Abstract
New variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continue to arise and prolong the coronavirus disease 2019 (COVID-19) pandemic. Here we used a cell-free expression workflow to rapidly screen and optimize constructs containing multiple computationally designed miniprotein inhibitors of SARS-CoV-2. We found the broadest efficacy with a homo-trimeric version of the 75-residue angiotensin converting enzyme 2 (ACE2) mimic AHB2 (TRI2–2) designed to geometrically match the trimeric spike architecture. In the cryo-electron microscopy structure, TRI2 formed a tripod on top of the spike protein which engaged all three receptor binding domains (RBDs) simultaneously as in the design model. TRI2–2 neutralized Omicron (B.1.1.529), Delta (B.1.617.2), and all other variants tested with greater potency than that of monoclonal antibodies used clinically for the treatment of COVID-19. TRI2–2 also conferred prophylactic and therapeutic protection against SARS-CoV-2 challenge when administered intranasally in mice. Designed miniprotein receptor mimics geometrically arrayed to match pathogen receptor binding sites could be a widely applicable antiviral therapeutic strategy with advantages over antibodies and native receptor traps. By comparison, the designed proteins have resistance to viral escape and antigenic drift by construction, precisely tuned avidity, and greatly reduced chance of autoimmune responses.
ONE SENTENCE SUMMARY
Computationally designed trivalent minibinders provide therapeutic protection in mice against emerging SARS-CoV-2 variants of concern.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to cause a global pandemic with more than 300 million infections and 5.5 million deaths as of January 2022 (https://covid19.who.int/). Monoclonal antibodies (mAbs) targeting the SARS-CoV-2 spike (S) glycoprotein (1) have been an effective treatment for improving outcomes for patients with coronavirus disease 2019 (COVID-19) (2–5), but many are sensitive to viral escape through point mutations in their epitopes on the S trimer (6, 7), and producing mAbs in sufficient quantities for population scale use during a global pandemic is technically and financially challenging (8). Indeed, the continued emergence of variants of concern (VOCs) jeopardizes the effectiveness of currently approved mAb treatments and vaccines (9–14). In particular mutations in the rapidly spreading B.1.1.529 (Omicron) variant disrupt binding of most receptor binding motif-targeted mAbs, and have been shown to reduce neutralization potency more than 100-fold for five of the seven clinical mAbs used for the prophylactic or therapeutic treatment of COVID-19 (15–18). Thus, there is an urgent need for interventions whose efficacy is not disrupted by the observed ongoing antigenic drift, as is the case for a few mAbs (19–24).
As an alternative to mAbs, we previously computationally designed two classes of minibinder proteins that block the SARS-CoV-2 receptor binding domain (RBD) interaction with its host receptor, angiotensin converting enzyme 2 (ACE2) (25). The first class, exemplified by AHB2, adopts a similar binding mode to and incorporates residues from the main RBD-interacting helix of ACE2 in a custom designed 3-helix bundle that has low overall sequence similarity with ACE2 (fig. S1). The second class, exemplified by LCB1 and LCB3, contain an entirely new designed RBD binding interface. These minibinders neutralize the WA1/2020 SARS-CoV-2 virus with half maximal inhibitory concentration (IC50) values in the range of 23 pM (LCB1) to 15 nM (AHB2) (25). The designs express at high concentrations in Escherichia coli and are highly thermostable (25), which could considerably streamline manufacturing and decrease the cost of goods for clinical development. LCB1 has demonstrated protective activity as both pre-exposure prophylaxis and post-exposure therapy in human ACE2 (hACE2)-expressing transgenic mice, but mutations in the B.1.351 (Beta) and P.1 (Gamma) VOCs were shown to reduce binding potency (26, 27).
Here, we sought to develop constructs containing three minibinder domains that could simultaneously engage all three RBDs on a single S protein, and by virtue of this multivalent binding, potently neutralize SARS-CoV-2 variants. Multivalency can increase the apparent affinity for target antigens (28–30), including against SARS-CoV-2 (31–36). We considered two classes of constructs. The first contain multiple distinct minibinder domains linked together to maximize RBD binding avidity; these constructs have the advantages that LCB1 and LCB3 are very high affinity binders on their own, and the three domains contain different sets of contacts with the RBD, making escape in principle more difficult (32, 37). The second consists of self-assembling homotrimers of minibinders geometrically matched to the 3 RBDs on a single spike; although AHB2 is lower affinity than LCB1 and LCB3, and the sites targeted are less diverse than the first class, homotrimers of AHB2 have the advantage that the ACE2 binding site is inherently less mutable as the virus must bind ACE2 to infect cells (24, 38). We describe the design, optimization, and escape resistance of both classes of constructs. We find that the top constructs have considerable promise as potential countermeasures in the ongoing COVID-19 pandemic.
RESULTS
RBD mutations impact minibinder binding.
To determine the potential for mutations to arise that disrupt LCB1 and AHB2 binding to the RBD, we performed deep mutational scans using site saturation mutagenesis of the RBD (38). We found that for LCB1, the widely observed K417N mutation results in a greater than 10-fold reduction in affinity and the E406W and Y453K/R mutations result in a greater than 100-fold reduction in affinity (fig. S2). For AHB2, we similarly observed several mutations, including K417N, E406W, and Y453K/R, that reduce the affinity of the minibinder for the RBD.
Multivalent minibinders bind to SARS-CoV-2 RBDs.
To improve the ability of the minibinders to neutralize circulating SARS-CoV-2 variants, we developed multivalent versions with geometries enabling simultaneous engagement of all 3 RBDs in a single S trimer (1) to increase binding avidity. Multivalent minibinders might be less sensitive to mutations that would escape binding of the monovalent minibinders; a 100x reduction in binding affinity of a sub-picomolar binder would still result in an affinity in a therapeutic range in a multivalent construct (39). We also hypothesized that constructs with binding domains containing different sets of contacts with the target epitope could prevent escape (32, 37). To design multivalent constructs, we started from optimized versions of the previously described LCB1, AHB2, and LCB3 minibinders (hereafter referred to as monomers MON1, MON2, and MON3, respectively; table S1) (25).
To rapidly prototype multivalent minibinder designs, we developed a cell-free protein synthesis (CFPS) workflow which combines an in vitro DNA assembly step followed by polymerase chain reaction (PCR) to generate linear expression templates that are used to drive CFPS and enable rapid prototyping of new minibinder designs (fig. S3). The workflow enables assembly and translation of synthetic genes and generation of purified protein in as little as 6 hours, is compatible with high-throughput, automated experimentation using an acoustic liquid handler (Echo 525), and is easily scaled for the production of mg quantities of protein (40, 41). To assess multivalent binding, we coupled the workflow to an AlphaLISA protein-protein interaction (PPI) competition assay to enable comparison of dissociation rates of the designed proteins against either the monomeric RBD or the trimeric HexaPro SARS-CoV-2-S-glycoprotein (S6P) (42).
Because multivalency largely impacts dissociation rate constants of protein-protein interactions, we reasoned that an in-solution off-rate screen could distinguish differences between mono- and multivalent binding (43). Multivalent minibinders were allowed to fully associate with the target protein, then reactions were split in two and either 100-fold molar excess of untagged competitor (to prevent reassociation) or buffer was added. MON1, MON2, and MON3 target overlapping epitopes (25), and thus mono- or multivalent versions of these minibinders were selected as competitors. The ratio of the competitor to no-competitor condition measurements were calculated to determine the fraction of the complex dissociated (44).
Paralleling previous work where trimeric binders were targeted to the sialic acid-binding site on influenza hemagglutinin (30), we first designed self-assembling homotrimeric versions of the MON1, MON2, and MON3 miniproteins geometrically matched to the three RBDs in the S trimer (hereafter referred to as TRI; for example, TRI1–1 represents a homotrimer of MON1 with homotrimerization domain 1, table S1, data file S1). We designed and screened more than 100 different homotrimeric minibinders, with varied linker lengths and homotrimzeriation domains, using the CFPS workflow. We observed that many of the homotrimeric constructs exhibited slower dissociation rates than the corresponding monomers; much larger effects were observed with dissociation from the S trimer than monomeric RBD, consistent with multivalent binding (Fig. 1 and fig. S4). In total, we tested eleven different oligomerization domains and found that nine of these domains yielded at least one design with a linker length that improved dissociation rates on par with the top binders (fig. S4). Designs with domains four and eleven exhibited slower dissociation rates compared to their monomeric counterpart, but faster than the top designs (fig. S4E); this is likely indicative of an inability to simultaneously engage all three target epitopes or dissociation of the oligomerization domains themselves. The top binders exhibited little to no dissociation from S trimer after 7 days of incubation with competitor, indicating a likely apparent dissociation rate constant of 1×10−7 s−1 or slower (Fig. 1B). This is a marked improvement, more than four orders of magnitude for the TRI2 proteins, over the dissociation rate constants of the corresponding monomeric minibinders (fig. S5). We selected two trimeric scaffolds, the designed two ring helical bundle SB175 (domain 2) and the T4 foldon (domain 1) (45) (table S2), to proceed with based on the screening results and previous experience with these scaffolds.
Fig 1. Multivalent minibinders exhibit very slow dissociation rates upon binding to the prefusion SARS-CoV-2-S glycoprotein trimer.
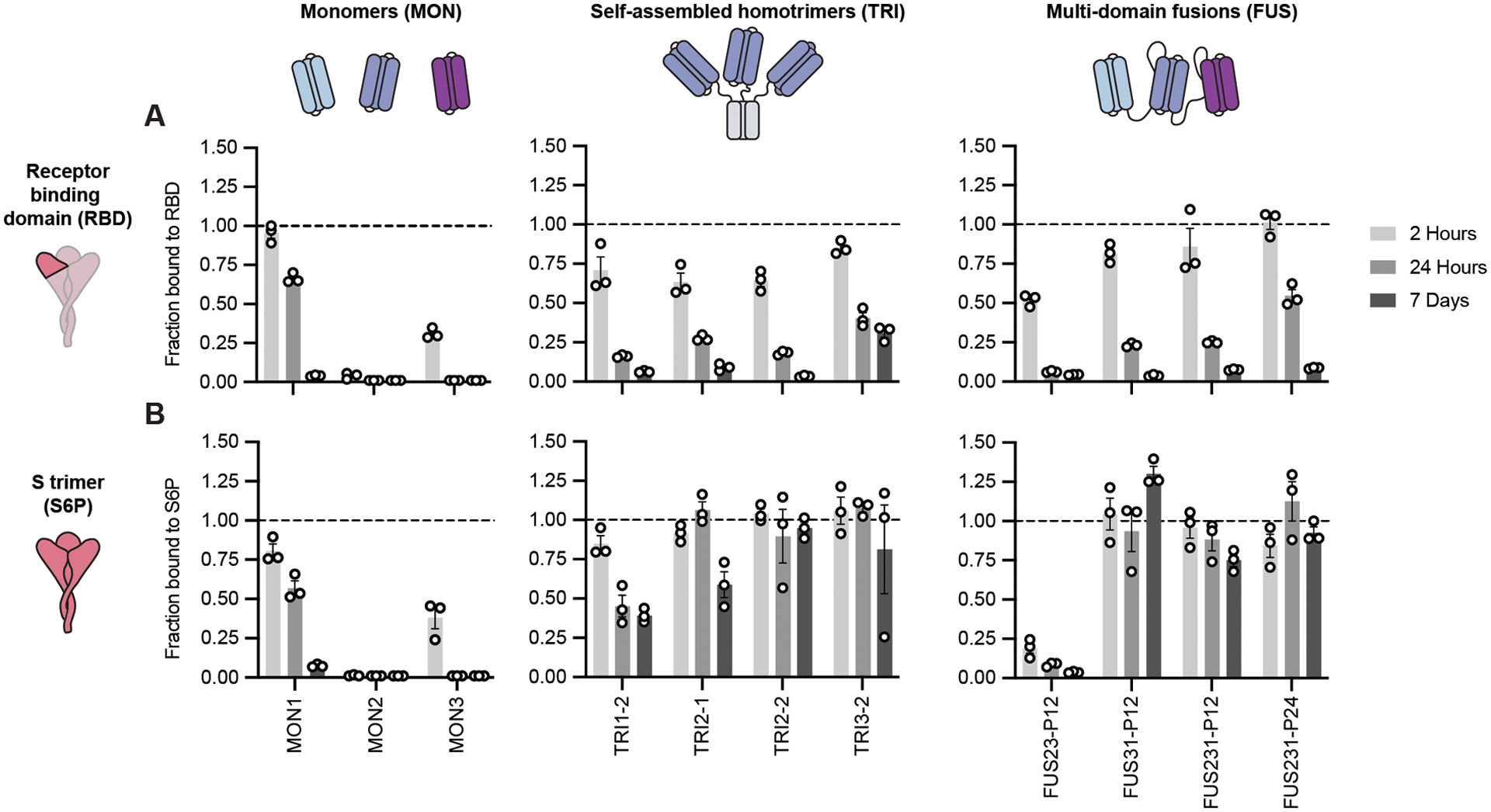
Dissociation of the minibinder construct was monitored by competition with 100-fold molar excess of untagged MON1 using AlphaLISA (Mean ± SEM, n = 3 technical replicates from a single experiment). (A) Dissociation was measured for indicated minibinder constructs complexed with the receptor-binding domain of SARS-CoV-2 (RBD). (B) Dissociation was measured for the indicated minibinder constructs complexed with the S trimer (S6P).
