Abstract
The transcription factor Forkhead box M1 (FoxM1) is overexpressed in breast cancers and correlates with poor prognosis. Mechanistically, FoxM1 associates with CBP to activate transcription and with Rb to repress transcription. While the activating function of FoxM1 in breast cancer has been well documented, the significance of its repressive activity is poorly understood. Using CRISPR-Cas9 engineering, we generated a mouse model that expresses FoxM1 harboring point mutations that block binding to Rb while retaining its ability to bind CBP. Unlike FoxM1-null mice, mice harboring Rb-binding mutant FoxM1 did not exhibit significant developmental defects. The mutant mouse line developed PyMT-driven mammary tumors that were deficient in lung metastasis, which was tumor cell-intrinsic. Single-cell RNA-seq of the tumors revealed a deficiency in pro-metastatic tumor cells and an expansion of differentiated alveolar-type tumor cells, and further investigation identified that loss of the FoxM1/Rb interaction caused enhancement of the mammary alveolar differentiation program. The FoxM1 mutant tumors also showed increased Pten expression, and FoxM1/Rb was found to activate Akt signaling by repressing Pten. In human breast cancers, expression of FoxM1 negatively correlated with Pten mRNA. Furthermore, the lack of tumor-infiltrating cells in FoxM1 mutant tumors appeared related to decreases in pro-metastatic tumor cells that express factors required for infiltration. These observations demonstrate that the FoxM1/Rb-regulated transcriptome is critical for the plasticity of breast cancer cells that drive metastasis, identifying a pro-metastatic role of Rb when bound to FoxM1.
INTRODUCTION
Metastasis to distal organ is the main cause of mortality from breast cancer. A greater understanding of the molecular events that drive metastatic progression will be important for development of more effective management strategies. The fork-head box transcription factor FoxM1 is significant in that regards because it is overexpressed in breast cancer and its overexpression coincides with aggressive progression and metastasis (1). Moreover, FoxM1 synergizes with a number of pro-oncogenic signaling pathways, including, PDGF (2), Wnt/b-catenin (3), TGF-b (4), STAT3 (5), and Hedgehog pathways (6) to drive aggressive progression. High expression of FoxM1 has been shown also to correlate with drug resistance in breast cancers. For example, FoxM1 over-expression correlates with herceptin-resistance (7), as well as resistance to cytotoxic chemotherapeutics (8). A recent study provided evidence that resistance to PI3-kinase-inhibitors correlates with increased accumulation of FoxM1 (9).
The cancer-related studies on FoxM1 thus far focused mainly on the transcriptional stimulatory activities of FoxM1 that increase expression of a variety of genes important for tumor progression. FoxM1 also possesses a transcriptional repressor function. FoxM1 associates with the retinoblastoma (Rb)(10) protein in G1 phase of the cell cycle (11). We showed that the FoxM1/Rb interaction is important for repression of the mammary differentiation gene Gata3 (1). The repression involves recruitment of DNMT3B and methylation of the CpG-islands in the Gata3 promoter (1). FoxM1/Rb also represses the luminal differentiation gene FoxA1 in breast cancer cells and FoxA2 in liver cancer cells through a similar mechanism (12,13). However, the roles of the repressor activity of FoxM1 in cancer development/progression is poorly understood.
FoxM1 is phosphorylated by Cyc-Cdks, and that phosphorylation serves as a primer for phosphorylation by Plk1(14,15). The phosphorylation by Cyc-Cdks or Plk1 inhibited FoxM1’s interaction with Rb (12). The Plk1-mediated phosphorylation of FoxM1 serves as a molecular switch in deciding interactions with the activation partner CBP versus the repression partner Rb. That allowed the two activities of FoxM1 to be separated from each other using targeted mutagenesis. For example, a phospho-mimetic mutant of FoxM1 (FoxM1DD), at two S-residues (715 and 724) in the Plk1-sites, binds to CBP and activates transcription, but fails to bind Rb and is deficient in the repressor activity (12).
To investigate the role of the repression function of FoxM1 in mammary tumors, we introduced knock-in Plk1-sites phospho-mimetic FoxM1DD mutations into the MMTV-PyMT mouse mammary tumor model. We show that the repression function of FoxM1 is dispensable for primary tumor growth, but it is remarkably important for metastasis. A variety of mechanistic analyses demonstrate that the repression mechanism of FoxM1/Rb is necessary for regulating tumor differentiation and support the pro-metastatic tumor cells that create a microenvironment with abundant infiltrating cells for metastasis.
MATERIALS AND METHODS
Mouse studies
All mice were generated and maintained in C57BL/6J background. The C57BL/6J mice were purchased from Charles River Laboratories. FoxM1DD/DD mice were generated in this study. The MMTV-PyMT mice were purchased from the Jackson Laboratory (stock#022974), as well as mT/mG mice, stock #007576.
FoxM1DD/DD MMTV-PyMT mice were generated by crossing FoxM1DD/DD mice with MMTV- PyMT mice. FoxM1DD/DD mT/mG mice were generated by crossing FoxM1DD/DD mice with mT/mG mice. FoxM1DD/DD MMTV-PYMT mT/mG mice were generated by crossing FoxM1DD/DD mT/mG mice with MMTV-PYMT mice. Only female mice were used for the experiments. Primary tumors reaching diameter of 2 cm marked end-point of the experiments. Total of 22 FoxM1+/+ PYMT+, 34 FoxM1+/DD PYMT+ and 39 FoxM1DD/DD PYMT+ female mice were examined for primary tumors.
For orthotopic implantation, primary tumor cells (for isolation, please look below) were resuspended in full medium (10%FBS, 1% Pen/Strep in DMEM) and Matrigel (Corning) in a 1:1 ratio with a final concentration of 5x105 cells/ml. One hundred microliters of cells were transplanted to the left fourth mammary gland fat pad of recipient mice. Mice were sacrificed and lungs harvested when orthotopic tumors reached 2 cm in size. For tail vein injection, primary tumor cells were resuspended in PBS at final concentration of 106 cells/ml. One hundred microliters of cell suspension were injected into wild-type female mouse through tail vein. Eight recipient mice were injected with FoxM1+/+ PYMT+ and 6 mice were injected with FoxM1DD/DD PYMT+ tumor cells. Four weeks after tail vein injection mice were sacrificed and lungs analyzed. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Illinois at Chicago (UIC).
Primary tumor cell isolation
Mouse tumors were harvested, washed in PBS and cut into small pieces (diameter around 2 mm). After centrifugation at 400 g for 5 minutes, supernatant was discarded and pellet was transferred to plates containing DMEM supplemented with 10% FBS and 1% antibiotics (Pen/Strep) in an incubator at 37°C with 5% CO2. Fresh primary tumor cells were used only up to second passage.
Cell Culture
Human breast cancer cell lines, MCF7 (ATCC Cat# HTB-22, RRID:CVCL_0031) and MDA-MB-231 (ATCC Cat# CRM-HTB-26, RRID:CVCL_0062) were obtained from American Type Culture Collection (ATCC), and used within six months after resuscitation, within 5th to 10th passages. They were authenticated by ATCC using short tandem repeat profiling. No testing for Mycoplasma was performed in the laboratory. Primary Mouse Embryonic Fibroblasts, MEFs, were isolated from 13.5 days old embryos following standard procedure. All cells were cultured in DMEM supplemented with 10% fetal bovine serum and 1% antibiotics (Pen/Strep). MEFs medium was also supplemented with 2% glutamine.
Sample preparation for scRNA-Seq
To prepare cells for scRNAseq, tumors were harvested from 2 independent female FoxM1+/+ PYMT+ (10 tumor nodules) and 2 FoxM1DD/DD PYMT+ mice (6 tumor nodules). All visible necrotic or cystic areas were excluded. Tumors were washed in PBS and dissociated into single-cell suspension. Shortly, tumors were mechanically chopped and digested with 0.1% Collagenase IV and 0.01% DNase in DMEM at 37°C for 30 min. Cells were washed twice with PBS followed by lysis of RBCs with ACK Lysing Buffer (Gibco), and filtered using 40 μm sterile cell strainers. After two washes, cells were resuspended in 0.04% BSA-PBS, and trypan blue staining was performed to asses for the viability of the cells.
