Abstract
Besides mRNA, rRNA, and tRNA, cells contain many other noncoding RNA that display critical roles in the regulation of cellular functions. Human genome sequencing revealed that the majority of non-protein-coding DNA actually codes for non-coding RNAs. The dynamic nature of RNA results in its motile and deformative behavior. These conformational transitions such as the change of base-pairing, breathing within complemented strands, and pseudoknot formation at the 2D level as well as the induced-fit and conformational capture at the 3D level are important for their biological functions including regulation, translation, and catalysis. The dynamic, motile and catalytic activity has led to a belief that RNA is the origin of life. We have recently reported that the deformative property of RNA nanoparticles enhances their penetration through the leaky blood vessel of cancers which leads to highly efficient tumor accumulation. This special deformative property also enables RNA nanoparticles to pass the glomerulus, overcoming the filtration size limit, resulting in fast renal excretion and rapid body clearance, thus low or no toxicity. The biodistribution of RNA nanoparticles can be further improved by the incorporation of ligands for cancer targeting. In addition to the favorable biodistribution profiles, RNA nanoparticles possess other properties including self-assembly, negative charge, programmability, and multivalency; making it a great material for pharmaceutical applications. The intrinsic negative charge of RNA nanoparticles decreases the toxicity of drugs by preventing nonspecific binding to the negative charged cell membrane and enhancing the solubility of hydrophobic drugs. The polyvalent property of RNA nanoparticles allows the multi-functionalization which can apply to overcome drug resistance. This review focuses on the summary of these unique properties of RNA nanoparticles, which describes the mechanism of RNA dynamic, motile and deformative properties, and elucidates and prepares to welcome the RNA therapeutics as the third milestone in pharmaceutical drug development.
Keywords: Ribonucleic acid (RNA), RNA nanotechnology, RNA dynamics, Deformative property, Drug development, Drug delivery, Cancer treatment
1. Introduction
Ribonucleic acid (RNA) plays an important role in biological processes. It includes coding RNA (mRNA) for protein translation, and non-coding RNA (ncRNA) for regulation of biological functions. The ncRNA has two subcategories, housekeeping ncRNAs such as transfer RNA (tRNA) and ribosomal RNA (rRNA) as well as regulatory ncRNAs such as small interfering RNA (siRNA) and microRNA (miRNA) [1,2]. The malfunction and misregulation of natural RNAs can cause disease, which makes them promising targets for therapeutic purposes [1]. RNA can be targeted by oligonucleotides such as patisiran siRNA and small molecules such as risdiplam for disease treatment [3–7]. In addition to the natural RNAs, artificial RNAs have been generated for the therapeutic purposes, that can be used as therapeutic agents directly such as siRNA or as drug delivery platforms such as RNA nanoparticles [3,8]. With the approval of numerous RNA drugs including antisense oligonucleotide (ASO), siRNA, and mRNA vaccine, RNA drugs have become the third milestone in drug development after small molecule drugs and protein-based drugs [8–12].
RNA is a polymeric molecule that is composed of four types of nucleotides with four nitrogenous bases including adenine (A), guanine (G), cytosine (C), and uracil (U). The canonical Watson-Crick (A-U, G-C) and non-canonical (e. g., G-U) bases pairs allow RNA molecules to form diverse secondary and tertiary structures [13]. The secondary structure of RNA is formed mainly by WC base-pairing and can be predicted using nearest neighbor parameters. The tertiary structure contains the long-range interactions of secondary motifs, and the quaternary structure refers to the higher-order architectures formed via RNA-RNA interaction or RNA interaction with other molecules such as proteins. The precise structure of RNA and its dynamics are essential for RNA’s functional versatility. RNA molecules in solution possess a set of conformations whose distribution is proven to change with environmental triggers [14]. RNA dynamics covers motions at a wide range of timescales including secondary structure transition, tertiary interaction change, and “jittering” motions [15]. RNA dynamics are closely related to their biological functions such as catalysis in ribozyme, translation in the ribosome, and gene regulation in riboswitch [14–16].
RNA nanotechnology is the study of nanoparticles whose main composition is RNA [8]. It includes the construction of RNA nanoparticles with various complexity and the application of RNA nanoparticles such as their biomedical usage [9]. RNA nanoparticles are designed by naturally derived RNA motifs or from scratch with the assistance of computation modeling [17–19]. RNA nanoparticles can be constructed through different approaches, including one-pot assembly, rolling circle transcription (RCT) and RNA origami [20–24]. The deformative property of RNA, in combination of their advantageous size (10–40 nm) to avoid nonspecific diffusion and nonspecific entry to organs, allows RNA nanoparticles to penetrate leaky blood vessels and accumulate at the tumor site as well as bypass renal filtration and reduce toxicity [25]. This favorable biodistribution of RNA nanoparticles could be further improved by the incorporation of ligand or aptamer for active cancer-targeting [26,27]. In addition to the unique deformative property, there are many other advantages of RNA nanotechnology for pharmaceutical applications. RNA naturally carries a negative charge which minimizes its interaction with negative charged cell membrane without the need to coat nanoparticles with polymers, thus preventing interactions with healthy organs. RNA nanoparticles have high programmability including shape, size, stability, and stoichiometry that self-assemble with high stability from shorter oligonucleotides [9]. Due to the high hydrophilic nature of RNA, RNA nanoparticles could significantly increase the water solubility of hydrophobic drugs thus eliminating the use of toxic formulation [28,29]. RNA is multivalent which allows them to be multi-functionalized and can be synthesized with defined structure and stoichiometry, which is beneficial for overcoming drug resistance by achieving combination therapy [30,31]. RNA nanoparticles are commonly synthesized to include targeting ligands that enhance local nanoparticle concentrations and allow for specific cell entry through receptor-mediated endocytosis [32]. In combination with extracellular vesicles, RNA nanoparticles could improve the delivery efficiency of therapeutic oligos such as siRNA by targeting delivery and avoiding endosome trapping [32,33]. Together, RNA nanoparticles have delivered small molecule drugs such as doxorubicin (DOX) and paclitaxel (PTX) as well as oligonucleotide drugs such as siRNA and anti-miRNA for disease treatment [28,34–36]. Furthermore, the immunogenicity of RNA nanoparticles is size, shape, and sequence-dependent leading to an adjustable immune response; and the lack of proteins eliminates issues with antibody induction and allows for repeatable intravenous injections without resulting toxicities [37]. They showed promises for the treatment of various types of cancer, including breast, prostate, colorectal cancer, and other disease such as central nervous system (CNS) and eye diseases (Table 1) [28,36,38–41].
Table 1.
RNA nanoparticles functionalized with different therapeutic agents for disease treatment.
| Therapy type | Therapeutic group | Targeting group | Nanoparticle | Construction | Disease type | Ref |
|---|---|---|---|---|---|---|
| Small molecule drug | DOX | Annexin A2apt | 3WJ | Self-assembly | Ovarian cancer | [34] |
| CPT | FA | 3WJ | Self-assembly | Cancer | [29] | |
| PTX | EGFRapt | 4WJ | Self-assembly | TNBC | [28] | |
| Therapeutic oligo | BRCAA1 siRNA | FA | 3WJ | Self-assembly | Gastric cancer | [42] |
| Survivin siRNA | FA/PSMAapt/EGFRapt | 3WJ & EV | Self-assembly | CRC, prostate and breast cancer | [33] | |
| BIRC5 siRNA | EGFRapt | 3WJ & EV | Self-assembly | NSCLC | [43] | |
| VEGF siRNA | N/A | siRNA nano ball/RAPSI | RCT | AMD, cancer | [44,45] | |
| USE1 siRNA | N/A | BRC | RCT | Lung cancer | [46] | |
| Anti-miR17 | PSMAapt | 3WJ | Self-assembly | Prostate cancer | [38] | |
| Anti-miR21 | EGFRapt /CD133apt /PSMAapt | 3WJ | Self-assembly | TNBC, prostate cancer | [35,38,47] | |
| Thrombin aptamer | N/A | 2-helix RNA origami | RNA origami | Blood coagulation | [48] | |
| Combination | PTX, miR122 | GalNAc | 6WJ | Self-assembly | HCC | [31] |
| DGLA, D5D siRNA | EpCAMapt | 3WJ | Self-assembly | Colon and lung cancer | [49,50] | |
| TAM, MED1 siRNA | HER2apt | 3WJ | Self-assembly | Breast cancer | [30] |
2. RNA as a dynamic, deformative, and motile being
2.1. RNA hierarchical structure in relationship to motion
As classical biological studies have elucidated the expansive roles of RNA within cells, the understanding of the complex folding of RNA structures have also been demonstrated and predicted by experimental and computational approaches. Determination of RNA structure is important for understanding its biological functions and designing therapeutics to target RNA for disease treatment [7]. The hierarchy of RNA structures includes primary, secondary and tertiary structure. The primary structure of RNA is the nucleotide sequence which contains the ribose sugar, nucleobases, and phosphodiester bond. The secondary structure of RNA is the local folds created through Watson-Crick base pairing, which contains both the base-paired regions and non-paired regions. The base-paired region refers to helices and the non-paired region includes apical loops, internal loops, and bulges (Fig. 1A) [51,52]. Current RNA secondary structure prediction methods are mainly based on free energy minimization using nearest-neighbor parameters [53]. Recently, new methods such as SPOT-RNA combine the nearest neighbor model with machine learning which improves the prediction accuracy by covering all base pairings including lone, pseudoknotted, and noncanonical base pairs as well as triplet interactions [54]. The tertiary structure is composed of secondary structural motifs that are brought together through long distance interactions (Fig. 1B) [51]. The long-range intramolecular interactions that drive the folding of RNA tertiary structures includes interactions between helical regions such as coaxial stacking, between helical regions and unpaired regions such as tetraloop motif, and between unpaired regions such as pseudoknots and loop-loop interactions [55,56]. RNAs such as ribozyme and riboswitch require complex tertiary structure to exhibit biological activities. Furthermore, the quaternary structure forms when RNA molecules participate in interactions with other molecules such as proteins and nucleic acids. RNAs can form ribonucleoproteins with RNA-binding proteins such as ribosome and spliceosome, RNAs can also form higher order assemblies with other RNAs such as bacteriophage Phi29 prohead RNA (pRNA) which forms hexamer with six RNA subunits (Fig. 1C) [57,58].
Fig. 1.
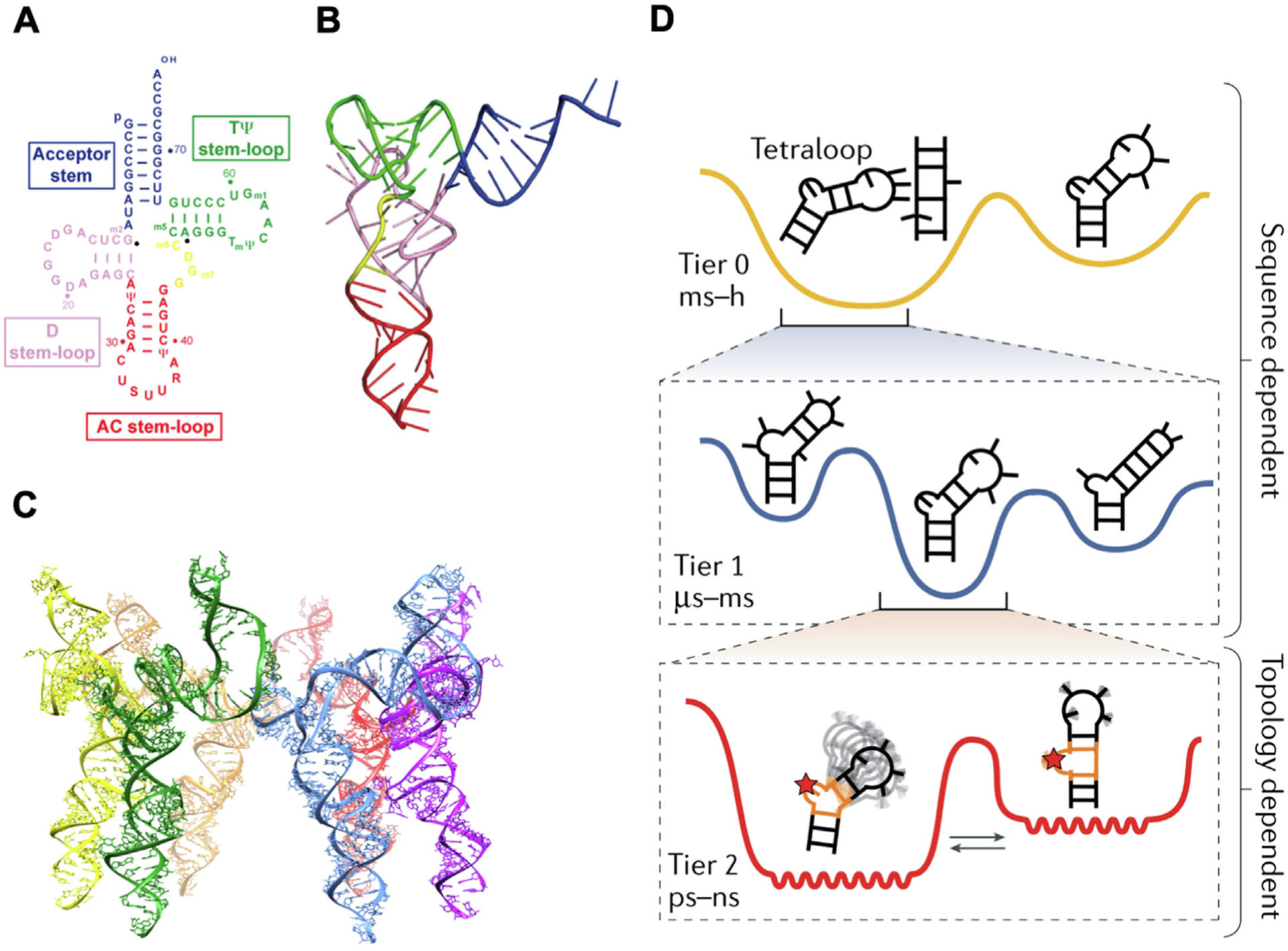
RNA hierarchical structure and dynamics. (A) Secondary and (B) tertiary structures of tRNA (Lys, 3). Reprinted with permission from [51]. Copyright 2010 Molcular Diversity Preservation International; (C) Quaternary structure of phi29 pRNA hexamer. Adapted with permission from [57]. Copyright 2021 American Chemical Society; (D) Three tiers of RNA dynamics in hierarchical free-energy landscape. Adapted with permission from [14]. Copyright 2019 Springer Nature Limited.
Instead of remaining static, RNA relies on their dynamic properties which is encoded by the inherent properties of RNA structure and the external trigger factors to achieve complex functionality [16,59]. An RNA molecule in a biological condition forms a statistical distribution of many interconverting conformations, which can be described using a continuous and rugged free-energy landscape [15]. The population of each conformation within this statistical ensemble depends on its free energy [14]. Environmental triggers such as variation in temperature, concentrations, and addition or presence of other interacting molecules such as proteins or RNA substrates can change the relative populations of different conformations. The rate and difficulty of transformation between two conformations depends on their energy barriers. The similar internal energies of two conformations allows for faster and more prevalent transitions. And the conformational change can occur at secondary structural level such as riboswitch during gene regulation and tertiary structural level such as ribozyme during catalysis. RNA dynamics includes both internal motions and external triggered rearrangement that covers many motional modes with a large range of amplitudes and timescales. Due to the hierarchal structuring of RNA (primary, secondary, tertiary), each level of structuring acts independently of other levels. Similarly, RNA motion is divided into three independent modes based on their locations in a hierarchical free-energy landscape. These levels of motion not only describe the level of motion in the RNA but can each independently happen without requiring motion from other tiers. Tier 0 refers to the secondary structural transitions at larger than millisecond timescales. These are the largest RNA dynamics that involve restructuring of RNA formations and global base pairing. Tier 0 motion comes at high energy costs due to the unfavorable energies of breaking several base pairs. Tier 1 includes base-pair changes and tertiary dynamics at timescale ranging from microsecond to millisecond. These smaller interactions are the breaking and reforming of singular base pairs that can be described as nucleic acid breathing that is divided into four categories: base-pair melting, base-pair reshuffling, base-pair isomerization, and long-range tertiary interaction. Each of these dynamic types works to optimize internal thermodynamic stabilities of the RNA molecules. Finally, Tier 2 contains “jittering” motions including interhelical and loop dynamics which happens at picosecond to nanosecond timescales (Fig. 1D) [15]. These rather rapid dynamics of an RNA results in the “vibration” of the molecule when observed in set space and include unpaired bulge and internal loop residues, sugar repuckering, and phosphate-backbone reorientation. Several studies have undertaken the examination of the structure and motion of RNA using nuclear magnetic resonance (NMR) [60–65], X-ray crystallography [66,67], single-molecule imaging microscopy [68–70], cryogenic electron microscopy (cryo-EM), small-angle scattering (SAS), atomic force microscopy (AFM), and fluorescence resonance energy transfer (FRET) [71–73]. The increasing amount of experimental data acquired from these approaches has significantly improved the computational modeling and simulation of the RNA structural dynamics [73–76].
