Summary:
We show that myriad, disparate mechanisms that diversify genomes and transcriptomes can be captured by a unifying principle: sample-dependent sequence variation. This variation occurs in both RNA and DNA and functions to regulate transcript expression and adaptation. Using this insight, we develop a novel highly efficient algorithm – NOMAD – that performs inference on raw reads without any genomic reference or sample metadata. NOMAD unifies data-scientifically driven discovery with previously unattainable speed and generality. Examples include SARS-CoV-2, humans, and non-model animals and plants with both bulk and single cell RNA-sequencing data. A snapshot of its novel discoveries include missing variants in SARS-CoV-2, gene regulation in diatoms epiphytic to eelgrass, an oceanic plant critical to the carbon cycle and significantly impacted by climate change, and in octopus where it identifies isoform regulation in genes missing from the reference. NOMAD is a new unifying approach to sequence analysis that enables expansive discovery.
One-sentence summary:
We present a unifying, reference-free formulation of disparate genomic problems bypassing reference genomes.
Introduction
Sequence variation – mutation, reassortment or rearrangement of nucleic acids – is fundamental to regulating gene expression, and to evolution and adaptation across the tree of life. Consider the sequence composition of two recently diverged genomes, such as the emergence of two viral strains. In a typical example, the vast majority of the two emergent strains’ genomic sequence is shared, and the strains only differ in sequence at a few strain-defining locations. These sequence differences could be point mutations, small or large insertions or deletions, or other rearrangements, including those as a consequence of mobile elements. This toy example illustrates a fundamental feature of a set of genomes that have shared ancestral genetic material and have since undergone selection. Their genomes have regions of sample-dependent sequence diversification: a shared identical sequence neighbors one that is diverse between samples. This feature characterizes genomes in much more generality than two viral strains. It includes any number of polymorphic members of a population including where genetic variation associated with disease or phenotype, mobile elements themselves (Abante, Wang and Salzman, 2022) or their genomic targets, loci that are rearranged or hypermutated (Medhekar and Miller, 2007) within an organism such as V(D)J recombination and somatic hypermutation, among many others.
Now consider the sequence composition of an alternatively spliced RNA transcript with a cassette exon, exon 2, that is differentially included in different cells (samples). Again, these transcripts have the property that the sequence of exon 1 will be a constant common sequence to all transcripts, but the sequence composition downstream of it will differ depending on if exon 2 is skipped: if it is, the sequence downstream will represent exon 3, and if not, exon 2. Thus, the set of sequences of these transcripts fit within the sequence signal that NOMAD will detect.
Again, this feature of a constant sequence juxtaposed next to one with a sequence composition that is sample-dependent characterizes a regulated transcriptome more generally, and can detect any sequences sample-specifically impacted by RNA editing, RNA splicing, alternative polyadenylation or allele-specific expression. Moreover, many other genomic problems across different ‘omic measurement designs can be mapped to a feature NOMAD will detect: one where a constant sequence flanks a set of variable sequences with sequence-dependent biological function (see Supplement).
Mapping the detection of disparate biological problems to a common model is both conceptually appealing but also allows us to design a single algorithm that solves disparate genomic problems including those described above, called NOMAD. NOMAD 1) provides a single unifying solution to disparate problems in genomics; 2) enables statistical inference for sequences of candidate biological function that bypasses reference genomes completely, operating directly on raw reads; 3) prioritizes and identifies biologically regulated sequences enabling biologists to focus on sequences with differential regulation across samples. Here we explain the concepts behind NOMAD and apply them to biological examples where reference genomes are well curated and domains where they are not. In both cases, NOMAD expands the scope of inference possible today and promises to be of broad use for genomic discovery in most domains where it is applied. Practitioners of genomics are familiar with how time consuming, and many times ad hoc, these procedures can be. Three main problems with the existing paradigm in genomics are highlighted below.
The first problem is fundamental: specifying and relying on a reference genome can be highly limiting for biological inference, and generating and or aligning to a reference is foundational in the field of genomics today. Alignment-based methods struggle when sequences can map to multiple or repetitive locations, and or reference genomes are incorrect or incomplete (Shi et al., 2022; Zhao, Shi and Pollard, 2022). These regions comprise ~54 % of the human genome (Nurk et al., 2022) and are sometimes among the most important to analyze. This means users must supervise running and make subjective interpretation at many steps, including subjective decisions about which statistical tests to perform on the output: eg. whether to run a statistical test for detecting SNPs, or alternative splicing, or some other test. This creates several major impediments to discovery.
In human genomics, workflows beginning with reference alignment miss important variation absent from assemblies, even with pangenomic approaches. We call methods that begin with alignment, “reference-first”. It is well-known that genetic variants associated with ancestries of under-studied populations are poorly represented in databases (Sherman et al., 2019), resulting in health disparities (West, Blacksher and Burke, 2017). In disease genomics (Wang et al., 2022), sequences of pathogenic cells may be missing from reference genomes, or no reference genome exists, as in genomically unstable tumors (C.-Z. Zhang et al., 2015). Many species do not have reference genomes even when they in principle could be attained due to logistical and computational overheads. In these situations, alignment-first approaches fail. Practitioners of human genomics can see more detailed and further examples of the insights from NOMAD’s application to human single cell RNA-seq data as well as its improved precision compared to state of the art algorithms (Dehghannasiri et al., 2022).
In viral surveillance, reference-first approaches are even more problematic (The Nucleic Acid Observatory Consortium, 2021). Viral reference genomes cannot capture the complexity of viral quasispecies (Kirkegaard, van Buuren and Mateo, 2016) or the vast extent of polymorphism (Kim et al., 2020). New viral assemblies are constantly being added to reference databases (Edgar et al., 2022; Zayed et al., 2022). In the microbial world, pre-specifying a set of reference genomes is infeasible due to its inherent rapid genomic changes. References also cannot capture insertional diversity of mobile elements, which have significant phenotypic and clinical impact (Evans et al., 2020) and are only partially cataloged in references (Wright, Comeau and Langille, 2022). Finally, reliance on mapping is also statistically problematic as it reduces raw, observed sequence data to a set of positions in the genome, and introduces errors both due to intrinsically randomized procedure of modern mappers (Langmead et al., 2009; Dobin et al., 2013) and at the very least, and computational procedure introduces some uncertainty.
A second issue with the existing paradigm in genomics is that the practitioner must choose a specific bioinformatic procedure: in an RNA-seq experiment, is the analyst interested in V(D)J recombination, RNA editing or splicing (or others)? No current single algorithm can detect all events. Consider transposable elements, known to drive evolution and cause an unascertained number of human diseases (Pascarella et al., 2022). A custom and multi-step workflow is required to detect these insertions because they are highly polymorphic, and the algorithm must address the issues of multi-mapping and the high degree of repetitive sequence (Shi et al., 2022; Zhao, Shi and Pollard, 2022). Similarly, if V(D)J recombination is present in a sample, but a custom workflow is not specified to detect these rearrangements, they will not be reported. Further examples abound in plant and microbial genomics (Michael and VanBuren, 2020).
A final issue with genomic algorithms today is that the statistical inference they provide is based on summaries generated from alignment, which necessarily introduce noise. Technically, statistical inference is conditional on aligners, resulting in poorly calibrated p-values. More than a purely statistical issue, this problem can significantly impact biological discovery. Because statistical analysis in reference-first approaches is conditional on the output of the noisy alignment step, complicated null distributions for test statistics must be derived, which is sometimes infeasible in closed-form. It is then difficult and in some cases impossible (Chung and Romano, 2013) to provide valid statistical significance levels in downstream analyses, such as in differential expression analysis. When null distributions are not available, computationally intensive resampling is required, increasing the computational burden by orders of magnitude. Even when employed, these procedures report discoveries based on a nominal FDR, typically based on permutation distributions, there is no guarantee that these values are accurate. In fact have been repeatedly shown to be wildly inaccurate, sometimes yielding an FDR of .5 when an FDR of .05 was desired (Romano, Sesia and Candès, 2019). Together, these three points, along with the extensive manual time modern bioinformatics consumes, there is a convincing argument to attempt to develop an alternative to the way genomics is approached today.
NOMAD makes important inroads in overcoming these three significant problems in genomics. Moreover, it presents a unified way to address one of the foremost challenges for biological discovery in genomics: to identify sample-dependent sequence variation, the variation most likely to be regulated and functional. NOMAD both recovers such true positive variation in diverse biological contexts, from viral genomes to human transcripts, and discovers new biology. We illustrate a downstream computational procedure for interpretation and novel discovery on NOMAD’s outputs in a set of selected examples ranging from highly scrutinized (SARS-CoV-2 and human) to studies eelgrass, an ocean-dwelling plant of great importance to carbon cycling which have comparatively little genomic study (Jueterbock et al., 2021), and of Octopus bimaculoides, which has a recently assembled genome where we use NOMAD to illuminate transcriptional regulation missing from the reference. As a snapshot of these new discoveries, NOMAD predicts un-annotated genomic variants in the well-studied delta and Omicron strains of SARS-CoV-2, uncovers novel cell-type regulation of gene paralogs and cell-type specific expression of alleles in the major histocompatibility locus. In mouse lemur, NOMAD discovers T cell receptor rearrangements impossible to identify with other tools. Finally, we illustrate further NOMAD’s novel regulatory biological discovery using reads with no partial mapping to reference genomes in Octopus bimaculoides and the eelgrass Zostera marina. While we present specific examples in this paper, NOMAD’s approach is general and should be of wide interest for theorists and practitioners across biological disciplines.
NOMAD is a statistics-first approach to identify sample-dependent sequence variation
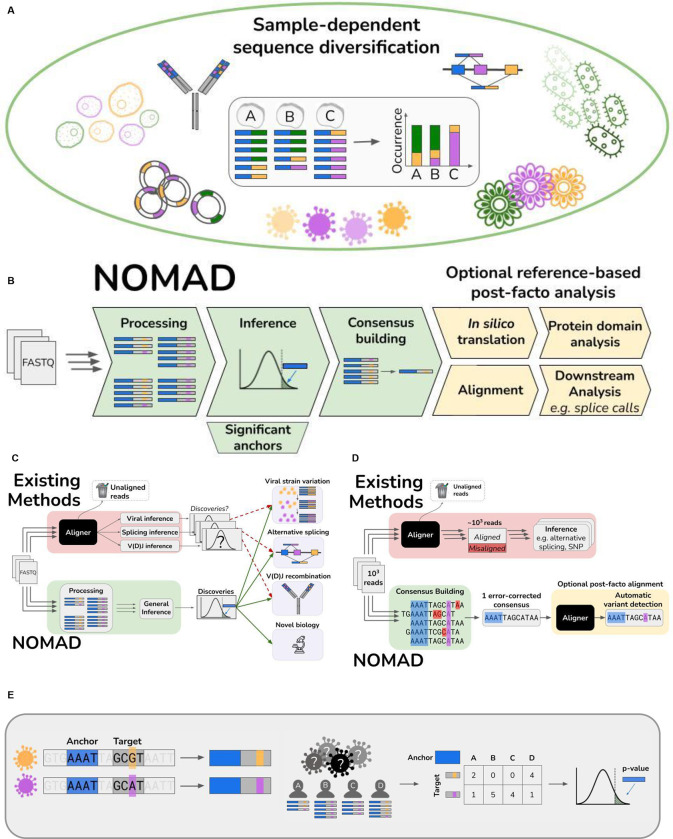
We first show that multitudes of genomic analyses reduce to detecting sample-dependent sequence variation, and admit natural probabilistic formulations. History suggests that formulating scientific problems mathematically can radically improve the precision of inference, discovery and computational efficiency (ref. eg population genetics or compressive sensing) (Donnelly and Tavaré, 1995; Candes and Wakin, 2008). Thus, approaching genomics through this fundamental probabilistic formulation is naturally expected to yield more general, efficient and powerful solutions. Here, we leverage the probabilistic formulation above to derive a statistical test on raw sequencing read data (e.g. FASTQ files) to discover sample-dependent sequence variation completely bypassing references or ontology. We implement the test in a highly efficient algorithmic workflow, providing novel discovery across disparate biological disciplines (Fig. 1B,C).
Figure 1.
A. Generality of NOMAD model. The study of sample-dependent sequence variation unifies problems in disparate areas of genomics which are currently studied with application-specific models and algorithms. Viral genome mutations, alternative splicing, and V(D)J recombination all fit under this framework, where sequence variation depends on the sample (through cell-type or infection strain type). Myriad problems in plant genomics, metagenomics and biological adaptation are subsumed by this framework.
B. Overview of NOMAD pipeline. NOMAD takes as input raw FASTQ files for any number of samples >1 and processes them in parallel, counting (anchor, target) pairs per sample. NOMAD performs inference on these aggregated counts, outputting statistically significant anchors. For each significant anchor, a denoised per-sample consensus sequence is built (Fig. 1D). NOMAD also enables optional reference-based post-facto analysis. If a reference genome is available, NOMAD can align the consensus sequences to the reference, enabling denoised downstream analysis (e.g. SNPs, indels, or splice calls). In silico translation of consensuses can optionally be used to study relationships of anchors to protein domains by mapping to databases such as Pfam (Methods).
C. Overview of NOMAD (green) versus existing workflows (red). Existing workflows (red) discard low-quality reads during FASTQ processing and alignment, only then performing statistical testing after algorithmic bias is introduced; p-values are then not unconditionally valid. Further, for every desired inferential task, a different inference pipeline must be used. NOMAD (green) performs direct statistical inference on raw FASTQ reads, bypassing alignment and enabling data-scientifically driven discovery. Due to its generality, NOMAD can simultaneously detect myriad biological examples of sample-dependent sequence variation.
D. NOMAD consensus building. NOMAD constructs a per-sample consensus sequence for every significant anchor by taking all reads in which the anchor (blue) appears, and recording plurality votes for each nucleotide, denoising reads while preserving the true variant; sequencing errors in red and biological mutations in purple. Existing approaches require alignment of all reads to a reference prior to error correction, requiring orders of magnitude more computation, discarding reads in both processing and alignment, and potentially making erroneous alignments due to sequencing error. They further require inferential steps, e.g. to detect if there is a SNP or alternatively spliced variant.
E. Construction of NOMAD anchor, target pairs. A stylized expository example of viral surveillance: 4 individuals A-D are infected with one of two variants (orange and purple), differing by a single basepair (orange and purple). NOMAD anchor k-mers are blue (k=4), followed by a lookahead distance of L=2, and the corresponding k-mer targets. Given sequencing reads from the 4 individuals as shown, NOMAD generates a target by sample contingency table for this blue anchor, and computes a p-value to test if this anchor has sample-dependent sequence diversity.
