Abstract
BACKGROUND
Hepatic steatosis is a major cause of chronic liver disease. Two-dimensional (2D) ultrasound is the most widely used non-invasive tool for screening and monitoring, but associated diagnoses are highly subjective.
AIM
To develop a scalable deep learning (DL) algorithm for quantitative scoring of liver steatosis from 2D ultrasound images.
METHODS
Using multi-view ultrasound data from 3310 patients, 19513 studies, and 228075 images from a retrospective cohort of patients received elastography, we trained a DL algorithm to diagnose steatosis stages (healthy, mild, moderate, or severe) from clinical ultrasound diagnoses. Performance was validated on two multi-scanner unblinded and blinded (initially to DL developer) histology-proven cohorts (147 and 112 patients) with histopathology fatty cell percentage diagnoses and a subset with FibroScan diagnoses. We also quantified reliability across scanners and viewpoints. Results were evaluated using Bland-Altman and receiver operating characteristic (ROC) analysis.
RESULTS
The DL algorithm demonstrated repeatable measurements with a moderate number of images (three for each viewpoint) and high agreement across three premium ultrasound scanners. High diagnostic performance was observed across all viewpoints: Areas under the curve of the ROC to classify mild, moderate, and severe steatosis grades were 0.85, 0.91, and 0.93, respectively. The DL algorithm outperformed or performed at least comparably to FibroScan control attenuation parameter (CAP) with statistically significant improvements for all levels on the unblinded histology-proven cohort and for “= severe” steatosis on the blinded histology-proven cohort.
CONCLUSION
The DL algorithm provides a reliable quantitative steatosis assessment across view and scanners on two multi-scanner cohorts. Diagnostic performance was high with comparable or better performance than the CAP.
Keywords: Ultrasound, Liver steatosis, Deep learning, Screening, Computer-aided diagnosis
Core Tip: Ultrasound is widely used to evaluate liver steatosis, but it is subjective. We developed a deep learning algorithm for quantitative steatosis scoring from ultrasound. The algorithm was trained on > 200000 images and composed of different scanners and viewpoints from both hepatic lobes. High diagnostic performance was measured across all viewpoints in separate histology proven groups, which was comparable to or better than the control attenuation parameter. We demonstrated high agreement across scanners and viewpoints. Thus, our deep learning algorithm provides a quantitative assessment with high performance and reliability.
INTRODUCTION
Liver steatosis, or fatty liver disease, is a major cause of chronic liver disease worldwide. It is estimated that nonalcoholic fatty liver disease (NAFLD) affects 20%-30% of the global population[1-3] and is associated with increased risks of cardiovascular disease, type 2 diabetes, and metabolic risk factors[4]. Unfortunately, it is an undertreated and underdiagnosed disease[5]. For those patients with more aggressive nonalcoholic steatohepatitis, the risks of liver cirrhosis, liver failure, and hepatocellular carcinoma are higher[6-8]. Liver steatosis is also associated with chronic viral hepatitis. For instance, hepatitis C can impair glucose metabolism, resulting in the accumulation of fat droplets in the liver and increasing hepatocarcinogenesis[9]. There have been reports on the interaction between liver steatosis and hepatitis B. Previous reports have also suggested that liver steatosis is associated with delayed hepatitis B surface antigen seroclearance[10]. Therefore, assessing liver steatosis is also important for chronic viral hepatitis.
Liver needle biopsy is the gold standard diagnosis at present. However, even with machine learning-based interpretation[11,12], the invasiveness of the biopsy severely limits its clinical applicability as a screening and assessment tool, and at the same time it is prone to sampling error. Thus, with the growing prevalence of NAFLD and nonalcoholic steatohepatitis, accurate, reliable, and accessible non-invasive screening tools are increasingly important to quantify liver steatosis and provide follow-up monitoring[4].
Such tools include magnetic resonance imaging with derived proton density fat fraction (MRI-PDFF), quantitative ultrasound (US), and two-dimensional (2D) US diagnoses[13]. MRI-PDFF is the best non-invasive test and can be used as a gold standard in clinical trials, but it is costly for routine clinical care[5,14]. As for quantitative US, its most popular variant is FibroScan with its control attenuation parameter (CAP) scores[15]. CAP is more available than MRI-PDFF but still requires dedicated equipment. Finally, 2D US exams have been widely used for the diagnosis of liver disease for 5 decades[13], which is partly driven by the prevalence of US equipment and its low cost. In clinical practice, 2D US is also the first-line screening modality for the detection of liver cancer[16]. Therefore, steatosis can also be assessed from studies with a primary aim of liver tumor screening. Given these considerations, it is not surprising that 2D US is the most common tool for assessing liver steatosis[4].
A recent NAFLD epidemiology meta-analysis revealed that 90.6% of 392 studies in China used 2D US as the diagnostic modality of choice[3]. Unfortunately, US steatosis scores are considered a subjective diagnosis. A meta-analysis in 2011 reported that the kappa statistics for inter- and intra-observer reliability showed poor numbers[4]. A 2014 analysis[17], focusing on US steatosis assessment derived from routine clinical care, concluded that intra- and inter-observer agreement of binary assessment was only 51%-68% and 39%-40%, respectively, and it also noted that there was a lack of reported reliability measurements of categorical assessments. Both studies attributed the low reliability to different image acquisition practices across institutions and the subjective and variable nature of US image interpretation. More recently, Hong et al[18] investigated the reliability of categorical US assessments of different features and reported only moderate inter-rater agreements (intra-class correlation coefficients of 0.54) for the overall steatosis impression.
A promising alternative is to apply machine learning algorithms, e.g., deep learning (DL), on 2D US liver scans. The goal is to provide a quantitative measure of liver steatosis directly from 2D US images for clinical decision support. Efforts toward this end have been reported. However, limitations in the analysis, i.e., small training set sizes[19,20], only binary assessments[20-24], and single-scanner data[23-27], restrict the conclusions that can be drawn. Moreover, the reliability of these assessments, either across scanners or across different US views of the liver, have not been assessed. Relatedly, the specific number of images needed for a reliable diagnosis, and of which liver viewpoints, has also not been well articulated or characterized. This is crucial for an at-large adoption of any imaging-based diagnostic tool.
To address these limitations, we developed a scalable DL algorithm to quantitatively assess liver steatosis from 2D US using a retrospectively mined big data cohort. Cross-scanner and cross-view reliability was measured in addition to diagnostic performance against gold standard histopathological diagnoses on two clinical cohorts. Direct comparisons against CAP were also performed. The DL algorithm might serve as an effective tool for liver steatosis screening and monitoring.
MATERIALS AND METHODS
Patient cohorts and image collection
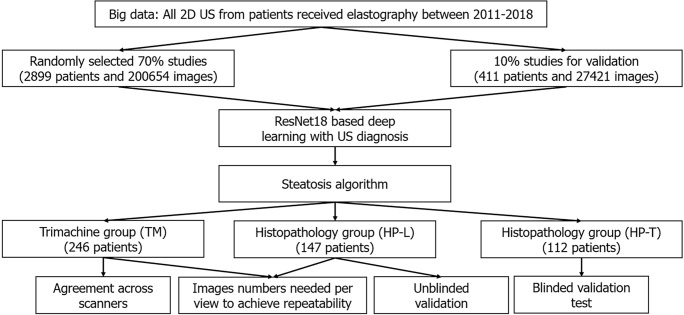
Multiple patient cohorts were collected for our study, as shown in Table 1 and Figure 1. The clinicopathological makeup of each dataset can be found in Supplementary Table 1. All US images underwent the same automatic cropping and resampling preprocessing, which is described in the Supplementary material. This study was approved by the Institutional Review Board of the Chang Gung Medical Foundation (CGMH IRB No. 201801283B0).
Table 1.
