Abstract
Background
Metastasis and chemoresistance are major culprits of cancer mortality, but factors contributing to these processes are incompletely understood.
Methods
Bioinformatics methods were used to identify the relations of Smyca expression to clinicopathological features of human cancers. RNA-sequencing analysis was used to reveal Smyca-regulated transcriptome. RNA pull-down and RNA immunoprecipitation were used to examine the binding of Smyca to Smad3/4 and c-Myc/Max. Chromatin immunoprecipitation and chromatin isolation by RNA purification were used to determine the binding of transcription factors and Smyca to various gene loci, respectively. Real-time RT-PCR and luciferase assay were used to examine gene expression levels and promoter activities, respectively. Xenograft mouse models were performed to evaluate the effects of Smyca on metastasis and chemoresistance. Nanoparticle-assisted gapmer antisense oligonucleotides delivery was used to target Smyca in vivo.
Results
We identify lncRNA Smyca for its association with poor prognosis of many cancer types. Smyca potentiates metabolic reprogramming, migration, invasion, cancer stemness, metastasis and chemoresistance. Mechanistically, Smyca enhances TGF-β/Smad signaling by acting as a scaffold for promoting Smad3/Smad4 association and further serves as a Smad target to amplify/prolong TGF-β signaling. Additionally, Smyca potentiates c-Myc-mediated transcription by enhancing the recruitment of c-Myc/Max complex to a set of target promoters and c-Myc binding to TRRAP. Through potentiating TGF-β and c-Myc pathways, Smyca synergizes the Warburg effect elicited by both pathways but evades the anti-proliferative effect of TGF-β. Targeting Smyca prevents metastasis and overcomes chemoresistance.
Conclusions
This study uncovers a lncRNA that coordinates tumor-relevant pathways to orchestra a pro-tumor program and establishes the clinical values of Smyca in cancer prognosis and therapy.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13045-022-01306-3.
Keywords: LncRNA, TGF-β, Smad, c-Myc, EMT, Metastasis, Chemoresistance
Background
Metastasis and therapy resistance are two leading causes of death for cancer patients, and epithelial–mesenchymal transition (EMT) is tightly associated with these two culprits [1, 2]. Among the molecular pathways that promote EMT to worsen the prognosis of cancer patients, TGF-β/Smad pathway plays a potent and prevalent role [3, 4]. Mechanistically, TGF-β binds its cell surface receptors, the type II and type I receptor complex, to induce the recruitment and phosphorylation of Smad2/3. The phosphorylated Smad2 and/or Smad3 forms an oligomer with Smad4 for translocation into the nucleus [5–7]. In the nucleus, Smad complex elicits multiple mechanisms to induce EMT. For instance, Smad activates the transcription of several EMT transcription factors, such as SNAI1, SNAI2, ZEB and Twist through direct or indirect mechanisms and increases the activity of Twist [8–11]. Smad also activates the expression of mesenchymal markers Fibronectin, Vimentin and Collagen αI [12, 13] and represses epithelial markers E-cadherin and occludin [14]. Through promoting EMT and EMT-associated malignant phenotypes such as invasion, metastasis, stem cell-like properties and immunosuppression/inflammation [3], TGF-β/Smad pathway functions as a key driver for cancer progression.
In addition to the potent tumor-promoting effects, TGF-β/Smad signaling also induces cytostatic effects, such as arresting cell cycle at G1 phase and inducing apoptosis [15]. By possessing both tumor-promoting and tumor-suppressive functions, the effects of TGF-β/Smad signaling on cancer are complex and often depend on the types and stages of cancer and the contexts of tumor cells and tumor microenvironment [16, 17]. For instance, TGF-β signaling usually prevents the generation of hyperplastic/premalignant lesions in normal tissues through its cytostatic effects. However, once tumors are progressed to an aggressive stage, the tumor-promoting effects become dominant. This functional switch of TGF-β/Smad signaling has been attributed to the activation of a number of oncogenic proteins that modulate Smad posttranslational modifications or Smad-controlled transcriptional programs [18]. For instance, c-Myc is known to counteract the growth inhibitory effect of TGF-β/Smad by reverting Smad-mediated transcriptional activation of CDK inhibitors CDKN2B and CDKN1A, also known as p15INK4B and p21Cip1, respectively [19–21]. Nevertheless, our understanding on the factors that dictate the net outcome of TGF-β/Smad signaling in cancer remains incomplete. Furthermore, several anti-TGF-β therapies showed poor clinical outcomes in cancer patients despite the success in the in vitro and preclinical models [22]. This points out the need for a better understanding of the cross talk between TGF-β pathway and other cancer pathways for stratifying patients that may benefit from the anti-TGF-β therapy or for designing more effective treatment regimens.
LncRNAs are known to elicit profound effects on cancer and can serve as prognostic/diagnostic biomarkers and therapeutic targets in many cancer types [23]. To date, lncRNAs have been shown to regulate a broad spectrum of cancer hallmarks and numerous cancer pathways [24]. TGF-β/Smad and c-Myc are among the “hot spots” for lncRNA-centered regulations. A large number of lncRNAs are reported to regulate c-Myc gene expression in cis or trans, influence on c-Myc protein stability or modulate c-Myc transcriptional activity [25, 26]. Similarly, a plethora of lncRNAs are linked to TGF-β/Smad signaling [3, 27]. These lncRNAs function as effectors to impact on a specific function of TGF-β signaling or act as regulators to modulate a specific step in the TGF-β/Smad signaling. Moreover, certain lncRNAs control a positive or negative feedback regulation of TGF-β/Smad signaling to amplify or dampen the signaling output, respectively. Despite the diverse modes of lncRNAs in regulating TGF-β/Smad signaling, lncRNAs that coordinate TGF-β signaling with other cancer pathways to alter the dichotomous roles of TGF-β in cancer have not been reported.
Smyca (Smad/Myc coactivator; also known as LOC284454) was originally identified as a p68-interacting RNA by RNA immunoprecipitation followed by high throughput sequencing. This 1.7 kb lncRNA is conserved only in primates and is generated by Drosha-mediated cleavage of a primary transcript for separating it from the precursor of miR-23a ~ 27a ~ 24–2 cluster [28]. Previous studies indicated that Smyca is upregulated in nasopharyngeal carcinoma and hepatocellular carcinoma (HCC) and associated with poor prognosis of these cancer types [29, 30]. Furthermore, serum level of Smyca is upregulated in nasopharyngeal carcinoma, oral cancer and thyroid cancer [31]. In this study, we identified Smyca based on its association with aggressive progression and poor prognosis of multiple cancer types. Smyca binds and activates Smad3 and Smad4 to enhance their interaction and promoter targeting. Additionally, Smyca binds c-Myc to enhance c-Myc-mediated transcription. Through potentiating both TGF-β/Smad and c-Myc pathways, Smyca promotes EMT and multiple EMT-associated malignant features, but avoids TGF-β-induced growth inhibition. Furthermore, Smyca acts through TGF-β and c-Myc signaling to synergize glycolysis. Consistent with the multifaceted tumor-promoting roles, targeting Smyca suppresses metastasis and sensitizes tumors to chemotherapy. Thus, Smyca is a lncRNA that turns the dichotomous roles of TGF-β toward tumor promotion and represents a potential therapeutic target for cancers with aberrant activation of TGF-β and/or c-Myc pathways.
Materials and methods
Cell culture and transfection
LM6 is a subclone of MDA-MB-231 cells derived from six-round enrichment of lung metastatic cells via an experimental metastasis model [32]. 293FT, MDA-MB-231, MDA-MB-468, Hs578T, MCF7, BT-549, BT-474, ZR75-1, LM6 cells and normal mammary epithelial cell line M10 were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin. The HCC cell lines Malaru and NTU-BL were grown in high-glucose DMEM supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 1 × non-essential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin. Transfection was performed using Lipofectamine 2000 reagent (Invitrogen) or TransIT-X2 transfection reagent (Mirus) according to the manufacturers’ instructions.
Plasmids
The full-length Smyca cDNA was amplified by RT-PCR from HT29 cells and subcloned to pLAS5w, pRK5-Flag and pcDNA. To generate the Smyca (1-500), Smyca (501-1000), Smyca (1001-1500), Smyca (1501-1772) and Smyca (△1-500) constructs, the corresponding cDNA fragments were amplified via PCR and subcloned to pLAS5w and pcDNA. Smyca (△1001-1500) mutant was generated using the NEBuilder HiFi DNA Assembly Kit (New England Biolabs). To generate the Smad7 reporter construct, a DNA fragment corresponding to nucleotides − 584 to + 160 of the Smad7 gene containing an SBE sequence (5’-GTCTAGAC) was amplified from the genomic DNA of MDA-MB-231 cells and inserted to pGL3-Basic plasmid. The SNAI2 reporter construct was described previously [33]. The 3TP-Luc and 4 × SBE-Luc constructs were obtained from Rik Derynck (University of California at San Francisco) and the SERPINE1-Luc construct was from Xin-Hua Feng (Zhejiang University, Zhejiang, China). The Myc-responsive reporter construct was purchased from Qiagen. The full-length cDNAs for Smad3 and Smad4 were amplified by PCR from pRK5F-Smad3 and pRK5F-Smad4 [34], respectively, and subcloned to pGEX4T-1 and pVL1392. To generate Smad3△MH1 and Smad4△MH1 mutants, Smad3 cDNA fragment containing nucleotides 409 to 1278 and Smad4 cDNA fragment containing nucleotides 427 to 1659 were amplified via PCR and subcloned to pVL1392. To generate GFP fusion constructs for full-length c-Myc, c-Myc TAD (1-150), c-Myc△TAD (151-439) and c-Myc△DBD (1–319), the corresponding cDNA fragments were amplified by PCR from pLAS3w-c-Myc and subcloned to pEGFP-N3.
Antibodies and reagents
Antibodies used in this study were obtained from commercial sources and are described in Additional file 1: Table S1. Recombinant human proteins TGF-β, EGF and bFGF were obtained from R&D Systems, whereas c-Myc was purchased from Abcam. TβRI (type I TGF-β receptor) inhibitor SB431542, c-Myc inhibitor 10058-F4, cisplatin, doxorubicin (Dox), protamine sulfate and calf thymus DNA were purchased from Sigma-Aldrich (St Louis, MO). DOTAP, cholesterol and DSPE-PEG2000 were purchased from Avanti Polar Lipids (Alabaster, AL).
RNA interference
Smyca shRNAs predicted via BLOCK-iT™ RNAi Designer (https://rnaidesigner.thermofisher.com/) were generated by Purigo Biotechnology, Inc. (Taiwan), and cloned to pLKO.1. siRNA targeting c-Myc was purchased from Dharmacon (Lafayette, CO, USA). Other shRNAs were from RNA Technology Platforms and Gene Manipulation Core Facility, Taiwan. The sequences of siRNAs and shRNAs are listed in Additional file 1: Table S2.
Migration and invasion
Transwell migration and invasion assays were performed as described [35]. At the end of incubation, cells that had migrated onto the lower membrane surface were fixed with 4% formaldehyde, stained with 0.5% DAPI and photographed at least five random fields for each well under a Nikon fluorescence microscope. The migrated cells in each field were counted automatically via the “Analyze Particles” function of Image J.
Spheroid formation
Cells were plated on 96-well ultra-low attachment plates at a tenfold serial dilution from 1 to 1000 cells per well and cultured in tumor sphere medium (DMEM/F12 medium supplemented with B27 (Invitrogen), 20 ng/ml EGF, 10 ng/ml bFGF and 5 μg/ml insulin (Invitrogen)). After 3 weeks, tumor spheres were imaged by an Olympus Scan^R high-content screen station (Olympus). Spheres with a diameter > 20 μm were scored and the percentage of wells containing tumor spheres was measured. The sphere-forming frequency of each group was analyzed via the extreme limiting dilution assay (ELDA) model (http://bioinf.wehi.edu.au/software/elda/).
Flow cytometry
Cells were trypsinized by dissociation buffer (Invitrogen) and resuspended in blocking solution (Ca2+, Mg2+-free HBSS containing 2% goat serum). Next, cells were incubated with APC-conjugated anti-CD44 antibody (BD Bioscience) and PE-conjugated anti-CD24 antibody (BD Bioscience) for 1 h at 4 °C. After washes, the labeled cells were analyzed by a Beckman CytoFLEX flow cytometer. The breast cancer stem cell population (CD44high and CD24low/−) was calculated via the CytExpert software.
Quantitative RT-PCR (qRT-PCR)
Total RNAs were extracted by TRIZOL reagent (Invitrogen) and quantified by NanoDrop (Thermo Scientific). Reverse transcription was performed using iScript™ cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s instructions. Real-time PCR was performed on a LightCycler® 480 System with SYBR Green I Master Kit (Roche). GAPDH was used as an internal control. Smyca copy number was calculated by the equation of “Number of copies = Amount (ng) × 6.022 × 1023/Length (bp)x1 × 109x660” in which “Amount” was determined by a “Ct value to amount” standard curve generated from known concentrations of a Smyca-containing plasmid. The PCR primers used in this study are listed in Additional file 1: Table S3.
Luciferase assay
Cells transfected with the pGL3-based firefly luciferase construct together with the pRK5F-based Renilla luciferase plasmid were used for luciferase assay with the Dual-Glo Luciferase Reporter Assay System (Promega) followed by the manufacturer’s instructions. The firefly luciferase activity was normalized to that of Renilla luciferase activity.
In situ hybridization
In situ hybridization was performed with the ViewRNA ISH Tissue 2-Plex Assay Kit (Thermo Fisher) according to the manufacturer’s instructions. Briefly, the paraformaldehyde-fixed cells were permeablized with protease and hybridized with the commercial Smyca tilling Type 1 probe set (Thermo Fisher, Catalog #VA1-3016541) at 40 °C for 2 h. After washing out the unlabeled probes, the hybridization signal was amplified by 2 steps of branch DNA hybridization at 40℃ for 40 min. These branch DNA amplifiers were then labeled by alkaline phosphatase, and fluorescence was developed by incubating with Fast Red substrates at 40 °C for 60 min. The slide was then counterstained with DAPI and examined by an Olympus FV3000RS inverted confocal microscope equipped with 60x/1.40 oil objective lens (Olympus Objective Lens, PlanApo N). Images were collected by an Olympus FV3000 FV31S-SW (v 2.40) software.
Subcellular fractionation
Cells were trypsinized and passed through a 70 µm cell strainer (BD Falcon) to remove cell clumps. After centrifugation, the cell pellet was resuspended in 200 µl ice-cold cytoplasmic lysis buffer (10 mM Tris pH 7.5, 150 mM NaCl and 0.15% NP-40), chilled on ice for 5 min, transferred to a tube containing 500 µl ice-cold sucrose buffer (10 mM Tris pH 7.5, 150 mM NaCl and 24% sucrose) and centrifuged at 13,000 rpm for 10 min. The supernatant was collected as the cytoplasmic fraction. The pellet was resuspended in 200 µl of cytoplasmic lysis buffer without NP-40 and centrifuged again with 500 µl sucrose buffer. The pellet was washed and resuspended in 200 µl ice-cold glycerol buffer (20 mM Tris pH 7.5, 75 mM NaCl, 0.5 mM EDTA, 50% glycerol and 0.85 mM DTT) and then lysed by 200 µl of ice-cold nuclei lysis buffer (20 mM HEPES pH 7.5, 7.5 mM MgCl2, 0.2 mM EDTA, 300 mM NaCl, 1 M urea, 1% NP-40 and 1 mM DTT). The nuclear lysate was vortexed vigorously for 5 s, incubated on ice for 1 min and centrifuged at 14,000 rpm for 2 min. The supernatant was transferred to a fresh tube and saved as the nuclear fraction. 10% of the cytoplasmic and nuclear fractions were used for RNA extraction and qRT-PCR analysis.
