Abstract
The small nucleolar RNA (snoRNA) is a type of small non-coding RNA widely distributed in the nucleoli of eukaryotic cells, promoting cancer development. The aim of this study was to assess box C/D snoRNA 89 (SNORD89) dysregulations in endometrial cancer. According to the TCGA database as well as the International Federation of Gynecology and Obstetrics (FIGO), higher SNORD89 expression is found in endometrial cancer tissues. In addition, the SNORD89 expression level was higher in endometrial carcinoma with lymph node metastasis than in endometrial carcinoma without lymph node metastasis. By interacting with the conservative chaperone protein methylase fibrillarin (Fbl), SNORD89 inhibits the translation process of the Bim gene, leading to a decrease in Bim protein. Cancer-promoting effect of SNORD89 can be reversed by Fbl knockdown or Bim overexpressing. What’s more, ASO-mediated silencing of SNORD89 could inhibit endometrial cancer cell proliferation and migration ability. Taken together, SNORD89 can modify Bim through 2′-O-methylation and affect downstream signaling pathways to promote endometrial cancer occurrence and development. The role of methylation modification in the prevention and treatment of endometrial cancer provides a new understanding and SNORD89 may be a new diagnostic and therapeutic target for endometrial cancer.
Subject terms: Endometrial cancer, Tumour biomarkers
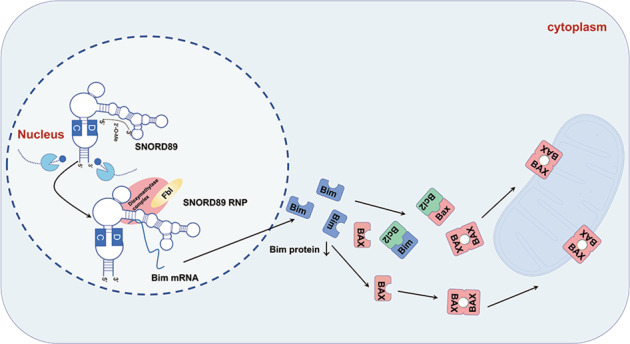
Mechanism of action of SNORD89
Introduction
Endometrial cancer is the most common gynecological malignant tumor in developed countries. Its morbidity and mortality also continue to soar [1]. The 5-year relative survival rate of early endometrial cancer is high (96%). However, late diagnosis reduces the 5-year relative survival rate dramatically (18%) [2]. Endometrial cancers with high malignancy and poor prognosis are characterized by the loss of tumor suppressor genes or amplification of multiple oncogenes. Therefore, an in-depth investigation of the molecular mechanism of the occurrence and development of endometrial cancer is essential for the early detection, diagnosis, and treatment of endometrial cancer. The small nucleolar RNA (snoRNA) is a non-coding RNA (ncRNA) [3, 4]. At present, ncRNA is generally believed to potentially regulate various cellular processes, including chromosome remodeling, transcription, translation, and signal transduction [5].
The abnormal expression and dysfunction of ncRNA are also involved in the occurrence and development of multiple malignant tumors, such as liver cancer [6], endometrial carcinoma [7], and colorectal cancer [8]. snoRNA is a type of small ncRNA widely distributed in the nucleus of eukaryotic cells, with a length of 60–300 nt. It can bind to four nucleolar ribonucleoproteins (Fbl/Nop1p, Nop56, Nop58/Nop5p, and P15.5 KD/Snu13p) to form a snoRNP complex and participate in ribosomal RNA (rRNA) processing, RNA shearing, and translation to process regulation and oxidative stress response [9]. Several recent studies have reported that snoRNAs are closely related to cancer occurrence and development. For example, snoRNA42 overexpression is related to the invasive biological behavior of non-small cell lung cancer (NSCLC), and its expression level is inversely proportional to the overall survival rate of patients, suggesting that snoRNA42 may become a new prognostic marker and potential therapeutic target of NSCLC [10]. In addition, snoRNA73A, snoRNA73B, and snoRNA74A can activate poly [ADP-ribose] polymerase (PARP-1) to regulate downstream signaling pathways and participate in breast cancer development by combining with the DNA repair enzyme PARP-1 in the absence of DNA damage stimulation [11]. Moreover, SNORD126 activates the PI3K–AKT pathway in liver and colorectal cancer by combining with hnRNPK to promote tumor cell proliferation in vitro and tumor formation in xenograft models in vivo [12]. Ravo et al. conducted high-throughput genetic analysis on paired normal, hyperplastic, and cancerous endometrial tissues obtained by endometrial biopsy. They found increased SNORD116 and SNORD2 expression, while SNORD3 was decreased in cancerous endometrial tissues [13]. This suggests that snoRNA is closely related to the occurrence and development of endometrial cancer. However, the specific mechanism of action and possible signaling pathways of snoRNA in endometrial cancer has not yet been reported.
SNORD89 belongs to the Box C/D snoRNAs in snoRNA, located at 2q11.2, also known as HBII-289. It is characterized by highly conserved boxes, called C (typical motif RUGAUGA, where R is purine) and D (typical motif CUGA). Box C/D snoRNAs could combine with Fbl/Nop1p, Nop56, Nop58/Nop5p, and P15.5 KD/Snu13p to form a snoRNP complex and 2′-O-methylate modification of rRNA, transfer RNA (tRNA), and other RNAs, among which Fbl is the key 2′-O-methylase of Box C/D snoRNA [14]. Besides, studies have shown that some snoRNAs can be processed into miRNA precursors, forming snoRNA-like microRNAs (sno-miRNAs) similar to snoRNA that plays a miRNA-like role [15]. miRNA-like roles are the same functions played by miRNAs and can silence the expression of target genes. AGO2 is the core element of the RNA-induced silencing complex. It not only promotes the degradation of the target mRNA and inhibits its protein translation in the miRNA/siRNA pathway, but also regulates the biosynthesis and maturation of miRNA, playing a key role in the regulatory pathway [16–18].
According to the function, members of the Bcl-2 family can be divided into three groups: two groups of antiapoptotic members and one group of proapoptotic members. Bcl-2 members mostly contain Bcl-2 tumor domains (BH domains), ranging from BH1 to BH4. Bim belongs to the only BH3 proapoptotic group of the Bcl-2 family, promoting many tumors and cell disorders, such as lung cancer [19], breast cancer [20, 21], osteosarcoma [22], prostatic cancer [23], and melanoma [24]. Various chemotherapeutic drugs contain Bim as an intermediary executor of cell death [25, 26]. Bim is located in the cytoplasm and can induce apoptosis by directly activating the proapoptotic Bax/Bak or by neutralizing the antiapoptotic Bcl-2 protein (Bcl-2 and Bcl-XL) when activated by foreign signals. When activated directly or indirectly by Bim, Bax will be inserted into the mitochondrial outer membrane (MOM) and oligomerized with Bax/Bak into the pore, enhancing the permeability of MOM and promoting the release of mitochondrial interstitial membrane proteins such as Cytc into the cytoplasm. After Cytc enters the cytoplasm, it can bind to apoptotic protease activator 1 (Apaf-1) to activate Apaf-1 and its oligomer into a sevenfold caspase activation complex, known as Apaf-1 apoptotic body. Apaf-1 can successively recruit and activate caspase-9 and caspase-3 to participate in the hydrolysis of various proteins and finally induce cell apoptosis [27]. In addition to directly activating Bax and promoting apoptosis, Bim can exert an antiapoptotic effect by antagonizing the antiapoptotic protein Bcl-2, which can not only bind Bax directly to inhibit Bax activation but also bind the Apaf-1 apoptotic body to inhibit the apoptosis process.