Overview of development and testing datasets
|
Stage
|
Name
|
Purpose
|
Labels
|
Patients
|
Studies
|
Images
|
| DL learning | BD-L | Big data, to train the neural network | 2D US Dx | 2899 | 17149 | 200654 |
| DL validation | BD-V | Big data, to tune model performance | 2D US Dx | 411 | 2364 | 27421 |
| Testing | HP-U | Histopathology-proven group, to (a) measure the trend between DL predictions and histology (b) measure reliability across 2D US liver viewpoints | Histology | 147 | 147 | 1647 |
| TM | Tri-machine data US Dx group, to (a) measure reliability across 2D US liver viewpoints and (b) measure reliability across scanners | - | 246 | 733 | 9215 | |
| HP-T1 | Histology proven group to measure the trend between DL predictions and histology | Histology, CAP | 112 | 112 | 1996 |
Labels blind to deep learning researchers during course of algorithmic development.
DL: Deep learning; US: Ultrasound; BD-L: Big data learning group; BD-V: Big data validation group; HP-U: Histopathology unblinded test group; TM: Trimachine group; HP-T: Histopathology blinded test group; CAP: Control attenuation parameter; 2D: Two-dimensional; Dx: Diagnosis.
Figure 1.
Flowchart. 2D-US: Two-dimensional ultrasound.
Big data groups (big data learning group and big data validation group)
We retrospectively collected a big data dataset from the picture archiving and communication system of CGMH, a major hospital in Taiwan (over 4 million outpatient visits/year). All patients who received elastography (a quantitative US technology), specifically acoustic radiation force impulse imaging and FibroScan, between January 3, 2011 and September 28, 2018 represented the index patients. From the index patients, we extracted all 2D US studies that were acquired within the same 2011-2018 period, resulting in multiple studies per patient. Seventy percent of the patients and their corresponding US studies were randomly selected as training data (big data learning group, BD-L), totaling 2899 patients and 200654 images. We used another 10% of the patients for validating and tuning our DL algorithm (big data validation group, BD-V), totaling 411 patients and 27421 images. The remaining 20% of the patients were not used as part of this study. In addition, any patients also found in the other datasets were also moved to this excluded cohort. The collected images were generated from 13 known scanners, which are listed in Supplementary Table 2. Each US study was accompanied by a 2D US diagnosis of steatosis severity from visual assessment using established guidelines[13,28], generated through the course of routine clinical care. These labels are used to train the DL algorithm. We enrolled the maximum number of patients that fit our inclusion criteria to provide as much data as possible for the DL algorithm.
Unblinded test group (histopathology unblinded test group)
We used a collection and selection protocol reported in prior work[29-32]. Specifically, since 2011 it has been CGMH policy to obtain elastography measurements for all patients undergoing a liver biopsy, i.e., acoustic radiation force impulse and FibroScan once the latter became available. The histopathology unblinded test group (HP-U) dataset (n = 147) consists of patients with FibroScan diagnoses between the dates November 27, 2014 to September 26, 2019. Identical to a prior study[29], we included patients with chronic hepatitis B virus (HBV), chronic hepatitis C virus (HCV), and non-hepatitis B/C virus (NBNC) liver diseases. We excluded those with decompensated liver disease, toxic hepatitis, alcoholism, or autoimmune liver diseases. After exclusion of these etiologies, the majority of the NBNC group were NAFLD patients (90.3% in HP-U group, Supplementary Table 1B). Histopathological analysis was performed by the same clinician (S.F.H.), and any retrospectively collected histopathological analysis not originally performed by S.F.H. was redone. Liver biopsies were collected mainly at the CGMH outpatient center and were performed to evaluate fibrosis, steatosis, or an unknown etiology[29-32]. Percutaneous liver biopsy was performed under US guidance with an 18-gauge core needle and an automatic pistol device (Magnum; Bard Peripheral Vascular, Inc, Tempe, AZ, United States). Under this standard procedure a liver specimen greater than 1.2 cm was obtained. However, a portion of the histology data was obtained from tumor resection. Following Kleiner et al[33], the histology diagnoses were graded into normal (5%), mild (5% and 33%), moderate (33% and 66%) and severe (66%) based on the liver fat cell fraction.
Because we were interested in US analysis, additional selection criteria were also applied. A patient must have a 2D US study that was: (1) Acquired within 3 mo of the biopsy; and (2) acquired with one of the Siemens Acuson S2000, Philips IU22, or Toshiba Aplio 300 scanners. We also excluded patients with tumors > 3 cm and with multiple cysts. Finally, US studies must have ≥ 10 images of the viewpoints depicted in Figure 1. If more than one US study qualified, we randomly selected one. All labels in HP-U were unblinded to the DL researchers of this work, but the data was treated as a test set, meaning it was only analyzed after development was complete.
Tri-machine group
We prospectively collected this cohort (n = 246) from patients that had both an US and a FibroScan study ordered and if D.I.T., Y.C.C., T.H.H., or C.J.C. conducted the image study. With the agreement of these patients, they were scanned by Siemens Acuson S2000, Philips IU22, and Toshiba Aplio 300 scanners on the same day. Studies were collected over a period from August 30, 2018 to August 27, 2019, and we only included patients diagnosed with the HBV, HCV, and NBNC criteria used in HP-U. The tri-machine (TM) cohort allowed for an assessment of agreement across scanners.
Blind testing group (histopathology blinded test group)
Finally, we included histopathology blinded test group (HP-T, n = 112), a clinical testing dataset whose labels were blind to the DL researchers involved in this project during development of the algorithm. HP-T was collected from patients that had received a liver biopsy between April 18, 2011 to May 5, 2015 and September 1, 2018 to January 29, 2021. Associated FibroScan diagnoses were only available for the later subset of patients. Also, unlike HP-U, US studies were not restricted to only the Siemens, Philips, and Toshiba scanners of TM and HP-U. In particular, 18 studies were acquired with the Aloka SSD 5500 or ATL: HDI 5000 scanners, which are not currently considered premium scanners.
Image selection
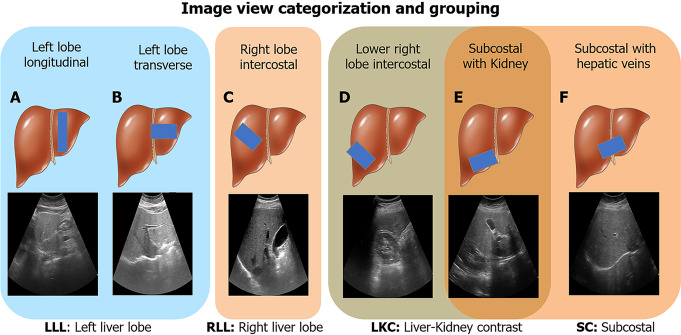
We were interested in investigating performance and reliability across viewpoints. Thus, we only included US images from the viewpoints shown in Figure 2, which can be categorized into four view groups: Left liver lobe (LLL), right liver lobe (RLL), liver/kidney contrast (LKC), and subcostal (SC). For HP-U, HP-T, and BWC, we only included studies that had ≥ 10 images of any of the studied viewpoints. BD-L and BD-V were automatically filtered with an algorithm explained in the Supplementary material.
Figure 2.
Image view categorization and grouping. Six ultrasound image viewpoints were used in this study. A: Left lobe longitudinal; B: Left lobe transverse; C: Right lobe intercostal; D: Lower right lobe intercostal (depicting liver/kidney contrast); E: Subcostal depicting liver/kidney contrast; F: Subcostal with hepatic veins. These views were further categorized into four groups: Left liver lobe (A and B), right liver lobe (C), liver/kidney contrast (D and E), and subcostal (E and F). LLL: Left liver lobe; RLL: Right liver lobe; LKC: Liver/kidney contrast; SC: Subcostal. Liver cartoons adapted from the DataBase Center for Life Science (https://commons.wikimedia.org/wiki/File:201405_liver.png), licensed under the Creative Commons Attribution 4.0 International[51] (Copyright permission see Supplementary material).
