Abstract
Single nucleotide polymorphism (SNP) of long noncoding RNA (lnc)RNA has been reported to be an important factor in cancer development. Recently, lncRNA homeobox transcript antisense intergenic RNA (HOTAIR) was indicated to induce tumorigenesis of several cancer types, but the association between the SNP of lncRNA HOTAIR and lung cancer susceptibility has remained undetermined. The present meta-analysis aimed to investigate the effect of HOTAIR polymorphism on susceptibility to lung cancer. The PubMed, Ovid Medline, Embase and Cochrane Library databases were thoroughly searched. Studies containing data on the incidence of lung cancer in patients with different HOTAIR SNPs were included. The Hardy-Weinberg equilibrium was analyzed to determine genotype distribution and allele frequencies. The odds ratio (OR) was pooled to evaluate the association of different SNPs with the susceptibility to lung cancer. A total of six studies comprising 1,715 patients with lung cancer and 2,745 healthy controls were finally included. A total of 4 SNPs (rs12826786, rs1899663, rs920778 and rs4759314) were reported. Analyses for all of these SNPs individually indicated that the lncRNA HOTAIR rs1899663 C>A polymorphism was a risk factor for lung cancer (dominant mode, AA+CA vs. CC: OR=0.816, 95% CI=0.707-0.942, P=0.005). The present study was the first meta-analysis investigating the association between lncRNA HOTAIR and lung cancer susceptibility. The results indicated that the lncRNA HOTAIR rs1899663 C>A polymorphism is a risk factor for lung cancer. LncRNA HOTAIR may be of value in lung cancer screening, particularly for populations with high-risk factors, as well as prognosis prediction. Future investigations are required to further clarify the intrinsic mechanism of the role of HOTAIR in the oncogenesis of lung cancer.
Keywords: lncRNA, HOTAIR, SNP, lung cancer, susceptibility
Introduction
Lung cancer has brought a heavy burden to human society and families, with high morbidity and mortality rates among all types of malignant tumors (1). The statistical report on cancer incidence and mortality published by the International Cancer Center (GLOBOCAN 2018) suggests that lung cancer is a leading cause of death of patients with cancer in China and worldwide (2). Globally, it is estimated that there are 2.1 million new cases of lung cancer and 1.8 million lung cancer-related deaths each year (2). In China, the incidence of lung cancer is reported to be 5.7 per million and the total number of new cases is ~787,000, accounting for 20% of the total malignant tumor cases (3).
The occurrence and development of lung cancer are closely related to the tumor micro-environment, personal living habits and genetic differences (4,5). Smoking, history of lung-related diseases, air pollutants, family history of tumors, ethnic differences, obesity, occupational exposure and poor eating habits may increase the risk of lung cancer (6). Although smoking is a well-recognized risk factor for lung cancer, lung cancer also occurs in 10-25% of non-smokers. The influence of personal genetic factors on the pathogenesis of lung cancer should not be underestimated (7,8).
With the rise of molecular epidemiology, the ENCODE project indicated that up to 75% of human genome nucleotides may be transcribed into the original transcripts, producing a series of common RNAs including non-coding RNAs (ncRNA) (9). A large number of studies have indicated that most ncRNAs do not encode proteins but have specific functions (10). At the level of apparent modification, long ncRNA (lncRNA) affects numerous aspects of chromosome biology by remodeling chromatin structure and regulating a large number of genes. Based on its various characteristics and regulatory mechanisms, the abnormal expression of lncRNA may also be involved in the occurrence, development and metastasis of tumors, thus affecting tumor staging and the occurrence of various cancers (11,12). The potential impact of lncRNA on the occurrence, development, treatment and prognosis of lung cancer is worthy of further investigation (13).
Single nucleotide polymorphism (SNP) refers to the polymorphism in the DNA sequence caused by the variation of a single nucleotide in the genome. It is a heritable variation that exists widely in the human genome (14). It is estimated that there is one SNP per 1,000 bases on average, which is recognized as the third-generation genetic marker after restriction fragment length polymorphism and microsatellites. SNP is involved in the transcription of coding genes by influencing the binding of transcription factors, thereby affecting gene expression. Researchers have discovered a large number of SNPs associated with diseases through genome-wide association studies and about one-third of the SNPs are located in non-protein coding regions, indicating the SNPs of lncRNA may also have an important role in disease (15). Emerging evidence suggested that the SNPs of lncRNAs are related to the susceptibility to and prognosis of various types of malignant tumor, such as breast cancer, gastric cancer and colorectal cancer (16).
