Abstract
The emergence of SARS-CoV-2 in December 2019 resulted in the COVID-19 pandemic. Recurring disease outbreaks repeatedly overloaded the public health sector and severely affected the global economy. We developed a candidate COVID-19 vaccine based on a recombinant Newcastle disease virus (NDV) vaccine vector, encoding a pre-fusion stabilized full-length Spike protein obtained from the original SARS-CoV-2 Wuhan isolate. Vaccination of hamsters by intra-muscular injection or intra-nasal instillation induced high neutralizing antibody responses. Intranasal challenge infection with SARS-CoV-2 strain Lelystad demonstrated that both vaccination routes provided partial protection in the upper respiratory tract, and almost complete protection in the lower respiratory tract, as measured by suppressed viral loads and absence of histological lung lesions. Activity wheel measurements demonstrated that animals vaccinated by intranasal inoculation rapidly recovered to normal activity. NDV constructs encoding the spike of SARS-CoV-2 may be attractive candidates for development of intra-nasal COVID-19 booster vaccines.
Keywords: COVID-19, SARS-CoV-2, Spike protein, NDV, Vaccine
1. Introduction
In December 2019 a cluster of patients with severe respiratory tract disease was reported in Wuhan, China. The disease was caused by a novel zoonotic coronavirus, closely related to severe acute respiratory syndrome (SARS)-coronavirus. The virus was efficiently transmitted between humans, resulting in the COVID-19 pandemic. More than two years later, several COVID-19 vaccines have been developed and authorized for use. The majority of those are based on the SARS-CoV-2 Spike protein (S), and are administered by injection. Whereas most of these vaccines are effective at preventing moderate to severe COVID-19, their effectiveness at preventing infection of the upper respiratory tract (URT) and inhibiting virus transmission is limited.
We set out to develop a candidate COVID-19 vaccine based on a live-attenuated strain of Newcastle Disease virus (NDV), the type species of the genus Orthoavulavirus of the family Paramyxoviridae [1]. NDV is an avian virus that is prevalent in all species of birds, but in wild birds usually does not cause disease. However, in commercial poultry some NDV strains cause respiratory and neurologic clinical signs known as Newcastle disease, which can lead to severe economic losses [2]. Naturally occurring lentogenic (low-virulent) NDV strains, such as LaSota, are used worldwide as live-attenuated vaccines to control Newcastle disease in poultry. The establishment of reverse genetics for NDV [2] allowed insertion of an additional transcription unit (ATU) encoding foreign proteins, thus making it possible to use recombinant NDV as vaccine vector [3]. Potential strengths include the absence of pre-existing immunity, the induction of robust humoral and cellular immune responses, and the possibility of delivery via the respiratory route [4]. Importantly, the virus grows to high titers in either fertilized chicken eggs or in FDA-approved cell lines such as Vero cells [5].
Here, we describe the generation of a recombinant NDV (rNDV) expressing a prefusion-stabilized S protein of SARS-CoV-2, and show its immunogenicity and protective capacity in a vaccination and challenge model in hamsters.
2. Methods
2.1. Cells and plasmids
DF-1 (ATCC CRL-12203) cells were cultured in DMEM supplemented with Glutamax (Gibco), antibiotics and 10% fetal calf serum (Sigma). The cDNA clone of the lentogenic NDV strain LaSota (pNDFL2) and the helper plasmids (pCIneo-NP, pCIneo-P and pCIneo-L) have been described previously [2], [6]. The sequence of the S gene of the SARS-CoV-2 Wuhan strain [7] was modified to remove the polybasic cleavage site and stabilize the protein in its pre-fusion conformation by 2P modification of the S2 unit as described elsewhere [8], rendering the S protein non-fusogenic. The human codon-optimized gene (Genscript) was introduced into the cDNA clone as additional transcription unit (ATU) between the NDV phosphoprotein (P) and matrix (M) genes by using the pGEM-PM cassette [6].