Western blot and immunoprecipitation
Cells were lysed with RIPA buffer containing 50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 1 mM DTT, 1 mM phenylmethylsulphonyl fluoride (PMSF), 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mM sodium vanadate, 4 mM sodium pyrophosphate and 20 mM NaF. Immunoprecipitation and Western blot using cell lysates with equal amounts of proteins were performed as described [36].
In silico expression-based analysis and prediction
The RNA-seq data and survival information derived from patients of various cancer types were downloaded from The Cancer Genome Atlas (TCGA) via the UCSC Xena platform (http://xena.ucsc.edu) [37]. Microarray data from human breast, liver and colon cancer patients were retrieved from the NCBI’s Gene Expression Omnibus (GEO) database. The association between Smyca expression and cancer patient survival was conducted by Graphpad Prism using the Kaplan–Meier survival analysis along with the log-rank test. The correlation analysis for the expression of two genes was conducted by Pearson’s correlation analysis using the Python Statistical package scipy stats.
RNA-sequencing (RNA-seq) analysis
Total RNAs were extracted by the NucleoSpin RNA Kit (Macherey-Nagel, Duren, Germany). Only RNA samples with a clear peak of 25–200 nt on the electrophenogram (small RNA fraction) and an RNA integrity number of at least 8 were used for the subsequent library construction and sequencing. After removing rRNAs from the RNA samples, RNA libraries were generated by the Agilent Sure Select Strand Specific RNA Library Prep Kit (Illumina). Next-generation sequencing was performed in HiSeq4000 (Illumina) for 150 bp paired-end reads by Novogene (Biotools). Reads were mapped against the human reference genome (GRCh38/hg38) using TopHat v2.0.12 to generate align reads for each sample. The align reads were counted and normalized to per million bases by HTSeq to obtain gene abundance. Differentially expressed genes (DEGs) were determined using DESeq v1.10.1.
Pathway and gene set enrichment analyses
The DEGs identified by RNA-seq were subjected to the Ingenuity Pathway Analysis (IPA, Qiagen) or a web-based gene set analysis toolkit WebGestalt, 2019 (http://www.webgestalt.org/). To confirm the enrichment between Smyca-induced signature and published malignancy-related signatures from the MSigDB database, Smyca-induced transcriptome was submitted to the Gene Set Enrichment Analysis (GSEA) v4.1.0 Java Web Start. Gene sets with a false discovery rate (FDR) < 0.05 by comparing the enrichment score to the enrichment results generated from 1000 random permutations were considered as statistically significant.
Chromatin immunoprecipitation (ChIP)
Cells were treated with 1% formaldehyde for 15 min for cross-linking the chromatins followed by quenched with 125 mM glycine for 5 min. Next, cells were lysed with ChIP lysis buffer (3 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 0.5% N-lauroylsarcosine, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mM sodium vanadate, 4 mM sodium pyrophosphate and 20 mM NaF) and sonicated to achieve the majority of DNA fragments within 200–500 bp. The lysate was precleared with Protein A and incubated with the desired antibodies at 4 °C for overnight, followed by the addition of 100 μl Protein-A magnetic beads with rotation for 1 h. The immunocomplexes were washed 4 times with ChIP wash buffer (50 mM HEPES pH 7.5, 1 mM EDTA, 1% NP-40, 500 mM LiCl, 0.7% sodium deoxycholate, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mM sodium vanadate, 4 mM sodium pyrophosphate and 20 mM NaF), followed by TE buffer (10 mM Tris pH 8.0 and 1 mM EDTA) containing 50 mM NaCl. The protein–DNA complex was eluted twice from beads with 150 μl elution buffer (1% SDS, 200 mM NaCl, 100 µg/ml RNase A, 100 U/ml RNase H in TE buffer) at 65 °C for 20 min. The immunoprecipitated DNA fragments were incubated with 1 mg/ml proteinase K at 65 °C for 2 h to remove the cross-linked proteins, purified by a PCR purification kit (Qiagen) and analyzed by quantitative PCR (qPCR). The sequences of qPCR primers are listed in Additional file 1: Table S3.
In silico analysis of ChIP-seq data
The Smad2/3 ChIP-seq data derived from TGF-β-treated Hs578T and BT-549 cells were downloaded from GSE83788. The aligned BAM files of ChIP-seq datasets were then subjected to MACS2 (Galaxy Version 2.1.1.20160309.6) to identify the Smad2/3 binding sites with a threshold of FDR < 0.05. The binding sites were mapped to the nearest genes by Galaxy platform (https://usegalaxy.org/) using the definition of promoter region as a segment between 2 kb upstream and 10 kb downstream of the transcriptional starting site (12 kb in length) in the human reference sequence assembly (NCBI Build 37/hg19, February 2009). The Smad2/3 occupied genes were then converted to “Gene Symbol” by BioDBnet (https://biodbnet-abcc.ncifcrf.gov/).
Chromatin isolation by RNA purification (ChIRP)
ChIRP was performed by adapting a protocol described previously [38]. Briefly, tiling 20-mer antisense oligonucleotide probes targeting Smyca and the negative control probes for lacZ RNA were designed using the online probe designer at singlemoleculefish.com (http://www.singlemoleculefish.com/designer.html), synthesized and biotinylated by Genomics, Inc. The sequences of these probes are listed in Additional file 1: Table S4. Cells were treated with 1.25% glutaraldehyde for 15 min and then with 125 mM glycine for 5 min. The cross-linked chromatins were sheared into 200–500 bp by sonication and hybridized with each of the probe sets, followed by streptavidin magnetic beads capturing and wash/elution steps. The ChIRP captured chromatins were treated with Protease/RNase and analyzed by qPCR. The sequences of qPCR primers are listed in Additional file 1: Table S3.
RNA pull-down analysis
Biotin-labeled RNAs were generated by in vitro transcription using the AmpliScribe T7-Flash Biotin-RNA Transcription Kit (Epicentre Biotechnologies) and purified by the NucleoSpin RNA Kit (Macherey-Nagel, Duren, Germany). To form the proper secondary structure, 30 pmol biotinylated RNA was heated to 90 °C in the RNA structure buffer (10 mM Tris pH 7.0, 100 mM KCl and 10 mM MgCl2) for 2 min, chilled on ice for 2 min and incubated at room temperature for 20 min. The RNA was mixed with nuclear extracts or purified proteins and incubated at room temperature for 1 h, followed by incubating with Streptavidin magnetic beads (GE Healthcare) at room temperature for 1 h. After washes, the pull-down complexes were analyzed by Western blot. Alternatively, the folded RNA was incubated with recombinant protein conjugated on beads at room temperature for 1 h. After washes, the pull-down RNA was extracted by TRIZOL reagent and analyzed by qRT-PCR.
For preparing nuclear extracts, cells were harvested by 2 ml PBS and mixed with 8 ml nuclear isolation buffer (0.32 M sucrose, 10 mM Tris pH 7.5, 5 mM MgCl2 and 1% Triton X-100) on ice for 20 min with frequent mixing. After centrifugation at 2,500 × g for 15 min, the pellet was resuspended with 1 ml ice-cold RIP buffer (150 mM KCl, 25 mM Tris pH 7.4, 5 mM EDTA, 0.5% NP-40, 0.5 mM DTT, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mM sodium vanadate, 4 mM sodium pyrophosphate, 20 mM NaF and 100 U/ml SUPERaseIN). The nuclear lysate was sonicated using a Qsonica Q700 system according to the manufacturer’s protocol, followed by centrifugation at 13,000 rpm for 20 min. The supernatant was recovered as the nuclear extract.
RNA immunoprecipitation (RIP)
Cells were lysed with polysome lysis buffer containing 15 mM Tris pH 7.5, 300 mM NaCl, 1% Triton X-100, 1 mM DTT, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mM sodium vanadate, 4 mM sodium pyrophosphate, 20 mM NaF and 100 U/ml SUPERaseIN. The lysates were precleared with Protein-A magnetic beads (Millipore, Bedford, MA, USA) at 4 °C for 60 min. After adding 2 μg antibody into the precleared lysates, the immunoprecipitated protein-RNA complex was captured by Protein-A magnetic beads at 4 °C for 2 h. For experiment involving sequential immunoprecipitations, the bound proteins were eluted twice with 150 μg/ml M2 peptide in TBS buffer (50 mM Tris pH 7.5 and 150 mM NaCl). The eluent was diluted to 1 ml with the adjustment of salts and detergent to 50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 5% glycerol and 5 mM EDTA, followed by the second immunoprecipitation. The immunoprecipitated proteins and coprecipitated RNAs were extracted from the beads with sample buffer and TRIZOL reagent and analyzed by qRT-PCR and immunoprecipitation, respectively.
Cell proliferation and viability assays
To determine the proliferation rate, cells were seeded on 96-well plates at a density of 2000 cells/well, cultured overnight and treated with various inhibitors. Alternatively, transfected cells were seeded at a density of 8000 cells/well. Then, cells were pulse labeled with 10 μM BrdU for 2 h. After fixation, BrdU incorporation was determined by the BrdU Cell Proliferation Assay Kit (Merck Millipore) according to the manufacturer's instructions. To test cell viability in response to chemotherapeutic agents, cells were seeded at a density of 2000 cells/well on 96-well plates. After attachment, cells were cultured in medium containing Dox or cisplatin for 2 or 3 days, respectively. Cell viability was determined by incubating with 0.4 mg/ml methyl thiazolyl diphenyl tetrazolium bromide (MTT, Sigma-Aldrich) for 2 h, followed by cell lysis with DMSO and absorbance measurement at 590 nm.
Measurement of glucose and lactate levels
Cells were seeded on 6-well plates at a density of 2 × 105 cells/well and treated with various inhibitors. The conditioned media were harvested at 0 and 24 h (for measuring glucose) or 0 and 48 h (for measuring lactate) post-treatment. The concentrations of glucose and lactate were determined by commercial ELISA-based kits (BioVision, Inc.) according to the manufacturer's instructions.
Measurement of extracellular acidification rate (ECAR)
ECAR was determine by the Seahorse Extracellular Flux analyzer (XFe24, Seahorse Bioscience, Billerica, MA, USA) with the Seahorse XF Glycolysis Stress Test Kit according to the manufacturer’s instructions. Briefly, cells were seeded at a density of 40,000 cells/well in 24-well plates and allow to attach. Three hours later, the cells were treated with various pathway inhibitors for 24 h. The glycolytic metabolic profiles were determined by sequential injections of glucose (10 mM), oligomycin (1 uM) and 2-deoxy-D-glucose (75 mM). The Seahorse Wave 2.3 software was used for data analyses, and ECAR was expressed in mpH/min. The maximum glucose response was referred to as glycolysis, whereas the glycolytic reserve was calculated as: maximum oligomycin response—maximum glucose response.
Preparation of PEGylated liposome–polycation DNA nanoparticle (LPD-NP) formulations
LPD-NPs were prepared according to a previously method [39, 40] with slight modifications. Briefly, liposomes composed of DOTAP and cholesterol (1:1 molar ratio) were prepared by thin film hydration followed by sonication for 3 min (5 s on/5 s off, Amp 30%). To prepare LPD-NPs, 22 µl of protamine (2 mg/ml), 120 µl of deionized water, and 24 µl of 1:1 weight ratio of gapmer (Qiagen, 2 mg/ml) and calf thymus DNA (2 mg/ml) were mixed and kept at room temperature for 10 min before adding 60 µl of liposome (20 mmol/l). LPD-NPs were stood at room temperature for 10 min before the addition of DSPE-PEG. LPD-NPs were then mixed with 33.6 µl of DSPE-PEG2000 (10 mg/ml) and kept at 50–60 °C for 10 min. The sequences of gapmers are listed in Additional file 1: Table S2.
Animal studies
For experimental metastasis model, MDA-MB-231 cells tagged with luciferase were resuspended (1 × 106 cells/0.1 ml HBSS) and injected into the tail vein of 7-week-old female NOD/SCID (NOD.CB17-Prkdcscid/NcrCrlBltw) mice (BioLASCO Taiwan Co., Taipei, Taiwan) by a 27-gauge needle. Lung metastasis was monitored by bioluminescence imaging using the PerkinElmer In Vivo Imaging System (Waltham, MA, USA). The mice were then killed for histological analysis of the lung.
To monitor chemoresistance, 2 × 106 MDA-MB-231 cells were orthotopically injected into the fat pad of 7-week-old female Nu/Nu (Bltw:NU-Foxn1nu) mice (BioLASCO Taiwan Co., Taipei, Taiwan). When tumors grew to about 30 mm3 in volume, Dox (6 mg/kg) diluted with 0.9% NaCl was administered via intraperitoneal injection every week. The sizes of tumors were measured every 3 or 4 days, and their volumes were calculated using the equation mm3 = 1/2 × length (mm) × (width (mm))2.
To evaluate the anti-tumor effect of LPD-NPs containing Smyca gapmer ASO, LM6 tumor-bearing NOD/SCID mice were injected intratumorally with 30 µg LPD-NPs and intraperitoneally with Dox (6 mg/kg) every week. The sizes of tumors were measured every 3 or 4 days. At the end of experiment, the tumors were removed to visualize their sizes. To evaluate the anti-metastasis effect of LPD-NPs containing Smyca gapmer ASO, LM6 tumor-bearing NOD/SCID mice were intratumorally injected with 30 µg LPD-NPs every week. Lung metastasis was monitored by bioluminescence imaging. All mouse experiments were conducted with the approval from the Experimental Animal Committees of Academia Sinica and Taipei Medical University.
Human specimens
Snap-frozen primary breast cancer tissues stored in liquid nitrogen were obtained from Taipei Medical University BioBank and mRNAs extracted from HCC tissues and paired noncancerous adjacent tissues were obtained from National Health Research Institutes BioBank. Written informed consents were obtained from all patients. The research design, study protocols and information security were approved by the Institutional Review Boards of Taipei Medical University and Academia Sinica.
Results
Smyca high expression correlates with poor prognosis and aggressive progression of multiple cancer types
In an attempt to identify cancer-relevant lncRNAs, we searched for lncRNAs associated with adverse prognosis of cancer. By querying TCGA data sets from a variety of cancer types, we found that Smyca high expression correlated with poor overall survival and disease-free survival of several cancer types, including kidney clear cell carcinoma, lower grade glioma, adrenocortical cancer and mesothelioma (Additional file 1: Fig. S1A, B). Furthermore, the association of Smyca high expression with poor overall survival was also observed from two breast cancer cohorts and one colon cancer cohort downloaded from the GEO database as well as an in house HCC cohort (Additional file 1: Fig. S1C–F). In the HCC cohort, although Smyca expression did not differ significantly between tumor and adjacent non-tumor tissues, its high expression was modestly but significantly associated with higher tumor stages and invasive phenotypes (Additional file 1: Fig. S1G–I). Similar associations were observed from an in house breast cancer cohort (Additional file 1: Fig. S1J, K). In this cohort, Smyca expression was higher in the basal-like subtype, compared with the less malignant HER2-enriched and luminal subtypes (Additional file 1: Fig. S1L). These findings collectively support Smyca as a prognostic marker for several cancer types and suggest its role in tumor promotion.