In this study, to explore the possible molecular mechanism of SNORD89 in endometrial cancer we performed in vitro and in vivo experiments.
Results
SNORD89 expression is increased in endometrial carcinoma
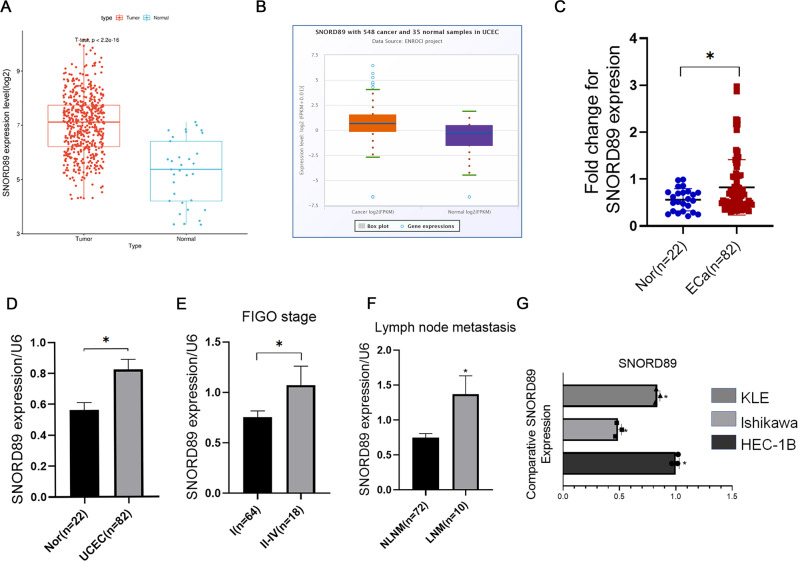
In the TCGA database and starbase database analysis, the expression of SNORD89 in endometrial cancer tissues was higher than that in normal endometrial tissues (Fig. 1A, B, *P < 0.05). qRT-PCR detected significantly increased expression of SNORD89 in endometrial cancer tissues compared with its expression in normal endometrial tissues (Fig. 1C, D, *P < 0.05). In the analysis of the relationship between SNORD89 expression level and clinicopathological features of endometrial cancer, the SNORD89 expression level was higher in the International Federation of Gynecology and Obstetrics (FIGO) stage II-IV diseases than in FIGO stage I diseases (Fig. 1E, *P < 0.05). In addition, the SNORD89 expression level was higher in endometrial carcinoma with lymph node metastasis than in endometrial carcinoma without lymph node metastasis (Fig. 1F, *P < 0.05). qRT-PCR analyses revealed that SNORD89 expression was the lowest in Ishikawa cells, but the highest in HEC-1B cells (Fig. 1G, *P < 0.05). Therefore, Ishikawa cells were used for transfection of SNORD89 overexpression plasmid, and HEC-1B cells were used for SNORD89 ASO transfection. qRT-PCR was used to confirm the transfection efficiency of plasmid- or ASO-transfected cells (Fig. 2A, E, *P < 0.05).
Fig. 1. Gene expression levels.
A Data from TCGA database; B Data from starBase; C, D SNORD89 in tumoral (n = 82) and nontumoral (n = 22) endometrial tissue; E SNORD89 in stage I (n = 64) and stage II-IV (n = 18); F SNORD89 in tissues with lymph node metastasis, negative (n = 72), positive (n = 10). G SNORD89 in EC cell lines (n = 3). Values are presented as the mean ± SD. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
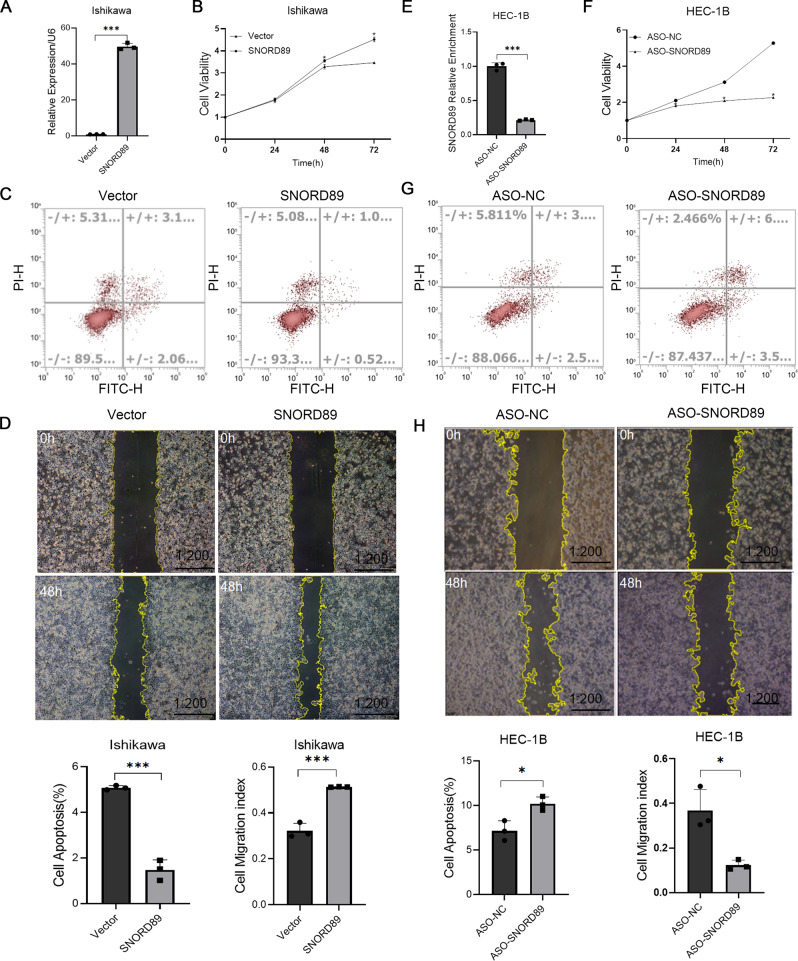
Fig. 2. Effect of SNORD89 on EC cells in vitro.
