Abstract
Proteins that catalyse homologous recombination have been identified in all living organisms and are essential for the repair of damaged DNA as well as for the generation of genetic diversity. In bacteria homologous recombination is performed by the RecA protein, whereas in the eukarya a related protein called Rad51 is required to catalyse recombination and repair. More recently, archaeal homologues of RecA/Rad51 (RadA) have been identified and isolated. In this work we have cloned and purified the RadA protein from the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus and characterised its in vitro activities. We show that (i) RadA protein forms ring structures in solution and binds single- but not double-stranded DNA to form nucleoprotein filaments, (ii) RadA is a single-stranded DNA-dependent ATPase at elevated temperatures, and (iii) RadA catalyses efficient D-loop formation and strand exchange at temperatures of 60–70°C. Finally, we have used electron microscopy to visualise RadA-mediated joint molecules, the intermediates of homologous recombination. Intriguingly, RadA shares properties of both the bacterial RecA and eukaryotic Rad51 recombinases.
INTRODUCTION
Homologous recombination is the fundamental process by which all living organisms repair damage to their DNA, restart stalled replication forks and generate genetic diversity in their progeny. The mechanism of homologous recombination involves bringing two DNA molecules together, searching for homology in the sequences and then exchanging the DNA strands (1,2). In prokaryotes the RecA protein is responsible for catalysing homologous recombination and recombinational DNA repair (3–5). Homologues of RecA (named Rad51) have been identified in eukaryotes and have been shown to utilise similar mechanisms of recombination (1,2,6).
The archaea constitute the third primary kingdom of living organisms and are evolutionarily distinct from the bacteria and the eukarya (7,8). RecA/Rad51 homologues (named RadA) from various archaea have recently been identified (9–13). These archaeal recombinase proteins appear to perform the same essential roles in the archaea as in other organisms since deletion of the radA gene in Haloferax volcanni leads to UV hypersensitivity, the absence of detectable recombination and slow growth (14).
The archaeal RadA proteins share ∼40% amino acid identity with the eukaryotic Rad51 proteins but only ∼20% identity with bacterial RecA (9). Furthermore, the eukaryotic Rad51 proteins and the archaeal RadA proteins share an extended N-terminal domain that is outside the RecA core domain and is likely to be involved in species-specific interactions with other proteins (13,15). Intriguingly, both RadA and Rad51 proteins lack the C-terminal domain of RecA which is thought to inhibit binding of double-stranded DNA (dsDNA) by RecA (16–19).
Many archaeal proteins have sequences that are similar to their eukaryotic counterparts and it seems that the eukarya share a more common ancestor with the archaea than with the bacteria (8,20,21). Therefore, the archaea may provide useful model systems for the complex eukaryotic machinery of recombination and replication. For example, the archaea possess homologues of the eukaryotic single-stranded DNA (ssDNA)-binding protein RPA (22–24) and the DNA double-strand break processing protein Mre11 (23,25). Although the archaeal genomes sequenced thus far have not revealed homologues of the Rad52, Rad54 or Rad59 repair proteins (23,25), a protein named RadB has been identified that has some homology to RadA (33%) with which it interacts (13). RadB also has some similarity to the Rad55/57 proteins from yeast (11), which stimulate Rad51-mediated recombination (26,27). It has also been demonstrated that the RadB protein from a thermophilic archaeon can complement the UV sensitivity of a recA deletion in Escherichia coli suggesting a possible role in homologous recombination for RadB (28). Furthermore, the RadB protein from Pyrococcus furiosis has been shown to interact with Hjc protein, an archaeal Holliday junction resolvase (13).
Although transcription of the recA gene is induced by agents that damage DNA, such as UV irradiation or methyl methane sulfonate (29), UV irradiation does not increase the transcription of radA genes (9,13). Since many of the archaeons are hyperthermophiles and flourish in extreme environments (70–100°C and hundreds of atmospheres pressure) it is possible that constitutive expression of DNA repair proteins is essential for the maintenance of genome stability in the presence of continual DNA damage. The thermophilic RadA proteins have evolved and adapted to exist and function at high temperatures. The RadA protein from Pyrobaculum islandicum remains structurally stable at 87°C and in highly denaturing conditions (4.5 M guanidine hydrochloride) (30). The thermoresistance of RadA proteins is due to key amino acid substitutions (e.g. serine to threonine), which increase the hydrophobicity of individual α-helices leading to an overall increase in the thermal stability of the protein (31).
The recent biochemical characterisation of various archaeal RadA proteins has confirmed that these proteins exhibit many characteristics of homologous recombination proteins (10,13,32). It has been demonstrated that RadA from Sulfolobus solfataricus has DNA-dependent ATPase activity, forms nucleoprotein filaments on DNA and catalyses a weak strand-exchange activity (10). Similarly, RadA from the hyperthermophilic archaeon Desulfurococcus amylolyticus catalyses ATP hydrolysis and strand exchange at temperatures of 80–95°C, the optimal temperature for growth (32).
Given the remarkable properties of these hyperthermophilic recombination proteins we were interested in examining an archaeal RadA protein in further detail. We report here that (i) RadA protein from Archaeoglobus fulgidus forms ring structures in solution and filaments upon binding ssDNA, (ii) RadA does not appear to bind or form filaments on dsDNA, and (iii) RadA catalyses DNA-stimulated ATP hydrolysis, D-loop formation and DNA strand exchange. The optimal reaction temperature is 60–70°C. We have also used electron microscopy to visualise joint molecules formed by RadA, the intermediates of homologous recombination. Interestingly, we find that the RadA protein catalyses D-loop formation and strand exchange reactions with high efficiency, similar to the bacterial RecA protein, but that the overall reaction kinetics are slow, and more akin to those shown by eukaryotic Rad51 proteins.
MATERIALS AND METHODS
Cloning and overexpression of RadA
RadA protein was expressed in 1 l cultures of Luria broth containing 100 µg/ml ampicillin and 34 µg/ml chloramphenicol, each inoculated with a 5 ml culture of E.coli B834(DE3) ‘codon plus’ (Stratagene) containing the radA gene cloned into the NdeI and NotI sites of pET22b vector (Novagen). The cultures were grown whilst shaking at 37°C until the OD600 reached 0.5–0.6, and then RadA synthesis was induced by the addition of 1 mM isopropyl-β-d-thiogalactoside. Growth was continued for 3 h before the cells were harvested by centrifugation at 5000 g. The cell pellets were resuspended in 20 ml of 50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 mM dithiothreitol (DTT) and 10% sucrose, and frozen at –80°C until required.
