Abstract
We have developed a novel technology that makes it possible to detect simple nucleotide polymorphisms directly within a sample of total genomic DNA. It allows, in a single Southern blot experiment, the determination of sequence identity of genomic regions with a combined length of hundreds of kilobases. This technology does not require PCR amplification of the target DNA regions, but exploits preparative size-fractionation of restriction-digested genomic DNA and a newly discovered property of the mismatch-specific endonuclease CEL I to cleave heteroduplex DNA with a very high specificity and sensitivity. We have used this technique to detect various simple mutations directly in the genomic DNA of isogenic pairs of recombinant Pseudomonas aeruginosa, Escherichia coli and Salmonella isolates. Also, by using a cosmid DNA library and genomic fractions as hybridization probes, we have compared total genomic DNA of two clinical P.aeruginosa clones isolated from the same patient, but exhibiting divergent phenotypes. The mutation scan correctly detected a GA insertion in the quorum-sensing regulator gene rhlR and, in addition, identified a novel intragenomic polymorphism in rrn operons, indicating very high stability of the bacterial genomes under natural non-mutator conditions.
INTRODUCTION
Spontaneous acquisition of simple mutations (i.e. single or few nucleotide substitutions, insertions/deletions or inversions) by structural or regulatory genes can have pronounced effects on various cellular processes (1,2). In pathogenic bacteria, simple mutations that alter gene expression or modify gene products can provide a selective advantage during the course of a single infection or epidemic spread (3). Such pathogenicity-adaptive (or pathoadaptive) mutations can help bacteria to, for example, avoid immune recognition, optimize host tissue tropism or acquire antibiotic resistance (4). They occur spontaneously in a great variety of genes and in regions that are located in different parts of the genome. Here, we describe an effective method for screening large regions of genomic DNA of bacterial strains to identify the presence and location of simple genetic mutations.
A prominent example of a bacterial pathogen capable of adaptive (micro)evolution in the course of infection is Pseudomonas aeruginosa, the causative agent of chronic lung infections in patients with cystic fibrosis and urinary tract and burn infections in compromised patients (5). Several pathoadaptive mutations have been described in P.aeruginosa including loss of function mutations in the mucA gene that result in the overproduction of an alginate capsule and increased bacterial resistance to host clearance mechanisms and anti-microbial drugs (6,7). Additional mutations are responsible for the loss of flagellar motility, auxotrophy or altered lipopolysaccharide (LPS) structure (8–10) and, in some cases, the adaptive microevolution might be facilitated by a mutator phenotype of the infecting clones (11,12). However, in most cases the nature of the underlying mutations remains unclear. As the pathoadaptive phenotype can result from simple mutations in a great variety of structural genes or regulatory regions, it is very difficult to define the location of such subtle alterations on a genomic level. Genome-wide comparative methods, such as restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA (RAPD) analysis or genomic microarrays are not suited to search for simple mutational differences between whole genomes. Single nucleotide substitutions and small insertions/deletions can be identified by several methods, including gene sequencing, differential gel mobility of heterologous DNA, oligonucleotide hybridization, and the use of DNA mismatch-cleaving, -modifying or -binding compounds (13–16). However, these techniques, even with automation, usually require PCR amplification of a DNA region containing the mutation and, therefore, are limited to the analysis of a relatively small number of specific regions.
In this study, we have developed and tested a method that allows screening of large regions of genomic DNA for the detection of various types of nucleotide alterations.
MATERIALS AND METHODS
Bacterial strains and plasmids
Pseudomonas aeruginosa strains UPA204 and UPA205 were isolated concurrently from the urine of a patient with a catheterized urinary bladder and symptoms of an acute urinary tract infection. Pyocyanin, elastase and pyoverdine production of these and other clinical isolates of P.aeruginosa were determined as described previously (17). Pseudomonas aeruginosa PAO1 is a standard reference strain for studies on pathogenesis, genetics and gene mapping, and it expresses most of the virulence factors commonly associated with clinical P.aeruginosa isolates (18). Pseudomonas aeruginosa PAO1seq is an isolate of PA01 of which the genome has been recently sequenced and annotated (19). Genomic sequencing has revealed that PAO1seq differs from PA01 by a spontaneously occurred CTGA tandem duplication in pilC. Pseudomonas aeruginosa PDO300 is PA01 derivative with a G deletion in a poly G strip and an A→G substitution in mucA that were introduced by a homologous exchange (gift of Dr S. Suh, Virginia Commonwealth University, VA, USA). Escherichia coli C1845 is a clinical strain isolated from a patient with diarrhea (20). Escherichia coli G1148 strain is a derivative of C1845 with an AAT→TAAC replacement in daaP and a downstream C→G substitution that were introduced by a homologous recombination (S.Moseley, unpublished data). Salmonella enteritidis TH1966 is a derivative (K.Hughes, unpublished data) of a standard reference strain Salmonella enterica serovar Typhimurium LT2 (21). TH1967 is a derivative of TH1966 with a spontaneously occurring A→C substitution in nadC.
