Abstract
Diffusion magnetic resonance imaging (dMRI) tractography is an advanced imaging technique that enables in vivo reconstruction of the brain’s white matter connections at macro scale. It provides an important tool for quantitative mapping of the brain’s structural connectivity using measures of connectivity or tissue microstructure. Over the last two decades, the study of brain connectivity using dMRI tractography has played a prominent role in the neuroimaging research landscape. In this paper, we provide a high-level overview of how tractography is used to enable quantitative analysis of the brain’s structural connectivity in health and disease. We focus on two types of quantitative analyses of tractography, including: 1) tract-specific analysis that refers to research that is typically hypothesis-driven and studies particular anatomical fiber tracts, and 2) connectome-based analysis that refers to research that is more data-driven and generally studies the structural connectivity of the entire brain. We first provide a review of methodology involved in three main processing steps that are common across most approaches for quantitative analysis of tractography, including methods for tractography correction, segmentation and quantification. For each step, we aim to describe methodological choices, their popularity, and potential pros and cons. We then review studies that have used quantitative tractography approaches to study the brain’s white matter, focusing on applications in neurodevelopment, aging, neurological disorders, mental disorders, and neurosurgery. We conclude that, while there have been considerable advancements in methodological technologies and breadth of applications, there nevertheless remains no consensus about the “best” methodology in quantitative analysis of tractography, and researchers should remain cautious when interpreting results in research and clinical applications.
1. Introduction
Diffusion magnetic resonance imaging (dMRI) tractography is an imaging method that uniquely enables in vivo reconstruction of the brain’s white matter connections at macro scale (Basser et al., 2000). Since the first dMRI tractography methods were proposed in 1998–2000 (Basser, 1998; Basser et al., 2000; Conturo et al., 1999; Mori et al., 1999; Westin et al., 1999), tractography has enabled mapping of the brain’s structural connectivity in many neurological applications such as aging, development and disease (Assaf and Pasternak, 2008; Ciccarelli et al., 2008; Essayed et al., 2017; Nucifora et al., 2007; Shi and Toga, 2017; Yamada et al., 2009). Initial applications of tractography enabled the visualization of white matter tracts, a primarily qualitative approach that remains highly relevant today both in the clinic (Abhinav et al., 2015; Essayed et al., 2017; Pujol et al., 2015) and for the study of neuroanatomy (Catani et al., 2002; Ciccarelli et al., 2003; Makris et al., 2005). Recently, quantitative approaches have become popular tools for studying the brains connectivity and tissue microstructure using tractography. In this paper, we provide a high-level overview of how tractography is used to enable quantitative analysis of the brain’s structural connectivity. This review is intended to be useful to researchers studying the white matter, developers of quantitative analysis methods, and clinicians interpreting results related to tractography.
Due to the large number of proposed quantitative approaches and the evolution of the field over two decades, there is a proliferation of terminology related to the quantitative analysis of dMRI tractography. Therefore, we begin with a listing of common terms used throughout this paper and their definitions, as provided in Table 1. We also provide a visualization of many key concepts that will be used in the paper (Fig. 1).
Table 1.
Common terms used throughout this paper and their definitions. We note that there are traditional terms that are widely used, but are not technically or biologically precise; in this table, we emphasize such terms and encourage the avoidance of their usage in future studies.
| Term | Definition |
|---|---|
| Tractography / Fiber tracking | Any computational process that estimates the anatomical trajectories of the white matter fiber pathways from dMRI data. Note: The term “Fiber tracing” and “Fiber tracking” are also widely used. However, we strongly suggest avoiding this term because “tracing” is frequently used in the context of ex vivo tracer injections, and “tracking” refers to following a moving object while white matter fibers do not move. |
| Streamline | A set of ordered points in 3D space, encoding a trajectory estimated through performing tractography (see Fig. 1(b)). Note: The term “fiber ” is also widely used. However, we strongly suggest avoiding this term because “fiber” is in reference to biology, while “streamline” implies the digital reconstruction of such. |
| Tractogram | A set of streamlines, often generated in such a way as to cover the entire white matter of the brain in order to capture any possible white matter connections. This can be referred to as a “whole-brain tractogram” (see Fig. 1(c)). |
| Steamline cluster/ Streamline connection/ Streamlines within an edge/ White matter parcel | A set of streamlines resulting from the subdivision of a tractogram. Such sets can result from a variety of methods for dividing the tractogram; hence there are several names in use for them. While such a set is often called a “fiber bundle,” “fiber tract,” or “fiber cluster” in the literature, to clarify that a set of streamlines is a computational data representation, we avoid using the word “fiber” in this paper. |
| Fiber Bundle / Fiber Tract / Fiber Fasciculus / Fiber Pathway | These terms have biological meanings as a set of white matter fibers (axons) forming a corticocortical or corticosubcortical connection in the brain (Schmahmann, Schmahmann, Pandya, 2009). In the neuroimaging literature, these terms are commonly used instead to refer to white matter connections reconstructed using tractography. Following this practice, in this paper we will use the terms “fiber tract,” “fiber bundle,” “fiber fasciculus,” or “fiber pathway” to refer to a white matter structure that corresponds to known anatomy with a traditional name (e.g., the corpus callosum or the corticospinal tract) (see Fig. 1(d)). |
| Brain Connectivity | In neuroimaging, the somewhat elusive and ambiguous concept of brain connectivity refers to measures of the structural and/or functional relationship between different brain regions (Horwitz, 2003; Rossini et al., 2019; Sakkalis, 2011; Uddin, 2013). |
| Structural Connectivity | A specific type of brain connectivity. Two brain regions are structurally connected if a fiber tract physically interconnects them. This is typically measured in vivo in humans using dMRI. However, there is no consensus on how this should be best quantified. Structural connectivity measures (also called “weights” or “strengths”) can include a variety of quantitative connectivity measures computed from a specific set of streamlines corresponding to a pathway of interest (e.g. those connecting two specific endpoints). The goal is often to approximate the true underlying fiber density or number of axons (Jones, 2010; Smith et al., 2020a). |
| Connectome / Brain Connectivity Matrix | A two-dimensional matrix wherein the rows and columns correspond to specific brain gray matter regions of interest (ROIs), and the value stored within each element of the matrix is the computed connectivity “ strength” between those regions corresponding to that row & column (Sporns, Tononi, Kötter, 2005) (see Fig. 1(e)). Such data are described mathematically as a graph. This matrix representation is directly inspired by invasive axonal tract-tracing experiments in animals, where the results of multiple studies are expressed as a quantitative connectivity matrix (Wakker et al., 2012). Without further specification, we use “connectome” throughout to implicitly refer to the structural connectome constructed using dMRI tractography, as opposed to those derived through other imaging modalities (e.g., functional MRI). |
| Diffusion Model | A theoretical model that connects the dMRI signal to salient features of tissue microstructure at the cellular level (Novikov et al., 2018; Panagiotaki et al., 2012; Yablonskiy and Sukstanskii, 2010). This includes tissue microstructure or biophysical models such as Neurite Orientation Dispersion and Density Imaging (NODDI) (Zhang et al., 2012) and Free Water (FW) (Pasternak et al., 2012, 2009, as well as diffusion signal representations that include the diffusion tensor (Basser and Pierpaoli, 1996), diffusion kurtosis (Jensen and Helpern, 2010) and many others (Afzali et al., 2020; Alexander, 2006). |
| Microstructural Measure | Any parameter extracted from a diffusion model fit in each voxel that provides information regarding the underlying tissue microstructure (e.g., the fractional anisotropy (FA) that describes water diffusion anisotropy (Basser and Pierpaoli, 2011)). |
| Tractometry / Profilometry / Tract-based morphometry | An along-tract profiling analysis technique to investigate the distribution of the microstructural measures along the fiber pathway (Bells et al., 2011b; O’Donnell et al., 2009; Yeatman et al., 2012). |
Fig. 1.
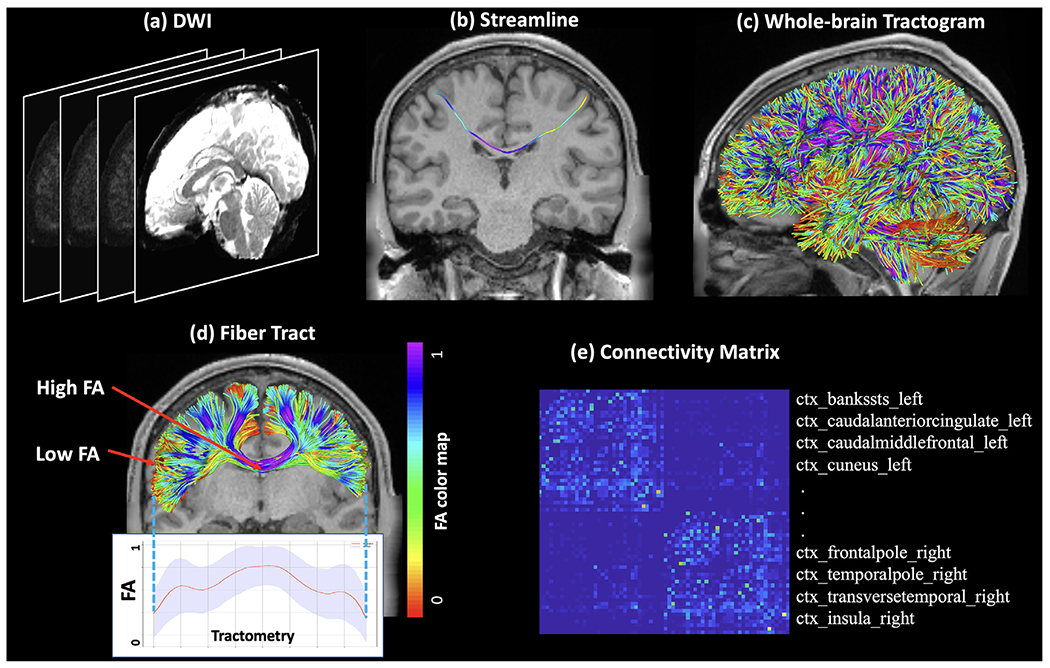
Graphic illustration of tractography. (a) Example dMRI data, also known as diffusion weighted imaging (DWI) data. (b) An individual streamline computed after performing tractography. (c) An example whole-brain tractogram that consists of streamlines covering the entire white matter. (d) A fiber bundle that consists of a set of streamlines, representing an anatomical fiber tract named the corpus callosum. The streamlines are colored by a microstructural measure, i.e., fractional anisotropy (FA) that measures the anisotropy of water diffusion. The distribution of FA along the fiber tract is provided to show the tractometry of the tract. A low FA can be seen at the endpoint region of the streamlines (near the cortex) and a high FA can be seen in the middle of the streamlines (in the deep white matter). (e) An example brain structural connectivity matrix, which is constructed based on white matter tractography from the whole brain. Each row (and column) represents a brain gray matter ROI (See Fig. 3(c) for an example brain gray matter parcellation), and the value in an element of the matrix is the strength of the white matter connection between the two corresponding ROIs (quantified as the number of streamlines in this case).
Many quantitative tractography approaches can be applied to study the brain’s structural connectivity in health and disease. The fundamental goal of these analyses is to estimate quantitative measures of connectivity (or microstructure) of some pathway (or pathways) of interest. Quantitative analyses of tractography can be categorized into two main categories or styles: tract-specific analyses and connectome-based analyses (this categorization is helpful but imperfect, as some approaches blend aspects of both analysis styles). Tract-specific analysis refers to research that is typically hypothesis-driven and studies particular anatomical fiber tracts (Alexander et al., 2007; Levitt et al., 2020; Shany et al., 2017; Yeo et al., 2014). Tract-specific analysis has been increasingly adopted, particularly for the study of local white matter regions in health and disease (Alexander et al., 2007; Shany et al., 2017; Yeo et al., 2014). Connectome-based analysis refers to research that is more data-driven and generally studies the structural connectivity of the entire brain (Bastiani et al., 2012; Ingalhalikar et al., 2014; Sporns et al., 2005; Zalesky et al., 2012). This type of analysis aims to understand patterns of whole-brain anatomical connectivity, and therefore relies on tractography performed across the entire white matter.
In this paper, we first provide a brief introduction to tractography (Section 2), then a review of methodology for quantitative tractography analysis (Sections 3–5), followed by a review of studies that use quantitative tractography analysis to study the brain in health and disease (Section 6). For the methodology review, we organize the paper according to three main processing steps that are common across most approaches for quantitative analysis of tractography. The first common step corrects potential biases in tractography reconstruction that may otherwise be detrimental to, or outright preclude, quantitative analysis of such data; we refer to this as tractography correction (Section 3). The second common processing step identifies white matter pathways (e.g., subdivisions of the tractogram) that are meaningful for quantification of brain connectivity; we refer to this as tractography segmentation (Section 4). The third common step extracts quantitative indices that describe the microstructure and/or the “strength” of the brain connections; we refer to this as tractography quantification (Section 5). In each section, we aim to describe methodological choices, their popularity, and their potential pros and cons. Finally, we review studies that have used quantitative tractography approaches to study the brain’s white matter, including applications in development, aging, neurological disorders, mental disorders, and neurosurgery (Section 6).
2. Brief introduction to tractography
“Tractography” can refer to any computational process that estimates the anatomical trajectories of white matter fiber pathways from dMRI data. There are many methods available to perform tractography, including traditional streamline tractography based on the diffusion tensor model (Basser et al., 2000; Basser and Pierpaoli, 1996), which was originally proposed (as “hyperstreamlines”) for the visualization of tensor fields in the computer graphics community in 1993 (Delmarcelle and Hesselink, 1993). More recently, many streamline tractography methods using advanced diffusion models have been proposed and are under active development (Behrens et al., 2007; Descoteaux et al., 2008; Feng and He, 2020; Fernandez-Miranda et al., 2012; Friman et al., 2006; Jackowski et al., 2005; Jeurissen et al., 2014; Lazar and Alexander, 2005; Malcolm et al., 2010; Reddy and Rathi, 2016; Tournier et al., 2010; 2003; Wasserthal et al., 2019). Furthermore, many alternative tractography algorithms have been proposed, such as front evolution (Pichon et al., 2005; Sepasian et al., 2012; Tournier et al., 2003), geodesic (identifying likely connection trajectories based on assumed endpoints) (Jbabdi et al., 2008; 2007; Li et al., 2014; O’Donnell et al., 2002; Schreiber et al., 2014; Wu et al., 2009), atlas-based (Wasserthal et al., 2019; Yendiki et al., 2011), differential tractography (Yeh et al., 2019b) and various forms of “global” tractography that simultaneously fit the entire tractography reconstruction to all image data (either by progressively forming linked chains from a large set of randomly-initialized short segments (Christiaens et al., 2015; Fillard et al., 2009; Kreher et al., 2008; Mangin et al., 2013; Mangin et al., 2002; Reisert et al., 2011; Teillac et al., 2017)or iteratively perturbing a set of candidate trajectories (Battocchio et al., 2020; Close et al., 2015; Lemkaddem et al., 2014; Wu et al., 2012b). Work is additionally underway to improve tractography using machine learning (Neher et al., 2017; Poulin et al., 2019; Sarwar et al., 2020; Wasserthal et al., 2019; Wegmayr et al., 2018).
While a detailed introduction of tractography methods is out of the scope of this review, we refer the readers to these review papers (Jeurissen et al., 2019; Neher et al., 2015; Rheault et al., 2020b; Smith et al., 2020c) that are specific to tractography algorithms.
Many software packages are available to perform tractography research, including ANIMA (Descoteaux et al., 2008), BrainSUITE (Shattuck and Leahy, 2002), Camino (Cook et al., 2006), COMMIT (Daducci et al., 2013; 2014; Ocampo-Pineda et al., 2021; Schiavi et al., 2020a), Diffusion toolkit (Wang et al., 2007), Dipy (Garyfallidis et al., 2014), DMIPY (Fick et al., 2019), DSI studio (Yeh et al., 2013), DTIstudio (Jiang et al., 2006), ExploreDTI (Leemans et al., 2009), Fiber-Navigator (Chamberland et al., 2014), FSL (Jenkinson et al., 2012), MITK (Wolf et al., 2005), MRtrix3 (Tournier et al., 2012; 2019), PANDA (Cui et al., 2013), SlicerDMRI (Norton et al., 2017; Zhang et al., 2020b), TractSeg (Wasserthal et al., 2018), and Tracula (Yendiki et al., 2011). It is important to note that tractography is known to be sensitive to the choice of the underlying fiber tracking algorithms, which can further affect any subsequent quantitative analyses using the tractography data. On one hand, tractography results may vary across the large set of available diffusion models (Afzali et al., 2020; Alexander, 2006; Panagiotaki et al., 2012; Yablonskiy and Sukstanskii, 2010) and tractography methods (Basser et al., 2000; Behrens et al., 2007; Descoteaux et al., 2008; Feng and He, 2020; Fernandez-Miranda et al., 2012; Friman et al., 2006; Jackowski et al., 2005; Jeurissen et al., 2014; Lazar and Alexander, 2005; Malcolm et al., 2010; Reddy and Rathi, 2016; Tournier et al., 2010; 2003; Wasserthal et al., 2019). On the other hand, tractography results can also be sensitive to the choice of parameters (such as seeding and stopping thresholds) within a certain algorithm (Chamberland et al., 2014; Côté et al., 2013; Gong et al., 2018; Moldrich et al., 2010; Xie et al., 2020). While many research studies have been performed to compare different tractography algorithms (Bastiani et al., 2012; Fillard et al., 2011; Petrov et al., 2017; Pujol et al., 2015; Sinke et al., 2018; Wilkins et al., 2015; Zhan et al., 2015), there is no consensus on “the best algorithm.”
3. Improving tractography quality: methods for tractography correction
The goal of tractography correction methods is to tackle potential biases and errors that are produced in conventional or raw tractography algorithms. Where such an issue can be clearly demonstrated in exemplar data, and a tailored mechanism proposed that intrinsically addresses the issue, it is expected that the employment of such a mechanism in the reconstruction of empirical image data will lead to overall improved biological accuracy of the reconstruction (even in the absence of direct validation of such), which is important for the extraction of anatomical fiber tracts (Section 4.1) and the construction of quantitative structural connectivity matrices (Section 5.5). In this section, we consider the near-ubiquitous “streamlines” paradigm for dMRI tractography (Basser, 1998; Basser et al., 2000; Behrens et al., 2007; Conturo et al., 1999; Descoteaux et al., 2008; Feng and He, 2020; Fernandez-Miranda et al., 2012; Friman et al., 2006; Jackowski et al., 2005; Lazar and Alexander, 2005; Malcolm et al., 2010; Mori et al., 1999; Reddy and Rathi, 2016; Tournier et al., 2010; 2003; Wasserthal et al., 2019; Westin et al., 1999), and the ways in which specific undesirable behaviors or biases may be ameliorated through specific targeted mechanisms; see Fig. 2 for a visual summary of the relevant tractography biases included.
Fig. 2.

Demonstrations of various forms of errors and biases in tractography, as described in Section 3. Images are exemplars only; therefore there exist many other possible manifestations of such biases and errors. 1. Low-order integration methods under-estimate bundle curvature, leading to reconstructed paths overshooting the underlying curved trajectory. 2. In the absence of tailored constraints, streamlines may terminate in locations other than those known to contain axon synapses. 3. The number of streamlines generated within a bundle may not be proportional to the underlying axonal density of that bundle. 4. Because of the low resolution of diffusion MRI relative to the complexity of gyral folding, streamline termination may accumulate at gyral crowns rather than being distributed uniformly across the cortical ribbon. 5. Where macroscopic WM bundles intersect, streamlines may erroneously traverse part of one bundle and part of another, producing trajectories that are not present in the underlying structure (Schilling et al., 2021).
3.1. Curvature overshoot bias
Possibly the earliest bias established for dMRI tractography was the fact that if the streamline algorithm simply takes a step of finite size in the direction of the local fiber orientation at the current vertex (a so-called “first-order” method), streamlines in curved bundles will tend to under-estimate the curvature, resulting in erroneous trajectories (Tournier et al., 2002). While use of a step size that is small relative to the image voxel size mitigates the issue, use of a higher-order integration method during tracking that directly accounts for such curvature is a more direct solution (Basser et al., 2000). However, doing so in a manner that is compatible with diffusion models that account for crossing fibers can be technically difficult (Aydogan and Shi, 2020; Cherifi et al., 2018; Tournier et al., 2010).
3.2. Termination bias
“Termination” refers to the location at which the propagation of a streamline is ceased. Just as a tractography algorithm should produce streamlines whose tangents are faithful to the underlying fiber orientations, those streamlines should also terminate at locations corresponding to the endpoints of the underlying fiber tracts. Biases or inadequacies in such can result in partial fiber streamlines that stop prematurely in the white matter — despite the fact that white matter fibers synapse in the gray matter (Daducci et al., 2016) — or even that enter fluid-filled regions or cross sulcal banks. Such issues can be prevalent as tractography algorithms often operate using only the fitted diffusion model (e.g., diffusion tensor (Basser and Pierpaoli, 1996) or constrained spherical deconvolution (CSD) (Jeurissen et al., 2014; Tournier et al., 2007) in each image voxel. While these models provide strong evidence regarding fiber orientations to inform the direction of streamline propagation, the evidence they provide regarding where such fibers stop (and hence where streamlines should ideally be terminated) is weak (indeed, the diffusion-weighted signal itself does not provide direct evidence of fiber terminations (Smith et al., 2020b). Most fiber tracking algorithms exploit heuristic thresholds on features such as diffusion model anisotropy and streamlines curvature to serve as tractography termination criteria. However, the indirect nature of these metrics for this task, combined with the inferior spatial resolution of diffusion-weighted imaging, leads to such errors being highly prevalent (Smith et al., 2012). These common termination criteria are therefore not adequate to ensure biologically plausible streamline generation, resulting in a substantial proportion of streamlines from whole-brain tractography being unreasonable for quantification of white matter fiber connectivity (Yeh et al., 2016a).
The ill-posed nature of streamline terminations can be addressed by utilizing anatomical reference data to impose relevant prior information. This can ensure that the reconstructed streamlines fulfill some fundamental assumptions we could make, given an a priori understanding of how neurons are organized in the brain. For instance, axons emanate from cell bodies within the gray matter and connect to other cells elsewhere in either the brain or body, but they do not synapse in the middle of white matter or propagate into ventricles. Unlike conventional tractography algorithms, in which termination points can be distributed almost anywhere in the brain, the Anatomically-Constrained Tractography (ACT) framework (Smith et al., 2012) and similar methods (Girard and Descoteaux, 2012; Lemkaddem et al., 2014; Yeh et al., 2017) ensure that both streamline propagation and termination are constrained based on knowledge of where neuronal fibers locate, e.g., connecting between gray matter areas via the white matter. Although these methods cannot guarantee that every streamline accurately traces the complete trajectory of an underlying fiber connection, they do prevent the generation of streamlines that cannot possibly represent biological connectivity within the brain.
3.3. Connection density biases
While streamline tractography provides estimates of structural connection trajectories that are consistent with the underlying fiber orientations, it does not provide any guarantees regarding consistency between the number of such reconstructed connections with the density of those underlying fibers (i.e., the actual number of axons in a white matter region) (Jones, 2010; Jones et al., 2013). This limitation is often expressed as the mantra “streamline count is not quantitative.” There are myriad factors that may modulate the empirical streamline count between pathways, or within a pathway across individuals. Many are emergent phenomena from the nuances of the operation of any particular streamlines algorithm and therefore difficult to characterise. Nevertheless, the fact it can be trivially demonstrated that both that streamline count can be erroneously modulated in the absence of a difference in connection strength and erroneously constant in the presence of a difference in connection strength (Figs. 2, 3) highlights the inappropriateness of applying such a strong biological interpretation to raw streamline count.
Fig. 3.

Tractography segmentation: (a) Example whole-brain tractogram computed by performing tractography in DWI data; (b) Example anatomical tracts extracted from the tractogram; and (c) Example structural connectivity matrix constructed by performing whole brain tractography segmentation between all pairs of FreeSurfer cortical regions.
What would give greater confidence to the interpretation of tractography-based connection density as a quantitative measure is if, throughout the white matter, there were correspondence between the local density of reconstructed connections and the local density of fibers as evidenced by the diffusion image data (Smith et al., 2020a). Initially, aspiration for achieving this was limited to “global” tractography algorithms as mentioned in Section 2. More recently, methods have been devised to address this task by instead utilizing a pre-generated whole-brain streamline tractogram and modulating the contribution of each streamline toward the reconstructed white matter fiber density. Initial methods achieved this by selecting a subset of streamlines that best fit the diffusion image data, i.e., BlueMatter (Sherbondy et al., 2009), MicroTrack (Sherbondy et al., 2010) and SIFT (Smith et al., 2013), while later methods estimate a multiplier to be applied to each streamline, i.e., COMMIT (Daducci et al., 2013; 2014), LiFE (Pestilli et al., 2014), SIFT2 (Smith et al., 2015b), COMMIT2 (Schiavi et al., 2020a) and COMMIT2tree (Ocampo-Pineda et al., 2021); these “weights” represent an effective cross-sectional area of each streamline, which can be utilized as a direct measure of connection density (Smith et al., 2020a) (or indeed to modulate streamline contributions toward other aggregate measures of the connectivity “strength”; see Section 5).
3.4. Gyral bias
Gyral bias in tractography is the bias towards generating streamlines that terminate in gyri rather than sulci (Schilling et al., 2018a; Wu et al., 2020b), which reduces the agreement of the initiations or terminations of fiber streamlines with known ground truth from tracer studies and histology (Aydogan et al., 2018; Budde and Annese, 2013). The source of the gyral bias is multifactorial, such as the complexity of axon arrangement at the junction of cortical grey matter and superficial white matter (Van Essen et al., 2014). The partial volume effect from the limited MRI spatial resolution introduces difficulties in distinguishing complex fiber configurations based on the reconstructed fiber orientation distributions (FODs). Increasing the image resolution is beneficial to mitigate the gyral bias (Heidemann et al., 2012; Sotiropoulos et al., 2016), but is constrained by the signal-to-noise ratio of the acquired dMRI data. Even operating on high-resolution dMRI data, current tractography algorithms have been shown to deviate from the ground-truth fiber projections derived from histological staining, with greater streamline densities at gyral crowns than sulcal banks (Schilling et al., 2018b). The complex folding and convolutions of cortical gyri also pose challenges for the reconstruction of long-range connections (Reveley et al., 2015).
A surface flow approach has been proposed to model the arrangement of superficial axonal fibers at gyral crowns, thereby improving the consistency between streamlines’ initiations or terminations and the expectations from histological data (St-Onge et al., 2018). More recently, the gyral bias has been shown to be alleviated using an asymmetric FOD technique (Wu et al., 2020c), which can depict the highly-curved fiber geometry appearing at the superficial white matter (Bastiani et al., 2017; Wu et al., 2020c).
3.5. False positive connections
Several orders of magnitude separate the resolution of dMRI acquisitions (cubic millimeters) from the typical size of the axons (few micrometers). This discrepancy introduces ambiguities in the white matter that can be compatible with multiple streamline configurations and are difficult for tractography to resolve (Girard et al., 2020; Guevara et al., 2012; Maier-Hein et al., 2017). This ill-posed nature of tractography has received a renewed interest in the past few years, and several studies have raised serious concerns about the anatomical accuracy of the reconstructions (Maier-Hein et al., 2017; Thomas et al., 2014). In particular, it has been shown that tractography techniques suffer from a large number of false positives, i.e., streamlines that are reconstructed but do not correspond to real anatomical bundles, and that these erroneous connections can severely bias the estimation of connectivity (Drakesmith et al., 2015; Maier-Hein et al., 2017; Zalesky et al., 2016b). The anatomical accuracy of the reconstructions can be improved by manually filtering the streamlines with inclusion/exclusion ROIs, as seen in Section 4.1, but this requires prior knowledge of “where white matter pathways start, where they end, and where they do not go” (Schilling et al., 2020a). However, more automated procedures are desirable especially when analyzing large cohorts of subjects.
A widely-used approach to attempt to alleviate false positive connections when building a structural connectivity matrix is thresholding (Fornito et al., 2016), which refers to the removal of edges whose strength is smaller than a certain cut-off value. This strategy implicitly assumes a correlation between the strength of a reconstructed edge and its validity, which can be violated in a range of ways due to the errors produced by streamlines tractography not being simply random variance. The capacity for thresholding to differentiate between true and spurious connections remains a debated topic in the field.
Alternative solutions have been proposed, which are either knowledge- or data-driven. The former are described in Section 4.1, and typically use fully- or semi-automated algorithms, e.g., clustering, as a means to detect and discard streamlines considered outliers based on anatomical priors or geometrical properties, e.g., length and shape. On the other hand, the methods described in Section 3.3 for addressing density biases can also be used as filtering procedures, in which the streamlines that are not supported by the acquired dMRI data, i.e., whose estimated weight is zero, are discarded. While these approaches achieve good results in terms of removing duplicate streamlines or isolated ones that are likely spurious, actually none proved effective in discriminating between true and spurious connections in the connectome networks (Maier-Hein et al., 2017; Schiavi et al., 2020a).
It was recently demonstrated that the performance of filtering methods can be significantly boosted by combining knowledge- and data-driven strategies. These new formulations (Ocampo-Pineda et al., 2021; Schiavi et al., 2020a) allow taking explicitly into account two fundamental assumptions about the connections in the brain: (i) fibers are naturally organized in bundles (Mandonnet et al., 2018; Udin and Fawcett, 1988), and (ii) the number of bundles should be low to minimize the overall wiring cost (Bullmore and Sporns, 2012). This prior knowledge helps resolve some of the ambiguities present in the data and, as a consequence, allows significant improvement of the anatomical accuracy of the connectome (Ocampo-Pineda et al., 2021; Schiavi et al., 2020a). This is achieved by organizing the streamlines into groups and seeking for solutions which explain the measured signal with the minimum number of bundles, using the Group Lasso regularization (Yuan and Lin, 2006), thus promoting sparsity in the space of connections, rather than in the space of individual streamlines as implicitly done in previous filtering methods.
