Abstract
Cancer cells can be conceived as “living organisms” interacting with cellular or non‐cellular components in the host internal environment, not only the local tumor microenvironment but also the distant organ niches, as well as the immune, nervous and endocrine systems, to construct a self‐sustainable tumor ecosystem. With increasing evidence for the systemic tumor‐host interplay, we predict that a new era of cancer therapy targeting the ecosystemic vulnerability of human malignancies has come. Revolving around the tumor ecosystem scoped as different hierarchies of primary, regional, distal and systemic onco‐spheres, we comprehensively review the tumor‐host interaction among cancer cells and their local microenvironment, distant organ niches, immune, nervous and endocrine systems, highlighting material and energy flow with tumor ecological homeostasis as an internal driving force. We also substantiate the knowledge of visualizing, modelling and subtyping this dynamically intertwined network with recent technological advances, and discuss ecologically rational strategies for more effective cancer therapies.
Keywords: ecological therapy, neuroendocrine system, onco‐sphere, tumor‐host interplay, pre‐metastatic niche, tumor ecosystem, tumor immunity, tumor microenvironment
Abbreviations
- ANGPTL4
angiopoietin‐like 4
- APCs
antigen‐presenting cells
- ASO
antisense oligonucleotide
- ATP
adenosine triphosphate
- BCS
breast‐conserving surgery
- BMDCs
bone marrow‐derived cells
- CAFs
cancer‐associated fibroblasts
- CAR T
chimeric antigen receptor CAR T cell
- CCDC25
coiled‐coil domain containing 25
- CCL
C‐C motif chemokine ligand
- CCR
C‐C motif chemokine receptor
- cGAS
cyclic GMP‐AMP synthase
- CSCs
cancer stem cells
- CSF‐1
colony‐stimulating factor‐1
- CSF‐1R
colony‐stimulating factor‐1 receptor
- CTL
cytotoxic T lymphocyte
- CTLA4
cytotoxic T‐lymphocyte‐associated protein 4
- CXCL
C‐X‐C motif chemokine ligand
- CXCR
C‐X‐C motif chemokine receptor
- DC
dendritic cell
- ECM
extracellular matrix
- EGF
epidermal growth factor
- GM‐CSF
granulocyte‐macrophage colony‐stimulating factor
- HER2
human epidermal growth factor receptor 2
- HIF‐1α
hypoxia inducible factor‐1 α
- HLA
human leukocyte antigen
- HPA
hypothalamic‐pituitary‐adrenal
- ICB
immune checkpoint blockade
- ICIs
immune checkpoint inhibitors
- IFN
interferon
- IL‐1R8
interleukin‐1 receptor 8
- IL‐6
interleukin‐6
- IL‐8
interleukin‐8
- KIRs
killer cell immunoglobulin‐like receptors
- LGALS3
Galactoside‐binding soluble 3
- LOX
lysyl oxidases
- mAb
monoclonal antibody
- MDSCs
myeloid‐derived suppressor cells
- MEK
mitogen‐activated extracellular signal‐regulated kinase
- MHC
major histocompatibility complex class
- MMPs
matrix metalloproteinases
- mTOR
mammalian target of rapamycin
- NAC
neoadjuvant chemotherapy
- NK
natural killer
- NKG2A
natural killer group 2A
- NSCLC
non‐small cell lung cancer
- OXPHOS
oxidative phosphorylation
- PARP
poly ADP‐ribose polymerase
- PCa
Prostate cancer
- PD 1
programmed cell death 1
- PDGFR
platelet‐derived growth factor receptor
- PD‐L1
programmed cell death 1 ligand 1
- PDX
patient‐derived xenograft
- PERK
protein‐kinase‐R‐like endoplasmic reticulum kinase
- PitNET
pituitary neuroendocrine tumor
- PMNs
pre‐metastatic niches
- PPP
pentose phosphate pathway
- ROCK
Rho‐associated kinase
- ROS
reactive oxygen species
- RT
radiation therapy
- RTK
receptor tyrosine kinase
- SCLC
small‐cell lung cancer
- SNS
sympathetic nervous system
- STING
stimulator of interferon genes
- TAMs
tumor‐associated macrophages
- TCA
tricarboxylic acid
- TGFβ
transforming growth factor β
- Th1/2
T helper 1/2
- TIGIT
T cell immunoreceptor with immunoglobulin and ITIM domains
- TIL
tumor‐infiltrating lymphocyte
- TKIs
tyrosine kinase inhibitors
- TME
tumor microenvironment
- TNFα
tumor necrosis factor α
- Tregs
Regulatory T cells
- VEGF
vascular endothelial growth factor
- VEGFR
vascular endothelial growth factor receptor
- VGCC
voltage‐gated calcium channels
1. BACKGROUND
Cancer is a heterogeneous disease of clonal evolution, as the tumor cells with genetic drift may acquire selective advantages that allow for rapid adaptation to external stress and survive, termed as “survival of the fittest” [1]. This evolution‐like process selects for more potent tumor clones to expand in an alien habitat, which are also prone to be therapy‐resistant. Generally, the fitness of neoplastic cells is shaped by their interactions with the surrounding niche and the host systemic environment respectively, which, in essence, is analogous to the interplay between living organisms and the ecosystem. Adopting an ecological viewpoint, most researchers believe that cancer is a complex cellular ecosystem in which malignant cells co‐exist and collaborate with other host cells within their microenvironment [2, 3]. Moreover, single‐cell genomic has been widely applied for direct mapping of these localized “tumor ecosystem” [4, 5]. With a more holistic view, cancer ecosystem can also be perceived as a broad and intercrossing host‐tumor biosphere, in which the “living organisms”, as tumor cells, and their local/distal “living habitats”, as tumor microenvironment (TME) and distant organ niches, along with the internal/external stimuli (e.g., cytokines, hormones, neural mediators, nutrition) work in concert to promote malignant evolvement of cancer cells (Table 1) [6, 7]. Since this concept has extended beyond the TME, we herein introduce the term onco‐sphere, which differs from “oncosphere” ‐ a tapeworm embyo. In this way, onco‐spheres can be classified among niches where tumor initiates, expands and spreads, namely the primary, regional (lymph nodes), distal (pre/post‐metastatic) as well as systemic onco‐spheres accordingly (Figure 1).
TABLE 1.
Tumor ecosystem paralleling to the Earth's biosphere
| Ecology | Natural ecosystem | Artificial ecosystem | Tumor ecosystem |
|---|---|---|---|
| Biosphere | Earth | Earth | Patient |
| Ecosystem | Aquatic and terrestrial ecosystem | Urban ecosystem | Tissue and organ system |
| Metacommunities | Zoobenthos | Cities | Metastases |
| Species | Animals and plants | Humans | Cancer cells and host cells |
| Biotope | Water or land | Land | Extracellular matrix |
| Biogas | Swamp gas | Smoke and greenhouse gases | Cytokines, hormones and neurotransmitters |
| Abiotic factors | Land or water | Technology, transportation and infrastructure | Oxygen, acidity and therapeutic intervention |
| Energy source | Solar energy | Natural ecosystems | Food |
| Nutrient cycling | Biogeochemical cycling | Material supply and waste disposal | Intercellular signaling and metabolic interplay |
| Ecosystem collapse | Mass extinction | Urban pollution or mass migration | Organ failure and death |
FIGURE 1.
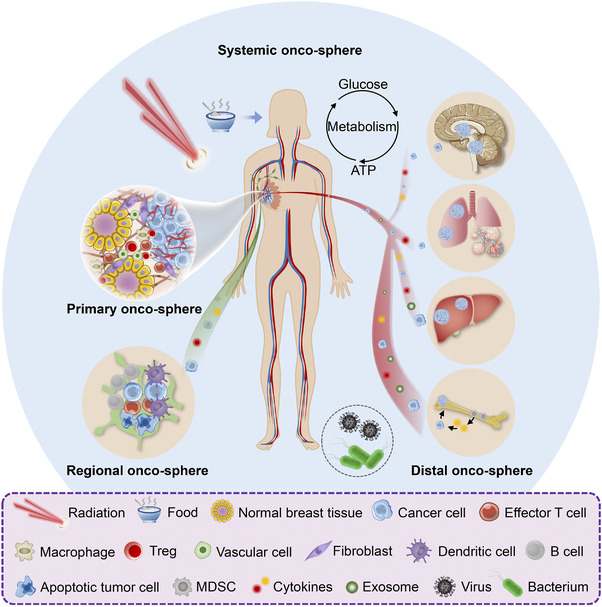
The hierarchy of tumor ecosystem. Taking breast cancer as an example, the patient's systemic environment can be perceived as an integral ecosystem and scoped at three different levels: primary (cancerous breast), regional (metastatic lymph node) and distal (metastatic brain, lung, liver or bone) onco‐spheres. The material and energy flow throughout the systemic onco‐sphere are largely ascribed to its metabolic function within the host macroenvironment. Besides internal factors, food and nutrient, microorganisms (e.g., viruses and bacteria) and therapeutic interventions (e.g., irradiation) serve as external stimuli that also shape the tumor ecosystem. Abbreviations: Tregs, Regulatory T cells; MDSCs, myeloid‐derived suppressor cells.
Employing the principles of ecology, cancer cells act as the emerging species that employ different metabolic and reproductive strategies to hijack resources and space from the existing host cells, evade/defense predation by the host immunity, and cooperate to disperse throughout the circulation, followed by co‐evolution with new onco‐spheres and end up colonizing as macroscopic outgrowths [8]. Three essential factors are critical for the onco‐sphere formation: energy source, microenvironment, and interactive network. Energy is vital for cells to survive and thrive, sourcing from biomolecules such as glucose, proteins and lipids, or elements like iron and phosphorus. Of note, the material circulation and energy flow, largely dependent on the metabolic interplay among cells, serves as a driving force to maintain the tumor ecological homeostasis, resembling the biogeochemical cycle to the natural ecosystem. In the onco‐spheres, the extracellular space can be invaded, colonized and remodeled by neoplastic cells and their accomplices. In return, multifarious environmental selective pressures are reshaping the malignant traits of tumor cells. The cellular interactions that either benefit cancer growth (e.g., commensalism, mutualism) or counteract cancer development (e.g., predation, parasitism and competition) largely determine their ecological roles in different onco‐spheres. Evolutionarily, neoplastic cells cooperate by sharing diffusible factors which enable individual cell to bypass the need to gather all the cancer hallmarks one by one [9]. The mutualistic relationship between tumor cells and activated stromal cells also confers fitness advantages for both of them to co‐evolve [10, 11]. Moreover, the model of tumor and antitumor immunity may resemble that of prey and predator. In nature, predators grow and thrive by feeding on the prey population, while in the tumor ecosystem, cancer cells, the prey, can develop a wide range of counter‐adaptations to evade from immune predation. Unlike predation with an abrupt gain or loss, parasitism is characterized by propagation without causing immediate death to the host. Similarly, cancer cells hijack nearby tissues for spatial and metabolic benefits, as they can exploit growth factors, command neo‐angiogenesis and break down the extracellular matrix (ECM) for collective invasion. Competition exists not only within heterogeneous cancer populations but also among cancer cells and stromal cells, while fighting for living resources like oxygen and energy. Besides, miscellaneous selective pressures (e.g., nutrient deprivation, hypoxia, oxidative stress, augmented stiffness, lifestyle deterioration and therapeutic interventions) might trigger metabolic rewiring in cancerous and stromal cells to compensate for the hostile extracellular milieu, further reprogramming the onco‐sphere more hospitable for malignant growth [12].
The concept of cancer ecosystem extends and deepens our understanding of cancer pathobiology beyond its local interplay with the TME. Indeed, tumor cells interact with the host macroenvironment via systemic nervous and endocrine signaling, manifesting as paraneoplastic syndromes. Herein, we focus on systemic tumor‐host interaction among cancer cells and their microenvironment, distal organ niches, immune, nervous and endocrine system, scoping tumor ecosystem as different hierarchies of onco‐spheres. With successful visualizing, modelling and subtyping of the tumor ecosystem, we also discuss ecologically rational strategies to develop novel targeted therapies for cancer treatment.
