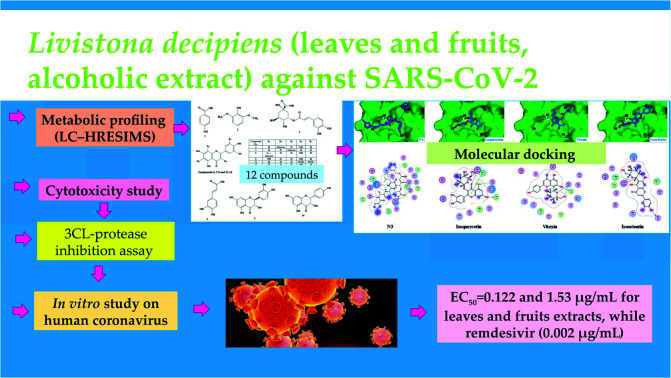
Abstract
In late December 2019, a pandemic coronavirus disease 2019 (COVID-19) emerged in Wuhan, China and spread all over the globe. One of the promising therapeutic techniques of viral infection is to search for enzyme inhibitors among natural phytochemicals using molecular docking to obtain leads with the least side effects. The COVID-19 virus main protease (Mpro) is considered as an attractive target due to its pivotal role in controlling viral transcription and replication. Metabolic profiling of the crude extract of Livistona decipiens Becc. (Arecaceae) leaves and fruit dereplicated twelve metabolites using LC-HRESIMS. Molecular docking simulation and in silico ADME profiling of these annotated compounds proposed that tricin is a promising lead against COVID-19 virus Mpro. The alcoholic extract was shown to inhibit SARS-CoV-2 through in vitro culture and RT-PCR testing with EC50 = 0.122 and 1.53 μg mL−1 for leaves and fruit extracts, respectively, when compared with that of the FDA-approved anti-COVID-19 remdesivir (0.002 μg mL−1). Preliminary steps were also performed including the 3CL-protease inhibition assay and cytotoxicity study. It is worthwhile to find a cheap, safe, natural source for promising anti-SARS-CoV-2 agents that can be further tested in vivo against the COVID-19 virus Mpro. This study provides scientific basis for demonstrating beneficial effects of L. decipiens application on human health during the corona pandemic.
1. Introduction
A pneumonia outbreak of unknown etiology emerged in Wuhan, China in December 2019 and rapidly became an epidemic, ravaging China and subsequently reached a pandemic threat to the whole world. The World Health Organization (WHO) has called this novel coronavirus SARS-CoV-2 disease “COVID-19” and declared it a Public Health Emergency of International Concern (PHEIC).1 SARS-CoV-2 is an RNA-enveloped virus that encodes four structural proteins.2 Exceptionally, the S protein has attracted great attention because of its earlier role during attachment of the virus to the host cell surface receptor called the Angiotensin Converting Enzyme-2 (ACE-2) receptor.3 Upon infection of the host cell, the viral genome is translated into a large polyprotein which is cleaved by viral-encoded proteases (Mpro), releasing several non-structural proteins responsible for replicating the viral genome and generating nested transcripts that are used in the synthesis of viral proteins.4 Various treatment strategies were tested such as azithromycin, chloroquine derivatives, and convalescent plasma. No specific clinically effective antiviral drugs are currently approved for the treatment of SARS-CoV-2 infections.5 There is a global urgent need to find an efficient safe antiviral therapy for this pandemic disease.
Natural products are considered as viable lead candidates that have a highly significant role in drug discovery.6–9Livistona decipiens Becc. mostly known as ribbon fan palm is one of the palm species belonging to the Arecaceae family.10 It is rich in important metabolites including flavonoid glycosides such as vitexin, vicenin II, orientin, and aglycones such as quercetin and apigenin.11,12 Biological reports on L. decipiens showed that it has antihyperlipidemic, antioxidant, cytotoxic, antimicrobial and ulceroprotective activities.11 To the best of the authors' knowledge, there have been no previous studies about its effect on SARS-CoV-2. Interestingly, it was reported that another species, L. chinensis, possesses antiviral activity against human immunodeficiency virus type 1 (HIV-1).12
Therefore, the objectives of this study were to prepare the alcoholic extract of L. decipiens aerial parts, identify its components using the LC-HRESIMS technique and investigate its anti-COVID-19 activity using the preliminary steps of a molecular docking study, 3CL-protease inhibition assay, and cytotoxic activity study. Finally, to assess the potential ability of the alcoholic extract to affect human SARS-CoV-2, an in vitro assay was used with the aid of remdesivir as reference.