Next, we generated two- and three-domain fusions of the MON1, MON2, and MON3 minibinders separated by flexible linkers (hereafter referred to as FUS; for example, FUS31-P12 represents a fusion of MON3 to MON1 separated by a 12 amino acid proline-alanine-serine (P12) linker, table S1, data file S1). We screened more than 100 different fusions using the CFPS workflow, evaluating different minibinder orderings and a range of linker compositions and lengths that span the distances between the termini of the domains when bound to the “open” and “closed” states of the RBD (Fig. 1, and fig. S4, A, B, and F) (1). We evaluated both glycine-serine (denoted as G) and proline-alanine-serine (denoted as P) linkers (46) and observed similar binding characteristics (Fig. 1 and fig. S4). We observed occasional truncation of the G linkers during expression and purification by E. coli proteases; however, this was less frequent for the P linkers than for the G linkers. FUS31 and FUS231 constructs showed slower dissociation against S6P than RBD, and exhibited slower dissociation than all monomeric minibinders tested, consistent with multivalent S6P engagement (Fig. 1). The top binders exhibited little dissociation from S6P after 7 days, indicating a likely apparent dissociation rate constant of 1×10−7 s−1 or slower, representing one order of magnitude or greater improvement over the corresponding monomeric minibinder dissociation rate constant (fig. S5). Finally, to determine the potential for low-cost purification by heat treatment, we recombinantly expressed MON1, FUS231-P12, and TRI2–2 in E. coli. The heat-treated soluble fraction was enriched with the expressed minibinder and contaminating background proteins were largely precipitated (fig. S6).
Structural studies of minibinders in complex with SARS-CoV-2 S.
We next determined how the designed multivalent proteins engage multiple RBDs on a single S trimer; multivalent engagement on a virion typically requires binding of a single S trimer due to the relatively sparse S distribution (47–49). For some designs, FUS31-G8 and TRI1–5-G2 for example (table S1), initial screening using negative stain EM revealed considerable cross-linking and aggregation of S trimers upon addition of the constructs (fig. S7), consistent with binding to RBDs on different S trimers. In contrast, for constructs TRI2–2, FUS231-G10, FUS231-P24 and FUS31-G10, we observed less cross-linking, consistent with multivalent engagement of a single S trimer for each minibinder. To determine the binding modes of these compounds to the S trimer and characterize the structure of the MON2 and RBD interactions at high resolution, we carried out cryogenic electron microscopy (cryoEM) characterization of these complexes (Fig. 2).
Fig 2. CryoEM structures of multivalent minibinders in complex with the SARS-CoV-2 S6P glycoprotein.
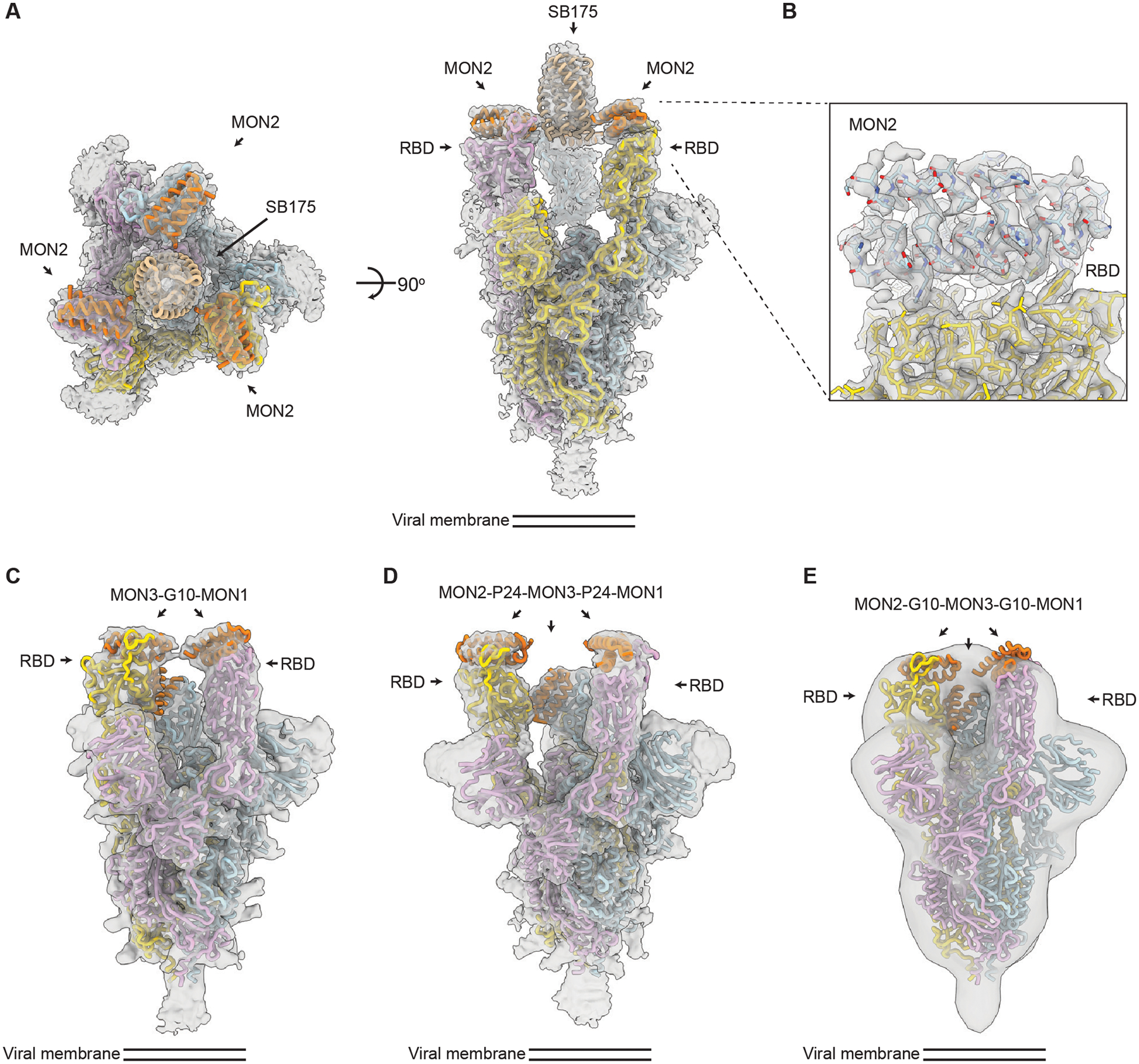
TRI2–2 is a homotrimer of MON2 using the SB175 homotrimerization domain, FUS31-G10 is a fusion of MON3 to MON1 with a 10 amino acid glycine-serine linker, FUS213-P24 is a fusion of MON2 to MON1 to MON 3 with a 24 amino acid proline-alanine-serine linker, and FUS213-G10 is a fusion of MON2 to MON1 to MON 3 with a 10 amino acid glycine-serine linker. (A) A CryoEM map of TRI2–2 in complex with the S6P in two orthogonal orientations is shown. (B) A zoomed-in view of the TRI2–2 and RBD complex was obtained using focused 3D classification and local refinement. The RBD and MON2 built in the 2.9 Å resolution cryoEM map are shown in yellow and blue, respectively. (C) A cryoEM map of FUS31-G10 bound to S6P is shown. (D) A cryoEM map of FUS231-P24 bound to S6P is shown. (E) A negative-stain EM map of FUS231-G10 in complex with S6P is shown. S and minibinder models were docked in the whole map by rigid body fitting for visualization. In all panels, the EM density is shown as a transparent gray surface, S protomers (PDB 7JZL) are rendered in yellow, cyan, and pink and minibinders (PDB 7JZU, 7JZM, and MON2 structure was determined in this study) are shown in orange.
The cryoEM structures of the TRI2–2, FUS31-G10, and the FUS231-P24 constructs in complex with S6P were determined at resolutions of 2.8, 4.6, and 3.9 Å respectively (Fig. 2A to D, fig. S8 to S11, and table S3), and a negative stain reconstruction was obtained with FUS231-G10 in complex with S6P (Fig. 2E). The TRI2–2/S6P cryoEM structure closely matched the TRI2–2 trimer design, with all three RBDs in the open state bound to MON2 (Fig. 2A and B, fig. S8 and S9). In the FUS31-G10 and S6P complex, FUS31-G10 is bound to two RBDs adopting an open conformation (Fig. 2C, fig. S8 and S10). The distance between the two RBDs in the open conformation is shorter in the FUS31-G10 than in the FUS231-P24 structure (Fig. 2C and D), suggesting that the bound minibinder holds the RBDs together, in agreement with the shorter linkers used in the former minibinder construct. In the structure, two molecules of FUS31-G10 are bound to a single S trimer with the third RBD being occupied by a second FUS31-G10 molecule. In the structure of FUS231-P24 bound to S6P, the three RBDs are participating in complex formation (Fig. 2D, fig. S8 and S11). The limited resolution in the region comprising the minibinder-bound RBDs and linkers precludes definitive assignment of minibinder identity at each binding site and relative connectivity between each minibinder module. The distances between the termini of the minibinder domains, however, is compatible with the computational design models and suggestive of engagement of either 2 (FUS31-G10) or 3 of the RBDs (FUS231-P24) in a single S trimer by the multivalent minibinders.
The structure of MON2 in complex with the S trimer has not previously been determined. Starting from the TRI2–2/S6P cryoEM data, we improved the RBD/MON2 densities using focused classification and local refinement, yielding a map at 2.9 Å resolution enabling visualization of the interactions formed by MON2 with the RBD (Fig. 2B). Superimposition of the design MON2 model to the corresponding cryoEM structure, using the RBD as reference, shows that the MON2 minibinder closely matched the design model with backbone Cɑ RMSD of 1.3 Å (fig. S8E and F). Together with previous structures of MON1 and MON3 (25), these data illustrate the accuracy with which both protein scaffolds and binding interfaces can now be computationally designed.
Multivalent minibinders enable rapid detection of SARS-CoV-2 S protein.
Having confirmed the binding mode of the FUS231 proteins by cryoEM, we designed an S trimer sensor, reasoning that the high affinity binding of the FUS231 proteins to the S trimer could make a useful diagnostic (50). We hypothesized that it would be possible to construct a bioluminescence resonance energy transfer (BRET) sensor for S trimer, where simultaneous engagement of all three minibinders in FUS231 with the S trimer would bring the N- and C-termini close enough together to enable efficient energy transfer. Towards this goal, we designed a BRET sensor based on FUS231-P12 with teLuc and mCyRFP3 fused to the N- and C-terminus of FUS231-P12 respectively (Fig. 3A) (51, 52). Upon binding of the sensor protein to a stabilized S protein with 2 proline mutations (S2P) (1, 50), we observed a 350% increase in the 590 nm:470 nm BRET ratio, which was not observed when bound to the RBD alone, and determined the limit of detection to be 11 pM S2P (Fig. 3B and C, and fig. S12). Furthermore, these results support the proposed multivalent binding mode for the FUS231 proteins.
Fig 3. FUS231-P12 enables detection of SARS-CoV-2 S trimer through BRET.