Training steatosis assessment DL algorithm
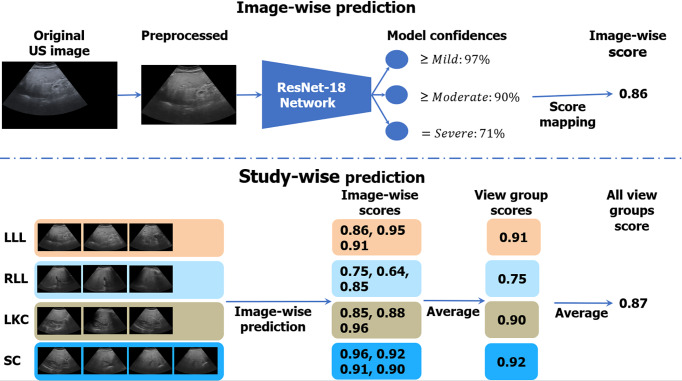
Figure 3 illustrates the algorithmic workflow of our DL algorithm. Using the images from BD-L, we trained a multi-class deep ResNet18[34] DL classifier using the 2D US diagnoses, which were ordinal labels ranging from 0 to 3 corresponding to None; Mild; Moderate; and Severe steatosis[35]. We treated each image independently in training and followed the well-known binary decomposition approach to ordinal classification of Frank and Hall[36]. After training, a simple transformation produced a continuous score[37] for each image that ranged from 0 to 1, with higher scores corresponding to more severe steatosis. As Figure 3 indicates, during inference, we took the mean of the image-wise scores within and across each view group. The view group scores were further averaged to produce an “All View Groups” score. If one or several view groups were missing, the “All View Groups” score was calculated with what view groups were available. More details on the training strategy can be found in the Supplementary material, and a listing of hyper-parameters is given in Supplementary Table 5.
Figure 3.
Algorithmic workflow. Images were first comprehensively preprocessed to remove regions outside the ultrasound beam. A deep learning neural network, called ResNet-18, was trained on individual ultrasound images in the big data learning group. The model predicted confidences in three binary cutoffs: “≥ mild”, “≥ moderate”, or “= severe” steatosis. The confidences were mapped to a continuous image-wise score in the range of [0, 1]. View-group scores were produced by averaging each image within the group. An “All View Groups” score was produced by averaging all available view group scores. In the figure’s example, the gold standard histopathology diagnosis was a fatty cell percentage of 90%. LLL: Left liver lobe; RLL: Right liver lobe; LKC: Liver/kidney contrast; SC: Subcostal.
Statistical analysis
A listing of all experiments can be found in Supplementary Table 3. We evaluated the reliability and diagnostic performance of the DL algorithm. We judged P values < 0.05 as significant and corrected for multiple comparisons using the Holm-Bonferroni procedure[38].
Reliability studies
Experiment 1: We used TM and HP-U to assess how many images were needed per view group to achieve repeatability. Note, for the TM dataset we randomly selected only one US study for each patient to avoid sampling the same patient more than once. As advocated by Bland and Altman[39,40], we graphed the within-subject standard deviations across different view-group steatosis scores and measured the repeatability coefficient (RC)[40]. The difference between two repeated measurements should be within the RC value for 95% of the US studies. However, because the within-subject standard deviation was not uniform across our data (typically greater variability in repeated measurements at moderate steatosis levels), we regressed a non-uniform RC and used the worst-case RC value as a summary statistic, with 95% confidence intervals computed using percentile bootstrap (1000 bootstrap samples)[41]. More details on this calculation can be found in the Supplementary material.
Experiment 2: We also evaluated agreement across scanners using TM. We conducted a Bland-Altman analysis[39] on the difference values across view-group scores and calculated the limits of agreement (LOAs[40]), which are the limits by which 95% of the disagreements fall under[40]. Like Experiment 1, variability tended to be higher at moderate levels of severity. Therefore, we regressed non-uniform LOAs and used the maximum upper LOAs and minimum lower LOAs as summary statistics. More details can be found in the Supplementary material. We also calculated the percentage agreement, using the cutoff levels determined in Experiment 3 below.
Diagnostic testing
Experiment 3a: We validated the DL model diagnostic performance using the images and histopathological labels of HP-U. Although the HP-U labels were not blinded to the DL researchers during algorithmic development, it was treated like a test set, i.e. evaluation was only performed once model development was complete. Histopathological diagnoses were separated into four ordinal labels[27]. For three separations of fatty percentages, i.e., grade ≥ 5%, grade ≥ 33%, and grade ≥ 66%, we used receiver operating characteristic (ROC) curve analysis and measured the area under the curve of the ROC (AUCROC). Trend tests between the DL assessment and histopathological grades were conducted using the non-parametric Jonckheere-Terpstra test[42]. The 95% confidence intervals of all AUCROCs were calculated based on DeLong’s non-parametric test[43]. When needed, we determined cutoff values using the values that maximized the Youden index[44].
Experiment 3b: We compared the performance of FibroScan with that of the DL assessment, and statistical significance of any differences in AUCROCs were assessed using the StAR[45] implementation of DeLong’s non-parametric test[43].
Experiment 4a: Finally, we tested our DL assessment on HP-T, whose histopathological labels were blind to the DL researchers during model development.
Experiment 4b: For patients with FibroScan diagnoses, we also compared its performance with that of the DL assessment. Experiments 4a and b used the same statistical analyses as Experiments 3a and b.
RESULTS
Repeatable measurements with a moderate number of images
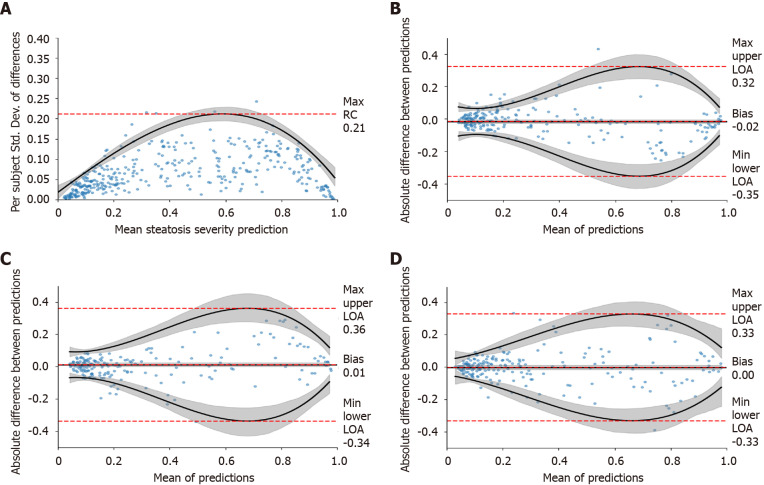
We first determined how many images were needed for each view group to reach a repeatable measurement (Experiment 1 in Supplementary Table 3) using TM (one random study per patient) (n = 246) and HP-U (n = 147). The results when using three images per view group can be found in Table 2, and the complete set of results from one-to-four images can be found in Supplementary Table 6. As can be seen, depending on the view group, the max RC value was equal to or less than 30% of the DL assessment scale, which ranged from 0 to 1. In the worst case, 95% of the differences between repeated measurements should be within this max RC value. As the repeatability graph of Figure 4A demonstrates, this max RC value occurred at moderate levels of steatosis severity, which have a wide tolerance (e.g., moderate fatty cell content is commonly determined as being anything between 33% and 66%). The RC values for mild and severe were much lower, suggesting that a 30% RC value was highly conservative for these ranges of severity. The repeatability graphs for all other view groups can be found in Supplementary Figure 1. Together, these data demonstrate the DL assessment can attain repeatable measurements with a moderate number of images for each view group. For the remainder of evaluations, we will only include assessments that have a minimum of three images for each view group.
Table 2.