In recent years, researchers have discovered that the homeobox (HOX) gene family participates in embryonic development (17). In their study from 2007, Rinn et al (18) used high-resolution microarray technology to identify and analyze hundreds of ncRNAs transcribed from the HOX gene cluster from 11 types of fibroblasts and discovered HOX transcript antisense intergenic RNA (HOTAIR) at the HOXC gene locus located on chromosome 12. HOTAIR was recently considered a novel oncogene, but it is controversial whether the SNP of lncRNA HOTAIR has a risk effect to influence lung cancer susceptibility (19). LncRNA HOTAIR is located at 12q13.13 and the length is 2.1 kb. There are four gene clusters in the genome: HOXA, HOXB, HOXC and HOXD (20). The transcription of these four gene clusters produces several long-chain ncRNAs and their expression has a temporal and spatial specificity (21). Although HOTAIR is produced from the HOXC sequence, it has the function of inhibiting the expression of the HOXD gene. The secondary structure of HOTAIR contains four independent folding units, two of which are evolutionarily conserved protein-binding domains (22). HOTAIR is produced and enriched in the nucleus and expressed in the cytoplasm. The HOTAIR-mediated epigenetic silencing function works by acting as a scaffold for the polycomb repressive complex 2 (PRC2) and lysine specific demethylase 1/RE1-silencing transcription factor (REST) co-repressor 1/REST complex. PRC2 binds to the 5' end of HOTAIR and regulates chromosome occupancy through the subunit EZH2 of the PRC2 complex, leading to the methylation of histone H3 lysine in the chromosome sequence region of the HOXD gene (23). Studies have indicated that at least 16 types of malignant tumor have increased expression of HOTAIR (24-26). Furthermore, it was reported that overexpressed HOTAIR combines with PRC2 to regulate gene expression patterns by changing the methylation status of H3K27, thus increasing the ability of cancer cells to metastasize and invade. Knockdown of HOTAIR or PRC2 is able to inhibit the invasion and metastasis ability of cancer cells (27). A number of studies focusing on lung cancer have reported that HOTAIR has a proto-oncogene function in lung cancer. For instance, compared with adjacent tissues, the expression of HOTAIR in lung cancer tissue is significantly higher (28). The increased HOTAIR expression in non-small cell lung cancer is similar to that of lung adenocarcinoma and lung squamous cells (29). Another study determined that elevated HOTAIR expression was associated with the brain metastasis rate of lung cancer (30). The association between the SNP of lncRNA HOTAIR and lung cancer susceptibility remains to be further investigated.
Thus, the present systematic review and meta-analysis was performed to investigate the association between lncRNA HOTAIR SNP and lung cancer susceptibility.
Materials and methods
Literature search and search strategy
This systematic review and meta-analysis was performed following the preferred reporting items for systematic review and meta-analysis (PRISMA) guidelines (31). A computerized literature search was performed in the PubMed (https://pubmed.ncbi.nlm.nih.gov/), Ovid Medline (https://www.wolterskluwer.com/en/solutions/ovid/ovid-medline-901), Embase (https://www.embase.com) and Cochrane Library databases (https://www.cochranelibrary.com/) up to May 2nd 2021. Relevant studies were also searched in Chinese databases, including Chinese National Knowledge Infrastructure (CNKI; https://www.cnki.net/) and Wanfang (https://www.wanfangdata.com.cn/index.html). Google scholar (https://scholar.google.com/) was searched for gray literature that is not formally published in sources such as books or journal articles.
Regarding the search strategy, terms included the following items: (‘lncRNA’ or ‘long non-coding RNA’) and (‘SNP’ or ‘polymorphism’ or ‘variants’) and (‘lung cancer’ or ‘lung tumor’ or ‘lung neoplasm’ or ‘adenocarcinoma’ or ‘lung carcinoma’) and (‘HOTAIR’ or ‘HOX transcript antisense intergenic RNA’). The items were searched for keywords and medical sub-headings. All results were downloaded and imported into Endnote X6 (Thomson Reuters) for further literature screening.
Inclusion and exclusion criteria
All of the studies evaluating and assessing the association between lncRNA HOTAIR polymorphism and the susceptibility to lung cancer were included in the present study. The inclusion criteria were as follows: i) Patients with lung cancer; ii) case-control studies on the polymorphism of lncRNA HOTAIR; iii) number of patients >50. The exclusion criteria were as follows: i) Reviews, observational studies, comments, case series and case reports; ii) lack of data on lncRNA HOTAIR; iii) lack of information on lung cancer risk; and iv) lack of available data that were able to be extracted from the text/table/graphs or obtained from the authors on request. There was no limitation in terms of the language of the studies included and the reference lists of reviews were also screened for the retrieval and further inclusion of studies.
Literature screening, data extraction and quality assessment
A total of two investigators (CK and XF) independently screened titles and abstracts according to the inclusion and exclusion criteria. The full-text was further screened for data extraction or assessed for the inclusion criteria. In the case of any disagreements, a third investigator (XH) was consulted to resolve the divergence.
The data were extracted by two reviewers independently (CK and XF). All the data were imported into a standard form, including the name of the first author of the study, publication year, recruitment years and country, genotyping methods and the common polymorphism of HOTAIR. In addition, the sample size, as well as the age, sex, smoking history and alcohol use of healthy participants (control group) and patients with lung cancer (case group) were recorded. The raw data are provided as supplemental material (Table SI).
The study quality was evaluated according to Newcastle-Ottawa Scale (NOS), with a score of 7-9 considered to indicate high quality, whereas a score of 0-6 was indicative of low quality (32).
Statistical analysis
The meta-analysis was performed with Stata 15.0 software (StataCorp LP). The genotype frequencies of lncRNA HOTAIR polymorphisms for the Hardy-Weinberg equilibrium (HWE) were calculated for the controls using the χ2 test and P<0.05 was considered to indicate a significant disequilibrium. The risk was pooled by the odds ratio (OR) and 95% confidence intervals (CI). The result of the pooled OR was visualized through the forest plot by using the metan module of Stata. The heterogeneity existing among studies was evaluated by the I2 statistic and χ2 test, with I2 ≥50% indicating the presence of heterogeneity. The random-effects model was used if heterogeneity was present among studies, while otherwise, the fixed-effects model was used. The risks of the heterozygote and variant homozygote compared with the wild-type homozygote were estimated. Furthermore, the dominant and recessive mode of the variant allele were evaluated. The sensitivity analysis was performed by reperforming the OR calculation while omitting one individual study at a time. Sensitivity analysis was performed for rs12826786, rs1899663, rs4759314 and rs920778 for all modes, including the dominant mode, recessive mode, additive mode, heterozygote mode, homozygote mode and allele mode. Egger's linear regression method was used to statistically assess the publication bias by Stata 15.0. The funnel plot for exploring the missed-reported articles was also analysed with the metafunnel module of Stata to visually display the outcomes of publication bias assessment. Subgroup analysis was performed using the metan module of Stata. The further heterogeneity analysis was performed based on the results of the subgroup analysis. P<0.05 was considered to indicate statistical significance for all analyses.
