino acids of the N-terminus or 336 amino acids of the C-terminus were clearly detectable in crude extracts prepared from the respective transformants. As expected, these truncated genes failed to complement a fas1 null mutation due to the lack of acetyl transferase and malonyl transferase catalytic domains, respectively. However, both truncation variants lacking distinct functional domains were also unable to stimulate FAS2–lacZ expression in a fas1 mutant background. Thus, these results do not yet allow us to map a regulatory domain within a defined region of Fas1.
Although Fas1 clearly influences the steady-state concentration of the FAS2 transcript (Fig. 1), biosynthesis of the mRNA or its stability may be affected. However, preliminary data (based on inhibition of de novo transcription in the presence of 1,10-phenanthroline) argue against an increased rate of decay of FAS2 mRNA in the absence of Fas1. Thus, we favor a mechanism based on DRS-affected biosynthesis of FAS2 mRNA, possibly mediated by a factor interacting with the DRS. In contrast with downstream binding sites described so far, sequence comparisons gave no evidence for an interaction of pleiotropic factors such as Rap1 or Med8 with the FAS2 DRS. Gel retardation assays with total protein extract from a fas1 deletion strain and a FAS1 multi-copy transformant did not reveal a different binding pattern to the FAS2 DRS (1/150 fragment used as a probe; not shown). We also attempted to relieve DRS-dependent repression of an FAS2–lacZ reporter gene by an unproductive increase of DRS copy number (multiple copies of FAS2 1/150 fragment on an episomal plasmid). However, competition of binding for a presumed negative factor by an excess of individual DRS elements did not stimulate FAS2–lacZ expression (data not shown). Thus, it remains to be shown whether the FAS2 DRS does in fact act at the DNA level. Alternatively, the DRS could function as a regulator of transcriptional elongation in the initiated FAS2 mRNA, similar to what has been shown for the tat-responsive element TAR in the HIV mRNA (45). Nuclear run-on assays may provide evidence for such a mechanism. In contrast with TAR, which is a positive element, the FAS2 DRS may reduce processivity of transcript elongation, possibly mediated by an unknown RNA-binding factor. An excess of Fas1 could increase processivity of elongation by deactivation of the repressor. Sequence modelling with nt 22–66 of the FAS2 reading frame, which represents the core of the DRS, allowed us to propose a potential stem–loop structure comprising nt 24–60, containing 10 conventional AU and GC base pairs together with 2 GU base pairs and a loop of 8 nt (Fig. 5B). With data supporting regulation of transcriptional elongation, the significance of this secondary structure may be investigated by RNA/protein interaction assays as well as by the introduction of mutations into the DRS that specifically interfere with the stem–loop but maintain its coding potential.
Acknowledgments
ACKNOWLEDGEMENTS
We thank E. Schweizer for stimulating discussions and valuable suggestions. D. Wolf and his group kindly provided strains and plasmids. We also thank B. Förtsch, S. Knab and I. Korakianitou for support in the initial phase of this work. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 473).
REFERENCES
- 1.Struhl K. (1989) Molecular mechanisms of transcriptional regulation in yeast. Annu. Rev. Biochem., 58, 1051–1077. [DOI] [PubMed] [Google Scholar]
- 2.Schweizer M., Roberts,L.M., Höltke,H.-J., Takabayashi,K., Höllerer,E., Hoffmann,B., Müller,G., Köttig,H. and Schweizer,E. (1986) The pentafunctional FAS1 gene of yeast: its nucleotide sequence and order of the catalytic domains. Mol. Gen. Genet., 203, 479–486. [DOI] [PubMed] [Google Scholar]
- 3.Schweizer E., Müller,G., Roberts,L.M., Schweizer,M., Rösch,J., Wiesner,P., Beck,J., Stratmann,D. and Zauner,I. (1987) Genetic control of fatty acid synthetase biosynthesis and structure in lower fungi. Fat Sci. Technol., 89, 570–577. [Google Scholar]
- 4.Chirala S.S., Kuziora,M.A., Spector,D.M. and Wakil,S.J. (1987) Complementation of mutations and nucleotide sequence of FAS1 gene encoding β subunit of yeast fatty acid synthase. J. Biol. Chem., 262, 4231–4240. [PubMed] [Google Scholar]
- 5.Mohamed A.H., Chirala,S.S., Mody,N.H., Huang,W.-Y. and Wakil,S.J. (1988) Primary structure of the multifunctional α subunit protein of yeast fatty acid synthase derived from FAS2 gene sequence. J. Biol. Chem., 263, 12315–12325. [PubMed] [Google Scholar]
- 6.Schweizer E. (1989) Biosynthesis of fatty acids and related compounds. In Ratledge,C. and Wilkinson,S.G. (eds), Microbial Lipids. Academic Press, London and New York, Vol. 2, pp. 3–50.