Smyca promotes EMT, cancer stemness, migration and invasion
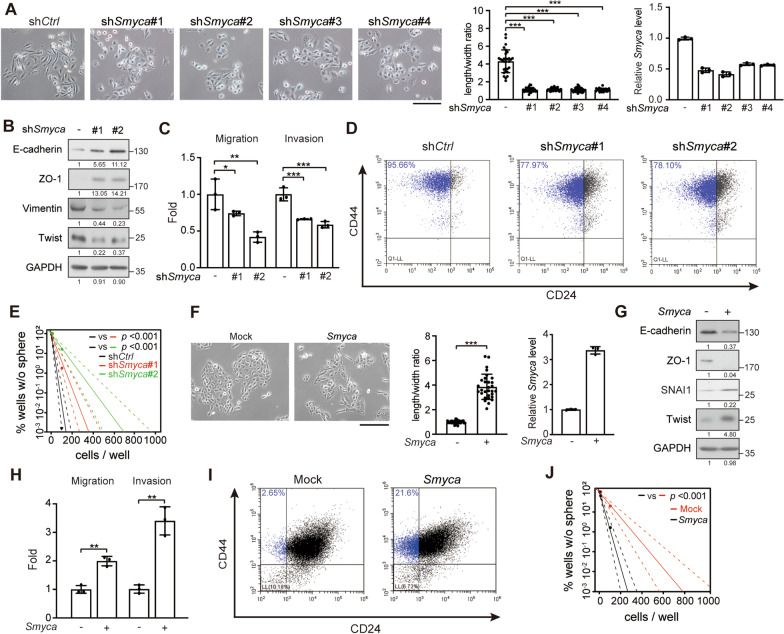
In line with the findings derived from breast cancer patients, Smyca showed higher expression levels in mesenchymal/basal-like than epithelial-like breast cancer cell lines (Additional file 1: Fig. S2A). We therefore investigated whether Smyca promotes EMT, a feature of mesenchymal/basal-like subtype. Four shRNAs were individually introduced to mesenchymal/basal-like breast cancer cell line MDA-MB-231. These shRNAs reduced Smyca expression without affecting the expression of miRNAs in the miR-23a ~ 27a ~ 24–2 cluster (Fig. 1A and Additional file 1: Fig. S2B). Smyca knockdown switched the mesenchymal morphology of MDA-MB-231 cells to epithelial, as indicated by the reduction in cell length/width ratio (Fig. 1A). Furthermore, Smyca knockdown elevated the expression of epithelial markers E-cadherin and ZO-1 and reduced the mesenchymal markers Vimentin and Twist (Fig. 1B). These findings are consistent with an induction of mesenchymal-to-epithelial transition (MET). Smyca knockdown in another mesenchymal/basal-like breast cancer cell line Hs578T similarly upregulated epithelial markers and downregulated mesenchymal markers (Additional file 1: Fig. S2C). In line with the MET induction, Smyca knockdown in MDA-MB-231 cells inhibited migration and invasion (Fig. 1C). Next, we evaluated the effect of Smyca on cancer stemness, a feature tightly linked to EMT. By monitoring the breast cancer stem-like cell markers CD44high/CD24low/−, we found a high stem-like population in MDA-MB-231 cells, consistent with previous reports [41, 42]. This stem-like population was reduced by Smyca knockdown (Fig. 1D). Furthermore, Smyca knockdown decreased the ability of MDA-MB-231 cells to form mammary spheres (Fig. 1E). In the reciprocal sets of experiments, we chose to overexpress Smyca in mammary epithelial cell M10 and epithelial-like breast cancer cell line MCF7, which expressed lower levels of Smyca comparing to MDA-MB-231 cells (Additional file 1: Fig. S2A). We found that Smyca overexpression in these cells promoted EMT (Fig. 1F, G and Additional file 1: Fig. S2D). Smyca overexpression in M10 cells also promoted migration and invasion and enhanced stemness features (Fig. 1H–J). Notably, Smyca overexpression or knockdown did not affect cell proliferation and survival (Additional file 1: Fig. S2E–H). Thus, our study revealed a critical role of Smyca in mediating EMT and EMT-associated malignant features, such as migration, invasion and tumor stemness.
Fig. 1.
Smyca promotes EMT, migration, invasion and stemness. (A) Morphological assessment of MDA-MB-231 cells stably expressing four different Smyca shRNAs. Bar, 100 μm. Cell length/width ratios and Smyca expression levels (normalized to the control group) are shown in the middle and right panels, respectively. Data are mean ± SD, n = 30 (middle) or 3 (right). P values are determined by one-way ANOVA with Tukey’s post hoc test, *** P < 0.001. (B, G) Western blot analysis of EMT markers in MDA-MB-231 cells stably expressing Smyca shRNAs (B) or M10 cells stably overexpressing Smyca (G). The amounts of each protein in relation to the control cells are indicated under the bands. (C, H) Migration and invasion assays of MDA-MB-231 cells stably expressing Smyca shRNAs (C) or M10 cells stably overexpressing Smyca (H). Data are mean ± SD from three independent experiments. P values are determined by one-way ANOVA with Tukey’s post hoc test (C) or unpaired t test (H), *P < 0.05, **P < 0.01, *** P < 0.001. (D, I) Flow cytometry analysis of the expression of breast cancer stem cell markers in MDA-MB-231 cells stably expressing Smyca shRNAs (D) or M10 cells stably overexpressing Smyca (I). Stem cell populations are marked by blue and the percentages are indicated. (E, J) Sphere formation assay of MDA-MB-231 cells stably expressing Smyca shRNAs (E) or M10 cells stably overexpressing Smyca (J). Solid and dashed lines are derived from means and standard deviations, respectively. Data are mean ± SD from three independent experiments. P values are determined by Chi-square test. (F) Morphological assessment of M10 cells stably overexpressing Smyca. Bar, 100 μm. Cell length/width ratios and Smyca expression levels (normalized to the control group) are shown in the middle and right panels, respectively. Data are mean ± SD, n = 30 (middle) or 3 (right). P values are determined by unpaired t test, *** P < 0.001
Smyca promotes TGF-β/Smad signaling
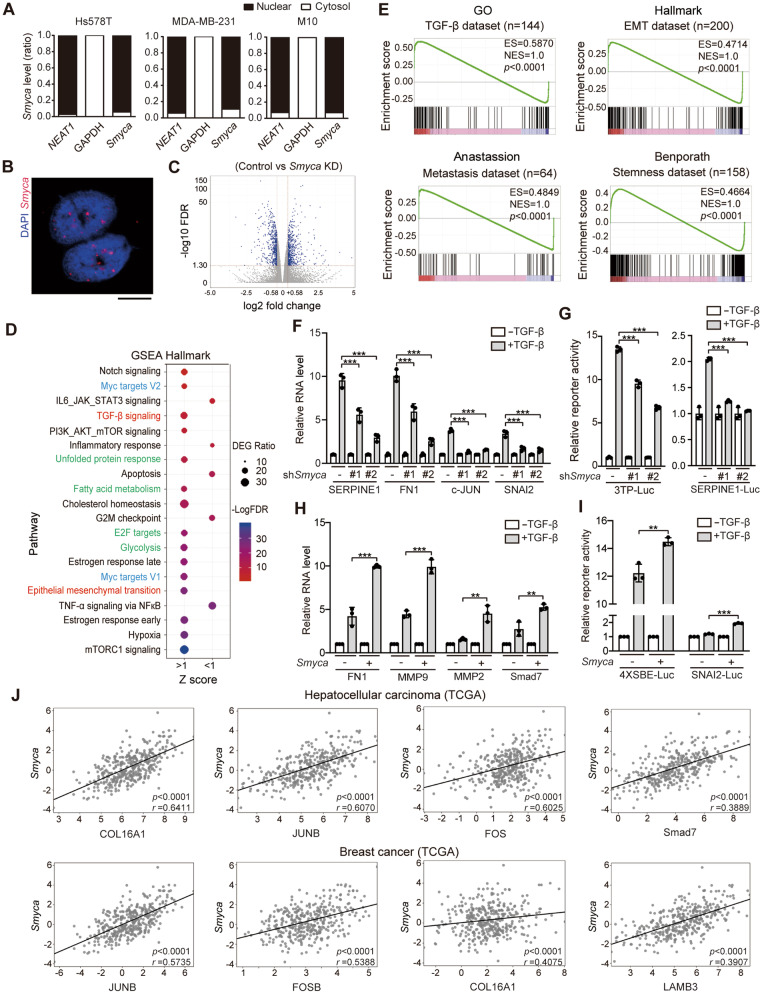
Next, we explored the mechanism by which Smyca potentiates EMT. Cell fractionation analysis and fluorescence in situ hybridization assay revealed that Smyca was mainly distributed to the nucleus (Fig. 2A, B). Since nuclear lncRNAs often regulate gene expression, we determined the impact of Smyca on transcriptome by RNA-seq analysis of MDA-MB-231 cells expressing control or Smyca shRNA. DEGs were defined with the criteria: control/Smyca knockdown ≥ 1.5 or ≤ 0.66 and FDR < 0.05 and a total of 2201 DEGs were recovered in three biological repeats (Fig. 2C). GSEA Hallmark Pathway analysis of these DEGs revealed that EMT and TGF-β signaling were among the upregulated hallmarks (Fig. 2D, red). Furthermore, GSEA analysis found that Smyca-induced transcriptome correlated significantly and positively with TGF-β, stemness, metastasis and EMT signatures from multiple sources (Fig. 2E and Additional file 1: Fig. S3A). Comparison of Smyca-induced DEGs with the Smad2/3 ChIP-seq data derived from Hs578T or BT-549 cells [43] revealed a subset of overlapped genes (Additional file 1: Fig. S3B and Table S5). These findings prompted us to investigate the impact of Smyca on TGF-β/Smad signaling. Importantly, Smyca knockdown in MDA-MB-231 cells diminished TGF-β-induced expression of multiple downstream genes, such as FN1 (Fibronectin 1), SERPINE1 (encoding PAI-1), c-JUN and SNAI2 and TGF-β-mediated transcriptional activation of Smad-responsive reporters 3TP-Luc and SERPINE1-Luc (Fig. 2F, G). Smyca knockdown in Hs578T cells similarly attenuated TGF-β-induced expression of downstream genes and activation of Smad-responsive reporters 4 × SBE-Luc and SERPINE1-Luc (Additional file 1: Fig. S3C, D). A similar reduction in TGF-β-induced 4 × SBE-Luc reporter activity was found by Smyca knockdown in HCC cell line Malaru (Additional file 1: Fig. S3E), which expressed a higher level of Smyca than another HCC cell line NTU-BL (Additional file 1: Fig. S2A). Conversely, Smyca overexpression in M10 cells increased TGF-β-induced downstream gene expression and Smad-responsive reporter activities (Fig. 2H, I). These data identify Smyca as a positive regulator of TGF-β/Smad signaling, which is in line with its function in promoting EMT and EMT-associated tumor malignancies.
Fig. 2.
Smyca enhances TGF-β signaling. (A) qRT-PCR analysis of the ratios of nuclear and cytoplasmic Smyca from indicated cells. NEAT1 and GAPDH were used as controls. (B) Representative image for Smyca subcellular distribution analyzed by in situ hybridization on MDA-MB-231 cells. Bar, 10 μm. (C) Comparison of RNA-seq data derived from MDA-MB-231 cells expressing control shRNA and Smyca shRNA #1. DEGs are marked by blue dots. (D) GSEA Hallmark Pathway analysis of DEGs shown in (C). The top enriched and depleted hallmarks are shown by the order of FDR (bottom to top). (E) Representative GSEA plots for the match of Smyca signature with the indicated signatures. Enrichment score (ES) and normalized enrichment score (NES) are indicated. The full set of GSEA data is shown in Additional file 1: Fig. S3A. (F, H) qRT-PCR analysis of indicted genes in MDA-MB-231 cells stably expressing Smyca shRNAs (F) or M10 cells stably expressing Smyca (H) and treated with or without 5 ng/ml TGF-β for 24 h. Data are normalized with that of untreated group in each cell. (G, I) Luciferase reporter assay on MDA-MB-231 cells stably expressing Smyca shRNAs (G) or M10 cells stably expressing Smyca (I), transfected with indicated reporters and treated with or without 5 ng/ml TGF-β for 24 h. Data in (F), (G), (H), and (I) are normalized with that of untreated control and expressed as mean ± SD from three independent experiments. P values are determined by one-way ANOVA with Tukey’s post hoc test (F, G) or unpaired t test (H, I), **P < 0.01, ***P < 0.001. (J) Representative correlation plots of Smyca expression with the expression of indicated TGF-β target genes by analyzing HCC or breast cancer data sets from TCGA (n = 369 for HCC and 1099 for breast cancer). Pearson’s coefficients and P values are indicated. Additional correlative data are shown in Additional file 1: Fig. S3F
To validate the clinical relevance of Smyca to TGF-β-induced transcriptional program, we queried RNA-seq data from TCGA data sets. Remarkably, Smyca expression correlated positively (Pearson’s correlation r > 0.3) with the expression of a large set of TGF-β/Smad target genes in the tumor tissues of HCC and breast cancer patients (Fig. 2J and Additional file 1: Fig. S3F). Similar findings were obtained by analyzing the microarray data of breast cancer and HCC patients from the GEO data sets (Additional file 1: Fig. S3G). Importantly, most of these genes are of tumor-promoting functions and contribute to invasion and metastasis. GO analysis of this set of genes revealed that response to TGF-β, cell migration/chemotaxis, cell adhesion mediated by integrin signaling and collagen metabolic process are among the enriched GO terms (Additional file 1: Fig. S3H). Thus, these data revealed the association of Smyca with TGF-β-induced transcription of a set of tumor-promoting genes in human tumor tissues, highlighting the clinical relevance of Smyca-promoted TGF-β signaling to tumor progression.