RadA purification
For RadA purification, the cells were thawed and lysed by sonication (on ice) in the presence of 0.2 mg/ml phenylmethylsulphonyl fluoride. The supernatant was clarified by centrifugation at 20 000 g and then heated at 60°C for 5 min followed by cooling on ice. The precipitate was removed by centrifugation at 20 000 g. The cleared supernatant was diluted with 0.1 M K2HPO4 pH 6.8 and 1 mM DTT until the conductivity approached that of the same buffer. This supernatant was then applied to a 5 ml hydroxyapatite column (Bio-Rad) pre-equilibrated with 0.1 M K2HPO4 pH 6.8 and 1 mM DTT. The column was washed with 3 column vol of 0.1 M K2HPO4 pH 6.8 and 1 mM DTT, and then the RadA was eluted with a 50 ml linear gradient of 0.2–1.0 M K2HPO4 pH 6.8 and 1 mM DTT. The peak fractions were pooled and loaded onto a 5 ml heparin column (Pharmacia) pre-equilibrated with 50 mM Tris–HCl pH 7.5, 1 mM DTT and 1 mM EDTA (TDE buffer). The RadA was again diluted to a conductivity approaching that of the column buffer before loading onto the heparin column. The heparin column was then washed with 3 column vol of TDE, and RadA was eluted with a 100 ml linear gradient of 0–1.0 M NaCl in the same buffer. The peak fractions were pooled and dialysed against TDE and concentrated to 50 ml. The protein was applied to a MonoQ 10/10 column (Pharmacia) pre-equilibrated in TDE and washed with 3 column vol of buffer. RadA was eluted using a 10 column vol linear gradient of 0–1.0 M NaCl. Peak fractions containing RadA were pooled and treated with DNaseI and RNaseA in the presence of 2 mM MgCl2 at 37°C for 30 min. The RadA was diluted 3-fold with 1 M (NH4)2SO4, 100 mM Tris–HCl pH 7.0, 1 mM DTT and 1 mM EDTA, loaded onto an 8 ml octyl–Sepharose column (Pharmacia) and pre-equilibrated in the same buffer. The column was washed with 3 column vol of buffer and then the RadA was eluted by applying a 20 ml linear gradient of 1.0–0 M (NH4)2SO4. Prior to storage of the RadA protein at –80°C, glycerol was added to a final concentration of 10% (v/v). The purity of the RadA protein was monitored throughout on 12% SDS–PAGE. The yield of RadA was ∼20–40 mg from 1 l of culture.
Escherichia coli RecA protein was prepared by a modification of published procedures (33) and stored in 20 mM Tris–HCl pH 7.5, 0.1 mM EDTA, 10% (v/v) glycerol, 0.5 mM DTT. Concentrations of protein are expressed in moles of monomer. Restriction enzymes (New England Biolabs) were used as recommended by the suppliers.
DNA substrates
Supercoiled plasmid pPB4.3 DNA (34,35) was prepared using Qiagen Plasmid Mega Kits followed by equilibrium centrifugation on caesium chloride/ethidium bromide gradients. Linear DNA (400 bp) was prepared by treating pPB4.3 with HindIII and NdeI, and the 400 bp fragment was excised from an agarose gel followed by purification using QIAquick gel extraction columns (Qiagen). Linear fragments [1.2 kb (HindIII + BamHI), 3 kb (HindIII + BamHI) and 4.3 kb (HindIII only)] were prepared in a similar manner to the 400 bp fragment. Circular single-stranded pPB4.3 DNA was prepared as described previously (34). φX174 dsDNA was purchased from New England Biolabs. Nicked plasmid DNA for electron microscopy was prepared by treating 10 µg supercoiled plasmid φX174 DNA with 5 U of DNaseI in 10 mM Tris–HCl pH 7.9, 10 mM MgCl2, 50 mM NaCl and 1 mM DTT for 30 min at 37°C. The nicked DNA was then purified by phenol/chloroform extraction and ethanol precipitation followed by dialysis against Tris–HCl pH 7.5, 0.1 mM EDTA.
The oligonucleotide was prepared using standard procedures on an Applied Biosystems 394 DNA Synthesiser. The oligonucleotide used in this study was 5′-GGGCGAATTGGGCCCGACGTCGCATGCTCCTCTAGACTCGAGGAATTCGGT-ACCCCGGGTTCGAAATCGATAAGCTTACAGTCTCCATTTAAAGGACAAG. The oligonucleotide was gel-purified using denaturing PAGE as described previously (36). DNA was 5′-32P-end-labelled using polynucleotide kinase (USB) and [γ-32P]ATP. Unless stated otherwise, DNA concentrations are expressed in moles of nucleotides.
Electron microscopy
Binding reactions contained 2.5 µM RadA protein in 50 mM triethanolamine-acetate (TEA) pH 7.5, 1 mM MgCl2, 2 mM ATP (or ATP-γ-S) and 1 mM DTT. Reactions carried out in the presence of DNA contained 2.5 µM RadA and 5 µM ssDNA (circular single-stranded pPB4.3) or 5 µM dsDNA (nicked plasmid φX174) in the same buffer. Following incubation at 60°C for 10 min, the complexes were fixed by the addition of glutaraldehyde to 0.2% followed by 15 min of incubation at 37°C. Strand exchange reactions (for electron microscopy) were performed in 50 mM TEA pH 7.5, 2 mM MgCl2, 2 mM ATP and 1 mM DTT. Reactions (10 µl) contained single-stranded pPB4.3 DNA (10 µM) and 6 µM RadA in buffer. After 5 min at 55°C, 7 µM 3 kb linear DNA was added and incubation continued for 5 min followed by fixation with 0.2% glutaraldehyde for 15 min at 37°C. Samples were diluted and washed in 5 mM Mg(OAc)2 before uranyl acetate staining (37). Complexes were visualised at a magnification of 25 500× using a Philips CM100 electron microscope.