Results
Study selection and characteristics of included studies
A total of 46 studies were retrieved by searching the databases as described above, as well as 8 studies via other methods. Thus, a total of 54 studies focusing on the association of lncRNA HOTAIR with lung cancer were identified based on the search strategy. After excluding duplicates and other studies based on the inclusion and exclusion criteria, six studies were finally included in the present study (33-38). The literature screening process is presented in Fig. 1.
Figure 1.
Flowchart of the literature screening. Records were searched via databases (n=46) and other sources (n=8). Finally, a total of 6 records were considered eligible and included in the present meta-analysis. SNP, single nucleotide polymorphism; lncRNA, long noncoding RNA; HOTAIR, homeobox transcript antisense intergenic RNA.
The characteristics of the included studies are described in Table I. The year of publication ranged from 2016 to 2020 and the recruitment period ranged from 1995 to 2017. Of the six studies, four were from China and the other two studies were from Turkey and Japan, respectively. Furthermore, two studies used MassArray to detect the SNPs of lncRNA, while the other four studies used the TaqMan assay instead. A total of 1,715 patients with lung cancer and 2,745 healthy participants were included in the analysis of the present study. The average age of the lung cancer group was 57.9-64.3 years, while it was 56.6-64.8 years for the healthy participants (control group). The proportion of male subjects ranged from 29.9 to 96.9% in the control group, as compared to 35.4-81.6% in the lung cancer group. A history of smoking was present in 20.1-96.6% of the healthy participants and in 29.9-99.4% of the patients with lung cancer. The results of the quality assessment are listed in Table I. Of the studies, four scored as >6 and were considered to be of high quality.
Table I.
Characteristics of the included studies.
| Control group | Case group | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author (year) | Country | Recruitment year | Cancer type | Ethnicity | Genotyping method | Quality score | Sample, n | Age, years | Male sex, n (%) | Smoking, n (%) | Alcohol consumption, n (%) | Sample, n | Age, years | Male sex, n (%) | Smoking, n (%) | Alcohol consumption, n (%) | (Refs.) |
| Ren (2020) | China | 2015-2017 | All lung cancer | Asian | MassArray system | 8 | 196 | 64.82±10.00 | 139 (70.9) | 190 (96.9) | 188 (95.9) | 196 | 64.32±10.26 | 136 (69.4) | 195 (99.4) | 196(100) | (33) |
| Minn (2020) | Japan | 1995-2006 | All lung cancer | Asian | TaqMan Assay-PCR | 8 | 1,241 | NG | NG | NG | NG | 129 | NG | NG | NG | NG | (34) |
| Wang (2018) | China | 2006-2010 | All lung cancer | Asian | TaqMan Assay-PCR | 7 | 451 | NG | 280 (62.1) | 159 (35.2) | NG | 262 | NG | 188 (71.8) | 169 (64.5) | NG | (35) |
| Li (2018) | China | NG | All lung cancer | Asian | TaqMan Assay-PCR | 7 | 551 | 56.69±15.65 | 165 (29.9) | 111 (20.1) | NG | 543 | 57.99±11.51 | 192 (35.4) | 165 (30.4) | NG | (36) |
| Dadaş (2018) | Turkey | NG | NSCLC | Caucasian | TaqMan Assay-PCR | 6 | 93 | 61.77±12.00 | 67 (72.0) | 61 (65.6) | NG | 87 | 59.27±10.55 | 71 (81.6) | 26 (29.9) | NG | (37) |
| Gong (2016) | China | 2011-2013 | All lung cancer | Asian | MassARRAY | 5 | 213 | NG | 80 | NG | NG | 498 | NG | 394 (79.1) | NG | NG | (38) |
NG, not given; NSCLC, non-small cell lung cancer.
Genotype frequency distributions
Table II displays the genotype frequency distributions of the five SNPs of lncRNA HOTAIR. Regarding HOTAIR rs128261786 and HOTAIR rs1899663, PHWE<0.05 was calculated for the study by Wang et al (35), indicating unbalanced data in terms of the two SNPs.
Table II.
Common gene polymorphisms of homeobox transcript antisense intergenic RNA in the included studies.
| A, rs920778 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | |||||||||
| Author (year) | Total | AA | AG | GG | Total | AA | AG | GG | PHWE | (Refs.) |
| Ren (2020) | 183 | 104 | 69 | 10 | 184 | 114 | 60 | 10 | 0.7416 | (33) |
| Minn (2020) | 1,241 | 698 | 473 | 70 | 129 | 75 | 53 | 1 | 0.3844 | (34) |
| Wang (2018) | 451 | 233 | 192 | 26 | 262 | 110 | 132 | 20 | 0.0947 | (35) |
| Li (2018) | 551 | 326 | 203 | 22 | 543 | 324 | 197 | 22 | 0.1640 | (36) |
| B, rs12826786 | ||||||||||
| Controls | Cases | |||||||||
| Author (year) | Total | AA | AG | GG | Total | AA | AG | GG | PHWE | (Refs.) |
| Wang (2018) | 451 | 355 | 85 | 11 | 262 | 200 | 49 | 13 | 0.0047 | (35) |
| Li (2018) | 551 | 381 | 156 | 14 | 543 | 340 | 185 | 18 | 0.6767 | (36) |
| Dadaş (2018) | 93 | 38 | 41 | 14 | 87 | 33 | 41 | 13 | 0.5934 | (37) |
| C, rs1899663 | ||||||||||
| Controls | Cases | |||||||||
| Author (year) | Total | AA | AG | GG | Total | AA | AG | GG | PHWE | (Refs.) |
| Ren (2020) | 188 | 123 | 60 | 5 | 187 | 131 | 52 | 4 | 0.4656 | (33) |
| Wang (2018) | 452 | 254 | 189 | 9 | 262 | 178 | 66 | 18 | 0.0001 | (35) |
| Dadaş (2018) | 93 | 41 | 40 | 12 | 87 | 39 | 37 | 11 | 0.6495 | (37) |
| D, rs4759314 | ||||||||||
| Controls | Cases | |||||||||
| Author (year) | Total | AA | AG | GG | Total | AA | AG | GG | PHWE | (Refs.) |
| Wang (2018) | 451 | 412 | 37 | 2 | 262 | 238 | 21 | 3 | 0.2463 | (35) |
| Ren (2020) | 184 | 168 | 16 | 0 | 175 | 158 | 17 | 0 | 0.5375 | (33) |
| Li (2018) | 551 | 489 | 61 | 1 | 543 | 526 | 16 | 1 | 0.5268 | (36) |
HWE, Hardy-Weinberg equilibrium.
