Abstract
Although antibodies are well developed and widely used in cancer therapy and diagnostic fields, some defects remain, such as poor tissue penetration, long in vivo metabolic retention, potential cytotoxicity, patent limitation, and high production cost. These issues have led scientists to explore and develop novel antibody alternatives. Protein scaffolds are small monomeric proteins with stable tertiary structures and mutable residues, which emerged in the 1990s. By combining robust gene engineering and phage display techniques, libraries with sufficient diversity could be established for target binding scaffold selection. Given the properties of small size, high affinity, and excellent specificity and stability, protein scaffolds have been applied in basic research, and preclinical and clinical fields over the past two decades. To date, more than 20 types of protein scaffolds have been developed, with the most frequently used being affibody, adnectin, ANTICALIN®, DARPins, and knottin. In this review, we focus on the protein scaffold applications in cancer therapy and diagnosis in the last 5 years, and discuss the pros and cons, and strategies of optimization and design.
Although antibodies are well developed and widely used in cancer therapy and diagnostic fields, some defects remain, such as poor tissue penetration, long in vivo metabolic retention, potential cytotoxicity, patent limitation, and high production cost.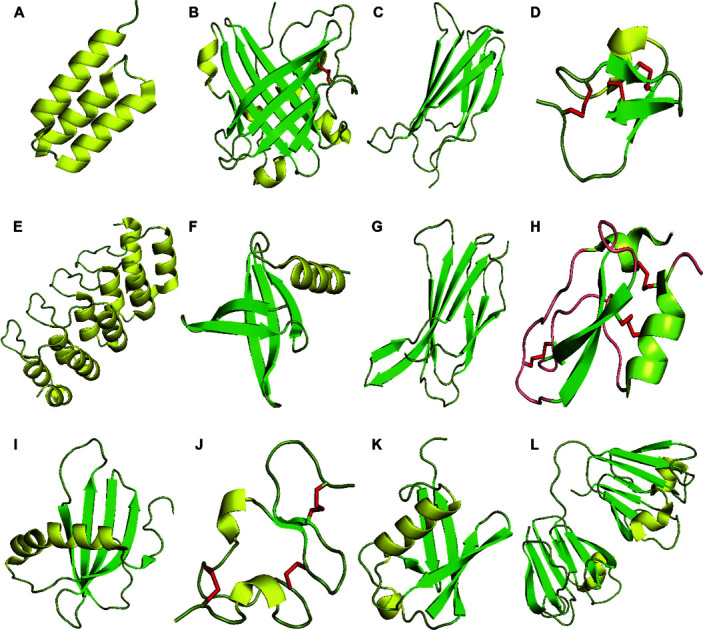
Introduction
Cancer is a severe disease that threatens human health. Traditional cancer treatments were designed as one-size-fits-all approaches; however, not all patients benefited from the same method owing to inter- and intra-tumor heterogeneity between individuals.1 To deal with these issues, precision medicine using multi-omics information analyses has been proposed to identify the crucial tumor markers for classifying patients and tailoring precisely targeted treatments.2–4
Monoclonal antibodies against tumor markers are the most widely investigated targeting elements for precise cancer diagnosis and treatment.5,6 Generally, antibodies are large size proteins, which comprise two pairs of heavy and light chains that form a stable Y-shape structure via disulfide bond linking. Antibodies can recognize biomarkers on cell surfaces, thereby activating immune responses or suppressing protumoral biological activities.7,8 However, several defects deter the further application of antibodies. For instance, the large molecular weight leads to poor tissue penetration for solid tumors, and hinders the binding of inside antigenic epitopes and the internalization of antibody-conjugated drugs.9 Posttranslational modifications are indispensable in facilitating the production of antibodies with complete functionality; however, these require the use of a high-cost and time-consuming eukaryotic expression system, which limits the large-scale production and access to most patients.10 Moreover, it may be difficult to introduce free residues and chemical linkers for further drug conjugation. In addition, in some situations, the antibody constant fragment (Fc) might incur immune-related cytotoxicity, resulting in damage to normal tissues and organs.11 While truncated antibodies such as antigen-binding fragments (Fab/scFvs) and alpaca nanobodies have been developed to remedy the shortcomings of full-size antibodies, some defects remained, such as lower avidity after modification and the requirement of humanization processing for reducing immunogenicity.12,13
Hence, urgent demands for addressing these limitations have motivated scientists to explore novel antibody alternatives. Based on the discoveries of small molecule proteins with binding function and the gradual development of biotechnology, it is feasible to confer targeting ability to small size proteins by mutating the variable region.14 Technically, a protein scaffold (Fig. 1)† is a small single-domain molecule (1–20 kDa), composed of two parts, which are analogous to antibodies. The first component is the constant region, which comprises a couple of α-helixes or beta-sheets constituting a rigid tertiary structure, while the second is the variable region, which is formed by a few exposed loops or several residues in the rigid secondary structures. The constant regions sustain inherent conformational stability of protein scaffolds, and variable sites provide a specific binding capability to various target molecules via structural ligand–receptor pairing or chemical forces. Generally, 10–20 amino acid residues in the protruded loops or rigid structures can be randomized to obtain novel affinity molecules. Protein scaffolds often lack disulfide bonds and require no translational modifications, allowing production using cost-effective prokaryotic expression systems. Furthermore, unnatural and free residues can be introduced into protein scaffolds via chemical synthesis for drug or diagnostic reagent conjugation. Small protein scaffolds are usually soluble with good physicochemical stability which favors their in vivo application.9,15 Despite the many advantages of protein scaffolds, they are more suitable for short-term cancer imaging, not therapy. Table 1 summarizes the major properties and developed targets of protein scaffolds.
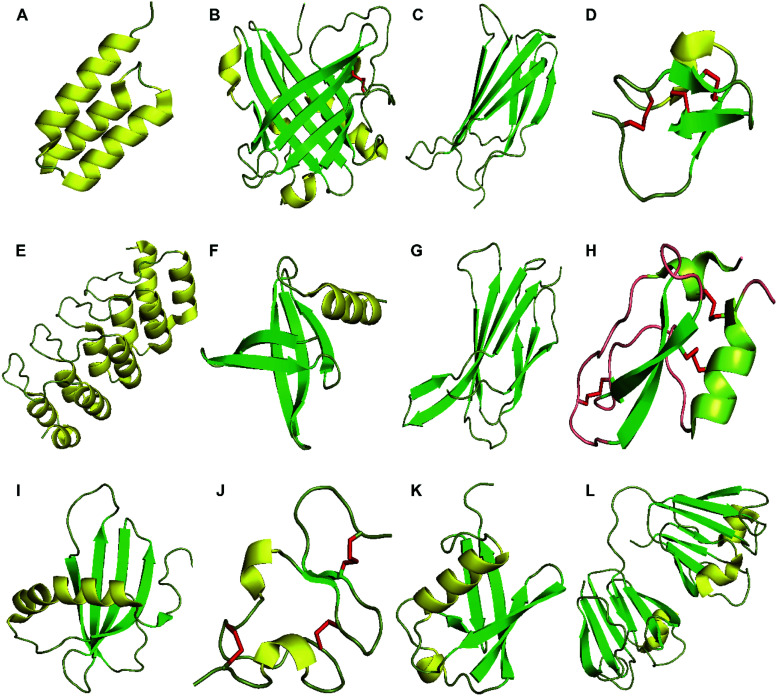
Fig. 1. Structures of proteins scaffolds. Notes: cartoon-illustrations are generated using PyMol, β-sheets are shown in green, α-helix structures are displayed in yellow and disulfide bonds are colored in red. (A) Affibody (PDB ID:3MZW); (B) ANTICALIN® (PDB ID: 4GH7); (C) adnectin or monobody (PDB ID: 1TTG); (D) Knottin (PDB ID: 2IT7); (E) designed ankyrin repeat proteins (PDB ID: 1MJ0); (F) Nanofitin or Affitin (PDB ID: 4CJ2); (G) centyrins (PDB ID: 5L2H); (H) Kunitz (PDB ID: 1KTH); (I) affimer (PDB ID: 1NB5); (J) avimer (PDB ID:1AJJ); (K) Affilin based on the ubiquitin (PDB ID:1UBI); and (L) affilin based on the γ-B-crystalin (PDB ID: 2JDG).
The overview of protein scaffolds properties and molecular targets.
| Protein scaffolds | Parental protein | Molecular size (kDa) | Affinity (Kd) | Structural feature | Targets |
|---|---|---|---|---|---|
| Affibody | B-domain of staphylococcal protein A | 6.5 | μM–pM | α-Helix | RSV-G protein, Taq DNA polymerase, human factor VIII, human IgA, Staphylococcal protein A domain, apolipoprotein A1, CD28, GP120, transferrin, CD25, c-Jun, H-Ras, Raf-1, VEGFR, HER2, murine lgG1, insulin, IAPP, IL-6, Tau, IGF1R, Aβ, TNF-α, IL-17, VEGFR-2, LOV, HER3, C5, CAIX, PDGFRβ, PD-L1, FcRn, HPV16 E7, EGFR, CTX-M15, CD276, EBV-LMP-2, fibrinogen, IgE, IgM, IL-1, IL-8, HAS, transthyretin |
| ANTICALIN® | Lipocalin | 18–20 | nM–pM | β-Barrel and terminal α-helix | Fluorescein, digoxigenin, hemoglobin, nonsymmetric phthalic acid ester, CTLA-4-Fc, estradiol-2, MET, hepcidin, VEGF-A, fibronectin extra-domain B, CD137, IL4Rα, PSMA, Aβ peptide, VEGFR-3, monosaccharide, HSP70, PCSK9, petrobactin, colchicine, CD98, IL-17, IL-23 |
| Adnecin/monobody | 10th type III domain of human fibronectin | 10 | μM–pM | β-Sandwich | Ubiquitin, estrogen receptor α, leptin, TNF-α, αvβ3 integrin, c-Src, VEGFR-2, hen egg-white lysozyme, MBP, hSUMO4, phosphor-IκBα, SARS-N protein, Abl kinase SH2 domain, EGFR, IGF-1R, IL-6, Fyn-SH3 domain, human IgG, Pak1 kinase, IL-23, Erk-2 CD domain, SHP2 SH2 domains, GFP, PCSK9, hEphA2, β-galactosidase, CDC34, COPS5, MAP2K5, SF3A1, USP11, human pregnane X receptor, KEAP1, GPR56, myostatin, H-Ras, CD4, C258S, methyltransferase, mesothelin, Candida rugosa lipase 1, MAPK, WDR5, PD-L1, gp41, glypican-3, VEGFR2, CD80, oncoprotein aurora A kinase, SET, Bcr-Abl p210, MLKL N-terminal domain |
| DARPins | Human ankyrin repeat proteins | 14–21 | nM–pM | α-Helix and β-turn | MBP, JNK2 kinase, NIa(pro) proteinase, AcrB, Na+ citrate symporter, caspase-2, apo Plk-1, CD4, TNF-α, EGFR, HER2, HER4, human IgG-Fc, IgE, neurotensin receptor 1, FcεRIα, EpCAM, NEMO-CC2-LZ, BCL-2, adenoviruses fiber protein knob domain, MsbA, JNK, LmrCD, tubulin, ERK2, BCL-W, VEGF-A, caspase-3, gp120, antigen FVIII, PDGF-BB, BCL-XL, caspase-7, human telomere quadruplex, Aβ, GFP, IL-4, CDCP1, lamin A/C, CD23, tubulin, MCL-1, IL-13, ribosomal protein S6 kinase 2, FccRIIB, K-Ras, cathepsin B, HSA, GluA4, CD105, NKp46, TNFR2, mTFP1, MET, EpoR, TcdB, ESAT-6, listeriolysin O, CD8, HER3, VEGF, HGF, CD137, CD40, fibroblast activation protein, Wnt enhanceosome ChiLS, CD8, KRAS |
| Knottin | Cystine kont from agatoxin, chlorotoxin, EETI, kalata B1, MCoTI-I/II, SOTI, AgRP | <4 | Sub-nM–nM | β-Strands and 3 S–S formed macrocycle | Matrix metalloproteinase-2, thrombopoietin, FMDV-3C protease, VEGF-A, human thrombopoietin receptor, β-tryptase, NS2B–NS3 protease, MC4R, CXCR4, neuropilin-1, neuropilin-2, human matriptase-1, Hdm2/X, CTLA-4, BCR-ABL, α-synuclein, thrombospondin-1, SET protein, factor XIIa, angiotensin, IL-10, ebvIL-10, human fibronectin ED-B domain, αvβ3, αvβ5, α5β1, α(IIb)β(3), α9β1,α4β1, αvβ6 integrin |
| Kunitz | Serine protease inhibitor from BPTI/APPI/ TFPI | 7 | nM–pM | α-Helix, β-sheet and 3 S–S | Human plasma kallikrein, mesotrypsin, KLK-related peptidase 6, factor VIIa (TF-FVIIa), factor X (FXa), thrombin, IL-6R, human plasmin, neutrophil elastase, matriptase-2, Rho-GTPases, uPA receptor |
| Affimer | Human stefin A protease inhibitor | 11 | μM–nM | α-Helix, β-strands | CDK2,Src-Homology 2 (SH2) domains, CRP, SUMO, VEGFR2, HER4, AGR2, TRPV1 ion channel, PEDF, tenascin C, GFP, osteonectin, HTV-derived protein UL49, NS1 protein, tubulin, Myc, CDK4, PI-3K-P85 subunit, PD-L1 and LAG-3 |
| Nanofitin/affitin | DNA binding protein Sac7d | 7 | μM–sub-nM | β-Barrel capped with an α-helix | Human IgG, EGFR, GFP, PulD protein, hrEpCAM, chicken egg lysozyme, staphylococcal protein A, thermophilic CelD |
| Avimer | A-domain of cell membrane receptors (LRP,VLDLR) | 4 | Sub-nM | β-Strands and 3 S-S | IL-6, endocrine hormone FGF21, MET, human type II collagen, BAFF |
| Centyrins | Fn III domain human tenascin C | 10 | nM–pM | β-Sheet | C-MET, FcγRIIB, murine IL-17A EGFR, S. aureus leukocidins, rat TNFα |
| Affilin | Human γ-B-crystallin | 20 | μM–pM | β-Sheet | HER2, human IgG-Fc, EGFR, fibronectin ED-B domain, human proNGF protein, human papillomavirus E7 protein, estradiol/testosterone |
| Ubiquitin | 10 | α/β | |||
| ADAPT | Albumin-binding domain | 5 | Sub-nM–nM | α-Helix | HER2, HER3, IL-23, IL-17, hIFNγ, TNF-α |
Conjugation strategies for protein scaffolds
Conjugating proteins with diagnostic or therapeutic compounds is necessary for functional probes or nanomedicine preparation.16,17
Generally, there are four ways to realize bioconjugation. Cysteine-based conjugation uses the thiol group within free cysteine to react with maleimide or iodoacetamide-modified compounds and form stable covalent bonds.18 Besides, reporters or drugs can be directly conjugated to the disulfide bonds using next-generation maleimides.19 Lysine-based conjugation is built upon the good nucleophilicity of the reactive amine side chain from the solvent-accessible lysine. Normally, N-hydroxysuccinimidyl (NHS)- or sulfo-NHS ester-modified compounds react with the amine group of lysines to produce amide or amidine bonds,20 and unnatural amino acid (UAA)-based biorthogonal reaction and enzymatic conjugation are also feasible.21,22 For instance, 5-hydroxytryptophan (5 HTP) can be incorporated into specific sites of proteins via tryptophanyl-tRNA synthetase, following which compounds carrying aromatic diazonium groups quickly interact with 5 HTP via the azo-coupling reaction and realize conjugation.23 Cycloaddition-conjugation using l-azidohomoalanine (Aha) has replaced the l-methionine of the protein in methionine-auxotrophic E. coli strains to generate stable triazole groups via azide amino acid reacting with the alkyne or dibenzocyclooctyne of payloads.24 Photocrosslinking conjugation replaces the stop codon of protein with benzoylphenylalanine (BPA), while compounds with photoreactive antibody-binding domains (pAbBDs) can be conjugated with the BPA group under ultraviolet irradiation.25 The enzymatic conjugation strategies take advantage of enzymes that selectively recognize and catalyze the protein-tag and compound interaction. Enzyme-recognition tag combinations, such as sortaseA-LPXTG tag, trypsiligase-YRH tag, N-myristoyltransferase-(M) GXXXS/TXXX tag, tubulin tyrosine ligase-tub tag, lipoic acid ligase-LAP tag, farnesyltransferase-C-terminal CaaX tag, and transglutaminase–glutamine–lysine, have also been applied for enzymatic conjugation.26
To date, cysteine-based conjugation remains the universal site-specific conjugation method for most protein scaffolds that have no cysteine, including affibody, centyrin, DARPins, and ADPTA. However, for cysteine-rich scaffolds such as knottin, kuntiz, ANTICALIN®, and avimer, thioether bond-based conjugation may result in inhomogeneous conjugates and disulfide scrambling of scaffolds without a proper re-oxidation environment. UAA and enzymatic catalysis are more appropriate conjugation approaches for disulfide bond-based scaffolds. Via the incorporation of an azide residue or a polypeptide-tag into the N-terminus, protein scaffolds can easily realize site-specific conjugation and produce homogeneous conjugates with little impact on the affinity and original structure. However, recognition tag sequence introduction and the slow reaction of cycloaddition-conjugation may be inevitable.
