SUMMARY.
Gulls are the known reservoir for H13 and H16 influenza A viruses (IAV) but also host a diversity of other IAV subtypes. Gulls also share habitats with both ducks and shorebirds, increasing the potential for cross-species IAV transmission. We serologically tested laughing gulls (Leucophaeus atricilla) collected at Delaware Bay during May when they were in direct contact with IAV-infected shorebirds; both species feed on horseshoe crab (Limulus polyphemus) eggs on beaches during this month. From 2010 to 2014, antibody prevalence as determined by competitive blocking enzyme-linked immunosorbent assay ranged from 25%–72%. Antibodies to H13 and H16 were detected by hemagglutination inhibition (HI) tests in 12% and 24% of tested gulls, respectively. Results from virus microneutralization (MN) tests for antibodies to H1–H12, H14, and H15 varied among years but the highest prevalence of neutralizing antibodies was detected against H1 (24%), H5 (25%), H6 (35%), H9 (33%), and H11 (42%) IAV. The subtype diversity identified by serology in gulls was dominated by Group 1 HA subtypes and only partially reflected the diversity of IAV subtypes isolated from shorebirds.
Keywords: antibodies, influenza A virus, laughing gull, Leucophaeus atricilla microneutralization, serology
RESUMEN.
Anticuerpos contra el virus de la influenza A en gaviotas en la Bahía de Delaware, en los Estados Unidos. Las gaviotas son el reservorio conocido de los virus de la influenza H13 y H16 (IAV), pero también son los hospederos de una diversidad de otros subtipos del virus de la influenza. Las gaviotas también comparten hábitat con patos y aves costeras, lo que aumenta la posibilidad de que transmisión cruzada de virus de la influenza entre las especies. Se analizaron de manera serológica las gaviotas reidoras (Leucophaeus atricilla) recolectadas en la bahía de Delaware durante el mes de Mayo cuando estaban en contacto directo con aves playeras infectadas por el IAV; ambas especies se alimentan de los huevos de cangrejos de herradura (Limulus polyphemus) en las playas durante este mes. De 2010 a 2014, la prevalencia de anticuerpos determinada por un ensayo de inmunoabsorción con enzimas ligadas de tipo competitivo, osciló entre de 25% a 72%. Los anticuerpos para H13 y H16 fueron detectados por pruebas de inhibición de la hemaglutinación (HI) en el 12% y 24% de las gaviotas ensayadas, respectivamente. Los resultados de pruebas de microneutralización viral (MN) para anticuerpos contra H1-H12, H14 y H15 varió entre años, pero la mayor prevalencia de anticuerpos neutralizantes se detectó en contra virus de influenza H1 (24%), H5 (25%), H6 (35%), H9 (33%) y H11 (42%). La diversidad de subtipo identificada por serología en gaviotas estaba dominado por subtipos HA grupo 1 y refleja sólo parcialmente la diversidad de subtipos de virus de influenza aislados de aves playeras.
Gulls are a recognized reservoir for influenza A viruses (IAV) and, worldwide, most avian HA subtypes of IAV (HA1-12, H13, H16) have been detected from more than 20 gull species (1). Some HA subtypes, such as H13 and H16, are commonly isolated from gulls and are maintained exclusively within gull populations (5); other subtypes are shared with shorebird and waterfowl species and are reported from gulls less frequently (19). With the exception of the H13 and H16 viruses in fledgling gulls (5,20,21), the reported prevalence of IAV based on virus isolation is generally low (<5%) but can vary with season (1,11,13). In contrast, IAV antibody prevalence estimates in numerous gull specieshave consistently been high, generally exceeding 50% (2,18).
Like many gull species, laughing gulls (Leucophaeus atricilla) can be infected with numerous IAV HA subtypes and are long-distance intercontinental migrants that can range from the northeastern United States to northern South America. Laughing gulls also are an extremely common species at Delaware Bay where a high prevalence of IAV is annually observed in shorebirds that share feeding habitats with this species (9). The source(s) of the viruses that annually infect shorebirds at this site, and the extent of IAV sharing between laughing gulls and shorebirds during this period, are not well described.