2.2. Rescue of recombinant NDV-S
DF-1 cells were infected with recombinant fowlpox virus-T7 [9] and after two hours co-transfected with pNDFL-Wuhan-SARS2-S, pCIneo-NP, pCIneo-P and pCIneo-L using X-tremeGENE™ HP DNA Transfection Reagent (Roche). After five days, the culture supernatant was harvested, filtered and subsequently inoculated into 11-day-old embryonated SPF chicken eggs. After two passages in SPF chicken eggs the allantoic fluid was harvested, clarified, aliquoted and stored at −80 °C. The stock had a titer of 3.2 × 108 TCID50/ml in DF-1 cells.
2.3. Immuno peroxidase monolayer assay
Monolayers infected with rNDV-S were washed with PBS and frozen at −20 °C. After thawing, the monolayers were fixed with 4% formaldehyde for 10 min and washed with 0.05% Tween-80 in PBS. The NDV fusion protein (NDV F) and SARS-CoV2 Spike protein were detected using mouse monoclonal antibody (8E12A8C3, generated by WBVR) and human monoclonal antibody S309 [10], respectively. After three washes (0.05% Tween-80 in PBS), Rabbit Anti-Mouse IgG HRP (Dako) or Goat Anti-Human IgG HRP (Southern Biotech) were used as secondary antibodies, and AEC (3-amino-9-ethyl-carbazole; Sigma) as substrate.
2.4. Animal study
The animal study was performed under legislation of the Dutch Central Authority for Scientific procedures on Animals (CCD license no. AVD4010020209446), the experimental plan (2020.D-0007.013) was approved by the Animal Welfare Body of Wageningen University and Research.
Twenty-four female SPF Syrian Golden hamsters (Mesocricetus auratus), strain RjHan:AURA, were obtained from Janvier (Le Genest-Saint-Isle, France). The animals were 8 weeks (juvenile) at first vaccination, and were individually housed as detailed elsewhere [11]. Animals were randomized into three groups of eight animals, and allowed an acclimatization period of 11 days. Vaccinations were performed on days 0 and 21, delivering rNDV-S (107 TCID50 per dose) by injection (group 1: intra-muscular, 100 µl), intra-nasal instillation (group 2, 100 µl, divided over 50 µl per nostril) or intra-nasal inoculation with PBS (mock control, group 3, 100 µl). Three weeks after the second vaccination all animals were challenged by intra-nasal inoculation of SARS-CoV-2 (104.5 TCID50) strain Lelystad, an early isolate containing the D614G mutation, as described [11]. As clinical readout, activity of the hamsters was monitored by introducing activity tracking wheels (Tecniplast, Buguggiate, Italy) as described [11].
All animal handlings (except if only oropharyngeal swabs had to be collected) were performed under injection anesthesia using ketamine and medetomidine, as detailed elsewhere [11]. Blood samples were collected by retro-orbital vein puncture (under anesthesia) at 0, 21 and 42 days post vaccination. Four days post challenge, four out of eight animals in each group were euthanized and necropsies, histopathological and immunohistochemical analyses were performed as described [11].
2.5. Viral load measurements
RNA was isolated from oropharyngeal swabs and quantitative PCRs for total or subgenomic SARS-CoV-2 E gene were performed as described [11]. Primers and probes used were Fw:ACAGGTACGTTAATAGTTAATAGCGT, Rev:ATATTGCAGCAGTACGCACACA, Pr:FAM-ACACTAGCCATCCTTACTGCGCTTCG-BHQ and Fw:CGATCTCTTGTAGATCTGTTCTC, Rev:ATATTGCAGCAGTACGCACACA, Pr:FAM-ACACTAGCCATCCTTACTGCGCTTCG-BHQ, respectively.
2.6. Neutralization assays
A pseudovirus neutralization assay was performed as described elsewhere [12], using SARS-CoV-2 S pseudotyped vesicular stomatitis virus bearing the firefly luciferase reporter gene in Vero E6 cells. The SARS-CoV-2 live virus neutralization assay was performed as described [11]. In the latter assay, the WHO International Standard for anti-SARS-CoV-2 immunoglobulin (human, NIBSC code: 20/136) resulted in a VN titer of 300.