Affibody
The B domain of Staphylococcal protein A is the parental scaffold of the affibody (ABY) protein scaffold originally responsible for Fc binding of immunoglobulin G.27 The B-domain contains 58 amino acid residues (6.5 kDa), constituting a three α-helix bundle motif without disulfide bonds. The B-domain was then artificially synthesized and denoted as the Z domain or affibody, which was used to construct a combinatorial library by randomizing 13 soluble surface residues located in the first two helixes.28 In theory, a specific affibody against any given target can be selected via an affibody-display phage library. As the third helix contributes to the structural integrity of the affibody but not binding function, a 2-helix affibody was developed. Compensating stability with disulfide bridges or site mutation offered advantages including a smaller size, quicker clearance, and lower imaging background.29 A homodimeric affibody ZAβ3 has also been used against amyloid peptides to screen the aggregation inhibitor for Tau protein.30 The Swedish company “Affibody AB,” founded in 1998, has explored affibodies and owns almost 300 patents.31 Affibodies modified with imaging contrast agents, fluorescent labels, chemical drugs, and radiolabels have been applied in therapeutic and diagnostic cancer research.32,33
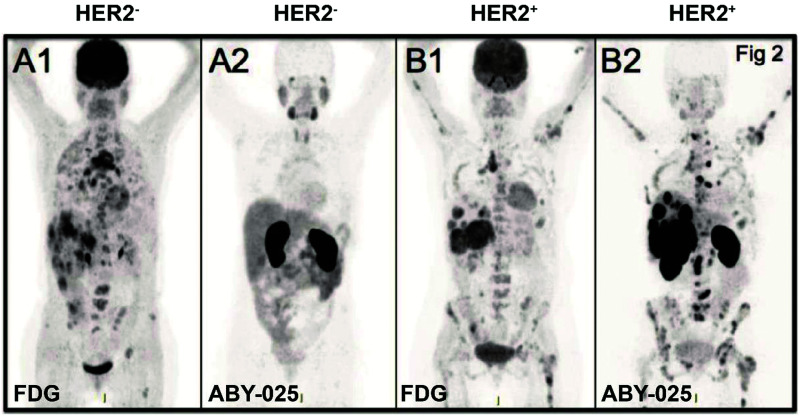
The anti-HER2 (human epidermal growth factor receptor 2) affibody is the most intensively investigated affibody thus far, with ABY-002(ZHER2:342) being the first to be applied in clinical computed tomography (CT) imaging for patients with breast cancer.34 However, owing to its partial high hepatic accumulation, ABY-002 was optimized to improve its specificity, stability, and hydrophilicity to produce ABY-025. A clinical phase II study of 68Ga-ABY-025 and 111In-ABY-025 demonstrated rapid accumulation in the tumor site within 1 h and showed clearance from the kidneys. The imaging results provided the metastatic details of HER2-positive primary tumors for several patients (NCT01858116, NCT01216033) (Fig. 2).‡ 35 A recently reported second generation HER2 affibody, ZHER2:41071, replaced the –NDA– with an –SES– sequence at the C-terminal. Single photon emission computed tomography (SPECT) imaging of the [99mTc]Tc-ZHER2:41071 tracer showed a significantly lower renal uptake by 25–30 fold (5.9 ± 2.1% ID g−1), as well as a tumor-to-blood ratio of 363 ± 84 in SKOV-3 xenografts in contrast to [99mTc]Tc-ZHER2:2395 (renal uptake and tumor to blood ratios are 183.8 ± 27.3% ID g−1 and 121 ± 24, respectively).36 Given the precise quantification for HER2 expression and no obvious systemic side effects, it was favorable to adjust therapeutic schemes and monitor tumor progress.37–39 More recently, a positron emission tomography (PET) imaging study performed in patients with advanced gastric cancer with low dose 68Ga-NOTA-MALM-ZHER2:342 showed a 3-fold higher lesion uptake compared with the HER2-negative group (SUVmax 10.5 ± 12.5 vs. 3.5 ± 1.7, p = 0.005) 2 h post-injection, without interference from simultaneous HER2 therapy.40 Another bivalent affibody, ABY-027, was generated by fusing ABY-025 with an albumin-binding domain to improve the retention time and kinetic profile. Nevertheless, tumor tolerance to the HER2 targeted treatments may occur as a result of upregulation of HER3 (human epidermal growth factor receptor 3) receptors. 89Zr-DFO-ZHER3:8698 has been used to track the expression of HER3 after heat shock protein 90 kDa (Hsp90) inhibitor-AUY922 treatment. The results demonstrated favorable HER3 quantification by PET imaging, making it possible to evaluate and predict HER2-targeted therapy efficacy.41 Moreover, the optimized HER3-specific probe [57Co] Co-(HE)3-ZHER3-DOTA significantly improved imaging contrast and prolonged tumor retention for up to 24 h in BxPC-3 xenografts via SPECT imaging.42 Another epidermal growth factor receptor (EGFR)-binding affibody ABY-029 (Z03115-Cys), labeled with an NIR (near-infrared ray) dye (IRDye® 800CW), provided an excellent in vivo imaging effect for glioma in nude mice. Meanwhile, early phase I studies of ABY-029 for gliomas, sarcomas, and head and neck cancers had completed (NCT02901925, NCT03282461, NCT03154411).43–46 Detecting the insulin-like growth factor 1 receptor (IGF-1R) expression level is essential for assessing therapeutic efficacy and prognosis. To overcome the non-specific accumulation of the first anti-IGF-1R affibody imaging probe 111In-DOTA-ZIGF1R:4551, a novel IGF-1R-specific affibody ZIGF-1R:4:40 labeled 64Cu was used. The results showed significantly reduced non-specificity, low background imaging interference, and the majority being excreted from the kidneys and urinary systems in a glioma mice model.47 Volunteers were recruited for tumor PET imaging with 68Ga-NODAGA-ZIGF-1R:4:40 (NCT02916394). The radiolabeled tracer 18F-NOTA-ZPD-L1_1 specific for programmed death-ligand 1 (PD-L1) exhibited high specificity to PD-L1, quick clearance, and good imaging contrast for mixed tumor-bearing mice via PET imaging, which is consistent with the results of immunohistochemistry; therefore, this tracer may have potential for use in clinical PD-L1 detection.48 Recently, ZCAIX:2 and ZCAIX:4 affibody molecules were screened against a prognostic tumor marker, carbonic anhydrase IX (CAIX). The radionuclide imaging probes 99mTc-(HE)3-ZCAIX:2 and 125I-ZCAIX:4 were evaluated in a mouse model, and demonstrated a high tumor tissue intake and a favorable tumor–blood ratio for clearly delineating the tumor.49 Additionally, the biomarker B7-H3 (CD276), which is highly expressed in breast cancer, was combined with indocyanine green (ICG) and microbubbles (MB) to generate ABYB7-H3-ICG and MBABY-B7-H3 for photoacoustic imaging, which demonstrated enhanced contrast signal and deep imaging effects in a mammary tumor model.50,51
Fig. 2. HER2-specific-affibody imaging in the clinical trial. Notes: PET imaging of HER2-negative patients (A) and HER2-positive patient (B) with 18F-FDG (A1, B1) and 68Ga-ABY-025 (A2, B2); standard uptake value (SUV) is normalized to 10 for all images; darker colors indicate higher uptake. Specific uptake and high contrast image is shown with 68Ga-ABY-025 compared to 18F-FDG-PET image. Adapted from ref. 35. Abbreviations: HER2, human epidermal growth factor receptor 2; ABY, affibody; SUV, standard uptake value; PET, positron emission tomography; and FDG, fluorodeoxyglucose.
Although no affibodies have been tested for cancer therapy in the clinic alone, many ongoing preclinical cancer studies for drug delivery or theranostics combined with affibodies have achieved promising results. For instance, the (ZHER2:2891)2-ABD-MC-DM1 conjugates efficiently delivered the cytotoxic maytansine to HER2 over-expressed tumor tissue and demonstrated high uptake rates and potent ablation efficacy, which prolonged the survival time of mice.52 A triple-modal (MRI/MSOT/CT) theranostic platform was built with Au–Fe2C Janus nanoparticles and ZHER2:342, which achieved deeper penetration and selective accumulation in mice tumor tissues, thus conferring high contrast imaging and efficient tumor ablation via photothermal therapy.53 Furthermore, NIR-830-ZHER2:342-IONP-Cisplatin theranostic nanoparticles exhibited substantial inhibition of primary and peritoneal metastatic tumors in an ovarian cancer mouse model, highlighting their potential to solve the defects of traditional debulking surgery.54 TAM-HER3, a bivalent HER3-specific affibody, drastically suppressed heregulin-induced phosphorylation, effectively blocked receptors in HER3-positive tumor, and presented equal therapeutic efficacy and safety compared to seribantumab.55 Besides, to overcome off-target effects triggered by nanoparticle cytotoxicity, a bioactivatable GSH-responsive nanogel decorated with anti-EGFR affibody was shown to specifically accumulate and internalize in the tumor tissue, ultimately realizing controllable photosensitizer release under a low GSH concentration intratumor environment for photodynamic therapy.56 Liposome-coated doxorubicin coupled with ZEGFR:1907 also provided a safe drug delivery platform with low cytotoxicity and improved antitumor efficacy.57 Moreover, ZPDGFRβ affibody was fused to the N-terminus of mouse TNFα and conjugated with doxorubicin; it efficiently permeated the pericytes of PDGFRβ (platelet-derived growth factor receptor β) positive tumors and accumulated in the tumor lesion. This method of drug delivery showed desirable antitumor efficacy and tumor vessel reshape, potentially due to the reduced secretion of VEGF and increased level of intercellular cell adhesion molecule-1.58 The ZPDGFRβ-IR700 probe has also been used for photodynamic therapy for colorectal cancer.59 ZHPV16E7 is the recently discovered affibody specific to HPV16 positive tumors; combined with exotoxin A, ZHPV16E7 affitoxin384 exhibited significant tumor ablation in a cervical cancer mice model.60 More recently, an affibody against the latent membrane protein 2 (LMP-2) associated with Epstein-Barr virus-induced tumors was fused with exotoxin for toxin delivery. ZEBVLMP-2142–Z142X affitoxin showed potent cytotoxicity and favorable antitumor efficacy in LMP-2-expressed xenograft mice.61
ANTICALIN®
ANTICALIN® scaffolds are a class of affinity molecules engineered from small extracellular proteins, lipocalins, which recognize small hydrophobic molecules and cell surface receptors for transportation, mediating the immune response, or allergens and hormones.62 Cup-shaped lipocalins consist of eight antiparallel β-strands, forming a β-barrel rigid framework, a single α-helix linked to the c-terminal, and four variable loops exposed on the surface, which constitute the binding pocket. The sequence and length diversity of loops provide the possibility for engineering new affinity ligands, the so-called anticalins.63 The first reported ANTICALIN® was based on the butterfly-derived bilin-binding protein (BBP), which was reshaped to combine fluorescein.64 To improve the safety and decrease the immunogenicity for human disease study, tear lipocalin (Tcl/Lcn1) and the neutrophil-gelatinase associated lipocalin (NGAL/Lcn2), with high compatibility and flexibility, were applied to construct human ANTICALIN® libraries.65 The “Pieris” company, which has nine ongoing ANTICALIN® projects, has committed to realizing their commercialization since 2001, and many anticalins have also been reported in preclinical research.66–68 However, only a few anticalins that are specific to biomarkers, including MET, HSP70, VEGF-A, VEGFR-3, PSMA, CD137, CD98, and ED-B, have been investigated for detecting or treating cancer.