Although reports of high IAV antibody prevalence in gulls suggest that serologic data could be utilized to better understand the natural history of IAV in gull populations and provide insight into possible cross-species interactions, recent serologic testing has centered on detecting antibodies to IAV-reactive antigens such as the nucleoprotein (NP). The objectives of this study were to document the presence of IAV neutralizing antibodies in laughing gull populations at Delaware Bay and to determine if subtype diversity, as determined by serology, reflects the IAV subtype diversity present in laughing gulls and cohabiting shorebirds, primarily ruddy turnstones (Arenaria interpres), as determined by virus isolation.
MATERIALS AND METHODS
During May 2010–2014, 199 adult laughing gulls (Larus argentatus) and six herring gulls (Scientific name) were captured with cannon nets on the beaches of Delaware Bay in New Jersey. Combined cloacal and oropharyngeal swabs were collected and tested by virus isolation in chicken eggs (7). Additional virus isolation results from laughing gulls from 1986–2009 were obtained through the Influenza Research Database (IRD) (16). Comparison virus isolation data from cohabiting shorebirds during 2010–2014 were compiled from data provided by St. Jude Children’s Research Hospital and the Southeastern Cooperative Wildlife Disease Study; these birds were sampled and tested as previously described (17).
Serum samples were collected via jugular venipuncture. Serum samples were tested by commercial blocking enzyme-linked immunosorbent assay (bELISA; IDEXX Laboratories, Westbrook, ME) for IAV antibodies (2). Samples testing positive by bELISA were also tested by virus microneutralization (MN). Antigens for MN were prepared in Madin Darby canine kidney cells (MDCK; American Type Culture Collection, Manassas, VA). During virus propagation, and in all MN test procedures, cells were maintained in minimal essential media (MEM; Sigma-Aldrich, St. Louis, MO) containing L-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK)-trypsin (final concentration of 1 μg/ml; Worthington Biochemical Corporation, Lakewood, NJ) and antibiotics (final concentration of 100 units penicillin, 0.1 mg streptomycin, and 0.25 mg amphotericin B/ml; Sigma-Aldrich). Antigens were stored at −80 C until used. For antibody testing, sera were diluted 1:10 in MEM and heat inactivated at 57 C for 30 min. Serum samples were screened at a 1:20 dilution against all antigens. For the screen, 25 μl of the diluted serum (1:10) were placed into a single well of a 96-well, v-bottom plate corresponding to each antigen. An additional well for each serum sample served as a serum control to determine potential toxicity. A positive control well using chicken antisera to each antigen (provided by the National Veterinary Services Laboratory [APHIS, USDA]) and a negative control well using MEM were also included. Each antigen (25 μl containing 100 median tissue culture infective doses [102.0 TCID50]) was added to each well, not including the serum control wells which received 25 μl of MEM. Plates were incubated for 2 hr at room temperature after which 25 μl from each well was transferred to a second 96-well tissue culture plate with a confluent monolayer of MDCK cells. Prior to transfer, the tissue culture plate containing the MDCK cells was washed two times with Dulbecco’s phosphate-buffered saline (Sigma-Aldrich), and 150 μl of trypsinsupplemented MEM was added to each well. The inoculated tissue culture plate was incubated at 5% CO2 at 37 C and was visually read at 48–72 hr. For the test result to be considered valid, all controls (serum, positive, and negative) had to meet their expected negative or positive status. In addition, based on back titration in MDCK cells (four replicates per dilution), the viral titer of the antigen had to fall within 101.5 and 102.5 TCID50/25 μl. Sera were considered positive on the screen if complete neutralization (no cytopathic effect [CPE]) was observed. All positive serum samples were titrated. Each positive serum sample was diluted 2-fold in MEM on a 96-well, v-bottom plate (final volume of 25 μl/well at dilutions 1:20 to 1:640) and tested as described above. If CPE was observed at the minimum 1:20 dilution, the sample was classified as negative; if not, the positive titer was recorded as the highest dilution at which no CPE was observed. Viruses used as antigens in the MN assays included A/mallard/NJ/AI10-4263/2010 (H1N1), A/mallard/MN/AI08-2755/2008 (H2N3), A/mallard/MN/AI10-2593/2010 (H3N8), A/mallard/MN/AI10-3208/2010 (H4N6), A/mallard/MN/AI11-3933/2011 (H5N1), A/mallard/MN/AI08-2721/2008 (H6N1), A/mallard/MN/ AI08-3770/2009 (H7N9), A/mallard/MN/SG-01048/2008 (H8N4), A/ RUTU/DE/AI11-809/2011 (H9N2), A/mallard/MN/SG-00999/2008 (H10N7), A/mallard/MN/SG-00930/2008 (H11N9), A/mallard/MN/SG-3285/2007 (H12N5), A/blue-winged teal/TX/AI13-1028/2013 (H14N5), and A/wedge-tailed shearwater/Western Australia/2327/1983 (H15N6).
