Abstract
Recent biochemical, biophysical, and genetic studies have shown that heparan sulfate, a major component of the cellular glycocalyx, participates in infection of SARS-CoV-2 by facilitating the so-called open conformation of the spike protein, which is required for binding to ACE2. This review highlights the involvement of heparan sulfate in the SARS-CoV-2 infection cycle and argues that there is a high degree of coordination between host cell heparan sulfate and asparagine-linked glycans on the spike in enabling ACE2 binding and subsequent infection. The discovery that spike protein binding and infection depends on both viral and host glycans provides insights into the evolution, spread and potential therapies for SARS-CoV-2 and its variants.
Graphical abstract
Introduction
SARS-CoV-2 is a single-stranded RNA, positive-sense, enveloped virus that primarily targets the respiratory epithelial cells lining the upper and lower airways with evidence of spread to other organs. The spike (S) protein studs the outer membrane of the virus and mediates the initial steps of viral infection and spread. It acts by engaging cellular host receptors, such as angiotensin-converting enzyme 2 (ACE2) and neuropilin-1 (NRP1), and it undergoes processing via the transmembrane protease serine 2 (TMPRSS2) or other proteases [1]. Additionally, infection is facilitated by heparan sulfate (HS), a long negatively charged polysaccharide expressed by all animal cells [2, ∗∗3, 4, ∗∗5]. Here we review current literature demonstrating the involvement of HS in SARS-CoV-2 infection. We discuss the mechanism of action from a structural perspective and explore the coordination between HS binding and asparagine-linked glycans.
The glycocalyx and viral infection
All cells are surrounded by a glycocalyx composed of various types of glycoconjugates, including asparagine-linked (N-linked) and serine/threonine-linked (O-linked) glycoproteins, glycosphingolipids, and proteoglycans. Given their location at the interface between cells and the extracellular milieu, it is not surprising that many viruses exploit glycoconjugates as attachment factors and receptors for infection, including influenza virus, herpes simplex virus (HSV), human immunodeficiency virus (HIV), and coronaviruses (SARS-CoV and MERS) [6, 7, 8, 9]. Often the dependence of viral attachment on host glycans is obscured in vitro by the high–affinity interaction with protein receptors, for example ACE2 for HCoV-NL63 and HVEM for HSV, which may be more exposed in cultured cells compared to their exposure in host tissues, such as the lung [10,11]. In fact, MERS binding to sialic acids was only described years after the discovery of its interaction with dipeptidyl peptidase 4 (DPP4) [12]. Glycan-binding may act as the initial step for cellular attachment, bringing the virus close to the epithelial cell membrane where it can interact with its protein receptor(s) (Figure 1 ). This general mechanism appears conserved among coronaviruses (CoVs) such as MERS, SARS-CoV, and HCoV-NL63, which are related to SARS-CoV-2 and bind via the viral spike protein to HS and other glycans [12, 13, 14, 15, 16]. The conservation of this mechanism across CoVs suggests that glycan attachment plays a role in CoV infectivity. Unfortunately, the glycocalyx is often ignored in studies of viral infection that focus only on protein receptors. This review focuses on the participation of cell surface HS and spike protein glycans in the interaction of the SARS-CoV-2 spike protein with cells and infection.
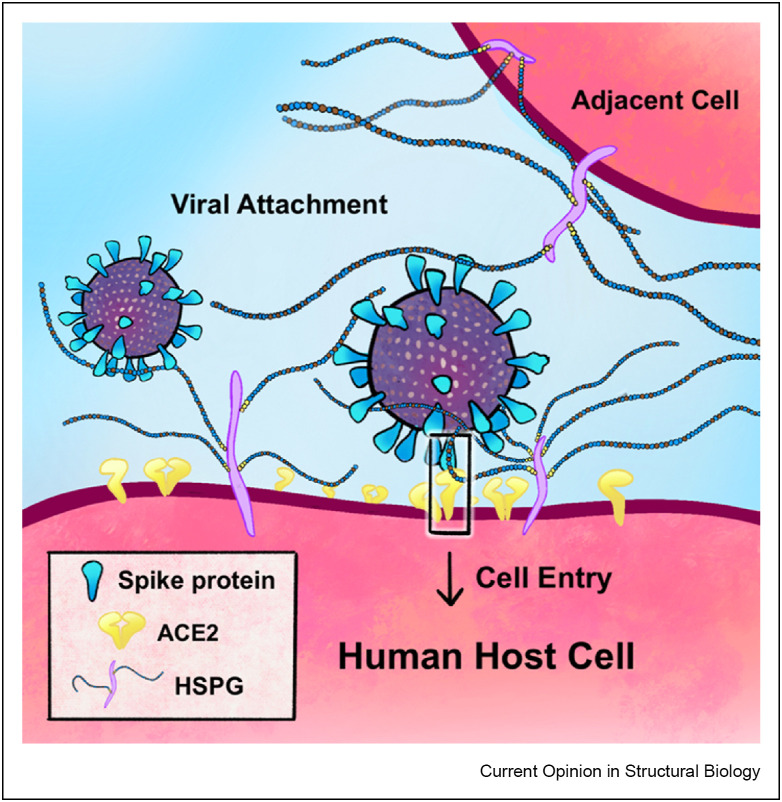
Figure 1.
Cartoon illustration depicting SARS-CoV-2 viral attachment on a host cell via initial binding to heparan sulfate proteoglycans (HSPGs), followed by ternary complex formation between SARS-CoV-2 spike protein, HSPG and ACE2 (highlighted with a rectangular box).