2. Materials and methods
2.1. Plant material and preparation of the alcoholic extract of L. decipiens
The fruit and leaves of Livistona decipiens Becc. were separately collected on Jan. 2018 from the El-Orman Botanical Garden, Giza, Egypt. They were kindly identified by Eng. Trease Labib, former-Head of El Orman Botanical Garden and Dr Reem Samir Hamdy, Lecturer of Plant Taxonomy, Botany Department, Faculty of Science, Cairo University, Giza, Egypt. A voucher specimen (BUPD-72-2018) was deposited at the Department of Pharmacognosy, Faculty of Pharmacy, Beni-Suef University, Egypt. The air-dried and finely powdered leaves (30 g) and fruit (50 g) of L. decipiens were exhaustively macerated separately using ethanol 70% (3 × 30 mL, each) at room temperature and concentrated under vacuum to afford 230 and 450 mg crude extracts, respectively.
2.2. Metabolic profiling
The alcoholic extracts were separately subjected to LC-HRESIMS analysis according to Owis et al. 2020.5 LC-HRESIMS analysis was performed on an Acquity Ultra-Performance Liquid Chromatography system coupled to a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, MA, USA). Chromatographic separation was carried out using a BEH C18 column of 2.1 × 100 mm and 1.7 μm particle size (Waters, Milford, USA) in addition to a guard column of 2.1 × 5 mm, 1.7 μm particle size. A linear binary gradient solvent system was used consisting of 0–100% eluent B for 6 min, at a flow rate of 0.3 mL min−1 and 0.1% formic acid in water (v/v) as solvent A in addition to acetonitrile as solvent B. The column temperature was set at 40 °C and the injection volume was adjusted at 2 μL. After chromatographic separation, the metabolites were detected using mass spectrometry by electrospray ionization (ESI). The source was set at 120 °C. The ESI capillary voltage was operated at 0.8 kV and the sampling cone voltage was adjusted to 25 V. Nitrogen at a flow rate of 800 L h−1, at 350 °C was used as the desolvation gas while the cone gas was at a flow rate of 30 L h−1. The mass range was set at m/z 50–1200 for TOF-MS. In MZmine, the RAW data is imported by selecting the ProteoWizard-converted negative files in mzML format. The peaks were detected using a chromatogram builder and chromatogram deconvolution. A local minimum search algorithm was applied, and the isotopes were also identified via the isotopic peaks' grouper. The missing peaks were detected by a gap-filling peak finder. An adduct search and a complex search was performed. The processed data set was subjected to a molecular formula prediction as well as peak identification. The negative ionization mode data sets from the plant extract were dereplicated against the Dictionary of Natural Products database (DNP).
2.3. Molecular docking simulation and in silico ADME profiling
Molecular modeling and visualization processes were performed using Molecular Operating Environment (MOE) 2019.01 software (Chemical Computing Group, Montreal, QC, Canada). First, the X-ray structure of COVID-19 Mpro protease (PDB code 7BQY) was retrieved from the protein data bank. Then, the MOE QuickPrep protocol was used to prepare the protein structure, followed by removing all the water molecules. Docking calculations were performed using the induced fit method, where the co-crystallized ligand N3 was considered as the center of the binding site.13 All other parameters, such as the force field and the number of docking poses, were left untouched as in their MOE default settings. To assess our docking results, two structurally similar COVID-19 virus Mpro inhibitors, namely cinanserin and shikonin, were also docked with our ligands. The generated docking poses were visually inspected, where the binding free energy (ΔG) in kcal mol−1 of all ligands was calculated for the best docking pose with the top-score.14 The Swiss ADME online database was used to predict the drug-likeness and ADME properties of all ligands in the present study.15
2.4. 3CL-protease inhibition assay
The 3CL-protease inhibition assay was used to assess the effect of L. decipiens leaves and fruit extracts on the inhibition of the main protease (Mpro) enzyme which is responsible for viral replication and transcription and consequently affects human SARS-CoV-2. The assay was performed according to the manufacturer's protocol (3CL Protease (SARS-CoV-2) Assay Kit, BPS Bioscience). The fluorescence intensity was measured in a microtiter plate-reading fluorimeter (TECAN reader) capable of excitation at a wavelength of 360 nm and detection of emission at a wavelength of 460 nm.
