Abstract
Physical cues in the extracellular microenvironment regulate cancer cell metastasis. Functional microRNA (miRNA) carried by cancer derived exosomes play a critical role in extracellular communication between cells and the extracellular microenvironment. However, little is known about the role of exosomes loaded miRNAs in the mechanical force transmission between cancer cells and extracellular microenvironment. Herein, our results suggest that stiff extracellular matrix (ECM) induced exosomes promote cancer cell migration. The ECM mechanical force regulated the exosome miRNA cargo of prostate cancer cells. Exosome miRNAs regulated by the ECM mechanical force modulated cancer cell metastasis by regulating cell motility, ECM remodeling and the interaction between cancer cells and nerves. Focal adhesion kinase mediated-ECM mechanical force regulated the intracellular miRNA expression, and F-actin mediate-ECM mechanical force regulated miRNA packaging into exosomes. The above results demonstrated that the exosome miRNA cargo promoted cancer metastasis by transmitting the ECM mechanical force. The ECM mechanical force may play multiple roles in maintaining the microenvironment of cancer metastasis through the exosome miRNA cargo.
Keywords: Extracellular environment mechanical forces, Exosomes, Prostate cancer, Cancer metastasis, MicroRNA
Highlights
-
•
ECM mechanical force-induced exosomes regulate cancer cell migration.
-
•
ECM mechanical forces regulate the cancer cell exosomes miRNA cargo.
-
•
ECM mechanical forces regulated exosomes miRNAs modulate cancer metastasis by remodeling extracellular microenvironment.
1. Introduction
Physical cues modulate cancer metastasis [1,2]. Studies have demonstrated that the mechanical force of the extracellular microenvironment plays an important role in the process of cancer cell epithelial-mesenchymal transition (EMT) [3], migration [4], and immune cell infiltration [1]. The extracellular matrix (ECM) mechanical force transmits through adhesion into the cell-ECM interface mediated by integrin, and then through the cytoskeleton into cells to regulate cell behavior by affecting the tension of the cytoskeleton [[5], [6], [7]]. Cell junctions formed into the cell-cell interface can transmit mechanical force to contact cells and regulate the behavior of neighboring cells by affecting the contact cytoskeleton [8,9]. However, the long-distance transmission of the mechanical force between non-contact cells remains unclear. Exosomes, as intercellular communication signalosomes secreted by cells [10], provide the possibility of long-distance transmission of ECM mechanical force between non-contact cells.
Previous studies have shown that cancer cell exosomes regulate cancer cell proliferation [11,12], metastasis [13], angiogenesis [14], extracellular matrix remodeling [15], immune escape [16], and drug resistance [17]. The microRNA (miRNA) in exosomes plays a key role in cancer metastasis [[18], [19], [20], [21]]. Some previous studies have demonstrated that the ECM mechanical force can regulate exosome secretion [[22], [23], [24]]. However, whether exosomes can transmit the mechanical force to regulate cell behavior via miRNA remains unclear.
Here, substrates with different stiffness were prepared as described in our previous work to simulate the mechanical force changes in the extracellular microenvironment during cancer metastasis [25]. Prostate cancer (PCa) cells were selected to discuss the possibility and mechanism of cancer cell transmission of mechanical force signals via exosome miRNA. Our results showed that substrate stiffness-induced exosomes regulated cell migration. Furthermore, substrate stiffness also regulated exosome miRNA. The substrate stiffness-regulated exosome miRNA modulated cancer metastasis by regulating cell motility, ECM remodeling and the interaction between cancer cells and nerves. Focal adhesion kinase (FAK) and F-actin played a key role in the process of substrate stiffness regulating the PCa exosome miRNA. The above results imply that cancer cell exosome miRNA can transmit the ECM mechanical force to distant cells and participate in the construction of the extracellular microenvironment necessary for cancer cell metastasis.
2. Materials and methods
Preparation of substrates with different stiffness: Substrates with stiffness of 46 KPa and 0.7 KPa were prepared using PDMS based on the previous work [25,26].
Cell culture: Mycoplasma-negative PCa LNCaP cells (Changhai Hospital, Shanghai, China) were cultured in 1640 medium (11879020; Gbico) supplemented with 10% v/v exosome-free fetal bovine serum (FBS; 10091148; Gbico), 100 U mL−1penicillin, and 100 μg mL−1 streptomycin (10378016; Gibco) at 37 °C with 5% CO2.
Exosome-free FBS: FBS was deleted of exosomes by ultracentrifugation at 160000 g g for 6 h. Then the supernatant was collected and sterilized via a 0.22 filter membrane (SLGP033RB, MF-Millipore) [27].
Cell migration assays: The cells grown on different substrates or treated with different exosomes were collected. 1 × 105 cells were added to the Transwell (353097; Corning) chamber to assess cell migration efficiency as previously described [25].