Plasmids and cosmids
To test for restoration of pyocyanin production, the clinical isolates were transformed with pMB5, a pLAFR1-based plasmid encoding rhlABRI’ genes (17). As a nadC-specific probe we have used plasmid pKH385 incorporating the 3′ portion of the gene and the downstream region (K.Hughes, unpublished data). Cosmids used in this study as probes for Southern blot hybridization were obtained by cloning 35–45 kb chromosomal regions of P.aeruginosa PA01 and were provided by Dr M. Olson, University of Washington (WA, USA).
Pulse-field gel electrophoresis
Pulse-field gel electrophoresis (PFGE) profiles of the UPA204 and UPA205 strains were determined by digesting the chromosomal DNA with SpeI endonuclease for 16 h followed by electophoretic separation in 0.8% agarose under pulse-field conditions.
PCR amplification
The 1678 bp fragment containing the 730 bp coding sequence of rhlR was amplified by using high-fidelity Herculase™ DNA Polymerase (Stratagene) and the following primer pair: 5′-CCTGAACGGTGCTGGCATAAC-3′ (top) and 5′-CGAAACGGCTGACGACCTCAC-3′ (bottom).
Heteroduplex formation and CEL I treatment
Heteroduplex DNA was obtained by heat-denaturing a sample of either PCR-amplified or fractionated genomic DNA fragments (from two strains being compared) at 95°C for 10 min followed by fast cooling to 85°C at a rate 2°C/s and then a gradual cooling to 40°C at a rate of 3°C/h to allow random re-annealing of the denatured DNA strands. CEL I is a 30 kDa plant nuclease purified from celery as described previously (22). CEL I treatment of the re-annealed DNA samples (10 ng for the purified PCR products and 200–1000 ng for the genomic fractions) was performed at 1:1 to 1:4 w/w enzyme/DNA ratio in a volume of 20 µl of reaction buffer containing 10 mM Tris–HCl, 150 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 10% formamide, 9% glycerol, 100 µg/ml BSA (pH 7.8). The reaction mixture was incubated at 40°C for 60 min, put on ice, mixed 1:10 with TBE agarose-loading buffer followed by electrophoretic separation in 0.8% agarose gel.
DNA fractionation
Purified genomic DNA from corresponding bacterial clones (500 µg each) was combined and digested to completion with an appropriate restriction endonuclease followed by size-fractionation in 0.5% agarose using a preparative gel electrophoresis apparatus (Model 491 Prep Cell from Bio-Rad Laboratories) under conditions described previously (23).
Membrane blot hybridization
Alkaline-denatured DNA was transferred from a liquid sample (for a slot blot hybridization) or from the agarose gel (for a Southern blot hybridization) to Nytran Plus nylon membranes (Schleicher & Schuell) under neutral conditions and crosslinked to the membranes by UV light. 32P-labeled hybridization probes were prepared from either PCR-amplified DNA, plasmids, cosmids or genomic fractions (see above) using the Random Primers DNA Labeling System (Gibco BRL). Membrane blot hybridization was performed under high-stringency conditions according to the manufacturer’s instructions.
RESULTS
Characterization of rhlR variants of P.aeruginosa clinical isolates
The mutation scan was developed using pathogenic isolates of P.aeruginosa with a novel type of phenotypic variation. It was found that 15% (50 out of 334 tested) of urinary tract infection isolates of P.aeruginosa, when cultured on pseudomonas isolation agar plates, produce a fluorescent pyoverdin pigment but not a blue pyocyanin pigment (M.Brint, unpublished data; presented at the 100th General Meeting of the American Society for Microbiology, Los Angeles, CA, May 2000). This phenotype has also been found among isolates from certain other sources. A urine specimen from one patient with a catheterized bladder yielded a mixture of a pyocyanin-positive isolate, UPA204, and a pyocyanin-negative isolate, UPA205. Each strain showed the same PFGE profile (Fig. 1A). It is known that production of pyocyanin in P.aeruginosa, but not of pyoverdin, requires a transcriptional regulator, RhlR, which is part of a cell density-dependent quorum-sensing cascade (17). When a plasmid encoding rhlR was introduced into UPA205, it resumed production of the pigment (Fig. 1B). Similar results were obtained by introducing the rhlR plasmid into eight different pyocyanin-negative clinical isolates (not shown). Therefore, the pyocyanin-negative phenotype might be due to a structural mutation of the RhlR, it’s down regulation, or functional inhibition.