Single-cell RNA-Seq
For Drop-seq we followed the protocol version 3.1 [31] with following modifications. Samples and beads were diluted to approximately 110 cells/μl and 120 beads/μl, respectively, and loaded onto a microfluidic chip. The lysis buffer contained 0.4% Sarkosyl (Sigma). The number of cycles in the PCR step post-exonuclease is 16. The cDNA post-PCR was purified twice with 0.6x Agencourt AMPure XP (Beckman Coultier). The tagmented DNA for sequencing was purified twice: first with 0.6× and then with 1× Agencourt AMPure XP (Beckman Coultier). Quality control of amplified cDNA and sequencing-ready library was determined using both Qubit and Agilent TapeStation 4200 instrument.
High-throughput sequencing and processing raw datasets
Drop-seq libraries were sequenced on a NextSeq 500 instrument (Illumina) at the University of Illinois at Chicago Sequencing Core (UICSQC). The Drop-seq samples were processed for read alignment and gene expression quantification following Drop-seq cook-book (version 1.2 Jan 2016)7 (http://mccarrolllab.com/dropseq/; [31]). STAR (RRID:SCR_004463) aligner was used to align the reads against Mus musculus mm10 genome version GRCm38.p5 (Ensembl 90: Aug 2017). The DNA sequences for both Cre and PYMT elements were added to the genome as chromosomes. The program FastQC (RRID:SCR_014583, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to check quality of reads and mapping. Uniquely mapped reads were grouped and counted to generate digital expression matrices.
scRNA-Seq data analysis
For the dataset analysis, we used packages Seurat (version 3.0.0) and R (version 3.5.3) and followed standard tutorial instructions from the Seurat website (https://satijalab.org/seurat/).
We performed quality control analysis using gene expression matrices. Low-quality cells were filtered out using 400 and 2,000 genes per cell as a low and top cutoff, respectively, and min. cell = 5. To exclude dying cells, content of mitochondrial genes was set to less than 5% reads per cell. We performed integrative analysis by determining the anchor points in each dataset (16). To run linear dimensional reduction, top 2000 variable genes were used as input for PCA analysis. The first 40 principal components were used for non-linear dimensional reduction. Graph-based method was used to cluster cells at resolution of 1.0. UMAP was used to visualize clusters.
Tissue staining and immunohistochemistry
Breast tumors and lungs were collected and fixed in 10% formalin (Fisher Chemical) for 24 hours. After fixation, tissues were washed with PBS and left in 75% ethanol until processing, when tissues were passed through grades of alcohols and embedded in paraffin. Sections 5μm thick were prepared, baked overnight at 55°C and stained following standard procedures. Staining was performed with hematoxylin and eosin (H&E) or with specific antibodies. Antigen retrieval was done using sodium citrate buffer at 95°C for 20 minutes, after which slides were allowed to cool down to room temperature. Sections were treated with 3% H2O2 (in methanol) for 5 minutes to quench endogenous peroxidases. After blocking with normal serum, appropriate primary antibodies were incubated overnight at 4°C. Secondary antibodies from Vectastain ABC Kit (Vector Labs) were incubated for 30 minutes at room temperature, and 3, 3-diaminobenzidine [60] kit (Vector Labs) was used to visualize the signal. The sections were counterstained with hematoxylin. The images were acquired and analyzed using Leica DMi8 microscope, LAS X Version: 3.4.2.18368 software. At least 10 fields of tumor nodules per mouse, harvested from 4 different FoxM1+/+ PYMT+ or 6 different FoxM1 DD/DD PYMT+ mice were quantified. Following primary antibodies have been used for IHC staining: anti-CD68 (Abcam, Cat# ab125212, RRID:AB_10975465), anti-CD206 (Cell Signaling Technology, Cat# 24595, RRID:AB_2892682), anti-Ly6G (BioLegend, Cat# 127602, RRID:AB_1089180), anti-CD31 (Santa Cruz Biotechnology, Cat# sc-1506, RRID:AB_2161037), anti-F4/80 (BioRad, Cat # MCA497G, RRID:AB_872005), anti-CD3 antibody (Abcam, Cat# ab16669, RRID:AB_443425) and anti-ki67 (Abcam, Cat# ab16667, RRID:AB_302459).
Protein analysis by immunoblotting
The cells were harvested and washed with cold PBS and lysed in ice-cold lysis buffer containing 0.4 mM NaCl, 20 mM Tris-HCl, pH 7.5, 0.1% NP40, 5% (V/V) glycerol, 1 mM NaF, 1 mM Na-orthovanadate and protease inhibitor cocktail. After 60 minutes incubation on ice, released proteins were collected by centrifugation and the protein concentration was measured via Bio-Rad Bradford protein assay. Extracts (100 μg of total cell protein) were subjected to polyacrylamide-SDS gel electrophoresis followed by blotting to nitrocellulose. The membranes were blocked with 5% skim milk in TBS with 0.1% Tween-20 for 1 hour at room temperature and incubated with specific primary antibodies at 4°C overnight, followed by incubation with appropriate secondary antibody at room temperature for 1 hour. Enhanced chemiluminescence western blot substrate was used for film development, and ImageJ (ImageJ, RRID:SCR_003070) was used for data quantification.
Tumors collected from 4 FoxM1+/+ and 5 FoxM1DD/DD PYMT+ mice were snap frozen in a dry ice-ethanol mixture and preserved at −80°C until required for the experiment. The tissue samples for western blotting were homogenized in ice-cold lysis buffer containing 50mM Hepes-KOH (pH 7.5), 300mM NaCl, 1mM EDTA, 1 mM EGTA, 1 mM DTT, 0.1% Tween-20, 10% glycerol, 10mM β-glycerolphosphate, 1 mM NaF, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 0.2 mM PMSF, and processed as stated above. Following primary antibodies were used: anti-FoxM1 (Santa Cruz Biotechnology Cat# sc-500, RRID:AB_631521 ), anti-α-tubulin (Sigma-Aldrich Cat# T9026, RRID:AB_477593 ), anti-CBP (Santa Cruz Biotechnology Cat# sc-7300, RRID:AB_626817 ), anti-Rb (Cell Signaling Technology Cat# 9309, RRID:AB_823629 ), anti-Pten (Cell Signaling Technology Cat# 9552, RRID:AB_10694066 ), anti-pan Akt (Cell Signaling Technology Cat# 4691, RRID:AB_915783 ) and anti-pAktS308 (Cell Signaling Technology Cat# 9275, RRID:AB_329828). Quantification has been performed using ImageJ (RRID:SCR_003070).
RT-PCR analysis
RNA was Trizol extracted (Invitrogen) and cDNA was synthesized using Bio-Rad reverse transcriptase. cDNA was amplified using SYBR Green (Bio-Rad) and analyzed via iCycler software (Biorad MyIQ single-channel QPCR instrument, RRID:SCR_019736). All primer sequences are shown in Table S2. RT-PCR analysis was performed on RNA isolated from 3 different pairs of FoxM1+/+ and FoxM1DD/DD PYMT+ primary tumors or isolated from 3 different pairs of FoxM1+/+ and FoxM1DD/DD PYMT+ primary tumor cell lines. Each biological replicate was assayed in technical triplicate.