A Transfection efficiency after overexpression of SNORD89 (n = 3); B SNORD89 overexpression induced cell viability (n = 3) and D cell migration (n = 3), C while reduced cell apoptosis (n = 3). E Transfection efficiency after downregulation of SNORD89 (n = 3); SNORD89 downregulation reduced F cell viability (n = 3), H cell migration (n = 3), while increased G cell apoptosis (n = 3). Values are presented as the mean ± SD. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
SNORD89 overexpression promoted the proliferation and migration of endometrial cancer cells and inhibited apoptosis
Ishikawa cells were transfected with plasmids-overexpressing SNORD89. To evaluate the role of SNORD89 in the proliferation, apoptosis, and migration of endometrial cancer cells, cell count, apoptosis, and wound-healing experiments were performed. The results revealed that SNORD89 overexpression could promote cell proliferation (Fig. 2B, *P < 0.05), reduce apoptosis (Fig. 2C, *P < 0.05), and induce cell migration (Fig. 2D, *P < 0.05).
The knockdown of SNORD89 inhibited the proliferation and migration of endometrial cancer cells and promoted cell apoptosis
We transfected HEC-1B cells with ASO SNORD89 and knocked down the expression level of SNORD89 for the cell function experiment. As shown in Fig. 2F–H, SNORD89 knockdown can inhibit cell proliferation and migration and promote cell apoptosis compared with the negative control group.
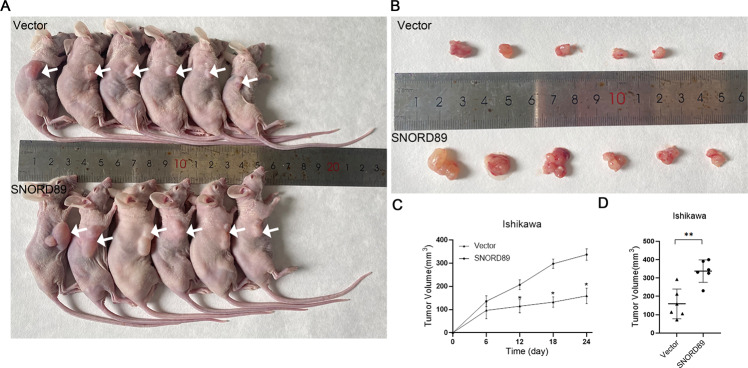
In vivo effect of SNORD89 overexpression
Following the experiments detailed in the Materials and methods section, a subcutaneous tumor formation experiment was performed in nude mice to detect the in vivo effect of SNORD89. The results showed that compared with the control group, the tumor formation rate was faster, and the tumor volume was larger in mice injected with SNORD89 Ishikawa-overexpressed cells (Fig. 3A–D, *P < 0.05).
Fig. 3. Effect of SNORD89 overexpression on xenograft tumors in nude mice.
A–C Tumorigenicity was greater in mice injected with the SNORD89-overexpressing cells than in vector; D The tumor volume was greater (n = 6). Values are presented as the mean ± SD. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
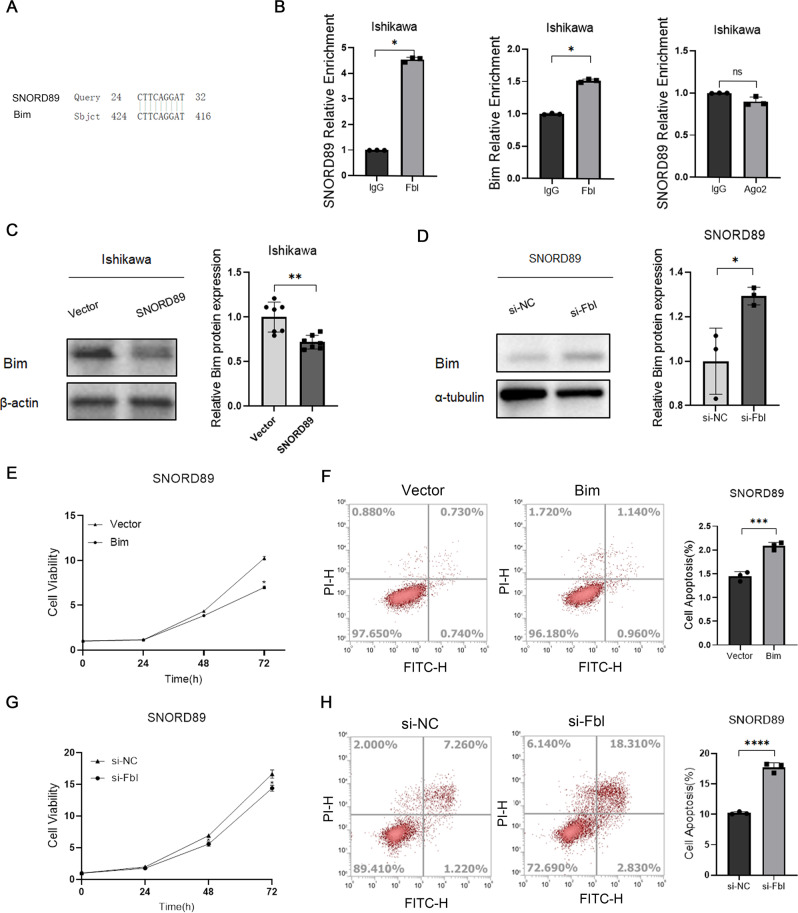
SNORD89 downregulates Bim gene expression via Fbl
A possible binding site between the tumor suppressor gene Bim and SNORD89 was found through the gene complementary pairing with SNORD89 sequence base by bioinformatics prediction (NCBI database) (Fig. 4A). RIP experiments confirmed that Bim could combine with SNORD89, and the expression level of Bim protein decreased after SNORD89 was overexpressed in Ishikawa cells (Fig. 4B, C, *P < 0.05). Meanwhile, we found that overexpression of Bim in SNORD89-overexpressed Ishikawa cells reverses the cell survival effect of SNORD89. (Fig. 4E, F, *P < 0.05). Therefore, it was hypothesized that SNORD89 might play an oncogenic role in endometrial cancer by regulating Bim. SNORD89 performs 2′-O-methylation modification mainly by binding to RNA-methylated proteins such as Fibrillarin (Fbl) or competitive endogenous RNA by binding to proteins such as Argonaute (AGO2). RIP experimental results showed that SNORD89 could not bind AGO2 but could bind the Fbl protein (Fig. 4B, *P < 0.05). To further verify that SNORD89 regulates Bim expression through Fbl, we knocked down Fbl in Ishikawa cells with overexpressed SNORD89, Fbl knockdown reversed the antiapoptotic effect of SNORD89 (Fig. 4G, H, *P < 0.05), suggesting that in combination with Fbl SNORD89 suppresses Bim gene expression.
Fig. 4. The relationship between SNORD89, Bim, and Fbl.