2.7. Statistical analysis
Statistical analysis was performed by fitting generalized (linear) regression models (GLM) or -mixed models (GLMM) to account for repeated measures in time. The analysis was performed using the statistical package R. GLMM were fitted using the library lme4. Variables such as activity and body weight were measured daily during the course of the experiment. To account for deviation of linearity in time (weight loss/gain or increased/decreased activity), natural splines were introduced to reflect different phases in the experiment (arrival, first and second vaccination, introduction of wheels, challenge and recovery). A Bonferroni correction was applied when performing multiple comparisons.
3. Results
Recombinant NDV-S was generated by introducing an ATU encoding the prefusion-stabilized S protein of SARS-CoV-2 into the genome of the NDV vaccine strain LaSota, between the viral P and M genes (Fig. 1 A). Serial dilution of the virus, which was harvested after the second passage in embryonated chicken eggs, in chicken fibroblast cell-line DF-1 resulted in comparable expression of NDV F (Fig. 1B) and SARS-CoV-2 S (Fig. 1C).
Fig. 1.
(A) Schematic design of rNDV-S, a recombinant NDV (strain LaSota) containing an ATU between the P and M genes encoding a codon-optimized open reading frame expressing the Spike (S) protein of the Wuhan strain of SARS-CoV-2 (containing the D614G mutation). The S protein was stabilized in its pre-fusion conformation by addition of two prolines in the S2 unit and removing the polybasic cleavage site, as described elsewhere [8]. (B, C):DF-1 cells were incubated with serial dilutions of the rescued virus (10−3, 10−4,10−5 or 10-6) or with culture medium (control), followed by immunoperoxidase monolayer staining with monoclonal antibodies to the NDV fusion (F) protein (B) or the SARS-CoV-2 S protein (C).
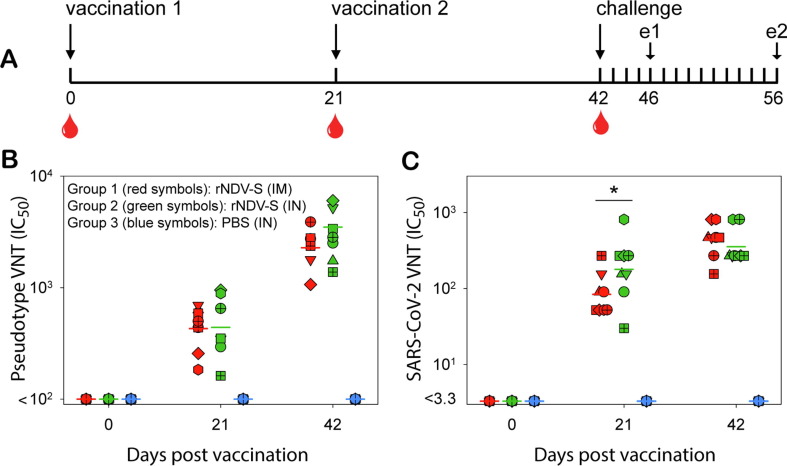
To assess the immunogenicity and protective potential of the rNDV-S construct, a vaccination and challenge study was performed in Syrian hamsters (Fig. 2 A). Two sequential vaccinations by either IM injection (group 1) or IN inoculation (group 2) induced high S-specific antibody responses as demonstrated by either a VSV-pseudovirus assay (Fig. 2B) or a SARS-CoV-2 neutralization assay (Fig. 2C). At 21 days post vaccination (DPV) levels of VN antibodies measured in the SARS-CoV-2 assay were significantly higher in animals of group 2 as compared to group 1 (p = 0.035), but no differences were observed at 42 DPV (p = 0.55).
Fig. 2.
