Abstract
Aims
The potential signaling pathways and core genes in ulcerative colitis (UC) were investigated in this study. Furthermore, potential mechanisms of BBR in treating UC were also explored.
Methods
Expression profiling by array of UC patients were obtained from Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) were determined with the differential analysis. The biological functions of DEGs were analyzed through the Database for Annotation, Visualization and Integrated Discovery (DAVID). The Gene Set Enrichment Analysis (GSEA) was applied to analyze the expression differences between two different phenotype sample sets. Dextran sulfate sodium (DSS) was applied to establish UC model of mice and lipopolysaccharide (LPS) was utilized to induce inflammatory damage of NCM460 cells. Therapeutic effects of berberine (BBR) on disease performance, pathologic changes and serum supernatant indices were analyzed in vivo. To further investigate the potential mechanisms of BBR in treating UC, the expression of genes and proteins in vivo and in vitro were examined by RT-qPCR, immunohistochemical staining and western blotting.
Results
Immune-inflammatory genes were identified and up-regulated significantly in UC patients. In addition, IFN-γ signaling pathway and its core genes were significantly up-regulated in the phenotype of UC. All disease performance and the pathologic changes of UC in mice were evidently ameliorated by BBR treatment. The pro-inflammatory cytokines of serum, including CXCL9, CXCL1, IL-17 and TNF-α, in UC mice were significantly reduced by treatment of BBR. In terms of mechanisms of BBR in treating UC, the pro-inflammatory and immune-related genes, encoding IFN-γ, IRF8, NF-κB and TNF-α decreased significantly in UC mice followed by BBR treatment. Meanwhile, the expression of IFN-γ and its initiated targets, including IRF8, Ifit1, Ifit3, IRF1, were suppressed significantly by BBR treatment in vivo. The blocking of IFN-γ in vitro led to the silence of IFN-γ signaling pathway after exposure to BBR. Furthermore, the blocking of IFN-γ in vitro led to the silence of IFN-γ signaling pathway after exposure to BBR.
Conclusion
BBR holds anti-inflammatory activity and can treat UC effectively. The anti-inflammatory property of BBR is tightly related to the suppression of IFN-γ signaling pathway, which is crucial in immune-inflammatory responses of the colon mucosa.
Keywords: Ulcerative colitis, Berberine, IFN-γ, Inflammation
1. Introduction
Ulcerative colitis (UC) is an extremely refractory disease in the intestine. UC has aroused concern over the past few decades due to its increasing incidence and fatality rate worldwide. The prevalence in Asian populations ranges from 5.3 to 63.6 per 100,000 (Niriella et al., 2010), whereas in North America, it ranges from 37.5 to 238 per 100,000 (Cosnes et al., 2011). The mean percentage of deaths ascribed to UC was 17% (range 11% to 30%) (Jess et al., 2013). The diffuse and idiopathic inflammation in the rectal and colonic mucosa is the pivotal pathology of UC (Ordás et al., 2012). 95% cases of UC mainly originate from the rectum and could be extended constantly and circumferentially to more proximal parts of the whole intestine (da Silva et al., 2014). Typical clinical symptoms of UC patients include abdominal pain, bloody diarrhea, urgency, and tenesmus (Feuerstein and Cheifetz, 2014). UC has decreased quality of life (QOL) evidently, and it may lead to death if not treated properly. Unfortunately, there is no cure for UC at present (Eisenstein, 2018). Instead, patients with UC turn to anti-inflammatory therapies (such as 5-aminosalicylate [5-ASA], corticosteroids) to manage mild to moderate symptoms or to immunosuppressants or anti-tumor necrosis factor (TNF) therapies to control more severe UC (Rubin et al., 2019). However, many patients do not obtain lasting relief of symptoms and improvement of QOL. Thus, the therapeutic regimes at present still cannot meet the clinical treatment needs of UC and more effective drugs or therapies are badly required to treat UC efficiently.
The basic pathological changes of UC are inflammatory lesions of the colonic mucosa (Kaur and Goggolidou, 2020, Porter et al., 2020). Inflammation is an adaptive response of the body, which can be triggered by harmful stimuli and conditions such as microbial infection and tissue damage (Medzhitov, 2008). Inflammation is coordinated by a complex network of inflammatory mediators. The basic inflammation pathway consists of four links: (1) inducer; (2) sensor; (3) mediator; (4) effector (Medzhitov, 2008, Medzhitov, 2010). Interferon (IFN), as the main member of cytokines (mediator), plays an important role in the inflammatory cascade. IFN was first discovered as an antiviral cytokine, which mainly interferes with viral replication in mammalian cells (Isaacs and Lindenmann, 1957). Based on different structural homology and related specific receptors, IFN can be divided into types I, II and III (de Weerd and Nguyen, 2012, Ivashkiv and Donlin, 2014). Among them, type II IFN only includes IFN-γ in acid-labile form, which mainly binds to receptors IFN-γR1 and IFN-γR2. The signal transduction of IFN-γ mainly via the JAK/STAT pathway (Rauch et al., 2013, Majoros et al., 2017). In addition, STAT homodimer (such as STAT-1, STAT-2, STAT-3 homodimer) or heterodimer (such as STAT1/2, STAT1/3, STAT2/6 heterodimer) through combine with interferon regulatory factor (IRF) to activate target genes. In terms of inflammation regulation, IFN-γ mainly activates STAT1 homodimer and IRF (such as IRF1, IRF8) through JAK1 and JAK2 (Hu and Ivashkiv, 2009, Majoros et al., 2017). Meanwhile, as a Th1 type effector cytokine, IFN-γ regulates inflammation through adaptive immune response and innate immune response. IFN-γ promotes Th1-type pro-inflammatory response and inhibits Th2-type anti-inflammatory response (Teixeira et al., 2005). Another way for IFN-γ to aggravate the effect of inflammation is to enhance the response of macrophages to inflammatory stimuli. IFN-γ can enhance the signal transduction of TLRs, promote the activation of NF-κB, and inhibit the transmission of IL-10-STAT3 signal by increasing the activity of GSK3-β, resulting in a significant decrease in the expression of the anti-inflammatory factor IL-10 (Hu et al., 2006, Chan et al., 2009). Despite IFN-γ and its related pathway playing a critical role in regulating inflammation, relevant researches of IFN-γ signaling pathway in UC are still vacant to some extent.
Berberine (BBR) is a natural isoquinoline alkaloid and has been widely applied in clinics for many centuries. Many studies have revealed BBR and its derivatives possess high activity to break the cycle of inflammation in UC (Li et al., 2016, Cui et al., 2018, Li et al., 2020). Meanwhile, our previous study has revealed that BBR could suppress chronic inflammation initiated by IFN-γ in the stomach (Yang et al., 2020), which indicated BBR could inhibit IFN-γ and its related pathway potentially. In this study, we further investigated the role of IFN-γ signaling pathway in UC. Furthermore, we also explored potential mechanisms of BBR in treating UC, contributing to a better understanding of the treatment roles of BBR in UC.
2. Materials and methods
2.1. Bioinformatics of UC
The Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo) database was applied to obtain clinical data and RNA expression of UC patients. Differentially expressed genes (DEGs) were determined with an empirical Bayesian approach using the Bioconductor “limma” package in R software (Ritchie et al., 2015). LogFC (fold change) > 1.5 and false discovery rate (FDR) < 0.05 were set as significantly up-regulated differential expression genes (DEGs), logFC < −1.5 and FDR < 0.05 were set as significantly down-regulated DEGs. Then, the relevant signaling pathways of up-regulated or down-regulated DEGs were explored via the Database for Annotation, Visualization, and Integrated Discovery database (DAVID, https://david.ncifcrf.gov, version 6.8). Kyoto Encyclopedia of Genes and Genome (KEGG) enrichment analysis and Gene Ontology (GO) analysis in the DAVID database were applied to indicate the relevant signaling pathways and biological process (BP) of DEGs. The Gene Set Enrichment Analysis (GSEA) was applied to analyze the expression differences between two different phenotype sample sets. The normalized enrichment score (NES) ≥ 1.5, NOM p-value ≤ 0.05, and FDR q-value ≤ 0.25 were considered as significant gene sets.
2.2. Reagents
BBR was purchased from Chroma Biotechnology Co. Ltd. Purity ≥ 98% BBR (Cat. No. CHB181028) was dissolved in pure dimethyl sulfoxide (DMSO). Purity ≥ 95% BBR (Cat. No. CHB180606) was dissolved in sodium chloride injection (NS 0.9%). Mesalazine was obtained from Hengcheng pharmaceutical group Huainan Co., Ltd., (Cat. No. H20020211). Dextran sulfate sodium (DSS, Cat. No. 216011080) was purchased from MP Biomedicals, Inc (Irvine, CA, USA). Lipopolysaccharide (LPS, Cat. No. SMB00610) was purchased from Sigma-Aldrich (St Louis, MO, USA). Cell counting kit-8 (CCK-8, Lot. PG658) was obtained from Dojindo Molecular Technologies, Inc (Tokyo, Japan). Fecal occult blood test paper (Cat. No. B201201) was purchased from Zhuhai Besso Biotechnology Co., Ltd. Selective inhibitor of IFN-γ (Cat. No. AX-024) was purchased from MedChem Express Co., Ltd. (Shanghai, China).