Research is still underway to determine the best method(s) for filtering, with some disagreement on the beneficial versus detrimental effect of these methods on the structural connectome (Buchanan et al., 2020; Civier et al., 2019; Drakesmith et al., 2015; Frigo et al., 2020; Schiavi et al., 2020a; Yeh et al., 2016a).
4. Defining regions for quantitation: methods for tractography segmentation
The goal of white matter tractography segmentation is to identify white matter pathways (e.g., subdivisions of the tractogram) that are meaningful for quantification of the brain’s structural connectivity. Critically, this step enables cross-subject quantitative comparison of the segmented white matter pathways; see (Smith et al., 2014) for a review of general considerations in multi-subject dMRI studies. (Tractography segmentation is also critical for qualitative applications such as visualization of fiber tracts, e.g., for neurosurgical white matter mapping; see Section 6.4.) Tractography segmentation methods can be generally grouped into two categories, related to the tract-specific and connectome-based analysis approaches, as illustrated in Fig. 3. The first category of methods identify anatomical white matter fiber tracts and label them with traditional names (such as the arcuate fasciculus or the corticospinal tract) (Section 4.1). The second category of methods parcellate, or subdivide, the entire white matter into many white matter parcels based on information about the streamline trajectories and/or endpoints (Section 4.2).
4.1. Anatomical tract identification
Anatomical tract identification aims to identify tractography streamlines that correspond to anatomically named white matter fiber tracts. This task is non-trivial, given the complex structures of the white matter anatomy and the large number of streamlines in the tractogram.
Conventional anatomical tract identification methods rely on manual streamline selection, also referred to as virtual dissection, where experts in anatomy interactively select tractography streamlines using manually drawn ROIs in the brain (Catani et al., 2002; de Schotten et al., 2011; Stieltjes et al., 2001; Wakana et al., 2007). Usually, inclusion ROIs are placed in the gray matter (cortical and subcortical) to define where the streamlines should terminate and in the white matter to define where the streamlines should pass, and exclusion ROIs are placed in other regions to exclude undesired streamlines (Rheault et al., 2020a; Wakana et al., 2007). Manual streamline selection is considered to be the gold standard to delineate anatomical fiber tracts in tractography and has been widely used to validate other anatomical tract identification techniques (Poulin et al., 2019; Pujol et al., 2015; Xie et al., 2020). Manual selection is also used in studies where specific clinical expertise is needed, e.g., for presurgical white matter mapping in tumor patients where the tumor and lesion can largely displace white matter fiber tracts (Radmanesh et al., 2015).
Due to the fact that manual tract selection is time-consuming and has high clinical and expert labor costs, modern studies increasingly focus on automated tract identification methods. These methods can be categorized into three categories: ROI-based, streamline labeling, and direct segmentation.
The ROI-based methods are the most commonly used. They automate ROI computation and perform streamline selection based on the ROIs a streamline terminates in and/or passes through. The majority of the ROI-based methods leverage a brain ROI atlas and use an image registration to automatically align the ROIs in the atlas space to the subject space (Allan et al., 2020; Astolfi et al., 2020; Hua et al., 2008; Lawes et al., 2008; Schurr et al., 2019; Verhoeven et al., 2010; Wassermann et al., 2016; Yeatman et al., 2012; Zhang et al., 2008; 2010). Currently, popularly used brain ROI atlases include those provided in Freesurfer (Desikan et al., 2006; Fischl, 2012), MNI–ICBM152 (Mazziotta et al., 2001; Mori et al., 2008; Oishi et al., 2009; 2008), and JHU-DTI (Wakana et al., 2007; 2004) (see (Hansen et al., 2020) for a review of more atlases). There are also ROI-based methods that directly predict ROIs in subject space using machine learning (Astolfi et al., 2020; Li et al., 2020b).
The second category of automated tract identification methods, streamline labeling, assigns an anatomical label to each individual streamline. Often, streamline labeling is done by computing the geometric distance of each streamline to labeled streamlines in a reference tract segmentation, and then assigning a streamline label based on the closest reference tract (Bertò et al., 2020; Clayden et al., 2007; Gupta et al., 2018; 2017b; Kumar et al., 2019; Labra et al., 2017; Lam et al., 2018; Liu et al., 2019; Maddah et al., 2005; Xu et al., 2019; Yang et al., 2020; Zhang et al., 2020a). There are also learning-based segmentation approaches that train a model from the reference tract segmentation data and predict an anatomical label for each streamline in a new subject (Gupta et al., 2018; 2017b; Kumar et al., 2019; Lam et al., 2018; Liu et al., 2019; Xu et al., 2019; Yang et al., 2020; Zhang et al., 2020a). To reduce the amount of labeling for each streamline, many streamline labeling methods first group streamlines into clusters (known as fiber clustering), followed by assigning an anatomical label for each cluster, thus labeling each streamline (Avila et al., 2019; Chekir et al., 2014; Garyfallidis et al., 2018; Guevara et al., 2012; Li et al., 2010; O’Donnell and Westin, 2007; Román et al., 2017; Ros et al., 2013; Siless et al., 2018; 2020; Tunç et al., 2014; Vázquez et al., 2020; Wu et al., 2020a; Yeh et al., 2018; Yoo et al., 2015; Zhang et al., 2018c; Ziyan et al., 2009).
The third category of automated tract identification is called direct segmentation in the literature (Bazin et al., 2011; Dong et al., 2019; Eckstein et al., 2009; Li et al., 2020a; Lu et al., 2020; Ratnarajah and Qiu, 2014; Reisert et al., 2018; Rheault et al., 2019; Wasserthal et al., 2018; 2019; Yendiki et al., 2011; Zöllei et al., 2019). Unlike the aforementioned methods that work on precomputed tractography streamline data, the direct segmentation methods operate on DWI data (Yendiki et al., 2011; Zöllei et al., 2019) or derived volumetric image data (e.g., FOD maps (Wasserthal et al., 2018) and streamline density maps (Lu et al., 2020)) to predict the location for multiple tracts of interest, followed by performing tractography for only these tracts. Although the direct segmentation methods are relatively new and are relatively less used than the ROI-based and streamline labeling methods, the success of methods such as TRACULA (Yendiki et al., 2011), Tract-Seg (Wasserthal et al., 2018) and Bundle-specific tractography (BST) (Rheault et al., 2019) has shown highly promising tract segmentation performance.
There are several points that should be considered when selecting a method for anatomical tract identification. First, given the fact that there is no ground truth in tractography, validation of a tract identification method is difficult. One commonly used strategy to assess a tract identification method is to evaluate its reproducibility (in terms of intra- and inter-rater as well as test-retest reproducibility) (Rheault et al., 2020a; Tong et al., 2019; Zhang et al., 2019). Methods using manual streamline selection have been shown to be highly reproducible for specific tracts and segmentation protocols using the diffusion tensor tractography (Wakana et al., 2007), but with more modern multi-fiber tractography, reproducibility is lower, potentially due to the increase in spurious streamlines (Rheault et al., 2020a). However, manual streamline selection is still considered to be the gold standard for benchmarked comparisons and algorithm training (Kreilkamp et al., 2019; Wasserthal et al., 2018; Zhang et al., 2010). For automated tract identification, one study indicates that streamline labeling has higher test-retest reproducibility than ROI-based segmentation (Zhang et al., 2019); however, no consensus has been reached given that many existing tract identification methods have not been compared. The second point is that a good tract identification method should be highly consistent across different populations and acquisitions (Sydnor et al., 2018; Wasserthal et al., 2018; Yendiki et al., 2011; Zhang et al., 2018c). This is particularly important for automated tract identification methods, which are essential to perform across-lifespan and multi-site studies, which usually involve a large number of datasets (Alexander et al., 2017; Casey et al., 2018; Cetin-Karayumak et al., 2020; Harms et al., 2018; Thompson et al., 2017). One factor that can affect the consistency of methods that segment tractography streamline data is the underlying tractography method that is employed. In general, studies indicate that modern multi-fiber tractography is more sensitive when performing tractography and thus is more consistent across subjects and timepoints than traditional single-fiber DTI tractography (Bucci et al., 2013; Prčkovska et al., 2016). The third point is that anatomical tract identification results can be highly variable across studies due to a lack of consistent fiber tract definitions, where different methods may give different results for the same target anatomical fiber tracts (Dick and Tremblay, 2012; Rheault et al., 2020a; Schilling et al., 2020b). Work is underway to provide sets of consistent rules for fiber tract definitions (e.g. the White Matter Query Language (Wassermann et al., 2016) and tractography-based fiber tract atlases (Mori et al., 2008; Yeh et al., 2018; Zhang et al., 2018c) that are provided in an open fashion for use and possible extension or modification by the community. The fourth point is that a method for performing tractography must be chosen: tract-specific tractography may be more computationally efficient if the number of tracts of interest is small, while whole-brain tractography can be computationally expensive but enables tract identification or parcellation of the whole white matter. (Note that a whole-brain tractogram is required for filtering (Sections 3.3 and 5.3) and analyses of the brain as a network using a connectivity matrix (Sections 5.5 and 5.6)). However, computation of whole-brain tractography considers the entire white matter simultaneously, estimating all fiber tracts using the same set of tractography parameters; this may not be ideal for fiber tracts that are difficult to compute using tractography, e.g., those requiring a lower anisotropy threshold, more lenient curvature threshold, or a dedicated tracking algorithm (such as the optic radiation (Sherbondy et al., 2008; Tax et al., 2014)). Recent approaches propose to better reconstruct white matter anatomy by exploring the tractography parameter space, e.g., by combining results from multiple tractography algorithms (“ensemble tractography”) (Takemura et al., 2016) and by exploring millions of parameter combinations (“augmented fiber tracking”) (Yeh, 2020). The fifth point is that selection of different tract identification methods may require different image registration methods. The ROI-based methods usually require volumetric registration (e.g., between a population T1-weighted template and individual dMRI data) to align ROIs to the dMRI space. While there are many sophisticated tools to compute these registrations such as FSL (Jenkinson et al., 2012) and ANTs (Avants et al., 2009), the performance is limited by nontrivial factors. For example, the intermodality registration between dMRI and T1-weighted images can be affected by differences in image resolutions (Malinsky et al., 2013) and echo-planar imaging (EPI) distortion in dMRI data (Albi et al., 2018). Many streamline labeling methods use streamline-based registration to directly align individual tractography data to a tractography atlas where a reference tract segmentation is available. Most widely used approaches perform a linear registration (Garyfallidis et al., 2015; O’Donnell et al., 2012), while many studies have proposed to perform non-linear approaches to enable deformations of the streamlines (Benou et al., 2019; Chandio and Garyfallidis, 2020; Olivetti et al., 2016). While using streamline-based registration can avoid the intermodality registration issue in ROI-based methods, streamline registration (especially a nonlinear registration) may be computationally heavier than volumetric registration. Most of the direct segmentation approaches use a training tract segmentation dataset and thus may not require image registration or require a small amount of work for registration (e.g., a rigid volumetric registration); however, we note that curation of a training tract segmentation dataset may require extensive work compared to performing image registration. Lastly, deep learning techniques have been increasingly applied for anatomical tract identification, showing high potential for future work and improvements in computational speed (Chen et al., 2021; Gupta et al., 2017a; 2018; 2017b; Lam et al., 2018; Reisert et al., 2018; Wasserthal et al., 2018; 2019; Xu et al., 2019; Zhang et al., 2020a).
4.2. Parcellation of the whole-brain tractogram
Parcellation of the entire white matter (as represented by a whole-brain tractogram) aims to enable quantitative analysis of all possible white matter connections in the whole brain. There are generally two categories of methods: cortical-parcellation-based methods and fiber clustering methods (O’Donnell et al., 2013). The cortical-parcellation-based methods are more widely used as they enable construction of a connectivity matrix and its subsequent analysis using techniques from graph theory (as described in Section 5.6) (Bassett and Bullmore, 2017; Bullmore and Sporns, 2009a; Gong et al., 2009a; Ingalhalikar et al., 2014; Sporns et al., 2005; Yeh et al., 2016b; Zalesky et al., 2012). Though relatively less used, fiber clustering tractography parcellation methods are increasingly applied to study the brain’s structural connectivity in applications such as disease classification and between-population statistical analysis (Feng et al., 2020; Ji et al., 2019; Wu et al., 2018; Zhang et al., 2018a; 2018b). (Note that these methods could perhaps more correctly be called “streamline clustering,” but the term “fiber clustering” is very established in the literature.)
The cortical-parcellation-based methods work from a gray-matter-centric perspective. They parcellate tractography according to a cortical (and sometimes a subcortical) gray matter parcellation, focusing on the structural connectivity among different gray matter ROIs (Bassett and Bullmore, 2017; Bullmore and Sporns, 2009a; Gong et al., 2009a; Ingalhalikar et al., 2014; Sporns et al., 2005; Yeh et al., 2016b; Zalesky et al., 2012). Specifically, tractography segmentation is performed by extracting streamlines that connect pairs of ROIs. Therefore the resulting tractography segmentation is mainly determined by the selection of a cortical parcellation scheme. The majority of methods adopt a cortical parcellation that is computed from T1-weighted or T2-weighted MRI. The most popularly used cortical parcellation is the Freesurfer Desikan-Killiany cortical parcellation (Desikan et al., 2006; Fischl, 2012), while many other cortical parcellation schemes have also been widely used (Destrieux et al., 2010; Shattuck et al., 2008; Tzourio-Mazoyer et al., 2002). Many cortical-parcellation-based methods have also used a functional cortical parcellation computed using functional MRI data (Eickhoff et al., 2005; Glasser et al., 2016; Schaefer et al., 2018; Yeo et al., 2011). Finally, an alternative approach is to use a vertex-wise cortical “parcellation,” i.e., by identifying streamlines that connect pairs of vertices in the cortical surface (Besson et al., 2014; Fellner et al., 2020; Tian et al., 2021). Currently, there is no consensus about which brain parcellation technique could be most useful (de Reus and Van den Heuvel, 2013; Yeh et al., 2020; Zalesky et al., 2012).
The fiber clustering methods work instead from a white-matter-centric perspective. They group tractography streamlines according to their geometric trajectories, describing the structural connectivity according to the white matter anatomy (Avila et al., 2019; Chekir et al., 2014; Garyfallidis et al., 2018; Guevara et al., 2012; Li et al., 2010; O’Donnell and Westin, 2007; Román et al., 2017; Ros et al., 2013; Siless et al., 2018; 2020; Tunç et al., 2014; Vázquez et al., 2020; Wu et al., 2020a; Yeh et al., 2018; Yoo et al., 2015; Zhang et al., 2018c; Ziyan et al., 2009). In general, fiber clustering methods start with a computation of pairwise streamline geometric similarities, followed by a computational clustering method to group similar streamlines into clusters. Compared to the cortical-parcellation-based methods that focus on streamline terminal regions, fiber clustering methods leverage the full lengths of the streamline trajectories. As a result, the cortical-parcellation-based methods can erroneously group streamlines that follow completely different trajectories through the white matter but nevertheless end up at the same gray matter endpoints, whereas fiber clustering can be insensitive to streamlines diverging at the cortex if they pass through the same deep white matter regions. In addition, unlike the cortical-parcellation-based methods that leverage additional cortical parcellation information, the majority of fiber clustering methods work on tractography data only, such that there is no need for an inter-MR-modality registration, e.g., an image registration between dMRI and T1w images that can be affected by differences in image resolution (Malinsky et al., 2013) and echo-planar imaging (EPI) distortion in dMRI data (Albi et al., 2018). Due to these factors, several studies have demonstrated advantages of fiber clustering methods over the cortical-parcellation-based methods, including more consistent parcellation across subjects (Sydnor et al., 2018; Zhang et al., 2017) and a higher reproducibility between test-retest scans (Zhang et al., 2019).
When choosing a method to perform parcellation of the entire white matter, the previously described points related to reproducibility and consistency of anatomical tract identification across different populations and acquisitions (as described in Section 4.1) are also important facts to consider (Buchanan et al., 2014; Smith et al., 2015a; Zhang et al., 2019; 2018d).
An additional point to be highlighted for parcellation of the entire white matter is the scale or the granularity of the parcellation, i.e., how many white matter parcels are to be obtained after parcellation. For constructing the connectome, this is related to the choice of the cortical parcellation (Hagmann et al., 2008a; Messé, 2020), while for fiber clustering this is related to the number of fiber clusters obtained by the clustering algorithms (Wu et al., 2020a; Zhang et al., 2018c). Various parcellation scales have been applied in different studies, ranging from tens to millions of white matter parcels (Besson et al., 2014; Liu et al., 2017; Osmanlıoğlu et al., 2020; Rodrigues et al., 2013; Zalesky et al., 2010; Zhang et al., 2018a). Choosing parcellation scales depends on the target application, e.g., studies have suggested a fine scale white matter parcellation (over 2000 parcels) can be beneficial for machine learning and statistical analysis (Liu et al., 2017; Zhang et al., 2018a) while a coarse scale parcellation (fewer than 200 parcels) can improve the connectome consistency across individuals (Osmanlıoğlu et al., 2020; Rodrigues et al., 2013). We note that inter-subject connectome consistency may also be improved using other approaches, e.g., consistency-based thresholding (Baum et al., 2018; Roberts et al., 2017).
5. Performing quantitative analysis: methods for tractography quantification
The goal of tractography quantification is to extract quantitative measures that are useful to assess the structural connectivity of the brain’s white matter pathways. In this section, we will first introduce the quantitative measures that can be computed from tractography (Section 5.1). Then we will focus on how to extract these measures within an individual white matter fiber pathway (Section 5.2) and how to perform filtering techniques to reduce potential biases in the extracted measures (Section 5.3). Next, we introduce how the extracted measures can be used for tract-specific analysis of anatomical white matter tracts (Section 5.4) and for computing the edge weights to construct a connectivity matrix (Section 5.5). Finally, we will introduce topological analyses of whole-brain connectivity based on graph theoretical measures and more advanced connectome measurements (Section 5.6).
5.1. Quantitative measures computed from tractography
There are many quantitative measures that can be computed from tractography, which can be subdivided into two categories based on the source of data used to compute the measures. The first category of quantitative measures are based on the tractography data alone, which are often used to define the connectivity strength (Jones, 2010; Smith et al., 2020a; Sotiropoulos and Zalesky, 2019; Yeh et al., 2020). The number of streamlines (NOS) is one widely used measure to quantify the connectivity strength (Hagmann et al., 2008b; Ingalhalikar et al., 2014; Roberts et al., 2016; Sporns et al., 2005; van Dellen et al., 2018). Several studies conducted on animals using invasive tract-tracing techniques have shown good agreement between the NOS and the biological connectivity as measured using ex vivo tract-tracing data (Delettre et al., 2019; Girard et al., 2020; van den Heuvel et al., 2015a). However, because of the difference in resolution between the measured dMRI signal and actual axon dimensions (as described in Section 3.3: “Connection density biases”), many studies have emphasized that NOS does not provide a truly quantitative measure of connection strength (Jones, 2010; Jones et al., 2013; Sotiropoulos and Zalesky, 2019; Yeh et al., 2020). Recent work aims to provide alternative, more quantitative measures of connectivity that better estimate the underlying connection density (Smith et al., 2020a) or modulate streamline contributions toward aggregate measures of connectivity (see Section 5.3). Other quantitative measures can be computed directly from tractography, such as the volume and the length of a fiber tract (Bajada et al., 2019; Behrman-Lay et al., 2015; Catani et al., 2007; Chandio et al., 2020a; Colby et al., 2012a; Grinberg et al., 2018; Rheault et al., 2020a; Song et al., 2015; Yeatman et al., 2012), have also been used.
The second category of quantitative measures computed from tractography leverages microstructural measures that are usually computed from a diffusion model (e.g., diffusion tensor model) or another quantitative imaging modality (e.g., myelin imaging). In this category of measures, tractography is generally used to define the locations in which microstructural measures are sampled. (This sampling may be done after performing tractography, or during performing tractography as part of sampling or computing a diffusion model). The most widely used microstructural measures are based on computational modeling of the dMRI signal. Popularly used diffusion-modeling-based microstructural measures include the fractional anisotropy, axial, radial and mean diffusivities (FA, AD, RD and MD respectively) derived from traditional diffusion tensor model (Basser and Pierpaoli, 1996). More advanced measures include those reflecting inter- and intra-cellular signal fractions derived from Diffusion Kurtosis Imaging (DKI) (Jensen and Helpern, 2010), Free Water modeling (Pasternak et al., 2012; 2009), Neurite Orientation Dispersion and Density Imaging (NODDI) (Zhang et al., 2012), Spherical Mean Technique (SMT) (Kaden et al., 2016), or Apparent Fiber Density (AFD) (Raffelt et al., 2012). It is important to be aware that changes in dMRI microstructural measures can be non-specific, in part due to the difference in scales between voxel-averaged dMRI signals (mm scale) and the scale of the individual axons and cells that are probed by the diffusing water (micrometer scale). For example, many factors (e.g., cell death, edema, gliosis, inflammation, change in myelination, increase in connectivity of crossing fibers, increase in extracellular or intracellular water, etc.) may cause changes in FA (Assaf and Pasternak, 2008; Jones et al., 2013; Le Bihan and Johansen-Berg, 2012). More details about sensitivity, specificity and interpretability of these microstructural measures are out of the scope of the present review but the reader can refer to the following studies (Alexander et al., 2019; Jelescu and Budde, 2017; Novikov et al., 2019; 2018). Other approaches for tractography quantification leverage information from imaging modalities other than dMRI, such as T1-weighted (T1w) imaging (Boshkovski et al., 2020a; Yeatman et al., 2014a), myelin sensitive maps (Lee et al., 2020; Mancini et al., 2020; Piredda et al., 2021) and g-ratio (Campbell et al., 2018). For example, researchers have used the longitudinal relaxation rate (R1), a measure sensitive to myelin, along the fiber tracts (Boshkovski et al., 2020a; Yeatman et al., 2012).
These microstructural measures can be computed along fiber pathways using several approaches. Usually, this is done by computing a model in each voxel to derive a 3D microstructural map (e.g., an FA image) and then using tractography to define the locations from which the values of this microstructural measure should be sampled. This sampling may be done by first generating a binary mask that defines the voxels through which streamlines pass and then sampling from an underlying microstructure image within this mask (Ciccarelli et al., 2003; Heiervang et al., 2006; Kunimatsu et al., 2004; Voineskos et al., 2010). The sampling may alternatively be done by sampling microstructural measures at each point of the streamlines from a pre-calculated microstructure image (Ciccarelli et al., 2003; Heiervang et al., 2006; Kunimatsu et al., 2004; Voineskos et al., 2010). Instead of using a pre-calculated microstructure image, the diffusion or fiber model can be simultaneously estimated when performing tractography so that the microstructural measures are directly computed at points along each streamline (Girard et al., 2017; Gong et al., 2018; Malcolm et al., 2010; Olszewski et al., 2017; Reddy and Rathi, 2016).
5.2. Domain of analysis: extraction of quantitative measures within white matter fiber pathways
After choosing a quantitative measure of interest (Section 5.1) and a method for tractography segmentation (Section 4), then there are many methods available to extract the measure within an individual white matter fiber pathway. This is important to enable tract-specific analysis (Section 5.4) and construction of a connectivity matrix (Section 5.5). Two main strategies can be employed: using a scalar value as a summary statistic, or using data along the length of the fiber pathway.
The most commonly adopted strategy for extraction of quantitative measures is to compute a scalar value as a summary statistic of the fiber pathway. (This is required for connectome-based analysis and is a popular approach for tract-specific analysis). While some quantitative measures intrinsically provide a single scalar value per pathway, e.g. NOS or tract volume, others (e.g. sampled values of a quantitative metric) necessitate calculation of some statistic in order to produce such a scalar. For microstructural measures, the mean measure within the fiber pathway is the most widely used. Other summary statistics such as the median, maximum and minimum have also been employed (Boshkovski et al., 2020b; Zhang et al., 2018a; 2018b). A summary statistic can be obtained in several ways according to how microstructural measures are computed along fiber pathways (as described in Section 5.1). It can be computed within a binary mask defining a fiber pathway in a microstructure image (e.g. the mean FA within the mask) (Ciccarelli et al., 2003; Heiervang et al., 2006; Jolly et al., 2021; Kunimatsu et al., 2004; Voineskos et al., 2010; Wilson et al., 2021). It can also be computed by averaging microstructural measures sampled along each point of the streamlines (Gong et al., 2018; Jones et al., 2006; Olszewski et al., 2017; Zhang et al., 2018a; 2019). The choice of the best summary statistic for data measured within a fiber pathway is still open. For example, compared to the mean statistic, studies have shown that the median can be more robust against outliers and does not rely on the normality assumption for the distribution of the microstructure parameter along a streamline (Boshkovski et al., 2020b; Zhang et al., 2018b). One study has suggested that the maxima and minima are more discriminative than the mean value in machine-learning-based disease classification (Zhang et al., 2018a).
The second strategy to quantify an individual fiber pathway is to measure the distribution of the microstructural measures along the fiber pathway. This can enable the study of the tissue microstructure in local regions along the fiber pathway. This approach requires definition of a coordinate system, sampling framework, or surface-based representation to define how to sample the microstructural data of interest (Bells et al., 2011a; Chen et al., 2016b; Colby et al., 2012b; Corouge et al., 2006; Goodlett et al., 2009; Irimia et al., 2020; Jones et al., 2005; O’Donnell et al., 2009; Yeatman et al., 2012; Yeh, 2020). Most often, data are averaged across corresponding points along each streamline in the pathway, such that data can be analyzed (e.g., an along-tract or along-pathway plot) versus streamline arc length or other parameterization (Colby et al., 2012b; Corouge et al., 2006; Yeatman et al., 2012). Other approaches fit a medial surface representation to the pathway, such that sampled data can be represented in two dimensions and can be analyzed not only along the pathway but also across its cross-section (Chen et al., 2016b; Qiu et al., 2010; Yushkevich et al., 2009).
5.3. Quantitative measures using filtering techniques
The so called filtering methods discussed in Section 3.3 can also be used to perform “microstructure-informed” tractography with the aim to extract quantitative measures of white matter fiber pathways and reduce potential bias in the estimation of connectivity. Assuming that the microstructure properties corresponding to a single streamline remain constant along its path, these methods assign a specific microstructural property to each reconstructed pathway. The underling assumption comes from the fact that a streamline reconstructed from tractography cannot represent a single axon, but rather a group of axons following the same trajectory, and thus we can suppose that on average the microstructure properties the magnetic resonance signal is sensitive to at achievable resolution remain constant. While in its original implementation SIFT (Smith et al., 2013) provides only the NOS adjusted based on voxel-wise spherical deconvolution, its evolution SIFT2 (Smith et al., 2015b) optimizes per-streamline cross-section multipliers to match a whole-brain tractogram to fixel-wise fiber densities and thus provides a value for each streamline that can be used (for example) to define the edge weights of the connectome. Similarly, LiFE (Pestilli et al., 2014), COMMIT (Daducci et al., 2013; 2014; Schomburg and Hohage, 2019), COMMIT2 (Schiavi et al., 2020a) and COMMIT2tree (Ocampo-Pineda et al., 2021) deconvolve the measured dMRI signal on the streamlines and assign a single contribution, or “weight”, to each one of them using classical multi-compartment models (Panagiotaki et al., 2012). These methods have been used to investigate properties of healthy and pathological brains, showing better performance than the standard NOS (Schiavi et al., 2020b; Smith et al., 2015a; Yeatman et al., 2014b). In particular, because of their flexible implementations, COMMIT, COMMIT2 and COMMIT2tree allow complementing tractography with biophysical models of the tissue microstructure and, thus, enable one to access more quantitative and biologically informative features of individual bundles, such as average axon diameter (Barakovic et al., 2018), myelin content (Schiavi et al., 2019) and bundle-specific T2 (Barakovic et al., 2021b).
5.4. Tract-specific analysis using statistical or machine learning techniques
Once a quantitative measure has been extracted for individual fiber pathways, several options exist for tract-specific analysis, which may be hypothesis-driven using statistical analysis or data-driven using machine learning. These methods may rely on a summary statistic per tract, or analyze data along a tract.
One widely used analysis is based on a hypothesis-driven strategy to assess if there are statistical differences of tract(s)-of-interest between groups (e.g., between health and disease, or between different subtypes of a disease). Usually, given a quantitative measure of interest, a selected summary statistic (e.g. mean of the chosen microstructural measure) is extracted from the tract(s)-of-interest and is used to compute the level of group differences (e.g., the p-value) using statistical group-wise comparison methods such as Student’s t-test, ANOVA, or other more advanced statistical analysis methods (Habeck and Stern, 2010; Martí-Juan et al., 2020; Ombao et al., 2016; Zhang et al., 2018b). Another hypothesis-driven analysis uses a regression model (e.g., a generalized linear model (Dobson and Barnett, 2018) or support vector regression (Zhang and O’Donnell, 2020)) to assess correlation of the tract quantitative measurement and a behavioral or a disease symptom score. Such analyses have been applied to study, e.g., how white matter fiber tracts are affected in individuals with different disease severity (Cha et al., 2015; Chiang et al., 2016; Price et al., 2007), how tracts relate to cognitive and socio-emotional function (Zekelman et al., 2021), and how tracts are developing during human neurodevelopment and aging (Gertheiss et al., 2013; Hasan et al., 2009a; Michielse et al., 2010). One important point to note is that correction for multiple comparisons is needed if there are multiple tracts and/or multiple quantitative measurements analyzed. Commonly used multiple comparison correction methods include false discovery rate (FDR) (Benjamini and Hochberg, 1995) and Bonferroni (Holm, 1979) methods. However, it is important to highlight that such multiple comparison correction methods, which assume independence across data samples, can be overly conservative when applied to tests performed on multiple tracts and/or quantitative measurements that are not independent. For example, MD and FA are not mutually mathematically orthogonal measures (Ennis and Kindlmann, 2006).