Because representative H13 and H16 IAVs could not be propagated in MDCK cells supplemented with trypsin, hemagglutination inhibition (HI) tests were used to test for antibodies against these viruses, as described (14). Viruses used for HI tests included A/AWPE/MN/AI07-1819/2007 (H13N9) and A/ring-billed gull/DE/AI10-1144/2010 (H16N3).
RESULTS
From 2009–2014, 13 IAV were isolated from laughing gulls including H5N3 (n = 1), H10N2 (n = 2), H10N7 (n = 2), H10N8 (n = 2), H11N2 (n = 1), H11N4 (n = 1), H11N9 (n = 2), H12N2 (n = 1), and H16N3 (n = 1). Additionally, 24 IAV isolates reported from laughing gulls from 1986–2008 included H1N9 (n = 2), H2N7 (n = 1), H3N6 (n = 1), H4N6 (n = 1), H6N3 (n = 1), H6N8 (n = 4), H7N3 (n = 1), H9N1 (n = 1), H9N5 (n = 1), H9N9 (n = 1), H10N2 (n = 1), H11N1 (n = 2), H11N2 (n = 1), H11N9 (n = 1), H13N2 (n = 1), H13N9 (n = 1), and H16N3 (n = 4) (16). Overall, of the 1296 virus isolation attempts from laughing gulls reported in the IRD that were sampled in Delaware, New Jersey, and New York from 1986 to 2014, 40 IAV were isolated representing a prevalence of 3.1%.
Antibodies to IAV were detected in 61% of 199 laughing gulls sampled from 2010–2014 (Table 1). Antibody prevalence estimates varied by year ranging from 25%–72%. Of the six herring gulls sampled during 2012 and 2014, three (50%) tested positive for IAV antibodies. Results from MN (n = 104) and HI (n = 82) testing of bELISA-positive serum samples from laughing gulls are shown in Table 2. Positive MN results were detected in 82 of the 104 bELISA-positive samples. Of these 25, 18, 22, 5, 7, 3, and 2 samples tested positive to 1, 2, 3, 4, 5, 6, and 7 HA subtypes by MN, respectively. Antibody prevalence varied by subtype and was highest for H1, H5, H6, H9, H11, H13, and H16 (Table 2; Fig. 1). As shown in Figure 1 where HA subtypes are arranged based on their phylogenetic relatedness (6), all of these HA subtypes are included in Group 1.
Table 1.
Prevalence of antibodies to influenza A viruses as detected by bELISA in laughing gulls sampled at Delaware Bay, New Jersey, U. S. A.
| Year | Number tested | No. positive | % positive |
|---|---|---|---|
| 2010 | 32 | 23 | 72 |
| 2011 | 28 | 7 | 25 |
| 2012 | 14 | 9 | 64 |
| 2013 | 80 | 54 | 68 |
| 2014 | 45 | 28 | 62 |
| Total | 199 | 121 | 61 |
Table 2.