2.3. DSS-induced mice with UC
The model using DSS-induced mice with UC was previously reported (Li et al., 2019, Cui et al., 2021). Specific pathogen-free (SPF), male C57BL/6 mice (n = 50, weight: 20 ± 2 g) were purchased from Beijing Sibeifu Animal Breeding Center (Permission No. SCXK-(jing) 2019–0010). Mice were maintained under SPF conditions in a shelter sustained facility and provided with sterile food and water. All breeding and experiments were undertaken with review and approval from the Animal Ethical and Experimental Committee of the Fifth Medical Center of PLA General Hospital (Approval ID: IACUC-2018-010).
3% DSS (w/v) was applied to induce UC in mice. Mice in the control group received deionized drinking water for 14 days. Model group of UC was given to drink 3% DSS (w/v) for 7 consecutive days. Experimental mice were randomly divided into the control group, UC group, mesalazine group, BBR with low-dose and high-dose groups (n = 10 per group) respectively. Mice in the positive group received 0.364 g/kg·d−1 mesalazine. Mice in the low-dose BBR group and high-dose BBR group received 20 mg/kg·d−1 and 40 mg/kg·d−1 BBR respectively. Mesalazine and BBR were orally administered once daily for 7 days. On the 14th day, all mice were sacrificed. The blood samples were obtained from the orbital sinus, colon tissue was kept at −80 °C condition and then taken for further experiments.
2.4. Cell culture
Human-originated colon epithelial cells (NCM460, Lot. CL0393) line were purchased from Fenghui Biological Cell Center (Hunan, China). NCM460 cells were cultured in DMEM (Gibco, Life) medium, containing 10% FBS (Gibco, Life), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, Life). The cells were cultured in a humidified incubator with 5% CO2 at 37 °C. NCM460 cells were processed with 0.8 μg·ml−1 LPS in the absence or presence of BBR.
2.5. Disease performance and disease activity index (DAI)
The severity of colitis was evaluated by monitoring disease performance daily, including body weight, food consumption, stool consistency and fecal occult blood. The DAI was calculated based on the scoring system as previously described by Wirtz, S, et al (Wirtz et al., 2017). Fecal occult blood was assessed using a fecal occult blood test paper. DAI values were calculated as the sum of the weight loss score, diarrhea score and occult blood score.
2.6. Hematoxylin-eosin (HE) staining of colon tissue
Mice were sacrificed at the end of the experiment. Parts of the colons were fixed in 4% paraformaldehyde cleared in xylene, embedded in paraffin, and cut into 5 mm thick slices. Colon tissue sections were stained with hematoxylin-eosin (HE). Furthermore, tissue damage index (TDI) was applied to evaluate the degree of inflammatory cell infiltration based on HE results.
2.7. Measurement of specific serum biomarkers
The Synergy H1 Hybrid Reader (Biotech, USA) was employed to measure serum biochemical indices, including chemokines 9 (CXCL9), chemokines 1 (CXCL1), interleukin-17 (IL-17), and Tumor necrosis factor-α (TNF-α). The enzyme-linked immunosorbent assay (ELISA) kits of CXCL9 (Cat No.: JYM1038Mo), CXCL1 (Cat No.: JYM0924Mo), IL-17 (Cat No.: JYM0554Mo) and TNF-α (Cat No.: JYM0218Mo) were purchased from JYMBIO Biotechnology Co., Ltd. (Wuhan, China). All assays were performed rigorously according to the manufacturer's instructions.
2.8. Immunohistochemical (IHC) staining of colon tissue
Colon tissue samples were fixed and preserved in 4% paraformaldehyde, and cut into 5 mm thick slices by using a microtome. Paraffin sections of colon tissues were stained with a polyclonal anti-IFN-γ Ab (Cat No.: NBP2-66900, Novusbio, 1:200) and rabbit anti-IRF8 Ab (Cat No.: 18977-1-AP, Proteintech, 1:200) were utilized. NIS Elements Imaging Software Version 4.0 at 200× magnification (Olympus, Japan) were applied to photograph images.
2.9. Real-time quantitative PCR for mRNA expressions
Microarray analysis was applied by using RNA extracts from colon tissue. TRIzol reagent (Nordic Bioscience, Beijing, China) was applied and converted into cDNA by reverse transcription kit (Promega, Madison, USA) according to manufacturer's protocol. The primers sequences of IFN-γ, IRF8, NF-κB, and TNF-α are listed in Table 1. Quantitative Real-time PCR for these mRNA was performed and analyzed using cDNA and SYBR Green PCR Master Mix (Nordic Bioscience, Beijing, China). RT-qPCR was performed on a 7500 fast real-time PCR system (Applied Biosystems, Foster City, CA, USA).β-actin as an endogenous reference, and the relative amounts of mRNA were determined based on 2−ΔΔCt calculations.
Table 1.
Primers sequences of IFN-γ, IRF8, NF-κB, and TNF-α.
| Genes | Primer | Sequences (5′–3′) |
|---|---|---|
| IFN-γ | Forward | GCACAGTCATTGAAAGCCTAGAAAGTC |
| Reverse | GCCAGTTCCTCCAGATATCCAAGAAG | |
| IRF8 | Forward | GCAGGATTACAATCAGGAGGTGGATG |
| Reverse | TCTTGTTCAGAGCACAGCGTAACC | |
| NF-κB | Forward | TAAATGGGAAACCGTATGAGCCTGTG |
| Reverse | GTTGTAGCCTCGTGTCTTCTGTCAG | |
| TNF-α | Forward | CGCTCTTCTGTCTACTGAACTTCGG |
| Reverse | GTGGTTTGTGAGTGTGAGGGTCTG | |
| β-actin | Forward | TATGCTCTCCCTCACGCCATCC |
| Reverse | GTCACGCACGATTTCCCTCTCAG |
2.10. Western blotting (WB) analysis for protein expression
The following main antibodies were applied: anti-IFN-γAb (Novusbio, Cat No.: NBP2-66900, dilution: 1:500), anti-IRF8 Ab (Proteintech, Cat No.: 18977-1-AP, dilution: 1:1000), anti-Ifit1 Ab (Proteintech, Cat No.: 23247-1-AP, dilution: 1:500), anti-Ifit3 Ab (BIOSS, Cat No.: bs-15515R, dilution: 1:500), anti-IRF1 Ab (BIOSS, Cat No.: bsm-52114R, dilution: 1:1000), anti-TNF-α Ab (Proteintech, Cat No.: 17590-1-AP, dilution: 1:1000), GAPDH monoclonal antibody (Proteintech, Cat No.: 60004-1-Ig, dilution: 1:8000) and HRP-conjugated secondary antibody (goat anti-mouse IgG (H + L) (Abcam, Cat No.: ab6721, dilution, 1:15000). To quantify protein levels, the grey values of the blots in scanned images were measured using ImageJ Pro Plus software (National Institute of Health, Bethesda, MD, USA), and the GAPDH was set as a loading control for the grey value of each target protein.
2.11. Statistics analysis
All data were presented as mean ± standard deviation (SD) and analyzed with the SPSS software program (version 21.0; SPSS Inc., Chicago, IL, USA). Data were presented using one-way ANOVA followed by LSD test. For the data of repeated measurement, repeated measures analysis of variance was applied, and the LSD method was used for multiple comparisons. P < 0.05 was considered statistically significant and P < 0.01 was highly significant. GraphPad Prism software for Windows (version 6.02; Inc., San Diego, USA) and R software (version 4.0.4; https://www.r‑project.org/) were utilized for visible presentation of all results.