ATPase assay
Reactions (50 µl) contained 300 µM single- or double-stranded φX174 DNA, 5 µM RadA or RecA in 50 mM triethanolamine–HCl pH 7.5, 15 mM Mg(OAc)2, 0.2 mM ATP, supplemented with 50 nCi [γ-32P]ATP (3000 Ci/mmol), 1 mM DTT and 100 µg/ml BSA. Incubations were at 60°C for RadA and 37°C for RecA unless indicated otherwise. At the indicated times, aliquots (5 µl) were removed, stopped by addition of 2.5 µl 0.5 M EDTA, and analysed using CEL 300 PEI/UV254 (Polygram) thin layer chromatography plates developed in 1 M formic acid/0.5 M LiCl. The percentage of [γ-32P]ATP hydrolysed to [γ-32P]ADP was quantified with a Molecular Dynamics Storm 860 PhosphorImager. Reactions carried out at varying temperatures were incubated for 120 min.
D-loop assay
Standard reactions (10 µl) contained 5′-32P-end-labelled 100mer ssDNA (3 µM), and the indicated concentration of RadA or RecA in standard buffer (50 mM triethanolamine–HCl pH 7.5, 15 mM MgCl2, 2 mM ATP, 1 mM DTT, 100 µg/ml BSA, 20 mM creatine phosphate and 2 U/ml creatine phosphokinase). After 5 min at the indicated temperatures, excess supercoiled pPB4.3 DNA (0.3 mM) was added and incubation was continued for 10 min for RadA and 1 min for RecA. Reaction products were deproteinised by the addition of one-fifth vol of stop buffer (0.1 M Tris–HCl pH 7.5, 0.1 M MgCl2, 3% SDS and 10 mg/ml proteinase K) followed by 30 min incubation at 37°C. Labelled DNA products were analysed by electrophoresis through 0.8% agarose gels run in TEA buffer at 4 V/cm for 3 h, dried onto filter paper and visualised by autoradiography. Quantification was carried out using a Molecular Dynamics Storm 860 PhosphorImager.
For the time-course studies, reactions (80 µl) contained 100mer ssDNA (3 µM) and RadA or RecA (4 µM) in standard buffer. After 5 min at 60°C (RadA) or 37°C (RecA), supercoiled pPB4.3 DNA (0.3 mM) was added and incubation was continued. At the times indicated, aliquots (10 µl) were taken, deproteinised and analysed on agarose gels as described.
Strand exchange (gel assay)
Reactions (10 µl) contained single-stranded pPB4.3 DNA (15 µM) with the indicated amounts of RadA or RecA in standard buffer. After 5 min at 60°C (RadA) or 37°C (RecA), either 1.5 µM 400 bp linear DNA, 4.2 µM 1.2 kb linear DNA or 15 µM 4.3 kb linear DNA was added and incubation continued for 90 min. Reaction products were deproteinised by the addition of one-fifth vol of stop buffer (0.1 M Tris–HCl pH 7.5, 0.1 M MgCl2, 3% SDS, 5 µg/ml ethidium bromide and 10 mg/ml proteinase K) followed by 30 min incubation at 37°C. Labelled DNA products were analysed on agarose gels as before.
RESULTS AND DISCUSSION
RadA protein forms rings and nucleoprotein filaments
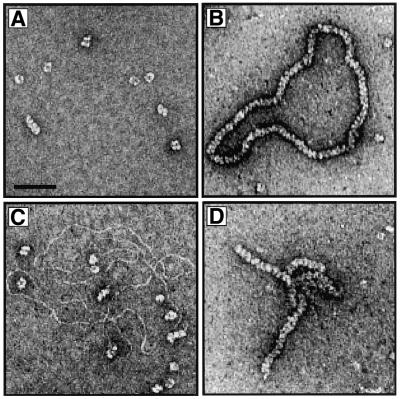
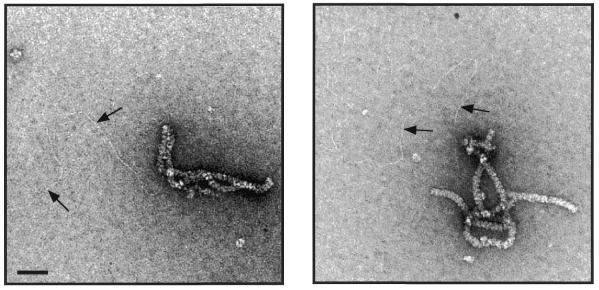
The RadA protein from A.fulgidus was cloned and purified to near homogeneity as described in Materials and Methods. A sample of purified RadA was analysed by SDS–PAGE and is shown in Figure 1. In previous studies it was demonstrated that RecA and Rad51 proteins can form nucleoprotein filaments with ssDNA or dsDNA (18,38–40). To determine whether purified RadA protein forms filaments upon binding DNA, we analysed RadA alone (Fig. 2A) or complexes formed between RadA and ssDNA (Fig. 2B) or dsDNA (Fig. 2C) using electron microscopy after negative staining with uranyl acetate.
Figure 1.
Purification of RadA protein. Purified RadA protein (1 µg) was analysed by 10% SDS–PAGE followed by Coomassie Brilliant Blue staining. Numbers correspond to the size of indicated proteins. The RadA protein is 37.2 kDa.
Figure 2.
Four images showing either RadA protein alone or RadA interacting with ssDNA or dsDNA. (A) Electron microscopic visualisation of RadA protein in the presence of ATP showing the formation of ring-like structures. The complexes were formed and analysed by electron microscopy as described in Materials and Methods. The scale bar represents 50 nm. (B) Image of complex formed between RadA and ssDNA in the presence of ATP-γ-S. (C) Image of RadA incubated with dsDNA in the presence of ATP. The results were identical in the presence of ATP-γ-S. (D) Image of the complex formed between RadA and ssDNA in the presence of ATP.
In the absence of DNA, RadA showed a striking tendency to form ring-like structures (Fig. 2A). Rings were seen in the presence of ATP or ATP-γ-S. The finding that RadA protein forms ring-like structures in vitro is interesting in relation to recent findings with other recombination proteins and enzymes involved in DNA recombination and repair (41). Similar ring-like structures have been observed with RecA (42,43), Rad51 (44,45), the meiosis-specific recombinase Dmc1 (46), Rad52 (47,48) and RuvB (49). Although ring structures present a large surface area for binding DNA and this may be the mechanism of action for proteins involved in ssDNA annealing such as Rad52 (50,51), it is not clear what function rings perform for proteins that catalyse strand invasion such as RecA, Rad51 or RadA. Ring structures may, in fact, be the inactive storage form of the recombinase that are transported to the sites of DNA repair and then converted into functional helical nucleoprotein filaments when loaded onto ssDNA.