Smyca enhances Smad3/Smad4 complex formation and recruitment to its target promoters
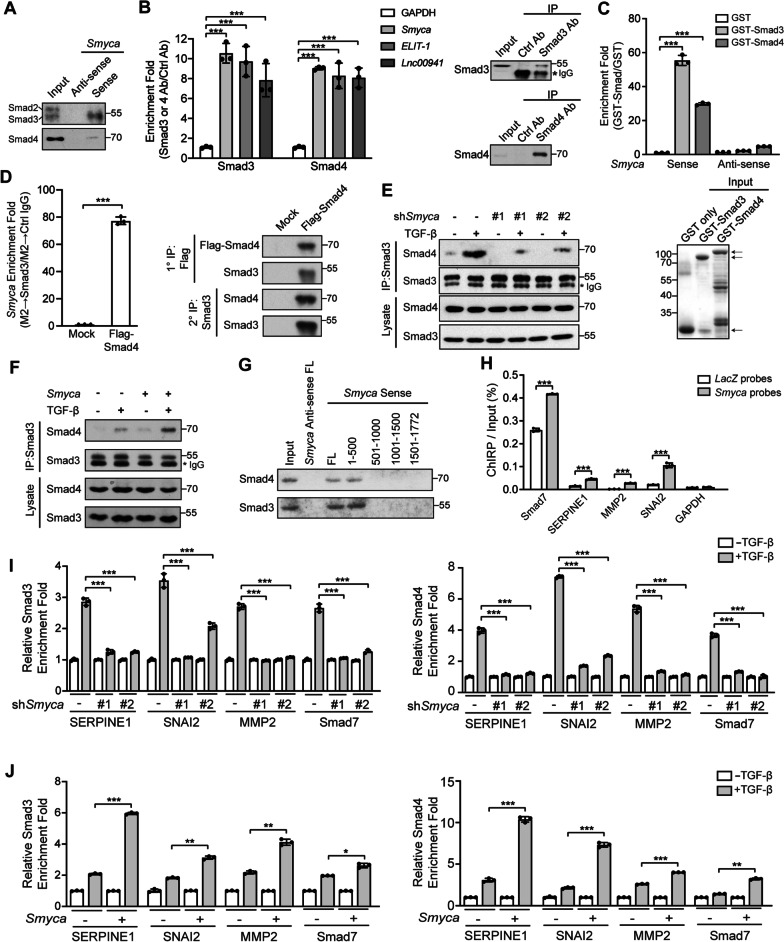
We next investigated how Smyca promotes TGF-β signaling. Given the enrichment of Smyca in the nucleus, we explored its impact on Smads. RNA pull-down assay by incubating in vitro transcribed Smyca with MDA-MB-231 nuclear lysates revealed that Smyca interacted specifically with Smad3 and Smad4, but not Smad2 (Fig. 3A). RIP analysis further detected a significant enrichment of Smyca in the Smad3 and Smad4 immunoprecipitates, and the enrichment folds were comparable to that of other Smad-binding lncRNAs, such as ELIT-1 and Lnc00941 (Fig. 3B). To determine whether Smyca binds Smad3 and Smad4 directly, bacterially purified GST-Smad3 or GST-Smad4 was incubated with in vitro transcribed Smyca or its antisense RNA. We observed a robust enrichment of Smyca, but not its antisense RNA, in beads containing GST-Smad3 or GST-Smad4, compared with GST alone (Fig. 3C). To interrogate whether Smyca, Smad3 and Smad4 form a tertiary complex in vivo, we introduced Flag-Smad4 into MDA-MB-231 cells, stimulated cells with TGF-β, and performed two-step immunoprecipitation analysis to isolate the Smad3/Smad4 complex. We found that Smyca was highly enriched in the Smad3/Smad4 complex (Fig. 3D). Because Smyca assembles a tripartite complex with Smad3 and Smad4, we determined whether Smyca enhances the association between the two Smads. Smyca knockdown in MDA-MB-231 cells attenuated TGF-β-induced Smad3/Smad4 association, without affecting the expression of Smad3 and Smad4 and the phosphorylation of Smad3 (Fig. 3E and Additional file 1: Fig. S4A, B). Conversely, Smyca overexpression in M10 or MDA-MB-231 cells enhanced TGF-β-induced Smad3/Smad4 complex formation (Fig. 3F and Additional file 1: Fig. S4C). To investigate the structural determinants for the association among Smyca, Smad3 and Smad4, we mapped their interaction regions. Using a panel of deletion mutants, we found that Smyca (1–500) segment bound Smad3 and Smad4 as efficiently as the full-length Smyca in a RNA pull-down assay, whereas other fragments bound neither Smad3 nor Smad4 (Fig. 3G). Accordingly, removal of the 1–500 segment abrogated the potentiating effects of Smyca on TGF-β-induced Smad3/Smad4 association and Smad-responsive reporter activity (Additional file 1: Fig. S4C, D). Deletion mapping analysis further showed a critical role of the MH1 domains of both Smad3 and Smad4 in their interactions with Smyca (Additional file 1: Fig. S4E). These findings support that Smyca (1-500) segment binds the MH1 domains of Smad3 and Smad4, which do not overlap with the MH2 domains responsible for the p-Smad3/Smad4 interaction [44]. Thus, Smyca functions as a scaffold to provide an additional binding surface for enhancing the association of Smad3 with Smad4.
Fig. 3.
Smyca enhances Smad3/Smad4 complex formation and promoter recruitment. (A, G) RNA pull-down assay using MDA-MB-231 nuclear extracts and biotinylated sense or antisense Smyca or Smyca deletion fragments. Antibodies that recognize Smad2/3 and Smad3 only were used in (A) and (G), respectively. (B) RIP analysis for the enrichment of indicated lncRNAs in Smad3 or Smad4 immunoprecipitates derived from MDA-MB-231 cells. Data are normalized with that from the control antibody. The presence of Smad3 or Smad4 in the immunoprecipitates is shown on the right. (C) Bacterially purified GST-Smad3 or GST-Smad4 bound on beads was incubated with sense or antisense Smyca. The bound Smyca was analyzed by qRT-PCR and normalized with that from the GST only group. The input GST fusion proteins are shown on the bottom and marked by arrows. (D) Smad4-associated complex was immunoprecipitated from MDA-MB-231 cells transfected with Flag-Smad4 and treated with 5 ng/ml TGF-β for 2 h. The immunocomplex was eluted and further precipitated with anti-Smad3 antibody to isolate the Smad3/Smad4 complex. Smyca enrichment in this complex was analyzed by qRT-PCR. (E, F) Immunoprecipitation analysis of Smad3 and Smad4 interaction in MDA-MB-231 cells stably expressing Smyca shRNAs (E) or M10 cells stably expressing Smyca (F) and treated with or without 5 ng/ml TGF-β for 1 h. (H) ChIRP assay for Smyca occupancy on the indicated Smad target loci. GAPDH was used as a control. Tilling biotinylated oligonucleotides complementary to LacZ or Smyca were used to pull down the RNA-associated chromatins from MDA-MB-231 cells treated with 5 ng/ml TGF-β for 2 h, followed by qRT-PCR analysis. Data are normalized with the inputs. (I, J) ChIP analysis of the recruitment of Smad3 or Smad4 to the indicated promoters in MDA-MB-231 cells stably expressing Smyca shRNAs (I) or Smyca (J) and treated with or without 5 ng/ml TGF-β for 2 h. The enrichment folds are normalized with that from untreated group. Data in (B), (C), (D), (H), (I), and (J) are mean ± SD, n = 3. P values are determined by one-way ANOVA with Tukey’s post hoc test (B, C, I) or unpaired t test (D, H, J), *P < 0.05, **P < 0.01, ***P < 0.001
To determine whether Smyca acts directly on the Smad3/Smad4 target chromatins, we conducted ChIRP assay using a set of probes complementary to the Smyca sequence for pulling down Smyca from TGF-β-stimulated MDA-MB-231 cells. Subsequent qPCR analysis demonstrated the recruitment of Smyca to promoters of a set of Smad targets (Fig. 3H). Furthermore, ChIP assay showed that Smyca knockdown reduced the recruitment of Smad3 and Smad4 to the promoter regions of these Smad targets, whereas Smyca overexpression elicited an opposite effect (Fig. 3I, J). Thus, our findings suggest that the Smyca/Smad3/4 complex acts on chromatins to promote the transcription of Smad target genes.
Smyca governs a positive feedback regulation of TGF-β signaling
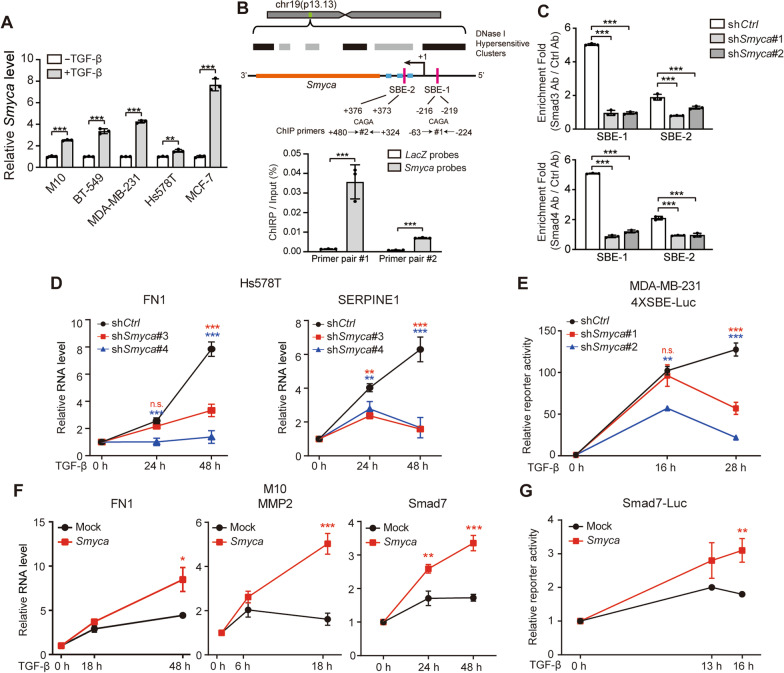
Having identified an effect of Smyca on promoting Smad-mediated transcription, we next explored the conditions that regulate Smyca expression. Interestingly, Smyca expression was induced by TGF-β in multiple cell lines and Smad3 or Smad4 knockdown abolished TGF-β-induced Smyca expression (Fig. 4A and Additional file 1: Fig. S5A). Furthermore, two Smad-binding elements (SBEs) were found in 5’ regulatory and intra-gene regions of the Smyca gene. ChIRP and ChIP assays demonstrated the recruitment of Smyca and Smad3/Smad4 to the two regions, respectively (Fig. 4B, C). Furthermore, Smyca knockdown diminished the binding of Smad3 and Smad4 to these SBEs (Fig. 4C). These findings not only uncovered an autoregulatory mode of Smyca for its own transcription, but also suggested a role of Smyca in mediating a positive feedback regulation of TGF-β/Smad signaling. Accordingly, we showed that Smyca knockdown in Hs578T or MDA-MB-231 cells decreased the amplitude and duration of TGF-β-induced gene expression (Fig. 4D and Additional file 1: Fig. S5B). Smyca knockdown also shortened the responsive period for TGF-β-induced Smad-responsive reporter activity (Fig. 4E). Conversely, Smyca overexpression in M10 cells increased the amplitude and duration of TGF-β-induced gene expression and reporter activity (Fig. 4F, G). Thus, Smyca governs a positive feedback regulation of TGF-β/Smad signaling to enhance and prolong the signal output.
Fig. 4.
Smyca governs a positive feedback regulation of TGF-β signaling. (A) qRT-PCR analysis of Smyca expression in indicated cell lines treated with 5 ng/ml TGF-β for 6 h. Data are normalized with that of untreated group. (B) Upper panel: Smyca gene architecture. The transcription starting site (+ 1), regions corresponding to miR-23a ~ 27a ~ 24–2 clusters (blue), Smyca (orange) and locations of the two SBEs (red) and two sets of PCR primers covering the SBEs are indicated. The corresponding chromosome location of Smyca gene (green) and the DNase I hypersensitive clusters (gray and black) are shown on the top. Bottom panel: ChIRP assay for Smyca occupancy on its own promoter. Tilling biotinylated oligonucleotides complementary to LacZ or Smyca were used to pull down the RNA-associated chromatins from MDA-MB-231 cells treated with 5 ng/ml TGF-β for 2 h, followed by qRT-PCR analysis. Data are normalized with the inputs. The locations of two sets of PCR primers covering the two SBE regions of Smyca promoter are shown on the top. (C) ChIP analysis of the recruitment of Smad3 and Smad4 to the indicated SBEs in MDA-MB-231 cells stably expressing control or Smyca shRNAs and treated with 5 ng/ml TGF-β for 2 h. (D, F) qRT-PCR analysis of indicated genes in Hs578T cells stably expressing Smyca shRNAs (D) or M10 cells stably expressing Smyca (F) and treated with or without 5 ng/ml TGF-β for indicated time points. Data are normalized with that of untreated group (0 h). (E, G) Luciferase reporter assay on MDA-MB-231 cells stably expressing Smyca shRNAs (E) or M10 cells stably expressing Smyca (G) and treated with or without 5 ng/ml TGF-β for indicated time points. Data are normalized with that of untreated group (0 h). Data in all panels are mean ± SD, n = 3. P values are determined by unpaired t test (A, B, F, G) or one-way ANOVA with Tukey’s post hoc test (C, D, E), *P < 0.05, **P < 0.01, ***P < 0.001
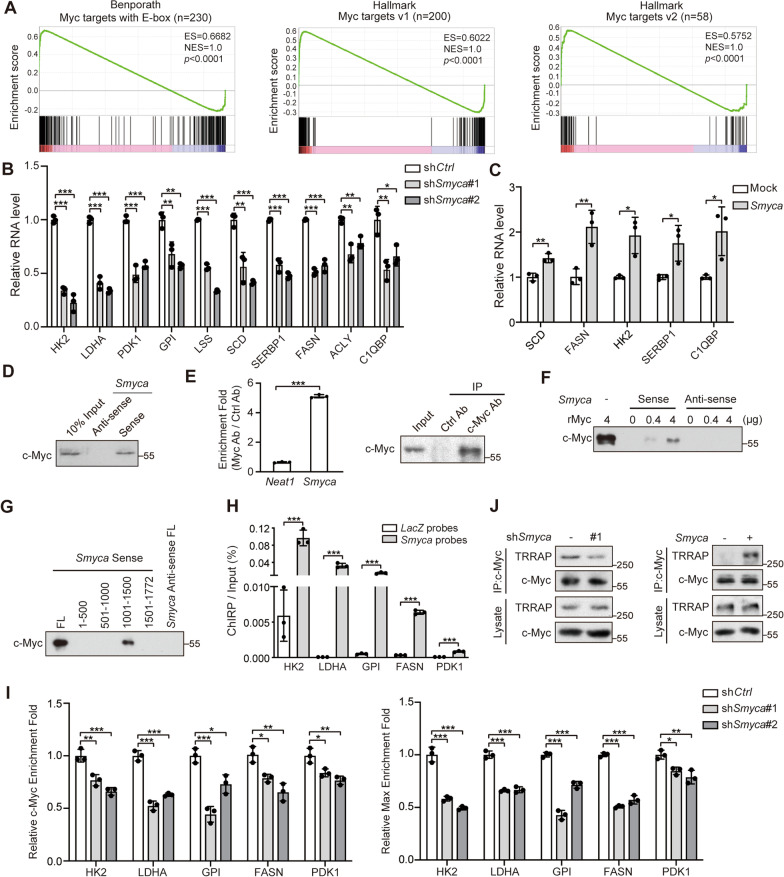
Smyca activates c-Myc signaling by promoting chromatin recruitment of c-Myc/Max complex and c-Myc/TRRAP interaction
Besides TGF-β signaling, GSEA Hallmark Pathway analysis revealed that Smyca-regulated gene signature correlated significantly with Myc target signatures (Fig. 2D, blue). Furthermore, Smyca-regulated genes are enriched with genes in glycolysis and fatty acid metabolism (Fig. 2D, green), many of which are c-Myc targets [45]. In addition, Smyca-regulated gene signature correlated with the signature of E2F targets (Fig. 2D, green), which act downstream of c-Myc [46], as well as unfolded protein response (Fig. 2D, green), which is often induced by an unconstraint Myc activity in cancer cells [47]. Accordingly, GSEA analysis showed significant correlations of Smyca-induced gene signature with c-Myc target signature from multiple sources (Fig. 5A). These findings prompted us to investigate the impact of Smyca on c-Myc signaling. Importantly, Smyca knockdown in MDA-MB-231 or Malaru cells downregulated the expression of a set of c-Myc target genes (Fig. 5B and Additional file 1: Fig. S6A), most of which involve in cancer metabolic reprograming. In contrast, Smyca overexpression in MCF7 cells showed an opposite effect (Fig. 5C). Using a c-Myc-responsive luciferase reporter, we showed that Smyca overexpression increased the reporter activity, whereas Smyca knockdown diminished this activity (Additional file 1: Fig. S6B, C). Furthermore, by retrieving RNA-seq data from TCGA data sets, we found that Smyca expression levels positively correlated with the expression of a set of c-Myc target genes in the tumor tissues of breast cancer and HCC patients (Additional file 1: Fig. S6D), thus supporting the clinical relevance of Smyca-activated c-Myc signaling to human cancers. Together, our study identified a role of Smyca in promoting c-Myc signaling.