To begin, we define an “anchor” k-mer in a read, and define an anchor to have sample-dependent diversity if the distribution of k-mers starting R base pairs downstream of it (called “targets”) depends on the sample (Fig. 1E). This definition can be generalized to different constructions of anchors and targets, and can be applied to diverse data sources including DNA, RNA and protein sequence. The length of the anchor and target (k) are general. For intuition, suppose k is long enough an anchor of length k is found in a unique position in a genome, though this is not necessary and in fact sometimes not desired. In this case, suppose the anchor is found in a sequence adjacent to one which has a strain-specific mutation (Fig. 1C). Then, target counts following this anchor quantify the sequence variants, up to errors in sampling such as introduced in library preparation or sequencing. NOMAD’s null states that each sample has the same distribution of variants immediately following this anchor. This framework detects sample-specific sequence variation including a broad range of biological features from mutations or indels (an anchor immediately preceding these variants, as in Fig. 1C), differentially regulated alternative splicing (an anchor in the constitutive exon), V(D)J recombination, RNA editing and many other features, for example CRISPR repeats (Supplement, work in preparation). While anchors were chosen of length k=27 for biological inference in this manuscript, genomic inference is similar under a range of parameter choices (Methods, Kokot et al, in preparation).
The above is a unifying formulation that allows expansive numbers problems to be studied (Supplement) and allows us to develop a novel statistics-first approach, NOMAD (Novel multi-Omics Massive-scale Analysis and Discovery), that is reference-free and operates directly on raw sequencing data. It is an extremely computationally efficient algorithm to detect sample-dependent sequence variation, through the use of a novel statistic of independent interest that provides closed form p-values, meaning no permutation, resampling or numerical solutions to complex likelihoods are required for significance tests (Methods). Arguably, the most important reason NOMAD can reveal new biology is that all sequences predicted to have sample-dependent diversity are called without access to any reference and annotation information, though they can be optionally used for post-facto interpretation. This key property makes NOMAD fundamentally different from existing methods which critically rely on reference mapping prior to statistical inference. To illustrate, as a special case NOMAD automatically detects differential isoform expression. In this domain, the closest approach to NOMAD is Kallisto (Bray et al., 2016). However, Kallisto still requires a reference transcriptome and statistical resampling for inference, and is further challenged to provide accurate expression estimates for more than a handful of paralogous genes and isoforms due to model limitations, specifically parameter identifiability. Further, unlike NOMAD, Kallisto cannot discover spliced isoforms de novo.
Summary of NOMAD’s technical innovation
NOMAD is by default an unsupervised algorithm that does not require any sample identity metadata, though user-defined metadata can be optionally used (Methods). Significance testing is detailed in the Methods. At a high level, NOMAD tests whether sequences have evidence of sample-dependent sequence variation: an anchor is called significant if our statistical test rejects the hypothesis that for a sequence a, the conditional distribution of observing a target sequence t a distance R downstream of a is sample-independent (Fig. 1E). Significance testing is performed by finding approximate best splits of the samples into two groups (Methods). If a user desires, the test can be restricted to user-defined groups. Anchors are reported with an effect size taking values in [0,1]. This value measures how well partitioned target sequences are between sample groups: 0 if the groups have no difference in target distributions and increasing to 1 when the target distributions of the two groups are disjoint. NOMAD has significant power in detecting two-group type deviations, e.g. mutations between one group of Omicron samples and one group of Delta samples, but also has substantial power against more complex alternatives that involve combinatorial expression, as we show with detection of V(D)J recombination. The statistical basis for this power is discussed in greater detail in a separate statistical work (Baharav, Tse and Salzman, 2023).
NOMAD has multiple major technical innovations: 1) a parallelizable, fully containerized, and computationally efficient approach to parse FASTQ files into tables that summarize joint kmer composition, 2) a novel statistical analysis of the derived tables, using concentration inequalities to derive closed form p-values, 3) a micro-assembly-based consensus sequence representing the dominant error-corrected sequence, similar to (Magoč and Salzberg, 2011; Motahari et al., 2013; Edgar, 2016), downstream of the anchor for post-facto interpretation and identification of SNPs, indels or isoforms, to name a few (Fig. 1D). If post anchor-identification inference is desired, the consensus, rather than the raw reads, is aligned. This reduces the number of sequences to align by ~1000x in real data.
NOMAD’s novel approach yields an extremely computationally efficient implementation without code optimization. We ran NOMAD on a 2015 Intel laptop with an Intel(R) Core(TM) i7–6500U CPU @ 2.50GHz processor, generating significance calls for single cell RNA-seq totaling over 10 million reads in only 1 hour 45 min. When performed on a compute cluster, the same analysis is completed in an average of 22.8 minutes with 750 MB of memory for 10 million reads. NOMAD provides a dramatic speed up over existing methods for de novo splicing detection and significance calls because it reduces the number of reads needed for optional alignment by orders of magnitude. Further, NOMAD bypasses resampling-based approaches for significance testing, such as permutation testing, which create further computational overhead.
NOMAD discovers sequence variation in proteins at the host-viral interface without a genomic reference
We first show that NOMAD automatically detects viral strain evolution without any reference genome, or knowledge of the sample origin. Existing approaches are computationally-intensive, require genome assemblies and rely on heuristics. It is still difficult to call strain variation with them, and impossible to know how many strains are currently missed, though estimates suggest orders of magnitude remain to be discovered (Edgar et al., 2022).
Yet, emergent viral threats or variants of concern, e.g. within SARS-CoV-2, will necessarily be absent from reference databases. Because virus’ genomes are under selective pressure to diversify when infecting a host, NOMAD should prioritize anchors near genome sequences that are under selection, in theory, based purely on their statistical features: sequences flanking variants that distinguish strains have consistent sample-dependent sequence diversity. When NOMAD is run on patients infected with varying relative titers of Omicron and Delta strains of SARS-CoV-2, significant anchors are expected to be called adjacent to strain-specific mutations (Fig. 2A); they should be, and as we show are, discoverable without any knowledge of a reference. NOMAD rediscovers known variants without a reference genome, overcoming significant challenges in the field of viral genomics.
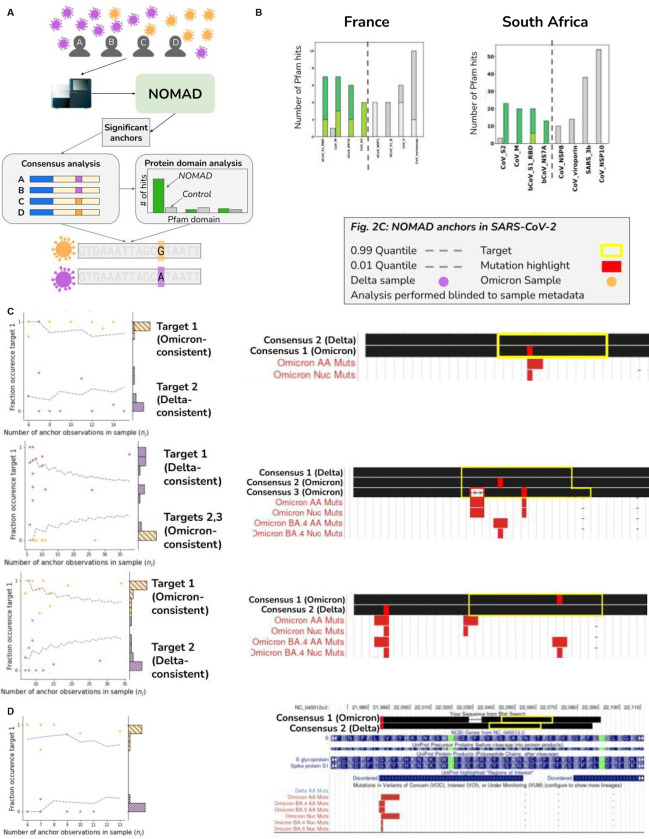
Figure 2.
A. Stylized example representing NOMAD workflow for viral strain identification. Patients with varying viral strains are sampled; two representative strains with differentiating mutations are depicted in orange and purple. NOMAD is run on raw FASTQs generated from sequencing patient samples. Significant anchors are called without a reference genome or clinical metadata. Optional post-facto analysis quantifies domain enrichment via in silico translation of consensus sequences derived from NOMAD-called anchors versus controls. Consensuses can also be used to call variants de novo and can be compared to annotated variants e.g. in SARS-CoV-2, Omicron.
B. NOMAD SARS-CoV-2 protein profile hits (anchor effect size >.5) to the Pfam database (greens) and control (greys) for France and South Africa datasets; ordered by enrichment in NOMAD hits compared to control showing large distributional differences (chi-squared test p-values France: < 3.7E-7, SA: <2.5E-39). Spike protein domains are highly enriched in NOMAD calls versus controls. In the France data, the most NOMAD-enriched domain is the betacoronavirus S1 receptor binding domain (hypergeometric p=2.3E-2, corrected) followed by Orf7A (hypergeometric p=6.4E-2, corrected), known to directly interact with the host innate immune defense. In the South Africa data, the most enriched NOMAD profiles are CoV S2 (p=2.9E-6) and the coronavirus membrane protein (p=8.4E-8).
C. Examples of NOMAD-detected anchors in SARS-CoV-2 (France data) classified as strain-defining. Scatterplots (left) show the fraction of each sample’s observed fraction of target 1 (the most abundant target) for three representative anchors, binomial confidence intervals: (.01,.99), p=empirical fraction occurrence of target 1 (Supplement). y-axis shows histogram of the fraction occurrence of target 1. Mutations (right) found in the targets are highlighted in purple, BLAT shows single nucleotide mutations match known Omicron mutations. Binomial p-values of 1.6E-21, 1.0E-13, and 1.8E-16 respectively (Methods). These uncorrected p-values, and are not used for any significance testing, but rather to quantify the visually separated nature of the plots. The anchor in (top) maps to the coronavirus membrane protein; anchors in (middle and bottom) map to the spike protein. One sample (out of 26) depicted in the bottom plot has a consensus mapping perfectly to the Wuhan reference; 3 other consensuses contain annotated Omicron mutations, some designated as VOC in May of 2022, 3 months after these samples were collected.
D. Example of a NOMAD-detected anchor in SARS-CoV-2 that is not classified as strain-defining. Same format as Fig. 2C, showing a 6 basepair deletion in the spike protein that only occurs in patients infected with Omicron. Binomial p-value of 1.3E-19. The highlighted deletion is not annotated as a VOC yet it is present only in samples classified as Omicron.
To test this prediction, we analyzed oropharyngeal swabs from patients with SARS-CoV-2 from 2021–12-6 to 2022–2-27 in France, a period of known Omicron-Delta coinfection (Bal et al., 2022). We ran NOMAD and analyzed anchors with high degree of target partitioning by sample measured by a per-anchor quantity we call effect size: high effect sizes are predicted if samples can be approximately partitioned by strain, although results are similar without this threshold (here, effect size>0.5 was used). NOMAD’s statistical test identifies biologically significant anchors, as shown by comparing NOMAD’s significant anchors against a set of control anchors, which are generated as the most abundant anchor sequences (Methods). We perform protein domain analysis independent of any reference genome to illustrate reference-genome-free interpretation of NOMAD’s calls. For each anchor, we assigned a protein domain label based on in silico translation of its consensus sequence (Mistry et al., 2021). The protein domain with the best mapping to the Pfam database is assigned to the anchor, producing a set of “protein profiles” (Methods, Supplementary Table 1). NOMAD protein profiles are significantly different from controls (p=3.7E-7, chi-squared test, Fig. 2B). The most NOMAD-enriched domains are the receptor binding domain of the betacoronavirus spike glycoprotein (7 NOMAD vs 0 control hits, p=2.3E-2 hypergeometric p-value, corrected) and Orf7A a transmembrane protein that inhibits host antiviral response (6 NOMAD vs 0 control hits, p=6.4E-2 hypergeometric p-value, corrected). All analysis is blind to the data origin (SARS-CoV-2).
We further analyzed patient samples from the original South African genomic surveillance study that identified the Omicron strain during the period 2021–11-14 to 2021–11-23 (Viana et al., 2022), again without metadata or reference genomes and directly on input FASTQ files (Methods). NOMAD protein profiles were significantly different from controls (chi-squared test, p=2.5E-39, Fig. 2B). NOMAD-enriched domains in France and South African are highly consistent: the top four domains in both datasets are permutations of each other. The most NOMAD-enriched domains versus controls were the betacoronavirus S2 subunit of the spike protein involved in eliciting the human antibody response (Poh et al., 2020) (23 NOMAD vs 2 control hits, p=2.9E-6 hypergeometric p-value, corrected), the matrix glycoprotein which interacts with the spike (20 NOMAD vs 0 control hits, p=8.4E-8 hypergeometric p-value, corrected), and the receptor binding domain of the spike protein to which human antibodies have been detected (Jörrißen et al., 2021) (20 NOMAD vs 0 control hits, p=8.4E-8 hypergeometric p-value, corrected). All domains are biologically predicted to be under strong selective pressure, and thus exhibit sample-specific sequence diversity; NOMAD discovers this de novo.
We aligned NOMAD anchors with high effect size (Supplement) to the Wuhan, Delta, and Omicron BA.1 and BA.2 reference strains to test if NOMAD anchors rediscovered known strain-defining mutations. We defined an anchor to recover a strain defining mutation if any of its 2 targets (>5% of total anchor counts) mapped to different strains (Supplement). NOMAD anchors are significantly enriched for being mutation-consistent: in the French data, 252 anchors are significant and have effect size >.5. Of the 83 anchors that also map to a reference and have a target mapping to a reference, 69.9% (58/83) of NOMAD’s calls were strain defining versus 5.6% (14/252) in the control (p=1.3E-31, hypergeometric test). Anchors that are not classified as strain-defining include examples of unannotated mutations (Fig). For the South African data, 201 anchors are significant and have high effect size. 26% (34/130) of NOMAD’s called anchors were mutation consistent vs 3.0% (6/201) for the control (p=3.1E-10, hypergeometric test). This is due to the low prevalence of Omicron in the dataset, where we see that further restricting the set of anchors to those occurring in at least 5 samples, 93% (14/15) of the anchors are strain-defining.
Examples of strain-defining anchors detected by NOMAD are presented in Fig. 2 where metadata is used post-facto to visualize sample expression of each strain-defining target. Examples include anchors in the membrane (Fig. 2C, top) and spike protein (Fig. 2C, middle and bottom). Differences between consensus sequences and Wuhan reference illustrate NOMAD’s unsupervised rediscovery of annotated strain-specific variation, including co-detection of a deletion and a mutation (Fig. 2C). While the Wuhan reference genome was used for post-facto interpretation, for emphasis: no alignment or sample metadata was used to generate NOMAD’s calls, only to interpret them (Fig. 2). NOMAD consensuses also extend discovery (Fig. 2C), identifying strain-specific variation beyond the target: both are annotated Omicron variants (Fig. 2C). NOMAD’s statistical approach automatically links discovered variants within patients de novo: one consensus contains the Omicron Variant of Concern (VOC) T22882G; a second consensus has a single VOC T22917G identified in Omicron strains BA.4 and BA.5 in May of 2022, 3 months after the analyzed samples were collected; a third consensus contains the VOC as well as the VOC G22898A; a single further consensus shows no mutations, consistent with Delta infection. Together, this suggests that mutations in BA.4 and BA.5 were circulating well before the VOC was called in May 2022. This implies that the VOC could have been automatically detected by NOMAD before it was present in reference annotations. A large fraction of called genetic variants exist outside of currently annotated SARS-CoV-2 variants, a subject of future work (Supplement). One such example is a 6 basepair deletion in the spike protein whose presence is perfectly predicted by whether a patient (sample) was infected with Delta or Omicron (Fig. 2D).