ChIP assay
MCF7 cells were fixed for 10 min in 1% formaldehyde and quenched with glycine (125 nM). Sonicated cell lysate was incubated with either IgG (Santa Cruz Biotechnology Cat#sc-2025, RRID:AB_737182 (mouse) or Santa Cruz Biotechnology Cat#sc-2027, RRID:AB_737197, rabbit )), GFP (Abcam, Cat# ab290, RRID:AB_303395), FoxM1 (Santa Cruz Biotechnology Cat# sc-500, RRID:AB_631521) or Rb (Cell Signaling Technology Cat# 9309, RRID:AB_823629) antibody and collected with protein-A and protein-G beads with salmon sperm DNA. DNA was extracted from beads and PCR amplification was done with primers listed in Table S2.
FACS analysis
Tumor nodules from 4 FoxM1+/+ PYMT+ and 5 FoxM1DD/D PYMT+ mice were harvested, washed in PBS, chopped into small pieces (around 2mm) and dissociated by collagenase type IV for 30 min at 37°C. Cell debris and red blood cells were removed with centrifugation and ACK (Gibco) lysis buffer. The cell pellets were washed with PBS and passed through a 40 μm cell strainer to form a single cell suspension. Lastly, cells were counted with trypan blue to determine cell number and viability. Cells (resuspended in PBS with 2%FBS and 2mM EDTA) were incubated with antibodies for 20 minutes on ice. Samples were analyzed using Cyan ADP flow cytometer (Beckman Coulter) and Summit software. Following antibodies were used: anti-CD90-FITC (HIS51, eBiosciences, Thermo Fisher Scientific Cat# 11-0900-81, RRID:AB_465151), anti-CD24-PE (M1/69, BD Biosciences Cat# 553262, RRID:AB_394741) and CD45-APC (30-F11, eBiosciences, Thermo Fisher Scientific Cat# 17-0451-82, RRID:AB_469392).
Statistical analysis
Statistical significance was calculated by the unpaired Student’s t test (2 tailed). Statistically significant changes were indicated with asterisks (*p < 0.05; **p < 0.01, ***p<0.001).
Data Availability
Drop-seq scRNA-seq data are available in the NCBI Gene Expression Omnibus database (GEO), RRID:SCR_005012, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178612 Accession token: mlgpsiokrnmdfkz) ChIP-Seq data are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE176383 Accession token: svkpwakmznazfqv.
RESULTS
Mammary tumors expressing a repression-deficient mutant of FoxM1, FoxM1DD, are deficient in metastatic progression
Previously, we used point mutations to separate the transcription activator function from the transcriptional repressor function of FoxM1 (12). A phospho-mimetic mutant, FoxM1DD, in which the two Plk1 phosphorylation sites were replaced by aspartic acid, retains the transcriptional activator function, but fails to repress the mammary differentiation genes Gata3 and FoxA1 (12). We employed CRISPR/Cas9 technology to alter the corresponding residues in mouse genome to aspartic acid (Fig.S1A–B). To screen for the mutant allele, we design PCR primers that specifically recognize desired mutations (FoxM1S727 mutated to aspartic acid where S727 corresponds to human S715, and FoxM1S736 mutated to aspartic acid where S736 corresponds to human S724, Fig.S1A). The mutation introduced a Cla1-site that offered additional screening method for the wanted substitutions (Fig.S1B and D). From 36 mice, we obtained two mouse lines in C57BL6 background (Fig.S1C–D, founder 10 and founder 18), both heterozygous for the DD mutations. Next generation sequencing confirmed desired mutation and found that one of them (founder 18) harbored additional substitution in FoxM1 gene (Serine729 into Cytosine, Fig.S1E), hence was excluded from further experiments. We crossed the founder mouse with wild-type C57BL6 mice to expand colony, and subsequently crossed the heterozygotes to obtain the homozygous FoxM1DD/DD line. In those crosses, homozygous FoxM1DD/DD pups were born in expected Mendelian ratio (Table S1). Extracts from the embryonic fibroblasts (MEFs) were assayed by western blots to detect expression of FoxM1 (Fig.1A). Moreover, immunoprecipitation of FoxM1 from the extracts of FoxM1DD/DD expressing MEFs co-immunoprecipitated CBP, but not Rb (Fig.1B). Surprisingly, unlike the FoxM1−/− line, which is embryonic lethal (17), there was no gross developmental defect in the FoxM1DD/DD line. Examination of 2 male and 2 female adult FoxM1DD/DD mice by necropsies did not reveal any significant abnormality in any of the mouse organs or tissues (Supplemental pathologist’s report). Together, these observations indicate that FoxM1’s interaction with the Rb-family of proteins is not essential for mouse embryonic development.
Figure 1. FoxM1DD/DD PYMT+ mouse mammary tumors are deficient in metastasis.

(A) FoxM1 protein expression in MEFs. (B) FoxM1 interaction with CBP and Rb in FoxM1+/DD and FoxM1DD/DD MEFs. IP was performed using FoxM1 antibody. (C) Number of tumor nodules at the end-point of the experiment. (D) Percentage of mice that did not reach the end-point at indicated time point. (E) Quantification showing percentage of mice with visible lung metastatic nodules at the end-point of the experiment. (F) Lungs of FoxM1+/+ PYMT+ and FoxM1DD/DD PYMT+ female mice. Arrows are pointing at metastatic nodules. (G) Metastatic area in the lungs. p value was calculated using Student’s unpaired t test (*p < 0.05).
Since over-expression of FoxM1 coincides with aggressive mammary tumor progression (1) we sought to investigate effects of the mutations in mammary tumors. We generated the MMTV-PyMT FoxM1DD/DD strain, and compared mammary tumors with MMTV-PyMT females that are in the FoxM1+/+ and FoxM1+/DD background. As shown in Fig.1C and Fig.1D, there was no significant difference in the development of PyMT-driven mammary tumors among the three genotypes. All mice reached the end-point (diameter equal of 2 cm) at comparable times and developed similar number of tumor nodules that were comparable in histology and proliferation (Fig.S2). Interestingly, however, there were significant differences when metastasis to the lung was examined (Fig.1E–G). Majority of the FoxM1+/+ PyMT+ (61.5%) and FoxM1+/DD PyMT + (59.5%) female mice developed lung metastasis at the end-point, whereas a significantly lower number of the FoxM1DD/DD PYMT+ female mice developed lung metastasis at the end-point of the experiment (27.3%, Fig.1E). More importantly, the lung nodules in the FoxM1DD/DD background were much smaller (Fig.1F). Quantification of the metastatic areas in the lung showed even more robust difference (Fig.1G). These observations suggest that while dispensable for primary tumor development, the interactions between FoxM1 and Rb is important for mammary tumor metastasis.
Deficiency in metastasis is tumor cell-intrinsic
Because FoxM1DD/DD is a whole-body knock-in line, we investigated whether the tumor cells themselves are deficient in metastatic progression. We crossed our FoxM1+/+ PYMT+ and FoxM1DD/DD PYMT+ mice with mT/mG reporter mouse line (detailed description of the mouse lines can be found in the Methods section), harvested tumors when they reached end-point, and isolated tumor cells. The tumor cells were orthotopically implanted in wild-type C57BL6 mammary glands. In those experiments, 13 out of 14 FoxM1+/+ (92.86%) orthotopic implantations developed mammary tumors, and 8 out of 11 (72.72%) FoxM1DD/DD implantations developed tumors. Interestingly, whereas 6 out of 13 mice (46.15%) implanted with FoxM1+/+ cells exhibited lung metastasis, only 1 out of 8 mice (12.5%) implanted with FoxM1DD/DD cells that developed mammary tumors had lung metastasis (albeit microscopic only) (Fig.2A–B), indicating that the deficiency is tumor-cell intrinsic. Moreover, FoxM1+/+ cells orthotopically implanted in FoxM1DD/DD recipient female mice developed primary tumors at frequency of 80% (4 out of 5 mice) and 50% of the mice also developed lung metastasis, solidifying the notion that metastatic deficiency of FoxM1DD/DD PYMT+ tumors is tumor cell-intrinsic (Fig.2A–B). Also, we observed evidence that lentiviral-mediated transduction of FoxM1-cDNA into FoxM1DD/DD tumor cells restored lung metastasis upon orthotopic implantation into mammary fat pad of FoxM1DD/DD females (Fig.S3).