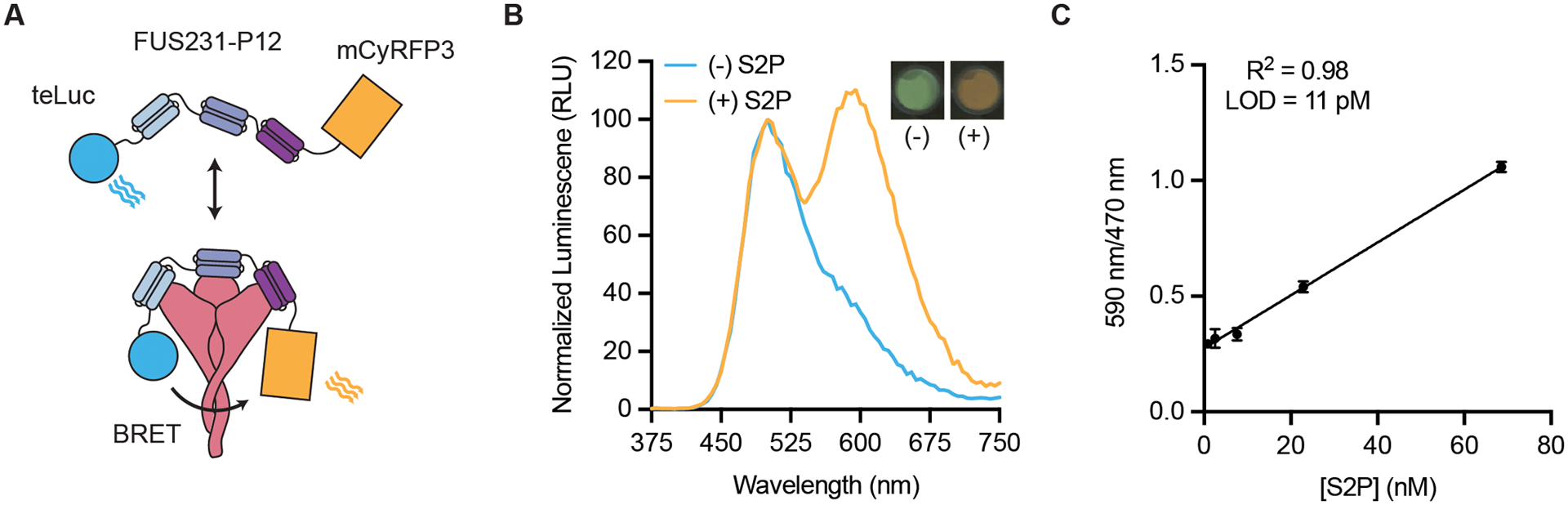
(A) A schematic representation of the BRET sensor, teluc-FUS231-P12-mCyRFP3, to detect S trimer is shown. (B) Luminescence emission spectra and image of the BRET sensor (100 pM) in the presence (orange trace, 100 pM) and absence (blue trace) of S2P are shown. Emission color change was observed using a mobile phone camera (inset top right). RLU, relative light units. (C) Titration of S2P with 100 pM sensor protein is shown. LOD indicates limit of detection. R2 value is shown on the graph. Data are presented as mean ± SEM with n = 3 technical replicates from a single experiment.
Multivalent minibinders bind tightly to SARS-CoV-2 variants.
We next evaluated the resiliency of the binding of multivalent minibinders to the previously identified MON1 and MON2 escape mutants as well as mutations present in the B.1.1.7 (Alpha), B.1.351 (Beta), and P.1 (Gamma) SARS-CoV-2 VOCs. We first measured the off-rate of the best multivalent minibinders using competition AlphaLISA with TRI2–1 against a panel of mutant S glycoproteins (Fig. 4A). Multivalent minibinders were allowed to fully associate with mutant S trimers and subsequently were competed with 100-fold molar excess of untagged TRI2–1 to measure dissociation of the complex. The two-domain fusions (FUS23 and FUS31) did not show improved binding to the tested point mutants. The three-domain fusions (FUS231) retained binding to the tested mutants, indicating that they are more resistant to mutations than their monomeric counterparts, although E406W, Y453R, and the combination of K417N, E484K, and N501Y mutations (present in the B.1.351 S trimer) increased the dissociation rate more than 100-fold. Consistent with these results, we also observed increased dissociation rates for the FUS231 proteins against the B.1.351 and P.1 spikes by surface plasmon resonance (SPR) (fig. S13). The TRI1 and TRI3 homotrimers showed similar mutational tolerance in the competition experiment, with the same E406W, Y453R, and B.1.351 mutations causing increased dissociation rates. Strikingly, the TRI2 designs showed little dissociation after 24 hours against any of the tested S trimer mutants.
Fig 4. Multivalency enhances both the breadth and potency of neutralization against SARS-CoV-2 variants by minibinders.
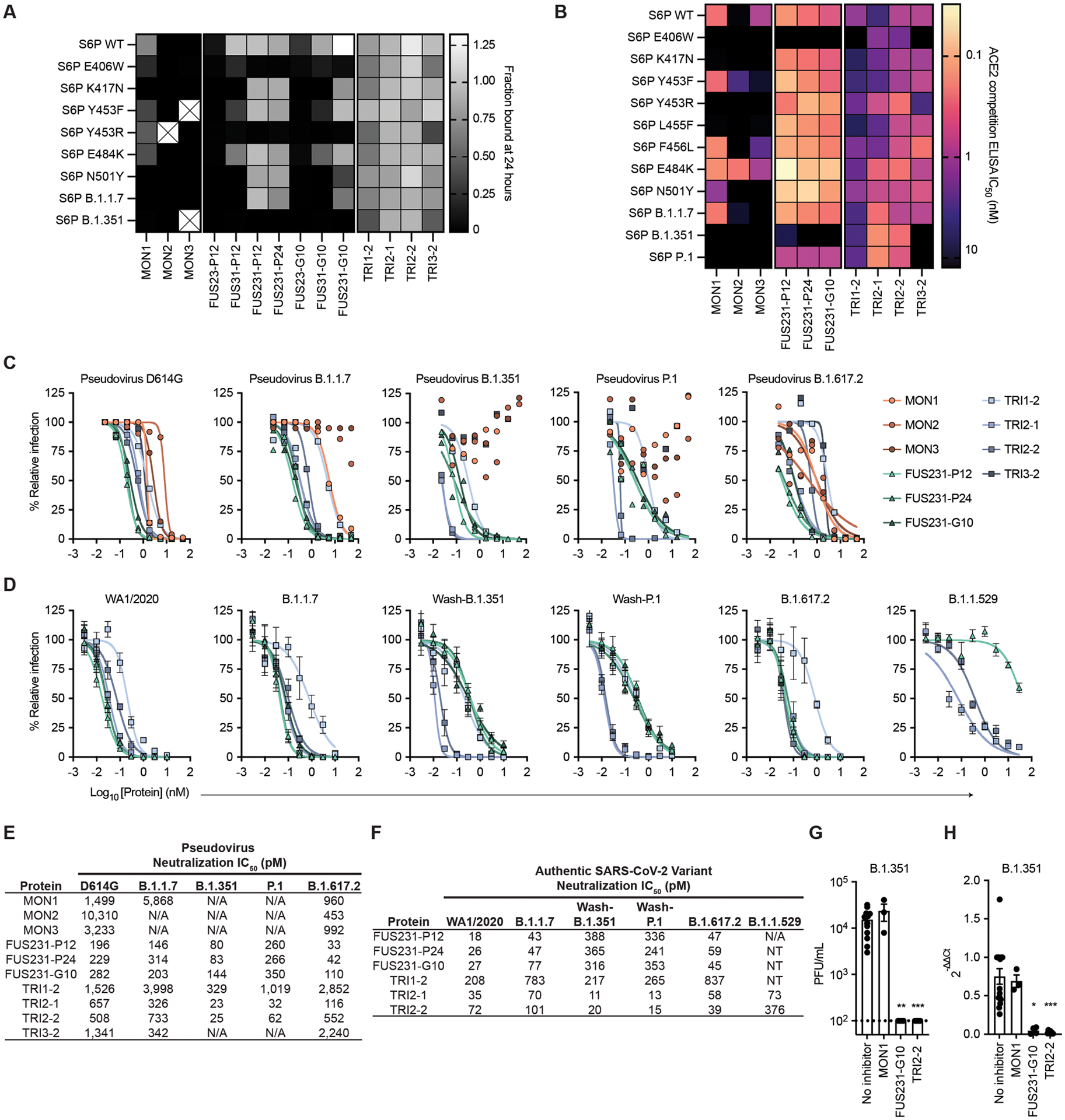
(A) Dissociation of minibinder constructs from S6P variants after 24 hours was measured by competition with untagged TRI2–1 using AlphaLISA. Means are shown with n = 3 technical replicates from a single experiment. Cells containing an X indicate insufficient signal in the absence of a competitor to quantify the fraction of protein bound. (B) Competition of minibinder constructs with ACE2 for binding S6P were measured by ELISA. Data are presented as mean values for n = 2 technical replicates representative of two independent experiments. (C) Neutralization of SARS-CoV-2 pseudovirus variants by minibinder constructs are shown. Data are presented as means of n = 2 technical replicates representative of two independent experiments. (D) Neutralization of authentic SARS-CoV-2 by minibinder constructs was measured. Data are presented as mean ± SEM with n = 4 technical replicates from two independent experiments for all but B.1.1.529, where n = 8 technical replicates from four independent experiments. (E) Summary of neutralization potencies of multivalent minibinder constructs against SARS-CoV-2 pseudovirus variants are shown. N/A indicates an IC50 value above the tested concentration range and an IC50 greater than 50,000 pM. (F) Summary of neutralization potencies of multivalent minibinder constructs against authentic SARS-CoV-2 variants are shown. N/A indicates an IC50 value above the tested concentration range and an IC50 greater than 30,000 pM. NT indicates not tested. (G) Replicating authentic B.1.351 virus in the presence of minibinder constructs (0.3 μM) was quantified in human kidney organoids. Data are presented as mean ± SEM, n = 4 biological replicates with 2 to 3 technical replicates per experiment. Data were compared to the no inhibitor control by a Kruskal-Wallis test with Dunn’s post-hoc analysis; ** P < 0.01, *** P < 0.001. Dashed line indicates lower limit of detection of plaque assay. (H) Relative gene expression of SARS-CoV-2 envelope protein (SARS-CoV2-E) was measured in kidney organoids post viral infection with and without multivalent minibinders (0.3 μM). Data are presented as mean ± SEM of n = 4 biological replicates with 2 to 3 technical replicates per experiment. Data were compared to the no inhibitor control by a Kruskal-Wallis test with Dunn’s post-hoc analysis; * P < 0.05, *** P < 0.001.
We subsequently screened the top multivalent minibinders for binding to mutant S trimers by an ACE2 competition enzyme-linked immunosorbent assay (ELISA), which correlates with neutralization potency (53). The minibinders were pre-incubated with the S6P variants before binding to immobilized ACE2 (Fig. 4B and fig. S14). In line with deep mutational scanning data, we observed impaired binding to the E406W, K417N, and Y453R mutants in addition to several other mutants. Two mutations, Y453F and E484K, improved MON2 binding, consistent with MON2 mimicry of the ACE2 interaction interface (38). Compared to the monovalent minibinders, we observed reduced effects of mutations in the competition IC50 values of the FUS231 and TRI2 minibinders and, to a lesser extent, of the TRI1 and TRI3 minibinders against the tested S6P variants, except for E406W (Fig. 4B and fig. S14D).
Multivalent minibinders potently neutralize circulating SARS-CoV-2 variants.
To investigate the efficacy of the multivalent minibinders for preventing viral infection, we performed neutralization assays with the inhibitors using both pseudotyped lentiviruses and authentic SARS-CoV-2 variants (Fig. 4C to F, fig. S15). Against pseudoviruses displaying S proteins corresponding to the B.1.1.7, B.1.351, P.1, B.1.617.1, B.1.617.2 (Delta), and B.1.617.2.1 (Delta plus, AY.1) variants, all three monomer minibinders showed reduced neutralization capacity as compared to the Wuhan-Hu-1 D614G strain; in contrast, many of the multivalent minibinders were less affected in an ACE2 overexpressing cell line (Fig. 4C and E, and fig. S15A and C). The same proteins were also evaluated against pseudoviruses containing the E406W, L452R, and Y453F mutations, which again had little impact on neutralization for most multivalent minibinders tested (fig. S15A and C). This suggests that the increase in affinity from multivalency improved neutralization breadth. The top neutralizing minibinders from this screen were tested for neutralization of a panel of authentic SARS-CoV-2 viruses including a historical WA1/2020 strain, B.1.1.7, B.1.526 (Iota), B.1.1.529 (Omicron), B.1.617.1, B.1.617.2, and B.1.617.2.1 natural isolates, and chimeric WA1/2020 strains encoding spike genes corresponding to those of B.1.351 (Wash-B.1.351), and P.1 (Wash-P.1) variants. Again, the top candidates maintained pM-range IC50 values (Fig. 4D and F, and fig. S15B and D), except for the FUS231 proteins, which did not fully neutralize the B.1.1.529 variant in the tested concentration range. The TRI2 proteins maintained potent neutralization across all tested variants, notably including the B.1.1.7, Wash-B.1.351, Wash-P.1, B.1.617.2, and B.1.1.529 variants. Impressively, the TRI2 proteins potently neutralized the B.1.1.529 variant whereas many clinical mAbs for the treatment of COVID-19 do not (table S4) (15–17).