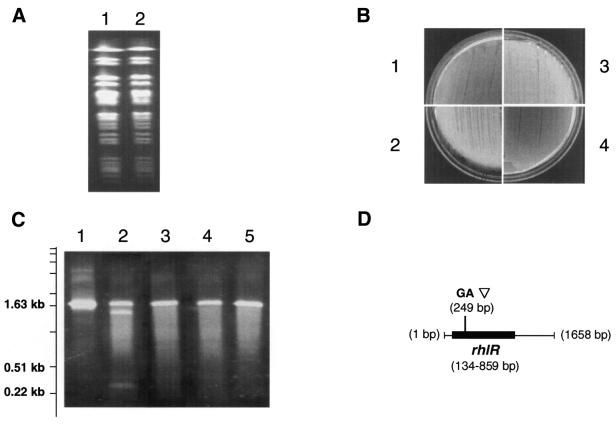
Figure 1.
(A) PFGE profiles of the UPA204 and UPA205 strains. Track 1, UPA204; track 2, UPA205. The chromosomal digest was performed with SpeI endonuclease for 16 h. The top band is at 533.5 kb, the bottom of the first cluster of bands is at 194.0 kb and the bottom band in the second band cluster is at 97.0 kb. (B) Pyocyonin production on pseudomonas isolation agar: plate 1, UPA204; plate 2, UPA205; plate 3, UPA205 transformed with pLARF1 vector; plate 4, UPA205 transformed with an rhlR-encoding plasmid pMB5. (C) Agarose gel analysis of CEL I treatment of PCR-amplified rhlR gene regions of UPA204 and UPA205. Track 1, the amplified rhlR fragments of UPA204 and -205 strains are combined, heat-denatured and re-annealed without CEL I treatment; track 2, same as track 1, but treated with CEL I; track 3, the amplified rhlR fragments of UPA204 only, heat-denatured, re-annealed and treated with CEL I; track 4, the rhlR fragments of UPA205 only, heat-denatured, re-annealed and treated with CEL I; track 5, the rhlR fragments of UPA204 and -205 strains are combined and treated with CEL I without heat-denaturation/re-annealing. On the left the gel is a scale corresponding to a partial 1 kb ladder (Gibco BRL; shown marks correspond to 6.11, 5.09, 4.07, 3.05, 2.04, 1.64, 1.02, 0.51 and 0.22 kb, respectively). (D) rhlR and the GA insertion within the PCR-amplified region. Total length of the PCR fragment is 1658 bp (from UPA204). Position of the gene and mutation is marked from the beginning of the PCR fragment.
To determine whether UPA205 possesses an altered rhlR, the corresponding regions from UPA204 and UPA205 were amplified, the DNA fragments combined, heat-denatured and slowly cooled to allow heteroduplex formation. If the rhlR sequences were not identical, the DNA heteroduplexes would contain a mispaired region that could be recognized by a mismatch-specific enzyme. Re-annealed DNA samples were treated with the mismatch-specific endonuclease CEL I (22), then analyzed by agarose gel electrophoresis (Fig. 1C). In samples containing re-annealed DNA from UPA204 and UPA205, two CEL I-cleavage fragments were observed, indicating the presence of a mismatched region in the rhlR heteroduplex. [Note: When the amount of enzyme is limiting, CEL I is known only to nick one of the two DNA strands on the 3′ side of the mismatch nucleotide (22). The double-strand breaking activity is due to the use of an ∼100-fold higher concentration of CEL I relative to that described previously (22) as well as modified reaction buffer conditions. The mechanism of mismatch-specific DNA cleavage by CEL I is currently under investigation.] Sequence analysis of the PCR products revealed that the rhlR of UPA205a has GA insertion in position 140 that leads to a frame-shift and inactivation of the 730 bp long gene (Fig. 1D). Therefore, the pyocyanin-negative phenotype of the UPA205 strain is likely to be a result of the rhlR mutation. If the rhlR mutation was acquired by UPA205 during evolution within the host, this strain and UPA204 might represent clonal variants of the same strain and, in this case, there should be minimal differences between the genomes.
Design of the genomic mutation scan
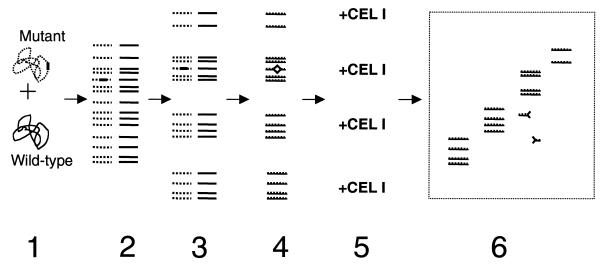
Although both strains demonstrated identical PFGE profiles, this or other currently available methods of genetic analysis are not suited to examine their isogenicity. To screen large portions of the bacterial genome for the presence of nucleotide polymorphisms, we have designed a strategy for a mutational scan that further exploits the mismatch-cleavage property of the CEL I endonuclease. However, instead of using a PCR-amplified DNA product, the heteroduplex DNA fragments are formed directly from genomic DNA of two bacterial strains. Importantly, before DNA denaturation/re-annealing and CEL I treatment, total genomic DNA of both strains is digested with a restriction enzyme and size-fractionated (Fig. 2). A CEL I-cleaved heteroduplex within a certain size range would produce at least one DNA fragment of a size smaller than the DNA fragments within that particular fraction, and could be separated from the non-cleaved DNA by gel electrophoresis. We have designated the proposed technology Genomic Identity Review by Annealing of Fractionated Fragments (GIRAFF).