In the presence of ssDNA and ATP-γ-S, helical nucleoprotein filament structures were observed (Fig. 2B). The RadA filaments appeared very similar to those formed by RecA, since they exhibited the characteristic striated appearance of RecA–DNA filaments (38,52). This extended and regular filament structure has also been observed with Rad51 protein (18,39).
The binding of ATP transforms the bacterial RecA protein to a DNA-binding state with a high affinity for ssDNA (1,53). In contrast to RecA protein’s preference for ssDNA, Rad51 protein associates equally well with ssDNA or dsDNA to form nucleoprotein filaments (18,39,40). Since both Rad51 and RadA proteins lack the C-terminal domain of RecA, which inhibits binding of dsDNA by RecA (16–19), we were eager to examine whether RadA also binds to ssDNA and dsDNA equally. Surprisingly, when RadA was incubated with dsDNA in the presence of ATP or ATP-γ-S, we observed only protein rings and naked DNA (Fig. 2C). To confirm RadA’s preference for binding ssDNA, band-shift experiments were performed with RadA and ssDNA or dsDNA in the presence of ATP, ATP-γ-S, various Mg2+ concentrations and at several temperatures. RadA formed filaments on ssDNA under all conditions (except in the absence of ATP/ATP-γ-S), but we did not observe protein–DNA complexes on dsDNA under any of the conditions examined (data not shown).
Such a preference for binding ssDNA may allow targeting of RadA protein to the ssDNA tails at the ends of processed double-strand breaks in vivo. If bacterial RecA and eukaryotic Rad51 are at opposite ends of the spectrum with regard to the binding to DNA, it appears that archaeal RadA may be more similar to RecA and perhaps shows an even greater affinity for ssDNA.
In the presence of ATP, we observed that nucleoprotein filaments formed on ssDNA were often aligned and intertwined (Fig. 2D). Since reaction conditions containing ATP/Mg2+ are closest to the physiological state and are most likely suitable for recombination as shown, intertwined nucleoprotein filaments may be indicative of ATP-dependent DNA alignment and homologue search, the initiating step of homologous recombination.
RadA protein is a ssDNA-dependent ATPase
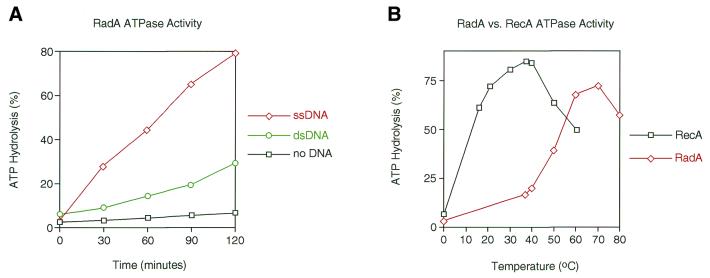
A fundamental property of the RecA and Rad51 proteins is their ability to hydrolyse ATP to ADP in a DNA-dependent manner (17,40,54,55). To examine the ability of purified RadA protein to hydrolyse ATP, we assayed its ATPase activity in the presence of ssDNA and dsDNA. RadA-dependent ATPase activity was found to be most efficient in the presence of ssDNA (Fig. 3A). The rate of ATP hydrolysis was considerably reduced in the presence of dsDNA and virtually no ATP was hydrolysed in the absence of DNA. The rate of RadA-mediated ATP hydrolysis was calculated to be in the order of 0.26–0.37 ATP/min in the presence of ssDNA. While this rate is ∼100-fold lower than that of RecA (kcat of 18–30 min–1; 56), it is similar to the rates reported for yeast Rad51 (kcat of 0.73 min–1; 57) and human Rad51 (kcat of 0.16 min–1; 55). ATP hydrolysis was not detected in the absence of magnesium (data not shown).
Figure 3.
The ATPase activity of RadA protein. (A) Graph showing the percentage of ATP hydrolysed to ADP by RadA in the presence of ssDNA, dsDNA or no DNA. (B) Graph showing the percentage of ATP hydrolysed to ADP by RadA or RecA incubated with ssDNA at a range of temperatures. ATPase assays were carried out as described in Materials and Methods.
To measure the thermal dependence of RadA, the ATPase activity of the protein was examined over a wide range of temperatures. RadA was found to exhibit the highest levels of ATP hydrolysis at temperatures of ∼60–70°C, whereas ATPase activity was dramatically reduced at temperatures of 37–40°C (Fig. 3B). In contrast, E.coli RecA protein showed optimal ATP hydrolysis at 37–40°C, the optimal growth temperature for E.coli (Fig. 3B). These studies indicate that the optimal reaction conditions of these proteins are similar to the normal growth conditions of the host organism. Other groups have also reported increased ATPase activity at high temperatures using RadA proteins from different hyperthermophiles (10,13,30,32). The RadA protein from D.amylolyticus is particularly unusual in that it displayed optimal ATPase activity at 95°C and hydrolysed very little ATP at 70°C (32).
RadA protein catalyses high temperature D-loop formation
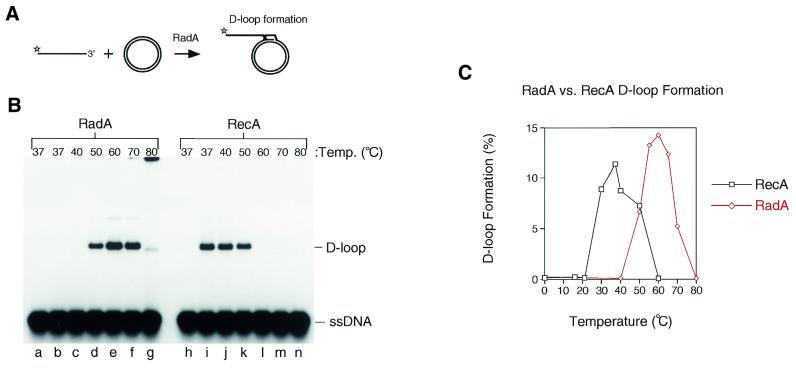
The definitive test for a homologue of RecA/Rad51 is the ability of the protein to catalyse ATP-dependent homologous pairing and strand exchange. To study A.fulgidus RadA catalysing homologous pairing and strand invasion, we used a D-loop assay. In this assay 5′-32P-labelled linear ssDNA (100mer) was incubated with varying concentrations of RadA at a range of temperatures, and then an excess of homologous supercoiled plasmid was added (Fig. 4A). After a short incubation period, the products were deproteinised and analysed by agarose gel electrophoresis. RecA protein was used as a control. RadA protein formed efficient D-loops at 50–70°C (Fig. 4B, lanes d–f). At 80°C the DNA products were found to form aggregates that failed to enter the gel due to the expected DNA denaturation that occurs at elevated temperatures (Fig. 4B, lane g). As expected, RecA protein formed D-loop complexes at temperatures from 37–50°C (Fig. 4B, lanes i–k). In both cases, D-loop formation required the presence of magnesium, ATP and homologous DNA substrates (data not shown).