Another tract-specific analysis strategy is data-driven and uses machine learning techniques to perform tasks such as disease classification and prediction (Deng et al., 2019; O’Dwyer et al., 2012; Payabvash et al., 2019; Zhang et al., 2018a), as well as prediction of behavioral measures and traits (Seguin et al., 2020a; Tian et al., 2021). In machine learning analysis, the quantitative measures computed from individual fiber pathways are treated as feature descriptors, which are input to a machine learning algorithm (e.g., a support vector machine) to train a model from a set of training samples with known information (e.g., labels such as disease or healthy control). Then the trained model can be used to predict new samples. Unlike statistical analysis methods that usually aim to find white matter structures with statistical group differences, the machine-learning-based methods aim to offer predictive relevance. This can be advantageous in situations where the within-group heterogeneity is large (e.g., in psychiatric diseases) and thus a statistical group comparison, which essentially compares the “average patient” to the “average control,” is not informative. Another advantage of machine learning is that it can be easily applied to multimodal data. For example, the combination of structural and functional connectivity measures has been shown to increase predictive power (Whitfield-Gabrieli et al., 2016).
Instead of using a single scalar value to summarize the entire pathway, many methods perform along-tract analysis to investigate the distribution of the microstructural measures along the fiber pathway (variously called “tractometry,” “profilometry,” and “tract-based morphometry”) (Bells et al., 2011b; O’Donnell et al., 2009; Yeatman et al., 2012). Along-tract analysis enables the study of local regions along the tract. After mapping microstructural measures along each point of the streamlines along the fiber pathway, a widely used strategy is to analyze data along the tract profile. One popular approach is to average data across corresponding points along each streamline in the pathway and leverage a prototype, core, or mean streamline to enable statistical analysis comparing subjects or groups (Colby et al., 2012b; Wang et al., 2016; Yeatman et al., 2012). Other approaches can employ streamlines of the entire tract in the statistical analyses (Chandio et al., 2020b). Additional studies have analyzed tracts represented as surfaces (Chen et al., 2016b; Qiu et al., 2010; Yushkevich et al., 2009) or performed data reduction techniques to reduce the dimensionality of the point-wise measurements along tracts (Ceschin et al., 2015; Chamberland et al., 2019; Geeraert et al., 2020). Several studies have shown that along-tract analysis can be more sensitive to detect differences that are not apparent in the mean value within the tract (Colby et al., 2012b; O’Donnell et al., 2009; St-Jean et al., 2019).
5.5. Construction of a structural connectivity matrix
Quantitative measures extracted for individual fiber pathways can also be used to construct a structural connectivity matrix to map the whole-brain network of structural connectivity between all pairs of gray matter regions (Hagmann et al., 2008a; Sporns et al., 2005). This approach is motivated by a general shift of focus away from the roles of isolated regions towards understanding the brain as a networked system (Barch et al., 2016; Friston, 2002). Construction of the connectivity matrix has been described in several reviews (Sotiropoulos et al., 2016; Yeh et al., 2020) and includes two major steps. First, parcellation of the whole-brain tractogram is needed to compute all possible fiber pathways in the brain, i.e., white matter parcels connecting all pairs of gray matter regions (as described in Section 4.2). Then, a metric of “connectivity” is needed to define quantitative edge weights in the connectivity matrix (see Fig. 1(e) for a graphic illustration). This is usually done by computing a scalar value for each white matter parcel (as described in Section 5.2). The most popular scalar value for connectivity matrix analysis is the NOS (Hagmann et al., 2008b; Ingalhalikar et al., 2014; Roberts et al., 2016; Sporns et al., 2005; van Dellen et al., 2018), where higher values are considered to indicate stronger connectivity between pairs of gray matter regions. Other scalar values that are generally interpreted as measures of connectivity “strength” include microstructural measures, e.g., FA (Bathelt et al., 2017) and R1 (Boshkovski et al., 2020a). For scalar values that are interpreted as inversely related to connectivity “strength,” such as RD, MD, or maps reflecting extracellular or isotropic/free water components of the signal, a transformation of the connectivity matrix may be performed using the reciprocal or the log function prior to any subsequent connectivity analysis (Fornito et al., 2016).
While the construction of the connectivity matrix is straightforward, multiple factors that need careful consideration have been pointed out by researchers. Selection of a gray matter parcellation scheme (as described in Section 4.2) is the first essential consideration when constructing a structural connectivity matrix, and it has been shown to be highly consequential in connectome analysis (de Reus and Van den Heuvel, 2013; Yeh et al., 2020; Zalesky et al., 2012). Myriad components of the processing pipeline can also influence the resulting connectome matrix data, even down to the level of detail of the mechanism by which streamlines are assigned to those parcels (Yeh et al., 2019a). In addition, connectomes are usually post-processed to reduce potential bias in the estimation of graph measures. This step is typically done by simply thresholding according to some rule or to reach a predefined density, or using filtering techniques described in Section 5.3. Selection of tractography filtering strategies is important and has been shown to affect the analysis of the topology of brain networks (Civier et al., 2019; Frigo et al., 2020; Yeh et al., 2016a) and affect sensitivity and specificity when analyzing the connectomes (Zalesky et al., 2016a). Moreover, as concluded in several studies (Bastiani et al., 2012; Qi et al., 2015; Yeh et al., 2016a), several other factors (such as q-space sampling schemes, fiber orientation models, and tractography algorithms) that are not specifically reviewed in our paper, have also been shown to affect most network measures derived from the connectivity matrix.
5.6. Graph theoretical measures and advanced structural connectome measurements
A structural connectivity matrix defines a network (or graph) of whole-brain anatomical connectivity, where the nodes are typically gray matter regions or ROIs, and the edges represent the connectivity between these regions (Hagmann et al., 2008a; Sporns et al., 2005) (Fig. 4a). The disciplines of graph theory and network science provide a framework to quantify network properties of brain connectivity (Rubinov and Sporns, 2010) and investigate whole-brain models of structural wiring (Betzel et al., 2016). Over the last two decades, the study of the brain as a network has become increasingly popular in the neuroscience and neuroimaging communities, giving rise to the new fields of connectomics and network neuroscience (Bassett and Sporns, 2017; Fornito et al., 2013; 2015; 2016).
Fig. 4.

Graph theoretical analysis of the human connectome. (a) Network diagram of structural connectivity, where nodes represent gray matter ROIs and connections depict white matter fiber bundles. (b) Schematics of key graph theory concepts applied to the analysis of structural connectivity. The thickness of connections indicates the strength of structural connectivity between two ROIs (e.g., NOS or integrated FA). Region-level graph measures such as degree, strength, and centrality identify ROI B as an important hub in the network. At the mesocale, two modules or communities are observed. Intra-module connectivity is dense and inter-module connectivity in sparse. At the global scale, communication paths delineate multi-step sequences linking anatomically unconnected ROIs. Two possible paths connecting ROIs A and C are highlighted in green (shortest path) and orange (alternative path).
A daunting number of graph theoretical measures have been proposed to study the network properties of the brain’s connectome (Bullmore and Sporns, 2009b; Iturria-Medina et al., 2008; Rubinov and Sporns, 2010). The choice of which measures to focus on is dependent on the research questions and hypotheses at hand. Graph measures can be classified based on their resolution or scope—from local measures that quantify properties of individual ROIs (nodes), to mesoscale measures describing clusters of interconnected ROIs, to global measures that describe whole-brain connectivity properties (Betzel and Bassett, 2017; Fornito et al., 2016) such as network communication in the brain (Graham et al., 2020) (Fig. 4b). In the following paragraphs, we give examples of these popular graph measures that quantify aspects of brain networks at each level, from local to global.
At the local level, node centrality measures provide a useful characterization of the integrative importance of individual brain regions (Sporns et al., 2007). Degree and strength are the simplest and most popular node centralities, quantifying, respectively, the number of connections and the sum of connection weights of individual regions (see Oldham et al., 2019 for a review on advanced node centrality measures). Centrality measures find particular utility in identifying hub regions that may play a central role in integrative brain function (van den Heuvel and Sporns, 2013). Indeed, hubs identified with these measures have been found to entail high metabolic expenditures and be disproportionately implicated in brain disorders (Crossley et al., 2014; Fornito et al., 2015).
At the mesocale, structural connectivity can be partitioned into modules (also known as communities) that reflect the presence of clusters embedded in the brain’s wiring (Betzel, 2020; Sporns and Betzel, 2016). The practice of uncovering such clusters, called community detection or modular decomposition, helps to summarize the complexity of structural networks into coarse-grained blocks, aiding the identification of meaningful sub-networks and connectivity motifs relevant to specific research questions. Typically, ROIs belonging to the same module are densely and strongly interconnected, while connectivity between modules is sparse. This modular structure constrains the flow of information through the network in a manner thought to promote functional segregation and clusters of specialized information processing (Betzel et al., 2013). Popular methods for community detection in structural brain networks include the Louvain algorithm and stochastic block models, though determining best practices around the utilization of these and other approaches remain an active topic of research (Fortunato and Hric, 2016).
At a global scale, graph measures can be used to describe how the brain’s structural wiring supports the communication of information between distant ROIs and cognitive systems (Avena-Koenigsberger et al., 2018; Mišić et al., 2015). While structurally connected regions can directly communicate, signal propagation between unconnected ROIs requires a sequence of one or more intermediate connections to enable communication. Graph measures of network communication presume that the strength of structural connections reflects the capacity of fiber bundles to support fast and reliable propagation of electrical signals. Connections with, e.g., large NOS, high integrated FA, or short average streamline length are typically considered to support efficient network communication. Traditionally, connectome communication has been quantified using graph measures based on the notion that neural information transfer occurs via topological shortest paths. Shortest-path-based measures include, e.g., the popular characteristic path length and network efficiency (Rubinov and Sporns, 2010). However, algorithmically, the identification of shortest paths demands individual regions to possess global knowledge of structural connectivity, a requirement unlikely to be met in decentralized systems such as the brain (Avena-Koenigsberger et al., 2019; Goñi et al., 2014; Seguin et al., 2018). In addition, connectome networks are known to strike a balance between wiring minimization (shortest paths) and formation of connections between brain regions with similar connectivity profiles (homophily) (Betzel et al., 2016).
In light of these shortcomings, recent work has stated to focus on alternative models of connectome communication, such as network diffusion (Estrada and Hatano, 2008; Goñi et al., 2014), linear transmission models (Mišić et al., 2015), and navigation (Seguin et al., 2018) (see Avena-Koenigsberger et al., 2018 for a review). Determining which approaches most accurately reflect underlying biological processes remains a crucial open question (Seguin et al., 2020b). Nonetheless, recent work has shown that advanced communication measures can elucidate how the organization of structural connectivity shapes the brains functional dynamics (Goñi et al., 2014; Imms et al., 2021; Mišic et al., 2018; Wang et al., 2019). For instance, despite the inherent limitations of dMRI tractography in resolving axonal fiber directionality, graph measures computed on the undirected human connectome can reveal patterns of asymmetric communication, as evidenced by significant associations to the directionality of effective connectivity computed using dynamic causal modeling (Seguin et al., 2019). Therefore, the use of advanced communication measures enables the study of directional and asymmetric functional interactions from structural connectivity mapped with dMRI.
6. Applications of quantitative tractography analysis
In the last two decades, modern dMRI-based tractography that offers quantitative measurements of fiber tracts has extended neuroanatomy methods and become a key technique for the study of the brain’s white matter in a wide range of applications. In this section, we focus on several essential areas including development, aging, neurological disorders, mental disorders, and neurosurgery. These studies can be organized into two types that correspond to the above-mentioned tract-specific and connectome-based analyses. The first category concerns major fiber pathways or localized structural connectivity in order to capture fine-grained details of brain circuits. The second category regards the entire white matter as a complex system and thus adopts clustering analysis for feature reduction or utilizes graph theoretical analysis to quantify topological characteristics with respect to information integration and segregation.
6.1. Developmental tractography of the brain
Brain white matter undergoes highly ordered changes during the ontogeny from a neural tube to a complex connectome. In the past decade, researchers have implemented dMRI tractography investigations across broad brain developmental stages ranging from the middle fetal stage to adulthood (Cao et al., 2017; Dubois et al., 2014; Gilmore et al., 2018; Huang, 2010; Lebel and Deoni, 2018; Lebel et al., 2019; Qiu et al., 2015; Zhao et al., 2019b).
For the prenatal stage, most tractography studies aim to identify the migration pathways of neurons and emergence of key tracts of the brain as important developmental milestones (Dubois et al., 2014; Gilmore et al., 2018; Huang, 2010; Vasung et al., 2019). For instance, using ex vivo dMRI scans on fetal brain samples, researchers are able to identify the major tract parts of the fornix and cingulum bundles at 13 weeks, but a part of the corpus callosum at 15 weeks (Huang et al., 2009). Similar studies also show that the radial and tangential pathways organization are prominent at 17 weeks but gradually disappear at later gestational ages, and all major structural pathways are already identifiable by term (Takahashi et al., 2002). These results are consistent with classical histological studies (Mitter et al., 2015; Vasung et al., 2010). Due to recent advances in methodological progress such as motion correction algorithms, in vivo tractography on the fetal brain has become available and exhibit highly agreement with ex vivo and neuroanatomical studies (Christiaens et al., 2021; Khan et al., 2019), which indicates strong potential for future prenatal brain investigations.
In the postnatal stage, in vivo tractography offers a superior tool to chart the spatiotemporal maturation patterns of the reconstructed tracts by employing quantitative dMRI metrics from the infancy period (Dubois et al., 2014; Gilmore et al., 2018; Lebel and Deoni, 2018; Qiu et al., 2015; Vasung et al., 2019) to the adolescent stage (Lebel et al., 2019; Tamnes et al., 2018). Despite focusing on different developmental ranges, these studies consistently observe significant age-related increases of FA and MD in widespread fiber tracts during postnatal growth in both cross-sectional and longitudinal population designs (Lebel and Deoni, 2018). Studies also characterize several typical nonlinear trajectories (piecewise, exponential and quadratic) of diffusion metrics for major tracts across large developmental ranges such as infancy to childhood (Pecheva et al., 2017; Reynolds et al., 2019; Stephens et al., 2020) or adolescence to adulthood (Chang et al., 2015; Lebel and Beaulieu, 2011). Researchers can further estimate the growth of tracts at the subsystem level by multivariate analysis; for example, one study investigated the maturational calendars of linguistic bundles and found that the dorsal pathway development falls behind the ventral pathway at birth and catches up later at the first postnatal months (Dubois et al., 2016).
Using the newly emerging developmental connectomics framework, researchers are able to seek the wiring principles of brain white matter tracts (Cao et al., 2017; 2016; Collin and Van Den Heuvel, 2013; Gilmore et al., 2018; Ouyang et al., 2019; Tymofiyeva et al., 2014; Vértes and Bullmore, 2015; Zhao et al., 2019b). Many studies have discovered largely adult-like topological structures including significant small-world, modular, and rich-club organization during the middle to final trimester of gestation (Brown et al., 2014; Song et al., 2017; Tymofiyeva et al., 2013; Van Den Heuvel et al., 2015b; Zhao et al., 2019b), which reveals that the whole brain tract network is already highly efficient and specialized around term age. During prenatal development, the whole network becomes increasingly efficient and segregated with typical system-level refinements such as increased small-worldness (Brown et al., 2014) and increased modularity (Van Den Heuvel et al., 2015b). After birth until adulthood, the brain network reshapes to improve integration capacity with the reconfigurations of decreased modularity and decreased small-worldness (Chen et al., 2013; Dennis et al., 2013; Hagmann et al., 2010; Huang et al., 2015; Tymofiyeva et al., 2013). In terms of regional changes, although neonatal brains already show similar structural hub distribution as adult brains, many refinements occur for the topological roles for each node (Yap et al., 2011). The nodal efficiency of provincial hubs and the strength of within-module connections (Zhao et al., 2019a) develop fast during the prenatal stage, and hubs expand into the inferior frontal and insula regions at term age (Ball et al., 2013; Van Den Heuvel et al., 2015b). After birth, the left anterior cingulate gyrus and left superior occipital gyrus become hubs in toddler brains (Huang et al., 2015). The centrality of the pre-cuneus and cuneus still increases before pre-adolescence (Huang et al., 2015; Yap et al., 2011).
The interpretation of the above tract-related changes largely depends on the corresponding biological changes during development. In the fetal brain, it is probably caused by pre-myelination phases such as proliferation and maturation of oligodendrocyte and progenitor cells, the glia that form the myelin sheath (Jakovcevski et al., 2009). In the early postnatal stage, ongoing myelination is a clear cause (Paydar et al., 2014) while axonal packing and decreasing water content (Neil et al., 2002) may also make a contribution. In later childhood and adolescence, results seem to be attributed to changes of neurite density, particularly axonal packing (Lebel and Deoni, 2018).
6.2. Aging and lifespan tractography of the brain
Aging is associated with broad alterations of axon density and axonal structure. Researchers have performed many tractography investigations to trace the normal aging patterns and lifespan trajectories of brain connectivity across wide age ranges, spanning childhood until old age (Collin and Van Den Heuvel, 2013; Madden et al., 2009; Moseley, 2002).
Early tractography studies usually estimated the aging effect on specific fiber bundles such as the corpus callosum (Sullivan et al., 2006), uncinate fasciculus (Hasan et al., 2009a), and fornix (Stadlbauer et al., 2008b), which show consistent changes including decreased FA, increased MD and decreased tract volume. By subdividing across or along certain tracts (as described in Section 5.4), researchers have been able to delineate the elaborate aging patterns within a tract (Davis et al., 2009; Michielse et al., 2010; Sullivan et al., 2006). For instance, FA values of the corpus callosum declined with aging only in the genu part while not in the body and splenium parts in participants aged 22–84 years (Michielse et al., 2010). Using various tract identification methods, many studies have investigated a wide range of tracts (Davis et al., 2009; Stadlbauer et al., 2008a; Sullivan et al., 2010; Westlye et al., 2010), revealing a typical spatial degeneration pattern across brain systems in which prefrontal association tracts are the most vulnerable to aging, while cingulum, temporal and parietal-occipital commissural connections are relatively preserved. From a lifespan view (Hasan et al., 2010; 2009b; Lebel et al., 2010; 2012; Yeatman et al., 2014a), although most tracts show increased FA during childhood and adolescence and decreased FA at older ages, the age when FA reaches its highest peak varies across different white matter tracts. In general, the anterior and posterior parts of tracts peak earlier than the central part of tracts, around 20 to 40 years old (Lebel et al., 2010), supporting a classical “last-in-first-out” theory. Recently, with increasing numbers of adult imaging datasets and development of automated tract identification pipelines (as described in Section 4.1), researchers have began to assess the effect of aging on the brain’s tracts by employing thousands of participants (Cox et al., 2016; de Groot et al., 2015; Tseng et al., 2021), which brings us into an unprecedented “big data” era (Xia and He, 2017).
From a connectomics perspective (as described in Section 5.6), tractography studies show that the basic layouts of the brain’s structural network are largely preserved until old age, such as the economical small-world character, the modular organization and rich club organization (Gong et al., 2009b; Li et al., 2020c; Wen et al., 2011; Wu et al., 2012a). Meanwhile, both global and local alterations of the brain’s structural topology occur with aging, including decreased network efficiency (Bi et al., 2021; Gong et al., 2009b; Li et al., 2020c; Madden et al., 2020; Shu et al., 2018b; Wu et al., 2012a) and decreased inter-/intra-modularity strength that may be due to the widespread degeneration of whole brain tracts (Li et al., 2020c; Wu et al., 2012a) and loss of frontal hubs that may be driven by the vulnerable prefrontal tracts (Zhao et al., 2015). These tractography-based network changes may offer potential imaging markers for individualized cognition. As strong evidence, a recent large sample study implicates the global dimensions of variation in the human structural connectome in aging-related cognitive decline (Madole et al., 2020). Lifespan changes of the connectome also show good consistency with cognitive maturation (Zhao et al., 2015). For instance, following an inverted U-shaped trajectory, the peak ages of network efficiency and small-world measures (30 years) fall within the range of peak ages on general cognitive performance (Schaie, 2005; Schroeder and Salthouse, 2004), and brain regions within the default mode network, which is involved broadly in high-level cognition (Raichle, 2015; Vatansever et al., 2018; 2017), display the latest maturation age of peak.
These various tract-related changes could be attributable to different physiological brain changes at older ages. For instance, decreased axonal density and loss of fibers may cause a decline of FA values, while an increased amount of water content along with healthy atrophy may cause increased MD (Moseley, 2002). Results from a study that employ an advanced diffusion model (NODDI) indicate that the age-related declines in FA are primarily induced by declines in neurite density rather than changes in tract complexity (Cox et al., 2016). Using multi-modal dMRI and PET imaging analysis, researchers further interpreted the metabolic mechanisms underlying aging-related changes of brain tractography, in which they found that the overall connectome efficiency-metabolism coupling across brain regions significantly increased with aging (Bi et al., 2021).
6.3. Tractography in neurological and mental brain disorders
dMRI tractography has become an indispensable tool for studies on various brain disorders including, but not limited to, multiple sclerosis (MS) (Fleischer et al., 2019; Sbardella et al., 2013), amnestic mild cognitive impairment (aMCI) (Bai et al., 2009; Zhao et al., 2017a), Alzheimer’s disease (Jack and Holtzman, 2013; Lo et al., 2010; Mito et al., 2018; Toga and Thompson, 2013), stroke (Mukherjee, 2005), schizophrenia (Collin et al., 2016; Goldsmith et al., 2018; Wheeler and Voineskos, 2014), depression (De Witte and Mueller, 2017; Korgaonkar et al., 2014), obsessive-compulsive disorder (OCD) (Cao et al., 2021; Chiu et al., 2011; Gan et al., 2017; Gruner et al., 2012; Koch et al., 2014; Widge et al., 2021), attention-deficit hyperactivity disorder (ADHD) (Cao et al., 2014; 2013; Damatac et al., 2020; Hong et al., 2014) and autism (Ikuta et al., 2014; Langen et al., 2012; Thomas et al., 2011; Zhang et al., 2018a). In this section, we are condensing the explanation using two exemplar disorders, multiple sclerosis and schizophrenia, as the methods utilized in the study of such are representative of the work done across a breadth of disorders. Compared to traditional voxel-based analysis, quantitative tractography not only offers an irreplaceable tool to accurately quantify the abnormalities along local trajectories of fiber bundles in vivo but also provides a unique opportunity to investigate brain diseases at a network level (Collin et al., 2016; Fleischer et al., 2019; Wheeler and Voineskos, 2014).
6.3.1. Multiple sclerosis
Multiple sclerosis is a demyelinating disease with multiple focal white matter lesions (Fleischer et al., 2019). Accurate quantification of tract-specific damage is especially important for patients in MS because researchers have found that the location and surrounding damages of lesions contribute more than the number and volume of lesions (Lipp et al., 2020). Accurate anatomical localization of affected tracts is also essential to understand the symptomatology, disease evolution, and intervention effects (Barkhof, 1999; 2002; Barkhof et al., 2009; Charil et al., 2003; Kolind et al., 2012; Lin et al., 2005). However, traditional voxel-based analysis of brain white matter is usually performed in a standard space and seems sensitive to registration error (Colby et al., 2012b). Therefore, tractography in native space becomes suitable for MS patients because it is free of registration and allows inter-subject variability of tract location and shape (Lipp et al., 2020). Since the pathological lesions of MS hinder basic tract reconstruction, clinical studies frequently employ advanced tractography methods to reconstruct individual tract streamlines (Sbardella et al., 2013) and obtain feasible results even when brain lesions are present (Lipp et al., 2020). After segmenting a tract of interest, specific dMRI metrics can be estimated within the tract (as described in Section 5.2). Tractography studies in MS have found that decreased FA or increased MD value in specific tracts, such as the corticospinal tract, the corpus callosum and cerebellar peduncles, are related to individual motor disabilities (Anderson et al., 2011; Kern et al., 2011; Lin et al., 2005; Wilson et al., 2003) and longitudinal changes of diffusion metrics of these tracts in patients are associated with the rehabilitation of motor function during training (Filippi et al., 2019; Prosperini et al., 2014). Importantly, quantitative tractography analysis in MS further highlights that white matter tracts exhibiting normal appearance are also affected pathologically (Droby et al., 2015; Fleischer et al., 2019) and may be influenced by different pathological factors such as iron or myelin (Fang et al., 2019). The FA abnormalities in these tracts are found to be significantly correlated with cognitive abilities in pediatric patients at the early phase of the disease (Sbardella et al., 2013). These individual tract-based quantitative metrics are also promising in early clinical trials. For instance, researchers may use this approach to study specific rehabilitation interventions within relevant white matter tracts (Bonzano et al., 2014). By combining along-tract analysis and intra-cortical measurements, researchers further found that although the abnormalities in white matter tracts of MS are concomitant with intra-cortical injuries, this relationship is not strictly following a tract-cortex-specific pattern (Louapre et al., 2016).
MS is considered to be a typical disconnection syndrome due to the massive lesions observed (Rocca et al., 2015). Researchers have observed consistent disrupted network integration including reduction of global efficiency and network strength (Liu et al., 2018; Schiavi et al., 2020b; Shu et al., 2016; 2011), breakdown of long-range connections that account for remote communication (Muthuraman et al., 2016), decreased frontal network communicability that represents the parallel communication capacity (Li et al., 2013) and impairments of rich-club connectivity that serve as core highways (Shu et al., 2018a) in MS patients compared with healthy controls. Meanwhile, studies find increased clustering and modular connectivity patterns at 6 and 12 months after the first clinical event, which indicates a disease-related abnormality of network segregation ability (Fleischer et al., 2017; Muthuraman et al., 2016). Addressing such topological network abnormalities in MS benefits the tracking of high-level behavior disruptions and complex disease state at individual level. The disruption of structural networks is associated with impaired cognitive performance, especially involving attention and executive function (Llufriu et al., 2017). The decrease in global efficiency is significantly correlated with Expanded Disability Status Scale (EDSS) scores and disease duration (Shu et al., 2011). Researchers have further used network metrics to differentiate clinical subtypes of MS and achieved promising accuracy (Kocevar et al., 2016).
6.3.2. Schizophrenia
Schizophrenia has been considered to be a dysconnectivity disorder with a complex nature since the late 19th century (Collin et al., 2016; Goldsmith et al., 2018). Modern tractography techniques provide in vivo evidence for this hypothesis, indicating that schizophrenia may be seen in terms of affected pathways between cortical regions (Goldsmith et al., 2018; Griffa et al., 2013). For instance, the corpus callosum exhibits impairments of decreased FA in anatomical subdivisions in schizophrenia. These impairments are found reproducible across studies (Ohoshi et al., 2019; Price et al., 2007; Shahab et al., 2018) and are predictive for multiple cognitive domains (Sui et al., 2018) in both health and patients across datasets. A general question about schizophrenia is whether it is a disorder of whole brain dysfunction or one of selectively affected neural systems (Fornito et al., 2017). Studies frequently find consistent decreases of FA in tracts among fronto-temporal systems such as the superior/inferior longitudinal fasciculi, uncinate fasciculi, arcuate fasciculi, and cingulum bundles (Peters and Karlsgodt, 2015; Wheeler and Voineskos, 2014) in both chronic and first episode schizophrenia patients. These tract-specific impairments exhibit significant symptom associations in deficits of working memory (Karlsgodt et al., 2008), hallucinations (Shergill et al., 2007) and attention (Abdolalizadeh et al., 2020). However, increasing evidence suggests that the impairments of tracts in schizophrenia are more widespread than classical frontotemporal regions (Fitzsimmons et al., 2013; Kelly et al., 2018; Klauser et al., 2017; Mamah et al., 2019; Zalesky et al., 2011), which emphasizes the need for whole brain investigation. There is evidence that tractography analysis showed higher specificity and sensitivity for detecting white matter impairments in schizophrenia, compared to the voxel-based analysis approach (Kanaan et al., 2006). Meanwhile, by combining along-tract profiles of whole brain fiber pathways (which cannot be obtained by voxel-based analysis) with an unsupervised clustering framework, researchers were able to identify biologically defined subtypes of schizophrenia rather than symptom-based divisions, which shows great promise for future studies (Sun et al., 2015).
Using the graph-theoretical framework (as described in 5.6), structural connectome studies (Collin et al., 2016; Fornito et al., 2017) have shown altered global network topology such as reduced network efficiency and increased characteristic path length in chronic (van den Heuvel et al., 2010; Wheeler and Voineskos, 2014; Zalesky et al., 2011) or first episode patients (Zhang et al., 2015), unaffected siblings (Collin et al., 2014b) and high-risk infants (Shi et al., 2012), compared with healthy controls. These results support that the impaired network integration may be foundational to schizophrenia’s etiology and possibly reflect genetic vulnerability. Meanwhile, the disproportionate deficits of long-distance association edges, disruptions of anatomical brain hubs and rich club connections (Collin et al., 2014a; Crossley et al., 2017; Klauser et al., 2017; Van Den Heuvel et al., 2013; Yeo et al., 2016) in both patients and their unaffected siblings also suggest damage of topological infrastructures for inter-domain functional integration in schizophrenia (Collin et al., 2016). Studies also find evidence of abnormally enhanced network segregation such as increased modularity in schizophrenia patients and individuals with high-risk syndrome (Schmidt et al., 2017; Van Den Heuvel et al., 2013). These deficiencies of network topology are generally associated with the cognitive impairments and treatment responses during disease. The reduced global efficiency of patients is correlated with Positive and Negative Syndrome Scale (PANSS) scores (Wang et al., 2012) while higher global efficiency trend to be found in first-onset patients who subsequently respond to treatment, compared to non-responders (Crossley et al., 2017). The disruption of rich-club organization is also associated with PANSS scores and shows potential for classification between schizophrenia patients and healthy controls (Cui et al., 2019; Zhao et al., 2017b). Tractography-based networks are also a valuable framework to enable cross-species comparisons for pathogenesis; for instance, by comparing brain network connectivity between humans and chimpanzees, researchers find evidence of evolutionary modifications of human white matter connectivity to significantly overlap with the cortical pattern of schizophrenia-related dysconnectivity (Van Den Heuvel et al., 2019).
