Abstract
Deoxynucleoside triphosphate (dNTP) triphosphohydrolases (dNTPases) are important enzymes that may perform multiple functions in the cell, including regulating the dNTP pools and contributing to innate immunity against viruses. Among the homologs that are best studied are human sterile alpha motif and HD domain–containing protein 1 (SAMHD1), a tetrameric dNTPase, and the hexameric Escherichia coli dGTPase; however, it is unclear whether these are representative of all dNTPases given their wide distribution throughout life. Here, we investigated a hexameric homolog from the marine bacterium Leeuwenhoekiella blandensis, revealing that it is a dGTPase that is subject to allosteric activation by dATP, specifically. Allosteric regulation mediated solely by dATP represents a novel regulatory feature among dNTPases that may facilitate maintenance of cellular dNTP pools in L. blandensis. We present high-resolution X-ray crystallographic structures (1.80–2.26 Å) in catalytically important conformations as well as cryo-EM structures (2.1–2.7 Å) of the enzyme bound to dGTP and dATP ligands. The structures, the highest resolution cryo-EM structures of any SAMHD1-like dNTPase to date, reveal an intact metal-binding site with the dGTP substrate coordinated to three metal ions. These structural and biochemical data yield insights into the catalytic mechanism and support a conserved catalytic mechanism for the tetrameric and hexameric dNTPase homologs. We conclude that the allosteric activation by dATP appears to rely on structural connectivity between the allosteric and active sites, as opposed to the changes in oligomeric state upon ligand binding used by SAMHD1.
Keywords: cryo-EM, crystallography, enzyme structure, enzyme mechanism, allosteric regulation, nucleoside/nucleotide metabolism, structure–function
Abbreviations: AS1, primary allosteric site; AS2, secondary allosteric site; CTF, contrast transfer function; dG, 2′-deoxyguanosine; dNTP, deoxynucleoside triphosphate; dNTPase, dNTP triphosphohydrolase; HD, histidine–aspartate; hSAMHD1, human SAMHD1; PDB, Protein Data Bank; PNP, purine nucleoside phosphorylase; RNR, ribonucleotide reductase; SAMHD1, sterile alpha motif and HD domain–containing protein 1; XOD, xanthine oxidase
Proper control of the cellular nucleotide pools is critically important, particularly during DNA replication when both the absolute and relative concentrations of the four deoxynucleoside triphosphates (dNTPs) must be carefully balanced and maintained (1, 2). Disturbances in the dNTP levels have been shown to lead to defective DNA replication or strongly elevated mutation rates (3, 4, 5). Control of dNTP levels is exerted at several points, most prominently their synthesis. At this stage, the enzyme ribonucleotide reductase (RNR) is responsible for reducing the ribonucleotides to the corresponding 2′-deoxynucleotides, usually at the diphosphate level. To achieve the proper level and balance, RNR employs two separate types of allosteric sites: an “activity site” that functions as an on/off switch and a “specificity site” that uses feedback information from the existing dNTP mix to regulate the relative ratios of the four DNA precursors (6). In addition to regulation at synthesis, dNTP levels are controlled by activities that degrade triphosphates. Among these activities are a group of enzymes, the dNTP triphosphohydrolases (dNTPases), which hydrolyze dNTPs into the corresponding 2′-deoxynucleosides and inorganic triphosphate. These dNTPases belong to the histidine–aspartate (HD) domain superfamily, whose members share a common fold surrounding the HD motif (H…HD…D) that coordinates the active-site divalent metal (7).
The earliest discovered example of a dNTPase is the dGTP-specific dGTP triphosphohydrolase (dGTPase) from the bacterium Escherichia coli (8, 9, 10). Its role in promoting genome stability is supported by the observed mutator effect associated with deletion of the corresponding dgt gene (11, 12). Many other examples of triphosphohydrolases in other organisms have since been revealed. The human enzyme, sterile alpha motif and HD domain–containing protein 1 (SAMHD1), is a general dNTPase, hydrolyzing all four dNTPs (13); it, too, plays an important role in determining the cellular dNTP pools (14). Furthermore, deficiency in human SAMHD1 (hSAMHD1) activity leads to an autoimmune disease, termed Aicardi–Goutières syndrome, signifying the importance of its function in human health (15, 16). It also serves an antiviral role against HIV and other viruses through its dNTPase activity by depleting the dNTP pools necessary for viral replication. The significance of the antiviral function is further indicated by the fact that several viruses encode dNTPase inhibitor molecules, such as the Vpx protein of HIV-2 (17) and the Gp1.2 protein of E. coli bacteriophage T7 (18, 19).
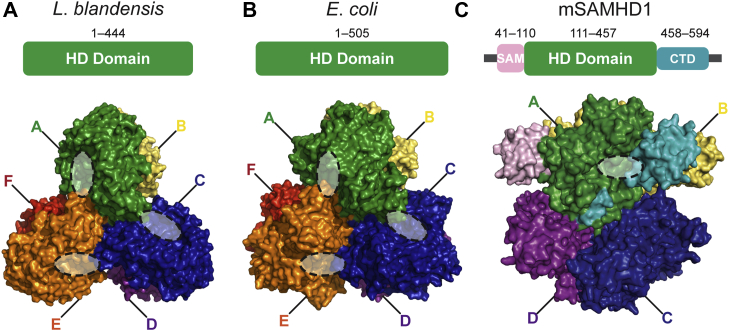
Among the dNTPases, differences in domain architecture result in distinct monomer–monomer interactions that produce varied oligomeric states and opportunities for allosteric regulation (Fig. 1). E. coli dGTPase is a homohexamer, which can be viewed as a set of three parallel dimers (12). Compared with the E. coli enzyme, mammalian SAMHD1 includes an N-terminal SAM, an HD domain with a number of insertions and deletions, and an additional C-terminal domain. These extensions alter the monomer–monomer interaction surfaces and ultimately produce a homotetramer.
Figure 1.
Crystal structures of SAMHD1 homologs from three species. Generated in PyMOL. A, Leeuwenhoekiella blandensis homolog (PDB ID: 3BG2). B, Escherichia coli dGTPase (PDB ID: 4XDS). C, mouse SAMHD1 isoform 1 (PDB ID: 6BRG). A domain architecture diagram is shown for each crystal structure, with monomer A colored accordingly. All other monomers are labeled and colored for each homolog. The visible active-site cavities for each homolog are indicated by dashed ovals. We define the HD domain as the entire protein for E. coli dGTPase (505 amino acids in total) and L. blandensis dGTPase homolog (444 amino acids in total). For mouse SAMHD1, we define the HD domain boundaries by structural conservation with the hexameric homologs, whereas the start of the SAM domain and end of the CTD are defined here by the residues that are ordered in the crystal structure shown. The domain numbering is listed above the diagram (626 amino acids in total). CTD, C-terminal domain; HD, histidine–aspartate; PDB, Protein Data Bank; SAM, sterile alpha motif; SAMHD1, sterile alpha motif and HD domain–containing protein 1.
dNTPases are regulated by the binding of activators at allosteric sites located at the interface between monomers, which allows for responsiveness to cellular conditions. The hexameric E. coli dGTPase is allosterically activated by ssDNA, which binds to the monomer–monomer interface within one dimer of the overall hexamer (12, 20). This is the only reported example of a hexameric homolog bound to its allosteric activator (12); however, structural evidence for its precise activation mechanism was limited to side-chain conformational changes observed in the active site. The tetrameric hSAMHD1 dNTPase is activated by GTP or dGTP at a primary allosteric site (AS1) along with dNTP binding to a secondary allosteric site (AS2) (13, 21, 22, 23), whereas the tetrameric dNTPase from Enterococcus faecalis, EF1143, is activated by dGTP binding to AS1 with nanomolar affinity in vitro (24, 25). Crystal structures of tetrameric dNTPases bound to a variety of allosteric activators have indicated that allosteric activation in vitro is driven by changes in oligomerization state accompanied by conformational changes in the active site (23, 24, 25, 26, 27, 28). In contrast, the bacterial hexameric homologs are found as hexamers even in the absence of allosteric ligands (12, 29), so the precise mode of allosteric activation has been less clear.
hSAMHD1 and mouse SAMHD1 and the related E. faecalis EF1143 have been crystallized bound to dNTP substrates using a variety of biochemical means to limit turnover, including inactivating mutations to the HD motif (23, 26), employing nonhydrolyzable substrate analogs (26, 30), or replacing important catalytic metal ions with inhibitory Ca2+ or Zn2+ (24, 27, 31). These structures have revealed all four dNTP substrates bound to the active site. Barnes et al. (32) recently reported the first substrate-bound structures of a hexameric homolog, the WT E. coli dGTPase bound to dGTP or α-thio-dGTP with a catalytically competent HD motif. This was achieved by crystal crosslinking, which reduced turnover and trapped the dGTP substrate. The observed substrate-binding conformation differed from that of the tetrameric homologs because of the differences in active-site structure yielding differences in substrate-binding surfaces. However, these conformational differences might not necessarily imply differences in mechanism.
The impetus of the current work was to obtain deeper insight into the structural and mechanistic features of this class of enzymes, particularly the mechanisms that drive allosteric activation and active-site and allosteric-site ligand specificity. We noted the presence in the Protein Data Bank (PDB) of a crystal structure of a hexameric homolog from the bacterium Leeuwenhoekiella blandensis, with a structure ∼15% smaller than the E. coli dGTPase, deposited by the New York SGX Research Center for Structural Genomics (PDB ID: 3BG2; Fig. 1). L. blandensis is a member of Flavobacteriaceae (33), a family that is collectively important for ecosystem restoration following algal blooms (34, 35). In this work, we identify the L. blandensis SAMHD1 homolog as a hexameric dGTPase that is activated specifically by dATP. This represents a novel mode of allosteric regulation amongst the SAMHD1 homologs. X-ray crystallographic structures of enzyme variants indicate the importance of loop movement near the dGTP substrate–binding site. Importantly, we obtained high-resolution cryo-EM structures of the enzyme (i) in the absence of nucleotides (apo) or in complexes that included (ii) dGTP in the active site, (iii) dATP in both the allosteric and active sites, and (iv) dGTP in the active site and dATP in the allosteric site. Together, these structures reveal the structural basis for nucleotide specificity at the dGTP substrate and the dATP allosteric sites. Conformational changes between the ligand-free and ligand-bound states suggest that the dATP binding at the allosteric site increases turnover and affinity for the dGTP substrate by driving a conformational change of an active-site loop that recognizes the substrate 2′-deoxyribose moiety.