2.2. The dynamics of RNA at energy, structure, and function level
2.2.1. Energy landscape: Melting temperature and nearest neighbor dynamics
RNA thermodynamics refers to RNA structural changes affected by temperature. The melting temperature (Tm) is the temperature at which half of the total RNA complexes in the system are denatured into unfolded single strands. The Tm of RNA is length, sequence, and concentration dependent; longer strand, more GC pairs, and higher concentration leads to higher Tm. It also depends on counter ion because RNA is negative charged, thus, cations such as sodium (Na+) and magnesium (Mg2+) bind to RNA and form a shield which neutralize the overall negative charge. The measurement of Tm provides a convenient way to evaluate the thermodynamic parameters such as Gibbs free energy (G), enthalpy (H) and entropy (S) from experimental data. By adjusting the composing strand and overall structure, RNA nanoparticles with desirable thermodynamic properties have been constructed to meet the requirement of various applications. Phi29 3WJ has been proven to be a thermostable RNA motif, however, the multiple drug conjugation significantly decreases its thermostability due to increased steric constrains and physical hindrance. The Tm of Phi29 3WJ decreased to 32° C after conjugation with 10 copies of PTX, which makes it unstable at physiological condition (37° C) [28] (Fig. 2A). To improve the drug loading capacity, RNA nanoparticles with increased thermostability need to be constructed. It is generally believed that increasing GC content of RNA nanoparticles would result in increased Tm value and therefore stabilize RNA nanoparticles. However, unrestricted increase of GC percentage can result in misfolding as the chance of self-dimerization and the formation of quadruplex also increases. Thus, a new 4WJ RNA nanoparticle has been designed with more base pairing resulting from more and longer branches. The annealing temperature (Ta) of 4WJ (80.9° C) itself is much higher than 3WJ (58.4° C) which indicates its high thermostability. This ultra-high thermostability allows 4WJ RNA nanoparticles to have high drug loading capacity without diminishing its stability. After conjugation with 24 copies of PTX, 4WJ remains its high thermostability with the Tm of 79° C, which enables its in vivo application without stability concerns [28] (Fig. 2A).
Fig. 2.
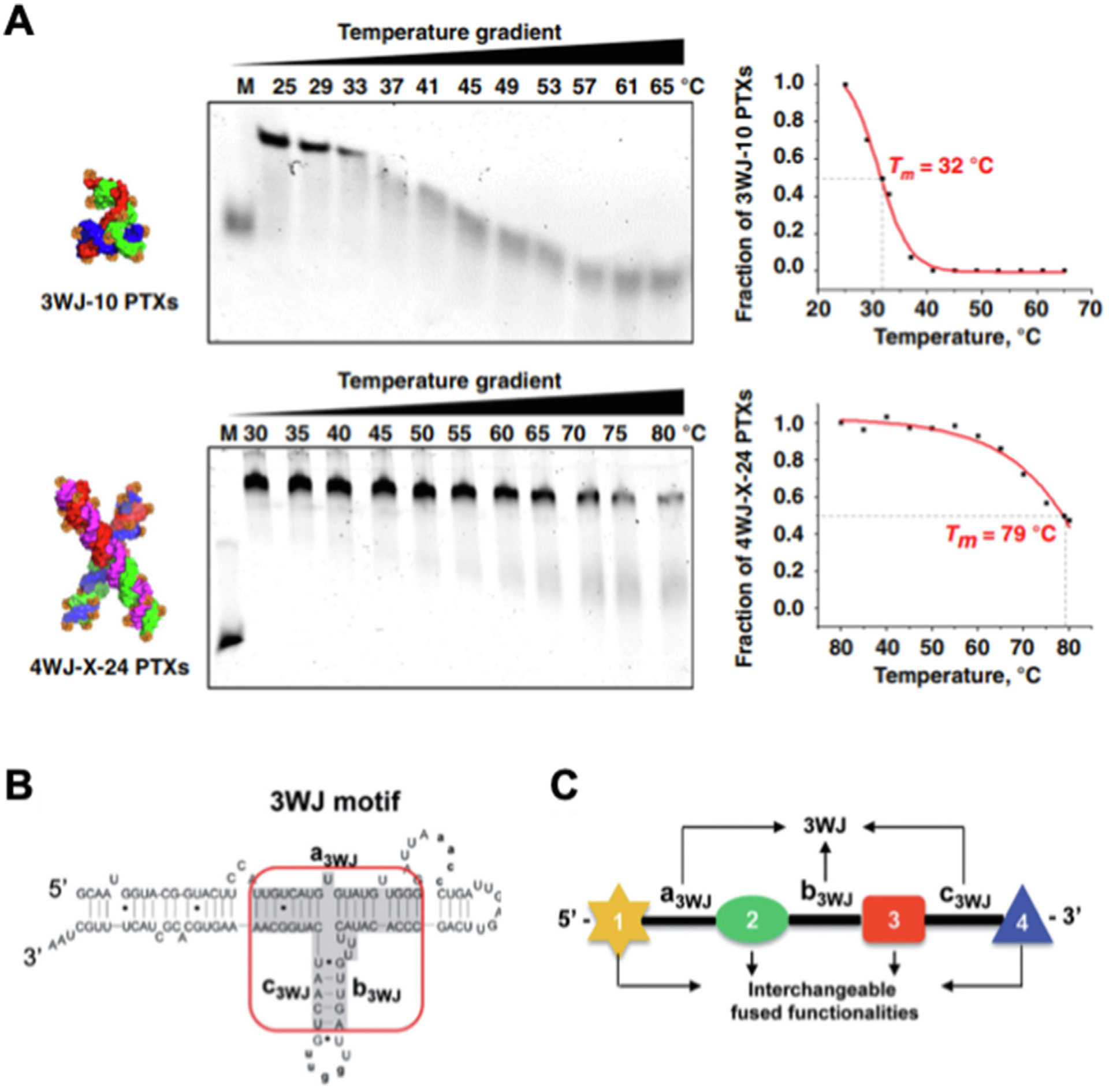
Melting temperature and nearest neighbor dynamics of RNA nanoparticles. (A) TGGE gels and their quantification curves of 3WJ-10 PTXs and 4WJ-X-24 PTXs RNA nanoparticles. Reprinted with permission from [28]. Copyright 2020 The Author(s); (B) Secondary structure of pRNA with central 3WJ motif (red circle) composing a3WJ, b3WJ, and c3WJ strands. Reprinted with permission from [78]. Copyright 2013 Oxford University Press; (C) The strategy of constructing RNA complexes harboring multiple functional groups (1–4) driven by the formation of 3WJ core. Reprinted with permission from [78]. Copyright 2013 Oxford University Press.
In addition to the interaction within base pairs, the interaction between base pairs in nucleic acid strands also affect the free energy of RNA molecules. The nearest neighbor model is used for predicting RNA secondary structure using thermodynamic parameters by minimizing the Gibbs free energy computed with stacking energy associated costs between matching with complementary nucleotides of not just single base pairs but ‘neighboring’ base pairs. The nearest neighbor principle also plays an important role during the folding of RNA structures. Phi29 pRNA has been reported to have a highly thermodynamically stable 3WJ core (Tm 58° C), which is resistant to 8 M urea denaturation and serum degradation [77,78] (Fig. 2B). During the folding process, the formation of stable 3WJ core facilitates the correct folding of other attaching modules, such as aptamer, siRNA, and ribozyme, attached to the pRNA strand, which allows them to remain functional [78] (Fig. 2C). Mechanism study of pRNA with malachite green (MG) aptamer demonstrated the role of nearest neighbor principle in RNA folding process [78]. Studies have shown pRNA-MG aptamer with interfering sequences at 5′ showed different fluorescent intensity during transcription and after refolding. During transcription, as the 5′ interfering sequence is synthesized first, it complements to the MG aptamer sequence which disrupt its structure leading to decreased fluorescent intensity. However, the fluorescent intensity is regained after refolding, during which the highly stable 3WJ core forms first at high temperature leading to the correct folding of the MG aptamer. The presence of 5′ interfering sequence pairs with the MG aptamer leading to the kinetical trap of RNA in the misfolding structure during transcription, while the denaturation and annealing process allows the thermostable 3WJ core to form which results in correct formation of the MG aptamer.
2.2.2. Structure transition: Pseudoknot formation and RNA breathing
Structure transition via RNA breathing and pseudoknot formation governed by the nearest neighbor principle resulted in a conformational dynamic process that causes passive motion.
RNA pseudoknots are conformations of folded knot-shaped 3D configurations that are not true topological knots [79]. It is a secondary structure containing two stem loops, or one stem and one stem loop. Hairpin type (H-type) pseudoknots, consisting two helical stems and two loops, are the most common type and is formed by base-pairing between a hairpin loop and the single-stranded region of the second hairpin [80] (Fig. 3A). In H-type pseudoknot, two helical stems stack coaxially to form a quasi-continuous structure and the coaxial stacking stabilizes the overall RNA structure [80]. Pseudoknots play an indispensable role in the structure and function of many RNAs such as ribozyme and ribonucleoproteins (RNPs) such as ribosome and telomerase [79,81]. The formation of pseudoknot is essential in a variety of biological processes such as the regulation of translation initiation by inducing ribosomal frameshifting [81,82].
Fig. 3.
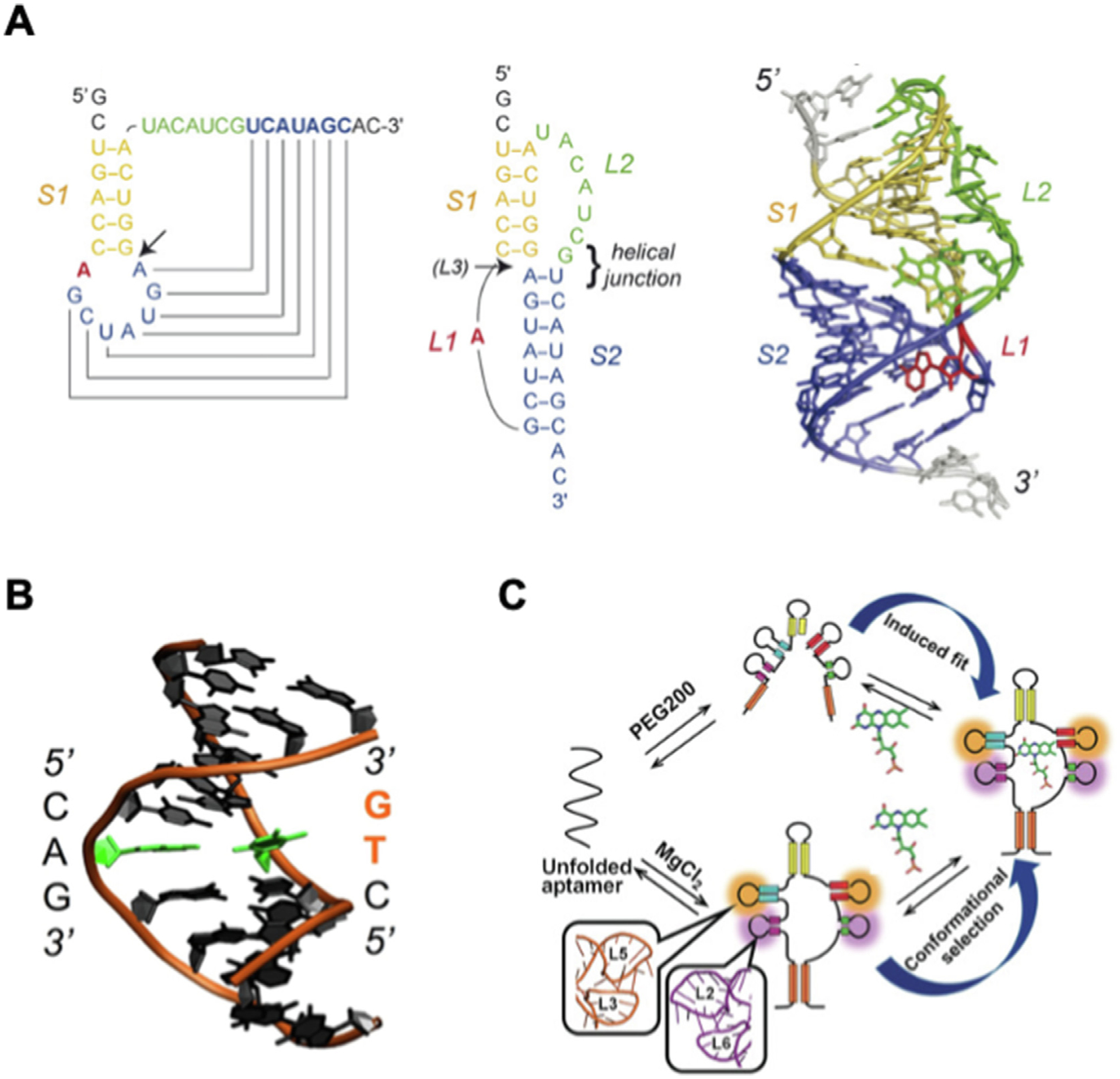
RNA dynamics at structure and function level. (A) Folding topology and 3D structural model of the phage T2 gene 32 autoregulatory pseudoknot (PBD 2TPK). Adapted with permission from [79]. Copyright 2008 Elsevier B.V.; (B) DNA breathing demonstrated by dsDNA with the breaking A-T base pair (green) generated from atomistic MD simulation. Reprinted with permission from [84]. Copyright 2016 Oxford University Press; (C) Mechanism of FMN recognition by induced fit under crowding conditions with PEG200 and by conformational capture under dilute condition with MgCl2. Reprinted with permission from [89]. Copyright 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinhem.
Double-stranded DNA (dsDNA) has spontaneous conformational fluctuation, also called DNA breathing, which leads to the breaking of base pairs and transit single stranded region (ssDNA), also called a DNA bubble (Fig. 3B) [83–85]. Due to the similar composition with DNA, RNA also processes the breathing process. However, RNA breathing is not widely recognized as it does not generate of an observable bubble at double strand region as seen in DNA [83]. Nevertheless, RNA breathing has great impact on the dynamic property of RNA as it exposes the nucleotides within the base pairs to the environment causing the formation of motifs such as pseudoknot that leads to a cascade of structural changes at the secondary and tertiary level.
2.2.3. Configuration adjustment: Induced fit and conformational capture
RNA interacts with proteins and small molecules to exhibit biological functions such as riboswitch regulation and ribozyme cleavage. The dynamics of RNA interacting with other molecules fall under induced fit and conformational selection models. The induced fit model proposes that RNA actively adapts and changes its shape upon binding to substrate and continually reform until reaching a final stable state. The interaction between tRNA and MiaA transferase involves a mutually induced fit mechanism with the movements of large domain in MiaA leading to the partial unfolding of anticodon loop in tRNA [86]. The conformational selection model assumes that the RNA confirmation required for stable binding preexists, and the binding of substrate stabilizes this confirmation which makes it predominant in the conformational population. The SAM-I riboswitch aptamer has confirmations with both open and closed binding pocket in the ligand-free state despite the energetic differences [87]. RNA systems participating intermolecular interactions utilizing both mechanisms have also been reported. U1A-RNA molecular recognition is mediated by the contact of U1A with RNA through conformational selection and the stabilization of U1A-RNA complex by induced fit [88]. In another study, FMN riboswitch aptamer showed FMN binding through different mechanism in different environment (Fig. 3C). The FMN recognition mechanism of riboswitch aptamer is conformational selection in the presence of Mg2+ ion and is induced fit in the presence of molecular crowding agent such as PEG200 without Mg2+ [89].
2.3. RNA dynamics before, during, and after intrinsic equilibrium
RNA dynamics can be classified into three statuses, before, during, and after intrinsic equilibrium. RNA itself has an intrinsic equilibrium that aims to fold the nucleic acids populations into the lowest energy conformation possible. RNA populations exist as ensembles of folding conformations, with the population as a whole is in equilibrium with defined percentages of the population remaining in each conformation. However, individual RNA molecules are in transient motion between each conformational state. This ensemble of RNA conformations provides a stable set of RNA folded states and a level of equilibrium. Before intrinsic equilibrium refers to the stage of RNA cotranscriptional folding, during means the status of native RNA reaches equilibrium of possible conformations, after refers to the transition of RNA conformations triggered by external changes.