Although Vero-hACE2-TMPRSS2 (transmembrane serine protease 2) cells are useful for neutralization studies, they likely do not fully reflect the human cell infectivity. Recent findings highlight the relevance of using non-transformed human organoid models for SARS-CoV-2 research (54). SARS-CoV-2 can infect and replicate in human kidney organoids, specifically targeting kidney tubular epithelial cells expressing ACE2 receptors, responsible for viral entry (55, 56). Therefore, we generated kidney organoids from the H9 human embryonic stem cell line (57) (fig. S16) and evaluated the ability of the multivalent minibinders to prevent SARS-CoV-2 viral entry and replication. Replication of the B.1.351 variant was inhibited when the virus was pre-incubated with designed multivalent minibinders FUS231-G10 and TRI2–2, but not with MON1 (Fig. 4G). Quantitative reverse transcription PCR (RT-qPCR) analysis of viral RNA from the kidney organoids also showed reduced SARS-CoV-2 envelope protein (SARS-CoV2-E) gene expression in the presence of either FUS231-G10 or TRI2–2 (Fig. 4H). These data show that designed multivalent minibinders are potent neutralizers of the B.1.351 variant in a human organoid system.
Multivalent minibinders resist viral escape.
Given the promising data showing that multivalent minibinders can neutralize SARS-CoV-2 VOCs, we tested the multivalent minibinders for resistance against viral escape mutations in the S trimer (Fig. 5A and B) (6). Plaque assays were performed with a VSV-SARS-CoV-2 S chimera on Vero CCL-81 cells with minibinders included in the overlay to halt spread of non-resistant viruses. In positive control wells, inclusion in the overlay of 2B04, a potent neutralizing antibody targeting the RBD (6, 58–60), resulted in multiple escape mutants in each plate similar to previously reported escape mutants (Fig. 5A) (6). In contrast, for both FUS231-P12 and TRI2–2, escape mutants were not isolated in 36 replicate wells for each protein (fig. S17). These data indicate that both the FUS231-P12 and TRI2–2 proteins are more difficult to escape than 2B04. Given the known mutation rate of the VSV RNA polymerase L (61) and the number of viral particles screened, we estimated (table S5) that, for the multivalent minibinders, the screened pool of viral mutants contains a large fraction of the possible single amino acid substitutions (34% to 88%) and a small fraction of the possible double amino acid substitutions (0.4% to 9.6%) within the region of the RBD that contacts the minibinders. Taken together with the results of the single site saturation mutagenesis studies for the monovalent minibinders (fig. S2) these findings indicate that at least two or more mutations in the RBD are likely necessary to escape binding of the multivalent minibinders.
Fig 5. Top multivalent minibinder candidates are resistant to viral escape.
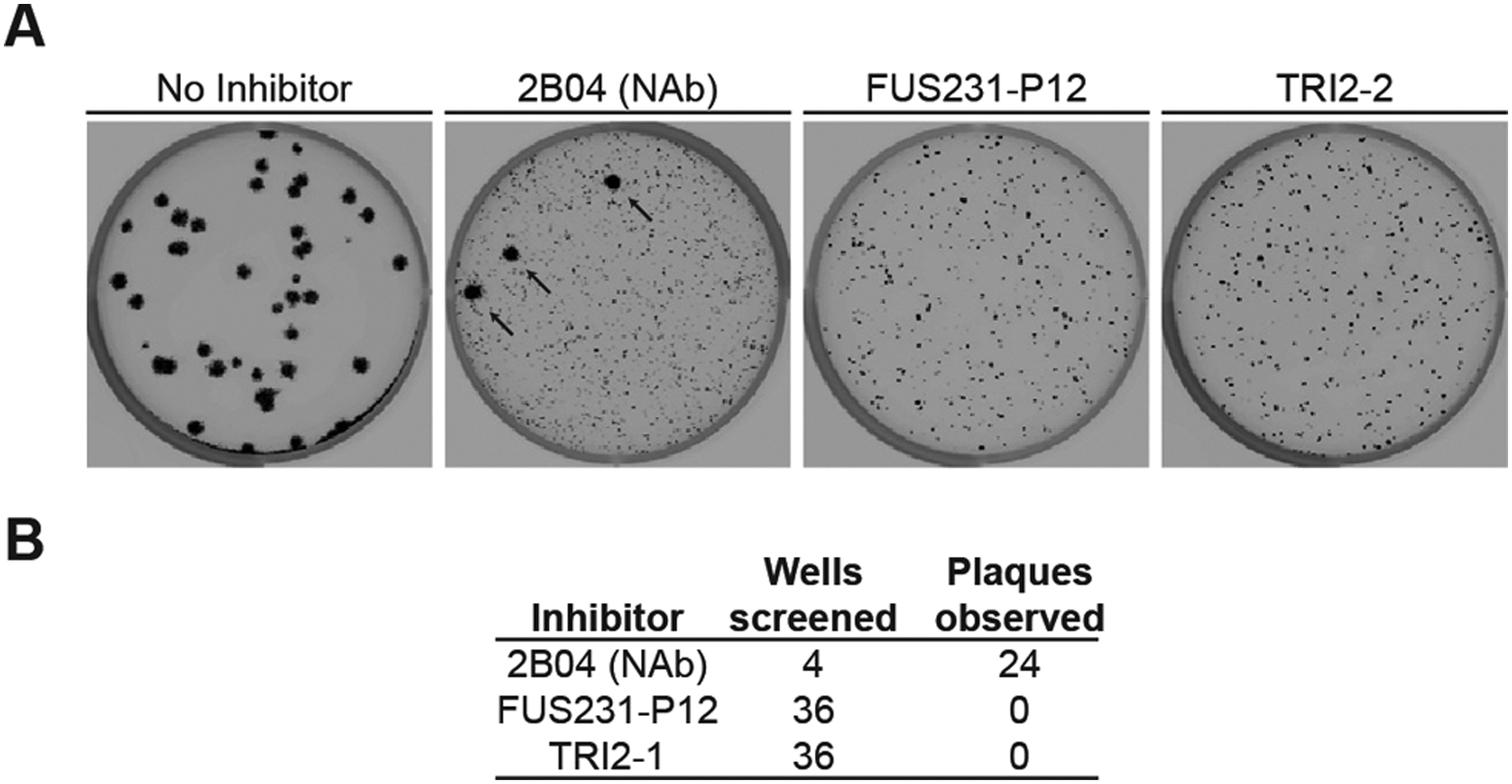
(A) Plaque assays were performed to isolate VSV-SARS-CoV-2 S chimera virus escape mutants against a control neutralizing antibody (2B04) and the FUS231-P12 and TRI2–2 multivalent minibinders. For each inhibitor tested, Vero CCL-81 cells were incubated with VSV-SARS-CoV-2 S chimera virus for one hour, followed by addition of the inhibitor protein at a fully neutralizing concentration and further incubation to allow for replication and spread of resistant viruses. Thirty-six independent selections were carried out for each minibinder compound in a single experiment; representative examples are shown in the images. Small plaques are indicative of inhibited viral spreading and large plaques, highlighted by black arrows, are indicative of viral escape mutants spreading. (B) A summary of the results of the viral escape screen are shown. NAb, neutralizing antibody.
Multivalent minibinder confers protection in human ACE2-expressing transgenic mice.
To determine whether the multivalent minibinders can prevent or treat SARS-CoV-2 infection in vivo, we performed pre-exposure prophylaxis or post-exposure therapy studies in highly susceptible K18-hACE2 transgenic mice (62) with TRI2 multivalent minibinders, which retained the most consistent binding to all S trimer variants tested. For prophylaxis, a single 50 μg dose (about 2.5 mg/kg) of TRI2–1 or TRI2–2 was administered directly to the nasal cavity (intranasal administration) one day prior to inoculation with 103 focus forming units (FFU) of the indicated SARS-CoV-2 VOCs (Fig. 6A). In all cases, intranasal administration of TRI2–1 or TRI2–2 protected mice against SARS-CoV-2-induced weight loss (Fig. 6B). At 6 days post infection, viral burden in tissues was reduced in almost all primary (lung and nasal wash) and secondary sites (heart, spleen, brain) of viral replication in TRI2–1 and TRI2–2 treated animals (Fig. 6C). To determine the therapeutic potential of TRI2–2, we inoculated K18-hACE2 mice with 103 FFU of Wash-B.1.351 or B.1.617.2 and one day later, administered a single 50 μg dose of minibinder intranasally (Fig. 6D). Treatment with TRI2–2 protected against weight loss and reduced viral burden in all tissues except nasal washes (Wash-B.1.351) or the spleen (B.1.617.2) (Fig. 6E and F). TRI2–2 therapy at D+1 reduced infectious virus titers in the lungs of Wash-B.1.351- and B.1.617.2-infected mice (fig. S18). We determined the pharmacokinetics of TRI2–2 after intranasal administration by quantitative competition ELISA. Substantial concentrations of TRI2–2 were detected in the lung lysate and serum 48 hours after administration (fig. S19) but was too low for confident quantification in nasal turbinates after the first time point and for confident quantification in nasal washes at all time points. These results indicate that intranasal administration of TRI2–1 or TRI2–2 confer protection against SARS-CoV-2 infection as both pre-exposure prophylaxis and post-exposure therapy in a stringent model of disease.
Fig 6. Top multivalent minibinder candidates protect mice from SARS-CoV-2 challenge.
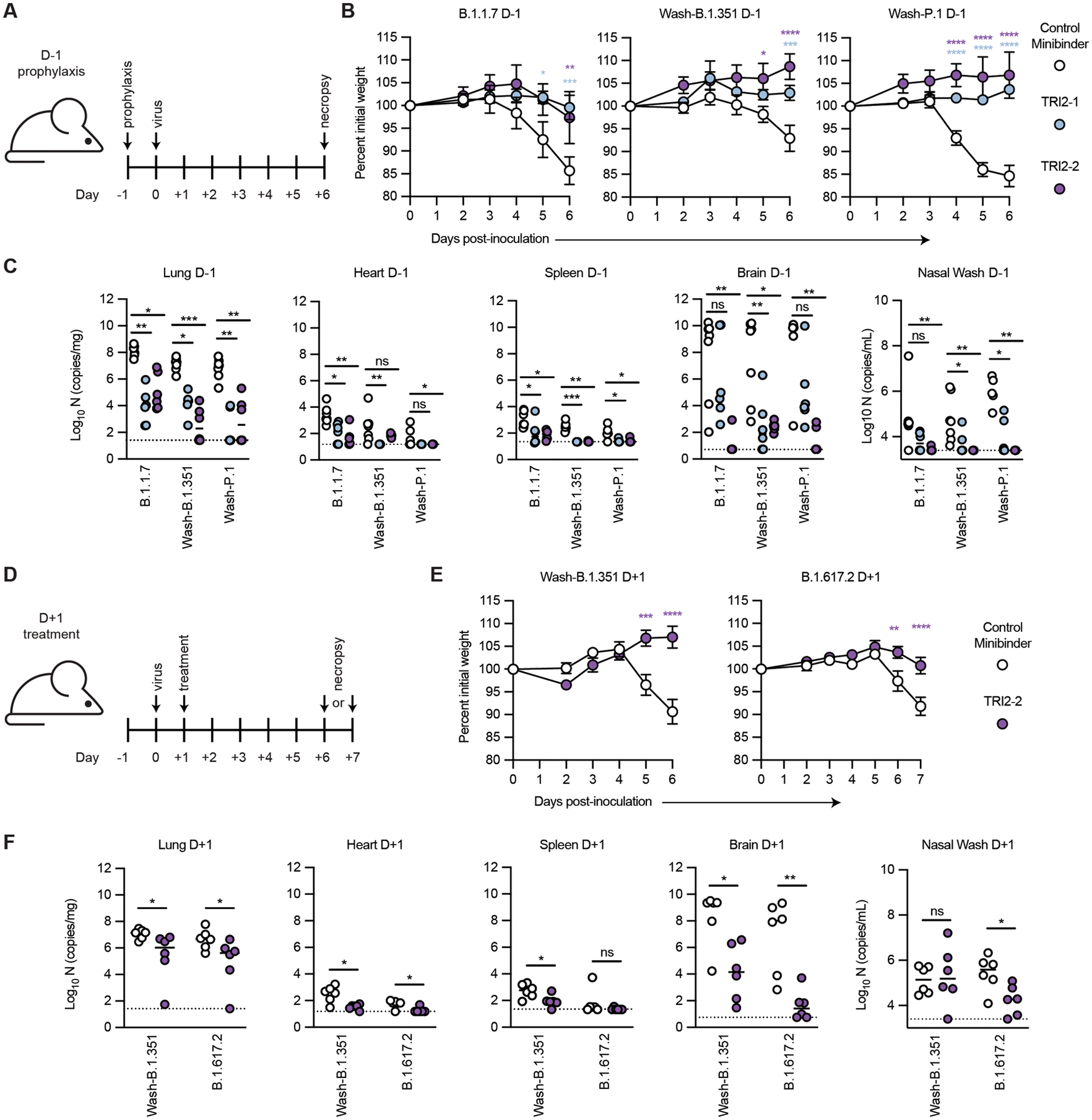
(A) K18-hACE2-transgenic mice (n = 6 from two independent experiments) were dosed with 50 μg of the indicated minibinder by intranasal (i.n.) administration (50 μl total) 24 hours prior (D-1) to infection with 103 focus forming units (FFU) of SARS-CoV-2 variants B.1.1.7, Wash-B.1.351, or Wash-P.1 i.n. on Day 0. (B) Daily weight change following inoculation was measured. Data are presented as mean ± SEM. Data were analyzed by a two-way ANOVA with Sidak’s post-test; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001 as compared to the control minibinder. (C) At 6 days post infection (dpi), animals (n = 6 from two independent experiments) were euthanized and analyzed for SARS-CoV-2 viral RNA by RT-qPCR in the lung, heart, spleen, brain, and nasal wash. Horizontal bars indicate median; dashed lines represent the limit of detection. Data were analyzed by a Kruskal-Wallis test with Dunn’s post-hoc analysis; ns, not significant, * P < 0.05, ** P < 0.01, *** P < 0.001. (D) K18-hACE2-transgenic mice (n = 6 from two independent experiments) were dosed with 50 μg of the indicated minibinder by i.n. administration (50 μl total) 24 hours after (D+1) infection with 103 FFU of the SARS-CoV-2 Wash-B.1.351 or B.1.617.2 variant on Day 0. (E) Daily weight change following inoculation was measured. Data are presented as mean ± SEM. Data were analyzed by two-way ANOVA with Sidak’s post-test; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001). (F) At 6 dpi (B.1.351) or 7 dpi (B.1.617.2), animals (n = 6 from two independent experiments) were euthanized and analyzed for SARS-CoV-2 viral RNA by RT-qPCR in the lung, heart, spleen, brain, and nasal wash. Horizontal bars indicate median; dashed lines represent the limit of detection. Data were analyzed by a two-tailed Mann-Whitney test; ns, not significant, * P < 0.05, ** P < 0.01.