A There are pairing points between SNORD89 and Bim; B Fbl can bind Bim mRNA and SNORD89, but SNORD89 cannot bind Ago2 (n = 3). C Overexpression of SNORD89 in Ishikawa cells can reduce the protein level of Bim (n = 6); D Downregulation of Fbl can increase the overexpression of Bim protein in Ishikawa cells (n = 3); E–H downregulation of Fbl or overexpression of Bim can reverse the oncogenic effect of SNORD89 (n = 3); Values are presented as the mean ± SD. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
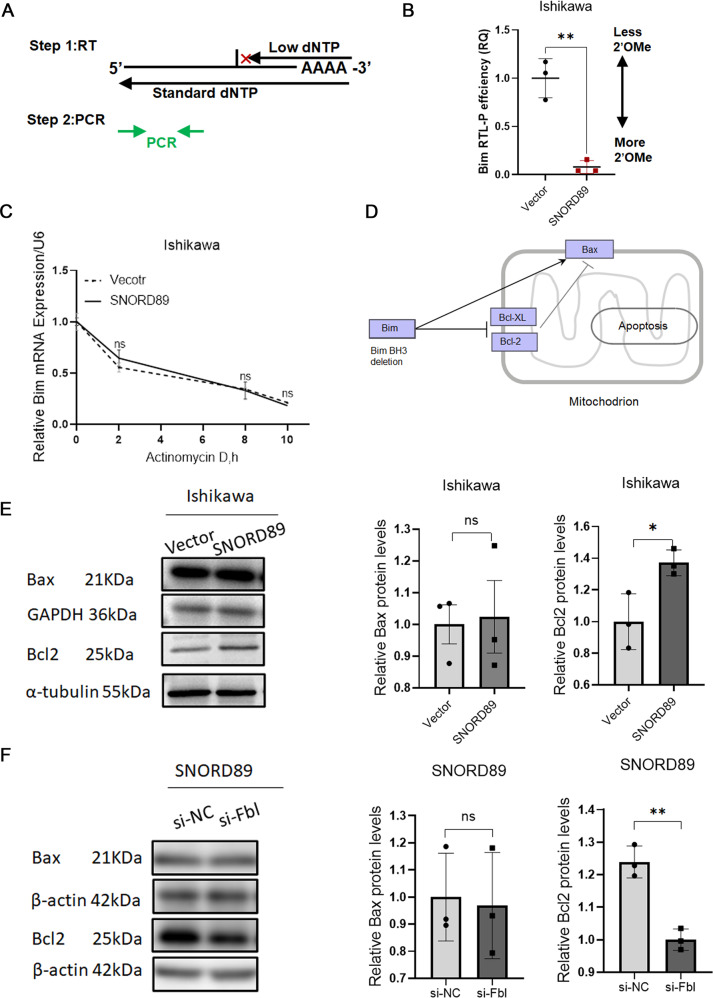
The relationship between SNORD89 and Bim 2′-O-methylation was detected by the RTL-P experiment
In order to verify whether SNORD89 combined with FBL regulates Bim expression by affecting the 2′-O-methylation modification level, we used the RTL-P experiment for detection, and the specific experimental method was obtained from existing literature. Figure 5A illustrates the principle of the RTL-P experiment, in which reverse transcriptase immediately stops cDNA synthesis before encountering 2′-O-methylated nucleotides at low concentrations of dNTPs. RTL-P experimental results showed that the Bim products of SNORD89 overexpressed were significantly reduced at low concentration, suggesting higher 2′-O-methylation activity and more 2′-O-methylation sites (Fig. 5B, *P < 0.05).
Fig. 5. Changes of Bim mRNA and downstream pathways.
A Illustration of the RTL-P method. The amount of qPCR product from the low dNTP reactions of two different samples can be compared to establish the relative “RTL-P efficiency (RQ)”. RQ relative quantity. See Methods for detail. B Under the low concentration of dNTPs, the Bim product was significantly reduced after overexpression of SNORD89 and showed higher RTL-P efficiency. C Bim mRNA stability had no significant change compared with the vector group (n = 3). D Bim/Bcl-2/Bax was an important signaling pathway that mediated apoptosis. E Overexpression of SNORD89 increased Bcl2 protein level and Bcl2 / Bax ratio (n = 3). F In Ishikawa cells overexpressing SNORD89, silencing Fbl reduced Bcl2 protein and Bcl2/Bax ratio (n = 3). Values are presented as the mean ± SD. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
Relationship between SNORD89 overexpression and Bim mRNA stability
We performed RNA stability tests in SNORD89-overexpressed Ishikawa cells, and the results confirmed that SNORD89 overexpression did not affect the stability of Bim mRNA (Fig. 5C, *P < 0.05). The above results indicated that SNORD89 combined with Fbl increased the level of 2′-O-methylation modification of Bim mRNA, affecting the complementary base pairing of Bim mRNA, thus reducing Bim protein.
Changes in protein levels in endometrial cancer cell lines after SNORD89 overexpression
The above results indicated that SNORD89 regulated Bim 2′-O-methylation modification by binding Fbl to form a snoRNP complex, thus reducing the Bim protein level. Through the KEGG database (Fig. 5D), we found that Bim/Bcl-2/Bax was an important signaling pathway that mediated apoptosis. Western blotting showed that after SNORD89 overexpression, the Bcl-2 protein level and Bcl-2/Bax ratio increased (Fig. 5E, *P < 0.05). The results of the rescue experiment revealed that Bcl-2 protein level and Bcl-2/Bax ratio were decreased after Fbl silencing in Ishikawa cells with overexpressed SNORD89 (Fig. 5F, *P < 0.05). The above results indicated that SNORD89 was involved in the occurrence and development of endometrial cancer by combining Fbl to form a snoRNP complex and regulating the Bim/Bcl-2/Bax signaling pathway.
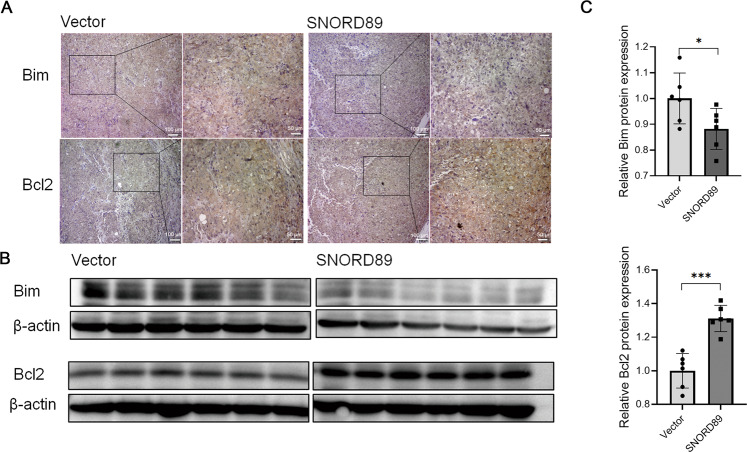
SNORD89 overexpression can decrease Bim protein level and increase Bcl-2 protein level in vivo
Immunohistochemical analysis of tumor tissues collected in vivo from subcutaneous tumorigenesis experiment in nude mice showed decreased expression of the Bim protein and increased expression of the Bcl-2 protein in tumor tissues generated by Ishikawa cells with overexpressed SNORD89 (Fig. 6A). By extracting tumor tissue proteins and conducting Western blotting experiments, the Bcl-2 protein increased and Bim protein levels decreased in tumor tissues with overexpressed SNORD89 (Fig. 6B, C, *P < 0.05). The above results further verified that SNORD89 was involved in the occurrence and development of endometrial cancer by regulating the Bim/Bcl-2/Bax signaling pathway.