Results
The L. blandensis SAMHD1 homolog is a dGTPase allosterically activated by dATP
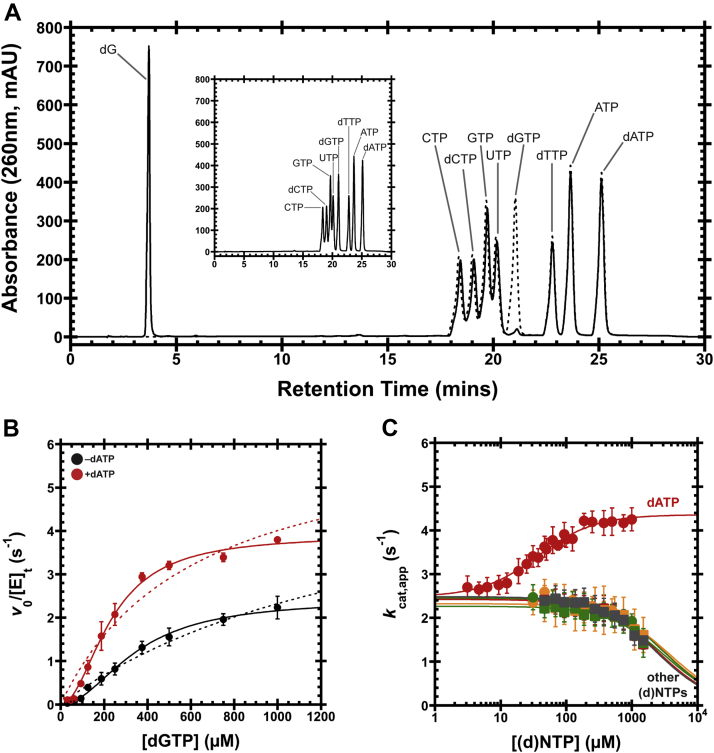
We determined the substrate specificity of the L. blandensis SAMHD1 homolog and established that it is highly specific for dGTP (Fig. 2). We incubated the enzyme with a mixture of the eight standard dNTPs and NTPs and found that it readily hydrolyzed dGTP without hydrolyzing the other (d)NTPs (Fig. 2A). These analyses also confirmed that the L. blandensis dGTPase is a triphosphohydrolase as expected, releasing 2′-deoxyguanosine (dG) as a product. We next measured kinetic parameters for the dGTPase and found that the kcat was 2.6 ± 0.1 s−1, the KM was 390 ± 30 μM, and the Hill coefficient was close to 2 (1.7 ± 0.1) (Fig. 2B and Table S1). The KM value is significantly higher than the expected cellular concentration of dGTP (50–100 μM) (5, 36), indicating that the in vivo dGTPase turnover rate will be fairly low. However, a high KM might be beneficial for this enzyme because an enzyme’s turnover rate is maximally responsive to changes in the substrate concentration when operating below the KM, allowing the cell to respond meaningfully to fluctuations in dGTP concentration. Furthermore, the hydrolysis activity displays a sigmoidal dependence on concentration (nH near 2) (Fig. 2B, solid versus dashed lines), similar to what has been observed for the E. coli dGTPase (12, 37), further increasing the potential for responsiveness to in vivo dGTP concentration fluctuations. In a multimeric protein like dGTPase, a sigmoidal dependence on substrate concentration may indicate intersubunit cooperativity during catalysis, enzyme hysteresis, or that dGTP is itself an activator at the allosteric site. Although the precise mechanistic basis for this cooperativity has not been delineated, we can likely rule out self-activation by substrate binding to the allosteric site because dGTP is not found in the allosteric site even when supplied at millimolar concentrations (see the cryo-EM section later).
Figure 2.
Leeuwenhoekiella blandensis dGTPase activity and allosteric activation.A, 1-h hydrolysis of a mixture containing all eight standard dNTPs and NTPs by the dNTPase homolog from L. blandensis. The eight (d)NTPs were reacted together (1 mM each), with the substrates and products separated by HPLC (solid line). A control reaction at time 0 is shown as a dashed line and separately in the inset, for clarity. B, v0/[E]tversus [dGTP] plots with or without 500 μM dATP. Data are presented as the mean and SEM of three independent experiments. The solid lines are the fit of Equation 1 to the data with nH = 2, whereas the dashed lines are the fit for nH = 1. For dGTP-only, kcat = 2.6 ± 0.1 s−1, KM = 390 ± 30 μM, and nH = 1.7 ± 0.1. With 500 μM dATP, kcat = 3.8 ± 0.1 s−1, KM = 224 ± 9 μM, and nH = 2.2 ± 0.1. C, dGTPase activity at 1 mM dGTP, titrating the remaining seven standard (d)NTPs. We refer to this substrate concentration as an apparent kcat (kcat,app) because it is above the threshold required to bring the turnover within 20% of the kcat, thereby approximating conditions with saturating substrate. Data are plotted as the mean and SEM of four independent experiments. Activation was observed for dATP (red circles), with K1/2 = 36 ± 3 μM and nH = 1.3 ± 0.1. Inhibitory effects are observed for dTTP (orange circles), dCTP (green circles), ATP (red squares), UTP (orange squares), CTP (green squares), and GTP (gray squares). The IC50s are estimated to be >2 mM. dNTP, deoxynucleoside triphosphate; dNTPase, dNTP triphosphohydrolase.
We next characterized the L. blandensis dGTPase activity in more detail and found that it is allosterically activated by dATP. To probe any allosteric specificity, we fixed the dGTP concentration at 1 mM (kcat,app) and titrated in each of the other seven standard (d)NTPs (Fig. 2C). The enzyme showed robust activation with dATP, with an activating K1/2 of 36 ± 3 μM and a Hill coefficient near 1 (1.3 ± 0.1). The remaining six nucleotides showed inhibitory effects in the millimolar range (IC50 >2 mM). These weaker effects might indicate competition for the active site by a nucleotide with lower affinity than dGTP. We then fixed the dATP concentration at 500 μM in order to approximate a saturating dATP activator concentration and repeated the dGTP substrate titration (Fig. 2B, red). Under these conditions, the dGTPase kcat was 3.8 ± 0.1 s−1, the KM was 224 ± 9 μM, and nH was again close to 2 (2.2 ± 0.1). These results indicate that dATP enhances enzyme activity by both increasing kcat (by ∼50%) and decreasing KM (by ∼70%). While these effects appear modest, and the activated KM remains above the expected in vivo dGTP concentration, the relative effect of the activation will again be most effective when the substrate concentration is below the KM. For instance, if we calculate the fold activation using nH = 2 (Fig. 2B and Table S1), the turnover rate is enhanced by dATP by more than threefold whenever dGTP is less than ∼160 μM, but the activation drops to ∼1.6-fold as the dGTP concentration increases to saturation.
Crystal structures of L. blandensis dGTPase identify a putative dATP allosteric site and a loop critical for enzymatic activity
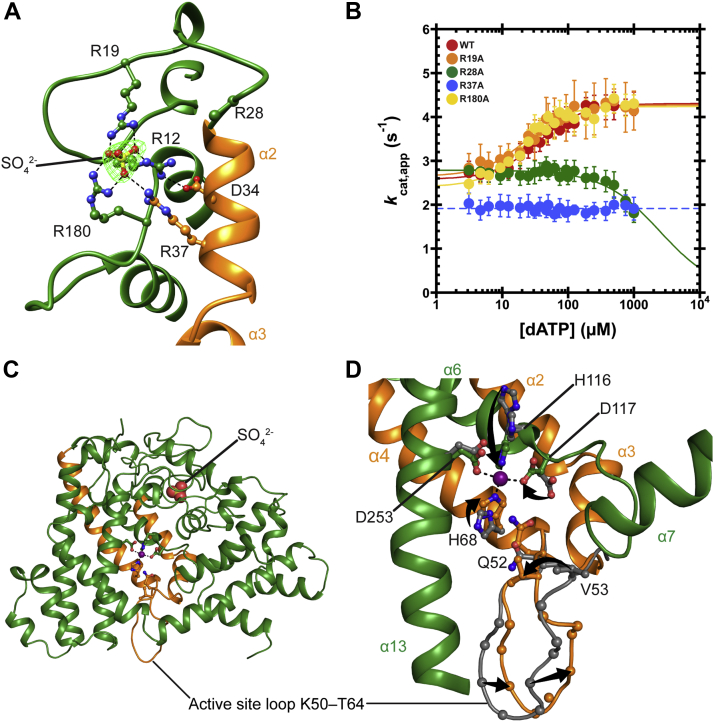
To explore the structural basis for the dGTPase activity and its allosteric activation, we determined several crystal structures of the enzyme in the absence of dNTP ligands (Table S2). These structures yielded a nearly complete model of the dGTPase peptide chain including several regions that were disordered in the previous structure (PDB ID: 3BG2). The hexameric structure is best described as a set of three parallel dimers with >1500 Å2 buried surface area between the two monomers within each dimer, compared with <600 Å2 between monomers from different dimers. We examined the structures for the possible dATP allosteric site by focusing on these intradimer interfaces, since in the case of the E. coli dGTPase, this interface contained the ssDNA activator (12) (Fig. S1, A and B). Interestingly, we found sulfate ions bound in the corresponding locations, coordinated by several arginine residues (Figs. 3A and S1, A and B). We reasoned that this arginine-rich region may play a role in enzyme activation by dATP. To probe this hypothesis, we mutated three arginine residues to alanine: R19, R37, and R180, which directly interact with the sulfate ion in our crystal structures (Fig. S2). We also mutated the disordered R28, which is on a loop near the sulfate ion site but does not interact with the sulfate. Importantly, when we measured dATP activation of the mutant enzymes, we found that R28A and R37A eliminated activation by dATP (Fig. 3B). We concluded that the L. blandensis dATP allosteric site, like the ssDNA-activating site in the E. coli dGTPase, is likely at the monomer–monomer interface.
Figure 3.
Crystal structures of Leeuwenhoekiella blandensis dGTPase.A, close-up of a sulfate-binding site showing the position of nearby arginine residues. Sulfate Fo–Fc omit map contoured to 6σ (green mesh). Alpha helices α2 and α3 are important for our proposed allosteric activation model (discussed later), so they are colored orange and labeled for reference. Generated in Chimera. B, dATP titrations at 1 mM dGTP. Data are plotted as the mean and SEM of four independent experiments. Relative to the WT (red; data are replotted from Fig. 2C for reference), R19A (orange) and R180A (purple) have minimal effect on activation by dATP (K1/2 = 36 ± 3 μM, 23 ± 3 μM, and 21 ± 2 μM, respectively). R28A (green) and R37A (blue) eliminate activation by dATP, with dATP seemingly inhibiting R28A at higher concentrations and R37A activity entirely independent of dATP in the concentration range we tested. C, overview of monomer A. The orange helices (α2, α3, and α4) are colored as such for reference. Generated in PyMOL. D, the active site of R37A dGTPase. We use the R37A crystals for this analysis because loop residues 57 to 60 are disordered in the WT crystals, whereas we have a complete picture of the active-site loop in the R37A structures. Crystals were soaked with Mn2+ prior to freezing (solid, orange, and green) or frozen without soaking Mn2+ (gray). Structures were aligned by the HD motif in PyMOL (RMSD = 0.347 over the four α-carbon atoms). C-alpha atoms on the loop are shown as spheres. Arrows are highlighting residue displacement between the structures. Generated in PyMOL. HD, histidine–aspartate.