(A) Study design of rNDV-S vaccination and challenge study in hamsters. Three groups of eight Syrian golden hamsters received two rNDV-S vaccinations (dose 107 TCID50) by intra-muscular (IM) injection (group 1), intra-nasal (IN) inoculation (group 2) or received an IN inoculation with PBS as mock control (group 3). Three weeks after the second immunization, all animals were IN inoculated with the Lelystad strain of SARS-CoV-2. Animals were euthanized four (n = 4) or fourteen (n = 4) days post challenge, indicated by e1 and e2, respectively. All animals in groups 1 and 2 developed virus neutralizing antibody levels, as measured by pseudovirus (B) or SARS-CoV-2 (C) neutralization assays. Data are presented as individual values, horizontal lines indicate geometric means. Symbol shapes and colors are used uniformly throughout the manuscript. Statistical analysis of the differences between groups 1 and 2 showed a significant difference in the titers as determined by live virus VNT in sera collected 21DPV (p = 0.035).
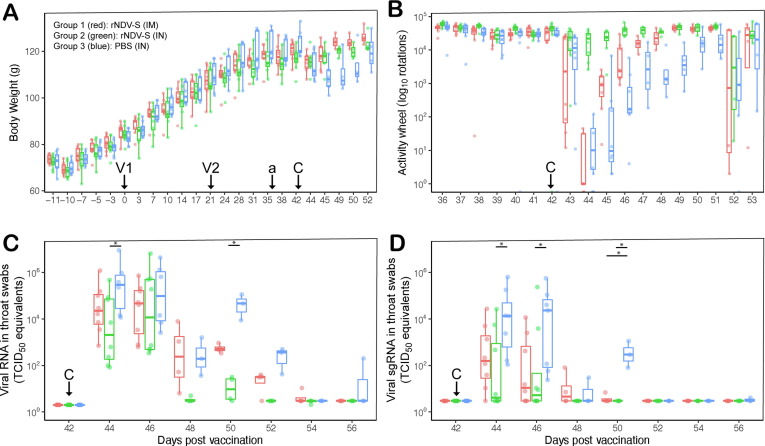
Three weeks after the second vaccination (42 DPV), all hamsters were challenged with SARS-CoV-2 strain Lelystad by intra-nasal inoculation. All animals demonstrated loss of body weight (BW), but the vaccinated animals lost less weight than the mock control animals (Fig. 3 A). In the activity measurements, animals in groups 1 returned to pre-challenge activity levels earlier than the mock controls, but animals in group 2 only showed a minor transient decrease in activity (Fig. 3B). Based on the GLMM analysis, the body weights of animals in group 3 were significantly lower than those of animals in groups 1 and 2 during the time period after challenge virus infection (42 – 52 DPV) (interaction group:time p < 0.001). Similarly, a GLMM showed that activity measurements after challenge virus infection were significantly less reduced in animals of group 2 as compared to those in groups 1 and 3. The mock control animals of group 3 showed the most substantial drop of activity, which also lasted significantly longer than in animals of groups 1 and 2 to recover (interaction group:time p < 0.001). Measurement of viral genome loads in oropharyngeal swabs detected by quantitative RT-PCR using primers targeting genomic (Fig. 3C) or subgenomic (Fig. 3D) RNA (sgRNA) showed that viral shedding was significantly more reduced in the animals of group 2 than the animals of group 3, particularly in the first four days post challenge. No significant differences were observed between groups 1 and 3 or groups 1 and 2.
Fig. 3.
(A) Body weights of hamsters were not affected by vaccination, but showed a transient drop after challenge infection, with the strongest decrease in the mock-vaccinated animals of group 3. Arrows indicate vaccinations (V1, V2), placement of activity wheel (a) or challenge infection (C). Measurements are shown as box plots. (B) Activity patterns of hamsters showed a transient drop after challenge infection, with the strongest decrease in the mock-vaccinated animals of group 3 followed by the IM vaccinated animals of group 1. Intranasally vaccinated hamsters showed the least activity reduction post challenge. SARS-CoV-2 loads in oropharyngeal swabs of hamsters as measured by RT-PCR based on primer sets directed to genomic (C) or subgenomic (D) viral RNA. Using generalized mixed model analysis, BW of group 3 was significantly lower than groups 1 and 2 during the time period 42 – 52 DPV. Activity measurements during the same time period were significantly different in group 2 as compared to groups 1 and 3, but also in group 1 as compared to group 3.
