Abstract
Objectives:
Automated whole brain segmentation from magnetic resonance images is of great interest for development of clinically relevant volumetric markers for various neurological diseases. While deep learning methods have demonstrated remarkable potential in this area, they may perform poorly in non-optimal conditions, such as with limited training data. Manual whole brain segmentation is an incredibly tedious process, so minimizing the dataset size required for training segmentation algorithms may be of wide interest. The purpose of this study was to compare the performance of the prototypical deep learning segmentation architecture (U-Net) with a previously published atlas-free traditional machine learning method, Classification using Derivative-based Features (C-DEF) for whole brain segmentation, in the setting of limited training data.
Materials and Methods:
C-DEF and U-Net models were evaluated after training on manually curated data from 5, 10, and 15 participants in two research cohorts – (i) people living with clinically diagnosed HIV infection and (ii) relapsing-remitting Multiple Sclerosis (MS), each acquired at separate institutions, and between 5 and 295 participant data using a large, publicly available and annotated dataset of glioblastoma and lower grade glioma (BraTS). Statistics was performed on Dice Similarity Coefficient (DSC) using repeated-measures ANOVA and Dunnett-Hsu pairwise comparison.
Results:
C-DEF produced better segmentation than U-Net in lesion (29.2 – 38.9%) and cerebrospinal fluid classes (5.3% – 11.9%) when trained with data from 15 or fewer participants. Unlike C-DEF, U-Net showed significant improvement when increasing the size of the training data (24 – 30% higher than baseline). In the BraTS dataset, C-DEF produced equivalent or better segmentations than U-Net for enhancing tumor and peritumoral edema regions across all training data sizes explored. However, U-Net was more effective than C-DEF for segmentation of necrotic tumor tissue when trained on 10 or more participants, probably due to the inconsistent signal intensity of the tissue class.
Conclusions:
These results demonstrate that classical machine learning methods can produce more accurate brain segmentation than the far more complex deep learning methods when only small or moderate amounts of training data are available (n ≤ 15). The magnitude of this advantage varies by tissue and by cohort, while U-Net may be preferable for deep grey matter and necrotic tumor segmentation, particularly with larger training datasets (n ≥ 20). The magnitude of this advantage varies by tissue and by cohort. Given that segmentation models often need to be re-trained for application to novel imaging protocols or pathology, the bottleneck associated with large-scale manual annotation could be avoided with classical machine learning algorithms such as C-DEF.
Keywords: Brain segmentation, Deep learning, Machine learning, MRI
1. Introduction
In recent years, imaging markers of atrophy and inflammation derived from brain segmentation have been widely used to inform disease status and progression in neurological disorders (1–3). These volumetric markers, particularly with regards to white matter lesions (4, 5), are of great interest as potential endpoints for clinical trials. Although labels manually drawn by an expert rater remain the gold standard for segmentation accuracy, this is prohibitively tedious and costly to do on a large scale. Whole brain segmentation is particularly time-consuming due to the large number of detailed contours that must be delineated across many individual slices. Automation of brain segmentation can therefore help by delivering fast and reproducible imaging markers (6).
Among automated brain segmentation algorithms, supervised learning methods have demonstrated strong capability to model the signal intensity profiles and spatial context of brain tissues to produce robust brain segmentations. Atlas-free segmentations offer ability to segment the brain in a disease-independent manner, using signal intensity signatures of various tissue types from multiple contrasts and derived image filters (7). Recently, deep learning has risen to the forefront by demonstrating state-of-the-art results on a range of medical image segmentation benchmarks, including the MICCAI Brain Tumor Segmentation challenge, Medical Segmentation Decathlon, and others (8–11). The U-Net framework, introduced in 2015 by Ronneberger et al., used a convolutional neural network with skip connections to capture complex spatial information at multiple scales (12). Since then, much research has focused on improvement of this framework using various strategies for multiscale feature aggregation (9, 13, 14) and localization (15, 16). Despite the proliferation of U-Net-style architectures, many of the same challenges still persist with regards to model development and deployment. Model training requires specialized GPU resources that are of little use in other aspects of clinical informatics workflows. Due to the high number of parameters (often greater than a million) involved in training for large 3D or 4D (if multi-contrast) volumes, GPU memory limitations often dictate that input size be reduced, with patch extraction being the most common approach (17, 18). With this in mind, the relative performance of these highly complex deep learning algorithms as opposed to less resource-intensive machine learning algorithms needs to be investigated.
Perhaps more importantly, deep-learning models still frequently rely on extensive training datasets and demonstrate inconsistent generalizability across imaging protocols and pathologies. This is significant because a model trained on hundreds of well-labeled public datasets may still find itself to be of limited use while segmenting naïve images. Bias introduced by factors such as scanner type and imaging parameters can be great enough to significantly confound segmentation tasks (19). These factors can make transfer learning or domain adaptation with few labeled examples challenging, particularly if the disease or scanner protocol of interest is rare or novel, making previously learned features less relevant. Therefore, real-world applications of automated segmentation often rely on training models from scratch with manually curated labels from each dataset. Such manual annotation requires expert knowledge and therefore places a substantial time demand on qualified experts. Manual annotation is also prone to significant inter- and intra-operator variability requiring consensus readings (20), further adding to the cost of curation. This can create a significant bottleneck for model development in many cases (21), so reducing the size of training dataset that is needed may be of wide interest. There is therefore a need for algorithms that are robust to training on limited data and a greater understanding of the behavior of supervised segmentation methods under limited training conditions. Several previous studies have observed effect of training dataset size on the performance of deep learning models for medical segmentation tasks, including for head-and-neck CT (22) and carotid ultrasound (23). However, this line of inquiry remains underexplored in the domain of whole brain segmentation. In addition, these studies typically do not consider alternative segmentation methods such as classical machine learning for comparison.
This study aims to address these gaps in knowledge by comparison of a deep-learning method (U-Net) and a classical machine learning method (C-DEF) for segmentation on several clinical datasets under limited training conditions. Each model was evaluated in parallel on two moderately sized cohorts (n=20 for each cohort) composed of patients diagnosed with HIV infection and relapsing-remitting MS, respectively. Algorithms were assessed solely for tissue segmentation performance within each cohort in order to assess the reliability of each method. The amount of training data provided for training was varied in order to assess the relationship between training data abundance and segmentation performance. This comparison was then extended to a publicly available segmentation benchmark, the MICCAI BraTS 2020 Training dataset, modified to classify only the manually delineated pathological tissue subtypes, to validate the observed trends over a larger range of training dataset sizes.
