Objectives
Most previous studies have analyzed bacteria in tumors using resected pancreatic cancer (PC) tissues, because it is difficult to obtain tissue samples from unresectable advanced PC. We aimed to determine whether minimal tissue obtained by endoscopic ultrasound–guided fine-needle aspiration is useful for microbiome analysis.
Methods
Thirty PC and matched duodenal and stomach tissues (N = 90) were prospectively collected from 30 patients who underwent endoscopic ultrasound–guided fine-needle aspiration. Bacterial DNA was extracted, and 16S rRNA sequencing was performed. The primary outcome was the success rate of bacterial detection in tumors. Bacterial diversity and structure were investigated.
Results
The bacterial detection rates were 80%, 100%, and 97% in PC, gastric, and duodenal samples, respectively. Pancreatic cancer tissues showed a lower α-diversity and a significantly different microbial structure than stomach and duodenal tissues. Proteobacteria were more abundant, whereas Firmicutes, Bacteroidetes, and Fusobacteria were less abundant in PC tissues than in stomach and duodenal tissues. Acinetobacter was more abundant in PC tissues than in stomach and duodenal tissues, and Delftia was more frequently detected in resectable PC.
Conclusions
Endoscopic ultrasound–guided fine-needle aspiration samples were valuable for PC microbiome analysis, revealing that the bacterial composition of PC is different from that of the stomach and duodenum.
Key Words: endoscopic ultrasound–guided fine-needle aspiration, bacteria, microbiome, pancreas, pancreatic cancer
Pancreatic cancer (PC) is the seventh leading cause of cancer-related deaths worldwide.1 In recent years, more than 80% of patients have been diagnosed with unresectable PC due to locally advanced or distant metastasis,2 and the 5-year survival rate of patients with unresectable PC is reportedly approximately 8% in the United States3 and 6.5% in Japan.4 Of the cancers diagnosed as localized and resectable, the 5-year survival rate is reportedly 25% in the United States5,6 and 18.8% in Japan.7 Hence, along with early diagnosis, the development of new treatment strategies is needed.
There are numerous studies regarding the human microbiome and its potential role in various diseases, including cancer.8–10 The gut microbiome is associated with the response to immune-checkpoint inhibitors11,12 and chemotherapeutic agents.13 Various tumors harbor unique bacterial assemblages,14 with the presence of tumor bacteria being related to prognosis in various cancers.15–17 The pancreas was once regarded as a sterile organ18; however, studies have shown the presence of more bacteria in pancreatic tumor tissue than in noncancerous pancreatic tissue.19,20 Tumor bacteria in PC are associated with the progression of PC,21 chemoresistance to gemcitabine,19 and the prognosis of patients with resected PC.17 These findings suggest that the PC microbiome might play an important role in cancer development and, therefore, represent a potential therapeutic target. However, most previous studies have analyzed the PC microbiome using resected tumor tissues only,17 with most patients with advanced PC being excluded from the analysis because of the diagnosis of having an unresectable tumor and the difficulty in obtaining resected PC tissue. Pancreatic cancer tissue collected before treatment would be needed to use information about bacteria to select better neoadjuvant therapies for patients with resectable PC.
Endoscopic ultrasound–guided fine-needle aspiration (EUS-FNA) is a safe procedure for histologic diagnosis in patients with suspected PC.22 Previous studies have reported the utility of genetic analysis of various cancers using specimens obtained via EUS-FNA.23–25 Therefore, this technology may be valuable for assessing the PC microbiome. We aimed to examine the PC tumor microbiome by analyzing samples obtained through EUS-FNA.
MATERIALS AND METHODS
Patients and Study Design
This was a single-center, prospective, observational study. Patients clinically diagnosed with PC were recruited from May 2019 to April 2020. Those 20 years or older who had undergone EUS-FNA for histologic or cytologic diagnosis of PC were included in the study. Bacterial analysis was performed using samples from patients who were diagnosed by a pathologist as having pancreatic ductal adenocarcinoma. Resectability was defined according to the Japanese Classification of Pancreatic Cancer.4 All procedures were conducted in accordance with the ethical standards of the Helsinki Declaration of 1964 and its later versions. All patients received information about the trial in written form, and provided informed consent before enrolment. The study design and protocol were approved by the institutional review board of the Hokkaido University Hospital (approval no. 018-0271).
Assessment of Outcomes
The primary study outcome was the success rate of the detection of bacteria in pancreatic tissues collected by EUS-FNA, which was defined as the success of the amplicon sequence analysis. The secondary outcomes were the comparison of bacterial diversity and composition between normal tissues (stomach and duodenal mucosa) and resectable and unresectable PC.