Fig. 5.
Smyca promotes c-Myc/Max complex recruitment to its target promoters and c-Myc/TRRAP binding. (A) GSEA plots for the match of Smyca signature with Myc signature from indicated sources. (B, C) qRT-PCR analysis of indicated c-Myc targets in MDA-MB-231 cells stably expressing Smyca shRNAs (B) or MCF7 cells stably expressing Smyca (C). Data are normalized with that derived from control cells. (D, F, G) RNA pull-down analysis by incubating MDA-MB-231 nuclear extracts (D, G) or indicated amounts of recombinant c-Myc protein (F) with biotinylated sense or antisense Smyca (D, F) or its deletion mutants (G). (E) RIP assay for the enrichment of Smyca in c-Myc immunoprecipitates derived from MDA-MB-231 cells. Data are normalized with that derived from Neat1. The presence of c-Myc in the immunoprecipitates is shown on the right panel. (H) ChIRP assay for detecting Smyca occupancy on indicated c-Myc target loci. Tilling biotinylated oligonucleotides complementary to LacZ or Smyca were used to pull down the RNA-associated chromatins from MDA-MB-231 cells, followed by qRT-PCR analysis. Data are normalized with that of inputs. (I) ChIP analysis of the recruitment of c-Myc or Max to indicated promoters in MDA-MB-231 cells stably expressing Smyca shRNAs. The enrichment folds are normalized with that from control cells. Data in (B), (C), (E), (H) and (I) are mean ± SD, n = 3. P values are determined by one-way ANOVA with Tukey’s post hoc test (B, I) or unpaired t test (C, E, H), *P < 0.05, **P < 0.01, ***P < 0.001. (J) Immunoprecipitation analysis of c-Myc binding to TRRAP in MDA-MB-231 cells stably expressing Smyca shRNA or Smyca
Next, we determined the mechanism underlying Smyca-promoted c-Myc signaling. RNA pull-down and RIP assays demonstrated a specific interaction of Smyca with c-Myc (Fig. 5D, E). Furthermore, purified recombinant c-Myc (rMyc) was readily pulled down by in vitro transcribed Smyca (Fig. 5F), indicating their direct interaction. We further showed that deletion of the transcriptional activating domain (TAD), but not DNA-binding domain, of c-Myc greatly reduced its binding to Smyca, whereas TAD alone was sufficient in binding Smyca (Additional file 1: Fig. S6E). Reciprocally, Smyca (1001-1500) region was responsible for binding c-Myc (Fig. 5G). Furthermore, deletion of this region abrogated the ability of Smyca to upregulate c-Myc-responsive reporter activity (Additional file 1: Fig. S6B). Thus, our study revealed a role of Smyca in binding c-Myc to enhance c-Myc-mediated gene expression. Notably, Smyca overexpression or knockdown affected neither the expression of c-Myc nor the ability of c-Myc to form a complex with Max (Additional file 1: Fig. S6F, G). Nevertheless, ChIRP assay demonstrated the occupancy of Smyca to a number of c-Myc target promoters (Fig. 5H). Furthermore, Smyca knockdown compromised the recruitment of both c-Myc and Max to these promoters (Fig. 5I). These findings suggest that Smyca guides c-Myc/Max complex to its target promoter. In addition to this effect, Smyca knockdown decreased c-Myc binding to TRRAP, which is required for c-Myc-mediated transcriptional activation by acting as a scaffold for bringing the STAGA and NuA4 histone acetyltransferase complexes [48]. In contrast, Smyca overexpression elevated c-Myc/TRRAP complex (Fig. 5J). Thus, our data identified dual roles of Smyca in activating c-Myc-mediated transcription, i.e., promoting the recruitment of c-Myc/Max complex to its target promoters and enhancing c-Myc binding to TRRAP.
c-Myc and Smad3/4 form separate complexes with Smyca and compete for binding Smyca
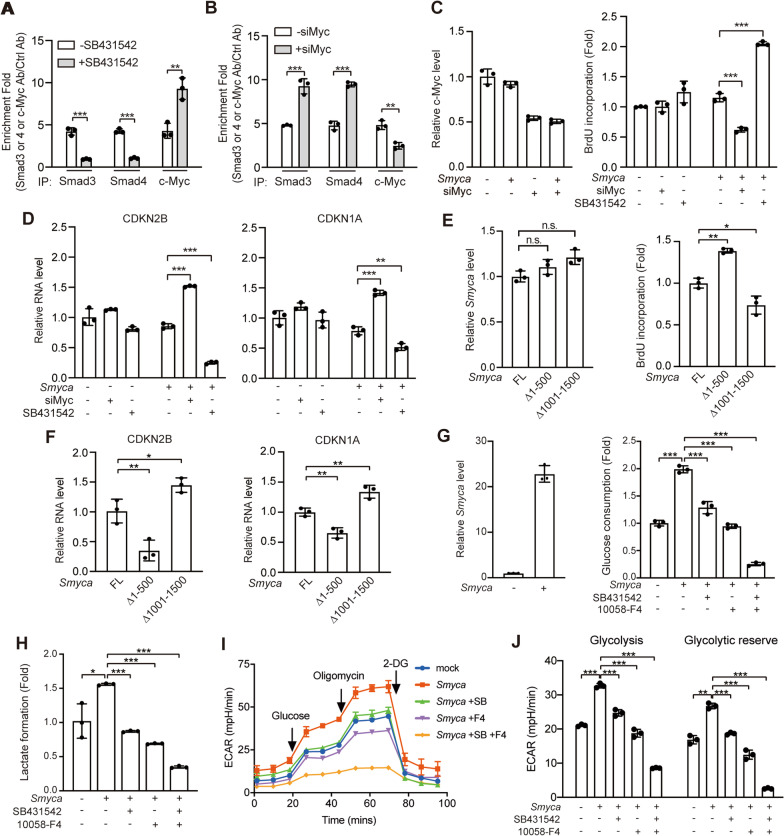
A previous study reported the ability of c-Myc to form a complex with Smad3 [19]. Since Smyca uses different regions for binding Smad3/Smad4 and c-Myc, we determined whether Smyca could function as a bridge to enhance the interaction of c-Myc with Smad3. However, Smyca overexpression did not affect the interaction of c-Myc with Smad3 in TGF-β-stimulated cells (Additional file 1: Fig. S7A). Furthermore, while expression of Smyca (1-500) or Smyca (1001-1500) segment alone, i.e., the Smad-binding or c-Myc-binding region, induced the activities of Smad- or c-Myc-responsive reporters, respectively, combined expression of these two segments showed no additive effect (Additional file 1: Fig. S7B, C). This finding suggests that the two binding regions function independently. To further determine the role of Smyca in the interplay between Smad and c-Myc pathways, we investigated whether Smad3/4 and c-Myc compete for binding Smyca. Importantly, administration of TβRI inhibitor SB431542, which blocks Smad3 phosphorylation and Smad3/4 nuclear entry, increased Smyca binding to c-Myc, whereas c-Myc depletion by siRNA enhanced Smyca binding to Smad3/4 (Fig. 6A, B). Together, these findings suggest that Smad3/4 and c-Myc form separate complexes with Smyca to independently activate Smad and c-Myc signaling, respectively. However, Smad3/4 and c-Myc can compete for binding Smyca and thus blocking one axis shifts the balance to promote the other axis.
Fig. 6.
Smyca coordinates with TGF-β and c-Myc pathways for stimulating glycolysis and preventing growth inhibition. (A, B) RIP assays for the enrichment of Smyca in Smad3, Smad4 or c-Myc immunoprecipitates derived from MCF7 cells stably expressing Smyca and treated with or without 5 µM SB431542 for 24 h (A), or MCF7 cells stably expressing Smyca and transfected with c-Myc siRNA (B). (C, D) Cell proliferation (C) and qRT-PCR analysis (D) of MCF7 cells stably expressing Smyca, transfected with c-Myc siRNA, and/or treated with 5 µM SB431542 for 24 h. Validation of c-Myc knockdown efficiency is shown on the left panel in (C). (E, F) Cell proliferation (E) and qRT-PCR analysis (F) of MCF7 cells transfected with indicated Smyca constructs. The expression levels of Smyca and mutants are shown on the left panel in (E). (G, H) Glucose consumption (G) and lactate formation (H) in BT-549 cells transfected with Smyca and treated with 10 µM SB431542 and/or 150 µM 10058-F4 for 24 h (G) or 48 h (H). Validation of Smyca overexpression is shown on the left panel in (G). (I, J) Glycolysis stress profile (I) and glycolysis and glycolytic reserve rates (J) were measured using MDA-MB-231 cells transfected and treated as in (G). Data in all panels are mean ± SD, n = 3. P values are determined by unpaired t test (A, B), one-way (C, D, E, F) or two-way (G, H, J) ANOVA with Tukey’s post hoc test, *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant
Smyca coordinates c-Myc and TGF-β pathways to regulate tumor proliferation and glycolysis
The identification of Smyca as a coactivator for Smad- and c-Myc-mediated transcription is of a particular importance in cancer biology. Since the two pathways control cell proliferation in opposite directions, we propose that the Smyca/c-Myc axis would induce an effect to neutralize the growth inhibitory function of Smyca/Smad axis. Consistent with this hypothesis, while stable overexpression of Smyca in MCF7 cells only modestly affected proliferation, suppression of c-Myc expression by an siRNA or c-Myc activity by a chemical inhibitor (10058-F4) in Smyca-overexpressing MCF7 cells greatly decreased cell proliferation (Fig. 6C and Additional file 1: Fig. S7D). Conversely, blockage of TGF-β pathway by SB431542 stimulated proliferation of Smyca-overexpressing cells (Fig. 6C). Previous studies indicated that Smad and c-Myc are both recruited to the promoters of CDK inhibitors CDKN2B and CDKN1A. While Smad activates their transcription, c-Myc elicits a repressive effect [19–21, 49, 50]. Consistently, ChIP assay revealed that Smyca enhanced the recruitment of Smad3, Smad4 and c-Myc to the promoters of CDKN2B and CDKN1A and these effects were compromised by blocking TGF-β or c-Myc signaling with SB431542 or 10058-F4, respectively (Additional file 1: Fig. S7E, F). Furthermore, inhibiting or silencing c-Myc in Smyca-overexpressing MCF7 cells elevated the expression of these CDK inhibitors, whereas blocking TGF-β pathway showed opposite effects (Fig. 6D and Additional file 1: Fig. S7G). Besides breast cancer cells, blockage of c-Myc and TGF-β pathways in Smyca-overexpressing HCC cell line NTU-BL also showed contrasting effects on cell proliferation and the expression of cell cycle regulators CDKN1A and CCNA1 (also known as cyclin A1) (Additional file 1: Fig. S7H, I). In line with the opposite roles of Smyca/Smad and Smyca/c-Myc axes in proliferation, the Smad-binding defective mutant Smyca (△1-500) induced a higher proliferation and lower expression of CDKN2B and CDKN1A, compared with full-length Smyca, whereas the c-Myc-binding defective mutant Smyca (△1001-1500) elicited opposite effects (Fig. 6E, F). Together, our data revealed that Smyca activates c-Myc signaling to avoid the growth inhibitory effect of Smyca/Smad axis.
Besides proliferation, c-Myc plays a key role in rewiring tumor cell metabolism and Warburg effect is among the best characterized metabolic alterations induced by c-Myc [45]. Notably, evidence has emerged that TGF-β pathway also promotes glycolysis [51]. Thus, we tested whether Smyca could act through TGF-β and c-Myc pathways to synergize glycolysis. Smyca overexpression in BT-549 cells greatly potentiated glucose consumption and lactate formation, the initial step and the final product of glycolysis, respectively, and these effects were attenuated by inhibiting either TGF-β or c-Myc pathway. Combined inhibition of both pathways further decreased glucose consumption and lactate formation (Fig. 6G, H). Furthermore, Seahorse analysis revealed a significant induction of ECARs (including glycolysis and glycolytic reserve) by Smyca overexpression. Again, blockage of TGF-β or c-Myc pathway alone diminished such effect, whereas blockage of both pathways showed a synergistic reduction effect (Fig. 6I, J). Thus, Smyca coactivates c-Myc and TGF-β pathways to synergize glycolysis.
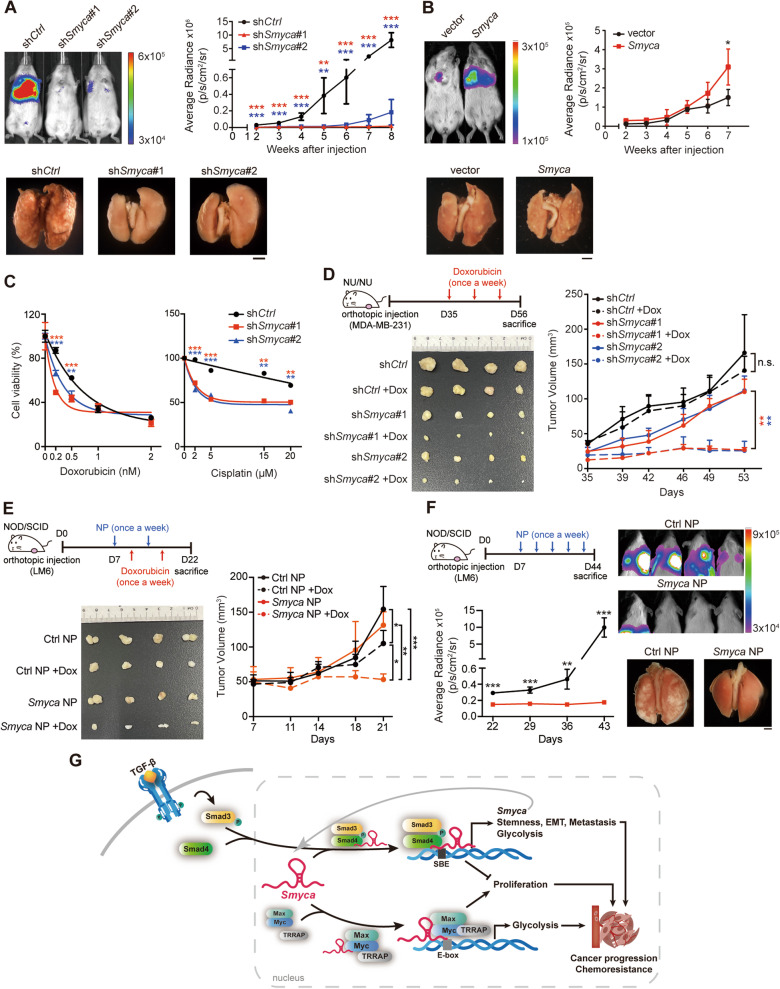
Targeting Smyca overcomes metastasis and chemoresistance
Our findings for the function of Smyca as a coactivator of TGF-β/Smad and c-Myc pathways strongly suggest its potential as a target for cancer therapy. Metastasis and drug resistance are the two leading causes of cancer mortality and both are associated with TGF-β-induced EMT [1, 2]. In addition, Warburg effect-induced acidification of tumor microenvironment potentiates metastasis [52]. We therefore tested the effects of Smyca on metastasis and chemoresistance. Using an experimental metastasis model by injecting tumor cells into the circulation of NOD/SCID mice, we found that Smyca knockdown in MDA-MB-231 cells greatly impaired lung metastasis (Fig. 7A). Conversely, Smyca overexpression in MDA-MB-231 cells enhanced metastasis (Fig. 7B). Next, we tested the effect of Smyca on chemoresistance. Using MTT assay, we showed that Smyca knockdown enhanced the sensitivity of MDA-MB-231 cells to Dox and cisplatin (Fig. 7C), two commonly used chemotherapeutic drugs in treating triple-negative breast cancer (TNBC) [53]. Similarly, in a breast cancer orthotopic model, Dox treatment almost completely blocked the growth of tumors derived from Smyca knockdown cells. However, tumor formed by parental cells showed little response to Dox (Fig. 7D). These findings collectively demonstrated the promoting effects of Smyca on metastasis and chemoresistance.