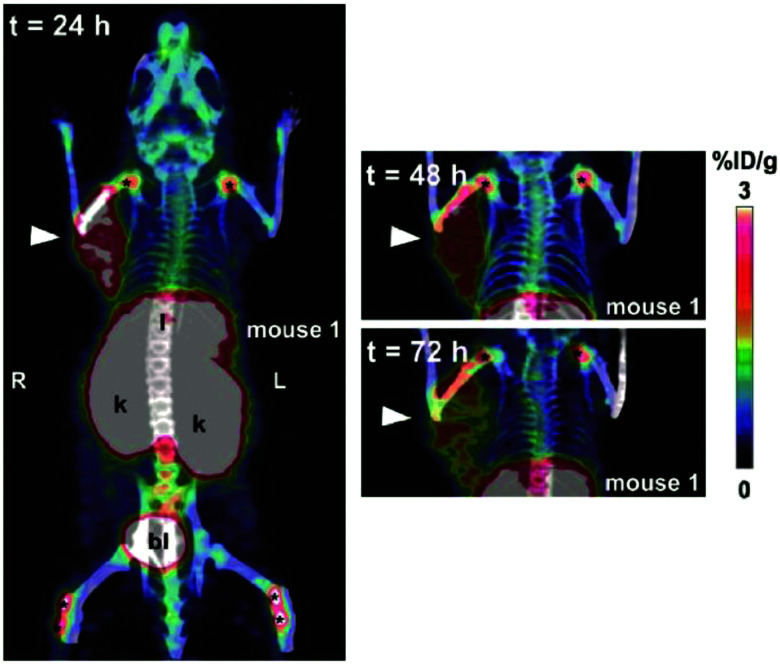
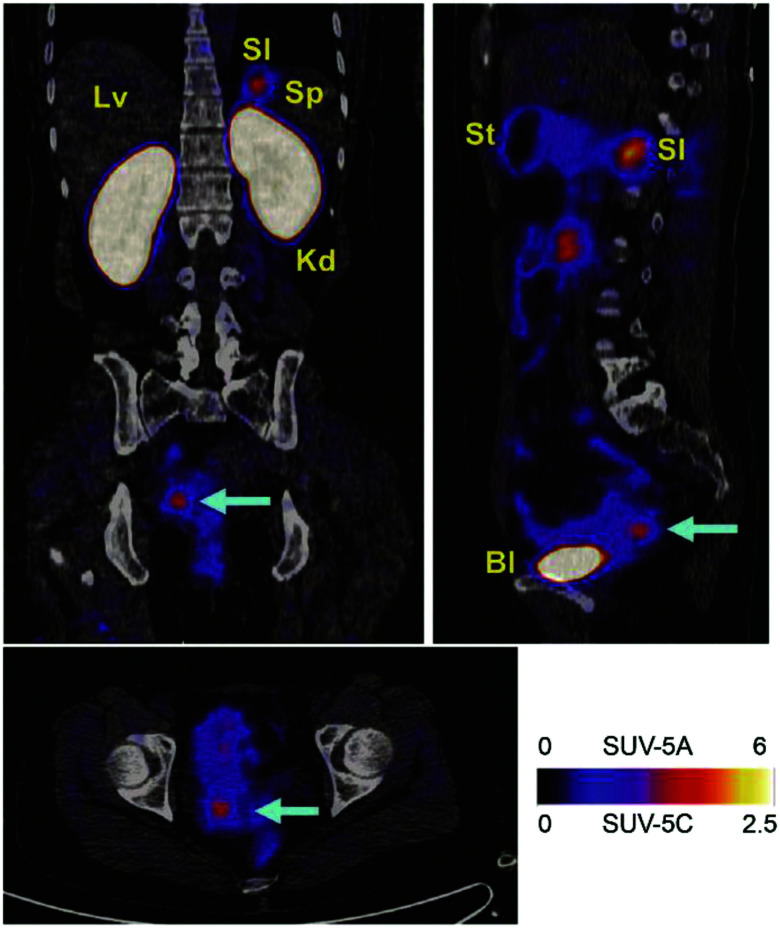
An early study of PET tracer 89Zr-PRS-110 for MET (hepatocyte growth factor receptor) imaging showed specific and low background imaging effects in the MET-expressing tumor models, which demonstrated the feasibility of ANTICALIN® as a targeting moiety for in vivo diagnosis.69 Anticalins (N7A, N7E, and N9B) against fibronectin ED-B (extra domain B), which is uniquely expressed in neogenic blood vessels, labeled with a γ-emitter 123I, presented high specificity and affinity to over-vascularized glioblastoma multiforme, as verified by histochemistry and in vitro autoradiography.70 Later, a ketone group was introduced into ANTICALIN® N7A for conjugation with Alexa 488, which realized safe ED-B detection at the cellular level and demonstrated its potential as an in vivo imaging probe.71 Meanwhile, prostate-specific membrane antigen (PSMA) and vascular endothelial growth factor receptor-3 (VEGFR-3)-specific anticalins combined with immunofluorescence staining were shown to exhibit high binding affinity and selectivity to PSMA- and VEGFR-3-positive cells, demonstrating the potential of these anticalins for further in vivo imaging and therapy studies of solid tumor.72,73 The 70 kDa heat-shocking protein (HSP70) is commonly expressed in primary and metastatic tumors and is commonly upregulated after radiochemotherapy treatment. HSP70 PET imaging with the 89Zr-labeled HSP70-binding ANTICALIN® BBG10C/I revealed deep tissue penetration and selective enrichment in mouse tumor sites. Moreover, strong in vitro tumor inhibition was realized by conjugating BBG10C/I with the toxin gelonin.74 The latest published ANTICALIN® P3D11, which targets a malignant tumor marker, human CD98hc ectodomain (hCD98hcED), was examined in the first CD98 in vivo imaging study by labeling with 89Zr and PASylation modification. The PET imaging performance was investigated in a prostate carcinoma xenograft-bearing mouse model and showed picomolar affinity, high tumor lesion accumulation (8.6 ± 1.1% ID g−1), and a good tumor-to-blood ratio of 11.8 (Fig. 3).75,76
Fig. 3. In vivo PET study of CD98hc-binding ANTICALIN® tracer. Notes: prostate cancer xenografts mice were injected intravenously with ANTICALIN® probes D11vs-PAS200-Dfo 89Zr; PET imaging data are acquired at 24 h, 48 h, and 72 h post-injection. Strong imaging signal is shown at 24 h (arrowhead) and gradually reduced at 48 h and 72 h; * indicated joints. Adapted from ref. 75. Abbreviation: l, liver; k, kidneys; and bl, bladder.
The most widely studied ANTICALIN® for cancer treatment is PRS-343 against a crucial costimulatory immune receptor 4-1BB/CD137, which plays a critical role in mediating the immune response and is typically expressed on tumor-infiltrating lymphocytes.77 To safely activate T cells and reduce damage to the peripheral tissue of the tumor, the anti-HER2 monoclonal antibody trastuzumab was fused to the 4-1BB-specific ANTICALIN®; this dual-targeting fusion protein selectively accumulated in the tumor tissues and site-specifically activated tumor-localized T cells to achieve excellent tumor regression efficacy in a SKOV3 mouse model. The peripheral toxicity and pharmacological profile of PRS-343 have been verified in cynomolgus monkeys and in a mouse toxicology model.78 A phase I trial of PRS-343 for patients with advanced HER2-positive tumors has been completed, and phase II studies of the combination with tucatinib, ramucirumab, and paclitaxel are currently recruiting (NCT03330561, NCT05190445).79–82 Meanwhile, PRS-342 consisting of α-Glypican 3 antibodies fused to 4-1BB-specific ANTICALIN® also showed strong suppression of tumor growth and activation of T cells in a hepatocellular carcinoma mouse model.83 Moreover, the 4-1BB-specific ANTICALIN® combined with PD-L1 inhibitor (atezolizumab) achieved excellent co-stimulation efficacy for immune system activation in the tumor microenvironment and successfully ablated tumor cells in a mouse model. A phase II trial (NCT05159388) for combination therapy of this fusion protein (also called PRS-344) is currently recruiting. Additionally, the phase I study (NCT01141257) of ANTICALIN® PRS-050-PEG40 (Angiocal) against VEGF-A was completed in 2013. Clinical data have shown safety and efficient inhibition of angiogenesis by reducing MMP-2 in patients with advanced solid tumors.84 Later, PEGylated PRS-050 was evaluated in A673 sarcoma xenografts, and was shown to strongly block vascular permeability and reduce microvessel density to inhibit tumor growth. This compound provided favorable safety with no side effects of platelet aggregation and thrombosis resulting from bevacizumab therapy.85
Adnectin/monobody
Fibronectins are a type of macro-glycoprotein that widely exist in animals and are responsible for intercellular communication and constitute the extracellular matrix. One of the major components of human fibronectins is the tenth fibronectin type 3 domain (10FN3), a subfamily of immunoglobulin superfamily composed of seven β-strands, which forms a β-sandwich framework connected by six loops, three of which are variable.86 As their structural features resemble those of the variable heavy chains of antibodies, 10FN3 was engineered as a novel protein binder, termed monobody (10 kDa).87 The first devised 10FN3 scaffold against ubiquitin was generated by mutating residues in the BC and FG loops, while the DE loop was introduced later for screening the TNF-α binding ligand.88 The commercialized monobody was named Trinectin by the “Adnexus” company, which was then acquired by “Bristol-Myers Squibb” and renamed Adnectin in 2007.89 A conventional loop-based adnectin library was designed by randomizing residues within the BC, DE, and FG loops of 10FN3. Later, side-and-loop-based libraries were constructed by reshaping the second CD-FG β-sheets, which formed a concave binding pocket suitable for convex receptor epitopes, such as globular macromolecules.90 Currently, more than 50 adnectin molecules have been selected and used in biological or medical research.91,92
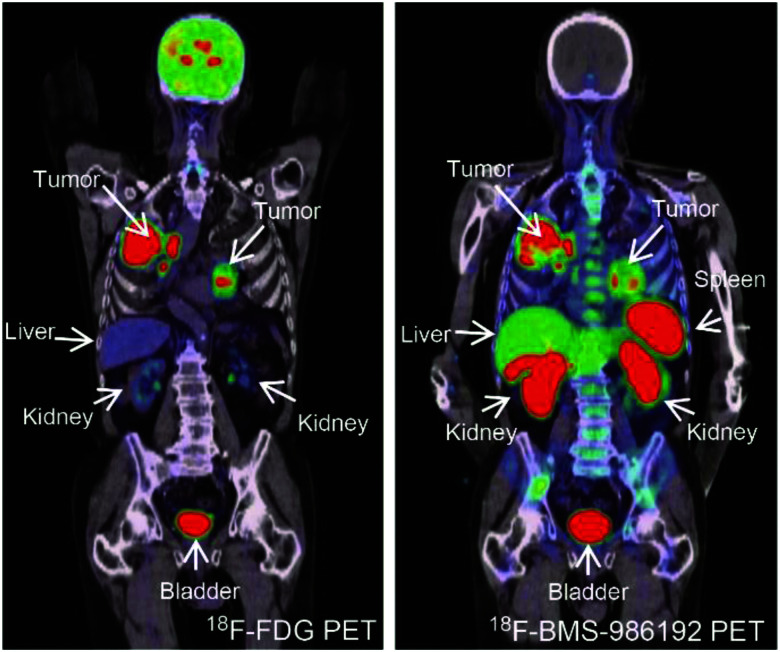
Anti-vascular endothelial growth factor receptor 2 (VEGFR2) adnectin CT-322/BMS-844203/FN3VEGFR2 is the first one to enter the clinic trial, showing a good pharmacokinetic profile and safety (NCT00374179).93 Functionalizing with molecularly targeted MB generated an ultrasound imaging probe, MB-FN3VEGFR2, which showed high affinity, selective accumulation, and strong ultrasound signal in a breast cancer mouse model.94 Another adnectin E1 against human EphA2, which is aberrantly expressed in diverse cancers, was used in an in vivo imaging study of the fluorescent probe E1-Rluc8 and a radiolabeled tracer 64Cu-NOTA-E1 in a subcutaneous prostate tumor model, with results showing a strong signal, selective accumulation, and rapid clearance.95,96 So far, two anti-PD-L1 adnectins, FN3hPD-L1-01 and BMS-986192, have been investigated for PD-L1 imaging, and the latter has shown good results in the clinic. 64Cu-FN3hPD-L1 exhibited high specificity and a tumor-to-muscle ratio of 13 after 4 h in mice for PD-L1 expression quantification.97,98 Moreover, the PET imaging probe 18F-BMS-986192 showed picomolar binding affinity, spleen accumulation, and rapid clearance from the kidneys and bladder in healthy cynomolgus monkey, and favorable dosimetry results provided an estimated safe dose for human use of 2.20 × 10−1 mSv/MBq. The in vivo study of 18F-BMS-986192 in HT-29 and L2987 xenograft models presented peak radioactivity of tumor tissue 25 min post-injection and low background imaging for PD-L1 expression evaluation.99 Human study of PD-L1 whole body quantification for patients with terminal non-small cell lung cancer (NSCLC) was completed with 18F-BMS-986192, which exhibited safe pharmacokinetic performance and accurately detected the heterogeneity of PD-L1 distribution between individuals and diverse tumor lesions compared to the results of conventional biopsies. A phase I study on 18F-BMS-986192 PET imaging in patients with oral cavity squamous cell carcinoma is ongoing (NCT03843515) (Fig. 4).100,101 A PD-L1 PET imaging pilot study (NCT03520634) also demonstrated that 18F-BMS-986192 provided a baseline uptake for predicting the lesion volume recession effect after immune checkpoint inhibitor treatments.102
Fig. 4. PD-L1-targeting adnectin probes for clinical PET imaging. Notes: a patient with advanced NSCLC (PD-L1 expression >50%) was administered with 18F-FDG PET (left) and 18F-BMS-986192 adnectin PET tracer (right); compared to the FDG tracer, the PET image of adnectin showed high tumor-to-background contrast and heterogeneous uptake at tumor sites. Adapted from ref. 100. Abbreviations: NSCLC, non-small-cell lung carcinoma; and PD-L1, programmed death-ligand 1.
In terms of cancer therapy, a phase I trial of CT-322 demonstrated significant blocking of VEGFR2 receptors and efficient inhibition of angiogenesis and metastasis in solid tumors, although it failed to treat recurrent glioblastoma in a phase II study.103 Later, CT-322 modified by PASylation demonstrated a significant boost in bioactivity, pharmacokinetic properties, and half-life extension in a mouse model than PEGylation, thus providing a safe and effective drug delivery system for further clinical study.104 EGFR-specific adnectin-E3 has also been used to engineer a new chimeric antigen receptor T cell system; this adnectin-based CAR-T, which showed lower immunogenicity and improved selectivity and safety, offered equivalent tumor ablation efficacy in an immunodeficient NSG mouse model in contrast with the conventional scFv-based CAR system.105 Another adnectin, Mb(S4), against WDR5 (necessary for the mixed-lineage leukemia 1 [MLL1] methylation), was genetically encoded with a TMP-inducible expression system as an inhibitor for WDR5–MLL1 interaction, and functioned to reduce methyltransferase activity, downregulate MLL1 relative gene expression, and extend survival in the MLL-AF9 leukemia model.106 The RAS binding adnectin NS1 could intervene in the α4–α5 subunit interface to inhibit RAS activation, and combined with a doxycycline-inducible expression system, presented a good antitumor effect in a mouse model.107 Glypican-3-specific adnectin A1 conjugated with the microtube inhibitor tubulysin showed effective tumor regression in Hep3B xenografts, which persisted for 2 weeks.108 More recently, a mesothelin-binding adnectin was reported with the ability to induce apoptosis and, combined with mitomycin C, showed a strong tumor inhibition efficacy in A431 cells.109
Designed ankyrin repeat proteins (DARPins)
Ankyrin repeat (AR) proteins with helix–loop–helix conformations are present in virtually all phyla and are exclusively involved in mediating protein–protein interactions, including transcription, ion transportation, and cell cycle modulation.110 Crystal structure and consensus sequence analyses of the ankyrin motif revealed that the conserved hydrophobic core of the α-helix constituted rigid elements, while the less conserved loop region of solvent-accessible surface contributed to molecular recognition, which forms the basis of the protein binder design.111 DARPin scaffolds comprise 2–3 human AR motifs flanked by capping repeats at both the N- and C-terminals, which protect them from aggregation during the folding process. Each AR unit (33 amino acids) is composed of two antiparallel α-helices with a loop and a β-turn extended from each end for binding to the next AR, and 6–7 residues within each repeat are available for randomization.112 Based on the consensus design strategy, a biopharmaceutical company called “Molecular Partners AG” founded in 2004 has developed new DARPin molecules as antibody alternatives for clinical translation.113 Moreover, a loopDARPin library was constructed by introducing the CDR-H3 domain of antibodies to solve the limitations of traditional DARPins, including the less convex and somewhat rigid binding surface.114 Until now, more than 60 DARPin molecules have been reported, and owing to their small size (14–18 kDa), physicochemical stability, and ease for producing multivalent or multispecific targeting elements, DARPins have become attractive for diagnostic and therapeutic applications.115,116
The Cy5.5-labeled DARPin 8h6 was the first reported DARPin-based fluorescent probe for in vivo imaging study. This probe is specific for cathepsin B, which is overexpressed in the tumor microenvironment, and shows a strong long-term signal ratio up to 72 h in a breast cancer model, despite partial liver accumulation.117 Moreover, HER2-specific DARPins G3, 9_29 labeled with fluorescein and a radionuclide, respectively, exhibited a good tumor-tissue ratio and high contrast SPECT imaging effect in a HER2-positive malignant tumor model. Moreover, the superparamagnetic nanoparticle probe SPIO-G3-5MF and upconversion nanoparticle UCNP-PMAO-DARPin-9_29 were effective in diagnosing SKBR-3 xenograft tumor models, with selective accumulation and long-term retention.118,119 Furthermore, DARPin G3 was genetically displayed on the surface of exosomes and radiolabeled with 99mTc to take advantage of the DARPin specificity and the homing ability of exosomes. Planar imaging has shown that these probes rapidly accumulated in breast cancer, with a high contrast signal detected for visualizing tumor lesions.120 The phase I SPECT imaging study in patients with primary breast cancer (NCT04277338) proved that the 99mTc-(HE)3-G3 tracer had good tolerability, biodistribution, and ability to discern HER2 positive or negative tumor.121 Moreover, the newly reported imaging probe 125I-PIB-H6-Ec1 selectively binds to the biomarker EpCAM (epithelial cell adhesion molecules) with picomolar affinity based on the DARPin molecule Ec1. In an in vivo xenograft mouse model, this probe demonstrated lower non-specific accumulation and favorable tumor-to-organ ratios for tumor imaging (Fig. 5).122,123
Fig. 5. In vivo SPECT imaging of EpCAM-specific DARPins probes. Notes: OVCAR-3 (A) and SKOV-3(B) xenografts models; mice are injected with the DARPin probe [125I] I-PIB-Ec1 and imaged at 6 hours post-injection. High contrast tumor imaging and mainly cleared through the kidneys has shown. Adapted from ref. 123. Abbreviations: SPECT, Single-photon emission computed tomography; EpCAM, epithelial cell adhesion molecules; DARPins, designed ankyrin repeat proteins; K, kidneys; and T, tumors.