Reliability studies in different views
|
|
Repeatability study
|
Cross-scanner agreement study
|
|||||
|
View
|
n
|
Max RC
|
n
|
Bias
|
Min lower LOA
|
Max upper LOA
|
Agreement
|
| LLL | 342 | 0.27 (0.24, 0.29) | 237 | 0.00 (-0.01, 0.01) | -0.37 (-0.32, -0.42) | 0.37 (0.32, 0.42) | 92% |
| RLL | 370 | 0.21 (0.19, 0.23) | 232 | 0.00 (-0.01, 0.01) | -0.37 (-0.33, -0.42) | 0.37 (0.33, 0.42) | 92% |
| LKC | 267 | 0.30 (0.27, 0.34) | 183 | 0.00 (-0.01, 0.01) | -0.35 (-0.29, -0.41) | 0.35 (0.29, 0.41) | 93% |
| SC | 297 | 0.26 (0.24, 0.29) | 182 | 0.00 (-0.01, 0.01) | -0.36 (-0.31, -0.42) | 0.36 (0.31, 0.42) | 94% |
| All view groups | - | - | 237 | 0.00 (-0.01, 0.01) | -0.25 (-0.21, -0.28) | 0.24 (0.21, 0.28) | 94% |
On the left, the max repeatability coefficient was tabulated when using three images for each view group (Histopathology unblinded test group and Trimachine group datasets). On the right, the bias, worst-case limits of agreement, and agreement (%) were tabulated across different view groups for the Trimachine dataset. The results for all scanner pairs were combined. Parentheses enclose bootstrapped 95% confidence intervals. RC: Repeatability coefficient; LOA: Limits of agreement; LLL: Left liver lobe; RLL: Right liver lobe; LKC: Liver/kidney contrast; SC: Subcostal.
Figure 4.
Repeatability study. A: A repeatability coefficient plot for right liver lobe when using three images; B-D: Cross-scanner Bland-Altman plots for Siemens-Toshiba (B), Toshiba-Philips (C), and Philips-Siemens (D), respectively. Cross-scanner plots were depicted for “All View Groups” when using ≥ three images per view group. Grey-shaded areas indicate 95% confidence intervals. RC: Repeatability coefficient; LOA: Limit of agreement.
Agreement across scanners was high
We used the TM dataset (n = 246) to measure agreement across measurements taken on the same day but with three different scanners (Experiment 2 in Supplementary Table 3). As the Bland-Altman plots for “All View Groups” of Figure 4B-D demonstrate, the LOAs are worse at moderate levels of severity, but they are much tighter at milder and more severe levels. This mirrors the repeatability results. Table 2 presents the Bland-Altman summary statistics for all view groups, demonstrating that the cross-scanner agreement is roughly equivalent for all view groups. For brevity we combined the results across all scanner pairs together. As can be seen, the agreement across scanners is high. The bias for “All View Groups” was close to zero, and the worst case LOAs were around 35% for every view group. The LOAs for “All View Groups” fell to 25%, suggesting that examining all view groups together can increase reliability. The percentage agreement numbers are all 92% or higher, further underscoring the high agreement across the tested scanners.
High diagnostic performance on clinical test sets
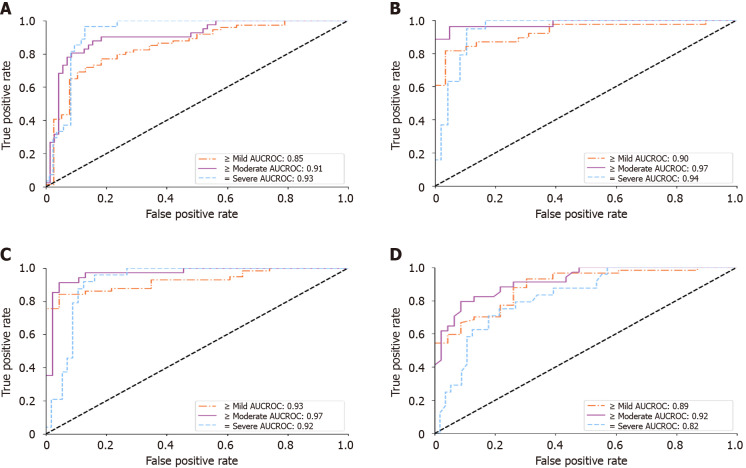
With the reliability studies completed, we validated the DL score against a histopathological gold standard for diagnosing mild-to-severe (≥ 5%), moderate-to-severe (≥ 33%), and severe (≥ 66%) fatty percentages. The results on HP-U and HP-T, Experiments 3 and 4 in Supplementary Table 3, respectively, are presented in Table 3. Focusing primarily on the HP-T results, the “Complete 4 view group study” of Table 3 only selects studies that had three or more images for every view group. This allowed an “apples-to-apples” comparison across view groups. As can be seen, the performance was comparable across view groups, suggesting that each view group provided similar diagnostic value, with all providing AUCROCs ≥ 0.84. The “Individual view group study,” on the other hand, presented results when examining each view group individually, meaning only the view group in question required three or more images. This allowed for a larger sample size. In this setting “All View Groups” denoted the mean score across all view groups with three or more images, which can comprise different view groups from study to study. The ROC curves for “All View Groups” for the individual view group study can be found in Figure 5A. As can be seen in Table 3, the results for the individual view groups were broadly like those of the complete view groups study with AUCROCs ≥ 0.81. Comparing the results of HP-U to HP-T, the AUCROCs were generally similar, reinforcing the above results. However, the HP-U results were generally better, especially for diagnosing mild-to-severe steatosis. This is due to HP-T including non-premium Aloka and ATL HDI scanners in its cohort, which make it harder to accurately assess steatosis. Indeed, as Figure 5B indicates, when only selecting for the Siemens, Philips, and Toshiba premium scanners the AUCROC scores for HP-T were much improved, indicating that scanner choice does make an impact. Finally, the “FibroScan comparison study” compared the performance of FibroScan directly with the DL assessment. As can be seen, the DL assessment AUCROC values were better than FibroScan, and statistical significance was reached for all levels on the HP-U dataset and for “= severe” on the HP-T dataset (Figure 5).
Table 3.