Figure 2. Lack of metastasis in FoxM1DD/DD PYMT-driven tumors is tumor cell-intrinsic.

(A) Images of orthotopic tumors and corresponding lungs. (B) Table summarizing incidences of mammary tumors and lung metastasis after orthotopic transplantation. (C-D) Images of lungs (C) and quantification (D) after tail vein injection of FoxM1+/+ or FoxM1DD/DD PYMT+ cells into c57/bl6 recipient female mice. Arrows are pointing at metastatic nodules (E). CD90+CD24+CD45- cancer stem cells were analyzed by FACS. Quantification is shown in the bottom panel. p value was calculated using Student’s unpaired t test.
To further investigate tumor cell-autonomous deficiency, tumor cells derived from FoxM1+/+ and FoxM1DD/DD backgrounds were injected in female C57BL6 mice via the tail vein. All mice injected with the FoxM1+/+ tumor cells exhibited robust lung metastasis. The mice injected with FoxM1DD/DD tumor cells exhibited significantly lower number of visible tumor nodules in the lung (Fig.2C). Quantification further confirmed a greatly reduced metastatic potentials of the FoxM1DD/DD tumor cells (Fig.2D). Previous studies on lung metastasis of the tail vein injected MMTV-PyMT tumor cells indicated a role of the CD90+/CD24+/CD45- cancer stem-like cells in growth of the mammary tumor cells in the lung (18). We analyzed the CD45-depleted tumor cells for the frequency of the CD90+/CD24+ population by FACS using fluorescent tagged CD90-ab and CD24-ab. As shown in Fig.2E, tumor cells derived from the FoxM1DD/DD mice contained significantly lower number of the CD90+CD24+ cells. Together the results clearly indicate that the deficiency in metastasis of the FoxM1DD/DD tumors is tumor cell-intrinsic, and that it is likely related to fewer cancer stem-like cells.
Single-cell RNA-Seq revealed expansion of the differentiated alveolar-type tumor cells and reduction in pro-metastatic tumor cells in tumors expressing repression-deficient FoxM1
Recent studies using single-cell RNA-Seq of the MMTV-PyMT tumors by our colleagues Chen et al. (19) and others (20, 21) demonstrated extensive heterogeneity of the mammary tumor cells. To further explore the deficiencies of the FoxM1DD tumors, we carried out single-cell RNA-Seq analyses. 10 mammary tumor nodules isolated from 2 independent FoxM1+/+ mice and 6 tumor nodules from 2 independent FoxM1DD/DD mice were harvested, dissociated into single-cell suspensions and subjected to Drop-Seq strategy for single-cell RNA-Seq (22). After quality control filtering to remove data from poor quality cells (cells with low gene detection (<400), and cells with high proportion of mitochondrial genes (>5%)), we obtained high quality RNA-Seq data from a total of 9849 cells, consisting of 6431 FoxM1+/+ tumor-derived cells and 3418 FoxM1DD/DD tumor-derived cells. Seurat was used for integrated analyses to cluster the cells and identify cell-types in the combined data sets (16). The tumor-derived cells clustered in 19 discrete clusters at resolution 1.0 (Fig.3A). Using well-established cell markers (23), we identified the three main cell lineages in PyMT-driven tumors (Fig.3A–B, Fig.S4) including epithelial, immune and stromal cells. Consistent with previous publications (19), we observed significant heterogeneity in the tumor cells, and identified 11 different clusters of epithelial cells (Fig.3B and Fig.S5). In addition, we detected clusters that express specific markers of tumor associated macrophages (clusters 0, 12, 14 and 15), T lymphocytes (cluster10), neutrophils (cluster17), myCAFs (cluster13), iCAFs (cluster18) and endothelial cells (cluster19) (Fig.3A–B, and Fig.S5).
Figure 3. Analysis of FoxM1+/+ PYMT+ and FoxM1DD/DD PYMT+ primary mammary tumors using single-cell RNAseq.

(A) UMAP plot of pooled FoxM1+/+ and FoxM1DD/DD cells isolated from primary mammary tumors. (B-C) Dot plots showing expression of epithelial, stromal, immune (B) and metastatic (C) marker genes. (D) UMAP plot of total cells separated by genotype. (E) Pie charts showing percentage of each cell populations in FoxM1+/+ and FoxM1DD/DD PYMT+ tumors.
Cluster 11 in our analyses is equivalent to cluster 13 of Chen et al (19) that was shown to contain pro-metastatic cells. For example, cluster 11 tumor cells express Krt14, Vim, E-cadherin, Ereg, Jag1, Adamts1 and Cav1 that were shown to mark metastatic cells (19), as well as other epithelial cell markers including Krt7, Krt8, Krt17 and Krt18 (Fig.3B–C) that were demonstrated to be expressed in pro-metastatic cell cluster. The cells in cluster 11 also expressed several well-known pro-metastatic genes, including Vegf, Egfr, Pgf, Adamts1 and Pdgfα (Fig.3C). In our single-cell RNA-Seq analyses, Cd90-mRNA (Thy1) was very low in epithelial cells and not reliable. However, expression of Cd44, another marker of cancer stem-like cells (24), is clearly detected in the cluster 11 population (Fig.3C). Interestingly, when the UMAPs for the FoxM1DD/DD and FoxM1+/+ clusters were plotted separately, a significant difference in the number of cells corresponding to cluster 11 was observed. Consistent with deficiency in metastasis, there were fewer cluster 11 cells in the FoxM1DD/DD tumors (Fig.3D). Cluster 11 in FoxM1+/+ tumor represented 4.82% of the total tumor cells, whereas the same cluster in FoxM1DD/DD tumor represented 1.42% of the total tumor cells. The difference between the two genotypes was validated further by assaying several metastasis genes and Cd44-mRNA in the tumor cells (shown later in Fig.7).
Figure 7. FoxM1DD/DD PYMT+ tumors are deficient in tumor cells involved in recruitment of tumor-infiltrating cells.

(A) Dot plot showing expression of Csf1, Ccl2, Mif, Il1β, Vegfα, Pdgfα and Ptges across epithelial cell clusters. (B) Feature plots showing expression of Mfge8, Il1β, Ccl5 and Ccl4 in FoxM1+/+ and FoxM1DD/DD epithelial cells separately. (C-D) Validation of single-cell RNAseq data by RT-PCR. p value was calculated using Student’s unpaired t test. *p<0.05, ***p<0.001.
We investigated the cell-types in the FoxM1+/+ and FoxM1DD/DD tumors using previously defined markers of mouse mammary epithelium (23). As shown in Figs.4A and B, distinct cell populations for luminal progenitors, alveolar, basal/myoepithelial and hormone-sensing lineages were identified. Interestingly, the majority of the tumor cells in FoxM1DD/DD tumors corresponded to differentiated alveolar cells (77.53% of epithelial cells in FoxM1DD/DD compared to 41.82% of epithelial cells in FoxM1+/+ tumors), and they were clustered in clusters 1, 2, 3, 8, and 9 that expressed the alveolar markers (Csn3, Lalba, and Wap). We used well defined luminal progenitor marker genes Aldh1a3, Cd14, Kit, Ltf and Spp1 (21,25) to identify luminal progenitor-type cells in FoxM1+/+ and FoxM1DD/DD tumors and found that these cells cluster in cluster 6. Interestingly, while 12.78% of FoxM1+/+ tumor cells corresponded to cluster 6, only 4.0% FoxM1DD/DD cells belonged to the same cluster (Fig.4A–B and Fig.S6). Overall, analyses of the tumor cell-types indicated fewer progenitor-like and basal/myoepithelial cells, but more differentiated alveolar cells in the FoxM1DD/DD tumors, compared to those in the FoxM1+/+ tumors (Fig.4B). Consistent with that, a greater number of FoxM1DD/DD tumor cells expressed the milk protein k-casein mRNA at the higher levels compared to FoxM1+/+ tumor cells (Fig.4C). The higher expression of k-casein, Lalba, and Wap in FoxM1DD/DD tumors were confirmed further using RT-PCR in three tumor nodules isolated from three different females of each genotype (Fig.4D).