6.4. Tractography in neurosurgery
For neurosurgeons, in vivo tractography is invaluable for the planning and implementation of surgery, allowing for the visualization and localization of white matter tracts that are displaced or otherwise affected by the tumor (Jellison et al., 2004; O’Donnell et al., 2017). Although there remain technical considerations (Essayed et al., 2017), in-vivo tractography offers essential spatial information about individual anatomical pathways for brain surgery planning and implementation (though quantitative metrics of local tracts are relatively less used in clinical care) in the fields of lesion resection and deep brain stimulation (DBS).
During tumor resection surgeries, in-vivo tractography, especially in intraoperative MRI, allows for the accurate visualization and localization of white matter tracts that are displaced or affected by tumors (Chen et al., 2016a; Jellison et al., 2004), which is important for the quality of life and overall survival of neurosurgical patients (Costabile et al., 2019; Essayed et al., 2017; Fortin et al., 2012; Panesar et al., 2019). In neoplastic lesion surgery, balancing the trade-off between function preservation and maximized resection is highly significant. The classical gold standard is the intraoperative identification of eloquent areas by direct electrical stimulation (DES) (Duffau, 2005; 2015; Essayed et al., 2017; Szelényi et al., 2010). Recent applications of in vivo tractography from intraoperative MRI have shown a relatively high correlation between distances to reconstructed tracts and positive DES intensities (Essayed et al., 2017). Using intraoperative MRI has also shown to provide a more comprehensive surgical white matter mapping by identifying fiber tracts that were not detected in preoperative tractography (Javadi et al., 2017). Studies have shown that when combined with navigated transcranial magnetic stimulation (nTMS), intraoperative tractography aids in preserving speech for patients with tumors adjacent to the superior longitudinal fasciculus (Negwer et al., 2017; Sollmann et al., 2018). Additionally, in language area surgery, when awake surgery is impossible or unavailable, tractography-based estimation may become the only available solution for assessing involved white matter tracts (D’Andrea et al., 2016). The applications of in vivo tractography have been widely used in various lesions located in both supratentorial and infratentorial areas (Ellis et al., 2012; Fernandes-Cabral et al., 2016; Kovanlikaya et al., 2011), involving not only the major tracts but also smaller fiber structures (Meola et al., 2016).
In psychiatric surgery, tractography results are used to improve targeting during DBS surgery for patients with refractory psychiatric diseases such as depression, obsessive-compulsive disorder (OCD), or posttraumatic stress disorder (PTSD) (Davidson et al., 2020; Fenoy, 2020). For instance, the ventral tegmental area projection pathway (VTApp), which is the potential target of stimulation in many translational cases during psychiatric DBS, cannot be appreciated on standard structural MRI images. In-vivo tractography can be used to maximize the contact with the VTApp of stimulation electrode by optimizing the invasive depth and trajectory after the visualization of fiber streamlines (Coenen et al., 2019; Davidson et al., 2020; Schlaepfer et al., 2013). Such a DBS approach brings rapid improvements on clinical responses and remissions in several open-label trials, although it needs to be further studied through large randomized trials (Coenen et al., 2019). Similar efforts have also been pursued for major targets including the subcallosal cingulum (SCC) and ventral capsule/ventral striatum (VC/VS) in clinical practice (For a review, see Davidson et al., 2020).
The brain connectomic concept benefits traditional surgical strategy by a potential shift from focusing on local topography to a network-guided “oncological disconnection surgery” (Duffau, 2021; Hart et al., 2020). In a classical neurosurgical view, the inflexible definition of “eloquent” brain regions represented by the sensorimotor, language and visual cortices serves as an irrefutable principle: tumors involving “eloquent” cortex would not be selected for resection while tumors locating in “non-eloquent” areas could be operated without considerations (Spetzler and Martin, 1986). However, considering the potential for functional compensation and the presence of individual variation, a common location of “eloquent” areas across patients seems inaccurate (Southwell et al., 2017). By combining tractography atlases with intraoperative electrical stimulation for functional mapping, researchers have constructed probabilistic atlases of white matter pathways and structural hubs that define a minimal common brain connectome with a low inter-individual variability and a low potential of post-lesional compensation (Herbet et al., 2016; Sarubbo et al., 2020). Such prior “structural and functional skeletons” provide a whole brain knowledge map of essential brain sub-networks, which may be helpful in informing the resection of certain neuronal circuits during tumor surgery (Duffau, 2021). For instance, connectomic studies have shown that the lexical access speed, which is related to the return to professional work after awake surgery for patients with low-grade gliomas, may be completely disrupted only when the left inferior longitudinal fasciculus (ILF) is damaged (Moritz-Gasser et al., 2012). An elaborate surgical management can now take into account the white matter circuits involved with functional reconfiguration that affected by tumors/lesion (Van Horn et al., 2012). Researchers also have tried to develop a “connectomic risk signature” for lesion surgery using brain function simulation based on tractography results (Aerts et al., 2018). In this study, the authors employed neural dynamic models on empirical tractography data of a cohort of patients with gliomas and meningiomas to create a virtual brain functional network and observed distinct individual signatures depending whether the brain regions were directly affected by tumors. In psychiatric surgery, advanced tractography facilitates the combination between brain connectomic analyses and neuromodulation interventions (Davidson et al., 2020; Horn et al., 2017; Riva-Posse et al., 2018), which fuels the emerging concept of “Connectomic DBS” (Baldermann et al., 2021; Horn and Fox, 2020). This simulation approach aims to modulate an optimal interconnected network instead of targeting local sites. It may partly reduce the ambiguity about the best intervention target and is quite suitable for brain disorders that involve several abnormal circuits, e.g., OCD (For a review, see (Baldermann et al., 2021)). Furthermore, connectomic neuromodulation may contribute to identifying specific networks that lead to symptom-specific treatments, paving the way for personalized therapy (Horn and Fox, 2020). For instance, in Parkinson’s disease, researchers found that DBS on sub-thalamic nucleus linking to the supplementary motor area is associated with improvement in bradykinesia and rigidity, while DBS on nucleus structurally commenting to M1 is associated with improvement in tremor (Akram et al., 2017). These studies indicate the potential of using individual quantitative tractography to generate neurosurgical prognostic markers at the brain system level in the future.
7. Discussion and conclusion
In this review paper, we have provided a high-level overview of how tractography can be used to enable quantitative analysis of the brain’s structural connectivity in health and disease. We reviewed methods that are involved in the main processing steps for quantitative analysis of tractography. We also reviewed studies that have used quantitative tractography to study the brain’s white matter.
For research and clinical applications using tractography, we note that researchers should be extremely cautious when making biological interpretations about quantitative results. The reconstructed streamlines are only simulated entities that do not correspond directly to nerve fibers (Jeurissen et al., 2019; Yeh et al., 2020), and basic diffusion metrics are only inferences based on local diffusion properties, which are not direct measures of tissue properties (Assaf et al., 2019). Multiple review papers illustrate pitfalls to be avoided when performing quantitative dMRI analysis and studying the brains connectivity using tractography (Daducci et al., 2016; Jones, 2010; O’Donnell and Pasternak, 2015; Rheault et al., 2020c). Research is underway to improve biological specificity to the type of tissue change, by improving the information that is obtained at the acquisition level (Barakovic et al., 2021a; Henriques et al., 2020; Hutter et al., 2018; Ning et al., 2019; Shemesh et al., 2016; Westin et al., 2016), and by proposing advanced mathematical modeling and machine learning techniques (Nath et al., 2021; Ning et al., 2021; Pizzolato et al., 2020; Wu and Miller, 2017).
The anatomical accuracy of tractography is an ongoing issue in quantitative tractography analysis, where both false positive and false negative tractography results pose challenges (Maier-Hein et al., 2017; Thomas et al., 2014). While technical improvements (e.g., using fiber filtering techniques as introduced in Section 3.5) can improve anatomical accuracy, researchers should be cautious when interpreting results in research and clinical applications by considering prior anatomical knowledge learned from studies in histology (Huttenlocher, 1984; Miller et al., 2012; Sidman and Rakic, 1973; Yakovlev and Lecours, 1967) and animals (Song et al., 2003; 2002).
Tractography has enabled the study of the brain’s white matter connections across the lifespan in health and disease. Overall, we conclude that, while there have been considerable advancements in methodological technologies and breadth of applications, there nevertheless remains no consensus about the “best” methodology in quantitative analysis of tractography, and researchers should remain cautious when interpreting results in research and clinical applications.
Acknowledgements
We acknowledge the following funding. FZ and LJO acknowledge funding provided by the following National Institutes of Health (NIH) grants: P41EB015902, R01MH074794, R01MH125860 and R01MH119222. FZ also acknowledges a BWH Radiology Research Pilot Grant Award. YH and TDZ are supported by the Natural Science Foundation of China (Grant Nos. 81620108016, 81801783). RS is supported by fellowship funding from the National Imaging Facility (NIF), an Australian Government National Collaborative Research Infrastructure Strategy (NCRIS) capability. CHY is grateful to the Ministry of Science and Technology of Taiwan (MOST 109-2222-E-182-001-MY3) for the support.
Footnotes
Credit Author Statement
Fan Zhang: Conceptualization, Project administration, Visualization, Writing – original draft, Writing – review & editing. Alessandro Daducci: Conceptualization, Writing – original draft, Writing – review & editing. Yong He: Conceptualization, Writing – original draft, Writing – review & editing. Simona Schiavi: Conceptualization, Writing – original draft, Writing – review & editing. Caio Seguin: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. Robert Smith: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. Chun-Hung Yeh: Conceptualization, Writing – original draft, Writing – review & editing. Tengda Zhao: Conceptualization, Writing – original draft, Writing – review & editing. Lauren J. O’Donnell: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
References
- Abdolalizadeh A, Ostadrahimi H, Mohajer B, Darvishi A, Sattarian M, Ershadi ASB, Abbasi N, 2020. White matter microstructural properties associated with impaired attention in chronic schizophrenia: a multi-center study. Psychiatry Res. Neuroimaging 302, 111105. [DOI] [PubMed] [Google Scholar]
- Abhinav K, Yeh FC, Mansouri A, Zadeh G, Fernandez-Miranda JC, 2015. High-definition fiber tractography for the evaluation of perilesional white matter tracts in high-grade glioma surgery. Neuro. Oncol 17, 1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts H, Schirner M, Jeurissen B, Van Roost D, Achten E, Ritter P, Marinazzo D, 2018. Modeling brain dynamics in brain tumor patients using the virtual brain. eNeuro 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzali M, Pieciak T, Newman S, Garifallidis E, Özarslan E, Cheng H, Jones DK, 2020. The sensitivity of diffusion MRI to microstructural properties and experimental factors. J. Neurosci. Methods 108951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akram H, Sotiropoulos SN, Jbabdi S, Georgiev D, Mahlknecht P, Hyam J, Foltynie T, Limousin P, De Vita E, Jahanshahi M, et al. , 2017. Subthalamic deep brain stimulation sweet spots and hyperdirect cortical connectivity in Parkinson’s disease. Neuroimage 158, 332–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albi A, Meola A, Zhang F, Kahali P, Rigolo L, Tax CM, Ciris PA, Essayed WI, Unadkat P, Norton I, et al. , 2018. Image registration to compensate for EPI distortion in patients with brain tumors: an evaluation of tract-specific effects. J. Neuroimaging 28, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander AL, Lee JE, Lazar M, Boudos R, DuBray MB, Oakes TR, Miller JN, Lu J, Jeong EK, McMahon WM, et al. , 2007. Diffusion tensor imaging of the corpus callosum in autism. Neuroimage 34, 61–73. [DOI] [PubMed] [Google Scholar]
- Alexander DC, 2006. An introduction to computational diffusion MRI: the diffusion tensor and beyond. In: Visualization and Processing of Tensor Fields. Springer, pp. 83–106. [Google Scholar]
- Alexander DC, Dyrby TB, Nilsson M, Zhang H, 2019. Imaging brain microstructure with diffusion MRI: practicality and applications. NMR Biomed. 32, e3841. [DOI] [PubMed] [Google Scholar]
- Alexander LM, Escalera J, Ai L, Andreotti C, Febre K, Mangone A, Vega-Potler N, Langer N, Alexander A, Kovacs M, et al. , 2017. An open resource for transdiagnostic research in pediatric mental health and learning disorders. Sci. Data 4, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan PG, Briggs RG, Conner AK, O’Neal CM, Bonney PA, Maxwell BD, Baker CM, Burks JD, Sali G, Glenn CA, et al. , 2020. Parcellation-based tractographic modeling of the ventral attention network. J. Neurol. Sci 408, 116548. [DOI] [PubMed] [Google Scholar]
- Anderson V, Wheeler-Kingshott C, Abdel-Aziz K, Miller D, Toosy A, Thompson A, Ciccarelli O, 2011. A comprehensive assessment of cerebellar damage in multiple sclerosis using diffusion tractography and volumetric analysis. Multiple Scler. J 17, 1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf Y, Johansen-Berg H, Thiebaut de Schotten M, 2019. The role of diffusion MRI in neuroscience. NMR Biomed. 32, e3762. [DOI] [PubMed] [Google Scholar]
- Assaf Y, Pasternak O, 2008. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: a review. J. Mol. Neurosci 34, 51–61. [DOI] [PubMed] [Google Scholar]
- Astolfi P, De Benedictis A, Sarubbo S, Bertó G, Olivetti E, Sona D, Avesani P, 2020. A stem-based dissection of inferior fronto-occipital fasciculus with a deep learning model. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI). IEEE, pp. 267–270. [Google Scholar]
- Avants BB, Tustison N, Song G, 2009. Advanced normalization tools (ANTS). Insight J 2, 1–35. [Google Scholar]
- Avena-Koenigsberger A, Misic B, Sporns O, 2018. Communication dynamics in complex brain networks. Nat. Rev. Neurosci 19, 17–33. [DOI] [PubMed] [Google Scholar]
- Avena-Koenigsberger A, Yan X, Kolchinsky A, van den Heuvel M, Hagmann P, Sporns O, 2019. A spectrum of routing strategies for brain networks. PLoS Comput. Biol 15, e1006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila NL, Lebenberg J, Rivière D, Auzias G, Fischer C, Poupon F, Guevara P, Poupon C, Mangin JF, 2019. Inference of an extended short fiber bundle atlas using sulcus-based constraints for a diffeomorphic inter-subject alignment. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 323–333. [Google Scholar]
- Aydogan DB, Jacobs R, Dulawa S, Thompson SL, Francois MC, Toga AW, Dong H, Knowles JA, Shi Y, 2018. When tractography meets tracer injections: a systematic study of trends and variation sources of diffusion-based connectivity. Brain Struct. Funct 223, 2841–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydogan DB, Shi Y, 2020. Parallel transport tractography. IEEE Trans. Med. Imaging 40, 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Zhang Z, Watson DR, Yu H, Shi Y, Yuan Y, Qian Y, Jia J, 2009. Abnormal integrity of association fiber tracts in amnestic mild cognitive impairment. J. Neurol. Sci 278, 102–106. [DOI] [PubMed] [Google Scholar]
- Bajada CJ, Schreiber J, Caspers S, 2019. Fiber length profiling: a novel approach to structural brain organization. Neuroimage 186, 164–173. [DOI] [PubMed] [Google Scholar]
- Bakker R, Wachtler T, Diesmann M, 2012. CoCoMac 2.0 and the future of tract-tracing databases. Front. Neuroinform 6, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldermann JC, Schüller T, Kohl S, Voon V, Li N, Hollunder B, Figee M, Haber SN, Sheth SA, Mosley PE, et al. , 2021. Connectomic deep brain stimulation for obsessive-compulsive disorder. Biol. Psychiatry [DOI] [PubMed] [Google Scholar]
- Ball G, Srinivasan L, Aljabar P, Counsell SJ, Durighel G, Hajnal JV, Rutherford MA, Edwards AD, 2013. Development of cortical microstructure in the preterm human brain. Proc. Natl. Acad. Sci 110, 9541–9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakovic M, Girard G, Romascano DPR, Patino Lopez JR, Descoteaux M, Innocenti G, Jones DK, Thiran JP, Daducci A, 2018. Assessing feasibility and reproducibility of a bundle-specific framework on in vivo axon diameter estimates at 300mT/m. 26th Annual Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). [Google Scholar]
- Barakovic M, Tax CM, Rudrapatna U, Chamberland M, Rafael-Patino J, Granziera C, Thiran JP, Daducci A, Canales-Rodríguez EJ, Jones DK, 2021. Resolving bundle-specific intra-axonal T2 values within a voxel using diffusion-relaxation tract-based estimation. Neuroimage 227, 117617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakovic M, Tax CMW, Rudrapatna U, Chamberland M, Rafael-Patino J, Granziera C, Thiran JP, Daducci A, Canales-Rodríguez EJ, Jones DK, 2021. Resolving bundle-specific intra-axonal T2 values within a voxel using diffusion-relaxation tract-based estimation. Neuroimage 227, 117617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barch DM, Verfaellie M, Rao SM, 2016. Introduction to JINS special issue on human brain connectivity in the modern era: relevance to understanding health and disease. J. Int. Neuropsychol.Soc 22, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkhof F, 1999. MRI in multiple sclerosis: correlation with expanded disability status scale (EDSS). Multiple Scler. J 5, 283–286. [DOI] [PubMed] [Google Scholar]
- Barkhof F, 2002. The clinico-radiological paradox in multiple sclerosis revisited. Curr. Opin. Neurol 15, 239–245. [DOI] [PubMed] [Google Scholar]
- Barkhof F, Calabresi PA, Miller DH, Reingold SC, 2009. Imaging outcomes for neuroprotection and repair in multiple sclerosis trials. Nat. Rev. Neurol 5, 256–266. [DOI] [PubMed] [Google Scholar]
- Basser PJ, 1998. Fiber-tractography via diffusion tensor MRI (DT-MRI). In: Proceedings of the 6th Annual Meeting ISMRM, Sydney, Australia, p. 1226. [Google Scholar]
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A, 2000. In vivo fiber tractography using DT-MRI data. Magn. Reson. Med 44, 625–632. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C, 1996. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J. Magn. Reson. Ser. B 111, 209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C, 2011. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J. Magn. Reson 213, 560–570. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET, 2017. Small-world brain networks revisited. Neuroscientist 23, 499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Sporns O, 2017. Network neuroscience. Nat. Neurosci 20, 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani M, Cottaar M, Dikranian K, Ghosh A, Zhang H, Alexander DC, Behrens TE, Jbabdi S, Sotiropoulos SN, 2017. Improved tractography using asymmetric fibre orientation distributions. Neuroimage 158, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani M, Shah NJ, Goebel R, Roebroeck A, 2012. Human cortical connectome reconstruction from diffusion weighted MRI: the effect of tractography algorithm. Neuroimage 62, 1732–1749. [DOI] [PubMed] [Google Scholar]
- Bathelt J, Barnes J, Raymond FL, Baker K, Astle D, 2017. Global and local connectivity differences converge with gene expression in a neurodevelopmental disorder of known genetic origin. Cereb. Cortex 27, 3806–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battocchio M, Schiavi S, Descoteaux M, Daducci A, 2020. Improving tractography accuracy using dynamic filtering. CDMRI. accepted to appear [Google Scholar]
- Baum GL, Roalf DR, Cook PA, Ciric R, Rosen AF, Xia C, Elliott MA, Ruparel K, Verma R, Tunç B, et al. , 2018. The impact of in-scanner head motion on structural connectivity derived from diffusion MRI. Neuroimage 173, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin PL, Ye C, Bogovic JA, Shiee N, Reich DS, Prince JL, Pham DL, 2011. Direct segmentation of the major white matter tracts in diffusion tensor images. Neuroimage 58, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW, 2007. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? Neuroimage 34, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman-Lay AM, Usher C, Conturo TE, Correia S, Laidlaw DH, Lane EM, Bolzenius J, Heaps JM, Salminen LE, Baker LM, et al. , 2015. Fiber bundle length and cognition: a length-based tractography MRI study. Brain Imaging Behav. 9, 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bells S, Cercignani M, Deoni S, Assaf Y, Pasternak O, Evans C, Leemans A, Jones D, 2011. Tractometry–comprehensive multi-modal quantitative assessment of white matter along specific tracts. In: Proc. ISMRM, p. 1. [Google Scholar]
- Bells S, Cercignani M, Deoni S, Assaf Y, Pasternak O, Evans CJ, Leemans A, Jones DK, 2011. Tractometry–comprehensive multi-modal quantitative assessment of white matter along specific tracts. In: Proc. ISMRM, p. 1. [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. [Google Scholar]
- Benou I, Veksler R, Friedman A, Raviv TR, 2019. Combining white matter diffusion and geometry for tract-specific alignment and variability analysis. Neuroimage 200, 674–689. [DOI] [PubMed] [Google Scholar]
- Bertò G, Bullock D, Astolfi P, Hayashi S, Zigiotto L, Annicchiarico L, Corsini F, De Benedictis A, Sarubbo S, Pestilli F, et al. , 2020. Classifyber, a robust streamline-based linear classifier for white matter bundle segmentation. BioRxiv. [DOI] [PubMed] [Google Scholar]
- Besson P, Lopes R, Leclerc X, Derambure P, Tyvaert L, 2014. Intra-subject reliability of the high-resolution whole-brain structural connectome. Neuroimage 102, 283–293. [DOI] [PubMed] [Google Scholar]
- Betzel RF, 2020. Community detection in network neuroscience. arXiv preprint arXiv:2011.06723. [Google Scholar]
- Betzel RF, Avena-Koenigsberger A, Goñi J, He Y, de Reus MA, Griffa A, Vértes PE, Mišic B, Thiran JP, Hagmann P, van den Heuvel M, Zuo XN, Bullmore ET, Sporns O, 2016. Generative models of the human connectome. Neuroimage 124, 1054–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel RF, Bassett DS, 2017. Multi-scale brain networks. Neuroimage 160, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel RF, Griffa A, Avena-Koenigsberger A, Goñi J, Thiran JP, Hagmann P, Sporns O, 2013. Multi-scale community organization of the human structural connectome and its relationship with resting-state functional connectivity. Netw. Sci 1, 353–373. [Google Scholar]
- Bi Q, Wang W, Niu N, Li H, Wang Y, Huang W, Chen K, Xu K, Zhang J, Chen Y, et al. , 2021. Relationship between the disrupted topological efficiency of the structural brain connectome and glucose hypometabolism in normal aging. Neuroimage 226, 117591. [DOI] [PubMed] [Google Scholar]
- Bonzano L, Tacchino A, Brichetto G, Roccatagliata L, Dessypris A, Feraco P, De Carvalho MLL, Battaglia MA, Mancardi GL, Bove M, 2014. Upper limb motor rehabilitation impacts white matter microstructure in multiple sclerosis. Neuroimage 90, 107–116. [DOI] [PubMed] [Google Scholar]
- Boshkovski T, Kocarev L, Cohen-Adad J, Mišić B, Lehéricy S, Stikov N, Mancini M, 2020. The R1-weighted connectome: complementing brain networks with a myelin-sensitive measure. Netw. Neurosci 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshkovski T, Kocarev L, Cohen-Adad J, Mišić B, Lehéricy S, Stikov N, Mancini M, 2020. The R1-weighted connectome: complementing brain networks with a myelin-sensitive measure. Netw. Neurosci 1–34. doi: 10.1101/2020.08.06.237941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Miller SP, Booth BG, Andrews S, Chau V, Poskitt KJ, Hamarneh G, 2014. Structural network analysis of brain development in young preterm neonates. Neuroimage 101, 667–680. [DOI] [PubMed] [Google Scholar]
- Bucci M, Mandelli ML, Berman JI, Amirbekian B, Nguyen C, Berger MS, Henry RG, 2013. Quantifying diffusion MRI tractography of the corticospinal tract in brain tumors with deterministic and probabilistic methods. Neuroimage Clin. 3, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan CR, Bastin ME, Ritchie SJ, Liewald DC, Madole JW, Tucker-Drob EM, Deary IJ, Cox SR, 2020. The effect of network thresholding and weighting on structural brain networks in the uk biobank. Neuroimage 211, 116443. doi: 10.1016/j.neuroimage.2019.116443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan CR, Pernet CR, Gorgolewski KJ, Storkey AJ, Bastin ME, 2014. Test–retest reliability of structural brain networks from diffusion MRI. Neuroimage 86, 231–243. [DOI] [PubMed] [Google Scholar]
- Budde MD, Annese J, 2013. Quantification of anisotropy and fiber orientation in human brain histological sections. Front. Integr. Neurosci 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O, 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci 10, 186–198. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O, 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci 10, 186–198. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O, 2012. The economy of brain network organization. Nat. Rev. Neurosci 13, 336–349. [DOI] [PubMed] [Google Scholar]
- Campbell JSW, Leppert IR, Narayanan S, Boudreau M, Duval T, Cohen-Adad J, Pike GB, Stikov N, 2018. Promise and pitfalls of g-ratio estimation with MRI. Neuroimage 182, 80–96. doi: 10.1016/j.neuroimage.2017.08.038. [DOI] [PubMed] [Google Scholar]
- Cao M, Huang H, He Y, 2017. Developmental connectomics from infancy through early childhood. Trends Neurosci. 40, 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Huang H, Peng Y, Dong Q, He Y, 2016. Toward developmental connectomics of the human brain. Front. Neuroanat 10, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Shu N, Cao Q, Wang Y, He Y, 2014. Imaging functional and structural brain connectomics in attention-deficit/hyperactivity disorder. Mol. Neurobiol 50, 1111–1123. [DOI] [PubMed] [Google Scholar]
- Cao Q, Shu N, An L, Wang P, Sun L, Xia MR, Wang JH, Gong GL, Zang YF, Wang YF, et al. , 2013. Probabilistic diffusion tractography and graph theory analysis reveal abnormal white matter structural connectivity networks in drug–naive boys with attention deficit/hyperactivity disorder. J. Neurosci 33, 10676–10687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Yang X, Luo J, Wang P, Meng F, Xia M, He Y, Zhao T, Li Z, 2021. The effects of cognitive behavioral therapy on the whole brain structural connectome in unmedicated patients with obsessive-compulsive disorder. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 104, 110037. [DOI] [PubMed] [Google Scholar]
- Casey B, Cannonier T, Conley MI, Cohen AO, Barch DM, Heitzeg MM, Soules ME, Teslovich T, Dellarco DV, Garavan H, et al. , 2018. The adolescent brain cognitive development (ABCD) study: imaging acquisition across 21 sites. Dev. Cogn. Neurosci 32, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catani M, Allin MP, Husain M, Pugliese L, Mesulam MM, Murray RM, Jones DK, 2007. Symmetries in human brain language pathways correlate with verbal recall. Proc. Natl. Acad. Sci 104, 17163–17168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catani M, Howard RJ, Pajevic S, Jones DK, 2002. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. Neuroimage 17, 77–94. [DOI] [PubMed] [Google Scholar]