Fig. 6. Effect of SNORD89 overexpression on xenograft tumors in nude mice.
A–C SNORD89 overexpression can reduce the expression of Bim protein and increase the expression of Bcl2 protein in vivo (n = 6). All results are representative of at least three independent experiments. Values are presented as the mean ± SD. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
Discussion
To search for diagnostic and therapeutic markers of endometrial cancer, we screened various snoRNAs from the TCGA database and found a significantly increased expression of SNORD89 in endometrial cancer tissues compared with normal endometrial tissues. We further examined the expression of SNORD89 in our clinical samples, the result of which was still the case. We speculated that it was related to the FIGO stage and lymph node metastasis and that SNORD89 might be involved in the occurrence and progression of endometrial cancer and affect the prognosis.
To evaluate the function of SNORD89 in endometrial cancer cells, we performed in vitro cell experiments and in vivo subcutaneous tumorigenesis experiments in nude mice. The transfection of SNORD89-overexpressing plasmids into Ishikawa cells with low SNORD89 expression promoted cell proliferation, inhibited cell apoptosis, and induced cell migration. However, the cell experiment results were reversed after the expression level of SNORD89 was knocked down in HEC-1B cells. In vivo subcutaneous tumorigenesis experiments in nude mice also confirmed that SNORD89 overexpression can induce tumor growth in vivo. These results indicated that SNORD89 had an oncogenic effect in endometrial cancer. However, the specific mechanism of action of SNORD89 in endometrial cancer remained unclear.
We proved through the RIP experiment that SNORD89 does not combine with AGO2, but with Fbl. Fbl silencing inhibited the proliferation of ovarian cancer cells with stable overexpression of SNORD89. Therefore, we believe that SNORD89 may promote the development and progression of endometrial cancer by influencing the 2′-O-methylation modification of target genes.
BLAST search for downstream genes complementary to the SNORD89 sequence revealed a binding point might exist between SNORD89 and Bim. The RIP experiment confirmed that Fbl could bind with Bim and that SNORD89 overexpression significantly reduced the expression of Bim protein. Therefore, we believe that SNORD89 may be involved in the progression of endometrial cancer by regulating Bim. In order to verify that SNORD89 plays its carcinogenic role through 2′-O-methylation modification of Bim mRNA, we designed corresponding primers and detected whether there was 2′-O-methylation modification at SNORD89 and Bim pairing site by the RTL-P method. The results confirmed that Bim methylation modification was enhanced after overexpression of SNORD89.
2′-O-methylation modification refers to the addition of methyl at the 2′ positions of nucleic acid ribose so that the 2′-OH group is replaced by 2′-O-methyl [28]. This modification is mainly mediated by snoRNAs and has been confirmed to be involved in various cancers. For example, long ncRNA ZFAS1 promotes snoRNA-mediated 2′-O-methylation through Nop58 recruitment and is involved in the development and prognosis of colorectal cancer [29]. Methyltransferase HENMT1 inhibits apoptosis of lung cancer cells by enhancing its stability by 2′-O-methylation of the 3′-terminal of Mir-21-5p [30]. Several studies have demonstrated that 2′-O-methylation modification was mainly located in rRNA [31]. Recently, some studies have shown that snoRNA could perform 2′-O-methylation modification on the mRNA of the target gene, and these modification sites could regulate mRNA and protein expression of the target gene [32].
2′-O-methylation modification can protect mRNA from hydrolysis, thus increasing the mRNA stability. However, this modification limits RNA chain conformation and suppleness, interferes with the ribosome monitoring base on the space and the homologous codon-anticodon spiral interaction, causes the excessiveness of homologous aminoacylation tRNAs in the initial selection and calibration to refuse, and inhibits the expression of target genes. Further, through RNA stability and Western blotting experiments, we confirmed that SNORD89 do not alter the stability of Bim mRNA, but reduces the expression level of Bim protein. This suggests that SNORD89 may promote the occurrence and development of endometrial cancer by binding Fbl to form a snoRNP complex 2′-O-methylation to modify Bim mRNA and reduce the expression level of Bim protein.
Bim is a member of the Bcl-2 family. Bcl-2 family proteins constitute key intracellular checkpoints and regulators of cell death and can inhibit or promote apoptosis. Irregularities in the members of the Bcl-2 family are found in various cancers and diseases, including breast cancer [33] and prostate cancer [34]. Some studies have suggested that the intracellular Bcl-2/Bax protein ratio is the key to cell survival [35]. We found that the Bcl-2/Bax protein ratio increased significantly after SNORD89 overexpression. Suggesting that SNORD89 may affect downstream signaling pathways through Bim. Meanwhile, the rescue experiment showed that Bim protein increased after Fbl knockdown in the Ishikawa cell line stably transfected with overexpressed SNORD89 plasmid. These results suggest that Fbl protein plays an important role in SNORD89’s regulation of Bim. Therefore, we believe that SNORD89 may modify Bim mRNA through 2′-O-methylation, reduce the expression level of Bim protein, increase the ratio of Bcl2 to Bax, and inhibit the activation of Bax, the release of mitochondrial Cytc, and subsequent apoptosis.
Finally, immunohistochemical analysis of tumor tissues collected from subcutaneous tumorigenesis experiment in nude mice confirmed that SNORD89 overexpression in vivo could decrease the expression of Bim protein and increase the expression of Bcl-2 protein in endometrial cancer cells. By extracting tumor tissue proteins and Western blotting experiments, the Bcl-2 protein were increased, and the levels of Bim proteins were decreased in tumor tissues overexpressing SNORD89. SNORD89 can be further verified by influencing Bim and its mediated Bcl-2/Bax signaling axis to participate in the occurrence and development of endometrial cancer.
Conclusions
SNORD89 can modify Bim through 2′-O-methylation and affect downstream signaling pathways to promote endometrial cancer occurrence and development. It enables snoRNA-mediated RNA 2′-O-methylation. The role of methylation modification in the prevention and treatment of endometrial cancer provides a new understanding and SNORD89 may be a new diagnostic and therapeutic target for endometrial cancer.
Materials and methods
Bioinformatics analysis
The TCGA database (https://portal.gdc.cancer.gov/) and starbase database (http://starbase.sysu.edu.cn/) are used to analyze the expression level of SNORD89 in normal endometrial tissues and endometrial cancer tissues. The NCBI database (https://www.ncbi.nlm.nih.gov/) was used to predict downstream target genes with complementary base pairs in SNORD89. The KEGG database (https://www.kegg.jp/) is used to query Bim mediated signal pathway.