Fig. 7.
Smyca serves as a therapeutic target to prevent metastasis and chemoresistance. (A, B) Experimental metastasis assay for MDA-MB-231 cells stably expressing Smyca shRNAs (A) or Smyca (B). Representative images of the bioluminescence analysis at 8 weeks (A) or 7 weeks (B) after injection and the kinetics of metastasis at indicated time points are shown on the top left and right panels, respectively. The lung images at 8 weeks (A) and 7 weeks (B) after injection are shown on the bottom. Bar, 2 mm. (C) Cell viability assay of MDA-MB-231 cells stably expressing Smyca shRNAs and treated with Dox or cisplatin at indicated doses. (D) Nude mice orthotopically implanted with MDA-MB-231 cells stably expressing control or Smyca shRNAs and treated with Dox or DMSO as indicated (top left panel). Tumor volumes were measured at indicated days and plotted on the right. Tumors were surgically removed at the killing day and their sizes are shown on the bottom left panel. (E) NOD/SCID mice orthotopically implanted with LM6 cells were injected with NPs and/or Dox as indicated (top left panel). Tumor volumes at indicated days are shown on the right. Tumors were surgically removed at the killing day and their sizes are shown on the bottom left panel. (F) NOD/SCID mice orthotopically implanted with LM6 cells were injected with NPs as indicated (top left panel). Representative images of the bioluminescence analysis at 6 weeks after transplantation and the kinetics of metastasis at indicated time points are shown on the top right and bottom left panels, respectively. The lung images at 6 weeks after transplantation are shown on the bottom right. Bar, 2 mm. Data in (A), (B), (C), (D), (E), and (F) are mean ± SD, n = 5 (A), n = 4 (B, D, E, F), or n = 3 (C). P values are determined by one-way (A, C) or two-way (D, E) ANOVA with Tukey’s post hoc test or unpaired t test (B, F), *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant. (G) Schematic presentation of the roles of Smyca in coactivating TGF-β/Smad and Myc pathways to promote tumor progression
Next, we thought to develop a clinically applicable approach for targeting Smyca. To this end, we used Smyca-specific gapmer antisense oligonucleotides (ASO), which efficiently downregulated Smyca expression upon transfecting into MDA-MB-231 cells (Additional file 1: Fig. S8A). The Smyca gapmer ASO and control gapmer were loaded into LPD-NPs [40]. As to the TNBC model, we used LM6, a highly metastatic variant of MDA-MB-231 cells [32]. Incubation of LM6 cells with LPD-NPs carrying Smyca gapmer ASO dose-dependently reduced Smyca expression, compared with cells receiving control gapmer LPD-NPs (Additional file 1: Fig. S8B). Using an orthotopic model, we found that injection of Smyca gapmer ASO-loaded LPD-NPs into the NOD/SCID mice bearing LM6-derived tumors greatly enhanced the anti-tumor effect of Dox (Fig. 7E). Administration of Smyca gapmer ASO LPD-NPs in mice carrying orthotopically implanted LM6 cells reduced the size and weight of primary tumors and prevented lung metastasis (Fig. 7F and Additional file 1: Fig. S8C). qRT-PCR analysis of the primary tumors found that Smyca gapmer ASO LPD-NPs downregulated the expression of not only Smyca, but also a set of tumor-promoting TGF-β/Smad and c-Myc targets in vivo (Additional file 1: Fig. S8D). These findings highlight a potential of Smyca as an anticancer target and Smyca gapmer ASO as a promising agent for combating cancer metastasis and chemoresistance.
Discussion
This study identifies Smyca as a lncRNA that coactivates two prominent pathways in controlling tumor malignancies, i.e., the TGF-β/Smad and c-Myc pathways, thereby driving tumor progression and therapy resistance. Mechanistically, the nuclear-residing Smyca functions as a scaffold to enhance the formation of Smad3/4 complex and its recruitment to a set of target promoters. Furthermore, Smyca binds c-Myc to promote the association of c-Myc with TRRAP and the recruitment of c-Myc/Max complex to some of target promoters. Through these mechanisms, Smyca not only potentiates multiple cancer hallmarks induced by the two pathways, including metabolic reprogramming, migration, invasion, cancer stemness, metastasis and chemoresistance, but also evades TGF-β-induced growth inhibition by stimulating the c-Myc proliferating signal (Fig. 7G). Consistent with these tumor-promoting effects, Smyca high expression correlates with poor prognosis and aggressiveness of multiple cancer types. Furthermore, targeting Smyca by the gapmer ASO approach efficiently blocks tumor metastasis and sensitizes tumor cells to chemotherapy. These findings highlight the prognostic and therapeutic values of Smyca in human cancers.
Smyca not only promotes TGF-β/Smad signaling but is itself a transcriptional target of Smad (Fig. 7G). This mutual reinforcement establishes a positive feedback loop to elevate the amplitude and duration of TGF-β signaling. Of note, a previous study identified the differential cellular responses to TGF-β treatment at early and late time points. At the late phase, R-Smad is redirected to the promoters of a set of invasion genes via a cooperation with JUNB [43], thus suggesting a role of sustained TGF-β signaling in favoring a tumor-promoting outcome. Similarly, HCC patients with a late TGF-β signature are associated with invasive phenotype, metastasis and poor prognosis [54]. Consistent with these previous studies, Smyca enhances TGF-β-induced expression of many invasion genes and Smyca expression correlates positively with the expression of a large set of invasion/metastasis genes in cancer patients. Thus, the ability of Smyca to govern a positive feedback regulation of TGF-β signaling may contribute in part to its ability to switch the dichotomous functions of TGF-β toward tumor promotion.
Intriguingly, Smyca binds Smad3 but not Smad2, which is in analogous to another lncRNA, ELIT-1 [55]. Notably, a number of studies have reported the distinct roles of these two Smads in cancer progression. While Smad3 elicits pro-tumor functions by enhancing EMT, invasion and angiogenesis, Smad2 displays opposite effects in many scenarios to suppress tumor progression [8, 56–58]. These differential roles of the two Smads may be attributed to the differences in the affinity to promoters of target genes or in the recruitment of partners or regulators [58, 59]. Our finding that Smyca binds Smad3 but not Smad2 to elevate the expression of pro-tumor effectors of Smad not only is consistent with these previous studies but also suggests Smyca as one factor that contributes to the differential effects of Smad2 and Smad3 on tumor progression. Through the ability to govern a positive feedback regulation of TGF-β signaling to prolong the signal duration, Smyca would endow its binding partner Smad3 with a preference to turn on the expression of a set of invasion genes. In this way, Smad3 could outcompete Smad2 for preferentially gaining a tumor-promoting function.
Consistent with its distribution to the chromatins [28], Smyca not only is recruited to the promoters of a set of Smad3/4 and c-Myc targets, but enhances the recruitment of Smad3/4 and c-Myc/Max complexes to these loci. Since Smyca also potentiates Smad3/4 complex formation, we cannot exclude the possibility that the latter is merely a consequence of the former. However, due to the lack of effect of Smyca on c-Myc/Max association, our findings do support a role of Smyca in guiding the c-Myc/Max complex to some of their target promoters. It remains unclear whether Smyca is recruited to c-Myc and Smad3/4 target loci directly or indirectly via other factors, such as modified histones or chromatin remodeling factors. Intriguingly, Smyca also enhances c-Myc binding to TRRAP, which recruits histone acetyltransferase complexes to promote H3/4 acetylation and transcription [48]. Future studies are needed to investigate the order and interdependency for the loading of Smyca, c-Myc/Max and TRRAP to c-Myc target promoters as well as the choreography of the Smyca/c-Myc/TRRAP complex.
Smyca binds Smad3/4 and c-Myc via non-overlapping regions, and each region is sufficient and necessary for activating Smad- or c-Myc-mediated transcription. Furthermore, the two regions act independently and no additive effect is observed. Accordingly, Smyca does not promote Smad3/4 interaction with c-Myc, suggesting that Smyca forms separate complexes with Smad3/4 and c-Myc. Nevertheless, depleting c-Myc results in an elevation of Smyca/Smad3/4 complex, while depleting nuclear Smad3/4 by TβRI inhibitor increases Smyca/c-Myc complex. These findings reveal the existence of a competition between Smad3/4 and c-Myc for binding Smyca. Such competition might be resulted from the following scenarios. First, Smyca may be a limiting factor in cells in relation to Smad3/4 and c-Myc. Therefore, when Smyca binds to one partner and is recruited to the target promoters of this partner, it would limit the chance of Smyca to be recruited to the target promoters of another partner. Additionally, although Smad3/4 and c-Myc bind to separate regions in Smyca, the binding of one partner may recruit additional factors and/or alter the conformation of Smyca, thereby masking or eliminating the binding site of another partner. Regardless of the underlying mechanisms, this competitive binding is consistent with our finding that blockage of one pathway readily shifts the effects of Smyca on cell proliferation and the expression of cell cycle regulators. Thus, Smyca forms separate complexes with Smad3/4 and c-Myc and their competitive binding could determine the effect of Smyca on cell proliferation.
It is unclear whether Smyca acts on all target loci of Smad3/Smad4 and c-Myc/Max complexes or particular subsets of them. Intersection of Smyca-induced DEGs with published Smad2/3 ChIP-seq data revealed partial overlaps, suggesting the latter possibility. However, one caveat is that the DEGs and ChIP-seq data were derived from different cell types and treatment conditions. It would require further omics studies, such as the comparative ChIRP-seq and ChIP-seq analyses, conducted on matched cellular contexts to address this important issue. Lastly, although p-Smad3 readily binds Smad4 via their MH2 domains, Smyca interacts with the MH1 domains of the two Smads to further enhance their interaction and thus functions as a RNA scaffold. Even though the Smad3- and Smad4-binding regions are both located to the Smyca (1–500) segment, we postulate that non-overlapping regions in this segment are responsible for binding Smad3 and Smad4, thereby fulfilling a scaffold function. Notably, the ability of Smyca to enhance the association of a protein complex by providing an additional binding surface is in analogous to another lncRNA GUARDIN, which binds both BRCA1 and BARD1 to potentiate their association [60].
LncRNAs have emerged as a new class of biomarkers for cancer diagnosis and prognosis [23]. We uncover a remarkable value of Smyca for the prognosis of a number of cancer types. Notably, a recent study reported an upregulation of Smyca level in the patient plasma of several cancer types [31], implying the utility of Smyca as a noninvasive diagnostic biomarker. Besides diagnosis and prognosis, lncRNAs are also great targets of cancer therapy, because drugs targeting RNAs are easier design and synthesis than drugs targeting proteins. The ability of Smyca to activate two cancer-relevant pathways for potentiating multiple cancer hallmarks highlights its potential as a target for cancer therapy. In support of this notion, we show that NP-mediated delivery of Smyca gapmer ASO to the tumor-bearing mice significantly blocks tumor metastasis and overcomes chemoresistance, two major causes of cancer mortality.
Conclusions
In conclusion, our study identifies Smyca as a lncRNA that coactivates TGF-β/Smad and c-Myc pathway to potentiate tumor progression, metastasis and chemoresistance. Furthermore, our findings reveal a great promise for the clinical applications of Smyca as a prognostic biomarker and a therapeutic target for certain cancer types.