Figure 4. FoxM1/Rb interaction is critical for maintenance of poorly-differentiated state of breast cancer cells.

(A) Dot plot showing expression of mammary epithelial lineage markers. (B) Frequency of identified tumor cell lineages. (C) Violin plot showing expression of kappa-casein (Csn3). (D) Validation of differential expression of milk protein mRNAs by RT-PCR. (E) Feature plot showing Gata3 and Elf5 expression. (F) Validation of differential expression of luminal differentiation factors Gata3 and FoxA1 by RT-PCR. p value was calculated using Student’s unpaired t test. ns=nonsignificant, p>0.05, *p<0.05, **p<0.01, ***p<0.001.
Expansion of the differentiated alveolar type tumor cells correlates with upregulation of the FoxM1/Rb-regulated mammary differentiation genes
Mammary tumor in the MMTV-PyMT model was shown to originate from the alveolar progenitors (20). The increase in the differentiated alveolar type tumor cells in the FoxM1DD/DD tumors suggests an activation of the differentiation program in the FoxM1DD/DD tumors. That is interesting because FoxM1/Rb represses expression of Gata3 (1), which was shown to promote alveolar morphogenesis (26). For example, expression of Gata3 in mammary stem cells induced maturation along the alveolar luminal lineage (26). Moreover, expression of Gata3 negatively regulates the stem and luminal progenitor populations in mammary gland (1). Therefore, we investigated expression of Gata3 in the FoxM1DD/DD tumors. Consistent with the repression of Gata3 by FoxM1/Rb, a significantly greater number of the tumor cells in FoxM1DD/DD background expressed Gata3, compared to those in the FoxM1+/+ tumors (Fig.4E). Moreover, a larger population of epithelial cells in the FoxM1DD/DD tumors expressed alveolar-differentiation gene Elf5 (Fig.4E). We confirmed increase in Gata3 expression in FoxM1DD/DD tumors using RT-PCR experiments (Fig.4F). In those experiments, we also observed increased expression of FoxA1 (Fig.4F), which is involved in ductal morphogenesis (27). Gata3 and FoxA1 are direct repression targets of FoxM1/Rb (12), Elf5 is not known to be a target of FoxM1. Increased expression of Gata3 in the FoxM1DD/DD tumors is significant also because Gata3 was shown to inhibit metastasis of mammary tumors (28).
FoxM1/Rb activates Akt-signaling by repressing expression of Pten
Our single-cell RNA-Seq analyses also revealed an increased number of tumor cells in the FoxM1DD/DD tumors expressed the tumor suppressor gene Pten at higher levels (Fig.5A). Increased Pten expression is interesting for the following reasons. First, PyMT-driven oncogenesis involves activation of PI3-kinase (29), and PTEN opposes the effects of PI3-kinase activation. Moreover, loss of Pten expression in tumor cells has been implicated in breast cancer metastasis (30) and in regulation of breast cancer stem cells (31). We validated the scRNA-Seq data by RT-PCR as well as by western blotting (Fig.5B–C). PI3-kinase activates Akt by recruiting it to the membrane where it is phosphorylated at residue 308 by PDK1. Increased Pten expression is expected to reduce membrane recruitment, leading to reduction in 308 phosphorylation of Akt. Indeed, FoxM1DD/DD tumors exhibited reduced Akt-308 phosphorylation compared to those in the extracts of FoxM1+/+ tumors (Fig.5C–D).
Figure 5. FoxM1/Rb complex is critical for Pten transcriptional repression in mouse and human breast cancers.

(A) Feature plot showing Pten expression. (B) Pten mRNA expression in 3 different pairs of FoxM1+/+ and FoxM1DD/DD PYMT+ primary tumors. (C) Western blot showing protein levels of Pten, Akt and pAktS308 in mammary gland tumors. (D) Quantification of phosphorylated Akt(S308) relative to total pan-Akt in tumor extracts. (E) Pten mRNA expression after FoxM1 silencing in MCF7 cells. (F) Pten mRNA expression after GFP-FoxM1 or GFP-FoxM1DD transient overexpression measured by RT-PCR. (G and H) ChIP assay performed on chromatin extract isolated from MCF7 cells transiently transfected with GFP, GFP-FoxM1 and GFP-FoxM1DD. Pull down was performed with GFP (G) or Rb (H) antibody and was normalized to the value from GFP transfected samples. (I) FoxM1 and Pten inverse correlation in human breast cancer samples. Analysis was performed using cBioportal (http://cbioportal.org). Pearson and Spearman correlations are indicated. p value was calculated using Student’s unpaired t test. **p<0.01, ***p<0.001.
Next, we investigated whether FoxM1/Rb directly regulates expression of Pten in mammary tumors. Using human breast cancer cell line MCF7, we observed that knockdown of FoxM1 increased mRNA expression of Pten, whereas over-expression of FoxM1 inhibited expression of Pten, indicating that Pten is a potential repression target of FoxM1 (Fig.5E–F). Moreover, consistent with FoxM1/Rb-mediated repression of Pten, expression of FoxM1DD, deficient in Rb-binding, failed to inhibit expression of Pten (Fig.5F). Recently, using liver cancer cell lines, we observed evidence for a ChIP-signal of FoxM1 on the Pten chromatin in a ChIP-Seq experiment (See Method). We carried out ChIP experiments using MCF7 cells, and confirmed occupancies of the endogenous FoxM1 and Rb at that site in the Pten chromatin (Fig.S7I and S7J). To further investigate recruitment of Rb by the FoxM1DD mutant, MCF7 cells were transfected with GFP-FoxM1 or GFP-FoxM1DD. The cells were then subjected to ChIP experiments using GFP- and Rb-antibody. Both GFP-FoxM1 and GFP-FoxM1DD bound to the Pten chromatin, however, FoxM1DD transfected cells, unlike the wild-type FoxM1 transfected cells, did not exhibit recruitment of Rb to the Pten chromatin (Fig.5G–H). Next, we analyzed the publicly available TCGA datasets to investigate evidence for regulation of Pten expression by FoxM1 in breast cancer samples. Consistent with the repression of Pten by FoxM1, we found significant inverse correlation between FoxM1 and Pten mRNAs in human breast cancer samples (Fig.5I).
FoxM1 DD/DD tumors are deficient in tumor-infiltrating cells
In addition to the differences in tumor cells, we observed huge differences in the populations of the tumor-infiltrating cells. The FoxM1DD/DD tumors contain much lower numbers of tumor associated macrophages (TAMs), neutrophils, T-lymphocytes, endothelial cells and CAFs (Fig.3D–E). The lack of tumor-infiltrating cells in FoxM1DD/DD tumors is interesting because those cells have been shown to be critical for lung metastasis of mammary tumors in the MMTV-PyMT model. TAMs have tumor-promoting phenotype (M2 polarization state) and they conduct tumor suppressive environment by inhibiting CD8+ T cell function. Moreover, TAMs aid metastatic progression by participating in ECM remodeling and degradation, by stimulating angiogenesis and supporting cancer stemness. Furthermore, CD4+ T cells have been shown to promote lung metastasis in PyMT-driven breast cancer by enhancing pro-tumorigenic properties of macrophages (32). And finally, neutrophils have been recognized in PyMT-driven breast cancers as critical for metastasis to the lungs (19). Therefore, we sought to validate lack of infiltrating cells in FoxM1DD/DD breast tumors by immunohistochemical (IHC) analyses of the tumor sections. We used F4/80 and CD68 antibodies to detect the macrophages, CD206 to identify M2-polarized (tumor promoting) macrophages, CD3 antibody to detect the T-cells, Ly6G-antibody to detect the neutrophils and CD31 to identify endothelial cells. Tumor sections from 3 to 5 mice were analyzed by IHC. Indeed, there were significant differences in the population of tumor-infiltrating macrophages, T-cells, neutrophils and endothelial cells (Figs.6A–D and Fig.S7). To investigate whether the difference in the infiltrating cells is related to deficiencies in the tumor cells, we carried out similar IHC analyses on tumor sections derived from orthotopic implantations of FoxM1+/+ or FoxM1DD/DD tumor cells in the wild type C57BL6 mice. The FoxM1DD/DD tumor cells implanted in the wild type background were deficient in recruiting the tumor-infiltrating and M2 macrophages, T-cells, neutrophils and endothelial cells (Fig.6E–H, Fig.S7).