MP0250 DARPin against HGF, VEGF-A, and human serum albumin is the first tri-specific DARPin. A clinical phase II program (NCT02194426) demonstrated the good tolerability and pharmacokinetic properties of MP0250 in patients with advanced solid tumors, in which 14 patients showed slight tumor shrinkage.124–126 Moreover, MP0250 has completed another phase II trial for patients with multiple myeloma with the combinational therapy of bortezomib and dexamethasone (NCT03136653).127,128
More recently, a MET antagonist of DARPins was shown to tightly bind two epitopes of the extracellular domain of MET and efficiently block MET-dependent signaling pathway activation and inhibit proliferation in gastric carcinoma cells.129 Moreover, the EGFR-specific DARPin-E01 fused with the TPD prodomain avoided EGFR–ligand shedding by inhibiting A-disintegrin and metalloproteinase. The fusion protein E01-GS-TPD potently blocked EGFR receptors and reduced their kinase activity, showing significant inhibition of tumor cell proliferation and migration in vitro.130 Another study genetically modified oncolytic measles virus with EGFR-binding DARPins E.01 and MMPA1 activated by tumor-associated MMPs. The bi-specific engineered virus MV-MMPA1-E.01 displayed reduced off-target effects and demonstrated favorable therapeutic efficacy in GBM xenograft models.131 HER2-specific DARPins have also been broadly studied for cancer treatment. For instance, the hybrid upconversion nanoparticles, UCNP-R-T, composed of radionuclide 90Y and exotoxin A, and anti-HER2 DARPin-PE40 generated nanotheranostic regents achieved background-free imaging and effective tumor ablation in a breast cancer mouse model.132 Patients with HER2-positive solid tumors were recruited for a phase I study with the HER2-binding DARPin drug MP0274 (NCT03084926).
EpCAM, EGFR, and HER2-targeting DARPins used to engineer monospecific and multispecific CAR-T cells showed desirable antitumor efficacy equivalent to scFv-based CAR-T cells, and the multispecific CAT-T cells reduced antigen escape and enhanced synergistic effect of immunotherapy in different NSG mouse xenograft models.133,134 The DARPin molecules TREG005 and TREG006 serve as agonists to TNFR2 (CD120b) of tumor-infiltrating T cells, and have been shown to strongly activate the NF-κB signaling pathway in vitro, and facilitate CD8+ T cell infiltration into tumor tissue and enhance IFN-γ expression in an in vivo model.135 MP0310 (AMG506) and MP0317 are bispecific DARPin scaffolds against 4-IBB (CD137) and CD40, respectively, and have been combined with the fibroblast activation protein to activate the tumor-localized immune response in an actively recruiting phase I study (NCT04049903, NCT05098405).
Knottin
Knottin molecules are a subclass of the cystine-knot mini-protein family, which broadly exists in many plants, animals, and fungi in the form of inhibitors that play essential roles in mediating signaling transduction, channel blocking, and proteolysis inhibition.136,137 The knottin scaffold consists of three antiparallel β-strands and three disulfide bonds (I–IV, II–V, and III–VI) with several exposed variable loops; generally, the third disulfide bond threads through a macrocycle, which was constituted by the first two disulfide bonds and then formed a constrained knot structure. This rigid knot fold confers its excellent structural stability to temperature, proteolysis, and chemical denaturation; besides, good sequence tolerance has been shown given that the sequence identity of the knottin motif is very low, excluding the six cysteines.138 It is worth noting that the knottin molecule can be produced in microbes, despite the existence of disulfide bonds. Aside from the above advantages, small size knottin scaffolds (<4 kDa) also show good pharmacological properties, targeting specificity and non-immunogenicity. Knottin scaffolds were engineered as new protein binders for devising targeting imaging probes and drug delivery systems.139,140 Knottin scaffolds are being explored by several companies, including “Jazz,” “Ironwood,” “Cyclogenix,” and “AstraZeneca,” and some FDA-approved knottin-drugs like “Ziconotide” and “Linaclotide” have been marketed although not for cancer.141–144 Currently, most reported knottin scaffolds originate from animals and plants, and a few have been used to construct knottin display libraries. While grafting with known binding motifs as agonists or antagonists is more prevalent, partial mainstream knottin scaffolds have also been reported.
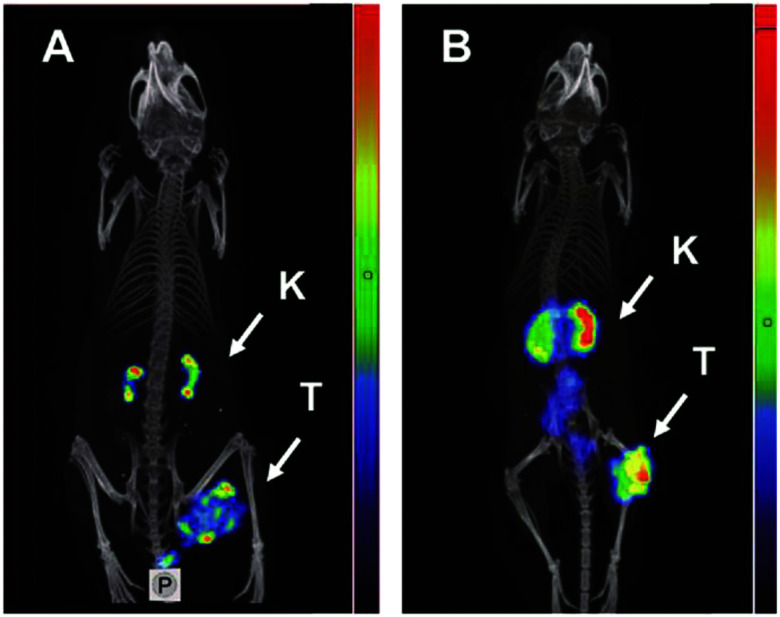
The most commonly investigated knottin scaffold for tumor imaging and therapy is integrin-targeting integrin αvβ6 binding knottin (R01-MG) labeled with the contrast agent A740. This scaffold has been used for photoacoustic and fluorescence imaging in A431 xenograft mice, in which maximal signal and significant contrast enhancement were observed in tumor tissues after 1 h, demonstrating rapid localization and good imaging effect.145 R01-MG-IRDye800 has also been evaluated in subcutaneous and orthotopic tumor models, with results showing a higher tumor-to-background ratio of 2.5–2.7. Furthermore, a strong contrast fluorescent signal under 750 nm laser exposure for 30 min proved its ability for imaging-guidance surgery and potential for further clinical application.146 More recently, three PET tracers based on R01-MG, [18F]FDG and [18F]FP-R01-MG-F2, and [68Ga]NODAGA-R01-MG have been evaluated in patients with pancreatic, lung, or cervical cancer and healthy volunteers, which demonstrated their safety, selective and quick accumulation in tumor tissues, and low background interference imaging for cancer diagnosis (NCT02683824) (Fig. 6).147 Another knottin scaffold (MC-FN-010) against the extra domain B of human fibronectin, which is associated with tumor angiogenesis, conjugated to Alexa Fluor 680, presented a significant uptake and favorable imaging capacity in glioblastoma mouse models.148
Fig. 6. αvβ6-Targeting Knottin tracer of PET/CT imaging in the clinical trial. Notes: a cervical cancer patient was administered with Knottin probe 68Ga-NODAGA-R01-MG; cervical tumor pointed by the cyan arrowhead and SUVmean reached 4.79 ± 0.37. Adapted from ref. 147. Abbreviations: Lv, liver; Sp, spleen; Bl, bladder; Kd, kidneys; Si, small intestine; and St, stomach.
EETI-2.5F knottin against integrin receptors (αvβ3, αvβ5, and α5β1) was dimerized through oxime-conjugation, the dimer 2.5_AO reached a 150-fold binding affinity improvement compared to the monomer and more effectively inhibited glioblastoma and breast cancer cell proliferation and migration in vitro compared to clinical cilengitide.149 EETI-2.5F modification helps the cell cycle inhibitor gemcitabine to evade its resistance in pancreatic cancer cells, and the drug conjugate was efficiently released intracellularly, which achieved potent suppression of tumor growth in vitro and demonstrated its potential for targeted drug delivery.150 EETI-2.5F combining with the tubulin inhibitor MMAF, EETI-2.5F-Fc-MMAF, efficiently suppressed tumors and extended the mice's survival time in the U-87 MG xenograft model when administered at 10 mg kg−1 twice a month.151 Furthermore, the oncolytic measles virus (MV) modified with EETI-2.5F was intravenously injected into a glioblastoma mouse model and showed excellent antitumor efficacy.152 Additionally, another natural knottin molecule, chlorotoxin/CTX, isolated from scorpion venom, can permeate through the blood–brain barrier easily and specifically accumulate in neuroectodermal tumors.153 Ribosome inhibitor gelonin fused with chlorotoxin significantly suppressed the translation process in U-87 MG cells, with lower toxicity and strongly ablated tumor tissue performance in the U-87 MG xenograft mice.154 The PET tracer 18F-FP-chlorotoxin is rapidly taken up by the MMP-2 expressed glioma at 90 min post-injection and showed a high contrast imaging effect.155 Chlorotoxin-ICG, also called BLZ-100/tozuleristide, was tested in spontaneous canine tumors, and exhibited a 20-fold tumor-to-tissue contrast for intraoperative imaging.156 Medulloblastoma imaging with BLZ-100 enhanced signal contrast between the tumor core and margins for clear tumor delineation.157 A phase I trial of BLZ-100 for recurrent gliomas has shown safe dose tolerability up to 30 mg in humans, while further phase II/III trials are ongoing (NCT02234297, NCT03579602).158
Other scaffolds
In addition to the scaffolds described above, some other less developed or emerging scaffolds have been reported for cancer research.
Kunitz
The kunitz scaffold (∼7 kDa) was engineered from protease inhibitors with a rigid α-helix and β-sheet folds that are structurally constrained by three disulfide bonds.15 The “Shire (Dyax)” company has explored kunitz molecules for commercialization.159 A few Kunitz-type scaffolds, angiopep-2 (ANG) for glioblastoma imaging, Amblyomin-X for tumor inhibition via regulating Rho-GTPases and uPAR signaling, and APPI-4M blocking the invasion of KLK6-dependent breast cancer in vitro, were reported.160–162
Affimer
Another scaffold, affimer (∼14 kDa), derives from the human stefin A protease inhibitors with an α-helix over four antiparallel β-sheets and two-variable loops.163 Affimer scaffolds are currently being developed by “Avacta Life Sciences”.164 Recently, HER4 binding affimer 5 conjugated with CF640R-maleimide fluorescent dyes have been tested for imaging of MCF7 cells in vitro. The VEGFR2-specific affimer B8 and A9 were labelled with biotin as probes and presented higher sensitivity in histochemical staining assay compared to the commercial VEGFR2 antibody.165,166
Nanofitin
The nanofitin scaffold (7 kDa), also called affitin, originated from the DNA binding protein Sac7d family and was first commercialized by the “Affilogic” company.167,168 Engineered EGFR-specific nanofitin B10 has been used to generate the radiolabeled probe 18F-FBEM-Cys-B10, which provided excellent imaging results in the A431 tumor model.169
Avimer
An avimer scaffold based on the A-domain of diverse membrane receptors is another antibody memetic that has been explored by the “Amgen” company.170,171 Recently, a previously reported C426 avimer scaffold has been designed as a novel C-MET reporter by introducing a gly3-cys sequence at the c-terminal. Although the computational model and ELISA showed good traceability, further in vivo study is needed.172
Centyrins
Centyrins are derived from the Fn3 domains of the tenascin scaffold, and the “Janseen” company is currently exploring the commercialization of centryins.173 The EGFR-specific centyrin 83V2 conjugated with the tubulin inhibitor MMAF exhibited potent cytotoxicity in NCI-H1573 cells. Simultaneously, 83v2 labeled with the NIR dye S0456 localized at the tumor lesion after 4 h, and showed a long retention time beyond 24 h, with a high contrast imaging effect observed in xenograft mice.174,175
Affilin
Ubiquitin molecules (∼76 amino acids) comprising an α/β secondary structure and molecular recognition function were engineered as affilin scaffolds and are currently being explored by the “Scil Proteins” company.176,177 Moreover, HER2 and EGFR binding affilins fused to the anti-EGFR monoclonal antibody cetuximab have been shown to achieve bi-specificity and strong inhibition of cancer cell proliferation in vitro.178
ADAPT
The albumin-binding domain with good solubility generally extends the half-life of drugs or probes. Based on the interaction energy calculation, 11 variable residues on the surface were identified by alanine scanning, which were then engineered to albumin-binding domain-derived affinity protein (ADAPT).179 An ADAPT6 scaffold specific to HER2 was labeled with a radionuclide and showed high contrast imaging in vivo and better delineation for small metastases. Besides, ADAPT6 conjugated with exotoxin A (ETA) demonstrated excellent efficacy as a cancer therapeutic in a preclinical study.180–182
Design of protein scaffolds
Although protein scaffolds have acquired promising achievements in different application fields, it remains vital to devise or discover novel scaffold molecules considering the limitations of the protein scaffolds, such as patent monopoly, short half-life, immunogenicity, and less accessibility to the flat area of targets. One should keep in mind that a suitable candidate scaffold should possess a compact structure with a rigid core and good tolerance to residue substitution or engraftment without losing its structural integrity and stability.