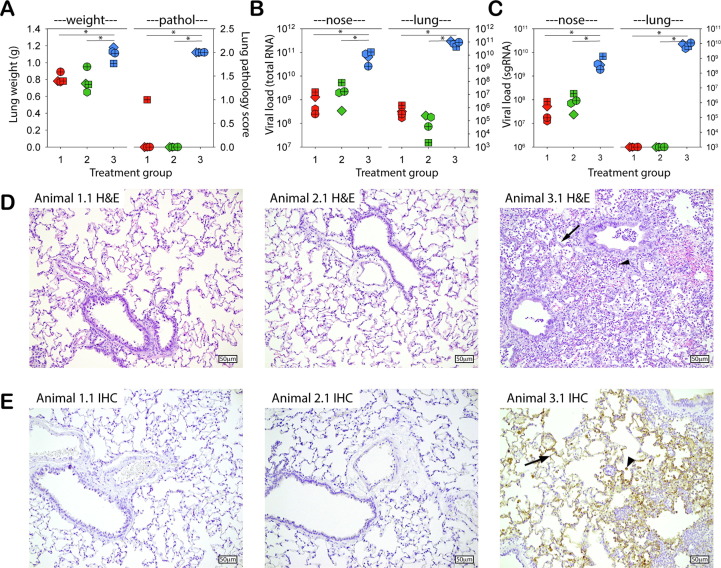
Four days post challenge, four out of eight animals in each group were euthanized and necropsies were performed. Animals in groups 1 and 2 had significantly lower lung weights and a lower macroscopic pathology score than those of the mock control animals in group 3 (Fig. 4 A). Vaccinated animals showed significantly reduced genomic RNA loads in conchae (1–2 log reduction) and lungs (5–6 log reduction), independent of vaccination route (Fig. 4B). A significant reduction of sgRNA was also detected in nasal conchae, while no sgRNA could be detected in the lungs of any of the vaccinated animals (Fig. 4C).
Fig. 4.
(A) Lung weights (left y-axis) and gross pathology score (0–4) (right y-axis) of hamsters euthanized 4 days post challenge infection (4 DPI). Data are shown as individual values of the animals euthanized 4 DPI. (B, C): SARS-CoV-2 genome loads in nasal conchae (left) or lungs suspensions (right) of hamsters euthanized 4 DPI, as measured by RT-PCR based on primer sets directed to genomic (B) or subgenomic (C) viral RNA. (D): Lung histopathology at DPI 4 (Hematoxylin and eosin (H&E) stain) of representative animals in group 1 (animal 1.1) and 2 (animal 2.1) without SARS-CoV-2 related histopathology and group 3 (animal 3.1) with thickened alveolar walls (arrow), increase in inflammatory cells (arrowhead) and type II pneumocyte proliferation, objective 20x. (E): Immunohistochemical (IHC) stain of the lungs of the same animals shown in panel (D), using an antibody to SARS-CoV-2; no expression of SARS-CoV-2 protein was found in group 1 and 2; group 3 showed extensive positive staining in pneumocytes (arrow), desquamated pneumocytes and/or alveolar macrophages (arrowhead), objective 20x. Color codes are identical to other figures: red symbols represent group 1 (rNDV-S, IM), green symbols group 2 (rNDV-S, IN) and blue symbols group 3 (PBS, IN).
Histopathological analysis showed extensive thickening of alveolar walls, type II pneumocyte proliferation and inflammatory cells in the alveoli of animals in group 3, representing interstitial pneumonia (see example in Fig. 4D). Immunohistochemical staining demonstrated widespread presence of cells expressing SARS-CoV-2 S protein (see example in Fig. 4E). In contrast, H&E staining of the lungs of the animals in groups 1 and 2 showed no histological lesions and IHC showed no evidence of cells expressing SARS-CoV-2 S (see examples in Fig. 4D and E).