During transcription, an RNA strand increases its length until reaching the final product. However, during the relatively slow transcription process RNA has been shown to fold into structures to conserve energy at a much more rapid rate. Recent studies revealed the rearrangement pathway of non-native RNA intermediates during cotranscriptional folding of E. coli signal recognition particle (SRP) RNA using molecular dynamics (MD) simulation (Fig. 4A) [90]. The cotranscriptional folding of RNA is driven by local interactions and distant interactions. Local interactions are mainly mediated by the base-pairing changes such as formation and melting (Tier 0 and 1) at the nearest neighbor region, which can be imaged as the RNA breathing process. Distant interactions are mainly mediated by the formation and deformation of tertiary motifs (Tier 1) such as pseudoknots. The base-pairing regions are continuously changing which leads to the formations of different tertiary motifs resulting in the formation of RNA structure with relatively high stability and low free energy. The structure is constantly changing during transcription as more nucleotides are added onto the RNA strands until it reaches the intrinsic equilibrium at full length.
Fig. 4.
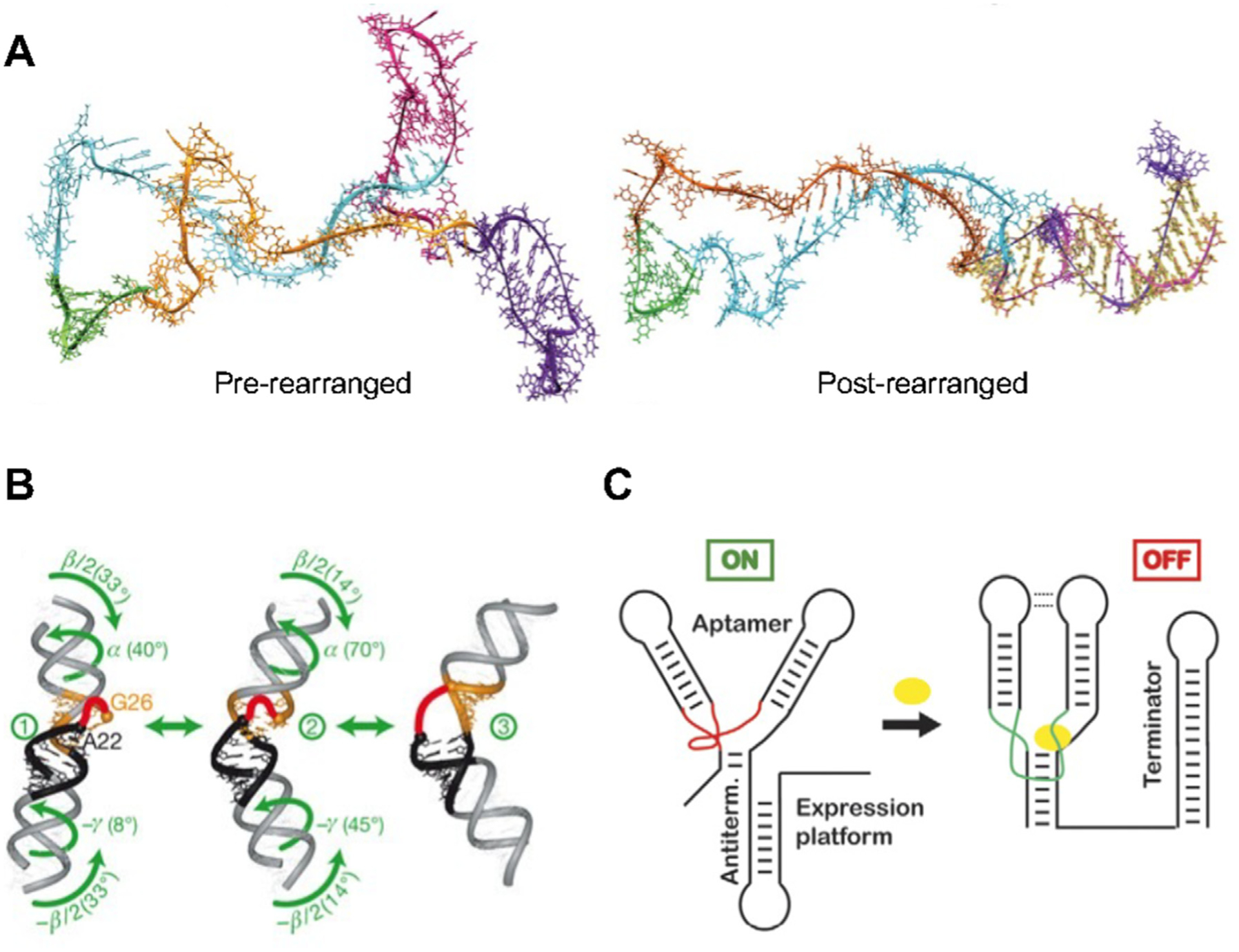
RNA dynamics before, during, and after intrinsic equilibrium. (A) Pre-rearranged structure and post-rearranged structure with the formation of extended helix (yellow) of E. coli SPR RNA during cotranscriptional folding. Reprinted with permission from [90]. Copyright 2020 Elsevier Inc.; (B) Three conformation of TAR with interhelical motions (Black: Helix I; Orange: Helix II). Reprinted with permission from [62]. Copyright 2007 Nature Publishing Group; (C) The transition of riboswitch from on to off status after ligand (yellow) binding. Reprinted with permission from [16]. Copyright 2008 Elsevier Ltd.
RNAs once folded into their equilibrium structure will maintain a relative level of stability and defined structure with small reorganization through intramolecular conformation changes. Intramolecular zipping/unzipping of base pairs and backbone conformational changes routinely occur throughout the RNA on a scale of < 0.1 μs. This reorganization of small regions of an RNA strand provides small vibrational movements in the RNA over time. Within RNA structures, frequent and rapid motion can be found on the 10−9 to 10−5 second time scale [16,60,62,91]. Using NMR, Zhang et al. were able to demonstrate the inter-helical bending and twisting, and axially twisting of transactivation response element (TAR) RNA from the HIV-1 virus without the presence of its ligand (Fig. 4B) [62]. These dynamics within the RNA provides an overall motion of the RNA molecule itself. Furthermore, RNA motion has been found within RNA tetraloops, including the smallest know RNA structure the UUCG tetraloop RNA hairpin [64,92]. This natural tetraloop demonstrates significant mobility and multiple conformations while retaining high stability [92]. RNAs intrinsically contain motion via intramolecular zipping/unzipping of base pairs, sugar puckering, and backbone conformational changes that can create a floppy or wiggling motion of the RNA over time.
Environmental changes can lead to movement of a region of the RNA or the entire RNA structure, resulting in redistribution of conformations in the previous dynamic equilibrium [14]. This motion and restructuring are due to the higher affinity and lower energy landscape to the rearranged interaction with the substrate. Riboswitches are able to change conformation to change gene expression upon sensing the presence of metabolites, vitamins, metal ions, and temperature changes [93–95]. The signaling off of genes is highly orchestrated with riboswitch conformational changes or motion in the RNA. The aptamers region of riboswitches can have motion required of the riboswitch to switch to an on or off state (Fig. 4C) [16]. In addition, intermolecular interactions with other proteins and RNAs can lead to restructuring of large regions of an RNA. Ribosome assembly is the most well studied case of RNA motion in this category as 16S ribosomal RNA (rRNA) associate with 20 ribosomal proteins [68,96]. These motions were probed by Kim et al. through fluorescence resonance energy transfer (FRET) studies to observe the motion in real-time [68]. It was found that the 5′ region of the 16S rRNA initiated interactions with the S4 ribosomal protein that led to stable non-native intermediate before changing the frequency and direction of RNA helix motions into a native complex allowing for completion of the 30S complex formation.
3. The Dynamic, deformative and motile properties of RNA nanoparticles enable their favorable biodistribution
It has been reported that RNA nanoparticles have a rubbery property which facilitate their favorable biodistribution, including efficient cancer accumulation and fast renal excretion [25]. This unique advantage is based on the dynamic property of RNA, which results in its versatile conformational changes within the biological environment using induced fit and configuration capture mechanisms.
3.1. Programmability over shape, size, stability, and stoichiometry
One of the advantages of RNA nanoparticle is that their shape, size, stability, and stoichiometry can be easily and precisely controlled by changing the sequence, length, and modification of their composing RNA strands, which makes RNA nanoparticle an idea drug delivery platform [9].
RNA has been shown to fold into nanocomplexes with stable structures from one or multiple RNA oligo strands. Myriads of RNA nanoarchitectures with various 2D structures, such as RNA triangle, square and pentagon (Fig. 5C) [97], as well as 3D structures, such as RNA polygons, polyhedrons, and dendrimers, have been rationally designed and assembled [98–101]. The approaches to design RNA with specific shape include utilization of existing RNA motifs with specific angles and de novo design assisted by computation [17–19]. Atomic force microscopy (AFM) and cryogenic electron microscopy (Cryo-EM) are two methods to confirm the shape of RNA nanoparticles [28,97]. The size of RNA nanoparticles can be easily controlled by changing the length of composing RNA strands. RNA squares with the size of 5, 10, and 20 nm have been constructed by modulating the length of the core and side strands accordingly (Fig. 5B) [21]. Gel electrophoresis and dynamic light scattering (DLS) are two methods to demonstrate the relative size and measure the hydrodynamic diameter of RNA nanoparticles, respectively [21]. There is a large repertoire of existing RNA nanoparticles with versatile structures covering a wide range of size which is continuously expanding [9]. Besides small RNA nanoparticle s constructed by bottom-up assembly, large RNA nanoparticles are also available using RNA origami (up to 6000 nt) and RCT (100–2000 nm) method [102,103]. The size and shape of RNA nanoparticles have great impact on their in vivo behavior including circulation time and organ accumulation [104]. Furthermore, RNA nanoparticles with special size and shape could provide protection to loaded cargos. RNA prism and RNA dendrimer have been used to protect the embedded RNA aptamer from degradation by ribonuclease (RNase) and protect the encapsulated drug from interaction with surrounding proteins, respectively [98,105]. More recently, reconfigurable RNA nanocubes with “open” and “closed” states were constructed which demonstrates the potential of developing RNA nanoparticles with changeable conformation in response to biological stimulus for advanced biomedical applications [106].
Fig. 5.
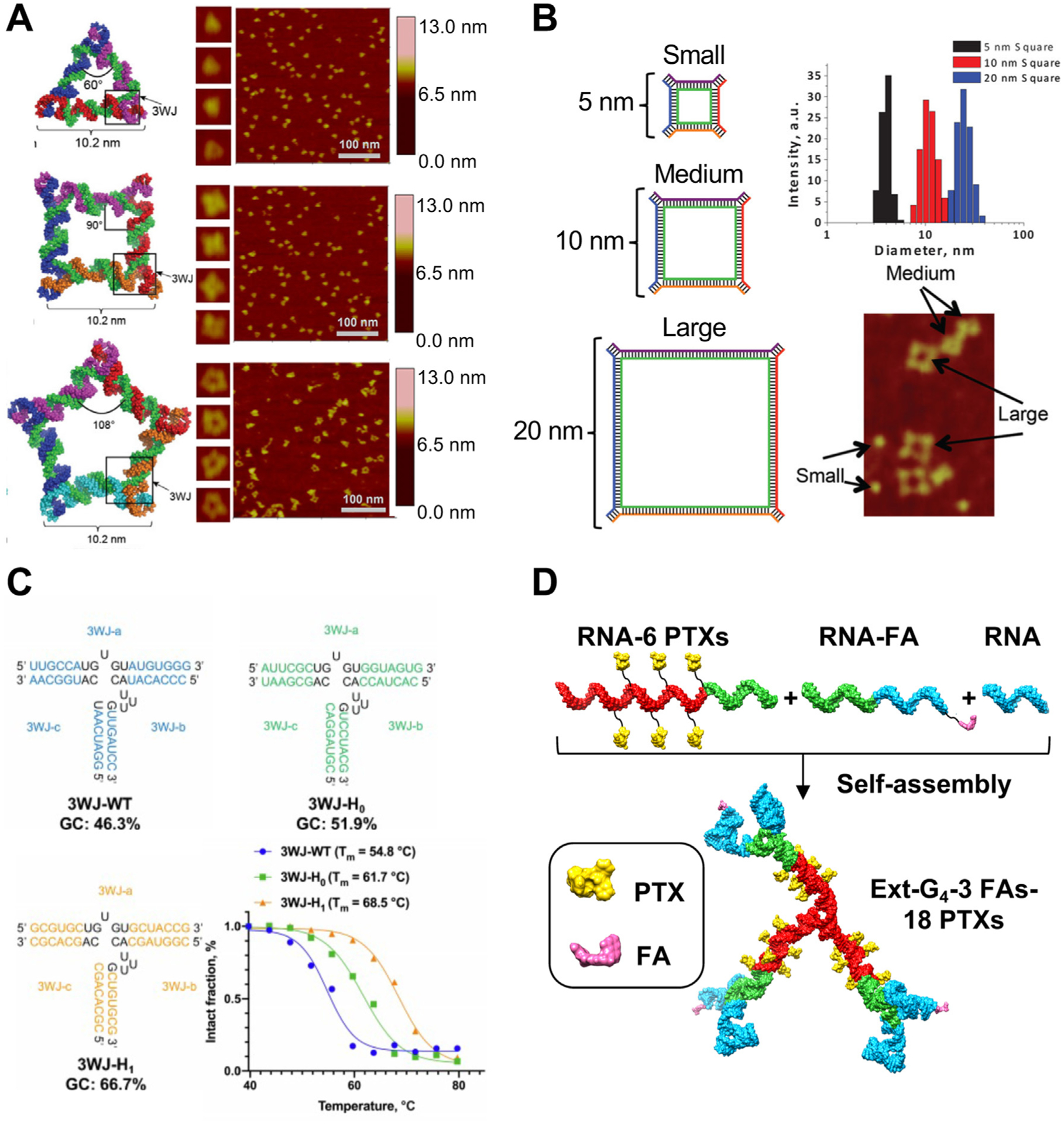
Programmability of RNA nanoparticles over (A) shape; (B) size; (C) stability; and (D) stoichiometry. (A) Schematic and AFM image of triangle, square, and pentagon RNA nanoparticles. Adapted with permission from [97]. Copyright 2014 Oxford University Press; (B) Schematic, size measurement, and AFM image of 5, 10, and 20 nm square RNA nanoparticles. Adapted with permission from [21]. Copyright 2014 American Chemical Society; (C) Secondary structure and melting profile of three 3WJs with different sequences. Adapted with permission from [105]. Copyright 2020 Royal Society of Chemistry; (D) Self-assembly of RNA dendrimers with 18 PTXs (yellow) and 3 FAs (magenta) using RNA strands with 6 PTXs, with 1 FA, and without modification. Reprinted with permission from [105]. Copyright 2020 Royal Society of Chemistry.
Besides shape and size, the thermal and enzymatic stability of RNA nanoparticles can also be controlled by changing the composition and sequence of composed RNA strands. Natural RNA molecules are sensitive to nuclease and are unstable in biological fluids, and this property leads to short half-life and unsatisfied therapeutic effect for unmodified RNA therapeutics [107]. Various kinds of chemical modification at the backbone, base, ribose have been generated and incorporated into RNA therapeutics and RNA nanoparticles to increase their stability and avoid the rapid degradation in serum [108]. Nucleotides with various base modifications, such as 2′-Fluoro (2′-F), 2′-O-methyl (2′-OMe), and 2′-O-methoxyethyl (2′-MOE) have been incorporated into RNA nanoparticles to improve their thermal and enzymatic stability and ensure their integrity for in vivo applications [109]. The most used modified phosphoramidites for the construction of RNA nanoparticles are 2′-F or 2′-OMe modified pyrimidines (2′-F/2′-OMe C and 2′-F/2′-OMe U) [110]. One approach to modulate thermal stability is to change the composition of RNA nanoparticles. Phi29 3WJs with the same sequence but different composition, including DNA, RNA, 2′-F RNA, and locked nucleic acid (LNA) showed different thermal stabilities which were measured by temperature gradient gel electrophoresis (TGGE) [111]. The Tm for 3WJDNA, 3WJRNA, 3WJ2′F RNA and 3WJLNA at 10μ M was 39.3 °C, 58.0 °C, 67.5 °C, and higher than 80 °C, respectively [111]. Furthermore, the sequence of RNA nanoparticles can also be adjusted to tune the stability of the nanoparticle. The thermal stability of 3WJs with the same composition but different sequences has also been measured by TGGE, which indicates higher guanine-cytosine (GC) contents leads to higher Tm. The Tm of three 2′-F 3WJs with 46.3%, 51.9%, and 66.7% GC content at 2.5μ M was 54.8 °C, 61.7 °C, and 68.5 °C, respectively (Fig. 5C) [105]. In addition to simple RNA structures such as 3WJ, the thermostability of sophisticated structures such as RNA dendrimers can also be controlled. RNA dendrimers utilizing 3WJs with different thermostability as the building block showed temperature-triggered layer-by-layer property, with the outer-layer dissociating at low temperature while the inner-layer remaining intact [105]. The pharmacokinetic (PK) profile of RNA nanoparticles is highly affected by their thermal and enzymatic stability. To improve the biodistribution of RNA nanoparticles, highly stable RNA nanoparticles have been developed by incorporating modified nucleotides and increasing GC percentage, which showed prolonged circulation time and retention at tumor site in mouse model [25].