LNCaP exosome isolation: exosome isolation: Cell supernatants were cultured on different substrates with the medium replaced every 24 h, centrifuged at 300 g for 10min, 2000g for 20min and 15000 g for 40min with High-Speed Refrigerated Centrifuge (CR21 N, HITACHI), and ultracentrifuged at 120000 g for 1 h with Optima XPN-100 Ultracentrifuge (Beckman Coulter). The pellets were collected and resuspended in PBS to a final concentration of 2 × 107/μl particles.
Exosome characterization: The diameter of exosomes was characterized by nanoparticle tracking analysis (NTA, ZetaView PMX 110, Particle Metrix). The exosome morphology was characterized by transmission electron microscopy (TEM, JEM-1230, JEOL) [11].
Western blotting: Cells or exosomes were lysed with RIPA buffer (PC101; Epizyme) supplemented with protease and phosphatase inhibitors (A32959; Thermo Scientific), separated by SDS-PAGE and then transferred to the PVDF membrane (ISEQ00010; MIillipore), which was then blocked, incubated with primary antibodies (TSG101, ab125011, Abcam; Calnexin, ab213243, Abcam; CD9, 13174, CST) for 14 h at 4 °C, and then with horseradish peroxidase (HRP)-conjugated secondary antibodies. Signals were collected using the ChemiScope system (Clinx Science Instruments).
High-throughput sequencing of miRNA in exosomes: Exosomes were collected and the high-throughput sequencing data were obtained by Illumina platform.
Differential expression analysis: The other ncRNA and duplicate data were filtered by Bowtie [28]. The remaining data were predicted to be known or new miRNAs by genome and miRBas data [29]. The DESeq2 R package (1.10.1) was used to perform differential analysis of miRNA [30]. |log2(FC)|≥0.58 and p value ≤ 0.05 (corrected by the Benjamini and Hochberg) were defined as differentially expressed. MiRanda and Targetscan were used to predict miRNA target genes and obtain the GO annotation information [31,32]. GOseq R package was used to analyze the GO enrichment of differential target genes [33]. The Fisher's exact test was used to calculate the significant enrichment of the target genes in the KEGG pathway [34]. miRNA sequencing and analysis of exosomes were completed by the biomarker platform (http://www.biomarker.com.cn/).
Quantitative real-time polymerase chain reaction (qRT-PCR): Total RNA was extracted with Total RNA Extraction Reagent (R401-01; Vazyme) from cultured cells or collected exosomes, reverse-transcribed with miRNA 1st Strand cDNA Synthesis Kit (MR101-02) and amplified with miRNA Universal SYBR qPCR Master Mix (MQ101-01; Vazyme) by Q-PCR (4485696, Thermo Scientific) [13]. U6 was used as an internal reference for normalization [35].
Inhibition assays: After 24 -h culture of LNCaP cells on different substrates, the F-actin inhibitor cytoskeleton B (HY-16928; MCE; 10 μM), FAK inhibitor PF-573288 (HY-10461; MCE; 10 μM), or ROCK inhibitor Y-27632 (HY-10071; MCE; 10 μM) was added to cell culture medium [25]. The cells were co-cultured with inhibitor-containing medium for 24 h. The supernatants were collected to isolate exosomes, and cells were collected to extract total RNA for qRT-PCR assay.
Data analysis: All data were analyzed using t-tests in Matlab R2014a software (MathWorks, Natick, MA, USA). Image-Pro Plus 6.0 (Media Cybernetics) was used for quantitative photometric analysis of the images.
3. Results and discussion
3.1. Exosomes transmit substrate stiffness information to regulate PCa cell migration
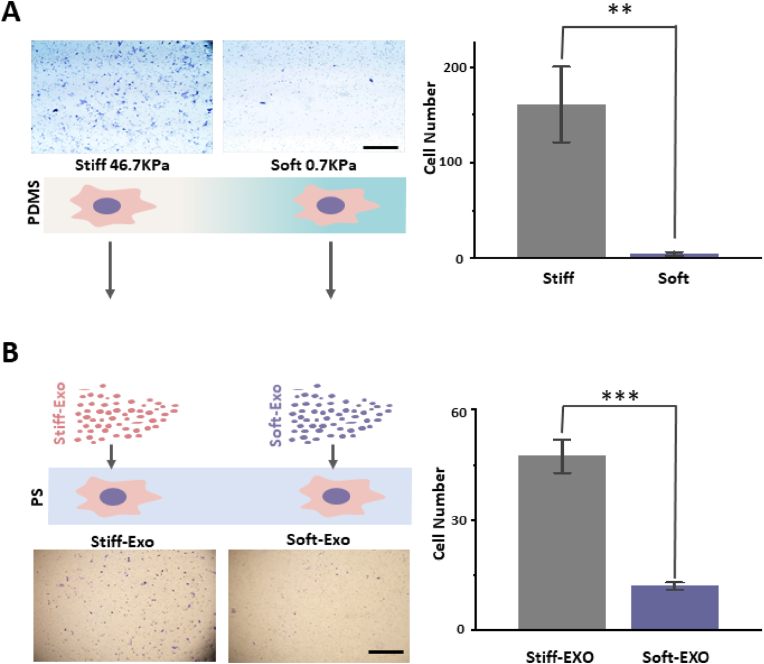
According to our previous work [25], LNCaP cells were cultured on stiff (46.7 KPa) or soft (0.7 KPa) substrates for 48 h. Then, the cells were collected and Transwell was used to measure the cell migration ability. The results showed that compared with soft substrates, LNCaP cells cultured on stiff substrates had significantly higher migration efficiency (Fig. 1 A, p = 0.00235). Next, LNCaP was cultured on different substrates for 48 h, and the cell supernatant was collected to isolate exosomes. The exosomes induced by the stiff substrate (Stiff-Exo) and the exosomes induced by the soft substrate (Soft-Exo) were added to the culture medium (2 × 109 particles/2 × 106 cells) and co-cultured with LNCaP cells grown in a Petri dish made of polystyrene (PS) for 48 h. Then, LNCaP cells treated with different exosomes were collected, and Transwell results show that compared with Soft-Exo treated LNCaP cells, Stiff-Exo treated LNCaP cells have a significantly higher migration efficiency (Fig. 1 B, p = 1.87488E-4). In summary, the exosomes can transmit extracellular matrix mechanical force to regulate PCa cell migration.