Subtype-specific antibodies detected from laughing gulls sampled at Delaware Bay, NJ as determined by microneutralization (MN) and hemagglutination inhibition (HI) tests.
| MN |
HI |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Year | H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | H9 | H10 | H11 | H12 | H14 | H15 | H13 | H16 |
| 2010 | 1/15 (6)A | 0/15 B | 0/15 | 0/15 | 4/15 (23) | 3/15C (17) | 0/15 | 0/15 | 5/15 (29) | 1/15 (6) | 5/15 (29) | 0/15 | 1/15 | 0/15 | NT | NT |
| 2011 | 1/5 (20) | 0/5 | 0/5 | 0/5 | 1/5 (20) | 1/5 (20) | 0/5 | 0/5 | 1/5 (20) | 0/5 | 1/5 (20) | 0/5 | 0/5 | 0/5 | NT | NT |
| 2012 | 2/9 (22) | 1/9 (11) | 0/9 | 0/9 | 1/9 (11) | 1/9 (11) | 0/9 | 0/9 | 2/9 (40) | 0/9 | 2 (40) | 0/9 | 0/9 | 0/9 | 2/9 (22) | 4/9 (44) |
| 2013 | 19/49 (39) | 0/49 | 2/49 (4) | 1/49 (4) | 14/49 (29) | 22/49 (45) | 3/49 (6) | 3/49 (4) | 27/49 (55) | 0/49 | 27/49 (55) | 3/49 (6) | 0/49 | 0/49 | 3/47 (6) | 9/47 (18) |
| 2014 | 2/26 (8) | 0/26 | 0/26 | 0/26 | 6/26 (23) | 9/26 (35) | 2/26 (4) | 0/26 | 9/26 (35) | 2/26 (4) | 9/26 (35) | 0/26 | 0/26 | 0/26 | 10/26 (38) | 7/26 (27) |
| Total (%) | 25/104 (24) | 1/104 (1) | 2/104 (2) | 1/104 (1) | 26/104 (25) | 36/104 (35) | 5/104 (5) | 4/104 (4) | 35/104 (33) | 3/104 (3) | 44/104 (42) | 3/104 (3) | 1/104 (1) | 0/104 (0) | 15/82 (12) | 20/82 (24) |
Number positive/number tested (% positive). All tested sera also tested positive on bELISA for antibodies to type A influenza virus.
Italicized data indicate the presence of that subtype in avian influenza virus isolates from shorebirds, primarily ruddy turnstones, sampled at the same time.
Boldface italicized data represent the predominant HA subtype detected that year in shorebirds.
Fig. 1.
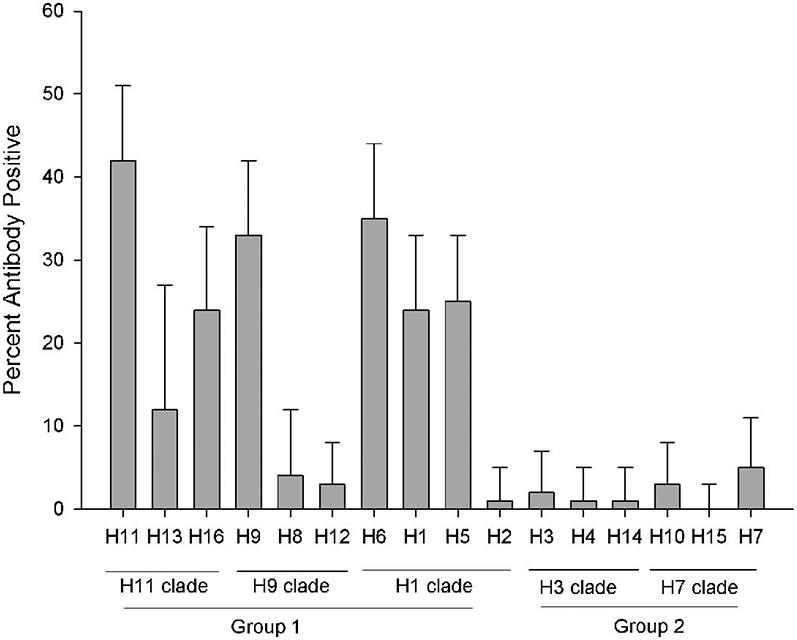
Hemagglutinin-specific antibody prevalence in laughing gulls as determined by MN (H1–12, 14, and 15) and HI (H13 and 16). Subtypes as arranged based on phylogenic relatedness. Error bars represent 95% confidence limits.