Moreover, the multivalent RNA strands can be modified with therapeutic, targeting, and imaging groups at internal and terminal sites which enables high loading capacity and ease of multi-functionalization of RNA nanoparticles [105]. The amount of cargo, including drug, ligand, and dye, loaded into RNA nanoparticles can be precisely controlled, which is not achievable for other commonly used nanoparticles [31]. Small molecule drugs such as PTX and ligands such as folate (FA) can be covalently conjugated to RNA strands via click reaction, which are then self-assembled into nanoparticles [28,112,113]. RNA dendrimers encapsulating 18 copies of PTX inside and displaying 3 copies of FA outside have been constructed for targeted cancer treatment (Fig. 5D) [105]. As for therapeutic oligos and RNA aptamers, they are normally attached to the 3′ or 5′ end of scaffold RNA strands during the chemical solid-phase synthesis [35]. RNA nanoparticles with siRNA, miRNA, anti-miRNA have been constructed, respectively and showed promising tumor suppression effects [31,35,50]. Immunomodulators such as unmethylated cytosine-phosphate-guanine oligodeoxynucleotides (CpG ODN) and specific RNA sequences (SEQs) have also been attached to RNA nanoparticles which exhibited cytokine induction in both cell and mouse model [37,97]. Fluorescent dyes are normally attached to the terminal end of RNA strands either by using specific phosphoramidites during the synthesis of RNA strands or by conjugation reactions after the synthesis of RNA strands [114,115]. Fluorescent labeling allows for evaluating the stability, cellular trafficking and in vivo biodistribution behavior of RNA nanoparticles [32,104,105].
3.2. The rubbery property of RNA facilitates passive tumor accumulation
Unfavorable pharmacokinetic profile is a challenge for both small molecule drug and therapeutic oligos. Improvement of delivery to disease site is a logical approach to increase their therapeutic effects and decrease undesirable side effects. RNA nanoparticles have been designed as a delivery platform to facilitate drug to pass biological barriers and reach tumor environment by strong passive accumulation and active targeting. The passive accumulation of RNA nanoparticles at tumor site is contributed by two factors: the enhanced permeability and retention (EPR) effect of tumor and the rubbery, deformative, and amoeba property of RNA nanoparticles [116,117]. The EPR effect is caused by the fast growth of tumor tissue which requires extra blood vessels to provide nutrients and oxygen [118]. This causes tumor vasculature to have relatively large pores (0.1 – 3μ m in diameter) due to incomplete endothelial lining in comparison to normal blood vessels, which allows the extravasation of nanoparticles in tumor tissue [119]. Artificial RNA nanoparticles with chemical modifications, like natural RNA molecules, possess dynamic property which leads to conformational changes through motions such as base pair changes while remaining intact nanocomplexes. The dynamics of RNA nanoparticles contains the interconversion of conformations with large diameter and small diameter, and the conformational equilibrium is constantly changing after entering the blood circulation. When RNA nanoparticles reach tumor microenvironment with hypertonic blood vessel and abnormal blood flow velocity, their conformational population changes as the structure with smaller diameters penetrate the blood vessel and accumulate at tumor site [90]. It has been reported that, 10 nm and 20 nm RNA square nanoparticles both showed stronger accumulation and longer retention time in tumor compared with other vital organs at 12 and 24 hr post intravenous (IV) injection, which indicates the abnormal tumor vasculature and lymphatic drainage plays an important role in passive tumor accumulation [25]. Furthermore, 4WJ RNA nanoparticles showed strong tumor accumulation at 8 hr post IV injection, while more stiff iron and gold nanoparticles without deformability showed no or less tumor accumulation, which demonstrates the deformative property enhances passive tumor accumulation (Fig. 6A) [25]. The role of this dynamic structure plays in the hemodynamic profile of RNA nanoparticle have not been fully investigated. There is an increasing recognition of the importance of understanding the physiological effect of the vasculature temperature, composition, and pressure on the biodistribution of RNA nanoparticles. For example, the influence of the interaction of RNA nanoparticles with proteins in the blood circulation and other mechanisms that might contribute to the tumor accumulation such as endocytosis should be investigated [120].
Fig. 6.
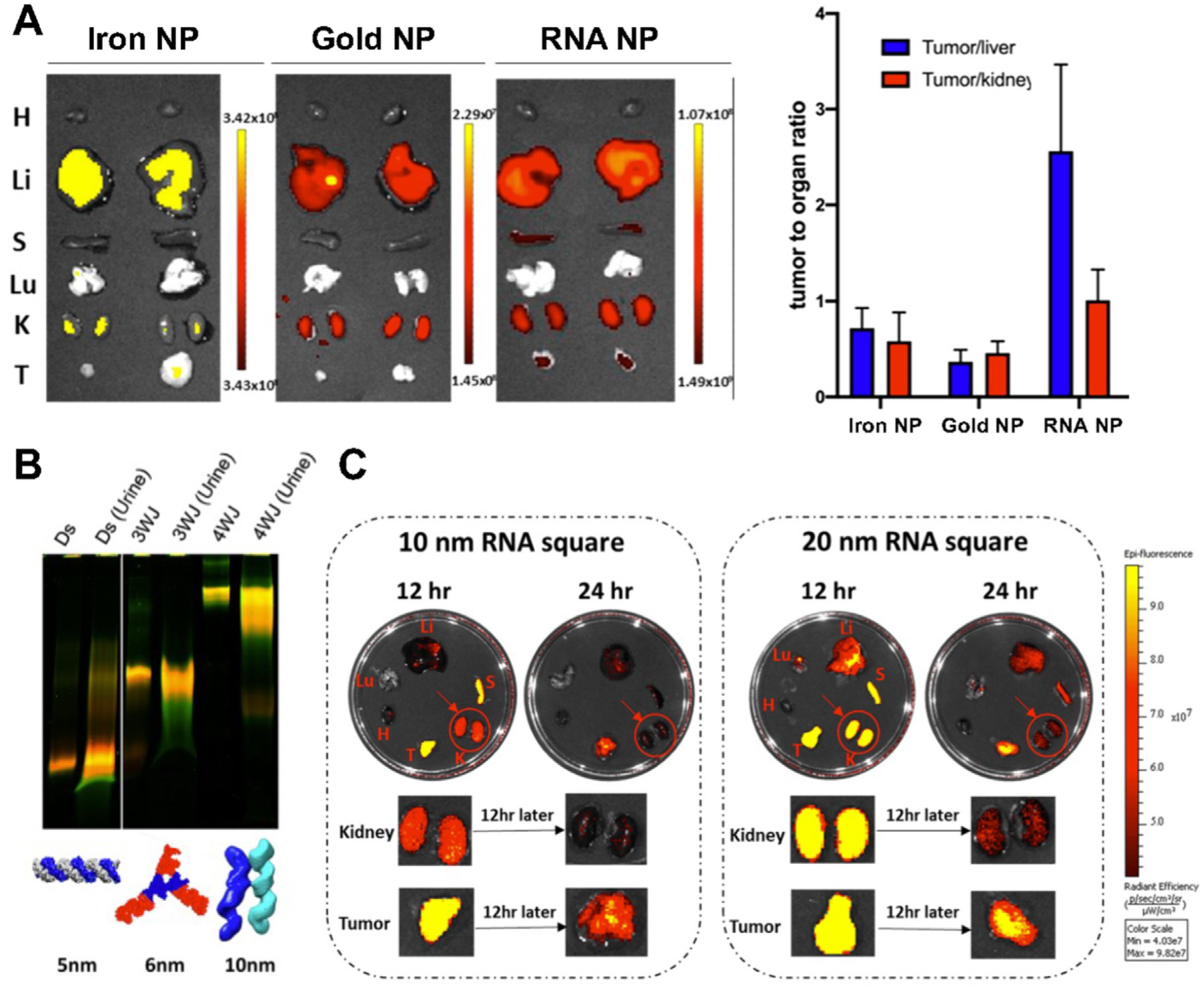
Deformative ability of RNA nanoparticles contributes to favorable biodistribution. (A) Biodistribution comparison of iron, gold, and RNA nanoparticles with IVIS images of tumors and organs, and the fluorescent quantification of tumor to organ (liver and kidney) ratio; (B) Gel image of urine samples at 0.5 hr post IV injection of dsRNA, 3WJ, and 4WJ RNA nanoparticles; (C) IVIS images of organs and tumors collected at 12 and 24 hr post IV injection of 10 nm and 20 nm RNA square nanoparticles. Adapted with permission from [25]. Copyright 2020 American Chemical Society.
3.3. The rubbery property of RNA enables passive renal excretion and quick body clearance
In addition to facilitating vasculature penetration, the deformative property can also help RNA nanoparticles to pass through renal excretion with similar mechanism. The limit of renal excretion is 5.5 nm, and proteins with the hydrodynamic diameter of ~5 – 6 nm can be cleared rapidly from the body by renal filtration and urinary excretion [121,122]. Studies have shown that dsRNA, 3WJ and 4WJ RNA nanoparticles with the size of 5, 6, and 10 nm were detected in urine samples at 0.5 h after IV injection (Fig. 6B) [25]. The excretion of intact 10 nm 4WJ RNA nanoparticles indicates the deformative property of RNA nanoparticles helps them to squeeze through the renal filtration. In another biodistribution study of RNA square with the size of 10 and 20 nm, the fluorescent intensity in kidney is significantly reduced from 12 hr to 24 hr post IV injection, which further confirms the fast renal clearance of deformative RNA nanoparticles. (Fig. 6C) [25]. The rapid renal excretion eliminates the concern of accumulation of RNA nanoparticles in vital organs which might lead to the long-term toxicity. The elements, such as size, shape, modification, that affect the biodistribution of RNA nanoparticles including tumor accumulation and renal filtration need to be investigated systematically like the immunogenicity evaluation by quantitative structure–activity relationship (QSAR) modeling [123].
3.4. Functionalization for active cancer targeting
The appropriate size and the deformative ability of RNA nanoparticles helps them to cross the leaky blood vessel and accumulate at tumor site. To be internalized into cancer cells, it requires the RNA nanoparticles to be functionalized with targeting groups such as small molecule ligands and RNA aptamers [124]. Certain epitopes or receptors have over- or exclusive-expression on cancer cells which has been used for active targeting using small molecule, antibody/peptide, and DNA/RNA aptamer [125–127]. Active targeting group allows RNA nanoparticles to specifically bind to cancer cells and minimize the interaction with normal cells thus decrease the non-specific toxicity [26,112]. This also has the potential to decrease the drug dosage and further reduce the side effects [31]. Small molecule targeting ligand, such as FA and N-Acetylgalactosamine (GalNAc), can be attached to RNA nanoparticles during the synthesis of RNA strands using special phosphoramidite or after the RNA synthesis via click reaction. FA has strong binding affinity to folate receptor which is overexpressed on the surface of various cancer cells, including ovarian, breast, colon, and lung cancer [128–130]. Phi29 3WJ-FA nanoparticle showed simultaneous targeting of colorectal cancer (CRC) cells in major sites of metastasis such as liver, lymph nodes, and lung, without accumulation in health cells in CRC metastasis mouse model (Fig. 7A, B, C) [112]. GalNAc, which binds to asialoglycoprotein receptor (ASGP-R) on hepatocyte, has been used intensively as the targeting ligand in the siRNA therapies for the treatment of liver diseases [131]. The conjugation of GalNAc to 6WJ RNA nanoparticle significantly increased its binding to ASGP-R overexpressing HepG2 cells in comparison to bare 6WJ RNA nanoparticles [31]. Furthermore, 6WJ-GalNAc RNA nanoparticle efficiently delivered both PTX and miR122 therapeutics to tumor site in hepatocellular carcinoma (HCC) xenograft mouse model [31]. In addition to small molecules, RNA aptamers can also be attached to RNA nanoparticles as the targeting group by extending the scaffold RNA strand with aptamer sequence during RNA synthesis. RNA aptamers are RNAs with defined structures that have high binding affinity to certain proteins such as receptors on cell membrane [132]. RNA aptamers have been shown to direct the nanoparticles into cells through endocytosis pathway [133,134]. RNA nanoparticles with the incorporation of prostate-specific membrane antigen (PSMA), epidermal growth factor (EGFR), and epithelial cell adhesion molecule (EpCAM) RNA aptamers for prostate cancer, TNBC, and CRC targeting have been developed [35,36,38]. Phi29 3WJ-PSMAapt RNA nanoparticles showed strong tumor accumulation in vivo in comparison to 3WJ without aptamer (Fig. 7D, E, F) [38]. And it was used to targeted deliver therapeutic anti-miRNA LNA in LNCaP C4-2 prostate cancer xenograft mouse model [38]. Currently, most active targeting RNA nanoparticles only possess one copy of ligand or aptamer, which showed good targeting effect to cancer cell both in vitro and in vivo. The targeting efficiency could be improved by manipulating the amount and position of ligand attached to RNA nanoparticles. Besides small molecule and RNA aptamer, antibodies and antibody fragments have been commonly attached to nanoparticles for active targeting [135]. Antibodies are Y-shaped glycoproteins which are used by the immune system to recognize and bind to antigens. Antibody have been conjugated with drugs as antibody-drug conjugates (ADCs) or with nanoparticles as antibody conjugated nanoparticles (ACNPs) [136,137]. So far, 11 ADCs have been approved by the FDA and the first antibody-oligonucleotide conjugate (AOC) entered clinically trial in 2021 [138]. The lessons and tricks learned from the development of ADCs and AOCs will contribute to the rational design of antibody conjugated RNA nanoparticles to facilitate the selective delivery of their entities.
Fig. 7.
