Abstract
Objective
To develop and validate a prognostic model for LEF discontinuation with abnormal blood test results.
Methods
Data from the Clinical Practice Research Datalink Gold and Aurum were used for model development and external validation, respectively. Participants prescribed LEF between 1 January 2007 and 31 December 2019 were followed up from 6 months after the first general practitioner prescription to the earliest of date of outcome, death, 5 year follow-up or 31 December 2019. Candidate prognostic factors were ascertained using theory and data-driven approaches. Penalized Cox regression was performed to develop the risk equation, followed by internal validation using 500 bootstraps to correct for optimism. Multiple imputation was applied to handle missing data. Model performance was assessed in terms of calibration and discrimination.
Results
Data for 1487 and 2329 participants contributing 3140 and 5246 person-years follow-up were included in the development and validation cohorts, respectively. Thirteen candidate predictors were included in the model. Epilepsy and either cytopenia or elevated liver enzymes during the first 6 months of shared-care LEF prescription were strong predictors of drug discontinuation with a hazard ratio of 4.39 (95% CI 1.74, 11.06) and 3.06 (2.15, 4.35), respectively. The unadjusted and optimism-adjusted calibration slope in development data was 1.00 (95% CI 0.75, 1.25) and 0.72 (95% CI 0.47, 0.97), respectively. The calibration slope in validation data was 0.91 (95% CI 0.74, 1.07). The model showed prognostic separation with an optimism-adjusted Royston D statistic of 0.73 (95% CI 0.44, 1.02).
Conclusion
We have developed and externally validated an easy-to-use prognostic model that may be used to risk stratify monitoring for LEF toxicity and to make informed choices about risks when choosing treatments.
Keywords: leflunomide, rheumatoid arthritis, psoriatic arthritis, drug toxicity, monitoring
Rheumatology key messages.
One in five patients established on long-term leflunomide discontinue treatment with abnormal monitoring blood tests.
This is the first prognostic model to discriminate patients at varying risk of leflunomide toxicity.
The developed tool may be used to risk stratify monitoring after successful stabilization on leflunomide.
Introduction
LEF is used in the treatment of inflammatory arthritis when low-dose weekly MTX is either contraindicated, ineffective or causes side effects [1]. Although head-to-head trials suggest comparable efficacy to MTX ≤15 mg/week, LEF is less well tolerated, with a higher risk of treatment discontinuation, mainly due to cytopenia and elevated liver enzymes [2–5]. For instance, up to 7.1% of patients commenced on LEF in clinical trials discontinued it by 12 months due to elevated liver enzymes [2, 4]. Real-world data indicate that 9.3% and 20.5% of patients initiated on long-term LEF discontinue treatment with abnormal blood test results by 1 and 5 years, respectively [5].
The risk factors for target organ damage from LEF are not well understood. In the absence of this information, those prescribed long-term LEF undergo monitoring blood tests every 3 months [6, 7]. This strategy of routine periodic testing may not be necessary for those at low risk. Additionally, better understanding of predictors for target organ damage will aid patients and rheumatologists when choosing DMARDs. Thus the aim of this study was to develop and externally validate a prognostic model for LEF discontinuation due to abnormal monitoring blood tests at 5 years.
Methods
Data source
Data from the Clinical Practice Research Datalink (CPRD) Gold and Aurum were used for model development and external validation, respectively [8, 9]. CPRD is an anonymized longitudinal database of electronic health records and its participants are representative of the UK population in terms of age, sex and ethnicity [8]. It includes information on demographic details, lifestyle factors (e.g. smoking, alcohol intake), diagnoses, results of investigations including blood tests and details of general practitioner (GP) prescriptions during clinical care.
CPRD Gold and Aurum complement each other in terms of nationwide coverage of general practice surgeries. The former uses Vision software while the latter uses EMIS. Some general practice surgeries have contributed data to both CPRD Gold and Aurum databases. Data from such surgeries were excluded from the validation cohort using a bridging file provided by the CPRD to allow for true independent external validation.
Ethical approval
Ethical approval was obtained from the Independent Scientific Advisory Committee of the Medicines and Healthcare Products Regulatory Agency (reference 19_275R).
Study design
This was a cohort study. The study period was 1 January 2007 to 31 December 2019. The study population comprised those who received a first shared-care LEF prescription from their GP in the study period.
In the UK, DMARDs are initiated in the hospital rheumatology clinic and prescriptions are initially issued by the rheumatologist until a stable, effective and well-tolerated dose is reached. During this period, the rheumatology team oversees monitoring blood tests. Once the patient is established on treatment, the responsibility for prescribing and monitoring is handed to the patient’s GP under a shared-care protocol endorsed by the BSR and the Royal College of General Practitioners [6]. The GP consults with the rheumatologist if there are abnormal blood test results or any side effects and changes in treatments are directed by the rheumatologist.