- Ceschin R, Lee VK, Schmithorst V, Panigrahy A, 2015. Regional vulnerability of longitudinal cortical association connectivity: associated with structural network topology alterations in preterm children with cerebral palsy. Neuroimage Clin. 9, 322–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin-Karayumak S, Di Biase MA, Chunga N, Reid B, Somes N, Lyall AE, Kelly S, Solgun B, Pasternak O, Vangel M, et al. , 2020. White matter abnormalities across the lifespan of schizophrenia: a harmonized multi-site diffusion MRI study. Mol. Psychiatry 25, 3208–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J, Fekete T, Siciliano F, Biezonski D, Greenhill L, Pliszka SR, Blader JC, Roy AK, Leibenluft E, Posner J, 2015. Neural correlates of aggression in medication-naive children with ADHD: multivariate analysis of morphometry and tractography. Neuropsychopharmacology 40, 1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberland M, Raven EP, Genc S, Duffy K, Descoteaux M, Parker GD, Tax CM, Jones DK, 2019. Dimensionality reduction of diffusion MRI measures for improved tractometry of the human brain. Neuroimage 200, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberland M, Whittingstall K, Fortin D, Mathieu D, Descoteaux M, 2014. Real–time multi-peak tractography for instantaneous connectivity display. Front. Neuroinform 8, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandio BQ, Garyfallidis E, 2020. StND: Streamline-based non-rigid partial-deformation tractography registration. Medical Imaging Meets NeurIPS. [Google Scholar]
- Chandio BQ, Risacher SL, Pestilli F, Bullock D, Yeh FC, Koudoro S, Rokem A, Harezlak J, Garyfallidis E, 2020. Bundle analytics, a computational framework for investigating the shapes and profiles of brain pathways across populations. Sci. Rep 10, 17149. doi: 10.1038/s41598-020-74054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandio BQ, Risacher SL, Pestilli F, Bullock D, Yeh FC, Koudoro S, Rokem A, Harezlak J, Garyfallidis E, 2020. Bundle analytics, a computational framework for investigating the shapes and profiles of brain pathways across populations. Sci. Rep 10, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YS, Owen JP, Pojman NJ, Thieu T, Bukshpun P, Wakahiro ML, Berman JI, Roberts TP, Nagarajan SS, Sherr EH, et al. , 2015. White matter changes of neurite density and fiber orientation dispersion during human brain maturation. PLoS ONE 10, e0123656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charil A, Zijdenbos AP, Taylor J, Boelman C, Worsley KJ, Evans AC, Dagher A, 2003. Statistical mapping analysis of lesion location and neurological disability in multiple sclerosis: application to 452 patient data sets. Neuroimage 19, 532–544. [DOI] [PubMed] [Google Scholar]
- Chekir A, Descoteaux M, Garyfallidis E, Côté MA, Boumghar FO, 2014. A hybrid approach for optimal automatic segmentation of white matter tracts in HARDI. In: 2014 IEEE Conference on Biomedical Engineering and Sciences (IECBES). IEEE, pp. 177–180. [Google Scholar]
- Chen Y, Zhang C, Song Y, Makris N, Rathi Y, Cai W, Zhang F, O’Donnell LJ, 2021. Deep fiber clustering: anatomically informed unsupervised deep learning for fast and effective white matter parcellation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 497–507. [Google Scholar]
- Chen Z, Liu M, Gross DW, Beaulieu C, 2013. Graph theoretical analysis of developmental patterns of the white matter network. Front. Hum. Neurosci 7, 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Tie Y, Olubiyi O, Zhang F, Mehrtash A, Rigolo L, Kahali P, Norton I, Pasternak O, Rathi Y, et al. , 2016. Corticospinal tract modeling for neurosurgical planning by tracking through regions of peritumoral edema and crossing fibers using two-tensor unscented Kalman filter tractography. Int. J. Comput. Assist. Radiol. Surg 11, 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang H, Yushkevich PA, Liu M, Beaulieu C, 2016. Maturation along white matter tracts in human brain using a diffusion tensor surface model tract-specific analysis. Front Neuroanat 10, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherifi D, Boudjada M, Morsli A, Girard G, Deriche R, 2018. Combining improved euler and Runge-Kutta 4th order for tractography in diffusion-weighted MRI. Biomed. Signal Process. Control 41, 90–99. [Google Scholar]
- Chiang H, Chen Y, Shang C, Tseng WI, Gau SS, 2016. Different neural substrates for executive functions in youths with ADHD: a diffusion spectrum imaging tractography study. Psychol. Med 46, 1225. [DOI] [PubMed] [Google Scholar]
- Chiu CH, Lo YC, Tang HS, Liu IC, Chiang WY, Yeh FC, Jaw FS, Tseng WYI, 2011. White matter abnormalities of fronto-striato-thalamic circuitry in obsessive—compulsive disorder: a study using diffusion spectrum imaging tractography. Psychiatry Res. Neuroimaging 192, 176–182. [DOI] [PubMed] [Google Scholar]
- Christiaens D, Cordero-Grande L, Pietsch M, Hutter J, Price AN, Hughes EJ, Vecchiato K, Deprez M, Edwards AD, Hajnal JV, et al. , 2021. Scattered slice shard reconstruction for motion correction in multi-shell diffusion MRI. Neuroimage 225, 117437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaens D, Reisert M, Dhollander T, Sunaert S, Suetens P, Maes F, 2015. Global tractography of multi-shell diffusion-weighted imaging data using a multi-tissue model. Neuroimage 123, 89–101. [DOI] [PubMed] [Google Scholar]
- Ciccarelli O, Catani M, Johansen-Berg H, Clark C, Thompson A, 2008. Diffusion-based tractography in neurological disorders: concepts, applications, and future developments. Lancet Neurol. 7, 715–727. [DOI] [PubMed] [Google Scholar]
- Ciccarelli O, Parker G, Toosy A, Wheeler-Kingshott C, Barker G, Boulby P, Miller D, Thompson A, 2003. From diffusion tractography to quantitative white matter tract measures: a reproducibility study. Neuroimage 18, 348–359. [DOI] [PubMed] [Google Scholar]
- Civier O, Smith RE, Yeh CH, Connelly A, Calamante F, 2019. Is removal of weak connections necessary for graph-theoretical analysis of dense weighted structural connectomes from diffusion MRI? Neuroimage 194, 68–81. doi: 10.1016/j.neuroimage.2019.02.039. [DOI] [PubMed] [Google Scholar]
- Clayden JD, Storkey AJ, Bastin ME, 2007. A probabilistic model-based approach to consistent white matter tract segmentation. IEEE Trans. Med. Imaging 26, 1555–1561. [DOI] [PubMed] [Google Scholar]
- Close TG, Tournier JD, Johnston LA, Calamante F, Mareels I, Connelly A, 2015. Fourier tract sampling (FouTS): a framework for improved inference of white matter tracts from diffusion MRI by explicitly modelling tract volume. Neuroimage 120, 412–427. [DOI] [PubMed] [Google Scholar]
- Coenen VA, Bewernick BH, Kayser S, Kilian H, Boström J, Greschus S, Hurlemann R, Klein ME, Spanier S, Sajonz B, et al. , 2019. Superolateral medial forebrain bundle deep brain stimulation in major depression: a gateway trial. Neuropsychopharmacology 44, 1224–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby JB, Soderberg L, Lebel C, Dinov ID, Thompson PM, Sowell ER, 2012. Along-tract statistics allow for enhanced tractography analysis. Neuroimage 59, 3227–3242. doi: 10.1016/j.neuroimage.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby JB, Soderberg L, Lebel C, Dinov ID, Thompson PM, Sowell ER, 2012. Along-tract statistics allow for enhanced tractography analysis. Neuroimage 59, 3227–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin G, Kahn RS, De Reus MA, Cahn W, Van Den Heuvel MP, 2014. Impaired rich club connectivity in unaffected siblings of schizophrenia patients. Schizophr. Bull 40, 438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin G, Sporns O, Mandl RC, Van Den Heuvel MP, 2014. Structural and functional aspects relating to cost and benefit of rich club organization in the human cerebral cortex. Cereb. Cortex 24, 2258–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin G, Turk E, Van Den Heuvel MP, 2016. Connectomics in schizophrenia: from early pioneers to recent brain network findings. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 1, 199–208. [DOI] [PubMed] [Google Scholar]
- Collin G, Van Den Heuvel MP, 2013. The ontogeny of the human connectome: development and dynamic changes of brain connectivity across the life span. Neuroscientist 19, 616–628. [DOI] [PubMed] [Google Scholar]
- Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, McKinstry RC, Burton H, Raichle ME, 1999. Tracking neuronal fiber pathways in the living human brain. Proc. Natl. Acad. Sci 96, 10422–10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PA, Bai Y, Hall MG, Nedjati-Gilani S, Seunarine KK, Alexander DC, 2006. Camino: diffusion MRI reconstruction and processing. In: International Society for Magnetic Resonance in Medicine (ISMRM), p. 2759. [Google Scholar]
- Corouge I, Fletcher PT, Joshi S, Gouttard S, Gerig G, 2006. Fiber tract-oriented statistics for quantitative diffusion tensor MRI analysis. Med. Image Anal 10, 786–798. [DOI] [PubMed] [Google Scholar]
- Costabile JD, Alaswad E, D’Souza S, Thompson JA, Ormond DR, 2019. Current applications of diffusion tensor imaging and tractography in intracranial tumor resection. Front. Oncol 9, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté MA, Girard G, Boré A, Garyfallidis E, Houde JC, Descoteaux M, 2013. Tractometer: towards validation of tractography pipelines. Med. Image Anal 17, 844–857. [DOI] [PubMed] [Google Scholar]
- Cox SR, Ritchie SJ, Tucker-Drob EM, Liewald DC, Hagenaars SP, Davies G, Wardlaw JM, Gale CR, Bastin ME, Deary IJ, 2016. Ageing and brain white matter structure in 3,513 UK biobank participants. Nat. Commun 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Marques TR, Taylor H, Chaddock C, Dell’Acqua F, Reinders AA, Mondelli V, DiForti M, Simmons A, David AS, et al. , 2017. Connectomic correlates of response to treatment in first-episode psychosis. Brain 140, 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Scott J, Carletti F, Fox PT, McGuire P, Bullmore ET, 2014. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain 137, 2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui LB, Wei Y, Xi YB, Griffa A, De Lange SC, Kahn RS, Yin H, Van den Heuvel MP, 2019. Connectome-based patterns of first-episode medication-naive patients with schizophrenia. Schizophr. Bull 45, 1291–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Zhong S, Xu P, Gong G, He Y, 2013. Panda: a pipeline toolbox for analyzing brain diffusion images. Front. Hum. Neurosci 7, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daducci A, Dal Palú A, Descoteaux M, Thiran JP, 2016. Microstructure informed tractography: pitfalls and open challenges. Front. Neurosci 10, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daducci A, Dal Palu A, Lemkaddem A, Thiran JP, 2013. A convex optimization framework for global tractography. In: 2013 IEEE 10th International Symposium on Biomedical Imaging. IEEE, pp. 524–527. [Google Scholar]
- Daducci A, Dal Palù A, Lemkaddem A, Thiran JP, 2014. COMMIT: convex optimization modeling for microstructure informed tractography. IEEE Trans. Med. Imaging 34, 246–257. [DOI] [PubMed] [Google Scholar]
- Damatac CG, Chauvin RJ, Zwiers MP, van Rooij D, Akkermans SE, Naaijen J, Hoekstra PJ, Hartman CA, Oosterlaan J, Franke B, et al. , 2020. White matter microstructure in attention-deficit/hyperactivity disorder: a systematic tractography study in 654 individuals. Biol. Psychiatry Cognit. Neurosci. Neuroimaging [DOI] [PubMed] [Google Scholar]
- D’Andrea G, Familiari P, Di Lauro A, Angelini A, Sessa G, 2016. Safe resection of gliomas of the dominant angular gyrus availing of preoperative FMRI and intraoperative DTI: preliminary series and surgical technique. World Neurosurg. 87, 627–639. [DOI] [PubMed] [Google Scholar]
- Davidson B, Lipsman N, Meng Y, Rabin JS, Giacobbe P, Hamani C, 2020. The use of tractography-based targeting in deep brain stimulation for psychiatric indications. Front. Hum. Neurosci 14, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SW, Dennis NA, Buchler NG, White LE, Madden DJ, Cabeza R, 2009. Assessing the effects of age on long white matter tracts using diffusion tensor tractography. Neuroimage 46, 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot M, Ikram MA, Akoudad S, Krestin GP, Hofman A, van der Lugt A, Niessen WJ, Vernooij MW, 2015. Tract-specific white matter degeneration in aging: the Rotterdam study. Alzheimer’s Dement. 11, 321–330. [DOI] [PubMed] [Google Scholar]
- de Reus MA, Van den Heuvel MP, 2013. The parcellation-based connectome: limitations and extensions. Neuroimage 80, 397–404. [DOI] [PubMed] [Google Scholar]
- de Schotten MT, Bizzi A, Dell’Acqua F, Allin M, Walshe M, Murray R, Williams SC, Murphy DG, Catani M, et al. , 2011. Atlasing location, asymmetry and inter-subject variability of white matter tracts in the human brain with mr diffusion tractography. Neuroimage 54, 49–59. [DOI] [PubMed] [Google Scholar]
- De Witte NA, Mueller SC, 2017. White matter integrity in brain networks relevant to anxiety and depression: evidence from the human connectome project dataset. Brain Imaging Behav. 11, 1604–1615. [DOI] [PubMed] [Google Scholar]
- Delettre C, Messé A, Dell LA, Foubet O, Heuer K, Larrat B, Meriaux S, Mangin JF, Reillo I, de Juan Romero C, Borrell V, Toro R, Hilgetag CC, 2019. Comparison between diffusion MRI tractography and histological tract-tracing of cortico-cortical structural connectivity in the ferret brain. Netw. Neurosci 3, 1038–1050. doi: 10.1162/netn_a_00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmarcelle T, Hesselink L, 1993. Visualizing second-order tensor fields with hyperstreamlines. IEEE Comput. Graph. Appl 13, 25–33. [Google Scholar]
- Deng Y, Hung KS, Lui SS, Chui WW, Lee JC, Wang Y, Li Z, Mak HK, Sham PC, Chan RC, et al. , 2019. Tractography-based classification in distinguishing patients with first-episode schizophrenia from healthy individuals. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 88, 66–73. [DOI] [PubMed] [Google Scholar]
- Dennis EL, Jahanshad N, McMahon KL, de Zubicaray GI, Martin NG, Hickie IB, Toga AW, Wright MJ, Thompson PM, 2013. Development of brain structural connectivity between ages 12 and 30: a 4-tesla diffusion imaging study in 439 adolescents and adults. Neuroimage 64, 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descoteaux M, Deriche R, Knosche TR, Anwander A, 2008. Deterministic and probabilistic tractography based on complex fibre orientation distributions. IEEE Trans. Med. Imaging 28, 269–286. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. , 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980. [DOI] [PubMed] [Google Scholar]
- Destrieux C, Fischl B, Dale A, Halgren E, 2010. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage 53, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick AS, Tremblay P, 2012. Beyond the arcuate fasciculus: consensus and controversy in the connectional anatomy of language. Brain 135, 3529–3550. [DOI] [PubMed] [Google Scholar]
- Dobson AJ, Barnett AG, 2018. An Introduction to Generalized Linear Models. CRC press. [Google Scholar]
- Dong X, Yang Z, Peng J, Wu X, 2019. Multimodality white matter tract segmentation using CNN. In: Proceedings of the ACM Turing Celebration Conference-China, pp. 1–8. [Google Scholar]
- Drakesmith M, Caeyenberghs K, Dutt A, Lewis G, David A, Jones D, 2015. Overcoming the effects of false positives and threshold bias in graph theoretical analyses of neuroimaging data. Neuroimage 118, 313–333. doi: 10.1016/j.neuroimage.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droby A, Fleischer V, Carnini M, Zimmermann H, Siffrin V, Gawehn J, Erb M, Hildebrandt A, Baier B, Zipp F, 2015. The impact of isolated lesions on white-matter fiber tracts in multiple sclerosis patients. Neuroimage Clin. 8, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J, Dehaene-Lambertz G, Kulikova S, Poupon C, Hüppi PS, Hertz-Pannier L, 2014. The early development of brain white matter: a review of imaging studies in fetuses, newborns and infants. Neuroscience 276, 48–71. [DOI] [PubMed] [Google Scholar]
- Dubois J, Poupon C, Thirion B, Simonnet H, Kulikova S, Leroy F, Hertz-Pannier L, Dehaene-Lambertz G, 2016. Exploring the early organization and maturation of linguistic pathways in the human infant brain. Cereb. Cortex 26, 2283–2298. [DOI] [PubMed] [Google Scholar]
- Duffau H, 2005. Lessons from brain mapping in surgery for low-grade glioma: insights into associations between tumour and brain plasticity. Lancet Neurol. 4, 476–486. [DOI] [PubMed] [Google Scholar]
- Duffau H, 2015. Stimulation mapping of white matter tracts to study brain functional connectivity. Nat. Rev. Neurol 11, 255. [DOI] [PubMed] [Google Scholar]
- Duffau H, 2021. Brain connectomics applied to oncological neuroscience: from a traditional surgical strategy focusing on glioma topography to a meta-network approach. Acta Neurochir. 1–13. [DOI] [PubMed] [Google Scholar]
- Eckstein I, Shattuck DW, Stein JL, McMahon KL, de Zubicaray G, Wright MJ, Thompson PM, Toga AW, 2009. Active fibers: matching deformable tract templates to diffusion tensor images. Neuroimage 47, T82–T89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff SB, Stephan KE, Mohlberg H, Grefkes C, Fink GR, Amunts K, Zilles K, 2005. A new spm toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage 25, 1325–1335. [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Rutka JT, Kulkarni AV, Dirks PB, Widjaja E, 2012. Corticospinal tract mapping in children with ruptured arteriovenous malformations using functionally guided diffusion-tensor imaging: report of 3 cases. J. Neurosurg. Pediatr 9, 505–510. [DOI] [PubMed] [Google Scholar]
- Ennis DB, Kindlmann G, 2006. Orthogonal tensor invariants and the analysis of diffusion tensor magnetic resonance images. Magn. Reson. Med 55, 136–146. [DOI] [PubMed] [Google Scholar]
- Essayed WI, Zhang F, Unadkat P, Cosgrove GR, Golby AJ, O’Donnell LJ, 2017. White matter tractography for neurosurgical planning: a topography-based review of the current state of the art. Neuroimage Clin. 15, 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada E, Hatano N, 2008. Communicability in complex networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys 77, 036111. [DOI] [PubMed] [Google Scholar]
- Fang FY, Chiang FL, Stephens N, Huang SY, Bilgic B, Tantiwongkosi B, Romero R, 2019. Characterization of normal-appearing white matter in multiple sclerosis using quantitative susceptibility mapping in conjunction with diffusion tensor imaging. Neuroradiology 61, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellner M, Varga B, Grolmusz V, 2020. The frequent complete subgraphs in the human connectome. PLoS ONE 15, e0236883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, He J, 2020. Asymmetric fiber trajectory distribution estimation using streamline differential equation. Med. Image Anal 63, 101686. [DOI] [PubMed] [Google Scholar]
- Feng Y, Yan W, Wang J, Song J, Zeng Q, Zhao C, 2020. Local white matter fiber clustering differentiates Parkinson’s disease diagnoses. Neuroscience 435, 146–160. [DOI] [PubMed] [Google Scholar]
- Fenoy A, 2020. Challenges in deep brain stimulation for depression. Braz. J. Psychiatry 42, 347–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Cabral DT, Zenonos GA, Hamilton RL, Panesar SS, Fernandez-Miranda JC, 2016. High-definition fiber tractography in the evaluation and surgical planning of Lhermitte-Duclos disease: a case report. World Neurosurg. 92, 587.e9–587.e13. [DOI] [PubMed] [Google Scholar]
- Fernandez-Miranda JC, Pathak S, Engh J, Jarbo K, Verstynen T, Yeh FC, Wang Y, Mintz A, Boada F, Schneider W, et al. , 2012. High-definition fiber tractography of the human brain: neuroanatomical validation and neurosurgical applications. Neurosurgery 71, 430–453. [DOI] [PubMed] [Google Scholar]
- Fick RH, Wassermann D, Deriche R, 2019. The dmipy toolbox: diffusion MRI multi–compartment modeling and microstructure recovery made easy. Front. Neuroinform 13, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi M, Preziosa P, Rocca MA, 2019. Brain mapping in multiple sclerosis: lessons learned about the human brain. Neuroimage 190, 32–45. [DOI] [PubMed] [Google Scholar]
- Fillard P, Descoteaux M, Goh A, Gouttard S, Jeurissen B, Malcolm J, Ramirez-Manzanares A, Reisert M, Sakaie K, Tensaouti F, et al. , 2011. Quantitative evaluation of 10 tractography algorithms on a realistic diffusion mr phantom. Neuroimage 56, 220–234. [DOI] [PubMed] [Google Scholar]
- Fillard P, Poupon C, Mangin JF, 2009. A novel global tractography algorithm based on an adaptive spin glass model. In: Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 927–934. [DOI] [PubMed] [Google Scholar]
- Fischl B, 2012. Freesurfer. Neuroimage 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons J, Kubicki M, Shenton ME, 2013. Review of functional and anatomical brain connectivity findings in schizophrenia. Curr. Opin. Psychiatry 26, 172–187. [DOI] [PubMed] [Google Scholar]
- Fleischer V, Gröger A, Koirala N, Droby A, Muthuraman M, Kolber P, Reuter E, Meuth SG, Zipp F, Groppa S, 2017. Increased structural white and grey matter network connectivity compensates for functional decline in early multiple sclerosis. Multiple Scler. J 23, 432–441. [DOI] [PubMed] [Google Scholar]
- Fleischer V, Radetz A, Ciolac D, Muthuraman M, Gonzalez-Escamilla G, Zipp F, Groppa S, 2019. Graph theoretical framework of brain networks in multiple sclerosis: a review of concepts. Neuroscience 403, 35–53. [DOI] [PubMed] [Google Scholar]
- Fornito A, Bullmore ET, Zalesky A, 2017. Opportunities and challenges for psychiatry in the connectomic era. Biol. Psychiatry Cogn. Neurosci.Neuroimaging 2, 9–19. [DOI] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Breakspear M, 2013. Graph analysis of the human connectome: promise, progress, and pitfalls. Neuroimage 80, 426–444. [DOI] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Breakspear M, 2015. The connectomics of brain disorders. Nat. Rev. Neurosci 16, 159–172. [DOI] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Bullmore ET, 2016. Fundamentals of brain network analysis. [Google Scholar]
- Fortin D, Aubin-Lemay C, Boré A, Girard G, Houde JC, Whittingstall K, Descoteaux M, 2012. Tractography in the study of the human brain: a neurosurgical perspective. Can. J. Neurol. Sci 39, 747–756. [DOI] [PubMed] [Google Scholar]
- Fortunato S, Hric D, 2016. Community detection in networks: a user guide. Phys. Rep 659, 1–44. [Google Scholar]
- Frigo M, Deslauriers-Gauthier S, Parker D, Ismail AAO, Kim JJ, Verma R, Deriche R, 2020. Diffusion MRI tractography filtering techniques change the topology of structural connectomes. J. Neural Eng 17, 065002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friman O, Farneback G, Westin CF, 2006. A Bayesian approach for stochastic white matter tractography. IEEE Trans. Med. Imaging 25, 965–978. [DOI] [PubMed] [Google Scholar]
- Friston K, 2002. Beyond phrenology: what can neuroimaging tell us about distributed circuitry? Annu. Rev. Neurosci 25, 221–250. [DOI] [PubMed] [Google Scholar]
- Gan J, Zhong M, Fan J, Liu W, Niu C, Cai S, Zou L, Wang Y, Wang Y, Tan C, et al. , 2017. Abnormal white matter structural connectivity in adults with obsessive–compulsive disorder. Transl. Psychiatry 7, e1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garyfallidis E, Brett M, Amirbekian B, Rokem A, Van Der Walt S, Descoteaux M, Nimmo-Smith I, 2014. Dipy, a library for the analysis of diffusion MRI data. Front. Neuroinform 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garyfallidis E, Côté MA, Rheault F, Sidhu J, Hau J, Petit L, Fortin D, Cunanne S, Descoteaux M, 2018. Recognition of white matter bundles using local and global streamline-based registration and clustering. Neuroimage 170, 283–295. [DOI] [PubMed] [Google Scholar]
- Garyfallidis E, Ocegueda O, Wassermann D, Descoteaux M, 2015. Robust and efficient linear registration of white-matter fascicles in the space of streamlines. Neuroimage 117, 124–140. [DOI] [PubMed] [Google Scholar]
- Geeraert BL, Chamberland M, Lebel RM, Lebel C, 2020. Multimodal principal component analysis to identify major features of white matter structure and links to reading. PLoS ONE 15, e0233244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertheiss J, Goldsmith J, Crainiceanu C, Greven S, 2013. Longitudinal scalar-on–functions regression with application to tractography data. Biostatistics 14, 447–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore JH, Knickmeyer RC, Gao W, 2018. Imaging structural and functional brain development in early childhood. Nat. Rev. Neurosci 19, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard G, Caminiti R, Battaglia-Mayer A, St-Onge E, Ambrosen KS, Eskildsen SF, Krug K, Dyrby TB, Descoteaux M, Thiran JP, Innocenti GM, 2020. On the cortical connectivity in the macaque brain: acomparison of diffusion tractography and histological tracing data. Neuroimage 221, 117201. [DOI] [PubMed] [Google Scholar]
- Girard G, Daducci A, Petit L, Thiran JP, Whittingstall K, Deriche R, Wassermann D, Descoteaux M, 2017. AxTract: toward microstructure informed tractography. Hum. Brain Mapp 38, 5485–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard G, Descoteaux M, 2012. Anatomical tissue probability priors for tractography. In: CDMRI, pp. 174–185. [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. , 2016. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goñi J, van den Heuvel MP, Avena-Koenigsberger A, Velez de Mendizabal N, Betzel RF, Griffa A, Hagmann P, Corominas-Murtra B, Thiran JP, Sporns O, 2014. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc. Natl. Acad. Sci. U.S.A 111, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith DR, Crooks CL, Walker EF, Cotes RO, 2018. An update on promising biomarkers in schizophrenia. Focus 16, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, He Y, Concha L, Lebel C, Gross DW, Evans AC, Beaulieu C, 2009. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb. Cortex 19, 524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, Rosa-Neto P, Carbonell F, Chen ZJ, He Y, Evans AC, 2009. Age- and gender-related differences in the cortical anatomical network. J. Neurosci 29, 15684–15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zhang F, Norton I, Essayed WI, Unadkat P, Rigolo L, Pasternak O, Rathi Y, Hou L, Golby AJ, et al. , 2018. Free water modeling of peritumoral edema using multi-fiber tractography: application to tracking the arcuate fasciculus for neurosurgical planning. PLoS ONE 13, e0197056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodlett CB, Fletcher PT, Gilmore JH, Gerig G, 2009. Group analysis of DTI fiber tract statistics with application to neurodevelopment. Neuroimage 45, S133–S142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D, Avena-Koenigsberger A, Mišić B, 2020. Editorial: network communication in the brain. Netw. Neurosci 4, 976–979. doi: 10.1162/netn_e_00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffa A, Baumann PS, Thiran JP, Hagmann P, 2013. Structural connectomics in brain diseases. Neuroimage 80, 515–526. [DOI] [PubMed] [Google Scholar]
- Grinberg F, Maximov II, Farrher E, Shah NJ, 2018. Microstructure-informed slow diffusion tractography in humans enhances visualisation of fibre pathways. Magn. Reson. Imaging 45, 7–17. [DOI] [PubMed] [Google Scholar]
- Gruner P, Vo A, Ikuta T, Mahon K, Peters BD, Malhotra AK, Uluğ AM, Szeszko PR, 2012. White matter abnormalities in pediatric obsessive-compulsive disorder. Neuropsychopharmacology 37, 2730–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara P, Duclap D, Poupon C, Marrakchi-Kacem L, Fillard P, Le Bihan D, Leboyer M, Houenou J, Mangin JF, 2012. Automatic fiber bundle segmentation in massive tractography datasets using a multi-subject bundle atlas. Neuroimage 61, 1083–1099. [DOI] [PubMed] [Google Scholar]
- Gupta T, Patil SM, Tailor M, Thapar D, Nigam A, 2017a. BrainSegNet: a segmentation network for human brain fiber tractography data into anatomically meaningful clusters. arXiv preprint arXiv:1710.05158. [Google Scholar]
- Gupta V, Thomopoulos SI, Corbin CK, Rashid F, Thompson PM, 2018. Fibernet 2.0: An automatic neural network based tool for clustering white matter fibers in the brain. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE, pp. 708–711. [Google Scholar]