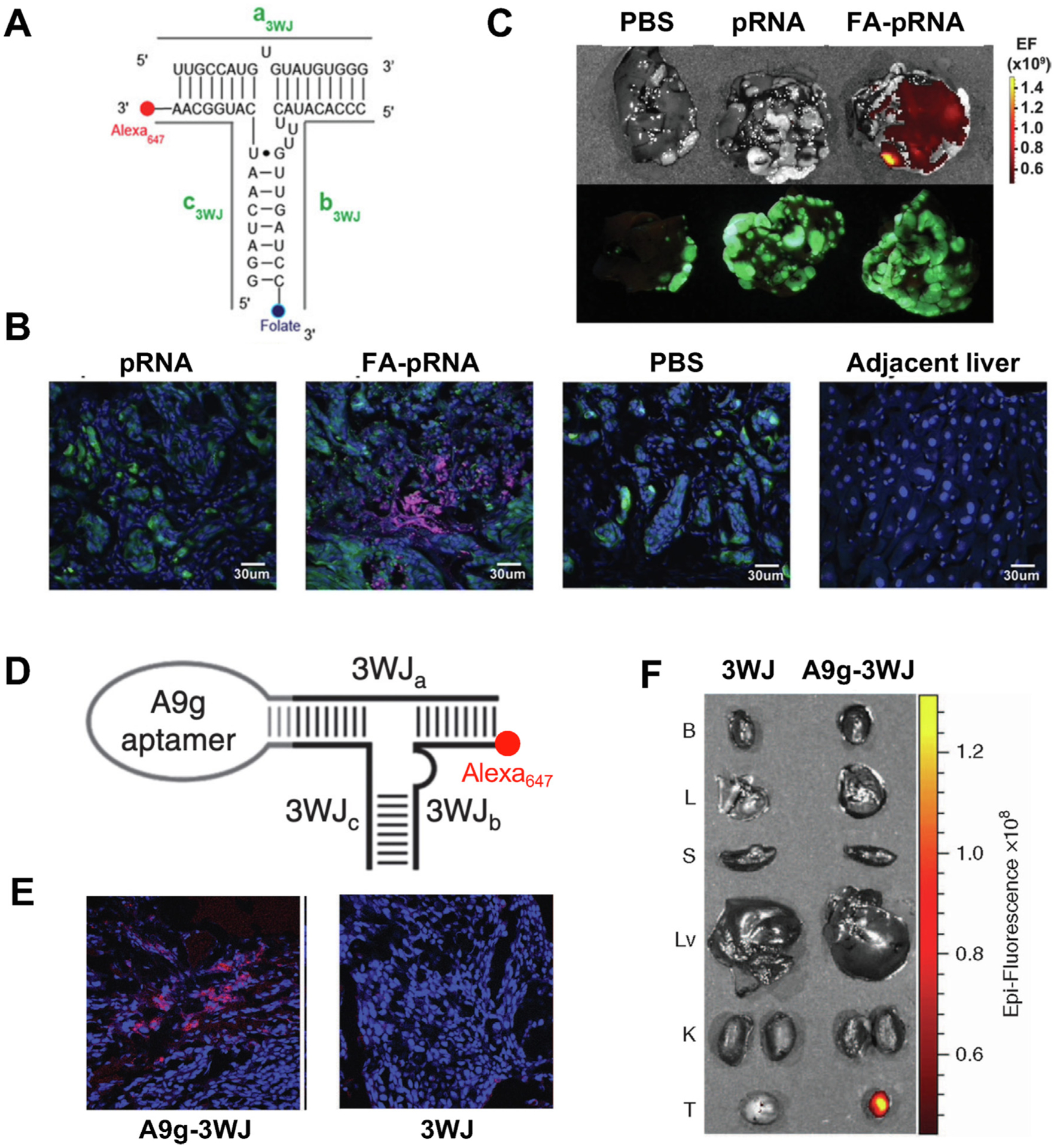
Active targeting of RNA nanoparticles with small molecule ligand (A, B, C) and RNA aptamer (D, E, F). (A) Design of Phi29 3WJ with Alexa647 fluorescent dye and FA targeting ligand; (B) Fluorescent confocal images of liver metastasis and adjacent healthy liver at 6 hr post IV injection of PBS and RNA nanoparticles. (Blue: DAPI; Green: GFP expressing cancer cells; Red: Alexa647); (C) Alexa (top panel) and GFP (bottom panel) fluorescent images of liver with metastasis at 6 hr post IV injection of PBS, pRNA and FA-pRNA. Adapted with permission from [112]. Copyright 2015 American Chemical Society; (D) Design of Phi29 3WJ with Alexa647 fluorescent dye and PSMA RNA aptamer; (E) Fluorescent confocal images of tumor samples at 8 hr post IV injection of RNA nanoparticles (Blue: DAPI; Red: Alexa647); (F) Biodistribution of A9g-3WJ and 3WJ in organs and tumors demonstrated by Alexa647 signal. Adapted with permission from [38]. Copyright 2016 American Society of Gene & Cell Therapy.
4. Advantages of RNA nanotechnology that facilitates drug development
4.1. Hydrophilic RNA enhances the solubility of hydrophobic drugs
The poor water solubility is a major issue for many hydrophobic anticancer drugs such as paclitaxel (PTX) and camptothecin (CPT) [139]. PTX is a tubulin targeting drug that has been widely used for cancer treatment [139,140]. However, it requires the usage of Cremophor EL, a formulation vehicle, for dissolution, which results in side effects such as hypersensitive reactions [141,142]. CPT is a topoisomerase inhibitor, and its clinical application is limited because of its low water solubility [143]. RNA nanotechnology offers a new strategy to solve this solubility issue utilizing its highly hydrophilic property. RNA nanoparticles loaded with multiple copies of anticancer drugs, such as 3WJ-7-CPT and 4WJ-24-PTX, have been constructed [28,29]. The preparation of RNA nanoparticles attached with hydrophobic drugs is achieved by synthesizing RNA strands with multiple alkyne modifications followed by conjugation with prodrug, such as PTX-azide and CPT-azide, via click reaction. After the conjugation to the RNA strands, the water solubility of hydrophobic anticancer drugs was significantly increased by 1,000-fold for CPT from 2.7μ g/mL to 2.8 mg/mL and 32,000-fold for PTX from 0.4μ g/mL to 12.8 mg/mL (Fig. 8A, B) [28,29]. This can not only solve the formulation problem, but also eliminate some side effects such as unfavorable immune response. In vivo studies of the production of cytokines and chemokines demonstrated the decreased immunostimulation for RNA-PTX nanoparticles dissolved in aqueous buffer compared to free PTX dissolved in Cremophor EL/ethanol [28]. Studies have shown that 40% of the drug candidates in the developmental stages have low or inconsistent bioavailability due to the poor water solubility which leads to unsatisfactory therapeutic response [144]. RNA nanoparticle provides a universal platform for the solubilization and delivery of these hydrophobic drugs. Besides solving the formulation issue, RNA nanoparticles could stabilize the drug, prolong the circulation, and improve the PK profile, which has the potential to reduce the side effects by lower the drug dosage. Studies showed that RNA nanoparticles loaded with drug showed tumor inhibition effect without causing toxicity to vital organs for short-term treatment [28]. More studies are required to investigate the acute and chronic toxicity of RNA-drug nanoparticle treatment and compare with the toxicity caused by free drug at the same dosage and/or with the same therapeutic outcome.
Fig. 8.
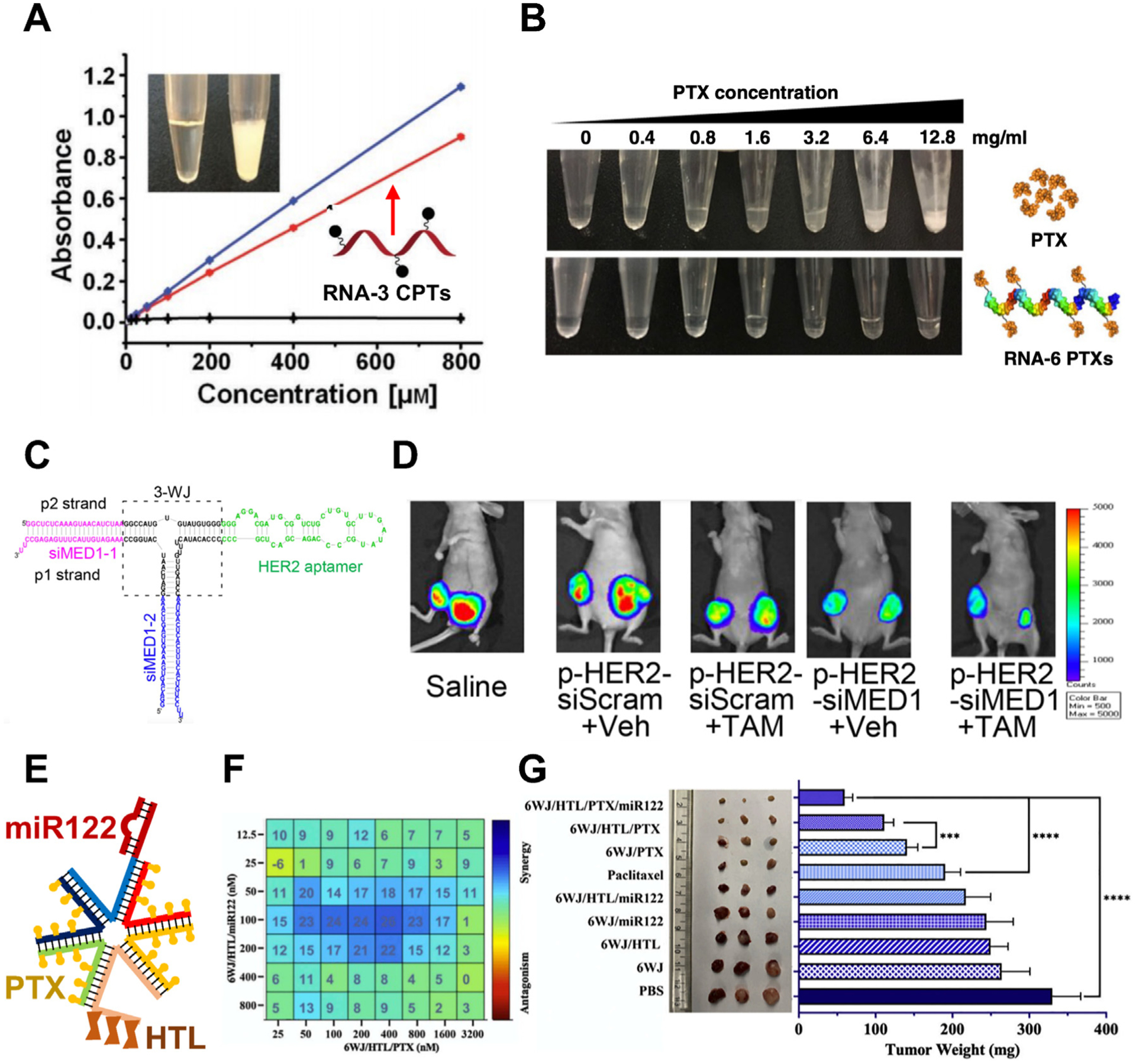
RNA nanoparticles improve water solubility of drug and overcome drug resistance. RNA increases water solubility of CPT (A) and PTX (B) Adapted with permission from [28,29]. Copyright 2019 The Authors and 2020 The Author(s); (C) Design of multifunctional 3WJ RNA nanoparticle with MED siRNA and HER2 aptamer; (D) IVIS lumina images of tumors in mice bearing breast tumor after treatment. Adapted with permission from [30]. Copyright 2016 American Chemical Society; (E) Design of multifunctional 6WJ RNA nanoparticle with PTX, miR122, and HTL; (F) Synergistic cytotoxic effect of PTX and miR122 using MTT assay and HSA synergy model; (G) Image and quantification of tumors harvest from mice bearing HCC tumor after treatment. Adapted with permission from [31]. Copyright 2020 Elsevier B.V.
4.2. Overcome drug resistance utilizing multivalent property
Another clinical challenge of anticancer drugs is the drug resistance, which is related to 90% of failures in the chemotherapy during the invasion and metastasis of cancers. There are several mechanisms of drug resistance in the cancer cells including decreased drug uptake, increased drug efflux, altered drug metabolism, enhanced DNA repair, etc [145]. The multivalent property of RNA molecules enables the convenient multi-functionalization of RNA nanoparticles. Multiple copies of different therapeutic agents could be loaded into RNA nanoparticles simultaneously. Each nucleotide of RNA nanoparticles is a potential position for the conjugation of small molecule drugs. And each strand of RNA nanoparticles offers two terminal ends for the extension of functional RNA sequences, such as siRNA, miRNA, and RNA aptamers. Phi29 3WJ attached with mediator subunit (MED1) siRNAs and human epidermal growth factor receptor 2 (HER2) RNA aptamer was designed for the treatment of estrogen receptor α (ER α) positive breast cancer with endocrine therapy resistance (Fig. 8C) [30]. 3WJ-HER2apt-siMED was showed to overcome the tamoxifen (TAM) resistance in xenograft mouse model by silencing the ER coactivator and targeting HER2 overexpressing cancer cells (Fig. 8D) [30]. A more recent study reported the development of 6WJ RNA nanoparticles attached with 24 copies of PTX, one copy of miR122, and one copy of GalNAc for the treatment of HCC (Fig. 8E) [31]. miR122 is highly expressed in the liver and could efficiently downregulate oncogenic proteins such as a disintegrin and metalloproteinase domain-containing protein 10 (ADAM10) and drug exporters such as P-glycoproteins (P-gp) [146]. The combination of PTX and miR122 leads to enhanced toxicity to HCC cells due to their synergistic cytotoxicity effect and drug efflux inhibition (Fig. 8F) [31]. In addition, GalNAc increases the targeting and accumulation of PTX and miR122 to tumor site. All these factors led to the best tumor suppression outcome in HCC xenograft mouse model compared to PTX only, miR122 only, and other control groups (Fig. 8G) [31]. Using RNA nanoparticles for the co-delivery of two or multiple types of drugs gives advantage over the traditional combination chemotherapy. RNA nanoparticles allow combined drugs with different PK parameters to reach cancer cells simultaneously with the defined ratio resulting in the desired synergistic effect [147]. Furthermore, RNA nanoparticles attached with 4-1BB aptamer showed T cell activation and antitumor function which demonstrates their potential to achieve the combination of chemotherapy and immunotherapy [148–150].
4.3. Deliver therapeutic oligos and facilitate endosome escape
Small interfering RNAs (siRNAs) are short double-stranded RNAs that cause targeted gene suppression by binding to targeted mRNA sequences and leading to the cleavage and degradation of mRNA [151]. Currently, three siRNA drugs, patisiran, givosiran, and lumasiran, for different liver diseases have been approved by FDA and more siRNA drugs for the treatment of eye, skin, kidney, liver, and cancer diseases are under clinical stage [152]. This shows the great potential of utilizing siRNA for a wide range of clinical applications. However, there are many challenges for the efficient siRNA delivery [153]. One is the lack of efficient delivery to the disease side after systemic administration, and another one is the endosome trapping, which limits the amount of siRNA delivered to the targeted site in cytosol [154]. To solve the delivery obstacle, siRNA has been loaded into extracellular vesicles (EVs) that are decorated with RNA nanoparticles on the surface for targeted delivery [155]. The decorative RNA nanoparticles contain one cholesterol for anchoring on EVs and one targeting group such as folate to guide EVs to cancer cells [32,33]. This strategy utilizes the decorative RNA nanoparticles for targeting to tumor cells and the EVs for fusing with cell membrane and avoiding endosome trapping, thus achieving the optimized delivery of siRNA. Ligand and aptamer displaying EVs loaded with survivin siRNA showed promises for the treatment of colon, prostate, breast, and lung cancer [33,43]. FA displaying EVs loaded with survivin siRNA decreased the tumor growth rate in patient-derived colorectal cancer xenograft (PDX-CRC) mouse model [33]. In addition to small molecule ligand, RNA aptamers such as PSMA aptamer (PMSAapt) and EGFR aptamer (EGFRapt), have also been displayed on the surface of EVs for targeted delivery of siRNA, respectively (Fig. 9A) [43]. EGFRapt displaying EVs loaded with survivin siRNA showed specific tumor accumulation and significant tumor inhibition effect in both breast cancer and lung cancer xenograft mouse model (Fig. 9B) [43].
Fig. 9.