Sample Collection
Pancreatic cancer tissues were collected from patients by EUS-FNA, which was performed using a linear echoendoscope (GF-UCT260; Olympus Medical Systems, Tokyo, Japan) and 22- or 25-gauge needles (Expect or Acquire; Boston Scientific Japan, Tokyo, Japan). The aspirated material was smeared onto microscope slides for on-site examination, and Diff-Quik staining (Kokusai Shiyaku, Kobe, Japan) of all specimens was performed to confirm malignancy. Endoscopic ultrasound–guided fine-needle aspiration white samples, which were considered to include malignant cells near the area of Diff-Quik staining, were stored at −80°C until DNA extraction. Hematoxylin/eosin staining was performed, and a pathologist reviewed the slides.
Duodenal and stomach mucosa were collected from the same patients using a pair of biopsy forceps (Radial Jaw 4; Boston Scientific), and all samples were immediately stored at −80°C until DNA extraction.
DNA Extraction From Samples and 16S rRNA Sequencing
DNA extraction from the samples, preparation of a 16S rRNA library, and sequencing were conducted at Takara Bio, Inc (Kusatsu, Japan) using the MiSeq system (Illumina, San Diego, Calif) according to the manufacturer's protocol. DNA was extracted from the samples using the NucleoSpin Soil kit (Macherey-Nagel, Düren, Germany). All extracted DNA samples were quantified by fluorescence detection using the Quant-iT dsDNA Assay Kit (Thermo Fisher Scientific, Waltham, Mass) and purified using Agencourt AMPure XP (Beckman Coulter, Brea, Calif). Sequencing libraries were prepared using the 16S (V3-V4) Metagenomic Library Construction Kit for NGS (Takara Bio, Kusatsu, Japan). The first polymerase chain reaction amplification was performed using the primer pair 341 F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) and 806 R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACHVGGGTWTCTAAT-3′) with Illumina adaptor overhang sequences. The second polymerase chain reaction amplification was performed using the Nextera XT Index Kit v2 (Illumina). Sequencing libraries were purified using Agencourt AMPure XP (Beckman Coulter) and quantified by fluorescence detection using the Quant-iT dsDNA Assay Kit (Thermo Fisher Scientific). Clonal clusters of the libraries were generated and sequenced on a MiSeq system (Illumina) with the MiSeq Reagent v3 kit (Illumina) in the 2 × 250-bp mode.
16S rRNA-Based Taxonomic Analysis and Statistical Analysis
Amplicon sequence analysis was performed using Quantitative Insights Into Microbial Ecology 2 (QIIME2 v.2020.8; https://qiime2.org/).26 Paired-end sequences were imported into QIIME2 and denoised using the DADA2 pipeline. The demultiplexed sequencing data were denoised using the DADA2 plugin with parameter settings of –p-trunc-len-f 245 –p-trunc-len-r 245 –p-trim-left-f 0 –p-trim-left-r 0. Sequences were then dereplicated, checked for sequence variants, merged, and finally chimera checked by DADA2. In total, 1,187,971 sequences were obtained after quality filtering and processing. We performed diversity analysis and differential abundance analysis. For the diversity analysis, the samples were rarefied to a sampling depth of 2404 sequences per sample. Core diversity metrics were analyzed, including the number of amplicon sequence variants, Shannon entropy for α-diversity, and permutational multivariate analysis of variance for β-diversity. In the differential abundance analysis, amplicon sequence variants were selected by aligning the sequences with those in the latest Silva 16S database (v.138; https://www.arb-silva.de/). We then performed linear discriminant analysis effect size (LEfSe)27 and analysis of composition of microbiomes (ANCOM)28 to determine whether there were significant differences in the relative abundance of taxa between the groups. Statistically significant differences (P < 0.05) between groups comprising categorical and continuous variables were compared using χ2 tests and nonparametric Mann-Whitney U test or Kruskal-Wallis test, respectively. All statistical analyses were performed using SPSS ver. 26 (IBM Corp, Armonk, NY).
RESULTS
Thirty patients were included in this study, and their major clinicopathological characteristics are summarized in Table 1. Of these patients, 19 had pancreatic head cancer and 11 had pancreatic body or tail cancer, with 11 displaying distant metastasis. Tumor tissues from 12 patients were aspirated through the stomach, whereas those from the remaining 18 patients were aspirated through the duodenum. Only one patient received antibiotics within 30 days before sample collection. Among the collected samples, 24 (80%) EUS-FNA samples, 30 (100%) stomach samples, and 29 (97%) duodenum samples could be analyzed by 16S rRNA sequencing. We found no significant differences between patients who provided EUS-FNA samples that were successfully analyzed and those whose samples could not be analyzed (Table 2).
TABLE 1.