Inclusion and exclusion criteria
Participants with autoimmune rheumatic disease (AIRD, e.g. RA, axial SpA, etc.), age ≥18 years, with ≥12 months of follow-up in CPRD Gold (or Aurum for validation) prior to a first-ever prescription of LEF were eligible [5]. Exclusion criteria included chronic liver disease, haematological malignancy, myelodysplasia, haemolytic anaemia, neutropenia, idiopathic thrombocytopenic purpura and chronic kidney disease (CKD) stage ≥4, as detailed previously [5].
Outcome
The outcome was drug discontinuation with abnormal blood test results, defined as a prescription gap of ≥90 days, with abnormal blood test results (or diagnostic code indicating an abnormal blood test result) within ±60 days of the last prescription [5]. See the Supplementary Methods (available at Rheumatology online) for thresholds used to define abnormal blood test results.
Start of follow-up
Participants were followed-up from 180 days after the first LEF prescription issued by the GP until the earliest of date of outcome, death, transfer out of the practice, date of last data collection from the practice, 5 years or 31 December 2019.
Predictors
Predictors were ascertained using theory and data-driven approaches.
Theory driven
Clinical members of the team comprising a hepatologist, nephrologist, haematologist, rheumatologist, gastroenterologist and GP suggested potential predictors. These were supplemented with drugs that increase the risk of LEF toxicity according to the British National Formulary (BNF).
Demographic or lifestyle factors including age, sex, BMI and alcohol intake were included because they increase the risk of drug-induced liver injury (DILI), and smoking was included because it increases the clearance of LEF [10, 11].
Drugs that increase the risk of LEF toxicity as per the BNF, specifically statins, paracetamol, MTX, 5-aminosalicylates, carbamazepine and sodium valproate.
Comorbidities: diabetes was included as it increases the risk of DILI [10].
Cytopenia (neutrophil count <2 × 109/l, total leucocyte count <4 × 109/l or platelet count <150 × 109/l) or liver enzyme elevation (alanine aminotransferase/aspartate aminotransferase levels >35 IU/l) during the first 6 months of shared-care LEF prescription were included. This is because blood test abnormalities predict cytopenia and/or transaminitis due to other DMARDs [12, 13].
The latest record of demographic and lifestyle factors prior to the start of follow-up, diagnostic code for comorbidities in the 2 years prior to the start of follow-up and prescription and blood test results in the 6 months prior to the start of follow-up were used to define the prognostic factors. A longer look-back was used to capture data on comorbidities, as GPs usually review patients with chronic illnesses annually.
Data driven
All diagnoses for study participants within 2 years of the start of follow-up were extracted and classified into chronic disease categories. Hypothesis-free logistic regression adjusted for age and gender was undertaken to identify potential prognostic factors that associate with the outcome of interest. Potential risk factors associated with outcome with P < 0.10 and present in ≥1% of the derivation cohort were included in the prognostic model. Uncommon prognostic factors were excluded to avoid model imbalance.
Sample size
To minimize model overfitting and ensure precise estimation of overall risk, the minimum sample size required for new model development was 1398 participants (189 events) based on a maximum of 20 parameters, a Cox–Snell R2 value of 0.12, estimated event rate of 0.057/person-year, a 5 year time horizon and a mean follow-up period of 2.36 years using the findings from our earlier work [5] (see Supplementary Methods, available at Rheumatology online, for Stata syntax).
Statistical analysis
Mean (s.d.) and n (%) were used for descriptive purposes. We applied multiple imputation to handle missing values using chained equations. We carried out 10 imputations in the development dataset, as there tends to be no additional benefit from using >5–10 imputations [14]. We used five imputations for the validation data—a pragmatic approach considering the large size of CPRD Aurum. The imputation model included all candidate predictors, the Nelson–Aalen cumulative hazard function and outcome variables.
Model development
All candidate predictors were included in the Cox model and coefficients of each predictor were estimated and combined using Rubin’s rule across the imputed datasets. We formed the risk equation for predicting an individual’s risk of LEF discontinuation due to abnormal blood test results at 5 years of follow-up using the developed model’s baseline survival function at = 5 years, a non-parametric estimate of the survival function when all predictor values are set to zero, which is equivalent to the Kaplan–Meier product-limit estimate, along with the estimated regression coefficients (β) and the individual’s predictor values (X). This process ultimately led to an equation for the predicted absolute risk over time t [15]:
Predicted event risk at 5 years = 1 − S0(t = 5)exp(βX), where S0(t = 5) is the baseline survival function at 5 years of follow-up and βX is the linear predictor, β1x1 + β2x2 + … + βpxp. Regression coefficients (β) are estimated from the developed model.