2.5. In vitro study on human coronavirus
The human lung adenocarcinoma epithelial cell line (A549, American Type Culture Collection, USA) and human coronavirus (COV 19) were kindly supplied from VACSERA, Egypt. Prior to assessment of the antiviral activity of L. decipiens leaves and fruit extracts, a cytotoxicity study was performed using the MTT assay as described by Mosmann, 1983 (ref. 16) and the absorbance was measured at 530 nm wavelength. To evaluate the antiviral potential of the L. decipiens extracts against SARS-CoV-2, a classical cell culture was performed.17 The percentage of inhibition of SARS-CoV-2 replication was determined through estimation of the viral RNA concentration using a quantitative RT-PCR (real time-polymerase chain reaction) assay performed according to the manufacturer's protocol (The genesig® Real-Time PCR Coronavirus COVID-19 (CE IVD), Primerdesign Ltd) and remdesivir was used as a reference standard.
2.6. Statistical analysis
The previous experiments were performed in triplicate and the data were presented as mean ± standard error (SE). For studying the significance of differences between results, a one-way analysis of variance (ANOVA) was used.
3. Results and discussion
3.1. Metabolic analysis
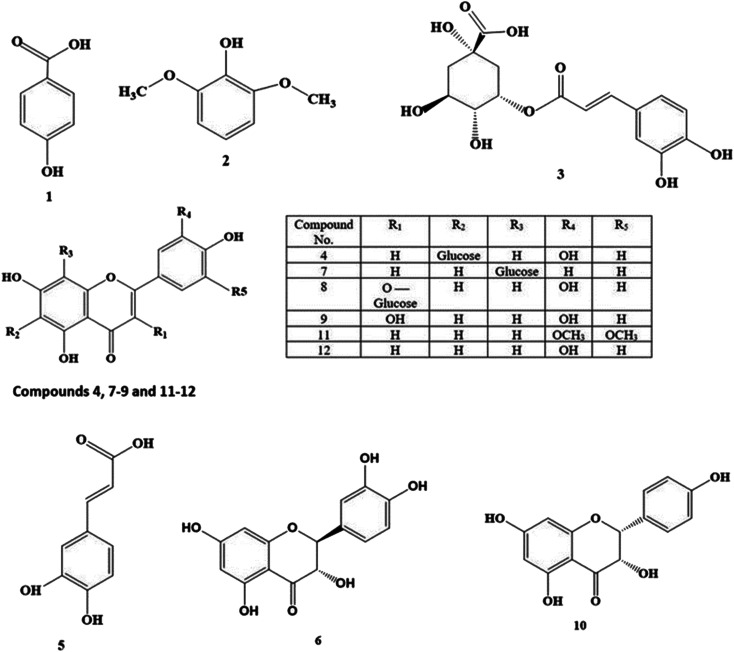
LC-HRESIMS is a useful tool for characterization of different secondary metabolites present in a complex system such as plants.18–20 Metabolic profiling of L. decipiens leaves and fruit, led to the identification of 12 compounds (Table 1, Fig. 1). The individual components belong to several classes including eight flavonoids (five aglycones and three glucosides), phenolic acids (two caffeic acid and one benzoic acid derivatives) and a methoxylated phenol.
LC-HR-ESIMS dereplication results of the alcoholic extract of Livistona decipiens leaves and fruita.