Serologic results from gulls only partially reflect the subtype diversity present in shorebirds. As shown on Table 2, H5, H6, and H11 IAV were isolated from the shorebird population during most years, and in 2010 and 2011, H6 and H9 represented the predominant subtypes isolated from shorebirds, respectively. In both of those years, neutralizing antibodies to these HA subtypes were detected in gulls. In contrast, although a high prevalence of H1 antibodies was detected in gulls, this subtype was poorly represented in shorebird isolates during this same period. With the H10 and H12 viruses which together represent predominant shorebird isolates in three of the five years (Table 2), antibody prevalence in gulls was consistently low, with neutralizing antibodies detected in less than 6% of the 104 bELISA-positive laughing gulls. During years when H10 IAV (2013) and H12 IAV (2012 and 2014) predominated in shorebirds, antibodies to these HA subtypes were not detected in gulls (Table 2).
DISCUSSION
The low prevalence of reported virus isolation-positive laughing gulls and the high prevalence of antibodies to NP were consistent with previous reports from gull studies. The observed annual variation in prevalence (25%–72%) cannot be explained but may have resulted from variations in the prevalence and subtype diversity present in the shorebird populations, variation in adult gull age structure during an individual year, or the timing of gull sampling. At Delaware Bay, subtype diversity varies by year, and peak prevalence associated with these annual outbreaks can be short-lived and limited to a single week (12).
Subtype diversity present in the reported virus isolates from laughing gulls from 1986–2014 included representatives of all HA subtypes except H8, H14, and H15. Antibodies to all HA subtypes except H15 were also detected in laughing gulls sampled at Delaware Bay from 2010–2014. Predominant subtypes identified by serologic results, however, were skewed toward Group 1 HAs, specifically H1, H5, H6, H9, H11, H13, and H16 (Table 2; Fig. 1). All of these subtypes, except H1, H9, and H13, were represented in recent (2009–2014) virus isolation data from laughing gulls sampled at Delaware Bay. The predominance of these Group 1 subtypes in laughing gulls also is consistent with virus isolation results reported for ring-billed gulls (Larus delawarensis) sampled in Maryland during 1977–1979; the 70 virus isolations reported in that study included H2, H5, H6, H9, H11, and H13 IAV (6). A total of 234 IAV isolates from species in the family Laridae worldwide (including Canada, Republic of Georgia, Iceland, Netherlands, and the United States) are reported in the IRD (16). Of these, 145 are H13 and H16 IAV. The remaining 89 isolates include H1–H7 and H9–H12 (Fig. 2); however, H1, H2, H6, and H11 IAVs represented 68% of these 89 isolates. Although the H1, H5, H6, H9, H11, and other Group 1 HA subtypes appear to be prevalent in gulls, they are not restricted to gulls and occur frequently in both waterfowl and shorebirds (10), and in some cases, viruses shared between waterfowl and gulls may be genetically identical (19). The high prevalence of antibodies to Group 1 IAV and their overrepresentation in reported isolates from gulls (Fig. 2) may reflect some degree of gull adaptation, as occurs with H13 and H16. Evidence for a potential gull lineage of H11 viruses was recently reported based on phylogenetic characterization of viruses isolated from gulls in Georgia (11).
Fig. 2.