Receiver operating characteristic analysis on the histopathology unblinded test group and histopathology blinded test group cohorts for diagnosing steatosis grades
|
|
HP-U
|
HP-T
|
||||||||
|
View
|
n
|
AUC ≥ 5% |
AUC ≥ 33% |
AUC ≥ 66% |
Acc
|
n
|
AUC ≥ 5% |
AUC ≥ 33% |
AUC ≥ 66% |
Acc
|
| Complete 4 view group study1 | ||||||||||
| LLL | 41 | 0.98 (0.93, 1.00) | 0.95 (0.89, 1.00) | 0.94 (0.87, 1.00) | 83% | 51 | 0.90 (0.82, 0.98) | 0.92 (0.81, 1.00) | 0.90 (0.82, 0.99) | 88% |
| RLL | 41 | 0.96 (0.90, 1.00) | 0.95 (0.89, 1.00) | 0.89 (0.79, 0.99) | 85% | 51 | 0.84 (0.73, 0.95) | 0.93 (0.84, 1.00) | 0.92 (0.85, 1.00) | 96% |
| LKC | 41 | 0.96 (0.90, 1.00) | 0.95 (0.89, 1.00) | 0.93 (0.84, 1.00) | 83% | 51 | 0.84 (0.73, 0.95) | 0.95 (0.90, 1.00) | 0.88 (0.79, 0.97) | 90% |
| SC | 41 | 0.96 (0.90, 1.00) | 0.92 (0.84, 1.00) | 0.89 (0.79, 0.99) | 83% | 51 | 0.88 (0.79, 0.97) | 0.93 (0.86, 1.00) | 0.88 (0.77, 0.99) | 90% |
| All view groups | 41 | 0.96 (0.90, 1.00) | 0.94 (0.88, 1.00) | 0.92 (0.83, 1.00) | 83% | 51 | 0.88 (0.79, 0.98) | 0.95 (0.88, 1.00) | 0.91 (0.83, 0.99) | 94% |
| Individual view group study1 | ||||||||||
| LLL | 103 | 0.95 (0.90, 0.99) | 0.93 (0.87, 0.98) | 0.91 (0.86, 0.97) | 80% | 96 | 0.84 (0.76, 0.93) | 0.92 (0.85, 0.99) | 0.93 (0.88, 0.98) | 90% |
| RLL | 138 | 0.94 (0.91, 0.98) | 0.91 (0.86, 0.98) | 0.85 (0.78, 0.92) | 83% | 109 | 0.82 (0.74, 0.90) | 0.89 (0.83, 0.96) | 0.92 (0.87, 0.97) | 92% |
| LKC | 88 | 0.96 (0.92, 1.00) | 0.92 (0.86, 0.98) | 0.84 (0.76, 0.92) | 80% | 71 | 0.81 (0.69, 0.93) | 0.93 (0.87, 0.99) | 0.89 (0.81, 0.96) | 90% |
| SC | 117 | 0.93 (0.89, 0.98) | 0.91 (0.85, 0.96) | 0.86 (0.79, 0.92) | 79% | 90 | 0.86 (0.77, 0.94) | 0.89 (0.82, 0.96) | 0.90 (0.83, 0.97) | 88% |
| All view groups | 147 | 0.95 (0.91, 0.98) | 0.92 (0.88, 0.96) | 0.87 (0.81, 0.92) | 76% | 112 | 0.85 (0.77, 0.93) | 0.91 (0.85, 0.97) | 0.93 (0.88, 0.98) | 90% |
| FibroScan comparison study1,2 | ||||||||||
| All view groups | 147 | 0.953 (0.92, 0.98) | 0.953 (0.92, 0.98) | 0.923 (0.88, 0.97) | 77% | 80 | 0.93 (0.87, 0.98) | 0.97 (0.93, 1.00) | 0.923 (0.86, 0.98) | 91% |
| FibroScan | 147 | 0.88 (0.81, 0.95) | 0.88 (0.81, 0.95) | 0.80 (0.73, 0.87) | 62% | 80 | 0.89 (0.82, 0.96) | 0.92 (0.86, 0.98) | 0.82 (0.73, 0.92) | 68% |
Filtered with a minimum of 3 images for each view group.
Selecting only studies with associated FibroScan control attenuation parameter (CAP) scores.
Area under the curve of the receiver operating characteristic significantly better than FibroScan CAP scores.
“Complete 4 view groups study” only selects studies where every view group is qualifying (three or more images), whereas “Individual view group study” examines the performance of each qualifying view group individually. Numbers in parentheses are 95% confidence intervals. All trends between the deep learning/CAP score and the histopathology grades were significant (P < 0.001). “Acc” is the classification accuracy when the threshold values calculated by optimizing the Youden index[44] are applied. LLL: Left liver lobe; RLL: Right liver lobe; LKC: Liver/kidney contrast; SC: Subcostal; HP-U: Histopathology unblinded test group; HP-T: Histopathology blinded test group; AUC: Area under the curve.
Figure 5.
Receiver operating characteristic analysis on histopathology blinded test group. A and B: Receiver operating characteristic curves of the deep learning model for diagnosing hepatic steatosis grades on histopathology blinded test group (HP-T) when using all scanners and only the Siemens/Toshiba/Philips premium scanners, respectively; C and D: Only select for histopathology blinded test group studies with FibroScan diagnoses, corresponding to the performance of the deep learning algorithm and FibroScan, respectively. All receiver operating characteristic curves were measured against a histopathological gold standard. AUCROC: Area under the curve of the receiver operating characteristic.
DISCUSSION
Incorporating different 2D US scanner models and brands, different liver viewpoints, and prospectively and retrospectively collected images, we demonstrated that a DL-based assessment can provide quantitative and reliable hepatic steatosis scores.
Unlike attenuation imaging (ATI)[46], FibroScan, or some other reported DL solutions[22,25,26], our algorithm accepts images taken from both hepatic lobes rather than a selected area of interest in a specific location. We categorized 2D US images into six major viewpoints (Figure 2), which we further grouped into four view groups. We found that for each view group three images were enough to reach acceptable max RC values of 21%-30% (Table 2, Figure 4A), where the best and worst max RC values corresponded to the RLL and LKC view group, respectively. The relatively poorer repeatability of the LKC view group was likely due to the heterogenous makeup of viewpoints, as it comprises both subcostal and right lobe intercostal images. To put these repeatability ranges in context, the RC for gold standard histopathology fatty percentage assessment has been reported to be 38%[47] with poor intraclass correlation agreements of 0.57[48]. Considering our DL model’s worst max RC value was 30% and that RC values tended to be much better than 30% at milder or more severe levels of steatosis (Figure 4 and Supplementary Figure 1), these repeatability measures compare well. It is also encouraging that the DL algorithm was more repeatable at milder and severe steatosis levels since such patients should indeed be less ambiguous to categorize.
We also demonstrated good cross-scanner agreement. Bland-Altman analysis suggested bias across scanners was near zero with acceptable LOAs (35%-37%) for individual view groups, with the LOAs falling to 25% when using “All View Groups.” When using categorical labels, these numbers correspond to agreements of 90% to 96%. An encouraging sign for generalizability is that cross scanner agreement is high for the Siemens: S2000 scanner, despite it being poorly represented in BD-L.
In terms of diagnostic performance, we validated on two different histology-proven cohorts (HP-U and HP-T). The “Complete view group study” of Table 3 indicated that diagnostic performance remained stable across view groups, with comparable and high AUCROC values (≥ 0.84). These results were reinforced by the “Individual view group study,” which allowed each view group to be investigated individually, with a corresponding larger sample size. Again, the DL model posted good AUCROC values (≥ 0.81) across view groups and histopathology grades. As highlighted in the results, the AUCROCs in HP-T tended to be lower than in HP-U, which was most pronounced for diagnosing > 5% fatty percentage. This is explained by the 18 earlier studies acquired by low resolution scanners in HP-T (Supplementary Table 2), which produced relatively lower quality images than the more recent premium scanners. Indeed, as Figure 4B shows, excluding the older scanners raised the HP-T AUCROC scores considerably so that they matched the HP-U group (from 0.85 to 0.90 AUCROC for ≥ 5% fatty percentage). Consequently, even though performance seemed to be stable across the tested premium scanners, the DL algorithm can be sensitive to scanner quality, which is a matter requiring further investigation.
The clinical diagnosis of our big data was generally based on the principle of Hemaguchi score, US-FLI score, and hepatorenal steatosis score[13]. Our DL algorithm demonstrated good quantitative performance despite being trained on subjective 2D US categorical labels with inter-observer variations. We speculate this is due to the exceptionally large training BD-L dataset, which included diagnoses from 63 clinicians. This allowed the DL model to “learn” and distill from a variety of clinical assessments. In addition, the DL model’s continuous output allowed for a calibration against accepted gold standards. Although the distribution of etiologies between the big data training cohort and histology-proven validation cohorts were unavoidably different, validation performance remained high. Thus, these distributional differences did not seem to impede performance on a representative sample from a clinical population.
Importantly, we compared the DL algorithm’s performance head-to-head with CAP of FibroScan. The DL algorithm’s “All View Groups” score reported higher AUCROCs (0.92-0.97 vs 0.80-0.92) and accuracies (77%-91% vs 62%-68%) than CAP (Table 3). Statistically significant improvements were achieved for all levels on the HP-U cohort, and only for “= severe” on the HP-T cohort. This is encouraging because, unlike CAP, our DL algorithm can be applied to many different liver viewpoints and does not require additional equipment outside of an US scanner. Our DL algorithm enjoys similar advantages in flexibility over other quantitative US techniques[25,49], which examines the attenuation, brightness, or echogenicity change within a small region of interest (ROI). Such restrictions may result in missing other useful information for steatosis diagnosis (for example, liver/kidney contrast and the loss of vessel walls). Quantitative US can also not be applied to patients where the specific ROI is unable to be imaged and is often limited to a specific scanner, e.g., ATI only works with Canon scanners.