2. Materials and Methods
2.1. Datasets and Preprocessing
Multi-modal brain MRI scans were obtained at 3T from study participants clinically diagnosed with relapsing-remitting multiple sclerosis using 2010 revised McDonald Criteria (24) (MS cohort) and people living with HIV infection (HIV cohort). MS cohort participants were recruited as part of the FutureMS study (https://future-ms.org). Study protocols were approved by the institutional review board of the National Institute of Neurological Disorders and Stroke (reference number NCT01875588) for the HIV cohort and the National Health Service South East Scotland Research Ethics Committee 02 (reference number 15/SS/0233) for the MS cohort, and all participants provided written informed consent. Images used in this analysis included MPRAGE (TR/TE/TI = 2500/2.26/1100 ms, 1 mm isotropic resolution), 3D FLAIR (TR/TE/TI = 5000/393/1800 ms, 1mm isotropic resolution), PD/T2 (2D FSE, TR/TE = 3630/9.6 and 96 ms, resolution = 0.7×0.7×3) acquired on a Siemens Prisma 3T for the MS cohort, and MPRAGE (TR/TE/TI = 6.9/3.2/900 ms, 1 mm isotropic resolution), FLAIR (TR/TE/TI = 4800/350/1650 ms, 1mm isotropic resolution), PD/T2 (2D TSE, TR/TE = 3418/15 and100 ms, 0.9×0.9×3 mm resolution) acquired on the Philips Achieva 3T (Philips Healthcare, Andover, MA) for the HIV cohort. All MR images were co-registered to the corresponding MPRAGE image and were skull-stripped, bias-field corrected, and normalized using AFNI tools (25) as described in (7). Whole brain segmentations were obtained using FreeSurfer (26) on the MPRAGE and FLAIR images, then converted into the following labels: cerebrospinal fluid (CSF), grey matter (GM), and white matter (WM). These labels were then manually edited for errors and lesions were manually drawn by either a trained neurologist (YM, SO) or a clinical neuroscientist (RM), each with more than 5 years of experience.
The multi-institutional MICCAI BraTS 2020 Training dataset (BraTS cohort, https://www.med.upenn.edu/cbica/brats2020/data.html) was used to check the applicability of our findings to a much larger dataset with widely accepted gold-standard masks (11). This dataset contains pre-processed multimodal input data and expert-drawn glioma region segmentations for 369 participants. The images were individually z-score normalized prior to modeling. For these experiments, the healthy tissue labels were omitted as it contained a mixture of signal intensity profiles from WM, GM, and CSF, and therefore was not relevant to the techniques being tested herein. The three remaining labels of the tumor regions were used for training and testing: enhancing tumor (ET), peritumoral edema (PE), and necrotic/non-enhancing core (NCR/NET).
2.2. Brain Segmentation Methods
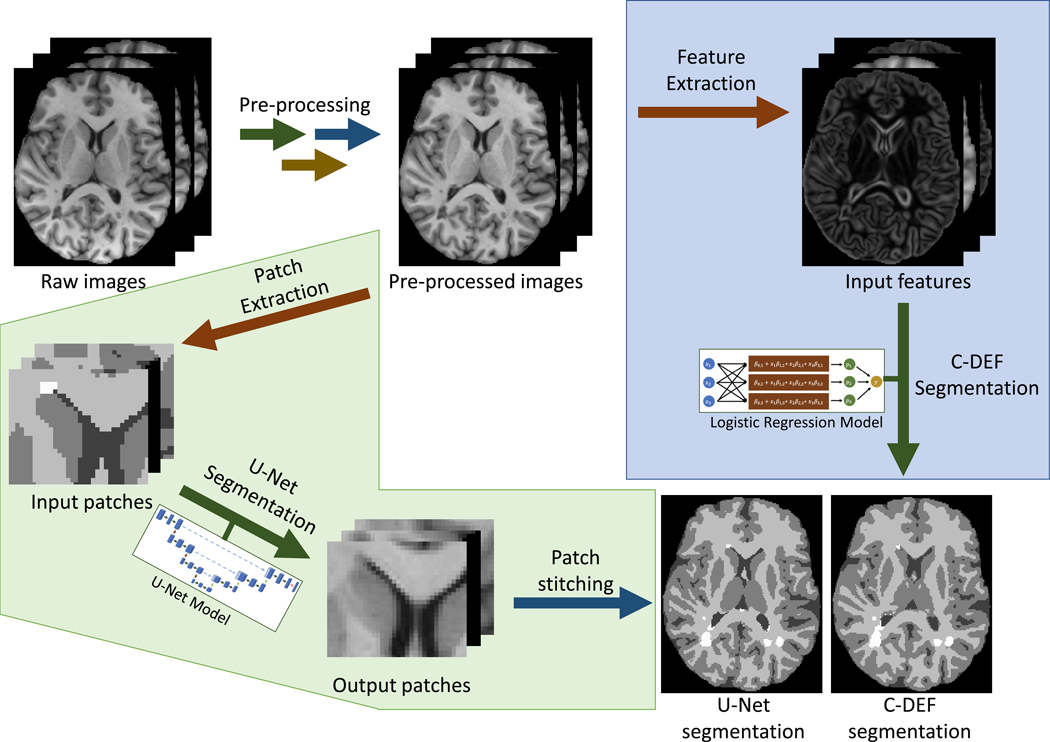
Figure 1 shows the inference pipeline for our parallel evaluation of C-DEF and U-Net. All models were trained from scratch on each cohort. In the C-DEF pathway (blue shading), image features were derived and used as inputs for a logistic regression classifier following the previously published method (7). Gaussian blur and Gaussian gradient filters with kernel sizes of 33, 53, 73, 93, 173, 333, and 653 were calculated for each contrast and concatenated with the unfiltered images (27). These features were z-score normalized based on the mean and standard deviation of the entire training dataset, then used to train a multinomial logistic regression classifier with a maximum of 200 iterations and L2 regularization (C=0.05) to prevent overfitting. For detailed methods, please see (7). C-DEF models were implemented on a computing cluster with 64x Intel® Xeon® CPUs (Intel Corporation, Santa Clara, CA) with 256 GB RAM running CentOS 7.7.
Figure 1: Analysis workflow:
Classification using DErivative-based Features (C-DEF, shaded in blue) and U-Net (shaded in green) algorithms for brain segmentation. Preprocessing consisted of co-registration, bias correction, and intensity normalization.