3. Results
3.1. Immune-inflammatory related genes were significantly up-regulated in UC patients
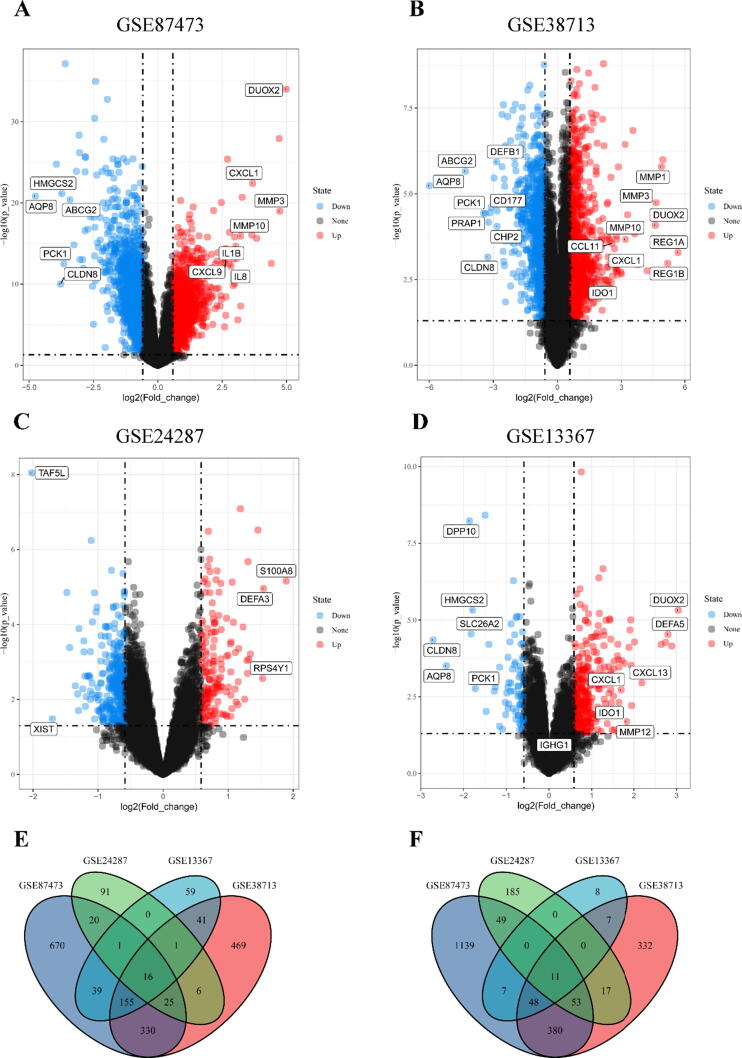
Four gene expression databases about UC patients and control cases, GSE87473 (Li et al., 2018a, Li et al., 2018b), GSE38713 (Planell et al., 2013), GSE24287 (Zhang et al., 2012), and GSE13367 (Bjerrum et al., 2010) were included in bioinformatics analysis. The main information of these four databases is presented in Table 2. In total, four databases involved 197 UC patients and 79 control cases. There were 5578 DEGs in these four databases followed by “limma” analysis. Among them, 298 DEGs were significantly up-regulated and 264 DEGs were significantly down-regulated (Supplement 1). In the database of GSE87473, a total of 2805 DEGs were obtained, of which 131 were up-regulated DEGs and 148 were down-regulated DEGs (Fig. 1A). The expression of DUOX2, MMP3, CXCL1, MMP10, IL1B, IL8 and CXCL9 were significantly up-regulated, ABCG2, HMGCS2, PCK1 CLDN8 and AQP8 were significantly down-regulated (Fig. 1A). In the database of GSE38713, a total of 1903 DEGs were obtained, of which 140 were up-regulated DEGs and 108 were down-regulated DEGs (Fig. 1B). In the database of GSE24287, a total of 473 DEGs were obtained, including 3 up-regulated DEGs and 2 down-regulated DEGs (Fig. 1C). In the database of GSE13367, a total of 397 DEGs were obtained, of which 24 were up-regulated DEGs and 6 were down-regulated DEGs. Among them, DUOX2, IGHG1, DEFA5, CXCL13, MMP12, CXCL1 and IDO1 were significantly up-regulated, PCK1, HMGCS2, SLC26A2, DPP10, AQP8 and CLDN8 were significantly down-regulated, respectively (Fig. 1D). Altogether, 16 up-regulated DEGs (CXCL3, DUOXA2, DUOX2, TIMP1, PI3, MMP1, PDZK1IP1, MMP3, CCL18 CXCL1, C4BPB, IL1B, LCN2, F3, S100A12, S100A9, Fig. 1E) and 11 down-regulated DEGs (HSD17B2, DHRS11, AQP8, UGT2A3, GUCA2A, MEP1B, EPHX2, ABCG2, GUCA2B, CHP2, CLDN8, Fig. 1F) were distributed simultaneously in these four databases. All these 16 up-regulated DEGs were immune-inflammatory related genes (Smallridge et al., 2014, Li et al., 2021), and up-regulated significantly in UC patients.
Table 2.
Main information of gene expression profiling by array.
| Database | Tissue | PlatformRef | Probe name and version | Sample in control group | Sample in UC group | Country |
|---|---|---|---|---|---|---|
| GSE87473 | colon | GPL13158 | Affymetrix HT HG-U133 + PM Array Plate | 21 | 106 | USA |
| GSE38713 | colon | GPL570 | Affymetrix Human Genome U133 Plus 2.0 Array | 13 | 30 | Spain |
| GSE24287 | proximal ileal margin |
GPL6480 | Agilent-014850 Whole Human Genome Microarray 4x44K G4112F | 25 | 27 | USA |
| GSE13367 | colon | GPL570 | Affymetrix Human Genome U133 Plus 2.0 Array | 20 | 34 | Denmark |
Fig. 1.
Identification of DEGs in GSE87473, GSE38713, GSE24287, and GSE13367 databases. A: Volcano plot of DEGs in GSE87473; B: Volcano plot of DEGs in GSE38713; C: Volcano plot of DEGs in GSE24287; D: Volcano plot of DEGs in GSE13367; E: Venn plot of up-regulated DEGs (logFC > 1.5 and FDR < 0.05); F: Venn plot of down-regulated DEGs (logFC < −1.5 and FDR < 0.05). All up-regulated genes marked as red points, down-regulated genes marked as blue points, and no significantly different genes marked as black points. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Up-regulated DEGs were significantly enriched in IFN-γ mediated pathway and biological process
Then, we applied the 298 up-regulated DEGs and 264 down-regulated DEGs for KEGG pathway enrichment analysis and GO analysis in DAVID to indicate the relevant signaling pathways and biological process of DEGs. KEGG pathway enrichment analysis revealed that up-regulated DEGs were significantly enriched in cytokine-cytokine receptor interaction, chemokine signaling pathway and TNF signaling pathway (Fig. 2A). Meanwhile, up-regulated DEGs were mainly enriched in the inflammatory response, immune response, neutrophil chemotaxis and cellular response to interferon-gamma (IFN-γ) followed by GO analysis (Fig. 2B). Conclusively, IFN-γ was particularly mediated in the pathways and biological process of the inflammatory response (Cordes et al., 2021), immune response (Gogokhia et al., 2019), and neutrophil chemotaxis (Bonecchi et al., 1999).
Fig. 2.
KEGG pathway enrichment analysis and GO analysis of up-regulated and down-regulated DEGs. A: KEGG pathway enrichment analysis of up-regulated DEGs; B: Biological process analysis of up-regulated DEGs; C: KEGG pathway enrichment analysis of down-regulated DEGs; D: Biological process analysis of down-regulated DEGs.
In addition, results revealed the down-regulated DEGs were significantly enriched in retinol metabolism and steroid hormone biosynthesis followed by KEGG pathway enrichment analysis (Fig. 2C). Furthermore, down-regulated DEGs were significantly enriched in digestion, drug metabolic process and steroid metabolic process after being analyzed by GO analysis (Fig. 2D).
3.3. IFN-γ signaling pathway was significantly up-regulated in the phenotype of UC
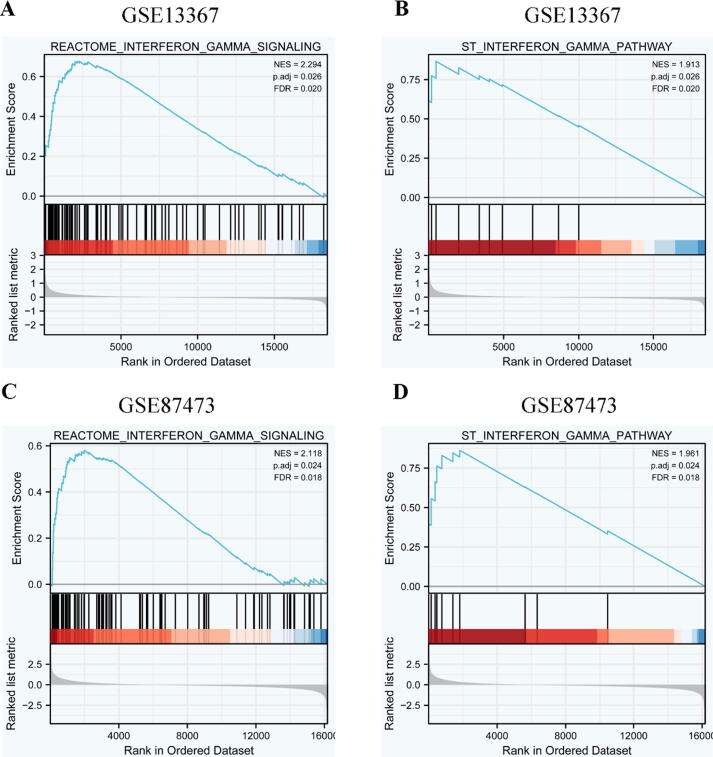
GSEA was applied to analyze the expression differences between two different phenotype sample sets (UC group and control group) in the databases of GSE13367 and GSE87473. All results were presented in Supplement 2. In the database of GSE13367, a total of 614 gene sets were enriched in the phenotype of the UC group, and 142 gene sets were enriched in the phenotype of the control group. IFN-γ signaling pathway was significantly up-regulated in the phenotype of UC, no matter in Reactome database (ES = 0.68, NES = 2.294, P.adj = 0.026, Fig. 3A), or in ST database (ES = 0.87, NES = 1.913, P.adj = 0.026, Fig. 3B). In the database of GSE87473, a total of 563 gene sets were enriched in the phenotype of the UC group, and 215 gene sets were enriched in the phenotype of the control group. IFN-γ signaling pathway was also significantly up-regulated in Reactome database (ES = 0.58, NES = 2.118, P.adj = 0.024, Fig. 3C), and in ST database (ES = 0.86, NES = 1.961, P.adj = 0.024, Fig. 3D). GSEA indicated IFN-γ signaling pathway and its core genes were significantly up-regulated in UC and were very likely involved in the occurrence and development of UC.