- 7.Schüller H.-J., Hahn,A., Tröster,F., Schütz,A. and Schweizer,E. (1992) Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J., 11, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopes J.M., Hirsch,J.P., Chorgo,P.A., Schulze,K.L. and Henry,S.A. (1991) Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipid precursors. Nucleic Acids Res., 19, 1687–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailis A.M., Lopes,J.M., Kohlwein,S.D. and Henry,S.A. (1992) Cis and trans regulatory elements required for regulation of the CHO1 gene of Saccharomyces cerevisiae. Nucleic Acids Res., 20, 1411–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasslacher M., Ivessa,A.S., Paltauf,F. and Kohlwein,S.D. (1993) Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J. Biol. Chem., 268, 10946–10952. [PubMed] [Google Scholar]
- 11.Hiesinger M., Wagner,C. and Schüller,H.-J. (1997) The acetyl-CoA synthetase gene ACS2 of the yeast Saccharomyces cerevisiae is coregulated with structural genes of fatty acid biosynthesis by the transcriptional activators Ino2p and Ino4p. FEBS Lett., 415, 16–20. [DOI] [PubMed] [Google Scholar]
- 12.Schüller H.-J., Richter,K., Hoffmann,B., Ebbert,R. and Schweizer,E. (1995) DNA binding site of the yeast heteromeric Ino2p/Ino4p basic helix-loop-helix transcription factor: structural requirements as defined by saturation mutagenesis. FEBS Lett., 370, 149–152. [DOI] [PubMed] [Google Scholar]
- 13.Lopes J.M. and Henry,S.A. (1991) Interaction of trans and cis regulatory elements in the INO1 promoter of Saccharomyces cerevisiae. Nucleic Acids Res., 19, 3987–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schüller H.-J., Schorr,R., Hoffmann,B. and Schweizer,E. (1992) Regulatory gene INO4 of yeast phospholipid biosynthesis is positively autoregulated and functions as a trans-activator of fatty acid synthase genes FAS1 and FAS2 from Saccharomyces cerevisiae. Nucleic Acids Res., 20, 5955–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambroziak J. and Henry,S.A. (1994) INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem., 269, 15344–15349. [PubMed] [Google Scholar]
- 16.Schwank S., Ebbert,R., Rautenstrauss,K., Schweizer,E. and Schüller,H.-J. (1995) Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res., 23, 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White M.J., Hirsch,J.P. and Henry,S.A. (1991) The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J. Biol. Chem., 266, 863–872. [PubMed] [Google Scholar]
- 18.Wagner C., Blank,M., Strohmann,B. and Schüller,H.-J. (1999) Overproduction of the Opi1 repressor inhibits transcriptional activation of structural genes required for phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Yeast, 15, 843–854. [DOI] [PubMed] [Google Scholar]
- 19.Wagner C., Dietz,M., Wittmann,J., Albrecht,A. and Schüller,H.-J. (2001) The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol., 41, 155–166. [DOI] [PubMed] [Google Scholar]
- 20.Schüller H.-J., Schütz,A., Knab,S., Hoffmann,B. and Schweizer,E. (1994) Importance of general regulatory factors Rap1p, Abf1p and Reb1p for the activation of yeast fatty acid synthase genes FAS1 and FAS2. Eur. J. Biochem., 225, 213–222. [DOI] [PubMed] [Google Scholar]
- 21.Santangelo G.M. and Tornow,J. (1990) Efficient transcription of the glycolytic gene ADH1 and three translational component genes requires the GCR1 product, which can act through TUF/GRF/RAP binding sites. Mol. Cell. Biol., 10, 859–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vignais M.L., Woudt,L.P., Wassenaar,G.M., Mager,W.H., Sentenac,A. and Planta,R.J. (1987) Specific binding of TUF factor to upstream activation sites of yeast ribosomal protein genes. EMBO J., 6, 1451–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egner R., Thumm,M., Straub,M., Simeon,A., Schüller,H.-J. and Wolf,D.H. (1993) Tracing intracellular proteolytic pathways: Proteolysis of fatty acid synthase and other cytoplasmic proteins in the yeast Saccharomyces cerevisiae. J. Biol. Chem., 268, 27269–27276. [PubMed] [Google Scholar]
- 24.Schüller H.-J., Förtsch,B., Rautenstrauss,B., Wolf,D.H. and Schweizer,E. (1992) Differential proteolytic sensitivity of yeast fatty acid synthetase subunits α and β contributing to a balanced ratio of both fatty acid synthetase components. Eur. J. Biochem., 203, 607–614. [DOI] [PubMed] [Google Scholar]