Structure of heparan sulfate
HS is a negatively charged, linear polysaccharide that assembles while attached to one or more of the 17 known transmembrane, GPI-anchored, or secreted proteoglycans (Figure 2 ). These proteoglycans are expressed in a tissue- and cell-type specific manner. HS is assembled by the addition of alternating residues of N-acetyl-d-glucosamine (GlcNAc) and d-glucuronic acid (GlcA) to a primer tetrasaccharide attached to the proteoglycan core protein. During polymerization, the chains undergo processing reactions in which subsets of N-acetyl-d-glucosamine residues undergo N-deacetylation and N-sulfation; adjacent glucuronic acid units undergo epimerization at C5 to alpha-l-iduronic acid (IdoA); and ester-linked sulfate groups are installed at C2 of the uronic acids, at C6 of the glucosamine residues, and occasionally at C3 of the glucosamine residues. These modification reactions do not go to completion, giving rise to domains with variably sulfated sugars and different contents of iduronic acids interspersed by domains lacking all or most of these modifications. The variable length of the sulfated domains, their pattern of sulfation, and their spacing create unique motifs to which a variety of HS-binding proteins interact. These motifs are expressed differentially across cells and tissues, allowing for modulation of specific cellular functions [17]. Protein interactions with HS are driven largely by electrostatic interactions of the sulfate and carboxyl groups in these motifs with positively charged amino acids in the HS-binding domains of HS binding proteins and by hydrogen bonding. The variation in structure of HS across different tissues and cell types may impact tissue tropism of viruses and other pathogens [18]. Importantly, the structural features of HS are well conserved between species and thus may play a role in zoonotic transmission of viruses. Heparin, a fractionated form of tissue HS isolated from bovine and porcine entrails, is highly sulfated and iduronic acid-rich, binds to antithrombin and acts as a potent anticoagulant. Heparin is available commercially and is often used as a surrogate for HS, which has only recently become available through scientific vendors.
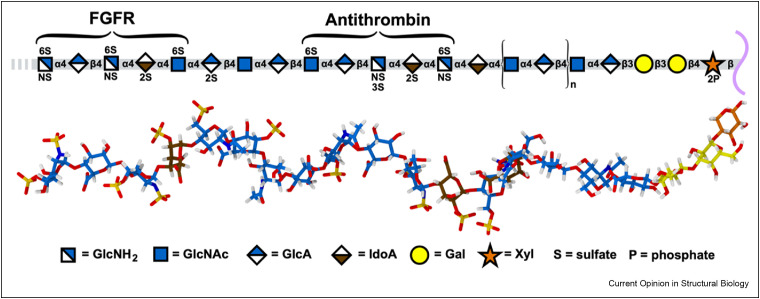
Figure 2.
Representative example of heparan sulfate (HS) structure, sulfation pattern and 3D spatial geometry. Top panel: The SNFG representation of an HS 18-mer connected to the tetrasaccharide (glucuronic acid-galactose-galactose-xylose) linker that is attached to HSPG core protein (HSPG core protein shown in purple) [19, 19]. A potential binding site for a fibroblast growth factor receptor and the high affinity binding site for antithrombin is indicated. Bottom panel: a molecular model, shown in stick form and in a biologically relevant conformation, reflecting the oligosaccharide in the top panel.
HS enhances SARS-CoV-2 infection
The impact of HS on SARS-CoV-2 infection has been demonstrated in vitro by different approaches: (i) genetic and enzymatic manipulation of cell surface HS [3,5]; (ii) molecular and biophysical interrogation of spike protein binding to HS [2,3,20,21]; (iii) structural characterization of putative HS binding sites in the spike protein as well as putative binding modes [3]; (iv) pharmacological intervention by inhibition of HS biosynthesis and competition by heparin and HS mimetics [3,5,21,22]; and (v) non-biased genome-wide CRISPR-Cas9 knockout screens aimed at the identification of critical host factors for infection [23, 24, 25, 25, 26]. HS enhances binding of recombinant SARS-CoV-2 receptor binding domain (RBD) and recombinant full-length trimeric spike protein to cells, as well as infection by SARS-CoV-2 spike pseudotyped viruses and authentic SARS-CoV-2 and other seasonal coronaviruses to cells [3,24,25]. Comparable in vivo studies have not yet been performed, but the ubiquitous distribution of HS across tissues suggests that attachment to HS and possibly other glycans in the glycocalyx helps facilitate infection.