Model validation
We assessed the performance of the model in terms of calibration (where 1.00 is the ideal) by plotting agreement between predicted and observed events. We performed internal validation to correct calibration for optimism (overfitting) by bootstrapping with 500 replacement samples of the development data in each imputed dataset. We fitted the full model in each bootstrap sample to quantify the performance in the bootstrap sample (apparent performance) and applied the same model to the original sample to test model performance and optimism (difference in test performance and apparent performance). The uniform shrinkage factor was then estimated as the average of calibration slopes from each of the bootstrap samples. This process was repeated in each imputed dataset and the final uniform shrinkage was calculated by averaging across the estimated shrinkage estimates from all imputations. To account for overfitting during the model development process, the original β coefficients were penalized by the final uniform shrinkage factor and the baseline hazard was re-estimated on the basis of the shrunken β coefficients to ensure that overall calibration was maintained, producing a final model. We calculated the D statistic, a measure of discrimination, interpreted as a log hazard ratio (HR), the exponential of which gives the HR comparing two groups defined by above/below the median of the linear predictor, and plotted Kaplan–Meier curves in risk groups to visually assess separation. The cut-points are the 16th, 50th and 84th centiles of the linear predictor (mean ± 1 s.d.) as determined by Cox’s method [16, 17].
External validation of the model
Independent external validation of the final model was performed using data from CPRD Aurum within the same start and end of follow-up periods. General practice surgeries that also contributed data to CRPD Gold were excluded from the validation cohort. The final developed model equation was applied to each individual in the validation dataset and then we examined calibration and discrimination as described above. In addition, we examined calibration at 5 years by plotting agreement between predicted risk and observed event rate.
We used Stata/MP version 16 (StataCorp, College Station, TX, USA) for all statistical analyses. This study was reported in line with the Transparent Reporting of a multivariate prediction model for Individual Prognosis or Diagnosis (TRIPOD) guidelines [18].
Results
Study participants
Data for 1487 and 2329 participants contributing 3140 and 5246 person-years follow-up were included in the development and validation cohorts, respectively (Table 1; Supplementary Figs S2 and S3, available at Rheumatology online). The majority of participants in both cohorts had RA, were female and the cohorts had similar prevalence of lifestyle factors, comorbidities and drug treatments.
Table 1.
Baseline characteristics of the study population
| Variables | Development cohort (CPRD Gold) (n = 1487) | Validation cohort (CPRD Aurum) (n = 2329) |
|---|---|---|
| Age, mean (s.d.), years | 57 (13) | 57 (13) |
| Sex (female), n (%) | 979 (65.8) | 1580 (67.8) |
| BMI (kg/m2), n (%) | ||
| <18.5 | 28 (1.9) | 28(1.2) |
| 18.5–24.9 | 426 (28.7) | 651 (28.0) |
| 25.0–29.9 | 470 (31.6) | 728 (31.3) |
| ≥30 | 495 (33.3) | 821(35.3) |
| Missing | 68 (4.6) | 101(4.3) |
| Current smoker, n (%) | ||
| No | 1168 (78.6) | 1878 (80.6) |
| Yes | 319 (21.5) | 451 (19.4) |
| Alcohol use (units/week), n (%) | ||
| Non-user | 329 (22.1) | 519 (22.3) |
| Low (1–14) | 805 (54.1) | 931 (40.0) |
| Moderate (15–21) | 43 (2.9) | 109 (4.7) |
| Hazardous (>21) | 76 (5.1) | 112 (4.8) |
| Ex-user | 88 (5.9) | 354 (15.2) |
| Missing | 146 (9.8) | 304 (13.1) |
| Autoimmune rheumatic disease, n (%) | ||
| RA | 970 (65.2) | 1518 (65.2) |
| PMR/GCA | 91 (6.1) | 201 (8.6) |
| SpA | 426 (28.7) | 610 (26.2) |
| Comorbidities, n (%) | ||
| Epilepsy or prescribed carbamazepine or valproate | 19 (1.3) | 26 (1.1) |
| Diabetes | 149 (10.2) | 278 (11.9) |
| CKD | 74 (5.0) | 57 (2.5) |
| Other DMARDs, n (%) | ||
| MTX or 5-aminosalicylates | 467 (31.4) | 758 (32.6) |
| Other drugs, n (%) | ||
| Statins | 341 (22.9) | 531 (22.8) |
| Paracetamol | 287 (19.3) | 464 (19.92) |
| Blood test abnormalities, n (%) | ||
| At-least mild cytopenia or liver enzyme elevation in 6 months preceding the start of follow-up | 325 (21.9) | 514 (22.1) |
On data-driven analyses in the derivation cohort, epilepsy, CKD and nutritional intolerances were associated with the outcome of interest with P < 0.10 (Supplementary Table S1, available at Rheumatology online). As nutritional intolerances were only present in 0.15% of the derivation cohort, it was not taken forward as a candidate predictor. A diagnosis of epilepsy and prescription of sodium valproate or carbamazepine was merged together to create a single candidate predictor (epilepsy) to avoid multicollinearity. We used fraction polynomials to model non-linear risk relationships with continuous predictors (BMI and age), but these were found to be no better than the linear terms, hence BMI and age were not transformed (data not shown). Thirteen candidate predictors (17 predictor parameters) were selected to be included in the model (Table 2).