DISCUSSION
Both strategies for generating multivalent S protein binders from miniproteins, self-assembling homotrimers (TRI) and multi-domain fusions (FUS), yielded designs with improved affinity, neutralization of current and historical VOCs, and resistance to escape mutants over their monovalent counterparts (25, 26). The TRI2 proteins maintained the strongest binding across all S trimer variants tested, likely because MON2 is an ACE2 mimic, similar to the recently reported S2K146 mAb (15, 24). This combination of trivalency and receptor mimicry could be a useful general approach for combating viral escape and antigenic drift (15, 24, 36, 53, 63, 64).
The designs also have potential advantages as therapeutics over ACE2 receptor traps and mAbs. When compared to receptor traps (55, 65–67), TRI2–2 has a low risk of eliciting host-directed anti ACE2 responses due to low sequence similarity between MON2 and ACE2 (fig. S1). On a per mass basis, the TRI2 proteins are more potent neutralizers than all currently authorized mAbs for the treatment of COVID-19 (15, 16), and, unlike most clinical mAbs, they maintain activity against Omicron (table S4). The multivalent minibinders are amenable to large-scale production in microorganisms like E. coli, making them more cost-effective to manufacture than mAbs (8). Furthermore, their small size and stability may enable direct nebulization into the human upper respiratory tract (3, 68–70), a strategy that could increase accessibility for patients over the typical intravenous or subcutaneous routes used for administering neutralizing mAbs.
The high potency of the multivalent constructs, in particular TRI2–2 against Omicron, Delta, and the other tested VOCs, makes them promising candidate SARS-CoV-2 therapeutics, and they are currently undergoing further preclinical development and investigational new drug (IND) enabling studies. These efforts will address limitations in our current study. First, anti-drug antibodies are a concern with non-human proteins and, although MON1 and other minibinders (26, 71) elicit little or no immune response, additional studies are required to determine the immunogenicity of the multivalent constructs. Second, it will be important to assess the pharmacokinetics following different modes of administration; in humans, it may be necessary to distribute the compound deeper into the respiratory system for post infection efficacy. Third, as with any new drug candidate going through the drug development pipeline, it will be necessary to assess its stability as well as its potency and toxicity after prolonged administration.
In summary, our integration of structure-guided computational protein design, cell-free DNA assembly, cell-free expression, and a competition-based off-rate screen enabled the rapid design and optimization of S trimer-engaging multivalent minibinders. Scaling cell-free expression to manufacture mg quantities of endotoxin-free protein for cell-based neutralization assays further reduced the time required to evaluate lead molecules. The developed pipeline has direct relevance to diagnostics as well; the FUS231-based BRET sensor is easy to use, fast, and has the potential to be less expensive than state-of-the-art lateral flow assay-based antigen tests (72, 73). Our integrated computational and experimental pipeline should enable the rapid generation of potent protein-based medical countermeasures and diagnostic reagents against newly emerging pathogens.
MATERIALS AND METHODS
Study design
The objective of this study was to design and evaluate multivalent minibinders that neutralize SARS-CoV-2 variants containing mutations within the RBD. At the outset, we hypothesized that multivalency would overcome mutations that reduce binding for individual monomeric minibinders. Designed proteins were evaluated in controlled laboratory experiments, first using biophysical methods with purified proteins (AlphaLISA and ELISA competition assays) followed by in vitro methods requiring cell culture (pseudovirus and authentic virus neutralization assays). The top candidates from neutralization assays were screened by electron microscopy for cross-linking multiple S trimers and the candidates that were found to minimally cross-link S trimers were subjected to structural analysis by cryoEM. The most promising proteins were evaluated in vivo in mice. In all studies where cell lines were used, the cell line is noted in the corresponding materials and methods section. The total number and type of experimental replicates is noted in each figure legend. Details on the in vivo mouse study compliance with best practices can be found in the corresponding materials and methods section. No sample-size calculations were performed to power each in vivo study. Instead, sample sizes and study endpoints were determined based on previous in vivo virus challenge experiments. For all other experiments, sample size was selected based on previous literature and previous experience. In the animal studies, mice were randomly assigned to the control and treatment groups. Animal caretakers and researchers were not blinded to the study groups or during the assessment of the outcomes. Data that underlie the results reported in this article can be found in data file S2, data file S3, and in the deposited data listed in the data and materials availability statement.
Statistical analysis
Statistical significance was determined by a P value < 0.05 using GraphPad Prism 9 software. Only non-parametric tests were used throughout this article. Analysis of mouse weight changes was performed using a two-way analysis of variance (ANOVA) with Sidak’s post-test for multiple comparisons. Statistical analysis of viral load between two groups was performed using either a Kruskal-Wallis test with Dunn’s post-hoc analysis for multiple comparisons or a two-tailed Mann-Whitney test as noted in the corresponding figure captions.
Supplementary Material
Acknowledgements:
We thank Ashty S. Karim and Lauren Clark for their laboratory support during COVID-19, Ali Ellebedy for providing antibodies, Sara Fernandez Dunne and Matt Clutter for assistance with AlphaLISA method development, Barry Lutz for advice on bioassay development for the detection of S trimer, Alyssa Blackstone for assistance in bioanalytical method development, and Jonathan Himmelfarb for helpful discussions.
Funding:
This work was supported by: the Defense Threat Reduction Agency contracts HDTRA1-15-10052 and HDTRA1-20-10004 (to M.C.J); the David and Lucile Packard Foundation (to M.C.J.); the Camille Dreyfus Teacher-Scholar Program (to M.C.J.); Department of Defense National Defense Science and Engineering Graduate (NDSEG) Fellowship Program (NDSEG-36373, to A.C.H.); Department of Defense Peer Reviewed Medical Research Program awards W81XWH-21-1-0006 and W81XWH-21-1-0007 (to H. R-B., D.B., M.G., and B.S.F.); DARPA Synergistic Discovery and Design (SD2) HR0011835403 contract FA8750-17-C-0219 (to I.G., L. Cao, L.Carter, L.K., N.E., R.R., D.V., and D.B.); DOD contract W81XWH-20-1-0270-2019, AI145296, and AI143265 (to M.G. and T.Y.S.); the Audacious Project at the Institute for Protein Design (to D.B. and H-.W.Y.); Eric and Wendy Schmidt by recommendation of the Schmidt Futures (to H-.W.Y., L.M., L.K, R.R., and I.G.); the Bill & Melinda Gates Foundation (OPP1156262 to D.V. and D.B.; INV-004949 to J.D.B.); The Open Philanthropy Project Improving Protein Design Fund (to D.B. and S.E.B.); a Career Award at the Scientific Interface Grant from the Burroughs Wellcome Fund (to S.E.B.); European Commission MSCA CC-LEGO 792305 (to A.L.); the Wu Tsai Translational Investigator Fund at the Institute for Protein Design (to G.U.); the National Institute of General Medical Sciences (R01GM120553 to D.V.) (NIH1P01GM081619, R01GM097372, R01GM083867 and the NHLBI Progenitor Cell Biology Consortium U01HL099997; UO1HL099993 to H.R-B.); the National Institute of Allergy and Infectious Diseases (DP1AI158186 to D.V.; HHSN272201700059C to D.V., L.S., and D.B., R37 AI1059371 to S.P.J.W.; R01 AI145486 to B.R.L. and N.P.); the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK117914, R01DK130386, and U01DK127553 to B.S.F.; F31DK130550 to L.H.); the National Center for Advancing Translational Sciences (UG3TR002158 to J.H. and B.S.F.); the United World Antiviral Research Network -UWARN- one of the Centers Researching Emerging Infectious Diseases “CREIDs”; U01 AI151698-01 (to D.B., L.S., M.G., and H-.W.Y.); R01 AI157155 (to M.S.D.), a Pew Biomedical Scholars Award (to D.V.); Investigators in the Pathogenesis of Infectious Disease Awards from the Burroughs Wellcome Fund (to D.V.); Fast Grants (to D.V. and A.A.); A Helen Hay Whitney Foundation postdoctoral fellowship (to J.B.C); T90 Training Grant (to Y.T.Z); an HHMI Fellowship from the Damon Runyon Cancer Research Foundation (to T.N.S); Howard Hughes Medical Institute (to J.D.B, D.V., and D.B.), the National Institute of Health Cellular and Molecular Biology Training Grant (T32GM007270 to A.A.), the University of Washington Arnold and Mabel Beckman cryoEM center and the National Institute of Health grant S10OD032290 (to D.V.). This project has also been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272201700059C (to D.V., L.S., and D.B.). J.D.B, D.V. and D.B. are investigators of the Howard Hughes Medical Institute.
Competing Interests:
Each contributor attests that they have no competing interests relating to the subject contribution, except as disclosed. A.C.H, L.Cao, I.G., J.B.C., L.M., L.K., L.Carter, K.W., G.U., J.B., N.E., A.C.W., B.V., R.R. Y-J.P, H-W.Y., L.S., D.V., M.D., M.C.J. and D.B. are co-inventors on a patent application that incorporate discoveries described in this article (Application #: PCT/US2021/034069, Title: SARS-COV-2 inhibitors). D.B. and H-W.Y. are co-inventors on a patent application describing the S protein sensors described in this article (Application #: 63/216,891, Title: Multivalent SARS-CoV-2 Spike Protein Biosensor). Z.L. and S.P.J.W. are co-inventors on a patent application covering the use of VSV-SARS-CoV-2 as a vaccine vector and screening tool (Application #: PCT/US2021/027275, Title: SARS CoV-2 Vaccines and High Throughput Screening Assays Based on Vesicular Stomatitis Virus Vectors). B.S.F. and L.H. are inventors on patent applications related to kidney organoid differentiation and application (Application #: US2021290632A1, Title: High-Throughput Automation of Organoids for Identifying Therapeutic Strategies; Application #: US10815460B2, Title: Three-dimensional differentiation of epiblast spheroids to kidney organoids models stage-specific epithelial physiology, morphogenesis, and disease). J.D.B. may receive a share of IP revenue as an inventor on a Fred Hutchinson Cancer Research Center-optioned technology/patent (Application #: WO2020006494, Title: Cell-Stored Barcoded Deep Mutational Scanning Libraries and Uses of the Same) related to deep mutational scanning of viral proteins.