Clausen et al. showed that spike protein trimers bind to heparin and to more relevant cell surface HS. Binding to HS and ACE2 occurs in a cooperative manner [3]. Negative stain-electron transmission microscopy and image analysis showed that the binding of recombinant trimeric spike protein to HS enhances the conversion to the “up RBD” or “open spike,” which is required for binding to ACE2 [3]. Moreover, binding studies using spike protein mutated to stabilize either the RBD “closed” or the RBD “open” confirmation showed that both bound to HS with similar affinities, but only the RBD “open” form bound to ACE2 [3]. These findings suggest a model in which the spike protein initially interacts with cell surface HS, resulting in an increase in the number of RBDs in the “up/open” conformation, which in turn leads to enhanced binding to ACE2 receptors and subsequent stabilization of the interaction of the trimer and the virion with cells [3]. When cells were genetically depleted of HS by CRISPR-Cas9 knock-out of the HS biosynthetic machinery, recombinant spike protein was significantly impaired in binding to the cell surface [3]. Similarly, a decrease in SARS-CoV-2 binding to the cell surface occurred when ACE2 was genetically depleted, and the residual ACE2-independent binding was shown to depend on HS. Other studies have confirmed that the spike protein can engage cells independently of ACE2 expression, although the efficiency of engagement was diminished [3,27,28]. Infection may occur independently of ACE2, possibly through micropinocytosis mediated by HSPG internalization [29]. These findings suggest that HS captures SARS-CoV-2 virions, increasing the local viral concentration and presentation of these virions to ACE2 and other proteinaceous receptors [27]. The interaction of virus with HS may facilitate tangential spread of viral progeny across tissues, even to tissues that do not express ACE2. In addition, HS is expressed across phylogeny, suggesting that the virus could, in a similar way, exploit HS in other susceptible animals, including bats, pangolins, mink, and others. The biochemical and cell culture-based evidence for the involvement of HS in SARS-CoV-2 infection is compelling, but additional experiments are needed to establish the involvement of HS in animal models.
Model of engagement of spike protein with heparan sulfate
Several groups have utilized computational methods to identify sites in the spike protein that engage HS [3,20,21]. These studies each identified different polybasic regions, consistent with the architecture of HS binding sites [17]. Taking a more holistic approach and looking at these proposed sites in sequence, suggests the presence of a long polyanion binding site starting at the RBD, running down between the RBD and the N-terminal domain (NTD), and down to the polybasic furin cleavage site (FCS) (Figure 3 a–d) [3,20,21,30]. Electrostatic potential maps of the SARS-CoV-2 spike surface indeed show that this proposed polyanion binding site is a positively charged channel connecting the RBD and FCS (Figure 3c). Furthermore, due to its trimeric nature, the native spike protein exhibits three of these polyanion binding sites creating the potential for highly flexible and multivalent spike–HS binding modes (Figure 3a–b). Interestingly, the loss of an N-linked glycan at position N370 in SARS-CoV spike leaves a vacant binding site on the RBD primed to receive another oligosaccharide. This idea aligns with data suggesting that the proposed HS binding region in the SARS-CoV RBD carries less positive charge compared to the same site in the SARS-CoV-2 RBD, corresponding to reduced binding affinity to heparin/HS [3].
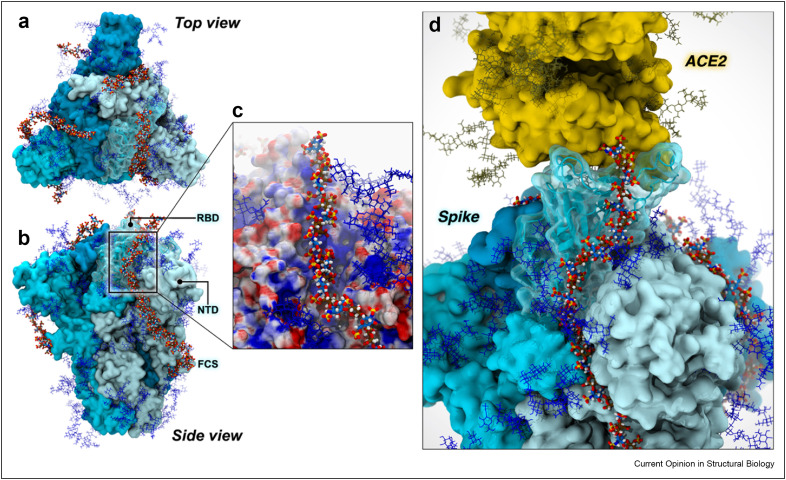
Figure 3.
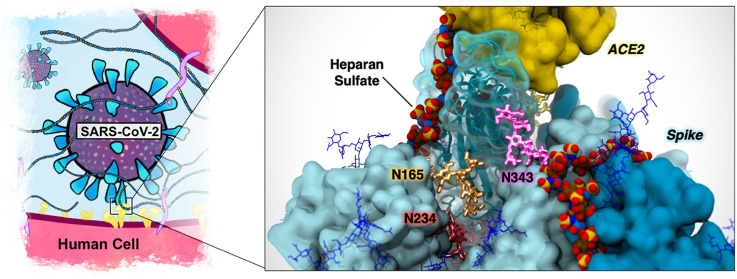
Spike/heparan sulfate binding. ACE2 and the SARS-CoV-2 spike protein are displayed with yellow and cyan surfaces, respectively. The RBD is depicted with a transparent cyan surface. Spike protein and ACE2 are surrounded by a glycan shield generated by N-linked glycans illustrated with blue and dark gold sticks, respectively. HS is depicted with a per-atom-colored, stick representation: oxygen, red; hydrogen, white; nitrogen, blue; sulfur, yellow. Carbon atoms of N-acetyl-d-glucosamine residues are colored in blue, and carbon atoms of l-iduronic acid residues are colored in brown. a–b, Top and side view of the SARS-CoV-2 spike protein bound to HS. c, Electrostatic potential projected onto the RBD HS-binding patch with heparin bound. The surface is colored from red (negative) to blue (positive), representing electrostatic potential values of −4 kbT/e to +4 kbT/e. d, HS contributes to the formation of a ternary complex with the SARS-CoV-2 spike and ACE2.