| No. | Metabolite name | L. decipiens | RT (min) | MF | m/z | Calculated m/z | |
|---|---|---|---|---|---|---|---|
| Leaves | Fruit | ||||||
| 1 | ρ-Hydroxybenzoic acid | ✗ | ✓ | 2.0135 | C7H6O3 | 137.0238 | 138.0317 |
| 2 | Syringol | ✗ | ✓ | 2.0486 | C8H10O3 | 153.0557 | 154.0629 |
| 3 | Neochlorogenic acid | ✓ | ✓ | 2.2486 | C16H18O9 | 353.0881 | 354.0951 |
| 4 | Isoorientin | ✓ | ✓ | 2.3678 | C21H20O11 | 447.0932 | 448.1006 |
| 5 | Caffeic acid | ✓ | ✓ | 2.3739 | C9H8O4 | 179.0346 | 180.0422 |
| 6 | (+)-Catechin | ✓ | ✓ | 2.4451 | C15H14O6 | 289.0713 | 290.0790 |
| 7 | Vitexin | ✓ | ✗ | 2.6408 | C21H20O10 | 431.0974 | 432.1056 |
| 8 | Isoquercetin | ✓ | ✓ | 2.7137 | C21H20O12 | 463.0876 | 464.0955 |
| 9 | Quercetin | ✗ | ✓ | 2.7605 | C15H10O7 | 301.0351 | 302.0427 |
| 10 | (−)-Epiafzelechin | ✗ | ✓ | 2.9733 | C15H14O5 | 273.0766 | 274.0841 |
| 11 | Tricin | ✗ | ✓ | 3.8475 | C17H14O7 | 329.0658 | 330.0739 |
| 12 | Luteolin | ✓ | ✓ | 4.2993 | C15H10O6 | 285.0763 | 286.0477 |
MF: molecular formula, RT: retention time, min: minute.
Fig. 1. Structures of the dereplicated metabolites from the alcoholic extract of Livistona decipiens leaves and fruit.
3.2. Molecular docking simulation and in silico ADME profiling
Recently, Jo et al. reported that isoquercetin has reasonable inhibitory activity against MERS-CoV 3C-like protease with an IC50 of 37.03 μM.21 Moreover, shikonin is a clinical trial antitumor drug candidate isolated from Lithospermum erythrorhizon that has recently been reported to have a potent inhibitory activity against COVID-19 virus Mpro with IC50 of 15.75 ± 8.22 μM.22 Shikonin is a naphthoquinone derivative that is structurally similar to flavonoids. Likewise, cinanserin is a cinnamic acid derivative that has moderate antiviral activity against COVID-19 virus with IC50 of 20.61 ± 0.97 μM.22 Our dereplicated compounds contain isoquercetin and other structurally related flavonoids as well as cinnamic acid derivatives, thus our ligands are more likely to have a comparable inhibitory activity against the COVID-19 virus. Subsequently, a flexible molecular docking simulation was performed to predict the binding affinities and orientations of our ligands within the substrate-binding site of the COVID-19 virus Mpro.
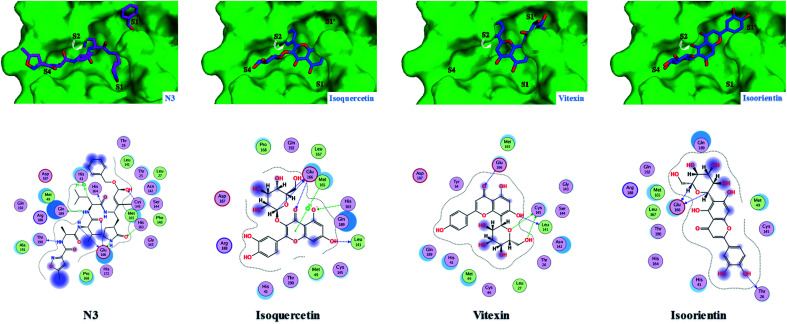
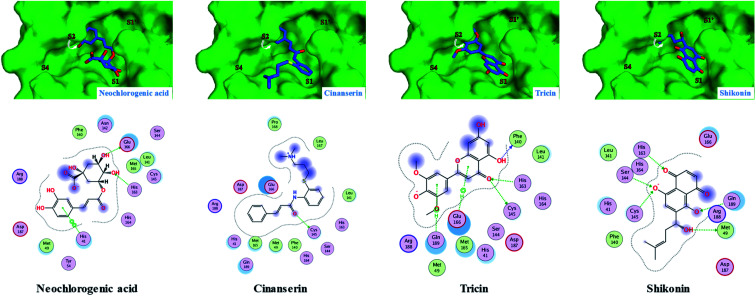
The top-ranked poses for our ligands bound to the substrate-binding site of the COVID-19 virus Mpro (PDB code: 7BQY)22 compared to two COVID-19 virus Mpro inhibitors (cinanserin and shikonin) and the co-crystallized inhibitor N3 are illustrated in Fig. 2–5 as predicted by Molecular Operating Environment (MOE) 2019.01 software (Chemical Computing Group, Montreal, QC, Canada). Besides, the reported IC50 of the two COVID-19 virus Mpro inhibitors (cinanserin and shikonin) and the predicted binding free energy (ΔG) of these two inhibitors as well as our ligands to COVID-19 virus Mpro protein are summarized in Table 2.