Figure 2.
Scheme of the proposed technology to screen for nucleotide polymorphism within genomic DNA. Step 1: total genomic DNA from two bacterial strains is purified and combined (a possible polymorphic site is indicated as a solid bar). Step 2: complete endonuclease restriction of the combined genomic DNA. Step 3: size-fractionation of the restricted DNA fragments. Step 4: DNA heteroduplex formation by heat-denaturation and re-annealing of the fractionated DNA fragments (the mutation-induced mispaired region is indicated by an open square). Step 5: CEL I treatment of the re-annealed DNA fragments. Step 6: agarose gel analysis of CEL I-treated DNA fractions (the mismatch-cleaved fragments are indicated as shortened fragments with halved squares on the ends).
Detection of the rhlR mutation directly in genomic DNA
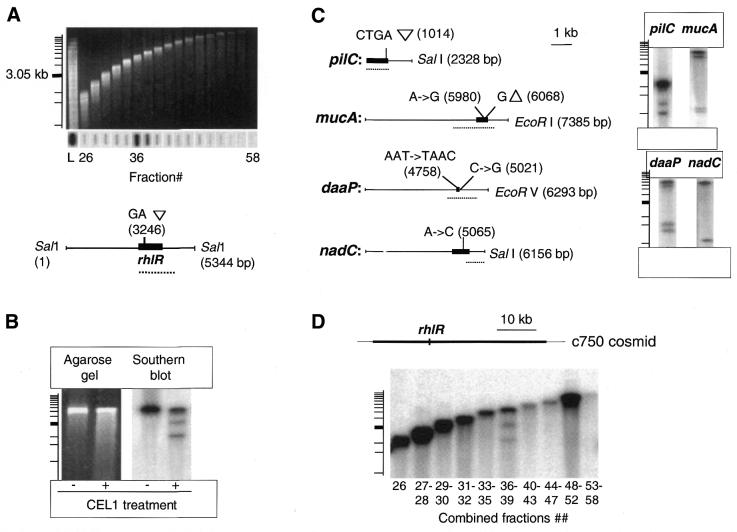
We have examined whether the rhlR mutation could be detected by GIRAFF directly in restricted genomic DNA of UPA204 and UPA205 strains. Purified genomic DNA from UPA204 and UPA205 (500 µg each) was digested with SalI endonuclease and separated into 30 fractions (Fig. 3A) by preparative gel electrophoresis. Using an rhlR-specific hybridization probe, fractions 36 and 37 were found to contain rhlR (Fig. 3A), which corresponded to the predicted location of the rhlR gene within a 5.2 kb restriction fragment (Fig. 3A) according to the genomic sequence of P.aeruginosa strain PAO1 (19). It is important to note, however, that due to the high complexity of these relatively large genomic fractions, the rhlR-containing fragment is expected to constitute only a minor portion of fractions 36 or 37. DNA from fraction 36 was heat-denatured, re-annealed and treated with CEL I under conditions described in the Materials and Methods and analyzed by agarose gel electrophoresis. Direct visualization of CEL I-cleaved fragments was not possible due to the high complexity of DNA in the fraction, and also because of the background created by non-specific 5′-exonuclease activity of CEL I which results from a high concentration of the enzyme being used (A.Yeung, unpublished observation). Therefore, the agarose-separated DNA was analyzed by Southern blot hybridization using a 32P-labeled rhlR-specific probe (Fig. 3B). Three fragments were detected that corresponded to the original 5.2 kb fragment and two fragments of ∼3.2 and 2 kb in size, which agreed with the expected location of the mutation between the sites flanking the rhlR gene. Therefore, CEL I was capable of specifically detecting the rhlR mutation within a highly complex DNA sample.
Figure 3.
(A) Agarose gel analysis of size-fractionated SalI-restricted genomic DNA of UPA204/205 strain pair and, underneath the gel, Southern slot blot hybridization of the corresponding fractions with a 32P-labeled rhlR gene-specific probe (even fractions only starting from the fraction 26; L, the loaded restricted DNA before fractionation). Marks of the 1 kb ladder scale on the left correspond to 12.22, 11.20, 10.18, 9.16, 8.14, 7.13, 6.11, 5.09, 4.07, 3.05 (in bold), 2.04, 1.64 and 1.02 kb. Below: the expected position of the GA insertion in rhlR (solid bar) within the SalI restriction fragment based on the genomic sequence of P.aeruginosa strain PA01 (dotted line indicates the PCR-amplified region used as the hybridization probe). (B) Agarose gel and Southern blot analysis of the denatured/re-annealed fraction 36 DNA, without (–) and with (+) CEL I treatment. The 1 kb ladder scale as described (A). (C) Expected position of the known mutations in pilC, mucA, daaP and nadC regions within corresponding restriction fragments. The genes are indicated by a solid bar; regions used as the hybridization probes are indicated by a dotted line. (The pilC-, mucA- and daaP-specific probes were based on the corresponding PCR-amplified gene regions, while as a nadC-specific probe we have used plasmid pKH385 incorporating only the 3′ portion of the gene and the downstream region, which explains detection only of the CEL I-cleaved fragment of the lower size). On the right, Southern blot analysis of the CEL I-treated genomic fractions that contain the corresponding restriction fragments. (D) Hybridization of the total genomic blot of UPA204/205 with the rhlR-containing cosmid c705. The thin line corresponds to the vector DNA, while the thick line corresponds to the 46 kb of cloned P.aeruginosa PA01 DNA.