This putative HS-binding site on the spike protein surface is maintained in the rapidly emerging SARS-CoV-2 variants of concern (VOCs), despite the multiple mutations that have occurred relative to the original circulating strain. In fact, mutations in the Delta (B.1.617.2) variant's spike protein sequence has resulted in three additional positively charged amino acids along the putative HS binding groove [31,32]. The most recent variant of concern, Omicron (B.1.1.529), was first identified by S gene dropout in PCR tests due to the more than 30 mutations per protomeric chain [33]. As in the case of the Delta variant spike, many of these mutations result in an overall increase in positive charge in the spike protein and four such mutations (mutations resulting in an increase in positive charge) lie along the putative HS-binding groove [34]. These observations suggest that the more infectious variants of SARS-CoV-2 have increased their potential for viral spread, possibly by increasing their affinity for HS. Additional modeling as well as biochemical studies are needed to confirm this hypothesis.
Coordination of heparan sulfate binding with spike N-linked glycans
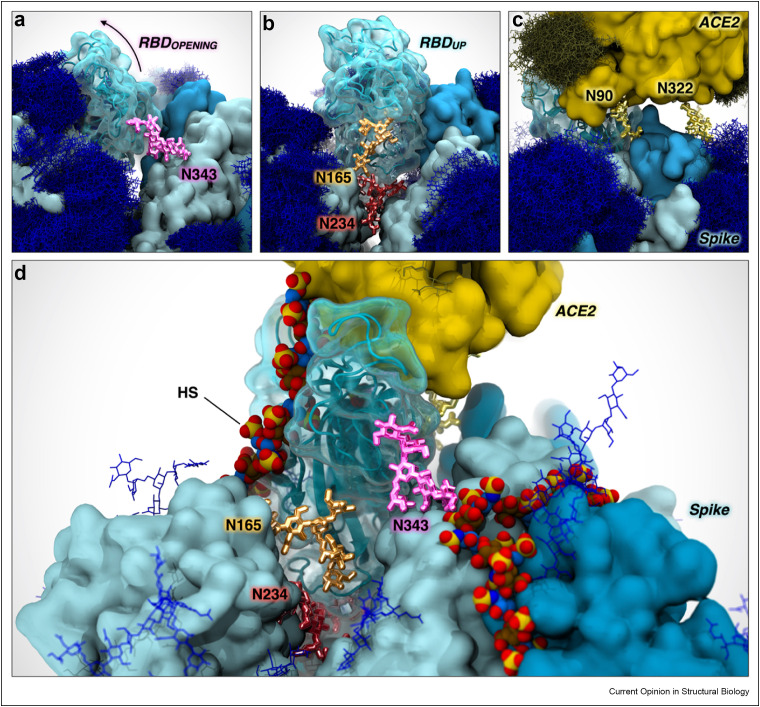
The spike protein contains 22 N-linked glycans per protomer, forming a glycan shield around much of the protein and masking antigenic sites from the immune system. The shield arises from the intrinsic flexibility of N-glycans due to rotations around the glycosidic bonds (phi, psi, and omega angles) which, when modeled over time, gives the spike a characteristic “furry” look (Figure 4 a–c) [35]. However, recent biophysical studies have demonstrated that the N-linked glycans on spike are also part of the weaponry utilized by the virus to infect host cells, exhibiting functional roles that go beyond shielding. Extensive weighted ensemble simulations of the SARS-CoV-2 spike protein revealed that the glycan at position N343 acts like a “molecular crowbar” to facilitate the RBD's down-to-up (closed-to-open) conformational change by wedging and pushing several RBD residues along the opening pathway [36]. When the spike is in the open state, it transitions to an “attacking” mode, where the Receptor Binding Motif (RBM) emerges from the glycan shield to bind to ACE2 [36]. Molecular dynamics simulations of the open spike protein revealed that glycans linked to N165 and N234 act as “kickstands” to lock the RBD in the up conformation, priming this sophisticated machinery for ACE2 binding [35]. This observation is the first to demonstrate a functional, active role of glycans in the context of viral host-cell attachment and infection. Although the N-linked glycans on spike have been shown to help drive the RBD conformational change to the “up” position, HS may also facilitate this process. During the RBD opening pathway, N165 interacts significantly with the RBD, engaging it as it moves up; in the final steps of this pathway, N165 swings under the RBD, taking its position beside N234 [37]. The HS binding patch on the spike RBD [2,3,20,21,32,34] overlaps significantly with the RBD binding site for N165 [35,36]. Thus, HS has the potential to replace N165 in this role, thereby modulating the well-timed RBD opening at the glycocalyx. Additionally, the handshake between the RBD and ACE2 is stabilized by two N-linked glycans at position N90 and N322 of ACE2 (Figure 4c) [38,39]. Interestingly, the N-glycan at N546 has also been shown to stabilize spike-ACE2 interactions via glycan–glycan interactions [39]. HS can also participate in this stabilization; as can be seen in Figure 4d, a ternary complex could easily be accommodated between the spike, ACE2 and HS.
Figure 4.
Interplay between N-glycans and heparan sulfate in priming the spike for infection. In all panels, ACE2 and the SARS-CoV-2 spike protein are displayed with yellow and cyan surfaces, respectively. The RBD is depicted with a transparent cyan surface. Glycans contributing to the glycan shield are illustrated with blue sticks. Hydrogen atoms have been hidden for clarity. a, The N-glycan at N343, highlighted in magenta, facilitates RBD opening, acting as a molecular crowbar. b, N-glycans at N165 and N234, depicted with orange and red sticks, respectively, stabilize the RBD in the “up” state, locking-and-loading the RBD for infection. c, N-glycans at N90 and N322 of ACE2, highlighted with yellow sticks, stabilize ACE2-RBD binding. d, HS, represented with per-atom-colored space filling spheres, modulate RBD opening and stabilization together with N-linked glycans (oxygen, red; hydrogen, white; nitrogen, blue; sulfur, yellow; carbons of N-acetyl-d-glucosamine residues, blue; and carbons of l-iduronic acid residues, brown).