Figure 6. FoxM1DD/DD PYMT+ mammary gland tumors are deficient in TAMs, neutrophils and endothelial cells.
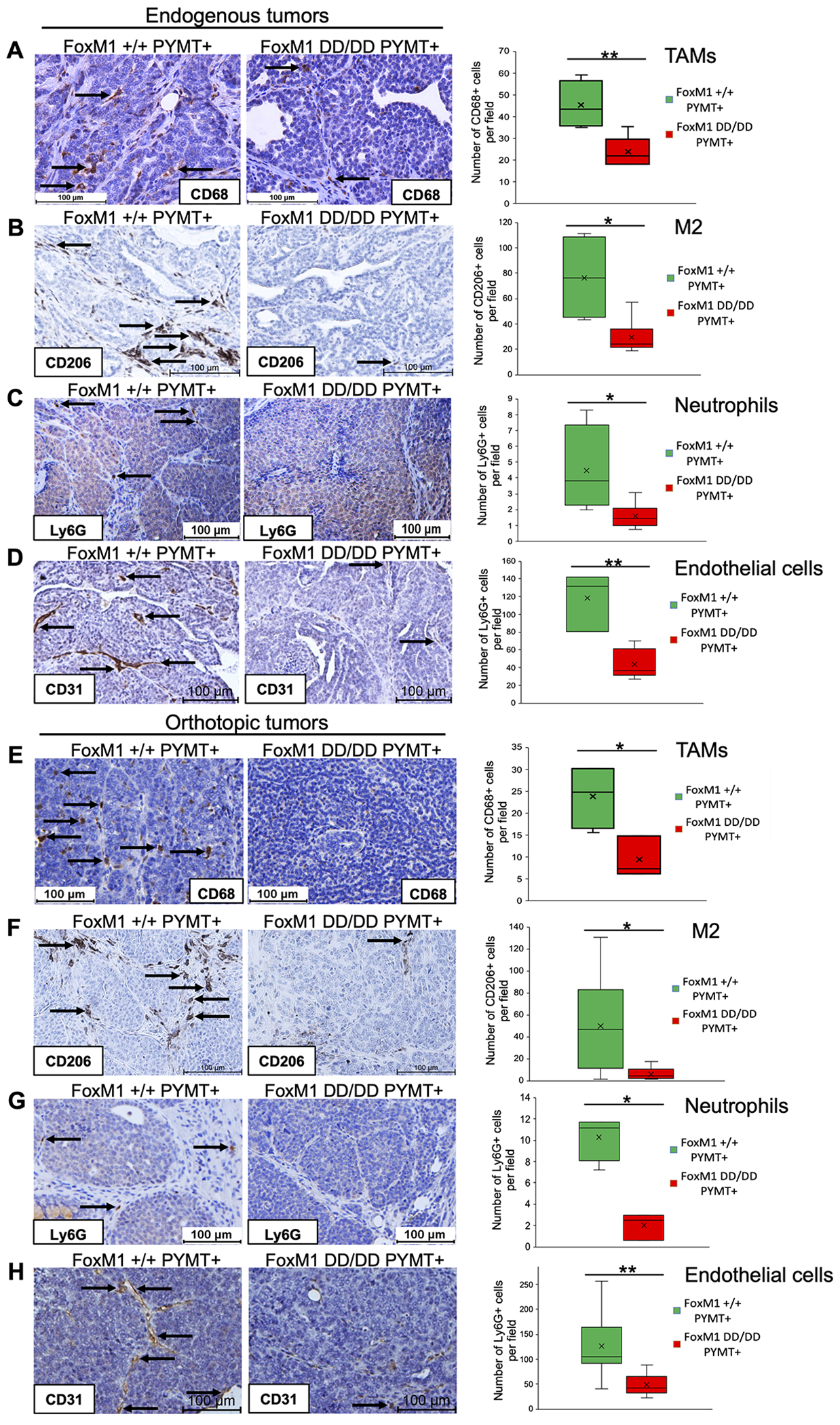
Immunohistochemical staining of FoxM1+/+ and FoxM1DD/DD PYMT-driven endogenous mammary gland tumors was performed with CD68 (A), CD206 (B), Ly6G (C) or CD31 antibody (D). IHC staining of mammary gland orthotopic tumors was carried out with CD68 (E), CD206 (F), Ly6G (G) or CD31 antibody (H). p value was calculated using Student’s unpaired t test. *p<0.05, **p<0.01.
Lack of tumor-infiltrating cells is related to fewer tumor cells that express the factors important for infiltration
Reduced number of the tumor-infiltrating cells in the FoxM1DD/DD tumor that was confirmed in orthotopic tumors derived from the FoxM1DD/DD tumor cells suggests that those cells are deficient in secretion of recruitment factors. Tumor cells have been shown to secrete growth factors and cytokines that are involved in recruitment of non-tumor cells that generate metastasis-permissive microenvironment (Reviewed in (33)). Our single-cell RNA-Seq analyses identified clusters of tumor cells that express cytokines involved in the recruitment of infiltrating cells. Cells in the pro-metastatic cluster 11 express Csf1, Mif, and Vegfa, (Fig.7A), all demonstrated to be important for recruitment of immune cells into tumors (34,35). As noted above, there were fewer cells of FoxM1DD/DD genotype in cluster 11 compared to FoxM1+/+ cells (1.42% vs. 4.82%). Interestingly, the cells in the same cluster exclusively expressed mRNAs for the fibroblast recruitment and reprograming factors, such as Vegfa, Pdgfa and Ptges (Fig.7A). Additionally, we identified recruitment factors, Ccl2, Il1b, Mfge8, Ccl5 and Ccl4 that were not associated with particular clusters, but were expressed in a “salt-and-pepper” manner throughout the epithelial cell clusters (Fig.7B). Importantly, those were specifically downregulated in the FoxM1DD/DD tumor cells.
We sought to validate the difference by assaying for the aforementioned mRNAs in tumor cells derived from FoxM1+/+ and FoxM1DD/DD tumors. Fewer cells in the cluster 11 predicted lower total expression of those genes in the FoxM1DD/DD tumors. Total RNAs from cultured tumor epithelial cells rather than total RNAs from the tumors were compared because many of those genes are expressed also by the non-tumor cells present in greater number in the FoxM1+/+ tumors. Indeed, RT-PCR assays showed reduced levels of those mRNAs in cells derived from the FoxM1DD/DD tumors compared to those from the FoxM1+/+ tumors (Fig.7C), whereas no significant difference in FoxM1 expression was observed (Fig.7D). The RT-PCR experiments were performed on biological and technical triplicates of both genotypes. Taken together, reduced expression of the recruitment factors by the tumor cells in the FoxM1DD/DD background explain why those tumors are deficient in tumor-infiltrating cells.