We further analyzed 499 samples collected in California (2020) before viral strain divergence in the spike had been reported (Gorzynski et al., 2020) as a negative control. No enrichment of NOMAD protein profiles related to the spike or Orf7a domains were observed (Supplementary Fig. 2B), supporting the idea that NOMAD calls are not false positives. To explore the generality of NOMAD for reference-free discovery in other viral infections, we additionally ran NOMAD on a study of influenza-A and of rotavirus breakthrough cases (Supplemental Fig. 2A,B, Methods). NOMAD Protein profile analysis showed enrichment in domains involved in viral suppression of the host response and regulated alternative splicing (Supplement). Together, these datasets suggest that NOMAD analysis could aid in viral surveillance, including detecting emergence of variants of concern directly from short read sequencing (Jacot et al., 2021).
NOMAD discovers paralog-specific expression in single cell RNA-seq
NOMAD is a general algorithm to discover sample-dependent sequence variation in disparate applications including RNA expression and beyond. To illustrate the former, we ran NOMAD without any parameter tuning on single cell RNA-seq datasets, testing if it could perform the fundamental but previously distinct tasks of identifying regulated expression of paralogous genes and alternative splicing (Fig. 3A). Each of these tasks is difficult, and each currently constitute sub-disciplines within bioinformatics. Algorithms to perform these tasks are time intensive to run and require significant intellectual resources for their application-specific design.
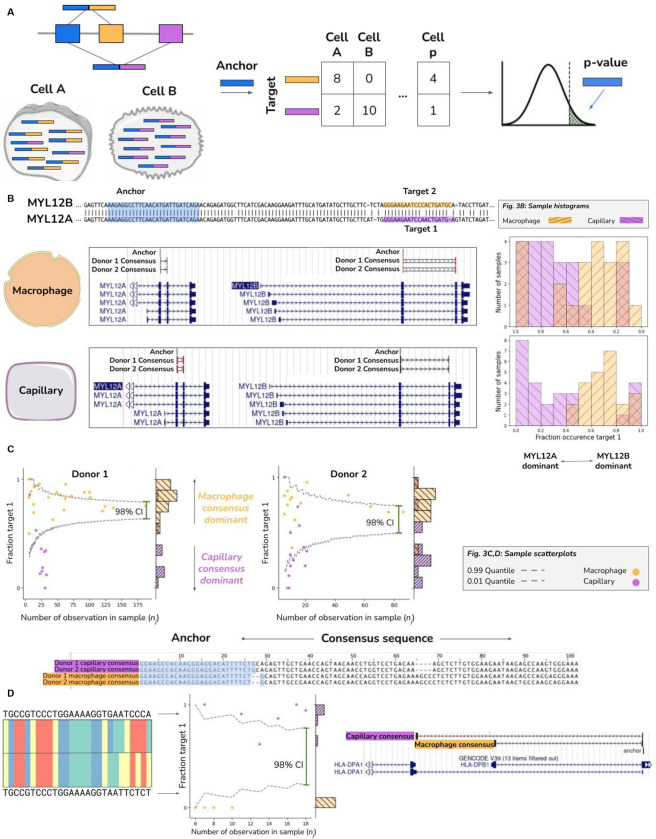
Figure 3.
A. Toy diagram depicting differentially regulated alternative splicing detection with 3 exons and 2 isoforms. Isoform 1 consists of exon 1 (blue) and exon 2 (orange), and is predominantly expressed in cell A. Isoform 2 consists of exon 1 (blue) and exon 3 (purple) and is primarily expressed in cell B.. An anchor sequence in exon 1 (blue), then generates target sequences in exon 2 (orange) or exon 3 (purple). Counts are used to generate a contingency table, and NOMAD’s statistical inference detects this differentially regulated alternative splicing.
B. Detection of differential regulation of MYL12A/B isoforms. (top-left) Shared anchor (q-value 2.5E-8, donor 1, 2.3E-42 for donor 2) highlighted in yellow, maps post fact to both MYL12 isoforms, highlighting the power of NOMAD inference: MYL12A and MYL12B isoforms share >95% nucleotide identity in coding regions. (bottom-left) NOMAD’s approach automatically detects target and consensus sequences that unambiguously distinguish the two isoforms. (right) In both donors, NOMAD reveals differential regulation of MYL12A and MYL12B in capillary cells (MYL12A dominant) and macrophages (MYL12B dominant).
C. NOMAD-identified anchor mapping to HLA-DRB1 (shared anchor, q-value of 4.0E-10 for donor 1, 1.2E-4 for donor 2). Scatter plots show cell-type regulation of different HLA-DRB1 alleles not explained by a null binomial sampling model (p<2E-16) for donor 1, (p<5.6E-8) for donor 2, finite sample confidence intervals depicted in gray (Methods). Each (donor, cell-type) pair has a dominant target, per-cell fractions represented as “fraction target 1” in scatterplots, and a dominant consensus mapping to the HLA-DRB1 3’ UTR (multiway alignment); donor 1 capillary consensus contains an insertion and deletion.
D. Donor 1 specific splice variant of HLA-DPA1. Anchor q-value: 7.9E-22. Detected targets are consistent with macrophages exclusively expressing the short splice isoform which excises a portion of the ORF and changes the amino acid composition and 3’ UTR compared to the dominant splice isoform in capillary cells; splice variants found de novo by NOMAD consensuses. Binomial hypothesis test as in 3C for cell-type specific target expression depicted in scatter plots (binomial p<2.8E-14).
First, we tested if NOMAD discovers alternatively spliced genes in single cell RNA-seq (Smart-seq 2) data from human macrophage versus capillary lung cells, chosen because they have a recently established positive control of alternative splicing, MYL6, a subunit of the myosin light chain (Olivieri et al., 2021). NOMAD rediscovered MYL6 and made new, reproducible discoveries not reported in the literature (Supplemental Table 3). For example, we discovered reproducible cell-type specific regulation of MYL12 isoforms, MYL12A and MYL12B. Like MYL6, MYL12 is a subunit of the myosin light chain. In humans (as in many species) two paralogous genes, MYL12A and MYL12B, sharing >95% nucleotide identity in the coding region, are located in tandem on chromosome 18 (Fig. 3B). Reference-first algorithms fail to quantify differential expression of MYL12A and MYL12B due to mapping ambiguity. NOMAD automatically detects targets that unambiguously distinguish the two paralogous isoforms, and demonstrates their clear differential regulation in capillary cells and macrophages (Fig. 3B). We confirmed MYL12 isoform specificity in pairwise comparisons of the same cell types in two further independent single cell sequencing studies of primary cells from the same cell-types (Supplement). MYL12 was recently discovered to mediate allergic inflammation by interacting with CD69 (Hayashizaki et al., 2016); while today little is known about differential functions of the two MYL12 paralogs, the distinct roles of highly similar actin paralogs provides a precedent (Perrin and Ervasti, 2010; Vedula et al., 2017).
Another significant challenge in human genomics is analyzing variation in the major histocompatibility (MHC) locus, including cell-type specific expression. The MHC is one of the most polymorphic regions of the human genome and carries many significant disease risk associations (Matzaraki et al., 2017). Despite its central importance in human immunity and complex disease, allotypes are difficult to quantify and have not and perhaps cannot be exhaustively annotated. Statistical methods to reliably distinguish allotype expression at single-cell level do not exist, while analysis of bulk data requires custom algorithms. Because the locus is polymorphic, standard RNA-seq alignment based methods consider reads from the MHC to be multimapped and may discard them (Dobin et al., 2013; Bray et al., 2016). NOMAD calls novel reproducible cell-type specific allelic expression and splicing in the MHC, discovering allele-specific expression of HLA-DRB1 (Fig. 3C) where macrophage and capillary cells preferentially express different alleles in donors 1 and 2, differing by an insertion. Across donors, the allele expressed is similar within the same cell-type. In another example of NOMAD’s findings in the MHC, NOMAD predicts novel cell-type specific splicing which would change the amino acid and 3’ UTR sequence of HLA-DPA1 in donor 2 (Methods, Fig. 3D).
These empirical results bear out a snapshot of NOMAD’s discovery power. They validate the theoretical prediction that NOMAD has high statistical power to simultaneously identify isoform expression variation and allelic expression, including that missed by existing algorithms (Supplementary Table 3). Finally, while in this manuscript we focus on anchors that have aligned to the human genome, NOMAD makes many predictions of cell-type specific RNA expression that include sequences that map to repetitive elements or do not map to the human reference (Supplemental Methods). In donor 1, 53% (in donor 2 61%), of 4010 (resp. 4603) anchors map to the human genome and no other reference; 35% (resp. 30%) map to both the Rfam and human genome (Supplemental Figure 5); 6% and 7% have no map to any reference used for annotation which includes repetitive and mobile elements (Supplemental Methods). As an example, 9 and 18 anchors (donor 1 and 2 respectively) BLAST to MHC alleles in the NCBI database (Supplementary Table 3). Together, this data suggests that NOMAD enables automatic discovery of biologically important sequences outside of the human reference.
Unsupervised discovery of B, T cell receptor diversity from single-cell RNA-seq
B and T cell receptors are generated through V(D)J recombination and somatic hypermutation, yielding sequences that are absent from any reference genome and cannot be cataloged comprehensively due to their diversity (>1012 sequences) (Briney et al., 2019). Detecting and quantifying V(D)J variation is critical to understanding the natural immune response from virus’ to tumors (Gee et al., 2018; Cao et al., 2020; Grant et al., 2021), (Gee et al., 2018; Grant et al., 2021), and can be used to engineer drugs. Existing methods to identify V(D)J rearrangement require specialized workflows that depend on receptor annotations and alignment; they thus fail when the loci are unannotated or when sequences have variation with high distance from annotated references (Canzar et al., 2017; Lindeman et al., 2018). In many organisms, T cell receptor loci are incompletely unannotated as they must be manually curated (The Tabula Microcebus Consortium et al., 2021). Further, V(D)J recombination will be missed in standard RNA-seq workflows. In summary, it remains difficult and time consuming to identify T cell receptor (TCR) and B cell receptor (BCR) variants, and algorithms still suffer from blindspots due to heuristics embedded in their pipelines.
To illustrate the power of NOMAD’s general and reference-free statistical approach, we tested if NOMAD could identify TCR and BCR rearrangements in the absence of annotations. We ran NOMAD with identical parameters used in analysis of other data on 111 natural killer T and 289 B cells isolated from the spleen of two mouse lemurs (Microcebus murinus) profiled by Smart-Seq2 (SS2) (The Tabula Microcebus Consortium et al., 2021), and performed the same analysis on a random choice of 50 naive B cells from the peripheral blood and 128 CD4+ T cells from two human donors profiled with SS2 (Tabula Sapiens Consortium* et al., 2022) for comparison (Fig. 4A, Methods). For emphasis, no sample metadata was used; NOMAD was not provided a reference genome or any knowledge of the samples’ biological origin or annotation. NOMAD’s novel statistical test has power to detect combinatorial target expression where each sample has a unique or nearly unique target deriving from recombination and hypermutation.
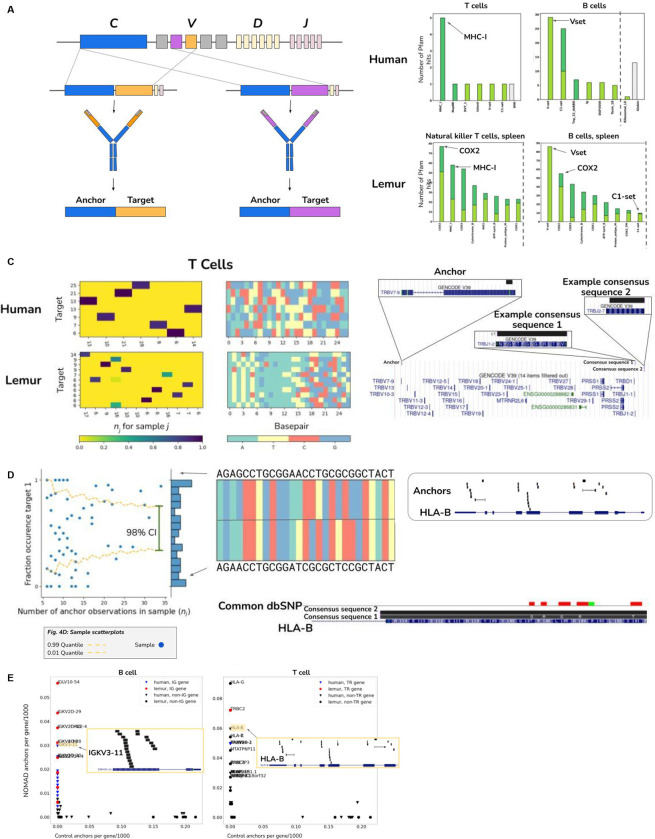
Figure 4.
A. Toy diagram depicting V(D)J recombination, specifically of different variable regions in the heavy chain. An anchor sequence in the constant region (blue), generates target sequences (orange and purple) during V(D)J recombination, in which immunoglobulins may receive different gene segments during rearrangement. NOMAD is able to rediscover and detect these recombination events by prioritizing sample-specific TCR and BCR variants.
B. Analysis of lemur and human B (left) and T (right) cells. Human genes are depicted as triangles; lemur as circles. Post facto alignments show variable regions in the kappa light chain in human B cells are most densely hit by NOMAD anchors and absent from controls; in T cells, the HLA loci and TRB including its constant and variable region are most densely hit, which are absent from controls. x-axis indicates the fraction of the 1000 control anchors (most abundant anchors) that map to the named transcript, y-axis indicates the fraction of NOMAD’s 1000 most significant anchors that map to the named transcript. Each inset depicts anchor density alignment in the IGKV region (left) and HLA-B in CD4+ T cells (top right) and TRBC-2 (bottom right), showing these regions are densely hit.
C. In human T cells (right), we show a NOMAD anchor in the TRVB7–9 gene, and two example consensuses which map to disjoint J segments, TRBJ1–2 and TRBJ2–7. Histograms of this anchor depict combinatorial single-cell (columns) by target (row) expression of targets detected by NOMAD. Histogram for lemur T cells depicted similarly; lemur T cell anchor maps to the human gene TBC1D14.
D. NOMAD-annotated anchors are enriched in HLA-B (top Fig. 4.D.1). Sample scatterplot (middle)Fig. 4.D.2 shows that T cells have allelic-specific expression of HLA-B, not explicable by low sampling depth (binomial test as in Fig. 3d,e described in Methods, p< 4.6E-24). Fig. 4.D.3: HLA-B sequence variants are identified de novo by the consensus approach (bottom), including allele-specific expression of two HLA-B variants, one annotated in the genome reference, the other with 5 SNPs coinciding with annotated SNPS.