Detection of various types of simple mutations in different bacterial genomes
To test the ability of the CEL I enzyme to recognize different nucleotide mismatches in the genomes of different bacterial species, we used four pairs of isogenic strains differing by mutations of known nature and location. These included (i) P.aeruginosa PAO1 and PAO1seq which differ by a CTGA tandem duplication in pilC; (ii) P.aeruginosa PAO1 and PDO300 with a G deletion in a poly G strip and an A→G substitution in mucA; (iii) E.coli G1310 and G1148 with an AAT→TAAC replacement in daaP and a downstream C→G substitution; and (iv) S.enteritidis TH1966 and TH1967 with an A→C substitution in nadC (Fig. 3C). Total genomic DNA of the strain pairs was combined, digested with an appropriate restriction enzyme and size-fractionated as described above. The restriction enzyme choice was defined to test the capability of the GIRAFF technology to detect mutations in the genomic restriction fragments of various size. Fractions containing mutated gene regions were identified by gene-specific hybridization probes and subjected to denaturing/re-annealing, CEL I treatment and Southern blot hybridization with specific probes (see Materials and Methods). In all DNA fractions tested the original and novel bands of predicted size were clearly identifiable (Fig. 3C) indicating the feasibility of GIRAFF for detecting various types of nucleotide substitutions or insertions/deletions directly in the genomic restriction fragments of different size and species.
Mutation probing of large genomic regions
To test the ability of GIRAFF to screen larger chromosomal regions, we examined whether it was possible to detect the rhlR mutation by probing with cosmid c750. This cosmid was derived from a cosmid library of P.aeruginosa strain PAO1 (provided by Dr Maynard Olson, University of Washington Genome Center, Seattle, WA, USA) and carries rhlR within a 46 kb chromosomal region. We also tested whether two to four adjacent genomic fractions can be combined for the analysis to facilitate the hybridization of all genomic fragments on a single membrane blot. We found that probing a blot of total UPA204/UPA205 genomic DNA with labeled cosmid c750 was as efficient in detecting the rhlR mutation as the probing of a single fraction with the rhlR-specific probe (Fig. 3D). Similarly, it was possible to detect the pilC and mucA mutations in genomic blots of the respective P.aeruginosa strains (see above) by substituting the gene-specific probes with corresponding cosmids (not shown). Therefore, CEL I is capable of recognizing mismatch-containing DNA in large genomic fractions. Each of the combined fractions contains, on average, 10% of the total genomic DNA of P.aeruginosa, a bacterial pathogen with one of the largest genomes (∼6 Mb). The successful use of cosmid DNA probes also illustrates that by using a single hybridization blot it is possible to interrogate at least 46 kb of chromosomal DNA for mutations. In fact, we have found that a genetic probe composed of several different cosmids (with a total size of inserted DNA up to 200 kb) could be used for hybridization of a genomic blot with detection of the cleavage-specific fragments (not shown). Therefore, the GIRAFF technology is capable of analyzing in a single genomic blot many dozens or even hundreds of kilobases of chromosomal DNA, validating its feasibility for genomic mutation analysis.
Other than the rhlR mutation-specific fragments, no cleavage bands were detected by cosmid c750 hybridization in any of the combined DNA fractions of the UPA204 and UPA205 strains. Because CEL I is highly sensitive to a variety of DNA mismatches, the lack of additional bands indicates that, within the probed regions of chromosomes, the sequences are identical. Overall, we have probed the UPA204/UPA205 genomic replicas by 26 non-overlapping cosmids (in six separate Southern blots) with a total size of the inserted DNA of 1.085 Mb (17.5% of the P.aeruginosa PAO1 genome) and did not detect any additional nucleotide polymorphism, strongly suggesting a clonal identity of the UPA204/UPA205 genomes. In contrast, using cosmids as probes for GIRAFF analysis of combined genomic DNA of two unrelated strains the presence of multiple polymorphic sites was always detected (not shown).