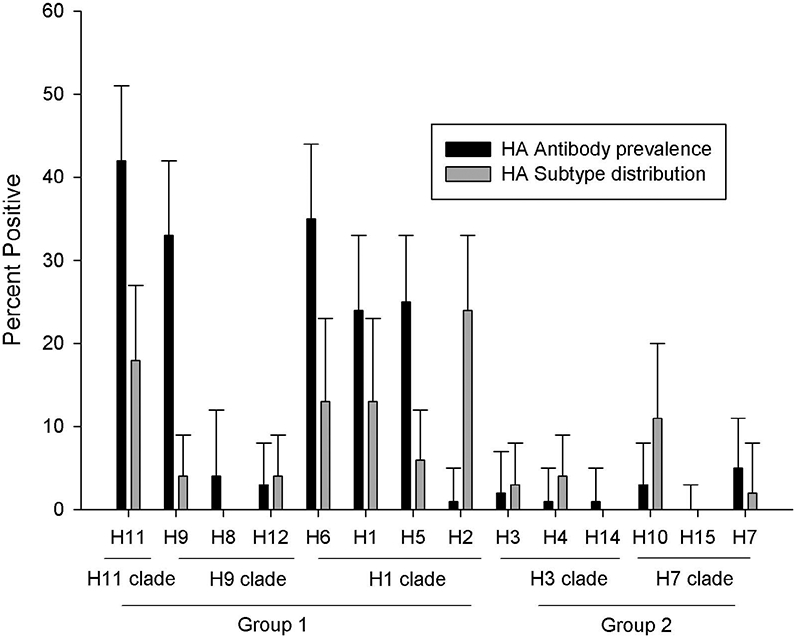
Antibody prevalence observed in laughing gulls (based on 104 and 82 samples tested by MN and HI, respectively) and HA subtype diversity represented in 234 reported IAV isolations from Laridae worldwide as included in the IRD (16). Error bars represent 95% confidence limits.
The inconsistency observed between HA subtype diversity, as indicated by neutralizing antibodies in gulls and virus isolation from shorebirds, may reflect host-related variation in gull vs. shorebird susceptibility; it may also relate to viral source. At present, the sources of IAV that infect shorebirds annually at Delaware Bay are not understood, but the multiple IAV subtypes annually present at this site probably represent viruses that migrate with shorebirds and viruses that are transmitted to shorebirds by gulls or ducks during spring migration or after arrival at Delaware Bay (3,15). The high prevalence of H11 antibodies in gulls and the occurrence of these viruses in shorebirds during most years at this site are consistent with a pattern of local interspecies transmission that could reflect either a gull or shorebird source. Isolates of H11 IAV from wintering ruddy turnstones in Brazil are most related genetically to isolates from ruddy turnstones from Delaware Bay (4); wintering areas in South America also are utilized by laughing gulls. In contrast, the frequent occurrence of H12 in shorebirds with limited evidence of infection and spillover in gulls is consistent with a pattern of local transmission that is more restricted within shorebird populations. The H12 viruses have been reported from laughing gulls at Delaware Bay during surveillance from 1986–2014 but represent only 3% of the observed HA diversity. Also, the H12 IAVs are not well represented in isolates reported from gulls in the IRD, where they represent only 2 of 234 (1%) isolates (16). Variation in the ability of IAV derived from shorebirds to replicate in ducks has been previously demonstrated (8). Based on results from our study, host differences may also exist between gulls and shorebirds.
Serologic data, especially related to IAV, are difficult to interpret due to potential cross-reactions between subtypes, lack of comparative data between different tests, and limited information related to host immune response. This is especially true with serologic data derived from wild birds such as gulls, which are long lived and can be infected numerous times with different IAV subtypes during their lifetime. It is possible that the high prevalence of antibodies to the Group 1 HAs was exaggerated by cross-reactive antibodies between related HAs. Although we cannot discount this possibility, the low antibody prevalence to other Group 1 HAs (H2 and H8) suggests that these effects were minimal. With regard to test format, it is likely that the low prevalence of H13 and H16 antibodies as compared to H1, H5, H6, and H11 reflected differences in HI and MN test sensitivity rather than true differences. Although HI antibody prevalence estimates for H13 and H16 viruses were low compared to neutralizing antibodies of H1–H12, H14, and H15, the H13 and H16 HA subtypes are the most common viruses isolated from gulls worldwide (1) and locally at Delaware Bay (10,17). It also is unknown how long an antibody response for a given subtype is detectable by either MN or HI. Without knowledge on the duration of the detectable immune response, it is difficult to directly compare serologic and virus isolation data. The potential long-term persistence of antibodies and the relatively short duration of IAV shedding may explain some of the inconsistency observed in this study, such as the detection of antibodies to H1 with few virus isolates from gulls or cohabiting shorebirds during the sampling period. Although additional work is needed to improve our ability to interpret serologic data from gulls and other wild birds, supporting subtype-specific serologic data can provide a unique perspective to better define IAV subtype diversity present in avian species or taxonomic groups and to identify potential species interactions that may facilitate IAV maintenance within the wild bird reservoir.