Figure 4.
Strand invasion and D-loop formation by RadA and RecA. (A) The in vitro system for the initiation of RadA-mediated strand invasion. Linear ssDNA is bound by RadA to form nucleoprotein filaments. These filaments then search for homology and invade the homologous circular duplex DNA to form a D-loop structure. Stars indicate 5′-32P-labels. (B) Strand invasion reactions were carried out between 32P-end-labelled linear ssDNA (100mer) and homologous covalently closed circular pPB4.3 DNA using RadA (lanes b–g) or RecA (lanes i–n) at the indicated temperatures. Lanes a and h, no protein. (C) Graph showing the percentage of D-loop formation obtained with RadA and RecA at a range of temperatures using the in vitro system described in (A) and (B).
To further refine the optimal temperature for strand invasion by RadA, further experiments were carried out and the results were quantified and are shown graphically in Figure 4C. The optimal temperature for RadA-mediated strand invasion was found to be 60°C with a dramatic reduction in D-loop formation below 40°C and above 80°C. In contrast, the optimal reaction temperature for RecA was found to be 37°C with product levels dropping off below 20°C and above 60°C (Fig. 4C). It was noticeable in these experiments that the RadA protein formed D-loops as efficiently as RecA protein (12–15%; Fig. 4C). Using the same conditions and substrates we had previously observed that human Rad51 protein formed only ∼0.6% D-loops (58).
Time-course of RadA-mediated strand invasion
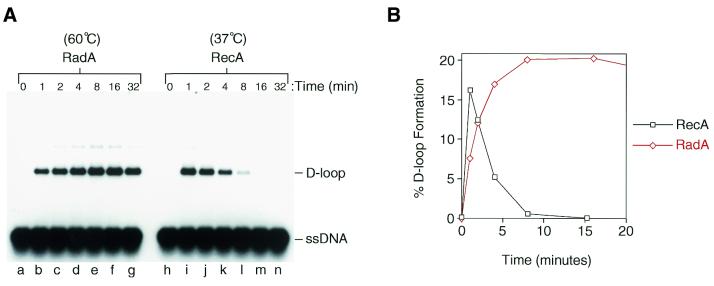
Strand invasion by RecA has been shown to be a rapid and reversible reaction in which the invading ssDNA is introduced into the homologous duplex DNA briefly (∼4 min) and is then removed in what is known as the ‘D-loop cycle’ (59). The human Rad51 protein also displays a D-loop cycle, but in this case the reaction is slower with a peak in D-loop formation seen at ∼16 min (58). To examine RadA-mediated strand invasion, time-course reactions were performed with both RadA and RecA (Fig. 5). Using this relatively short ssDNA substrate (100 nt), D-loop formation by RecA peaked after only 1 min (Fig. 5A, lane i) and by 16 min almost all complexes had dissociated back to the starting substrates (Fig. 5A, lane m). In contrast, RadA-mediated D-loop formation peaked at ∼8–16 min (Fig. 5A, lanes e and f), which is very similar to the kinetics displayed by the human Rad51 protein (58). Quantification of the data shown in Figure 5A revealed the dramatically different strand invasion profiles of the two proteins (Fig. 5B). These results indicate that A.fulgidus RadA protein is a bona fide hyperthermophilic recombinase that catalyses homologous pairing with an efficiency similar to that of RecA, but with reaction kinetics more resembling that of eukaryotic Rad51 protein.
Figure 5.
Time-course of strand invasion by RadA and RecA. (A) Strand invasion reactions were performed with RadA (lanes b–g) and RecA (lanes i–n), and samples removed and stopped at the times indicated as described in Materials and Methods. Lanes a and h, samples were taken prior to protein addition. (B) Graph showing the percentage of D-loop formation obtained at various times using the RadA and RecA reactions presented in (A).
RadA protein catalyses strand exchange
To measure the extent of strand exchange we used linear duplex DNA and homologous circular ssDNA (Fig. 6A) (60). Three linear substrates of varying lengths were prepared and 5′-32P-labelled: (i) a 400 bp linear duplex; (ii) a 1.2 kb linear duplex; and (iii) a full-length 4.3 kb linear duplex. Each substrate was incubated with 4.3 kb circular ssDNA and RadA in strand exchange buffer. In reactions between ssDNA and the 400 bp linear duplex, RadA protein formed products efficiently at 60°C. Unfortunately, the displaced 400 nt ssDNA was not observed by agarose gel electrophoresis since this product migrates at the same position as the 400 bp duplex substrate (Fig. 6B, lanes b–f). However, using the longer 1.2 kb substrate (Fig. 6B, lanes j-k), efficient RadA-mediated strand exchange was again observed and we were now able to visualise the displaced ssDNA (1.2 kb). These results demonstrate that A.fulgidus RadA is capable of promoting strand transfer through at least 1200 bp of duplex DNA.
Figure 6.
Strand exchange and joint molecule formation by RadA. (A) The in vitro system for RadA-mediated strand exchange. Single-stranded circular pPB4.3 is bound by RadA to form nucleoprotein filaments. These filaments then search for homology and invade the homologous linear duplex DNA to form a joint molecule. Complete strand transfer results in the release of 32P-end-labelled linear ssDNA. Stars indicate 5′-32P-labels. (B) Strand exchange reactions were carried out between single-stranded circular pPB4.3 and 32P-end-labelled 400 bp linear duplex DNA (lanes a–f) or 32P-end-labelled 1.2 kb linear duplex DNA (lanes g–l) using the indicated amounts of RadA. Lanes a and g, no protein. (C) Strand exchange reactions were carried out between single-stranded circular pPB4.3 and 32P-end-labelled 4.3 kb linear duplex DNA using the indicated amounts of RadA (lanes a–f) or RecA (lane g). Lane a, no protein.