Attempts to cocrystallize the enzyme with nucleotides or to soak the apo crystals with nucleotides did not yield any nucleotide-bound structures. However, Mn2+ soaking yielded informative results, revealing a reorganization of the HD-motif metal-binding site accompanied by a conformational change in a nearby active-site loop (K50–T64, see later). Specifically, the addition of Mn2+ filled the metal site, which was otherwise poorly occupied, and led to full coordination with the four HD motif residues: H68, H116, D117, and D253 (Fig. 3, C and D). The K50–T64 loop (Fig. 3D) contains the conserved residues Q52 and V53 (e.g., see Fig. 5C). The corresponding residues in hSAMHD1 (Q149/L150) (23) or E. coli dGTPase (Q53/V54) (32) recognize the 2′-deoxyribose of the substrate, so we reasoned that this loop movement is likely important for catalysis. In the metal-free conformation, the side chain of V53 is pointed “up” into the active site in a position that would sterically clash with the guanine ring of the substrate (as judged by aligning the HD motif with substrate-bound structures of homologs). In the Mn2+-bound conformation, the V53 side chain is rolled over into a position that is expected to be more amenable to substrate binding: a 4.7 Å displacement is measured from the V53 β-carbon. The next several residues in the loop move in the opposite direction by as much as 4.5 Å. We deleted four residues to shorten the loop (Δ55–58) and determined crystal structures of the mutant in the presence or the absence of Mn2+. We found that the deletion introduced structural rigidity, locking Q52 and V53 into their metal-free conformation, regardless of the metal content of the active site (Fig. S1C). Importantly, the Δ55 to 58 mutant displayed no measurable dGTPase activity (Fig. S1D), consistent with the notion that the ability of the loop to adopt alternate conformations is essential for catalysis.
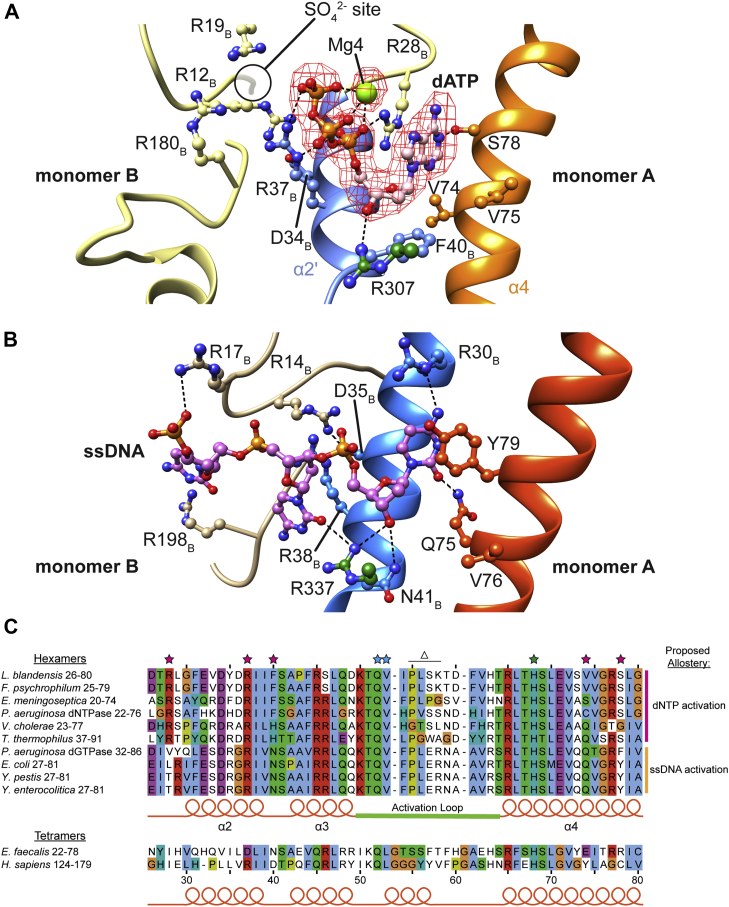
Figure 5.
Leeuwenhoekiella blandensis dGTPase allosteric site compared with Escherichia coli dGTPase. Generated in Chimera. A, dATP-binding residues from the H125A L. blandensis dGTPase + dGTP + dATP structure (th 0.11). Monomer A residues are colored green/orange, monomer B is colored blue/yellow, dATP is pink, M3 is green, and all are colored by heteroatom. There is additional density (at left) found in the same position as the sulfate ion from our crystal structures. The postprocessed map was generated using DeepEMhancer. B, the equivalent ssDNA-binding site in the E. coli dGTPase (Protein Data Bank [PDB] ID: 4X9E). C, alignment of dGTPase homologs, limited to the sequence containing the residues that we identified as important for allosteric activation in the L. blandensis homolog. The residues that directly interact with dATP are indicated by magenta stars. Residues that bind the dGTP 2′-deoxyribose are indicated by cyan stars. One of the two histidine–aspartate (HD) motif histidine residues is also present in this sequence, indicated by the green star. The residues that we deleted from the active-site loop in the Δ55 to 58 mutation are indicated by the Δ. Proposed allosteric site specificity for the hexameric homologs is labeled at right. The sequences included in the alignment are: L. blandensis dGTPase, EAQ51213; Flavobacterium psychrophilum dGTPase, WP_011962456; Enterococcus meningoseptica dNTPase, WP_078793617; Pseudomonas aeruginosa dNTPase (PA3043), NP_251733; Vibrio cholerae VC1979, Q9KQL9; Thermus thermophilus dGTPase (TT1383), BAC98488; P. aeruginosa dGTPase (PA1124), NP_249815.1; E. coli dGTPase, P15723; Yersinia pestis dGTPase, WP_002209369; Yersinia enterocolitica dGTPase, WP_075339176; Enterococcus faecalis dNTPase, WP_002357999; and Homo sapiens SAMHD1, Q9Y3Z3. SAMHD1, sterile alpha motif and HD domain–containing protein 1.
Cryo-EM structures of the L. blandensis dGTPase with dGTP and dATP
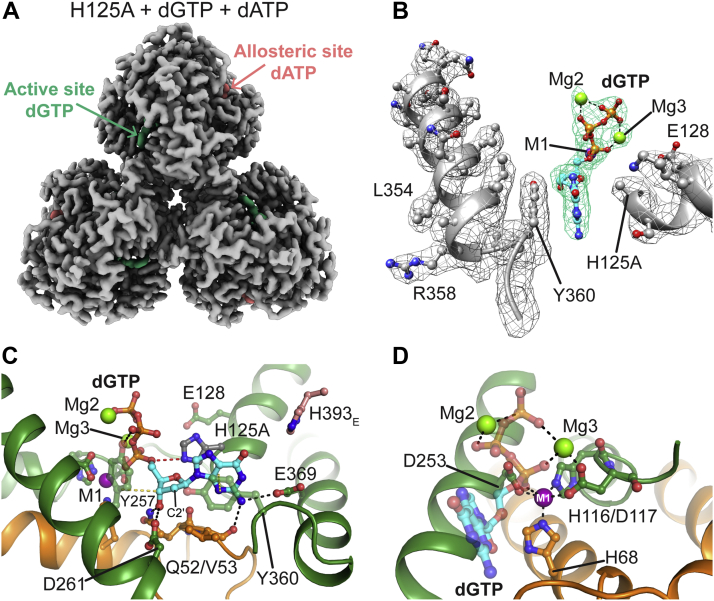
We then turned to cryo-EM to obtain structures of the dGTPase in ligand-bound states. Indeed, using this approach, we successfully obtained high-resolution cryo-EM structures of (i) WT dGTPase without nucleotide ligands (apo, 2.1 Å), (ii) WT bound to dATP activator (2.7 Å), (iii) a catalytically deficient mutant (H125A) bound to dGTP substrate (2.5 Å), and (iv) H125A bound to both dGTP and dATP (2.6 Å) (Figs. 4 and S2). The maps are all below 3 Å and of sufficient quality to yield complete density for each of the ligands (Fig. S3), many dGTPase side chains (Fig. 4B), and >97% of the dGTPase main chain. In order to be able to trap the dGTP substrate in a precatalytic state while maintaining a catalytically competent metal site, we mutated His125, which is the acid of a proposed catalytic dyad (His125/Glu128) (30, 32), to alanine. This histidine is a universally conserved residue amongst SAMHD1-related dNTPase homologs, whereas the glutamate is conserved as either Glu or Asp. When the equivalent histidine residues were mutated to alanine in E. coli (H126A) or hSAMHD1 (H215A), there were severe reductions in activity (30, 32). Indeed, when we tested the activity of the L. blandensis H125A mutant, we found that the rate constant for turnover (v0/[E]t) at 1 mM dGTP was reduced by approximately 16,000-fold (Fig. S4), which was sufficient to allow us to trap the substrate in the active site.
Figure 4.
Cryo-EM structures of dGTPase with dGTP bound in the active site.A, H125A dGTPase (gray) bound to both dGTP substrate (teal) and dATP activator (pink) (th 0.11). The postprocessed map was generated using DeepEMhancer, whereas the image was generated in ChimeraX. B, electron density for alpha helices in the active site (gray) and the dGTP substrate (teal) in monomer A in the H125A + dGTP structure (th 0.1). The postprocessed map was generated using DeepEMhancer, whereas the image was generated in Chimera. C, monomer A from the H125A + dGTP structure highlighting the guanine- and 2′-deoxyribose-binding residues. The point of view is from the left side of B. dGTPase residues are colored green/orange as for previous figures, dGTP is cyan, and each is colored by heteroatom; M1 is purple, whereas Mg2 and Mg3 are green. Hydrogen bonds are black dashed lines, whereas van der Waals/stacking interactions with Y257 and Y360 are yellow dashed lines. H125 (gray) was added back in silico using the Mutagenesis Wizard in PyMOL with backbone-dependent rotamers imposed. The H125 nitrogen NE2 would be 3.1 Å from the 5′-oxygen atom and in line to protonate it during catalysis (red dashed line). D, monomer A from the H125A + dGTP structure highlighting the positions of the three metal ions. The point of view is approximately the position of the helix containing H125 and E128 in C. Black dashed lines are metal-ligand interactions.