In the U-Net pathway (green shading), a 3D U-Net model was used to train and predict on 3D image patches, then the output patches were concatenated to produce the final segmentation. U-Net architecture was adapted from the original 3D U-Net method (28) with 32 filters in the first convolution layer but with the use of padded convolution layers and randomly sampled input patches for training (18). Key hyperparameters including patch size, batch size, learning rate, and number of epochs were tuned by 5-fold cross-validation grid search on the MS cohort (data not shown), and the configuration with the highest average DSC was chosen. Based on this, the Adam optimizer with categorical cross-entropy loss and a learning rate of 1 × 10−3 was trained for a maximum of 50 epochs with a batch size of 60, and early stopping was conditioned on validation loss to prevent overfitting. For each participant, 1000 random nonzero voxels were selected and patches of size 323 were extracted centered at each voxel. Data augmentation consisted of random patch-wise reflections, rotations, and elastic deformations during training. The U-Net model was implemented in Keras with Tensorflow backend on an NVIDIA® v100-SXM2 GPU (NVIDIA, Santa Clara, CA) with 32 GB VRAM and 8 Intel® Xeon® Gold 6410 CPUs (Intel Corporation, Santa Clara, CA) with 64 GB RAM.
2.3. C-DEF vs U-Net Comparison
The optimized C-DEF and U-Net methods were then applied to the full MS and HIV cohorts. Models were evaluated by 5-fold cross-validation using an 80/20 training/testing split for C-DEF and U-Net, with 25% of the training data used for validation in the latter case. For each cross-validation fold, the training (including validation, in the case of U-Net) dataset was then subsampled to include either 5, 10, or 15 participants for model fitting to simulate limited training data availability. This subsampling was randomly generated but kept consistent between corresponding C-DEF and U-Net runs (e.g., C-DEF-5 and U-Net-5) to provide a fair comparison. The same comparison was also applied to the BraTS cohort with additional models trained with data from 20, 40, 80, 160, and 295 (maximum available) participants as well. Mean computational cost statistics were calculated from three-fold repeated measurements.
2.4. Qualitative Evaluation
Output segmentations for the HIV and MS cohorts were first visually inspected, then qualitatively scored by an experienced neurologist (SO) or trained clinical neuroscientist (RM), respectively, while blinded to the method and model used to generate each segmentation. A 5-point rating scale was used ranging from 1 = ‘very bad’ to 5 =‘ very good’ for each tissue class. Net score for each segmentation was calculated as the mean of the ratings for all tissue classes, with lesion segmentations weighted double due to their importance as a potential marker for neurological disease progression.
2.5. Quantitative Evaluation and Statistical Analysis
Dice Similarity Coefficient (DSC) scores were calculated according to the following formula:
where TP, FP, and FN are the number of true positive, false positive, and false negative voxels, respectively, when segmentation results were compared to labels manually drawn by an expert neurologist. Statistical analyses were performed using SAS® version 9.4 (SAS Institute Inc, Cary, NC). Box-Cox data transformation was applied to the two data sets with minimum observations <0.1. The Shapiro–Wilk (sample size<50) or Kolmogorov–Smirnov test was used to check the normality assumption. For each tissue class, repeated-measures analysis of variance (RM-ANOVA) was conducted to evaluate the effect of the scoring method on mean DSC. Intraclass correlation coefficient (ICC) and 95% confidence intervals were calculated to examine the correlation and agreement of the different scoring methods (treated as raters). Bland-Altman agreement analysis was performed using segmentation tissue volumes of model pairs to examine potential biases between C-DEF and U-Net models trained on the same data. The Bonferroni-corrected p-value <0.05 was considered to be statistically significant.
2.6. Code and Data Availability
BraTS is a publicly available dataset. Code and data for C-DEF and prototypical U-Net models used herein will be made available via GitHub upon acceptance of manuscript. The MS and HIV data in this project are confidential and obtained via a natural history study, but may be obtained with Material Transfer Agreements, subject to NIH’s policy on data sharing.
3. Results
3.1. Participant Cohorts
The Institutional Review Board approved the study protocols and all participants provided written informed consent. Images acquired from participants diagnosed with relapsing-remitting MS (MS cohort, n = 20, 13 women, age 36 ± 8 years, time from diagnosis 0.2 ± 0.1 years, and Expanded Disability Status Scale (EDSS) 2.5 ± 1.8), and people living with HIV (HIV cohort, n = 18, 10 women, age 56 ± 4 years, time from diagnosis 20 ± 8 years) were used in this study (mean ± SD). Label masks were derived from FreeSurfer segmentations (26), which were converted to white matter, grey matter, and CSF labels. These were carefully edited by one of three neurologists and/or clinical neuroscientists (RM, YM, SO), who then manually added the lesion class. In addition, a collection of multimodal MRI scans of glioblastoma (n=293) and lower grade glioma (n=76) patients with manually drawn tumor segmentation masks were downloaded from the MICCAI BraTS 2020 online portal and modified to exclude normal brain regions (11, 29) (https://www.med.upenn.edu/cbica/brats2020/data.html).
3.2. Whole Brain Segmentation
Qualitative examination of segmentation results from C-DEF trained on data from 5, 10, and 15 MS cohort participants revealed no obviously visible improvement (Figure 2A, top row; mean ratings: 3.77, 3.79, and 3.79, respectively), whereas significant improvements (yellow arrows) were seen, especially for lesion segmentation, by increasing the training data supplied to U-Net (Figure 2A, middle row; mean ratings: 3.55, 3.90, and 3.93, respectively). U-Net-5 had low lesion sensitivity, especially for small punctate lesions (red arrows), and higher occurrence of false positives from artifactual hyperintensities (not shown). These results were mostly replicated in the HIV cohort (Figure 2B), with the exception that U-Net-15 (4.12) rated worse than U-Net-10 (4.58) due to errors in the brainstem.