Supplementary Information
Additional file 1. Figure S1. Smyca expression correlates with poor prognosis. (A-F) Kaplan-Meier analysis of Smyca expression in relation to the overall survival (A, C, E, F) and disease-free survival (B, D) of indicated cancer types. Data were retrieved from TCGA data sets (A, B) or GEO data sets (C, D, E), or generated from an in house cohort (F). Patients were grouped into high and low expression based on the median expression level. Patients without survival information were omitted. Hazard ratio (HR) and P values are determined by log-rank test. (G) Smyca expression in HCC tumor tissues and adjacent normal tissues analyzed by qRT-PCR. (H–K) The correlation of Smyca expression with the stage (H, J) and invasiveness (I, K) of indicated cancer types. Data in (G-K) are presented by Whiskers boxplot, whiskers: min to max, bound of box: lower and upper quartiles, center line: median. Data points derived from basal-like subgroup of patients are marked in blue in (K). P values in (G), (H), (I), (J), (K) are determined by unpaired t-test. (L) Smyca expression in different subtypes of breast cancer patients analyzed by qRT-PCR. Data are expressed as mean ± SD. P values are determined by oneway ANOVA with Tukey’s post hoc test. Figure S2. Smyca promotes EMT. (A) Smyca expression in different breast cancer and HCC cell lines analyzed by qRT-PCR and represented as copy numbers. Data are expressed as mean ± SD from three independent experiments. (B) qRT-PCR analysis of indicated miRNAs in MDA-MB-231 cells stably expressing various Smyca shRNAs. Data are normalized with that of control cells and are expressed as mean ± SD from three independent experiments, ns, not significant by one-way ANOVA with Tukey’s post hoc test. (C, D) Western blot analysis of EMT markers in Hs578T cells stably expressing Smyca shRNAs (C) or MCF7 cells stably overexpressing Smyca (D). The amounts of each protein in relation to the control cells are indicated under the bands. Smyca expression levels in these stable lines are shown on the left panels. Data are mean ± SD from three independent experiments. (E-H) Cell proliferation (E, F) and cell viability (G, H) assays of M10 cells stably expressing Smyca (E, G) or MDA-MB-231 cells stably expressing Smyca shRNAs (F, H). Data are mean ± SD, n=3. P values are determined by unpaired t-test (E, G) or one-way ANOVA with Tukey’s post hoc test (F, H), ns, not significant. Figure S3. Bioinformatics and cell-based analysis for the relation of Smyca to TGF-β signaling. (A) Summary of the GSEA analysis for the match of Smyca signature with the indicated signatures. Data origin indicates the source database or the first author identifying the gene signature. (B) Intersection of Smyca-induced DEGs derived from MDA-MB-231 cells with Smad2/3 ChIP-seq data derived from Hs578T cells and BT-549 cells. The full list of overlapped genes is shown in Table S5. (C) qRT-PCR analysis for the expression of indicated TGF-β target genes in Hs578T cells stably expressing Smyca shRNAs and treated with or without 5 ng/ml TGF-β for 48 hr. Data are normalized with that of untreated group in each cell. (D, E) Luciferase assay for the Smad-target reporters transfected into Hs578T cells (D) or Malaru cells (E) stably expressing Smyca shRNAs and treated with or without 5 ng/ml TGF-β for 24 hr. The Smyca knockdown efficiencies are shown on the left panel in (E). Data in (C), (D), (D) are normalized with that of untreated control and presented as mean ± SD, n=3. P values are determined by one-way ANOVA with Tukey’s post hoc test, **P < 0.01, ***P < 0.001. (F, G) Summary of the correlations of Smyca expression with the expression of indicated TGF-β target genes by analyzing HCC or breast cancer data sets from TCGA (F) or GEO (G) databases. Pearson’s coefficients and P values are indicated. (H) GO analysis using the set of TGF-β target genes with expression levels showing positive correlations with Smyca expression in HCC and/or breast cancers. Figure S4. Smyca binds MH1 domains of Smad3 and Smad4 without affecting their expression and Smad3 phosphorylation. (A) Western blot analysis of Smad3 and Smad4 expression in MDA-MB-231 cells stably expressing Smyca shRNAs. (B) Western blot analysis for Smad3 phosphorylation in MDA-MB-231 cells stably expressing Smyca shRNAs and treated with or without 5 ng/ml TGF-β for 30 min. The blots are representative of three independent experiments and quantitative data are shown on the right. (C) Immunoprecipitation analysis of Smad3 and Smad4 interaction in MDA-MB-231 cells stably expressing indicated Smyca constructs and treated with or without 5 ng/ml TGF-β for 1 hr. (D) Luciferase assay for the Smad-responsive reporter in MDA-MB-231 cells transfected with indicated Smyca constructs and treated with or without 5 ng/ml TGF-β for 24 hr. The expression levels of Smyca are shown on the left panel. (E) Baculovirally purified Smad3, Smad4, and their MH1 deletion mutants bound on beads were incubated with biotinylated sense or antisense Smyca. The bound Smyca was analyzed by qRT-PCR. The equal inputs of recombinant proteins are shown on or antisense Smyca. The bound Smyca was analyzed by qRT-PCR. The equal inputs of recombinant proteins are shown on the right. Data in (B), (D) and (E) are mean ± SD, n=3. Pvalues are determined by one-way ANOVA with Tukey’s post hoc test (B, D) or unpaired t-test (E), *** P < 0.001; ns., not significant. Figure S5. Smyca is a Smad target and mediates a positive feedback control of TGF-β signaling. (A) qRT-PCR analysis of Smyca expression in MDA-MB-231 cells stably expressing Smad3 or Smad4 shRNAs and treated with or without 5 ng/ml TGF-β for 24 hr. The knockdown efficiencies of Smad3 and Smad4 shRNAs are shown on the left and middle panels, respectively. (B) qRT-PCR analysis of TGF-β target gene expression in MDA-MB-231 cells stably expressing Smyca shRNAs and treated with or without 5 ng/ml TGF-β for indicated time points. Data are normalized with that of untreated group (0 h). Data in (A) and (B) are mean ± SD, n=3. P values are determined by unpaired t-test (A) or one-way ANOVA with Tukey’s post hoc test (B), *** P < 0.001. Figure S6. Smyca promotes c-Myc transcription activity without affecting its expression or interaction with Max. (A) qRT-PCR analysis of the expression of indicated c-Myc target genes in Malaru cells stably expressing control or Smyca shRNA. Data are normalized with that of control cells. (B, C) Luciferase assay for a c-Myc-reponsive reporter transfected into MDA-MB-231 cells stably expressing Smyca or mutant (B) or Smyca shRNAs (C). The expression levels of Smyca and its mutant are shown on the left panel in (B). (D) Summary of the correlations of Smyca expression with the expression of indicated c-Myc target genes by analyzing breast cancer and HCC data sets from TCGA. Pearson’s coefficients and P values are indicated. (E) Maping the Smyca-binding region in c-Myc. Top: Schematic presentaton of c-Myc domains. The various GFP-c-Myc truncated proteins were purified from transfected 293T cells and incubated with biotinylated sense or antisense Smyca. The bound Smyca was analyzed by qRT-PCR. Data are normalized with that from GFP only control. The input levels of various GFP fusion proteins are shown on the right and marked by arrows. (F) Western blot analysis of c-Myc expression in indicated cells stably expressing Smyca or Smyca shRNAs. (G) Immunoprecipitation analysis of c-Myc/Max complex formation in MDA-MB-231 cells stably expressing Smyca or Smyca shRNA. Data in (A), (B), (C), (E) are mean ± SD, n=3. P values are determined by unpaired t-test (A), or one-way ANOVA with Tukey’s post hoc test (B, C, E), *P < 0.05, **P < 0.01, *** P < 0.001. Figure S7. Smyca-promoted c-Myc singaling neutralizes the growth inhibitory effect of Smyca-promoted TGF-β signaling. (A) Immunoprecipitation analysis of c-Myc/Smad complex formation in MDA-MB-231 cells stably expressing Smyca and treated with 5 ng/ml TGF-β for 2 hr. (B, C) Luciferase assay for a Smad-responsive reporter (B) or c-Myc-responsive reporter (C) transfected into MDA-MB-231 cells together with indicated Smyca constructs and treated with or without 5 ng/ml TGF-β for 24 hr. The expression levels of Smyca (1-500) and (1001-1500) fragments are shown on the left and middle panels in (B), respectively. (D, H) Cell proliferation assay of MCF7 cells stably expressing Smyca and treated with 75 μM 10058-F4 for 24 hr (D) or NTU-BL cells stably expressing Smyca and treated with 5 μM SB431542 or 75 μM 10058-F4 for 24 hr (H). (E, F) ChIP analysis for Smad3, Smad4, and c-Myc binding to the promoter regions of CDKN2B (E) and CDKN1A (F) genes in MDA-MB-231 cells stably expressing Smyca and treated with 5 μM SB431542 and/or 150 μM 10058-F4 for 2 hr. (G, I) qRTPCR analysis of the expression of indicated genes in MCF7 (G) or NTU-BL (I) cells stably expressing Smyca and treated with 5 μM SB431542 or 75 μM 10058-F4 for 24 hr. Smyca expression levels are shown on the left panel of (I). Data in (B), (C), (D), (E), (F), (G), (H), (I) are mean ± SD, n=3. P values are determined by one-way ANOVA with Tukey’s post hoc test, *P < 0.05, **P < 0.01, ***P < 0.001. Figure S8. Downregulation of the expression of Smyca, Smad targets, and c-Myc targets by Smyca gapmer ASO. (A, B, D) qRT-PCR analysis of Smyca or indicated mRNAs in MDA-MB-231 cells transfected with Smyca gapmer ASO (A), LM6 cells treated with indicated doses of NPs carrying Smyca gapmer ASO or control gapmer (B), or LM6 tumor-bearing mice treated with NPs carrying Smyca gapmer ASO or control NPs (D). (C) The morphology, size and weight of primary tumors taken from the sacrifice day for experiment shown in Fig. 7F. Data in all panels are mean ± SD, n=3 (A, B, D) or 4 (C). P values are determined by unpaired t-test, *P < 0.05, **P < 0.01, ***P < 0.001. Table S1. Antibody details. Table S2. siRNA, shRNA and Gapmer sequences. Table S3. Sequences of PCR primers. Table S4. ChIRP probe sequences. Table S5. List of genes that are regulated by Smyca and bound by Smad2/3.
Acknowledgements
We thank Rik Derynck and Xin-Hua Feng for constructs, RNA Technology Platforms and Gene Manipulation Core Facility for shRNA constructs, TMU Core Facility for guiding the Seahorse analysis, Chin-Chun Hung for assisting confocal and flow cytometry analyses and Hsin-Jien Kung for valuable discussions.
Abbreviations
- ASO
Antisense oligonucleotides
- CCNA1
Cyclin A1
- ChIP
Chromatin immunoprecipitation
- ChIRP
Chromatin isolation by RNA purification
- DEGs
Differentially expressed genes
- DMEM
Dulbecco’s modified Eagle’s medium
- Dox
Doxorubicin
- ECAR
Extracellular acidification rate
- ELDA
Extreme limiting dilution assay
- EMT
Epithelial–mesenchymal transition
- FBS
Fetal bovine serum
- FDR
False discovery rate
- FN1
Fibronectin1
- GEO
Gene expression omnibus
- GSEA
Gene set enrichment analysis
- HCC
Hepatocellular carcinoma
- IPA
Ingenuity pathway analysis
- LncRNA
Long noncoding RNA
- LPD-NPs
Liposome–polycation–DNA nanoparticles
- MET
Mesenchymal-to-epithelial transition
- MTT
Methyl thiazolyl diphenyl tetrazolium bromide
- PMSF
Phenylmethylsulphonyl fluoride
- RIP
RNA immunoprecipitation
- rMyc
Recombinant c-Myc
- RNA-seq
RNA-sequencing
- qRT-PCR
Real-time quantitative PCR
- qPCR
Quantitative PCR
- SBE
Smad-binding elements
- Smyca
Smad/Myc coactivator
- TAD
Transcriptional activating domain
- TβRI
Type I TGF-β receptor
- TCGA
The cancer genome atlas
- TNBC
Triple-negative breast cancer
Author contributions
H.Y.C. and R.H.C. conceived this study. H.Y.C., S.J.C., X.L., A.C.W., R.I.J., C.C.L. and S.Y.C. performed the experiments and analyzed data. H.Y.C., J.H.S., C.L.L. and Y.T.K. performed bioinformatics analyses. K.W.H. prepared NP and Y.C. designed and supervised NP studies, and Y.D.L. P.W.H. and Y.S.J. contributed to reagents and materials. R.H.C. directed and coordinated study, designed the research and oversaw the project. H.Y.C., S.J.C., Y.C. and R.H.C. prepared the manuscript. All authors read and approved the final manuscript.
Funding
This work is supported by NHRI Innovative Research Grant NHRI-EX111-11127BI (to R.H.C.), an intramural fund from Institute of Biological Chemistry, Academia Sinica (to R.H.C.), Taipei Medical University Research Grant TMU104-AE1-B33 (to H.Y.C.) and Ministry of Science and Technology Grant 109-2320-B-038-019-MY3 (to H.Y.C.).
Availability of data and materials
All data needed to evaluate the conclusions in the paper are present in the paper and/or Additional file 1. The RNA-seq data were deposited to the GEO database with the accession number GSE181028. Other data and materials are available upon reasonable request.
Declarations
Ethic approval and consent to participate
The procedures related to human subjects were approved by the Institutional Review Boards of Taipei Medical University and Academia Sinica. Animal experiments were performed according to the protocols approved by the Experimental Animal Committees of Academia Sinica and Taipei Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Derynck R, Weinberg RA. EMT and cancer: more than meets the eye. Dev Cell. 2019;49:313–316. doi: 10.1016/j.devcel.2019.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell. 2019;49:361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao Y, Baker D, Ten Dijke P. TGF-beta-mediated epithelial-mesenchymal transition and cancer metastasis. Int J Mol Sci. 2019;20:2767. doi: 10.3390/ijms20112767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 6.Kang JS, Liu C, Derynck R. New regulatory mechanisms of TGF-beta receptor function. Trends Cell Biol. 2009;19:385–394. doi: 10.1016/j.tcb.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 8.Hoot KE, Lighthall J, Han G, Lu SL, Li A, Ju W, et al. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest. 2008;118:2722–2732. doi: 10.1172/JCI33713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang Y, Chen CR, Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003;11:915–926. doi: 10.1016/S1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 10.Morita T, Mayanagi T, Sobue K. Dual roles of myocardin-related transcription factors in epithelial mesenchymal transition via slug induction and actin remodeling. J Cell Biol. 2007;179:1027–1042. doi: 10.1083/jcb.200708174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell. 2007;18:3533–3544. doi: 10.1091/mbc.e07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaimori A, Potter J, Kaimori JY, Wang C, Mezey E, Koteish A. Transforming growth factor-beta1 induces an epithelial-to-mesenchymal transition state in mouse hepatocytes in vitro. J Biol Chem. 2007;282:22089–22101. doi: 10.1074/jbc.M700998200. [DOI] [PubMed] [Google Scholar]
- 13.Nawshad A, Medici D, Liu CC, Hay ED. TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex. J Cell Sci. 2007;120:1646–1653. doi: 10.1242/jcs.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol. 2009;11:943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Alexander PB, Wang XF. TGF-beta family signaling in the control of cell proliferation and survival. Cold Spring Harb Perspect Biol. 2017;9:a022145. doi: 10.1101/cshperspect.a022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David CJ, Massague J. Contextual determinants of TGFbeta action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19:419–435. doi: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta. 2007;1775:21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Feng XH. SMAD-oncoprotein interplay: potential determining factors in targeted therapies. Biochem Pharmacol. 2020;180:114155. doi: 10.1016/j.bcp.2020.114155. [DOI] [PubMed] [Google Scholar]
- 19.Feng XH, Liang YY, Liang M, Zhai W, Lin X. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B) Mol Cell. 2002;9:133–143. doi: 10.1016/S1097-2765(01)00430-0. [DOI] [PubMed] [Google Scholar]
- 20.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 21.Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 22.Teixeira AF, Ten Dijke P, Zhu HJ. On-target anti-TGF-beta therapies are not succeeding in clinical cancer treatments: what are remaining challenges? Front Cell Dev Biol. 2020;8:605. doi: 10.3389/fcell.2020.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 24.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iaccarino I. lncRNAs and MYC: an intricate relationship. Int J Mol Sci. 2017;18:1497. doi: 10.3390/ijms18071497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang JF, Yang L, Chen LL. The long noncoding RNA regulation at the MYC locus. Curr Opin Genet Dev. 2015;33:41–48. doi: 10.1016/j.gde.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Papoutsoglou P, Moustakas A. Long non-coding RNAs and TGF-beta signaling in cancer. Cancer Sci. 2020;111:2672–2681. doi: 10.1111/cas.14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das M, Renganathan A, Dighe SN, Bhaduri U, Shettar A, Mukherjee G, et al. DDX5/p68 associated lncRNA LOC284454 is differentially expressed in human cancers and modulates gene expression. RNA Biol. 2018;15:214–230. doi: 10.1080/15476286.2017.1397261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan C, Tang Y, Wang J, Wang Y, Xiong F, Zhang S, et al. Long non-coding RNA LOC284454 promotes migration and invasion of nasopharyngeal carcinoma via modulating the Rho/Rac signaling pathway. Carcinogenesis. 2019;40:380–391. doi: 10.1093/carcin/bgy143. [DOI] [PubMed] [Google Scholar]