Main findings
The association between different HOTAIR SNPs and lung cancer susceptibility is presented in Table III. No significant association with the susceptibility for lung cancer was obtained for SNPs rs12826786, rs4359714 and rs920778 (P>0.05 for all modes). However, the lncRNA HOTAIR rs1899663 C>A polymorphism was a risk factor for lung cancer (dominant mode, AA+CA vs. CC: OR=0.816, 95% CI=0.707-0.942, P=0.005). However, for the recessive mode, additive mode, heterozygote mode, homozygote mode and allele mode, no significant association with lung cancer was observed.
Table III.
Association of homeobox transcript antisense intergenic RNA gene polymorphisms and lung cancer risk.
| SNP/mode | Comparison | N | OR (95% CI) | P-value | I2 (%) | Model for meta-analysis |
|---|---|---|---|---|---|---|
| rs12826786 | ||||||
| Dominant | TT+CT vs. CC | 3 | 0.964 (0.793-1.172) | 0.716 | 56.30 | Random-effects |
| Recessive | TT vs. CC+CT | 3 | 1.101 (0.732-1.657) | 0.643 | 42.50 | Fixed-effects |
| Additive | TT+CC vs. TC | 3 | 1.043 (0.987-1.102) | 0.134 | 26.40 | Fixed-effects |
| Heterozygote | CT vs. CC | 3 | 0.899 (0.783-1.032) | 0.131 | 29.60 | Fixed-effects |
| Homozygote | TT vs. CC | 3 | 1.085 (0.730-1.613) | 0.686 | 49.90 | Fixed-effects |
| Allele | T vs. C | 3 | 0.990 (0.782-1.254) | 0.934 | 66.50 | Random-effects |
| rs1899663 | ||||||
| Dominant | AA+CA vs. CC | 3 | 0.816 (0.707-0.942) | 0.005 | 37.70 | Fixed-effects |
| Recessive | AA vs. CC+CA | 3 | 1.527 (0.552-4.225) | 0.415 | 72.10 | Random-effects |
| Additive | AA+CC vs. AC | 3 | 1.135 (0.969-1.331) | 0.117 | 69.90 | Random-effects |
| Heterozygote | CA vs. CC | 3 | 0.801 (0.607-1.056) | 0.116 | 64.50 | Random-effects |
| Homozygote | AA vs. CC | 3 | 1.382 (0.588-3.247) | 0.457 | 62.30 | Random-effects |
| Allele | A vs. C | 3 | 0.886 (0.762-1.030) | 0.115 | 0.00 | Fixed-effects |
| rs4759314 | ||||||
| Dominant | GG+AG vs. AA | 3 | 1.630 (0.720-3.690) | 0.241 | 85.00 | Random-effects |
| Recessive | GG vs. AA+AG | 2 | 1.933 (0.447-8.350) | 0.377 | 0.00 | Fixed-effects |
| Additive | AA+GG vs. AG | 3 | 0.966 (0.906-1.030) | 0.285 | 83.60 | Random-effects |
| Heterozygote | AG vs. AA | 3 | 1.612 (0.670-3.878) | 0.286 | 86.20 | Random-effects |
| Homozygote | GG vs. AA | 2 | 1.983 (0.456-8.617) | 0.361 | 0.00 | Fixed-effects |
| Allele | G vs. A | 3 | 1.649 (0.769-3.537) | 0.198 | 82.90 | Random-effects |
| rs920778 | ||||||
| Dominant | GG+AG vs. AA | 4 | 1.029 (0.902-1.173) | 0.670 | 53.60 | Random-effects |
| Recessive | GG vs. AA+AG | 4 | 0.914 (0.640-1.304) | 0.620 | 43.80 | Fixed-effects |
| Additive | AA+GG vs. AG | 4 | 0.967 (0.908-1.029) | 0.289 | 36.90 | Fixed-effects |
| Heterozygote | AG vs. AA | 4 | 1.050 (0.957-1.151) | 0.305 | 42.90 | Fixed-effects |
| Homozygote | GG vs. AA | 4 | 0.984 (0.555-1.747) | 0.957 | 53.60 | Random-effects |
| Allele | G vs. A | 4 | 1.008 (0.865-1.176) | 0.917 | 57.70 | Random-effects |
SNP, single nucleotide polymorphism; OR, odds ratio.
The sensitivity analysis suggested that the omission of any single study did not significantly alter the overall pooled ORs in dominant mode, recessive mode, additive mode, heterozygote mode, homozygote mode and allele mode (Fig. 2).
Figure 2.
Sensitivity analysis for each single nucleotide polymorphism. The sensitivity was performed by omitting one study at a time. In each row of the forest plot, one study was omitted. Sensitivity analysis was performed for rs12826786, rs1899663, rs4759314 and rs920778 for all modes including (A) dominant mode, (B) recessive mode, (C) additive mode, (D) heterozygote mode, (E) homozygote mode and (F) allele mode.