Figure 2: Qualitative assessment of segmentation at various training data sizes:
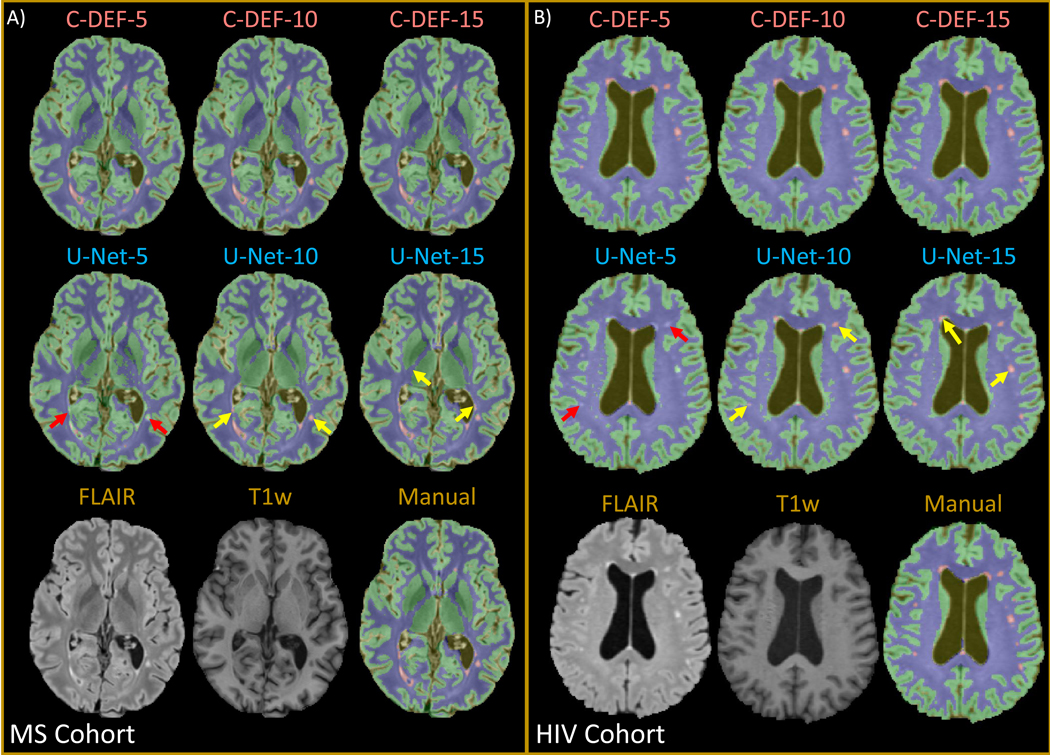
Output of segmentation performed with varied numbers of training data (5, 10, 15) using C-DEF and U-Net algorithms shown on a representative slice from a participant in (A) (Male, 34 years old) the MS cohort and (B) (Female, 56 years old) the HIV cohort. Bottom row shows pre-processed input scans and the manually drawn mask for reference. Red arrows indicate segmentation errors, while yellow arrows indicate areas of improved segmentation with more training data.
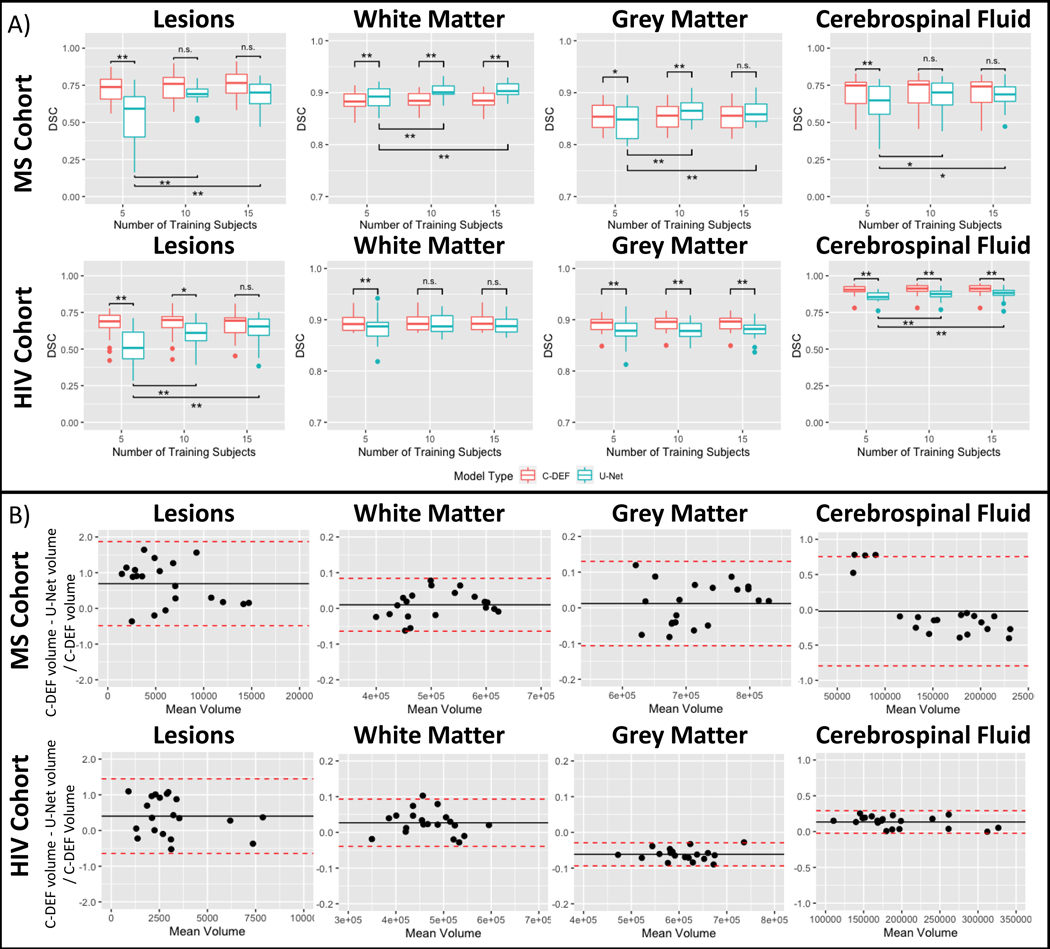
Quantitative results found that for both cohorts, DSC from C-DEF segmentations were unchanged with additional training data, but DSC from U-Net segmentation improved significantly with more training data (p<0.05, slice test applied to DSC from each tissue class, Figure 3A). In the MS cohort, DSC from the U-Net segmentation improved on average by 30% in the lesion class (p<0.001), 8% in the CSF class (p<0.01), 2% in the GM class (p<0.001), and 1% in WM class (p<0.001) when using training data from 15 participants compared to 5. U-Net segmentation in the HIV cohort showed similar DSC increases with a 24% improvement in the lesion class (p<0.001) and 2% improvement in the CSF class (p<0.01). It should be noted that the DSC from WM and GM segmentation was not significantly changed in the HIV cohort. Consistent with qualitative examination, quantitative results from U-Net-5 segmentation had worse DSC across all tissues than C-DEF-5 for both cohorts, except for WM in the MS cohort. Meanwhile, U-Net-15 segmentation had similar lesion, GM, and CSF DSC as C-DEF-15 segmentation in the MS cohort and similar lesion and WM DSC in the HIV cohort.