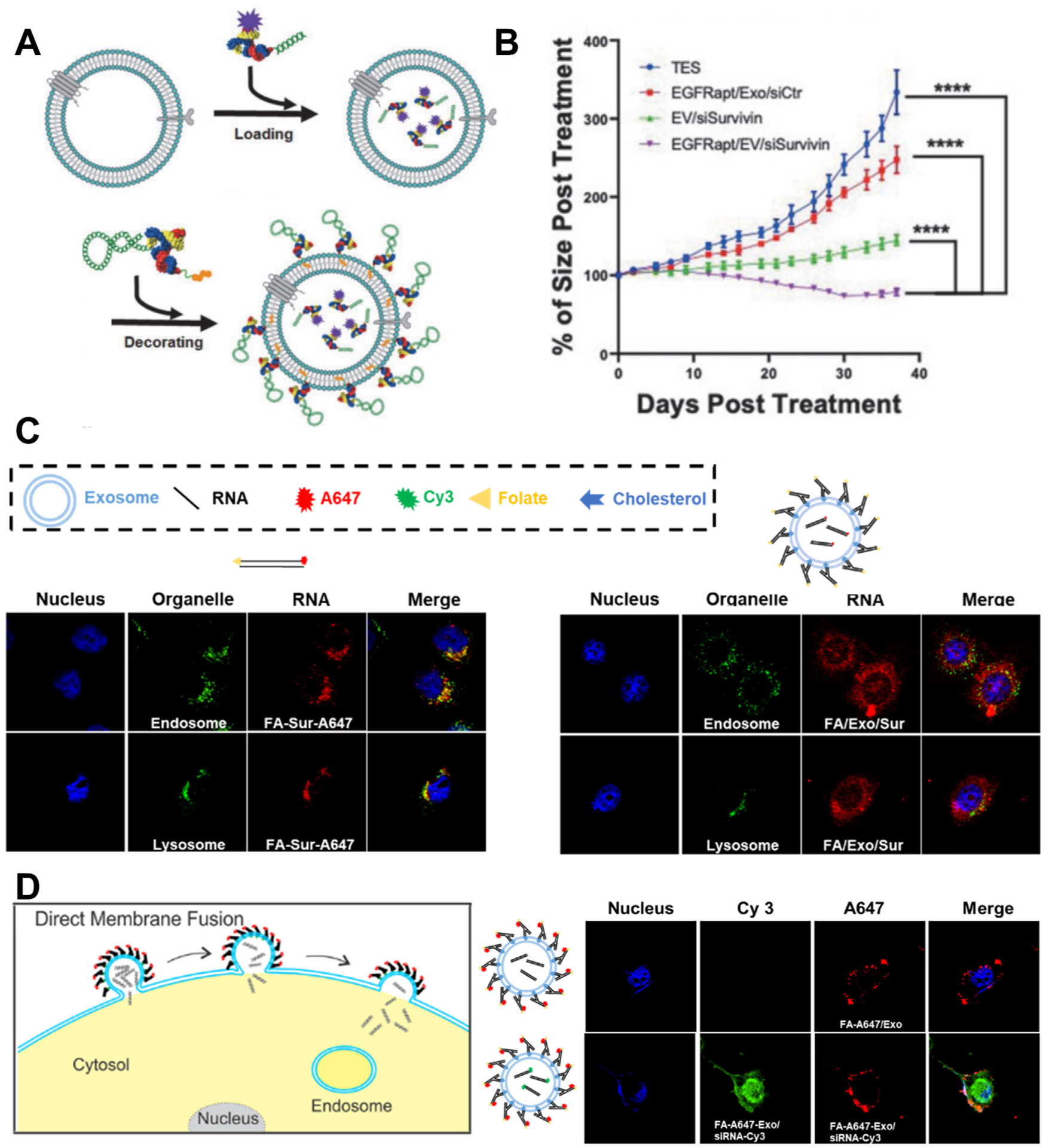
EVs decorated with RNA nanoparticles increase delivery efficiency and therapeutic effect of siRNA. (A) Schematic of EVs loaded with 3WJ-siRNA and decorated with 3WJ-EGFRapt; (B) Tumor regression curve of mice bearing NSCLC tumor after treatment. Adapted with permission from [43]. Copyright 2021 Mary Ann Liebert, Inc.; (C) Confocal images of cellular internalization of siRNA and siRNA loaded in RNA decorated EVs (Blue: DAPI; Green: endosome/lysosome; Red: RNA); (D) Schematic of direct membrane fusion mechanism of endosome escape pathway and cellular distribution of fluorescent 3WJ RNA and siRNA after cellular internalization of EVs (Blue: DAPI; Green: Cy3 labeled siRNA; Red: A647 labeled 3WJ RNA). Adapted with permission from [32]. Copyright 2020 Elsevier B.V.
More recently, the mechanism of how the combination of RNA nontechnology and EVs achieves the efficient therapeutic effect of siRNA has been elucidated. The intracellular behavior of siRNA and FA/EV/siRNA has been studied by confocal microscopy and colocalization analysis [32]. Free siRNA showed colocalization with endosome and lysosome while FA/EV/siRNA showed even distribution in the cytosol without specific accumulation in endosome or lysosome (Fig. 9C) [32]. More detailed confocal studies using EVs decorated with fluorescent labeled 3WJ-FA RNA nanoparticles showed this efficient cytosolic delivery of siRNA is likely mediated by the direct membrane fusion mechanism (Fig. 7D) [32]. Furthermore, FA/EV/siRNA showed increased tumor suppression effect compared to siRNA and EV/siRNA in KB cell derived cancer xenograft mouse model [32]. These results indicated that ligand displaying EVs could targeted deliver siRNA payload to tumor cell by receptor-mediated cytosolic delivery without endosome trapping, which leads to the improved therapeutic effect. So far, the RNA nanoparticle decorated exosomes have only been used for the targeted delivery of siRNA. Due to the relative large size and high drug loading capacity of EV, it can also be used to deliver large nucleic acid therapeutics such as mRNA and CRISPR/Cas 9 system for disease treatment [156,157].
5. Conclusion and perspectives
As more and more functional RNAs are discovered, they draw a lot of attention for disease treatment and pharmaceutical development, as the disease target, as the therapeutic agent, and as the delivery platform. Various RNA therapeutics such as ASO, siRNA, mRNA, and RNA aptamer have been approved for treatment of diseases containing genetic diseases such as spinal muscular atrophy and infectious diseases such as COVID-19 (Table 2). In addition to being used as the drug, RNA is also a great material to construct RNA nanoparticles for biomedical applications due to their unique properties such as deformability, programmability, and multi-functionality. Different methods such as bottom-up assembly, RNA origami, and RCT have been used to construct RNA nanoparticles with various size and shape. Despite the major achievements in predicting the structure and dynamic of biological RNA molecules, the modeling of the 3D structure RNA nanoparticles with multistrand, modification, and complicated structure as well as the simulation of their interaction with other molecules still have a long way to go.
Table 2.
FDA-approved therapeutic RNA.
| RNA type | Name | Target | Disease type | Company | Approved year |
|---|---|---|---|---|---|
| ASO | Nusinersen | Survival of motor neuron gene | Spinal muscular atrophy | Ionis | 2016 |
| Eteplirsen | Exon 51 of dystrophin pre-mRNA | Duchenne muscular dystrophy | Sarepta | 2016 | |
| Inotersen | 3′ untranslated region of TTR mRNA | Hereditary transthyretin-mediated amyloidosis | Ionis | 2018 | |
| Volanesoren | mRNA for apo-CIII | Familial chylomicronemia syndrome | Ionis | 2019 | |
| Golodirsen | Exon 53 of dystrophin pre-mRNA | Duchenne muscular dystrophy | Sarepta | 2019 | |
| siRNA | Parisiran | Transthyretin | Polyneuropathy caused by hATTR | Alnylam | 2018 |
| Givosiran | ALAS1 | Acute hepatic porphyria | Alnylam | 2019 | |
| Lumasiran | HAO1 mRNA | Primary hyperoxaluria type 1 | Alnylam | 2020 | |
| mRNA | BNT162b2 | Spike protein of SARS-CoV-2 | COVID-19 | BioNTech & Pfizer | 2020 |
| mRNA-1273 | Spike protein of SARS-CoV-2 | COVID-19 | Moderna | 2020 | |
| RNA aptamer | Pegatanib | VEGF-165 | Neovascular age-related macular degeneration | Bausch & Lomb | 2014 |
RNA nanoparticles have been functionalized with small molecule drug, oligonucleotide drug, fluorescent dye, targeting ligand, RNA aptamer, and immunomodulator for diagnosis and treatment of diseases. RNA nanoparticles have been shown to facilitate drug development by enhancing the therapeutic effect and minimizing the toxicity for both small molecule and nucleic acid drugs. For the incorporation of small molecule drugs to RNA nanoparticles, more linkers should be explored to expand the applicable drugs for conjugation. With attachment of different types of drugs to RNA becoming possible, the combinational chemotherapy can be easily achieved by the assembly of RNA nanoparticles using RNA strands with different drug conjugation. Furthermore, the combination of chemotherapy and immunotherapy can be achieved by RNA nanoparticles to improve the therapeutic outcome. Immunotherapeutic agents such as RNA vaccine, immune check point inhibitor and chimeric antigen receptor (CAR) T cell, are used for activating the immune system to attack cancer cells [158]. In addition to 4-1BB RNA aptamers, the possibility of attaching small molecule, aptamer, and antibody to RNA nanoparticles to inhibit immune checkpoint antagonist such as programmed death-1 (PD-1) and cytotoxic T-lymphocyte antigen-4 (CTLA-4) should be evaluated [159–161]. Currently, RNA nanoparticles are mainly designed for cancer treatment, but this platform is highly alterable and is proven for other medical usage such as anti-coagulation [48].
Significant progress has been made in the construction, functionalization, and application of RNA nanoparticles, but substantial work remains to make them a clinical reality. One challenge is the cost-efficient large scale production of RNA nanoparticles with high purity [162]. Long RNA strands are mainly produced by in vitro transcription, and this process is significantly improved due to the mRNA vaccine requirement during the pandemic. Short RNA strands are normally produced by solid-phase chemical synthesis using phosphoramidite, which has significantly improved over the past decade by increasing the nucleotide incorporation efficiency and the yield of production. This in turn resulted in the synthesis of RNA oligos up to 120 bases in length. Additionally, fermentation for the industrial scale production by using implantation of plasmids into bacteria can be used. Large-scale RNA complexes produced in bacteria escorted by a tRNA vector or by RCT has been reported [163]. To reduce the cost of storage and transportation of RNA nanoparticles, different preparation methods such as lyophilization and light-assisted drying have been studied [164].
Building a standard characterization protocol for the evaluation of RNA nanoparticles is also essential for their clinical usage. Although many studies have been done to evaluate the biodistribution of RNA nanoparticles using fluorescent and radioisotope labeling, more systematic and detailed studies are required to determine the influence of various parameters such as size, shape, and cargo on the PK parameters of RNA nanoparticles [26]. Multiple animal trials have verified the therapeutic effect of RNA nanoparticles in terms of suppressing tumor growth, however standard pharmacodynamic (PD) studies, such as dose response study as well as acute and chronic toxic studies, are lacking. The PK/PD profile of the drug loaded in RNA nanoparticles also needs to be evaluated and compared with the free drug to demonstrate the advantage of using RNA nanoparticles as drug delivery platform. As for the immunogenicity, the effect of contributors such as molecular weight (MW) and Tm on the immune response activities of nucleic acid nanoparticles has been studied and QSAR model has been built based on these results, but more RNA nanostructures with more diverse parameters are needed to refine the model [123]. In addition to the phenomenological study, more mechanism studies should be conducted to demonstrate the cellular uptake pathway, cellular trafficking and localization as well as tumor accumulation routes of RNA nanoparticles.
In conclusion, RNA nanoparticles demonstrated its great potential for pharmaceutic application, more systematic evaluation of RNA nanoparticles will provide more insights of the rational design and directions of their optimized usage.
Acknowledgements
The work was partially supported by NIH grant U01CA207946 to P.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation.
Footnotes
Declaration of Competing Interest
The authors declare the following competing financial interests: P.G. is the consultant, licensor, and grantee of Oxford Nanopore Technologies; as well as the cofounder, chairman of the board of directors, and president of the ExonanoRNA, LLC.
References
- [1].Winkle M, El-Daly SM, Fabbri M, Calin GA, Noncoding RNA therapeutics - challenges and potential solutions, Nat Rev Drug Discov 20 (2021) 629–651, 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Losko M, Kotlinowski J, Jura J, Long Noncoding RNAs in Metabolic Syndrome Related Disorders, Mediators Inflamm 2016 (2016) 5365209, 10.1155/2016/5365209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Roberts TC, Langer R, Wood MJA, Advances in oligonucleotide drug delivery, Nat Rev Drug Discov 19 (2020) 673–694, 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kristen AV, Ajroud-Driss S, Conceicao I, Gorevic P, Kyriakides T, Obici L, Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis, Neurodegener, Dis Manag 9 (2019) 5–23, 10.2217/nmt-2018-0033. [DOI] [PubMed] [Google Scholar]
- [5].Baranello G, Darras BT, Day JW, Deconinck N, Klein A, Masson R, Mercuri E, Rose K, El-Khairi M, Gerber M, Gorni K, Khwaja O, Kletzl H, Scalco RS, Seabrook T, Fontoura P, Servais L, Group FW, Risdiplam in Type 1 Spinal Muscular Atrophy, N Engl J Med 384 (2021) 915–923, 10.1056/NEJMoa2009965. [DOI] [PubMed] [Google Scholar]
- [6].Warner KD, Hajdin CE, Weeks KM, Principles for targeting RNA with drug-like small molecules, Nat Rev Drug Discov 17 (2018) 547–558, 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Falese JP, Donlic A, Hargrove AE, Targeting RNA with small molecules: from fundamental principles towards the clinic, Chem Soc Rev 50 (2021) 2224–2243, 10.1039/d0cs01261k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jasinski D, Haque F, Binzel DW, Guo P, Advancement of the Emerging Field of RNA Nanotechnology, ACS Nano 11 (2017) 1142–1164, 10.1021/acsnano.6b05737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Binzel DW, Li X, Burns N, Khan E, Lee WJ, Chen LC, Ellipilli S, Miles W, Ho YS, Guo P, Thermostability, Tunability, and Tenacity of RNA as Rubbery Anionic Polymeric Materials in Nanotechnology and Nanomedicine-Specific Cancer Targeting with Undetectable Toxicity, Chem Rev 121 (2021) 7398–7467, 10.1021/acs.chemrev.1c00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dhuri K, Bechtold C, Quijano E, Pham H, Gupta A, Vikram A, Bahal R, Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development, J Clin Med 9 (2020), 10.3390/jcm9062004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hou XC, Zaks T, Langer R, Dong YZ, Lipid nanoparticles for mRNA delivery, Nat Rev Mater 6 (2021) 1078–1094, 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang F, Zuroske T, Watts JK, RNA therapeutics on the rise, Nat Rev Drug Discov 19 (2020) 441–442, 10.1038/d41573-020-00078-0. [DOI] [PubMed] [Google Scholar]
- [13].Zwieb C, The principles of RNA structure architecture, Methods Mol Biol 1097 (2014) 33–43, 10.1007/978-1-62703-709-9_2. [DOI] [PubMed] [Google Scholar]
- [14].Ganser LR, Kelly ML, Herschlag D, Al-Hashimi HM, The roles of structural dynamics in the cellular functions of RNAs, Nat Rev Mol Cell Biol 20 (2019) 474–489, 10.1038/s41580-019-0136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mustoe AM, Brooks CL, Al-Hashimi HM, Hierarchy of RNA functional dynamics, Annu Rev Biochem 83 (2014) 441–466, 10.1146/annurev-biochem-060713-035524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Al-Hashimi HM, Walter NG, RNA dynamics: it is about time, Curr Opin Struct Biol 18 (2008) 321–329, 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Geary C, Chworos A, Verzemnieks E, Voss NR, Jaeger L, Composing RNA Nanostructures from a Syntax of RNA Structural Modules, Nano Lett 17 (2017) 7095–7101, 10.1021/acs.nanolett.7b03842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L, In vitro assembly of cubic RNA-based scaffolds designed in silico, Nat Nanotechnol 5 (2010) 676–682, 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yu J, Liu Z, Jiang W, Wang G, Mao C, De novo design of an RNA tile that self-assembles into a homo-octameric nanoprism, Nat Commun 6 (2015) 5724, 10.1038/ncomms6724. [DOI] [PubMed] [Google Scholar]
- [20].Severcan I, Geary C, Verzemnieks E, Chworos A, Jaeger L, Square-shaped RNA particles from different RNA folds, Nano Lett 9 (2009) 1270–1277, 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jasinski DL, Khisamutdinov EF, Lyubchenko YL, Guo P, Physicochemically tunable polyfunctionalized RNA square architecture with fluorogenic and ribozymatic properties, ACS Nano 8 (2014) 7620–7629, 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT, Self-assembled RNA interference microsponges for efficient siRNA delivery, Nat Mater 11 (2012) 316–322, 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Geary C, Grossi G, McRae EKS, Rothemund PWK, Andersen ES, RNA origami design tools enable cotranscriptional folding of kilobase-sized nanoscaffolds, Nat Chem 13 (2021) 549–558, 10.1038/s41557-021-00679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Geary C, Rothemund PW, Andersen ES, A single-stranded architecture for cotranscriptional folding of RNA nanostructures, Science 345 (2014) 799–804, 10.1126/science.1253920. [DOI] [PubMed] [Google Scholar]