Patient Characteristics
| Characteristic | Patients, n (n = 30) |
|---|---|
| Sex, male/female | 11/19 |
| Age, median (range), y | 70 (54–86) |
| Performance status, 0/1 | 18/12 |
| Primary location, head/body/tail | 19/5/6 |
| Resectability,* R or BR/UR | 15/15 |
| Drainage stent, present/absent | 6/24 |
| Initial treatment, CTX/CRT/resection/BSC | 25/1/1/3 |
| Aspiration, stomach/duodenum | 12/18 |
| Diabetes, yes/no | 10/20 |
| Use of antacids, yes/no | 14/16 |
| Use of antibiotics 1-month prior, yes/no | 1/29 |
*Defined according to the Japanese Classification of Pancreatic Cancer.4
BR indicates borderline resectable; BSC, best supportive care; CRT, chemoradiotherapy; CTX, chemotherapy; R, resectable; UR, unresectable.
TABLE 2.
Comparison Between Patients Who Provided Successfully Versus Unsuccessfully Analyzed EUS-FNA Samples
| Patients, n (n = 30) | |||
|---|---|---|---|
| Unsuccessful (n = 6) | Successful (n = 24) | P* | |
| Sex, male/female | 2/4 | 9/15 | 1.00 |
| Primary location, head/BT | 2/4 | 17/7 | 0.15 |
| Resectability,† R‡/UR | 3/3 | 12/12 | 1.00 |
| Drainage stent, present/absent | 1/5 | 5/19 | 1.00 |
| Aspiration, stomach/duodenum | 4/2 | 8/16 | 0.18 |
| Diabetes, yes/no | 1/5 | 9/15 | 0.63 |
| Use of antacids, yes/no | 1/5 | 13/11 | 0.18 |
*χ2 test.
†Defined according to the Japanese Classification of Pancreatic Cancer.4
‡BR was included in R.
BT indicates body or tail; R, resectable; UR, unresectable.
We then compared the α-diversity of bacteria between PC, stomach, and duodenum tissues. Using several indices, PC tissue was found to have a lower microbial diversity than stomach and duodenum tissues (Fig. 1). The β-diversity analysis revealed that the unweighted UniFrac distance differed significantly between PC tissues and stomach and duodenum tissues (P < 0.05), whereas the weighted UniFrac distance revealed a significant structural difference between PC and duodenum tissues (P < 0.05), but not between PC and stomach tissues (P = 0.051; Supplemental Fig. 1, http://links.lww.com/MPA/A949). These data showed that the bacterial composition of the pancreatic tumor differed from that of the gastrointestinal tract.
FIGURE 1.
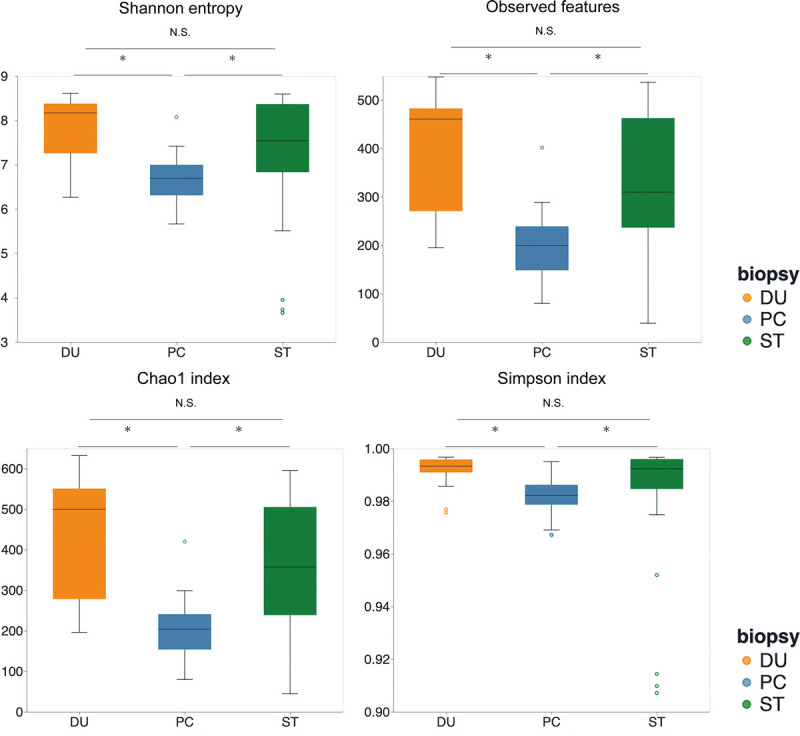
α-Diversity box plots among PC, ST, and DU tissues. *P < 0.05. DU, duodenum; NS, not significant; ST, stomach.
We then evaluated differences in the microbial taxonomic structure at the phylum level (Fig. 2). Firmicutes, Bacteroidetes, and Fusobacteria were significantly less abundant in PC tissues than in the gastrointestinal tract, whereas Proteobacteria were significantly more abundant in PC samples (Fig. 2B). We then conducted high-dimensional class comparisons using LefSe, which revealed considerable differences in the predominance of bacterial communities between PC, stomach, and duodenal tissues at the genus level. Evaluation of the relative frequency of the bacteria indicated that PC tissue exhibited a predominance of Acinetobacter and Pseudomonas compared with the gastrointestinal tract (Figs. 3A, B). The ANCOM also revealed a trend that Acinetobacter was abundant in PC (Fig. 3C).