Ligand mimicking is a mainstream design method, which requires engrafting known binding motifs onto scaffolds and reshaping scaffolds derived from a protein data bank (PDB) or in silico. Ligand mimicking consists of several parts: binding motif analysis, rational scaffold selection, computer simulation motif grafting, molecular docking, and further optimization of folding efficiency stability, and solubility.183
The motif sequence is based on the analysis of the ligand–receptor co-crystallization structure; normally, a consecutive sequence containing binding site is preferred. The flexible motifs are well accepted by most scaffolds; in contrast, one should consider whether the scaffold's conformational folding path matches with rigid motifs.184 Searching scaffolds from the PDB databases or de novo design library, and comparing similarity via PDBeFold or DALI is a good option; meanwhile, small protein inhibitors also require attention, such as typical kunitz, affimer, and knottin scaffolds.185 It should be noted that some de novo scaffolds may be difficult to be expressed in E. coli due to their evolutionary preference inconsistence.186 Combining with molecular modeling (Rosetta) and thermodynamic simulation calculation, it is possible to produce rational motif–scaffold structures. Moreover, solubility and folding efficacy could be optimized by replacing soluble residues and employing the fold-from-loops strategy.187,188 Candidate structures with high-affinity rates after multiple molecular docking comparisons are suitable for further expression and surface plasmon resonance (SPR) to verify their actual effects. Sometimes, low stability may affect correct folding, and it is worth trying to introduce disulfide-bridges with appropriate distances to improve the overall stability of the scaffolds.184
Tetsuya's group grafted two CD25-binding residues GGGV-D of daclizumab and YGY-RS-Y of basiliximab onto a zinc-finger peptide scaffold to construct a small antibody mimetic library, three CD25-binding ligands with 30 nM affinity were successfully selected.189 Cassie's group designed PD-1 agonist by inserting three binding motifs “WDYKY”, “ADYKR”, “WDYKR” into 34840 de novo-designed scaffolds, after Rosetta, fold-from-loopsm, flow cytometry sorting and affinity maturation processed, an agonist PD-MP1 were selected, monomeric PD-MP1 showed 6 μM affinity to mouse PD-1, and trimeric PD-MP1 strongly inhibited murine T cell activation in vitro.190 A helix-kinked-helix motif of PfEMP1 molecules which forms core binding site of endothelial protein C receptor was integrated to a three-helical bundle scaffold as a vaccine immunogen, the final binder could elicit antibodies, although not as effective as the whole CIDRalpha1 domain.184
Another approach scaffold reshaping is commonly involved in scaffold selection, solvent-accessible surface area calculation, stability evaluation, library construction, and binder panning verification.
First, existing scaffolds can be screened from the PDB by setting different criteria, such as the size range, human or microbe derived, cysteine-free or not, high-resolution, solubility (based on literature data), nontoxicity, and the number of loops, β-sheets or α-helices. The filtered results can be further refined by similarity alignments. Based on the FoldX platform, it is possible to calculate the energy difference of mutated surface residues to select diversified sites. Often loops with no specified structures tend to present a high solvent-accessible surface area that is suitable for randomization.191 Next, stability assessment can be achieved via the Eris platform, disulfide-bridge, or other slat-bridge introductions, and should be considered in some cases.192 Ideally, the diversified residues should be close in sequential and structural space, and libraries with good diversity require at least ten randomized sites in theory. Finally, it is possible to select designed scaffold binders against targets via experiments to evaluate their performance.193,194
Hackel's group searched ideal scaffolds from the PDB that conform to a series of criteria, 259 candidates were further evaluated via solvent accessible surface area calculation and destabilization after randomization; the T7 phage gene 2 protein (Gp2) with best rates was selected for library construction. Gp2 binder GaEGFR2.2.3 (18 ± 8 nM) and GP2-PDL1E4 (3–13 nM) against EGFR and PD-L1 were screened.193,195 Schneider's group screened scaffold candidates from the PDB databases, after sequence alignment, in silico saturation mutagenesis, structural evaluation, the 4PSF scaffold was finally selected to build the ProBi library, ProBi binder against human interleukin-10 with nanomolar affinity was screened.194
To sum up, with the development of various technologies, such as computer-aided design, protein prediction (Alphafold, Rosetta), molecular docking, molecular dynamics simulation, crystallographic structural analysis, deep sequencing, and learning, designing novel protein scaffolds will be easier and more feasible.
Conclusions
Protein scaffolds as antibody alternatives have been widely investigated for cancer diagnosis and therapy in preclinical and clinical studies over the past two decades.
In comparison, some outstanding advantages of protein scaffolds are irreplaceable, except for the high affinity and specificity. For example, the smaller molecular size (<20 kDa) facilitates tissue infiltration and deeper tumor lesion penetration.196 Protein scaffolds with small steric obstacles can easily access hidden epitopes located in the deep groove of some undruggable targets, such as Ras and Myc, which is impossible to approach using conventional antibodies.197–199 Moreover, the favorable physicochemical stability of scaffold binders is also appealing; generally, they can refold to their original structure after experiencing extreme temperature changes over 60 °C or following a broad range pH change (2.5–11). Some of them, such as knottin scaffolds, are able to withstand even higher temperatures (over 100 °C), making efficient coupling and photothermal reaction possible.9 Besides, the knottin scaffolds also show fantastic stability after being exposed to protease from the gastrointestinal tract, which demonstrates their potential for oral administration.200 No apparent affinity loss of protein scaffolds was found after modification with fluorescent dyes, radionuclides, and drugs compared to the antibody conjugates.13 In addition, small scaffolds of <200 amino acids, library reshaping, site-directed mutation, and specific residue introduction are easier than antibodies via gene engineering. Meanwhile, considerable and homogeneous modified products can be well controlled by introducing cysteine residues or azide groups to provide a single coupling site.201 Most importantly, protein scaffolds can be mass-produced via the prokaryotic expression system are more cost-effective and affordable for research and patients in the future.
Challenges
Despite some protein scaffolds showing promising performance in different fields, few can realize successful clinical translation for cancer imaging or therapy, partially due to their short half-life, immunogenicity, somewhat unspecific biodistribution, or other less good clinical effects during long-term different dose administration.
Small size scaffolds with a short half-life generally accelerate the extravasation rate, are rapidly cleared from the circulating blood system, and impair the tissue penetration efficacy. Therefore, strategies of retention time extension, such as PEGylation, PASylation, fusing with Fc fragments, or albumin-binding domain, should be considered.202 Furthermore, immunogenicity resulting from non-human-derived scaffolds must also be resolved. Previous studies have demonstrated that constant region residue substitution (N23T, S33K, and N43E) of the HER2 affibody dramatically lowers the interaction with Ig and IgM, while some other residue replacements (D36A, D37S, E47A) have been shown to reduce the HER2 binding activity or thermostability.203 Additionally, the randomized residue (Q32) of the affibody scaffold was directly involved in both the Fc and the VH3 interaction; by systematically comparing the relationship between Q32 amino acid usage frequencies and the actual immunogenicity effect, it is possible to find the least immunogenic residues for this position. Biodistribution is another shortcoming; except in the metabolic organs, liver and kidneys, some scaffolds may cause unspecific accumulation, which results in a high imaging background or cytotoxicity, along with an increased residency time and dose. According to previous studies, hydrophilicity and charges seem to be the crux for biodistribution.204,205 By replacing hydrophilic amino acids, the ZHER2:34 affibody (A42S, A46S, A54S) showed significantly improved hydrophilicity, reduced renal retention by nearly threefold, and lowered the hepatic uptake. ZHER2:V2 (N52S, D53E, A54S) also lowered the renal uptake by 25- to 30-fold compared to the original affibody.36,206
Prospects
Overall, protein scaffolds are promising alternatives with many favorable properties superior to antibodies. Besides, protein scaffolds have been widely applied not only in cancer imaging and therapy but also in basic research, such as detection and elimination of microbes, intracellular delivery, blocking of the signal pathway, crystallization analysis, and in vitro detecting probes. With the development of computer simulation, molecular docking, and experimental technologies, the inherent flaws of scaffold molecules are expected to be gradually reduced, and safer, more effective, and affordable novel scaffolds will emerge. We expect that protein scaffolds will provide us with more options for cancer diagnosis and therapy in the future.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was partially supported by the Shanghai Municipal Science and Technology Major Project (ZC).
Footnotes
Crystal structures for the protein scaffolds shown in Fig. 1 were obtained from the PDB (https://www.rcsb.org/): (A) affibody (PDB ID: 3MZW);207 (B) ANTICALIN® (PDB ID: 4GH7);208 (C) adnectin or Monobody (PDB ID: 1TTG);209 (D) Knottin (PDB ID: 2IT7);210 (E) designed ankyrin repeat proteins (PDB ID: 1MJ0);211 (F) nanofitin or affitin (PDB ID: 4CJ2);212 (G) centyrins (PDB ID: 5L2H);213 (H) Kunitz (PDB ID: 1KTH);214 (I) affimer (PDB ID: 1NB5);215 (J) avimer (PDB ID: 1AJJ);216 (K) affilin based on the ubiquitin (PDB ID: 1UBI);217 and (L) affilin based on the γ-B-crystalin (PDB ID: 2JDG).218
Fig. 2 is reproduced from ref. 35, annotation for HER2 positive or negative and different tracers used in each group has been added to the top and bottom of the cited figure (CC BY-NC link (https://creativecommons.org/licenses/by-nc/4.0/)); Fig. 3 is reproduced from ref. 75, the missing background part of the bottom right corner was filled without any information interference to the original picture (CC BY link (https://creativecommons.org/licenses/by/4.0/)); Fig. 4 is reproduced from ref. 100 (CC BY link (http://creativecommons.org/licenses/by/4.0/)); Fig. 5 is reproduced from ref. 123 (CC BY link (http://creativecommons.org/licenses/by/4.0/)); and Fig. 6 is reproduced from ref. 147 (CC BY link (http://creativecommons.org/licenses/by/4.0/)).
References
- Senft D. Leiserson M. D. M. Ruppin E. Ronai Z. A. Trends Mol. Med. 2017;23:874–898. doi: 10.1016/j.molmed.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone E. R. Oliva M. Sabatini P. J. B. Stockley T. L. Siu L. L. Genome Med. 2020;12:8. doi: 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig I. R. Fuchs O. Hansen G. von Mutius E. Kopp M. V. Eur. Respir. J. 2017;50:1700391. doi: 10.1183/13993003.00391-2017. [DOI] [PubMed] [Google Scholar]
- Olivier M. Asmis R. Hawkins G. A. Howard T. D. Cox L. A. Int. J. Mol. Sci. 2019;20(19):4781. doi: 10.3390/ijms20194781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau C. H. Steeg P. S. Figg W. D. Lancet. 2019;394:793–804. doi: 10.1016/S0140-6736(19)31774-X. [DOI] [PubMed] [Google Scholar]
- Zhao X. Ning Q. Mo Z. Tang S. Artif. Cells Nanomed. Biotechnol. 2019;47:3621–3630. doi: 10.1080/21691401.2019.1657875. [DOI] [PubMed] [Google Scholar]
- Butterfield L. H., Schoenberger S. P. and Lyczak J. B., Immunology, Infection, and Immunity, 2004, pp. 573–591 10.1128/9781555816148.ch24 [DOI] [Google Scholar]
- Corley R. B., Immunology, Infection, and Immunity, 2004, pp. 111–143 10.1128/9781555816148.ch6 [DOI] [Google Scholar]
- Yu X. Yang Y. P. Dikici E. Deo S. K. Daunert S. Annu. Rev. Anal. Chem. 2017;10:293–320. doi: 10.1146/annurev-anchem-061516-045205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. J. Jeong K. J. J. Biosci. Bioeng. 2015;120:483–490. doi: 10.1016/j.jbiosc.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Ochoa M. C. Minute L. Rodriguez I. Garasa S. Perez-Ruiz E. Inoges S. Melero I. Berraondo P. Immunol. Cell Biol. 2017;95:347–355. doi: 10.1038/icb.2017.6. [DOI] [PubMed] [Google Scholar]