2. ADAPTATIONS TO THE LOCAL MILIEU
The TME is composed of a variety of non‐tumoral cells as well as ECM. Herein, we termed the organization of tumor cells and their surrounding milieu as primary onco‐sphere. Among all the abnormal biological events that constitute cancer hallmarks, dysregulated tumor‐stroma interplay stands out. Cytokine shuttling, exosome transmission, metabolite diffusion together with direct cell‐cell contact enable constant intercellular communications, eliciting a wide array of malignant behaviors [13, 14]. Generally, information and energy flowing among various cell types in the onco‐spheres is fulfilled by signal transduction mediated by receptor ligation. Analogous to the commensal behaviors among different species, cancer‐derived mediators can recruit, activate or educate stromal cells, endowing them with tumor‐promoting phenotypes that favor angiogenesis, inflammation, and invasion. For example, Rho‐associated kinase (ROCK) released by breast tumor epithelia can induce fibroblast reprogramming and promote tumor progression through selective activation of protein‐kinase‐R‐like endoplasmic reticulum kinase (PERK) signaling [15].
2.1. Cancer‐stromal cell interaction
In many solid malignancies, cancer‐associated fibroblasts (CAFs) are the most abundant stromal cells that populate the onco‐sphere. With diverse cellular origins, these phenotypically heterogeneous fibroblasts evolve highly plastic functions during multistep cancer development, which we have comprehensively reviewed elsewhere [16]. For instance, a pro‐tumorigenic CAF subset (CD10+GPR77+) provides a protecting niche for cancer stem cells (CSCs) against chemotherapy in breast and lung cancers [17]. Besides interleukin‐6 (IL‐6) and IL‐8, chemokines and their cognate receptors stand at the crossroads of tumor‐CAF crosstalk, as CAFs activated by cancer‐releasing factors can secret a significant level of chemokines to act reciprocally on the cancer cells, promoting their malignancy [18]. Notably, CAFs release C‐X‐C motif chemokine ligand (CXCL)12 to exert a proliferation‐promoting effect on cancer cells expressing C‐X‐C motif chemokine receptor (CXCR)4 [19].
Tumor‐associated macrophages (TAMs) orchestrate tumor‐promoting inflammation, as they preferentially reside in the poorly vascularized tumor‐host tissue interface, in regions characteristic of hypoxia and where invasive events emerge [20]. Tumor cells often interact with macrophages by escaping phagocytosis and by polarizing them toward M2‐like phenotype via chemokines and cytokines such as C‐C motif chemokine ligand (CCL)2, transforming growth factor (TGF)β and IL‐10. Upon TAM activation, a vast diversity of growth factors, proteolytic enzymes, and other inflammatory mediators strongly involved in the cancerous program are released into the onco‐sphere. Among them, vascular endothelial growth factor (VEGF)‐A stimulates blood vessel formation for rapid tumor growth [21]. Reciprocal secretion of epidermal growth factor (EGF) by perivascular TAMs and colony‐stimulating factor (CSF)‐1 by cancer cells enhances intravasation of tumor cells [22]. Likewise, mesenchymal‐like cancer cells secrete granulocyte‐macrophage colony‐stimulating factor (GM‐CSF) to activate TAMs, which, in turn, induce the epithelial‐mesenchymal transition in cancer cells through CCL18‐mediated nuclear factor kappa‐B (NF‐κB) signaling, in a positive‐feedback loop [23, 24].
Additionally, neutrophils, mast cells and lymphocytes are also listed on the roster of tumor‐associated inflammatory cells, which coordinate with cancer cells to contribute greatly to the inflammatory and immunosuppressive state of the primary onco‐sphere [25, 26, 27]. Inflammation has been linked to tumorigenesis in a positive feed‐forward manner, as activated inflammatory cells produce reactive oxygen species and reactive nitrogen intermediates to induce mutations in cancer cells, and the resultant DNA damage reroutes the inflammatory response toward a pro‐tumor direction [28, 29].
2.2. Cancer‐vasculature interaction
Tumor angiogenesis is a fine‐tuned process that balances the pro‐ and anti‐angiogenic factors, beginning with the chemotaxis of endothelial cells and pericytes [21]. While VEGF‐A serves as a major angiogenic factor, thrombospondin‐1 is a key angiogenesis inhibitor, whose expression is tightly controlled either by overexpression of ras oncogene in the epithelial cells or by inactivation of tumor suppressors (e.g., tumor protein p53 and prepro‐retinoblastoma‐associated protein) in fibroblasts [30]. Of note, commensalism occurs between tumor cells and other cells in their vicinity, as neo‐angiogenesis brings ample nourishing factors (an abundant supply of blood, oxygen and diffusible factors) to the shared onco‐sphere. Being the blood vessel‐supporting cells on one hand, tumor vascular mural cells also crosstalk with tumor cells via paracrine secretion to modulate cancer growth, development and drug resistance [31, 32].
2.3. Cancer‐ECM interaction
As a principal non‐cellular component, the ECM not only provides cells with architectural and mechanical support within the onco‐sphere but also serves as a diverse reservoir of regulators involved in multiple cellular processes [33]. Malignant and stromal cells can deposit, break down and remodel the ECM through production of multitudinous ECM proteins, including collagens, fibronectins, laminins and proteolytic enzymes, which reciprocally fine‐tune the abnormal behaviors of these cells [34]. Specifically, biochemical changes within the ECM composition such as hydroxylation and enzymatic crosslinking can restructure the spatial compartments to release locally sequestered factors (e.g., VEGF‐A) into the primary onco‐sphere [35]. In addition, the biophysical properties of ECM, including stiffness, density, rigidity and tension, pose indispensable effects on the migratory phenotype and collective invasion of cancer cells [36]. Matrix degradation and turnover can be orchestrated by matrix‐degrading proteinases, notably matrix metalloproteinases (MMPs), allowing neo‐angiogenesis and further tumor invasion [37].
As such, tumor‐associated stromal cells and extracellular cues interacting with cancer cells synergistically construct a heterogeneous community that supports carcinogenesis. As the tumor grows, malignant cells compete with the surrounding milieu for spaces and metabolic benefits, creating a hypoxic, acidic, and nutrient‐poor onco‐sphere that favors clonal selection of more aggressive cancer cells to spread outward. Notably, accumulated extracellular lactate and concomitant acidification has profound impacts on the TME by hindering dendritic cell (DC) activation, inhibiting cytotoxic T lymphocyte (CTL) function [38, 39], promoting TAM polarization [40], stimulating endothelial cell‐induced angiogenesis [41], as well as facilitating fibroblast activation and MMP‐governed matrix remodeling [42, 43], which can ultimately lead to tumor progression. Furthermore, external stimuli like therapeutic interventions may reprogram the evolvement of primary onco‐spheres. Strikingly, hypoxia generated by hepatic surgery imposes an invasive CSC phenotype on the residual tumor cells [44]. Besides, neoadjuvant chemotherapy for breast cancer plays as a double‐edged sword, which on one hand, enhances antitumor immunity by inducing interferon (IFN) response, and on the other hand, polarizes macrophages towards a proinflammatory phenotype conferring chemoresistance [6].
Taken together, both internal and external factors of the primary onco‐spheres harness tumor cells and their accomplice in the local milieu with fitness advantage as well as aggressive potency.
3. PRE‐METASTATIC NICHES IN ORGANOTROPIC METASTASIS
In nature, new species of organisms may invade into an established ecosystem, closely resembling metastases of tumor cells to distant organs. The stepwise malignant events of cancer metastasis begin with cells invading the basal membrane of blood vessels, followed by cell intravasation into blood circulation, ending with cell extravasation and dissemination in target organs (invasive species survival in transport). The forerunners of metastatic cancer cells often lay dormant at the secondary site for a period of time (lag period) prior to forming detectable tumor outgrowth (invasive spread). Using the ecological principle, these newcomers start as a native population within a primary community, but somehow transport to certain new organ niches, impacting upon the native host cells along with the original ecosystem [45]. Accumulating studies have demonstrated that the pre‐metastatic niches (PMNs) preconditioned in the distant organs usher organotropic metastasis for the tumors cells that possess intrinsic metastatic propensity [46]. Interplay between tumor cells and pre/post‐metastatic niches in distant organs conceives the distal onco‐spheres.
3.1. Organ‐specific PMNs
Preceding the arrival of metastatic tumor cells, combined systemic effects of tumor‐derived factors, exosome cargos, metabolic flux and the like act in concert to assist the formation of pre‐metastatic onco‐spheres at specific organ sites. The sequential construction steps involve vascular leakage, awakening of stromal residents, ECM remodeling, recruitment of inflammatory and immune cells, etc [46].
The lung is one of the most frequent sites of cancer metastasis, in which PMN formation has been well documented. Vessel barrier breakdown is the earliest event of lung PMN, and enhanced pulmonary vascular permeability is attributed to TGFβ‐induced angiopoietin‐like 4 (ANGPTL4) expression by the disseminated cancer cells [47]. Factors secreted by primary tumors, including epiregulin, angiopoietin 2, cyclooxygenase 2 and MMPs, can cooperate to destabilize lung capillaries and empower circulating tumor cell extravasation [48, 49]. Moreover, tumor‐shed exosomes expressing integrins α6β4 and α6β1 fuse preferentially with pulmonary fibroblasts and further upregulate pro‐inflammatory S100 proteins to facillitate lung metastasis [50]. Stromal fibroblasts in the lung PMN contribute to lung metastasis as well. The pulmonary resident fibroblasts in the PMN can secrete fibronectin, a integrin very late antigen 4 (VLA4; α4β1) ligand, and recruit bone marrow‐derived hematopoietic progenitor cells that express VLA4, and thus foster a growth‐permissive niche for the incoming metastatic cancer cells [51]. Also, crosslinking of collagen due to hypoxia‐induced lysyl oxidases (LOX) not only provides a platform for the recruitment and adhesion of bone marrow‐derived cells (BMDCs) [52] but also increases tissue stiffness that enhances metastatic outgrowth [53]. Infiltration of proinflammatory cells is also involved in PMN formation. Exosomal RNAs released by primary cancer cells can stimulate pulmonary epithelia to produce chemokines for neutrophil recruitment that specifically supports metastatic initiation [54, 55]. Furthermore, tumor‐conditioned alveolar macrophages impose immunosuppression in the lung PMN by dampening the antitumor T cell responses orchestrated by antigen‐presenting cells (APCs) and tumoricidal T helper 1 (Th1) cells [56]. Additionally, tumor‐derived exosomes can activate NF‐κB signaling to polarize macrophages toward immunosuppressive phenotype in the lung PMN by reducing mitochondrial oxidative phosphorylation and enhancing glycolysis [57].
Regarding liver metastasis, colorectal cancer‐derived exosomes carrying miR‐25‐3p may upregulate vascular endothelial growth factor receptor (VEGFR)2, zonula occludens‐1, occludin and Claudin5 expression in endothelial cells, conferring vascular permeability and angiogenesis for the establishment of hepatic PMN [58]. In contrast to α6β4 and α6β1 that mediate lung metastasis, exosomes expressing integrin αvβ5 can be taken up by Kupffer cells, the tissue‐resident macrophages in the liver, which orchestrates organotropic metastasis to the liver [50]. Furthermore, Kupffer cells conditioned by pancreatic cancer exosomes can induce TGFβ secretion and fibronectin production by stellate cells, the tissue‐resisdent fibroblasts in the liver, producing a fibrotic niche permissive for tumor metastasis [59]. Another systemic response mouted by primary pancreatice cancer cells is critical to their liver metastasis, in which myeloid cell accumulation and hepatic fibrosis in the liver facillitate metastatic colonization by inducing IL‐6‐dependent production of serum amyloid A1 and A2 [60].
The metastatic organotropism of tumor cells to the bone and their tendency to induce a bone‐forming (osteoblastic) or bone‐lysing (osteolytic) phenotype largely depend on the interplay between neoplastic cells and osteoblasts/osteoclasts, which differ in various cancer types. Galactoside‐binding soluble 3 (LGALS3) secreted by hepatocellular carcinoma directly activates osteoclast fusion to establish a bone PMN for osteolytic metastases [61]. Hypoxic breast cancer secretome also confers focal pre‐metastatic bone lesion formation by inducing osteoclastogenesis via the ECM‐shaping enzyme LOX [62]. Prostate cancer (PCa) is inclined to bone metastasis since PCa‐derived matrix protein mindinand exosomes can stimulate osteoblast proliferation and differentiation to induce bone remodeling for the colonization of metastatic tumor cells [63, 64].
Brain metastases often arise from lung cancers, breast cancers and melanoma [65]. Although several studies have shown that cancer secretome (e.g., MMPs, exosomal microRNAs)‐mediated ECM remodeling, blood‐brain barrier permeability as well as activation of M2 microglia and astrocytes are involved in brain metastatic cascades, the detailed mechanisms underlying brain PMN formation remain less informed [66, 67, 68, 69]. Thus, whether putative molecules directly prime the brain‐tropism cancer metastasis requires further investigations.