Figure 3: Comparison of U-Net and C-DEF segmentation:
(A) Dice Similarity Coefficient (DSC) of the output of the C-DEF and U-Net models with manually annotated mask, when trained with varied numbers of training data (5, 10, 15). Asterisks indicate statistically significant differences (* p < 0.05, ** p < 0.005). Note that DSC (y-axis) for white and grey matter is scaled 0.7–1.0 for better visualization. (B) Bland-Altman analysis of tissue volumes of C-DEF and U-Net trained on data from 5 participants from the MS (top) and HIV (bottom) cohort. Bias (black line) and 95% CI (red dotted line) are indicated.
ICC between C-DEF models trained with data from 5, 10, and 15 participants were >0.92 and >0.96 in all tissue classes in the MS cohort and the HIV cohort, respectively. Between U-Net models it varied between 0.72 (CI: [0.52, 0.87]) in the WM class to less than 0.1 (CI: [−0.15, 0.39]) in the lesion class in the MS cohort and between 0.60 (CI: [0.35, 0.81]) in the CSF class to less than 0.4 (CI: [0.07, 0.65]) in the lesion class in the HIV cohort. Furthermore, Bland-Altman analysis (Figure 3B) found that U-Net-5 consistently gave much smaller lesion volumes compared to C-DEF-5 segmentation in both cohorts (MS cohort mean bias: 0.69, CI: [−0.49, 1.87], HIV cohort mean bias: 0.40, CI: [−0.64, 1.45]). The same comparison with models trained on 15 data points (not shown) found a 67–70% reduction in lesion volume bias (MS: 0.20 [−0.35, 0.75], HIV: 0.13 [−0.47, 0.73]) compared to the 5 participant models. In the MS cohort, C-DEF-5 produced smaller CSF volumes compared to U-Net-5 for most participants, with a few exceptions. Visual inspection of the four U-Net segmentations that deviated from this trend (not shown) revealed abnormally low sulcal CSF sensitivity. For each tissue class, the MS cohort had a greater range of relative volume differences, which is consistent with qualitative observations of greater similarity between C-DEF and U-Net segmentations in the HIV cohort. In addition, Bland-Altman analysis of C-DEF compared to the manually edited masks (not shown) confirmed that MS cohort CSF volumes for C-DEF were consistently lower than those present in the manual labels regardless of amount of training data used.
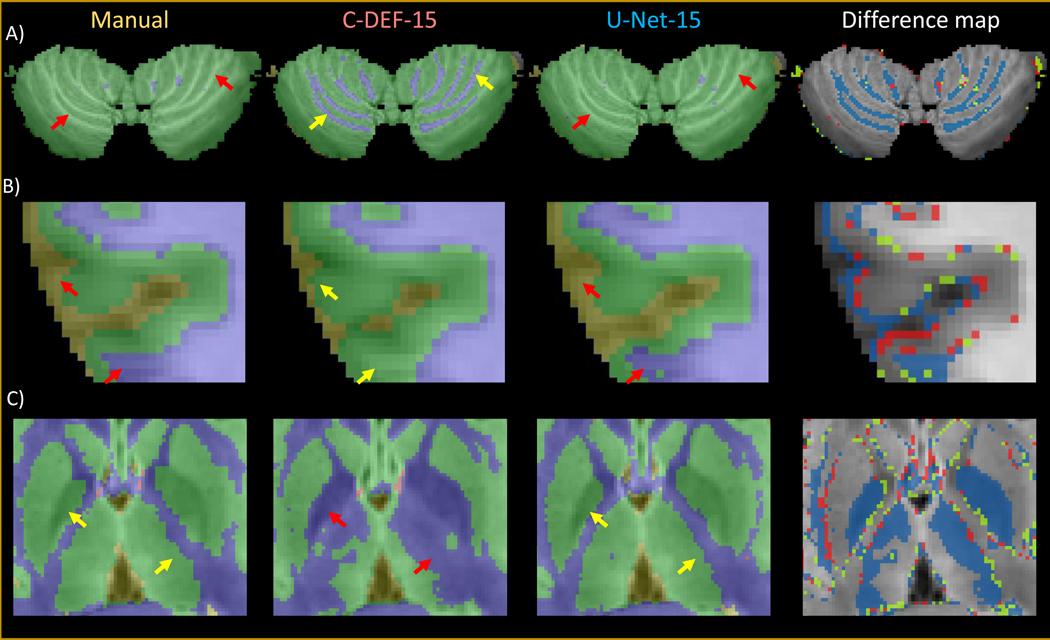
Difference map calculations of MS cohort segmentations revealed that C-DEF was better able to detect fine details of the cerebellar GM/WM folds, while U-Net typically failed to do so (Figure 4A). C-DEF also produced more plausible cortical grey matter boundaries than U-Net in many cases (Figure 4B). Qualitative inspection of the FreeSurfer-derived manually edited training labels found that these two areas were particularly prone to labeling noise and subtle errors, which were more readily replicated by U-Net than by C-DEF. Finally, C-DEF more often misclassified parts of subcortical grey matter structures, including the globus pallidus and thalamus, as white matter, while U-Net generally labeled them correctly (Figure 4C).
Figure 4: Effect of errors in training labels:
Zoomed-in details of segmentation outputs from a representative MS cohort participant (Male, 30 years old) using C-DEF-15, U-Net-15, and their difference maps compared to the manually annotated mask (blue: mismatched segmentation by C-DEF-15; red: mismatched segmentation by U-Net-15; green: mismatched segmentation by both C-DEF-15 and U-Net-15) for: (A) cerebellum, (B) cortical grey matter boundary, and (C) subcortical grey matter structures. Red arrows indicate segmentation errors, while yellow arrows indicate correctly labeled segmentation.
3.3. Tumor Segmentation of Glioma Dataset
In order to validate and extrapolate these findings, we downloaded a large publicly available dataset and modified it to simulate tissue segmentation using signal intensity profiles. Table 1 summarizes the performance of C-DEF and U-Net models on the modified BraTS cohort. C-DEF outperformed U-Net by mean DSC on the enhancing tumor (ET) and peritumoral edema (PE) classes when trained on data from 5 participants (p < 0.001, Dunnett-Hsu test), while the necrotic/non-enhancing tumor (NCR/NET) DSC was not significantly different between C-DEF-5 and U-Net-5. While no difference in mean DSC for any class was seen between C-DEF-5 and C-DEF-10, it did increase significantly for all classes between U-Net-5 and U-Net-10. When trained on much more data (>20 participants), C-DEF mean DSC did eventually increase significantly, with average improvement of 12% (ET), 7.6% (PE), and 190% (NCR/NET) from minimum to maximum amount of training data. Over the same range, U-Net scores increased 43% (ET), 62% (PE), and 200% (NCR/NET). C-DEF was better or statistically equivalent to U-Net by DSC for PE and ET tissue classes for every training regime tested, except for PE with 160 training data. However, U-Net did demonstrate a significant advantage on the NCR/NET tissue class for all except 5 and 15 training data models. While C-DEF models collectively produced ICCs of 0.81 [0.78, 0.83] and 0.89 [0.88, 0.91] for ET and PE DSC, indicating highly similar segmentation results, it had a far lower ICC of 0.62 [0.58, 0.66] for NCR/NET DSC, indicating only mild consistency in segmentation results for C-DEF models trained on different amounts of data.