- Wang Y. Fan Z. Shao L. Kong X. Hou X. Tian D. Sun Y. Xiao Y. Yu L. Int. J. Nanomed. 2016;11:3287–3303. doi: 10.2147/IJN.S107194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyer L. Hruska K. Veterinarni Medicina. 2018;57:439–513. doi: 10.17221/6336-VETMED. [DOI] [Google Scholar]
- Skerra A. J. Mol. Recognit. 2000;13:167–187. doi: 10.1002/1099-1352(200007/08)13:4<167::AID-JMR502>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Simeon R. Chen Z. Protein Cell. 2018;9:3–14. doi: 10.1007/s13238-017-0386-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher D. Helma J. Schneider A. F. L. Leonhardt H. Hackenberger C. P. R. Angew. Chem., Int. Ed. 2018;57:2314–2333. doi: 10.1002/anie.201708459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa S. Xavier C. Muyldermans S. Devoogdt N. Exp. Opin. Drug Delivery. 2016;13:1149–1163. doi: 10.1080/17425247.2016.1178235. [DOI] [PubMed] [Google Scholar]
- Dickgiesser S. Kellner R. Kolmar H. Rasche N. Methods Mol. Biol. 2019;2033:1–14. doi: 10.1007/978-1-4939-9654-4_1. [DOI] [PubMed] [Google Scholar]
- Morais M. Forte N. Chudasama V. Baker J. R. Methods Mol. Biol. 2019;2033:15–24. doi: 10.1007/978-1-4939-9654-4_2. [DOI] [PubMed] [Google Scholar]
- Chaubet G. Thoreau F. Wagner A. Drug Discovery Today Technol. 2018;30:21–26. doi: 10.1016/j.ddtec.2018.09.002. [DOI] [PubMed] [Google Scholar]
- Milczek E. M. Chem. Rev. 2018;118:119–141. doi: 10.1021/acs.chemrev.6b00832. [DOI] [PubMed] [Google Scholar]
- Adumeau P. Sharma S. K. Brent C. Zeglis B. M. Mol. Imaging Biol. 2016;18:153–165. doi: 10.1007/s11307-015-0920-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addy P. S. Erickson S. B. Italia J. S. Chatterjee A. Methods Mol. Biol. 2019;2033:239–251. doi: 10.1007/978-1-4939-9654-4_16. [DOI] [PubMed] [Google Scholar]
- Merten H. Schaefer J. V. Brandl F. Zangemeister-Wittke U. Plückthun A. Methods Mol. Biol. 2019;2033:253–273. doi: 10.1007/978-1-4939-9654-4_17. [DOI] [PubMed] [Google Scholar]
- Zappala F. Tsourkas A. Methods Mol. Biol. 2019;2033:275–286. doi: 10.1007/978-1-4939-9654-4_18. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Park K. Y. Suazo K. F. Distefano M. D. Chem. Soc. Rev. 2018;47:9106–9136. doi: 10.1039/C8CS00537K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson B. Moks T. Jansson B. Abrahmsen L. Elmblad A. Holmgren E. Henrichson C. Jones T. A. Uhlen M. Protein Eng. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- Nord K. Nilsson J. Nilsson B. Uhlen M. Nygren P. A. Protein Eng. 1995;8:601–608. doi: 10.1093/protein/8.6.601. [DOI] [PubMed] [Google Scholar]
- Webster J. M. Zhang R. Gambhir S. S. Cheng Z. Syud F. A. ChemBioChem. 2009;10:1293–1296. doi: 10.1002/cbic.200900062. [DOI] [PubMed] [Google Scholar]
- Gruning C. S. R. Mirecka E. A. Klein A. N. Mandelkow E. Willbold D. Marino S. F. Stoldt M. Hoyer W. J. Biol. Chem. 2014;289:23209–23218. doi: 10.1074/jbc.M114.560920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affibody, https://www.affibody.se, (accessed 14 November, 2020)
- Stahl S. Graslund T. Karlstrom A. E. Frejd F. Y. Nygren P. Lofblom J. Trends Biotechnol. 2017;35:691–712. doi: 10.1016/j.tibtech.2017.04.007. [DOI] [PubMed] [Google Scholar]
- Frejd F. Y. Kim K. T. Exp. Mol. Med. 2017;49:e306. doi: 10.1038/emm.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum R. P. Prasad V. Muller D. Schuchardt C. Orlova A. Wennborg A. Tolmachev V. Feldwisch J. J. Nucl. Med. 2010;51:892–897. doi: 10.2967/jnumed.109.073239. [DOI] [PubMed] [Google Scholar]
- Sörensen J. Velikyan I. Sandberg D. Wennborg A. Feldwisch J. Tolmachev V. Orlova A. Sandström M. Lubberink M. Olofsson H. Carlsson J. Lindman H. Theranostics. 2016;6:262–271. doi: 10.7150/thno.13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroujeni M. A.-O. Rinne S. A.-O. Vorobyeva A. A.-O. Loftenius A. Feldwisch J. Jonasson P. Chernov V. Orlova A. A.-O. Frejd F. Y. Tolmachev V. A.-O. Int. J. Mol. Sci. 2021;22(5):2770. doi: 10.3390/ijms22052770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen J. Velikyan I. Sandberg D. Wennborg A. Feldwisch J. Tolmachev V. Orlova A. Sandstrom M. Lubberink M. Olofsson H. Carlsson J. Lindman H. Theranostics. 2016;6:262–271. doi: 10.7150/thno.13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstrom M. Lindskog K. Velikyan I. Wennborg A. Feldwisch J. Sandberg D. Tolmachev V. Orlova A. Sorensen J. Carlsson J. Lindman H. Lubberink M. J. Nucl. Med. 2016;57:867–871. doi: 10.2967/jnumed.115.169342. [DOI] [PubMed] [Google Scholar]
- Sandberg D. Tolmachev V. Velikyan I. Olofsson H. Wennborg A. Feldwisch J. Carlsson J. Lindman H. Sorensen J. Eur. J. Nucl. Med. Mol. Imaging. 2017;44:1337–1346. doi: 10.1007/s00259-017-3650-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N. Liu C. Guo X. Xu Y. Gong J. Qi C. Zhang X. Yang M. Zhu H. Shen L. Yang Z. Eur. J. Nucl. Med. Mol. Imaging. 2020;48:161–175. doi: 10.1007/s00259-020-04898-5. [DOI] [PubMed] [Google Scholar]
- Martins C. D. Da Pieve C. Burley T. A. Smith R. Ciobota D. M. Allott L. Harrington K. J. Oyen W. J. G. Smith G. Kramer-Marek G. Clin. Cancer Res. 2018;24:1853–1865. doi: 10.1158/1078-0432.CCR-17-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne S. S. Dahlsson Leitao C. Saleh-Nihad Z. Mitran B. Tolmachev V. Ståhl S. Löfblom J. Orlova A. Int. J. Mol. Sci. 2020;21(6):1972. doi: 10.3390/ijms21061972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza A. L. Marra K. Gunn J. Samkoe K. S. Hoopes P. J. Feldwisch J. Paulsen K. D. Pogue B. W. Mol. Imaging Biol. 2017;19:41–48. doi: 10.1007/s11307-016-0980-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samkoe K. S. Sardar H. S. Gunn J. Feldwisch J. Linos K. Henderson E. Pogue B. Paulsen K. Proc. SPIE Int. Soc. Opt. Eng. 2019:10862. doi: 10.1117/12.2510935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J. T. Dsouza A. V. Marra K. Pogue B. W. Roberts D. W. Paulsen K. D. Biomed. Opt. Express. 2016;7:3280–3288. doi: 10.1364/BOE.7.003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samkoe K. S. Sardar H. S. Bates B. D. Tselepidakis N. N. Gunn J. R. Hoffer-Hawlik K. A. Feldwisch J. Pogue B. W. Paulsen K. D. Henderson E. R. J. Surg. Oncol. 2019;119:1077–1086. doi: 10.1002/jso.25468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. Cheng K. Liu Y. Hu X. Meng S. Cheng Z. Amino Acids. 2015;47:1409–1419. doi: 10.1007/s00726-015-1975-4. [DOI] [PubMed] [Google Scholar]
- Gonzalez Trotter D. E. Meng X. McQuade P. Rubins D. Klimas M. Zeng Z. Connolly B. M. Miller P. J. O'Malley S. S. Lin S. A. Getty K. L. Fayadat-Dilman L. Liang L. Wahlberg E. Widmark O. Ekblad C. Frejd F. Y. Hostetler E. D. Evelhoch J. L. J. Nucl. Med. 2017;58:1852–1857. doi: 10.2967/jnumed.117.191718. [DOI] [PubMed] [Google Scholar]
- Garousi J. Honarvar H. Andersson K. G. Mitran B. Orlova A. Buijs J. Lofblom J. Frejd F. Y. Tolmachev V. Mol. Pharm. 2016;13:3676–3687. doi: 10.1021/acs.molpharmaceut.6b00502. [DOI] [PubMed] [Google Scholar]
- Bam R. Laffey M. Nottberg K. Lown P. S. Hackel B. J. Wilson K. E. Bioconjugate Chem. 2019;30:1677–1689. doi: 10.1021/acs.bioconjchem.9b00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bam R. Lown P. S. Stern L. A. Sharma K. Wilson K. E. Bean G. R. Lutz A. M. Paulmurugan R. Hackel B. J. Dahl J. Abou-Elkacem L. Clin. Cancer Res. 2020;26:2140–2150. doi: 10.1158/1078-0432.CCR-19-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altai M. Liu H. Ding H. Mitran B. Edqvist P. H. Tolmachev V. Orlova A. Graslund T. J. Controlled Release. 2018;288:84–95. doi: 10.1016/j.jconrel.2018.08.040. [DOI] [PubMed] [Google Scholar]
- Ju Y. Zhang H. Yu J. Tong S. Tian N. Wang Z. Wang X. Su X. Chu X. Lin J. Ding Y. Li G. Sheng F. Hou Y. ACS Nano. 2017;11:9239–9248. doi: 10.1021/acsnano.7b04461. [DOI] [PubMed] [Google Scholar]
- Satpathy M. Wang L. Zielinski R. J. Qian W. Wang Y. A. Mohs A. M. Kairdolf B. A. Ji X. Capala J. Lipowska M. Nie S. Mao H. Yang L. Theranostics. 2019;9:778–795. doi: 10.7150/thno.29964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova A. Bass T. Z. Rinne S. S. Leitao C. D. Rosestedt M. Atterby C. Gudmundsdotter L. Frejd F. Y. Lofblom J. Tolmachev V. Stahl S. Mol. Pharmaceutics. 2018;15:3394–3403. doi: 10.1021/acs.molpharmaceut.8b00393. [DOI] [PubMed] [Google Scholar]
- He H. Nieminen A. L. Xu P. Biomater. Sci. 2019;7:5143–5149. doi: 10.1039/C9BM01237K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D. Yang Y. Yuan F. Fan Q. Wang F. Huang Y. Song H. Hu P. Wang R. Li G. Liu R. Li J. Int. J. Pharm. 2020;586:119541. doi: 10.1016/j.ijpharm.2020.119541. [DOI] [PubMed] [Google Scholar]
- Fan Q. Tao Z. Yang H. Shi Q. Wang H. Jia D. Wan L. Zhang J. Cheng J. Lu X. J. Controlled Release. 2019;302:63–78. doi: 10.1016/j.jconrel.2019.03.018. [DOI] [PubMed] [Google Scholar]
- Shi Q. Tao Z. Yang H. Fan Q. Wei D. Wan L. Lu X. Drug Delivery. 2017;24:1818–1830. doi: 10.1080/10717544.2017.1407011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P. Wang L. Hou B. Zhu J. Zhou M. Jiang J. Wang L. Chen S. Zhu S. Chen J. Zhang L. Theranostics. 2018;8:3544–3558. doi: 10.7150/thno.24607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S. Chen J. Xiong Y. Kamara S. Gu M. Tang W. Chen S. Dong H. Xue X. Zheng Z. M. Zhang L. PLoS Pathog. 2020;16:e1008223. doi: 10.1371/journal.ppat.1008223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower D. R. Biochem. J. 1996;318(Pt 1):1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer M. Skerra A. Methods Enzymol. 2012;503:157–188. doi: 10.1016/B978-0-12-396962-0.00007-0. [DOI] [PubMed] [Google Scholar]
- Beste G. Schmidt F. S. Stibora T. Skerra A. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1898–1903. doi: 10.1073/pnas.96.5.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C. Skerra A. BioDrugs. 2018;32:233–243. doi: 10.1007/s40259-018-0278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar A. Ahmad E. Zia Q. Rauf M. A. Owais M. Ashraf G. M. Int. J. Biol. Macromol. 2017;102:630–641. doi: 10.1016/j.ijbiomac.2017.04.045. [DOI] [PubMed] [Google Scholar]
- Gebauer M. Skerra A. Curr. Opin. Biotechnol. 2019;60:230–241. doi: 10.1016/j.copbio.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Pieris, https://www.pieris.com/platform, (accessed 14 November 2020)
- Van Scheltinga A. G. T. T. Hooge M. N. L. Hinner M. Verheijen R. B. Allersdorfer A. Hulsmeyer M. Nagengast W. B. Schroder C. P. Kosterink J. G. W. De Vries E. G. E. J. Nucl. Med. 2014;55:665–671. doi: 10.2967/jnumed.113.124941. [DOI] [PubMed] [Google Scholar]
- Albrecht V. Richter A. Pfeiffer S. Gebauer M. Lindner S. Gieser E. Schuller U. Schichor C. Gildehaus F. J. Bartenstein P. Tonn J. C. Skerra A. Glass R. Int. J. Cancer. 2016;138:1269–1280. doi: 10.1002/ijc.29874. [DOI] [PubMed] [Google Scholar]
- Reichert A. J. Poxleitner G. Dauner M. Skerra A. Protein Eng., Des. Sel. 2015;28:553–565. doi: 10.1093/protein/gzv048. [DOI] [PubMed] [Google Scholar]
- Barinka C. Ptacek J. Richter A. Novakova Z. Morath V. Skerra A. Protein Eng., Des. Sel. 2016;29:105–115. doi: 10.1093/protein/gzv065. [DOI] [PubMed] [Google Scholar]
- Richter A. Skerra A. Biol. Chem. 2017;398:39–55. doi: 10.1515/hsz-2016-0195. [DOI] [PubMed] [Google Scholar]
- Friedrich L. Kornberger P. Mendler C. T. Multhoff G. Schwaiger M. Skerra A. Biol. Chem. 2018;399:235–252. doi: 10.1515/hsz-2017-0207. [DOI] [PubMed] [Google Scholar]
- Deuschle F. C. Morath V. Schiefner A. Brandt C. Ballke S. Reder S. Steiger K. Schwaiger M. Weber W. Skerra A. Theranostics. 2020;10:2172–2187. doi: 10.7150/thno.38968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlapschy M. Binder U. Börger C. Theobald I. Wachinger K. Kisling S. Haller D. Skerra A. Protein Eng., Des. Sel. 2013;26:489–501. doi: 10.1093/protein/gzt023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Paulete A. R. Labiano S. Rodriguez-Ruiz M. E. Azpilikueta A. Etxeberria I. Bolaños E. Lang V. Rodriguez M. Aznar M. A. Jure-Kunkel M. Melero I. Eur. J. Immunol. 2016;46:513–522. doi: 10.1002/eji.201445388. [DOI] [PubMed] [Google Scholar]
- Hinner M. Aiba R. B. Jaquin T. Berger S. Durr M. Schlosser C. Allersdorfer A. Rothe C. Matis L. Olwill S. Cancer Res. 2017;77:3673. [Google Scholar]
- Hinner M. J. Bel Aiba R. S. Schlosser C. Jaquin T. Allersdorfer A. Berger S. Wiedenmann A. Matschiner G. Schüler J. Moebius U. Rothe C. Olwill S. A. Eur. J. Cancer. 2016;69:S99. doi: 10.1016/S0959-8049(16)32894-5. [DOI] [Google Scholar]
- Hinner M. J. Aiba R. S. B. Jaquin T. J. Berger S. Durr M. C. Schlosser C. Allersdorfer A. Wiedenmann A. Matschiner G. Schuler J. Moebius U. Rothe C. Matis L. Olwill S. A. Clin. Cancer Res. 2019;25:5878–5889. doi: 10.1158/1078-0432.CCR-18-3654. [DOI] [PubMed] [Google Scholar]