DISCUSSION
Results presented here are significant in several ways. First, we show that the transcriptional repression function of FoxM1 is important for metastasis of mammary tumor, which is related to the abundance of pro-metastatic and poorly differentiated type of tumor cells. Moreover, FoxM1 represses expression of the tumor suppressor Pten along with the mammary differentiation genes, and that mechanism is important for the accumulation of pro-metastatic, poorly differentiated tumor cells. Further, the pro-metastatic tumor cells in primary tumors, which are dependent on the repression mechanism of FoxM1, express cytokines and chemokines that are critical for recruitment of tumor-infiltrating cells. Since the repression mechanism of FoxM1 involves Rb, the results identify a pro-metastatic function of the tumor suppressor Rb when bound to FoxM1. Together, these results provide new insights into the tumor mechanisms of FoxM1/Rb that are important for evolution of metastatic cancer cells (Fig.8).
Figure 8. Schematic diagram depicting the role of FoxM1/Rb in breast cancer progression and metastasis.

In the breast tumor cells, high FoxM1 expression and intact retinoblastoma protein generate FoxM1/Rb repression complex that plays critical role in accumulation of the poorly differentiated cells, cancer stem-like and pro-metastatic cells. These cells in turn secrete recruitment factors required for establishment of pro-metastatic tumor microenvironment (TME) and subsequent metastasis. If FoxM1/Rb interaction is disrupted, tumors fail to accumulate poorly differentiated and pro-metastatic cells, resulting in reduced expression of chemokines and cytokines required for recruitment of tumor-infiltrating cells, altered TME and decreased metastatic potential.
Mutations of the Rb gene are common in the basal-type of breast cancers that are aggressive (36). How do those Rb-mutant tumors undergo metastatic progression? Rb has been shown to participate in controlling proliferation through its inhibitory effects on the E2F-target genes important for G1/S progression (37). Importantly, Rb is involved also in cellular differentiation. For example, Rb−/− embryos, rescued from lethality with wild-type placenta, exhibit skeletal muscle defects, resulting from deficiency in muscle differentiation (38–40). Moreover, a number of studies implicated roles of Rb in differentiation of cancer and normal cells (41–43). The differentiation function of Rb is conserved in Drosophila (44). Loss of Drosophila Rb and hippo pathway were shown to induce de-differentiation (45). Therefore, in the absence of Rb, as in Rb-mutations, tumor cells would maintain the progenitors more efficiently to support evolution of the metastatic cancer cells.
Our observations suggest that, in the Rb+ve tumors, the FoxM1/Rb interaction plays a significant role in evolution of the metastatic cells by suppressing differentiation pathways. In the absence of that suppression in the FoxM1DD/DD tumor, tumor cells undergo extensive differentiation to the alveolar lineage. We provided evidence that FoxM1/Rb, in addition to repressing the mammary differentiation gene Gata3 (Fig.4), represses the tumor suppressor Pten (Fig.5). Both Gata3 and Pten were shown to regulate breast cancer stem cells and metastasis (28,30,31). Interestingly, Gata3 was shown also to have profound effects on regulating the tumor microenvironment (46). Therefore, it is likely that higher expression of Gata3, Pten, and others, in the FoxM1DD/DD tumor caused expansion of the differentiated alveolar cell population limiting the number of the pro-metastatic type tumor cells, leading to the deficiencies in the tumor-infiltrating cells, important for metastasis. It is important to note that we did not rule out a possibility that the FoxM1DD protein has additional functions unrelated to its lack of interaction with Rb. But those possible unrelated functions of FoxM1DD are suppressed by the wild-type FoxM1. As shown in Fig.1E, the FoxM1DD allele is recessive, and expression of FoxM1 in FoxM1DD/DD tumor cells rescues metastasis (Fig.S3).
The pro-metastatic tumor cells in cluster 11, which express Csf1, Ccl2, Vegf and others, also express cancer stem cell marker Cd44 (24) and luminal progenitor marker Aldh1a3 (Figs.3C and 4A). Moreover, our FACS analyses also indicated significantly lower number of the CD90+/CD24+ mammary cancer stem-like cells in the FoxM1DD/DD tumors (Fig.2E). Therefore, it is tempting to speculate that the cells in the pro-metastatic clusters are derived from the stem/progenitor type of tumor cells. Also, the alveolar progenitor cells, which are more abundant in the FoxM1+/+ tumors, express Mfge8 that is involved in mammary gland involution (25), also participates in macrophage recruitment and M2 polarization (47), and associates with poor prognosis in breast cancer patients (48). Therefore, the differences in the stem/progenitor tumor cell populations and pro-metastatic cell population would explain why implantation of the FoxM1DD/DD tumor cells in the wild-type FoxM1 background failed to recruit the tumor-infiltrating cells and to undergo metastasis.
The observation that, in Rb+ve tumor, FoxM1/Rb is important for aggressive progression is significant with regards to breast cancer therapy. Notably, Palbociclib, which is being used to treat ER+ metastatic breast cancer, in addition to activating Rb, decreases the levels of FoxM1 (49). It has strong inhibitory effects on the protein levels of FoxM1 also in breast cancer cells (Fig.S8). Therefore, it is likely that mechanisms that cause accumulation of FoxM1 would contribute to development of Palbociclib-resistance. Conversely, tumors over-expressing FoxM1 may not respond to Palbociclib. That could potentially explain why not all Rb+ve patients benefit from Palbociclib therapies (50,51). Furthermore, increased accumulation of FoxM1 in ER+ breast cancer correlates with resistance to PI3-kinase inhibitors (9). Our observation that FoxM1 represses expression of Pten is significant in that regard because loss of Pten was shown to mediate cross-resistance to CDK4/6 and PI3-kinase inhibitors (52). Together, our findings highlight the importance of looking at the expression-levels of FoxM1 before treating patients with drugs that generate active Rb, as high FoxM1 partnering with active Rb would support evolution of aggressive type of tumor cells that are also resistant to the available therapeutics.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
This work provides new insights into how the interaction between FoxM1 and Rb facilitates the evolution of metastatic breast cancer cells by altering the transcriptome.
ACKNOWLEDGMENTS
This work was supported by a grant from the NCI (5 RO1 CA243247) to P. Raychaudhuri. P. Raychaudhuri is supported also by a grant (I01 BX000131) from the Department of Veterans Affair (Biomedical Laboratory Research Development Service). The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government. D. Kopanja is supported by the Gilead Sciences Research Scholars Program. M. Frolov acknowledges support from NIH grants (R35GM131707).
Financial Support:
This work was supported by a grant from the NCI (5 RO1 CA243247) to PR. PR is supported also by a grant (I01 BX000131) from the Department of Veterans Affair. DK is supported by the Gilead Sciences Research Scholars Program. MVF is supported by NIH grant (R35GM131707).