D.B. is a cofounder of Neoleukin Therapeutics. The Diamond laboratory has received unrelated sponsored research support from Moderna, Vir Biotechnology, and Emergent BioSolutions. The Veesler laboratory received an unrelated sponsored research agreement from Vir Biotechnology. M.C.J. is a cofounder of SwiftScale Biologics, Stemloop, Inc., Design Pharmaceuticals, and Pearl Bio. The interests of M.C.J. are reviewed and managed by Northwestern University in accordance with their conflict-of-interest policies. L.G., W.M. and C.T. are current employees of Amgen and own Amgen stock. Z.L. and S.P.J.W. received unrelated sponsored research agreements from Vir Biotechnology, AbbVie, and SAB therapeutics.
A.C.H. has consulted for SwiftScale Biologics and L.E.K. Consulting. M.S.D. is a consultant for Inbios, Vir Biotechnology, and Carnival Corporation and is on the Scientific Advisory Board of Moderna and Immunome. H.R-B. is a Scientific Advisor of CuriBio. J.D.B. consults for Moderna on viral evolution and epidemiology and Flagship Labs 77 on deep mutational scanning. All other authors declare no competing interests.
Footnotes
Data and Materials Availability:
All data associated with this study are in the paper or supplementary materials. Structural models and density maps have been deposited in the Protein Data Bank (PDB) (SARS-CoV-2/TRI2–2: 7UHC and SARS-CoV-2/TRI2–2 (local refinement): 7UHB) and Electron Microscopy Data Bank (EMDB) (SARS-CoV-2/TRI2–2: EMD-26512, SARS-CoV-2/TRI2–2 (local refinement): EMD-26511, SARS-CoV-2/FUS31-G10 (2RBD-open): EMD-26509, SARS-CoV-2/FUS31-G10 (3RBD-open): EMD-26510, SARS-CoV-2/FUS231-P24 (2RBD-open): EMD-26507, and SARS-CoV-2/FUS231-P24 (3RBD-open): EMD-26508). Illumina sequencing data for the deep mutational scanning experiments are available on NCBI SRA, BioSample SAMN19925005. Code for the analysis of the deep mutational scanning experiments are available on Zenodo https://doi.org/10.5281/zenodo.6377268. Requests for reagents (antibodies, viruses, and other proteins) should be directed to the corresponding authors and will be made available after completion of a Materials Transfer Agreement with the University of Washington. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using this material.
References:
- 1.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D, Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dougan M, Nirula A, Azizad M, Mocherla B, Gottlieb RL, Chen P, Hebert C, Perry R, Boscia J, Heller B, Morris J, Crystal C, Igbinadolor A, Huhn G, Cardona J, Shawa I, Kumar P, Adams AC, Van Naarden J, Custer KL, Durante M, Oakley G, Schade AE, Holzer TR, Ebert PJ, Higgs RE, Kallewaard NL, Sabo J, Patel DR, Dabora MC, Klekotka P, Shen L, Skovronsky DM, BLAZE-1 Investigators, Bamlanivimab plus Etesevimab in Mild or Moderate Covid-19. N. Engl. J. Med 385, 1382–1392 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta A, Gonzalez-Rojas Y, Juarez E, Crespo Casal M, Moya J, Falci DR, Sarkis E, Solis J, Zheng H, Scott N, Cathcart AL, Hebner CM, Sager J, Mogalian E, Tipple C, Peppercorn A, Alexander E, Pang PS, Free A, Brinson C, Aldinger M, Shapiro AE, COMET-ICE Investigators, Early Treatment for Covid-19 with SARS-CoV-2 Neutralizing Antibody Sotrovimab. N. Engl. J. Med 385, 1941–1950 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Xiao J, Hooper AT, Hamilton JD, Musser BJ, Rofail D, Hussein M, Im J, Atmodjo DY, Perry C, Pan C, Mahmood A, Hosain R, Davis JD, Turner KC, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Roque-Guerrero L, Acloque G, Aazami H, Cannon K, Simón-Campos JA, Bocchini JA, Kowal B, DiCioccio AT, Soo Y, Geba GP, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, Trial Investigators REGEN -COV Antibody Combination and Outcomes in Outpatients with Covid-19. N. Engl. J. Med 385, e81 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corti D, Purcell LA, Snell G, Veesler D, Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 184, 4593–4595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z, VanBlargan LA, Bloyet L-M, Rothlauf PW, Chen RE, Stumpf S, Zhao H, Errico JM, Theel ES, Liebeskind MJ, Alford B, Buchser WJ, Ellebedy AH, Fremont DH, Diamond MS, Whelan SPJ, Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29, 477–488.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, Li JZ, Bloom JD, Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 371, 850–854 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crook ZR, Nairn NW, Olson JM, Miniproteins as a Powerful Modality in Drug Development. Trends Biochem. Sci 45, 332–346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCallum M, Walls AC, Sprouse KR, Bowen JE, Rosen LE, Dang HV, De Marco A, Franko N, Tilles SW, Logue J, Miranda MC, Ahlrichs M, Carter L, Snell G, Pizzuto MS, Chu HY, Van Voorhis WC, Corti D, Veesler D, Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 374, 1621–1626 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCallum M, Bassi J, De Marco A, Chen A, Walls AC, Di Iulio J, Tortorici MA, Navarro M-J, Silacci-Fregni C, Saliba C, Sprouse KR, Agostini M, Pinto D, Culap K, Bianchi S, Jaconi S, Cameroni E, Bowen JE, Tilles SW, Pizzuto MS, Guastalla SB, Bona G, Pellanda AF, Garzoni C, Van Voorhis WC, Rosen LE, Snell G, Telenti A, Virgin HW, Piccoli L, Corti D, Veesler D, SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 373, 648–654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dejnirattisai W, Zhou D, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, Wang B, López-Camacho C, Slon-Campos J, Walter TS, Skelly D, Costa Clemens SA, Naveca FG, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert SC, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Paterson NG, Williams MA, Hall DR, Hulswit RJG, Bowden TA, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR, Antibody evasion by the P.1 strain of SARS-CoV-2. Cell 184, 2939–2954.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, Liu J, Errico JM, Xie X, Suryadevara N, Gilchuk P, Zost SJ, Tahan S, Droit L, Turner JS, Kim W, Schmitz AJ, Thapa M, Wang D, Boon ACM, Presti RM, O’Halloran JA, Kim AHJ, Deepak P, Pinto D, Fremont DH, Crowe JE Jr, Corti D, Virgin HW, Ellebedy AH, Shi P-Y, Diamond MS, Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med 27, 717–726 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen RE, Winkler ES, Case JB, Aziati ID, Bricker TL, Joshi A, Darling TL, Ying B, Errico JM, Shrihari S, VanBlargan LA, Xie X, Gilchuk P, Zost SJ, Droit L, Liu Z, Stumpf S, Wang D, Handley SA, Stine WB Jr, Shi P-Y, Davis-Gardner ME, Suthar MS, Knight MG, Andino R, Chiu CY, Ellebedy AH, Fremont DH, Whelan SPJ, Crowe JE Jr, Purcell L, Corti D, Boon ACM, Diamond MS, In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature 596, 103–108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collier DA, De Marco A, Ferreira IATM, Meng B, Datir R, Walls AC, Kemp SSA, Bassi J, Pinto D, Fregni CS, Bianchi S, Tortorici MA, Bowen J, Culap K, Jaconi S, Cameroni E, Snell G, Pizzuto MS, Pellanda AF, Garzoni C, Riva A, Elmer A, Kingston N, Graves B, McCoy LE, Smith KGC, Bradley JR, Temperton N, Lourdes Ceron-Gutierrez L, Barcenas-Morales G, Harvey W, Virgin HW, Lanzavecchia A, Piccoli L, Doffinger R, Wills M, Veesler D, Corti D, Gupta RK, Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 593, 136–141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, Pinto D, VanBlargan LA, De Marco A, di Iulio J, Zatta F, Kaiser H, Noack J, Farhat N, Czudnochowski N, Havenar-Daughton C, Sprouse KR, Dillen JR, Powell AE, Chen A, Maher C, Yin L, Sun D, Soriaga L, Bassi J, Silacci-Fregni C, Gustafsson C, Franko NM, Logue J, Iqbal NT, Mazzitelli I, Geffner J, Grifantini R, Chu H, Gori A, Riva A, Giannini O, Ceschi A, Ferrari P, Cippà PE, Franzetti-Pellanda A, Garzoni C, Halfmann PJ, Kawaoka Y, Hebner C, Purcell LA, Piccoli L, Pizzuto MS, Walls AC, Diamond MS, Telenti A, Virgin HW, Lanzavecchia A, Snell G, Veesler D, Corti D, Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 602, 664–670 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanBlargan L, Errico J, Halfmann P, Zost S, Crowe J, Purcell L, Kawaoka Y, Corti D, Fremont D, Diamond M, An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nature Medicine 28, 490–495 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCallum M, Czudnochowski N, Rosen LE, Zepeda SK, Bowen JE, Walls AC, Hauser K, Joshi A, Stewart C, Dillen JR, Powell AE, Croll TI, Nix J, Virgin HW, Corti D, Snell G, Veesler D, Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 375, 864–868 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Case JB, Mackin S, Errico J, Chong Z, Madden EA, Guarino B, Schmid MA, Rosenthal K, Ren K, Jung A, Droit L, Handley SA, Halfmann PJ, Kawaoka Y, Crowe JE Jr, Fremont DH, Virgin HW, Loo Y-M, Esser MT, Purcell LA, Corti D, Diamond MS, Resilience of S309 and AZD7442 monoclonal antibody treatments against infection by SARS-CoV-2 Omicron lineage strains. bioRxiv, 2022.03.17.484787 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D, Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Pinto D, Sauer MM, Czudnochowski N, Low JS, Tortorici MA, Housley MP, Noack J, Walls AC, Bowen JE, Guarino B, Rosen LE, di Iulio J, Jerak J, Kaiser H, Islam S, Jaconi S, Sprugasci N, Culap K, Abdelnabi R, Foo C, Coelmont L, Bartha I, Bianchi S, Silacci-Fregni C, Bassi J, Marzi R, Vetti E, Cassotta A, Ceschi A, Ferrari P, Cippà PE, Giannini O, Ceruti S, Garzoni C, Riva A, Benigni F, Cameroni E, Piccoli L, Pizzuto MS, Smithey M, Hong D, Telenti A, Lempp FA, Neyts J, Havenar-Daughton C, Lanzavecchia A, Sallusto F, Snell G, Virgin HW, Beltramello M, Corti D, Veesler D, Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science 373, 1109–1116 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starr TN, Czudnochowski N, Liu Z, Zatta F, Park Y-J, Addetia A, Pinto D, Beltramello M, Hernandez P, Greaney AJ, Marzi R, Glass WG, Zhang I, Dingens AS, Bowen JE, Tortorici MA, Walls AC, Wojcechowskyj JA, De Marco A, Rosen LE, Zhou J, Montiel-Ruiz M, Kaiser H, Dillen JR, Tucker H, Bassi J, Silacci-Fregni C, Housley MP, di Iulio J, Lombardo G, Agostini M, Sprugasci N, Culap K, Jaconi S, Meury M, Dellota E Jr, Abdelnabi R, Foo S-YC, Cameroni E, Stumpf S, Croll TI, Nix JC, Havenar-Daughton C, Piccoli L, Benigni F, Neyts J, Telenti A, Lempp FA, Pizzuto MS, Chodera JD, Hebner CM, Virgin HW, Whelan SPJ, Veesler D, Corti D, Bloom JD, Snell G, SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jette CA, Cohen AA, Gnanapragasam PNP, Muecksch F, Lee YE, Huey-Tubman KE, Schmidt F, Hatziioannou T, Bieniasz PD, Nussenzweig MC, West AP Jr, Keeffe JR, Bjorkman PJ, Barnes CO, Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies. Cell Rep 36, 109760 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.VanBlargan LA, Adams LJ, Liu Z, Chen RE, Gilchuk P, Raju S, Smith BK, Zhao H, Case JB, Winkler ES, Whitener BM, Droit L, Aziati ID, Bricker TL, Joshi A, Shi P-Y, Creanga A, Pegu A, Handley SA, Wang D, Boon ACM, Crowe JE Jr, Whelan SPJ, Fremont DH, Diamond MS, A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope. Immunity 54, 2399–2416.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park Y-J, De Marco A, Starr TN, Liu Z, Pinto D, Walls AC, Zatta F, Zepeda SK, Bowen JE, Sprouse KR, Joshi A, Giurdanella M, Guarino B, Noack J, Abdelnabi R, Foo S-YC, Rosen LE, Lempp FA, Benigni F, Snell G, Neyts J, Whelan SPJ, Virgin HW, Bloom JD, Corti D, Pizzuto MS, Veesler D, Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science 375, 449–454 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls AC, Park Y-J, Strauch E-M, Stewart L, Diamond MS, Veesler D, Baker D, De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370, 426–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Case JB, Chen RE, Cao L, Ying B, Winkler ES, Johnson M, Goreshnik I, Pham MN, Shrihari S, Kafai NM, Bailey AL, Xie X, Shi P-Y, Ravichandran R, Carter L, Stewart L, Baker D, Diamond MS, Ultrapotent miniproteins targeting the SARS-CoV-2 receptor-binding domain protect against infection and disease. Cell Host Microbe 29, 1151–1161.