Fig. 1.
Exosomes induced by different substrate stiffnesses regulate cell migration. A. LNCaP cells were cultured on different substrates. Cell migration was analyzed after 48 h using Transwell (left, scale bar: 1 mm). LNCaP cells grown on stiff substrates had a higher migration efficiency (p = 0.00235). B. The exosomes of LNCaP cells grown on different mechanical substrates were collected and co-cultured with LNCaP grown on polystyrene (PS) petri dishes (stiff substrate-induced exosomes: Stiff-Exo; soft substrate-induced exosomes: Soft-Eox). Cell migration was analyzed after 48-h co-culture by Transwell (left, scale bar: 1 mm). LNCaP cells co-cultured with Stiff-Exo had higher migration efficiency (p = 1.87488E-4). n = 3, *p < 0.05; **p < 0.01; ***p < 0.001, t-test.
An interesting opposite trend was observed in our study. It was reported that soft substrates promoted LNCaP cell migration at 72 h [25], but we found that they inhibited cell migration at 48 h (Fig. 1 A). The time-dependent regulatory effect of the mechanical force on cell migration will be discussed hereafter.
3.2. Characterization of PCa cell exosomes induced by different stiffness substrates
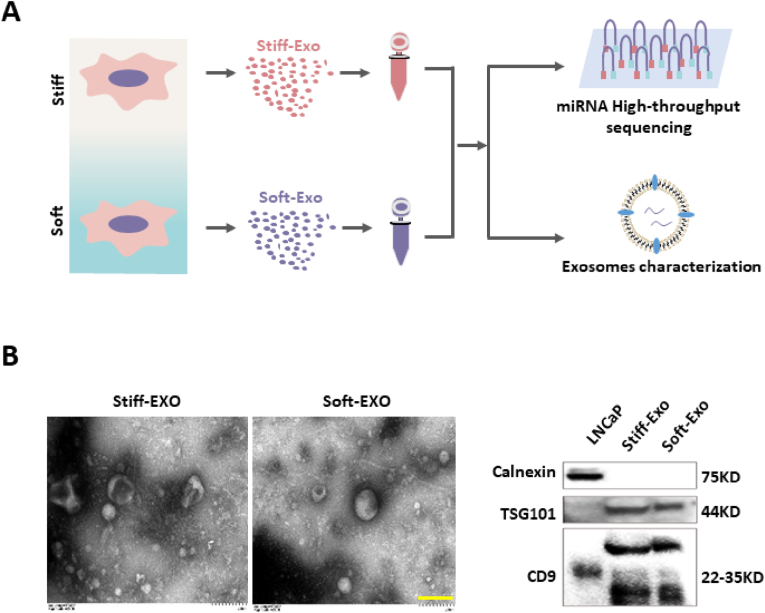
Stiff-Exo and Soft-Exo were collected for miRNA sequencing and characterization (Fig. 2 A). The results showed that substrate stiffness had no effect on the exosome morphology (Fig. 2 B) and size (Fig. S1) [11]. Both Stiff-Exo and Soft-Exo showed positive for CD9 and TSG101, and negative for Calnexin (Fig. 2 B), indicating that the collected exosomes were free of cellular debris contamination [36,37].
Fig. 2.
Collection and characterization of Stiff-Exo and Soft-Exo. A. Schematic diagram of collection and characterization of exosomes induced by different substrates stiffness. B. Characterization of Stiff-Exo and Soft-Exo morphology by transmission electron microscopy (left, bar = 200 nm). Characterization of exosome-related and cell-related protein markers by Western blot (right).