Footnotes
Conflict of Interest: None
REFERENCES
- 1.Carr JR, Kiefer MM, Park HJ, Li J, Wang Z, Fontanarosa J, et al. FoxM1 regulates mammary luminal cell fate. Cell reports 2012;1:715–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu G, Zhou A, Xue J, Huang C, Zhang X, Kang SH, et al. FoxM1 promotes breast tumorigenesis by activating PDGF-A and forming a positive feedback loop with the PDGF/AKT signaling pathway. Oncotarget 2015;6:11281–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer cell 2011;20:427–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. The Journal of clinical investigation 2014;124:564–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong AH, Wei P, Zhang S, Yao J, Yuan Y, Zhou AD, et al. FoxM1 Drives a Feed-Forward STAT3-Activation Signaling Loop That Promotes the Self-Renewal and Tumorigenicity of Glioblastoma Stem-like Cells. Cancer Res 2015;75:2337–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue J, Zhou A, Tan C, Wu Y, Lee HT, Li W, et al. Forkhead Box M1 Is Essential for Nuclear Localization of Glioma-associated Oncogene Homolog 1 in Glioblastoma Multiforme Cells by Promoting Importin-7 Expression. The Journal of biological chemistry 2015;290:18662–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr JR, Park HJ, Wang Z, Kiefer MM, Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res 2010;70:5054–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao S, Fan LY, Lam EW. The FOXO3-FOXM1 axis: A key cancer drug target and a modulator of cancer drug resistance. Semin Cancer Biol 2018;50:77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ros S, Wright AJ, D’Santos P, Hu DE, Hesketh RL, Lubling Y, et al. Metabolic Imaging Detects Resistance to PI3Kalpha Inhibition Mediated by Persistent FOXM1 Expression in ER(+) Breast Cancer. Cancer Cell 2020;38:516–33 e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wierstra I, Alves J. Transcription factor FOXM1c is repressed by RB and activated by cyclin D1/Cdk4. Biological chemistry 2006;387:949–62 [DOI] [PubMed] [Google Scholar]
- 11.Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Molecular and cellular biology 2004;24:2649–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukhopadhyay NK, Chand V, Pandey A, Kopanja D, Carr JR, Chen YJ, et al. Plk1 Regulates the Repressor Function of FoxM1b by inhibiting its Interaction with the Retinoblastoma Protein. Scientific Reports 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chand V, Pandey A, Kopanja D, Guzman G, Raychaudhuri P. Opposing Roles of the Fork-head box genes FoxM1 and FoxA2 in Liver Cancer. Mol Cancer Res 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YJ, Dominguez-Brauer C, Wang Z, Asara JM, Costa RH, Tyner AL, et al. A conserved phosphorylation site within the forkhead domain of FoxM1B is required for its activation by cyclin-CDK1. J Biol Chem 2009;284:30695–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive Integration of Single-Cell Data. Cell 2019;177:1888-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krupczak-Hollis K, Wang X, Kalinichenko VV, Gusarova GA, Wang IC, Dennewitz MB, et al. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Developmental biology 2004;276:74–88 [DOI] [PubMed] [Google Scholar]
- 18.Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2012;481:85–9 [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Ariss MM, Ramakrishnan G, Nogueira V, Blaha C, Putzbach W, et al. Cell-Autonomous versus Systemic Akt Isoform Deletions Uncovered New Roles for Akt1 and Akt2 in Breast Cancer. Mol Cell 2020;80:87–101 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao L, van Bragt MP, Li Z. A Long-Lived Luminal Subpopulation Enriched with Alveolar Progenitors Serves as Cellular Origin of Heterogeneous Mammary Tumors. Stem Cell Reports 2015;5:60–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valdes-Mora F, Salomon R, Gloss BS, Law AMK, Venhuizen J, Castillo L, et al. Single-cell transcriptomics reveals involution mimicry during the specification of the basal breast cancer subtype. Cell Rep 2021;35:108945. [DOI] [PubMed] [Google Scholar]
- 22.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015;161:1202–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bach K, Pensa S, Grzelak M, Hadfield J, Adams DJ, Marioni JC, et al. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat Commun 2017;8:2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009;11:1487–95 [DOI] [PubMed] [Google Scholar]
- 25.Yeo SK, Zhu X, Okamoto T, Hao M, Wang C, Lu P, et al. Single-cell RNA-sequencing reveals distinct patterns of cell state heterogeneity in mouse models of breast cancer. Elife 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nature cell biology 2007;9:201–9 [DOI] [PubMed] [Google Scholar]
- 27.Bernardo GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, et al. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development 2010;137:2045–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouros-Mehr H, Bechis SK, Slorach EM, Littlepage LE, Egeblad M, Ewald AJ, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer cell 2008;13:141–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dankort DL, Muller WJ. Signal transduction in mammary tumorigenesis: a transgenic perspective. Oncogene 2000;19:1038–44 [DOI] [PubMed] [Google Scholar]
- 30.Li K, Li GD, Sun LY, Li XQ. PTEN and SHIP: Impact on lymphatic metastasis in breast cancer. J Cancer Res Ther 2018;14:S937–S41 [DOI] [PubMed] [Google Scholar]
- 31.Bahena-Ocampo I, Espinosa M, Ceballos-Cancino G, Lizarraga F, Campos-Arroyo D, Schwarz A, et al. miR-10b expression in breast cancer stem cells supports self-renewal through negative PTEN regulation and sustained AKT activation. EMBO Rep 2016;17:1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, et al. CD4(+) T Cells Regulate Pulmonary Metastasis of Mammary Carcinomas by Enhancing Protumor Properties of Macrophages. Cancer Cell 2009;16:91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noe JT, Mitchell RA. MIF-Dependent Control of Tumor Immunity. Front Immunol 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol 2015;15:73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–52 [DOI] [PubMed] [Google Scholar]
- 37.Nevins JR. The Rb/E2F pathway and cancer. Human molecular genetics 2001;10:699–703 [DOI] [PubMed] [Google Scholar]
- 38.Zacksenhaus E, Jiang Z, Chung D, Marth JD, Phillips RA, Gallie BL. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev 1996;10:3051–64 [DOI] [PubMed] [Google Scholar]
- 39.de Bruin A, Wu L, Saavedra HI, Wilson P, Yang Y, Rosol TJ, et al. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci U S A 2003;100:6546–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 2003;421:942–7 [DOI] [PubMed] [Google Scholar]
- 41.Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG Jr., Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 2005;18:623–35 [DOI] [PubMed] [Google Scholar]
- 42.Berman SD, Yuan TL, Miller ES, Lee EY, Caron A, Lees JA. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol Cancer Res 2008;6:1440–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iavarone A, King ER, Dai XM, Leone G, Stanley ER, Lasorella A. Retinoblastoma promotes definitive erythropoiesis by repressing Id2 in fetal liver macrophages. Nature 2004;432:1040–5 [DOI] [PubMed] [Google Scholar]
- 44.Zappia MP, Rogers A, Islam A, Frolov MV. Rbf Activates the Myogenic Transcriptional Program to Promote Skeletal Muscle Differentiation. Cell Rep 2019;26:702–19 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicolay BN, Bayarmagnai B, Moon NS, Benevolenskaya EV, Frolov MV. Combined inactivation of pRB and hippo pathways induces dedifferentiation in the Drosophila retina. PLoS Genet 2010;6:e1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol 2013;15:201–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soki FN, Koh AJ, Jones JD, Kim YW, Dai JL, Keller ET, et al. Polarization of Prostate Cancer-associated Macrophages Is Induced by Milk Fat Globule-EGF Factor 8 (MFG-E8)-mediated Efferocytosis. Journal of Biological Chemistry 2014;289:24560–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu LF, Zhao L, Jia Z, Bi J, Wei Q, Song XY, et al. MFG-E8 overexpression is associated with poor prognosis in breast cancer patients. Pathol Res Pract 2019;215:490–8 [DOI] [PubMed] [Google Scholar]
- 49.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, et al. A Systematic Screen for CDK4/6 Substrates Links FOXM1 Phosphorylation to Senescence Suppression in Cancer Cells. Cancer Cell 2011;20:620–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spring LM, Wander SA, Andre F, Moy B, Turner NC, Bardia A. Cyclin-dependent kinase 4 and 6 inhibitors for hormone receptor-positive breast cancer: past, present, and future. Lancet 2020;395:817–27 [DOI] [PubMed] [Google Scholar]
- 51.Turner NC, Slamon DJ, Ro J, Bondarenko I, Im SA, Masuda N, et al. Overall Survival with Palbociclib and Fulvestrant in Advanced Breast Cancer. N Engl J Med 2018;379:1926–36 [DOI] [PubMed] [Google Scholar]
- 52.Costa C, Wang Y, Ly A, Hosono Y, Murchie E, Walmsley CS, et al. PTEN Loss Mediates Clinical Cross-Resistance to CDK4/6 and PI3Kalpha Inhibitors in Breast Cancer. Cancer Discov 2020;10:72–85 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Drop-seq scRNA-seq data are available in the NCBI Gene Expression Omnibus database (GEO), RRID:SCR_005012, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178612 Accession token: mlgpsiokrnmdfkz) ChIP-Seq data are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE176383 Accession token: svkpwakmznazfqv.