- 25.Fischer M., Hilt,W., Richter-Ruoff,B., Gonen,H., Ciechanover,A. and Wolf,D.H. (1994) The 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett., 355, 69–75. [DOI] [PubMed] [Google Scholar]
- 26.Gietz R.D. and Sugino,A. (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed]
- 27.Hill J.E., Myers,A.M., Koerner,T.J. and Tzagoloff,A. (1986) Yeast/E.coli shuttle vectors with multiple unique restriction sites. Yeast, 2, 163–167. [DOI] [PubMed] [Google Scholar]
- 28.Mumberg D., Müller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soni R., Carmichael,J.P. and Murray,J.A.H. (1993) Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr. Genet., 24, 455–459. [DOI] [PubMed] [Google Scholar]
- 30.Lynen F. (1973) Fatty acid synthetase in yeast. Methods Enzymol., 14, 17–33. [Google Scholar]
- 31.Werkmeister K., Johnston,R.B. and Schweizer,E. (1981) Complementation in vitro between purified mutant fatty acid synthetase complexes of yeast. Eur. J. Biochem., 116, 303–399. [DOI] [PubMed] [Google Scholar]
- 32.Mellor J., Dobson,M.J., Kingsman,A.J. and Kingsman,S.M. (1987) A transcriptional activator is located in the coding region of the yeast PGK gene. Nucleic Acids Res., 15, 6243–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purvis I.J., Loughlin,L., Bettany,A.J. and Brown,A.J. (1987) Translation and stability of an Escherichia coliβ-galactosidase mRNA expressed under the control of pyruvate kinase sequences in Saccharomyces cerevisiae. Nucleic Acids Res., 15, 7963–7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fantino E., Marguet,D. and Lauquin,G.J. (1992) Downstream activating sequence within the coding region of a yeast gene: specific binding in vitro of RAP1 protein. Mol. Gen. Genet., 236, 65–75. [DOI] [PubMed] [Google Scholar]
- 35.Fulton A.M., Rathjen,P.D., Kingsman,S.M. and Kingsman,A.J. (1988) Upstream and downstream transcriptional control signals in the yeast retrotransposon, TY. Nucleic Acids Res., 16, 5439–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farabaugh P., Liao,X.B., Belcourt,M., Zhao,H., Kapakos,J. and Clare,J. (1989) Enhancer and silencerlike sites within the transcribed portion of a Ty2 transposable element of Saccharomyces cerevisiae. Mol. Cell. Biol., 9, 4824–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farabaugh P.J., Vimaladithan,A., Turkel,S., Johnson,R. and Zhao,H. (1993) Three downstream sites repress transcription of a Ty2 retrotransposon in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinclair D.A., Kornfeld,G.D. and Dawes,I.W. (1994) Yeast intragenic transcriptional control: activation and repression sites within the coding region of the Saccharomyces cerevisiae LPD1 gene. Mol. Cell. Biol., 14, 214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrero P., Ramirez,M., Martinez-Campa,C. and Moreno,F. (1996) Identification and characterisation of two transcriptional repressor elements within the coding sequence of the Saccharomyces cerevisiae HXK2 gene. Nucleic Acids Res., 24, 1822–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarokin L. and Carlson,M. (1986) Short repeated elements in the upstream regulatory region of the SUC2 gene of Saccharomyces cerevisiae. Mol. Cell. Biol., 6, 2324–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaves R.S., Herrero,P. and Moreno,F. (1999) Med8, a subunit of the mediator CTD complex of RNA polymerase II, directly binds to regulatory elements of SUC2 and HXK2 genes. Biochem. Biophys. Res. Commun., 254, 345–350. [DOI] [PubMed] [Google Scholar]
- 42.Moreno-Herrero F., Herrero,P., Colchero,J., Baro,A.M. and Moreno,F. (1999) Analysis by atomic force microscopy of Med8 binding to cis-acting regulatory elements of the SUC2 and HXK2 genes of Saccharomyces cerevisiae. FEBS Lett., 459, 427–432. [DOI] [PubMed] [Google Scholar]
- 43.Kirkpatrick C.R. and Schimmel,P. (1995) Detection of leucine-independent DNA site occupancy of the yeast Leu3p transcriptional activator in vivo. Mol. Cell. Biol., 15, 4021–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q. and Johnston,S.A. (2001) Are all DNA binding and transcription regulation by an activator physiologically relevant? Mol. Cell. Biol., 21, 2467–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marciniak R.A. and Sharp,P.A. (1991) HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J., 10, 4189–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]