- Piha-Paul S. Bendell J. Tolcher A. Hurvitz S. Patnaik A. Shroff R. Pohlmann P. Zettl M. Hahn N. Krishnamurthy A. Duerr M. Mei J. Aviano K. Yusuf R. Matis L. Olwill S. Bruns I. Ku G. J. ImmunoTher. Cancer. 2020;8:A1. [Google Scholar]
- Hinner M. J. Aiba R.-S. B. Wiedenmann A. Schlosser C. Allersdorfer A. Matschiner G. Rothe C. Moebius U. Kohrt H. E. Olwill S. A. J. ImmunoTher. Cancer. 2015;3:P187. doi: 10.1186/2051-1426-3-S2-P187. [DOI] [Google Scholar]
- Bossenmaier B. Schlosser C. Aiba R. S. B. Barthels C. Weiche B. Eichner T. Yegres M. Olwill S. Immunology. 2019;79:3268. [Google Scholar]
- Mross K. Richly H. Fischer R. Scharr D. Buchert M. Stern A. Gille H. Audoly L. P. Scheulen M. E. PLoS One. 2013;8:e83232. doi: 10.1371/journal.pone.0083232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H. Hulsmeyer M. Trentmann S. Matschiner G. Christian H. J. Meyer T. Amirkhosravi A. Audoly L. P. Hohlbaum A. M. Skerra A. Angiogenesis. 2016;19:79–94. doi: 10.1007/s10456-015-9490-5. [DOI] [PubMed] [Google Scholar]
- Dickinson C. D. Veerapandian B. Dai X. P. Hamlin R. C. Xuong N. H. Ruoslahti E. Ely K. R. J. Mol. Biol. 1994;236:1079–1092. doi: 10.1016/0022-2836(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Chandler P. G. Buckle A. M. Cells. 2020;9:610. doi: 10.3390/cells9030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A. Bailey C. W. Huang X. Koide S. J. Mol. Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- Bristol-Myers Squibb, https://www.bms.com/, (accessed 14 November 2020)
- Koide A. Wojcik J. Gilbreth R. N. Hoey R. J. Koide S. S. J. Mol. Biol. 2012;415:393–405. doi: 10.1016/j.jmb.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A. Abou-Elkacem L. Methods Mol. Biol. 2019;2033:301–313. doi: 10.1007/978-1-4939-9654-4_20. [DOI] [PubMed] [Google Scholar]
- Sha F. Salzman G. Gupta A. Koide S. Protein Sci. 2017;26:910–924. doi: 10.1002/pro.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolcher A. W. Sweeney C. Papadopoulos K. P. Patnaik A. Chiorean E. G. Mita A. C. Sankhala K. K. Furfine E. Gokemeijer J. Iacono L. Clin. Cancer Res. 2011;17:363–371. doi: 10.1158/1078-0432.CCR-10-1411. [DOI] [PubMed] [Google Scholar]
- Abouelkacem L. Wilson K. E. Johnson S. M. Chowdhury S. M. Bachawal S. V. Hackel B. J. Tian L. Willmann J. K. Theranostics. 2016;6:1740–1752. doi: 10.7150/thno.15169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. A. Yoon H. S. Park S. H. Kim D. Y. Pyo A. Kim H. S. Min J. J. Hong Y. PLoS One. 2017;12:e0180786. doi: 10.1371/journal.pone.0180786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo A. You S. H. Sik Kim H. Young Kim J. Min J. J. Kim D. Y. Hong Y. Bioorg. Med. Chem. Lett. 2020;30:127262. doi: 10.1016/j.bmcl.2020.127262. [DOI] [PubMed] [Google Scholar]
- Natarajan A. Patel C. B. Ramakrishnan S. Panesar P. S. Long S. R. Gambhir S. S. Clin. Cancer Res. 2019;25:1774–1785. doi: 10.1158/1078-0432.CCR-18-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan S. Natarajan A. Chan C. T. Panesar P. S. Gambhir S. S. Protein Eng., Des. Sel. 2019;32:231–240. doi: 10.1093/protein/gzz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly D. J. Smith R. A. Morin P. Lipovsek D. Gokemeijer J. Cohen D. Lafont V. Tran T. Cole E. L. Wright M. Kim J. Pena A. Kukral D. Dischino D. D. Chow P. Gan J. Adelakun O. Wang X. T. Cao K. Leung D. Bonacorsi, Jr. S. J. Hayes W. J. Nucl. Med. 2018;59:529–535. doi: 10.2967/jnumed.117.199596. [DOI] [PubMed] [Google Scholar]
- Niemeijer A. N. Leung D. Huisman M. C. Bahce I. Hoekstra O. S. van Dongen G. Boellaard R. Du S. Hayes W. Smith R. Windhorst A. D. Hendrikse N. H. Poot A. Vugts D. J. Thunnissen E. Morin P. Lipovsek D. Donnelly D. J. Bonacorsi S. J. Velasquez L. M. de Gruijl T. D. Smit E. F. de Langen A. J. Nat. Commun. 2018;9:4664. doi: 10.1038/s41467-018-07131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman M. C. Niemeijer A. N. Windhorst A. D. Schuit R. C. Leung D. Hayes W. Poot A. Bahce I. Radonic T. Oprea-Lager D. E. Hoekstra O. S. Thunnissen E. Hendrikse N. H. Smit E. F. de Langen A. J. Boellaard R. J. Nucl. Med. 2020;61:1455–1460. doi: 10.2967/jnumed.119.240895. [DOI] [PubMed] [Google Scholar]
- Nienhuis P. H. Antunes I. F. Glaudemans A. Jalving M. Leung D. Noordzij W. Slart R. de Vries E. F. Hospers G. A. P. J. Nucl. Med. 2021;63(6):899–905. doi: 10.2967/jnumed.121.262368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff D. Kesari S. de Groot J. Mikkelsen T. Drappatz J. Coyle T. Fichtel L. Silver B. Walters I. Reardon D. Invest. New Drugs. 2015;33:247–253. doi: 10.1007/s10637-014-0186-2. [DOI] [PubMed] [Google Scholar]
- Aghaabdollahian S. Ahangari Cohan R. Norouzian D. Davami F. Asadi Karam M. R. Torkashvand F. Vaseghi G. Moazzami R. Latif Dizaji S. Sci. Rep. 2019;9:2978. doi: 10.1038/s41598-019-39776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. Cinay G. E. Zhao Y. Guo Y. Zhang X. Wang P. Mol. Ther. 2017;25:2466–2476. doi: 10.1016/j.ymthe.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. Xu J. Lee S. Tsai S. T. Zhou B. Kurosawa K. Werner M. S. Koide A. Ruthenburg A. J. Dou Y. Koide S. Nat. Chem. Biol. 2018;14:895–900. doi: 10.1038/s41589-018-0099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I. Spencer-Smith R. O'Bryan J. P. Oncogene. 2019;38:2984–2993. doi: 10.1038/s41388-018-0636-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovsek D. Carvajal I. M. Allentoff A. Barros A. Brailsford J. Cong Q. Cotter P. Z. Gangwar S. Hollander C. Lafont V. Protein Eng., Des. Sel. 2018;31:159–171. doi: 10.1093/protein/gzy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirois A. R. Deny D. A. Li Y. Fall Y. D. Moore S. J. Biotechnol. Bioeng. 2020;117:330–341. doi: 10.1002/bit.27204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P. Proteins. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- Mosavi L. K. Minor, Jr. D. L. Peng Z. Y. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz H. K. Stumpp M. T. Forrer P. Amstutz P. Pluckthun A. J. Mol. Biol. 2003;332:489–503. doi: 10.1016/S0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- Molecular Partners Allergan, https://www.molecularpartners.com/, (accessed 14 November 2020)
- Schilling J. Schoppe J. Pluckthun A. J. Mol. Biol. 2013;426:691–721. doi: 10.1016/j.jmb.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Rasool M. Malik A. Hussain M. Haq K. A. Butt K. Ashraf M. A. B. Naseer M. I. Asif M. Shaikh R. Mustafa M. Z. Alam Q. Rasool G. Ahmad W. Haque A. Kamal M. A. Curr. Pharm. Des. 2017;23:1610–1615. doi: 10.2174/1381612822666161208121829. [DOI] [PubMed] [Google Scholar]
- Boersma Y. L. Methods Mol. Biol. 2018;1798:307–327. doi: 10.1007/978-1-4939-7893-9_23. [DOI] [PubMed] [Google Scholar]
- Kramer L. Renko M. Zavrsnik J. Turk D. Seeger M. A. Vasiljeva O. Grutter M. G. Turk V. Turk B. Theranostics. 2017;7:2806–2821. doi: 10.7150/thno.19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. L. Tan J. E. Tian Y. Huang S. Sun P. H. Wang M. Han Y. J. Li H. S. Wu H. B. Zhang X. M. Xu Y. K. Wang Q. S. Biomaterials. 2017;147:86–98. doi: 10.1016/j.biomaterials.2017.09.010. [DOI] [PubMed] [Google Scholar]
- Guryev E. L. Shilyagina N. Y. Kostyuk A. B. Sencha L. M. Balalaeva I. V. Vodeneev V. A. Kutova O. M. Lyubeshkin A. V. Yakubovskaya R. I. Pankratov A. A. Ingel F. I. Novik T. S. Deyev S. M. Ermilov S. A. Zvyagin A. V. Toxicol. Sci. 2019;170:123–132. doi: 10.1093/toxsci/kfz086. [DOI] [PubMed] [Google Scholar]
- Molavipordanjani S. Khodashenas S. Abedi S. M. Moghadam M. F. Mardanshahi A. Hosseinimehr S. J. Eur. J. Pharm. Sci. 2020;148:105312. doi: 10.1016/j.ejps.2020.105312. [DOI] [PubMed] [Google Scholar]
- Bragina O. Chernov V. Schulga A. Konovalova E. Garbukov E. Vorobyeva A. Orlova A. Tashireva L. Sorensen J. Zelchan R. Medvedeva A. Deyev S. Tolmachev V. J. Nucl. Med. 2021;63(4):528–535. doi: 10.2967/jnumed.121.262542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyev S. M. Vorobyeva A. Schulga A. Abouzayed A. Gunther T. Garousi J. Konovalova E. Ding H. Graslund T. Orlova A. Tolmachev V. Int. J. Biol. Macromol. 2020;145:216–225. doi: 10.1016/j.ijbiomac.2019.12.147. [DOI] [PubMed] [Google Scholar]
- Vorobyeva A. Konovalova E. Xu T. Schulga A. Altai M. Garousi J. Rinne S. S. Orlova A. Tolmachev V. Deyev S. Int. J. Mol. Sci. 2020;21(9):3310. doi: 10.3390/ijms21093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler U. Ekawardhani S. Cornelius A. Gilboy P. Bakker T. R. Dolado I. Stumpp M. T. Dawson K. M. Oncotarget. 2017;8:98371–98383. doi: 10.18632/oncotarget.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azaro A. Rodon J. Middleton M. R. Baird R. D. Herrmann R. Fiedler U. Haunschild J. Hauptle M. Hermann F. Schreiner S. J. Clin. Oncol. 2018;36:2520. [Google Scholar]
- Baird R. D. Linossi C. Middleton M. Lord S. Harris A. Rodón J. Zitt C. Fiedler U. Dawson K. M. Leupin N. Stumpp M. T. Harstrick A. Azaro A. Fischer S. Omlin A. J. Clin. Oncol. 2021;39:145–154. doi: 10.1200/JCO.20.00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton M. R. Azaro A. Kumar S. Niedermann P. Rodón J. Herbschleb K. H. Steiner J. Zitt C. Feurstein D. Schreiner S. Turner D. Dawson K. Tadjalli-Mehr K. Baur E. Stumpp M. Harstrick A. Baird R. Omlin A. Ann. Oncol. 2016;27:vi115. doi: 10.1093/annonc/mdw368.04. [DOI] [Google Scholar]
- Rao L. De Veirman K. Giannico D. Saltarella I. Desantis V. Frassanito M. A. Solimando A. G. Ribatti D. Prete M. Harstrick A. Fiedler U. De Raeve H. Racanelli V. Vanderkerken K. Vacca A. Oncotarget. 2018;9:13366–13381. doi: 10.18632/oncotarget.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres F. Iamele L. Meyer T. Stuber J. C. Kast F. Gherardi E. Niemann H. H. Pluckthun A. J. Mol. Biol. 2019;431:2020–2039. doi: 10.1016/j.jmb.2019.03.024. [DOI] [PubMed] [Google Scholar]
- Soto-Gamez A. Chen D. Nabuurs A. G. E. Quax W. J. Demaria M. Boersma Y. L. Cancers. 2020;12(2):411. doi: 10.3390/cancers12020411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanauer J. R. H. Koch V. Lauer U. M. Muhlebach M. D. Mol. Ther. Oncolyt. 2019;15:186–200. doi: 10.1016/j.omto.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev E. L. Volodina N. O. Shilyagina N. Y. Gudkov S. V. Balalaeva I. V. Volovetskiy A. B. Lyubeshkin A. V. Sen A. V. Ermilov S. A. Vodeneev V. A. Petrov R. V. Zvyagin A. V. Alferov Z. I. Deyev S. M. Proc. Natl. Acad. Sci. U. S. A. 2018;115:9690–9695. doi: 10.1073/pnas.1809258115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegler E. Li S. Kim Y. J. Wang P. Hum. Gene Ther. 2017;28:726–736. doi: 10.1089/hum.2017.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan A. Rajan A. Salter A. I. Kosasih P. L. Wu Q. Voutsinas J. Jensen M. C. Pluckthun A. Riddell S. R. Clin. Cancer Res. 2019;25:7506–7516. doi: 10.1158/1078-0432.CCR-19-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. S. Mistry B. Guillard S. Ulrichsen J. C. Sandercock A. M. Wang J. Gonzalez-Munoz A. Parmentier J. Black C. Soden J. Freeth J. Jovanovic J. Leyland R. Al-Lamki R. S. Leishman A. J. Rust S. J. Stewart R. Jermutus L. Bradley J. R. Bedian V. Valge-Archer V. Minter R. Wilkinson R. W. Oncotarget. 2016;7:68278–68291. doi: 10.18632/oncotarget.11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S. Darbon H. Dyason K. Verdonck F. Tytgat J. FASEB J. 2003;17:1765–1767. doi: 10.1096/fj.02-1044fje. [DOI] [PubMed] [Google Scholar]
- Molesini B. Treggiari D. Dalbeni A. Minuz P. Pandolfini T. Br. J. Clin. Pharmacol. 2017;83:63–70. doi: 10.1111/bcp.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintzing J. R. Cochran J. R. Curr. Opin. Chem. Biol. 2016;34:143–150. doi: 10.1016/j.cbpa.2016.08.022. [DOI] [PubMed] [Google Scholar]
- Kolmar H. FEBS J. 2008;275:2684–2690. doi: 10.1111/j.1742-4658.2008.06440.x. [DOI] [PubMed] [Google Scholar]
- Avrutina O. Adv. Exp. Med. Biol. 2016;917:121–144. doi: 10.1007/978-3-319-32805-8_7. [DOI] [PubMed] [Google Scholar]
- AstraZeneca, https://www.astrazeneca.com/, (accessed 14 November)
- Jazz, https://www.jazzpharma.com/, (accessed 14 November 2020)
- Cyclogenix, https://www.cyclogenix.com/, (accessed 14 November 2020)
- Ironwood, https://ironwoodpharma.com/, (accessed 14 November 2020)
- Zhang C. Kimura R. H. Abouelkacem L. Levi J. Xu L. Gambhir S. S. J. Nucl. Med. 2016;57:1629–1634. doi: 10.2967/jnumed.115.169383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummers W. S. Kimura R. H. Abouelkacem L. Beinat C. Vahrmeijer A. L. Swijnenburg R. Willmann J. K. Gambhir S. S. Clin. Cancer Res. 2018;24:1667–1676. doi: 10.1158/1078-0432.CCR-17-2491. [DOI] [PubMed] [Google Scholar]
- Kimura R. H. Wang L. Shen B. Huo L. Tummers W. S. Filipp F. V. Guo H. H. Haywood T. Abouelkacem L. Baratto L. Nat. Commun. 2019;10:4673. doi: 10.1038/s41467-019-11863-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui B. G. Salomon N. Wustehube-Lausch J. Daneschdar M. Schmoldt H. U. Tureci O. Sahin U. Nat. Commun. 2020;11:295. doi: 10.1038/s41467-019-13948-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. W. Cochran F. V. Cochran J. R. J. Am. Chem. Soc. 2015;137:6–9. doi: 10.1021/ja508416e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox N. Kintzing J. R. Smith M. Grant G. A. Cochran J. R. Angew. Chem. 2016;55:9894–9897. doi: 10.1002/anie.201603488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier N. V. Ackerman S. E. Kintzing J. R. Chen R. Interrante M. F. Steiner A. R. Sato A. K. Cochran J. R. Mol. Cancer Ther. 2016;15:1291–1300. doi: 10.1158/1535-7163.MCT-15-0881. [DOI] [PubMed] [Google Scholar]