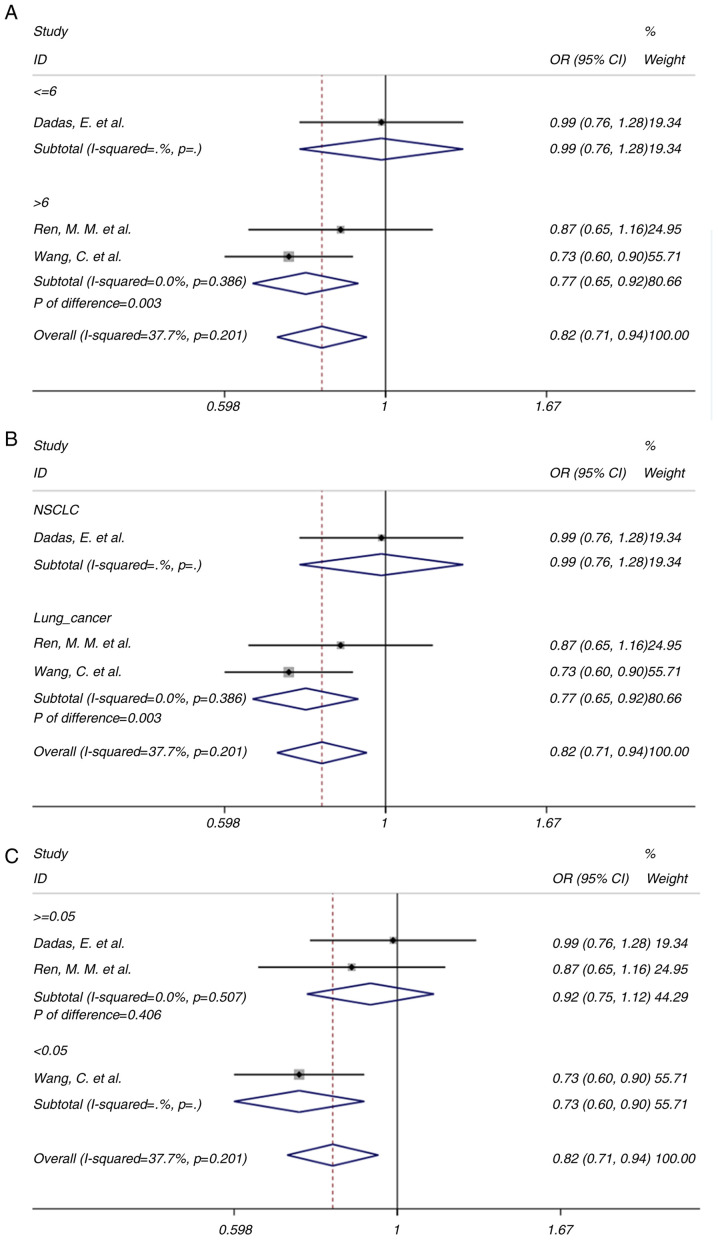
Since the lncRNA HOTAIR rs1899663 C>A polymorphism was identified as a risk factor for lung cancer (OR=0.816; Fig. 3A), a further analysis was performed, including a publication bias analysis and subgroup analysis in terms of rs1899663 (dominant mode, AA+CA vs. CC). The P-value as calculated using Egger's test (Fig. 3B) was 0.466 and the funnel plot indicated a good symmetry (Fig. 3C). These results suggested that no significant publication bias was present. In the further subgroup analysis, the studies with higher quality (NOS score >6) also suggested a significant association of the rs1899663 C>A polymorphism with the risk of lung cancer (OR=0.77, 95% CI=0.65-0.92, P=0.003; Fig. 4A). Similar results were obtained in the subgroup of cancer type (lung cancer) (Fig. 4B). As for the studies with PHWE≥0.05 (Fig. 4C), the trend for the risk of lung cancer was similar (OR=0.92, 95% CI=0.75-1.12, P=0.406). As for the study by Wang et al (35) with PHWE<0.05, a significant association was observed (OR=0.73, 95% CI=0.60-0.90, P=0.003). Of note, in each of the subgroups, no heterogeneity was detected (I2=0 for all subgroups with the number of studies >1), indicating that the NOS quality, type of cancer and gene frequency disequilibrium may be potential sources of heterogeneity. However, the overall heterogeneity for the rs1899663 C>A polymorphism was minor (I2=37.7%, P=0.201).
Figure 3.

Analysis for the rs1899663 C>A polymorphism. (A) The forest plot indicates the pooled OR for the effect of the rs1899663 C>A polymorphism on the risk of lung cancer. (B) Egger's test plot and (C) funnel plot for assessment of publication bias. OR, odds ratio; SND, standard normal distribution; se, standard error.
Figure 4.
Subgroup analysis. Subgroup analysis was performed based on (A) Newcastle-Ottawa Scale score quality assessment, (B) type of cancer and (C) P-value of Hardy-Weinberg equilibrium. OR, odds ratio; NSCLC, non-small cell lung cancer.
Discussion
The present study indicated that the lncRNA HOTAIR rs1899663 C>A polymorphism is a risk factor for lung cancer. In previous studies on the topic of HOTAIR-associated lung cancer susceptibility, two or three SNPs were usually investigated in an individual study. However, at least four SNPs (rs128261786, rs1899663, rs920778 and rs4759314) of HOTAIR have been reported in the lung cancer population. The present meta-analysis included all of these SNPs into the analysis and therefore arrived at a more comprehensive conclusion. The previous results remain controversial in that each SNP has been reported to be related to lung cancer in one or two of these previous studies. However, in the present meta-analysis, these data were integrated and according to the results, only rs1899663 was a significant risk factor. To the best of our knowledge, the present study was the first systematic review and meta-analysis focusing on the association between lncRNA HOTAIR and lung cancer susceptibility that has integrated all these four SNPs into its analysis, which is the novelty of this work.