The L. blandensis dGTPase active site reveals an intact metal site with three metal ions coordinating the substrate
The active site of our structures is well resolved and allows us to visualize a complete set of interactions between dGTP, the dGTPase, and three active-site metal ions (Fig. 4, B and C). A comparison of the active sites of L. blandensis, E. coli, and hSAMHD1 is presented in Fig. S5, whereas the electron densities and proposed coordination spheres for each metal ion are shown in Fig. S6. In L. blandensis, the dGTP guanine moiety is specifically recognized via hydrophobic and hydrogen bond interactions with the enzyme. Y360 stacks with the guanine ring, and the 2-amino group hydrogen bonds with the E369 side chain and the backbone carbonyl of V53 (Fig. 4C). Hydrogen bond and hydrophobic interactions favor the 2′-deoxyribose over a possible ribose. The 3′-OH of the 2′-deoxyribose is within hydrogen bonding distance to Q52 and D261, and the side chain of V53 accomplishes steric exclusion of a 2′-OH by a van der Waals interaction with the 2′-deoxyribose (Fig. 4C). These interactions are conserved among SAMHD1 homologs and explain why ribose-containing nucleotides are poor substrates (Figs. 2A and S5) (21, 23, 31, 38). The importance of V53 for substrate recognition is also supported by a V53T mutation, which impairs dGTPase activity in the absence of dATP, reducing the kcat sevenfold relative to WT (Fig. S1D and Table S1). The dGTP-bound conformation of V53 and the active-site loop is similar but not identical to the Mn2+-soaked crystal conformation. The universally conserved Y257 completes the sugar interaction surface with a van der Waals interaction on the opposite face of the 2′-deoxyribose ring (Fig. 4C).
The triphosphate moiety of dGTP is bound in an apparently catalytically competent conformation (Fig. 4, C and D). Y257, which also interacts with the 2′-deoxyribose ring, forms a hydrogen bond with the pro-SP nonbridging oxygen of the β-phosphate (Fig. 4C), whereas the γ-phosphate interacts with K191, Y192, and K209 (Fig. S7A). The pro-RP oxygen of the α-phosphate is bound to the catalytic metal, M1, which is itself coordinated by the HD motif (H68, H116/D117, and D253) (Fig. 4D). These five ligands form a nearly optimal trigonal bipyramidal coordination sphere (Fig. S6A) or a suboptimal octahedral coordination (S6b) for which the last ligand is a water that is not resolved in the electron density. The M1 observed in our structures is likely the Mg2+ supplied in excess during grid preparation and is built into each of our structures as such.
We noted additional density bridging the β-phosphates and γ-phosphates, where we have modeled a second metal ion (Mg2). There are two putative second-shell ligands for this site, E250 and D254, whereas the inner sphere ligands are probably water molecules that would be easily accommodated in this pocket (modeled into the site in Fig. S6C). We also noted additional spherical density bridging between the dGTP γ-phosphate and HD motif residue D117. We modeled here a third divalent metal (Mg3), which could assist catalysis by forming productive substrate–protein interactions to position the phosphates for nucleophilic attack (Fig. 4D). There are three observable ligands for Mg3 (Fig. 4D) including the D117 side chain, the pro-SP oxygen of the α-phosphate, and the γ-phosphate. The remaining sites required to complete an octahedral coordination sphere are not accounted for in the electron density, but these would be easily accommodated within the active site as water molecules (modeled into the site in Fig. S6D). One of the putative waters would be in position for an in-line nucleophilic attack on the scissile bond and is equivalent to the proposed nucleophilic water in the SAMHD1 active site (30). The other waters fit into cavities in the protein where one would be bound by N120 (H210 in hSAMHD1) and the other would be bound by N161 (E234 in hSAMHD1) and the H116 backbone carbonyl (Fig. S6D). Each of these protein ligands could therefore act as second-shell ligands for Mg3.
The three metal sites are found with different ligand compositions: M1 has a mixture of nitrogen and oxygen ligands, whereas Mg2 and Mg3 have only oxygen ligands. This could explain why the E. coli dGTPase displayed optimal activity when both MnCl2 and MgCl2 were present (18, 19, 32). The physiological M1 is likely a tightly bound transition metal like Mn2+, Co2+, or Fe2+, which would bind to the active-site nitrogen ligands better than a hard Lewis acid like Mg2+. Given their apparently octahedral coordination and the fact that their proposed ligands are all oxygen atoms, the Mg2 and Mg3 sites are probably filled by Mg2+ ions that bind in a substrate-dependent manner. Either one could feasibly enter the active site as Mg-dGTP, whereas the other probably fills the site from bulk solution.
dATP is not used as a substrate by the L. blandensis dGTPase (Fig. 2A), and it was therefore a surprise that it was found in the active sites when present as the sole ligand (WT + dATP structure, Fig. S3B). In contrast, when dGTP was the sole ligand in the H125A + dGTP structure, it was bound only to the active site with the allosteric site remaining unoccupied (Fig. S3C). At the active site, the sugar and phosphate moieties of dATP and dGTP are bound in similar positions, although they are not identical (Fig. S7A). However, the catalytic acid H125 is rotated away from the dATP substrate in a conformation that is not productive for the proposed reaction mechanism. Likewise, the dATP α-phosphate is bound farther from M1 and out of alignment for a nucleophilic attack, and there is no density for Mg2 or Mg3, all consistent with the lack of detectable dATPase activity for the enzyme (Fig. 2A). While this structure suggests that dATP could act as a competitive inhibitor by binding to the active site, this is unlikely to be the case under physiological conditions. The data in Figure 2B show a stimulatory effect for 500 μM dATP, even when it is in excess of the dGTP substrate in the low micromolar range.
dATP is specifically recognized at the L. blandensis allosteric site
Our cryo-EM structures confirm that the allosteric dATP-binding site of the L. blandensis dGTPase is located at the monomer–monomer interaction surfaces within each enzyme dimer (Figs. 5A and S2, B and D), where two dATP molecules are bound symmetrically per dimer. This location is near to where the ssDNA is bound to the E. coli dGTPase (Fig. 5), as also correctly predicted from the sulfate location in our crystal structures (Fig. 3). Adenine specificity is provided by the interaction of S78 with the Watson–Crick edge of the base and a stacking interaction with R28B (Fig. 5A). The loop containing R28 is partially disordered from Q24 to R28 in structures lacking dATP but becomes fully ordered when dATP is bound (e.g., Fig. S7B, cyan versus yellow). The adenine moiety is recognized through a hydrogen bond between S78 and the adenine N1 atom, a hydrogen bond acceptor (Fig. 5A). Conversely, a guanine base presents a hydrogen bond donor at this position, suggesting that S78 plays a role in base selectivity. Indeed, an S78G mutant was only weakly activated by dATP, the K1/2 being increased from 36 ± 3 μM to > 1.5 mM (Fig. S8A), confirming the importance of S78 for adenine binding.
V74 and V75 form a hydrophobic pocket below the plane of the adenine ring where the exocyclic 2-amino group of guanine would sterically clash (Fig. 5A). Thus, they likely also play a role in selecting against dGTP in the allosteric site. A V74Q mutant displayed lower overall dGTPase activity, but the residual dGTPase activity was still activated by dATP (Fig. S8B). However, dATP apparently binds to the mutant enzyme with lower affinity: K1/2 is increased from 36 ± 3 μM (WT) to 180 ± 16 μM (V74Q). The 2′-deoxyribose moiety of the dATP is bound by R307, which hydrogen bonds with the 3′-OH (Fig. 5A). F40B stacks with the sugar ring and would exclude a 2′-OH, thus preventing activation of the dGTPase activity by ATP. An F40N mutant eliminated allosteric activation by dATP and also displayed reduced dGTPase activity in its absence (Fig. S8C).
The dATP triphosphate moiety is bound by an ionic hydrogen bond between R28B and the α-phosphate pro-SP oxygen and by interactions between R37B and the α-phosphate pro-RP oxygen and the γ-phosphate (Fig. 5A). These interactions provide a structural explanation for why the R28A and R37A mutant enzymes were not activated by dATP. In contrast, R19B and R180B are more than 6 Å from the nearest atom in dATP (Fig. 5A), explaining why R19A and R180A mutations did not affect dATP allosteric activation (Fig. 3B). We identified additional density adjacent to the triphosphate moiety and modeled a fourth metal ion (Mg4) as Mg2+ (Fig. 5A).
Discussion
We report here high-resolution crystallographic and cryo-EM structures of a hexameric SAMHD1 homolog from the marine bacterium L. blandensis. The results are the highest resolution cryo-EM structures of any HD domain protein, to date, including a 2.1 Å resolution cryo-EM structure of the WT apo dGTPase (previous structures are >3 Å, e.g., (39, 40)). The near-atomic resolution structures provide new details of the intact HD motif’s metal coordination and reveal a catalytically competent conformation of the substrate at the metal site, which for the first time includes additional metal ions (Mg2 and Mg3) in a structure of a hexameric homolog. We propose that Mg3 is important for catalysis, positioning the substrate’s phosphates as well as aligning the nucleophilic water/hydroxide with the scissile bond for hydrolysis. We show that this homolog is specifically a dGTP triphosphohydrolase, further characterized by allosteric regulation using dATP as the sole activator in vitro. We describe the first ligand-bound structures of a SAMHD1/dGTPase homolog determined by cryo-EM, including several different biologically relevant nucleotide-bound states.