3.2. Homing and colonization of metastatic tumor cells
A successful metastasis necessitates tumor cells responding to chemotactic signals from PMNs hospitable for their seeding and colonization [70]. In organ‐specific metastases, directional migration of metastatic tumor cells towards chemokines occurs in a gradient‐dependent manner, while the chemokine gradient can be preestablished by distinct cell populations in target organs [71, 72]. Chemokine ligand‐receptor interactions, such as CXCL12/CXCR4, CCL21/C‐C motif chemokine receptor (CCR)7 and CCL25/CCR9, induce chemotactic and invasive responses in cancer cells and direct them towards metastatic destinations including the lymph nodes, lungs, liver or small intestine [73, 74]. Intriguingly, DNA of neutrophil extracellular traps in the liver or lungs can also act as a chemotactic factor to trap metastatic cancer cells that express a transmembrane DNA sensor, coiled‐coil domain containing 25 (CCDC25) [75]. Moreover, E‐selectin‐enriched discrete foci induced by endothelial cell‐focal adhesion kinase within the hyperpermeable lung vasculature results in preferential homing of metastatic tumor cells [76]. Different integrin expression patterns on tumor exosomes may determine metastatic tumor cell homing to either the lung or the liver PMNs, depending on their affinities to pulmonary fibroblasts or hepatic Kupffer cells, respectively [50].
As to the post‐seeding process, metastatic colonization is initially inefficient, especially when lacking the support of distant niches. However, upregulation of an ECM component periostin in stromal fibroblasts at distant organs may help to maintain the stemness as well as colonization of metastatic tumor cells [77]. Also, netrin‐4‐mediated basement membrane stiffness may enhance cancer metastasis in distant organs [78]. In addition, the dormant cancer cells residing in distant organ sites contribute to metastasis upon reactivation by environmental stimuli, which often leads to tumor relapse even following successful anticancer treatment for the primary tumor. Although the mechanisms for cancer dormancy remains obscure, it has been suggested that dormant tumor cells evade immunosurveillance to ensure long‐term latency and become re‐activated in response to vascular and ECM remodeling in the specialized immuno‐privileged niches, followed by a manifest metastatic relapse [79, 80]. Interestingly, a recent study has demonstrated that dormant breast cancer bone micrometastases could be remotivated and nourished by the osteogenic microenvironment to form macrometastases and even invigorate multi‐organ secondary metastases [81]. This finding underscored the dual effect of cancer dormancy and pre‐/post‐metastatic distal onco‐spheres on cancer metastasis.
In summary, combined actions by the establishenment of organ‐specific metastatic niches and the successefful homing/colonization of metastatic cancer cells precipitate the most malignant process.
4. INTERPLAY BETWEEN TUMORS AND IMMUNE SYSTEM
The concept of cancer immunoediting is an extension of cancer immunosurveillance that highlights the interaction between the tumor and host immune system throughout tumor initiation and progression [82, 83]. There are three phases in which the immune system shapes the tumor fate: elimination, equilibrium, and escape [83].
In the first phase, the tumor‐immune system interplay can be portrayed as a prey‐predator mode, whereby the malignant cells (preys) are largely eliminated under immunological surveillance orchestrated by both innate and adaptive immune systems (predators). Due to special anatomic location and immunologic characteristics, brain tumors were considered immune‐privileged. Nevertheless, ectopic expression of VEGF‐C enables lymphatic drainage and primes CD8+ T cell function, leading to immunosurveillance of glioblastoma [84]. On the other hand, antitumor γδ T cells function in an oxygen‐dependent way, while tumor hypoxia dampens γδ T cell‐mediated immunity against brain tumors [85]. In terms of the immune surveillance of cancer metastases, conventional natural killer (NK) cells and tissue‐resident type 1 innate lymphoid cells cooperate to constrain liver metastasis [86]. Interleukin‐1 receptor 8 (IL‐1R8) emerges as a new checkpoint for NK cell activation and antimetastatic defense in the liver and lung [87]. Moreover, a recent study revealed the mechanical dimension of immunosurveillance, in which overexpression of myocardin‐related transcription factors could sensitize cancer cells to CTL‐mediated killing by rigidifying the filamentous actin cytoskeleton [88].
However, if the immune system fails to perform a successful surveillance, its interplay with the tumor enters the second phase of dynamic equilibrium, in which the Darwinian selection of tumor variants endowed with immune tolerance occurs. As a result, tumor cells become inert, dormant or even evolve under the immunosurveillance pressure, while T regulatory cell responses escalate at this stage and create immune balance with the alleviated effector responses.
Upon tumor progression comes the third phase ‐ immune evasion, whereby the tumor variant survivors develop a variety of counter adaptations to escape the immune predation and expand in a runaway manner, resulting in macroscopic malignancy.
4.1. Loss of clonal neoantigen
Reduced neoantigen expression and impaired antigen presentation conduce to T cell tolerance. Mechanistically, chromosomal instability, promoter hypermethylation and neoantigen mutations may cause copy number loss or clonal neoantigen depletion at either DNA or RNA level [89]. On the other hand, mutation or dysregulation of the genes encoding major histocompatibility complex class (MHC)‐I antigen and/or proteins related to antigen‐processing machinery can result in aberrant MHC‐I expression, which impairs tumor antigen presentation [90]. Also, loss of heterozygosity in the human leukocyte antigen (HLA) complex, mutations weakening MHC stability, and decreased neoantigen peptide production may lead to deficient antigen presentation [89, 91].
4.2. Expression of NK cell inhibitory receptors
Self‐tolerance of NK cells is finely tuned by the expression of MHC‐I‐specific inhibitory receptors, such as killer cell immunoglobulin‐like receptors (KIRs) and natural killer group 2A (NKG2A), which discriminate normal cells expressing cognate MHC‐I from virus‐infected cells or tumor cells with MHC‐I downregulation [92]. Besides, NK cells also express non‐HLA‐specific inhibitory receptors. Among them, the T cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) could diminish IFN‐γ production and thus blunt NK cell immune control of the poliovirus receptor family ligand (e.g., CD155 and CD112)‐expressing tumors [93].
4.3. Upregulation of immune checkpoints
Both cytotoxic T‐lymphocyte‐associated protein (CTLA)4 and programmed cell death (PD)‐1 are major T cell inhibitory receptors, referred to as checkpoint regulators, that negatively modulate T cell receptor (TCR)‐dependent antitumor T cell responses. During the early stage of T cell activation, the upregulated CTLA4 on T cell surface competes with CD28 that binds to costimulatory molecules CD80 and CD86 expressed on APCs, thereby blocking the sustained activation of T cells during primary immune response [94]. Contrary to CTLA4, PD‐1 overexpression occurs in the late stage of T cell activation and poses inhibitory effects on T cell response through receptor ligation with programmed cell death‐ligand 1 (PD‐L1) [94]. It has been reported that oncogenic activation of the Janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) pathway confers high PD‐L1 expression and mediates immune escape in cancers [95, 96]. It is noteworthy that in head and neck squamous cell carcinoma, CSCs upregulate another immune checkpoint molecule CD276 to evade host immune surveillance [97]. Nowadays, attempts to target these checkpoint pathways have shown clinical benefits, rendering immune checkpoints as promising targets for cancer immunotherapy [98].
4.4. Induction of immunosuppressive cells
Regulatory T cells (Tregs) are instrumental in conferring T cell tolerance and alleviating anticancer immunity by targeting both effector T cells and APCs [99]. Besides being recruited from the periphery, locally differentiated or expanded tumor‐infiltrating Tregs (CD4+CD25+) could also arise from naive CD4+ T cells (CD4+CD25–) in situ [27]. Of note, deregulated metabolism in tumor cells leads to a metabolite‐depleted and acidic milieu that deprives effector T cells of nutrients. However, intratumoral Tregs can flexibly draw metabolic support from the glycolytic by‐product lactic acid to sustain their suppressive identity [100]. A recently uncovered cell‐programed nutrient partitioning mechanism also revealed the preferential uptake of glucose and glutamine by myeloid cells and cancer cells, respectively [101]. In human hepatoma, innate monocyte inflammation could induce a specific subset of IL‐21‐secreting T follicular helper‐like cells, which, in turn, prime M2b macrophage polarization and foster immune privilege [102]. Another notable immunosuppressive cell type is myeloid‐derived suppressor cells (MDSCs), which consist of the pathologically activated monocytes and relatively immature neutrophils with potent immune‐inhibitory activity [103, 104].
4.5. Generation of immunosuppressive cytokines
For adaptive immunity, excessive TGFβ represents a primary mechanism of immune evasion that hinders DC maturation and T cell activation but promotes Treg expansion and T cell exclusion [105]. On the other hand, for innate immunity, TGFβ controls the behaviors of macrophages, neutrophils and NK cells, forming a network of negative immune regulatory inputs [106]. IL‐10 similarly exerts immunosuppressive activities on APC‐induced antigen presentation and production of proinflammatory cytokines (e.g., IL‐12 and IL‐23) and alleviates effector T cell response as well as chronic inflammation‐mediated protumoral effects [107]. Additionally, cancer cells express a panel of pro‐apoptotic factors, especially Fas ligand, triggering T cell apoptosis and less infiltration into tumor [108]. NKILA, an NF‐κB‐interacting long non‐coding RNA (lncRNA) [109], shows potency in sensitizing tumor‐infiltrating T cells to activation‐induced cell death through NF‐κB inactivation [110]. As for T cell homing, upregulation of regulator of G protein signaling (RGS)1, rather than chemokines, in circulating CTLs and Th1 cells reduces their trafficking to and survival in breast and lung tumors [111]. Of note, STAT3 serves as a converging point of multiple oncogenic pathways, whose constitutive activation bridges tumor cells and the host immunity via both inhibition of immune mediators (e.g., IFN‐γ, IL‐12, CD80 and CD86) and promotion of immunosuppressive factors (e.g., IL‐6, IL‐10, TGFβ and VEGF) [112, 113]. The latter, in turn, prolong STAT3 signaling in a feed‐forward loop between cancer and immune cells [112, 113].
As such, decreased T cell trafficking to the tumors, increased apoptosis, or dysfunction of effector T cells, together with increased tumor infiltration by immunosuppressive cells, synergistically contribute to the formation of a “cold” (non‐T cell‐inflamed) onco‐sphere featuring immunologic anergy, tolerance or indifference, which favors cancer invasion and expansion (Figure 2). Therefore, deciphering the mechanisms underlying tumor immune evasion paves new paths for designing novel immunotherapy to convert “cold” cancers into “hot” (T cell‐inflamed) ones.
FIGURE 2.
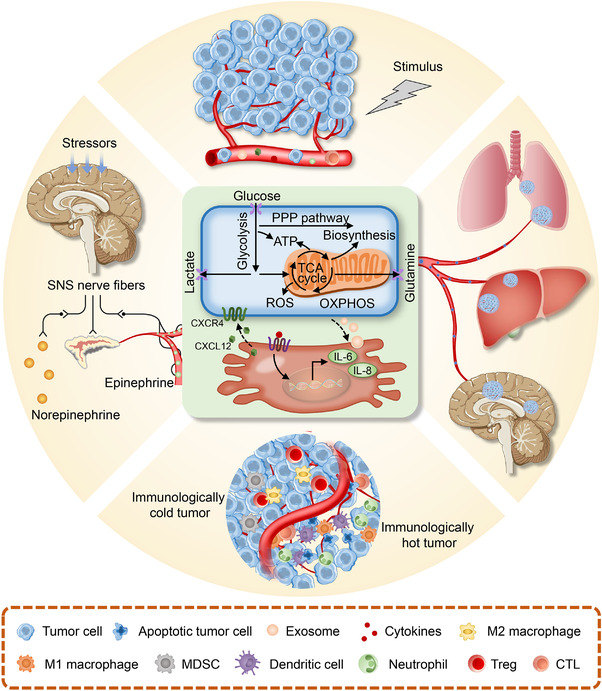
Intertwined interactions within the tumor ecosystem. The material and energy flow arising from tumor metabolism and intercellular communication (central) runs through the host macroenvironment, linking tumor microenvironment (upper) to distant organ niches (right), immune (bottom) as well as the nervous and the endocrine systems (left). Such tumor‐specific metabolic and other non‐metabolic hallmarks are intertwined to form a dual cause‐effect relationship that constitutes an evolving systemic onco‐sphere. Abbreviations: ATP, adenosine triphosphate; CXCL12, C‐X‐C motif chemokine ligand 12; CXCR4, C‐X‐C motif chemokine receptor4; IL‐6, interleukin‐6; IL‐8, interleukin‐8; MDSCs, myeloid‐derived suppressor cells; OXPHOS, oxidative phosphorylation; PPP, pentose phosphate pathway; ROS, reactive oxygen species; SNS, sympathetic nervous system; TCA, tricarboxylic acid; Treg, Regulatory T cells.