E. NOMAD protein profile analysis shows that NOMAD recovers domains known to be diversified in adaptive immune cells, bypassing any genome reference or alignment; control hits computed from the most abundant anchors have no such enrichment. In B cells, hits in the V set, IG like domains resembling the antibody variable region, are at a relatively high E-value, as predicted by protein diversification generated during V(D)J, making matching to reference domains imperfect. The third most hit domain is Tnp_22_dsRBD, a double stranded RNA binding domain, suggesting potential activation of LINE elements in B cells. COX2, known to be involved in immune response, is highly ranked in both lemur T and B cells. Plots were truncated for clarity of presentation as indicated by dashed grey line (Fig. S2 D–F).
NOMAD protein profiles (Fig. 4B) revealed that the most frequent hits in lemur B cell were IG-like domains resembling the antibody variable domain (86 hits), and COX2 (55 hits) a subunit of cytochrome c oxidase, known to be activated in the inflammatory response (Groeger et al., 2010). NOMAD’s top hits for Lemur T cell were COX2 and MHC_I (77 and 58 hits, respectively). Similar results were obtained for the human samples (Fig. 4B): even without pre-specifying a search for V(D)J rearrangement in B and T cells, NOMAD reidentified these loci. In addition, NOMAD generated novel predictions of cell-type specific allelic expression of HLA-B in T cells (Supplement) find statistical evidence that cells preferentially express a single allele of HLA-B (p<4,6E-24); consensus analysis shows SNPs found by NOMAD de novo in HLA-B are in fact concordant with known positions of polymorphism (Fig. 4D).
We further predicted that BCR and TCR rearrangements would also be discovered by investigating the transcripts most hit by NOMAD anchors. We mapped NOMAD-called lemur B and T cell anchors to an approximation of its transcriptome: that from humans which diverged from lemur ~60–75 million years ago (Ezran et al., 2017). Lemur B cell anchors most frequently hit the immunoglobulin light and kappa variable regions; lemur T cell anchors most frequently hit the HLA and T cell receptor family genes (Methods, Fig. 4C). Similar results are found in human B and T cells (Fig. 4E, Supplement). Transcripts with the most hits in the control were unrelated to immune function, showing that NOMAD identifies a signal not explained by k-mer abundance. To illustrate NOMAD’s statistical power and capacity for discovery, consider its comparison to existing pipelines. They cannot be run without the annotation for the lemur TCR locus; for assembling BCR sequences, pipelines e.g. BASIC (Canzar et al., 2017) cannot always identify V(D)J rearrangement, including in some cells profiled in the lemur dataset (The Tabula Microcebus Consortium et al., 2021). We selected the 35 B / plasma cells where BASIC could not programmatically identify variable gene families on the light chain variable region. NOMAD automatically identified anchors mapping to the IGLV locus, with consensus sequences that BLAST to the light chain variable region (Supplement). Together, NOMAD identifies sequences with adaptive immune function including V(D)J in both B and T cells de novo, using either no reference genome (protein profile analysis) or only an annotation guidepost from a related organism (human). In addition to being simple and unifying, NOMAD can extend discovery beyond what is available today with custom pipelines.
NOMAD uncovers novel biology in octopus and eelgrass
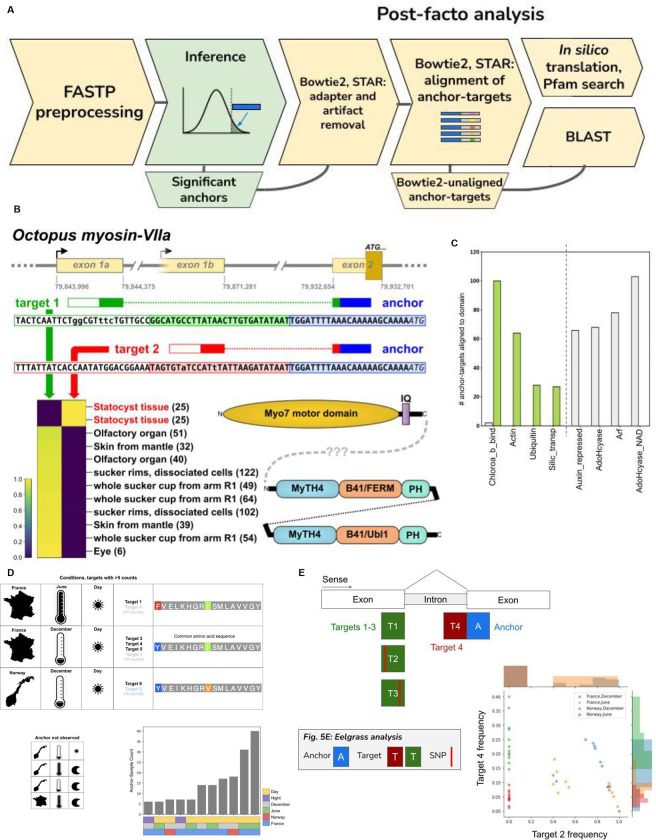
We applied NOMAD to two understudied domains of life to illustrate the breadth and generality of NOMAD’s discovery potential as well as its broad potential interest to biologists. First, we analyzed transcriptomic data from the nervous system of the non-model organism Octopus bimaculoides to illustrate transcript regulation missed by reference-first analyses. Second, we analyzed transcriptomic data from an ecological study of the seagrass species Zostera marina which is critical to carbon cycling and has a rapidly changing habitat due to climate change (Wilson and Lotze, 2019). While adaptation to climate change is critical, the mechanisms underlying its adaptation to dynamic temperatures and daylight ranges are unknown. We outline a general workflow that shows new discoveries made by NOMAD which would have been missed if the available reference had been used (Fig. 5a). Thus, while conventional mapping strategies are possible for these non-model organisms, they limit inference and discovery. We ran NOMAD in unsupervised mode with a lookahead distance of 0 for ease of interpretation (Methods). We mapped concatenated anchor-target pairs to available references, which include the reference genome of the species under study, as well as mobile genetic elements, adapter sequences and RFAM, among others, using both bowtie2 and STAR. We selected anchors where none of the anchors or anchor-target pairs mapped to the reference genome, even partially (Methods). While many follow-up analyses are possible after this stringent selection step, we concentrated on anchor-targets which mapped to protein domains in Pfam, again to showcase interpretable discoveries that would be missed by conventional genome alignment strategies (Fig. 5a).
Figure 5:
A. To showcase novel biology, anchors were concatenated with their adjacent targets (lookahead distance 0) and aligned with STAR and bowtie to reference genomes. Anchors where at most one anchor-target aligned by any method were selected for BLAST or in silico translation and Pfam mapping.
B. Anchor-targets have BLAST hits to O. sinensis chromosome LG8 (NC_043004.1) but fail alignment to the O. bimaculoides genome, are shown as reverse-complements, to give the mRNA sense-strand. The anchor is depicted in blue and targets are depicted in red or green; white boxes are sequences extending beyond the targets (from raw read data). Bases in lowercase are positions where the O. bimaculoides RNA-Seq data differs from the O. sinensis reference. Target 2 represents an unannotated alternative first exon, expressed in statocyst tissue; its 5’ extent is unknown so is grayed-out. The genomic coordinates shown are for O. sinensis chromosome LG8 (NC_043004.1); the sequence of exon 2 is missing from the O. bimaculoides genome assembly (whereas exon 1a and 1b are present but unannotated). The matching O. sinensis gene (LOC115214860) encodes a protein with only the motor and IQ domains. Adjacent to it in head-to-head orientation is a gene also annotated as myosin-VIIa (LOC115214969) which lacks a motor domain but has two repeats of other domains typical of myosin-VIIa. B41 = Band 4.1 homolog; FERM = (Four.1 protein, Ezrin, Radixin, Moesin) domain 1, F1 sub-domain; Ubl1 = first ubiquitin-like (Ubl) domain located at the N-terminus of coronavirus SARS-CoV non-structural protein 3 (Nsp3) and related proteins; PH = Pleckstrin homology-like domain.
C. In eelgrass SRP327909, the 4 domains with the most anchor-targets mapping to the domain (e-value <= 0.01) among NOMAD calls (green) and controls (gray). Highest NOMAD counts are in Chloroa_b_bind, part of the photosystem II light harvesting complex.
D. (L) Conditions in which anchor-targets were observed at least 5 times illustrate condition-specific target expression in daylight conditions. Targets expressed in distinct conditions produce distinct substitutions in the protein sequences, and in France December Day, 3 distinct targets produce a shared amino acid sequence. Amino acid sequences have best Pfam hits to Chlorophyll A-B binding protein (all e-values < 2.3e-07). BLAST (blastp) AA sequence 1 has 100% query cover and 100% identity to a transcript in 3 diatom species, AA sequence 2 has 100% query cover and 100% identity to a transcript in 4 diatom species, and AA sequence 3 has a single substitution to a transcript in 4 diatom species (100% query cover, 94.12% identity). BLAST (blastn) anchor-targets (AT) all best hit diatoms: AT1 best hits F. solaris (92% query cover, 92% identity), AT2 best hits E. pelagica (92% query cover, 94.23% identity), AT3 best hits E. pelagica (96% query cover, 92.31% identity), AT4 best hits 4 P. tricornutum (94% query cover, 96.08% identity), AT5 best hits E. pelagica and P. tricornutum (94% query cover, 94.12% identity), and AT6 best hits F. solaris (96% query cover, 92.31% identity). (R): anchor-sample counts (barplot) are highest in samples harvested during the day.
E. In eelgrass, NOMAD identifies season-specific splicing and location-specific polymorphism in anchor-targets. Pfam analysis assigns the sequences to NADH dehydrogenase transmembrane subunit. The top 3 targets in terms of abundance BLAST to splice junctions, and differ from one another by SNPs in two positions. The 4th most abundant target represents intron retention. In this anchor, only samples from France express target 2, partitioning perfectly by country. Within the samples from Norway, the collection month (December or June) can be perfectly predicted by fractional expression of target 4. Full sequences and additional discussion are presented in the supplemental text.
First, we used NOMAD to identify tissue-specific transcriptional regulation that would be missed by genome alignment, using RNA-Seq data from a single California two-spot octopus (Octopus bimaculoides) generated in a study on chemotactile sensation (van Giesen et al. 2020; personal communication). For each of the 254,066 significant anchors, we aligned the anchor concatenated to its most abundant target (Methods), 17,366 (6.8%) did not Bowtie2-align to any database, 26,199 (10.3%) did not STAR-align to the available reference. STAR alignments include sequences that are partially mapped through soft-clipping as well as mismatches and indels. To illustrate missing regulatory biology from genome alignments, we focused on a highly conservative list of 8,908 anchors which had at most one target aligning to a reference with either Bowtie2 or STAR (Methods). We used BLAST to query NOMAD-called anchor-targets that were completely unaligned by any method (Methods); matches to other Octopus species provide evidence that the assembled O. bimaculoides genome (Albertin et al., 2015) is incomplete.
Several sequences had BLAST matches to Octopus sinensis (Methods, Table 6), a species closely related to O. bimaculoides. One example maps to an unconventional Myosin VIIa (LOC115214860), also called MYO7A in humans; mutations in MYO7A cause Usher syndrome, leading to deafness and blindness (Wu et al., 2011). The anchor and both targets (as well as their extended consensuses) map to the O. sinensis genome. The more abundant target 1 and anchor also map to the 5’ end of an annotated myosin-VIIa transcript (XM_036505518.1), and represent the first and second exons, respectively. The more restricted target 2, expressed only in statocyst tissue, represents a novel alternative first exon (Figure 5B). The matches of target 1 and 2 consensus sequences in the O. sinensis genome are imperfect (but ≥92% identity), whereas they have perfect matches in the O. bimaculoides genome. The anchor has a perfect match in O. sinensis but is not found in the O. bimaculoides genome (Supplemental Text). We believe the annotated myo-VIIa gene and transcript in O. bimaculoides (LOC106880717; XM_052969897.1) are incomplete based on our evidence from RNA-Seq, as they lack sequences corresponding to target 1 and the anchor. While the O. sinensis gene model is more complete in this regard, it is likely that it is incorrect in other regards (Supplemental Text). The restriction of target 2 expression to statocyst tissue is intriguing given MYO7A’s association with Usher syndrome, as octopus statocyst is responsive to sound (Solé et al., 2013; Y. Zhang et al., 2015).
We highlight several other anchor-target pairs that do not map to the O. bimaculoides genome or transcriptome yet do map to annotated transcripts in O. sinensis: carboxypeptidase D, Upf2, and netrin (Supplemental Text). An anchor and its two targets map to the 3’ UTR of carboxypeptidase D; target 2 is a close match to the annotated transcript, whereas target 1 contains a 13 nt deletion with respect to target 2 and the genome. The two variants have surprisingly dichotomous expression: target 1 is expressed in sucker rims, the olfactory organ, and statocyst tissue; only target 2 is all other tissues sampled. A second anchor and its two targets map to the 3’ UTR of Upf2 (“regulator of nonsense transcripts 2”), involved in nonsense-mediated decay. Target 1 and 2 differ only in the number of CAG repeats -- either six or five, respectively, and show mutually exclusive expression in the tissues sampled. For both carboxypeptidase D and Upf2, O. bimaculoides has an annotated transcript yet the anchor or target sequences are missing, suggesting the O. bimaculoides annotation is incomplete. A third anchor-target set matches netrin, a protein involved in axon-guidance: an anchor-target pair differentiated by a single T to C variation, consistent with canonical RNA-editing was found having isoform usage specific to either eye, whole sucker cup, statocyst tissue, and olfactory organ, or to sucker rims (Supplemental Table 6, Supp. Figure 6C).
In this analysis, we focus on the strongest evidence that NOMAD discovers biology that is completely missed by using a reference genome. We have also not investigated novel proteins or transcripts among the thousands of anchors that are unmapped, and are not partially mapped to the Octopus genome by STAR or BLAST. Further, we only include discussion of examples where all anchor-targets are not mapped by STAR, and conjecture that relaxing this will reveal additional examples of transcript variation missed by reference-first methods. We anticipate that investigating these events in octopus, and in other organisms, will enable researchers to prioritize tissue-specific and tissue-regulated splicing and editing, among other (post-)transcriptional regulatory events.
To further demonstrate new biological discoveries from NOMAD, we analyzed RNA-seq data from a completely different domain of life: a seagrass species Zostera marina, also called eelgrass, which is under intense selection due to climate change and is critical to carbon sequestration in the ocean (Röhr et al., 2018). A reference genome for this diploid organism and its companion chloroplast and mitochondrial genome were assembled in 2016 (Olsen et al., 2016) and 2021 (Ma et al., 2021). Eelgrass is critical to global ocean ecosystems (Yu et al., 2020, 2022). As global oceans warm, eelgrass has established habitats in low-daylight arctic conditions, a requirement for the species as global temperatures force it to acclimate to high latitudes. Genetic and post-transcriptional adaptation that allow eelgrass to survive at low and high latitudes with greatly varying dynamic ranges of daylight exposure are unknown (Rock and Daru, 2021), but ecological studies have shown that eelgrass is extensively colonized by epiphytic diatoms (Jacobs and Noten, 1980) which have significant impacts on global carbon pumps (Tréguer et al., 2018), and regulate seagrass biomass and growth (Jacobs and Noten, 1980; Prazukin et al., 2022).