4. Discussion
In this study, we have generated a live-attenuated recombinant NDV expressing the SARS-CoV-2 S protein stabilized in its prefusion conformation (rNDV-S). The candidate COVID-19 vaccine was shown to induce S expression in cell culture, and was subsequently used in a vaccination and challenge study in hamsters. We directly compared vaccination by IM injection or IN instillation, which both induced high VN antibody responses. Three weeks after the second vaccination all animals were IN inoculated with SARS-CoV-2, and a partial protection in the URT and almost complete protection in the LRT was observed.
Despite the availability of safe and effective COVID-19 vaccines, there is still need for stable and low-cost vaccines that can inhibit transmission of SARS-CoV-2 and provide improved mucosal protection. NDV is an attractive vector [3], and can be produced in fertilized chicken eggs or cell culture [5]. NDV has previously been used to express the S protein of SARS-CoV [4], and already in 2020 generation of an NDV-based COVID-19 vaccine was described [13], [14], [15]. Two formulations were developed: one as live-attenuated and one as inactivated candidate COVID-19 vaccine. More recently, others have also reported immunogenicity and protective capacity of recombinant NDV-S vaccine candidates [16], [17], [18].
Our studies were performed in parallel, and confirm published immunogenicity and protective efficacy of recombinant NDV-S-based candidate COVID-19 vaccines. The major novelty of our results is that we show that the IN route of inoculation resulted in superior protection against disease, as measured by activity in a running wheel. However, it should be noted that IN inoculation of 100 µl per hamster results in both URT and LRT deposition. A study in non-human primates demonstrated that LRT delivery of the vaccine was crucial for immunogenicity [19], and this was recently corroborated in a human clinical trial that included both IN and IM delivery as vaccination routes [20]. The vaccine showed a good safety profile in humans, but required relatively high dosages for adequate immunogenicity [20].
Recombinant NDV-based COVID-19 vaccines may be attractive candidates for development of low-cost booster vaccines.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Bregtje Smit, Vera Nijman, Tony Smits, José Harders-Westerveen, Joukje Siebenga, Lisette Ruuls, Matthijn de Boer, Rianka Vloet, Sophie van Oort, Tjeerd van Dijk, Tom van Aalst and all members of the WBVR Department of Experimental Animal Research (DEAR) for their contributions to this study. This project was funded by Health ∼ Holland, Top Sector Life Sciences & Health (https://www.health-holland.com, project number LSHM20059). The authors declare no conflict of interest.
References
- 1.Rima B., Balkema-Buschmann A., Dundon W.G., Duprex P., Easton A., Fouchier R., et al. ICTV virus taxonomy profile: paramyxoviridae. J Gen Virol. 2019;100(12):1593–1594. doi: 10.1099/jgv.0.001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peeters B.P.H., de Leeuw O.S., Koch G., Gielkens A.L.J. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J Virol. 1999;73(6):5001–5009. doi: 10.1128/jvi.73.6.5001-5009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bello M.B., Yusoff K., Ideris A., Hair-Bejo M., Jibril A.H., Peeters B.P.H., et al. Exploring the prospects of engineered newcastle disease virus in modern vaccinology. Viruses. 2020;12(4):451. doi: 10.3390/v12040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiNapoli J.M., Kotelkin A., Yang L., Elankumaran S., Murphy B.R., Samal S.K., et al. Newcastle disease virus, a host range-restricted virus, as a vaccine vector for intranasal immunization against emerging pathogens. Proc Natl Acad Sci U S A. 2007;104(23):9788–9793. doi: 10.1073/pnas.0703584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fulber J.P.C., Farnós O., Kiesslich S., Yang Z., Dash S., Susta L., et al. Process development for newcastle disease virus-vectored vaccines in serum-free vero cell suspension cultures. Vaccines (Basel) 2021;9(11):1335. doi: 10.3390/vaccines9111335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kortekaas J., Dekker A., de Boer S.M., Weerdmeester K., Vloet R.P.M., Wit A.A.C.d., et al. Intramuscular inoculation of calves with an experimental Newcastle disease virus-based vector vaccine elicits neutralizing antibodies against Rift Valley fever virus. Vaccine. 