- Gupta V, Thomopoulos SI, Rashid FM, Thompson PM, 2017. FiberNET: an ensemble deep learning framework for clustering white matter fibers. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 548–555. [Google Scholar]
- Habeck C, Stern Y, 2010. Multivariate data analysis for neuroimaging data: overview and application to Alzheimer’s disease. Cell Biochem. Biophys 58, 53–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O, 2008. Mapping the structural core of human cerebral cortex. PLoS Biol. 6, e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O, 2008. Mapping the structural core of human cerebral cortex. PLoS Biol. 6, e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Sporns O, Madan N, Cammoun L, Pienaar R, Wedeen VJ, Meuli R, Thiran JP, Grant P, 2010. White matter maturation reshapes structural connectivity in the late developing human brain. Proc. Natl. Acad. Sci 107, 19067–19072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CB, Yang Q, Lyu I, Rheault F, Kerley C, Chandio BQ, Fadnavis S, Williams O, Shafer AT, Resnick SM, et al. , 2020. Pandora: 4-D white matter bundle population-based atlases derived from diffusion MRI fiber tractography. Neuroinformatics 1. doi: 10.1007/s12021-020-09497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MP, Somerville LH, Ances BM, Andersson J, Barch DM, Bastiani M, Bookheimer SY, Brown TB, Buckner RL, Burgess GC, et al. , 2018. Extending the human connectome project across ages: imaging protocols for the lifespan development and aging projects. Neuroimage 183, 972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MG, Romero-Garcia R, Price SJ, Santarius T, Suckling J, 2020. Connections, tracts, fractals, and the rest: aworking guide to network and connectivity studies in neurosurgery. World Neurosurg. 140, 389–400. [DOI] [PubMed] [Google Scholar]
- Hasan KM, Iftikhar A, Kamali A, Kramer LA, Ashtari M, Cirino PT, Papanicolaou AC, Fletcher JM, Ewing-Cobbs L, 2009. Development and aging of the healthy human brain uncinate fasciculus across the lifespan using diffusion tensor tractography. Brain Res. 1276, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan KM, Kamali A, Abid H, Kramer LA, Fletcher JM, Ewing-Cobbs L, 2010. Quantification of the spatiotemporal microstructural organization of the human brain association, projection and commissural pathways across the lifespan using diffusion tensor tractography. Brain Struct. Funct 214, 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan KM, Kamali A, Iftikhar A, Kramer LA, Papanicolaou AC, Fletcher JM, Ewing-Cobbs L, 2009. Diffusion tensor tractography quantification of the human corpus callosum fiber pathways across the lifespan. Brain Res. 1249, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann RM, Anwander A, Feiweier T, Knösche TR, Turner R, 2012. K-space and q-space: combining ultra-high spatial and angular resolution in diffusion imaging using zooppa at 7t. Neuroimage 60, 967–978. doi: 10.1016/j.neuroimage.2011.12.081. [DOI] [PubMed] [Google Scholar]
- Heiervang E, Behrens T, Mackay CE, Robson MD, Johansen-Berg H, 2006. Between session reproducibility and between subject variability of diffusion mr and tractography measures. Neuroimage 33, 867–877. [DOI] [PubMed] [Google Scholar]
- Henriques RN, Jespersen SN, Shemesh N, 2020. Correlation tensor magnetic resonance imaging. Neuroimage 211, 116605. [DOI] [PubMed] [Google Scholar]
- Herbet G, Maheu M, Costi E, Lafargue G, Duffau H, 2016. Mapping neuroplastic potential in brain-damaged patients. Brain 139, 829–844. [DOI] [PubMed] [Google Scholar]
- Holm S, 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat 65–70. [Google Scholar]
- Hong SB, Zalesky A, Fornito A, Park S, Yang YH, Park MH, Song IC, Sohn CH, Shin MS, Kim BN, et al. , 2014. Connectomic disturbances in attention-deficit/hyperactivity disorder: a whole-brain tractography analysis. Biol. Psychiatry 76, 656–663. [DOI] [PubMed] [Google Scholar]
- Horn A, Fox MD, 2020. Opportunities of connectomic neuromodulation. Neuroimage 221, 117180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn A, Reich M, Vorwerk J, Li N, Wenzel G, Fang Q, Schmitz-Hübsch T, Nickl R, Kupsch A, Volkmann J, et al. , 2017. Connectivity predicts deep brain stimulation outcome in Parkinson disease. Ann. Neurol 82, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz B, 2003. The elusive concept of brain connectivity. Neuroimage 19, 466–470. [DOI] [PubMed] [Google Scholar]
- Hua K, Zhang J, Wakana S, Jiang H, Li X, Reich DS, Calabresi PA, Pekar JJ, van Zijl PC, Mori S, 2008. Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. Neuroimage 39, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, 2010. Delineating neural structures of developmental human brains with diffusion tensor imaging. Sci. World J 10, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Shu N, Mishra V, Jeon T, Chalak L, Wang ZJ, Rollins N, Gong G, Cheng H, Peng Y, et al. , 2015. Development of human brain structural networks through infancy and childhood. Cereb. Cortex 25, 1389–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Xue R, Zhang J, Ren T, Richards LJ, Yarowsky P, Miller MI, Mori S, 2009. Anatomical characterization of human fetal brain development with diffusion tensor magnetic resonance imaging. J. Neurosci 29, 4263–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher PR, 1984. Synapse elimination and plasticity in developing human cerebral cortex. Am. J. Ment. Defic 88, 488–496. [PubMed] [Google Scholar]
- Hutter J, Slator PJ, Christiaens D, Teixeira RPA, Roberts T, Jackson L, Price AN, Malik S, Hajnal JV, 2018. Integrated and efficient diffusion-relaxometry using zebra. Sci. Rep 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T, Shafritz KM, Bregman J, Peters BD, Gruner P, Malhotra AK, Szeszko PR, 2014. Abnormal cingulum bundle development in autism: a probabilistic tractography study. Psychiatry Res. Neuroimaging 221, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imms P, Domínguez D JF, Burmester A, Seguin C, Clemente A, Dhollander T, Wilson PH, Poudel G, Caeyenberghs K, 2021. Navigating the link between processing speed and network communication in the human brain. Brain Struct. Funct 226, 1281–1302. [DOI] [PubMed] [Google Scholar]
- Ingalhalikar M, Smith A, Parker D, Satterthwaite TD, Elliott MA, Ruparel K, Hakonarson H, Gur RE, Gur RC, Verma R, 2014. Sex differences in the structural connectome of the human brain. Proc. Natl. Acad. Sci 111, 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia A, Fan D, Chaudhari NN, Ngo V, Zhang F, Joshi SH, O’Donnell LJ, 2020. Mapping cerebral connectivity changes after mild traumatic brain injury in older adults using diffusion tensor imaging and Riemannian matching of elastic curves. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI). IEEE, pp. 1690–1693. [Google Scholar]
- Iturria-Medina Y, Sotero RC, Canales-Rodríguez EJ, Alemán-Gómez Y, Melie-García L, 2008. Studying the human brain anatomical network via diffusion-weighted MRI and graph theory. Neuroimage 40, 1064–1076. [DOI] [PubMed] [Google Scholar]
- Jack CR Jr, Holtzman DM, 2013. Biomarker modeling of Alzheimer’s disease. Neuron 80, 1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski M, Kao CY, Qiu M, Constable RT, Staib LH, 2005. White matter tractography by anisotropic wavefront evolution and diffusion tensor imaging. Med. Image Anal 9, 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski I, Filipovic R, Mo Z, Rakic S, Zecevic N, 2009. Oligodendrocyte development and the onset of myelination in the human fetal brain. Front. Neuroanat 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javadi SA, Nabavi A, Giordano M, Faghihzadeh E, Samii A, 2017. Evaluation of diffusion tensor imaging–based tractography of the corticospinal tract: a correlative study with intraoperative magnetic resonance imaging and direct electrical subcortical stimulation. Neurosurgery 80, 287–299. [DOI] [PubMed] [Google Scholar]
- Jbabdi S, Bellec P, Toro R, Daunizeau J, Pelegrini-Issac M, Benali H, 2008. Accurate anisotropic fast marching for diffusion-based geodesic tractography. J. Biomed. Imaging 2008, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbabdi S, Woolrich M, Andersson J, Behrens T, 2007. A Bayesian framework for global tractography. Neuroimage 37, 116–129. [DOI] [PubMed] [Google Scholar]
- Jelescu IO, Budde MD, 2017. Design and validation of diffusion MRI models of white matter. Front. Phys 5, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellison BJ, Field AS, Medow J, Lazar M, Salamat MS, Alexander AL, 2004. Diffusion tensor imaging of cerebral white matter: a pictorial review of physics, fiber tract anatomy, and tumor imaging patterns. Am. J. Neuroradiol 25, 356–369. [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM, 2012. Fsl. Neuroimage 62, 782–790. [DOI] [PubMed] [Google Scholar]
- Jensen JH, Helpern JA, 2010. MRI quantification of non-gaussian water diffusion by kurtosis analysis. NMR Biomed. 23, 698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeurissen B, Descoteaux M, Mori S, Leemans A, 2019. Diffusion MRI fiber tractography of the brain. NMR Biomed. 32, e3785. [DOI] [PubMed] [Google Scholar]
- Jeurissen B, Tournier JD, Dhollander T, Connelly A, Sijbers J, 2014. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 103, 411–426. [DOI] [PubMed] [Google Scholar]
- Ji E, Guevara P, Guevara M, Grigis A, Labra N, Sarrazin S, Hamdani N, Bellivier F, Delavest M, Leboyer M, et al. , 2019. Increased and decreased superficial white matter structural connectivity in schizophrenia and bipolar disorder. Schizophr. Bull 45, 1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Van Zijl PC, Kim J, Pearlson GD, Mori S, 2006. DTIStudio: resource program for diffusion tensor computation and fiber bundle tracking. Comput. Methods Programs Biomed 81, 106–116. [DOI] [PubMed] [Google Scholar]
- Jolly AE, Bălăeţ M, Azor A, Friedland D, Sandrone S, Graham NS, Zimmerman K, Sharp DJ, 2021. Detecting axonal injury in individual patients after traumatic brain injury. Brain 144, 92–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, 2010. Challenges and limitations of quantifying brain connectivity in vivo with diffusion MRI. Imaging Med. 2, 341. [Google Scholar]
- Jones DK, Catani M, Pierpaoli C, Reeves SJ, Shergill SS, O’Sullivan M, Golesworthy P, McGuire P, Horsfield MA, Simmons A, et al. , 2006. Age effects on diffusion tensor magnetic resonance imaging tractography measures of frontal cortex connections in schizophrenia. Hum. Brain Mapp 27, 230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, Knösche TR, Turner R, 2013. White matter integrity, fiber count, and other fallacies: The do’s and don’ts of diffusion MRI. 10.1016/j.neuroimage.2012.06.081, arXiv:9905108 [DOI] [PubMed] [Google Scholar]
- Jones DK, Travis AR, Eden G, Pierpaoli C, Basser PJ, 2005. Pasta: pointwise assessment of streamline tractography attributes. Magn. Reson. Med 53, 1462–1467. [DOI] [PubMed] [Google Scholar]
- Kaden E, Kelm ND, Carson RP, Does MD, Alexander DC, 2016. Multi-compartment microscopic diffusion imaging. Neuroimage 139, 346–359. doi: 10.1016/j.neuroimage.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaan RA, Shergill SS, Barker GJ, Catani M, Ng VW, Howard R, McGuire PK, Jones DK, 2006. Tract-specific anisotropy measurements in diffusion tensor imaging. Psychiatry Res. Neuroimaging 146, 73–82. [DOI] [PubMed] [Google Scholar]
- Karlsgodt KH, van Erp TG, Poldrack RA, Bearden CE, Nuechterlein KH, Cannon TD, 2008. Diffusion tensor imaging of the superior longitudinal fasciculus and working memory in recent-onset schizophrenia. Biol. Psychiatry 63, 512–518. [DOI] [PubMed] [Google Scholar]
- Kelly S, Jahanshad N, Zalesky A, Kochunov P, Agartz I, Alloza C, Andreassen O, Arango C, Banaj N, Bouix S, et al. , 2018. Widespread white matter microstructural differences in schizophrenia across 4322 individuals: results from the enigma schizophrenia DTI working group. Mol. Psychiatry 23, 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern KC, Sarcona J, Montag M, Giesser BS, Sicotte NL, 2011. Corpus callosal diffusivity predicts motor impairment in relapsing–remitting multiple sclerosis: a TBSS and tractography study. Neuroimage 55, 1169–1177. [DOI] [PubMed] [Google Scholar]
- Khan S, Vasung L, Marami B, Rollins CK, Afacan O, Ortinau CM, Yang E, Warfield SK, Gholipour A, 2019. Fetal brain growth portrayed by a spatiotemporal diffusion tensor MRI atlas computed from in utero images. Neuroimage 185, 593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser P, Baker ST, Cropley VL, Bousman C, Fornito A, Cocchi L, Fullerton JM, Rasser P, Schall U, Henskens F, et al. , 2017. White matter disruptions in schizophrenia are spatially widespread and topologically converge on brain network hubs. Schizophr. Bull 43, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocevar G, Stamile C, Hannoun S, Cotton F, Vukusic S, Durand-Dubief F, Sappey–Marinier D, 2016. Graph theory-based brain connectivity for automatic classification of multiple sclerosis clinical courses. Front. Neurosci 10, 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K, Reeß TJ, Rus OG, Zimmer C, Zaudig M, 2014. Diffusion tensor imaging (DTI) studies in patients with obsessive-compulsive disorder (OCD): a review. J. Psychiatr. Res 54, 26–35. [DOI] [PubMed] [Google Scholar]
- Kolind S, Matthews L, Johansen-Berg H, Leite MI, Williams SC, Deoni S, Palace J, 2012. Myelin water imaging reflects clinical variability in multiple sclerosis. Neuroimage 60, 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar MS, Fornito A, Williams LM, Grieve SM, 2014. Abnormal structural networks characterize major depressive disorder: a connectome analysis. Biol. Psychiatry 76, 567–574. [DOI] [PubMed] [Google Scholar]
- Kovanlikaya I, Firat Z, Kovanlikaya A, Uluğ AM, Cihangiroglu MM, John M, Bingol CA, Ture U, 2011. Assessment of the corticospinal tract alterations before and after resection of brainstem lesions using diffusion tensor imaging (DTI) and tractography at 3T. Eur. J. Radiol 77, 383–391. [DOI] [PubMed] [Google Scholar]
- Kreher BW, Mader I, Kiselev VG, 2008. Gibbs tracking: a novel approach for the reconstruction of neuronal pathways. Magn. Reson. Med 60, 953–963. [DOI] [PubMed] [Google Scholar]
- Kreilkamp BA, Lisanti L, Glenn GR, Wieshmann UC, Das K, Marson AG, Keller SS, 2019. Comparison of manual and automated fiber quantification tractography in patients with temporal lobe epilepsy. Neuroimage Clin. 24, 102024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K, Siddiqi K, Desrosiers C, 2019. White matter fiber analysis using kernel dictionary learning and sparsity priors. Pattern Recognit. 95, 83–95. [Google Scholar]
- Kunimatsu A, Aoki S, Masutani Y, Abe O, Hayashi N, Mori H, Masumoto T, Ohtomo K, 2004. The optimal trackability threshold of fractional anisotropy for diffusion tensor tractography of the corticospinal tract. Magn. Reson. Med. Sci 3, 11–17. [DOI] [PubMed] [Google Scholar]
- Labra N, Guevara P, Duclap D, Houenou J, Poupon C, Mangin JF, Figueroa M, 2017. Fast automatic segmentation of white matter streamlines based on a multi-subject bundle atlas. Neuroinformatics 15, 71–86. [DOI] [PubMed] [Google Scholar]
- Lam PDN, Belhomme G, Ferrall J, Patterson B, Styner M, Prieto JC, 2018. Trafic: fiber tract classification using deep learning. In: Medical Imaging 2018: Image Processing. International Society for Optics and Photonics, p. 1057412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen M, Leemans A, Johnston P, Ecker C, Daly E, Murphy CM, dell’Acqua F, Durston S, Murphy DG, Consortium A, et al. , 2012. Fronto-striatal circuitry and inhibitory control in autism: findings from diffusion tensor imaging tractography. Cortex 48, 183–193. [DOI] [PubMed] [Google Scholar]
- Lawes INC, Barrick TR, Murugam V, Spierings N, Evans DR, Song M, Clark CA, 2008. Atlas-based segmentation of white matter tracts of the human brain using diffusion tensor tractography and comparison with classical dissection. Neuroimage 39, 62–79. [DOI] [PubMed] [Google Scholar]
- Lazar M, Alexander AL, 2005. Bootstrap white matter tractography (BOOT-TRAC). Neuroimage 24, 524–532. [DOI] [PubMed] [Google Scholar]
- Le Bihan D, Johansen-Berg H, 2012. Diffusion MRI at 25: exploring brain tissue structure and function. Neuroimage 61, 324–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C, Beaulieu C, 2011. Longitudinal development of human brain wiring continues from childhood into adulthood. J. Neurosci 31, 10937–10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C, Caverhill-Godkewitsch S, Beaulieu C, 2010. Age-related regional variations of the corpus callosum identified by diffusion tensor tractography. Neuroimage 52, 20–31. [DOI] [PubMed] [Google Scholar]
- Lebel C, Deoni S, 2018. The development of brain white matter microstructure. Neuroimage 182, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C, Gee M, Camicioli R, Wieler M, Martin W, Beaulieu C, 2012. Diffusion tensor imaging of white matter tract evolution over the lifespan. Neuroimage 60, 340–352. [DOI] [PubMed] [Google Scholar]
- Lebel C, Treit S, Beaulieu C, 2019. A review of diffusion MRI of typical white matter development from early childhood to young adulthood. NMR Biomed. 32, e3778. [DOI] [PubMed] [Google Scholar]
- Lee J, Hyun JW, Lee J, Choi EJ, Shin HG, Min K, Nam Y, Kim HJ, Oh SH, 2020. So you want to image myelin using MRI: an overview and practical guide for myelin water imaging. J. Magn. Reson. Imaging n/a doi: 10.1002/jmri.27059. [DOI] [PubMed] [Google Scholar]
- Leemans A, Jeurissen B, Sijbers J, Jones D, 2009. Exploredti: a graphical toolbox for processing, analyzing, and visualizing diffusion MR data. In: Proc. Intl. Soc. Mag. Reson. Med., p. 3537. [Google Scholar]
- Lemkaddem A, Skiöldebrand D, Dal Palú A, Thiran JP, Daducci A, 2014. Global tractography with embedded anatomical priors for quantitative connectivity analysis. Front. Neurol 5, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt JJ, Nestor PG, Kubicki M, Lyall AE, Zhang F, Riklin-Raviv T, O’Donnell LJ, McCarley RW, Shenton ME, Rathi Y, 2020. Miswiring of frontostriatal projections in schizophrenia. Schizophr. Bull 46, 990–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, de Groot M, Steketee RM, Meijboom R, Smits M, Vernooij MW, Ikram MA, Liu J, Niessen WJ, Bron EE, 2020. Neuro4neuro: a neural network approach for neural tract segmentation using large-scale population-based diffusion imaging. Neuroimage 116993. [DOI] [PubMed] [Google Scholar]
- Li H, Xue Z, Guo L, Liu T, Hunter J, Wong ST, 2010. A hybrid approach to automatic clustering of white matter fibers. Neuroimage 49, 1249–1258. [DOI] [PubMed] [Google Scholar]
- Li M, Ratnanather JT, Miller MI, Mori S, 2014. Knowledge-based automated reconstruction of human brain white matter tracts using a path-finding approach with dynamic programming. Neuroimage 88, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chen Z, Guo W, Zeng Q, Feng Y, 2020. Two parallel stages deep learning network for anterior visual pathway segmentation. CDMRI. pp. 1–1 [Google Scholar]
- Li X, Wang Y, Wang W, Huang W, Chen K, Xu K, Zhang J, Chen Y, Li H, Wei D, et al. , 2020. Age-related decline in the topological efficiency of the brain structural connectome and cognitive aging. Cereb. Cortex 30, 4651–4661. [DOI] [PubMed] [Google Scholar]
- Li Y, Jewells V, Kim M, Chen Y, Moon A, Armao D, Troiani L, Markovic-Plese S, Lin W, Shen D, 2013. Diffusion tensor imaging based network analysis detects alterations of neuroconnectivity in patients with clinically early relapsing-remitting multiple sclerosis. Hum. Brain Mapp 34, 3376–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Tench CR, Morgan PS, Niepel G, Constantinescu CS, 2005. Importance sampling’in MS: use of diffusion tensor tractography to quantify pathology related to specific impairment. J. Neurol. Sci 237, 13–19. [DOI] [PubMed] [Google Scholar]
- Lipp I, Parker GD, Tallantyre EC, Goodall A, Grama S, Patitucci E, Heveron P, Tomassini V, Jones DK, 2020. Tractography in the presence of multiple sclerosis lesions. Neuroimage 209, 116471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Feng J, Chen G, Wu Y, Hong Y, Yap PT, Shen D, 2019. Deepbundle: fiber bundle parcellation with graph convolution neural networks. In: International Workshop on Graph Learning in Medical Imaging. Springer, pp. 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lauer KK, Ward BD, Roberts CJ, Liu S, Gollapudy S, Rohloff R, Gross W, Xu Z, Chen G, et al. , 2017. Fine-grained parcellation of brain connectivity improves differentiation of states of consciousness during graded propofol sedation. Brain Connect. 7, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Duan Y, Dong H, Barkhof F, Li K, Shu N, 2018. Disrupted module efficiency of structural and functional brain connectomes in clinically isolated syndrome and multiple sclerosis. Front. Hum. Neurosci 12, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llufriu S, Martinez-Heras E, Solana E, Sola-Valls N, Sepulveda M, Blanco Y, Martinez-Lapiscina EH, Andorra M, Villoslada P, Prats-Galino A, et al. , 2017. Structural networks involved in attention and executive functions in multiple sclerosis. Neuroimage Clin. 13, 288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CY, Wang PN, Chou KH, Wang J, He Y, Lin CP, 2010. Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer’s disease. J. Neurosci 30, 16876–16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louapre C, Govindarajan ST, Giannì C, Cohen-Adad J, Gregory MD, Nielsen AS, Madigan N, Sloane JA, Kinkel RP, Mainero C, 2016. Is the relationship between cortical and white matter pathologic changes in multiple sclerosis spatially specific? A multimodal 7-T and 3-T MR imaging study with surface and tract-based analysis. Radiology 278, 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Li Y, Ye C, 2020. White matter tract segmentation with self-supervised learning. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 270–279. [Google Scholar]
- Maddah M, Mewes AU, Haker S, Grimson WEL, Warfield SK, 2005. Automated atlas-based clustering of white matter fiber tracts from DTMRI. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 188–195. [DOI] [PubMed] [Google Scholar]
- Madden DJ, Bennett IJ, Song AW, 2009. Cerebral white matter integrity and cognitive aging: contributions from diffusion tensor imaging. Neuropsychol. Rev 19, 415–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden DJ, Jain S, Monge ZA, Cook AD, Lee A, Huang H, Howard CM, Cohen JR, 2020. Influence of structural and functional brain connectivity on age-related differences in fluid cognition. Neurobiol. Aging 96, 205–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madole JW, Ritchie SJ, Cox SR, Buchanan CR, Hernández MV, Maniega SM, Wardlaw JM, Harris MA, Bastin ME, Deary IJ, et al. , 2020. Aging-sensitive networks within the human structural connectome are implicated in late-life cognitive declines. Biol. Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier-Hein KH, Neher PF, Houde JC, Côtë MA, Garyfallidis E, Zhong J, Chamberland M, Yeh FC, Lin YC, Ji Q, Reddick WE, Glass JO, Chen DQ, Feng Y, Gao C, Wu Y, Ma J, Renjie H, Li Q, Westin CF, Deslauriers-Gauthier S, González JOO, Paquette M, St-Jean S, Girard G, Rheault F, Sidhu J, Tax CMW, Guo F, Mesri HY, Dávid S, Froeling M, Heemskerk AM, Leemans A, Boré A, Pinsard B, Bedetti C, Desrosiers M, Brambati S, Doyon J, Sarica A, Vasta R, Cerasa A, Quattrone A, Yeatman J, Khan AR, Hodges W, Alexander S, Romascano D, Barakovic M, Auría A, Esteban O, Lemkaddem A, Thiran JP, Cetingul HE, Odry BL, Mailhe B, Nadar MS, Pizzagalli F, Prasad G, Villalon-Reina JE, Galvis J, Thompson PM, Requejo FDS, Laguna PL, Lacerda LM, Barrett R, Dell’Acqua F, Catani M, Petit L, Caruyer E, Daducci A, Dyrby TB, Holland-Letz T, Hilgetag CC, Stieltjes B, Descoteaux M, 2017. The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun 8, 1349. doi: 10.1038/s41467-017-01285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makris N, Kennedy DN, McInerney S, Sorensen AG, Wang R, Caviness VS Jr., Pandya DN, 2005. Segmentation of subcomponents within the superior longitudinal fascicle in humans: a quantitative, in vivo, DT-MRI study. Cereb. Cortex 15, 854–869. [DOI] [PubMed] [Google Scholar]
- Malcolm JG. Shenton ME. Rathi Y. 2010. Filtered multitensor tractography. IEEE Trans. Med. Imaging 29, 1664–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M. Peter R. Hodneland E. Lundervold AJ. Lundervold A. Jan J. 2013. Registration of fa and t1-weighted MRI data of healthy human brain based on template matching and normalized cross-correlation. J. Digit. Imaging 26, 774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamah D, Ji A. Rutlin J. Shimony JS. 2019. White matter integrity in schizophrenia and bipolar disorder: tract-and voxel-based analyses of diffusion data from the connectom scanner. Neuroimage Clin. 21, 101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini M, Karakuzu A, Cohen-Adad J, Cercignani M, Nichols TE, Stikov N, 2020. An interactive meta-analysis of MRI biomarkers of myelin. Elife 9, e61523. doi: 10.7554/eLife.61523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandonnet E, Sarubbo S, Petit L, 2018. The nomenclature of human white matter association pathways: proposal for a systematic taxonomic anatomical classification. Front. Neuroanat 12, 94. doi: 10.3389/fnana.2018.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin JF. Fillard P, Cointepas Y. Le Bihan D. Frouin V. Poupon C. 2013. Toward global tractography. Mapp. Connectome 80, 290–296. [DOI] [PubMed] [Google Scholar]
- Mangin JF. Poupon C. Cointepas Y. Riviére D. Papadopoulos-Orfanos D. Clark CA. Régis J. Le Bihan D, 2002. A framework based on spin glass models for the i nference of anatomical connectivity from diffusion-weighted MR data - a technical review. NMR Biomed. 15, 481–492. [DOI] [PubMed] [Google Scholar]
- Martí-Juan G. Sanroma-Guell G. Piella G. 2020. A survey on machine and statistical learning for longitudinal analysis of neuroimaging data in Alzheimer’s disease. Comput. Methods Programs Biomed 189, 105348. [DOI] [PubMed] [Google Scholar]
- Mazziotta J. Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, et al. , 2001. A probabilistic atlas and reference system for the human brain: international consortium for brain mapping (ICBM). Philos. Trans. R. Soc.London Ser. B 356, 1293–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola A, Yeh FC, Fellows-Mayle W, Weed J, Fernandez-Miranda JC, 2016. Human connectome-based tractographic atlas of the brainstem connections and surgical approaches. Neurosurgery 79, 437–455. [DOI] [PubMed] [Google Scholar]
- Messé A. 2020. Parcellation influence on the connectivity-based structure–function relationship in the human brain. Hum. Brain Mapp 41, 1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse S, Coupland N, Camicioli R, Carter R, Seres P, Sabino J, Malykhin N, 2010. Selective effects of aging on brain white matter microstructure: a diffusion tensor imaging tractography study. Neuroimage 52, 1190–1201. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, Fobbs AJ. Sousa AM. Šestan N, Wildman DE. et al. ,. 2012. Prolonged myelination in human neocortical evolution. Proc. Natl. Acad. Sci 109, 16480–16485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito R, Raffelt D, Dhollander T, Vaughan DN, Tournier JD, Salvado O, Brodtmann A, Rowe CC, Villemagne VL, Connelly A, 2018. Fibre-specific white matter reductions in Alzheimer’s disease and mild cognitive impairment. Brain 141, 888–902. [DOI] [PubMed] [Google Scholar]
- Mitter C, Jakab A, Brugger PC, Ricken G, Gruber GM, Bettelheim D, Scharrer A, Langs G, Hainfellner JA, Prayer D, et al. , 2015. Validation of in utero tractography of human fetal commissural and internal capsule fibers with histological structure tensor analysis. Front. Neuroanat 9, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mišic B. Betzel RF. Griffa A. de Reus MA. He Y. Zuo XN. van den Heuvel MP. Hagmann P, Sporns O, Zatorre RJ, 2018. Network-based asymmetry of the human auditory system. Cereb Cortex 28, 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mišić B. Betzel RF. Nematzadeh A. Goñi J. Griffa A. Hagmann P, Flammini A. Ahn YY, Sporns O, 2015. Cooperative and competitive spreading dynamics on the human connectome. Neuron 86, 1518–1529. [DOI] [PubMed] [Google Scholar]