We ran NOMAD on a matched collection of RNA-seq from eelgrass shoots collected in coastal regions of France, a mediterranean climate, and of Norway, a near-arctic climate, sampled in summer and winter and during day and night (Methods). Of the most abundant target per anchor, 33855/254991 (13.2%) anchor-target pairs failed alignment to any database (Bowtie2), 29,714 (11.6%) failed STAR alignment to a Zostera marina nuclear, chloroplast, and mitochondrial index, and 14,680 (5.7%) failed alignment by both methods.
In-silico translation and Pfam search of anchor-target pairs failing primary sequence alignment yielded hits to 118 protein domains (e value <.05, Supplemental Table 7). The largest enrichment of NOMAD-called Pfam domains versus controls hit Chlorophyll A-B binding protein domain, part of the light-harvesting complex of photosystem II (Fig. 5C). We selected an anchor with a large effect size (0.66) mapping to this domain (Fig. 5D). The most- and third-most abundant anchor-targets had respective best BLAST (blastn) hits with 92% sequence identity to transcripts covering 46/54 and 52/54 bases, to two diatoms (Fistulifera solaris and Epithemia pelagica, resp.). The second-and fourth-most abundant anchor-targets had marginally better BLAST hits with > 94% sequence identity to two different diatom genomes (Epithemia pelagica and Phaeodactylum tricornutum, respectively). The distribution of anchor-target counts is affected by all three main variables: geographic location, time of year, and time of day. The most frequently observed target dominates samples from France in June, daytime. This anchor is only detected in Norway December day samples. Targets detected only in France December samples, are more abundant in samples collected during day: all targets from sampling at night have fewer than 5 counts. Together, this suggests the hypothesis that diatom gene expression is impacted by circadian cycles– and upregulated during moderate daylight– and also that the abundance of the host diatom varies seasonally and by location.
Other domains enriched in NOMAD anchor-targets include those annotated as silicon transport, a known biochemical pathway in diatoms (Shrestha and Hildebrand, 2015), and HMG (high mobliy group) box protein domains (Supp. Fig. 7A). In investigated anchors with Pfam hits to Chlorophyll A-B binding (Fig. 5D), HMG box, and 2Fe-2S iron-sulfur cluster binding domain (Supp Figure 7A, Supp Figure 7B) the top hits from Protein BLAST are to diatom species. The two anchor-targets annotated as HMG-box had best BLAST (blastp) hits to a single diatom species with e-values of 0.007 and 1e-05. This suggests that these discoveries by NOMAD derive from co-associated diatoms (most likely species not specifically represented in the current database), rather than eelgrass per se, an inference not possible by restricting to reads mapping to the Zostera marina reference genome. For each example, target variation is strongly associated with collection location and or season (Fig. 5D). By bypassing reference alignments, NOMAD identifies differentially expressed sequence variation outside of the Zostera genome – likely within the associated epiphyte population. This sequence variation (Fig. 5D) tracks with sample metadata suggesting that genes and proteins expressed by diatoms may have important ecological and or regulatory features missed by past analyses. This highlights that NOMAD can automatically identify regulated sequence expression of co-associated species, whether known or unknown.
While we focused above on NOMAD’s discoveries that fail to map to the reference genome, NOMAD also provides insight and joint inference for anchor-targets mapping to the eelgrass reference genome. One example (Fig. 5E) is a gene encoding the NdhL subunit of chloroplast NADPH dehydrogenase complex (NDH), involved in cyclic electron transport (Laughlin, Savage and Davies, 2020). NOMAD detects a SNP discriminating samples collected in Norway from those in France and additionally, intron retention (Supplemental Text) that is regulated in June compared to December, completely discriminating these times in samples from Norway. This form of splicing regulation will be interesting for further study as modulation of NDH function may affect photosynthetic efficiency and oxidative stress under varying light conditions (M. Ma et al., 2021). Together, these highlighted examples show biology found by NOMAD would be missed by conventional pipelines and illustrate NOMAD’s ability to identify sequences completely missing from the genome, or unmappable, revealing new biology of tissue-specific, environmental, and metagenomic regulation. The analytic approach followed here is general and can be followed for any organism and can identify multiple mechanisms of regulation including editing to splicing and sequences missing from the reference genome.
Conclusion
Genomic analysis today is performed with complex computer scientific workflows in a highly domain-specific manner. Here, we have shown that a simple, direct analytic framework with transparent statistics can unify the approaches taken in disparate subfields of genomic data-science and enable novel discovery missed in current workflows. We have achieved this by stating their goal to discover sample-dependent sequence variation in a unified probabilistic model, and used this model to develop NOMAD. NOMAD is an efficient statistics-first algorithm that formulates and efficiently solves tasks in genome science with great generality.
We provided a snapshot of NOMAD’s discoveries in currently siloed subfields of genome science. When run on SARS-CoV-2 patient samples during the emergence of the Omicron variant, NOMAD finds that the spike protein is highly enriched for sequence variation, bypassing genome alignment completely. NOMAD provides evidence that Variants of Concern can be detected well before they are flagged as such or added to curated databases. This points to a broader impact for NOMAD in viral and other genomic surveillance, since emerging pathogens will likely have sequence variation missing from any reference.
NOMAD finds novel cell-type specific isoform expression in homologous genes missed by reference-guided approaches, such as in MYL12A/B and in the MHC (HLA) locus, even in a small sample of single cell data. Highly polymorphic and multicopy human genes have been recalcitrant to current genomic analyses and are critical to susceptibility to infectious and complex diseases, e.g. the MHC. NOMAD could shed new light on other polymorphic loci including non-coding RNAs, e.g. spliceosomal variants (Kuo et al., 2017; Buen Abad Najar, Yosef and Lareau, 2019). In addition, NOMAD unifies detection of many other examples of transcriptional regulation: intron retention, alternative linear splicing, allele-specific splicing, gene fusions, and circular RNA. Further, NOMAD prioritizes V(D)J recombination as the most sample-specific sequencing diversifying process in B and T cells of both human and mouse lemur, where inference in lemur is made using only an approximate genomic reference (human) which diverged from lemur ~60 million years ago.
Disparate data – DNA and protein sequence, or any “-omics” experiment, from Hi-C to spatial transcriptomics – can be analyzed through NOMAD’s framework (“Generality of NOMAD”, Supplement). NOMAD may also be impactful in analysis of plants, microbes, and mobile elements, including transposable elements and retrotransposons, which are far less well annotated, and are so diverse that references may never capture them. Beyond these discoveries, our work also shows that NOMAD provides a more efficient and powerful solution in domains where competing methods exist, such as the detection of differentially-regulated alternative-splicing.
NOMAD illustrates the power of statistics-first genomic analysis, where references and annotations are only optionally used for post-facto interpretation. References are valuable for interpretation of sequence data, including for example, in curating variants associated with disease, such as for genome-wide association studies. However, to be useful, references need not be used to filter raw data by reducing it to a set of positions and variants with respect to the reference alignment. This step itself introduces statistical bias in quantification, blindspots and errors. NOMAD provides a new approach to the use of references by translating the field’s “reference-first” approach to “statistics-first”, performing direct statistical hypothesis tests on raw sequencing data, enabled by its probabilistic modeling of raw reads rather than of alignment outputs. By design, NOMAD is highly efficient: it enables direct, large-scale study of sample-dependent sequence variation, completely bypassing the need for references or assemblies. NOMAD promises data-driven biological study with scope and power previously impossible.
Limitations of the study
Naturally, some problems cannot be easily formulated in the manner posed, such as cases where quantification of sample-specific RNA or DNA abundance alone is desired (e.g. differential gene expression analysis). However, the problems that can be addressed using this formulation span diverse fields which are of great current importance (Supplement), including those previously discussed. Further, NOMAD’s statistical test can be applied to any count matrices, including tables of gene expression by samples.
Materials and Methods
Anchor preprocessing and parameter choices
Anchors and targets are defined as contiguous subsequences of length k positioned at a distance R=max(0, (L - 2 * k) / 2) apart (rounded), where L is the length of the first read processed in the dataset. If L=100 and k=27, then R=23. Anchor sequences can be extracted as adjacent, disjoint sequences or as tiled sequences that begin at a fixed step size, to reduce computational burden. For this manuscript NOMAD was run with 1M reads per FASTQ file, anchor sequences tiled by 5bp, and k=27. To satisfy the independence assumption for computing p-values in the NOMAD statistics, only one read is used if the sequencing data is paired end. For this manuscript, we use FASTQ files from read 1; for HLCA datasets, both read 1 and read 2 were used. Extracted anchor and target sequences are then counted for each sample with the UNIX command, `sort | uniq -c`, and anchor-target counts are then collected across all samples for restratification by the anchor sequence. This stratification step allows for user control over parallelization. To reduce the number of hypotheses tested and required to correct for, we discard anchors that have only one unique target, anchors that appear in only 1 sample, and (anchor, sample) pairs that have fewer than 6 counts. Then, we retain only anchors having more than 30 total counts after the above thresholds were applied. This approach efficiently constructs sample by target counts matrices for each anchor. We note that for a fixed number of anchor-target pairs, under alternatives such as differential exon skipping, larger choices of R have provably higher power than smaller choices, following the style of analysis in (Salzman, Jiang and Wong, 2011). We give examples of how choices of k, R, and tiling length impact results in France SARS-CoV-2 data as follows, showing that NOMAD yields similar results for a range of parameter choices. Default parameters shown in bold: we tested K = [25, 27, 30]; Tile = [3, 5, 7]; Lookahead = [0, 15, 23]. For K = 25, 94.4% of anchors with default parameters contain at least one of the K=25 anchors as a substring. For K = 30, 93.8% of anchors with K=30 contain at least one of the anchors with default parameters a substring. For tile size of 3, 85% of the anchors from the default run can be found in the significant anchors of tile size of 3. For tile size of 7, 85% of the anchors from the default run can be found in the significant anchors of tile size of 5. For lookahead distance of 0, 37% of the anchors from the default run can be found in the significant anchors of tile size of 3; for lookahead distance of 15, 76% of the anchors from the default run can be found in the significant anchors. Overall, as tile size decreases, anchor calls increase (4715, 5522, 7891 for [7,5,3] respectively). As k varies, anchor calls stay essentially the same (5875, 5522, and 5958 for k=[25,27,30] respectively). Finally, for lookahead distance, the total number of calls decrease as lookahead distance increases (13239, 8295, 5522 for [0,15,23] respectively).
NOMAD p-values
While contingency tables have been widely analyzed in the statistics community (Agresti, 1992; Diaconis and Sturmfels, 1998; Chen et al., 2005), to our knowledge no existing tests provide closed form, finite-sample valid statistical inference with desired power for the application at hand (Supplement). We develop a test statistic S that has power to detect sample-dependent sequence diversity and is designed to have low power when there are a few outlying samples with low counts as follows. First, we randomly construct a function f, which maps each target independently to {0,1}. We then compute the mean value of targets with respect to this function. Next, we compute the mean within each sample of this function. Then, an anchor-sample score is constructed for sample j, Sj, as a scaled version of the difference between these two. Finally, the test statistic S is computed as the weighted sum of these Sj, with weights cj (which denote class-identity in the two-group case with metadata and are chosen randomly without metadata, see below). In the below equations, Dj,k denotes the sequence of the k-th target observed for the j-th sample, and M denotes the total number of observations of this anchor.
Statistically valid p-values are computed as:
by applying Hoeffding’s inequality on these sums of independent random variables (under the null). The derivation is detailed in the Supplement.
This statistic is computed for K different random choices of f, and in the case where sample group metadata is not available, jointly for each of the L random choices of c, here with K=10 and L=50. We call the random choice of cj’s “random c’s” below. The choice of f and c that minimize the p-value are reported, and are used for computing the p-value of this anchor. To yield valid p-values we apply Bonferroni correction over the L*K multiple hypotheses tested (just K when sample metadata is used and randomization on c is not performed). Then, to determine the significant anchors, we apply Benjamini-Yekutieli (BY) correction (Benjamini Hochberg (BH) with hypotheses with positive dependence) to the list of p-values (for each anchor), yielding valid FDR controlled q-values reported throughout the manuscript (Benjamini and Yekutieli, 2001) implemented with the statsmodels.api.stats.multipletests functionality in python.
NOMAD Effect size
NOMAD provides a measure of effect size when the cj’s used are +/− 1, to allow for prioritization of anchors with large inter-sample differences in target distributions. Effect size is calculated based on the split c and function f that yield the most significant NOMAD p-value. Fixing these, the effect size is the absolute value of the difference between the mean function value over targets (with respect to f) across those samples with cj = +1 denoted A+, and the mean over targets (with respect to f) across those samples with cj = −1 denoted A−.
This effect size has natural relations to a simple 2 group alternative hypothesis. It can also be shown to relate to the total variation distance between the empirical target distributions of the two groups. These connections are discussed further in the Supplement.
Consensus sequences
A consensus sequence is built for each significant anchor for the sequence downstream of the anchor sample. A separate consensus is built for each sample by aggregating all reads from this sample that contain the given anchor. Then, NOMAD constructs the consensus as the plurality vote of all these reads; concretely, the consensus at basepair i is the plurality vote of all reads that contain the anchor, i basepairs after the anchor appears in the read (a read does not vote for consensus base i if it has terminated within i basepairs after the anchor appeared). The consensus base as well as the fraction agreement with this base among the reads is recorded.
The consensus sequences can be used for both splice site discovery and other applications, such as identifying point mutations and highly diversifying sequences, e.g. V(D)J rearrangements. The statistical properties of consensus building make it an appealing candidate for use in short read sequencing (Ståhlberg et al., 2016), and may have information theoretic justification in de novo assembly (Motahari et al., 2013) (Supplement).
To provide intuition regarding the error correcting capabilities of the consensus, consider a simple probabilistic model where our reads from a sample all come from the same underlying sequence. In this case, under the substitution only error model, we have that the probability that our consensus for n reads makes a mistake at a given location i under independent sequencing error rate epsilon (substitution only) is at most
We can see that even for n=10, this probability is less than 1.3E-7 for a given basepair, which we can union-bound over the length of the consensus to yield a vanishingly small probability of error. Thus, for a properly aligned read, if a basepair differs between the consensus and reference it is almost certainly a SNP.
Element annotations
To identify false positive sequences or contextualize mobile genetic elements, anchors and targets are aligned with bowtie2 to a set of indices, corresponding to databases of sequencing artifacts, transposable elements, and mobile genetic elements. In these alignments, using bowtie2, the best hit is reported, relative to an order of priority (Abante, Wang and Salzman, 2022). The reference used are: UniVec, Illumina adapters, grass carp GCA_019924925 (Wu et al., 2022), Escherichia phage phiX174, Rfam (Kalvari et al., 2021), Dfam (Storer et al., 2021), TnCentral (Ross et al., 2021), ACLAME (Leplae et al., 2004), ICEberg (Bi et al., 2012), CRISPR direct repeats (Couvin et al., 2018), ITSoneDB (Santamaria et al., 2018), ITS2 (Selig et al., 2008), WBcel235, TAIR10, grch38_1kgmaj, Octopus bimaculoides (Albertin et al., 2015), Octopus bimaculoides transcriptome (van Giesen et al., 2020), Zostera marina (X. Ma et al., 2021), and Zostera marina chloroplast and mitochondrion (Olsen et al., 2016). Grass carp and other indices unrelated to the biological study are used as controls. To perform these annotations, bowtie2 indices were built from the respective reference fastas, using bowtie2-build with default parameters. Anchors and targets were then aligned to each index, using bowtie2-align with default parameters. For each sequence, we report the alignment to the reference and the position of that alignment for each reference in the prespecified set. Anchors and targets, and their respective element annotations, are reported in the element annotation summary files.