A general catalytic mechanism for dGTPase/SAMHD1 homologs
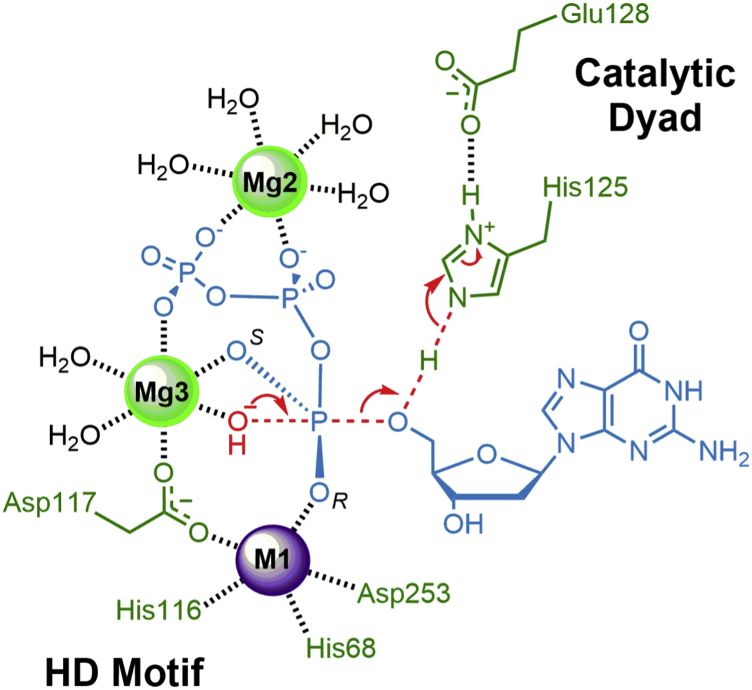
A recently proposed model for the catalytic mechanism of SAMHD1 homologs includes the nucleophilic water bound to three metal ions coordinated to the HD motif and the substrate, whereas a universally conserved His–Asp catalytic dyad protonates the leaving group (30, 32). Our data support this catalytic mechanism for the L. blandensis dGTPase (Fig. 6). Our metal M1 is equivalent to the Fe from the study by Morris et al. (30), whereas the Mg2/Mg3 ions are bound in roughly equivalent positions (compare Fig. S5, A and C). In the mechanism of Figure 6, the active-site metals interact with the substrate phosphates to align the nucleophilic water/hydroxide for an in-line attack on the scissile bond between the 5′-oxygen and the α phosphorous atom, whereas the catalytic dyad (H125 and E128 in L. blandensis) catalyzes the reaction by protonating the deoxyguanosine leaving group. The hexameric dGTPases and SAMHD1 include domain differences that result in the substrate β-phosphates and γ-phosphates binding in alternate conformations near Mg2 (Fig. S5A versus S5C). However, in both cases, the α-phosphates are positioned identically and align the scissile bond with the proposed H2Onuc for catalysis. Thus, this model provides a conserved mechanism among the distantly related SAMHD1/dGTPase homologs for generating the nucleophilic hydroxide and protonating the deoxynucleoside leaving group. In the case of hSAMHD1, the model is supported by differences in the ability of various divalent metals to support hydrolysis of SP-αS dNTP substrate analogs (41). In these experiments, replacing Mg2+ with the more thiophilic Co2+ enhanced the hydrolysis of SP-dTTPαS but not RP-dTTPαS, supporting the structural data showing the pro-SP oxygen bound to Mg3. In the present case, the model of Figure 6 is supported by the 1.6 × 104-fold catalytic defect of the L. blandensis H125A mutant (Fig. S4), commensurate with mutations to critical catalytic acid/base residues found in other enzymes (e.g., (42, 43)).
Figure 6.
Transition state of the proposed reaction mechanism for SAMHD1-related dNTPase homologs, with residues numbered for the Leeuwenhoekiella blandensis dGTPase. The α-phosphate pro-RP and pro-SP oxygens are labeled R and S, accordingly. SAMHD1, sterile alpha motif and HD domain–containing protein 1.
Sequence determinants of allosteric specificity
The allosteric activation of L. blandensis dGTPase by dATP is unique among the SAMHD1 homologs studied to date. The E. coli dGTPase is regulated by ssDNA (12), whereas the tetrameric hSAMHD1 and E. faecalis dNTPase (EF1143) are regulated by the twin binding of two distinct (d)NTPs (e.g., 13, 23, 24, 25). Nevertheless, a unifying feature is that all these allosteric effectors bind at the interface of two adjacent enzyme monomers near structural elements that are physically connected to the active sites (e.g., Fig. S5, orange helices). Indeed, the dATP-binding site is only slightly displaced from the site of the terminal nucleotide of the ssDNA fragment in the E. coli dGTPase (Figs. 5 and S9). Not surprisingly, given the differences in allosteric specificity, most of the residues that we identify for dATP binding are not conserved in the E. coli dGTPase. In E. coli, the L. blandensis S78 is replaced by Y79, which instead stacks with the terminal DNA base. L. blandensis V74 is instead Q75, which hydrogen bonds with the C2 carbonyl of a terminal pyrimidine and F40B is instead N41B, which interacts with the 3′-OH of the terminal nucleotide.
A sequence alignment of several relevant homologs shows distinct clusters of sequence homology for regions comprising the putative allosteric sites at their monomer–monomer interfaces (Fig. 5C). Likely, the differences in these sites coevolved with the distinct allosteries. Based on the sequence conservation and established in vitro allosteric specificities, we predict that homologs that encode allosteric sites like the L. blandensis enzyme will be activated by dNTPs (Fig. 5C). Homologs such as the Pseudomonas aeruginosa dNTPase (PA3043) (44) and some Flavobacterial homologs, including those from Flavobacterium psychrophilum and Enterococcus meningoseptica, have almost total conservation of the L. blandensis allosteric site and are likely also activated by dATP. Other homologs such as the Thermus thermophilus dNTPase and the putative Vibrio cholerae dGTPase (VC1979) are likely activated by other dNTPs, potentially dTTP or dCTP based on similarities to the nucleotide recognition side chains of the E. coli allosteric site. We also predict that the homologs that cluster with the E. coli enzyme are activated by ssDNA terminating in a pyrimidine nucleotide. This includes homologs from the closely related genus Yersinia as well as the P. aeruginosa dGTPase (PA1124) (Fig. 5C) (44, 45). The presence of the arginine at L. blandensis position 28 or E. coli position 30, which do not align with one another and are not in equivalent positions in their respective structures, could be a distinguishing feature between dNTP and ssDNA activation and might be used to predict these specificity differences in other homologs.
Model for activation by dATP
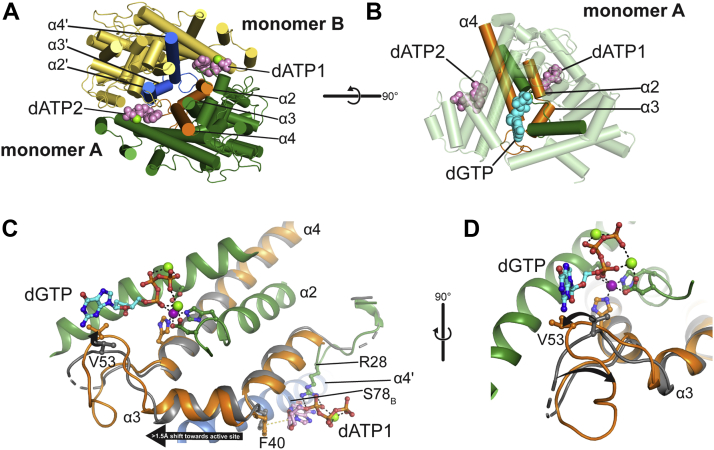
The present structural and biochemical data on the L. blandensis dGTPase allow us to propose a mechanistic model for its allosteric activation by dATP (Fig. 7). In this mechanism, the allosteric sites transmit dATP binding directly to a critical active-site substrate-binding loop (Fig. 7, orange/blue). At each monomer–monomer (intradimer) interaction surface, there are two dATP-binding sites (indicated as dATP1 and dATP2 for the A/B dimer, Fig. 7). dATP1, as defined here, makes contacts with monomer A residues R28A, R37A, F40A, and with monomer B residues V74B, S78B, and R307B. The reverse is true for dATP2. The two catalytic sites in the A/B dimer are located nearby and are physically connected to both dATP sites by way of each monomer’s helices α2, α3, and α4 (Fig. 7). Thus, in principle, either dATP molecule could influence either active site. However, the movements observed in the dATP-bound structures suggest different effects for each of the two dATPs. Specifically, it appears that only dATP1 affects monomer A’s active site, whereas dATP2 affects the active site of monomer B. Upon dATP binding, both F40 residues move by ∼1 Å upon interaction with the dATP 2′-deoxyribose, whereas the loops containing R28 become ordered (Figs. 5A and 7C). However, there is an associated >1.5 Å shift of each helix α3 downward along its respective helical axis, effectively “pushing” more than 1.5 Å further into each active site (Fig. 7C). Thus, dATP1 binding affects the active site of monomer A as shown in Figure 7C, whereas the complement is true for dATP2 and monomer B.
Figure 7.
Model for dGTPase activation by dATP. dATP binding to the allosteric site is proposed to activate dGTPase by way of the connections between the orange structural elements and the active-site loop. Generated in PyMOL. A, side-on view of the dimer containing monomers A and B. Monomer A is green, with helices α2, α3, and α4 colored orange. Monomer B is yellow, with helices α2', α3′, and α4′ colored blue. The dATP molecules are colored pink; the two M4 atoms are green spheres. B, overview of monomer A (transparent) highlighting the physical linkage between the active site and the two dATP sites (orange) and other important active-site structural elements (green), otherwise colored as in A with the dGTP colored cyan. C, displacement of helix α3 in dATP site 1 is coupled to the active-site loop movement in monomer A. dATP is pink, dGTP is cyan, and both are colored by heteroatom; M1 is shown as a purple sphere, whereas M2 and M3 are green spheres. R28 and F40 binding to dATP1 causes helix α3 to be displaced by >1.5 Å further into the active site (relative to the apo structure). There is a corresponding movement in the active-site loop displacing the V53 side chain (black arrow). The apo EM structure (gray) is aligned by the metal-coordinating residues of the monomer A histidine–aspartate (HD) motif. D, rotated 90° relative to C to highlight the loop movement in the active site (black arrows). V53 is “pushed” 3.6 Å further into the active site by α3 (as measured by the β-carbon), whereas P55 is “pulled” >5.5 Å in the opposite direction (as measured by the α-carbon).
This movement of α3 is accompanied by a “push–pull” displacement in the important active-site loop K50–T64. The critical residue V53 in the loop is “pushed” away from dATP1 by 3.6 Å (as measured from the β-carbon) into a more favorable interaction with dGTP substrate, whereas the next several residues in the loop are “pulled” in the opposite direction by more than 5.5 Å (as measured from the P55 α-carbon). As discussed earlier, the flexibility of this loop is essential for enzyme activity (Figs. 3, C and D and S1, C and D). The dGTP-free structures have the loop in a range of conformations that position the V53 side chain in a potential steric clash with the dGTP guanine ring (e.g., Figs. 3D and 7C, gray), whereas in the dGTP-bound conformation, the loop is rotated and the V53 side chain displaced to, instead, make a favorable van der Waals interaction with the 2′-deoxyribose (Fig. 7C, orange). Based on the aforementioned, we propose that dATP binding to the allosteric site either (1) prepositions the active-site loop favorably for dGTP binding or (2) alters the loop dynamics such that the energy required for dGTP binding to move the loop into the correct position is reduced. In either case, we propose that dATP binding improves dGTP binding by increasing the substrate affinity, thus enhancing catalysis.