- Moldrich RX, Pannek K, Hoch R, Rubenstein JL, Kurniawan ND, Richards LJ, 2010. Comparative mouse brain tractography of diffusion magnetic resonance imaging. Neuroimage 51, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S. Crain BJ. Chacko VP. Van Zijl PC. 1999. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann. Neurol 45, 265–269. [DOI] [PubMed] [Google Scholar]
- Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, et al. , 2008. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage 40, 570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz-Gasser S, Herbet G, Maldonado IL, Duffau H, 2012. Lexical access speed is significantly correlated with the return to professional activities after awake surgery for low-grade gliomas. J. Neurooncol 107, 633–641. [DOI] [PubMed] [Google Scholar]
- Moseley M. 2002. Diffusion tensor imaging and aging–a review. NMR Biomed. 15, 553–560. [DOI] [PubMed] [Google Scholar]
- Mukherjee P, 2005. Diffusion tensor imaging and fiber tractography in acute stroke. Neuroimaging Clin. 15, 655–665. [DOI] [PubMed] [Google Scholar]
- Muthuraman M, Fleischer V, Kolber P, Luessi F, Zipp F, Groppa S, 2016. Structural brain network characteristics can differentiate CIS from early RRMS. Front. Neurosci 10, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath V, Pizzolato M. Palombo M. Gyori N. Schilling KG. Hansen CH. Qi Y. Kanakaraj P, Landman BA, Chatterjee S, et al. , 2021. Resolving to super resolution multi-dimensional diffusion imaging (super-mudi). ISMRM & SMRT Annual Meeting. [Google Scholar]
- Negwer C, Sollmann N, Ille S, Hauck T, Maurer S, Kirschke JS, Ringel F, Meyer B, Krieg SM, 2017. Language pathway tracking: comparing nTMS-based DTI fiber tracking with a cubic ROIs-based protocol. J. Neurosurg 126, 1006–1014. [DOI] [PubMed] [Google Scholar]
- Neher PF. Côté MA. Houde JC. Descoteaux M. Maier-Hein KH. 2017. Fiber tractography using machine learning. Neuroimage 158, 417–429. [DOI] [PubMed] [Google Scholar]
- Neher PF, Descoteaux M, Houde JC, Stieltjes B, Maier-Hein KH, 2015. Strengths and weaknesses of state of the art fiber tractography pipelines–a comprehensive in-vivo and phantom evaluation study using tractometer. Med. Image Anal 26, 287–305. [DOI] [PubMed] [Google Scholar]
- Neil J. Miller J. Mukherjee P. Hüppi PS. 2002. Diffusion tensor imaging of normal and injured developing human brain-a technical review. NMR Biomed. 15, 543–552. [DOI] [PubMed] [Google Scholar]
- Ning L, Gagoski B, Szczepankiewicz F, Westin CF, Rathi Y, 2019. Joint relaxation-diffusion imaging moments to probe neurite microstructure. IEEE Trans. Med. Imaging 39, 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning L, Szczepankiewicz F, Nilsson M, Rathi Y, Westin CF, 2021. Probing tissue microstructure by diffusion skewness tensor imaging. Sci. Rep 11, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton I, Essayed WI, Zhang F, Pujol S, Yarmarkovich A, Golby AJ, Kindlmann G, Wassermann D, Estepar RSJ, Rathi Y, et al. , 2017. SlicerDMRI: open source diffusion MRI software for brain cancer research. Cancer Res. 77, e101–e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikov DS, Fieremans E, Jespersen SN, Kiselev VG, 2019. Quantifying brain microstructure with diffusion MRI: theory and parameter estimation. NMR Biomed. 32, e3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikov DS, Kiselev VG, Jespersen SN, 2018. On modeling. Magn. Reson. Med 79, 3172–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora PG, Verma R, Lee SK, Melhem ER, 2007. Diffusion-tensor MR imaging and tractography: exploring brain microstructure and connectivity. Radiology 245, 367–384. [DOI] [PubMed] [Google Scholar]
- Ocampo-Pineda M, Schiavi S, Rheault F, Girard G, Petit L, Descoteaux M, Daducci A. 2021. Hierarchical microstructure informed tractography. Brain Connect.. [DOI] [PubMed] [Google Scholar]
- O’Donnell L, Haker S, Westin CF, 2002. New approaches to estimation of white matter connectivity in diffusion tensor MRI: Elliptic PDEs and geodesics in a tensor-warped space. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 459–466. [Google Scholar]
- O’Donnell LJ, Golby AJ, Westin CF, 2013. Fiber clustering versus the parcellation-based connectome. Neuroimage 80, 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell LJ, Pasternak O, 2015. Does diffusion MRI tell us anything about the white matter? an overview of methods and pitfalls. Schizophr. Res 161, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell LJ, Suter Y, Rigolo L, Kahali P, Zhang F, Norton I, Albi A, Olubiyi O, Meola A, Essayed WI, et al. , 2017. Automated white matter fiber tract identification in patients with brain tumors. Neuroimage Clin. 13, 138–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell LJ. Wells WM. Golby AJ. Westin CF. 2012. Unbiased groupwise registration of white matter tractography. In: MICCAI, pp. 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell LJ, Westin CF, 2007. Automatic tractography segmentation using a high-dimensional white matter atlas. IEEE Trans. Med. Imaging 26, 1562–1575. [DOI] [PubMed] [Google Scholar]
- O’Donnell LJ, Westin CF, Golby AJ, 2009. Tract-based morphometry for white matter group analysis. Neuroimage 45, 832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dwyer L, Lamberton F, Matura S, Scheibe M, Miller J, Rujescu D, Prvulovic D, Hampel H, 2012. White matter differences between healthy young APOE4 carriers and non-carriers identified with tractography and support vector machines. PLoS ONE 7, e36024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohoshi Y, Takahashi S, Yamada S, Ishida T, Tsuda K, Tsuji T, Terada M, Shinosaki K, Ukai S, 2019. Microstructural abnormalities in callosal fibers and their relationship with cognitive function in schizophrenia: a tract-specific analysis study. Brain Behav. 9, e01357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Faria A, Jiang H, Li X, Akhter K, Zhang J, Hsu JT, Miller MI, van Zijl PC, Albert M, et al. , 2009. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and Alzheimer’s disease participants. Neuroimage 46, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Zilles K, Amunts K, Faria A, Jiang H, Li X, Akhter K, Hua K, Woods R, Toga AW, et al. , 2008. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. Neuroimage 43, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S. Fulcher B. Parkes L. Arnatkevic Iūte A, Suo C. Fornito A. 2019. Consistency and differences between centrality measures across distinct classes of networks. PLoS ONE 14, e0220061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivetti E, Sharmin N, Avesani P, 2016. Alignment of tractograms as graph matching. Front. Neurosci 10, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski AK, Kikinis Z, Gonzalez CS, Coman IL, Makris N, Gong X, Rathi Y, Zhu A. Antshel KM. Fremont W. et al. ,. 2017. The social brain network in 22q11. 2 deletion syndrome: a diffusion tensor imaging study. Behav. Brain Funct 13, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ombao H, Lindquist M, Thompson W, Aston J, 2016. Handbook of Neuroimaging Data Analysis. CRC Press. [Google Scholar]
- Osmanlioğlu Y. Alappatt JA. Parker D. Verma R. 2020. Connectomic consistency: a systematic stability analysis of structural and functional connectivity. J. Neural Eng 17, 045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M, Dubois J, Yu Q, Mukherjee P, Huang H, 2019. Delineation of early brain development from fetuses to infants with diffusion MRI and beyond. Neuroimage 185, 836–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotaki E, Schneider T, Siow B, Hall MG, Lythgoe MF, Alexander DC, 2012. Compartment models of the diffusion mr signal in brain white matter: a taxonomy and comparison. Neuroimage 59, 2241–2254. [DOI] [PubMed] [Google Scholar]
- Panesar SS, Abhinav K, Yeh FC, Jacquesson T, Collins M, Fernandez-Miranda J, 2019. Tractography for surgical neuro-oncology planning: towards a gold standard. Neurotherapeutics 16, 36–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak O, Shenton ME, Westin CF, 2012. Estimation of extracellular volume from regularized multi-shell diffusion MRI. Medical image computing and computer-assisted intervention: MICCAI … international conference on medical image computing and computer-assisted intervention 15, 305–312. doi: 10.1007/978-3-642-33418-4_38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak O, Sochen N, Gur Y, Intrator N, Assaf Y, 2009. Free water elimination and mapping from diffusion MRI. Magn. Reson. Med 62, 717–730. [DOI] [PubMed] [Google Scholar]
- Payabvash S, Palacios EM, Owen JP, Wang MB, Tavassoli T, Gerdes M, Brandes-Aitken A, Marco EJ, Mukherjee P, 2019. Diffusion tensor tractography in children with sensory processing disorder: potentials for devising machine learning classifiers. Neuroimage Clin. 23, 101831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paydar A, Fieremans E, Nwankwo J, Lazar M, Sheth H, Adisetiyo V, Helpern J, Jensen J, Milla S, 2014. Diffusional kurtosis imaging of the developing brain. Am. J. Neuroradiol 35, 808–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecheva D, Yushkevich P, Batalle D, Hughes E, Aljabar P, Wurie J, Hajnal JV, Edwards AD, Alexander DC, Counsell SJ, et al. , 2017. A tract-specific approach to assessing white matter in preterm infants. Neuroimage 157, 675–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestilli F, Yeatman JD, Rokem A, Kay KN, Wandell BA, 2014. Evaluation and statistical inference for human connectomes. Nat Meth 11, 1058–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BD, Karlsgodt KH, 2015. White matter development in the early stages of psychosis. Schizophr. Res 161, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D, Ivanov A, Faskowitz J, Gutman B, Moyer D, Villalon J, Jahanshad N, Thompson P, 2017. Evaluating 35 methods to generate structural connectomes using pairwise classification. In: International Conference on medical Image Computing and Computer-Assisted Intervention. Springer, pp. 515–522. [Google Scholar]
- Pichon E, Westin CF, Tannenbaum AR, 2005. A Hamilton-Jacobi-Bellman approach to high angular resolution diffusion tractography. Med. Image Comput. Comput.-Assist.Interv 8, 180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piredda GF, Hilbert T, Thiran JP, Kober T, 2021. Probing myelin content of the human brain with MRI: a review. Magn. Reson. Med 85, 627–652. doi: 10.1002/mrm.28509. [DOI] [PubMed] [Google Scholar]
- Pizzolato M, Palombo M, Bonet-Carne E, Tax CM, Grussu F, Ianus A, Bogusz F, Pieciak T, Ning L, Larochelle H, et al. , 2020. Acquiring and predicting multidimensional diffusion (MUDI) data: An open challenge. In: Computational Diffusion MRI. Springer, pp. 195–208. [Google Scholar]
- Poulin P, Jörgens D, Jodoin PM, Descoteaux M, 2019. Tractography and machine learning: current state and open challenges. Magn. Reson. Imaging 64, 37–48. [DOI] [PubMed] [Google Scholar]
- Price G, Cercignani M, Parker GJ, Altmann DR, Barnes TR, Barker GJ, Joyce EM, Ron MA, 2007. Abnormal brain connectivity in first-episode psychosis: a diffusion MRI tractography study of the corpus callosum. Neuroimage 35, 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosperini L, Fanelli F, Petsas N, Sbardella E, Tona F, Raz E, Fortuna D, De Angelis F, Pozzilli C, Pantano P, 2014. Multiple sclerosis: changes in microarchitecture of white matter tracts after training with a video game balance board. Radiology 273, 529–538. [DOI] [PubMed] [Google Scholar]
- Prčkovska V, Rodrigues P, Puigdellivol Sanchez A, Ramos M, Andorra M, Martinez-Heras E, Falcon C, Prats-Galino A, Villoslada P, 2016. Reproducibility of the structural connectome reconstruction across diffusion methods. J. Neuroimaging 26, 46–57. [DOI] [PubMed] [Google Scholar]
- Pujol S, Wells W, Pierpaoli C, Brun C, Gee J, Cheng G, Vemuri B, Commowick O, Prima S, Stamm A, et al. , 2015. The DTI challenge: toward standardized evaluation of diffusion tensor imaging tractography for neurosurgery. J. Neuroimaging 25, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Meesters S, Nicolay K, ter Haar Romeny BM, Ossenblok P, 2015. The influence of construction methodology on structural brain network measures: a review. J. Neurosci. Methods 253, 170–182. [DOI] [PubMed] [Google Scholar]
- Qiu A, Mori S, Miller MI, 2015. Diffusion tensor imaging for understanding brain development in early life. Annu. Rev. Psychol 66, 853–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Oishi K, Miller MI, Lyketsos CG, Mori S, Albert M, 2010. Surface-based analysis on shape and fractional anisotropy of white matter tracts in Alzheimer’s disease. PLoS ONE 5, e9811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmanesh A, Zamani AA, Whalen S, Tie Y, Suarez RO, Golby AJ, 2015. Comparison of seeding methods for visualization of the corticospinal tracts using single tensor tractography. Clin. Neurol. Neurosurg 129, 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffelt D, Tournier JD, Rose S, Ridgway GR, Henderson R, Crozier S, Salvado O, Connelly A, 2012. Apparent fibre density: a novel measure for the analysis of diffusion-weighted magnetic resonance images. Neuroimage 59, 3976–3994. doi: 10.1016/j.neuroimage.2011.10.045. [DOI] [PubMed] [Google Scholar]
- Raichle ME, 2015. The brain’s default mode network. Annu. Rev. Neurosci 38, 433–447. [DOI] [PubMed] [Google Scholar]
- Ratnarajah N, Qiu A, 2014. Multi-label segmentation of white matter structures: application to neonatal brains. Neuroimage 102, 913–922. [DOI] [PubMed] [Google Scholar]
- Reddy CP, Rathi Y, 2016. Joint multi-fiber NODDI parameter estimation and tractography using the unscented information filter. Front. Neurosci 10, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert M, Coenen VA, Kaller C, Egger K, Skibbe H, 2018. Hamlet: hierarchical harmonic filters for learning tracts from diffusion MRI. arXiv preprint arXiv:1807.01068. [Google Scholar]
- Reisert M, Mader I, Anastasopoulos C, Weigel M, Schnell S, Kiselev V, 2011. Global fiber reconstruction becomes practical. Neuroimage 54, 955–962. [DOI] [PubMed] [Google Scholar]
- Reveley C, Seth AK, Pierpaoli C, Silva AC, Yu D, Saunders RC, Leopold DA, Ye FQ, 2015. Superficial white matter fiber systems impede detection of long-range cortical connections in diffusion mr tractography. Proc. Natl. Acad. Sci 112, E2820–E2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JE, Grohs MN, Dewey D, Lebel C, 2019. Global and regional white matter development in early childhood. Neuroimage 196, 49–58. [DOI] [PubMed] [Google Scholar]
- Rheault F, De Benedictis A, Daducci A, Maffei C, Tax CMW, Romascano D, Caverzasi E, Morency FC, Corrivetti F, Pestilli F, Girard G, Theaud G, Zemmoura I, Hau J, Glavin K, Jordan KM, Pomiecko K, Chamberland M, Barakovic M, Goyette N, Poulin P, Chenot Q, Panesar SS, Sarubbo S, Petit L, Descoteaux M, 2020. Tractostorm: the what, why, and how of tractography dissection reproducibility. Hum. Brain Mapp 41, 1859–1874. doi: 10.1002/hbm.24917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheault F, Poulin P, Caron AV, St-Onge E, Descoteaux M, 2020. Common misconceptions, hidden biases and modern challenges of dMRI tractography. J. Neural Eng 17, 11001. doi: 10.1088/1741-2552/ab6aad. [DOI] [PubMed] [Google Scholar]
- Rheault F, Poulin P, Caron AV, St-Onge E, Descoteaux M, 2020. Common misconceptions, hidden biases and modern challenges of dMRI tractography. J. Neural Eng 17, 011001. [DOI] [PubMed] [Google Scholar]
- Rheault F, St-Onge E, Sidhu J, Maier-Hein K, Tzourio-Mazoyer N, Petit L, Descoteaux M, 2019. Bundle-specific tractography with incorporated anatomical and orientational priors. Neuroimage 186, 382–398. [DOI] [PubMed] [Google Scholar]
- Riva-Posse P, Choi K, Holtzheimer PE, Crowell AL, Garlow SJ, Rajendra JK, McIntyre CC, Gross RE, Mayberg HS, 2018. A connectomic approach for subcallosal cingulate deep brain stimulation surgery: prospective targeting in treatment-resistant depression. Mol. Psychiatry 23, 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Perry A, Lord AR, Roberts G, Mitchell PB, Smith RE, Calamante F, Breakspear M, 2016. The contribution of geometry to the human connectome. Neuroimage 124, 379–393. [DOI] [PubMed] [Google Scholar]
- Roberts JA, Perry A, Roberts G, Mitchell PB, Breakspear M, 2017. Consistency-based thresholding of the human connectome. Neuroimage 145, 118–129. [DOI] [PubMed] [Google Scholar]
- Rocca MA, Amato MP, De Stefano N, Enzinger C, Geurts JJ, Penner IK, Rovira A, Sumowski JF, Valsasina P, Filippi M, et al. , 2015. Clinical and imaging assessment of cognitive dysfunction in multiple sclerosis. Lancet Neurol. 14, 302–317. [DOI] [PubMed] [Google Scholar]
- Rodrigues P, Prats A, Gallardo-Pujol D, Villoslada P, Falcon C, Prčkovska V, 2013. Evaluating structural connectomics: the effect of the cortical parcellation scheme. In: Front. Neuroinform. Conference Abstract: Imaging the brain at different scales: How to integrate multi-scale structural information doi: 10.3389/conf.fninf.2013.10.00030. [DOI] [Google Scholar]
- Román C, Guevara M, Valenzuela R, Figueroa M, Houenou J, Duclap D, Poupon C, Mangin JF, Guevara P, 2017. Clustering of whole-brain white matter short association bundles using HARDI data. Front. Neuroinform 11, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros C, Güllmar D, Stenzel M, Mentzel HJ, Reichenbach JR, 2013. Atlas-guided cluster analysis of large tractography datasets. PLoS ONE 8, e83847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini P, Di Iorio R, Bentivoglio M, Bertini G, Ferreri F, Gerloff C, Ilmoniemi R, Miraglia F, Nitsche M, Pestilli F, et al. , 2019. Methods for analysis of brain connectivity: an IFCN-sponsored review. Clin. Neurophysiol 130, 1833–1858. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O, 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069. [DOI] [PubMed] [Google Scholar]
- Sakkalis V, 2011. Review of advanced techniques for the estimation of brain connectivity measured with EEG/MEG. Comput. Biol. Med 41, 1110–1117. [DOI] [PubMed] [Google Scholar]
- Sarubbo S, Tate M, De Benedictis A, Merler S, Moritz-Gasser S, Herbet G, Duffau H, 2020. Mapping critical cortical hubs and white matter pathways by direct electrical stimulation: an original functional atlas of the human brain. Neuroimage 205, 116237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarwar T, Seguin C, Ramamohanarao K, Zalesky A, 2020. Towards deep learning for connectome mapping: a block decomposition framework. Neuroimage 212, 116654. [DOI] [PubMed] [Google Scholar]
- Sbardella E, Tona F, Petsas N, Pantano P, 2013. DTI measurements in multiple sclerosis: evaluation of brain damage and clinical implications. Mult. Scler. Int 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo XN, Holmes AJ, Eickhoff SB, Yeo BT, 2018. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb. Cortex 28, 3095–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaie KW, 2005. Developmental Influences on Adult Intelligence: The Seattle Longitudinal Study. Oxford University Press. [Google Scholar]
- Schiavi S, Ocampo-Pineda M, Barakovic M, Petit L, Descoteaux M, Thiran JP, Daducci A, 2020. A new method for accurate in vivo mapping of human brain connections using microstructural and anatomical information. Sci. Adv 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavi S, Petracca M, Battocchio M, El Mendili MM, Paduri S, Fleysher L, Inglese M, Daducci A, 2020. Sensory-motor network topology in multiple sclerosis: structural connectivity analysis accounting for intrinsic density discrepancy. Hum. Brain Mapp. n/a doi: 10.1002/hbm.24989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavi S, Pizzolato M, Ocampo-Pineda M, Canales-Rodriguez EJ, Thiran JP, Daducci A, 2019. Is it feasible to directly access the bundle’s specific myelin content, instead of averaging? a study with microstructure informed tractography. ISMRM 27th Annual Meeting & Exhibition. [Google Scholar]
- Schilling K, Gao Y, Janve V, Stepniewska I, Landman BA, Anderson AW, 2018. Confirmation of a gyral bias in diffusion MRI fiber tractography. Hum. Brain Mapp 39, 1449–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling K, Gao Y, Janve V, Stepniewska I, Landman BA, Anderson AW, 2018. Confirmation of a gyral bias in diffusion MRI fiber tractography. Hum. Brain Mapp 39, 1449–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KG, Petit L, Rheault F, Remedios S, Pierpaoli C, Anderson AW, Landman BA, Descoteaux M, 2020. Brain connections derived from diffusion MRI tractography can be highly anatomically accurate’ if we know where white matter pathways start, where they end, and where they do not go. Brain Struct. Funct 225, 2387–2402. doi: 10.1007/s00429-020-02129-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KG, Rheault F, Petit L, Hansen CB, Nath V, Yeh FC, Girard G, Barakovic M, Rafael-Patino J, Yu T, et al. , 2020. Tractography dissection variability: what happens when 42 groups dissect 14 white matter bundles on the same dataset? Neuroimage 243, 118502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KG, Tax CM, Rheault F, Landman B, Anderson A, Descoteaux M, Petit L, 2021. Prevalence of white matter pathways coming into a single diffusion MRI voxel orientation: the bottleneck issue in tractography. bioRxiv doi: 10.1101/2021.06.22.449454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer TE, Bewernick BH, Kayser S, Mädler B, Coenen VA, 2013. Rapid effects of deep brain stimulation for treatment-resistant major depression. Biol. Psychiatry 73, 1204–1212. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Schmahmann J, Pandya D, 2009. Fiber Pathways of the Brain. OUP USA. [Google Scholar]
- Schmidt A, Crossley NA, Harrisberger F, Smieskova R, Lenz C, Riecher-Rössler A, Lang UE, McGuire P, Fusar-Poli P, Borgwardt S, 2017. Structural network disorganization in subjects at clinical high risk for psychosis. Schizophr. Bull 43, 583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg H, Hohage T, 2019. Formulation and efficient computation of l1 - and smoothness penalized estimates for microstructure-informed tractography. IEEE Trans. Med. Imaging 38, 1899–1909. [DOI] [PubMed] [Google Scholar]
- Schreiber J, Riffert T, Anwander A, Knösche TR, 2014. Plausibility tracking: a method to evaluate anatomical connectivity and microstructural properties along fiber pathways. Neuroimage 90, 163–178. [DOI] [PubMed] [Google Scholar]
- Schroeder DH, Salthouse TA, 2004. Age-related effects on cognition between 20 and 50 years of age. Pers. Individ. Dif 36, 393–404. [Google Scholar]
- Schurr R, Filo S, Mezer AA, 2019. Tractography delineation of the vertical occipital fasciculus using quantitative T1 mapping. Neuroimage 202, 116121. [DOI] [PubMed] [Google Scholar]
- Seguin C, van den Heuvel MP, Zalesky A, 2018. Navigation of brain networks. Proc. Natl. Acad. Sci. U.S.A 115, 6297–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin C, Razi A, Zalesky A, 2019. Inferring neural signalling directionality from undirected structural connectomes. Nat. Commun 10, 4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin C, Tian Y, Zalesky A, 2020. Network communication models improve the behavioral and functional predictive utility of the human structural connectome. Netw. Neurosci 4, 980–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin C, Tian Y, Zalesky A, 2020. Network communication models improve the behavioral and functional predictive utility of the human structural connectome. Netw. Neurosci [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepasian N, ten Thije Boonkkamp J, Ter Haar Romeny B, Vilanova Bartroli A, 2012. Multivalued geodesic ray-tracing for computing brain connections using diffusion tensor imaging. SIAM J. Imaging Sci 5, 483–504. [Google Scholar]
- Shahab S, Stefanik L, Foussias G, Lai MC, Anderson KK, Voineskos AN, 2018. Sex and diffusion tensor imaging of white matter in schizophrenia: a systematic review plus meta-analysis of the corpus callosum. Schizophr. Bull 44, 203–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shany E, Inder TE, Goshen S, Lee I, Neil JJ, Smyser CD, Doyle LW, Anderson PJ, Shimony JS, 2017. Diffusion tensor tractography of the cerebellar peduncles in prematurely born 7-year-old children. Cerebellum 16, 314–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM, 2002. Brainsuite: an automated cortical surface identification tool. Med. Image Anal 6, 129–142. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW, 2008. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 39, 1064–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh N, Jespersen SN, Alexander DC, Cohen Y, Drobnjak I, Dyrby TB, Finsterbusch J, Koch MA, Kuder T, Laun F, et al. , 2016. Conventions and nomenclature for double diffusion encoding NMR and MRI. Magn. Reson. Med 75, 82–87. [DOI] [PubMed] [Google Scholar]
- Sherbondy A, Rowe M, Alexander D, 2010. MicroTrack: an algorithm for concurrent projectome and microstructure estimation. In: Jiang T, Navab N, Pluim J, Viergever M (Eds.), Medical Image Computing and Computer-Assisted Intervention. Springer Berlin / Heidelberg, pp. 183–190. [DOI] [PubMed] [Google Scholar]
- Sherbondy AJ, Dougherty RF, Ananthanarayanan R, Modha DS, Wandell BA, 2009. Think global, act local; projectome estimation with BlueMatter. In: Medical Image Computing and Computer-Assisted Intervention. Springer-Verlag, London, UK, pp. 861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherbondy AJ, Dougherty RF, Ben-Shachar M, Napel S, Wandell BA, 2008. Contrack: finding the most likely pathways between brain regions using diffusion tractography. J. Vis 8. 15–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shergill SS, Kanaan RA, Chitnis XA, O’Daly O, Jones DK, Frangou S, Williams SC, Howard RJ, Barker GJ, Murray RM, et al. , 2007. A diffusion tensor imaging study of fasciculi in schizophrenia. Am. J. Psychiatry 164, 467–473. [DOI] [PubMed] [Google Scholar]
- Shi F, Yap PT, Gao W, Lin W, Gilmore JH, Shen D, 2012. Altered structural connectivity in neonates at genetic risk for schizophrenia: a combined study using morphological and white matter networks. Neuroimage 62, 1622–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Toga AW, 2017. Connectome imaging for mapping human brain pathways. Mol. Psychiatry 22, 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu N, Duan Y, Huang J, Ren Z, Liu Z, Dong H, Barkhof F, Li K, Liu Y, 2018. Progressive brain rich-club network disruption from clinically isolated syndrome towards multiple sclerosis. Neuroimage Clin. 19, 232–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu N, Duan Y, Xia M, Schoonheim MM, Huang J, Ren Z, Sun Z, Ye J, Dong H, Shi FD, et al. , 2016. Disrupted topological organization of structural and functional brain connectomes in clinically isolated syndrome and multiple sclerosis. Sci. Rep 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu N, Liu Y, Li K, Duan Y, Wang J, Yu C, Dong H, Ye J, He Y, 2011. Diffusion tensor tractography reveals disrupted topological efficiency i n white matter structural networks in multiple sclerosis. Cereb. Cortex 21, 2565–2577. [DOI] [PubMed] [Google Scholar]
- Shu N, Wang X, Bi Q, Zhao T, Han Y, 2018. Disrupted topologic efficiency of white matter structural connectome in individuals with subjective cognitive decline. Radiology 286, 229–238. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Rakic P, 1973. Neuronal migration, with special reference to developing human brain: a review. Brain Res. 62, 1–35. [DOI] [PubMed] [Google Scholar]
- Siless V, Chang K, Fischl B, Yendiki A, 2018. Anatomicuts: hierarchical clustering of tractography streamlines based on anatomical similarity. Neuroimage 166, 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siless V, Davidow JY, Nielsen J, Fan Q, Hedden T, Hollinshead M, Beam E, Bustamante CMV, Garrad MC, Santillana R, et al. , 2020. Registration-free analysis of diffusion MRI tractography data across subjects through the human lifespan. Neuroimage 116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinke MR, Otte WM, Christiaens D, Schmitt O, Leemans A, van der Toorn A, Sarabdjitsingh RA, Joëls M, Dijkhuizen RM, 2018. Diffusion MRI-based cortical connectome reconstruction: dependency on tractography procedures and neuroanatomical characteristics. Brain Struct. Funct 223, 2269–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Raffelt D, Tournier JD, Connelly A, 2020a. Quantitative streamlines tractography: methods and inter-subject normalisation.