Table 1:
Performance of C-DEF and U-Net models on BraTS cohort using limited training data
| Mean Dice Similarity Coefficient [95% CI] | |||
|---|---|---|---|
|
|
|||
| Model | Enhancing Tumor | Peritumoral Edema | Necrotic/Non-enhancing Tumor |
| C-DEF-5 | 0.74 [0.72, 0.76] | 0.81 [0.80, 0.83] | 0.15 [0.13, 0.18] |
| U-Net-5 | 0.58 [0.56, 0.60] | 0.55 [0.52, 0.57] | 0.17 [0.15, 0.20] |
| C-DEF-10 | 0.72 [0.70, 0.74] | 0.83 [0.81, 0.84] | 0.17 [0.15, 0.20] |
| U-Net-10 | 0.70 [0.68, 0.72] | 0.83 [0.82, 0.85] | 0.28 [0.25, 0.31] |
| C-DEF-15 | 0.76 [0.74, 0.77] | 0.85 [0.83, 0.86] | 0.24 [0.22, 0.27] |
| U-Net-15 | 0.73 [0.71, 0.75] | 0.82 [0.81, 0.84] | 0.27 [0.24, 0.30] |
| C-DEF-20 | 0.76 [0.74, 0.78] | 0.84 [0.82, 0.85] | 0.21 [0.19, 0.24] |
| U-Net-20 | 0.77 [0.75, 0.79] | 0.80 [0.79, 0.82] | 0.28 [0.25, 0.31] |
| C-DEF-40 | 0.80 [0.78, 0.81] | 0.85 [0.84, 0.87] | 0.28 [0.25, 0.31] |
| U-Net-40 | 0.78 [0.77, 0.80] | 0.85 [0.84, 0.87] | 0.34 [0.31, 0.38] |
| C-DEF-80 | 0.81 [0.80, 0.83] | 0.86 [0.85, 0.88] | 0.32 [0.29, 0.36] |
| U-Net-80 | 0.81 [0.80, 0.83] | 0.88 [0.86, 0.89] | 0.48 [0.44, 0.52] |
| C-DEF-160 | 0.83 [0.82, 0.85] | 0.87 [0.86, 0.89] | 0.39 [0.36, 0.43] |
| U-NET-160 | 0.82 [0.81, 0.84] | 0.89 [0.88, 0.91] | 0.51 [0.47, 0.55] |
| C-DEF-Alla | 0.83 [0.81, 0.84] | 0.88 [0.86, 0.89] | 0.43 [0.39, 0.46] |
| U-Net-Alla | 0.83 [0.81, 0.84] | 0.89 [0.88, 0.91] | 0.51 [0.47, 0.55] |
trained on data from 295 participants per fold
3.4. Computational Cost Comparison
Benchmark computational cost data for C-DEF and U-Net models were gathered during comparison on the MS cohort. When trained on the minimum 5 participants in the same CPU environment, U-Net (8400 ± 76 min) was more than two orders of magnitude slower than C-DEF (38.8 ± 0.7 min) in terms of overall training time per training participant. When deployed optimally on a v100 GPU, U-Net training time (33 ± 2 min) was comparable to C-DEF deployed on CPU, and inference was nearly 3 times faster (0.20 ± 0.01 min/participant compared to 0.55 ± 0.02 min/participant for C-DEF). It should be noted that C-DEF is currently only implemented on CPU. Both U-Net and C-DEF overall training time increased approximately linearly with the number of training participants in both the MS and HIV cohorts.
4. Discussion:
A logistic regression model using derived image textures (C-DEF) for brain segmentation performed equivalent or better by several key metrics than a prevalent deep learning algorithm (U-Net) when trained on manually edited masks from a small to moderate number of participants (n ≤ 15). In particular, C-DEF produced good lesion and CSF segmentations even when trained on the minimum 5 training participants, while U-Net did not. Performance of U-Net models for whole brain segmentation improved significantly with increasing amounts of training data, however, no significant improvements were seen in C-DEF models with increasing size of training data. U-Net produced better segmentation of certain structures, such as the thalamus and globus pallidus, with more subtle tissue intensity signatures. However, C-DEF was more robust to minor annotation errors in the training data, leading to better segmentation of certain structures, such as cerebellar folds and cortical grey matter boundaries. Moreover, deep learning algorithms such as U-Net require more compute resources than a classical machine learning method such as C-DEF to perform model training in a reasonable amount of time. Taken together, we have demonstrated that a classical machine learning algorithm such as C-DEF can produce equivalent or better whole brain segmentation based on signal intensity profiles than a much more resource intensive deep learning algorithm such as U-Net in the case of limited training data. Furthermore, accuracy of C-DEF segmentation is high even with limited (n=5) training data, and comparable to the accuracy of U-Net trained with larger (n=15) training data, making it a less labor intensive and more cost-effective option.
Minimizing the amount of manual label annotation required to obtain a reasonable model is an important goal often overlooked during model development. This step can have a large impact upon the time needed to implement a segmentation pipeline for a given application (30), as it is extremely tedious and requires expert knowledge of neuroanatomy and/or neuroradiology. This is particularly true for a task such as whole brain segmentation, as opposed to targeted quantification of a single structure, such as glioma segmentation. Given that the efficacy of any supervised learning model is highly dependent upon the quality of the training data, the threshold for acceptable annotations is generally quite high. It may therefore be advantageous to be able to obtain robust segmentations from only a few very carefully edited training data, rather than relying on a much larger pool of training data that may be more susceptible to large annotation errors due to its size. C-DEF achieves semantic segmentation by modeling individual voxels (along with their derived features), which means that a single annotated subject could be considered a source of millions of individual training data points. Meanwhile, U-Net dictates training on large image patches, which reduces the number of discrete training examples that can be obtained from each subject, even after data augmentation. Therefore, it is intuitive that C-DEF may be far less susceptible to overfitting and other penalties of insufficient training data.