Fig. 3.
GSEA of IFN-γ signaling pathway in the phenotype of UC. A-B: GSEA of IFN-γ signaling pathway in GSE13367 database; C-D: GSEA of IFN-γ signaling pathway in GSE87473 database.
3.4. BBR ameliorates disease performance of DSS-induced UC in mice
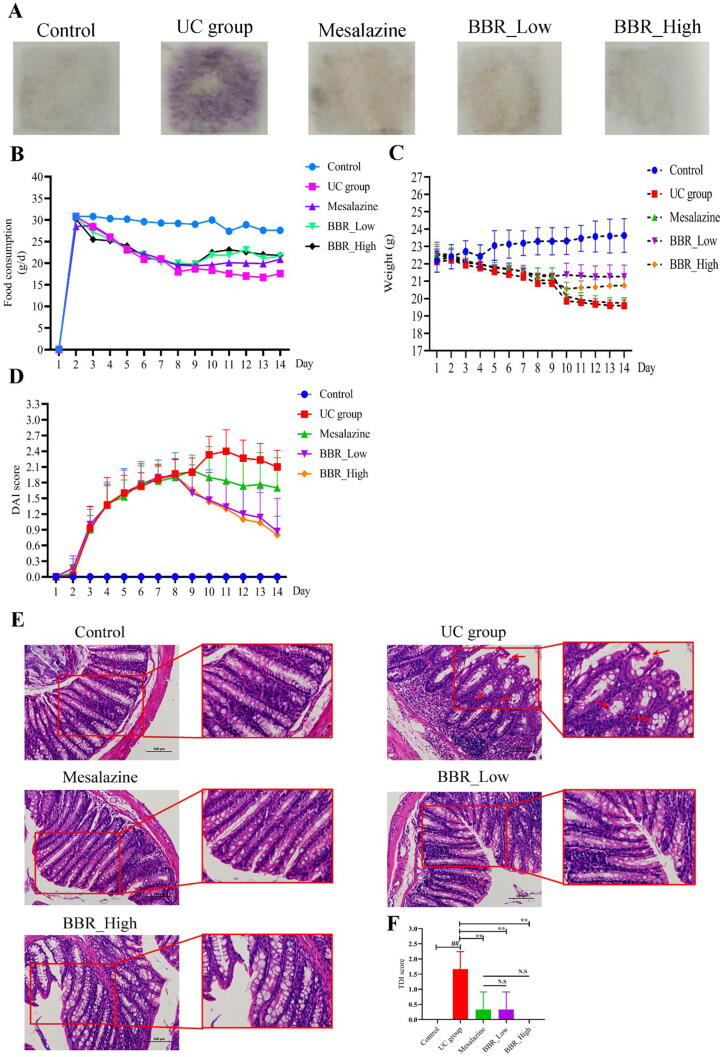
Then, the pharmacological effects of BBR on DSS-induced UC mice were evaluated. Fecal occult blood test in the UC group was obviously positive (++ to +++) since the drinking of 3% DSS from the third day (Fig. 4A). After being treated by mesalazine and BBR, the fecal occult blood test was changed into negative (Fig. 4A). Data of food consumption showed a visible downward trend in the UC group from the third day (Fig. 4B). On the contrary, there was a significant increasing trend followed by administration of mesalazine and BBR (Fig. 4B). Furthermore, weight loss was obvious in UC group from the fourth day (Fig. 4C). However, weight loss was not reversed significantly in the mesalazine group (Fig. 4C). In the BBR group, the weight of mice was increased significantly, especially in the group of BBR with low doses (20 mg/kg, Fig. 4C). Due to the severity of colitis, including weight loss, diarrhea and positive fecal occult blood test, was significantly increased in UC group, DAI values were also significantly increased in UC group (Fig. 4D). After treated by mesalazine and BBR, DAI values were decreased noticeably, especially in the group of BBR with high doses (40 mg/kg, Fig. 4D). HE staining of colon tissue indicated the integrity of the colon mucosa was lost in the UC group, the location of the crypts moved up and was irregular in shape, the number of goblet cells in the crypts was significantly reduced and crypts were swollen (indicated by the red arrow). The lamina propria infiltrated with neutrophils and lymphocytes, most of which can reach the mucosa and submucosa (Fig. 4E). Conversely, these pathologic changes in the mesalazine and BBR group were more slightly than in the UC group. In terms of the number of goblet cells in the crypts of the colon, administration of BBR at a high dose (40 mg/kg) increased the number of goblet cells in the crypts and decreased the infiltration of neutrophils and lymphocytes noticeably (Fig. 4E). Meanwhile, the TDI values were also decreased significantly followed by treatment of mesalazine and BBR (Fig. 4F).
Fig. 4.
Effects of BBR on disease performance of DSS-induced UC in mice. A: Fecal occult blood test of each group mice; B: Food consumption of each group mice; C: Bodyweight of each group mice; D: DAI values of each group mice; E: HE staining of each group mice; F: TDI values of each group mice. N.S: None statistically significant; ##P < 0.01 versus control group; ∗∗P < 0.01 versus UC group. BBR_Low: BBR with low dose; BBR_High: BBR with high dose.
3.5. BBR decreased pro-inflammatory cytokines of serum in UC mice
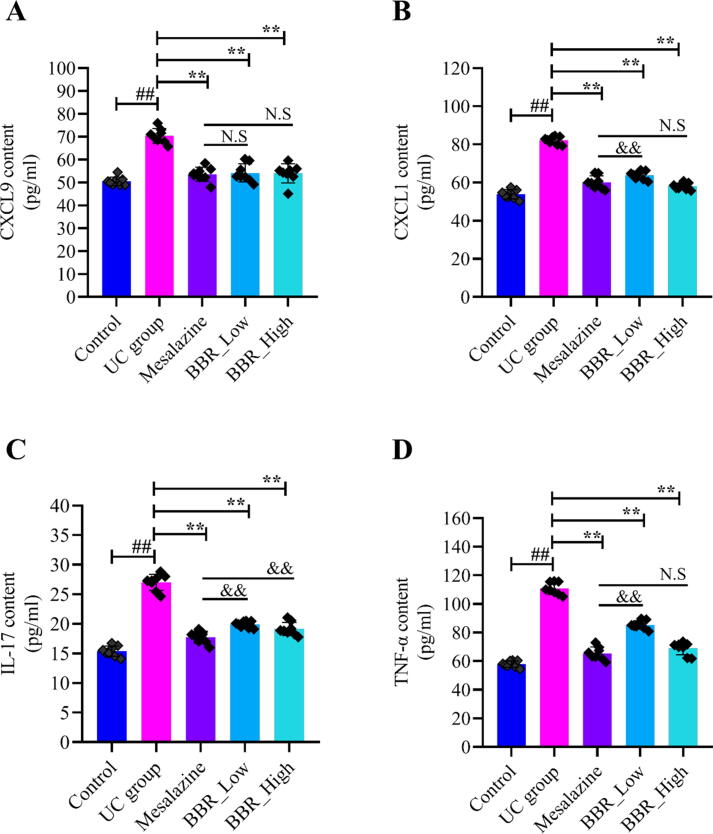
An ocean of studies reported the pro-inflammatory cytokines increased significantly when UC developed (Li et al., 2018a, Li et al., 2018b). The cytokines in serum, including CXCL9, CXCL1, IL-17 and TNF-α, were examined in this study. As presented in Fig. 5, all pro-inflammatory cytokines were increased noticeably in the UC group compared with the control group. On the contrary, CXCL9 in serum decreased distinctly in the mesalazine and BBR group compared with UC group (Fig. 5A). Meanwhile, CXCL1 (Fig. 5B), IL-17 (Fig. 5C) and TNF-α(Fig. 5D) were also significantly reduced followed by treatment of mesalazine and BBR. Altogether, these results implied that the activities of pro-inflammatory cytokines, including CXCL9, CXCL1, IL-17 and TNF-α got decreased by BBR intervention.