To test the ability of RadA to catalyse complete strand transfer using longer DNA substrates, similar reactions were carried out at 60°C using the 4.3 kb linear duplex (Fig. 6C). As a control, RecA reactions were performed at 37°C. We found that RadA catalysed efficient joint molecule formation, but no displaced ssDNA products were observed. In these reactions, a significant amount of DNA was trapped at the origin (Fig. 6C, lanes e and f) indicative of the formation of DNA networks. The RecA control reaction also had a similar amount of material trapped at the origin, but some displaced linear ssDNA was observed (Fig. 6C, lane g). However, this result was expected in view of the fact that the RecA reactions were carried out in the absence of the ssDNA-binding protein, which helps to stabilise the released ssDNA and prevent network formation (61). Extending the time of deproteinisation did not remove the material from the origin, although heating the samples at 60°C before loading the gels did release the retarded material suggesting it comprised of DNA networks (data not shown). From these experiments, we conclude that RadA catalyses efficient strand exchange reactions, but that the extent of strand transfer may be limited. Alternatively, RadA may be able to catalyse full-length strand exchange but the reactions require the presence of the archaeal RPA protein to stabilise the displaced ssDNA and prevent network formation.
The recent identification of archaeal homologues of the ssDNA-binding protein RPA may allow for more extensive strand exchange reactions to be performed (22–24). Indeed, the purification and characterisation of the recently discovered RadB proteins and Mre11 homologues in archaea will assist in the reconstitution of more complete systems of homologous recombination in these bizarre and fascinating organisms (13,23,25).
Visualisation of RadA-mediated joint molecule formation
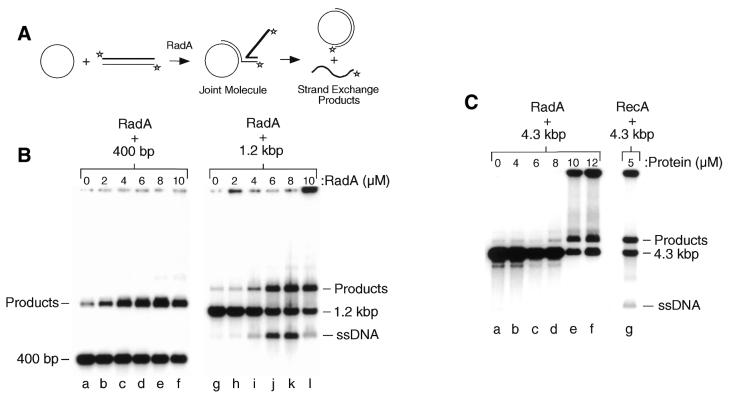
RadA protein has an unusually strong preference for binding and forming filaments on ssDNA compared to dsDNA (see above), and this presented us with an ideal opportunity to visualise joint molecules, since the ssDNA would be bound by RadA while the dsDNA would remain naked. A persistent problem with visualising recombination intermediates is that the recombinase often covers both ssDNA and dsDNA and forms protein–DNA aggregates. Conditions were optimised that reduced aggregate formation (2 mM Mg2+ and 5 min reactions) and used to visualise strand exchange intermediates between a 3 kb linear duplex and a 4.3 kb circular ssDNA by electron microscopy (Fig. 7). Two joint molecules are shown in Figure 7 in which the ssDNA is coated with RadA and has the collapsed, intertwined appearance of the molecule shown in Figure 2D. Each RadA–ssDNA nucleoprotein filament is attached to a naked linear dsDNA molecule (indicated by black arrows) and represents a joint molecule, the intermediate step of homologous strand exchange. No joint molecules were detected in the presence of ATP-γ-S using the gel assay indicating that RadA requires ATP for active filament formation and efficient strand exchange (data not shown).
Figure 7.
Two images showing RadA-mediated joint molecule formation. Electron microscopic visualisation of RadA strand exchange reactions showing the formation of joint molecules. The ssDNA is covered in RadA to form nucleoprotein filaments, whereas the dsDNA remains naked. The path of the dsDNA is indicated by black arrows. The scale bar represents 50 nm.
In light of the fact that the archaeal protein sequences are more similar to their eukaryotic counterparts than the bacterial sequences are, archaeal recombination reactions may serve as excellent simplified model systems for the more elaborate recombination machineries present in eukaryotic cells (2).
Acknowledgments
ACKNOWLEDGEMENTS
We thank the members of our laboratories for their comments and suggestions. We would also like to thank Professor Jacques Dubochet for his interest. We are grateful to the ICRF Oligonucleotide Synthesis Laboratory for DNA synthesis. This work was supported by the Imperial Cancer Research Fund, the Human Frontiers Science Program, the Swiss National Foundation and the Swiss–British Council Joint Research Program.