Endometrial cancer specimens
We collected 22 cases of normal endometrial tissues and 82 cases of endometrial cancer tissues from patients who underwent gynecological surgery in the Third Affiliated Hospital of Guangzhou Medical University. None of the patients received chemotherapy or radiotherapy before the operation. These tumor specimens were confirmed by two pathologists independently, then immediately frozen in liquid nitrogen and preserved at −80 °C until use. Informed consent was obtained from the subjects. The ethics committee of the Third Affiliated Hospital of Guangzhou Medical University approved the research (NO. 2020066), and all specimens were treated and anonymous according to ethical and legal standards.
Cell culture and transformation
SNORD89 expressions were screened from three human endometrial cancer cells (KLE, Ishikawa, and HEC-1B). Four human endometrial cancer cell lines were purchased from Jennio Biotech and cultured on Dulbecco’s Modified Eagle Medium (HEC-1B), Ham’s F-12 treatment medium (KLE), and RPMI 1640 medium (Ishikawa). All three media were supplemented with penicillin/streptomycin (100 U/mL) and 10% fetal bovine serum (FBS). All cell lines were recently STR identified and tested for mycoplasma contamination. Cells were cultured in an incubator with a constant temperature of 37 °C and 5% CO2. Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was used for plasmids, amido-bridged nucleic acid-flanked antisense oligonucleotides (ASOs), and siRNA transfection. An SNORD89-overexpressing plasmid was employed to upregulate SNORD89 expression (ACTGAGGAATGATGACAAGAAAAGGCCGAATTGCAGTGTCTCCATCAGCAGTTTGCTCTCCATGGGCACACGATGACAAAATATCCTGAAGCGAACCACTAGTCTGACCTCAGT). A Bcl-2 interacting mediator of cell death (Bim)-overexpressing plasmid was adopted to upregulate Bim expression (ATGGCAAAGCAACCTTCTGATGTAAGTTCTGAGTGTGACCGAGAAGGTAGACAATTGCAGCCTGCGGAGAGGCCTCCCCAGCTCAGACCTGGGGCCCCTACCTCCCTACAGACAGAGCCACAAGGTAATCCTGAAGGCAATCACGGAGGTGAAGGGGACAGCTGCCCCCACGGCAGCCCTCAGGGCCCGCTGGCCCCACCTGCCAGCCCTGGCCCTTTTGCTACCAGATCCCCGCTTTTCATCTTTATGAGAAGATCCTCCCTGCTGTCTCGATCCTCCAGTGGGTATTTCTCTTTTGACACAGACAGGAGCCCAGCACCCATGAGTTGTGACAAATCAACACAAACCCCAAGTCCTCCTTGCCAGGCCTTCAACCACTATCTCAGTGCAATGGCTTCCATGAGGCAGGCTGAACCTGCAGATATGCGCCCAGAGATATGGATCGCCCAAGAGTTGCGGCGTATTGGAGACGAGTTTAACGCTTACTATGCAAGGAGGTTAGAGAAATAG), and an SNORD89 ASO (CATGGGCACACGATGACAAA, Ruibo, Jiangsu, China) was applied to knockdown SNORD89 expression. The siRNA sequence for Fbl (si-Fbl) knockdown was GGGCTAAGGTTCTCTACCT.
Cell proliferation was detected by cell counting kit 8 (CCK8)
Cells were inoculated at 3000/well in 96-well plates, and transient transfection was performed after cell adherence. At specific time points (0, 24, 48, and 72 h), 10 μL of Cell Counting Kit 8 (CCK8, Yeasen, Shanghai, China) was added to each medium and cultured at 37 °C for 2 h. Then the absorbance was read at 450 nm. The experiment was repeated three times.
Wound-healing assay
In this experiment, 6 × 105 cells were seeded in 6-well plates. A vertical wound was marked in the Petri dish with the tip of a 200-μL pipette. Cells were cleaned three times with phosphate-buffered saline (PBS), and a 2-mL serum-free medium was added to the Petri dish. Wound healing was recorded at 0, 24, and 48 h. Image J software (National Institutes of Health, Bethesda, MD, USA) was used to determine the wound size and calculate the cell migration rate.
Flow cytometry analysis of apoptosis
Apoptosis was detected by Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) double staining according to the manufacturer’s protocol. Cells 3 × 105/well were inoculated in six-well plates and cultured to a cell density of 60–70% for transient transfection. At 48 h after transfection, the cells were washed with cold PBS twice, digested, suspended with trypsin without ethylenediaminetetraacetic acid (1 × 106 cells/mL), and cleansed with cold PBS three times. Cells were then cultured in 100 µL of binding buffer containing 5 µL of Annexin V-FITC and 5 µL of PI for 15 min in 4 °C darkness, with 400 µL of the binding buffer added to each tube. The cells were quantitatively analyzed by flow cytometry.
Real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
An RNA Quick Purification kit was used to extract the total RNA from cells and tissues, and a Fast Reverse Transcription kit was used to generate endogenous complementary DNA. The cDNA was used as a template for qRT-PCR using the SYBR Prime X Ex-TAQ Patent II Suite (Takara Bio Inc., Shiga, Japan). Finally, the relative expression of the gene was calculated by comparing the period threshold (Ct) value of the target gene with the control gene (U6, encoding glyceraldehyde-3-phosphate dehydrogenase or 18sRNA gene) according to the 2−ΔCt method.
RNA-binding protein immunoprecipitation (RIP) detection
TRIzol reagent (1 mL; Takamura, Shibuya, Japan) was used to extract the total RNA from cells or tissues. Then, 200 μL of chloroform was added to the extract, fully mixed, rested for 5 min, and centrifuged at 12,000 rpm for 20 min. The upper clarifying liquid was transferred to a new tube, and the RNA was precipitated with the same amounts of isopropyl alcohol. After centrifugation at 12,000 rpm for 20 min, the precipitate was retained and cleaned with 75% ethanol. Once fully dried at room temperature, sodium diethylpyrocarbonate (DEPC) water was used to dissolve the residue. The total RNA concentration was determined by spectrophotometry (Thermo Fisher Scientific, Shanghai, China). Then, using reverse transcriptase and random primers, 2 μg of RNA was reverse-transcribed into complementary DNA (cDNA) according to the manufacturer’s protocol (Yeasen). The cDNA was used as a template for qRT-PCR using the SYBR Prime X Ex-TAQ Patent II Suite (Takara Bio Inc., Shiga, Japan). Finally, the relative expression of the gene was calculated by comparing the period threshold (Ct) value of the target gene with the control gene (U6, encoding glyceraldehyde-3-phosphate dehydrogenase or 18sRNA gene) according to the 2−ΔCt method.
RNA stability test
After transfection with overexpressed SNORD89 plasmid, Ishikawa cells were treated with 5 μg/ml actinomycin D (ActD, CAS# A4262, Sigma). The total RNA was extracted from cells at 0, 2, 8, and 10 h after adding actinomycin D. qRT-PCR for analysis [36].