The advantages of C-DEF were more pronounced when training was performed on a smaller number of training data. C-DEF produced reasonably good segmentations after training on data from only 5 participants, while U-Net segmentations were significantly degraded when trained with so few data points. The latter is evidenced not only by raw DSC scores (Figure 3A) but also by the subpar CSF volumes produced in a single fold of U-Net-5 (Figure 3B, top rightmost plot), which are sufficiently aberrant as to indicate model instability. C-DEF was also more effective in segmentation of certain structures, such as cerebellar folds and sulcal grey matter boundaries, particularly in the MS cohort (Figures 4A, 4B). These regions were observed to be susceptible to labelling errors, which may indicate that C-DEF effectively lowers the quality threshold for training annotations by avoiding overfitting, provided that texture-enriched tissue intensity signatures are reasonably well-resolved. Indeed, we refrained from using the term gold-standard masks for manually annotated masks due to the presence of such errors. This capability presents a trade-off, since C-DEF was largely ineffective for segmentation of certain regions where tissue intensity signatures were not as well-resolved, such as thalamus and globus pallidus. As a result, U-Net demonstrated slightly better overall WM and GM DSC for some training regimes, but only on the MS cohort. Our training annotations were derived from FreeSurfer subcortical segmentation, which uses an atlas of annotated examples as priors (26). This mitigates the issue of overlapping tissue intensity signatures by utilizing a combination of local and global factors and their correlations to determine the most likely label for a given voxel. U-Net was given no such priors, but it was able to identify subcortical grey matter structures more reliably than C-DEF using only patch-wise spatial features. This difference could be due to a number of factors, but is likely related to the fact that the number and complexity of features captured by U-Net far surpasses the small (<50) number of localized image textures incorporated into a single-layer logistic regression model by C-DEF. It may also indicate that deep grey matter structures are not linearly distributed and therefore not easily separable by the linear decision boundaries defined by C-DEF, as opposed to the more complex boundaries available to a deep network such as U-Net.
In order to further explore the dependence of each algorithm on training data size, the MICCAI BraTS 2020 Training dataset was used as a supplementary comparison. This dataset was chosen based on its large, expert-curated set of gold-standard labels and its widespread acceptance as a public benchmark in the medical image segmentation community. Unfortunately, whole brain segmentation labels are not provided for this dataset, so the “healthy tissue” label encompasses regions of white matter, grey matter, and CSF, leading to an indeterminate tissue intensity signature. In order to adapt the tumor segmentation task to be more comparable to whole brain segmentation, we chose to omit the “healthy tissue” label from training and quantify segmentation of the three tumor regions (enhancing tumor, necrotic/non-enhancing tumor, and peritumoral edema) directly rather than using the nested anatomical labels (enhancing tumor, tumor core, whole tumor) of the official BraTS challenge. When evaluated on the enhancing tumor (ET) and peritumoral edema (PE) regions, C-DEF was better or equivalent to U-Net regardless of the number of participants used for training (up to 295). However, we observed that both C-DEF and U-Net struggled with segmentation of necrotic/non-enhancing tumor regions (NCR/NET), with U-Net producing significantly better results in this region with moderate (n=10) and large (n ≥ 20) training data sizes. The ET and NCR/NET labels were both modest minority classes, representing 19.8% and 22.2% of the data compared to 58.0% belonging to the peritumoral edema class. The NCR/NET label also had significant overlap with the PE class in terms of tissue intensity signatures (data not shown), which may have been exacerbated by the significant variation in intensity profiles in this multi-institutional dataset. These factors may have been at least in part responsible for the larger relative increase in NCR/NET DSC with more training.
MR images, especially at high fields, are prone to bias fields from radiofrequency excitation and receive profiles, which can confound segmentation using only local voxel intensities (20, 31). In addition, spatial context plays a critical role in tissue segmentation (7, 20). To address these issues, derived image textures offer a viable alternative to atlas-based strategies. Exhaustive optimization by searching all potential texture sets remains an intractably large task, which is one limitation of the C-DEF approach. U-Net offers one solution to this problem by effectively optimizing its own filters, albeit at great computational expense. The effects of changing the number of U-Net filters as well as spatial factors such as patch size and overall architecture depth are complicated, and exhaustive exploration of this space is beyond the scope of this work. Prior work has found evidence that deeper networks are not always better, depending on the complexity of the modeled dataset (14). For the comparison herein, we tried to match the maximum receptive field in the U-Net architecture (which depends on the size of the filters and depth of the architecture) to the maximum filter kernel used in the C-DEF model.
The scope of this inquiry was limited to comparison of one representative deep learning method and one classical machine learning algorithm (C-DEF) as “off-the-shelf” tools for whole brain segmentation. As such, we chose not to perform exhaustive hyperparameter optimization for either method. It is likely that such optimization could provide performance gains for the chosen datasets, but this remains difficult to predict a priori. Moreover, it is yet unclear if or when U-Net may definitively exceed overall C-DEF performance given additional training data, or if a different network architecture is required. Studies in recent years have produced numerous derivations of U-Net intended to boost performance in a variety of segmentation tasks (32–34). However, these methods remain largely developed and validated on large datasets, making it difficult to evaluate whether proposed modifications will enhance performance in a limited training context. Moreover, it is important to first establish the baseline performance characteristics of the U-Net model used herein in this context prior to investigation of more complicated approaches derived from this framework. For these reasons, we chose a 3D patch-based U-Net as our representative deep learning method. In the future, we plan to expand this line of inquiry by benchmarking some of these recent modifications to probe the impact of model architecture changes on training data dependence. Of particular interest for future examination is the recently proposed nnU-Net, which has shown impressive results on a wide range of medical image segmentation tasks (8).
This study demonstrates that there is an important niche for classical machine learning methods such as C-DEF to fill by providing robust models trained on only a few labeled examples. Avenues for future study include improvement of C-DEF segmentation of subcortical grey matter and comparison of recent U-Net-style methods in a larger whole brain segmentation dataset.
Acknowledgements
This work was supported by the Intramural Research Program at the NINDS. Thanks to Avindra Nath and Bryan Smith for providing the allHANDS (HIV cohort) dataset. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). In addition, FutureMS was funded by a grant from the Chief Scientist Office, Scotland, to Precision Medicine Scotland Innovation Centre (PMS-IC) and by Biogen Idec Ltd Insurance (combined funding under reference Exemplar SMS-IC010) to the University of Edinburgh. Further funding for authors included a grant from MS Society Edinburgh Centre for MS Research (RM; grant reference 133).
Sources of support:
This work was supported by the Intramural Research Program at the NINDS. Some images used in this study were sourced from FutureMS study, funded by a grant from the Chief Scientist Office, Scotland, to Precision Medicine Scotland Innovation Centre (PMS-IC) and by Biogen Idec Ltd Insurance (combined funding under reference Exemplar SMS-IC010) to the University of Edinburgh. Further funding for some of the authors included a grant from MS Society Edinburgh Centre for MS Research (RM; grant reference 133).