3.3. Substrate stiffness-modulated PCa exosome miRNA cargo participates in the establishment of the extracellular microenvironment in cancer metastasis
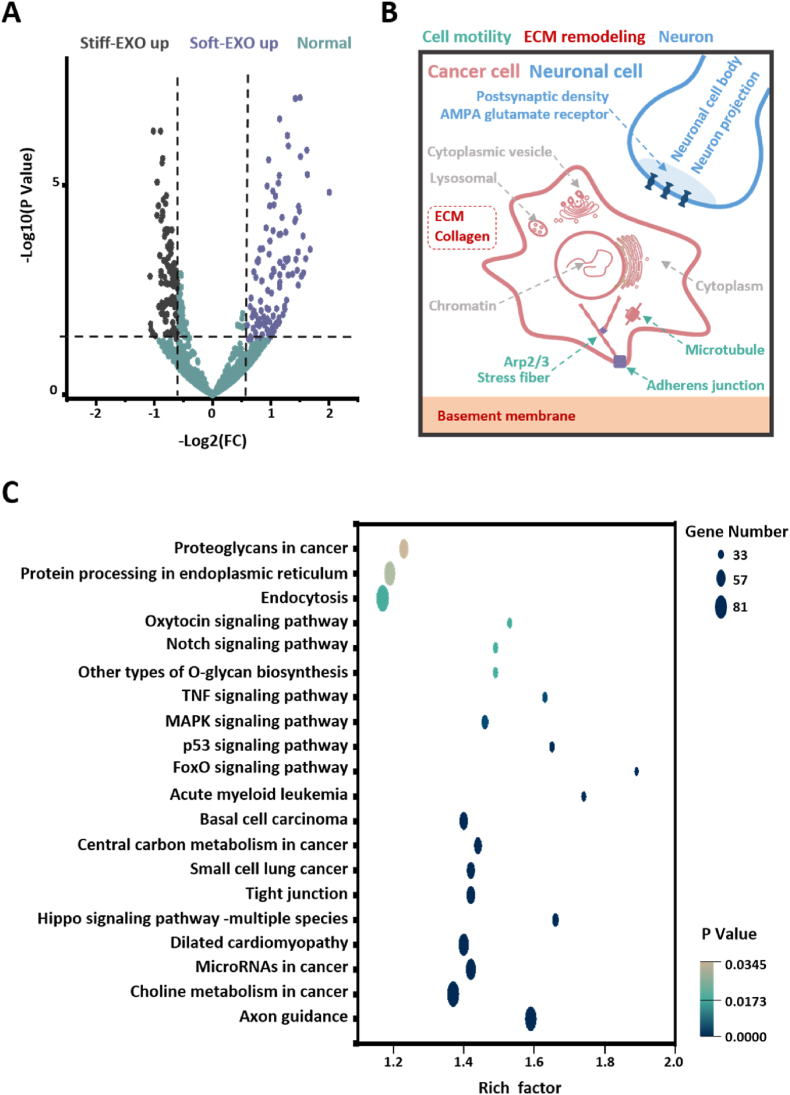
High-throughput sequencing results showed that there were 222 significantly different miRNAs between Stiff-Exo and Soft-Exo. Stiff-Exo had 101 upregulated miRNAs, and Soft-Exo had 121 upregulated miRNAs (Fig. 3 A, Table S1, |log2(FC)|≥0.58; p Value ≤ 0.05). The GO enrichment of cellular component (CC) showed that differential miRNAs mainly regulated genes associated with cell motility [8,9,[38], [39], [40]], ECM remodeling [[41], [42], [43]] and interaction between PCa cells and nerve cells (Fig. 3 B, Fig. S2, Table S3) [[44], [45], [46], [47], [48]]. The KEGG pathway enrichment results showed that, the differential miRNA target genes were enriched in pathways related to cancer progression, such as choline metabolism in cancer, microRNAs in cancer, small cell lung cancer (Fig. 3 C, Table S4). The above results indicate that the substrate stiffness played an important role in cancer progression by regulating miRNA. In summary, ECM mechanical forces regulated exosomes miRNA of PCa cells. The ECM mechanical force regulated exosomes miRNA is involved in the interaction between cancer cells and the extracellular microenvironment.
Fig. 3.
miRNA in LNCaP exosomes is regulated by substrate stiffness. A. Volcano map of differentially expressed miRNA in Stiff-Exo and Soft-Exo. |log2(FC)|≥0.58; p Value ≤ 0.05 is defined as difference. B. Schematic diagram of GO enrichment analysis (Cellular Component) of differential miRNAs target genes (top 19, sort according to p value). C. Enrichment analysis of differentially expressed miRNAs target genes in KEGG pathway (top 20, sort according to p value). n = 3.