FIGURE 2.
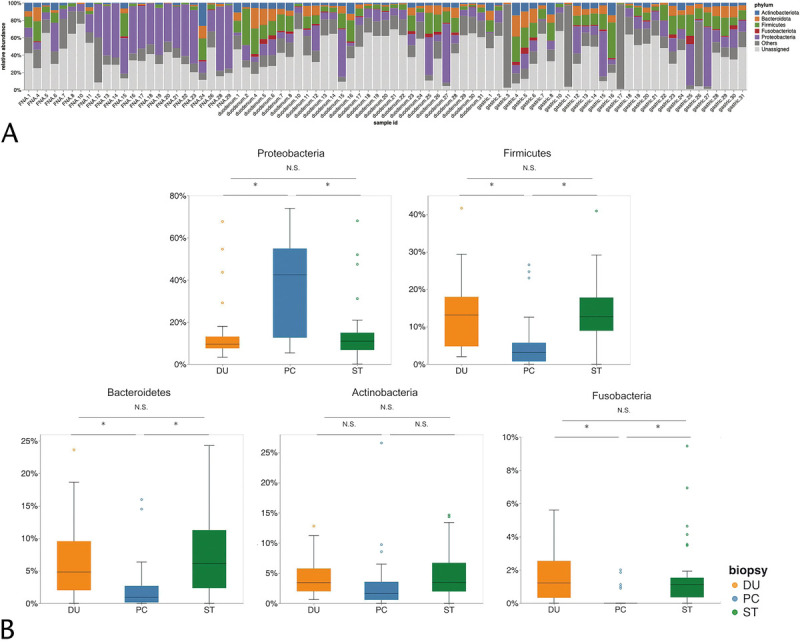
Microbiome communities among PC, ST, and DU tissues. A, Bar plots of the phylum level. B, Relative abundance of major phyla among PC, ST, and DU tissues. *P < 0.05. DU, duodenum; NS, not significant; ST, stomach.
FIGURE 3.
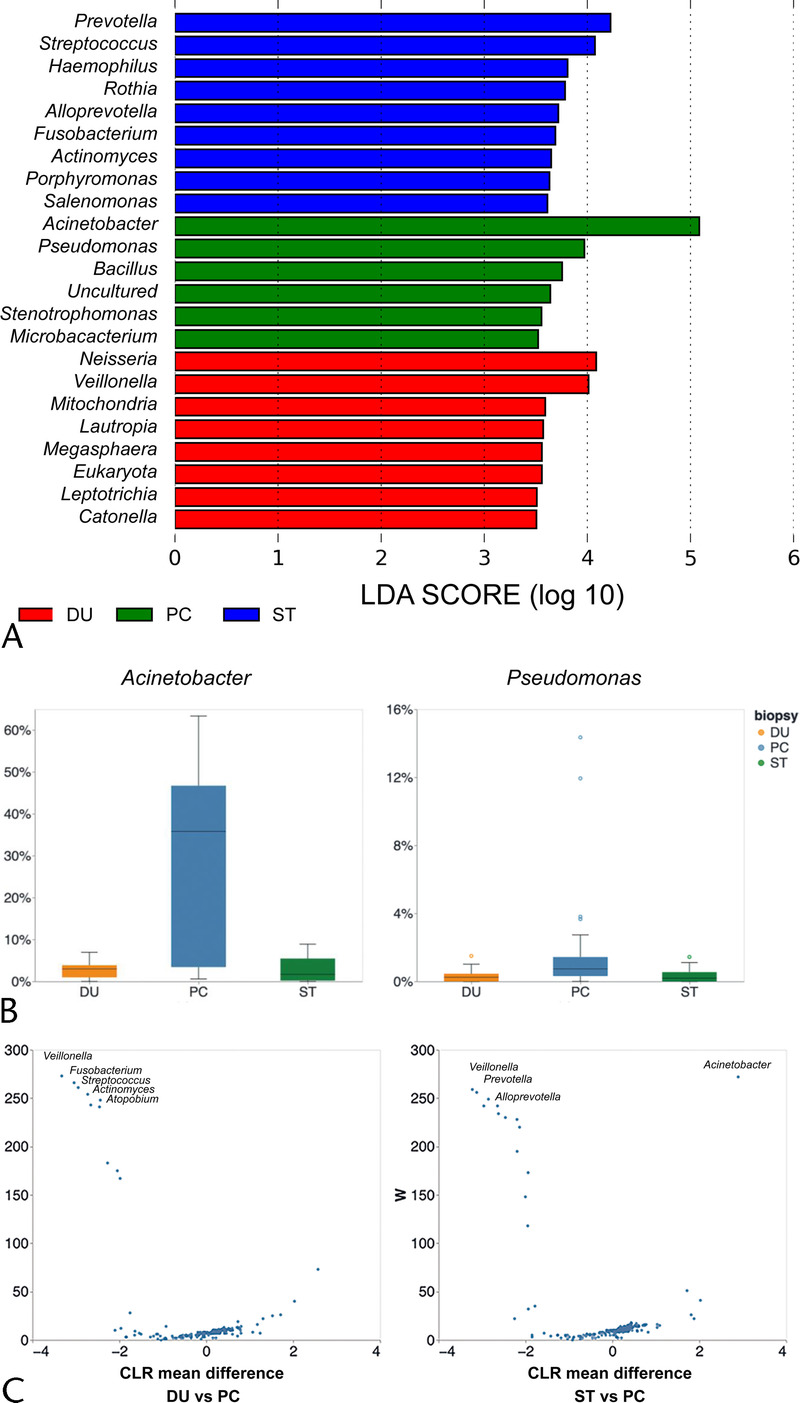
Identification of featured bacterial taxa. A, Bacterial taxa identified by LEfSe to be different among PC, ST, and DU tissues. The criterion for feature selection was log LDA score greater than 3.5. B, Relative abundance of specific bacteria among PC, ST, and DU tissues. C, Bacterial taxa identified by ANCOM to be different among PC, ST, and DU tissues. CLR, centered log ratio; DU, duodenum; LEfSe, linear discriminant analysis effect size; LDA, linear discriminant analysis; ST, stomach.
In the subgroup analysis, we analyzed patients with advanced PC who had been diagnosed as unresectable because of distant metastasis or invasion of major vessels. We found no significant difference in the Shannon index between resectable and unresectable PC tissues, although the Shannon index was lower in the stomach (P = 0.106) and duodenum (P < 0.05) samples