Western blotting
Bim antibody (22037-1-AP, diluted 1:500; Proteintech, IL, USA), Bcl-2 (12789-1-AP, diluted at 1:1000; Proteintech), Bax (50599-2-LG, diluted at 1:5000; Proteintech), α-tubulin (11224-1-AP, diluted at 1:2000; Proteintech), and endometrial cancer cells were washed with PBS, homogenized in lysis buffer with RIPA containing protease inhibitors, and centrifuged at 12,000 rpm at 4 °C for 20 min. Protein concentration was determined by a BCA Protein Concentration Determination Kit (No. P0009; Beyotime Biotech Inc.). Protein lysates were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to Hybond membranes under appropriate conditions (Amersham, Munich, Germany). They were sealed at room temperature with Tris-buffered saline with Tween (TBST) containing 3% bovine serum albumin for 2 h. After sealing, primary antibodies Bim, Bcl-2, Bax, and imprinting were incubated overnight at 4 °C. On day 2, the membrane was incubated with a secondary antibody (1:8,000, Proteintech) at room temperature for 2 h, and α-tubulin was selected as the internal reference. After incubation, cell membranes were washed with TBST three times, using a hypersensitive ECL Chemiluminescence Kit (NcmECL UItra; Sliding Biotech, Shanghai, China) for protein development. The film was scanned using a full-function gel imaging system. Western blotting was quantified using Image J software.
Subcutaneous tumorigenesis experiment in nude mice
The detection kit for subcutaneous tumor transmission was provided by Guangdong Medical Laboratory Animal Center (Guangdong, China) for BALB/C female nude mice, which were placed in a specific pathogen-free environment. The nude mice were randomly divided into groups according to the label. Ishikawa cells transfected with SNORD89/no-load plasmid (1 × 107 in 150 μL of FBS-free medium) were subcutaneously injected into 5-week-old female mice to establish a tumor-forming nude mice model. After tumor formation, the tumor volume was measured weekly with vernier calipers to understand tumor growth. When the tumor reached a certain size, the mice were euthanized, the tumor nodules were resected, and their volume was measured. No blinding was done.
Immunohistochemistry
The tumor tissue was collected by treating it with xylene and 100% ethanol and then decreasing the ethanol concentration. After antigen retrieval, endogenous peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 40 min, then the slides were washed three times with PBS, and incubated in 5% bovine serum albumin for 40 min, stained with anti-BIM antibody (1:100, Proteintech, USA) and BCL-2 antibody (1:100, Proteintech, USA), followed by enzyme-conjugated secondary antibody (Dako) incubation, washed in PBS for 5 min, developed using DAB reagent. Hematoxylin is used for redyeing and images taken with light microscopes are gained.
Reverse transcription at low dNTPs followed by PCR (RTL-P)
To detect the 2′-O-methylation (Nm) of SNORD89, RT-PCR was performed [32, 37]. The total reaction system was 20 μl. Containing 5 μg total RNA, a low (1 μM) or high (1 mM) concentration dNTPs (TaKaRa) and 1 μl specific RT primers (PF) were denatured at 65 °C for 5 min and then placed on ice. According to the instruction, 4 μl M-MLV RT 5×buffer (PROMEGA), 1 μl 200 u/μl M-MLV Reverse Transcriptase (PROMEGA), 1 μl 0.1 M DTT (PROMEGA), and 1 μl 40 u/μl RNase inhibitor (PROMEGA) were mixed at 50 °C for 1 h, and then heated at 70 °C for 15 min. cDNA is used as a template for QRT-PCR using the SYBR Prime X Ex-TAQ Patent II Suite (Takara). Finally, the relative expression of the gene was calculated by comparing the period threshold (Ct) of the target gene in the experimental group and the control group at low concentrations according to the 2−ΔCt method.
Ethical statement
The animal study was reviewed and approved by Guangdong Provincial Medical Experimental Animal Center (Approval no. S2021-101), with a license to use experimental animals [SYXK(Guangdong)2018–0002]. Written informed consent was obtained from the owners for the participation of their animals in this study.
Data analysis
GraphPad Prism (version 8.02(263)) was used for graphics and data analysis. Relevant data were expressed as mean ± standard mean error. Statistical analysis was performed using two-sample t-test and P < 0.05 was considered to indicate a significant difference.
Supplementary information
Acknowledgements
This work was supported by the National Nature Science Foundation of China (Grant No. 82072854 to YZ and 81872115 to SC); Guangzhou Medical Key Disciplines (2021–2023); and the Young S&T Talent Training Program of Guangdong Provincial Association for S&T(GDSTA), China (SKXRC202214).
Author contributions
SC and YZ conceived the study and participated in the study design. H-jB performed cell culture, RTL-P, qPCR, western blots, cell cycle, and flow cytometry apoptosis assay. XC and XL performed primer designs. Q-hL and J-yX analyzed the data. H-jB and WW organized Figures and wrote the manuscript. All authors read and approved the final manuscript.
Data availability
The data generated or analyzed during this study are included in this published article and its supplementary information files, Supplementary information is available at Cell Death Discovery’s website.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The Ethics Committee of Guangzhou Medical University approved the use of human tissue and experimental animals in this study (NO. 2020066). Informed consent was obtained from all patients.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41420-022-01102-5.