Genetic differences in DNA sequences lead to phenotypic variation, which affects changes in phenotypic characteristics, disease risks and responses to the environment (39). At the same time, human genetic diversity is not limited to individual polymorphisms, but also to the specific combinations of tightly linked alleles. The most common form of gene polymorphism is SNP, which refers to the polymorphism in the DNA sequence caused by the variation of a single nucleotide base in the genome. The types of SNP include conversion, transversion, insertion and deletion (40). SNP is associated with the function and expression level of the downstream protein by changing the protein-coding sequence and influencing the splicing changes of exons and introns, thereby affecting the individual's susceptibility to disease, prognosis and responsiveness to drugs (41).
To date, only a small number of studies have reported on the relationship between the SNP of the HOTAIR sequence and the susceptibility to malignancies. A study reported an enhancer-like sequence on the intron 1,719-2,353 bp downstream of the transcription start position of HOTAIR (42). There was a risk site for esophageal squamous cell carcinoma, rs920778, carrying the rs920778T allele. The expression level of the reported gene was higher than that of the rs920778C allele. This study also performed SNP typing and quantitative PCR using esophageal squamous cell carcinoma tissues and determined that individuals with the genotype rs920778TT had higher expression of HOTAIR than patients carrying rs920778CC (42). A study on papillary thyroid carcinoma indicated that in females, the rs920778TT and rs920778CT genotype increased the risk of developing papillary thyroid carcinoma by 0.75- and 0.46-fold, respectively (43). The study also verified the effect of the rs920778 locus polymorphism on HOTAIR expression in cells and tissues. It was indicated that cells carrying rs920778T alleles had higher HOTAIR expression than those with rs920778C alleles. In thyroid squamous cell carcinoma tissues and normal tissues, the expression of HOTAIR in cancer tissues and adjacent normal tissues of rs920778TT and rs920778CT genotype carriers was higher than that in rs920778CC genotype carriers (43). Previous results suggested that the expression of HOTAIR in individuals carrying the rs920778T allele was higher than that in individuals with rs920778C (44). Except for the proto-oncogene role in lung cancer, HOTAIR has also been indicated to be related to resistance to chemotherapy. Liu et al (45) reported that HOTAIR expression was significantly upregulated in cisplatin-resistant lung cancer cells and that knockdown of HOTAIR by RNA interference was able to resensitize the responses to cisplatin both in vitro and in vivo. On the other hand, overexpression of HOTAIR decreased the sensitivity of lung cancer cells to cisplatin. The HOTAIR-associated chemosensitivity regulation was indicated to involve the inhibition of cell proliferation, induction of G0/G1 cell-cycle arrest and apoptosis enhancement through regulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1 (p21) expression (45). Accordingly, another clinical investigation by the same group (28) suggested that high levels of HOTAIR expression were correlated with a more advanced pathological stage of non-small cell lung cancer (NSCLC), lymph-node metastasis, as well as a relatively poor prognosis. The upregulated HOTAIR may regulate NSCLC cell invasion and metastasis, partially via downregulation of HOXA5(28). In the present study, a critical literature review was performed to further understand the mechanisms of HOTAIR in cancer development. Apart from the p21WAF1/CIP1 and HOXA5 pathways, the most commonly acknowledged mechanism of HOTAIR is through the axis of microRNA (miR) and downstream signaling molecules. These axes include but are not limited to the following: HOTAIR/miR-222-3p/Cyclin Dependent Kinase 19 axis (46), HOTAIR/specificity protein 1/miR-199a axis (47), HOTAIR/miR-29b/phosphate and tension homology deleted on chromosome 10/PI3K axis (48), HOTAIR/miR-203/zinc finger E-box binding homeobox 1 axis (49), HOTAIR/estrogen receptor 1/miR-130b-3p axis (50), HOTAIR/miR-34a/Janus kinase 2/STAT3 axis (51), HOTAIR/miR-129-5p/ribosomal protein L14 axis (52), HOTAIR/miR-129-5p/frizzled class receptor 7 axis (53) and HOTAIR/miR-149-5p/doublecortin-like kinase 1 axis (54). Furthermore, HOTAIR also interacts with several classic signaling pathways, which are commonly involved in oncogenesis, such as CCL22 signaling (55), the Wnt/β-catenin signaling pathway (56), EZH2 and H3K27 methylation signaling (57). All of this evidence, as well as that provided by the present meta-analysis, suggests the importance of lncRNA HOTAIR SNP in the development of lung cancer.
Of note, there are certain limitations to the present study. First, due to the small sample of the included studies, it was not possible to analyze certain SNPs because only one study reported the respective relation. Furthermore, lung cancer susceptibility was indicated to be associated with the individual characteristics, such as lifestyle and family history, which was not possible to be extracted. In addition, the majority of the populations were from Asia and additional data are required for other populations. This work was not entered in registries such as the Cochrane Library or PROSPERO, yet the present meta-analysis was performed based on the PRISMA guidelines.
In conclusion, the present study was the first to systematically explore the association between lncRNA HOTAIR SNP and lung cancer susceptibility. It was indicated that populations carrying the lncRNA HOTAIR rs1899663 C>A polymorphism may have a high risk of developing lung cancer. LncRNA HOTAIR may be an important novel target in lung cancer prevention and prognosis prediction and future investigations are required to clarify the intrinsic mechanism of HOTAIR in lung cancer.
Supplementary Material
Acknowledgements
We thank our collaborator, Professor Jianjun Qiao (Department of Dermatology, the First Affiliated Hospital of Zhejiang University Medicine School, Hangzhou, China), for language and scientific editing support.