5. INTERTWINING WITH THE NEUROENDOCRINE SYSTEM
Benign or malignant tumors derived from the endocrine system are usually associated with hyper‐ or hypo‐hormonal activities, due to excessive or degenerative hormone secretion. A typical example among them is the pituitary neuroendocrine tumor (PitNET), as the name implies, manifesting hormone hypersecretion as well as various endocrine syndromes, in addition to headaches and visual deficits [114]. Among these syndromes, Cushing disease triggered by excessive pituitary adenomas‐derived adrenocorticotropin (ACTH) leads to a myriad of disabling and even lethal symptoms, such as diabetes, hypertension, central obesity, osteoporosis, muscle weakness, and psychological disturbances [115]. Moreover, PitNETs are associated with abnormal production of growth hormone, thyroid‐stimulating hormone, prolactin and follicle‐stimulating hormone and/or luteinizing hormone, resulting in hormonal‐excess disorders: acromegaly, thyrotoxicosis, reproductive and gonadal dysfunction, respectively [114].
Unlike PitNETs characterized with multiple neuroendocrine disorders, many solid tumors may also possess a neuroendocrine program (integrating neural and endocrine properties) and could even present paraneoplastic syndromes as the first clinical manifestation of the malignancy. Paraneoplastic syndromes are referred as anatomic and functional abnormalities caused by the remote or systemic biological effects of a tumor, which are irrelevant to the spreading or metastasis of cancer cells. A variety of tumor‐secreted factors may help to orchestrate the pathological process of paraneoplastic syndromes, which could be classified into two basic subtypes: the neurological symptoms largely due to tumor‐derived neural peptides or antineuronal antibodies, and the endocrine symptoms related to ectopic hormonal secretion by cancer cells with neuroendocrine features [116]. Among these tumors, small‐cell lung cancer (SCLC) is the most frequent malignancy associated with paraneoplastic syndromes [116], in which Lambert‐Eaton myasthenic syndrome featuring muscle weakness occurs more often than any other immune‐mediated neurological syndrome. In addition, pathogenic autoantibodies, raised against the voltage‐gated calcium channels (VGCCs) expressed by SCLC, may crossreact with VGCCs on the presynaptic nerve terminal of the neuromuscular junction, leading to neuromuscular disorders [117]. Furthermore, ACTH‐associated Cushing syndrome and inappropriate antidiuretic hormone secretion are other common endocrine manifestations of SCLC [116].
As an abnormal state of health in response to chronic wasting diseases, cachexia is commonly seen at late‐stage cancers and characterized by the gradual loss of muscles and fat tissues. Although cachectic patients are often anorexic due to the involuntary loss of appetite, anorexia is not the culprit for their weight loss [118]. Instead, tumor‐stimulated upregulation of gene expression and protein production raises the biochemical workload of the Cori cycle, accelerating the thermogenesis and basal metabolic rate. For instance, lipolysis which provides energy for both tumor cells and host cells often accounts for the loss of adipose tissue, while ubiquitin‐proteasome pathways and lysosomes are excessively activated to cause increased protein breakdown. Moreover, tumor‐secreted proteolysis‐inducing factors trigger the systemic production of tumor necrosis factor (TNF)‐α and glucocorticoids, resulting in severe muscle wasting in cancer patients [119].
On the other hand, deregulated neural or endocrine signaling within the host macroenvironment may also play a role in cancer initiation and development. Similar to an integrated organ, a solid tumor can be directly innervated by sympathetic nerve fibers, from which neurotransmitter norepinephrine is released and diffused to reach tumor cells [120]. Alternatively, adrenal gland‐releasing epinephrine is responsible for the hormonal regulation of tumor biology governed by the sympathetic nervous system [121, 122]. Contrary to the pro‐tumor roles of sympathetic (β‐adrenergic) nerves in PCa [122], the parasympathetic (cholinergic) division of the peripheral autonomic nervous system might eventually inhibit tumorigenesis and progression, as exemplified in pancreatic carcinomas [123].
All in all, a better understanding of the communication between tumor and neuroendocrine system through chemical messengers derived from neural cells, endocrine organs, the immune system and tumor cells within the systemic onco‐sphere is urgently needed for developing novel strategies toward effective cancer control.
6. FROM TUMOR ECOSYSTEM TO ANTITUMOR ECOLOGICAL THERAPY
As the tumor ecosystem is dynamically changing with cancer development, external stimuli such as therapeutic interventions also play a role in reshaping the local and systemic onco‐spheres. Allee effect is an ecological concept that manifests the association between population density or size and individual fitness in species, as individual fitness declines at low population density or size [124, 125]. Likewise, surgical removal of the tumor is supposed to reduce the cancer burden to a level below the Allee threshold, leaving the residual ones vulnerable to local or systemic adjuvant therapies such as radiotherapy and chemotherapy. Crucially, organ‐preserving surgery followed by adjuvant therapies might render the primary onco‐sphere susceptible to a new wave of therapeutic attack. For early‐stage breast cancer, equivalent clinical outcomes between breast‐conserving surgery (BCS) and mastectomy have been evidenced in a series of randomized controlled trials over the last three decades [126, 127, 128], while real‐world studies in recent years demonstrated improved 10‐year survival after BCS plus radiotherapy relative to mastectomy [129]. Similarly, bladder‐preserving tri‐modality therapy (transurethral resection of bladder tumor followed by concurrent chemoradiotherapy) has become a recognized alternative to radical cystectomy for patients with muscle‐invasive bladder cancer, due to comparable long‐term survival and high successful rate of bladder preservation [130]. Radiotherapy can induce DNA damage, not only causing direct tumor cell death but also triggering systemic antitumor immune responses as abscopal effect, featured by the release of inflammatory mediators, the increase of tumor‐infiltrating immune cells, and the upregulation of neoantigens. Mechanistically, radiation‐induced micronuclei in tumour cells activate cytosolic nucleic acid sensor pathways, notably cyclic GMP‐AMP synthase (cGAS)‐stimulator of interferon genes (STING), leading to type I IFN production in response to cytoplasmic DNA [131]. Further propagation of inflammatory signals potentially remodels the immune contexture of the onco‐spheres by stimulating DC maturation, effector T cell infiltrantion, as well as the recruitment of immunosuppressive populations such as Tregs, macrophages and MDSCs, depending on the cytokine milieu [132, 133, 134]. Despite that, the counterbalance of radiation‐mediated immunogenic and tolerogenic signalings could be broken by strategically combining radiotherapy with immunotherapy [135]. On the other hand, neoadjuvant therapies are initially devised to minize tumor burden and facilitate surgical eradication, but then, they might trigger systemic effects within the onco‐spheres. In patients with low‐risk distal rectal cancer, neoadjuvant chemoradiotherapy enables organ‐sparing transanal endoscopic microsurgery, resulting in a promising long‐term oncological and functional outcome [136]. Importantly, altered local and peripheral tumor immunity after neoadjuvant chemotherapy (NAC) could reshape the clinical outcomes in cancer patients with opposite ways: locally increased immune genes/sets associates with better long‐term outcome while high cytolytic T cell signatures in the peripheral blood tend to indicate persistent disease and recurrence [137]. As a coin has two sides, NAC might enhance IFN‐mediated antitumor immunity and simutaneously elicit protumoral inflammation by virtue of TAM polarization [6]. Additionally, neoadjuvant antibody (e.g., trastuzumab) treatment could induce antibody‐dependent cellular phagocytosis and lead to immunosuppression via upregulation of PD‐L1 and indoleamine 2,3‐dioxygenase in macrophages [138]. In this way, neoadjuvant therapies might reprogram the local or systemic onco‐spheres to call for combined immunotherapies or other targeted therapies. It is noteworthy that combined treatment with chemoradiotherapy plus immunomodulatory therapy favorably boosts the innate and adaptive immunity while reduces MDSCs and Tregs in the immune microenvironment [139]. As for immunotherapy, it has been reported that immune checkpoint blockade (ICB) leads to tumor and immune remodeling in metastaitic renal cell carcinoma, notably characteristic of upregulation of immunosuppressive programs [140]. Moreover, targeted inhibition of mitogen‐activated protein kinase kinase (MEK) can reprogram CD8+ T cells into regenerative stem cell‐like memory phenotypes that act as a reservoir for effector T cells with enhanced metabolic fitness and superior antitumor effects, without affecting TCR‐mediated cell priming [141].
As anticancer treatment may remodel the onco‐spheres into either tumor‐promoting or tumor‐suppressive, ecosystemic alterations during these processes also feed back to the therapeutic responses, in particular, rendering therapeutic resistance. For example, radiotherapy‐induced accumulation of immunosuppressive cells causes radiation resistance in tumors. Owing to the over‐production of CCL2 chemokine in irradiated tumor cells, a CCR2‐dependent recruitment of monocytes and Tregs contributes to TNFα‐mediated resistance [133]. Besides, MDSC mobilization and infiltration via activation of the STING/type I IFN/CCR2 pathway drives extrinsic radiation resistance in tumor, while anti‐CCR2 antibody treatment can alleviate suppressive inflammation and thus enhance the antitumor effects of STING agonists and radiotherapy [134]. Moreover, NAC‐induced IFN response upregulates a lncRNA in macrophages, endowing them with a proinflammatory phenotype that confers chemoresistance [6]. Concerning about the autonomous tumoral or microenvironmental changes, not only chromosomal alterations but also CD8+ T cell infiltration associate with response or resistance to PD‐1 blockade in advanced clear cell renal cell carcinoma [142]. Recently, the PD‐1/PD‐L1‐dependent expansion of a highly proliferative, overactivated, and apoptotic dysfunctional CD8+ tumor‐infiltrating lymphocyte (TIL) subset in the TME has been found to curb immunotherapy in advanced non‐small cell lung cancer (NSCLC) [143]. In addition, hepatic metastases co‐opt host peripheral tolerance mechanisms to create a systemic immune desert through macrophage‐mediated CD8+ T cell depletion, thereby dampening immunotherapy efficacy [144].
As such, both autonomous and non‐autonomous ecosystemic alterations impact the response to anticancer therapeutics. With the urgent need to overcome therapeutic resistance, more druggable targets have emerged. Recognizing tumor ecosystem as a network of dependencies facilitates the identification of therapeutically valuable elements created by the tumor‐host interplay. As tumor cells may mutate and evolve resistance in response to current cancer‐centric treatment paradigms, the host cells such as vascular cells, fibroblasts, macrophages and lymphocytes from local or systemic onco‐spheres might provide more stable targets for multitargeted cancer therapy.
For the whole tumor ecosystem, surgery, irradiation and chemotherapeutic agents represent three classic ecological ways of demographic perturbations of the cancer population, which can be the first strike to minimize the tumor burden below the Allee threshold as well. For more aggressive or resistant tumors with higher heterogeneity and capability of adaptation, a strategic second strike is indispensable to warrant further tumor control. As in the natural ecosystem, the most efficient way to kill a species is to alter its biosphere. Ecologically inspired second‐strike strategies could empirically involve intervention of its habitat (anti‐angiogenic therapy), disruption of the material/energy flow (metabolism modulators) and introduction of a predator (immunotherapy). Representative targets for above‐mentioned ecological therapies are summarized in Table 2. Specifically, targeting VEGF or its cognate receptor VEGFR by neutralizing monoclonal antibody (mAb) or tyrosine kinase inhibitors (TKIs), respectively, holds potential to inhibit tumor angiogenesis and has shown clinical benefits when combined with chemotherapy (or not) in treating advanced cancers [145]. The feedback loop between GM‐CSF and CCL18 can also be an appealing target to alleviate TAM‐mediated tumor‐promoting inflammation within the TME [23, 24]. Moreover, blocking chemokine ligand‐receptor interactions, notably CCL2/CCR2 and CXCL12/CXCR4, tends to hold back the recruitment of cancer‐associated stromal cells and thus prevent de novo niche formation. Due to the importance of hypoxia inducible factor (HIF)‐1α in metabolic adjustment, HIF‐1α inhibition becomes another attractive approach to disturb material and energy flow throughout the tumor ecosystem. The rapid development of cancer immunotherapy has recently led to new attempts to turn immunologically “cold” tumor “hot”. Building on the successes of checkpoint blockade antibodies against CTLA4 and PD‐1, multiple innovative immunotherapies are currently undergoing trial evaluation for cancer treatment, including ICB with novel targets (e.g., CD276), co‐stimulatory pathway (e.g., 4‐1BB, OX40 and CD40) agonists, cellular therapies typified by chimeric antigen receptor (CAR)‐T cell therapies, as well as therapeutic vaccination. Particularly, breast cancer patients may benefit from the human epidermal growth factor receptor 2 (HER2)/neu peptide (E75) vaccine alongside GM‐CSF (to improve T‐cell responses) treatment, while neoantigen‐based dendritic cell vaccines can potentially elicit specific T cell immunity and clinical therapeutic efficacy in advanced NSCLC [146]. Given the role of RGS1 in T cell exclusion, targeting RGS1 might also be promising to heat up the TME by increasing T cell trafficking and infiltration, further sensitizing originally “cold” tumors to concurrent or subsequent immunotherapies [111].