3.4. Substrate stiffness regulates cellular miRNA expression and sorting into exosomes through independent ways
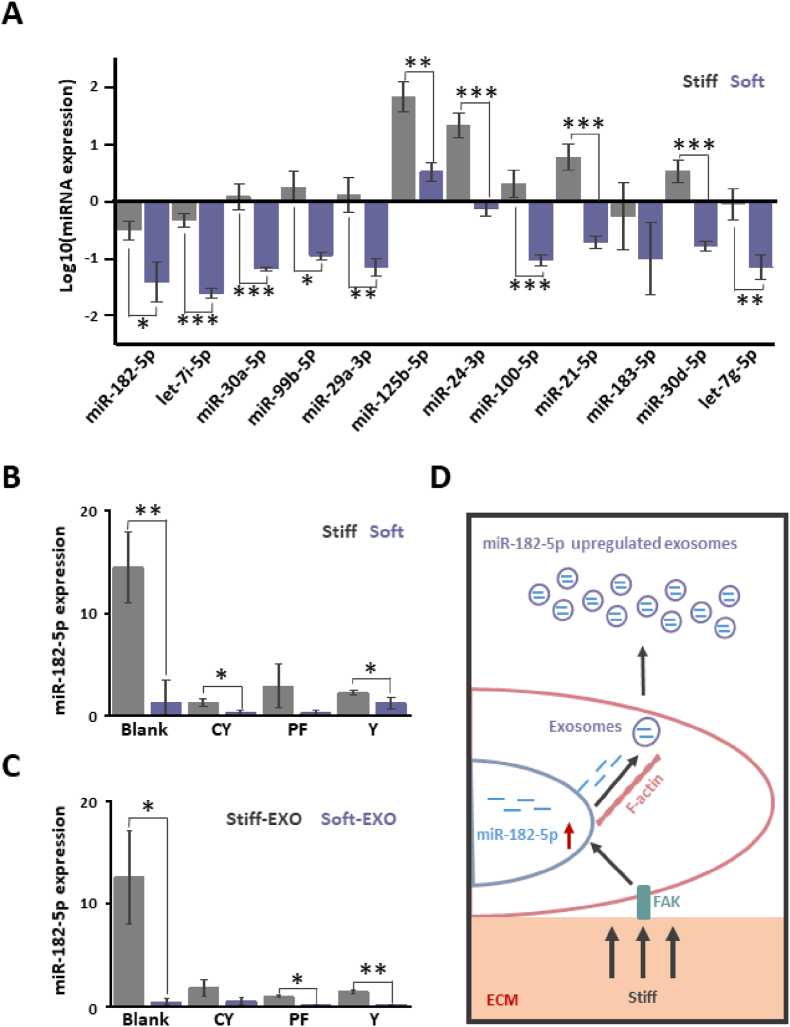
Then we explored the regulation process of exosomes miRNA by substrate stiffness. LNCaP cells were cultured on substrates with different stiffness for 48 h, collected and detected for the relative miRNA expression by qRT-PCR. The 12 upregulated miRNAs (FC > 1.8) in Stiff-Exo were selected to assess whether the substrate stiffness regulates its expression in the cell (Table S2). The results showed that except for miR-183-5p, the intracellular expression of all other miRNAs was significantly regulated by substrate stiffness (p < 0.05) (Fig. 4 A). The expression level of miRNAs in LNCaP cells grown on stiff substrates was significantly higher than those grown on soft substrates (p < 0.05) (Fig. 4 A). The above results demonstrate that the mechanism of ECM mechanical force to regulate the sorting of cancer cell miRNA cargo into exosomes is complex and different miRNAs depend on independent pathways.
Fig. 4.
Substrate stiffness regulates the expression of miRNA in cancer cells and transports miRNA to exosomes. A. LNCaP cells were cultured on different substrates for 48 h. Significantly different miRNA expressions in Stiff-Exo and Soft-Exo Q-PCR were detected by qRT-PCR. The results showed that, except for miR-183-5p, all other miRNAs were regulated by substrate stiffness (miR-182-5p: p = 0.0147; let-7i-5p: p = 1.24939E-4; miR-30a-5p: p = 6.96735E-4; miR-99b-5p: p = 0.0167; miR-29a-3p: p = 0.00302; miR-125b-5p: p = 0.00163; miR-24-3p: p = 6.32271E-4; miR-100-5p: p = 8.13899E-4; miR-21-5p: p = 5.60351E-4; miR-183-5p: p = 0.21238; miR-30d-5p: p = 4.79694E-4; let-7g-5p: p = 0.00562). B. LNCaP cells were cultured on different substrates for 24 h and then incubated with different inhibitors (F-actin inhibitor cytoskeleton B, CY; FAK inhibitor PF-573288, PF; ROCK inhibitor Y-27632, Y; 10 μM) for 24 h. The expression of miR-182-5p in cells was detected by qRT-PCR. The results showed that PF significantly inhibited the regulatory effect of substrate stiffness on miR-182-5p expression in cells (Blank: p = 0.00512; CY: p = 0.01741; PF: p = 0.10199; Y: p = 0.04764). C. LNCaP cells were cultured on different substrates for 24 h and then with different inhibitors for 24 h. The cell supernatant was collected to isolate Stiff-Exo and Soft-Exo. The expression of miR-182-5p in Stiff-Exo and Soft-Exo was detected by qRT-PCR. The results showed that CY significantly inhibited the regulatory effect of substrate stiffness on miR-182-5p transport to exosomes (Blank: p = 0.01; CY: p = 0.05816; PF: p = 0.01343; Y: p = 0.0031). D. Schematic diagram of the mechanism of ECM stiffness regulating miR-182-5p expression in cancer cells and transport to exosomes. The stiff ECM introduced information into LNCaP cells through FAK to promote miR-182-5p expression. LNCaP cells transferred stiff substrate-induced miR-182-5p to exosomes through F-actin. n = 3, *p < 0.05; **p < 0.01; ***p < 0.001, t-test.