Fig. 2. Predicted 2D/3D docking poses of three flavonoid glucosides (isoquercetin, vitexin, and isoorientin) occupying the same pocket as the co-crystallized ligand (N3) showing their binding interactions with the key amino acids in the active site of COVID-19 virus Mpro. Mpro is shown as the green background, N3 is in magenta and the flavonoids are in blue.
Fig. 3. Predicted 2D/3D docking poses of neochlorogenic acid and tricin compared to their corresponding structurally similar COVID-19 virus Mpro inhibitors, namely cinanserin and shikonin showing their binding interactions with the key amino acids in the active site of the COVID-19 virus Mpro. Mpro is shown as the green background and the ligands are in blue.
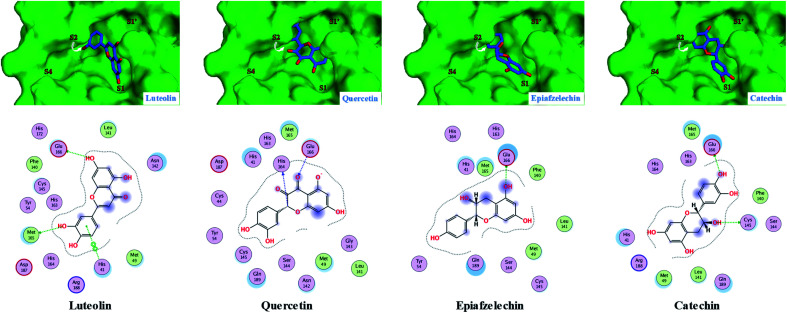
Fig. 4. Predicted 2D/3D docking poses of four flavonoids (luteolin, quercetin, epiafzelechin and catechin) showing their binding interactions with the key amino acids in the active site of the COVID-19 virus Mpro. Mpro is shown as the green background and the ligands are in blue.
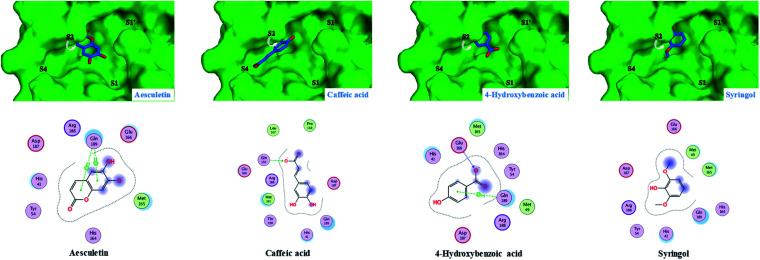
Fig. 5. Predicted 2D/3D docking poses of four dereplicated compounds (aesculetin, caffeic acid, 4-hydroxybenzoic acid and syringol) showing their binding interactions with the key amino acids in the active site of the COVID-19 virus Mpro. Mpro is shown as the green background and the ligands are in blue.
Predicted binding free energy (ΔG) in kcal mol−1 for dereplicated compounds with the active site of the COVID-19 virus Mpro (PDB 7BQY; co-crystallized with N3) compared to two structurally similar COVID-19 virus Mpro inhibitors, namely cinanserin and shikonin.
| Ligand | Predicted ΔG (kcal mol−1) COVID-19 virus Mpro | In vitro COVID-19 virus Mpro IC50a (μM) | Antiviral activity determined by qRT-PCRa (μM) |
|---|---|---|---|
| Isoquercetin (8) | −8.2 | NDb | NDb |
| Vitexin (7) | −7.6 | NDb | NDb |
| Isoorientin (4) | −7.6 | NDb | NDb |
| Cinanserin | −6.9 | 124.93 ± 7.89 | 20.61 ± 0.97 |
| Neochlorogenic acid (3) | −6.8 | NDb | NDb |
| Tricin (11) | −6.7 | NDb | NDb |
| Shikonin | −6.5 | 15.75 ± 8.22 | NDb |
| Quercetin (9) | −6.4 | NDb | NDb |
| Luteolin (12) | −6.2 | NDb | NDb |
| Epiafzelechin (10) | −6.2 | NDb | NDb |
| Catechin (6) | −5.8 | NDb | NDb |
| Caffeic acid (5) | −4.8 | NDb | NDb |
| Syringol (2) | −4.8 | NDb | NDb |
| Aesculetin | −4.7 | NDb | NDb |
| ρ-Hydroxybenzoic acid (1) | −4.4 | NDb | NDb |
In vitro COVID-19 virus Mpro IC50 and antiviral activity shown as reported (Jin et al. 2020).