Open questions
The discovery of HS as a key factor in SARS-CoV-2 infection raises many interesting questions. What are the molecular details of the SARS-CoV-2 binding and infection mechanism? How is the complex interplay between HS adhesion, spike N-glycans and ACE2 binding coordinated? If the virus binds to the glycocalyx, how does it escape its grip to spread across tissues? Do the additional positively charged amino acid residues present in Delta and Omicron enhance binding to HS and if so, does this enhance transmission? If HS binding increases viral fitness, is it then a driving factor in the emergence of novel variants? Is it also a factor important for zoonotic transmission?
There is conflicting data concerning the accessibility and abundance of HS in the lung. Studies utilizing the anti-HS mAb 10E4 in immunohistochemistry have suggested that lung tissue and monolayers of human airway epithelial cells grown at an air-liquid interface may not express abundant HS on the apical epithelium [40,41]. However, studies utilizing other fixation methods [42] or mAb 3G10 indicate that HS is expressed on lung apical epithelium [43,44]. These articles illustrate an important issue about methodology in determining HS accessibility and structure, which could potentially affect tissue tropism.
The trimeric spike protein can potentially bind to three HS chains and coupled with the presence of ∼24 spike trimers/virion [45], the avidity of the interaction between HS and the virion must be extremely high. Typical HSPGs contain more than one HS chain, but whether a single proteoglycan can act as a scaffold for docking a trimeric spike protein is unclear. Most cells express more than one type of HSPG, raising the question whether SARS-CoV-2 prefers specific HSPGs. The differential expression of the HSPGs across cell types is relevant because it could explain in part the tissue tropism of SARS-CoV-2 [18]. Finally, HS chains have polarity, with the non-reducing end pointing away from the core protein. Thus, engagement of spike protein with HS on a target cell would occur in a spatially organized manner, with the HS chains extending from the top of the spike towards the virion envelope. The location of the HS-binding site on the opposite side of the RBD where ACE2 binds would allow each spike protomer to form a ternary complex.
Can insights into the interaction of SARS-CoV-2 spike with HSPGs aid in the development of targeted therapies to control the spread of COVID-19 and future coronavirus outbreaks? Various types of heparin, such as unfractionated heparin, enoxaparin and split glycol heparin (lacking anticoagulant activity) were shown to block spike or RBD binding or abrogate SARS-CoV-2 infection [2,3,5,21,27,46,47]. Application of computational studies and binding studies to glycan arrays have defined unusual HS sequences with high affinity for spike protein [48]. These HS variants differ in structure and therefore vary in their capacity to block HS-spike interactions and infection by authentic virus. A polysulfated glycan analog, Pixatimod (PG545), was recently shown to potently block SARS-CoV-2 infection in a K18-hACE2 mouse model. Pixatimod markedly attenuated SARS-CoV-2 viral titer and COVID-19-like symptoms [49]. These findings strongly suggest that heparin or heparin analogs could prove efficacious for blocking infection.
Another approach is to identify small drug-like compounds that block HS formation or binding. A drug screen of FDA-approved drugs identified several inhibitors of HS-dependent endocytosis [5]. Notably, Mitoxantrone was the most potent inhibitor, almost completely blocking HS-dependent endocytosis of virus at 5 μM. Halofuginone, a prolyl tRNA synthetase inhibitor, reduces HS expression and attenuates spike binding and viral infection, in part due to inhibition of HSPGs [22]. Surfen a small molecule antagonist of HS-protein interactions, can also block spike protein binding and viral infection in vitro [50]. Clinical studies are needed now to establish the utility of these agents to block viral infection in human patients.
Conclusions
-
•
SARS-CoV-2 spike has high affinity for HS.
-
•
The SARS-CoV-2 spike protein is evolutionarily adapted to bind HS and evolution of the HS binding site may provide further advantage to recent SARS-CoV-2 variants of concern.
-
•
Modeling studies indicate that HS can bind an extended path along the spike protein surface, including regions of the receptor binding domain, the N-terminal domain, and the furin cleavage site
-
•
HS binding induces activation of the SARS-CoV-2 spike, working in concert with spike N-linked glycans, to induce the “RBD-up” or “open spike” conformation, which is required for ACE2 binding. HS can stabilize the spike-ACE2 interaction by participating in formation of a ternary complex.
-
•
Additional studies are needed to identify the relevant HSPGs that mediate binding and infection.
-
•
Anti-virals based on sulfated carbohydrates or heparin analogs have not yet been developed, but clinical trials are underway to explore this approach for treatment of SARS-CoV-2.
Credit author statement
Fiona L. Kearns: Formal analysis, Investigation methodology, Visualization, Writing - original draft, Writing - review & editing; Daniel R. Sandoval: Conceptualization, Investigation methodology, Visualization, Writing - original draft; Lorenzo Casalino: Formal analysis, Investigation methodology, Visualization, Visualization, Writing - original draft, Writing - review & editing; Thomas M. Clausen: Conceptualization, Funding acquisition, Visualization, Writing - original draft, Writing - review & editing; Mia A. Rosenfeld: Formal analysis, Investigation methodology, Visualization, Writing - original draft, Writing - review & editing; Charlotte B. Spliid: Conceptualization, Funding acquisition, Writing - original draft, Writing - review & editing; Rommie E. Amaro: Project administration, Supervision, Conceptualization, Funding acquisition, Writing - original draft, Writing - review & editing; Jeffrey D. Esko, Project administration, Supervision, Funding acquisition, Writing - original draft, Writing - review & editing.