Subsequently, the miR-182-5p was selected to discuss which mechanical elements were involved in the process of substrate stiffness in regulating the exosome miRNA, knowing that the miR-182-5p expression was regulated by the substrate stiffness. Mechanical forces in the ECM are usually transmitted into cells via FAK, F-actin and cytoskeleton tension to regulate PCa cell behavior [25,49]. Therefore, inhibitors of FAK, F-actin and cytoskeleton tension were selected to determine which mechanical elements were involved in the process of substrate stiffness regulating miRNA in exosomes. F-actin (CY), FAK (PF), or cytoskeleton tension (Y) inhibitors [25] were incubated with LNCaP cultured in different substrates for 24 h. The expression level of miR-182-5p was detected by qRT-PCR. The results showed that PF inhibited the regulation of miR-182-5p expression by substrate stiffness (Fig. 4 B). Meantime, the CY suppressed the difference of miR-182-5p between Stiff-Exo and Soft-Exo (Fig. 4 C). The stiff substrate transferred stiffness information into cells through FAK to regulate the expression of miR-182-5p, and sorted miR-182-5p into exosomes through F-actin (Fig. 4 D). Substrate stiffness regulated miR-182-5p expression in cells by sorting into exosomes through independent ways.
4. Conclusion
Our work has demonstrated that exosomes, as signalosomes, could transmit the extracellular matrix mechanical force on the regulation of PCa migration behavior to distant cells. The miRNA cargo in exosomes could be used as a mechanical force carrier to regulate cancer metastasis by regulating cell motility, ECM remodeling and the interaction between cancer cells and nerves. ECM mechanical force regulated the intracellular expression of miRNA and the sorting of miRNA into exosomes through two independent ways. FAK and F-actin played a key role in the process of packaging ECM mechanical force into exosome miRNA. The above results suggest that the mechanical force in the cancer cells ECM may participate in maintaining the microenvironment required for cancer metastasis through exosome miRNA.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank the biomarker platform for help with exosome miRNA sequencing and data analysis. We acknowledge the support of the China National Key Research and Development Program Stem Cell and Translational Research Key Projects (2018YFA0108300), the National Science Foundation of China (Grant No.31671545, 31971109), and Shanghai Key Laboratory of Cell Engineering (14DZ2272300).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101299.
Contributor Information
Huiwen Liu, Email: liuhw_11@126.com.
Yue Wang, Email: wangyuesmmu@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Nia H.T., Munn L.L., Jain R.K. Physical traits of cancer. Science. 2020;370 doi: 10.1126/science.aaz0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butcher D.T., Alliston T., Weaver V.M. A tense situation: forcing tumour progression. Nat. Rev. Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Q., Luo Q., Ju Y., Song G. Role of the mechanical microenvironment in cancer development and progression. Cancer Biol Med. 2020;17:282–292. doi: 10.20892/j.issn.2095-3941.2019.0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wirtz D., Konstantopoulos K., Searson P.C. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kechagia J.Z., Ivaska J., Roca-Cusachs P. Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 2019;20:457–473. doi: 10.1038/s41580-019-0134-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y., Ju L., Rushdi M., Ge C., Zhu C. Receptor-mediated cell mechanosensing. Mol. Biol. Cell. 2017;28:3134–3155. doi: 10.1091/mbc.E17-04-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens M.M., George J.H. Exploring and engineering the cell surface interface. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 8.Schwager S.C., Taufalele P.V., Reinhart-King C.A. Cell-cell mechanical communication in cancer. Cell. Mol. Bioeng. 2019;12:1–14. doi: 10.1007/s12195-018-00564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ladoux B., Mege R.M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 2017;18:743–757. doi: 10.1038/nrm.2017.98. [DOI] [PubMed] [Google Scholar]