ND, not determined.
It is clear from Fig. 2 that the top three hits; namely isoquercetin, vitexin, and isoorientin, are occupying the same pocket as the co-crystallized ligand (N3) in the active site of COVID-19 virus Mpro. Generally, all other docked ligands occupy mainly the S1 and S2 sites, except caffeic acid which occupies S2 and S4, as shown in Fig. 3–5.
As described in Table 2, the flexible docking results showed that the three flavonoid glucosides isoquercetin (−8.2), vitexin (−7.6), and isoorientin (−7.6) were the top ranked ligands followed by the five flavonoid aglycones such as tricin (−6.7), quercetin (−6.4), luteolin (−6.2), epiafzelechin (−6.2) and catechin (−5.8). The high binding affinity of these three flavonoid glucosides to COVID-19 virus Mpro can be explained by the ability of their saccharide moiety to make strong interactions with some key amino acids (Glu166 and Gln169) as represented in Fig. 2. This result is further emphasized in Fig. 2 where the glucoside moiety of both isoquercetin and isoorientin projects in S4 site, however it occupies S1′ in case of vitexin. Conversely, the lack of glucoside moiety in other docked flavonoid aglycones could illustrate their relative moderate affinity to the COVID-19 virus Mpro (Fig. 3 and 4). Small sized ligands such as caffeic acid, syringol, aesculetin and 4-hydroxybenzoic acid showed a binding free energy (ΔG) of less than 5 kcal mol−1 (Table 2). This weak binding affinity could be rationalized by the fact that their smaller surface area makes very few interactions with the amino acid residues in the protein active site as demonstrated in Fig. 5.
All ligands obey Lipinski's rule of five except the three flavonoid glucosides and neochlorogenic acid (Table 3). These four ligands have one or two violations to the Lipinski rule of drug likeness due to the presence of glucoside or the glucoside-like moiety, which decreases GI absorption and consequently results in poor bioavailability. It is very obvious that the glucoside moiety plays a key role in the binding affinity, but it has a negative impact on the drug-likeness and ADME properties (Table 3). Thus, flavonoid aglycones are more likely to be good leads for further optimization. For developing drugs after their optimization, pan-assay interference compounds (PAINS) are important features to be considered to remove false positive hits.23 All the compounds were found to have one or two structural alerts in PAINS structural alerts, except five ligands and the COVID-19 virus Mpro inhibitor, cinanserin. These five ligands are vitexin, tricin, epiafzelechin, syringol and 4-hydroxybenzoic acid. All these five ligands have some flaws in their ADME profiles except tricin. Although tricin has a moderate binding affinity to the COVID-19 virus Mpro, it has acceptable drug-like properties and an ADME profile with no PAINS alerts. These properties mean that tricin is a promising lead compound that can be further optimized as a potential antiviral candidate to eradicate the COVID-19 virus. Taking this all together, our in silico study sheds light on the importance of flavonoids as a promising scaffold to eradicate the COVID-19 virus, especially tricin.
Drug-likeness based on Lipinski's rule of five, ADME properties and medicinal chemistry parameters.
| Ligand | # of violations | Pgp substrate | GI absorption | Bioavailability score | PAINS alerts |
|---|---|---|---|---|---|
| Isoquercetin (8) | 2 | No | Low | 0.17 | 1 |
| Vitexin (7) | 1 | No | Low | 0.55 | 0 |
| Isoorientin (4) | 2 | No | Low | 0.17 | 1 |
| Cinanserin | 0 | No | High | 0.55 | 0 |
| Neochlorogenic acid (3) | 1 | No | Low | 0.11 | 1 |
| Tricin (11) | 0 | No | High | 0.55 | 0 |
| Shikonin | 0 | No | High | 0.55 | 2 |
| Quercetin (9) | 0 | No | High | 0.55 | 1 |
| Luteolin (12) | 0 | No | High | 0.55 | 1 |
| Epiafzelechin (10) | 0 | Yes | High | 0.55 | 0 |
| Catechin (6) | 0 | Yes | High | 0.55 | 1 |
| Caffeic acid (5) | 0 | No | High | 0.56 | 1 |
| Syringol (2) | 0 | No | High | 0.55 | 0 |
| Aesculetin | 0 | No | High | 0.55 | 1 |
| ρ-Hydroxybenzoic acid (1) | 0 | No | High | 0.56 | 0 |
3.3. Cytotoxicity, 3CL-protease inhibition assay and in vitro study on the human coronavirus
Safety and possible secondary effects are considered as major requirements during the search for a new antiviral lead. The natural L. decipiens extracts were screened for their cellular toxicity effect and for determination of the appropriate concentration for the in vitro inhibition assay. The estimated concentration associated with 50% cytotoxicity (CC50), was found to be 463.4 ± 26, 1451 ± 81.5 and 222.9 ± 12.5 μg mL−1 for L. decipiens leaves, fruit extracts and remdesivir, respectively. This means that the safest one is remdesivir followed by leaves and fruit extracts.