ACKNOWLEDGMENTS
We thank Larry Niles, Amanda Dey, Benjamin Wilcox, Morgan Slusher, and the many volunteers who captured the gull and shorebirds tested in this study. This work was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract HHSN272201400006C. The funding agencies were not involved in the design, implementation, or publishing of this study, and the research presented herein represents the opinions of the authors but not necessarily the opinions of the funding agencies.
Abbreviations:
- CPE
cytopathic effect
- HI
hemagglutination inhibition
- IAV
influenza A virus
- IRD
Influenza Research Database
- MEM
minimal essential media
- MDCK
Madin Darby canine kidney cells
- MN
microneutralization
- NP
nucleoprotein
- TCID50
median tissue culture infective doses
REFERENCES
- 1.Arnal A, Vittecoq M, Pearce-Duvet J, Gauthier-Clerc M, Boulinier T, and Jourdain E. Laridae: a neglected reservoir that could play a major role in avian influenza virus epidemiologic dynamics. Crit. Rev. Microbiol 41:508–519. 2015. [DOI] [PubMed] [Google Scholar]
- 2.Brown JD, Luttrell MP, Berghaus RD, Kistler W, Keeler SP, Howey A, Wilcox B, Hall J, Niles L, Dey A, Knutsen G, Fritz K, and Stallknecht DE. Prevalence of antibodies to type A influenza virus in wild avian species using two serologic assays. J. Wildl. Dis 46: 896–911. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown VL, Drake JM, Stallknecht DE, Pederson K, and Rohani PR. Dissecting a wildlife disease hotspot: the impact of multiple host species, environmental transmission and seasonality in migration, breeding and mortality. J. Royal Soc. Interface 2013 Feb 6,10(79): 20120804. doi: 10.1098/rsif.2012.0804. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Araujo J, de Azevedo Júnior SM, Gaidet N, Hurtado RF, Walker D, Thomazelli LM, Ometto T, Seixas MM, Rodrigues R, Galindo DB, da Silva AC, Rodrigues AM, Bomfim LL, Mota MA, Larrazábal ME, Branco JO, Serafini P, Neto IS, Franks J, Webby RJ, Webster RG, and Durigon EL. Avian influenza virus (H11N9) in migratory shorebirds wintering in the Amazon Region, Brazil. PLoS ONE 9:e110141. doi: 10.1371/journal.pone.0110141. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouchier RAM, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwann GF. Olsen B, Osterhaus ADME. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol 79:2814–2822. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graves IL Influenza viruses in birds of the Atlantic flyway. Avian Dis. 36:1–10. 1992. [PubMed] [Google Scholar]
- 7.Hanson BA, Luttrell MP, Goekjian VH, Beck JD, Niles L, Swayne DE, Senne DA, and Stallknecht DE. Is the occurrence of avian influenza virus in Charadriiformes species and location dependent? J. Wildl. Dis 44:351–361. 2008. [DOI] [PubMed] [Google Scholar]
- 8.Kawaoka Y, Chambers TM, Sladen WL, and Webster RG. Is the gene pool of influenza viruses in shorebirds and gulls different from that of ducks? Virology 163:247–250. 1988. [DOI] [PubMed] [Google Scholar]
- 9.Krauss S, Stallknecht DE, Negovetich NJ, Niles LJ, Webby RJ, and Webster RG. Coincident ruddy turnstone migration and horseshoe crab spawning creates an ecological “hot spot” for influenza viruses. Proc. Roy. Soc. B, Bio. Sci 277:3373–3793. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krauss S, Walker D, Paul-Pryor S, Niles L, Chenghong L, Hinshaw VS, and Webster RG. Influenza A viruses in migrating wild aquatic birds in North America. Vector-Borne Zoonotic Dis. 4:177–189. 2004. [DOI] [PubMed] [Google Scholar]