References
- 1.Makker V, Taylor MH, Aghajanian C, Oaknin A, Mier J, Cohn AL, et al. Lenvatinib plus pembrolizumab in patients with advanced endometrial cancer. J Clin Oncol. 2020;38:2981–92. doi: 10.1200/JCO.19.02627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdalla E, Troy R, Fall S, Elhussin I, Egiebor-Aiwan O, Nganwa D. Racial differences in 5-year relative survival rates of cervical cancer by stage at diagnosis, between African American (black) and white women, living in the state of Alabama, USA. BMC Cancer. 2020;20:830. doi: 10.1186/s12885-020-07338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Zhu S, Meng N, He Y, Lu R, Yan GR. ncRNA-encoded peptides or proteins and cancer. Mol Ther. 2019;27:1718–25. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu S, Wang J, He Y, Meng N, Yan GR. Peptides/proteins encoded by non-coding RNA: a novel resource bank for drug targets and biomarkers. Front Pharm. 2018;9:1295. doi: 10.3389/fphar.2018.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang XJ, Qi GT, Zhang XM, Wang L, Li FF. lncRNA RHPN1-AS1 promotes the progression of endometrial cancer through the activation of ERK/MAPK pathway. J Obstet Gynaecol Res. 2021;47:533–43. doi: 10.1111/jog.14548. [DOI] [PubMed] [Google Scholar]
- 7.Zhao D, Ren C, Yao Y, Wang Q, Li F, Li Y, et al. Identifying prognostic biomarkers in endometrial carcinoma based on ceRNA network. J Cell Biochem. 2020;121:2437–46. doi: 10.1002/jcb.29466. [DOI] [PubMed] [Google Scholar]
- 8.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–20. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 10.Mannoor K, Shen J, Liao J, Liu Z, Jiang F. Small nucleolar RNA signatures of lung tumor-initiating cells. Mol Cancer. 2014;13:104. doi: 10.1186/1476-4598-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DS, Camacho CV, Nagari A, Malladi VS, Challa S, Kraus WL. Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol Cell. 2019;75:1270–1285. doi: 10.1016/j.molcel.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barros-Silva D, Klavert J, Jenster G, Jerónimo C, Lafontaine DLJ, Martens-Uzunova ES. The role of oncoSnoRNAs and ribosomal RNA 2’-O-methylation in cancer. RNA Biol. 2021;18:61–74. doi: 10.1080/15476286.2021.1991167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravo M, Cordella A, Rinaldi A, Bruno G, Alexandrova E, Saggese P, et al. Small non-coding RNA deregulation in endometrial carcinogenesis. Oncotarget. 2015;6:4677–91. doi: 10.18632/oncotarget.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massenet S, Bertrand E, Verheggen C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017;14:680–92. doi: 10.1080/15476286.2016.1243646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–28. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–51. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Cui L, Nakano K, Obchoei S, Setoguchi K, Matsumoto M, Yamamoto T, et al. Small nucleolar noncoding RNA SNORA23, up-regulated in human pancreatic ductal adenocarcinoma, regulates expression of spectrin repeat-containing nuclear envelope 2 to promote growth and metastasis of xenograft tumors in mice. Gastroenterology. 2017;153:292–306. doi: 10.1053/j.gastro.2017.03.050. [DOI] [PubMed] [Google Scholar]
- 18.Monaco PL, Marcel V, Diaz JJ & Catez F. 2’-O-methylation of ribosomal RNA: towards an epitranscriptomic control of translation? Biomolecules 2018;8:106. [DOI] [PMC free article] [PubMed]
- 19.Tanaka K, Yu HA, Yang S, Han S, Selcuklu SD, Kim K, et al. Targeting Aurora B kinase prevents and overcomes resistance to EGFR inhibitors in lung cancer by enhancing BIM- and PUMA-mediated apoptosis. Cancer Cell. 2021;39:1245–1261. doi: 10.1016/j.ccell.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byerly JH, Port ER, Irie HY. PRKCQ inhibition enhances chemosensitivity of triple-negative breast cancer by regulating Bim. Breast Cancer Res. 2020;22:72. doi: 10.1186/s13058-020-01302-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Wang X, Zhu T, Zhang N, Wang L, Huang T, et al. Resistance to bortezomib in breast cancer cells that downregulate Bim through FOXA1 O-GlcNAcylation. J Cell Physiol. 2019;234:17527–37. doi: 10.1002/jcp.28376. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Xiao XQ, Qian YH, Zhou ZY. The CtBP1-p300-FOXO3a transcriptional complex represses the expression of the apoptotic regulators Bax and Bim in human osteosarcoma cells. J Cell Physiol. 2019;234:22365–77. doi: 10.1002/jcp.28802. [DOI] [PubMed] [Google Scholar]
- 23.Sur S, Steele R, Shi X, Ray RB. miRNA-29b inhibits prostate tumor growth and induces apoptosis by increasing Bim expression. Cells 2019;8:1455. [DOI] [PMC free article] [PubMed]
- 24.Dai DL, Wang Y, Liu M, Martinka M, Li G. Bim expression is reduced in human cutaneous melanomas. J Invest Dermatol. 2008;128:403–7. doi: 10.1038/sj.jid.5700989. [DOI] [PubMed] [Google Scholar]
- 25.Sun WL, He LY, Liang L, Liu SY, Luo J, Lv ML, et al. Ambra1 regulates apoptosis and chemosensitivity in breast cancer cells through the Akt-FoxO1-Bim pathway. Apoptosis 2022;27:329–41. [DOI] [PubMed]
- 26.Akiyama T, Dass CR, Choong PF. Bim-targeted cancer therapy: a link between drug action and underlying molecular changes. Mol Cancer Ther. 2009;8:3173–80. doi: 10.1158/1535-7163.MCT-09-0685. [DOI] [PubMed] [Google Scholar]
- 27.Shukla S, Saxena S, Singh BK, Kakkar P. BH3-only protein BIM: an emerging target in chemotherapy. Eur J Cell Biol. 2017;96:728–38. doi: 10.1016/j.ejcb.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Dimitrova DG, Teysset L, Carré C. RNA 2’-O-methylation (Nm) modification in human diseases. Genes 2019;10:117. [DOI] [PMC free article] [PubMed]
- 29.Wu H, Qin W, Lu S, Wang X, Zhang J, Sun T, et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2’-O-methylation via NOP58 recruitment in colorectal cancer. Mol Cancer. 2020;19:95. doi: 10.1186/s12943-020-01201-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang H, Jiao Z, Rong W, Qu S, Liao Z, Sun X, et al. 3’-terminal 2’-O-methylation of lung cancer miR-21-5p enhances its stability and association with Argonaute 2. Nucleic Acids Res. 2020;48:7027–40. doi: 10.1093/nar/gkaa504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xing YH, Chen LL. Processing and roles of snoRNA-ended long noncoding RNAs. Crit Rev Biochem Mol Biol. 2018;53:596–606. doi: 10.1080/10409238.2018.1508411. [DOI] [PubMed] [Google Scholar]
- 32.Elliott BA, Ho HT, Ranganathan SV, Vangaveti S, Ilkayeva O, Abou Assi H, et al. Modification of messenger RNA by 2’-O-methylation regulates gene expression in vivo. Nat Commun. 2019;10:3401. doi: 10.1038/s41467-019-11375-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Jiang H, Chen Y, Ren F. Resveratrol chemosensitizes adriamycin-resistant breast cancer cells by modulating miR-122-5p. J Cell Biochem. 2019;120:16283–92. doi: 10.1002/jcb.28910. [DOI] [PubMed] [Google Scholar]
- 34.Verdoodt B, Neid M, Vogt M, Kuhn V, Liffers ST, Palisaar RJ, et al. MicroRNA-205, a novel regulator of the anti-apoptotic protein Bcl2, is downregulated in prostate cancer. Int J Oncol. 2013;43:307–14. doi: 10.3892/ijo.2013.1915. [DOI] [PubMed] [Google Scholar]
- 35.Hashem KS, Elkelawy A, Abd-Allah S, Helmy NA. Involvement of Mfn2, Bcl2/Bax signaling and mitochondrial viability in the potential protective effect of Royal jelly against mitochondria-mediated ovarian apoptosis by cisplatin in rats. Iran J Basic Med Sci. 2020;23:515–26. doi: 10.22038/ijbms.2020.40401.9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CY, Ezzeddine N, Shyu AB. Messenger RNA half-life measurements in mammalian cells. Methods Enzymol. 2008;448:335–57. doi: 10.1016/S0076-6879(08)02617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong ZW, Shao P, Diao LT, Zhou H, Yu CH, Qu LH. RTL-P: a sensitive approach for detecting sites of 2’-O-methylation in RNA molecules. Nucleic Acids Res. 2012;40:e157. doi: 10.1093/nar/gks698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated or analyzed during this study are included in this published article and its supplementary information files, Supplementary information is available at Cell Death Discovery’s website.