2010;28(11):2271–2276. doi: 10.1016/j.vaccine.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broer R., Boson B., Spaan W., Cosset F.-L., Corver J. Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry. J Virol. 2006;80(3):1302–1310. doi: 10.1128/JVI.80.3.1302-1310.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britton P., Green P., Kottier S., Mawditt K.L., Penzes Z., Cavanagh D., et al. Expression of bacteriophage T7 RNA polymerase in avian and mammalian cells by a recombinant fowlpox virus. J Gen Virol. 1996;77(5):963–967. doi: 10.1099/0022-1317-77-5-963. [DOI] [PubMed] [Google Scholar]
- 10.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 11.Gerhards N.M., Cornelissen J.B.W.J., van Keulen L.J.M., Harders-Westerveen J., Vloet R., Smid B., et al. Predictive value of precision-cut lung slices for the susceptibility of three animal species for SARS-CoV-2 and validation in a refined hamster model. Pathogens. 2021;10(7):824. doi: 10.3390/pathogens10070824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun W., Leist S.R., McCroskery S., Liu Y., Slamanig S., Oliva J., et al. Newcastle disease virus (NDV) expressing the spike protein of SARS-CoV-2 as a live virus vaccine candidate. EBioMedicine. 2020;62:103132. doi: 10.1016/j.ebiom.2020.103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun W., McCroskery S., Liu W.-C., Leist S.R., Liu Y., Albrecht R.A., et al. A Newcastle disease virus (NDV) expressing a membrane-anchored spike as a cost-effective inactivated SARS-CoV-2 vaccine. Vaccines (Basel) 2020;8(4):771. doi: 10.3390/vaccines8040771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun W., Liu Y., Amanat F., González-Domínguez I., McCroskery S., Slamanig S., et al. A Newcastle disease virus expressing a stabilized spike protein of SARS-CoV-2 induces protective immune responses. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-26499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y., Shi W., Abiona O.M., Nazzari A., Olia A.S., Ou L.i., et al. Newcastle disease virus-like particles displaying prefusion-stabilized SARS-CoV-2 spikes elicit potent neutralizing responses. Vaccines (Basel) 2021;9(2):73. doi: 10.3390/vaccines9020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park J.-G., Oladunni F.S., Rohaim M.A., Whittingham-Dowd J., Tollitt J., Hodges M.D.J., et al. Immunogenicity and protective efficacy of an intranasal live-attenuated vaccine against SARS-CoV-2. iScience. 2021;24(9):102941. doi: 10.1016/j.isci.2021.102941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warner B.M., Santry L.A., Leacy A., Chan M., Pham P.H., Vendramelli R., et al. Intranasal vaccination with a Newcastle disease virus-vectored vaccine protects hamsters from SARS-CoV-2 infection and disease. iScience. 2021;24(11):103219. doi: 10.1016/j.isci.2021.103219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiNapoli J.M., Yang L., Samal S.K., Murphy B.R., Collins P.L., Bukreyev A. Respiratory tract immunization of non-human primates with a Newcastle disease virus-vectored vaccine candidate against Ebola virus elicits a neutralizing antibody response. Vaccine. 2010;29(1):17–25. doi: 10.1016/j.vaccine.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponce-de-Leon S, Torres M, Soto-Ramirez LE, Jose Calva J, Santillan-Doherty P, Carranza-Salazar DE, et al. Safety and immunogenicity of a live recombinant Newcastle disease virus-based COVID-19 vaccine (Patria) administered via the intramuscular or intranasal route: Interim results of a non-randomized open label phase I trial in Mexico. medRxiv. 2022. DOI: 10.1101/2022.02.08.22270676.