Fig. 5.
Effects of BBR on pro-inflammatory cytokines of serum in mice (n = 8). A: CXCL9 content in the serum of each group mice; B: CXCL1 content in the serum of each group mice; C: IL-17 content in the serum of each group mice; D: TNF-αcontent in serum of each group mice. N.S: None statistically significant; ##P < 0.01 versus control group; ∗∗P < 0.01 versus UC group. &&P < 0.01 versus mesalazine group. BBR_Low: BBR with low dose; BBR_High: BBR with high dose.
3.6. BBR suppressed IFN-γ and pro-inflammatory genes in DSS-induced UC mice
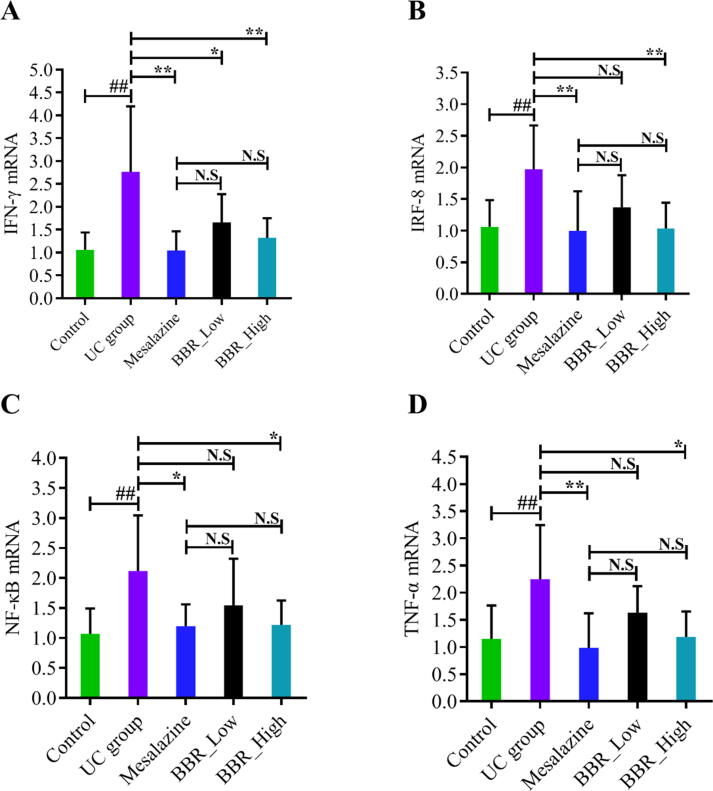
As consistent with the previous analysis results of bioinformatics, the expression of IFN-γ was significantly up-regulated in the UC group compared with the control group (Fig. 6A). Meanwhile, the expression of pro-inflammatory genes, including IRF8 (Fig. 6B), NF-κB (Fig. 6C) and TNF-α(Fig. 6D) were also up-regulated in the UC group. Conversely, all these up-regulated genes were suppressed by mesalazine and BBR intervention. Furthermore, administration of BBR at a high dose (40 mg/kg) expressed a lower level of IFN-γ (Fig. 6A), IRF8 (Fig. 6B), NF-κB (Fig. 6C) and TNF-α(Fig. 6D) noticeably compared with the UC group and low dose (20 mg/kg) group.
Fig. 6.
Effects of BBR on the mRNA expressions of IFN-γ, IRF8, NF-κB and TNF-α in DSS-induced UC mice. A: mRNA expressions of IFN-γ; B: mRNA expressions of IRF8; C: mRNA expressions of NF-κB; D: mRNA expressions of TNF-α. N.S: None statistically significant; ##P < 0.01 versus control group; ∗P < 0.05 versus UC group; ∗∗P < 0.01 versus UC group. BBR_Low: BBR with low dose; BBR_High: BBR with high dose.
3.7. BBR inhibited IFN-γ signaling pathway in vivo
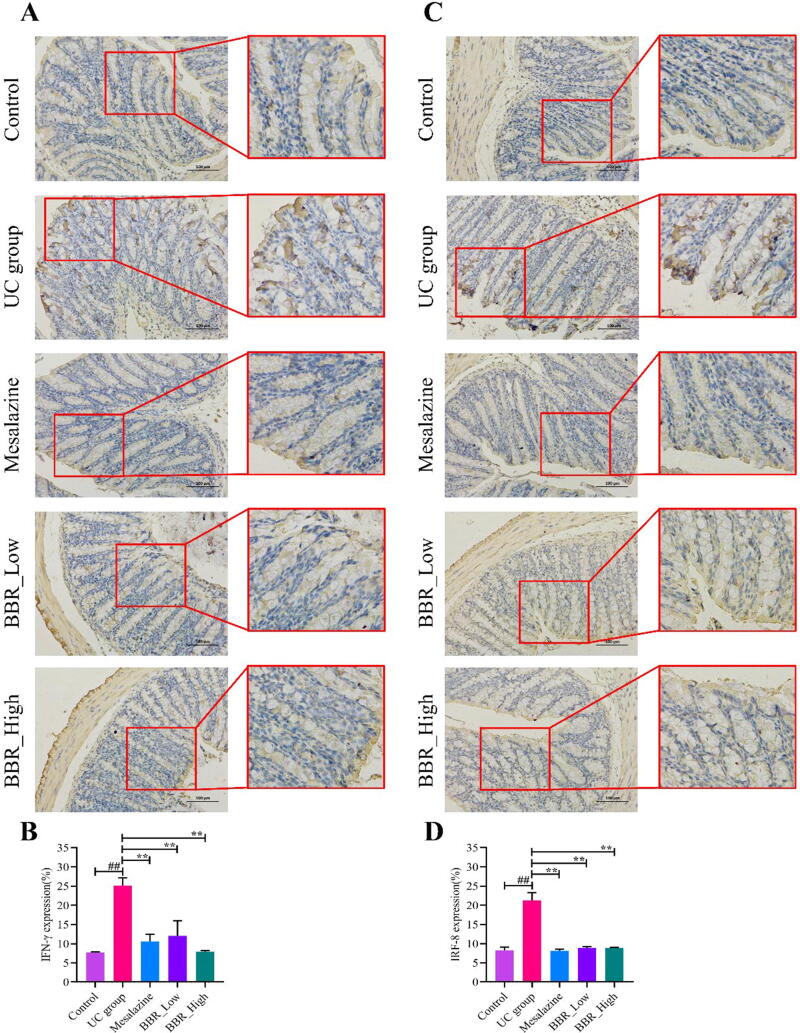
We reported previously that there existed feedback interaction between IFN-γ and IRF8 in inflammatory disease of the gastrointestinal tract (Yang et al., 2020). In this study, we measured the protein expression of IFN-γ and IRF8 in the colon via IHC staining. Consistent with the findings of mRNA, IHC analysis revealed that IFN-γ embraced a robust increase in the UC group (Fig. 7A, 7B). Correspondingly, IRF8 also holds a higher expression level in the UC group compared with the control group (Fig. 7C, 7D). Interfered by mesalazine and BBR, the expression of IFN-γ and IRF8 decreased significantly. Results of IHC are presented in Fig. 7.
Fig. 7.
IHC staining results of each group mice. A: IHC staining plot of IFN-γ in colon tissue; B: Quantitative analysis results of IFN-γ; C: IHC staining plot of IRF8 in colon tissue; D: Quantitative analysis results of IRF8. ##P < 0.01 versus control group; ∗∗P < 0.01 versus UC group. BBR_Low: BBR with low dose; BBR_High: BBR with high dose.