TABLE 2.
Selected therapeutic targets in the emerging and developing antitumor attempts
| Targets | Drugs | Biologic action | Target cells | Monotherapy or combinatorial therapies | Current status | Tumor types | Trial No. |
|---|---|---|---|---|---|---|---|
| Reprogram the TME | |||||||
| CSF‐1R | BLZ945 | Kinase inhibitor | Macrophages | Monotherapy or in combination with anti‐PD1 mAb (Spartalizumab) | Phase I/II ongoing | Advanced solid tumors | NCT02829723 |
| Pexidartinib | Kinase inhibitor | Macrophages | In combination with chemo‐radiotherapy (Temozolomide + RT) | Phase Ib/II completed | Glioblastoma | NCT01790503 | |
| CXCR1/2 | Reparixin | Allosteric inhibitor | Neutrophils | In combination with chemotherapy (Paclitaxel) | Phase II completed | Metastatic triple‐negative breast cancer | NCT02370238 |
| Hyaluronic acid | PEGPH20 | PEGylated enzyme | ECM | In combination with chemotherapy (Gemcitabine) | Phase I/II completed | Stage IV pancreatic cancer | NCT01453153 |
| Inhibit tumor angiogenesis | |||||||
| VEGF‐A | Bevacizumab | Anti‐VEGF mAb | Endothelial cells | In combination with standard chemotherapy (Taxanes, Gemcitabine, Capecitabine, or Vinorelbine) | Phase III completed | Metastatic breast cancer | NCT00281697 |
| VEGFR2 | Sorafenib | Kinase inhibitor | Endothelial cells | In combination with chemotherapy (Capecitabine) | Phase III completed | Advanced or metastatic HER2‐negative breast cancer | NCT01234337 |
| In combination with chemotherapy (Gemcitabine + Cisplatin) | Phase III completed | Advanced nonsquamous NSCLC | NCT00449033 | ||||
| Sunitinib | Kinase inhibitor | Endothelial cells | Monotherapy | Phase II/III ongoing | Glioblastoma | NCT03025893 | |
| Prevent PMN formation | |||||||
| CCR2 | MLN1202 | Anti‐CCR2 mAb | Macrophages | Monotherapy | Phase II completed | Metastatic cancer | NCT01015560 |
| CXCR4 | Plerixafor | CXCR4 antagonist | Hematopoietic progenitor cells | In combination with anti‐PD1 mAb (Cemiplimab) | Phase II ongoing | Metastatic pancreatic cancer | NCT04177810 |
| TGFβ | Fresolimumab | Anti‐TGFβ mAb | Fibroblasts | In combination with stereotactic ablative radiotherapy | Phase Ib/II ongoing | Stage IA/IB NSCLC | NCT02581787 |
| PDGFR | Imatinib | Kinase inhibitor | Fibroblasts; Endothelial cells | In combination with chemotherapy (Gemcitabine) | Phase II completed | Metastatic pancreatic cancer | NCT00161213 |
| Disrupt metabolic homeostasis | |||||||
| HIF‐1α | RO7070179 | ASO inhibitor | Tumor cells | Monotherapy | Phase I completed | HCC | NCT02564614 |
| Everolimus | mTOR inhibitor | Tumor cells, endothelial cells | In combination with chemotherapy (FOLFOX) and anti‐angiogenic therapy (Bevacizumab) | Phase I/II completed | Colorectal cancer | NCT01047293 | |
| EZN‐2968 | ASO inhibitor | Tumor cells | Monotherapy | Phase I completed | Advanced solid tumors | NCT01120288 | |
| Tirapazamine | Hypoxic cytotoxin | Hypoxic cancer cells | In combination with chemo‐radiation therapy (Cisplatin + 5‐fluorouracil + RT) | Phase II completed | Advanced squamous head and neck cancer | NCT00002774 | |
| Reboot antitumor immunity | |||||||
| CTLA4 | Ipilimumab | Anti‐CTLA4 mAb | T cells | In combination with anti‐PD1 mAb (Pembrolizumab) | Phase III ongoing | Stage IV, metastatic NSCLC | NCT03302234 |
| Monotherapy | Phase III completed | Metastatic melanoma | NCT01515189 | ||||
| Tremelimumab | Anti‐CTLA4 mAb | T cells | Monotherapy | Phase II completed | Colorectal cancer | NCT00313794 | |
| In combination with PARP inhibitor (Olaparib) | Phase I/II ongoing | Recurrent ovarian Cancer | NCT02571725 | ||||
| PD1 | Nivolumab | Anti‐PD1 mAb | T cells | In combination with anti‐CTLA4 mAb (Ipilimumab) | Phase III completed | Advanced melanoma | NCT03068455 |
| Pembrolizumab | Anti‐PD1 mAb | T cells | Monotherapy | Phase III completed | Advanced melanoma | NCT01866319 | |
| Monotherapy | Phase II completed | Metastatic high grade neuroendocrine tumors | NCT02939651 | ||||
| In combination with IDO1 inhibitor (Epacadostat) | Phase III completed | Cisplatin‐ineligible urothelial carcinoma | NCT03361865 | ||||
| In combination with chemotherapy (mFOLFOX6) | Phase II ongoing | Advanced colorectal cancer | NCT02375672 | ||||
| PD‐L1 | Atezolizumab | Anti‐PD‐L1 mAb | T cells | In combination with chemotherapy (Nab‐paclitaxel + Carboplatin) | Phase II ongoing | NSCLC | NCT02716038 |
| In combination with anti‐angiogenic therapy (Bevacizumab) | Phase III ongoing | Advanced or metastatic HCC | NCT03434379 | ||||
| In combination with chemo‐radiotherapy (Carboplatin + Paclitaxel + RT) | Phase II ongoing | NSCLC | NCT02525757 | ||||
| Durvalumab | Anti‐PD‐L1 mAb | T cells | In combination with MEK inhibitor (Trametinib) | Phase II ongoing | Microsatellite stable metastatic colon cancer | NCT03428126 | |
| After chemo‐radiotherapy | Phase II ongoing | Stage II‐IV rectal cancer | NCT03102047 | ||||
| In combination with anti‐CTLA4 mAb (Tremelimumab) | Phase II completed | Metastatic HER2 negative breast cancer | NCT02536794 | ||||
| In combination with anti‐CTLA4 mAb (Tremelimumab) and RT | Phase II ongoing | Invasive bladder cancer | NCT03702179 | ||||
ASO, antisense oligonucleotide; CCR2, C‐C motif chemokine receptor 2; CSF‐1R, colony‐stimulating factor‐1 receptor; CTLA4, cytotoxic T‐lymphocyte‐associated protein 4; CXCR, C‐X‐C motif chemokine receptor; ECM, extracellular matrix; IDO1, indoleamine 2,3 dioxygenase 1; HCC, hepatocellular carcinoma; HER2, human epidermal growth factor receptor 2; HIF‐1α, hypoxia inducible factor‐1 α; MEK, mitogen‐activated extracellular signal‐regulated kinase; mAb, monoclonal antibody; mTOR, mammalian target of rapamycin; NSCLC, non‐small cell lung cancer; PARP, poly ADP‐ribose polymerase; PD‐1, programmed cell death‐1; PDGFR, platelet‐derived growth factor receptor; PD‐L1, programmed cell death 1 ligand 1; RT, radiation therapy; RTK, receptor tyrosine kinase; TGFβ, transforming growth factor β; VEGF, vascular endothelial growth factor; VEGFR, vascular endothelial growth factor receptor.
In the long run, rational combination of these novel approaches with clinically approved cancer therapies will be valuable moving forward.
7. CHALLENGES AND PERSPECTIVES FOR TUMOR ECOSYSTEM RESEARCH
In brief, we propose that cancer cells (living organisms) dynamically interact with non‐tumoral cellular (other living organisms) and non‐cellular components (non‐living environmental factors) in the “host” internal environment (habitat) to construct a self‐sustainable cancer ecosystem as well as systemic onco‐sphere, which can be scoped at three different levels: primary, regional, and distal onco‐spheres. With the surge of big data (e.g., genomic data, radiomic data, environmental omic data), more features of tumor ecosystem in different dimensions are disclosed. The intertwined network constituted by these data remains difficult, due to the ecosystemic complexity, but more possible than ever, to be deciphered. For example, the burgeoning single‐cell RNA sequencing technique enables direct mapping of diverse immune phenotypes in the TME [147].
Recent advances in data management techniques, such as machine learning, parallel computing and artificial intelligence, also provide opportunities for visualizing the spatial‐temporal dynamics of different tumor ecosystems. Specifically, a convoluting neural network was exploited to reveal four structural patterns of the TIL map, which are associated with tumor subtypes and patient outcome [148]. Another deep‐learning technique MesoNet was further developed and could identify stromal regions linked to inflammation, cellular diversity and vacuolization, so as to predict the overall survival of mesothelioma patients [149]. On the other hand, the emergent mass cytometry imaging technologies enable synchronous analysis of multiple parameters at subcellular resolution using a single tissue slice, resulting in a high dimensional pathological image for direct visualization of the topography of tumor community. With Imaging Mass Cytometry, the single‐cell phenotypes and the spatial context of breast cancer have been revealed [150], while Multiplexed Ion Beam imaging employed in breast cancer could delineate three archetypical subtypes of tumor‐immune interactions: cold, mixed and compartmentalized, with distinct prognostic indications [151]. These technological advances for tumor pathology helps extracting hidden information from the spatial interactions within onco‐sphere networks, providing a new tool for diagnosis and precision oncology [152]. Other multi‐modal image modalities, such as intra‐vital two‐photon microscopy and positron emission tomography‐computed tomography, can also aid in monitoring the temporal dynamics of tumor ecosystem.
For modeling the tumor ecosystem, mathematical oncology, which combines mathematics, molecular modeling and simulation to study the uncontrolled proliferation and dissemination of cancer cells, has been used to parameterize the complexity of tumor ecosystem. Mathematical models provide a useful abstraction of clinical intuition to monitor tumor therapeutic response when combined with serial measurements of putative tumor burden biomarkers, such as prostate‐specific antigen for PCa, or L‐lactate dehydrogenase for melanoma [153]. More importanatly, these cell‐based models can simulate tumor behaviors and the intercellular interactions within the TME, predicting potential treatment strategies/targets [154]. By ecologically modeling the tumor growth dynamics with a matrix game model and considering the trade‐off between tumor burden and metastatic risk, West et al. [155] suggested that optimal tumor regression in postmenopausal estrogen receptor‐positive breast cancer patients might result from a 1‐month delay of aromatase inhibitor treatment combined with continuous immune checkpoint inhibitors (ICIs) against PD‐L1. Albeit optimizing combination therapy is an arduous task, the use of mathematical models helps expedite the generation of new treatment schedules superior to the traditional standard of care. Alternatively, the establishment of preclinical model systems, including patient‐derived xenografts (PDXs) and organoid cultures, allows in vivo and in vitro modeling of the onco‐spheres, accelerating the development of high‐throughput drug screening as well as treatment response monitoring for personalized medicine [156, 157]. As with other preclinical tools, PDXs and organoids also have limitations and several challenges need to be addressed. Specifically, PDX models based on immunocompromised mice cannot be used to study tumor‐associated immune cell function and assess immunotherapeutics. With gradual substitution of human stroma by murine components during serial passaging of tumors, the changing tumor ecological environment restrains drug response detection in PDXs [156]. Although organoid models of diverse cancer types have been reported, a set of unified standards and processes is still required in organoid generation. It is worth noting that patient‐derived organoids hold promise as a prospective clinical test for cancer patients, albeit costly and time‐consuming [157].