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Javanmardi K, Chou C-W, Terrace CI, Annapareddy A, Kaoud TS, Guo Q, Lutgens J, Zorkic H, Horton AP, Gardner EC, Nguyen G, Boutz DR, Goike J, Voss WN, Kuo H-C, Dalby KN, Gollihar JD, Finkelstein IJ, Rapid characterization of spike variants via mammalian cell surface display. Molecular Cell 81, P5099–5111.E8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverman J, Liu Q, Bakker A, To W, Duguay A, Alba BM, Smith R, Rivas A, Li P, Le H, Whitehorn E, Moore KW, Swimmer C, Perlroth V, Vogt M, Kolkman J, Stemmer WPC, Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nat. Biotechnol 23, 1556–1561 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Detalle L, Stohr T, Palomo C, Piedra PA, Gilbert BE, Mas V, Millar A, Power UF, Stortelers C, Allosery K, Melero JA, Depla E, Generation and Characterization of ALX-0171, a Potent Novel Therapeutic Nanobody for the Treatment of Respiratory Syncytial Virus Infection. Antimicrob. Agents Chemother 60, 6–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strauch EM, Bernard SM, La D, Bohn AJ, Lee PS, Anderson CE, Nieusma T, Holstein CA, Garcia NK, Hooper KA, Ravichandran R, Nelson JW, Sheffler W, Bloom JD, Lee KK, Ward AB, Yager P, Fuller DH, Wilson IA, Baker D, Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site. Nat. Biotechnol 35, 667–671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bracken CJ, Lim SA, Solomon P, Rettko NJ, Nguyen DP, Zha BS, Schaefer K, Byrnes JR, Zhou J, Lui I, Liu J, Pance K, QCRG Structural Biology Consortium, Zhou XX, Leung KK, Wells JA, Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2. Nat. Chem. Biol 17, 113–121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koenig P-A, Das H, Liu H, Kümmerer BM, Gohr FN, Jenster L-M, Schiffelers LDJ, Tesfamariam YM, Uchima M, Wuerth JD, Gatterdam K, Ruetalo N, Christensen MH, Fandrey CI, Normann S, Tödtmann JMP, Pritzl S, Hanke L, Boos J, Yuan M, Zhu X, Schmid-Burgk JL, Kato H, Schindler M, Wilson IA, Geyer M, Ludwig KU, Hällberg BM, Wu NC, Schmidt FI, Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science 371, eabe6230 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang Y, Nambulli S, Xiao Z, Liu H, Sang Z, Duprex WP, Schneidman-Duhovny D, Zhang C, Shi Y, Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science 370, 1479–1484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walser M, Rothenberger S, Hurdiss DL, Schlegel A, Calabro V, Fontaine S, Villemagne D, Paladino M, Hospodarsch T, Neculcea A, Cornelius A, Schildknecht P, Matzner M, Hänggi M, Franchini M, Kaufmann Y, S chaible D, Schlegel I, Iss C, Looser T, Mangold S, Herzog C, Schiegg D, Reichen C, Radom F, Bosshart A, Lehmann A, Haeuptle MA, Zürcher A, Vagt T, Sigrist G, Straumann M, Proba K, Veitonmäki N, Dawson KM, Zitt C, Mayor J, Ryter S, Lyoo H, Wang C, Li W, Drulyte I, Du W, Kaspar Binz H, de Waal L, Stittelaar KJ, Taplin S, Lewis S, Steiner D, van Kuppeveld FJM, Engler O, Bosch B-J, Stumpp MT, Amstutz P, Highly potent anti-SARS-CoV-2 multi-DARPin therapeutic candidates. bioRxiv, 2020.08.25.256339 (2020). [Google Scholar]
- 35.Schoof M, Faust B, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesbølle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Deshpande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Liu Y, Krogan NJ, Ralston CY, Swaney DL, García-Sastre A, Ott M, Vignuzzi M, QCRG Structural Biology Consortium, P. Walter, A. Manglik, An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 370, 1473–1479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linsky TW, Vergara R, Codina N, Nelson JW, Walker MJ, Su W, Barnes CO, Hsiang T-Y, Esser-Nobis K, Yu K, Reneer ZB, Hou YJ, Priya T, Mitsumoto M, Pong A, Lau UY, Mason ML, Chen J, Chen A, Berrocal T, Peng H, Clairmont NS, Castellanos J, Lin Y-R, Josephson-Day A, Baric RS, Fuller DH, Walkey CD, Ross TM, Swanson R, Bjorkman PJ, Gale M Jr, Blancas-Mejia LM, Yen H-L, Silva D-A, De novo design of potent and resilient hACE2 decoys to neutralize SARS-CoV-2. Science 370, 1208–1214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Copin R, Baum A, Wloga E, Pascal KE, Giordano S, Fulton BO, Zhou A, Negron N, Lanza K, Chan N, Coppola A, Chiu J, Ni M, Wei Y, Atwal GS, Hernandez AR, Saotome K, Zhou Y, Franklin MC, Hooper AT, McCarthy S, Hamon S, Hamilton JD, Staples HM, Alfson K, Carrion R, Ali S, Norton T, Somersan-Karakaya S, Sivapalasingam S, Herman GA, Weinreich DM, Lipsich L, Stahl N, Murphy AJ, Yancopoulos GD, Kyratsous CA, The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell 184, 3949–3961.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, Navarro MJ, Bowen JE, Tortorici MA, Walls AC, King NP, Veesler D, Bloom JD, Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 182, 1295–1310.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palomo C, Mas V, Detalle L, Depla E, Cano O, Vázquez M, Stortelers C, Melero JA, Trivalency of a Nanobody Specific for the Human Respiratory Syncytial Virus Fusion Glycoprotein Drastically Enhances Virus Neutralization and Impacts Escape Mutant Selection. Antimicrob. Agents Chemother 60, 6498–6509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun ZZ, Yeung E, Hayes CA, Noireaux V, Murray RM, Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth. Biol 3, 387–397 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Silverman AD, Karim AS, Jewett MC, Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet 21, 151–170 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Hsieh C-L, Goldsmith JA, Schaub JM, DiVenere AM, Kuo H-C, Javanmardi K, Le KC, Wrapp D, Lee AG, Liu Y, Chou C-W, Byrne PO, Hjorth CK, Johnson NV, Ludes-Meyers J, Nguyen AW, Park J, Wang N, Amengor D, Lavinder JJ, Ippolito GC, Maynard JA, Finkelstein IJ, McLellan JS, Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mammen M, Choi S-K, Whitesides GM, Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew. Chem. Int. Ed Engl 37, 2754–2794 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Pollard TD, De La Cruz EM, Take advantage of time in your experiments: a guide to simple, informative kinetics assays. Mol. Biol. Cell 24, 1103–1110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Güthe S, Kapinos L, Möglich A, Meier S, Grzesiek S, Kiefhaber T, Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J. Mol. Biol 337, 905–915 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Gräwe A, Stein V, Linker Engineering in the Context of Synthetic Protein Switches and Sensors. Trends Biotechnol 39, 731–744 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Ke Z, Oton J, Qu K, Cortese M, Zila V, McKeane L, Nakane T, Zivanov J, Neufeldt CJ, Cerikan B, Lu JM, Peukes J, Xiong X, Kräusslich HG, Scheres SHW, Bartenschlager R, Briggs JAG, Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turoňová B, Sikora M, Schürmann C, Hagen WJH, Welsch S, Blanc FEC, von Bülow S, Gecht M, Bagola K, Hörner C, van Zandbergen G, Landry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Mühlebach MD, Hummer G, Krijnse Locker J, Beck M, In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 370, 203–208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C, Zhang J, Weng T, Zhang Z, Wu Z, Cheng L, Shi D, Lu X, Lei J, Crispin M, Shi Y, Li L, Li S, Molecular Architecture of the SARS-CoV-2 Virus. Cell 183, 730–738.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeh HW, Karmach O, Ji A, Carter D, Martins-Green MM, Ai HW, Red-shifted luciferase-luciferin pairs for enhanced bioluminescence imaging. Nat. Methods 14, 971–974 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim BB, Wu H, Hao YA, Pan M, Chavarha M, Zhao Y, Westberg M, St-Pierre F, Wu JC, Lin MZ, A red fluorescent protein with improved monomericity enables ratiometric voltage imaging with ASAP3. Scientific Rep 12, 3678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, Silacci-Fregni C, Pinto D, Rosen LE, Bowen JE, Acton OJ, Jaconi S, Guarino B, Minola A, Zatta F, Sprugasci N, Bassi J, Peter A, De Marco A, Nix JC, Mele F, Jovic S, Rodriguez BF, Gupta SV, Jin F, Piumatti G, Lo Presti G, Pellanda AF, Biggiogero M, Tarkowski M, Pizzuto MS, Cameroni E, Havenar-Daughton C, Smithey M, Hong D, Lepori V, Albanese E, Ceschi A, Bernasconi E, Elzi L, Ferrari P, Garzoni C, Riva A, Snell G, Sallusto F, Fink K, Virgin HW, Lanzavecchia A, Corti D, Veesler D, Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 183, 1024–1042.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beumer J, Geurts MH, Lamers MM, Puschhof J, Zhang J, van der Vaart J, Mykytyn AZ, Breugem TI, Riesebosch S, Schipper D, van den Doel PB, de Lau W, Pleguezuelos-Manzano C, Busslinger G, Haagmans BL, Clevers H, A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat. Commun 12, 5498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Penninger JM, Mirazimi A, Montserrat N, Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 181, 905–913.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harder JL, Menon R, Otto EA, Zhou J, Eddy S, Wys NL, O’Connor C, Luo J, Nair V, Cebrian C, Spence JR, Bitzer M, Troyanskaya OG, Hodgin JB, Wiggins RC, Freedman BS, Kretzler M, European Renal cDNA Bank (ERCB), Nephrotic Syndrome Study Network (NEPTUNE), Organoid single cell profiling identifies a transcriptional signature of glomerular disease. JCI Insight 4 (2019), doi: 10.1172/jci.insight.122697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, Saad AF, Li MK, Hughes MR, Werff RV, Peters DT, Lu J, Baccei A, Siedlecki AM, Valerius MT, Musunuru K, McNagny KM, Steinman TI, Zhou J, Lerou PH, Bonventre JV, Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun 6, 8715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alsoussi WB, Turner JS, Case JB, Zhao H, Schmitz AJ, Zhou JQ, Chen RE, Lei T, Rizk AA, McIntire KM, Winkler ES, Fox JM, Kafai NM, Thackray LB, Hassan AO, Amanat F, Krammer F, Watson CT, Kleinstein SH, Fremont DH, Diamond MS, Ellebedy AH, A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. The Journal of Immunology 205, 915–922 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Errico JM, Zhao H, Chen RE, Liu Z, Case JB, Ma M, Schmitz AJ, Rau MJ, Fitzpatrick JAJ, Shi P-Y, Diamond MS, Whelan SPJ, Ellebedy AH, Fremont DH, Structural mechanism of SARS-CoV-2 neutralization by two murine antibodies targeting the RBD. Cell Rep 37, 109881 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su W, Sia SF, Schmitz AJ, Bricker TL, Starr TN, Greaney AJ, Turner JS, Mohammed BM, Liu Z, Choy KT, Darling TL, Joshi A, Cheng KM, Wong AYL, Harastani HH, Nicholls JM, Whelan SPJ, Bloom JD, Yen H-L, Ellebedy AH, Boon ACM, Neutralizing Monoclonal Antibodies That Target the Spike Receptor Binding Domain Confer Fc Receptor-Independent Protection against SARS-CoV-2 Infection in Syrian Hamsters. MBio 12, e0239521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wertz GW, Moudy R, Ball LA, Adding genes to the RNA genome of vesicular stomatitis virus: positional effects on stability of expression. J. Virol 76, 7642–7650 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hassan AO, Case JB, Winkler ES, Thackray L, Kafai M, Bailey AL, Mccune BT, Fox JM, Chen RE, Al WB, Turner JS, Schmitz AJ, Lei T, Shrihari S, Keeler P, Fremont DH, Greco S, Mccray PB, Perlman S, Holtzman MJ, Ellebedy AH, Diamond MS, A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 182, 744–753.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder J, Quispe J, Cameroni E, Gopal R, Dai M, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D, Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell 176, 1026–1039.