Funding Statement
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
Design of the meta-analysis: CK, XF and XH. Literature screening: CK and XF. Quality assessment: SC and XH. Check and approving the authenticity of the raw data: CK and XF. Statistical analysis: CK and XF and JL. Manuscript writing and revision: CK, XF, JL, SC and XH. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019;85(8) doi: 10.5334/aogh.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer. 2019;10:3–7. doi: 10.1111/1759-7714.12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taghvaee S, Sowlat MH, Hassanvand MS, Yunesian M, Naddafi K, Sioutas C. Source-specific lung cancer risk assessment of ambient PM2.5-bound polycyclic aromatic hydrocarbons (PAHs) in central Tehran. Environ Int. 2018;120:321–332. doi: 10.1016/j.envint.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Brasky TM, White E, Chen CL. Long-term, supplemental, one-carbon metabolism-related vitamin B use in relation to lung cancer risk in the vitamins and lifestyle (VITAL) cohort. J Clin Oncol. 2017;35:3440–3448. doi: 10.1200/JCO.2017.72.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hicks BM, Filion KB, Yin H, Sakr L, Udell JA, Azoulay L. Angiotensin converting enzyme inhibitors and risk of lung cancer: Population based cohort study. BMJ. 2018;363(k4209) doi: 10.1136/bmj.k4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin KF, Wu HF, Huang WC, Tang PL, Wu MT, Wu FZ. Propensity score analysis of lung cancer risk in a population with high prevalence of non-smoking related lung cancer. BMC Pulm Med. 2017;17(120) doi: 10.1186/s12890-017-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortunato O, Borzi C, Milione M, Centonze G, Conte D, Boeri M, Verri C, Moro M, Facchinetti F, Andriani F, et al. Circulating mir-320a promotes immunosuppressive macrophages M2 phenotype associated with lung cancer risk. Int J Cancer. 2019;144:2746–2761. doi: 10.1002/ijc.31988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The ENCODE (ENCyclopedia of DNA elements) project. Science. 2004;306:636–340. doi: 10.1126/science.1105136. ENCODE Project Consortium. [DOI] [PubMed] [Google Scholar]
- 10.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Shao A, Wang L, Hu K, Yu C, Pan C, Zhang S. The role of lncRNAs in the distant metastasis of breast cancer. Front Oncol. 2019;9(407) doi: 10.3389/fonc.2019.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Tang L. The application of lncRNAs in cancer treatment and diagnosis. Recent Pat Anticancer Drug Discov. 2018;13:292–301. doi: 10.2174/1574892813666180226121819. [DOI] [PubMed] [Google Scholar]
- 13.Lu T, Wang Y, Di Chen JL, Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther. 2018;11:8045–8052. doi: 10.2147/OTT.S178431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engle L, Simpson C, Landers J. Using high-throughput SNP technologies to study cancer. Oncogene. 2006;25:1594–1601. doi: 10.1038/sj.onc.1209368. [DOI] [PubMed] [Google Scholar]
- 15.Syvänen AC. Toward genome-wide SNP genotyping. Nat Genet. 2005;37 (Suppl):S5–S10. doi: 10.1038/ng1558. [DOI] [PubMed] [Google Scholar]
- 16.Andrew AS, Gui J, Sanderson AC, Mason RA, Morlock EV, Schned AR, Kelsey KT, Marsit CJ, Moore JH, Karagas MR. Bladder cancer SNP panel predicts susceptibility and survival. Hum Genet. 2009;125:527–539. doi: 10.1007/s00439-009-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- 18.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7(90) doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 22.Somarowthu S, Legiewicz M, Chillón I, Marcia M, Liu F, Pyle AM. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58:353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang Y, Wang X, Nguyen HT, Zhuo Y, Cui X, Fewell C, Flemington EK, Shan B. Induction of long intergenic non-coding RNA HOTAIR in lung cancer cells by type I collagen. J Hematol Oncol. 2013;6(35) doi: 10.1186/1756-8722-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Wang X, Zhang L, Yu J, Hao X. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 27.Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, Yu PF, Cheng XD. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13(464) doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao W, An Y, Liang Y, Xie XW. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:1930–1936. [PubMed] [Google Scholar]
- 30.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T, Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 31.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009;6(e1000097) doi: 10.1371/journal.pmed.1000097. PRISMA Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 33.Ren MM, Xu S, Wei YB, Yang JJ, Yang YN, Sun SS, Li YJ, Wang PY, Xie SY. Roles of HOTAIR in lung cancer susceptibility and prognosis. Mol Genet Genomic Med. 2020;8(e1299) doi: 10.1002/mgg3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minn AKK, Sato N, Mieno MN, Arai T, Muramatsu M. Association study of long non-coding RNA HOTAIR rs920778 polymorphism with the risk of cancer in an elderly Japanese population. Gene. 2020;729(144263) doi: 10.1016/j.gene.2019.144263. [DOI] [PubMed] [Google Scholar]