With a full‐scale understanding of the characteristics of tumor ecosystem based on aforementioned efforts, the next step is to subtype it, rendering it more clinically relevant. Current attempts in subtyping the tumor ecosystems generally involve two approaches: either in an unsupervised manner, e.g., using machine‐learning techniques, or exploiting our knowledge and understanding of its underlying mechanisms [148]. According to the presence or absence of PD‐L1 and TILs, immune microenvironment could be categorized into four types, which confer clinical implications [158]. For Type I (PD‐L1‐positive, with TILs), ICIs are recommended as the mitigation of immunosuppression, which could be translated to TIL‐induced antitumor response. For Types II and III (without TILs), ICIs can hardly be effective unless tumor‐reactive T cells are recruited into the tumors by additional approaches. For Type IV (PD‐L1‐negative, with TILs), immunosuppression might be ascribed to an alternative mechanism instead of PD‐L1/PD‐1 interaction, which results in insensitivity to ICIs. It is noteworthy that a consensus statement classifies tumor ecosystem based on the evo‐index and the eco‐index of each patient [159]. Determined by the spatial and temporal tumor heterogeneity, the evo‐index is an indicator of potential fitness of tumor in a given environment. Tumors with higher heterogeneity and mutation rate increase the likelihood to evolve resistant clones. The eco‐index, composed of both hazards and resources within the onco‐sphere, is a reflection of habitat hospitality towards cancer cells. Although novel and clinically relevant, this proposal is far from bedside due to the ambiguity and uncertainty in estimating the evo‐ or eco‐index. For example, which parameters are applicable to characterize the heterogeneity? What is the objective measurement of the environmental hazards or resources? Further studies are needed to addressed these questions.
Successful visualizing, modeling and subtyping of this ecosystem could lay the foundation for developing ecologically rational therapeutic strategies to improve cancer screening/detection, diagnosis/risk‐stratification, and treatment guidance. As tumors evolve, it remains challenging to steer tumor evolution towards a desired direction. However, a timely strategic blockade to tumor progression holds promise to achieve effective tumor control. For early‐stage cancers, it is important to tip the balance of tumor‐host competition by increasing the competitiveness of normal somatic cells as well as lowering the tissue support for malignant cells. As for metastatic cancers, malign stromal cells should be re‐educated or reprogrammed to prevent de novo niche formation. Strategies to block the tumor‐host interactions such as disturbance of energy flow and disruption of tumor vasculature could also pose negative effects on the neoplastic processes. Additionally, modulating the immunologically “cold” microenvironment towards an antitumor context would be a rational option, as the predator and the prey tend to vary in an opposite way according to the theory of ecology and Darwinian evolution. Last but not least, either psychological adjustment or pharmacological intervention to maintain mental health is of great significance in the clinical management of this systemic disease. Within the context of cancer ecosystem, it is tempting to expect that a new era of cancer ecotherapy targeting the ecosystemic vulnerability of human malignancies has come.
DECLARATIONS
COMPETING INTERESTS
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Erwei Song conceived the idea for the review article. Xueman Chen performed the literature search and drafted the manuscript. Both authors critically revised the work and approved the final version of the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
ACKNOWLEDGEMENTS
The authors thank Ping‐Pui Wong, Phei Er Saw and Kai Chen for proofreading the manuscript. This work was supported by grants from the Natural Science Foundation of China (81621004 and 81930081), the Guangdong Basic and Applied Basic Research Foundation (2021A1515010238) and the China Postdoctoral Science Foundation (2020M683106 and 2021T140763).
Chen X, Song E. The theory of tumor ecosystem. Cancer Commun. 2022;42:587–608. 10.1002/cac2.12316
REFERENCES
- 1. International Cancer Genome C , Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, et al. International network of cancer genome projects. Nature. 2010;464(7291):993–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, et al. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell. 2017;169(4):736–49.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Horning SJ. A new cancer ecosystem. Science. 2017;355(6330):1103. [DOI] [PubMed] [Google Scholar]
- 4. Slyper M, Porter CBM, Ashenberg O, Waldman J, Drokhlyansky E, Wakiro I, et al. A single‐cell and single‐nucleus RNA‐Seq toolbox for fresh and frozen human tumors. Nat Med. 2020;26(5):792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song X, Chang S, Seminario‐Vidal L, de Mingo Pulido A, Tordesillas L, Song X, et al. Genomic and single‐cell landscape reveals novel drivers and therapeutic vulnerabilities of transformed cutaneous T‐cell lymphoma. Cancer Discov. 2022;12:1294–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu J, Lao L, Chen J, Li J, Zeng W, Zhu X, et al. The IRENA lncRNA converts chemotherapy‐polarized tumor‐suppressing macrophages to tumor‐promoting phenotypes in breast cancer. Nature Cancer. 2021;2(4):457–73. [DOI] [PubMed] [Google Scholar]
- 7. Amend SR, Pienta KJ. Ecology meets cancer biology: the cancer swamp promotes the lethal cancer phenotype. Oncotarget. 2015;6(12):9669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu Q, Yu X, Li J, Sun S, Tu Y. Metabolic regulation in the immune response to cancer. Cancer Commun (Lond). 2021;41(8):661–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Axelrod R, Axelrod DE, Pienta KJ. Evolution of cooperation among tumor cells. Proc Natl Acad Sci U S A. 2006;103(36):13474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mueller MM, Fusenig NE. Friends or foes ‐ bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4(11):839–49. [DOI] [PubMed] [Google Scholar]
- 11. Ishiguro K, Yoshida T, Yagishita H, Numata Y, Okayasu T. Epithelial and stromal genetic instability contributes to genesis of colorectal adenomas. Gut. 2006;55(5):695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lyssiotis CA, Kimmelman AC. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017;27(11):863–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle‐packaged HIF‐1α‐stabilizing lncRNA from tumour‐associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21(4):498–510. [DOI] [PubMed] [Google Scholar]
- 14. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular Vesicles in Cancer: Cell‐to‐Cell Mediators of Metastasis. Cancer Cell. 2016;30(6):836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boyle ST, Poltavets V, Kular J, Pyne NT, Sandow JJ, Lewis AC, et al. ROCK‐mediated selective activation of PERK signalling causes fibroblast reprogramming and tumour progression through a CRELD2‐dependent mechanism. Nat Cell Biol. 2020;22(7):882–95. [DOI] [PubMed] [Google Scholar]
- 16. Chen X, Song E. Turning foes to friends: targeting cancer‐associated fibroblasts. Nat Rev Drug Discov. 2019;18(2):99–115. [DOI] [PubMed] [Google Scholar]
- 17. Su S, Chen J, Yao H, Liu J, Yu S, Lao L, et al. CD10(+)GPR77(+) Cancer‐Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell. 2018;172(4):841–56.e16. [DOI] [PubMed] [Google Scholar]
- 18. Mishra P, Banerjee D, Ben‐Baruch A. Chemokines at the crossroads of tumor‐fibroblast interactions that promote malignancy. J Leukoc Biol. 2011;89(1):31–9. [DOI] [PubMed] [Google Scholar]
- 19. Orimo A, Gupta PB, Sgroi DC, Arenzana‐Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF‐1/CXCL12 secretion. Cell. 2005;121(3):335–48. [DOI] [PubMed] [Google Scholar]
- 20. Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, et al. Direct visualization of macrophage‐assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67(6):2649–56. [DOI] [PubMed] [Google Scholar]
- 21. De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer. 2017;17(8):457–74. [DOI] [PubMed] [Google Scholar]
- 22. Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–6. [DOI] [PubMed] [Google Scholar]
- 23. Su S, Liu Q, Chen J, Chen F, He C, Huang D, et al. A positive feedback loop between mesenchymal‐like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25(5):605–20. [DOI] [PubMed] [Google Scholar]
- 24. Chen J, Yao Y, Gong C, Yu F, Su S, Liu B, et al. CCL18 from tumor‐associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19(4):541–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keeley T, Costanzo‐Garvey DL, Cook LM. Unmasking the Many Faces of Tumor‐Associated Neutrophils and Macrophages: Considerations for Targeting Innate Immune Cells in Cancer. Trends Cancer. 2019;5(12):789–98. [DOI] [PubMed] [Google Scholar]
- 26. Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, et al. A pan‐cancer single‐cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184(3):792–809.e23. [DOI] [PubMed] [Google Scholar]
- 27. Su S, Liao J, Liu J, Huang D, He C, Chen F, et al. Blocking the recruitment of naive CD4(+) T cells reverses immunosuppression in breast cancer. Cell Res. 2017;27(4):461–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sakurai T, He G, Matsuzawa A, Yu GY, Maeda S, Hardiman G, et al. Hepatocyte necrosis induced by oxidative stress and IL‐1 alpha release mediate carcinogen‐induced compensatory proliferation and liver tumorigenesis. Cancer Cell. 2008;14(2):156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watnick RS, Rodriguez RK, Wang S, Blois AL, Rangarajan A, Ince T, et al. Thrombospondin‐1 repression is mediated via distinct mechanisms in fibroblasts and epithelial cells. Oncogene. 2015;34(22):2823–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wong PP, Muñoz‐Félix JM, Hijazi M, Kim H, Robinson SD, De Luxán‐Delgado B, et al. Cancer Burden Is Controlled by Mural Cell‐β3‐Integrin Regulated Crosstalk with Tumor Cells. Cell. 2020;181(6):1346–63.e21. [DOI] [PubMed] [Google Scholar]
- 32. Zhang XN, Yang KD, Chen C, He ZC, Wang QH, Feng H, et al. Pericytes augment glioblastoma cell resistance to temozolomide through CCL5‐CCR5 paracrine signaling. Cell Res. 2021;31:1072–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cox TR. The matrix in cancer. Nat Rev Cancer. 2021;21:217–38. [DOI] [PubMed] [Google Scholar]
- 34. Mohan V, Das A, Sagi I. Emerging roles of ECM remodeling processes in cancer. Semin Cancer Biol. 2020;62:192–200. [DOI] [PubMed] [Google Scholar]
- 35. Lee S, Jilani SM, Nikolova GV, Carpizo D, Iruela‐Arispe ML. Processing of VEGF‐A by matrix metalloproteinases regulates bioavailability and vascular patterning in tumors. J Cell Biol. 2005;169(4):681–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu JS, Sheng SR, Liang XH, Tang YL. The role of tumor microenvironment in collective tumor cell invasion. Future Oncol. 2017;13(11):991–1002. [DOI] [PubMed] [Google Scholar]
- 37. Giraudo E, Inoue M, Hanahan D. An amino‐bisphosphonate targets MMP‐9‐expressing macrophages and angiogenesis to impair cervical carcinogenesis. J Clin Invest. 2004;114(5):623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, et al. Inhibitory effect of tumor cell‐derived lactic acid on human T cells. Blood. 2007;109(9):3812–9. [DOI] [PubMed] [Google Scholar]
- 39. Gottfried E, Kunz‐Schughart LA, Ebner S, Mueller‐Klieser W, Hoves S, Andreesen R, et al. Tumor‐derived lactic acid modulates dendritic cell activation and antigen expression. Blood. 2006;107(5):2013–21. [DOI] [PubMed] [Google Scholar]
- 40. Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, et al. Functional polarization of tumour‐associated macrophages by tumour‐derived lactic acid. Nature. 2014;513(7519):559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Végran F, Boidot R, Michiels C, Sonveaux P, Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF‐κB/IL‐8 pathway that drives tumor angiogenesis. Cancer Res. 2011;71(7):2550–60. [DOI] [PubMed] [Google Scholar]