The DL algorithm can be trained with large-scale retrospective datasets, which need neither costly machines nor annotation of regions of interest. Furthermore, in the inference stage, our algorithm does not require images from all hepatic views, e.g., the absence of the right hepatic lobe images is acceptable, as the performance of our algorithm with left hepatic lobe images is just as good. When using “All View Groups,” diagnostic AUCROCs did not improve. However, the improved cross-scanner agreement (Table 2) suggests using “All View Groups” may provide more reliability. Our algorithm could be used on different brands of scanners in different hospitals. However, each scanner or hospital may determine their cutoff values for different grades of steatosis based on their preferred gold standard (liver histology or MRI-PDFF). Its potential applications include improved steatosis screening and longitudinal tracking in clinical settings. Our trained algorithm may be shared with researchers for non-commercial use upon reasonable requests to the clinical principal investigator (D.I.T.) and subject to IRB approval. We expect it to have good generalizability given the diversity of the BD-L dataset. Because our algorithm is trained only on retrospective US-labeled data, which are readily available in most Picture Archiving and Communication Systems, we also believe the methodology we presented here to be highly generalizable and accessible to institutions wishing to train their own algorithms.
The use of machine learning technologies for non-invasive liver steatosis assessment has received attention for a number of years[19-27]. This work represents a significant step forward. Like this work, many prior solutions were built upon recent DL techniques[22,24-27], which Supplementary Table 7 summarizes. Apart from Gummadi et al[22], all other works only test on a single scanner. Moreover, our evaluation includes more than one etiology: HBV, HCV, and NBNC liver diseases. In terms of training, we use so-called “big-data,” which included 13 known different US scanner models and over 228000 images and 19000 US studies. This dwarfs the training set size of other works and should contribute to better model generalizability. Another important point is that most prior studies used one specific viewpoint[19,20,24] or manually defined area of interest[22,25,26], which can restrict their applicability. Like Byra et al[27], we investigated performance across different views. In addition, we investigated different viewpoints in both hepatic lobes. This advantage is most evident for patients with one resected hepatic lobe or a lobe whose space is occupied by a lesion. Furthermore, this study measured agreement across three different scanners and repeatability across different view groups and across different numbers of images per view group, which are analyses not found in other studies. Finally, we are the first to directly compare against a leading non-invasive alternative: CAP of FibroScan.
Our study has several limitations. First, we primarily used histology as the gold standard for assessing liver steatosis, which can suffer from sampling errors, processing variabilities, and intra- and inter-observer variability[48,50]. A histopathological gold standard also introduced patient selection bias. For example, CGMH histology is not required to initiate therapy in patients with HCV, whereas it is often required for patients with HBV or nonalcoholic steatohepatitis. In addition, we selected US studies completed within 3 mo of a liver histology study. If there were vigorous lifestyle changes within 3 mo, we may have a risk of deviation between fatty scores of US image and liver histology. Thus, investigating and validating against other non-invasive scores, both imaging and non-imaging based, remains an important task. Second, for assessing diagnostic performance we used retrospective data. To impose a degree of standardization, we required there to be at least 3 images in a view group and ≥ 10 images in the whole US study. Nonetheless, more controlled experimental settings may allow for more precise comparisons and follow-up prospective studies should implement an acquisition protocol to assess diagnostic performance in varied settings. In particular, the relatively poorer repeatability of the LKC view group should be investigated, and, if necessary, adjustments to the protocol should be made. Third, patients in HP-U and HP-T may also have co-occurring liver fibrosis, which likely has complex interactions with hepatic steatosis. The effect of this interaction on interpretations needs further study. Fourth, the scanner quality and model does seem to impact the DL model. Future work should better measure this impact, focusing on premium scanners not seen in the training data, which would better characterize generalizability. Fifth, the combined case numbers in HP-U and HP-T were more than 200, but the number with “complete view groups” was relatively small. Relatedly, the data collected in this study were all acquired from the CGMH institution, so any conclusions must be interpreted cautiously. Measuring performance across multiple centers, ideally using a prospective data collection protocol with larger evaluation cohorts, remains an important aspect of future work.
CONCLUSION
The DL algorithm provided a reliable quantitative steatosis assessment across view and scanners on two multi-scanner cohorts. Diagnostic performance was high with comparable or better performance than CAP. This algorithm could be applied to prospectively as well as retrospectively collected 2D US images.
ARTICLE HIGHLIGHTS
Research background
Two-dimensional (2D) ultrasound has been used for screening of liver steatosis for more than 5 decades. It is a cheap and non-invasive study.
Research motivation
Two-dimensional ultrasound is a subjective diagnosis that is not suitable for quantitative study.
Research objectives
To produce an objective steatosis diagnostic algorithm by deep learning from big data of 2D ultrasound images.
Research methods
Using multi-view ultrasound big data from a retrospective cohort of patients, we trained a deep learning algorithm to diagnose steatosis stages from clinical ultrasound diagnoses. Performance was validated on two multi-scanner unblinded and blinded histology-proven cohorts with histopathology diagnoses and a subset with FibroScan diagnoses. We also quantified reliability across scanners and viewpoints. Results were evaluated using Bland-Altman and receiver operating characteristic analysis.
Research results
The deep learning algorithm demonstrated repeatable measurements with a moderate number of images and high agreement across three premium ultrasound scanners. High diagnostic performance was observed across all viewpoints. Areas under the curve of the receiver operating characteristic to classify mild, moderate, and severe steatosis grades were 0.85, 0.91, and 0.93, respectively. This algorithm outperformed or performed at least comparably to FibroScan control attenuation parameter on the unblinded or blinded histology-proven cohort.
Research conclusions
This algorithm could give an objective diagnosis of steatosis from prospectively or retrospectively collected 2D ultrasound images.
Research perspectives
The cutoff values for different grades of steatosis would need future studies in different scanners and fibrosis status.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Institutional Review Board of the Chang Gung Medical Foundation (CGMH IRB No. 201801283B0).
Informed consent statement: Patients were not required to give informed consent to the study.
Conflict-of-interest statement: All authors declare no conflict of interest.
STROBE statement: The authors have read the STROBE Statement—checklist of items, and the manuscript was prepared and revised according to the STROBE Statement—checklist of items.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: The Gastroenterological Society of Taiwan, No. 398; American Association for the Study of Liver Diseases, No. 139352; Taiwan Association for study of Liver Disease, No. 0067; and Taiwan Medical Ultrasound Association, No. G355.
Peer-review started: December 18, 2021
First decision: January 23, 2022
Article in press: April 22, 2022
Specialty type: Radiology, nuclear medicine and medical imaging
Country/Territory of origin: Taiwan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cheng W, China; Rathnaswami A, India; Sagsoz ME, Turkey; Salvi M, Italy; Watanabe A, Japan S-Editor: Chen YL L-Editor: Filipodia P-Editor: Li X
Contributor Information
Bowen Li, Research and Development, PAII Inc., Bethesda, MD 20817, United States.
Dar-In Tai, Department of Gastroenterology and Hepatology, Chang Gung Memorial Hospital, Linkou Medical Center, Taoyuan 33305, Taiwan. tai48978@cgmh.org.tw.
Ke Yan, Research and Development, PAII Inc., Bethesda, MD 20817, United States.
Yi-Cheng Chen, Department of Gastroenterology and Hepatology, Chang Gung Memorial Hospital, Linkou Medical Center, Taoyuan 33305, Taiwan.