Cross-fraction probing of the entire bacterial genome and detection of a novel intragenomic polymorphism
Although the use of cosmid probing is a convenient method for examining genetic polymorphism within large genomic regions, the applicability of this approach for GIRAFF analysis of whole genomes of various bacterial species is obviously dependent on the availability of suitable cosmid clones. To avoid this limitation we tested whether it is feasible to hybridize genomic blots with restriction fractions of the same or related genomes, which were obtained using a different restriction enzyme. Because any two non-overlapping restriction sites are more or less randomly distributed relative to one another, DNA from restriction fractions generated by one enzyme would randomly hybridize across the genomic fractions generated by another restriction enzyme. Two alternative approaches could be used for obtaining hybridization probes for probing fractionated DNA. The first approach is to use the fractionated genomic DNA of a model fully sequenced strain P.aeruginosa PAO1 strain (19), which provides an opportunity to create a standard set of cross-fraction probes for GIRAFF analysis of various P.aeruginosa wild-type strains. The second approach uses genomic DNA of homologous origin, which allows probing of wild-type strain regions that could be unique to the tested isolates.
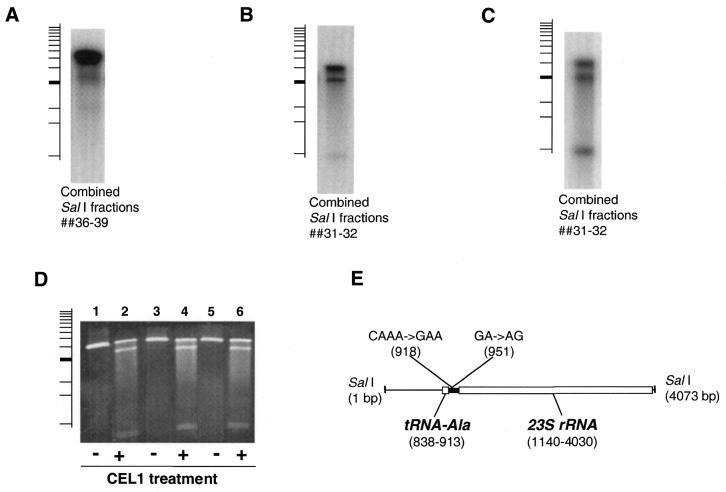
We have probed SalI-generated UPA204/UPA205 genomic fractions with different individual fractions obtained by BamHI restriction of genomic DNA of UPA205 or P.aeruginosa PAO1. The cross-fraction probing method detected the cleavage bands corresponding to the rhlR mutation (Fig. 4A) demonstrating the suitability of this approach for genome-wide mutation scanning. Using an entire set of the BamHI-generated fractions for cross-fraction probing of the genomic DNA of UPA204 and UPA205 strains resulted in detection of a novel pair of CEL I cleavage products in the combined fractions 31–32 (Fig. 4B). We have created a pBluescript library of the fraction 32 SalI fragments and, among 150 clones screened by blot hybridization, selected a plasmid detecting the novel cleavage products (Fig. 4C). It was determined that the restriction fragment susceptible to CEL I cleavage represented a 4073 bp internal region of the rrn operon, which is present in four nearly identical copies in the P.aeruginosa genome (19). In UPA204 and UPA205 strains the rrn operon is also present in four copies (data not shown). The corresponding DNA regions of each strain were amplified by PCR, denatured/annealed and treated with CEL I (Fig. 4D). The PCR-amplified fragments derived from either UPA204 or UPA205 formed identical mismatched heteroduplexes with or without combining with each other indicating the intragenomic origin of the rrn mismatch. Sequence analysis revealed that the rrn operon copies in both strains possess three adjacent polymorphic sites in a 227 bp Intergenic Transcribed Spacer between alanine tRNA and 23S rRNA genes—a C→G substitution in position 5, an A deletion in position 8 and GA→AG inversion in position 38 (Fig. 4E).
Figure 4.
(A) Detection of the rhlR mutation in the combined SalI fractions 36–39 by cross-fraction probing using an rhlR-containing BamHI fraction of P.aeruginosa PA01 genomic DNA. (B) Detection of novel CEL Icleavage products in the combined SalI fractions 31–32 by the cross-fraction probing using a BamHI fraction of the UPA204/205 genomic DNA. (C) Detection of the novel CEL I cleavage products in the combined SalI fractions 31–32 by probing with a pBluescript clone from the library of SalI fragments from fraction 31. (D) Agarose gel analysis of CEL I treatment of PCR-amplified rrn regions from UPA204 and UPA205, with and without treatment by CEL I. The PCR primers are internal to the SalI region of rrn operons and produce an amplified fragment of 3908 bp (the lower band is, therefore, of a smaller size than the corresponding band detected on the genomic fractions blot). Track 1, the amplified rrn regions of UPA204 and 205 strains are combined, heat-denatured and re-annealed without CEL I treatment; track 2, same as track 1, but treated with CEL I; track 3, the rrn regions of UPA204 only, heat-denatured and re-annealed without CEL I treatment; track 4, same as track 3, but treated with CEL I; track 5, the rrn regions of UPA205 only, heat-denatured and re-annealed without CEL I treatment; track 6, same as track 5, but treated with CEL I. (E) Position of polymorphic sites within the SalI fragment of rrn operons (Intergenic Transcribed Spacer region is indicated as a solid bar between the Ala-tRNA and 23S rRNA genes).