Possible role for dGTPases in regulating dNTP pools
dGTPases destroy the critical DNA precursor dGTP and therefore have the potential to affect the cellular dNTP concentrations available for DNA replication and repair. The dGTP concentration is expected to be affected directly, but other dNTPs could also be affected because of the regulatory features of the RNR, which shifts its production of 2′-deoxynucleotides to balance the individual dNTP concentrations. Recently, a new type of RNR was discovered in L. blandensis (46, 47, 48, 49), which includes a novel dimanganese center for radical generation as well as a novel domain architecture with the ATP cone residing on the small RNR subunit rather than being part of the large subunit. This novel RNR, like other RNRs previously described, is strongly inhibited by dATP binding to the cone domain. Given the importance of dATP for regulating both of these enzymes in vitro, wherein allosteric dATP binding activates dGTPase but inhibits RNR, L. blandensis may be an attractive model organism for future studies on dNTP pool balancing mechanisms and nucleotide metabolism regulatory circuits.
Experimental procedures
Cloning and mutagenesis
A pSGX3 plasmid encoding residues 3 to 442 of the L. blandensis MED217 dGTPase was purchased from the DNASU plasmid repository (plasmid ID: LbCD00297911). Expression plasmid pMCSG7-Lb_dgt was created by inserting dGTPase residues 3 to 442 into the pMCSG7 vector using LIC cloning (cloning primers can be found in Table S4). dGTPase variants were generated using the QuikChange II XL kit (Agilent) according to the vendor’s directions. Primers were designed using the Agilent QuickChange Primer Design tool (Table S4). The pMCSG7-Lb_dgt plasmid and mutations were verified by Sanger sequencing.
Expression and purification of WT and mutant L. blandensis dGTPase
E. coli strain BL21(DE3) was transformed with pMCSG7–Lb_dgt. Cultures were grown in 1 l terrific broth medium supplemented with 4% glycerol and 100 μg/ml ampicillin. Cultures were grown at 37 °C, shaking at 225 rpm to an absorbance of at least 1.5 at 600 nm, then cooled to 20 °C for at least 1 h, induced with 0.2 mM IPTG, and grown for an additional 14 to 18 h at 20 °C before harvesting by centrifugation. Cell pellets were stored at −20 °C until used for protein purification.
Cell pellets were resuspended in approximately 5 ml buffer A (100 mM Tris, pH 7.9, 500 mM NaCl, 30 mM imidazole, and 5% glycerol) per 1 g of cell paste. All purification steps were performed at 4 °C. Resuspended cells were incubated with bovine pancreas DNase (4 mg), lysozyme (40 mg), and MgCl2 (2 mM). Cells were lysed using a Branson 450 Digital Sonifier at 60% power, with three to four 1 min sonication cycles and at least 2 min between cycles. Lysate was clarified by centrifugation at 30,000g for 30 min. The soluble fraction was loaded onto a 5 ml HisTrap Ni–NTA column (Cytiva) in 95% buffer A and 5% buffer B (buffer A, but with 300 mM imidazole) and eluted with a gradient from 5% to 100% buffer B (45–300 mM imidazole). The sample was further purified by size-exclusion chromatography with a HiLoad 16/60 Superdex 200 column (Cytiva) in buffer C (50 mM Tris, pH 7.9, 100 mM NaCl, and 10% glycerol). The dGTPase eluted with an apparent molecular weight of 200 kDa. The purified protein samples were concentrated to 10 to 20 mg/ml (monomer), aliquoted, flash cooled with liquid nitrogen, and stored at −80 °C.
HPLC dGTPase activity assay
dGTPase was incubated with all four standard dNTPs and four standard NTPs at 1 mM, in a reaction buffer containing 100 mM Tris, pH 8, 10 mM MgCl2, and 125 μM MnCl2. The reaction was initiated by adding enzyme to substrate and then incubated at room temperature for 1 h. The timepoint was quenched with 1/10th volume 1 M NaOH (final concentration of 100 mM), diluted 1:4 with reaction buffer supplemented with 100 mM NaOH, spun on a 0.5 ml 30 kDa molecular weight cutoff Amicon centrifugal filter to separate the protein, and then the (d)NTP-containing flowthrough was neutralized with 1/10th volume 1 M HCl (final concentration of 90 mM NaCl). A zero-time control was made by quenching the enzyme with NaOH prior to adding substrate, then subsequently handled as with the 1 h timepoint. The neutralized flowthroughs were stored at −20 °C until they were thawed and loaded at 0.8 ml/min onto a Zorbax Eclipse XDB-C18 column (Agilent Technologies) on an Agilent 1200 Series HPLC instrument pre-equilibrated in 65% buffer HPLC-A (10 mM potassium phosphate, 5 mM PIC A [tetrabutylammonium phosphate; Waters], in 0.5% methanol, pH 7) and 35% buffer HPLC-B (50 mM potassium phosphate, 5 mM PIC A, in 30% methanol, pH 7). Products were eluted at 0.8 ml/min using a gradient from 35% to 60% HPLC-B from 0 to 10 min, then substrates were eluted with a second gradient from 60% to 80% HPLC-B with a parallel gradient from 0% to 5% of buffer HPLC-C (50% methanol) from 10 to 20 min. The column was washed in the final condition from 20 to 25 min, then returned to the starting condition over 1 min, and re-equilibrated for >5 min prior to injecting another sample. Absorbance traces were collected at 260 nm along with the absorbance spectra from 210 to 400 nm. The peak identities were confirmed using these spectra and by comparing the retention times with control samples containing the mixtures of only the four dNTPs or the four NTPs.
For the HPLC dGTP hydrolysis time-course assays, reactions were incubated for the indicated times. Timepoints were quenched and handled as described previously. The dGTP substrate and dG product were separated as described previously, absorbance traces were collected at 250 nm, the peak areas integrated, and the fraction product (dG/[dG + dGTP]) used to estimate product concentration at each timepoint, which were then used to determine initial velocities (v0). We corrected for differences in enzyme concentration between assays by dividing v0 by the total monomer (i.e., active site) concentration: ∼60 nM for WT and ∼600 nM for H125A. The resulting v0/[E]t values for the reactions at each timepoint are reported.
Enzyme-coupled dGTPase activity assay
The in vitro assay for dGTPase activity was adapted from our previously published enzyme-coupled assay (37) for use in a 96-well plate format. All reactions were carried out in a modified reaction buffer (100 mM Tris [pH 8], 5 mM MgCl2, 125 μM MnCl2, and 5 mM inorganic phosphate [pH 7]). Hydrolysis of dGTP to dG and inorganic triphosphate is coupled to a spectroscopic change by inclusion of purine nucleoside phosphorylase (PNP; Sigma) and xanthine oxidase (XOD; Sigma). PNP uses inorganic phosphate to convert the dG product into α-D-2′-deoxyribose 1-phosphate and guanine, and XOD then converts the guanine into 8-oxoguanine. This yields a different purine absorbance spectrum, with a maximum increase at 297 nm (37). Changes in absorbance at 297 nm were monitored in real time in a SpectraMax M5 microplate reader (Molecular Devices), with the reactions run in half-area, flat-bottom, UV-transparent Corning microplates (Sigma; catalog no.: CLS3679).
Given the potential for variance from a 1 cm pathlength in our plate assay, we used an experimental conversion factor to determine the product concentration from the change in absorbance. This conversion factor was determined by reacting a dilution series of guanine with XOD in the same reaction volume as our dGTPase reactions (40 μl). The endpoints of the Δ absorbance at 297 nm after the reactions reached completion were plotted against the starting concentrations of guanine. The conversion factor is the slope of this linear relationship (1.5 × 10−3 Δ absorbance at 297 nm/μM for a 40 μl reaction volume).
For dGTPase reactions, substrate stocks were serially diluted to yield two rows, each row also including a zero-substrate well. One row was diluted 1:1 with reaction buffer to be used as no-enzyme controls. The zero-substrate well from the no-enzyme control row was used as a plate blank. The reactions in the other row were initiated by 1:1 addition of a 2×-concentrated enzyme mixture (∼120 nM dGTPase monomer, 100 mU/ml PNP, and 1 U/ml XOD, in reaction buffer) using a multichannel pipette. The plate was shaken for 5 s, then the absorbance at 297 nm was read at 25 °C at the minimum interval allowed by the SoftMax Pro software (Molecular Devices; every 13 s when reading two rows). To normalize for the increase in baseline absorbance across the substrate dilution series, the absorbances for the substrate-only wells were subtracted from the corresponding reaction wells. Then, to correct for a slight upward drift in absorbance at 297 nm that was observed over long times in all enzyme-containing wells, the absorbance of the zero-substrate (enzyme-only) well in the reaction row was subtracted from the remaining reaction wells. This procedure was altered slightly for the v0 versus [dGTP] curves containing dATP, for which substrate stocks containing 500 μM dATP were serially diluted into reaction buffer also containing 500 μM dATP. The zero-substrate well in the no-enzyme control row (500 μM dATP in reaction buffer) was used as the plate blank, and the dATP-only well from the reaction row was subtracted from the dGTP titration wells. For the (d)NTP titrations, a stock of each (d)NTP containing 1 mM dGTP was serially diluted into reaction buffer also containing 1 mM dGTP. The zero-(d)NTP well in the no-enzyme control rows (1 mM dGTP in reaction buffer) were used as plate blanks, but the dGTP-only well from the reaction row was not subtracted from the dNTP titration wells.
Finally, all the data were converted into product concentrations at each timepoint using the experimental conversion factor described previously. A line was fit to the linear region of each concentration versus time curve to determine the initial velocity (v0), which was then divided by total dGTPase monomer concentration to yield v0/[E]t. For the v0/[E]t versus dGTP plots, a Michaelis–Menten equation with a Hill coefficient (n) was fit to the data (Equation 1). Equation 2 was fit to the kcat,app versus dATP activation curves, whereby kcat,0 is the rate constant for dGTPase activity without dATP, Δkcat is the amplitude of the activation, K1/2 is the concentration of dATP required for half-activation, and n is a Hill coefficient. Equation 3 was fit to the (d)NTP inhibition curves. In order to obtain an estimate of the IC50 from the incomplete inhibition data, we assumed the inhibition would reach completion and excluded a Hill coefficient.
| (1) |
| (2) |
| (3) |
Crystallization
To set trays, protein was thawed on ice, diluted to 10 mg/ml (monomer) with buffer C, and spun at 20,000g for 30 min at 4 °C. Crystals grew between 3 days and 2 weeks at 20 °C by vapor diffusion in 2 μl sitting drops that contained 1 μl of protein stock and 1 μl of reservoir solution (0.7–1 M potassium sodium tartrate, 0.14–0.20 M lithium sulfate, and 0.1 M Bis–Tris [pH 6.5]). Crystals were harvested in nylon loops, cryoprotected by soaking in well solution supplemented with 30% ethylene glycol, and flash cooled in liquid nitrogen. For structures of the metal complexes, the soaking solution also included 10 mM MnCl2.