- [25].Ghimire C, Wang H, Li H, Vieweger M, Xu C, Guo P, RNA Nanoparticles as Rubber for Compelling Vessel Extravasation to Enhance Tumor Targeting and for Fast Renal Excretion to Reduce Toxicity, ACS Nano 14 (2020) 13180–13191, 10.1021/acsnano.0c04863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang H, Guo P, Radiolabeled RNA Nanoparticles for Highly Specific Targeting and Efficient Tumor Accumulation with Favorable In Vivo Biodistribution, Mol Pharm 18 (2021) 2924–2934, 10.1021/acs.molpharmaceut.1c00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Guo S, Xu C, Yin H, Hill J, Pi F, Guo P, Tuning the size, shape and structure of RNA nanoparticles for favorable cancer targeting and immunostimulation, Wiley Interdiscip Rev Nanomed Nanobiotechnol 12 (2020), 10.1002/wnan.1582 e1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Guo S, Vieweger M, Zhang K, Yin H, Wang H, Li X, Li S, Hu S, Sparreboom A, Evers BM, Dong Y, Chiu W, Guo P, Ultra-thermostable RNA nanoparticles for solubilizing and high-yield loading of paclitaxel for breast cancer therapy, Nat Commun 11 (2020) 972, 10.1038/s41467-020-14780-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Piao X, Yin H, Guo S, Wang H, Guo P, RNA Nanotechnology to Solubilize Hydrophobic Antitumor Drug for Targeted Delivery, Adv Sci (Weinh) 6 (2019) 1900951, 10.1002/advs.201900951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang Y, Leonard M, Shu Y, Yang Y, Shu D, Guo P, Zhang X, Overcoming Tamoxifen Resistance of Human Breast Cancer by Targeted Gene Silencing Using Multifunctional pRNA Nanoparticles, ACS Nano 11 (2017) 335–346, 10.1021/acsnano.6b05910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang H, Ellipilli S, Lee WJ, Li X, Vieweger M, Ho YS, Guo P, Multivalent rubber-like RNA nanoparticles for targeted co-delivery of paclitaxel and MiRNA to silence the drug efflux transporter and liver cancer drug resistance, J Control Release 330 (2021) 173–184, 10.1016/j.jconrel.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zheng Z, Li Z, Xu C, Guo B, Guo P, Folate-displaying exosome mediated cytosolic delivery of siRNA avoiding endosome trapping, J Control Release 311–312 (2019) 43–49, 10.1016/j.jconrel.2019.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P, Li H, Haque F, Wang S, Croce CM, Guo B, Evers BM, Guo P, Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression, Nat Nanotechnol 13 (2018) 82–89, 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pi F, Zhang H, Li H, Thiviyanathan V, Gorenstein DG, Sood AK, Guo P, RNA nanoparticles harboring annexin A2 aptamer can target ovarian cancer for tumor-specific doxorubicin delivery, Nanomedicine 13 (2017) 1183–1193, 10.1016/j.nano.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shu D, Li H, Shu Y, Xiong G, Carson WE 3rd, Haque F, Xu R, Guo P, Systemic Delivery of Anti-miRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology, ACS Nano 9 (2015) 9731–9740, 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xu Y, Pang L, Wang H, Xu C, Shah H, Guo P, Shu D, Qian SY, Specific delivery of delta-5-desaturase siRNA via RNA nanoparticles supplemented with dihomo-gamma-linolenic acid for colon cancer suppression, Redox Biol 21 (2019), 10.1016/j.redox.2018.101085 101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Guo S, Li H, Ma M, Fu J, Dong Y, Guo P, Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles, Mol Ther Nucleic Acids, 9 (2017) 399–408. 10.1016/j.omtn.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Binzel DW, Shu Y, Li H, Sun M, Zhang Q, Shu D, Guo B, Guo P, Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology, Mol Ther 24 (2016) 1267–1277, 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Feng L, Li SK, Liu H, Liu CY, LaSance K, Haque F, Shu D, Guo P, Ocular delivery of pRNA nanoparticles: distribution and clearance after subconjunctival injection, Pharm Res 31 (2014) 1046–1058, 10.1007/s11095-013-1226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shi Z, Li SK, Charoenputtakun P, Liu CY, Jasinski D, Guo P, RNA nanoparticle distribution and clearance in the eye after subconjunctival injection with and without thermosensitive hydrogels, J Control Release 270 (2018) 14–22, 10.1016/j.jconrel.2017.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Smith JA, Braga A, Verheyen J, Basilico S, Bandiera S, Alfaro-Cervello C, Peruzzotti-Jametti L, Shu D, Haque F, Guo P, Pluchino S, RNA Nanotherapeutics for the Amelioration of Astroglial Reactivity, Mol Ther Nucleic Acids 10 (2018) 103–121, 10.1016/j.omtn.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cui D, Zhang C, Liu B, Shu Y, Du T, Shu D, Wang K, Dai F, Liu Y, Li C, Pan F, Yang Y, Ni J, Li H, Brand-Saberi B, Guo P, Regression of Gastric Cancer by Systemic Injection of RNA Nanoparticles Carrying both Ligand and siRNA, Sci Rep 5 (2015) 10726, 10.1038/srep10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li Z, Yang L, Wang H, Binzel DW, Williams TM, Guo P, Non-Small-Cell Lung Cancer Regression by siRNA Delivered Through Exosomes That Display EGFR RNA Aptamer, Nucleic Acid Ther 31 (2021) 364–374, 10.1089/nat.2021.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ryoo NK, Lee J, Lee H, Hong HK, Kim H, Lee JB, Woo SJ, Park KH, Kim H, Therapeutic effects of a novel siRNA-based anti-VEGF (siVEGF) nanoball for the treatment of choroidal neovascularization, Nanoscale 9 (2017) 15461–15469, 10.1039/c7nr03142d. [DOI] [PubMed] [Google Scholar]
- [45].Lee JH, Ku SH, Kim MJ, Lee SJ, Kim HC, Kim K, Kim SH, Kwon IC, Rolling circle transcription-based polymeric siRNA nanoparticles for tumor-targeted delivery, J Control Release 263 (2017) 29–38, 10.1016/j.jconrel.2017.03.390. [DOI] [PubMed] [Google Scholar]
- [46].Kim H, Lee YK, Han KH, Jeon H, Jeong IH, Kim SY, Lee JB, Lee PCW, BRC-mediated RNAi targeting of USE1 inhibits tumor growth in vitro and in vivo, Biomaterials 230 (2020), 10.1016/j.biomaterials.2019.119630 119630. [DOI] [PubMed] [Google Scholar]
- [47].Yin H, Xiong G, Guo S, Xu C, Xu R, Guo P, Shu D, Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133, Mol Ther 27 (2019) 1252–1261, 10.1016/j.ymthe.2019.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Krissanaprasit A, Key CM, Froehlich K, Pontula S, Mihalko E, Dupont DM, Andersen ES, Kjems J, Brown AC, LaBean TH, Multivalent Aptamer-Functionalized Single-Strand RNA Origami as Effective, Target-Specific Anticoagulants with Corresponding Reversal Agents, Adv Healthc Mater 10 (2021), 10.1002/adhm.202001826 e2001826. [DOI] [PubMed] [Google Scholar]
- [49].Yang X, Xu Y, Wang T, Shu D, Guo P, Miskimins K, Qian SY, Inhibition of cancer migration and invasion by knocking down delta-5-desaturase in COX-2 overexpressed cancer cells, Redox Biol 11 (2017) 653–662, 10.1016/j.redox.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pang L, Shah H, Wang H, Shu D, Qian SY, Sathish V, EpCAM-Targeted 3WJ RNA Nanoparticle Harboring Delta-5-Desaturase siRNA Inhibited Lung Tumor Formation via DGLA Peroxidation, Mol Ther Nucleic Acids 22 (2020) 222–235, 10.1016/j.omtn.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Isel C, Ehresmann C, Marquet R, Initiation of HIV Reverse Transcription, Viruses 2 (2010) 213–243, 10.3390/v2010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Benas P, Bec G, Keith G, Marquet R, Ehresmann C, Ehresmann B, Dumas P, The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop, RNA 6 (2000) 1347–1355, 10.1017/s1355838200000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Turner DH, Mathews DH, NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure, Nucleic Acids Res 38 (2010) D280–D282, 10.1093/nar/gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Singh J, Hanson J, Paliwal K, Zhou Y, RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning, Nat Commun 10 (2019) 5407, 10.1038/s41467-019-13395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Batey RT, Rambo RP, Doudna JA, Tertiary Motifs in RNA Structure and Folding, Angew Chem Int Ed Engl 38 (1999) 2326–2343, . [DOI] [PubMed] [Google Scholar]
- [56].Butcher SE, Pyle AM, The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks, Acc Chem Res 44 (2011) 1302–1311, 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- [57].Saeed A, Chan C, Guan H, Gong B, Guo P, Cheng X, Ouyang S, Structural Insights into gp16 ATPase in the Bacteriophage varphi29 DNA Packaging Motor, Biochemistry 60 (2021) 886–897, 10.1021/acs.biochem.0c00935. [DOI] [PubMed] [Google Scholar]
- [58].Jones CP, Ferre-D’Amare AR, RNA quaternary structure and global symmetry, Trends Biochem Sci 40 (2015) 211–220, 10.1016/j.tibs.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM, Functional complexity and regulation through RNA dynamics, Nature 482 (2012) 322–330, 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hansen AL, Al-Hashimi HM, Dynamics of large elongated RNA by NMR carbon relaxation, J Am Chem Soc 129 (2007) 16072–16082, 10.1021/ja0757982. [DOI] [PubMed] [Google Scholar]
- [61].Sun X, Zhang Q, Al-Hashimi HM, Resolving fast and slow motions in the internal loop containing stem-loop 1 of HIV-1 that are modulated by Mg2+ binding: role in the kissing-duplex structural transition, Nucleic Acids Res 35 (2007) 1698–1713, 10.1093/nar/gkm020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM, Visualizing spatially correlated dynamics that directs RNA conformational transitions, Nature 450 (2007) 1263–1267, 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- [63].Zhang Q, Sun X, Watt ED, Al-Hashimi HM, Resolving the motional modes that code for RNA adaptation, Science 311 (2006) 653–656, 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- [64].Ferner J, Villa A, Duchardt E, Widjajakusuma E, Wohnert J, Stock G, Schwalbe H, NMR and MD studies of the temperature-dependent dynamics of RNA YNMG-tetraloops, Nucleic Acids Res 36 (2008) 1928–1940, 10.1093/nar/gkm1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ilin S, Hoskins A, Ohlenschlager O, Jonker HR, Schwalbe H, Wohnert J, Domain reorientation and induced fit upon RNA binding: solution structure and dynamics of ribosomal protein L11 from Thermotoga maritima, Chembiochem 6 (2005) 1611–1618, 10.1002/cbic.200500091. [DOI] [PubMed] [Google Scholar]
- [66].Ding J, Swain M, Yu P, Stagno JR, Wang YX, Conformational flexibility of adenine riboswitch aptamer in apo and bound states using NMR and an X-ray free electron laser, J Biomol NMR 73 (2019) 509–518, 10.1007/s10858-019-00278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Spitale RC, Wedekind JE, Exploring ribozyme conformational changes with X-ray crystallography, Methods 49 (2009) 87–100, 10.1016/j.ymeth.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kim H, Abeysirigunawarden SC, Chen K, Mayerle M, Ragunathan K, Luthey-Schulten Z, Ha T, Woodson SA, Protein-guided RNA dynamics during early ribosome assembly, Nature 506 (2014) 334–338, 10.1038/nature13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Robb NC, Te Velthuis AJ, Wieneke R, Tampe R, Cordes T, Fodor E, Kapanidis AN, Single-molecule FRET reveals the pre-initiation and initiation conformations of influenza virus promoter RNA, Nucleic Acids Res 44 (2016) 10304–10315, 10.1093/nar/gkw884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Voith von Voithenberg L, Sanchez-Rico C, Kang HS, Madl T, Zanier K, Barth A, Warner LR, Sattler M, Lamb DC, Recognition of the 3’ splice site RNA by the U2AF heterodimer involves a dynamic population shift, Proc Natl Acad Sci U S A 113 (2016) E7169–E7175, 10.1073/pnas.1605873113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lee D, Walsh JD, Yu P, Markus MA, Choli-Papadopoulou T, Schwieters CD, Krueger S, Draper DE, Wang YX, The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes, J Mol Biol 367 (2007) 1007–1022, 10.1016/j.jmb.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mitra K, Frank J, Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps, Annu Rev Biophys Biomol Struct 35 (2006) 299–317, 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]
- [73].Li B, Cao Y, Westhof E, Miao Z, Advances in RNA 3D Structure Modeling Using Experimental Data, Front Genet 11 (2020), 10.3389/fgene.2020.574485 574485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Sponer J, Bussi G, Krepl M, Banas P, Bottaro S, Cunha RA, Gil-Ley A, Pinamonti G, Poblete S, Jurecka P, Walter NG, Otyepka M, RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview, Chem Rev 118 (2018) 4177–4338, 10.1021/acs.chemrev.7b00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Gao Y, Shi Y, Fu M, Feng Y, Lin G, Kong D, Jiang B, Simulation study of the effects of interstitial fluid pressure and blood flow velocity on transvascular transport of nanoparticles in tumor microenvironment, Comput Methods Programs Biomed 193 (2020), 10.1016/j.cmpb.2020.105493 105493. [DOI] [PubMed] [Google Scholar]
- [76].Vangaveti S, Ranganathan SV, Chen AA, Advances in RNA molecular dynamics: a simulator’s guide to RNA force fields, Wiley Interdiscip Rev RNA 8 (2017), 10.1002/wrna.1396. [DOI] [PubMed] [Google Scholar]
- [77].Shu D, Shu Y, Haque F, Abdelmawla S, Guo P, Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics, Nat Nanotechnol 6 (2011) 658–667, 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Shu D, Khisamutdinov EF, Zhang L, Guo P, Programmable folding of fusion RNA in vivo and in vitro driven by pRNA 3WJ motif of phi29 DNA packaging motor, Nucleic Acids Res 42 (2014), 10.1093/nar/gkt885 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Giedroc DP, Cornish PV, Frameshifting RNA pseudoknots: structure and mechanism, Virus Res 139 (2009) 193–208, 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Cao S, Chen SJ, Predicting RNA pseudoknot folding thermodynamics, Nucleic Acids Res 34 (2006) 2634–2652, 10.1093/nar/gkl346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lim NC, Jackson SE, Molecular knots in biology and chemistry, J Phys Condens Matter 27 (2015), 10.1088/0953-8984/27/35/354101 354101. [DOI] [PubMed] [Google Scholar]
- [82].Brierley I, Gilbert RJ, Pennell S, RNA pseudoknots and the regulation of protein synthesis, Biochem Soc Trans 36 (2008) 684–689, 10.1042/BST0360684. [DOI] [PubMed] [Google Scholar]
- [83].Fei J, Ha T, Watching DNA breath one molecule at a time, Proc Natl Acad Sci U S A 110 (2013) 17173–17174, 10.1073/pnas.1316493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sutthibutpong T, Matek C, Benham C, Slade GG, Noy A, Laughton C, JP KD, Louis AA, Harris SA, Long-range correlations in the mechanics of small DNA circles under topological stress revealed by multi-scale simulation, Nucleic Acids Res, 44 (2016) 9121–9130. 10.1093/nar/gkw815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].von Hippel PH, Johnson NP, Marcus AH, Fifty years of DNA “breathing”: Reflections on old and new approaches, Biopolymers 99 (2013) 923–954, 10.1002/bip.22347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Seif E, Hallberg BM, RNA-protein mutually induced fit: structure of Escherichia coli isopentenyl-tRNA transferase in complex with tRNA(Phe), J Biol Chem 284 (2009) 6600–6604, 10.1074/jbc.C800235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Liberman JA, Wedekind JE, Riboswitch structure in the ligand-free state, Wiley Interdiscip Rev, RNA 3 (2012) 369–384, 10.1002/wrna.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kurisaki I, Takayanagi M, Nagaoka M, Combined mechanism of conformational selection and induced fit in U1A-RNA molecular recognition, Biochemistry 53 (2014) 3646–3657, 10.1021/bi401708q. [DOI] [PubMed] [Google Scholar]
- [89].Rode AB, Endoh T, Sugimoto N, Crowding Shifts the FMN Recognition Mechanism of Riboswitch Aptamer from Conformational Selection to Induced Fit, Angew Chem Int Ed Engl 57 (2018) 6868–6872, 10.1002/anie.201803052. [DOI] [PubMed] [Google Scholar]
- [90].Yu AM, Gasper PM, Cheng L, Lai LB, Kaur S, Gopalan V, Chen AA, Lucks JB, Computationally reconstructing cotranscriptional RNA folding from experimental data reveals rearrangement of non-native folding intermediates, Mol Cell, 81 (2021) 870–883 e810. 10.1016/j.molcel.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Zhao L, Xia T, Direct revelation of multiple conformations in RNA by femtosecond dynamics, J Am Chem Soc 129 (2007) 4118–4119, 10.1021/ja068391q. [DOI] [PubMed] [Google Scholar]