Conflict of interest
J.D.E is a cofounder and T.M.C., and D.R.S. are consultants of Covicept Therapeutics, Inc. J.D.E. and the Regents of the University of California have licensed a university invention to and have an equity interest in TEGA Therapeutics, Inc., a vendor for heparan sulfate. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Acknowledgments
This work was supported by grants from the National Science Foundation RAPID IOS-20319 and National Institute of Health grant HL131474 (to J.D.E.), the Carlsberg Foundation (to C.B.S), National Institute of Health grant GM132826, National Science Foundation RAPID MCB 2032054, an award from the RCSA Research Corp., a UC San Diego Moore's Cancer Center 2020 SARS-CoV-2 seed grant and NIAID U19 AI171954 (to R.E.A). These sponsors were not involved in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
This review comes from a themed issue on Protein-Carbohydrate complexes and glycosylation
Edited by Sylvie Ricard-Blum and Joseph Zaia
References
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work was the defining paper describing ACE2 as a receptor for SARS-CoV-2 and that the serine protease TMPRSS2 was required for spike protein priming. A TMPRSS2 inhibitor approved for clinical use blocked entry and was suggested as a treatment option.
- 2.Liu L., Chopra P., Li X., Bouwman K.M., Tompkins S.M., Wolfert M.A., de Vries R.P., Boons G.J. Heparan sulfate proteoglycans as attachment factor for SARS-CoV-2. ACS Cent Sci. 2021;7:1009–1018. doi: 10.1021/acscentsci.1c00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study investigated the binding of the SARS-CoV-2 spike protein to cellular heparan sulfate and the impact of this interaction on spike protein mediated viral infection. Compelling genetic and biochemical data demonstrated that heparan sulfate is a required co-receptor for SARS-CoV-2 infection in addition to ACE2.
- 4.Chu H., Hu B., Huang X., Chai Y., Zhou D., Wang Y., Shuai H., Yang D., Hou Y., Zhang X., et al. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat Commun. 2021;12:134. doi: 10.1038/s41467-020-20457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Chen C.Z., Swaroop M., Xu M., Wang L., Lee J., Wang A.Q., Pradhan M., Hagen N., Chen L., et al. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020;6:80. doi: 10.1038/s41421-020-00222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that cellular heparan sulfate aids SARS-CoV-2 infection and that drugs that target heparan sulfate presentation are potential tools for SARS-CoV-2 treatment. Among the drugs characterized, Mitoxantrone was shown to be a potent HS inhibitor.
- 6.Cagno V., Tseligka E.D., Jones S.T., Tapparel C. Heparan sulfate proteoglycans and viral attachment: true receptors or adaptation bias? Viruses. 2019;11:596–620. doi: 10.3390/v11070596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stencel-Baerenwald J.E., Reiss K., Reiter D.M., Stehle T., Dermody T.S. The sweet spot: defining virus-sialic acid interactions. Nat Rev Microbiol. 2014;12:739–749. doi: 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connell B.J., Lortat-Jacob H. Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition. Front Immunol. 2013;4:385–397. doi: 10.3389/fimmu.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spear P.G., Shieh M.-T., Herold B.C., WuDunn D., Koshy T.I. In: Heparin and related polysaccharides. edn 1. Lane D.A., Lindahl U., editors. Plenum Press; 1992. Heparan sulfate glycosaminoglycans as primary cell surface receptors for herpes simplex virus; pp. 341–351. [DOI] [PubMed] [Google Scholar]
- 10.Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., DiMaio F., Bosch B.J., Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montgomery R.I., Warner M.S., Lum B.J., Spear P.G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 12.Li W., Hulswit R.J.G., Widjaja I., Raj V.S., McBride R., Peng W., Widagdo W., Tortorici M.A., van Dieren B., Lang Y., et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci U S A. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milewska A., Zarebski M., Nowak P., Stozek K., Potempa J., Pyrc K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J Virol. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milewska A., Nowak P., Owczarek K., Szczepanski A., Zarebski M., Hoang A., Berniak K., Wojarski J., Zeglen S., Baster Z., et al. Entry of human coronavirus NL63 into the cell. J Virol. 2018;92 doi: 10.1128/JVI.01933-17. e01933-01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang J., Yang N., Deng J., Liu K., Yang P., Zhang G., Jiang C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen L., McCord K.A., Bui D.T., Bouwman K.M., Kitova E.N., Elaish M., Kumawat D., Daskhan G.C., Tomris I., Han L., et al. Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2. Nat Chem Biol. 2021 doi: 10.1038/s41589-021-00924-1. [DOI] [PubMed] [Google Scholar]
- 17.Xu D., Esko J.D. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., Li Y., Liu Q., Yao Q., Wang X., Zhang H., Chen R., Ren L., Min J., Deng F., et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021;7:17. doi: 10.1038/s41421-021-00249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neelamegham S., Aoki-Kinoshita K., Bolton E., Frank M., Lisacek F., Lutteke T., O'Boyle N., Packer N.H., Stanley P., Toukach P., et al. Updates to the symbol nomenclature for glycans guidelines. Glycobiology. 2019;29:620–624. doi: 10.1093/glycob/cwz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S.Y., Jin W., Sood A., Montgomery D.W., Grant O.C., Fuster M.M., Fu L., Dordick J.S., Woods R.J., Zhang F., et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir Res. 2020;181 doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycroft-West C.J., Su D., Pagani I., Rudd T.R., Elli S., Gandhi N.S., Guimond S.E., Miller G.J., Meneghetti M.C.Z., Nader H.B., et al. Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the Spike S1 receptor-binding domain with heparin. Thromb Haemostasis. 2020;120:1700–1715. doi: 10.1055/s-0040-1721319. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that heparin and low molecular weight heparin binds and induce a conformational change in the spike protein receptor-binding domain of SARS-CoV-2. Binding to the RBD was dependent on the presence of 2-O or 6-O sulfate groups more than on N-sulfation and a hexasaccharide was the minimum size required for secondary structural changes to be induced in the RBD.