from patients with unresectable PC than in those with resectable PC (Fig. 4A). The β-diversity analysis showed no significant difference in unweighted and weighted UniFrac distances between resectable and unresectable PC tissues. There was no significant difference with respect to major taxa at the phylum level (Fig. 4B). Linear discriminant analysis effect size showed that Delftia was significantly more abundant in resectable than in unresectable PC tissues (Fig. 4C). Evaluation of the association between the aspiration site and bacteria in PC, to account for possible contamination of tumor tissue with stomach or duodenal bacteria, revealed no significant difference between the stomach- and duodenum-derived samples (Fig. 5).
FIGURE 4.
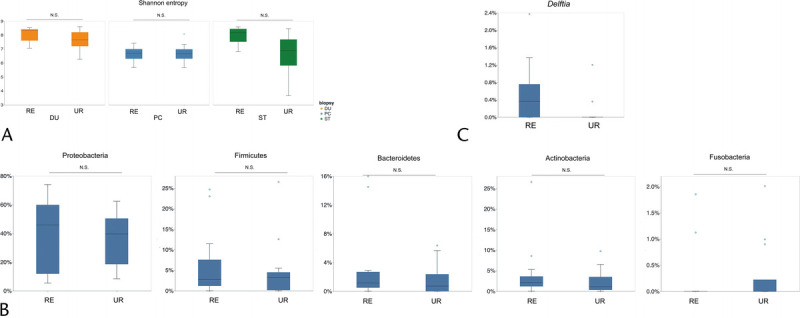
Comparison of bacterial diversity and composition between patients with resectable and unresectable PC. A, Shannon index between the RE and UR PC groups at each anatomical site. B, Relative abundance of major phyla and of (C) Delftia between the RE and UR PC groups. DU, duodenum; NS, not significant; PC, pancreatic cancer; RE, resectable; ST, stomach; UR, unresectable.
FIGURE 5.