- 11.Lewis N, Javakhishvili Z, Russell CA, Machablishvili A, Lexmond P, Verhagen JH, Vuong O, Onashvili T, Donduashvili M, Smith DJ, and Fouchier RAM. Avian influenza virus surveillance in wild birds in Georgia. PLoS ONE 8:e58534. doi: 10.1371/journal.pone.0058534. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maxted AM, Luttrell MP, Goekjian VH, Brown JD, Niles LJ, Dey AD, Kalasz KS, Swayne DE, and Stallknecht DE. Avian influenza virus dynamics in shorebird hosts. J. Wildl. Dis 48:322–334. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munster VJ, Baas C, Lexmond P, Waldenström J, Wallensten A, Fransson T, Rimmelzwann GF, Beyer WEP, Schetten M, Olsen B, Osterhaus ADME, and Fouchier RAM. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3:e61. doi: 10.1371/journal.ppat.0030061. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen JC Hemagglutination-inhibition test for avian influenza virus subtype identification and the detection and quantitation of serum antibodies to the avian influenza virus. In: Avian influenza virus, Spackman E, ed. Humana Press, Totowa, New Jersey. pp. 53–66. 2008. [DOI] [PubMed] [Google Scholar]
- 15.Ramey AM, Poulson RL, González-Reiche AS, Wilcox BR, Walther P, Link P, Carter DL, Newsome GM, Müller ML, Berghaus RD, Perez DR, Hall JS, and Stallknecht DE. Evidence for seasonal patterns in the relative abundance of avian influenza virus subtypes in North American waterfowl. J. Wildl. Dis 50:916–922. 2014. [DOI] [PubMed] [Google Scholar]
- 16.Squires RB, Noronha J, Hunt V, Garcia-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, Ramsey A, Zhou L, Zaremba S, Kumar S, Deitrich J, Klem E, and Scheuermann RH. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respir. Viruses doi: 10.1111/j.1750-2659.2011.00331.x. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stallknecht DE, Luttrell MP, Poulson R, Goekjian V, Niles L, Dey A, Krauss S, and Webster RG. Surveillance and testing approaches to detect and recover avian influenza viruses from shorebird populations. J. Wildl. Dis 48:382–393. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toennessen R, Kristoffersen AB, Jonassen CM, Hjortaas MJ, Hansen EF, Rimstad E, and Hauge AG. Virological and serological surveillance for type A influenza in the black-legged kittiwake (Rissa tridactyla). Virol. J 2013 Apr 10;10:112. doi: 10.1186/1743-422X-10-112. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tønnessen R, Kristoffersen AB, Jonassen CM, Hjortaas MJ, Hansen EF, Rimstad E, and Hauge AG. Molecular and epidemiological characterization of avian influenza viruses from gulls and dabbling ducks in Norway. Virol. J 2013. Apr 10;10:112. doi: 10.1186/1743-422X-10-112.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velarde R, Calvin SE, Ojkic D, Barker IK, and Nagy E. Avian influenza virus H13 circulating in ring-billed gulls (Larus delawarensis) in southern Ontario, Avian Dis. 54:411–419. 2010. [DOI] [PubMed] [Google Scholar]
- 21.Verhagen JH, Majoor F, Lexmond P, Vuong O, Kasemir G, Lutterop D, Osterhaus ADME, Fouchier RAM, and Kuiken T. Epidemiology of influenza A virus among black headed gulls, the Netherlands, 2006–2010. Emerg. Infect. Dis 20:138–141. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]