- 22.Sandoval D.R., Clausen T.M., Nora C., Cribbs A.P., Denardo A., Clark A.E., Garretson A.F., Coker J.K.C., Narayanan A., Majowicz S.A., et al. The prolyl-tRNA synthetase inhibitor halofuginone inhibits SARS-CoV-2 infection. bioRxiv. 2021 doi: 10.1101/2021.03.22.436522. [DOI] [Google Scholar]
- 23.Baggen J., Persoons L., Vanstreels E., Jansen S., Van Looveren D., Boeckx B., Geudens V., De Man J., Jochmans D., Wauters J., et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat Genet. 2021;53:435–444. doi: 10.1038/s41588-021-00805-2. [DOI] [PubMed] [Google Scholar]
- 24.Schneider W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A., et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184:120–132.e114. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184:106–119. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Israeli M., Finkel Y., Yahalom-Ronen Y., Paran N., Chitlaru T., Israeli O., Cohen-Gihon I., Aftalion M., Falach R., Rotem S., et al. Genome-wide CRISPR screens identify GATA6 as a proviral host factor for SARS-CoV-2 via modulation of ACE2. Nat Commun. 2022;13:2237. doi: 10.1038/s41467-022-29896-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bermejo-Jambrina M., Eder J., Kaptein T.M., van Hamme J.L., Helgers L.C., Vlaming K.E., Brouwer P.J.M., van Nuenen A.C., Spaargaren M., de Bree G.J., et al. Infection and transmission of SARS-CoV-2 depend on heparan sulfate proteoglycans. EMBO J. 2021;40 doi: 10.15252/embj.2020106765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puray-Chavez M., LaPak K.M., Schrank T.P., Elliott J.L., Bhatt D.P., Agajanian M.J., Jasuja R., Lawson D.Q., Davis K., Rothlauf P.W., et al. Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109364. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifed a lung adenocarcinoma cell line that did not express ACE2, but was infectable with SARS-CoV-2. They showed that these cells were infected through viral interaction with heparan sulfate, suggesting that heparan sulfate alone can support SARS-CoV-2 infection.
- 29.Chen K., Williams K.J. Molecular mediators for raft-dependent endocytosis of syndecan-1, a highly conserved, multifunctional receptor. J Biol Chem. 2013;288:13988–13999. doi: 10.1074/jbc.M112.444737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuurs Z.P., Hammond E., Elli S., Rudd T.R., Mycroft-West C.J., Lima M.A., Skidmore M.A., Karlsson R., Chen Y.H., Bagdonaite I., et al. Evidence of a putative glycosaminoglycan binding site on the glycosylated SARS-CoV-2 spike protein N-terminal domain. Comput Struct Biotechnol J. 2021;19:2806–2818. doi: 10.1016/j.csbj.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCallum M., Bassi J., De Marco A., Chen A., Walls A.C., Di Iulio J., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373:648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-H., Kearns F.L., Rosenfeld M.A., Casalino L., Papanikolas M.J., Simmerling C., Amaro R.E., Freeman R. GlycoGrip: cell surface-inspired universal sensor for betacoronaviruses. ACS Cent Sci. 2021 doi: 10.1021/acscentsci.1c01080. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work predicts GAG binding sites on the SARS-CoV-2 spike surface and provide mechanistic insight into how heparin or heparan sulfate may induce spike conformational changes at the cell surface. The authors present a new lateral-flow strip-based assay for capturing wildtype, Alpha, Beta, and Delta SARS-CoV-2 spike proteins.
- 33.Kannan S.R., Spratt A.N., Sharma K., Chand H.S., Byrareddy S.N., Singh K. Omicron SARS-CoV-2 variant: unique features and their impact on pre-existing antibodies. J Autoimmun. 2021;126 doi: 10.1016/j.jaut.2021.102779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiardi G., Richter S., Oreste P., Urbinati C., Rusnati M., Wade R.C. Three-fold mechanism of inhibition of SARS-CoV-2 infection by the interaction of the spike glycoprotein with heparin. J Biol Chem. 2021 doi: 10.1016/j.jbc.2021.101507. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study investigates a long chain heparin binding mode to the SARS-CoV-2 spike protein using docking and all-atom molecular dynamics simulations and reveals that long chain heparin can remain bound to spike over long timescales. They additionally propose that heparin inhibits S1/S2 cleavage by binding to positively charged patches located along the spike's head, from the RBD to the furin site.
- Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., Fogarty C.A., Barros E.P., Taylor B.C., McLellan J.S., et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study highlights the roles of glycans in the SARS-CoV-2 spike protein. Using all-atom molecular dynamics simulations, the authors characterize the glycan shield that allows the spike to escape the immune response. Beyond shielding, simulations and experimental studies reveal the functional role of N-linked glycans N165 and N234 in stabilizing the “up” RBD, priming the spike for infection.
- Sztain T., Ahn S.H., Bogetti A.T., Casalino L., Goldsmith J.A., Seitz E., McCool R.S., Kearns F.L., Acosta-Reyes F., Maji S., et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat Chem. 2021;13:963–968. doi: 10.1038/s41557-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reveals the opening mechanism of the RBD in the SARS-CoV-2 spike protein. Using weighted ensemble molecular dynamics simulations, the authors characterize the closed-to-open transition of the RBD and provide computational and experimental evidence of the key role of the N-linked glycan N343 in facilitating RBD opening.
- Harbison A.M., Fogarty C.A., Phung T., Satheesan A., Schulz B.L., Fadda E. Fine-tuning the Spike: role of the nature and topology of the glycan shield in the structure and dynamics of the SARS-CoV-2 S. Chem Sci. 2021 doi: 10.1039/d1sc04832e. in press:2021.2004.2001.438036. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study emphasizes the importance of glycan composition and size in the SARS-CoV-2 spike protein. The authors focus on the N-linked glycosylation site at position 370 that, at variance with SARS-CoV, was depleted in SARS-CoV-2. A glycan at this position would interact with the adjacent RBD, lying on a cleft that has a high affinity for charged oligosaccharides, such as heparan sulfate.
- 38.Barros E.P., Casalino L., Gaieb Z., Dommer A.C., Wang Y., Fallon L., Raguette L., Belfon K., Simmerling C., Amaro R.E. The flexibility of ACE2 in the context of SARS-CoV-2 infection. Biophys J. 2021;120:1072–1084. doi: 10.1016/j.bpj.2020.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H., et al. Virus-receptor Iinteractions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601. doi: 10.1016/j.chom.2020.08.004. e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson S.M., McNally B.A., Ioannidis I., Flano E., Teng M.N., Oomens A.G., Walsh E.E., Peeples M.E. Respiratory syncytial virus uses CX3CR1 as a receptor on primary human airway epithelial cultures. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shute J.K., Solic N., Shimizu J., McConnell W., Redington A.E., Howarth P.H. Epithelial expression and release of FGF-2 from heparan sulphate binding sites in bronchial tissue in asthma. Thorax. 2004;59:557–562. doi: 10.1136/thx.2002.002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Born J., Salmivirta K., Henttinen T., Ostman N., Ishimaru T., Miyaura S., Yoshida K., Salmivirta M. Novel heparan sulfate structures revealed by monoclonal antibodies. J Biol Chem. 2005;280:20516–20523. doi: 10.1074/jbc.M502065200. [DOI] [PubMed] [Google Scholar]
- 43.Allen B.L., Filla M.S., Rapraeger A.C. Role of heparan sulfate as a tissue-specific regulator of FGF-4 and FGF receptor recognition. J Cell Biol. 2001;155:845–858. doi: 10.1083/jcb.200106075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Izvolsky K.I., Zhong L., Wei L., Yu Q., Nugent M.A., Cardoso W.V. Heparan sulfates expressed in the distal lung are required for Fgf10 binding to the epithelium and for airway branching. Am J Physiol Lung Cell Mol Physiol. 2003;285:L838–L846. doi: 10.1152/ajplung.00081.2003. [DOI] [PubMed] [Google Scholar]
- 45.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tandon R., Sharp J.S., Zhang F., Pomin V.H., Ashpole N.M., Mitra D., McCandless M.G., Jin W., Liu H., Sharma P., et al. Effective Inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J Virol. 2021;95 doi: 10.1128/JVI.01987-20. e01987-01920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiwari V., Beer J.C., Sankaranarayanan N.V., Swanson-Mungerson M., Desai U.R. Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov Today. 2020;25:1535–1544. doi: 10.1016/j.drudis.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chittum J.E., Sankaranarayanan N.V., O'Hara C.P., Desai U.R. On the selectivity of heparan sulfate recognition by SARS-CoV-2 spike glycoprotein. ACS Med Chem Lett. 2021;12:1710–1717. doi: 10.1021/acsmedchemlett.1c00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimond S.E., Mycroft-West C.J., Gandhi N.S., Tree J.A., Le T.T., Spalluto C.M., Humbert M.V., Buttigieg K.R., Coombes N., Elmore M.J., et al. Synthetic heparan sulfate mimetic pixatimod (PG545) potently inhibits SARS-CoV-2 by disrupting the spike–ACE2 interaction. ACS Cent Sci. 2022;8:527–545. doi: 10.1021/acscentsci.1c01293. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe a potent sulfated glycoside (pixatimod) with anti-SARS-CoV-2 activity in vitro and in mice that targets the HS–Spike protein–ACE2 axis, providing a strong rationale for clinical investigation of pixatimod and other heparin analogs as a potential therapeutic for COVID-19
- 50.Yue J., Jin W., Yang H., Faulkner J., Song X., Qiu H., Teng M., Azadi P., Zhang F., Linhardt R.J., et al. Heparan sulfate facilitates spike protein-mediated SARS-CoV-2 host cell invasion and contributes to increased infection of SARS-CoV-2 g614 mutant and in lung cancer. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.649575. [DOI] [PMC free article] [PubMed] [Google Scholar]