REFERENCES
- 1.Bianco P.R., Tracy,R.B. and Kowalczykowski,S.C. (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci., 17, 570–603. [DOI] [PubMed] [Google Scholar]
- 2.Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox M.M. (2000) Recombinational DNA repair in bacteria and the RecA protein. Prog. Nucleic Acid Res. Mol. Biol., 63, 311–366. [DOI] [PubMed] [Google Scholar]
- 4.Kowalczykowski S.C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci., 25, 156–165. [DOI] [PubMed] [Google Scholar]
- 5.Cox M.M., Goodman,M.F., Kreuzer,K.N., Sherratt,D.J., Sandler,S.J. and Marians,K.J. (2000) The importance of repairing stalled replication forks. Nature, 404, 37–41. [DOI] [PubMed] [Google Scholar]
- 6.Baumann P. and West,S.C. (1998) Role of the human Rad51 protein in homologous recombination and double-stranded break repair. Trends Biochem. Sci., 23, 247–251. [DOI] [PubMed] [Google Scholar]
- 7.Woese C.R. and Fox,G.E. (1977) Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA, 74, 5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen G.J. and Woese,C.R. (1997) Archaeal genomics: an overview. Cell, 89, 991–994. [DOI] [PubMed] [Google Scholar]
- 9.Sandler S.J., Satin,L.H., Samra,H.S. and Clark,A.J. (1996) recA-like genes from three archaean species with putative protein products similar to Rad51 and Dmc1 proteins of the yeast Saccharomyces cerevisiae. Nucleic Acids Res., 24, 2125–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seitz E.M., Brockman,J.P., Sandler,S.J., Clark,A.J. and Kowalczykowski,S.C. (1998) RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev., 12, 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandler S.J., Hugenholtz,P., Schleper,C., DeLong,E.F., Pace,N.R. and Clark,A.J. (1999) Diversity of radA genes from cultured and uncultured archaea: comparative analysis of putative RadA proteins and their use as a phylogenetic marker. J. Bacteriol., 181, 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiRuggiero J., Brown,J.R., Bogert,A.P. and Robb,F.T. (1999) DNA repair systems in archaea: mementos from the last universal common ancestor? J. Mol. Evol., 49, 474–484. [DOI] [PubMed] [Google Scholar]
- 13.Komori K., Miyata,T., DiRuggiero,J., Holley-Shanks,R., Hayashi,I., Cann,I.K., Mayanagi,K., Shinagawa,H. and Ishino,Y. (2000) Both RadA and RadB are involved in homologous recombination in Pyrococcus furiosus. J. Biol. Chem., 275, 33782–33790. [DOI] [PubMed] [Google Scholar]
- 14.Woods W.G. and Dyall-Smith,M.L. (1997) Construction and analysis of a recombination-deficient (radA) mutant of Haloferax volcanii. Mol. Microbiol., 23, 791–797. [DOI] [PubMed] [Google Scholar]
- 15.Brendel V., Brocchieri,L., Sandler,S.J., Clark,A.J. and Karlin,S. (1997) Evolutionary comparisons of RecA-like proteins across all major kingdoms of living organisms. J. Mol. Evol., 44, 528–541. [DOI] [PubMed] [Google Scholar]
- 16.Benedict R.C. and Kowalczykowski,S.C. (1988) Increase in the DNA strand assimilation activity of RecA protein by removal of the C terminus and structure-function studies of the resulting protein fragment. J. Biol. Chem., 263, 15513–15520. [PubMed] [Google Scholar]
- 17.Roca A.I. and Cox,M.M. (1990) The RecA protein: structure and function. Crit. Rev. Biochem. Mol. Biol., 25, 415–456. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa T., Yu,X., Shinohara,A. and Egelman,E.H. (1993) Similarity of the yeast Rad51 filament to the bacterial RecA filament. Science, 259, 1896–1899. [DOI] [PubMed] [Google Scholar]
- 19.Sung P. and Robberson,D.L. (1995) DNA strand exchange mediated by a Rad51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell, 82, 453–461. [DOI] [PubMed] [Google Scholar]
- 20.Woese C.R. (1987) Bacterial evolution. Microbiol. Rev., 51, 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown J.R. and Doolittle,W.F. (1995) Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc. Natl Acad. Sci. USA, 92, 2441–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chedin F., Seitz,E.M. and Kowalczykowski,S.C. (1998) Novel homologs of Replication Protein A in archaea: implications for the evolution of ssDNA-binding proteins. Trends Biochem. Sci., 23, 273–277. [DOI] [PubMed] [Google Scholar]
- 23.Smith D.R., Doucettestamm,L.A., Deloughery,C., Lee,H.M., Dubois,J., Aldredge,T., Bashirzadeh,R., Blakely,D., Cook,R., Gilbert,K. et al. (1997) Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol., 179, 7135–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komori K. and Ishino,Y. (2001) Replication Protein A in Pyrococcus furiosusis involved in homologous DNA recombination. J. Biol. Chem., 276, 25654–25660. [DOI] [PubMed] [Google Scholar]
- 25.Klenk H.P., Clayton,R.A., Tomb,J.F., White,O., Nelson,K.E., Ketchum,K.A., Dodson,R.J., Gwinn,M., Hickey,E.K., Peterson,J.D. et al. (1997) The complete genome sequence of the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus. Nature, 390, 364–370. [DOI] [PubMed] [Google Scholar]
- 26.Johnson R.D. and Symington,L.S. (1995) Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol., 15, 4843–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung P. (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with Replication Protein-A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. [DOI] [PubMed] [Google Scholar]
- 28.Rashid N., Morikawa,M. and Imanaka,T. (1996) A RecA/RAD51 homologue from a thermophilic archaeon retains the major RecA domain only. Mol. Gen. Genet., 253, 397–400. [DOI] [PubMed] [Google Scholar]
- 29.Walker G.C. (1985) Inducible DNA repair systems. Annu. Rev. Biochem., 54, 425–458. [DOI] [PubMed] [Google Scholar]
- 30.Spies M., Kil,Y., Masui,R., Kato,R., Kujo,C., Ohshima,T., Kuramitsu,S. and Lanzov,V. (2000) The RadA protein from a hyperthermophilic archaeon Pyrobaculum islandicum is a DNA-dependent ATPase that exhibits two disparate catalytic modes, with a transition temperature at 75 degrees C. Eur. J. Biochem., 267, 1125–1137. [DOI] [PubMed] [Google Scholar]
- 31.Petukhov M., Kil,Y., Kuramitsu,S. and Lanzov,V. (1997) Insights into thermal resistance of proteins from the intrinsic stability of their alpha-helices. Proteins, 29, 309–320. [DOI] [PubMed] [Google Scholar]