Diffraction data collection and structure determination
Diffraction data were collected at 100 K on SER-CAT beamline ID22 at the Advanced Photon Source at Argonne National Laboratory. For each crystal, 200° of data were collected using 0.25° rotation and 0.1 s exposure per image. Data were processed using XDS (50) (Table S2). The structure of apo WT dGTPase was solved by molecular replacement with Phaser (51) in the PHENIX software suite (52) using the orphan L. blandensis dGTPase structure (PDB ID: 3BG2). Manual building and adjustment of the structure was completed using Coot (53). Refinement was performed using phenix.refine (54). Additional dGTPase structures were determined by isomorphous replacement with the apo WT structure. Structures were validated with MolProbity (55). Figures were prepared using PyMOL (Schrödinger, LLC).
Cryogenic-EM sample preparation and grid setting
About 1 mg of dGTPase was thawed on ice and then run over a 10/30 Superdex 200 Increase column (GE Healthcare) in buffer D (20 mM Tris [pH 7.5], 100 mM NaCl). The peak fraction (approximately 0.2 mg/ml hexamer) was used directly to set grids. The protein was mixed with appropriate substrates and MgCl2 (5 mM free concentration) at room temperature and used to set grids within 30 min (WT apo: 5 mM MgCl2; WT + dATP: 10 mM dATP and 15 mM MgCl2; H125A + dGTP: 10 mM dGTP and 15 mM MgCl2; H125A + dGTP + dATP: 10 mM dATP, 10 mM dGTP, and 25 mM MgCl2). Grids were prepared by applying 3 μl of the mixtures to glow-discharged UltrAuFoil R 1.2/1.3300 mesh grids (Quantifoil Micro Tools GmbH) followed by 2.5 to 3.5 s of blotting and vitrification in liquid ethane using an Automatic Plunge Freezer EM GP (Leica).
Cryo-EM data collection
Images of apo dGTPase were collected using a Titan Krios electron microscope (Thermo Fisher Scientific) operated at 300 keV and equipped with a K2 Summit direct electron detector (Gatan) placed behind a BioQuantum energy filter (Gatan) operated at the zero-loss peak with a 20 eV slit. Movies were collected in super-resolution mode (effective pixel size: 0.323 Å/pixel) at a nominal magnification of 2,15,000×, corresponding to a physical pixel size of 0.646 Å/pixel. About 3780 movies (50 frames/movie) were collected with defocus values ranging from −0.5 to −2.5 μm. Movies were recorded over 4 s resulting in a fluence of 58.6 e-/Å2 and flux of 6 e-/pixel/s.
Images of ligand-bound dGTPase were collected using a Talos Arctica electron microscope (Thermo Fisher Scientific) operated at 200 keV and equipped with a K2 Summit direct electron detector (Gatan, Inc). Movies were collected in counting mode at a nominal magnification of 450,00× corresponding to a physical pixel size of 0.932 Å/pixel. Overall, 361 (+dATP), 391 (+dGTP), or 279 (+dATP/dGTP) movies (60 frames/movie) were collected with defocus values ranging from −0.5 to −1.75 μm. Movies were recorded over 8.4 s resulting in a fluence of ∼54 e-/Å2 and flux of 5.6 e-/pixel/s.
Cryo-EM data processing
All data were processed in RELION (56) using normal procedures (Table S3 and Fig. S10). Briefly, movie frames were aligned using RELION’s own implementation of MOTIONCOR2 and the contrast transfer function (CTFs) estimated using CTFFIND-4.1 (57). CTF-corrected micrographs were manually inspected for quality, with 3239 (WT apo), 311 (WT + dATP), 367 (H125A + dGTP), and 256 (H125A + dGTP + dATP) used for the remaining steps. Particles were picked using Laplacian-of-Gaussian autopicking, extracted with 8× binning (for the apo dataset, effective pixel size of 5.168 Å/pix) or 4× binning (for the other datasets, 3.728 Å/pix), and submitted to 2D classification with alignment (T = 2). Good 2D classes were used to generate initial models at 15 Å (D3 symmetry), after which the particles were re-extracted with recentering and without downscaling and used for 3D autorefinement (apo dataset was still 2× binned at this step). These initial refined maps were submitted to 3D classification with alignment (k = 5, T = 4, D3 sym). For the apo dataset, the particles in each 3D class were unbinned prior to a refinement step to check for quality. A single class is included in the final dataset and was subjected to Bayesian polishing and CTF refinement to generate the final map. For the remaining datasets, the good 3D classes were combined, and the remaining particles were subjected to two rounds each of Bayesian polishing and CTF refinement. In all cases, the “good” classes carried forward were those for which the protein connectivity of the electron density agreed with our crystal structures and did not show excessive anisotropy. 3D autorefinement and postprocessing were run after each polishing and CTF refinement step to check the map quality improvements.
After the final postprocessing step, the resolution estimates, based on a Fourier shell correlation cutoff of 0.143, were 2.09 Å (WT apo), 2.74 Å (WT + dATP), 2.51 Å (H125A + dGTP), and 2.57 Å (H125A + dGTP + dATP). The half-maps from the final refinements were used to generate postprocessed maps in RELION, autosharpened maps in PHENIX, and postprocessed maps from the DeepEMhancer artificial neural network (these are the maps that are presented in our figures) (58). Each of these yielded Fourier shell correlation resolution estimates that are equivalent as judged by MTRIAGE (59). Our WT crystal structure was docked into each map for an initial real-space refinement in PHENIX (52). After this, iterative rounds of manual inspection and building in Coot (53) and real-space refinement in PHENIX were used to obtain the final models. Structures were validated with MolProbity (55). The interface analysis presented in the results was performed on the H125A + dATP + dGTP structure using the PISA server (60). Figures were prepared using PyMOL (Schrödinger, LLC), Chimera (61), or ChimeraX (62), as indicated in the figure legends.
Data availability
The structure factors and coordinates for the crystal structures have been deposited with the PDB under accession codes 7TU0 (WT dGTPase), 7TU1 (R37A dGTPase), 7TU2 (R37A dGTPase with Mn2+), 7TU3 (Δ55–58 dGTPase), and 7TU4 (Δ55–58 dGTPase with Mn2+). The cryo-EM maps and associated atomic models have been deposited in the Electron Microscopy Data Bank and PDB under accession codes EMD-26126/PDB ID: 7TU5 (WT apo), EMD-26127/PDB ID: 7TU6 (WT with dATP), EMD-26128/PDB ID: 7TU7 (H125A with dGTP), and EMD-26129/PDB ID: 7TU8 (H125A with dGTP and dATP).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Drs Niketa Bhawsinghka and Eric Sullivan for carefully reading this article. X-ray crystallographic datasets were collected by the Southeast Regional Collaborative Access Team at the Advanced Photon Source at Argonne National Laboratory. We thank Lars Pedersen and the Structural Biology Core facility at the National Institute of Environmental Health Sciences for their assistance. The apo dGTPase cryo-EM dataset was collected on a Krios instrument at the National Cancer Institute, whereas the other three cryo-EM datasets were collected using a Talos Arctica at the National Institute of Environmental Health Sciences. We thank Rick Huang and the staff of the National Institutes of Health IRP Cryo-EM Consortium for their assistance.
Author contributions
B. P. K., A. P. S., and M. J. B. methodology; B. P. K., A. P. S., A. L. H., and J. C. H. investigation; T. M. T. H. resources; B. P. K., A. P. S., and R. M. S. writing–original draft; R. M. S. supervision.
Funding and additional information
This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (project numbers: 1ZIAES050165, 1ZICES103326, and 1ZIAES101905 to T. M. T. H., M. J. B., and R. M. S., respectively). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by F. Peter Guengerich
Footnotes
Present address for Andrew P. Sikkema Research Department, New England Biolabs, Ipswich, MA 01938, USA. Present address for Allen L. Hsu Department of Physiology and Cellular Biophysics, Columbia University, New York, New York 10032, USA.