Genome annotations
Anchor, target, and consensus sequences can be aligned to reference genomes and transcriptomes, to provide information about the location of sequences relative to genomic elements.
For significant anchors, target, and consensus sequences, we report information regarding the anchor, target, and consensus sequences’ alignments to both a reference genome and transcriptome in the genome annotation summary files (table S4). All alignments reported below are run in two modes in parallel: bowtie2 end-to-end mode (the bowtie2 default parameters) and bowtie2 local mode (`-local`, in addition to the bowtie2 default parameters); the following columns are prefixed with “end_to_end” or “local”, for end-to-end mode and local mode, respectively.
To report alignments to the transcriptome, the sequences are aligned to the reference transcriptome with bowtie2, with `-k 1`, in addition to the above parameters, to report a maximum of one alignment per sequence. If there is a transcriptome alignment, we report the alignment to the reference and the MAPQ score of the alignment.
To report alignments to the genome, the sequences are aligned to the reference genome, with the same parameters above. If there is a genome alignment, we report the alignment to the reference, the strand of the alignment, and the alignment MAPQ score. To lend further context, we report any annotated gene intersection to the reference genome alignment, by first converting the genome alignments to BED format and then using `bedtools intersect` on the genome alignments BED file and a BED file of gene annotations (for this manuscript, we use hg38 RefSeq); for each sequence, we report the list of distinct gene intersections per sequence genome alignment. For each sequence genome alignment, we also report its distances to the nearest annotated exon junctions, by using `bedtools closest` with the sequence genome alignments and BED files of annotated exon start and end coordinates. To report distances of a sequence genome alignment to the nearest upstream exon starts and nearest upstream exon starts, we report the closest exon start and exon end, respectively, with the `bedtools closest` parameters `-D ref -id -t first`. To report distances of a sequence genome alignment to the nearest downstream exon starts and nearest downstream exon starts, we report the closest exon start and exon end, respectively, with the `bedtools closest` parameters `-D ref -iu -t first`.
NOMAD protein profiles
For each set of enriched anchors, homology-based annotation was attempted against an annotated protein database, the Pfam (Mistry et al., 2021). For each dataset, up to 1000 of the most significant anchors (q-value < 0.01) were retained for the following analysis: we first generated a substring of each downstream consensus by appending each consensus nucleotide assuming both conditions were met: a minimum observation count of 10 and a minimum agreement fraction of 0.8, until whichever metric first exhibited two consecutive failures at which point no further nucleotide was added. A limit of 1000 anchors was used due to computational constraints from HMMer3 (see below). Anchors that did not have any consensus nucleotides appended were kept as is. An extended anchor was generated for each experiment in which an anchor was found. Each extended anchor was then stored in a final concatenated multi FASTA file with unique seqID headers for each experiment’s extended anchors.
The number of matched anchors used for NOMAD and control analysis per dataset are as follows: 201 high effect size (.5) anchors in SARS-CoV-2 from South Africa, 252 high effect size (.5) anchors in SARS-CoV-2 from France; 1000 anchors (no effect size filter) were used for rotavirus, human T cells, human B cells, Microcebus natural killer T cells, and Microcebus B cells.
To assess these extended anchors for protein homology, this concatenated FASTA file was then translated in all six frames using the standard translation table using seqkit (Shen et al., 2016) prior to using hmmsearch from the HMMer3 package (Johnson, Eddy and Portugaly, 2010) to assess each resulting amino acid sequences against the Pfam35 profile Hidden Markov Model (pHMM) database.
All hits to the Pfam database were then binned at different E-value orders of magnitude and plotted. In each case, control assessments were performed by repeating the extension and homology searches against an equivalent number of control anchors, selected as the most frequent anchors from that dataset.
Lastly it is worth noting that while only counts of the best scoring Pfam hits were assessed in this study, other information is also produced by HMMer3. In particular, relative alignment positions are given for each hit which could be used to more finely pinpoint the precise locus at which sequence variation is detected.
We note that while the number of input anchors for NOMAD and control sets are matched, it is possible to have more control protein domains in the resulting barplots, as only high E-value hits to Pfam are reported in the visualizations.
Control analysis
To construct control anchor lists based on abundance, we considered all anchors input to NOMAD and counted their abundance, collapsing counts across targets. That is, an anchor receives a count determined by the number of times it appears at an offset of 5 in the read up to position R- max(0,R/2–2*k) where R is the length of the read, summed over all targets. The 1000 most abundant anchors were output as the control set. For analysis comparing control to NOMAD anchors, min(|NOMAD anchor list|,1000) most abundant anchors from the control set were used and the same number of NOMAD anchors were used, sorted by p-value.
SARS-CoV-2 analysis
The SARS-CoV-2 datasets used in this manuscript were analyzed with NOMAD’s unsupervised mode (no sample metadata provided). To identify high effect size anchors, a threshold of èffect_size_randCjs` > 0.5 was used (table S2). The Wuhan variant reference genome was downloaded from NCBI, assembly NC_045512.2. The Omicron and Delta mutation variants were downloaded as FASTA from the UCSC track browser in June 2022, with the following parameters: clade ‘Viruses’, assembly NC_045512.2, genome ‘SARS-CoV-2’, group ‘Variations and Repeats’, track ‘Variants of Concern’, and table ‘Omicron Nuc Muts (variantNucMuts_B_1_1_529)’ and ‘Delta Nuc Muts (variantNucMutsV2_B_1_617_2)’.
Variant genomes were downloaded in FASTA file format, and bowtie indices were built from these FASTA files, using default parameters. To determine alignment of anchors to the Wuhan genome, anchor sequences were converted to FASTA format and aligned to the Wuhan bowtie index with bowtie (default parameters). After mapping of NOMAD anchors, the number of control anchors were chosen to match the number of anchors mapped by bowtie to report comparable numbers.
Mutation consistency to the Omicron and Delta variants was reported as follows. For each anchor mapping to the Wuhan reference in the positive strand, an anchor at position x is called mutation-consistent if there is an annotated variant between positions x+k+D and x+R+2*D, where D=max(0, (L - 2 * k) / 2), L is the length of the first read processed in the dataset, and the factor of 2 reflects the bowtie convention of reporting the left-most base in the alignment. The reciprocal logic was used to define mutation-consistency for anchors mapping to the negative strand – e.g. a mutation had to occur between positions x-(R+D) and x. In total, we report: a) number of anchors mapping with bowtie default parameters to the Wuhan reference; b) number and fraction of mutation-consistent anchors as described above.
To determine what NOMAD calls (and control anchors) were strain defining we perform the following. To generate NOMAD’s calls, we filter for anchors that are significant (with a BY corrected p-value less than .05) and have large effect size (> .5), yielding a list of N NOMAD-called anchors. Control anchors are generated by taking the N anchors with the highest counts. For each of these anchors we construct their target x sample contingency table, first filtering out all anchors with fewer than 30 counts, only 1 unique target, or only 1 unique sample, and filtering out all samples with 5 or fewer counts. Then, we discard all targets that constitute less than 5% of the remaining counts for that anchor. These the remaining anchors and targets are then bowtie aligned to an index comprised of the Wuhan reference genome (NC_045512v2), and Delta (OK091006.1), Omicron BA.1 (OX315743.1), and Omicron BA.2 (OX315675.1) assemblies. For this alignment, options -a --best --strata were used. Then, for each set of anchors (NOMAD calls, and controls), the list is filtered to only anchors that align to one of the reference assemblies, further requiring that each anchor have at least one target that aligns to a reference assembly. Then for each anchor, we declare it to be strain defining if, for any of the reference assemblies, it has at least one target that maps to it and one target that does not.
Viral protein profile analysis
In influenza, NOMAD’s most frequently hit profiles were Actin (62 hits), and GTP_EFTU (23 hits), and the influenza-derived Hemagglutinin (17 hits), consistent with virus-induced alternative splicing of Actin (Thompson et al., 2020) and EF-Tu, further elucidating these proteins’ roles during infection (Sun and Whittaker, 2007; Kuo et al., 2017) (no such hits were found in the control). Similarly, in a study of metagenomics of rotavirus breakthrough cases, NOMAD protein profile analysis prioritized domains known to be involved in host immune suppression.
In rotavirus, the most enriched domain in NOMAD compared to control was the rotavirus VP3 (Rotavirus_VP3, 76 NOMAD hits vs 9 control hits), a viral protein known to be involved in host immune suppression (Song et al., 2020), and the rotavirus NSP3 (Rota_NSP3, 87 NOMAD vs 35 control hits), a viral protein involved in subverting the host translation machinery (Gratia et al., 2015), both proteins that might be expected to be under constant selection given their intimate host interaction.
Identifying cell-type specific isoforms in SS2
In the analysis of HLCA SS2 data, we utilize “isoform detection conditions” for alternative isoform detection. These conditions select for (anchor, target) pairs that map exclusively to the human genome, anchors with at least one split-mapping consensus sequence, mu_lev > 5, and M > 100. mu_lev is defined as the average target distance from the most abundant target as measured by Levenstein distance. To identify anchors and targets that map exclusively to the human genome, we included anchors and targets that had exactly one element annotation, where that one element annotation must be grch38_1kgmaj. To identify anchors with at least one split-mapping consensus, we selected anchors that had at least one consensus sequence with at least 2 called exons. The conditions on Levenshtein distance, designed to require significant across-target sequence diversity, significantly reduced anchors analyzed (excluding many SNP-like effects). We further restricted to anchors with M > 100, to account for the lower cell numbers in macrophage cells; note that the user can perform inference with a lower M requirement, based on input data. These isoform detection parameters were used to identify the SS2 examples discussed in this manuscript, MYL12. For HLA discussion, gene names were called using consensus_gene_mode.
Splice junction calls
To identify exon coordinates for reporting annotations in this manuscript, consensus sequences are mapped with STAR aligner (default settings) (Dobin et al., 2013). Gapped alignments are extracted and their coordinates are annotated with known splice junction coordinates using ‘bedtools bamtobed --split’; each resulting contiguously mapping segment is called a “called exon” (see below). From each consensus sequence, called exons are generated as start and end sites of each contiguously mapped sequence in the spliced alignment. These ‘called exons’ are then stratified as start sites and end sites. Note that the extremal positions of all called exons would not be expected to coincide with a splice boundary (see below); “called exon” boundaries would coincide with an exon boundary if they are completely internal to the set of called exon coordinates. Each start and end site of each called exon is intersected with an annotation file of known exon coordinates; it receives a value of 0 if the site is annotated, and 1 if it is annotated as alternative. The original consensus sequence and the reported alignment of the consensus sequence are also reported. Gene names for each consensus are assigned by bedtools intersect with gene annotations (hg38 RefSeq for human data by default), possibly resulting in multiple gene names per consensus.

Caption: Example of how spliced reads are converted to “called exons” (bottom) and are compared to annotated exons (top); right most and leftmost boundaries of called exons are not expected to coincide with annotated exon boundaries and are excluded from analysis of concordance between consensus called-exons and annotations.
HLA analysis in HLCA
NOMAD summary files were processed by restricting to anchors aligning to the human genome, and having at least 1 target with this characteristic. Further, mu_lev had to exceed 1.5. For HLA discussion, gene names were called using consensus_gene_mode.
B,T Cell Transcriptome Annotations
To determine the most frequent transcriptome annotation for a dataset, all significant anchors were mapped to the human transcriptome (GRCh38, Gencode) with bowtie2, using default parameters and `-k 1` to report at most one alignment per anchor. Then, the bowtie2 transcript hits are aggregated by counting over anchors. The transcript hits with the highest counts over all anchors were reported.
Further immune cell protein domain analysis
In human B and T cells, NOMAD blindly rediscovered the high degree of single-cell variability in the immunoglobulin (IG) in B cells: this locus was most highly ranked by anchor counts per transcript (Fig. 4E). In B cells, NOMAD anchor counts were highest in genes IGKV3–11, IGKV3D-20, IGKV3D-11, and IGKC, the first three being variable regions of the B cell receptor (Fig. 4E).
Parallel analysis of T cells showed similar rediscovery and extension of known biology: HLA-B, RAP1B, TRAV26–2, and TRBV20–1 were the highest-ranked transcripts in T cells measured by anchor counts. HLA-B is a major histocompatibility (MHC) class I receptor known to be expressed in T cells, and TRAV26–2 and TRBV20–1 are variable regions of the T cell receptor. T cell expression of HLA-B alleles has been correlated with T cell response to HIV (Kiepiela et al., 2004; Elahi et al., 2011). Fig. 4E shows many other genes known to be rearranged by V(D)J were also recovered. In the control sets for both B and T cells, enriched genes were unrelated to immune functions (Fig. 4E, Fig. S2 E,F).
HLA-B (Fig. 4E) is the most densely hit transcript in T cells. Mapping assembled consensuses shows two dominant alleles: one perfectly matches a reference allele, the other has 4 polymorphisms all corresponding with positions of known SNPs. NOMAD statistically identifies T cell variation in the expression of these two alleles, some T cells having only detectable expression of one but not the other (p< 4,6E-24) (Francis et al., 2022). Other HLA alleles called by NOMAD, including HLA-F, have similar patterns of variation in allele-specific expression (Supplement).
NOMAD comparison to BASIC analysis in lemur spleen B cells
To compare performance, we first ran BASIC on the lemur spleen B cells, with the following additional parameters: -a. We then ran NOMAD on cells where BASIC failed to identify the light chain variable gene family, by selecting cells annotated as “No BCR light chain” from the BASIC output. From the NOMAD output, we identified anchors which mapped to the IGL gene by bowtie; to do this, we used the command `grep IGL “$file”`, where “$file” corresponds to the NOMAD anchor genome annotations output file. This resulted in the following 5 anchors:
CCTCAGAGGAGGGCGGGAACAGCGTGA, CTCGGTCACTCTGTTCCCGCCCTCCTC, GCCCCCTCGGTCACTCTGTTCCCGCCC, GGGCGGGAACAGCGTGACCGAGGGGGC, TCACTCTGTTCCCGCCCTCCTCTGAGG.
We then fetched the consensus sequences associated with the above IGL-mapping anchors, and converted those consensus sequences into FASTA format. We ran the following command on that FASTA file (denoted by “$fasta”): `blastn -outfmt “$fmt” -query “$fasta” -remote -db nt -evalue 0.1 -task blastn -dust no -word_size 24 -reward 1 -penalty -3 -max_target_seqs 200`, where $fmt corresponds to “6 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue bitscore sseqid sgi sacc slen staxids stitle”.
From this BLAST output, we checked that light chain variable regions were identified, via grep for the term “light chain variable”, yielding 60 sequences. Each cell could have at most 5 contributions to this number, and thus at least 12 cells (conservatively) had NOMAD-identified partial light chain variable sequences.