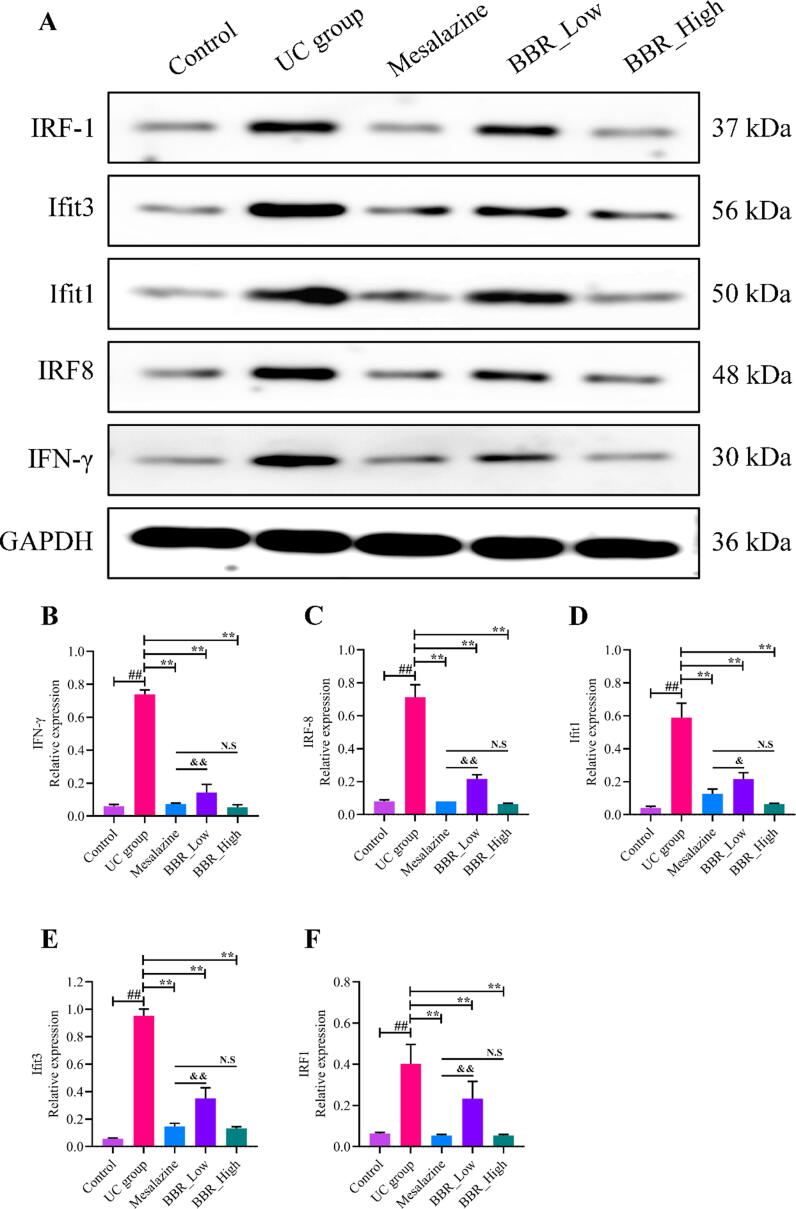
Subsequently, the expressions of IFN-γ and it's related targets, including IRF8, Ifit1, Ifit3, and IRF1 in colon tissues were estimated by WB analysis. As shown in Fig. 8, IFN-γ (Fig. 8A, 8B), IRF8 (Fig. 8A, 8C), Ifit3 (Fig. 8A, 8D), IRF1 (Fig. 8A, 8E) and Ifit1 (Fig. 8A, 8F) expressed at a relatively high level in UC group compared with the control group. After intervened by mesalazine and BBR, the expression of IFN-γ, IRF8, Ifit1, Ifit3, and IRF1 were significantly down-regulated. Noticeably, the expressions of these proteins were exceedingly significant differences in BBR low-dose group and BBR high-dose group. WB analysis findings provided stronger evidence that BBR inhibited IFN-γ signaling pathway in DSS-induced UC mice.
Fig. 8.
Proteins expression in each group mice estimated by WB analysis. A: WB images of IFN-γ, IRF8, Ifit1, Ifit3 and IRF1. B: Relative IFN-γ protein expression in colon tissue; C: Relative IRF8 protein expression in colon tissue; D: Relative Ifit1 protein expression in colon tissue; E: Relative Ifit3 protein expression in colon tissue; F: Relative IRF1 protein expression in colon tissue. N.S: None statistically significant; ##P < 0.01 versus control group; ∗∗P < 0.01 versus UC group. &P < 0.05 versus mesalazine group; &&P < 0.01 versus mesalazine group. BBR_Low: BBR with low dose; BBR_High: BBR with high dose.
3.8. BBR inhibited IFN-γ signaling pathway in vitro
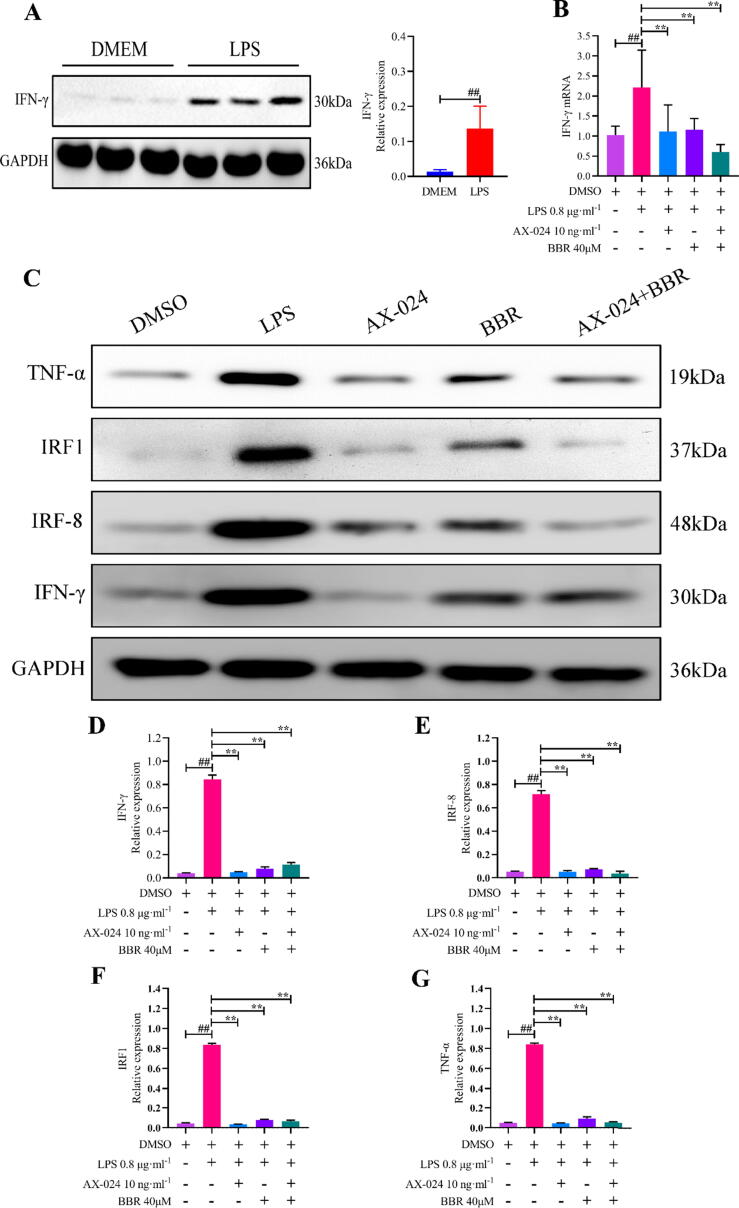
In addition, we further verified the effects of BBR on IFN-γ signaling pathway in vitro experiments. LPS with 0.8 μg·ml−1 was applied to induce NCM460 cells to inflammatory damage. Selective inhibitor of IFN-γ (AX-024, MedChem Express, Shanghai, China, 10 ng/ml) was applied to silence the expression of IFN-γ. As presented in Fig. 8A, the expression of IFN-γ protein was significantly up-regulated by exposure to LPS. Additionally, we observed that IFN-γ mRNA were decreased significantly with AX-024 blocking and BBR (with a concentration of 40 μM) intervention (Fig. 8B). Meanwhile, AX-024 plus BBR (combination group) was utilized to inhibit the expression of IFN-γ mRNA in NCM460 cells with LPS infection, result showed IFN-γ mRNA was further suppressed obviously (Fig. 8B). Furthermore, the expressions of IFN-γ (Fig. 9C, 9D) and its interactive proteins, including IRF8 (Fig. 9C, 9E), IRF1 (Fig. 9C, 9F) and TNF-α(Fig. 9C, 9G) were significantly inhibited by AX-024 and BBR intervention. Accordingly, these results suggested that BBR inhibited IFN-γ mediated signaling pathway in inflammatory cells.
Fig. 9.
Effects of BBR on the IFN-γ signaling pathway. A: Relative IFN-γ protein expression in NCM460 cells; B: mRNA expressions of IFN-γ in NCM460 cells; C: WB images of IFN-γ, IRF8, IRF1 and TNF-α. D: Relative IFN-γ protein expression in NCM460 cells; E: Relative IRF8 protein expression in NCM460 cells; F: Relative Ifit1 protein expression in NCM460 cells; G: Relative TNF-α protein expression in NCM460 cells; ##P < 0.01 versus control (DMEM/DMSO) group; ∗∗P < 0.01 versus LPS group.
4. Discussion
In this present study, results revealed that most of the immune-inflammatory genes were identified and up-regulated significantly in UC, which indicated immune-inflammatory response was the primary pathogenesis of colon mucosa in UC. In addition, IFN-γ signaling pathway was significantly up-regulated in the phenotype of UC, promoting to the aggravation of immune-inflammatory response in UC. Furthermore, BBR suppressed IFN-γ signaling pathway to exhibit anti-inflammatory effects in vivo and in vitro, contributing to the efficacious treatment of UC.