- 32.Kil Y.V., Baitin,D.M., Masui,R., Bonch-Osmolovskaya,E.A., Kuramitsu,S. and Lanzov,V.A. (2000) Efficient strand transfer by the RadA recombinase from the hyperthermophilic archaeon Desulfurococcus amylolyticus. J. Bacteriol., 182, 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eggleston A.K. and West,S.C. (2000) Cleavage of Holliday junctions by the Escherichia coliRuvABC complex. J. Biol. Chem., 275, 26467–26476. [DOI] [PubMed] [Google Scholar]
- 34.Baumann P. and West,S.C. (1997) The human Rad51 protein: polarity of strand transfer and stimulation by hRP-A. EMBO J., 16, 5198–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumann P. and West,S.C. (1999) Heteroduplex formation by human Rad51 protein: effects of DNA end-structure, hRP-A and hRad52. J. Mol. Biol., 291, 363–374. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook E.F., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Van Dyck, E., Hajibagheri,N.M.A., Stasiak,A. and West,S.C. (1998) Visualisation of human Rad52 protein and its complexes with hRad51 and DNA. J. Mol. Biol., 284, 1027–1038. [DOI] [PubMed] [Google Scholar]
- 38.Stasiak A., DiCapua,E. and Koller,T. (1981) Elongation of duplex DNA by RecA protein. J. Mol. Biol., 151, 557–564. [DOI] [PubMed] [Google Scholar]
- 39.Benson F.E., Stasiak,A. and West,S.C. (1994) Purification and characterisation of the human Rad51 protein, an analogue of E. coliRecA. EMBO J., 13, 5764–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast Rad51 protein. Science, 265, 1241–1243. [DOI] [PubMed] [Google Scholar]
- 41.Hingorani M.M. and O’Donnell,M. (1998) Toroidal proteins: running rings around DNA. Curr. Biol., 8, R83–R86. [DOI] [PubMed] [Google Scholar]
- 42.Heuser J. and Griffith,J. (1989) Visualization of RecA protein and its complexes with DNA by quick-freeze/deep etch electron microscopy. J. Mol. Biol., 210, 473–484. [DOI] [PubMed] [Google Scholar]
- 43.Yu X. and Egelman,E.H. (1997) The RecA hexamer is a structural homologue of ring helicases. Nature Struct. Biol., 4, 101–104. [DOI] [PubMed] [Google Scholar]
- 44.Baumann P., Benson,F.E., Hajibagheri,N. and West,S.C. (1997) Purification of human Rad51 protein by selective spermidine precipitation. Mutat. Res., 384, 65–72. [DOI] [PubMed] [Google Scholar]
- 45.Yu X., Jacobs,S.A., West,S.C., Ogawa,T. and Egelman,E.H. (2001) Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl Acad. Sci. USA, 98, 8419–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masson J.-Y., Davies,A.A., Hajibagheri,N., Van Dyck,E., Benson,F.E., Stasiak,A.Z., Stasiak,A. and West,S.C. (1999) The meiosis-specific recombinase Dmc1 forms rings structures and interacts with hRad51. EMBO J., 18, 6552–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinohara A., Shinohara,M., Ohta,T., Matsuda,S. and Ogawa,T. (1998) Rad52 forms ring structures and cooperates with RPA in single-strand DNA annealing. Genes Cells, 3, 145–156. [DOI] [PubMed] [Google Scholar]
- 48.Stasiak A.Z., Larquet,E., Stasiak,A., Müller,S., Engel,A., Van Dyck,E., West,S.C. and Egelman,E.H. (2000) The human Rad52 protein exists as a heptameric ring. Curr. Biol., 10, 337–340. [DOI] [PubMed] [Google Scholar]
- 49.Stasiak A., Tsaneva,I.R., West,S.C., Benson,C.J.B., Yu,X. and Egelman,E.H. (1994) The Escherichia coliRuvB branch migration protein forms double hexameric rings around DNA. Proc. Natl Acad. Sci. USA, 91, 7618–7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Passy S.I., Yu,X., Li,Z., Radding,C.M. and Egelman,E.H. (1999) Rings and filaments of β protein from bacteriophage λ suggest a superfamily of recombination proteins. Proc. Natl Acad. Sci. USA, 96, 4279–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parsons C.A., Baumann,P., Van Dyck,E. and West,S.C. (2000) Precise binding to single-stranded DNA termini by RAD52 protein. EMBO J., 19, 4175–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stasiak A. and DiCapua,E. (1982) The helicity of DNA in complexes with RecA protein. Nature, 299, 185–186. [DOI] [PubMed] [Google Scholar]
- 53.Roman L.J. and Kowalczykowski,S.C. (1986) Relationship of the physical and enzymatic properties of E. coli RecA protein to its strand exchange properties. Biochemistry, 25, 7375–7385. [DOI] [PubMed] [Google Scholar]
- 54.Ogawa T., Wabico,H., Tsurimoto,T., Horii,T., Masukata,H. and Ogawa,H. (1979) Characteristics of purified RecA protein and the regulation of its synthesis in vitro. Cold Spring Harb. Symp. Quant. Biol., 43, 909–915. [DOI] [PubMed] [Google Scholar]
- 55.Baumann P., Benson,F.E. and West,S.C. (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell, 87, 757–766. [DOI] [PubMed] [Google Scholar]
- 56.Brenner S.L., Mitchell,R.S., Morrical,S.W., Neuendorf,S.K., Schutte,B.C. and Cox,M.M. (1987) RecA protein promoted ATP hydrolysis occurs throughout RecA nucleoprotein filaments. J. Biol. Chem., 262, 4011–4016. [PubMed] [Google Scholar]
- 57.Sung P. and Stratton,S.A. (1996) Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem., 271, 27983–27986. [DOI] [PubMed] [Google Scholar]
- 58.McIlwraith M.J., Van Dyck,E., Masson,J.-Y., Stasiak,A.Z., Stasiak,A. and West,S.C. (2000) Reconstitution of the strand invasion step of double-strand break repair using human Rad51, Rad52 and RPA proteins. J. Mol. Biol., 304, 151–164. [DOI] [PubMed] [Google Scholar]
- 59.Shibata T., Ohtani,T., Iwabuchi,M. and Ando,T. (1982) D-loop cycle. A circular reaction sequence which comprises formation and dissociation of D-loops and inactivation and reactivation of superhelical closed circular DNA promoted by RecA protein of E. coli. J. Biol. Chem., 257, 13981–13986. [PubMed] [Google Scholar]
- 60.Cox M.M., Soltis,D.A., Livneh,Z. and Lehman,I.R. (1983) DNA strand exchange promoted by RecA protein and single-stranded DNA-binding protein of Escherichia coli. Cold Spring Harb. Symp. Quant. Biol., 47, 803–810. [DOI] [PubMed] [Google Scholar]
- 61.Lavery P.E. and Kowalczykowski,S.C. (1992) A postsynaptic role for single-stranded DNA-binding protein in RecA protein-promoted DNA strand exchange. J. Biol. Chem., 267, 9315–9320. [PubMed] [Google Scholar]