Cheng-Jen Chen, Department of Gastroenterology and Hepatology, Chang Gung Memorial Hospital, Linkou Medical Center, Taoyuan 33305, Taiwan.
Shiu-Feng Huang, Division of Molecular and Genomic Medicine, National Health Research Institute, Taoyuan 33305, Taiwan.
Tse-Hwa Hsu, Department of Gastroenterology and Hepatology, Chang Gung Memorial Hospital, Linkou Medical Center, Taoyuan 33305, Taiwan.
Wan-Ting Yu, Department of Gastroenterology and Hepatology, Chang Gung Memorial Hospital, Linkou Medical Center, Taoyuan 33305, Taiwan.
Jing Xiao, Research and Development, Ping An Insurance Group, Shenzhen 518001, Guangdong, China.
Lu Le, Research and Development, PAII Inc., Bethesda, MD 20817, United States.
Adam P Harrison, Research and Development, PAII Inc., Bethesda, MD 20817, United States.
Data sharing statement
No additional data are available.
References
- 1.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 2.Dietrich P, Hellerbrand C. Non-alcoholic fatty liver disease, obesity and the metabolic syndrome. Best Pract Res Clin Gastroenterol. 2014;28:637–653. doi: 10.1016/j.bpg.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Zhou F, Zhou J, Wang W, Zhang XJ, Ji YX, Zhang P, She ZG, Zhu L, Cai J, Li H. Unexpected Rapid Increase in the Burden of NAFLD in China From 2008 to 2018: A Systematic Review and Meta-Analysis. Hepatology. 2019;70:1119–1133. doi: 10.1002/hep.30702. [DOI] [PubMed] [Google Scholar]
- 4.Hernaez R, Lazo M, Bonekamp S, Kamel I, Brancati FL, Guallar E, Clark JM. Diagnostic accuracy and reliability of ultrasonography for the detection of fatty liver: a meta-analysis. Hepatology. 2011;54:1082–1090. doi: 10.1002/hep.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castera L. Non-invasive tests for liver fibrosis in NAFLD: Creating pathways between primary healthcare and liver clinics. Liver Int. 2020;40 Suppl 1:77–81. doi: 10.1111/liv.14347. [DOI] [PubMed] [Google Scholar]
- 6.Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol. 2019;16:411–428. doi: 10.1038/s41575-019-0145-7. [DOI] [PubMed] [Google Scholar]
- 7.Torres DM, Harrison SA. Diagnosis and therapy of nonalcoholic steatohepatitis. Gastroenterology. 2008;134:1682–1698. doi: 10.1053/j.gastro.2008.02.077. [DOI] [PubMed] [Google Scholar]
- 8.Singh S, Allen AM, Wang Z, Prokop LJ, Murad MH, Loomba R. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin Gastroenterol Hepatol. 2015;13:643–54.e1. doi: 10.1016/j.cgh.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupberger J, Croonenborghs T, Roca Suarez AA, Van Renne N, Jühling F, Oudot MA, Virzì A, Bandiera S, Jamey C, Meszaros G, Brumaru D, Mukherji A, Durand SC, Heydmann L, Verrier ER, El Saghire H, Hamdane N, Bartenschlager R, Fereshetian S, Ramberger E, Sinha R, Nabian M, Everaert C, Jovanovic M, Mertins P, Carr SA, Chayama K, Dali-Youcef N, Ricci R, Bardeesy NM, Fujiwara N, Gevaert O, Zeisel MB, Hoshida Y, Pochet N, Baumert TF. Combined Analysis of Metabolomes, Proteomes, and Transcriptomes of Hepatitis C Virus-Infected Cells and Liver to Identify Pathways Associated With Disease Development. Gastroenterology. 2019;157:537–551.e9. doi: 10.1053/j.gastro.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tai DI, Tsay PK, Chen WT, Chu CM, Liaw YF. Relative roles of HBsAg seroclearance and mortality in the decline of HBsAg prevalence with increasing age. Am J Gastroenterol. 2010;105:1102–1109. doi: 10.1038/ajg.2009.669. [DOI] [PubMed] [Google Scholar]
- 11.Salvi M, Molinaro L, Metovic J, Patrono D, Romagnoli R, Papotti M, Molinari F. Fully automated quantitative assessment of hepatic steatosis in liver transplants. Comput Biol Med. 2020;123:103836. doi: 10.1016/j.compbiomed.2020.103836. [DOI] [PubMed] [Google Scholar]
- 12.Munsterman ID, van Erp M, Weijers G, Bronkhorst C, de Korte CL, Drenth JPH, van der Laak JAWM, Tjwa ETTL. A Novel Automatic Digital Algorithm that Accurately Quantifies Steatosis in NAFLD on Histopathological Whole-Slide Images. Cytometry B Clin Cytom. 2019;96:521–528. doi: 10.1002/cyto.b.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferraioli G, Soares Monteiro LB. Ultrasound-based techniques for the diagnosis of liver steatosis. World J Gastroenterol. 2019;25:6053–6062. doi: 10.3748/wjg.v25.i40.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caussy C, Reeder SB, Sirlin CB, Loomba R. Noninvasive, Quantitative Assessment of Liver Fat by MRI-PDFF as an Endpoint in NASH Trials. Hepatology. 2018;68:763–772. doi: 10.1002/hep.29797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castera L, Friedrich-Rust M, Loomba R. Noninvasive Assessment of Liver Disease in Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2019;156:1264–1281.e4. doi: 10.1053/j.gastro.2018.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liaw YF, Tai DI, Chu CM, Lin DY, Sheen IS, Chen TJ, Pao CC. Early detection of hepatocellular carcinoma in patients with chronic type B hepatitis. A prospective study. Gastroenterology. 1986;90:263–267. doi: 10.1016/0016-5085(86)90919-4. [DOI] [PubMed] [Google Scholar]
- 17.Cengiz M, Sentürk S, Cetin B, Bayrak AH, Bilek SU. Sonographic assessment of fatty liver: intraobserver and interobserver variability. Int J Clin Exp Med. 2014;7:5453–5460. [PMC free article] [PubMed] [Google Scholar]
- 18.Hong CW, Marsh A, Wolfson T, Paige J, Dekhordy SF, Schlein AN, Housman E, Deiranieh LH, Li CQ, Wasnik AP, Jang HJ, Dietrich CF, Piscaglia F, Casola G, O'Boyle M, Richman KM, Valasek MA, Andre M, Loomba R, Sirlin CB. Reader agreement and accuracy of ultrasound features for hepatic steatosis. Abdom Radiol (NY) 2019;44:54–64. doi: 10.1007/s00261-018-1683-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byra M, Styczynski G, Szmigielski C, Kalinowski P, Michałowski Ł, Paluszkiewicz R, Ziarkiewicz-Wróblewska B, Zieniewicz K, Sobieraj P, Nowicki A. Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int J Comput Assist Radiol Surg. 2018;13:1895–1903. doi: 10.1007/s11548-018-1843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biswas M, Kuppili V, Edla DR, Suri HS, Saba L, Marinhoe RT, Sanches JM, Suri JS. Symtosis: A liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm. Comput Methods Programs Biomed. 2018;155:165–177. doi: 10.1016/j.cmpb.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Acharya UR, Sree SV, Ribeiro R, Krishnamurthi G, Marinho RT, Sanches J, Suri JS. Data mining framework for fatty liver disease classification in ultrasound: a hybrid feature extraction paradigm. Med Phys. 2012;39:4255–4264. doi: 10.1118/1.4725759. [DOI] [PubMed] [Google Scholar]
- 22.Gummadi S. Automated Machine Learning in the Sonographic Diagnosis of Non-alcoholic Fatty Liver Disease. AUDT 2020; 4: 176-182. [Google Scholar]