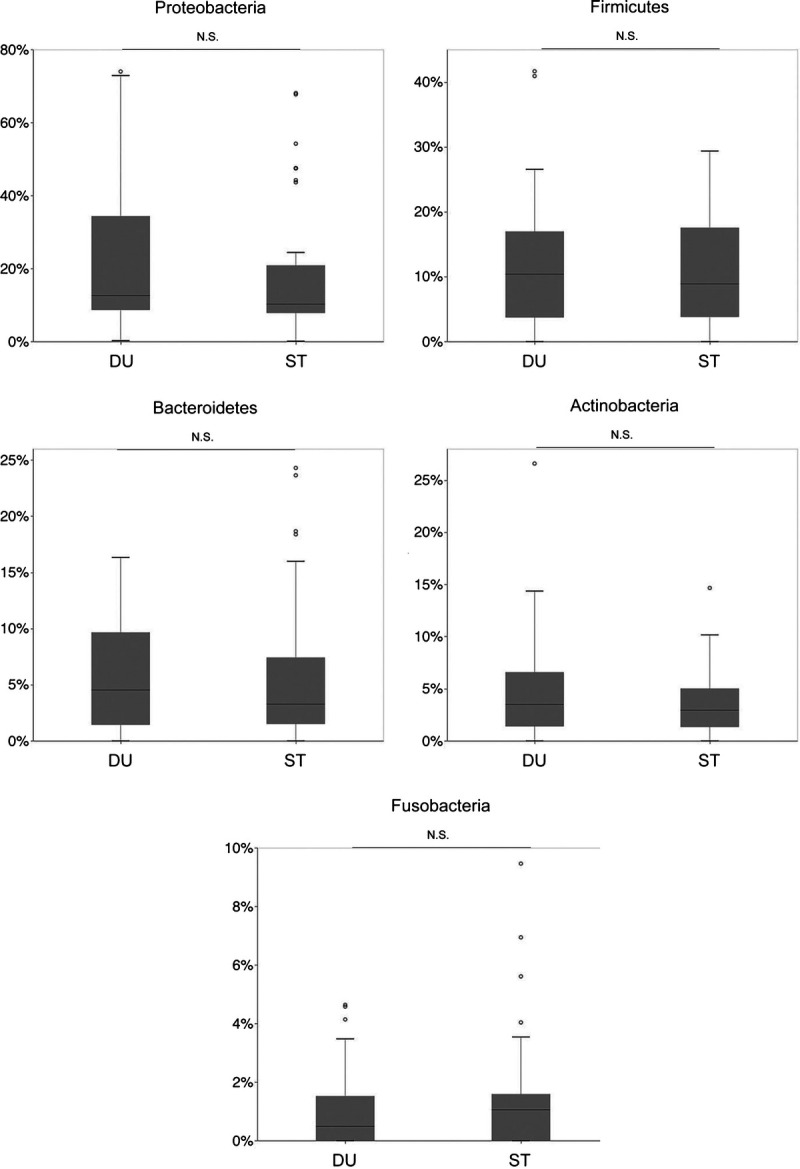
Relative abundance of major phyla in pancreatic cancer between ST-derived (pancreatic head cancer) and DU-derived (mostly pancreatic body/tail cancer) samples. DU, duodenum; NS, not significant; ST, stomach.
DISCUSSION
In this study, we demonstrated the efficacy of using minimal samples of PC tissue obtained by EUS-FNA for PC microbiome analysis. Recent studies have provided evidence related to the potential role of the microbiome in PC14,17,19,21; however, most of these previous studies have analyzed the PC microbiome using surgically resected tissue only. We were unable to obtain surgically resected PC tissue from patients with unresectable PC because tumor resection is not a treatment option for these patients. However, in the present study, we used tissue samples obtained by EUS-FNA, which is a standard diagnostic procedure for PC, to analyze the PC microbiome. Fewer tumor bacteria were detected when the tumor was located at the head or tail of the pancreas (Fig. 5), when tissue was aspirated through the stomach, or when antacids were not used. Tumor bacteria in PC are considered to originate from the duodenum,19,21 and our results reflect a lower number of bacteria in tumors located in the pancreatic body or tail. The use of antacids has been associated with the disturbance of the gut microbiome,29,30 which might affect the bacterial composition and abundance within the PC tissue. Several studies have reported that at the phylum level, Proteobacteria are abundant in PC tissue,19,21 and our findings agree with this observation. The duodenum microbiome is reportedly dominated by Firmicutes,31,32 although Proteobacteria are also abundant.32 Another study found a significant difference in microbial composition between duodenum and PC tissues.20 In the present study, we found a lower α-diversity in PC than in duodenum tissues, as well as a different bacterial composition. Considering that various tumors harbor unique bacteria,14 the PC microbiome might be specifically selected from duodenal bacteria.
Our results also revealed the taxonomic profile of unresectable PC, which had not been previously evaluated because of the difficulty in sample collection. We found no significant difference in tumor bacterial diversity and composition at the phylum level between patients with resectable and those with unresectable tumors. We also found that Delftia was more abundant in resectable PC, although the clinical implications of this finding remain unknown. Delftia is more frequently observed in the duodenum of patients with PC than in healthy controls,31 suggesting a possible relationship with the development of early phase PC. We observed a trend toward a lower α-diversity in the stomach and duodenal tissues from patients with unresectable PC than in the same tissues from patients with resectable PC. This trend might reflect gut dysbiosis, given that patients with unresectable PC often experience decreased appetite and diet changes.