IFN-γ is an endogenous cytokines of immunity and inflammation. It is primarily produced by immune cells, including natural killer (NK) cells, innate lymphoid cells (ILCs), T helper 1 (TH1) cells and CD8+ cytotoxic T lymphocytes (CTLs) (Ivashkiv, 2018). IFN-γ regulates the process of inflammation, tissue remodeling, macrophage activation, host defense and Th1 responses. Meanwhile, IFN-γ exerts regulatory functions to modulate Th and Treg differentiation (Hu and Ivashkiv, 2009). IFN-γ signaling has been conventionally defined as a cascade of tyrosine phosphorylation procedures triggered by the binding of IFN-γ to IFN-γ receptor (IFNγR, comprising the IFNγR1 and IFNγR2 subunits), which leads to the initiation of interferon-stimulated gene (ISG) transcription (Ivashkiv, 2018). Generally, IFN-γ utilizes the Janus kinases (JAKs) - signal transducer and activator of transcription (STAT) signaling pathway to the tyrosine phosphorylation of STAT1 (Hu and Ivashkiv, 2009, O'Connell et al., 2015, Cordes et al., 2020). Despite the fact that many biological functions of IFN-γ have been proved the induction of immune and inflammation effector genes were directly STAT1-mediated, it has become clear recently that IFN-γ is mediated by cross-regulation of cellular responses to other cytokines and inflammatory genes. Previous study has indicated the activation of IFN-γ mainly relied on a TLR9-dependent pathway and IFN-γ induction was required for transformation of immune responses and exacerbation of UC (Gogokhia et al., 2019). In addition, IFN-c activator factor (GAF), as a dimer of interferon, can bind to the IFN-c activation site (GAS) and then up-regulate interferon regulatory factor 8 (IRF8) (Nelson et al., 1996). When the expression of IFN-γ was suppressed, TNF-α production followed by activation of NF-κB and NO production were also decreased. In parallel, the inhibition of IFN-γ mediated cellular responses mainly via inhibiting the induction of IRF8 and IRF8-dependent genes (Takahashi et al., 2012). Previously, a genome-wide association study (GWAS) of IRF8 by employing ChIP-seq and RNA-seq indicated Ifit1, Ifit3 and IRF1 were the direct targets of IRF8 and the expressions were IRF8-dependent fashion (Yan et al., 2016). Meanwhile, IFN-γ drove IRF8 expression and IRF8 was identified as a major regulator for IFN-γ production, which amplified inflammatory responses (Yan et al., 2016, Salem et al., 2020). Obviously, the signaling pathway of IFN-γ in the immune-inflammatory responses can be summarized as IFN-γ initiated and exaggerated by IRF8-dependent genes.
In this study, we verified the role of IFN-γ signaling pathway in the occurrence and development of UC and further explored the potential effects of BBR, a natural isoquinoline alkaloid, on IFN-γ signaling pathway in treating UC. Results from this study indicated IFN-γ signaling pathway and its core genes were significantly up-regulated in UC. Accompanied by the increased expression of IFN-γ, its related genes and proteins (including NF-κB, TNF-α, IRF8, Ifit1, Ifit3 and IRF1) also up-regulated significantly in UC, which contributed to the exaggeration of immune-inflammatory responses in the colon mucosa. In addition, we observed that BBR possessed anti-inflammatory activity and could be a possible effective treatment to UC. All disease performance of UC, including fecal occult blood, food consumption and weight loss, in mice induced by DSS were ameliorated by BBR treatment. Furthermore, the pathologic changes of the colon mucosa, such as swollen crypts, infiltration of neutrophils and lymphocytes, were evidently ameliorated by BBR treatment. In terms of mechanisms of BBR in treating UC and regulating IFN-γ signaling pathway, the pro-inflammatory and immune-related genes, encoding IFN-γ, IRF8, NF-κB and TNF-α decreased significantly in UC mice followed by BBR treatment. These results supported the role of BBR as an anti-inflammatory agent in UC management. In addition, we also demonstrated the expression of IFN-γ and its initiated targets, including IRF8, Ifit1, Ifit3, IRF1, were suppressed significantly in UC mice followed by BBR treatment. Meanwhile, we revealed that the blocking of IFN-γ in vitro led to the silence of IFN-γ signaling pathway after exposure to BBR.
All in all, conclusions from this study suggested that BBR could be an anti-inflammatory agent in the UC treatment. The anti-inflammatory property of BBR is tightly related to the suppression of IFN-γ signaling pathway, which is crucial in immune-inflammatory responses of the colon mucosa. Findings from this study provided many novel targets for UC treatment and paved a theoretical cornerstone for BBR in treating UC at clinical practice.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors would like to thank all authors of references for their hard work.
Funding
This research was financially supported by the National key R & D project (No. 2018YFC1704500).
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2022.03.015.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Bjerrum J.T., Hansen M., Olsen J., et al. Genome-wide gene expression analysis of mucosal colonic biopsies and isolated colonocytes suggests a continuous inflammatory state in the lamina propria of patients with quiescent ulcerative colitis. Inflammatory Bowel Dis. 2010;16:999–1007. doi: 10.1002/ibd.21142. [DOI] [PubMed] [Google Scholar]
- Bonecchi R., Polentarutti N., Luini W., et al. Up-regulation of CCR1 and CCR3 and induction of chemotaxis to CC chemokines by IFN-gamma in human neutrophils. J. Immunol. (Baltimore, Md. : 1950) 1999;162:474–479. [PubMed] [Google Scholar]
- Chan M.M.P., Cheung B.K.W., Li J.C.B., Chan L.L.Y., Lau A.S.Y. A role for glycogen synthase kinase-3 in antagonizing mycobacterial immune evasion by negatively regulating IL-10 induction. J. Leukoc. Biol. 2009;86(2):283–291. doi: 10.1189/jlb.0708442. [DOI] [PubMed] [Google Scholar]
- Cordes F., Foell D., Ding J.N., Varga G., Bettenworth D. Differential regulation of JAK/STAT-signaling in patients with ulcerative colitis and Crohn's disease. World J. Gastroenterol. 2020;26(28):4055–4075. doi: 10.3748/wjg.v26.i28.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes F., Lenker E., Weinhage T., et al. Impaired IFN-γ-dependent STAT3 Activation Is Associated With Dysregulation of Regulatory and Inflammatory Signaling in Monocytes of Ulcerative Colitis Patients. Inflammatory Bowel Dis. 2021;27:887–901. doi: 10.1093/ibd/izaa280. [DOI] [PubMed] [Google Scholar]
- Cosnes J., Gower–Rousseau C., Seksik P., Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140(6):1785–1794.e4. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- Cui H., Cai Y., Wang L.i., Jia B., Li J., Zhao S., Chu X., Lin J., Zhang X., Bian Y., Zhuang P. Berberine Regulates Treg/Th17 Balance to Treat Ulcerative Colitis Through Modulating the Gut Microbiota in the Colon. Front. Pharmacol. 2018;9 doi: 10.3389/fphar.2018.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L.i., Guan X., Ding W., Luo Y.i., Wang W., Bu W., Song J., Tan X., Sun E., Ning Q., Liu G., Jia X., Feng L. Scutellaria baicalensis Georgi polysaccharide ameliorates DSS-induced ulcerative colitis by improving intestinal barrier function and modulating gut microbiota. Int. J. Biol. Macromol. 2021;166:1035–1045. doi: 10.1016/j.ijbiomac.2020.10.259. [DOI] [PubMed] [Google Scholar]
- Silva B.C.d., Lyra A.C., Rocha R., Santana G.O. Epidemiology, demographic characteristics and prognostic predictors of ulcerative colitis. World J. Gastroenterol. 2014;20(28):9458–9467. doi: 10.3748/wjg.v20.i28.9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerd N.A., Nguyen T. The interferons and their receptors–distribution and regulation. Immunol. Cell Biol. 2012;90(5):483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein, M., 2018. Ulcerative colitis: towards remission. Nature. 563, S33. 10.1038/d41586-018-07276-2. [DOI] [PubMed]