- 42. Martínez‐Ordoñez A, Seoane S, Avila L, Eiro N, Macía M, Arias E, et al. POU1F1 transcription factor induces metabolic reprogramming and breast cancer progression via LDHA regulation. Oncogene. 2021;40(15):2725–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baumann F, Leukel P, Doerfelt A, Beier CP, Dettmer K, Oefner PJ, et al. Lactate promotes glioma migration by TGF‐beta2‐dependent regulation of matrix metalloproteinase‐2. Neuro Oncol. 2009;11(4):368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Govaert KM, Emmink BL, Nijkamp MW, Cheung ZJ, Steller EJ, Fatrai S, et al. Hypoxia after liver surgery imposes an aggressive cancer stem cell phenotype on residual tumor cells. Ann Surg. 2014;259(4):750–9. [DOI] [PubMed] [Google Scholar]
- 45. Chen KW, Pienta KJ. Modeling invasion of metastasizing cancer cells to bone marrow utilizing ecological principles. Theor Biol Med Model. 2011;8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peinado H, Zhang H, Matei IR, Costa‐Silva B, Hoshino A, Rodrigues G, et al. Pre‐metastatic niches: organ‐specific homes for metastases. Nat Rev Cancer. 2017;17(5):302–17. [DOI] [PubMed] [Google Scholar]
- 47. Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin‐like 4. Cell. 2008;133(1):66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, et al. Mediators of vascular remodelling co‐opted for sequential steps in lung metastasis. Nature. 2007;446(7137):765–70. [DOI] [PubMed] [Google Scholar]
- 49. Huang Y, Song N, Ding Y, Yuan S, Li X, Cai H, et al. Pulmonary vascular destabilization in the premetastatic phase facilitates lung metastasis. Cancer Res. 2009;69(19):7529–37. [DOI] [PubMed] [Google Scholar]
- 50. Hoshino A, Costa‐Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, et al. VEGFR1‐positive haematopoietic bone marrow progenitors initiate the pre‐metastatic niche. Nature. 2005;438(7069):820–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Erler JT, Bennewith KL, Cox TR, Lang G, Bird D, Koong A, et al. Hypoxia‐induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15(1):35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cox TR, Bird D, Baker AM, Barker HE, Ho MW, Lang G, et al. LOX‐mediated collagen crosslinking is responsible for fibrosis‐enhanced metastasis. Cancer Res. 2013;73(6):1721–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, et al. Tumor Exosomal RNAs Promote Lung Pre‐metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell. 2016;30(2):243–56. [DOI] [PubMed] [Google Scholar]
- 55. Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis‐initiating breast cancer cells. Nature. 2015;528(7582):413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sharma SK, Chintala NK, Vadrevu SK, Patel J, Karbowniczek M, Markiewski MM. Pulmonary alveolar macrophages contribute to the premetastatic niche by suppressing antitumor T cell responses in the lungs. J Immunol. 2015;194(11):5529–38. [DOI] [PubMed] [Google Scholar]
- 57. Morrissey SM, Zhang F, Ding C, Montoya‐Durango DE, Hu X, Yang C, et al. Tumor‐derived exosomes drive immunosuppressive macrophages in a pre‐metastatic niche through glycolytic dominant metabolic reprogramming. Cell Metab. 2021;33(10):2040–58.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, et al. Cancer‐derived exosomal miR‐25‐3p promotes pre‐metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9(1):5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Costa‐Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, et al. Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee JW, Stone ML, Porrett PM, Thomas SK, Komar CA, Li JH, et al. Hepatocytes direct the formation of a pro‐metastatic niche in the liver. Nature. 2019;567(7747):249–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang S, Xu Y, Xie C, Ren L, Wu G, Yang M, et al. RNF219/α‐Catenin/LGALS3 Axis Promotes Hepatocellular Carcinoma Bone Metastasis and Associated Skeletal Complications. Adv Sci (Weinh). 2021;8(4):2001961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cox TR, Rumney RMH, Schoof EM, Perryman L, Høye AM, Agrawal A, et al. The hypoxic cancer secretome induces pre‐metastatic bone lesions through lysyl oxidase. Nature. 2015;522(7554):106–10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63. Borel M, Lollo G, Magne D, Buchet R, Brizuela L, Mebarek S. Prostate cancer‐derived exosomes promote osteoblast differentiation and activity through phospholipase D2. Biochim Biophys Acta Mol Basis Dis. 2020;1866(12):165919. [DOI] [PubMed] [Google Scholar]
- 64. Ardura JA, Álvarez‐Carrión L, Gutiérrez‐Rojas I, Friedman PA, Gortázar AR, Alonso V. MINDIN secretion by prostate tumors induces premetastatic changes in bone via β‐catenin. Endocr Relat Cancer. 2020;27(7):441–56. [DOI] [PubMed] [Google Scholar]
- 65. Steeg PS, Camphausen KA, Smith QR. Brain metastases as preventive and therapeutic targets. Nat Rev Cancer. 2011;11(5):352–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, et al. Microenvironment‐induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527(7576):100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lyle LT, Lockman PR, Adkins CE, Mohammad AS, Sechrest E, Hua E, et al. Alterations in Pericyte Subpopulations Are Associated with Elevated Blood‐Tumor Barrier Permeability in Experimental Brain Metastasis of Breast Cancer. Clin Cancer Res. 2016;22(21):5287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Carvalho R, Paredes J, Ribeiro AS. Impact of breast cancer cells´ secretome on the brain metastatic niche remodeling. Semin Cancer Biol. 2020;60:294–301. [DOI] [PubMed] [Google Scholar]
- 69. You H, Baluszek S, Kaminska B. Supportive roles of brain macrophages in CNS metastases and assessment of new approaches targeting their functions. Theranostics. 2020;10(7):2949–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ‐specific metastasis. Nat Rev Immunol. 2011;11(9):597–606. [DOI] [PubMed] [Google Scholar]
- 71. Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4(7):540–50. [DOI] [PubMed] [Google Scholar]
- 72. Murphy PM. Chemokines and the molecular basis of cancer metastasis. N Engl J Med. 2001;345(11):833–5. [DOI] [PubMed] [Google Scholar]
- 73. Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–6. [DOI] [PubMed] [Google Scholar]
- 74. Amersi FF, Terando AM, Goto Y, Scolyer RA, Thompson JF, Tran AN, et al. Activation of CCR9/CCL25 in cutaneous melanoma mediates preferential metastasis to the small intestine. Clin Cancer Res. 2008;14(3):638–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang L, Liu Q, Zhang X, Liu X, Zhou B, Chen J, et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature. 2020;583(7814):133–8. [DOI] [PubMed] [Google Scholar]
- 76. Hiratsuka S, Goel S, Kamoun WS, Maru Y, Fukumura D, Duda DG, et al. Endothelial focal adhesion kinase mediates cancer cell homing to discrete regions of the lungs via E‐selectin up‐regulation. Proc Natl Acad Sci U S A. 2011;108(9):3725–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Malanchi I, Santamaria‐Martínez A, Susanto E, Peng H, Lehr HA, Delaloye JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481(7379):85–9. [DOI] [PubMed] [Google Scholar]
- 78. Reuten R, Zendehroud S, Nicolau M, Fleischhauer L, Laitala A, Kiderlen S, et al. Basement membrane stiffness determines metastases formation. Nat Mater. 2021;20(6):892–903. [DOI] [PubMed] [Google Scholar]
- 79. Uhr JW, Scheuermann RH, Street NE, Vitetta ES. Cancer dormancy: opportunities for new therapeutic approaches. Nat Med. 1997;3(5):505–9. [DOI] [PubMed] [Google Scholar]
- 80. Phan TG, Croucher PI. The dormant cancer cell life cycle. Nat Rev Cancer. 2020;20(7):398–411. [DOI] [PubMed] [Google Scholar]
- 81. Zhang W, Bado IL, Hu J, Wan YW, Wu L, Wang H, et al. The bone microenvironment invigorates metastatic seeds for further dissemination. Cell. 2021;184(9):2471–86.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–8. [DOI] [PubMed] [Google Scholar]
- 83. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331(6024):1565–70. [DOI] [PubMed] [Google Scholar]
- 84. Song E, Mao T, Dong H, Boisserand LSB, Antila S, Bosenberg M, et al. VEGF‐C‐driven lymphatic drainage enables immunosurveillance of brain tumours. Nature. 2020;577(7792):689–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Park JH, Kim HJ, Kim CW, Kim HC, Jung Y, Lee HS, et al. Tumor hypoxia represses γδ T cell‐mediated antitumor immunity against brain tumors. Nat Immunol. 2021;22(3):336–46. [DOI] [PubMed] [Google Scholar]
- 86. Ducimetière L, Lucchiari G, Litscher G, Nater M, Heeb L, Nuñez NG, et al. Conventional NK cells and tissue‐resident ILC1s join forces to control liver metastasis. Proc Natl Acad Sci U S A. 2021;118(27):e2026271118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Molgora M, Bonavita E, Ponzetta A, Riva F, Barbagallo M, Jaillon S, et al. IL‐1R8 is a checkpoint in NK cells regulating anti‐tumour and anti‐viral activity. Nature. 2017;551(7678):110–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tello‐Lafoz M, Srpan K, Sanchez EE, Hu J, Remsik J, Romin Y, et al. Cytotoxic lymphocytes target characteristic biophysical vulnerabilities in cancer. Immunity. 2021;54(5):1037–54.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT, et al. Neoantigen‐directed immune escape in lung cancer evolution. Nature. 2019;567(7749):479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bukur J, Jasinski S, Seliger B. The role of classical and non‐classical HLA class I antigens in human tumors. Semin Cancer Biol. 2012;22(4):350–8. [DOI] [PubMed] [Google Scholar]
- 91. Jiang T, Shi T, Zhang H, Hu J, Song Y, Wei J, et al. Tumor neoantigens: from basic research to clinical applications. J Hematol Oncol. 2019;12(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, et al. Receptors for HLA class‐I molecules in human natural killer cells. Annu Rev Immunol. 1996;14:619–48. [DOI] [PubMed] [Google Scholar]
- 93. Sanchez‐Correa B, Valhondo I, Hassouneh F, Lopez‐Sejas N, Pera A, Bergua JM, et al. DNAM‐1 and the TIGIT/PVRIG/TACTILE Axis: Novel immune checkpoints for natural killer cell‐based cancer immunotherapy. Cancers (Basel). 2019;11(6):877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kubli SP, Berger T, Araujo DV, Siu LL, Mak TW. Beyond immune checkpoint blockade: emerging immunological strategies. Nat Rev Drug Discov. 2021;20:899–919. [DOI] [PubMed] [Google Scholar]
- 95. Song TL, Nairismägi ML, Laurensia Y, Lim JQ, Tan J, Li ZM, et al. Oncogenic activation of the STAT3 pathway drives PD‐L1 expression in natural killer/T‐cell lymphoma. Blood. 2018;132(11):1146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Prestipino A, Emhardt AJ, Aumann K, O'Sullivan D, Gorantla SP, Duquesne S, et al. Oncogenic JAK2(V617F) causes PD‐L1 expression, mediating immune escape in myeloproliferative neoplasms. Sci Transl Med. 2018;10(429):eaam7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang C, Li Y, Jia L, Kim JK, Li J, Deng P, et al. CD276 expression enables squamous cell carcinoma stem cells to evade immune surveillance. Cell Stem Cell. 2021;28(9):1597–613.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang Y, Wang M, Wu HX, Xu RH. Advancing to the era of cancer immunotherapy. Cancer Commun (Lond). 2021;41(9):803–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6(4):295–307. [DOI] [PubMed] [Google Scholar]
- 100. Watson MJ, Vignali PDA, Mullett SJ, Overacre‐Delgoffe AE, Peralta RM, Grebinoski S, et al. Metabolic support of tumour‐infiltrating regulatory T cells by lactic acid. Nature. 2021;591(7851):645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Reinfeld BI, Madden MZ, Wolf MM, Chytil A, Bader JE, Patterson AR, et al. Cell‐programmed nutrient partitioning in the tumour microenvironment. Nature. 2021;593(7858):282–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chen MM, Xiao X, Lao XM, Wei Y, Liu RX, Zeng QH, et al. Polarization of Tissue‐Resident TFH‐Like Cells in Human Hepatoma Bridges Innate Monocyte Inflammation and M2b Macrophage Polarization. Cancer Discov. 2016;6(10):1182–95. [DOI] [PubMed] [Google Scholar]