The PC microbiome might be a therapeutic target. Geller et al19 showed that tumor bacteria in PC, which are mainly Gammaproteobacteria, can inactivate gemcitabine. Gemcitabine-containing chemotherapies are standard treatments for PC,33,34 and retrospective studies have reported that the use of antibiotics might be associated with an increased efficacy of gemcitabine and gemcitabine-containing chemotherapy.35–37 Therefore, the PC microbiome may be a predictive marker for gemcitabine-containing therapy. In the present study, we showed that Gammaproteobacteria were dominant in PC tissue, an observation that is consistent with previous findings.19 Assessment of the PC microbiome using EUS-FNA before systemic therapy may lead to the selection of a more efficacious treatment. Modulating tumor bacteria using antibiotics or probiotics could represent a potential strategy for treating PC, although additional evidence is required.
Our study had some limitations. First, there was a risk of contamination. Using the EUS-FNA technique, PC tissues were collected through the stomach or duodenum, which may increase the risk of contamination. Our data showed that the composition of the PC microbiome did not include a major contamination by bacteria from the gastrointestinal tract. However, a minor contamination from stomach or duodenal bacteria may have occurred and could have affected our analyses. As opposed to percutaneous needle biopsy, there is no risk of skin contamination, which is another rich source of bacteria. Second, we did not compare the bacterial composition between EUS-FNA–obtained tissue and resected tumor tissue. As the amount of EUS-FNA–obtained tissue was small, it is possible that this tissue did not reflect the entire PC bacterial diversity and composition. However, a recent study showed that EUS-FNA–derived samples from formalin-fixed paraffin-embedded PC have bacterial profiles similar to those of samples derived from surgically resected PC,38 an observation which supports the efficacy of endoscopically collected samples for bacterial analysis.
In conclusion, we demonstrated the utility of EUS-FNA–derived PC tissue for analyzing the PC microbiome and identified differences in bacterial composition between PC tissue and stomach and duodenum mucosal tissue.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the patients and their families for participating in this study. We also thank Editage (http://www.editage.com) for English language editing.
Footnotes
This study was supported by Grants-in-Aid for Scientific Research (KAKENHI) of the Japan Society for the Promotion of Science (grant no. 19K17479).
The authors declare no conflicts of interest.
All authors contributed to the study conception and design. Material preparation and data collection were performed by S. Nakano, K.I., R.S., R. Saito, K.K., R. Sugiura, S.K., K.H., H.H., M.N., R.F., Y.T., K.N., and M.K. Data analyses were performed by S. Nakano, K.H.O., and S. Nakaoka. The first draft of the manuscript was written by St.N. All authors commented on previous versions of the manuscript. All authors read and approved the final version of the manuscript.
Supplemental digital contents are available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.pancreasjournal.com).
Contributor Information
Shintaro Nakano, Email: shintaro-n@frontier.hokudai.ac.jp.
Yasuyuki Kawamoto, Email: y-kawamoto0716@hotmail.co.jp.
Rika Saito, Email: trillium.kam@gmail.com.
Ken Ito, Email: captain0421@yahoo.co.jp.
Takahiro Yamamura, Email: taka_yama0218@yahoo.co.jp.
Kazuaki Harada, Email: haraharaccho0605@yahoo.co.jp.
Satoshi Yuki, Email: satoshi-yuuki175@joy.ocn.ne.jp.
Kazumichi Kawakubo, Email: kkawakubo-gi@umin.ac.jp.
Ryo Sugiura, Email: ryou99sugi@yahoo.co.jp.
Shin Kato, Email: shinchan1231@gmail.com.
Koji Hirata, Email: sugoina_sawaken@yahoo.co.jp.
Hajime Hirata, Email: h.hirapon@gmail.com.
Masahito Nakajima, Email: nakaji_saxophone@yahoo.co.jp.
Ryutaro Furukawa, Email: rfurukawa404@gmail.com.
Yunosuke Takishin, Email: ofmycloud@live.jp.
Kousuke Nagai, Email: n.kosuke0620@gmail.com.
Isao Yokota, Email: yokotai@pop.med.hokudai.ac.jp.
Keisuke H. Ota, Email: newkeisukeisuke413@eis.hokudai.ac.jp.
Shinji Nakaoka, Email: snakaoka@sci.hokudai.ac.jp.
Masaki Kuwatani, Email: mkuwatan@med.hokudai.ac.jp.
Naoya Sakamoto, Email: sakamoto@med.hokudai.ac.jp.