The 3CL protease inhibition assay was performed as a preliminary practical evaluation. L. decipiens exhibited a significant inhibitory action against 3CL-protease with IC50 = 15.58 ± 0.79 and 41.81 ± 2.12 μg mL−1 for leaves and fruit extracts. This result is consistent with the stabilized binding of compounds (1–12) at the binding site of Mpro.
The significant results of the effect of L. decipiens leaves and fruit extracts towards 3CL-protease highlighted the need to investigate their behavior on a cell culture basis. The prospective anti-human coronavirus effect of both extracts was investigated by an in vitro RT-PCR (real time-polymerase chain reaction) test which specifically detects SARS-CoV-2 RNA quantitatively. L. decipiens exhibited a significant inhibitory action against SARS-CoV-2 with EC50 = 0.122 and 1.53 μg mL−1 for leaves and fruit extracts, respectively, when compared with that of remdesivir (0.002 μg mL−1). This could be attributed to the presence of the flavonoid glucosides isoquercetin and isoorientin in addition to the flavonoid aglycones such as luteolin and catechin. It was worthwhile to find a natural product with the ability to inhibit SARS-CoV-2 replication at a value close to that of the FDA-approved anti-COVID-19 remdesivir.
Research shows that COVID-19 disease is characterized by an overexuberant inflammatory response leading to a major lung illness and consequent possible mortality. Anti-inflammatory agents are likely to be effective against the subsequent elevated cytokines levels, such as γ-interferon, typically associated with COVID-19 patients.24L. decipiens extract and its metabolites as anti-inflammatory natural agents12 are not only likely to reduce SARS-CoV-2 viral infectivity, but are also likely to reduce the host inflammatory response. Moreover, L. decipiens itself as a natural plant has the advantages of being a cheap, readily available, and safe drug source.
4. Conclusions
SARS-CoV-2 is the main causative agent of the COVID-19 pneumonia pandemic attack that emerged in Wuhan. Discovering known natural agents that can inhibit the SARS-CoV-2 main protease (Mpro) will lead to controlling viral transcription and replication. Twelve known compounds were identified from the alcoholic extract of L. decipiens leaves and fruit using LC-HRESIMS. The individual components belong to several classes including eight flavonoids (five aglycones and three glucosides), phenolic acids (two caffeic acid and one benzoic acid derivatives) and a methoxylated phenol. Identification of the annotated compounds enabled testing of the relationship between their interactions with the receptor in the N3 binding site in Mpro and their structure. The in silico structure-based screening and ADME study revealed that tricin is a potential anti-COVID lead compound. The alcoholic extract was shown to inhibit SARS-CoV-2 through the in vitro culture and RT-PCR test with EC50 = 0.122 and 1.53 μg mL−1 for leaves and fruit extracts, respectively, when compared with that of remdesivir (0.002 μg mL−1). This study provides scientific basis for demonstrating the health benefits of L. decipiens. In vivo testing is required to verify the observed effect of the active phytochemicals.
Conflicts of interest
The authors declare that there is no conflict of interest.