Supporting information
References
- 1.Coggins S.A., Mahboubi B., Schinazi R.F., Kim B. Mechanistic crosstalk between DNA/RNA polymerase enzyme kinetics and nucleotide substrate availability in cells: implications for polymerase inhibitor discovery. J. Biol. Chem. 2020;295:13432–13443. doi: 10.1074/jbc.REV120.013746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao N.Y., Schroeder J.W., Yurieva O., Simmons L.A., O’Donnell M.E. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12942–12947. doi: 10.1073/pnas.1309506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar D., Abdulovic A.L., Viberg J., Nilsson A.K., Kunkel T.A., Chabes A. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucl. Acids Res. 2011;39:1360–1371. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahluwalia D., Bienstock R.J., Schaaper R.M. Novel mutator mutants of E. coli nrdAB ribonucleotide reductase: insight into allosteric regulation and control of mutation rates. DNA Repair (Amst) 2012;11:480–487. doi: 10.1016/j.dnarep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahluwalia D., Schaaper R.M. Hypermutability and error catastrophe due to defects in ribonucleotide reductase. Proc. Natl. Acad. Sci. U. S. A. 2013;110:18596–18601. doi: 10.1073/pnas.1310849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofer A., Crona M., Logan D.T., Sjöberg B.M. DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 2012;47:50–63. doi: 10.3109/10409238.2011.630372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravind L., Koonin E.V. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 8.Kornberg S.R., Lehman I.R., Bessman M.J., Simms E.S., Kornberg A. Enzymatic cleavage of deoxyguanosine triphosphate to deoxyguanosine and tripolyphosphate. J. Biol. Chem. 1958;233:159–162. [PubMed] [Google Scholar]
- 9.Seto D., Bhatnagar S.K., Bessman M.J. The purification and properties of deoxyguanosine triphosphate triphosphohydrolase from Escherichia coli. J. Biol. Chem. 1988;263:1494–1499. [PubMed] [Google Scholar]
- 10.Beauchamp B.B., Richardson C.C. A unique deoxyguanosine triphosphatase is responsible for the OptA1 phenotype of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1988;85:2563–2567. doi: 10.1073/pnas.85.8.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gawel D., Hamilton M.D., Schaaper R.M. A novel mutator of Escherichia coli carrying a defect in the dgt gene, encoding a dGTP triphosphohydrolase. J. Bacteriol. 2008;190:6931–6939. doi: 10.1128/JB.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh D., Gawel D., Itsko M., Hochkoeppler A., Krahn J.M., London R.E., et al. Structure of Escherichia coli dGTP triphosphohydrolase, a hexameric enzyme with DNA effector molecules. J. Biol. Chem. 2015;290:10419–10429. doi: 10.1074/jbc.M115.636936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang S., Zhou X., Ahn J. Substrate specificity of SAMHD1 triphosphohydrolase activity is controlled by deoxyribonucleoside triphosphates and phosphorylation at Thr592. Biochemistry. 2016;55:5635–5646. doi: 10.1021/acs.biochem.6b00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franzolin E., Pontarin G., Rampazzo C., Miazzi C., Ferraro P., Palumbo E., et al. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14272–14277. doi: 10.1073/pnas.1312033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powell R.D., Holland P.J., Hollis T., Perrino F.W. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleoside triphosphohydrolase. J. Biol. Chem. 2011;286:43596–43600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belaglazova N., Flick R., Tchigvintsev A., Brown G., Popovic A., Nocek B., et al. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutiéres syndrome and HIV-1 restriction. J. Biol. Chem. 2013;288:8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaller T., Bauby H., Hué S., Malim M.H., Goujon C. New insights into an X-traordinary viral protein. Front. Microbiol. 2014;5:126. doi: 10.3389/fmicb.2014.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber H.E., Beauchamp B.B., Richardson C.C. Escherichia coli dGTP triphosphohydrolase is inhibited by gene 1.2 protein of bacteriophage T7. J. Biol. Chem. 1988;263:13549–13556. [PubMed] [Google Scholar]
- 19.Nakai H., Richardson C.C. The gene 1.2 protein of bacteriophage T7 interacts with the Escherichia coli dGTP triphosphohydrolase to form a GTP-binding protein. J. Biol. Chem. 1990;265:4411–4419. [PubMed] [Google Scholar]
- 20.Wurgler S.M., Richardson C.C. DNA binding properties of the deoxyguanosine triphosphate triphosphohydrolase of Escherichia coli. J. Biol. Chem. 1993;268:20046–20054. [PubMed] [Google Scholar]
- 21.Amie S.M., Bambara R.A., Kim B. GTP is the primary activator of the anti-HIV restriction factor SAMHD1. J. Biol. Chem. 2013;288:25001–25006. doi: 10.1074/jbc.C113.493619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen E.C., Seamon K.J., Cravens S.L., Stivers J.T. GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E1843–E1851. doi: 10.1073/pnas.1401706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji X., Tang C., Zhao Q., Wang W., Xiong Y. Structural basis of cellular dNTP regulation by SAMHD1. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4305–E4314. doi: 10.1073/pnas.1412289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vorontsov I.I., Minasov G., Kiryukhina O., Brunzelle J.S., Shuvalova L., Anderson W.F. Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex. J. Biol. Chem. 2011;286:33158–33166. doi: 10.1074/jbc.M111.250456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vorontsov I.I., Wu Y., DeLucia M., Minasov G., Mehrens J., Shuvalova L., et al. Mechanisms of allosteric activation and inhibition of the deoxyribonucleoside triphosphate triphosphohydrolase from Enterococcus faecalis. J. Biol. Chem. 2014;289:2815–2824. doi: 10.1074/jbc.M113.524207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji X., Wu Y., Yan J., Mehrens J., Yang H., DeLucia M., et al. Mechanism of allosteric activation of SAMHD1 by dGTP. Nat. Struct. Mol. Biol. 2013;20:1304–1309. doi: 10.1038/nsmb.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu C., Gao W., Zhao K., Qin X., Zhang Y., Peng X., et al. Structural insight into dGTP-dependent activtation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat. Comm. 2013;4:2722. doi: 10.1038/ncomms3722. [DOI] [PubMed] [Google Scholar]
- 28.Koharudin L.M.I., Wu Y., DeLucia M., Mehrens J., Gronenborn A.M., Ahn J. Structural basis of allosteric activation of Sterile α Motif and Histidine-Aspartate domain-containing protein 1 (SAMHD1) by nucleoside triphosphates. J. Biol. Chem. 2014;289:32617–32627. doi: 10.1074/jbc.M114.591958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo N., Nakagawa N., Ebihara A., Chen L., Liu Z.-J., Wang B.-C., et al. Structure of dNTP-inducible dNTP triphosphohydrolase: insight into broad specificity for dNTPs and triphosphohydrolase-type hydrolysis. Acta Crystallogr. D Biol. Crystallogr. 2007;63:230–239. doi: 10.1107/S0907444906049262. [DOI] [PubMed] [Google Scholar]
- 30.Morris E.R., Caswell S.J., Kunzelmann S., Arnold L.H., Purkiss A.G., Kelly G., et al. Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated hydrolysis. Nat. Commun. 2020;11:3165. doi: 10.1038/s41467-020-16983-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldstone D.C., Ennis-Adeniran V., Hedden J.J., Groom H.C.T., Rice G.I., Christodoulou E., et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 32.Barnes C.O., Wu Y., Song J., Lin G., Baxter E.L., Brewster A.S., et al. The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity. Proc. Natl. Acad. Sci. U. S. A. 2019;116:9333–9339. doi: 10.1073/pnas.1814999116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinhassi J., Bowman J.P., Nedashkovskaya O.I., Lekunberri I., Gomez-Consarnau L., Pedrós-Alió C. Leeuwenhoekiella blandensis sp. nov., a genome-sequenced marine member of the family Flavobacteriaceae. Int. J. Syst. Evol. Microbiol. 2006;56:1489–1493. doi: 10.1099/ijs.0.64232-0. [DOI] [PubMed] [Google Scholar]
- 34.Pinhassi J., Azam F., Hemphälä J., Long R.A., Martinez J., Zweifel U.L., et al. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 1999;17:13–26. [Google Scholar]
- 35.Riemann L., Steward G.F., Azam F. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 2000;66:578–587. doi: 10.1128/aem.66.2.578-587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaaper R.M., Mathews C.K. Mutational consequences of dNTP pool imbalances in E. coli. DNA Repair. 2013;12:73–79. doi: 10.1016/j.dnarep.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh D., Schaaper R.M., Hochkoeppler A. A continuous spectrophotometric enzyme-coupled assay for deoxynucleoside triphosphate triphosphohydrolases. Anal. Biochem. 2016;496:43–49. doi: 10.1016/j.ab.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondo N., Kuramitsu S., Masui R. Biochemical characterization of TT1383 from Thermus thermophilus identifies a novel dNTP triphosphohydrolase activity stimulated by dATP and dTTP. J. Biochem. 2004;136:221–231. doi: 10.1093/jb/mvh115. [DOI] [PubMed] [Google Scholar]
- 39.Gulati S., Palczewski K., Engel A., Stahlberg H., Kovacik L. Cryo-EM structure of phosphodiesterase 6 reveals insights into the allosteric regulation of type I phosphodiesterases. Sci. Adv. 2019;5:eaav4322. doi: 10.1126/sciadv.aav4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouvette J., Liu H.F., Du X., Zhou Y., Sikkema A.P., Mello J.F.R.E., et al. Beam image-shift accelerated data acquisition for near-atomic resolution single-particle cryo-electron tomography. Nat. Commun. 2021;12:1957. doi: 10.1038/s41467-021-22251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris E.R., Kunzelmann S., Caswell S.J., Purkiss A.G., Kelly G., Taylor I.A. Probing the catalytic mechanism and inhibition of SAMHD1 using the differential properties of RP- and SP-dNTPαS diastereomers. Biochemistry. 2021;60:1682–1698. doi: 10.1021/acs.biochem.0c00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Brien P.J., Ellenberger T. Human alkyladenine DNA glycosylase uses acid–base catalysis for selective excision of damaged purines. Biochemistry. 2003;42:12418–12429. doi: 10.1021/bi035177v. [DOI] [PubMed] [Google Scholar]
- 43.Dunican B.F., Hiller D.A., Strobel S.A. Transition state charge stabilization and acid–base catalysis of mRNA cleavage by the endoribonuclease RelE. Biochem. Biochem. 2015;54:7048–7057. doi: 10.1021/acs.biochem.5b00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mega R., Kondo N., Nakagawa N., Kuramitsu S., Masui R. Two dNTP triphosphohydrolases from Pseudomonas aeruginosa possess diverse substrate specificities. FEBS J. 2009;276:3211–3221. doi: 10.1111/j.1742-4658.2009.07035.x. [DOI] [PubMed] [Google Scholar]
- 45.Oh H.B., Lee K.-C., Park S.C., Song W.S., Yoon S.-I. Structural analysis of the dNTP triphosphohydrolase PA1124 from Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 2022;589:78–84. doi: 10.1016/j.bbrc.2021.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Grinberg I.R., Lundin D., Hasan M., Crona M., Jonna V.R., Loderer C., et al. Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit. eLIFE. 2018;7 doi: 10.7554/eLife.31529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grinberg I.R., Lundin D., Sahlin M., Crona M., Berggren G., Hofer A., et al. A glutaredoxin domain fused to the radical-generating subunit of ribonucleotide reductase (RNR) functions as an efficient RNR reductant. J. Biol. Chem. 2018;293:15889–15900. doi: 10.1074/jbc.RA118.004991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grinberg I.R., Berglund S., Hasan M., Lundin D., Ho F.M., Magnuson A., et al. Class Id ribonucleotide reductase utilizes a Mn2(IV,III) cofactor and undergoes large conformational changes on metal loading. J. Biol. Inorg. Chem. 2019;24:863–877. doi: 10.1007/s00775-019-01697-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hasan M., Banerjee I., Gringberg I.R., Sjöberg B.M., Logan D.T. Solution structure of the dATP-inactivated Class I Ribonucleotide Reductase from Leeuwenhoekiella blandensis by SAXS and cryo-electron microscopy. Front. Mol. Biosci. 2021;8:713608. doi: 10.3389/fmolb.2021.713608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Absch K.W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., et al. Phenix: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N., Mustyakimov M., et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J.H., Lindahl E., et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohou A., Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez-Garcia R., Gomez-Blanco J., Cuervo A., Carazo J.M., Sorzano C.O.S., Vargas J. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 2021;4:874. doi: 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afonine P.V., Klaholz B.P., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Biol. Crystallogr. 2018;74:814–840. doi: 10.1107/S2059798318009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krissinel E., Henrick K. Inference of macromolecular assemblies from a crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 61.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. USCF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure factors and coordinates for the crystal structures have been deposited with the PDB under accession codes 7TU0 (WT dGTPase), 7TU1 (R37A dGTPase), 7TU2 (R37A dGTPase with Mn2+), 7TU3 (Δ55–58 dGTPase), and 7TU4 (Δ55–58 dGTPase with Mn2+). The cryo-EM maps and associated atomic models have been deposited in the Electron Microscopy Data Bank and PDB under accession codes EMD-26126/PDB ID: 7TU5 (WT apo), EMD-26127/PDB ID: 7TU6 (WT with dATP), EMD-26128/PDB ID: 7TU7 (H125A with dGTP), and EMD-26129/PDB ID: 7TU8 (H125A with dGTP and dATP).