- 23.Owjimehr M, Danyali H, Helfroush MS, Shakibafard A. Staging of Fatty Liver Diseases Based on Hierarchical Classification and Feature Fusion for Back-Scan-Converted Ultrasound Images. Ultrason Imaging. 2017;39:79–95. doi: 10.1177/0161734616649153. [DOI] [PubMed] [Google Scholar]
- 24.Han A, Byra M, Heba E, Andre MP, Erdman JW Jr, Loomba R, Sirlin CB, O'Brien WD Jr. Noninvasive Diagnosis of Nonalcoholic Fatty Liver Disease and Quantification of Liver Fat with Radiofrequency Ultrasound Data Using One-dimensional Convolutional Neural Networks. Radiology. 2020;295:342–350. doi: 10.1148/radiol.2020191160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JR, Chao YP, Tsai YW, Chan HJ, Wan YL, Tai DI, Tsui PH. Clinical Value of Information Entropy Compared with Deep Learning for Ultrasound Grading of Hepatic Steatosis. Entropy (Basel) 2020;22 doi: 10.3390/e22091006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao W, An X, Cong L, Lyu C, Zhou Q, Guo R. Application of Deep Learning in Quantitative Analysis of 2-Dimensional Ultrasound Imaging of Nonalcoholic Fatty Liver Disease. J Ultrasound Med. 2020;39:51–59. doi: 10.1002/jum.15070. [DOI] [PubMed] [Google Scholar]
- 27.Byra M, Han A, Boehringer AS, Zhang YN, O'Brien WD Jr, Erdman JW Jr, Loomba R, Sirlin CB, Andre M. Liver Fat Assessment in Multiview Sonography Using Transfer Learning With Convolutional Neural Networks. J Ultrasound Med. 2022;41:175–184. doi: 10.1002/jum.15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saadeh S, Younossi ZM, Remer EM, Gramlich T, Ong JP, Hurley M, Mullen KD, Cooper JN, Sheridan MJ. The utility of radiological imaging in nonalcoholic fatty liver disease. Gastroenterology. 2002;123:745–750. doi: 10.1053/gast.2002.35354. [DOI] [PubMed] [Google Scholar]
- 29.Chen CJ, Tsay PK, Huang SF, Tsui PH, Yu WT, Hsu TH, Tai J, Tai DI. Effects of hepatic steatosis on non-invasive liver fibrosis measurements between hepatitis B and other etiologies. APPL SCI-BASEL. 2019;9:1961. [Google Scholar]
- 30.Lee CH, Wan YL, Hsu TH, Huang SF, Yu MC, Lee WC, Tsui PH, Chen YC, Lin CY, Tai DI. Interpretation US Elastography in Chronic Hepatitis B with or without Anti-HBV Therapy. APPL SCI-BASEL. 2017;7:1164. [Google Scholar]
- 31.Hsu TH, Tsui PH, Yu WT, Huang SF, Tai J, Wan YL, Tai DI. Cutoff Values of Acoustic Radiation Force Impulse Two-Location Measurements in Different Etiologies of Liver Fibrosis. J Med Ultrasound. 2019;27:130–134. doi: 10.4103/JMU.JMU_7_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai DI, Tsay PK, Jeng WJ, Weng CC, Huang SF, Huang CH, Lin SM, Chiu CT, Chen WT, Wan YL. Differences in liver fibrosis between patients with chronic hepatitis B and C: evaluation by acoustic radiation force impulse measurements at 2 locations. J Ultrasound Med. 2015;34:813–821. doi: 10.7863/ultra.34.5.813. [DOI] [PubMed] [Google Scholar]
- 33.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ Nonalcoholic Steatohepatitis Clinical Research Network. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 34.He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016: 770-778. [Google Scholar]
- 35.Scatarige JC, Scott WW, Donovan PJ, Siegelman SS, Sanders RC. Fatty infiltration of the liver: ultrasonographic and computed tomographic correlation. J Ultrasound Med. 1984;3:9–14. doi: 10.7863/jum.1984.3.1.9. [DOI] [PubMed] [Google Scholar]
- 36.Frank E, Hall M. A Simple Approach to Ordinal Classification. In: De Raedt L, Flach P, editors. Machine Learning: ECML 2001. Berlin, Heidelberg: Springer, 2001: 145-156. [Google Scholar]
- 37.Fürnkranz J, Hüllermeier E, Vanderlooy S. Binary Decomposition Methods for Multipartite Ranking. In: Buntine W, Grobelnik M, Mladenić D, Shawe-Taylor J, editors. Machine Learning and Knowledge Discovery in Databases. Berlin, Heidelberg: Springer, 2009: 359-374. [Google Scholar]
- 38.Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scand Actuar J . 1979;6:65–70. [Google Scholar]
- 39.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 40.Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8:135–160. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- 41.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Boca Raton, Florida, USA: CRC Press. [Google Scholar]
- 42.Higgins JJ. An Introduction to Modern Nonparametric Statistics. Pacific Grove, CA: Brooks/Cole, 2005: 101-104. [Google Scholar]
- 43.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 44.YOUDEN WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 45.Vergara IA, Norambuena T, Ferrada E, Slater AW, Melo F. StAR: a simple tool for the statistical comparison of ROC curves. BMC Bioinformatics. 2008;9:265. doi: 10.1186/1471-2105-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tada T, Iijima H, Kobayashi N, Yoshida M, Nishimura T, Kumada T, Kondo R, Yano H, Kage M, Nakano C, Aoki T, Aizawa N, Ikeda N, Takashima T, Yuri Y, Ishii N, Hasegawa K, Takata R, Yoh K, Sakai Y, Nishikawa H, Iwata Y, Enomoto H, Hirota S, Fujimoto J, Nishiguchi S. Usefulness of Attenuation Imaging with an Ultrasound Scanner for the Evaluation of Hepatic Steatosis. Ultrasound Med Biol. 2019;45:2679–2687. doi: 10.1016/j.ultrasmedbio.2019.05.033. [DOI] [PubMed] [Google Scholar]
- 47.St Pierre TG, House MJ, Bangma SJ, Pang W, Bathgate A, Gan EK, Ayonrinde OT, Bhathal PS, Clouston A, Olynyk JK, Adams LA. Stereological Analysis of Liver Biopsy Histology Sections as a Reference Standard for Validating Non-Invasive Liver Fat Fraction Measurements by MRI. PLoS One. 2016;11:e0160789. doi: 10.1371/journal.pone.0160789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El-Badry AM, Breitenstein S, Jochum W, Washington K, Paradis V, Rubbia-Brandt L, Puhan MA, Slankamenac K, Graf R, Clavien PA. Assessment of hepatic steatosis by expert pathologists: the end of a gold standard. Ann Surg. 2009;250:691–697. doi: 10.1097/SLA.0b013e3181bcd6dd. [DOI] [PubMed] [Google Scholar]
- 49.Paige JS, Bernstein GS, Heba E, Costa EAC, Fereirra M, Wolfson T, Gamst AC, Valasek MA, Lin GY, Han A, Erdman JW Jr, O'Brien WD Jr, Andre MP, Loomba R, Sirlin CB. A Pilot Comparative Study of Quantitative Ultrasound, Conventional Ultrasound, and MRI for Predicting Histology-Determined Steatosis Grade in Adult Nonalcoholic Fatty Liver Disease. AJR Am J Roentgenol. 2017;208:W168–W177. doi: 10.2214/AJR.16.16726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ratziu V, Charlotte F, Heurtier A, Gombert S, Giral P, Bruckert E, Grimaldi A, Capron F, Poynard T LIDO Study Group. Sampling variability of liver biopsy in nonalcoholic fatty liver disease. Gastroenterology. 2005;128:1898–1906. doi: 10.1053/j.gastro.2005.03.084. [DOI] [PubMed] [Google Scholar]
- 51.DataBase Center for Life Science. File:201405_liver.png. [cited 2 February 2022]. Available from: https://commons.wikimedia.org/wiki/File:201405_liver.png .
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.