DISCUSSION
Our study presents a proof-of-principle for a novel technological approach for the detection of unknown mutations and nucleotide polymorphisms on a level of hundreds of kilobases of genomic DNA. The technology is based on the finding that mismatch-specific endonuclease CEL I is able to recognize and cleave DNA heteroduplexes directly in a sample of genomic restriction fragments of a combined length of ∼0.6 Mb. This makes it feasible, for example, to compare the identity of nucleotide sequence of two entire bacterial genomes.
The technique was able to detect various simple mutations directly in the genomic DNA of isogenic pairs of recombinant P.aeruginosa, E.coli and Salmonella isolates. We have also compared total genomic DNA of two clinical P.aeruginosa clones isolated from the same patient, but exhibiting divergent quorum-sensing (RhlR-specific) phenotypes. The mutation scan correctly detected a GA insertion in the rhlR and, in addition, identified a novel intragenomic polymorphism in rrn operons. Identification of the rrn polymorphism illustrates the capability of GIRAFF technology to detect and, then, identify unknown nucleotide polymorphisms on a genomic scale. It also illustrated GIRAFF’s usefulness for detection of intragenomic polymorphisms that might occur within multicopy chromosomal operons, plasmids or phase-variable regions. It is necessary to note, however, that polymorphisms within chromosomal multicopy regions could be identified by GIRAFF as long as the polymorphism-containing restriction fragments are co-fractionated. Moreover, although phase-variable regions, such as genetic switches or tandem nucleotide repeats, might also contribute to the intragenomic polymorphism, their detection by GIRAFF will depend on the relative abundance of alternating genetic regions in the analyzed genomic DNA sample.
It is necessary to note some general pitfalls of the GIRAFF technology. GIRAFF is not suitable for the detection of duplications, insertions or deletions of relatively large genetic regions that might have occurred within the UPA204 or UPA205 genomes. Also, mutations would be missed that either alter the restriction sites used for the analysis or occur within close proximity to them. To detect these types of mutations it is necessary to repeat GIRAFF analysis using a different non-overlapping restriction enzyme for generating genomic fractions. Finally, the GIRAFF technology is most effective for finding a ‘needle-in-the-haystack’ type of mutation and not for comparative genomic analysis of highly diverged isolates, where a large number of polymorphic sites are present.
Despite the potential shortcomings of the GIRAFF technology, it is unlikely that even with additional testing, a significant number of novel mutational differences between the UPA204 and UPA205 genomes would be identified. Therefore, the two strains have obviously diverged from a common ancestor very recently. Because it is rather improbable that such closely related strains would colonize the same patient by chance, the rhlR mutation was most likely acquired during microevolution of UPA205 within the host during the course of infection. As the rhlR mutation is the only genetic difference detected, it is unlikely to be a neutral marker of other selective events, but, instead, could be of selective value itself for uropathogenic P.aeruginosa. RhlR function is critically important for the expression of a great variety of P.aerugonosa genes, including many virulence factors (24) and the selective nature of the mutation remains to be determined.
The finding of no additional polymorphisms between UPA204 and UPA205 indicates a very high stability of the bacterial genome under natural conditions. It is noteworthy, however, that these strains did not exhibit a mutator phenotype determined as described previously (12). With fully functional DNA repair systems, the rate of spontaneous mutations in bacteria is estimated to be ∼5 × 10–10 per bp per cell replication that, in P.aeruginosa, would result in a novel mutation in only ∼0.3% of bacterial population per generation (25). The UPA204 and UPA205 strains were isolated during a first-episode (i.e. non-recurrent) bladder infection, which would limit the number of the bacterial generations during host colonization. Taken together, these factors indicate a low probability for additional adaptive mutations to be co-selected or for neutral mutations to be hitchhiked in the genomes of tested clones.
With many genomic projects completed or underway, GIRAFF technology provides a valuable practical tool for studying intergenomic and intragenomic variability of pathogenic and other microorganisms that occurs under natural or experimental conditions. The preparative DNA fractionation and ability of CEL I to detect genetic alterations in highly complex DNA samples might also be applied to screening mutations and polymorphisms in the genomes of higher organisms (including human). Due to the significantly larger size, high complexity and diploidy, use of a long-range PCR or artificial chromosome constructs could be more practical for the latter task.
Acknowledgments
ACKNOWLEDGEMENTS
We thank D. Dykhuizen, M. Samadpour, D. Gottshling and S.-J. Suh for useful discussions and S.-J. Suh also for providing P.aeruginosa recombinant strains. Our work is supported by grants from the Cystic Fibrosis Foundation, National Institute of Health, National Science Foundation and Department for Veterans Affairs.