- Smith RE, Connelly A, Calamante F, 2020. Chapter 21 - diffusion MRI fiber tractography. In: Seiberlich N, Gulani V, Calamante F, Campbell-Washburn A, Doneva M, Hu HH, Sourbron S (Eds.), Advances in Magnetic Resonance Technology and Applications. In: Quantitative Magnetic Resonance Imaging, Vol. 1. Academic Press, pp. 533–569. [Google Scholar]
- Smith RE, Connelly A, Calamante F, 2020. Diffusion MRI fiber tractography. In: Advances in Magnetic Resonance Technology and Applications, Vol. 1. Elsevier, pp. 533–569. [Google Scholar]
- Smith RE, Tournier JD, Calamante F, Connelly A, 2012. Anatomically-constrained tractography: improved diffusion MRI streamlines tractography through effective use of anatomical information. Neuroimage 62, 1924–1938. [DOI] [PubMed] [Google Scholar]
- Smith RE, Tournier JD, Calamante F, Connelly A, 2013. SIFT: Spherical-deconvolution informed filtering of tractograms. Neuroimage 67, 298–312. [DOI] [PubMed] [Google Scholar]
- Smith RE, Tournier JD, Calamante F, Connelly A, 2015. The effects of SIFT on the reproducibility and biological accuracy of the structural connectome. Neuroimage 104, 253–265. [DOI] [PubMed] [Google Scholar]
- Smith RE, Tournier JD, Calamante F, Connelly A, 2015. SIFT2: enabling dense quantitative assessment of brain white matter connectivity using streamlines tractography. Neuroimage 119, 338–351. [DOI] [PubMed] [Google Scholar]
- Smith SM, Kindlmann G, Jbabdi S, 2014. Cross-subject comparison of local diffusion MRI parameters. In: Diffusion MRI. Elsevier, pp. 209–239. [Google Scholar]
- Sollmann N, Kelm A, Ille S, Schröder A, Zimmer C, Ringel F, Meyer B, Krieg SM, 2018. Setup presentation and clinical outcome analysis of treating highly language-eloquent gliomas via preoperative navigated transcranial magnetic stimulation and tractography. Neurosurg. Focus 44, E2. [DOI] [PubMed] [Google Scholar]
- Song JW, Mitchell PD, Kolasinski J, Ellen Grant P, Galaburda AM, Takahashi E, 2015. Asymmetry of white matter pathways in developing human brains. Cereb. Cortex 25, 2883–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Mishra V, Ouyang M, Peng Q, Slinger M, Liu S, Huang H, 2017. Human fetal brain connectome: structural network development from middle fetal stage to birth. Front. Neurosci 11, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SK, Sun SW, Ju WK, Lin SJ, Cross AH, Neufeld AH, 2003. Diffusion tensor imaging detects and differentiates axon and myelin degeneration in mouse optic nerve after retinal ischemia. Neuroimage 20, 1714–1722. [DOI] [PubMed] [Google Scholar]
- Song SK, Sun SW, Ramsbottom MJ, Chang C, Russell J, Cross AH, 2002. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage 17, 1429–1436. [DOI] [PubMed] [Google Scholar]
- Sotiropoulos SN, Hernández-Fernández M, Vu AT, Andersson JL, Moeller S, Yacoub E, Lenglet C, Ugurbil K, Behrens TE, Jbabdi S, 2016. Fusion in diffusion MRI for improved fibre orientation estimation: an application to the 3T and 7T data of the human connectome project. Neuroimage 134, 396–409. doi: 10.1016/j.neuroimage.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos SN, Zalesky A, 2019. Building connectomes using diffusion MRI: why, how and but. NMR Biomed. 32, e3752. doi: 10.1002/nbm.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell DG, Birk HS, Han SJ, Li J, Sall JW, Berger MS, 2017. Resection of gliomas deemed inoperable by neurosurgeons based on preoperative imaging studies. J. Neurosurg 129, 567–575. [DOI] [PubMed] [Google Scholar]
- Spetzler RF, Martin NA, 1986. A proposed grading system for arteriovenous malformations. J. Neurosurg 65, 476–483. [DOI] [PubMed] [Google Scholar]
- Sporns O, Betzel RF, 2016. Modular brain networks. Annu. Rev. Psychol 67, 613–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Honey CJ, Kötter R, 2007. Identification and classification of hubs in brain networks. PLoS ONE 2, e1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R, 2005. The human connectome: a structural description of the human brain. PLoS Comput. Biol 1, e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jean S, Chamberland M, Viergever MA, Leemans A, 2019. Reducing variability in along-tract analysis with diffusion profile realignment. Neuroimage 199, 663–679. [DOI] [PubMed] [Google Scholar]
- St-Onge E, Daducci A, Girard G, Descoteaux M, 2018. Surface-enhanced tractography (set). Neuroimage 169, 524–539. doi: 10.1016/j.neuroimage.2017.12.036. [DOI] [PubMed] [Google Scholar]
- Stadlbauer A, Salomonowitz E, Strunk G, Hammen T, Ganslandt O, 2008. Age-related degradation in the central nervous system: assessment with diffusion-tensor imaging and quantitative fiber tracking. Radiology 247, 179–188. [DOI] [PubMed] [Google Scholar]
- Stadlbauer A, Salomonowitz E, Strunk G, Hammen T, Ganslandt O, 2008. Quantitative diffusion tensor fiber tracking of age-related changes in the limbic system. Eur. Radiol 18, 130–137. [DOI] [PubMed] [Google Scholar]
- Stephens RL, Langworthy BW, Short SJ, Girault JB, Styner MA, Gilmore JH, 2020. White matter development from birth to 6 years of age: a longitudinal study. Cereb. Cortex 30, 6152–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieltjes B, Kaufmann WE, van Zijl PC, Fredericksen K, Pearlson GD, Solaiyappan M, Mori S, 2001. Diffusion tensor imaging and axonal tracking in the human brainstem. Neuroimage 14, 723–735. [DOI] [PubMed] [Google Scholar]
- Sui J, Qi S, van Erp TG, Bustillo J, Jiang R, Lin D, Turner JA, Damaraju E, Mayer AR, Cui Y, et al. , 2018. Multimodal neuromarkers in schizophrenia via cognition-guided MRI fusion. Nat. Commun 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan EV, Adalsteinsson E, Pfefferbaum A, 2006. Selective age-related degradation of anterior callosal fiber bundles quantified in vivo with fiber tracking. Cereb. Cortex 16, 1030–1039. [DOI] [PubMed] [Google Scholar]
- Sullivan EV, Rohlfing T, Pfefferbaum A, 2010. Quantitative fiber tracking of lateral and interhemispheric white matter systems in normal aging: relations to timed performance. Neurobiol. Aging 31, 464–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Lui S, Yao L, Deng W, Xiao Y, Zhang W, Huang X, Hu J, Bi F, Li T, et al. , 2015. Two patterns of white matter abnormalities in medication-naive patients with first-episode schizophrenia revealed by diffusion tensor imaging and cluster analysis. JAMA Psychiatry 72, 678–686. [DOI] [PubMed] [Google Scholar]
- Sydnor VJ, Rivas-Grajales AM, Lyall AE, Zhang F, Bouix S, Karmacharya S, Shenton ME, Westin CF, Makris N, Wassermann D, et al. , 2018. A comparison of three fiber tract delineation methods and their impact on white matter analysis. Neuroimage 178, 318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szelényi A, Bello L, Duffau H, Fava E, Feigl GC, Galanda M, Neuloh G, Signorelli F, Sala F, 2010. Intraoperative electrical stimulation in awake craniotomy: methodological aspects of current practice. Neurosurg. Focus 28, E7. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Hackney DB, Zhang G, Wehrli SL, Wright AC, O’Brien WT, Uematsu H, Wehrli FW, Selzer ME, 2002. Magnetic resonance microimaging of intraaxonal water diffusion in live excised lamprey spinal cord. Proc. Natl. Acad. Sci 99, 16192–16196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura H, Caiafa CF, Wandell BA, Pestilli F, 2016. Ensemble tractography. PLoS Comput. Biol 12, e1004692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamnes CK, Roalf DR, Goddings AL, Lebel C, 2018. Diffusion MRI of white matter microstructure development in childhood and adolescence: methods, challenges and progress. Dev. Cogn. Neurosci 33, 161–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax CM, Duits R, Vilanova A, ter Haar Romeny BM, Hofman P, Wagner L, Leemans A, Ossenblok P, 2014. Evaluating contextual processing in diffusion MRI: application to optic radiation reconstruction for epilepsy surgery. PLoS ONE 9, e101524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teillac A, Beaujoin J, Poupon F, Mangin JF, Poupon C, 2017. A novel anatomically-constrained global tractography approach to monitor sharp turns in gyri. In: International Conference on Medical Image Computing and Computer-assisted Intervention. Springer, pp. 532–539. [Google Scholar]
- Thomas C, Humphreys K, Jung KJ, Minshew N, Behrmann M, 2011. The anatomy of the callosal and visual-association pathways in high-functioning autism: a DTI tractography study. Cortex 47, 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoğlu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C, 2014. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc. Natl. Acad. Sci. U.S.A 111, 16574–16579. doi: 10.1073/pnas.1405672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Andreassen OA, Arias-Vasquez A, Bearden CE, Boedhoe PS, Brouwer RM, Buckner RL, Buitelaar JK, Bulayeva KB, Cannon DM, et al. , 2017. Enigma and the individual: predicting factors that affect the brain in 35 countries worldwide. Neuroimage 145, 389–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Yeo BT, Cropley V, Zalesky A, et al. , 2021. High-resolution connectomic fingerprints: mapping neural identity and behavior. Neuroimage 229, 117695. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM, 2013. Connectomics sheds new light on Alzheimer’s disease. Biol. Psychiatry 73, 390–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Q, He H, Gong T, Li C, Liang P, Qian T, Sun Y, Ding Q, Li K, Zhong J, 2019. Reproducibility of multi-shell diffusion tractography on traveling subjects: a multicenter study prospective. Magn. Reson. Imaging 59, 1–9. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A, 2007. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage 35, 1459–1472. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A, 2010. Improved probabilistic streamlines tractography by 2nd order integration over fibre orientation distributions. In: Proceedings of the International Society for Magnetic Resonance in Medicine, p. 1670. [Google Scholar]
- Tournier JD, Calamante F, Connelly A, 2012. Mrtrix: diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol 22, 53–66. [Google Scholar]
- Tournier JD, Calamante F, Gadian DG, Connelly A, 2003. Diffusion-weighted magnetic resonance imaging fibre tracking using a front evolution algorithm. Neuroimage 20, 276–288. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, King MD, Gadian DG, Connelly A, 2002. Limitations and requirements of diffusion tensor fiber tracking: an assessment using simulations. Magn. Reson. Med 47, 701–708. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Smith R, Raffelt D, Tabbara R, Dhollander T, Pietsch M, Christiaens D, Jeurissen B, Yeh CH, Connelly A, 2019. Mrtrix3: a fast, flexible and open software framework for medical image processing and visualisation. Neuroimage 202, 116137. [DOI] [PubMed] [Google Scholar]
- Tseng WYI, Hsu YC, Chen CL, Kang YJ, Kao TW, Chen PY, Waiter GD, 2021. Microstructural differences in white matter tracts across middle to late adulthood: a diffusion MRI study on 7167 UK biobank participants. Neurobiol. Aging 98, 160–172. [DOI] [PubMed] [Google Scholar]
- Tunç B, Parker WA, Ingalhalikar M, Verma R, 2014. Automated tract extraction via atlas based adaptive clustering. Neuroimage 102, 596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymofiyeva O, Hess C, Xu D, Barkovich A, 2014. Structural MRI connectome in development: challenges of the changing brain. Br. J. Radiol 87, 20140086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymofiyeva O, Hess CP, Ziv E, Lee PN, Glass HC, Ferriero DM, Barkovich AJ, Xu D, 2013. A DTI-based template-free cortical connectome study of brain maturation. PLoS ONE 8, e63310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M, 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15, 273–289. [DOI] [PubMed] [Google Scholar]
- Uddin LQ, 2013. Complex relationships between structural and functional brain connectivity. Trends Cogn. Sci 17, 600–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udin SB, Fawcett JW, 1988. Formation of topographic maps. Annu. Rev. Neurosci 11, 289–327. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Mandl RC, Stam CJ, Kahn RS, Pol HEH, 2010. Aberrant frontal and temporal complex network structure in schizophrenia: a graph theoretical analysis. J. Neurosci 30, 15915–15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O, 2013. Network hubs in the human brain. Trends Cogn. Sci 17, 683–696. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, de Reus MA, Feldman Barrett L, Scholtens LH, Coopmans FMT, Schmidt R, Preuss TM, Rilling JK, Li L, 2015. Comparison of diffusion tractography and tract-tracing measures of connectivity strength in rhesus macaque connectome. Hum. Brain Mapp 36, 3064–3075. doi: 10.1002/hbm.22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Heuvel MP, Kersbergen KJ, De Reus MA, Keunen K, Kahn RS, Groenendaal F, De Vries LS, Benders MJ, 2015. The neonatal connectome during preterm brain development. Cereb. Cortex 25, 3000–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Heuvel MP, Scholtens LH, De Lange SC, Pijnenburg R, Cahn W, Van Haren NE, Sommer IE, Bozzali M, Koch K, Boks MP, et al. , 2019. Evolutionary modifications in human brain connectivity associated with schizophrenia. Brain 142, 3991–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Heuvel MP, Sporns O, Collin G, Scheewe T, Mandl RC, Cahn W, Goñi J, Pol HEH, Kahn RS, 2013. Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiatry 70, 783–792. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Jbabdi S, Sotiropoulos SN, Chen C, Dikranian K, Coalson T, Harwell J, Behrens TE, Glasser MF, 2014. Chapter 16 - mapping connections in humans and non-human primates: Aspirations and challenges for diffusion imaging. In: Johansen-Berg H, Behrens TE (Eds.), Diffusion MRI (Second Edition). Academic Press, San Diego, pp. 337–358. doi: 10.1016/B978-0-12-396460-1.00016-0. [DOI] [Google Scholar]
- van Dellen E, Sommer IE, Bohlken MM, Tewarie P, Draaisma L, Zalesky A, Di Biase M, Brown JA, Douw L, Otte WM, et al. , 2018. Minimum spanning tree analysis of the human connectome. Hum. Brain Mapp 39, 2455–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn JD, Irimia A, Torgerson CM, Chambers MC, Kikinis R, Toga AW, 2012. Mapping connectivity damage in the case of phineas gage. PLoS ONE 7, e37454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasung L, Huang H, Jovanov-Milošević N, Pletikos M, Mori S, Kostović I, 2010. Development of axonal pathways in the human fetal fronto-limbic brain: histochemical characterization and diffusion tensor imaging. J. Anat 217, 400–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasung L, Turk EA, Ferradal SL, Sutin J, Stout JN, Ahtam B, Lin PY, Grant PE, 2019. Exploring early human brain development with structural and physiological neuroimaging. Neuroimage 187, 226–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatansever D, Manktelow A, Sahakian BJ, Menon DK, Stamatakis EA, 2018. Default mode network engagement beyond self-referential internal mentation. Brain Connect. 8, 245–253. [DOI] [PubMed] [Google Scholar]
- Vatansever D, Menon DK, Stamatakis EA, 2017. Default mode contributions to automated information processing. Proc. Natl. Acad. Sci 114, 12821–12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez A, López-López N, Houenou J, Poupon C, Mangin JF, Ladra S, Guevara P, 2020. Automatic group-wise whole-brain short association fiber bundle labeling based on clustering and cortical surface information. Biomed. Eng. Online 19, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven JS, Sage CA, Leemans A, Van Hecke W, Callaert D, Peeters R, De Cock P, Lagae L, Sunaert S, 2010. Construction of a stereotaxic DTI atlas with full diffusion tensor information for studying white matter maturation from childhood to adolescence using tractography-based segmentations. Hum. Brain Mapp 31, 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vértes PE, Bullmore ET, 2015. Annual research review: growth connectomics–the organization and reorganization of brain networks during normal and abnormal development. J. Child Psychol. Psychiatry 56, 299–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineskos AN, Lobaugh NJ, Bouix S, Rajji TK, Miranda D, Kennedy JL, Mulsant BH, Pollock BG, Shenton ME, 2010. Diffusion tensor tractography findings in schizophrenia across the adult lifespan. Brain 133, 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, Hua K, Zhang J, Jiang H, Dubey P, et al. , 2007. Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage 36, 630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakana S, Jiang H, Nagae-Poetscher LM, Van Zijl PC, Mori S, 2004. Fiber tract–based atlas of human white matter anatomy. Radiology 230, 77–87. [DOI] [PubMed] [Google Scholar]
- Wang D, Luo Y, Mok VC, Chu WC, Shi L, 2016. Tractography atlas-based spatial statistics: statistical analysis of diffusion tensor image along fiber pathways. Neuroimage 125, 301–310. [DOI] [PubMed] [Google Scholar]
- Wang Q, Su TP, Zhou Y, Chou KH, Chen IY, Jiang T, Lin CP, 2012. Anatomical insights into disrupted small-world networks in schizophrenia. Neuroimage 59, 1085–1093. [DOI] [PubMed] [Google Scholar]
- Wang R, Benner T, Sorensen AG, Wedeen VJ, 2007. Diffusion toolkit: a software package for diffusion imaging data processing and tractography. In: Proc. Intl. Soc. Mag. Reson. Med., Berlin, p. 3720.. [Google Scholar]
- Wang X, Seguin C, Zalesky A, Wong WW, Chu WCW, Tong RKY, 2019. Synchronization lag in post stroke: relation to motor function and structural connectivity. Netw. Neurosci 3, 1121–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann D, Makris N, Rathi Y, Shenton M, Kikinis R, Kubicki M, Westin CF, 2016. The white matter query language: a novel approach for describing human white matter anatomy. Brain Struct. Funct 221, 4705–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserthal J, Neher P, Maier-Hein KH, 2018. Tractseg-fast and accurate white matter tract segmentation. Neuroimage 183, 239–253. [DOI] [PubMed] [Google Scholar]
- Wasserthal J, Neher PF, Hirjak D, Maier-Hein KH, 2019. Combined tract segmentation and orientation mapping for bundle-specific tractography. Med. Image Anal 58, 101559. [DOI] [PubMed] [Google Scholar]
- Wegmayr V, Giuliari G, Holdener S, Buhmann J, 2018. Data-driven fiber tractography with neural networks. IEEE. 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), 1030–1033. [Google Scholar]
- Wen W, Zhu W, He Y, Kochan NA, Reppermund S, Slavin MJ, Brodaty H, Crawford J, Xia A, Sachdev P, 2011. Discrete neuroanatomical networks are associated with specific cognitive abilities in old age. J. Neurosci 31, 1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin CF, Knutsson H, Pasternak O, Szczepankiewicz F, Özarslan E, van Westen D, Mattisson C, Bogren M, O’Donnell LJ, Kubicki M, et al. , 2016. Q-space trajectory imaging for multidimensional diffusion MRI of the human brain. Neuroimage 135, 345–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin CF, Maier SE, Khidhir B, Everett P, Jolesz FA, Kikinis R, 1999. Image processing for diffusion tensor magnetic resonance imaging. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 441–452. [Google Scholar]
- Westlye LT, Walhovd KB, Dale AM, Bjørnerud A, Due-Tønnessen P, Engvig A, Grydeland H, Tamnes CK, Østby Y, Fjell AM, 2010. Life-span changes of the human brain white matter: diffusion tensor imaging (DTI) and volumetry. Cereb. Cortex 20, 2055–2068. [DOI] [PubMed] [Google Scholar]
- Wheeler AL, Voineskos AN, 2014. A review of structural neuroimaging in schizophrenia: from connectivity to connectomics. Front. Hum. Neurosci 8, 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S, Ghosh S, Nieto-Castanon A, Saygin Z, Doehrmann O, Chai X, Reynolds G, Hofmann S, Pollack M, Gabrieli J, 2016. Brain connectomics predict response to treatment in social anxiety disorder. Mol. Psychiatry 21, 680–685. [DOI] [PubMed] [Google Scholar]
- Widge AS, Zhang F, Gosai A, Papadimitrou G, Wilson-Braun P, Tsintou M, Palanivelu S, Noecker AM, McIntyre CC, O’Donnell L, et al. , 2021. Patient-specific connectomic models correlate with, but do not predict, outcomes in deep brain stimulation for obsessive-compulsive disorder. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins B, Lee N, Gajawelli N, Law M, Leporé N, 2015. Fiber estimation and tractography in diffusion MRI: development of simulated brain images and comparison of multi-fiber analysis methods at clinical b-values. Neuroimage 109, 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M, Tench C, Morgan P, Blumhardt L, 2003. Pyramidal tract mapping by diffusion tensor magnetic resonance imaging in multiple sclerosis: improving correlations with disability. J. Neurol. Neurosurg. Psychiatry 74, 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S, Pietsch M, Cordero-Grande L, Price AN, Hutter J, Xiao J, McCabe L, Rutherford MA, Hughes EJ, Counsell SJ, et al. , 2021. Development of human white matter pathways in utero over the second and third trimester. Proc. Natl. Acad. Sci 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf I, Vetter M, Wegner I, Böttger T, Nolden M, Schöbinger M, Hastenteufel M, Kunert T, Meinzer HP, 2005. The medical imaging interaction toolkit. Med. Image Anal 9, 594–604. [DOI] [PubMed] [Google Scholar]
- Wu K, Taki Y, Sato K, Kinomura S, Goto R, Okada K, Kawashima R, He Y, Evans AC, Fukuda H, 2012. Age-related changes in topological organization of structural brain networks in healthy individuals. Hum. Brain Mapp 33, 552–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Miller KL, 2017. Image formation in diffusion MRI: a review of recent technical developments. J. Magn. Reson. Imaging 46, 646–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Xie M, Zhou J, Anderson AW, Gore JC, Ding Z, 2012. Globally optimized fiber tracking and hierarchical clustering ’ a unified framework. Magn. Reson. Imaging 30, 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Xu Q, Xu L, Zhou J, Anderson AW, Ding Z, 2009. Genetic white matter fiber tractography with global optimization. J. Neurosci. Methods 184, 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Hong Y, Ahmad S, Lin W, Shen D, Yap PT, Consortium UBCP, et al. , 2020. Tract dictionary learning for fast and robust recognition of fiber bundles. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, 251–259. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Hong Y, Feng Y, Shen D, Yap PT, 2020. Mitigating gyral bias in cortical tractography via asymmetric fiber orientation distributions. Med. Image Anal 59, 101543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Hong Y, Feng Y, Shen D, Yap PT, 2020. Mitigating gyral bias in cortical tractography via asymmetric fiber orientation distributions. Med. Image Anal 59, 101543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang F, Makris N, Ning Y, Norton I, She S, Peng H, Rathi Y, Feng Y, Wu H, et al. , 2018. Investigation into local white matter abnormality in emotional processing and sensorimotor areas using an automatically annotated fiber clustering in major depressive disorder. Neuroimage 181, 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, He Y, 2017. Functional connectomics from a big data perspective. Neuroimage 160, 152–167. [DOI] [PubMed] [Google Scholar]
- Xie G, Zhang F, Leung L, Mooney MA, Epprecht L, Norton I, Rathi Y, Kikinis R, Al-Mefty O, Makris N, et al. , 2020. Anatomical assessment of trigeminal nerve tractography using diffusion MRI: a comparison of acquisition b-values and single-and multi-fiber tracking strategies. Neuroimage Clin. 25, 102160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Dong M, Lee MH, O’Hara N, Asano E, Jeong JW, 2019. Objective detection of eloquent axonal pathways to minimize postoperative deficits in pediatric epilepsy surgery using diffusion tractography and convolutional neural networks. IEEE Trans. Med. Imaging 38, 1910–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonskiy DA, Sukstanskii AL, 2010. Theoretical models of the diffusion weighted mr signal. NMR Biomed. 23, 661–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev P, Lecours A, 1967. The myelogenetic cycles of regional maturation of the brain. In: Regional Development of the Brain in Early Life, pp. 3–70. [Google Scholar]
- Yamada K, Sakai K, Akazawa K, Yuen S, Nishimura T, 2009. MR tractography: a review of its clinical applications. Magn. Reson. Med. Sci 8, 165–174. [DOI] [PubMed] [Google Scholar]
- Yang Z, Li X, Zhou J, Wu X, Ding Z, 2020. Functional clustering of whole brain white matter fibers. J. Neurosci. Methods 335, 108626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap PT, Fan Y, Chen Y, Gilmore JH, Lin W, Shen D, 2011. Development trends of white matter connectivity in the first years of life. PLoS ONE 6, e24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman JD, Dougherty RF, Myall NJ, Wandell BA, Feldman HM, 2012. Tract profiles of white matter properties: automating fiber-tract quantification. PLoS ONE 7, e49790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman JD, Wandell BA, Mezer AA, 2014. Lifespan maturation and degeneration of human brain white matter. Nat. Commun 5, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman JD, Weiner KS, Pestilli F, Rokem A, Mezer A, Wandell BA, 2014. The vertical occipital fasciculus: a century of controversy resolved by in vivo measurements. Proc. Natl. Acad. Sci 111, E5214–E5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CH, Jones DK, Liang X, Descoteaux M, Connelly A, 2020. Mapping structural connectivity using diffusion MRI: challenges and opportunities. J. Magn. Reson. Imaging doi: 10.1002/jmri.27188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CH, Smith RE, Dhollander T, Calamante F, Connelly A, 2019. Connectomes from streamlines tractography: assigning streamlines to brain parcellations is not trivial but highly consequential. Neuroimage 199, 160–171. [DOI] [PubMed] [Google Scholar]
- Yeh CH, Smith RE, Dhollander T, Connelly A, 2017. Mesh-based anatomically-constrained tractography for effective tracking termination and structural connectome construction. In: Proceedings of the ISMRM, p. 58. [Google Scholar]
- Yeh CH, Smith RE, Liang X, Calamante F, Connelly A, 2016. Correction for diffusion MRI fibre tracking biases: the consequences for structural connectomic metrics. Neuroimage 142, 150–162. [DOI] [PubMed] [Google Scholar]
- Yeh FC, 2020. Shape analysis of the human association pathways. Neuroimage 223, 117329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FC, Badre D, Verstynen T, 2016. Connectometry: a statistical approach harnessing the analytical potential of the local connectome. Neuroimage 125, 162–171. [DOI] [PubMed] [Google Scholar]
- Yeh FC, Panesar S, Fernandes D, Meola A, Yoshino M, Fernandez-Miranda JC, Vettel JM, Verstynen T, 2018. Population-averaged atlas of the macroscale human structural connectome and its network topology. Neuroimage 178, 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FC, Verstynen TD, Wang Y, Fernández-Miranda JC, Tseng WYI, 2013. Deterministic diffusion fiber tracking improved by quantitative anisotropy. PLoS ONE 8, e80713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FC, Zaydan IM, Suski VR, Lacomis D, Richardson RM, Maroon JC, Barrios–Martinez J, 2019. Differential tractography as a track-based biomarker for neuronal injury. Neuroimage 202, 116131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yendiki A, Panneck P, Srinivasan P, Stevens A, Zöllei L, Augustinack J, Wang R, Salat D, Ehrlich S, Behrens T, et al. , 2011. Automated probabilistic reconstruction of white-matter pathways in health and disease using an atlas of the underlying anatomy. Front. Neuroinform 5, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, et al. , 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo RA, Ryman SG, Van Den Heuvel MP, De Reus MA, Jung RE, Pommy J, Mayer AR, Ehrlich S, Schulz SC, Morrow EM, et al. , 2016. Graph metrics of structural brain networks in individuals with schizophrenia and healthy controls: group differences, relationships with intelligence, and genetics. J. Int. Neuropsychol.Soc 22, 240. [DOI] [PubMed] [Google Scholar]
- Yeo SS, Jang SH, Son SM, 2014. The different maturation of the corticospinal tract and corticoreticular pathway in normal brain development: diffusion tensor imaging study. Front. Hum. Neurosci 8, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SW, Guevara P, Jeong Y, Yoo K, Shin JS, Mangin JF, Seong JK, 2015. An example-based multi-atlas approach to automatic labeling of white matter tracts. PLoS ONE 10, e0133337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Lin Y, 2006. Model selection and estimation in regression with grouped variables. J. R. Stat. Soc. Ser. B 68, 49–67. [Google Scholar]
- Yushkevich PA, Zhang H, Simon TJ, Gee JC, 2009. Structure-specific statistical mapping of white matter tracts. In: Visualization and Processing of Tensor Fields. Springer, pp. 83–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Cocchi L, Fornito A, Murray MM, Bullmore E, 2012. Connectivity differences in brain networks. Neuroimage 60, 1055–1062. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Cocchi L, Gollo LL, van den Heuvel MP, Breakspear M, 2016. Connectome sensitivity or specificity: which is more important? Neuroimage 142, 407–420. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Cocchi L, Gollo LL, Heuvel M.P.v.d., Breaakspear M, 2016. Connectome sensitivity or specificity: which is more important? Neuroimage 142, 407–420. doi: 10.1016/j.neuroimage.2016.06.035. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Harding IH, Cocchi L, Yücel M, Pantelis C, Bullmore ET, 2010. Whole-brain anatomical networks: does the choice of nodes matter? Neuroimage 50, 970–983. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Seal ML, Cocchi L, Westin CF, Bullmore ET, Egan GF, Pantelis C, 2011. Disrupted axonal fiber connectivity in schizophrenia. Biol. Psychiatry 69, 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekelman LR, Zhang F, Makris N, He J, Chen Y, Xue T, Liera D, Drane DL, Rathi Y, Golby AJ, O’Donnell LJ, 2021. White matter association tracts underlying language and theory of mind: an investigation of 809 brains from the human connectome project. Neuroimage 118739. doi: 10.1016/j.neuroimage.2021.118739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L, Zhou J, Wang Y, Jin Y, Jahanshad N, Prasad G, Nir TM, Leonardo CD, Ye J, Thompson PM, et al. , 2015. Comparison of nine tractography algorithms for detecting abnormal structural brain networks in alzheimer’s disease. Front. Aging Neurosci 7, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Karayumak SC, Hoffmann N, Rathi Y, Golby AJ, O’Donnell LJ, 2020. Deep white matter analysis (DeepWMA): fast and consistent tractography segmentation. Med. Image Anal 65, 101761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Noh T, Juvekar P, Frisken SF, Rigolo L, Norton I, Kapur T, Pujol S, Wells W III Yarmarkovich A, et al. , 2020. SlicerDMRI: Diffusion MRI and tractography research software for brain cancer surgery planning and visualization. JCO Clin. Cancer Inf 4, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Norton I, Cai W, Song Y, Wells WM, O’Donnell LJ, 2017. Comparison between two white matter segmentation strategies: an investigation into white matter segmentation consistency. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). IEEE, pp. 796–799. [Google Scholar]
- Zhang F, O’Donnell LJ, 2020. Support vector regression. In: Machine Learning. Elsevier, pp. 123–140. [Google Scholar]
- Zhang F, Savadjiev P, Cai W, Song Y, Rathi Y, Tunç B, Parker D, Kapur T, Schultz RT, Makris N, et al. , 2018. Whole brain white matter connectivity analysis using machine learning: an application to autism. Neuroimage 172, 826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wu W, Ning L, McAnulty G, Waber D, Gagoski B, Sarill K, Hamoda HM, Song Y, Cai W, et al. , 2018. Suprathreshold fiber cluster statistics: leveraging white matter geometry to enhance tractography statistical analysis. Neuroimage 171, 341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wu Y, Norton I, Rathi Y, Golby AJ, O’Donnell LJ, 2019. Test–retest reproducibility of white matter parcellation using diffusion MRI tractography fiber clustering. Hum. Brain Mapp 40, 3041–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wu Y, Norton I, Rigolo L, Rathi Y, Makris N, O’Donnell LJ, 2018. An anatomically curated fiber clustering white matter atlas for consistent white matter tract parcellation across the lifespan. Neuroimage 179, 429–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC, 2012. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016. [DOI] [PubMed] [Google Scholar]
- Zhang R, Wei Q, Kang Z, Zalesky A, Li M, Xu Y, Li L, Wang J, Zheng L, Wang B, et al. , 2015. Disrupted brain anatomical connectivity in medication-naïve patients with first-episode schizophrenia. Brain Struct. Funct 220, 1145–1159. [DOI] [PubMed] [Google Scholar]
- Zhang W, Olivi A, Hertig SJ, Van Zijl P, Mori S, 2008. Automated fiber tracking of human brain white matter using diffusion tensor imaging. Neuroimage 42, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Oishi K, Faria AV, Jiang H, Li X, Akhter K, Rosa-Neto P, Pike GB, Evans A, et al. , 2010. Atlas-guided tract reconstruction for automated and comprehensive examination of the white matter anatomy. Neuroimage 52, 1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Descoteaux M, Zhang J, Girard G, Chamberland M, Dunson D, Srivastava A, Zhu H, 2018. Mapping population-based structural connectomes. Neuroimage 172, 130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Cao M, Niu H, Zuo XN, Evans A, He Y, Dong Q, Shu N, 2015. Age-related changes in the topological organization of the white matter structural connectome across the human lifespan. Hum. Brain Mapp 36, 3777–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Mishra V, Jeon T, Ouyang M, Peng Q, Chalak L, Wisnowski JL, Heyne R, Rollins N, Shu N, et al. , 2019. Structural network maturation of the preterm human brain. Neuroimage 185, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Sheng C, Bi Q, Niu W, Shu N, Han Y, 2017. Age-related differences in the topological efficiency of the brain structural connectome in amnestic mild cognitive impairment. Neurobiol. Aging 59, 144–155. [DOI] [PubMed] [Google Scholar]
- Zhao T, Xu Y, He Y, 2019. Graph theoretical modeling of baby brain networks. Neuroimage 185, 711–727. [DOI] [PubMed] [Google Scholar]
- Zhao X, Tian L, Yan J, Yue W, Yan H, Zhang D, 2017. Abnormal rich-club organization associated with compromised cognitive function in patients with schizophrenia and their unaffected parents. Neurosci Bull 33, 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziyan U, Sabuncu MR, Grimson WEL, Westin CF, 2009. Consistency clustering: a robust algorithm for group-wise registration, segmentation and automatic atlas construction in diffusion MRI. Int J Comput Vis 85, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllei L, Jaimes C, Saliba E, Grant PE, Yendiki A, 2019. TRActs constrained by underlying INfant anatomy (TRACULIna): an automated probabilistic tractography tool with anatomical priors for use in the newborn brain. Neuroimage 199, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]