- Feuerstein J.D., Cheifetz A.S. Ulcerative colitis: epidemiology, diagnosis, and management. Mayo Clin. Proc. 2014;89(11):1553–1563. doi: 10.1016/j.mayocp.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Gogokhia L., Buhrke K., Bell R., Hoffman B., Brown D.G., Hanke-Gogokhia C., Ajami N.J., Wong M.C., Ghazaryan A., Valentine J.F., Porter N., Martens E., O’Connell R., Jacob V., Scherl E., Crawford C., Stephens W.Z., Casjens S.R., Longman R.S., Round J.L. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host. Microbe. 2019;25(2):285–299.e8. doi: 10.1016/j.chom.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Ivashkiv L.B. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Paik P.K., Chen J., et al. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 2006;24:563–574. doi: 10.1016/j.immuni.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Isaacs A., Lindenmann J. Virus interference. I. The interferon. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1957;147:258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- Ivashkiv L.B. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018;18(9):545–558. doi: 10.1038/s41577-018-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14(1):36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jess T., Frisch M., Simonsen J. Trends in overall and cause-specific mortality among patients with inflammatory bowel disease from 1982 to 2010. Clin. Gastroenterol. Hepatol.: Off. Clin. Pract. J. Am. Gastroenterol. Associat. 2013;11(1):43–48. doi: 10.1016/j.cgh.2012.09.026. [DOI] [PubMed] [Google Scholar]
- Kaur A., Goggolidou P. Ulcerative colitis: understanding its cellular pathology could provide insights into novel therapies. J. Inflammat. (London, England). 2020;17:15. doi: 10.1186/s12950-020-00246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Feng C., Fan C., Yang Y., Yang X., Lu H., Lu Q., Zhu F., Xiang C., Zhang Z., He P., Zuo J., Tang W. Intervention of oncostatin M-driven mucosal inflammation by berberine exerts therapeutic property in chronic ulcerative colitis. Cell Death Dis. 2020;11(4) doi: 10.1038/s41419-020-2470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Strauss R., Ouahed J., Chan D., Telesco S.E., Shouval D.S., Canavan J.B., Brodmerkel C., Snapper S.B., Friedman J.R. Molecular comparison of adult and pediatric ulcerative colitis indicates broad similarity of molecular pathways in disease tissue. J. Pediatr. Gastroenterol. Nutr. 2018;67(1):45–52. doi: 10.1097/MPG.0000000000001898. [DOI] [PubMed] [Google Scholar]
- Li M.-Y., Luo H.-J., Wu X., Liu Y.-H., Gan Y.-X., Xu N., Zhang Y.-M., Zhang S.-H., Zhou C.-L., Su Z.-R., Huang X.-q., Zheng X.-B. Anti-inflammatory effects of huangqin decoction on dextran sulfate sodium-induced ulcerative colitis in mice through regulation of the gut microbiota and suppression of the ras-PI3K-Akt-HIF-1α and NF-κB pathways. Front. Pharmacol. 2019;10 doi: 10.3389/fphar.2019.01552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Cheng L., Wang Q.i., Zhou L. Comparative transcriptomic analysis reveals the immunosuppressive targets of mesalazine in dextran sulfate sodium-induced ulcerative colitis. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.698983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-H., Adam R., Colombel J.-F., Bian Z.-X. A characterization of pro-inflammatory cytokines in dextran sulfate sodium-induced chronic relapsing colitis mice model. Int. Immunopharmacol. 2018;60:194–201. doi: 10.1016/j.intimp.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Li Y.-H., Xiao H.-T., Hu D.-D., Fatima S., Lin C.-Y., Mu H.-X., Lee N.P., Bian Z.-X. Berberine ameliorates chronic relapsing dextran sulfate sodium-induced colitis in C57BL/6 mice by suppressing Th17 responses. Pharmacol. Res. 2016;110:227–239. doi: 10.1016/j.phrs.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Majoros A., Platanitis E., Kernbauer-Hölzl E., Rosebrock F., Müller M., Decker T. Canonical and Non-Canonical Aspects of JAK-STAT Signaling: Lessons from Interferons for Cytokine Responses. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454(7203):428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140(6):771–776. doi: 10.1016/j.cell.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Nelson N., Kanno Y., Hong C., et al. Expression of IFN regulatory factor family proteins in lymphocytes. Induction of Stat-1 and IFN consensus sequence binding protein expression by T cell activation. J. Immunol. (Baltimore, Md. : 1950) 1996;156:3711–3720. [PubMed] [Google Scholar]
- Niriella M.A., De Silva A.P., Dayaratne A.H., Ariyasinghe M.H., Navarathne M.MN., Peiris R.SK., Samarasekara D.N., Satharasinghe R.L., Rajindrajith S., Dassanayake A.S., Wickramasinghe A.R., de Silva H.J. Prevalence of inflammatory bowel disease in two districts of Sri Lanka: a hospital based survey. BMC Gastroenterol. 2010;10(1) doi: 10.1186/1471-230X-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell D., Bouazza B., Kokalari B., Amrani Y., Khatib A., Ganther J.D., Tliba O. IFN-γ-induced JAK/STAT, but not NF-κB, signaling pathway is insensitive to glucocorticoid in airway epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;309(4):L348–L359. doi: 10.1152/ajplung.00099.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordás I., Eckmann L., Talamini M., Baumgart D.C., Sandborn W.J. Ulcerative colitis. Lancet (London, England) 2012;380(9853):1606–1619. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- Planell N., Lozano J.J., Mora-Buch R., Masamunt M.C., Jimeno M., Ordás I., Esteller M., Ricart E., Piqué J.M., Panés J., Salas A. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut. 2013;62(7):967–976. doi: 10.1136/gutjnl-2012-303333. [DOI] [PubMed] [Google Scholar]
- Porter R.J., Kalla R., Ho G.-T. Ulcerative colitis: Recent advances in the understanding of disease pathogenesis. F1000 Res. 2020;9:294. doi: 10.12688/f1000research10.12688/f1000research.20805.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch I., Müller M., Decker T. The regulation of inflammation by interferons and their STATs. Jak-stat. 2013;2(1):e23820. doi: 10.4161/jkst.23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl. Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D.T., Ananthakrishnan A.N., Siegel C.A., Sauer B.G., Long M.D. ACG Clinical Guideline: Ulcerative Colitis in Adults. Am. J. Gastroenterol. 2019;114(3):384–413. doi: 10.14309/ajg.0000000000000152. [DOI] [PubMed] [Google Scholar]
- Salem S., Salem D., Gros P. Role of IRF8 in immune cells functions, protection against infections, and susceptibility to inflammatory diseases. Hum. Genet. 2020;139(6-7):707–721. doi: 10.1007/s00439-020-02154-2. [DOI] [PubMed] [Google Scholar]
- Smallridge R.C., Chindris A.-M., Asmann Y.W., Casler J.D., Serie D.J., Reddi H.V., Cradic K.W., Rivera M., Grebe S.K., Necela B.M., Eberhardt N.L., Carr J.M., McIver B., Copland J.A., Thompson E.A. RNA sequencing identifies multiple fusion transcripts, differentially expressed genes, and reduced expression of immune function genes in BRAF (V600E) mutant vs BRAF wild-type papillary thyroid carcinoma. J. Clin. Endocrinol. Metabol. 2014;99(2):E338–E347. doi: 10.1210/jc.2013-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Sugiyama T., Tokoro S., Neri P., Mori H. Inhibition of interferon-γ-induced nitric oxide production by 10-hydroxy-trans-2-decenoic acid through inhibition of interferon regulatory factor-8 induction. Cell. Immunol. 2012;273(1):73–78. doi: 10.1016/j.cellimm.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Teixeira L.K., Fonseca B.PF., Barboza B.A., Viola J.PB. The role of interferon-gamma on immune and allergic responses. Mem. Inst. Oswaldo Cruz. 2005;100(suppl 1):137–144. doi: 10.1590/s0074-02762005000900024. [DOI] [PubMed] [Google Scholar]
- Wirtz S., Popp V., Kindermann M., Gerlach K., Weigmann B., Fichtner-Feigl S., Neurath M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017;12(7):1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- Yan M., Wang H., Sun J., Liao W., Li P., Zhu Y., Xu C., Joo J., Sun Y., Abbasi S., Kovalchuk A., Lv N., Leonard W.J., Morse H.C. Cutting Edge: Expression of IRF8 in Gastric Epithelial Cells Confers Protective Innate Immunity against Helicobacter pylori Infection. J.I. 2016;196(5):1999–2003. doi: 10.4049/jimmunol.1500766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Wang R., Zhang J., Bao C., Zhang J., Li R., Chen X., Wu S., Wen J., Wei S., Li H., Cai H., Yang X., Zhao Y. Mechanism of berberine in treating Helicobacter pylori induced chronic atrophic gastritis through IRF8-IFN-γ signaling axis suppressing. Life Sci. 2020;248:117456. doi: 10.1016/j.lfs.2020.117456. [DOI] [PubMed] [Google Scholar]
- Zhang T., DeSimone R.A., Jiao X., Rohlf F.J., Zhu W., Gong Q.Q., Hunt S.R., Dassopoulos T., Newberry R.D., Sodergren E., Weinstock G., Robertson C.E., Frank D.N., Li E., Heimesaat M.M. Host genes related to paneth cells and xenobiotic metabolism are associated with shifts in human ileum-associated microbial composition. PLoS One. 2012;7(6):e30044. doi: 10.1371/journal.pone.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.