Supplementary Material
Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra01306a
References
- Team E. E. Eurosurveillance. 2020;25:200131e. [Google Scholar]
- Luan J., Lu Y., Jin X. and Zhang L., Biochemical and biophysical research communications, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V. Jung Y.-S. Liang P.-H. Expert Opin. Ther. Pat. 2013;23:1337–1348. doi: 10.1517/13543776.2013.823159. [DOI] [PubMed] [Google Scholar]
- Marra M. A. Jones S. J. Astell C. R. Holt R. A. Brooks-Wilson A. Butterfield Y. S. Khattra J. Asano J. K. Barber S. A. Chan S. Y. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Owis A. I. El-Hawary M. S. El Amir D. Aly O. M. Abdelmohsen U. R. Kamel M. S. RSC Adv. 2020;10:19570–19575. doi: 10.1039/D0RA03582C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owis A. I. El-Hawary M. S. El Amir D. Refaat H. Alaaeldin E. Aly O. M. Elrehany M. A. Kamel M. S. RSC Adv. 2021;11:13537–13544. doi: 10.1039/D1RA00142F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hawary S. S. Rabeh M. A. Raey M. A. E. El-Kadder E. M. A. Sobeh M. Abdelmohsen U. R. Albohy A. Andrianov A. M. Bosko I. P. Al-Sanea M. M. J. Biomol. Struct. Dyn. 2021:1–13. doi: 10.1080/07391102.2021.1885494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hawary S. S. Issa M. Y. Ebrahim H. S. Mohammed A. F. Hayallah A. M. El-Kadder E. M. A. Sayed A. M. Abdelmohsen U. R. Nat. Prod. Res. 2021:1–5. doi: 10.1080/14786419.2021.1917573. [DOI] [PubMed] [Google Scholar]
- El-Hawary S. S. Mohammed R. Bahr H. S. Attia E. Z. El-Katatny M. m. H. Abelyan N. Al-Sanea M. M. Moawad A. S. Abdelmohsen U. R. J. Appl. Microbiol. 2021;131:1193–1211. doi: 10.1111/jam.15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadry H. Shoala S. Gindi O. E. Sleem A. A. Mosharrafa S. Kassem M. Nat. Prod. Commun. 2009;4:1934578X0900400220. doi: 10.1177/1934578X0900400220. [DOI] [PubMed] [Google Scholar]
- Ibrahim H. Elshaarawy F. Haggag E. J. Adv. Pharm. Res. 2018;2:256–268. [Google Scholar]
- Ali R. F. A., CU theses, Cairo University, 2019 [Google Scholar]
- Abdelbaset M. S. Abdel-Aziz M. Ramadan M. Abdelrahman M. H. Bukhari S. N. A. Ali T. F. Abuo-Rahma G. E.-D. A. Bioorg. Med. Chem. 2019;27:1076–1086. doi: 10.1016/j.bmc.2019.02.012. [DOI] [PubMed] [Google Scholar]
- Ali T. F. Ciftci H. I. Radwan M. O. Koga R. Ohsugi T. Okiyama Y. Honma T. Nakata A. Ito A. Yoshida M. Bioorg. Med. Chem. 2019;27:1767–1775. doi: 10.1016/j.bmc.2019.03.003. [DOI] [PubMed] [Google Scholar]
- Daina A. Michielin O. Zoete V. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Günther S. Asper M. Röser C. Luna L. K. Drosten C. Becker-Ziaja B. Borowski P. Chen H.-M. Hosmane R. S. Antiviral Res. 2004;63:209–215. doi: 10.1016/j.antiviral.2004.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owis A. I. El-Naggar E.-M. B. Pharmacogn. Mag. 2016;12:S1. doi: 10.4103/0973-1296.176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag M. A. El-Din M. G. S. Selim M. A.-F. Owis A. I. Abouzid S. F. Food Biosci. 2021;39:100800. doi: 10.1016/j.fbio.2020.100800. [DOI] [Google Scholar]
- Owis A. I. Stud. Nat. Prod. Chem. 2018;59:509–530. [Google Scholar]
- Jo S. Kim H. Kim S. Shin D. H. Kim M. S. Chem. Biol. Drug Des. 2019;94:2023–2030. doi: 10.1111/cbdd.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. Y. Liu F. Jiang R. Yang X. You T. Liu X. Xiao C. Q. Shi Z. Jiang H. Rao Z. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Capuzzi S. J. Muratov E. N. Tropsha A. J. Chem. Inf. Model. 2017;57:417–427. doi: 10.1021/acs.jcim.6b00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J. Phelan A. Griffin I. Tucker C. Oechsle O. Smith D. Richardson P. Lancet Infect. Dis. 2020;20:400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.