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lempp FA, Soriaga LB, Montiel-Ruiz M, Benigni F, Noack J, Park Y-J, Bianchi S, Walls AC, Bowen JE, Zhou J, Kaiser H, Joshi A, Agostini M, Meury M, Dellota E Jr, Jaconi S, Cameroni E, Martinez-Picado J, Vergara-Alert J, Izquierdo-Useros N, Virgin HW, Lanzavecchia A, Veesler D, Purcell LA, Telenti A, Corti D, Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature 598, 342–347 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Guo L, Bi W, Wang X, Xu W, Yan R, Zhang Y, Zhao K, Li Y, Zhang M, Cai X, Jiang S, Xie Y, Zhou Q, Lu L, Dang B, Engineered trimeric ACE2 binds viral spike protein and locks it in “Three-up” conformation to potently inhibit SARS-CoV-2 infection. Cell Res 31, 98–100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glasgow A, Glasgow J, Limonta D, Solomon P, Lui I, Zhang Y, Nix MA, Rettko NJ, Zha S, Yamin R, Kao K, Rosenberg OS, Ravetch JV, Wiita AP, Leung KK, Lim SA, Zhou XX, Hobman TC, Kortemme T, Wells JA, Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A 117, 28046–28055 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Higuchi Y, Suzuki T, Arimori T, Ikemura N, Mihara E, Kirita Y, Ohgitani E, Mazda O, Motooka D, Nakamura S, Sakai Y, Itoh Y, Sugihara F, Matsuura Y, Matoba S, Okamoto T, Takagi J, Hoshino A, Engineered ACE2 receptor therapy overcomes mutational escape of SARS-CoV-2. Nat. Commun 12, 3802 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, Perry C, Pan C, Hosain R, Mahmood A, Davis JD, Turner KC, Hooper AT, Hamilton JD, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Kohli A, Sachdeva Y, Graber X, Kowal B, DiCioccio T, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med 384, 238–251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, Shawa I, Adams AC, Van Naarden J, Custer KL, Shen L, Durante M, Oakley G, Schade AE, Sabo J, Patel DR, Klekotka P, Skovronsky DM, SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med 384, 229–237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Brien MP, Forleo-Neto E, Musser BJ, Isa F, Chan K-C, Sarkar N, Bar KJ, Barnabas RV, Barouch DH, Cohen MS, Hurt CB, Burwen DR, Marovich MA, Hou P, Heirman I, Davis JD, Turner KC, Ramesh D, Mahmood A, Hooper AT, Hamilton JD, Kim Y, Purcell LA, Baum A, Kyratsous CA, Krainson J, Perez-Perez R, Mohseni R, Kowal B, DiCioccio AT, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, Weinreich DM, Covid-19 Phase 3 Prevention Trial Team, Subcutaneous REGEN-COV Antibody Combination to Prevent Covid-19. N. Engl. J. Med 385, 1184–1195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chevalier A, Silva D-A, Rocklin GJ, Hicks DR, Vergara R, Murapa P, Bernard SM, Zhang L, Lam K-H, Yao G, Bahl CD, Miyashita S-I, Goreshnik I, Fuller JT, Koday MT, Jenkins CM, Colvin T, Carter L, Bohn A, Bryan CM, Fernández-Velasco DA, Stewart L, Dong M, Huang X, Jin R, Wilson IA, Fuller DH, Baker D, Massively parallel de novo protein design for targeted therapeutics. Nature 550, 74–79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quijano-Rubio A, Yeh H-W, Park J, Lee H, Langan RA, Boyken SE, Lajoie MJ, Cao L, Chow CM, Miranda MC, Wi J, Hong HJ, Stewart L, Oh B-H, Baker D, De novo design of modular and tunable protein biosensors. Nature 591, 482–487 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elledge SK, Zhou XX, Byrnes JR, Martinko AJ, Lui I, Pance K, Lim SA, Glasgow JE, Glasgow AA, Turcios K, Iyer NS, Torres L, Peluso MJ, Henrich TJ, Wang TT, Tato CM, Leung KK, Greenhouse B, Wells JA, Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. Nat. Biotechnol 39, 928–935 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, Pettie D, King NP, Balazs AB, Bloom JD, Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 12, 513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hassan AO, Shrihari S, Gorman MJ, Ying B, Yaun D, Raju S, Chen RE, Dmitriev IP, Kashentseva E, Adams LJ, Mann C, Davis-Gardner ME, Suthar MS, Shi P-Y, Saphire EO, Fremont DH, Curiel DT, Alter G, Diamond MS, An intranasal vaccine durably protects against SARS-CoV-2 variants in mice. Cell Rep 36, 109452 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Case JB, Rothlauf PW, Chen RE, Liu Z, Zhao H, Kim AS, Bloyet L-M, Zeng Q, Tahan S, Droit L, Ilagan MXG, Tartell MA, Amarasinghe G, Henderson JP, Miersch S, Ustav M, Sidhu S, Virgin HW, Wang D, Ding S, Corti D, Theel ES, Fremont DH, Diamond MS, Whelan SPJ, Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe 28, 475–485.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fleishman SJ, Leaver-Fay A, Corn JE, Strauch E-M, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G, Meiler J, Baker D, RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One 6, e20161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hsia Y, Mout R, Sheffler W, Edman NI, Vulovic I, Park Y-J, Redler RL, Bick MJ, Bera AK, Courbet A, Kang A, Brunette TJ, Nattermann U, Tsai E, Saleem A, Chow CM, Ekiert D, Bhabha G, Veesler D, Baker D, Design of multi-scale protein complexes by hierarchical building block fusion. Nat. Commun 12, 2294 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang P-S, Ban Y-EA, Richter F, Andre I, Vernon R, Schief WR, Baker D, RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS One 6, e24109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin Y-R, Koga N, Tatsumi-Koga R, Liu G, Clouser AF, Montelione GT, Baker D, Control over overall shape and size in de novo designed proteins. Proc. Nat. Acad. Sci. U. S. A 112, E5478–E5485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rocklin GJ, Chidyausiku TM, Goreshnik I, Ford A, Houliston S, Lemak A, Carter L, Ravichandran R, Mulligan VK, Chevalier A, Arrowsmith CH, Baker D, Global analysis of protein folding using massively parallel design, synthesis, and testing. Science 357, 168–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dang B, Wu H, Mulligan VK, Mravic M, Wu Y, Lemmin T, Ford A, Silva D-A, Baker D, DeGrado WF, De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures. Proc. Nat. Acad. Sci. U. S. A 114, 10852–10857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khatib F, Cooper S, Tyka MD, Xu K, Makedon I, Popovic Z, Baker D, Players F, Algorithm discovery by protein folding game players. Proc. Natl. Acad. Sci. U. S. A 108, 18949–18953 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe JE Jr, Bloom JD, Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe 29, 44–57.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Starr TN, Greaney AJ, Dingens AS, Bloom JD, Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep Med 2, 100255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rabe BA, Cepko C, A Simple Enhancement for Gibson Isothermal Assembly. bioRxiv, 2020.06.14.150979 (2020). [Google Scholar]
- 87.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA 3rd, Smith HO, Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 88.Ojima-Kato T, Nagai S, Nakano H, Ecobody technology: rapid monoclonal antibody screening method from single B cells using cell-free protein synthesis for antigen-binding fragment formation. Sci. Rep 7, 13979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen Z, Kibler RD, Hunt A, Busch F, Pearl J, Jia M, VanAernum ZL, Wicky BIM, Dods G, Liao H, Wilken MS, Ciarlo C, Green S, El-Samad H, Stamatoyannopoulos J, Wysocki VH, Jewett MC, Boyken SE, Baker D, De novo design of protein logic gates. Science 368, 78–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daddona PE, Davis HM, Fogler WE, Klug TL, Ledonne NC, Mckinney JS, Method of removing endotoxin contaminants Patent, WO1989003885A1 (1989).
- 91.Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, Kong WP, Andres EL, Kettenbach AN, Denison MR, Chappell JD, Graham BS, Ward AB, McLellan JS, Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U. S. A 114, E7348–E7357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB, Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep 8, 15701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Russo CJ, Passmore LA, Electron microscopy: Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B, Automated molecular microscopy: the new Leginon system. J. Struct. Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 95.Tegunov D, Cramer P, Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH, New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Punjani A, Zhang H, Fleet DJ, Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 99.Zivanov J, Nakane T, Scheres SHW, A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, Henderson R, High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rosenthal PB, Henderson R, Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 102.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE, UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang RY-R, Song Y, Barad BA, Cheng Y, Fraser JS, DiMaio F, Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife 5, e17219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Frenz B, Walls AC, Egelman EH, Veesler D, DiMaio F, RosettaES: a sampling strategy enabling automated interpretation of difficult cryo-EM maps. Nat. Methods 14, 797–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Corman VM, Haage VC, Bleicker T, Schmidt ML, Mühlemann B, Zuchowski M, Jo WK, Tscheak P, Möncke-Buchner E, Müller MA, Krumbholz A, Drexler JF, Drosten C, Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study. Lancet Microbe 2, e311–e319 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bar-On YM, Flamholz A, Phillips R, Milo R, SARS-CoV-2 (COVID-19) by the numbers. Elife 9, 1–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Katsamba PS, Navratilova I, Calderon-Cacia M, Fan L, Thornton K, Zhu M, Bos TV, Forte C, Friend D, Laird-Offringa I, Tavares G, Whatley J, Shi E, Widom A, Lindquist KC, Klakamp S, Drake A, Bohmann D, Roell M, Rose L, Dorocke J, Roth B, Luginbühl B, Myszka DG, Kinetic analysis of a high-affinity antibody/antigen interaction performed by multiple Biacore users. Anal. Biochem 352, 208–221 (2006). [DOI] [PubMed] [Google Scholar]
- 109.Firth AE, Patrick WM, GLUE-IT and PEDEL-AA: new programmes for analyzing protein diversity in randomized libraries. Nucleic Acids Res 36, 281–285 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Case JB, Bailey AL, Kim AS, Chen RE, Diamond MS, Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology 548, 39–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R, The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boyken SE, Chen Z, Groves B, Langan RA, Oberdorfer G, Ford A, Gilmore JM, Xu C, DiMaio F, Pereira JH, Sankaran B, Seelig G, Zwart PH, Baker D, De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity. Science 352, 680–687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ueda G, Antanasijevic A, Fallas JA, Sheffler W, Copps J, Ellis D, Hutchinson GB, Moyer A, Yasmeen A, Tsybovsky Y, Park Y-J, Bick MJ, Sankaran B, Gillespie RA, Brouwer PJ, Zwart PH, Veesler D, Kanekiyo M, Graham BS, Sanders RW, Moore JP, Klasse PJ, Ward AB, King NP, Baker D, Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. Elife 9, e57659 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Boyken SE, Benhaim MA, Busch F, Jia M, Bick MJ, Choi H, Klima JC, Chen Z, Walkey C, Mileant A, Sahasrabuddhe A, Wei KY, Hodge EA, Byron S, Quijano-Rubio A, Sankaran B, King NP, Lippincott-Schwartz J, Wysocki VH, Lee KK, Baker D, De novo design of tunable, pH-driven conformational changes. Science 364, 658–664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Harbury PB, Kim PS, Alber T, Crystal structure of an isoleucine-zipper trimer. Nature 371, 80–83 (1994). [DOI] [PubMed] [Google Scholar]
- 116.Thomson AR, Wood CW, Burton AJ, Bartlett GJ, Sessions RB, Brady RL, Woolfson DN, Computational design of water-soluble α-helical barrels. Science 346, 485–488 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are in the paper or supplementary materials. Structural models and density maps have been deposited in the Protein Data Bank (PDB) (SARS-CoV-2/TRI2–2: 7UHC and SARS-CoV-2/TRI2–2 (local refinement): 7UHB) and Electron Microscopy Data Bank (EMDB) (SARS-CoV-2/TRI2–2: EMD-26512, SARS-CoV-2/TRI2–2 (local refinement): EMD-26511, SARS-CoV-2/FUS31-G10 (2RBD-open): EMD-26509, SARS-CoV-2/FUS31-G10 (3RBD-open): EMD-26510, SARS-CoV-2/FUS231-P24 (2RBD-open): EMD-26507, and SARS-CoV-2/FUS231-P24 (3RBD-open): EMD-26508). Illumina sequencing data for the deep mutational scanning experiments are available on NCBI SRA, BioSample SAMN19925005. Code for the analysis of the deep mutational scanning experiments are available on Zenodo https://doi.org/10.5281/zenodo.6377268. Requests for reagents (antibodies, viruses, and other proteins) should be directed to the corresponding authors and will be made available after completion of a Materials Transfer Agreement with the University of Washington. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using this material.