REFERENCES
- 1.Cairns J. (1975) Mutation selection and the natural history of cancer. Nature, 255, 197–200. [DOI] [PubMed] [Google Scholar]
- 2.Loeb L.A. and Christians,F.C. (1996) Multiple mutations in human cancers. Mutat. Res., 350, 279–286. [DOI] [PubMed] [Google Scholar]
- 3.Levin B.R. and Bull,J.J. (1994) Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol., 2, 76–81. [DOI] [PubMed] [Google Scholar]
- 4.Sokurenko E.V., Hasty,D.L. and Dykhuizen,D.E. (1999) Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol., 7, 191–195. [DOI] [PubMed] [Google Scholar]
- 5.Rainey B.P. and Moxon,E.R. (2000) Microbiology. When being hyper keeps you fit. Science, 288, 1186–1187. [DOI] [PubMed] [Google Scholar]
- 6.Boucher J.C., Yu,H., Mudd,M.H. and Deretic,V. (1997) Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun., 65, 3838–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H., Hanes,M., Chrisp,C.E., Boucher,J.C. and Deretic,V. (1998) Microbial pathogenesis in cystic fibrosis: pulmonary clearance of mucoid Pseudomonas aeruginosa and inflammation in a mouse model of repeated respiratory challenge. Infect. Immun., 66, 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahenthiralingam E., Campbell,M.E. and Speert,D.P. (1994) Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun., 62, 596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barth A.L. and Pitt,T.L. (1995) Auxotrophic variants of Pseudomonas aeruginosa are selected from prototrophic wild-type strains in respiratory infections in patients with cystic fibrosis. J. Clin. Microbiol., 33, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst R.K., Yi,E.C., Guo,L., Lim,K.B., Burns,J.L., Hackett,M. and Miller,S.I. (1999) Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science, 286, 1561–1565. [DOI] [PubMed] [Google Scholar]
- 11.LeClerc J.E., Li,B., Payne,W.L. and Cebula,T.A. (1996) High mutation frequencies among Escherichia coli and Salmonella pathogens. Science, 274, 1208–1211. [DOI] [PubMed] [Google Scholar]
- 12.Oliver A., Canton,R., Campo,P., Baquero,F. and Blazquez,J. (2000) High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science, 288, 1251–1254. [DOI] [PubMed] [Google Scholar]
- 13.Tawata M., Aida,K. and Onaya,T. (2000) Screening for genetic mutations. A review. Comb. Chem. High Throughput Screen, 3, 1–9. [DOI] [PubMed] [Google Scholar]
- 14.Nollau P. and Wagener,C. (1997) Methods for detection of point mutations: performance and quality assessment. IFCC Scientific Division, Committee on Molecular Biology Techniques. Clin. Chem., 43, 162–170. [PubMed] [Google Scholar]
- 15.Cotton R.G.H. (1997) Mutation Detection. Oxford University Press, Oxford.
- 16.Chakrabarti S., Price,B.D., Tetradis,S., Fox,E.A., Zhang,Y., Maulik,G. and Makrigiorgos,G.M. (2000) Highly selective isolation of unknown mutations in diverse DNA fragments: toward new multiplex screening in cancer. Cancer Res., 60, 3732–3737. [PubMed] [Google Scholar]
- 17.Brint J.M. and Ohman,D.E. (1995) Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR–RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR–LuxI family. J. Bacteriol., 177, 7155–7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holloway B.W. and Morgan,A.F. (1986) Genome organization in Pseudomonas. Annu. Rev. Microbiol., 40, 79–105. [DOI] [PubMed] [Google Scholar]
- 19.Stover C.K., Pham,X.Q., Erwin,A.L., Mizoguchi,S.D., Warrener,P. Hickey,M.J., Brinkman,F.S., Hufnagle,W.O., Kowalik,D.J., Lagrou,M. et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature, 406, 959–964. [DOI] [PubMed] [Google Scholar]
- 20.Bilge S.S., Clausen,C.R., Lau,W. and Moseley,S.L. (1989) Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J. Bacteriol., 171, 4281–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zinder N.D. and Lederberg,J. (1952) Genetic exchange in Salmonella. J. Bacteriol., 64, 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oleykowski C.A., Bronson Mullins,C.R., Godwin,A.K. and Yeung,A.T. (1998) Mutation detection using a novel plant endonuclease. Nucleic Acids Res., 26, 4597–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez H. and Akman,S.A. (1998) Large scale isolation of genes as DNA fragment lengths by continuous elution electrophoresis through an agarose matrix. Electrophoresis, 19, 646–652. [DOI] [PubMed] [Google Scholar]
- 24.Whiteley M., Lee,K.M. and Greenberg,E.P. (1999) Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA, 96, 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drake J.W. (1991) A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl Acad. Sci. USA, 88, 7160–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]