- 35.Wang C, Li Y, Li YW, Zhang HB, Gong H, Yuan Y, Li WT, Liu HY, Chen J. HOTAIR lncRNA SNPs rs920778 and rs1899663 are associated with smoking, male gender, and squamous cell carcinoma in a Chinese lung cancer population. Acta Pharmacol Sin. 2018;39:1797–1803. doi: 10.1038/s41401-018-0083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Yang Z, Li J, Lv X, Gao M, Bi Y, Zhang Z, Wang S, Li S, Li N, et al. Genetic variants in lncRNA HOTAIR are associated with lung cancer susceptibility in a Chinese Han population in China: A case-control study. Cancer Manag Res. 2018;10:5209–5218. doi: 10.2147/CMAR.S175961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dadaş E, Aydin M. Effect of HOTAIR rs12826786 and rs1899663 polymorphisms on lung cancer susceptibility and clinicopathological characteristics in a Turkish population: A hospital-based case-control study. Cell Mol Biol (Noisy-le-grand) 2018;64:97–102. [PubMed] [Google Scholar]
- 38.Gong WJ, Yin JY, Li XP, Fang C, Xiao D, Zhang W, Zhou HH, Li X, Liu ZQ. Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016;37:8349–8358. doi: 10.1007/s13277-015-4497-5. [DOI] [PubMed] [Google Scholar]
- 39.Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392:777–786. doi: 10.1016/S0140-6736(18)31268-6. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu Y, Sato S, Noguchi Y, Koyamatsu J, Yamanashi H, Higashi M, Nagayoshi M, Kadota K, Kawashiri SY, Nagata Y, et al. Impact of single nucleotide polymorphism on short stature and reduced tongue pressure among community-dwelling elderly Japanese participants: A cross-sectional study. Environ Health Prev Med. 2017;22(62) doi: 10.1186/s12199-017-0668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leaché AD, Oaks JR. The utility of single nucleotide polymorphism (SNP) data in phylogenetics. Annu Rev Ecol Evol Syst. 2017;48:69–84. [Google Scholar]
- 42.Zhang X, Zhou L, Fu G, Sun F, Shi J, Wei J, Lu C, Zhou C, Yuan Q, Yang M. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis. 2014;35:2062–2067. doi: 10.1093/carcin/bgu103. [DOI] [PubMed] [Google Scholar]
- 43.Zhu H, Lv Z, An C, Shi M, Pan W, Zhou L, Yang W, Yang M. Onco-lncRNA HOTAIR and its functional genetic variants in papillary thyroid carcinoma. Sci Rep. 2016;6(31969) doi: 10.1038/srep31969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen H, Zhou L, Yuan Q, Zhou C, Yang M. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog. 2016;55:90–96. doi: 10.1002/mc.22261. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, De W, Wang Z, Wang R. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One. 2013;8(e77293) doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan L, Lei H, Lin Y, Zhou Z, Li J, Wu A, Shu G, Ruger S, Yin G. Hotair promotes the migration and proliferation in ovarian cancer by miR-222-3p/CDK19 axis. Cell Mol Life Sci. 2022;79(254) doi: 10.1007/s00018-022-04250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Hou SF, Tang FJ, Liu DS, Chen ZZ, Zhang HL, Wang SH. HOTAIR/Sp1/miR-199a critically regulates cancer stemness and malignant progression of cutaneous squamous cell carcinoma. Oncogene. 2022;41:99–111. doi: 10.1038/s41388-021-02014-x. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Wu Q, Liu Y, Wang X, Ma C, Zhu W. LncRNA HOTAIR promotes chemoresistance by facilitating epithelial to mesenchymal transition through miR-29b/PTEN/PI3K signaling in cervical cancer. Cells Tissues Organs. 2022;211:16–29. doi: 10.1159/000519844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W, Liu J, Wu Q, Liu Y, Ma C. HOTAIR contributes to stemness acquisition of cervical cancer through regulating miR-203 interaction with ZEB1 on epithelial-mesenchymal transition. J Oncol. 2021;2021(4190764) doi: 10.1155/2021/4190764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang M, Wu K, Zhang P, Qiu Y, Bai F, Chen H. HOTAIR facilitates endocrine resistance in breast cancer through ESR1/miR-130b-3p axis: Comprehensive analysis of mRNA-miRNA-lncRNA network. Int J Gen Med. 2021;14:4653–4663. doi: 10.2147/IJGM.S320998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng S, Wang J, Zhang L, Li J, Jin Y. LncRNA HOTAIR promotes cancer stem-like cells properties by sponging miR-34a to activate the JAK2/STAT3 pathway in pancreatic ductal adenocarcinoma. Onco Targets Ther. 2021;14:1883–1893. doi: 10.2147/OTT.S286666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun N, Zhang W, Liu J, Yang X, Chu Q. Propofol inhibits the progression of cervical cancer by regulating HOTAIR/miR-129-5p/RPL14 axis. Onco Targets Ther. 2021;14:551–564. doi: 10.2147/OTT.S279942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu D, Zhu J, Fu Y, Li C, Wu B. LncRNA HOTAIR promotes breast cancer progression through regulating the miR-129-5p/FZD7 axis. Cancer Biomark. 2021;30:203–212. doi: 10.3233/CBM-190913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhan Y, Abuduwaili K, Wang X, Shen Y, Nuerlan S, Liu C. Knockdown of long non-coding RNA HOTAIR suppresses cisplatin resistance, cell proliferation, migration and invasion of DDP-resistant NSCLC cells by targeting miR-149-5p/doublecortin-like kinase 1 axis. Cancer Manag Res. 2020;12:7725–7737. doi: 10.2147/CMAR.S246299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang H, Peng J. LncRNA HOTAIR promotes proliferation, invasion and migration in NSCLC cells via the CCL22 signaling pathway. PLoS One. 2022;17(e0263997) doi: 10.1371/journal.pone.0263997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang Y, Song G, Liu H, Yang S, Yu X, Shi L. Silencing of long non-coding RNA HOTAIR alleviates epithelial-mesenchymal transition in pancreatic cancer via the Wnt/β-catenin signaling pathway. Cancer Manag Res. 2021;13:3247–3257. doi: 10.2147/CMAR.S265578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai ZY, Jin SM, Luo HQ, Leng HL, Fang JD. LncRNA HOTAIR regulates anoikis-resistance capacity and spheroid formation of ovarian cancer cells by recruiting EZH2 and influencing H3K27 methylation. Neoplasma. 2021;68:509–518. doi: 10.4149/neo_2021_201112N1212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.