- 103. Veglia F, Perego M, Gabrilovich D. Myeloid‐derived suppressor cells coming of age. Nat Immunol. 2018;19(2):108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Veglia F, Sanseviero E, Gabrilovich DI. Myeloid‐derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. 2021:21:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tauriello DVF, Palomo‐Ponce S, Stork D, Berenguer‐Llergo A, Badia‐Ramentol J, Iglesias M, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–43. [DOI] [PubMed] [Google Scholar]
- 106. Batlle E, Massagué J. Transforming Growth Factor‐β Signaling in Immunity and Cancer. Immunity. 2019;50(4):924–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ouyang W, O'Garra A. IL‐10 Family Cytokines IL‐10 and IL‐22: from Basic Science to Clinical Translation. Immunity. 2019;50(4):871–91. [DOI] [PubMed] [Google Scholar]
- 108. Strand S, Hofmann WJ, Hug H, Muller M, Otto G, Strand D, et al. Lymphocyte apoptosis induced by CD95 (APO‐1/Fas) ligand‐expressing tumor cells–a mechanism of immune evasion? Nat Med. 1996;2(12):1361–6. [DOI] [PubMed] [Google Scholar]
- 109. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF‐κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–81. [DOI] [PubMed] [Google Scholar]
- 110. Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation‐induced cell death. Nat Immunol. 2018;19(10):1112–25. [DOI] [PubMed] [Google Scholar]
- 111. Huang D, Chen X, Zeng X, Lao L, Li J, Xing Y, et al. Targeting regulator of G protein signaling 1 in tumor‐specific T cells enhances their trafficking to breast cancer. Nat Immunol. 2021;22:865–879. [DOI] [PubMed] [Google Scholar]
- 112. Wang Y, Shen Y, Wang S, Shen Q, Zhou X. The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 2018;415:117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X. Targeting STAT3 in Cancer Immunotherapy. Mol Cancer. 2020;19(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Heaney AP, Melmed S. Molecular targets in pituitary tumours. Nat Rev Cancer. 2004;4(4):285–95. [DOI] [PubMed] [Google Scholar]
- 115. Lacroix A, Feelders RA, Stratakis CA, Nieman LK. Cushing's syndrome. Lancet. 2015;386(9996):913–27. [DOI] [PubMed] [Google Scholar]
- 116. Gazdar AF, Bunn PA, Minna JD. Small‐cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer. 2017;17(12):725–37. [DOI] [PubMed] [Google Scholar]
- 117. Titulaer MJ, Lang B, Verschuuren JJ. Lambert‐Eaton myasthenic syndrome: from clinical characteristics to therapeutic strategies. Lancet Neurol. 2011;10(12):1098–107. [DOI] [PubMed] [Google Scholar]
- 118. Laviano A, Meguid MM, Rossi‐Fanelli F. Cancer anorexia: clinical implications, pathogenesis, and therapeutic strategies. Lancet Oncol. 2003;4(11):686–94. [DOI] [PubMed] [Google Scholar]
- 119. Tisdale MJ. Mechanisms of cancer cachexia. Physiol Rev. 2009;89(2):381–410. [DOI] [PubMed] [Google Scholar]
- 120. Shi M, Liu D, Yang Z, Guo N. Central and peripheral nervous systems: master controllers in cancer metastasis. Cancer Metastasis Rev. 2013;32(3‐4):603–21. [DOI] [PubMed] [Google Scholar]
- 121. Cole SW, Nagaraja AS, Lutgendorf SK, Green PA, Sood AK. Sympathetic nervous system regulation of the tumour microenvironment. Nat Rev Cancer. 2015;15(9):563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zahalka AH, Arnal‐Estapé A, Maryanovich M, Nakahara F, Cruz CD, Finley LWS, et al. Adrenergic nerves activate an angio‐metabolic switch in prostate cancer. Science. 2017;358(6361):321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Renz BW, Tanaka T, Sunagawa M, Takahashi R, Jiang Z, Macchini M, et al. Cholinergic Signaling via Muscarinic Receptors Directly and Indirectly Suppresses Pancreatic Tumorigenesis and Cancer Stemness. Cancer Discov. 2018;8(11):1458–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Courchamp F, Clutton‐Brock T, Grenfell B. Inverse density dependence and the Allee effect. Trends Ecol Evol. 1999;14(10):405–10. [DOI] [PubMed] [Google Scholar]
- 125. Johnson KE, Howard G, Mo W, Strasser MK, Lima E, Huang S, et al. Cancer cell population growth kinetics at low densities deviate from the exponential growth model and suggest an Allee effect. PLoS Biol. 2019;17(8):e3000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lichter AS, Lippman ME, Danforth DN, Jr. , d'Angelo T, Steinberg SM, deMoss E, et al. Mastectomy versus breast‐conserving therapy in the treatment of stage I and II carcinoma of the breast: a randomized trial at the National Cancer Institute. J Clin Oncol. 1992;10(6):976–83. [DOI] [PubMed] [Google Scholar]
- 127. van Dongen JA, Bartelink H, Fentiman IS, Lerut T, Mignolet F, Olthuis G, et al. Factors influencing local relapse and survival and results of salvage treatment after breast‐conserving therapy in operable breast cancer: EORTC trial 10801, breast conservation compared with mastectomy in TNM stage I and II breast cancer. Eur J Cancer. 1992;28a(4‐5):801–5. [DOI] [PubMed] [Google Scholar]
- 128. Fisher B, Anderson S, Bryant J, Margolese RG, Deutsch M, Fisher ER, et al. Twenty‐year follow‐up of a randomized trial comparing total mastectomy, lumpectomy, and lumpectomy plus irradiation for the treatment of invasive breast cancer. N Engl J Med. 2002;347(16):1233–41. [DOI] [PubMed] [Google Scholar]
- 129. van Maaren MC, de Munck L, de Bock GH, Jobsen JJ, van Dalen T, Linn SC, et al. 10 year survival after breast‐conserving surgery plus radiotherapy compared with mastectomy in early breast cancer in the Netherlands: a population‐based study. Lancet Oncol. 2016;17(8):1158–70. [DOI] [PubMed] [Google Scholar]
- 130. Giacalone NJ, Shipley WU, Clayman RH, Niemierko A, Drumm M, Heney NM, et al. Long‐term Outcomes After Bladder‐preserving Tri‐modality Therapy for Patients with Muscle‐invasive Bladder Cancer: An Updated Analysis of the Massachusetts General Hospital Experience. Eur Urol. 2017;71(6):952–60. [DOI] [PubMed] [Google Scholar]
- 131. McLaughlin M, Patin EC, Pedersen M, Wilkins A, Dillon MT, Melcher AA, et al. Inflammatory microenvironment remodelling by tumour cells after radiotherapy. Nat Rev Cancer. 2020;20(4):203–17. [DOI] [PubMed] [Google Scholar]
- 132. Chang MC, Chen YL, Lin HW, Chiang YC, Chang CF, Hsieh SF, et al. Irradiation Enhances Abscopal Anti‐tumor Effects of Antigen‐Specific Immunotherapy through Regulating Tumor Microenvironment. Mol Ther. 2018;26(2):404–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Mondini M, Loyher PL, Hamon P, Gerbé de Thoré M, Laviron M, Berthelot K, et al. CCR2‐Dependent Recruitment of Tregs and Monocytes Following Radiotherapy Is Associated with TNFα‐Mediated Resistance. Cancer Immunol Res. 2019;7(3):376–87. [DOI] [PubMed] [Google Scholar]
- 134. Liang H, Deng L, Hou Y, Meng X, Huang X, Rao E, et al. Host STING‐dependent MDSC mobilization drives extrinsic radiation resistance. Nat Commun. 2017;8(1):1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Marciscano AE, Ghasemzadeh A, Nirschl TR, Theodros D, Kochel CM, Francica BJ, et al. Elective Nodal Irradiation Attenuates the Combinatorial Efficacy of Stereotactic Radiation Therapy and Immunotherapy. Clin Cancer Res. 2018;24(20):5058–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Stijns RCH, de Graaf EJR, Punt CJA, Nagtegaal ID, Nuyttens J, van Meerten E, et al. Long‐term Oncological and Functional Outcomes of Chemoradiotherapy Followed by Organ‐Sparing Transanal Endoscopic Microsurgery for Distal Rectal Cancer: The CARTS Study. JAMA Surg. 2019;154(1):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Axelrod ML, Nixon MJ, Gonzalez‐Ericsson PI, Bergman RE, Pilkinton MA, McDonnell WJ, et al. Changes in Peripheral and Local Tumor Immunity after Neoadjuvant Chemotherapy Reshape Clinical Outcomes in Patients with Breast Cancer. Clin Cancer Res. 2020;26(21):5668–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Su S, Zhao J, Xing Y, Zhang X, Liu J, Ouyang Q, et al. Immune Checkpoint Inhibition Overcomes ADCP‐Induced Immunosuppression by Macrophages. Cell. 2018;175(2):442–57.e23. [DOI] [PubMed] [Google Scholar]
- 139. Hanoteau A, Newton JM, Krupar R, Huang C, Liu HC, Gaspero A, et al. Tumor microenvironment modulation enhances immunologic benefit of chemoradiotherapy. J Immunother Cancer. 2019;7(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Bi K, He MX, Bakouny Z, Kanodia A, Napolitano S, Wu J, et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell. 2021;39(5):649–61.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Verma V, Jafarzadeh N, Boi S, Kundu S, Jiang Z, Fan Y, et al. MEK inhibition reprograms CD8(+) T lymphocytes into memory stem cells with potent antitumor effects. Nat Immunol. 2021;22(1):53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Braun DA, Hou Y, Bakouny Z, Ficial M, Sant' Angelo M, Forman J, et al. Interplay of somatic alterations and immune infiltration modulates response to PD‐1 blockade in advanced clear cell renal cell carcinoma. Nat Med. 2020;26(6):909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Sanmamed MF, Nie X, Desai SS, Villaroel‐Espindola F, Badri T, Zhao D, et al. A Burned‐Out CD8(+) T‐cell Subset Expands in the Tumor Microenvironment and Curbs Cancer Immunotherapy. Cancer Discov. 2021;11(7):1700–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yu J, Green MD, Li S, Sun Y, Journey SN, Choi JE, et al. Liver metastasis restrains immunotherapy efficacy via macrophage‐mediated T cell elimination. Nat Med. 2021;27(1):152–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Liang P, Ballou B, Lv X, Si W, Bruchez MP, Huang W, et al. Monotherapy and Combination Therapy Using Anti‐Angiogenic Nanoagents to Fight Cancer. Adv Mater. 2021;33(15):e2005155. [DOI] [PubMed] [Google Scholar]
- 146. Ding Z, Li Q, Zhang R, Xie L, Shu Y, Gao S, et al. Personalized neoantigen pulsed dendritic cell vaccine for advanced lung cancer. Signal Transduct Target Ther. 2021;6(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single‐Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 2018;174(5):1293–308 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial Organization and Molecular Correlation of Tumor‐Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 2018;23(1):181–93 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Courtiol P, Maussion C, Moarii M, Pronier E, Pilcer S, Sefta M, et al. Deep learning‐based classification of mesothelioma improves prediction of patient outcome. Nat Med. 2019;25(10):1519–25. [DOI] [PubMed] [Google Scholar]
- 150. Jackson HW, Fischer JR, Zanotelli VRT, Ali HR, Mechera R, Soysal SD, et al. The single‐cell pathology landscape of breast cancer. Nature. 2020;578(7796):615–20. [DOI] [PubMed] [Google Scholar]
- 151. Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, et al. A Structured Tumor‐Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell. 2018;174(6):1373–87 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology ‐ new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16(11):703–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. West J, You L, Zhang J, Gatenby RA, Brown JS, Newton PK, et al. Towards Multidrug Adaptive Therapy. Cancer Res. 2020;80(7):1578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Kather JN, Poleszczuk J, Suarez‐Carmona M, Krisam J, Charoentong P, Valous NA, et al. In Silico Modeling of Immunotherapy and Stroma‐Targeting Therapies in Human Colorectal Cancer. Cancer Res. 2017;77(22):6442–52. [DOI] [PubMed] [Google Scholar]
- 155. West J, Robertson‐Tessi M, Luddy K, Park DS, Williamson DFK, Harmon C, et al. The Immune Checkpoint Kick Start: Optimization of Neoadjuvant Combination Therapy Using Game Theory. JCO Clin Cancer Inform. 2019;3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Byrne AT, Alférez DG, Amant F, Annibali D, Arribas J, Biankin AV, et al. Interrogating open issues in cancer precision medicine with patient‐derived xenografts. Nat Rev Cancer. 2017;17(4):254–68. [DOI] [PubMed] [Google Scholar]
- 157. Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364(6444):952–5. [DOI] [PubMed] [Google Scholar]
- 158. Smyth MJ, Ngiow SF, Ribas A, Teng MW. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13(3):143–58. [DOI] [PubMed] [Google Scholar]
- 159. Maley CC, Aktipis A, Graham TA, Sottoriva A, Boddy AM, Janiszewska M, et al. Classifying the evolutionary and ecological features of neoplasms. Nat Rev Cancer. 2017;17(10):605–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