References:
- 1.Marquez F, Yassa MA. Neuroimaging Biomarkers for Alzheimer’s Disease. Mol Neurodegener. 2019;14(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tur C, Moccia M, Barkhof F, et al. Assessing treatment outcomes in multiple sclerosis trials and in the clinical setting. Nat Rev Neurol. 2018;14(2):75–93. [DOI] [PubMed] [Google Scholar]
- 3.Cortese R, Collorone S, Ciccarelli O, Toosy AT. Advances in brain imaging in multiple sclerosis. Therapeutic advances in neurological disorders. 2019;12:1756286419859722-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jansen JFA, Vlooswijk MCG, Majoie HM, et al. White Matter Lesions in Patients With Localization-Related Epilepsy. Investigative Radiology. 2008;43(8):552–8. [DOI] [PubMed] [Google Scholar]
- 5.Sicotte NL, Voskuhl RR, Bouvier S, et al. Comparison of Multiple Sclerosis Lesions at 1.5 and 3.0 Tesla. Investigative Radiology. 2003;38(7):423–7. [DOI] [PubMed] [Google Scholar]
- 6.Hagiwara A, Fujita S, Ohno Y, Aoki S. Variability and Standardization of Quantitative Imaging: Monoparametric to Multiparametric Quantification, Radiomics, and Artificial Intelligence. Investigative Radiology. 2020;55(9):601–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selvaganesan K, Whitehead E, DeAlwis PM, et al. Robust, atlas-free, automatic segmentation of brain MRI in health and disease. Heliyon. 2019;5(2):e01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isensee F, Jaeger PF, Kohl SAA, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18(2):203–11. [DOI] [PubMed] [Google Scholar]
- 9.Ji Y, Zhang R, Li Z, et al. UXNet: Searching Multi-level Feature Aggregation for 3D Medical Image Segmentation. 2020. Cham. Springer International Publishing: 346–56. [Google Scholar]
- 10.Akkus Z, Galimzianova A, Hoogi A, et al. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J Digit Imaging. 2017;30(4):449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menze BH, Jakab A, Bauer S, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging. 2015;34(10):1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, 2015// 2015. Cham. Springer International Publishing: 234–41. [Google Scholar]
- 13.Ibtehaz N, Rahman MS. MultiResUNet : Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020;121:74–87. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans Med Imaging. 2020;39(6):1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlemper J, Oktay O, Schaap M, et al. Attention gated networks: Learning to leverage salient regions in medical images. Med Image Anal. 2019;53:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan Y. Automatic Brain Tumor Segmentation with Scale Attention Network. 2021. Cham. Springer International Publishing: 285–94. [Google Scholar]
- 17.Yuankai H, Zhoubing X, Yunxi X, et al. 3D whole brain segmentation using spatially localized atlas network tiles. NeuroImage. 2019;194:105–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao L, Feng X, Meyer CH, Alsop DC. Choroid Plexus Segmentation Using Optimized 3D U-Net. IEEE Int Symp Biomed Imaging, 3–7 April 2020 2020. 381–4. [Google Scholar]
- 19.Yan W, Huang L, Xia L, et al. MRI Manufacturer Shift and Adaptation: Increasing the Generalizability of Deep Learning Segmentation for MR Images Acquired with Different Scanners. Radiol Artif Intell. 2020;2(4):e190195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Despotović I, Goossens B, Philips W. MRI Segmentation of the Human Brain: Challenges, Methods, and Applications. Comput Math Methods Med. 2015;2015:450341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wichmann JL, Willemink MJ, De Cecco CN. Artificial Intelligence and Machine Learning in Radiology: Current State and Considerations for Routine Clinical Implementation. Investigative Radiology. 2020;55(9):619–27. [DOI] [PubMed] [Google Scholar]
- 22.Fang Y, Wang J, Ou X, et al. The impact of training sample size on deep learning-based organ auto-segmentation for head-and-neck patients. Physics in Medicine & Biology. 2021;66(18):185012. [DOI] [PubMed] [Google Scholar]
- 23.Lekadir K, Galimzianova A, Betriu A, et al. A Convolutional Neural Network for Automatic Characterization of Plaque Composition in Carotid Ultrasound. IEEE J Biomed Health Inform. 2017;21(1):48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Annals of neurology. 2011;69(2):292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162–73. [DOI] [PubMed] [Google Scholar]
- 26.Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–55. [DOI] [PubMed] [Google Scholar]
- 27.Dieckhaus HM R; Mina Y; Waldman AD; Parvathaneni P; Nair G. Expanding the Texture toolkit for Atlas-free Segmentation of Brain MRI. Organization for Human Brain Mapping Annual Meeting, 2021. Organization for Human Brain Mapping. [Google Scholar]
- 28.Çiçek Ö, Abdulkadir A, Lienkamp SS, et al. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. 2016. Cham. Springer International Publishing: 424–32. [Google Scholar]
- 29.Bakas S, Akbari H, Sotiras A, et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data. 2017;4:170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weikert T, Cyriac J, Yang S, et al. A Practical Guide to Artificial Intelligence–Based Image Analysis in Radiology. Investigative Radiology. 2020;55(1):1–7. [DOI] [PubMed] [Google Scholar]
- 31.Vrenken H, Jenkinson M, Horsfield MA, et al. Recommendations to improve imaging and analysis of brain lesion load and atrophy in longitudinal studies of multiple sclerosis. J Neurol. 2013;260(10):2458–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henry T, Carré A, Lerousseau M, et al. Brain Tumor Segmentation with Self-ensembled, Deeply-Supervised 3D U-Net Neural Networks: A BraTS 2020 Challenge Solution. 2021. Cham. Springer International Publishing: 327–39. [Google Scholar]
- 33.Rehman MU, Cho S, Kim J, Chong KT. BrainSeg-Net: Brain Tumor MR Image Segmentation via Enhanced Encoder-Decoder Network. Diagnostics (Basel). 2021;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-masni MA, Kim D-H. CMM-Net: Contextual multi-scale multi-level network for efficient biomedical image segmentation. Sci Rep. 2021;11(1):10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
BraTS is a publicly available dataset. Code and data for C-DEF and prototypical U-Net models used herein will be made available via GitHub upon acceptance of manuscript. The MS and HIV data in this project are confidential and obtained via a natural history study, but may be obtained with Material Transfer Agreements, subject to NIH’s policy on data sharing.