REFERENCES
- 1.International Agency for Research on Cancer . Global Cancer Observatory. Geneva, Switzerland: World Health Organization; 2018. [Google Scholar]
- 2.Cancer Facts & Figures 2020. Atlanta, GA: American Cancer Society; 2020. [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 4.Japanese Classification of Pancreatic Cancer. 7th ed. Tokyo, Japan: Japan Pancreas Society; 2016. [Google Scholar]
- 5.Geer RJ, Brennan MF. Prognostic indicators for survival after resection of pancreatic adenocarcinoma. Am J Surg. 1993;165:68–72; discussion 72-73. [DOI] [PubMed] [Google Scholar]
- 6.Yeo CJ Cameron JL Lillemoe KD, et al. Pancreaticoduodenectomy for cancer of the head of the pancreas. 201 patients. Ann Surg. 1995;221:721–731; discussion 731–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egawa S Toma H Ohigashi H, et al. Japan Pancreatic Cancer Registry; 30th year anniversary: Japan Pancreas Society. Pancreas. 2012;41:985–992. [DOI] [PubMed] [Google Scholar]
- 8.Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med. 2019;216:20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67:1716–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079–1089. [DOI] [PubMed] [Google Scholar]
- 11.Gopalakrishnan V Spencer CN Nezi L, et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Routy B Le Chatelier E Derosa L, et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science. 2018;359:91–97. [DOI] [PubMed] [Google Scholar]
- 13.Iida N Dzutsev A Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nejman D Livyatan I Fuks G, et al. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science. 2020;368:973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamura K Baba Y Nakagawa S, et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22:5574–5581. [DOI] [PubMed] [Google Scholar]
- 16.Yamaoka Y Suehiro Y Hashimoto S, et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J Gastroenterol. 2018;53:517–524. [DOI] [PubMed] [Google Scholar]
- 17.Riquelme E Zhang Y Zhang L, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178:795–806.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y Yang G You L, et al. Role of the microbiome in occurrence, development and treatment of pancreatic cancer. Mol Cancer. 2019;18:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geller LT Barzily-Rokni M Danino T, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noticewala SS Lin D Kouzy R, et al. Pancreatic cancer intratumoral microbiome and characteristics within paired patient samples. J Clin Oncol. 2020;38(4 suppl):744.abstract.31895608 [Google Scholar]
- 21.Pushalkar S Hundeyin M Daley D, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamada T Yasunaga H Nakai Y, et al. Severe bleeding and perforation are rare complications of endoscopic ultrasound–guided fine needle aspiration for pancreatic masses: an analysis of 3,090 patients from 212 hospitals. Gut Liver. 2014;8:215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirata K Kuwatani M Suda G, et al. A novel approach for the genetic analysis of biliary tract cancer specimens obtained through endoscopic ultrasound–guided fine needle aspiration using targeted amplicon sequencing. Clin Transl Gastroenterol. 2019;10:e00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kameta E Sugimori K Kaneko T, et al. Diagnosis of pancreatic lesions collected by endoscopic ultrasound–guided fine-needle aspiration using next-generation sequencing. Oncol Lett. 2016;12:3875–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds JP Zhou Y Jakubowski MA, et al. Next-generation sequencing of liquid-based cytology non-small cell lung cancer samples. Cancer Cytopathol. 2017;125:178–187. [DOI] [PubMed] [Google Scholar]
- 26.Bolyen E Rideout JR Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segata N Izard J Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandal S Van Treuren W White RA, et al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015;26:27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson MA Goodrich JK Maxan ME, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2016;65:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imhann F Bonder MJ Vich Vila A, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mei QX Huang CL Luo SZ, et al. Characterization of the duodenal bacterial microbiota in patients with pancreatic head cancer vs. healthy controls. Pancreatology. 2018;18:438–445. [DOI] [PubMed] [Google Scholar]
- 32.Ou G Hedberg M Hörstedt P, et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am J Gastroenterol. 2009;104:3058–3067. [DOI] [PubMed] [Google Scholar]
- 33.Von Hoff DD Ervin T Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unno M Motoi F Matsuyama Y, et al. Randomized phase II/III trial of neoadjuvant chemotherapy with gemcitabine and S-1 versus upfront surgery for resectable pancreatic cancer (Prep-02/JSAP-05). J Clin Oncol. 2019;37(4 suppl):189 .abstract. [DOI] [PubMed] [Google Scholar]
- 35.Nakano S Komatsu Y Kawamoto Y, et al. Association between the use of antibiotics and efficacy of gemcitabine plus nab-paclitaxel in advanced pancreatic cancer. Medicine (Baltimore). 2020;99:e22250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunakawa Y Arai H Izawa N, et al. Antibiotics may enhance the efficacy of gemcitabine treatment for advanced pancreatic cancer. Ann Oncol. 2018;29(suppl 8):viii205–viii270 .abstract 738P. [Google Scholar]
- 37.Imai H Saijo K Komine K, et al. Antibiotic therapy augments the efficacy of gemcitabine-containing regimens for advanced cancer: a retrospective study. Cancer Manag Res. 2019;11:7953–7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masi AC Oppong YEA Haugk B, et al. Endoscopic ultrasound (EUS)-guided fine needle biopsy (FNB) formalin fixed paraffin-embedded (FFPE) pancreatic tissue samples are a potential resource for microbiota analysis. Gut. 2021;70:999–1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