- 10.S E.L.A., Mager I., Breakefield X.O., Wood M.J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 11.Shang A., Gu C., Wang W., Wang X., Sun J., Zeng B., Chen C., Chang W., Ping Y., Ji P., Wu J., Quan W., Yao Y., Zhou Y., Sun Z., Li D. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-beta 1 axis. Mol. Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rezaie J., Akbari A., Rahbarghazi R. Inhibition of extracellular vesicle biogenesis in tumor cells: a possible way to reduce tumorigenesis. Cell Biochem. Funct. 2022;40:248–262. doi: 10.1002/cbf.3695. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H., Deng T., Liu R., Bai M., Zhou L., Wang X., Li S., Wang X., Yang H., Li J., Ning T., Huang D., Li H., Zhang L., Ying G., Ba Y. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 2017;8 doi: 10.1038/ncomms15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang M.K.S., Yue P.Y.K., Ip P.P., Huang R.L., Lai H.C., Cheung A.N.Y., Tse K.Y., Ngan H.Y.S., Wong A.S.T. Soluble E-cadherin promotes tumor angiogenesis and localizes to exosome surface. Nat. Commun. 2018;9:2270. doi: 10.1038/s41467-018-04695-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paggetti J., Haderk F., Seiffert M., Janji B., Distler U., Ammerlaan W., Kim Y.J., Adam J., Lichter P., Solary E., Berchem G., Moussay E. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood. 2015;126:1106–1117. doi: 10.1182/blood-2014-12-618025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricklefs F.L., Alayo Q., Krenzlin H., Mahmoud A.B., Speranza M.C., Nakashima H., Hayes J.L., Lee K., Balaj L., Passaro C., Rooj A.K., Krasemann S., Carter B.S., Chen C.C., Steed T., Treiber J., Rodig S., Yang K., Nakano I., Lee H., Weissleder R., Breakefield X.O., Godlewski J., Westphal M., Lamszus K., Freeman G.J., Bronisz A., Lawler S.E., Chiocca E.A. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu L., Ding J., Chen C., Wu Z.J., Liu B., Gao Y., Chen W., Liu F., Sun W., Li X.F., Wang X., Wang Y., Xu Z.Y., Gao L., Yang Q., Xu B., Li Y.M., Fang Z.Y., Xu Z.P., Bao Y., Wu D.S., Miao X., Sun H.Y., Sun Y.H., Wang H.Y., Wang L.H. Exosome-transmitted lncARSR promotes Sunitinib resistance in Renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Melo S.A., Sugimoto H., O'Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A., Lucci A., Ivan C., Calin G.A., Kalluri R. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita Y., Yoshioka Y., Ochiya T. Extracellular vesicle transfer of cancer pathogenic components. Cancer Sci. 2016;107:385–390. doi: 10.1111/cas.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thind A., Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles. 2016;5 doi: 10.3402/jev.v5.31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan Q., Yang L., Zhang X., Peng X., Wei S., Su D., Zhai Z., Hua X., Li H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018;414:107–115. doi: 10.1016/j.canlet.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 22.Schwager S.C., Bordeleau F., Zhang J., Antonyak M.A., Cerione R.A., Reinhart-King C.A. Matrix stiffness regulates microvesicle-induced fibroblast activation. Am. J. Physiol. Cell Physiol. 2019;317:C82–C92. doi: 10.1152/ajpcell.00418.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z., Maruyama K., Sakisaka Y., Suzuki S., Tada H., Suto M., Saito M., Yamada S., Nemoto E. Cyclic stretch force induces periodontal ligament cells to secrete exosomes that suppress IL-1 beta production through the inhibition of the NF-kappaB signaling pathway in macrophages. Front. Immunol. 2019;10:1310. doi: 10.3389/fimmu.2019.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patwardhan S., Mahadik P., Shetty O., Sen S. ECM stiffness-tuned exosomes drive breast cancer motility through thrombospondin-1. Biomaterials. 2021 doi: 10.1016/j.biomaterials.2021.121185. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z., Wang L., Xu H., Du Q., Li L., Wang L., Zhang E.S., Chen G., Wang Y. Heterogeneous responses to mechanical force of prostate cancer cells inducing different metastasis patterns. Adv. Sci. 2020;7 doi: 10.1002/advs.201903583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trappmann B., Gautrot J.E., Connelly J.T., Strange D.G., Li Y., Oyen M.L., Cohen Stuart M.A., Boehm H., Li B., Vogel V., Spatz J.P., Watt F.M., Huck W.T. Extracellular-matrix tethering regulates stem-cell fate. Nat. Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 27.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., Xiang J., Zhang T., Theilen T.M., Garcia-Santos G., Williams C., Ararso Y., Huang Y., Rodrigues G., Shen T.L., Labori K.J., Lothe I.M., Kure E.H., Hernandez J., Doussot A., Ebbesen S.H., Grandgenett P.M., Hollingsworth M.A., Jain M., Mallya K., Batra S.K., Jarnagin W.R., Schwartz R.E., Matei I., Peinado H., Stanger B.Z., Bromberg J., Lyden D. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedlander M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis B.P., Shih I.h., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian MicroRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 32.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Occhipinti G., Giulietti M., Principato G., Piva F. The choice of endogenous controls in exosomal microRNA assessments from biofluids. Tumour Biol. 2016;37:11657–11665. doi: 10.1007/s13277-016-5164-1. [DOI] [PubMed] [Google Scholar]