- Lal S. Raffel C. Mol. Ther. Oncolyt. 2017;7:57–66. doi: 10.1016/j.omto.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen G. Burks S. R. Frank J. A. Toxins. 2018;10:496. doi: 10.3390/toxins10120496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T. Min K. A. Cheong H. Moon C. Shin M. C. J. Drug Target. 2019;27:950–958. doi: 10.1080/1061186X.2018.1516221. [DOI] [PubMed] [Google Scholar]
- Zhao J. Wang Y. Li X. Gao S. Liu S. Song Y. Wang J. Xiong Y. Ma H. Jiang L. Contrast Media Mol. Imaging. 2018;2018:8439162. doi: 10.1155/2018/8439162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidel J. Kennedy K. C. Dernell W. S. Hansen S. Wiss V. Stroud M. R. Molho J. I. Knoblaugh S. E. Meganck J. A. Olson J. M. Cancer Res. 2015;75:4283–4291. doi: 10.1158/0008-5472.CAN-15-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. Girard E. J. Pakiam F. Seibel E. J. J. Med. Imaging. 2019;6:025005. doi: 10.1117/1.JMI.6.2.025005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil C. G. Walker D. G. Miller D. M. Butte P. Morrison B. Kittle D. S. Hansen S. J. Nufer K. L. Byrnes-Blake K. A. Yamada M. Lin L. L. Pham K. Perry J. Parrish-Novak J. Ishak L. Prow T. Black K. Mamelak A. N. Neurosurgery. 2019;85:E641–E649. doi: 10.1093/neuros/nyz125. [DOI] [PubMed] [Google Scholar]
- Dyax, https://www.takeda.com/, (accessed 14 November 2020)
- Tian T. Li J. Xie C. Sun Y. Lei H. Liu X. Xia J. Shi J. Wang L. Lu W. Fan C. ACS Appl. Mater. Interfaces. 2018;10:3414–3420. doi: 10.1021/acsami.7b17927. [DOI] [PubMed] [Google Scholar]
- Schmidt M. C. B. Morais K. L. P. Almeida M. E. S. Iqbal A. Goldfeder M. B. Chudzinski-Tavassi A. M. Cell Adhes. Migr. 2020;14:129–138. doi: 10.1080/19336918.2018.1516982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sananes A. Cohen I. Shahar A. Hockla A. De Vita E. Miller A. K. Radisky E. S. Papo N. J. Biol. Chem. 2018;293:12663–12680. doi: 10.1074/jbc.RA117.000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler L. K. Hoffmann T. Tomlinson D. C. Song Q. Lee T. Busby M. Nyathi Y. Gendra E. Tiede C. Flanagan K. Cockell S. J. Wipat A. Harwood C. Wagner S. D. Knowles M. A. Davis J. J. Keegan N. Ferrigno P. K. Protein Eng., Des. Sel. 2011;24:751–763. doi: 10.1093/protein/gzr019. [DOI] [PubMed] [Google Scholar]
- Avacta Life Sciences, https://avacta.com/, (accessed 14 November 2020)
- Wang W. Guo Y. Tiede C. Chen S. Kopytynski M. Kong Y. Kulak A. N. Tomlinson D. C. Chen R. Mcpherson M. J. ACS Appl. Mater. Interfaces. 2017;9:15232–15244. doi: 10.1021/acsami.6b13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiede C. Bedford R. Heseltine S. J. Smith G. Wijetunga I. Ross R. AlQallaf D. Roberts A. P. Balls A. Curd A. Hughes R. E. Martin H. Needham S. R. Zanetti-Domingues L. C. Sadigh Y. Peacock T. P. Tang A. A. Gibson N. Kyle H. Platt G. W. Ingram N. Taylor T. Coletta L. P. Manfield I. Knowles M. Bell S. Esteves F. Maqbool A. Prasad R. K. Drinkhill M. Bon R. S. Patel V. Goodchild S. A. Martin-Fernandez M. Owens R. J. Nettleship J. E. Webb M. E. Harrison M. Lippiat J. D. Ponnambalam S. Peckham M. Smith A. Ferrigno P. K. Johnson M. McPherson M. J. Tomlinson D. C. Elife. 2017;6:e24903. doi: 10.7554/eLife.24903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar G. Bellinzoni M. Maillasson M. Paillard-Laurance L. Alzari P. M. He X. Mouratou B. Pecorari F. Protein Eng., Des. Sel. 2013;26:267–275. doi: 10.1093/protein/gzs106. [DOI] [PubMed] [Google Scholar]
- Affilogic, https://www.affilogic.com/, (accessed 14 November 2020)
- Goux M. Becker G. Gorre H. Dammicco S. Desselle A. Egrise D. Leroi N. Lallemand F. Bahri M. A. Doumont G. Bioconjugate Chem. 2017;28:2361–2371. doi: 10.1021/acs.bioconjchem.7b00374. [DOI] [PubMed] [Google Scholar]
- Silverman J. Liu Q. Bakker A. To W. Duguay A. Alba B. M. Smith R. Rivas A. Li P. Le H. Whitehorn E. Moore K. W. Swimmer C. Perlroth V. Vogt M. Kolkman J. Stemmer W. P. Nat. Biotechnol. 2005;23:1556–1561. doi: 10.1038/nbt1166. [DOI] [PubMed] [Google Scholar]
- Amgen, https://www.amgen.com/science, (accessed 14 November 2020)
- Kohnehrouz B. B. Talischian A. Dehnad A. Nayeri S. Avicenna J Med Biotechnol. 2017;10:9–14. [PMC free article] [PubMed] [Google Scholar]
- Janseen, https://www.janssen.com, (accessed 14 November 2020)
- Goldberg S. Cardoso R. Lin T. Spinkadoms T. Klein D. Jacobs S. Dudkin V. Gilliland G. L. Oneil K. Protein Eng., Des. Sel. 2016;29:563–572. doi: 10.1093/protein/gzw054. [DOI] [PubMed] [Google Scholar]
- Mahalingam S. M. Dudkin V. Y. Goldberg S. Klein D. Yi F. Singhal S. O'Neil K. T. Low P. S. Bioconjug Chem. 2017;28:2865–2873. doi: 10.1021/acs.bioconjchem.7b00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorey S. Fiedler E. Kunert A. Nerkamp J. Lange C. Fiedler M. Bosse-Doenecke E. Meysing M. Gloser M. Rundfeldt C. Rauchhaus U. Hanssgen I. Gottler T. Steuernagel A. Fiedler U. Haupts U. J. Biol. Chem. 2014;289:8493–8507. doi: 10.1074/jbc.M113.519884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCIL proteins, https://www.scilproteins.com/, (accessed 14 November 2020)
- Kahl M. Settele F. Knick P. Haupts U. Bosse-Doenecke E. Protein Expr Purif. 2018;149:51–65. doi: 10.1016/j.pep.2018.04.013. [DOI] [PubMed] [Google Scholar]
- Ahmad J. N. Li J. Biedermannova L. Kuchař M. Sipova H. Semeradtova A. Cerný J. Petrokova H. Mikulecký P. Polinek J. Proteins. 2012;80:774–789. doi: 10.1002/prot.23234. [DOI] [PubMed] [Google Scholar]
- Liu H. Lindbo S. Ding H. Hober S. Graslund T. Int. J. Oncol. 2019;55:309–319. doi: 10.3892/ijo.2019.4814. [DOI] [PubMed] [Google Scholar]
- von Witting E. Garousi J. Lindbo S. Vorobyeva A. Altai M. Oroujeni M. Mitran B. Orlova A. Hober S. Tolmachev V. Eur. J. Pharm. Biopharm. 2019;140:109–120. doi: 10.1016/j.ejpb.2019.05.008. [DOI] [PubMed] [Google Scholar]
- Garousi J. Lindbo S. Mitran B. Buijs J. Vorobyeva A. Orlova A. Tolmachev V. Hober S. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva D. A. Correia B. E. Procko E. Methods Mol. Biol. 2016;1414:285–304. doi: 10.1007/978-1-4939-3569-7_17. [DOI] [PubMed] [Google Scholar]
- Barber N. M. Lau C. K. Y. Turner L. Watson G. Thrane S. Lusingu J. P. A. Lavstsen T. Higgins M. K. mSphere. 2021;6:e01081-20. doi: 10.1128/mSphere.01081-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. Sander C. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/S0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Pan X. Kortemme T. J. Biol. Chem. 2021;296:100558. doi: 10.1016/j.jbc.2021.100558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A. Tyka M. Lewis S. M. Lange O. F. Thompson J. Jacak R. Kaufman K. Renfrew P. D. Smith C. A. Sheffler W. Davis I. W. Cooper S. Treuille A. Mandell D. J. Richter F. Ban Y. E. Fleishman S. J. Corn J. E. Kim D. E. Lyskov S. Berrondo M. Mentzer S. Popović Z. Havranek J. J. Karanicolas J. Das R. Meiler J. Kortemme T. Gray J. J. Kuhlman B. Baker D. Bradley P. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia B. E. Bates J. T. Loomis R. J. Baneyx G. Carrico C. Jardine J. G. Rupert P. Correnti C. Kalyuzhniy O. Vittal V. Connell M. J. Stevens E. Schroeter A. Chen M. Macpherson S. Serra A. M. Adachi Y. Holmes M. A. Li Y. Klevit R. E. Graham B. S. Wyatt R. T. Baker D. Strong R. K. Crowe, Jr. J. E. Johnson P. R. Schief W. R. Nature. 2014;507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See K. Kadonosono T. Miyamoto K. Tsubaki T. Ota Y. Katsumi M. Ryo S. Aida K. Minegishi M. Isozaki T. Kuchimaru T. Kizaka-Kondoh S. Sci. Rep. 2021;11:22098. doi: 10.1038/s41598-021-01603-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan C. M. Rocklin G. J. Bick M. J. Ford A. Majri-Morrison S. Kroll A. V. Miller C. J. Carter L. Goreshnik I. Kang A. DiMaio F. Tarbell K. V. Baker D. Proc. Natl. Acad. Sci. U. S. A. 2021;118:e2102164118. doi: 10.1073/pnas.2102164118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J. Borg J. Stricher F. Nys R. Rousseau F. Serrano L. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S. Ding F. Dokholyan N. V. Nat. Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- Kruziki M. A. Bhatnagar S. Woldring D. R. Duong V. T. Hackel B. J. Chem. Biol. 2015;22:946–956. doi: 10.1016/j.chembiol.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P. N. Huličiak M. Biedermannová L. Černý J. Charnavets T. Fuertes G. Herynek Š. Kolářová L. Kolenko P. Pavlíček J. Zahradník J. Mikulecky P. Schneider B. Viruses. 2021;13(2):190. doi: 10.3390/v13020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruziki M. A. Sarma V. Hackel B. J. ACS Comb. Sci. 2018;20:423–435. doi: 10.1021/acscombsci.8b00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmachev V. Orlova A. Cancers. 2020;12(3):651. doi: 10.3390/cancers12030651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdine G. L. Walensky L. D. Clin. Cancer Res. 2007;13:7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- Grimm S. Lundberg E. Yu F. Shibasaki S. Vernet E. Skogs M. Nygren P. Gräslund T. N. Biotechnol. 2010;27:766–773. doi: 10.1016/j.nbt.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Shibasaki S. Karasaki M. Gräslund T. Nygren P. Sano H. Iwasaki T. AMB Express. 2014;4:82. doi: 10.1186/s13568-014-0082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K. Sugiura M. Kimura T. Int. J. Pept. 2015;2015:537508. doi: 10.1155/2015/537508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards D. A. Drug Discovery Today Technol. 2018;30:35–46. doi: 10.1016/j.ddtec.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Ahmadi M. K. B. Mohammadi S. A. Makvandi M. Mamouei M. Rahmati M. Dehghani H. Wood D. W. Curr. Pharm. Biotechnol. 2021;22:878–891. doi: 10.2174/1389201021999200824101035. [DOI] [PubMed] [Google Scholar]
- Feldwisch J. Tolmachev V. Lendel C. Herne N. Sjöberg A. Larsson B. Rosik D. Lindqvist E. Fant G. Höidén-Guthenberg I. Galli J. Jonasson P. Abrahmsén L. J. Mol. Biol. 2010;398:232–247. doi: 10.1016/j.jmb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Hackel B. J. Sathirachinda A. Gambhir S. S. Protein Eng., Des. Sel. 2012;25:639–647. doi: 10.1093/protein/gzs036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilia A. R. Piazzini V. Guccione C. Risaliti L. Asprea M. Capecchi G. Bergonzi M. C. Planta Med. 2017;83:366–381. doi: 10.1055/s-0043-102949. [DOI] [PubMed] [Google Scholar]
- Feldwisch J. Tolmachev V. Methods Mol. Biol. 2012;899:103–126. doi: 10.1007/978-1-61779-921-1_7. [DOI] [PubMed] [Google Scholar]
- Eigenbrot C. Ultsch M. Dubnovitsky A. Abrahmsén L. Härd T. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15039–15044. doi: 10.1073/pnas.1005025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer M. Schiefner A. Matschiner G. Skerra A. J. Mol. Biol. 2013;425:780–802. doi: 10.1016/j.jmb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Main A. L. Harvey T. S. Baron M. Boyd J. Campbell I. D. Cell. 1992;71:671–678. doi: 10.1016/0092-8674(92)90600-H. [DOI] [PubMed] [Google Scholar]
- Chiche L., Heitz A. and Le-Nguyen D., Solution structure of the squash trypsin inhibitor EETI-II, https://www.rcsb.org/structure/2IT7, (accessed 2 October 2007) [Google Scholar]
- Kohl A. Binz H. K. Forrer P. Stumpp M. T. Plückthun A. Grütter M. G. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1700–1705. doi: 10.1073/pnas.0337680100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa A. Pacheco S. Mechaly A. E. Obal G. Béhar G. Mouratou B. Oppezzo P. Alzari P. M. Pecorari F. PLoS One. 2014;9:e97438. doi: 10.1371/journal.pone.0097438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg S. D. Cardoso R. M. Lin T. Spinka-Doms T. Klein D. Jacobs S. A. Dudkin V. Gilliland G. O'Neil K. T. Protein Eng., Des. Sel. 2016;29:563–572. doi: 10.1093/protein/gzw054. [DOI] [PubMed] [Google Scholar]
- Arnoux B. Ducruix A. Prangé T. Acta Crystallogr. D: Biol. Crystallogr. 2002;58:1252–1254. doi: 10.1107/S0907444902007333. [DOI] [PubMed] [Google Scholar]
- Jenko S. Dolenc I. Guncar G. Dobersek A. Podobnik M. Turk D. J. Mol. Biol. 2003;326:875–885. doi: 10.1016/S0022-2836(02)01432-8. [DOI] [PubMed] [Google Scholar]
- Fass D. Blacklow S. Kim P. S. Berger J. M. Nature. 1997;388:691–693. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- Ramage R. Green J. Muir T. W. Ogunjobi O. M. Love S. Shaw K. Biochem. J. 1994;299(Pt 1):151–158. doi: 10.1042/bj2990151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach H. Fiedler E. Scheuermann T. Fiedler M. Stubbs M. T. Reimann C. Proetzel G. Rudolph R. Fiedler U. J. Mol. Biol. 2007;372:172–185. doi: 10.1016/j.jmb.2007.06.045. [DOI] [PubMed] [Google Scholar]