Timing for SS2
Because code was run on a server with dynamic memory, we report summary statistics as follows. For the steps parallelized by FASTQ file, such as anchor and target retrieval, total time for dataset run, as reported by Nextflow, was parsed per cell. Thus, the average time per cell is reported. For the steps parallelized by 64 files (q-value calculations), total extracted times were summed and divided by number of cells. For steps that consisted of aggregating files, total run time was divided by number of cells. Thus, the total time and memory should be multiplied by the total number of cells to achieve an estimate of the pipeline time for this dataset.
Laptop analysis details
Laptop specs:
An Intel(R) Core(TM) i7–6500U CPU @ 2.50GHz (launched in 2015)
2 cores, total of 4 threads, 3 of which NOMAD was allowed to use.
8 GB DDR3 RAM
SODIMM DDR3 Synchronous 1600 MHz (0.6 ns)
Laptop analysis dataset:
Ten B and T cells from donor 2 blood sequenced by Smart-Seq2 were used for the laptop benchmarking. These files totalled 43,870,027 reads, averaging 4.3M reads per cell. The fastq files for the Tabula Sapiens data were downloaded from https://tabula-sapiens-portal.ds.czbiohub.org/.
Files used:
TSP2_Blood_NA_SS2_B114581_B133053_Lymphocytes_A13_S73_R1_001.fastq.gz
TSP2_Blood_NA_SS2_B114581_B133053_Lymphocytes_A18_S78_R1_001.fastq.gz
TSP2_Blood_NA_SS2_B114581_B133053_Lymphocytes_A19_S79_R1_001.fastq.gz
TSP2_Blood_NA_SS2_B114581_B133053_Lymphocytes_A21_S81_R1_001.fastq.gz
TSP2_Blood_NA_SS2_B114581_B133053_Lymphocytes_A3_S63_R1_001.fastq.gz
TSP2_Blood_NA_SS2_B114581_B133053_Lymphocytes_A5_S65_R1_001.fastq.gz
TSP2_Blood_NA_SS2_B114581_B133053_Lymphocytes_A6_S66_R1_001.fastq.gz
TSP2_Blood_NA_SS2_B114581_B133053_Lymphocytes_A8_S68_R1_001.fastq.gz
TSP2_Blood_NA_SS2_B114581_B133053_Lymphocytes_A9_S69_R1_001.fastq.gz
TSP2_Blood_NA_SS2_B114581_B133053_Lymphocytes_B10_S94_R1_001.fastq.gz
Data and NOMAD runs for Zostera marina and Octopus bimaculoides._
Data was downloaded from SRP327909 (eelgrass), and SRP278619 (Octopus), using nf-core fetchngs run in default mode (Ewels et al., 2020) and preprocessed with FASTP (Chen et al., 2018) run in default mode to mitigate false positive calls due to adapter concatenation. NOMAD was run with gap length=0, anchor_unique_targets_threshold=1, anchor_count_threshold=50, anchor_samples_threshold=1, anchor_sample_counts_threshold=5, and excluding anchor-targets containing poly A / C / G / T run of length 8. 500 pairs of random c and f were chosen. Fastp v0.3.9 was installed on 2/23/23 using bioconda.
Supplemental Tables 6 and 7 – Anchor-target Bowtie and Pfam annotations
Anchor-targets are selected from the NOMAD calls if they do not contain a homopolymer length > 5, if effect size > 0.1, and if the corrected p value < 0.01. Element annotations are run as described on anchor-targets and all hits are reported; anchor-targets mapping to UniVec, Illumina adapters, or to the Ctenopharyngodon idella (grass carp which we used as a control as it contains many artifactual Illumina adapters, personal communication, GCA_019924925) genome are removed. Element annotations are run for anchors; anchor-targets are removed if the anchor maps to UniVec, Illumina adapters, or to the grass carp genome. Anchor-targets with no element annotation are in-silico translated in six frames (−3,−2,−1,1,2,3) to amino acid sequence and submitted to HMMer search of the Pfam database. Element annotations, NOMAD statistics, and the best Pfam hit by full sequence e-value across the 6 translation frames is reported for each of the selected anchor targets. Anchor-targets are aligned with STAR 2.7.5 to a reference index generated from a FASTA of grass carp and COVID-19; anchor-targets for which STAR reports a hit are removed. Anchor-targets are aligned with STAR 2.7.5 to a reference index generated from either the Octopus bimaculoides reference assembly or the Zostera marina nuclear, mitochondrial, and chloroplast genomic FASTA. Element annotations was run using NOMAD commit ID 728066b.
BLAST of anchor-targets in Octopus and Eelgrass
We selected anchors with no more than 1 target mapping with either STAR to the reference genome or Bowtie2 element annotations, thus cases where no sample-specific diversity would be detected if a reference genome were used. A target was selected if its fraction exceeded .5 and its anchor effect size exceeded .9. The 1808 anchor-target pairs satisfying this criterion were BLASTed and sequences mapping to an accession annotated as Octopus sinesis were further analyzed. BLAST hits were merged into table X with an indicator variable for whether the sequence was queried. X/Y had no BLAST hit. Selected anchor-targets are submitted to BLAST with parameters:
-db nt -evalue 0.1 -task blastn -dust no -word_size 24 -reward 1 -penalty -3 -max_target_seqs 4
Generation of Figure 6C.
Anchors are ordered by descending number of observations; the top 200,000 are concatenated with their anchor-targets and submitted to Pfam; these anchor-targets are defined as controls. The best Pfam hit by full sequence e-value is retained for each anchor-target. For each domain, the top 4 domains hit by anchor-targets in Supplemental table 7 and in the controls are plotted, where bar height is the number of anchor-targets that hit the domain with a full-sequence e-value < 0.01.
Generation of contingency table heatmaps.
To plot the anchor-target heatmaps, we exclude targets with low counts. Concretely, we filter out targets that occur fewer than 5 times, have less than 5% of the total counts of that anchor, and retain at most the top 10 targets, while ensuring that at least 2 targets are plotted. Then, all samples with fewer than 5 counts are discarded.
Supplementary Material
Acknowledgements
We thank Elisabeth Meyer for extensive edits, figure help, and assistance with exposition; Arjun Rustagi for extensive discussion and assistance in interpretation of viral biology; Roozbeh Dehghannasiri for selecting and collating data from HLCA/TSP/Tabula Microcebus analysis, running the SpliZ, and feedback on presentation; the Tabula Microcebus Consortium (Camille Ezran and Hannah Frank) for data sharing and discussion of B and T cell receptor detection algorithms; Jessica Klein for extensive figure assistance; Michael Swift for discussion of B and T cell receptor detection algorithms; Julia Olivieri for feedback on presentation and detailed comments on the manuscript; Andy Fire for useful discussions and feedback on the manuscript; Robert Bierman for figure assistance and laptop timing; Sebastian Deorwicz and Marek Kokot for early access to code, and Aaron Straight, Catherine Blish, Peter Kim, and all members of the Salzman lab for feedback and comments.
Funding
J.S. is supported by the Stanford University Discovery Innovation Award, National Institute of General Medical Sciences grant nos. R35 GM139517 and the National Science Foundation Faculty Early Career Development Program Award no. MCB1552196. T.Z.B. is supported by the National Science Foundation Graduate Research Fellowship Program and the Stanford Graduate Fellowship.
Footnotes
Competing interests
K.C., T.Z.B. and J.S. are inventors on provisional patents related to this work. The authors declare no other competing interests.
Data and materials availability
The code used in this work is available as a fully-containerized Nextflow pipeline (Di Tommaso et al., 2017) at https://github.com/salzmanlab/nomad, commit 1b73949d.
The human lung scRNA-seq data used here is accessible through the European Genome-phenome Archive (accession number: EGAS00001004344); FASTQ files from donor 1 and donor 2 were used. The FASTQ files for the Tabula Sapiens data (both 10X Chromium and Smart-seq2) were downloaded from https://tabula-sapiens-portal.ds.czbiohub.org/; B cells were used from donor 1 and T cells from donor 2. The mouse lemur single-cell RNA-seq data used in this study was generated as part of the Tabula Microcebus consortium; the FASTQ files were downloaded from https://tabula-microcebus.ds.czbiohub.org. Viral data was downloaded from the NCBI: SARS-CoV-2 from France (SRP365166), SARS-CoV-2 from South Africa (SRP348159), 2020 SARS-CoV-2 from California (SRR15881549), influenza (SRP294571), and rotavirus (SRP328899).
The sample sheets used as pipeline input, including individual sample SRA accession numbers, for all analyses are uploaded to pipeline GitHub repository. Similarly, scripts to perform supplemental analysis can be found on the pipeline repository.
FASTP v0.23.2 was installed on 2/15/23 using bioconda. R-NOMAD v0.3.9 was installed on 2/23/23, commit 5dafdc8.
Bibliography
- Abante J., Wang P.L. and Salzman J. (2022) “DIVE: a reference-free statistical approach to diversity-generating and mobile genetic element discovery,” BioRxiv [Preprint]. doi: 10.1101/2022.06.13.495703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti A. (1992) “A Survey of Exact Inference for Contingency Tables,” Statistical Science, 7(1), pp. 131–153. doi: 10.1214/ss/1177011454. [DOI] [Google Scholar]
- Albertin C.B. et al. (2015) “The octopus genome and the evolution of cephalopod neural and morphological novelties.,” Nature, 524(7564), pp. 220–224. doi: 10.1038/nature14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharav T.Z., Tse D. and Salzman J. (2023) “An interpretable, finite sample valid alternative to Pearson’s X2 for scientific discovery.” In preparation. [DOI] [PMC free article] [PubMed]
- Bal A. et al. (2022) “Detection and prevalence of SARS-CoV-2 co-infections during the Omicron variant circulation, France, December 2021 - February 2022,” medRxiv [Preprint]. doi: 10.1101/2022.03.24.22272871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. and Yekutieli D. (2001) “The control of the false discovery rate in multiple testing under dependency,” The Annals of Statistics, 29(4), pp. 1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- Bi D. et al. (2012) “ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria.,” Nucleic Acids Research, 40(Database issue), pp. D621–6. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L. et al. (2016) “Near-optimal probabilistic RNA-seq quantification.,” Nature Biotechnology, 34(5), pp. 525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Briney B. et al. (2019) “Commonality despite exceptional diversity in the baseline human antibody repertoire.,” Nature, 566(7744), pp. 393–397. doi: 10.1038/s41586-019-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buen Abad Najar C.F., Yosef N. and Lareau L.F. (2019) “Coverage-dependent bias creates the appearance of binary splicing in single cells,” BioRxiv [Preprint]. doi: 10.1101/2019.12.19.883256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candes E.J. and Wakin M.B. (2008) “An Introduction To Compressive Sampling,” IEEE signal processing magazine, 25(2), pp. 21–30. doi: 10.1109/MSP.2007.914731. [DOI] [Google Scholar]
- Canzar S. et al. (2017) “BASIC: BCR assembly from single cells.,” Bioinformatics, 33(3), pp. 425–427. doi: 10.1093/bioinformatics/btw631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. et al. (2020) “Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells.,” Cell, 182(1), pp. 73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. et al. (2018) “fastp: an ultra-fast all-in-one FASTQ preprocessor.,” Bioinformatics, 34(17), pp. i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. et al. (2005) “Sequential monte carlo methods for statistical analysis of tables,” Journal of the American Statistical Association, 100(469), pp. 109–120. doi: 10.1198/016214504000001303. [DOI] [Google Scholar]
- Chung E. and Romano J.P. (2013) “Exact and asymptotically robust permutation tests,” The Annals of Statistics, 41(2), pp. 484–507. doi: 10.1214/13-AOS1090. [DOI] [Google Scholar]
- Couvin D. et al. (2018) “CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins.,” Nucleic Acids Research, 46(W1), pp. W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghannasiri R. et al. (2022) “Unsupervised reference-free inference reveals unrecognized regulated transcriptomic complexity in human single cells,” BioRxiv [Preprint]. doi: 10.1101/2022.12.06.519414. [DOI] [Google Scholar]
- Diaconis P. and Sturmfels B. (1998) “Algebraic algorithms for sampling from conditional distributions,” The Annals of Statistics, 26(1). doi: 10.1214/aos/1030563990. [DOI] [Google Scholar]
- Di Tommaso P. et al. (2017) “Nextflow enables reproducible computational workflows.,” Nature Biotechnology, 35(4), pp. 316–319. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- Dobin A. et al. (2013) “STAR: ultrafast universal RNA-seq aligner.,” Bioinformatics, 29(1), pp. 15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P. and Tavaré S. (1995) “Coalescents and genealogical structure under neutrality.,” Annual Review of Genetics, 29, pp. 401–421. doi: 10.1146/annurev.ge.29.120195.002153. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2016) “UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing,” BioRxiv [Preprint]. doi: 10.1101/081257. [DOI] [Google Scholar]
- Edgar R.C. et al. (2022) “Petabase-scale sequence alignment catalyses viral discovery.,” Nature, 602(7895), pp. 142–147. doi: 10.1038/s41586-021-04332-2. [DOI] [PubMed] [Google Scholar]
- Elahi S. et al. (2011) “Protective HIV-specific CD8+ T cells evade Treg cell suppression.,” Nature Medicine, 17(8), pp. 989–995. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D.R. et al. (2020) “Systematic detection of horizontal gene transfer across genera among multidrug-resistant bacteria in a single hospital.,” eLife, 9. doi: 10.7554/eLife.53886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P.A. et al. (2020) “The nf-core framework for community-curated bioinformatics pipelines.,” Nature Biotechnology, 38(3), pp. 276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- Ezran C. et al. (2017) “The mouse lemur, a genetic model organism for primate biology, behavior, and health.,” Genetics, 206(2), pp. 651–664. doi: 10.1534/genetics.116.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.A. (1922) “On the Interpretation of X2 from Contingency Tables, and the Calculation of P,” Journal of the Royal Statistical Society, 85(1), p. 87. doi: 10.2307/2340521. [DOI] [Google Scholar]
- Francis J.M. et al. (2022) “Allelic variation in class I HLA determines CD8+ T cell repertoire shape and cross-reactive memory responses to SARS-CoV-2.,” Science Immunology, 7(67), p. eabk3070. doi: 10.1126/sciimmunol.abk3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee M.H. et al. (2018) “Antigen Identification for Orphan T Cell Receptors Expressed on Tumor-Infiltrating Lymphocytes.,” Cell, 172(3), pp. 549–563.e16. doi: 10.1016/j.cell.2017.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Giesen L. et al. (2020) “Molecular basis of chemotactile sensation in octopus.,” Cell, 183(3), pp. 594–604.e14. doi: 10.1016/j.cell.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzynski J.E. et al. (2020) “High-throughput SARS-CoV-2 and host genome sequencing from single nasopharyngeal swabs.,” medRxiv [Preprint]. doi: 10.1101/2020.07.27.20163147. [DOI] [Google Scholar]