- 30.Han L, Zhou W, Wu F. Long noncoding RNA LOC284454 promotes hepatocellular carcinoma cell invasion and migration by inhibiting Ecadherin expression. Oncol Rep. 2021 doi: 10.3892/or.2021.7956. [DOI] [PubMed] [Google Scholar]
- 31.Fan C, Wang J, Tang Y, Zhang S, Xiong F, Guo C, et al. Upregulation of long non-coding RNA LOC284454 may serve as a new serum diagnostic biomarker for head and neck cancers. BMC Cancer. 2020;20:917. doi: 10.1186/s12885-020-07408-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Apaya MK, Hsiao PW, Yang YC, Shyur LF. Deregulating the CYP2C19/epoxy-eicosatrienoic acid-associated FABP4/FABP5 signaling network as a therapeutic approach for metastatic triple-negative breast cancer. Cancers (Basel) 2020;12:199. doi: 10.3390/cancers12010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh HW, Hsu EC, Lee SS, Lang YD, Lin YC, Chang CY, et al. PSPC1 mediates TGF-beta1 autocrine signalling and Smad2/3 target switching to promote EMT, stemness and metastasis. Nat Cell Biol. 2018;20:479–491. doi: 10.1038/s41556-018-0062-y. [DOI] [PubMed] [Google Scholar]
- 34.Jang CW, Chen CH, Chen CC, Chen JY, Su YH, Chen RH. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nat Cell Biol. 2002;4:51–58. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- 35.Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 36.Chen YH, Huang TY, Lin YT, Lin SY, Li WH, Hsiao HJ, et al. VPS34 K29/K48 branched ubiquitination governed by UBE3C and TRABID regulates autophagy, proteostasis and liver metabolism. Nat Commun. 2021;12:1322. doi: 10.1038/s41467-021-21715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Sen J, Bathula SR, Yang Q, Fittipaldi R, Huang L. Novel cationic lipid that delivers siRNA and enhances therapeutic effect in lung cancer cells. Mol Pharm. 2009;6:696–705. doi: 10.1021/mp800136v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu JY, Chiang T, Liu CH, Chern GG, Lin TT, Gao DY, et al. Delivery of siRNA Using CXCR4-targeted nanoparticles modulates tumor microenvironment and achieves a potent antitumor response in liver cancer. Mol Ther. 2015;23:1772–1782. doi: 10.1038/mt.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricardo S, Vieira AF, Gerhard R, Leitao D, Pinto R, Cameselle-Teijeiro JF, et al. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J Clin Pathol. 2011;64:937–946. doi: 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- 43.Sundqvist A, Morikawa M, Ren J, Vasilaki E, Kawasaki N, Kobayashi M, et al. JUNB governs a feed-forward network of TGFbeta signaling that aggravates breast cancer invasion. Nucleic Acids Res. 2018;46:1180–1195. doi: 10.1093/nar/gkx1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chacko BM, Qin B, Correia JJ, Lam SS, de Caestecker MP, Lin K. The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat Struct Biol. 2001;8:248–253. doi: 10.1038/84995. [DOI] [PubMed] [Google Scholar]
- 45.Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beier R, Burgin A, Kiermaier A, Fero M, Karsunky H, Saffrich R, et al. Induction of cyclin E-cdk2 kinase activity, E2F-dependent transcription and cell growth by Myc are genetically separable events. EMBO J. 2000;19:5813–5823. doi: 10.1093/emboj/19.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolpaw AJ, Dang CV. MYC-induced metabolic stress and tumorigenesis. Biochim Biophys Acta Rev Cancer. 2018;1870:43–50. doi: 10.1016/j.bbcan.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 48.Lourenco C, Resetca D, Redel C, Lin P, MacDonald AS, Ciaccio R, et al. MYC protein interactors in gene transcription and cancer. Nat Rev Cancer. 2021;21:579–591. doi: 10.1038/s41568-021-00367-9. [DOI] [PubMed] [Google Scholar]
- 49.Feng XH, Lin X, Derynck R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J. 2000;19:5178–5193. doi: 10.1093/emboj/19.19.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/S0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 51.Hua W, Ten Dijke P, Kostidis S, Giera M, Hornsveld M. TGFbeta-induced metabolic reprogramming during epithelial-to-mesenchymal transition in cancer. Cell Mol Life Sci. 2020;77:2103–2123. doi: 10.1007/s00018-019-03398-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thews O, Riemann A. Tumor pH and metastasis: a malignant process beyond hypoxia. Cancer Metastasis Rev. 2019;38:113–129. doi: 10.1007/s10555-018-09777-y. [DOI] [PubMed] [Google Scholar]
- 53.Won KA, Spruck C. Triplenegative breast cancer therapy: current and future perspectives (Review) Int J Oncol. 2020;57:1245–1261. doi: 10.3892/ijo.2020.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–2067. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakai S, Ohhata T, Kitagawa K, Uchida C, Aoshima T, Niida H, et al. Long noncoding RNA ELIT-1 acts as a Smad3 cofactor to facilitate TGFbeta/Smad signaling and promote epithelial-mesenchymal transition. Cancer Res. 2019;79:2821–2838. doi: 10.1158/0008-5472.CAN-18-3210. [DOI] [PubMed] [Google Scholar]
- 56.Petersen M, Pardali E, van der Horst G, Cheung H, van den Hoogen C, van der Pluijm G, et al. Smad2 and Smad3 have opposing roles in breast cancer bone metastasis by differentially affecting tumor angiogenesis. Oncogene. 2010;29:1351–1361. doi: 10.1038/onc.2009.426. [DOI] [PubMed] [Google Scholar]
- 57.Singha PK, Pandeswara S, Geng H, Lan R, Venkatachalam MA, Dobi A, et al. Increased Smad3 and reduced Smad2 levels mediate the functional switch of TGF-beta from growth suppressor to growth and metastasis promoter through TMEPAI/PMEPA1 in triple negative breast cancer. Genes Cancer. 2019;10:134–149. doi: 10.18632/genesandcancer.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ying Z, Tian H, Li Y, Lian R, Li W, Wu S, et al. CCT6A suppresses SMAD2 and promotes prometastatic TGF-beta signaling. J Clin Invest. 2017;127:1725–1740. doi: 10.1172/JCI90439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aragon E, Wang Q, Zou Y, Morgani SM, Ruiz L, Kaczmarska Z, et al. Structural basis for distinct roles of SMAD2 and SMAD3 in FOXH1 pioneer-directed TGF-beta signaling. Genes Dev. 2019;33:1506–1524. doi: 10.1101/gad.330837.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD, et al. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. 2018;20:492–502. doi: 10.1038/s41556-018-0066-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Figure S1. Smyca expression correlates with poor prognosis. (A-F) Kaplan-Meier analysis of Smyca expression in relation to the overall survival (A, C, E, F) and disease-free survival (B, D) of indicated cancer types. Data were retrieved from TCGA data sets (A, B) or GEO data sets (C, D, E), or generated from an in house cohort (F). Patients were grouped into high and low expression based on the median expression level. Patients without survival information were omitted. Hazard ratio (HR) and P values are determined by log-rank test. (G) Smyca expression in HCC tumor tissues and adjacent normal tissues analyzed by qRT-PCR. (H–K) The correlation of Smyca expression with the stage (H, J) and invasiveness (I, K) of indicated cancer types. Data in (G-K) are presented by Whiskers boxplot, whiskers: min to max, bound of box: lower and upper quartiles, center line: median. Data points derived from basal-like subgroup of patients are marked in blue in (K). P values in (G), (H), (I), (J), (K) are determined by unpaired t-test. (L) Smyca expression in different subtypes of breast cancer patients analyzed by qRT-PCR. Data are expressed as mean ± SD. P values are determined by oneway ANOVA with Tukey’s post hoc test. Figure S2. Smyca promotes EMT. (A) Smyca expression in different breast cancer and HCC cell lines analyzed by qRT-PCR and represented as copy numbers. Data are expressed as mean ± SD from three independent experiments. (B) qRT-PCR analysis of indicated miRNAs in MDA-MB-231 cells stably expressing various Smyca shRNAs. Data are normalized with that of control cells and are expressed as mean ± SD from three independent experiments, ns, not significant by one-way ANOVA with Tukey’s post hoc test. (C, D) Western blot analysis of EMT markers in Hs578T cells stably expressing Smyca shRNAs (C) or MCF7 cells stably overexpressing Smyca (D). The amounts of each protein in relation to the control cells are indicated under the bands. Smyca expression levels in these stable lines are shown on the left panels. Data are mean ± SD from three independent experiments. (E-H) Cell proliferation (E, F) and cell viability (G, H) assays of M10 cells stably expressing Smyca (E, G) or MDA-MB-231 cells stably expressing Smyca shRNAs (F, H). Data are mean ± SD, n=3. P values are determined by unpaired t-test (E, G) or one-way ANOVA with Tukey’s post hoc test (F, H), ns, not significant. Figure S3. Bioinformatics and cell-based analysis for the relation of Smyca to TGF-β signaling. (A) Summary of the GSEA analysis for the match of Smyca signature with the indicated signatures. Data origin indicates the source database or the first author identifying the gene signature. (B) Intersection of Smyca-induced DEGs derived from MDA-MB-231 cells with Smad2/3 ChIP-seq data derived from Hs578T cells and BT-549 cells. The full list of overlapped genes is shown in Table S5. (C) qRT-PCR analysis for the expression of indicated TGF-β target genes in Hs578T cells stably expressing Smyca shRNAs and treated with or without 5 ng/ml TGF-β for 48 hr. Data are normalized with that of untreated group in each cell. (D, E) Luciferase assay for the Smad-target reporters transfected into Hs578T cells (D) or Malaru cells (E) stably expressing Smyca shRNAs and treated with or without 5 ng/ml TGF-β for 24 hr. The Smyca knockdown efficiencies are shown on the left panel in (E). Data in (C), (D), (D) are normalized with that of untreated control and presented as mean ± SD, n=3. P values are determined by one-way ANOVA with Tukey’s post hoc test, **P < 0.01, ***P < 0.001. (F, G) Summary of the correlations of Smyca expression with the expression of indicated TGF-β target genes by analyzing HCC or breast cancer data sets from TCGA (F) or GEO (G) databases. Pearson’s coefficients and P values are indicated. (H) GO analysis using the set of TGF-β target genes with expression levels showing positive correlations with Smyca expression in HCC and/or breast cancers. Figure S4. Smyca binds MH1 domains of Smad3 and Smad4 without affecting their expression and Smad3 phosphorylation. (A) Western blot analysis of Smad3 and Smad4 expression in MDA-MB-231 cells stably expressing Smyca shRNAs. (B) Western blot analysis for Smad3 phosphorylation in MDA-MB-231 cells stably expressing Smyca shRNAs and treated with or without 5 ng/ml TGF-β for 30 min. The blots are representative of three independent experiments and quantitative data are shown on the right. (C) Immunoprecipitation analysis of Smad3 and Smad4 interaction in MDA-MB-231 cells stably expressing indicated Smyca constructs and treated with or without 5 ng/ml TGF-β for 1 hr. (D) Luciferase assay for the Smad-responsive reporter in MDA-MB-231 cells transfected with indicated Smyca constructs and treated with or without 5 ng/ml TGF-β for 24 hr. The expression levels of Smyca are shown on the left panel. (E) Baculovirally purified Smad3, Smad4, and their MH1 deletion mutants bound on beads were incubated with biotinylated sense or antisense Smyca. The bound Smyca was analyzed by qRT-PCR. The equal inputs of recombinant proteins are shown on or antisense Smyca. The bound Smyca was analyzed by qRT-PCR. The equal inputs of recombinant proteins are shown on the right. Data in (B), (D) and (E) are mean ± SD, n=3. Pvalues are determined by one-way ANOVA with Tukey’s post hoc test (B, D) or unpaired t-test (E), *** P < 0.001; ns., not significant. Figure S5. Smyca is a Smad target and mediates a positive feedback control of TGF-β signaling. (A) qRT-PCR analysis of Smyca expression in MDA-MB-231 cells stably expressing Smad3 or Smad4 shRNAs and treated with or without 5 ng/ml TGF-β for 24 hr. The knockdown efficiencies of Smad3 and Smad4 shRNAs are shown on the left and middle panels, respectively. (B) qRT-PCR analysis of TGF-β target gene expression in MDA-MB-231 cells stably expressing Smyca shRNAs and treated with or without 5 ng/ml TGF-β for indicated time points. Data are normalized with that of untreated group (0 h). Data in (A) and (B) are mean ± SD, n=3. P values are determined by unpaired t-test (A) or one-way ANOVA with Tukey’s post hoc test (B), *** P < 0.001. Figure S6. Smyca promotes c-Myc transcription activity without affecting its expression or interaction with Max. (A) qRT-PCR analysis of the expression of indicated c-Myc target genes in Malaru cells stably expressing control or Smyca shRNA. Data are normalized with that of control cells. (B, C) Luciferase assay for a c-Myc-reponsive reporter transfected into MDA-MB-231 cells stably expressing Smyca or mutant (B) or Smyca shRNAs (C). The expression levels of Smyca and its mutant are shown on the left panel in (B). (D) Summary of the correlations of Smyca expression with the expression of indicated c-Myc target genes by analyzing breast cancer and HCC data sets from TCGA. Pearson’s coefficients and P values are indicated. (E) Maping the Smyca-binding region in c-Myc. Top: Schematic presentaton of c-Myc domains. The various GFP-c-Myc truncated proteins were purified from transfected 293T cells and incubated with biotinylated sense or antisense Smyca. The bound Smyca was analyzed by qRT-PCR. Data are normalized with that from GFP only control. The input levels of various GFP fusion proteins are shown on the right and marked by arrows. (F) Western blot analysis of c-Myc expression in indicated cells stably expressing Smyca or Smyca shRNAs. (G) Immunoprecipitation analysis of c-Myc/Max complex formation in MDA-MB-231 cells stably expressing Smyca or Smyca shRNA. Data in (A), (B), (C), (E) are mean ± SD, n=3. P values are determined by unpaired t-test (A), or one-way ANOVA with Tukey’s post hoc test (B, C, E), *P < 0.05, **P < 0.01, *** P < 0.001. Figure S7. Smyca-promoted c-Myc singaling neutralizes the growth inhibitory effect of Smyca-promoted TGF-β signaling. (A) Immunoprecipitation analysis of c-Myc/Smad complex formation in MDA-MB-231 cells stably expressing Smyca and treated with 5 ng/ml TGF-β for 2 hr. (B, C) Luciferase assay for a Smad-responsive reporter (B) or c-Myc-responsive reporter (C) transfected into MDA-MB-231 cells together with indicated Smyca constructs and treated with or without 5 ng/ml TGF-β for 24 hr. The expression levels of Smyca (1-500) and (1001-1500) fragments are shown on the left and middle panels in (B), respectively. (D, H) Cell proliferation assay of MCF7 cells stably expressing Smyca and treated with 75 μM 10058-F4 for 24 hr (D) or NTU-BL cells stably expressing Smyca and treated with 5 μM SB431542 or 75 μM 10058-F4 for 24 hr (H). (E, F) ChIP analysis for Smad3, Smad4, and c-Myc binding to the promoter regions of CDKN2B (E) and CDKN1A (F) genes in MDA-MB-231 cells stably expressing Smyca and treated with 5 μM SB431542 and/or 150 μM 10058-F4 for 2 hr. (G, I) qRTPCR analysis of the expression of indicated genes in MCF7 (G) or NTU-BL (I) cells stably expressing Smyca and treated with 5 μM SB431542 or 75 μM 10058-F4 for 24 hr. Smyca expression levels are shown on the left panel of (I). Data in (B), (C), (D), (E), (F), (G), (H), (I) are mean ± SD, n=3. P values are determined by one-way ANOVA with Tukey’s post hoc test, *P < 0.05, **P < 0.01, ***P < 0.001. Figure S8. Downregulation of the expression of Smyca, Smad targets, and c-Myc targets by Smyca gapmer ASO. (A, B, D) qRT-PCR analysis of Smyca or indicated mRNAs in MDA-MB-231 cells transfected with Smyca gapmer ASO (A), LM6 cells treated with indicated doses of NPs carrying Smyca gapmer ASO or control gapmer (B), or LM6 tumor-bearing mice treated with NPs carrying Smyca gapmer ASO or control NPs (D). (C) The morphology, size and weight of primary tumors taken from the sacrifice day for experiment shown in Fig. 7F. Data in all panels are mean ± SD, n=3 (A, B, D) or 4 (C). P values are determined by unpaired t-test, *P < 0.05, **P < 0.01, ***P < 0.001. Table S1. Antibody details. Table S2. siRNA, shRNA and Gapmer sequences. Table S3. Sequences of PCR primers. Table S4. ChIRP probe sequences. Table S5. List of genes that are regulated by Smyca and bound by Smad2/3.
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or Additional file 1. The RNA-seq data were deposited to the GEO database with the accession number GSE181028. Other data and materials are available upon reasonable request.