- [92].Hall KB, RNA in motion, Curr Opin Chem Biol 12 (2008) 612–618, 10.1016/j.cbpa.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR, Genetic control by a metabolite binding mRNA, Chem Biol 9 (2002) 1043, 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- [94].Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J, Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch, Nucleic Acids Res 35 (2007) 572–583, 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Ottink OM, Rampersad SM, Tessari M, Zaman GJ, Heus HA, Wijmenga SS, Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism, RNA 13 (2007) 2202–2212, 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Duss O, Stepanyuk GA, Puglisi JD, Williamson JR, Transient Protein-RNA Interactions Guide Nascent Ribosomal RNA Folding, Cell, 179 (2019) 1357–1369 e1316. 10.1016/j.cell.2019.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Khisamutdinov EF, Li H, Jasinski DL, Chen J, Fu J, Guo P, Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles, Nucleic Acids Res 42 (2014) 9996–10004, 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Khisamutdinov EF, Jasinski DL, Li H, Zhang K, Chiu W, Guo P, Fabrication of RNA 3D Nanoprisms for Loading and Protection of Small RNAs and Model Drugs, Adv Mater 28 (2016) 10079–10087, 10.1002/adma.201603180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Xu C, Zhang K, Yin H, Li Z, Krasnoslobodtsev A, Zheng Z, Ji Z, Guo S, Li S, Chiu W, Guo P, 3D RNA nanocage for encapsulation and shielding of hydrophobic biomolecules to improve the in vivo biodistribution, Nano Res 13 (2020) 3241–3247, 10.1007/s12274-020-2996-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Sharma A, Haque F, Pi F, Shlyakhtenko LS, Evers BM, Guo P, Controllable self-assembly of RNA dendrimers, Nanomedicine 12 (2016) 835–844, 10.1016/j.nano.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Li H, Zhang K, Binzel DW, Shlyakhtenko LS, Lyubchenko YL, Chiu W, Guo P, RNA nanotechnology to build a dodecahedral genome of single-stranded RNA virus, RNA Biol 18 (2021) 2390–2400, 10.1080/15476286.2021.1915620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Han D, Qi X, Myhrvold C, Wang B, Dai M, Jiang S, Bates M, Liu Y, An B, Zhang F, Yan H, Yin P, Single-stranded DNA and RNA origami, Science 358 (2017), 10.1126/science.aao2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Kim H, Kim D, Jeong J, Jeon H, Lee JB, Size-Controllable Enzymatic Synthesis of Short Hairpin RNA Nanoparticles by Controlling the Rate of RNA Polymerization, Polymers (Basel) 10 (2018), 10.3390/polym10060589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Jasinski DL, Li H, Guo P, The Effect of Size and Shape of RNA Nanoparticles on Biodistribution, Mol Ther 26 (2018) 784–792, 10.1016/j.ymthe.2017.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Li X, Vieweger M, Guo P, Self-assembly of four generations of RNA dendrimers for drug shielding with controllable layer-by-layer release, Nanoscale 12 (2020) 16514–16525, 10.1039/d0nr02614j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Blanco Carcache PJ, Guo S, Li H, Zhang K, Xu C, Chiu W, Guo P, Regulation of reversible conformational change, size switching, and immunomodulation of RNA nanocubes, RNA 27 (2021) 971–980, 10.1261/rna.078718.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Paunovska K, Loughrey D, Dahlman JE, Drug delivery systems for RNA therapeutics, Nat Rev Genet (2022), 10.1038/s41576-021-00439-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wan WB, Seth PP, The Medicinal Chemistry of Therapeutic Oligonucleotides, J Med Chem 59 (2016) 9645–9667, 10.1021/acs.jmedchem.6b00551. [DOI] [PubMed] [Google Scholar]
- [109].Khvorova A, Watts JK, The chemical evolution of oligonucleotide therapies of clinical utility, Nat Biotechnol 35 (2017) 238–248, 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Patra A, Paolillo M, Charisse K, Manoharan M, Rozners E, Egli M, 2’-Fluoro RNA shows increased Watson-Crick H-bonding strength and stacking relative to RNA: evidence from NMR and thermodynamic data, Angew Chem Int Ed Engl 51 (2012) 11863–11866, 10.1002/anie.201204946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Piao X, Wang H, Binzel DW, Guo P, Assessment and comparison of thermal stability of phosphorothioate-DNA, DNA, RNA, 2’-F RNA, and LNA in the context of Phi29 pRNA 3WJ, RNA 24 (2018) 67–76, 10.1261/rna.063057.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Rychahou P, Haque F, Shu Y, Zaytseva Y, Weiss HL, Lee EY, Mustain W, Valentino J, Guo P, Evers BM, Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration, ACS Nano 9 (2015) 1108–1116, 10.1021/acsnano.5b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Shu Y, Yin H, Rajabi M, Li H, Vieweger M, Guo S, Shu D, Guo P, RNA-based micelles: A novel platform for paclitaxel loading and delivery, J Control Release 276 (2018) 17–29, 10.1016/j.jconrel.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Jasinski DL, Yin H, Li Z, Guo P, Hydrophobic Effect from Conjugated Chemicals or Drugs on In Vivo Biodistribution of RNA Nanoparticles, Hum Gene Ther 29 (2018) 77–86, 10.1089/hum.2017.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Binzel DW, Guo S, Yin H, Lee TJ, Liu S, Shu D, Guo P, Rational design for controlled release of Dicer-substrate siRNA harbored in phi29 pRNA-based nanoparticles, Mol Ther Nucleic Acids 25 (2021) 524–535, 10.1016/j.omtn.2021.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Shi J, Kantoff PW, Wooster R, Farokhzad OC, Cancer nanomedicine: progress, challenges and opportunities, Nat Rev Cancer 17 (2017) 20–37, 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Maeda H, Toward a full understanding of the EPR effect in primary and metastatic tumors as well as issues related to its heterogeneity, Adv Drug Deliv Rev 91 (2015) 3–6, 10.1016/j.addr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- [118].Fang J, Nakamura H, Maeda H, The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect, Adv Drug Deliv Rev 63 (2011) 136–151, 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- [119].Nakamura Y, Mochida A, Choyke PL, Kobayashi H, Nanodrug Delivery: Is the Enhanced Permeability and Retention Effect Sufficient for Curing Cancer?, Bioconjug Chem 27 (2016) 2225–2238, https://doiorg/10.1021/acs.bioconjchem.6b00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sindhwani S, Syed AM, Ngai J, Kingston BR, Maiorino L, Rothschild J, MacMillan P, Zhang Y, Rajesh NU, Hoang T, Wu JLY, Wilhelm S, Zilman A, Gadde S, Sulaiman A, Ouyang B, Lin Z, Wang L, Egeblad M, Chan WCW, The entry of nanoparticles into solid tumours, Nat Mater 19 (2020) 566–575, 10.1038/s41563-019-0566-2. [DOI] [PubMed] [Google Scholar]
- [121].Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, Bawendi MG, Frangioni JV, Renal clearance of quantum dots, Nat Biotechnol 25 (2007) 1165–1170, 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Du BJ, Yu MX, Zheng J, Transport and interactions of nanoparticles in the kidneys, Nat Rev Mater 3 (2018) 358–374, 10.1038/s41578-018-0038-3. [DOI] [Google Scholar]
- [123].Johnson MB, Halman JR, Satterwhite E, Zakharov AV, Bui MN, Benkato K, Goldsworthy V, Kim T, Hong E, Dobrovolskaia MA, Khisamutdinov EF, Marriott I, Afonin KA, Programmable Nucleic Acid Based Polygons with Controlled Neuroimmunomodulatory Properties for Predictive QSAR Modeling, Small 13 (2017), 10.1002/smll.201701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC, Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology, Adv Drug Deliv Rev 66 (2014) 2–25, 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Gonzalez-Conchas GA, Rodriguez-Romo L, Hernandez-Barajas D, Gonzalez-Guerrero JF, Rodriguez-Fernandez IA, Verdines-Perez A, Templeton AJ, Ocana A, Seruga B, Tannock IF, Amir E, Vera-Badillo FE, Epidermal growth factor receptor overexpression and outcomes in early breast cancer: A systematic review and a meta-analysis, Cancer Treat Rev 62 (2018) 1–8, 10.1016/j.ctrv.2017.10.008. [DOI] [PubMed] [Google Scholar]
- [126].Torchilin VP, Multifunctional, stimuli-sensitive nanoparticulate systems for drug delivery, Nat Rev Drug Discov 13 (2014) 813–827, 10.1038/nrd4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Srinivasarao M, Galliford CV, Low PS, Principles in the design of ligand-targeted cancer therapeutics and imaging agents, Nat Rev Drug Discov 14 (2015) 203–219, 10.1038/nrd4519. [DOI] [PubMed] [Google Scholar]
- [128].Lu Y, Low PS, Folate-mediated delivery of macromolecular anticancer therapeutic agents, Adv Drug Deliv Rev 54 (2002) 675–693, 10.1016/s0169-409x(02)00042-x. [DOI] [PubMed] [Google Scholar]
- [129].Scaranti M, Cojocaru E, Banerjee S, Banerji U, Exploiting the folate receptor alpha in oncology, Nat Rev Clin Oncol 17 (2020) 349–359, 10.1038/s41571-020-0339-5. [DOI] [PubMed] [Google Scholar]
- [130].Sudimack J, Lee RJ, Targeted drug delivery via the folate receptor, Adv Drug Deliv Rev 41 (2000) 147–162, 10.1016/s0169-409x(99)00062-9. [DOI] [PubMed] [Google Scholar]
- [131].Springer AD, Dowdy SF, GalNAc-siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics, Nucleic Acid Ther 28 (2018) 109–118, 10.1089/nat.2018.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Panigaj M, Johnson MB, Ke W, McMillan J, Goncharova EA, Chandler M, Afonin KA, Aptamers as Modular Components of Therapeutic Nucleic Acid Nanotechnology, ACS Nano 13 (2019) 12301–12321, 10.1021/acsnano.9b06522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Zhou J, Rossi JJ, Cell-type-specific, Aptamer-functionalized Agents for Targeted Disease Therapy, Mol Ther Nucleic Acids 3 (2014), 10.1038/mtna.2014.21 e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Porciani D, Cardwell LN, Tawiah KD, Alam KK, Lange MJ, Daniels MA, Burke DH, Modular cell-internalizing aptamer nanostructure enables targeted delivery of large functional RNAs in cancer cell lines, Nat Commun 9 (2018) 2283, 10.1038/s41467-018-04691-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Richards DA, Maruani A, Chudasama V, Antibody fragments as nanoparticle targeting ligands: a step in the right direction, Chem Sci 8 (2017) 63–77, 10.1039/c6sc02403c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R, Engineering precision nanoparticles for drug delivery, Nat Rev Drug Discov 20 (2021) 101–124, 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Beck A, Goetsch L, Dumontet C, Corvaia N, Strategies and challenges for the next generation of antibody-drug conjugates, Nat Rev Drug Discov 16 (2017) 315–337, 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- [138].Mullard A, Antibody-oligonucleotide conjugates enter the clinic, Nat Rev Drug Discov 21 (2022) 6–8, 10.1038/d41573-021-00213-5. [DOI] [PubMed] [Google Scholar]
- [139].Singh S, Dash AK, Paclitaxel in cancer treatment: perspectives and prospects of its delivery challenges, Crit Rev Ther Drug Carrier Syst 26 (2009) 333–372, 10.1615/critrevtherdrugcarriersyst.v26.i4.10. [DOI] [PubMed] [Google Scholar]
- [140].Rowinsky EK, Donehower RC, Paclitaxel (taxol), N Engl J Med 332 (1995) 1004–1014, 10.1056/NEJM199504133321507. [DOI] [PubMed] [Google Scholar]
- [141].Singla AK, Garg A, Aggarwal D, Paclitaxel and its formulations, Int J Pharm 235 (2002) 179–192, 10.1016/s0378-5173(01)00986-3. [DOI] [PubMed] [Google Scholar]
- [142].Gelderblom H, Verweij J, Nooter K, Sparreboom A, Cremophor EL: the drawbacks and advantages of vehicle selection for drug formulation, Eur J Cancer 37 (2001) 1590–1598, 10.1016/s0959-8049(01)00171-x. [DOI] [PubMed] [Google Scholar]
- [143].Pommier Y, Topoisomerase I inhibitors: camptothecins and beyond, Nat Rev Cancer 6 (2006) 789–802, 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- [144].Kalepu S, Nekkanti V, Improved delivery of poorly soluble compounds using nanoparticle technology: a review, Drug Deliv, Transl Res 6 (2016) 319–332, 10.1007/s13346-016-0283-1. [DOI] [PubMed] [Google Scholar]
- [145].Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B, The Different Mechanisms of Cancer Drug Resistance: A Brief Review, Adv Pharm Bull 7 (2017) 339–348, 10.15171/apb.2017.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, Liu J, Cui Y, Bian X, Bie P, Qian C, MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest, Cancer Lett 310 (2011) 160–169, 10.1016/j.canlet.2011.06.027. [DOI] [PubMed] [Google Scholar]
- [147].Liang X, Gao C, Cui L, Wang S, Wang J, Dai Z, Self-Assembly of an Amphiphilic Janus Camptothecin-Floxuridine Conjugate into Liposome-Like Nanocapsules for More Efficacious Combination Chemotherapy in Cancer, Adv Mater 29 (2017), 10.1002/adma.201703135. [DOI] [PubMed] [Google Scholar]
- [148].Emens LA, Middleton G, The interplay of immunotherapy and chemotherapy: harnessing potential synergies, Cancer, Immunol Res 3 (2015) 436–443, 10.1158/2326-6066.CIR-15-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Mahoney KM, Rennert PD, Freeman GJ, Combination cancer immunotherapy and new immunomodulatory targets, Nat Rev Drug Discov 14 (2015) 561–584, 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- [150].Shu D, Zhang L, Bai X, Yu J, Guo P, Stoichiometry of multi-specific immune checkpoint RNA Abs for T cell activation and tumor inhibition using ultra-stable RNA nanoparticles, Mol Ther Nucleic Acids 24 (2021) 426–435, 10.1016/j.omtn.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, Liang XJ, Therapeutic siRNA: state of the art, Signal Transduct Target Ther 5 (2020) 101, 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Zhang MM, Bahal R, Rasmussen TP, Manautou JE, Zhong XB, The growth of siRNA-based therapeutics: Updated clinical studies, Biochem Pharmacol 189 (2021), 10.1016/j.bcp.2021.114432 114432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Juliano RL, The delivery of therapeutic oligonucleotides, Nucleic Acids Res 44 (2016) 6518–6548, 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Dowdy SF, Overcoming cellular barriers for RNA therapeutics, Nat Biotechnol 35 (2017) 222–229, 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- [155].Maas SLN, Breakefield XO, Weaver AM, Extracellular Vesicles: Unique Intercellular Delivery Vehicles, Trends Cell Biol 27 (2017) 172–188, 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO, RNA delivery by extracellular vesicles in mammalian cells and its applications, Nat Rev Mol Cell Biol 21 (2020) 585–606, 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Kim SM, Yang Y, Oh SJ, Hong Y, Seo M, Jang M, Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting, J Control Release 266 (2017) 8–16, 10.1016/j.jconrel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- [158].Riley RS, June CH, Langer R, Mitchell MJ, Delivery technologies for cancer immunotherapy, Nat Rev Drug Discov 18 (2019) 175–196, 10.1038/s41573-018-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Wu Q, Jiang L, Li SC, He QJ, Yang B, Cao J, Small molecule inhibitors targeting the PD-1/PD-L1 signaling pathway, Acta Pharmacol Sin 42 (2021) 1–9, 10.1038/s41401-020-0366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Pastor F, Berraondo P, Etxeberria I, Frederick J, Sahin U, Gilboa E, Melero I, An RNA toolbox for cancer immunotherapy, Nat Rev Drug Discov 17 (2018) 751–767, 10.1038/nrd.2018.132. [DOI] [PubMed] [Google Scholar]
- [161].Darvin P, Toor SM, Sasidharan Nair V, Elkord E, Immune checkpoint inhibitors: recent progress and potential biomarkers, Exp Mol Med 50 (2018) 1–11, 10.1038/s12276-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Chandler M, Panigaj M, Rolband LA, Afonin KA, Challenges to optimizing RNA nanostructures for large scale production and controlled therapeutic properties, Nanomedicine (Lond) (2020), 10.2217/nnm-2020-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Jasinski DL, Binzel DW, Guo P, One-Pot Production of RNA Nanoparticles via Automated Processing and Self-Assembly, ACS Nano 13 (2019) 4603–4612, 10.1021/acsnano.9b00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Tran AN, Chandler M, Halman J, Beasock D, Fessler A, McKeough RQ, Lam PA, Furr DP, Wang J, Cedrone E, Dobrovolskaia MA, Dokholyan NV, Trammell SR, Afonin KA, Anhydrous Nucleic Acid Nanoparticles for Storage and Handling at Broad Range of Temperatures, Small (2022) e2104814, 10.1002/smll.202104814. [DOI] [PMC free article] [PubMed] [Google Scholar]