- Grant R.A. et al. (2021) “Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia.,” Nature, 590(7847), pp. 635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratia M. et al. (2015) “Rotavirus NSP3 Is a Translational Surrogate of the Poly(A) Binding Protein-Poly(A) Complex.,” Journal of Virology, 89(17), pp. 8773–8782. doi: 10.1128/JVI.01402-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeger A.L. et al. (2010) “Cyclooxygenase-2 generates anti-inflammatory mediators from omega-3 fatty acids.,” Nature Chemical Biology, 6(6), pp. 433–441. doi: 10.1038/nchembio.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashizaki K. et al. (2016) “Myosin light chains 9 and 12 are functional ligands for CD69 that regulate airway inflammation.,” Science Immunology, 1(3), p. eaaf9154. doi: 10.1126/sciimmunol.aaf9154. [DOI] [PubMed] [Google Scholar]
- Jacobs R.P.W.M. and Noten T.M.P.A. (1980) “The annual pattern of the diatoms in the epiphyton of eelgrass (Zostera marina L.) at Roscoff, France,” Aquatic Botany, 8, pp. 355–370. doi: 10.1016/0304-3770(80)90065-0. [DOI] [Google Scholar]
- Jacot D. et al. (2021) “Assessment of SARS-CoV-2 Genome Sequencing: Quality Criteria and Low-Frequency Variants.,” Journal of Clinical Microbiology, 59(10), p. e0094421. doi: 10.1128/JCM.00944-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.S., Eddy S.R. and Portugaly E. (2010) “Hidden Markov model speed heuristic and iterative HMM search procedure.,” BMC Bioinformatics, 11, p. 431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörrißen P. et al. (2021) “Antibody Response to SARS-CoV-2 Membrane Protein in Patients of the Acute and Convalescent Phase of COVID-19.,” Frontiers in Immunology, 12, p. 679841. doi: 10.3389/fimmu.2021.679841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jueterbock A. et al. (2021) “Adaptation of temperate seagrass to arctic light relies on seasonal acclimatization of carbon capture and metabolism.,” Frontiers in plant science, 12, p. 745855. doi: 10.3389/fpls.2021.745855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I. et al. (2021) “Rfam 14: expanded coverage of metagenomic, viral and microRNA families.,” Nucleic Acids Research, 49(D1), pp. D192–D200. doi: 10.1093/nar/gkaa1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiepiela P. et al. (2004) “Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA.,” Nature, 432(7018), pp. 769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- Kim D. et al. (2020) “The Architecture of SARS-CoV-2 Transcriptome.,” Cell, 181(4), pp. 914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., van Buuren N.J. and Mateo R. (2016) “My Cousin, My Enemy: quasispecies suppression of drug resistance.,” Current opinion in virology, 20, pp. 106–111. doi: 10.1016/j.coviro.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S.-M. et al. (2017) “Inhibition of Avian Influenza A Virus Replication in Human Cells by Host Restriction Factor TUFM Is Correlated with Autophagy.,” mBio, 8(3). doi: 10.1128/mBio.00481-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. et al. (2009) “Ultrafast and memory-efficient alignment of short DNA sequences to the human genome.,” Genome Biology, 10(3), p. R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin T.G., Savage D.F. and Davies K.M. (2020) “Recent advances on the structure and function of NDH-1: The complex I of oxygenic photosynthesis.,” Biochimica et biophysica acta. Bioenergetics, 1861(11), p. 148254. doi: 10.1016/j.bbabio.2020.148254. [DOI] [PubMed] [Google Scholar]
- Leplae R. et al. (2004) “ACLAME: a CLAssification of Mobile genetic Elements.,” Nucleic Acids Research, 32(Database issue), pp. D45–9. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman I. et al. (2018) “BraCeR: B-cell-receptor reconstruction and clonality inference from single-cell RNA-seq.,” Nature Methods, 15(8), pp. 563–565. doi: 10.1038/s41592-018-0082-3. [DOI] [PubMed] [Google Scholar]
- Ma M. et al. (2021) “The significance of chloroplast NAD(P)H dehydrogenase complex and its dependent cyclic electron transport in photosynthesis.,” Frontiers in plant science, 12, p. 661863. doi: 10.3389/fpls.2021.661863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. et al. (2021) “Improved chromosome-level genome assembly and annotation of the seagrass, Zostera marina (eelgrass).,” F1000Research, 10, p. 289. doi: 10.12688/f1000research.38156.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T. and Salzberg S.L. (2011) “FLASH: fast length adjustment of short reads to improve genome assemblies.,” Bioinformatics, 27(21), pp. 2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzaraki V. et al. (2017) “The MHC locus and genetic susceptibility to autoimmune and infectious diseases.,” Genome Biology, 18(1), p. 76. doi: 10.1186/s13059-017-1207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhekar B. and Miller J.F. (2007) “Diversity-generating retroelements.,” Current Opinion in Microbiology, 10(4), pp. 388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P. and VanBuren R. (2020) “Building near-complete plant genomes.,” Current Opinion in Plant Biology, 54, pp. 26–33. doi: 10.1016/j.pbi.2019.12.009. [DOI] [PubMed] [Google Scholar]
- Mistry J. et al. (2021) “Pfam: The protein families database in 2021.,” Nucleic Acids Research, 49(D1), pp. D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motahari A. et al. (2013) “Optimal DNA shotgun sequencing: Noisy reads are as good as noiseless reads,” in 2013 IEEE International Symposium on Information Theory. 2013 IEEE International Symposium on Information Theory (ISIT), IEEE, pp. 1640–1644. doi:10.1109/ISIT.2013.6620505. [Google Scholar]
- Nurk S. et al. (2022) “The complete sequence of a human genome.,” Science, 376(6588), pp. 44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri J.E. et al. (2021) “RNA splicing programs define tissue compartments and cell types at single-cell resolution.,” eLife, 10. doi: 10.7554/eLife.70692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascarella G. et al. (2022) “Recombination of repeat elements generates somatic complexity in human genomes.,” Cell, 185(16), pp. 3025–3040.e6. doi: 10.1016/j.cell.2022.06.032. [DOI] [PubMed] [Google Scholar]
- Perrin B.J. and Ervasti J.M. (2010) “The actin gene family: function follows isoform.,” Cytoskeleton, 67(10), pp. 630–634. doi: 10.1002/cm.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh C.M. et al. (2020) “Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients.,” Nature Communications, 11(1), p. 2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prazukin A.V. et al. (2022) “Vertical distribution of epiphytic diatoms in relation to the eelgrass Zostera noltii canopy biomass and height,” Aquatic Botany, 176, p. 103466. doi: 10.1016/j.aquabot.2021.103466. [DOI] [Google Scholar]
- Rock B.M. and Daru B.H. (2021) “Impediments to understanding seagrasses’ response to global change,” Frontiers in Marine Science, 8. doi: 10.3389/fmars.2021.608867. [DOI] [Google Scholar]
- Röhr M.E. et al. (2018) “Blue carbon storage capacity of temperate eelgrass (Zostera marina) meadows,” Global Biogeochemical Cycles [Preprint]. doi: 10.1029/2018GB005941. [DOI] [Google Scholar]
- Romano Y., Sesia M. and Candès E. (2019) “Deep Knockoffs,” Journal of the American Statistical Association, pp. 1–27. doi: 10.1080/01621459.2019.1660174.34012183 [DOI] [Google Scholar]
- Ross K. et al. (2021) “Tncentral: a prokaryotic transposable element database and web portal for transposon analysis.,” mBio, 12(5), p. e0206021. doi: 10.1128/mBio.02060-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J., Jiang H. and Wong W.H. (2011) “Statistical Modeling of RNA-Seq Data.,” Statistical Science, 26(1). doi: 10.1214/10-STS343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria M. et al. (2018) “ITSoneDB: a comprehensive collection of eukaryotic ribosomal RNA Internal Transcribed Spacer 1 (ITS1) sequences.,” Nucleic Acids Research, 46(D1), pp. D127–D132. doi: 10.1093/nar/gkx855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selig C. et al. (2008) “The ITS2 Database II: homology modelling RNA structure for molecular systematics.,” Nucleic Acids Research, 36(Database issue), pp. D377–80. doi: 10.1093/nar/gkm827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. et al. (2016) “SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation.,” Plos One, 11(10), p. e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman R.M. et al. (2019) “Assembly of a pan-genome from deep sequencing of 910 humans of African descent.,” Nature Genetics, 51(1), pp. 30–35. doi: 10.1038/s41588-018-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z.J. et al. (2022) “Fast and accurate metagenotyping of the human gut microbiome with GT-Pro.,” Nature Biotechnology, 40(4), pp. 507–516. doi: 10.1038/s41587-021-01102-3. [DOI] [PubMed] [Google Scholar]
- Shrestha R.P. and Hildebrand M. (2015) “Evidence for a regulatory role of diatom silicon transporters in cellular silicon responses.,” Eukaryotic Cell, 14(1), pp. 29–40. doi: 10.1128/EC.00209-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé M. et al. (2013) “Ultrastructural damage of Loligo vulgaris and Illex coindetii statocysts after low frequency sound exposure.,” Plos One, 8(10), p. e78825. doi: 10.1371/journal.pone.0078825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. et al. (2020) “Reverse genetics reveals a role of rotavirus VP3 phosphodiesterase activity in inhibiting rnase L signaling and contributing to intestinal viral replication in vivo.,” Journal of Virology, 94(9). doi: 10.1128/JVI.01952-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhlberg A. et al. (2016) “Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing.,” Nucleic Acids Research, 44(11), p. e105. doi: 10.1093/nar/gkw224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer J. et al. (2021) “The Dfam community resource of transposable element families, sequence models, and genome annotations.,” Mobile DNA, 12(1), p. 2. doi: 10.1186/s13100-020-00230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. and Whittaker G.R. (2007) “Role of the actin cytoskeleton during influenza virus internalization into polarized epithelial cells.,” Cellular Microbiology, 9(7), pp. 1672–1682. doi: 10.1111/j.1462-5822.2007.00900.x. [DOI] [PubMed] [Google Scholar]
- Tabula Sapiens Consortium* et al. (2022) “The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans.,” Science, 376(6594), p. eabl4896. doi: 10.1126/science.abl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Nucleic Acid Observatory Consortium (2021) “A Global Nucleic Acid Observatory for Biodefense and Planetary Health,” arXiv [Preprint]. doi: 10.48550/arxiv.2108.02678. [DOI] [Google Scholar]
- The Tabula Microcebus Consortium et al. (2021) “Tabula Microcebus: A transcriptomic cell atlas of mouse lemur, an emerging primate model organism,” BioRxiv [Preprint]. doi: 10.1101/2021.12.12.469460. [DOI] [Google Scholar]
- Thompson M.G. et al. (2020) “Viral-induced alternative splicing of host genes promotes influenza replication.,” eLife, 9. doi: 10.7554/eLife.55500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tréguer P. et al. (2018) “Influence of diatom diversity on the ocean biological carbon pump,” Nature Geoscience, 11(1), pp. 27–37. doi: 10.1038/s41561-017-0028-x. [DOI] [Google Scholar]
- Vedula P. et al. (2017) “Diverse functions of homologous actin isoforms are defined by their nucleotide, rather than their amino acid sequence.,” eLife, 6. doi: 10.7554/eLife.31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R. et al. (2022) “Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa.,” Nature, 603(7902), pp. 679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. et al. (2022) “The Human Pangenome Project: a global resource to map genomic diversity.,” Nature, 604(7906), pp. 437–446. doi: 10.1038/s41586-022-04601-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West K.M., Blacksher E. and Burke W. (2017) “Genomics, health disparities, and missed opportunities for the nation’s research agenda.,” The Journal of the American Medical Association, 317(18), pp. 1831–1832. doi: 10.1001/jama.2017.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.L. and Lotze H.K. (2019) “Climate change projections reveal range shifts of eelgrass Zostera marina in the Northwest Atlantic,” Marine Ecology Progress Series, 620, pp. 47–62. doi: 10.3354/meps12973. [DOI] [Google Scholar]
- Wright R.J., Comeau A.M. and Langille M.G.I. (2022) “From defaults to databases: parameter and database choice dramatically impact the performance of metagenomic taxonomic classification tools,” BioRxiv [Preprint]. doi: 10.1101/2022.04.27.489753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-S. et al. (2022) “Chromosome-level genome assembly of grass carp (Ctenopharyngodon idella) provides insights into its genome evolution.,” BMC Genomics, 23(1), p. 271. doi: 10.1186/s12864-022-08503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. et al. (2011) “Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo.,” Science, 331(6018), pp. 757–760. doi: 10.1126/science.1198848. [DOI] [PubMed] [Google Scholar]
- Yu L. et al. (2020) “Somatic genetic drift and multilevel selection in a clonal seagrass.,” Nature Ecology & Evolution, 4(7), pp. 952–962. doi: 10.1038/s41559-020-1196-4. [DOI] [PubMed] [Google Scholar]
- Yu L. et al. (2022) “Ocean currents drive the worldwide colonization of the most widespread marine plant, eelgrass (Zostera marina),” BioRxiv [Preprint]. doi: 10.1101/2022.12.10.519859. [DOI] [Google Scholar]
- Zayed A.A. et al. (2022) “Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome.,” Science, 376(6589), pp. 156–162. doi: 10.1126/science.abm5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-Z. et al. (2015) “Chromothripsis from DNA damage in micronuclei.,” Nature, 522(7555), pp. 179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. (2015) “Hearing characteristics of cephalopods: modeling and environmental impact study.,” Integrative zoology, 10(1), pp. 141–151. doi: 10.1111/1749-4877.12104. [DOI] [PubMed] [Google Scholar]
- Zhao C., Shi Z.J. and Pollard K.S. (2022) “Pitfalls of genotyping microbial communities with rapidly growing genome collections,” BioRxiv [Preprint]. doi: 10.1101/2022.06.30.498336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code used in this work is available as a fully-containerized Nextflow pipeline (Di Tommaso et al., 2017) at https://github.com/salzmanlab/nomad, commit 1b73949d.
The human lung scRNA-seq data used here is accessible through the European Genome-phenome Archive (accession number: EGAS00001004344); FASTQ files from donor 1 and donor 2 were used. The FASTQ files for the Tabula Sapiens data (both 10X Chromium and Smart-seq2) were downloaded from https://tabula-sapiens-portal.ds.czbiohub.org/; B cells were used from donor 1 and T cells from donor 2. The mouse lemur single-cell RNA-seq data used in this study was generated as part of the Tabula Microcebus consortium; the FASTQ files were downloaded from https://tabula-microcebus.ds.czbiohub.org. Viral data was downloaded from the NCBI: SARS-CoV-2 from France (SRP365166), SARS-CoV-2 from South Africa (SRP348159), 2020 SARS-CoV-2 from California (SRR15881549), influenza (SRP294571), and rotavirus (SRP328899).
The sample sheets used as pipeline input, including individual sample SRA accession numbers, for all analyses are uploaded to pipeline GitHub repository. Similarly, scripts to perform supplemental analysis can be found on the pipeline repository.
FASTP v0.23.2 was installed on 2/15/23 using bioconda. R-NOMAD v0.3.9 was installed on 2/23/23, commit 5dafdc8.