- 36.Hoshino A., Kim H.S., Bojmar L., Gyan K.E., Cioffi M., Hernandez J., Zambirinis C.P., Rodrigues G., Molina H., Heissel S., Mark M.T., Steiner L., Benito-Martin A., Lucotti S., Di Giannatale A., Offer K., Nakajima M., Williams C., Nogués L., Pelissier Vatter F.A., Hashimoto A., Davies A.E., Freitas D., Kenific C.M., Ararso Y., Buehring W., Lauritzen P., Ogitani Y., Sugiura K., Takahashi N., Alečković M., Bailey K.A., Jolissant J.S., Wang H., Harris A., Schaeffer L.M., García-Santos G., Posner Z., Balachandran V.P., Khakoo Y., Raju G.P., Scherz A., Sagi I., Scherz-Shouval R., Yarden Y., Oren M., Malladi M., Petriccione M., De Braganca K.C., Donzelli M., Fischer C., Vitolano S., Wright G.P., Ganshaw L., Marrano M., Ahmed A., DeStefano J., Danzer E., Roehrl M.H.A., Lacayo N.J., Vincent T.C., Weiser M.R., Brady M.S., Meyers P.A., Wexler L.H., Ambati S.R., Chou A.J., Slotkin E.K., Modak S., Roberts S.S., Basu E.M., Diolaiti D., Krantz B.A., Cardoso F., Simpson A.L., Berger M., Rudin C.M., Simeone D.M., Jain M., Ghajar C.M., Batra S.K., Stanger B.Z., Bui J., Brown K.A., Rajasekhar V.K., Healey J.H., de Sousa M., Kramer K., Sheth S., Baisch J., Pascual V., Heaton T.E., La Quaglia M.P., Pisapia D.J., Schwartz R., Zhang H., Liu Y., Shukla A., Blavier L., DeClerck Y.A., et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–1061. doi: 10.1016/j.cell.2020.07.009. e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Apostolopoulou M., Mastrototaro L., Hartwig S., Pesta D., Strassburger K., de Filippo E., Jelenik T., Karusheva Y., Gancheva S., Markgraf D., Herder C., Nair K.S., Reichert A.S., Lehr S., Mussig K., Al-Hasani H., Szendroedi J., Roden M. Metabolic responsiveness to training depends on insulin sensitivity and protein content of exosomes in insulin-resistant males. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abi9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen M., Tremblay M.L., Yamada K.M. Phosphatases in cell-matrix adhesion and migration. Nat. Rev. Mol. Cell Biol. 2003;4:700–711. doi: 10.1038/nrm1199. [DOI] [PubMed] [Google Scholar]
- 39.Dogterom M., Koenderink G.H. Actin-microtubule crosstalk in cell biology. Nat. Rev. Mol. Cell Biol. 2019;20:38–54. doi: 10.1038/s41580-018-0067-1. [DOI] [PubMed] [Google Scholar]
- 40.SenGupta S., Parent C.A., Bear J.E. The principles of directed cell migration. Nat. Rev. Mol. Cell Biol. 2021;22:529–547. doi: 10.1038/s41580-021-00366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 42.Nissen N.I., Karsdal M., Willumsen N. Collagens and Cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J. Exp. Clin. Cancer Res. 2019;38:115. doi: 10.1186/s13046-019-1110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winkler J., Abisoye-Ogunniyan A., Metcalf K.J., Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020;11:5120. doi: 10.1038/s41467-020-18794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy M.B. The postsynaptic density at glutamatergic synapses. Trends Neurosci. 1997;20:264–268. doi: 10.1016/s0166-2236(96)01033-8. [DOI] [PubMed] [Google Scholar]
- 45.Kennedy M.B. Signal-processing machines at the postsynaptic density. Science. 2000;290:750–754. doi: 10.1126/science.290.5492.750. [DOI] [PubMed] [Google Scholar]
- 46.Okabe S. Molecular anatomy of the postsynaptic density. Mol. Cell. Neurosci. 2007;34:503–518. doi: 10.1016/j.mcn.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Amit M., Na'ara S., Gil Z. Mechanisms of cancer dissemination along nerves. Nat. Rev. Cancer. 2016;16:399–408. doi: 10.1038/nrc.2016.38. [DOI] [PubMed] [Google Scholar]
- 48.Zahalka A.H., Frenette P.S. Nerves in cancer. Nat. Rev. Cancer. 2020;20:143–157. doi: 10.1038/s41568-019-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagh K., Ishikawa M., Garcia D.A., Stavreva D.A., Upadhyaya A., Hager G.L. Mechanical regulation of transcription: recent advances. Trends Cell Biol. 2021;31:457–472. doi: 10.1016/j.tcb.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.