Table 2.
Final model HRs and β-coefficients
| Predictors | Adjusted HR (95% CI) | Coefficient |
|---|---|---|
| Age | 1.01 (0.99, 1.03) | 0.0094981 |
| Female sex | 1.24 (0.83, 1.83) | 0.2128283 |
| BMI (kg/m2) | 0.98 (0.95, 1.01) | −0.0171081 |
| Smoking status | ||
| Non-smoker/not recorded/ex-smoker | Reference | – |
| Current smoker | 0.90 (0.57, 1.42) | −0.1056694 |
| Alcohol consumption (units/week) | ||
| Non-drinker | Reference | – |
| Low (1–14) | 0.96 (0.63, 1.46) | −0.0400223 |
| Moderate (15–21) | 0.86 (0.26, 2.86) | −0.1474903 |
| Hazardous (>21) | 1.12 (0.47, 2.69) | 0.1171966 |
| Ex-drinker | 0.84 (0.37, 1.87) | −0.1774794 |
| AIRD type | ||
| RA | Reference | – |
| PMR or GCA | 1.03 (0.46, 2.30) | 0.026971 |
| SpA | 1.14 (0.76, 1.70) | 0.1266522 |
| Comorbidities | ||
| Epilepsya | 4.39 (1.74, 11.06) | 1.479007 |
| Diabetes | 0.88 (0.48, 1.60) | −0.1311263 |
| CKD | 1.72 (0.96, 3.06) | 0.5400153 |
| Other DMARDs | ||
| MTX or 5-aminosalicylates | 0.93 (0.64, 1.35) | −0.0733462 |
| Other drugs | ||
| Statins | 1.44 (0.94, 2.22) | 0.3666838 |
| Paracetamol | 1.45 (0.98, 2.16) | 0.3747208 |
| Blood test abnormalities | ||
| At-least mild cytopenia or liver enzyme elevation in the 6 months preceding the start of follow-up | 3.06 (2.15, 4.35) | 1.117226 |
Includes participants prescribed carbamazepine or valproate without a Read code for epilepsy.
Model development and identification of candidate predictors
In the development dataset, 136 outcome events occurred during the follow-up period at a rate of 43.32/1000 person-years (95% CI 36.62, 51.25). Epilepsy and presence of cytopenia or elevated liver enzymes during the first 6 months of shared-care LEF prescription were strong predictors of LEF discontinuation with an adjusted HR of 4.39 (95% CI 1.74, 11.06) and 3.06 (95% CI 2.15, 4.35), respectively (Table 2).
Apparent and internal validation performance statistics
As expected, the calibration slope in the development data was 1.00 (95% CI 0.75, 1.25). From the bootstrap, a uniform shrinkage factor of 0.73 was obtained and used to shrink predictor coefficients in the final model for optimism (Table 3); after re-estimation, the final model’s S0(5) was 0.914.
Table 3.
Model diagnosticsa
| Measure | Apparent performance (95% CI)b | Test performance (95% CI)c | Average optimism (95% CI)d | Optimism corrected performance (95% CI)e | External validation (CPRD Aurum) (95% CI) |
|---|---|---|---|---|---|
| Overall calibration slope | 1.00 (0.75, 1.25) | 0.72 (0.50–0.94) | 0.28 | 0.72 (0.47–0.97) | 0.91 (0.74–1.07) |
| Royston D statistic | 1.06 (0.77, 1.35) | 0.90 (0.63–1.17) | 0.33 | 0.73 (0.44–1.02) | 0.97 (0.89–1.05) |
| R 2 | 0.21 (0.12, 0.30) | 0.16 (0.08–0.24) | 0.10 | 0.11 (0.02–0.20) | 0.18 (0.16–0.21) |
Results from a single imputed dataset but similar across the other imputations (data not shown).
Refers to performance (95% CI) estimated directly from the data that was used to develop the model.
Determined by executing the full model in each bootstrap sample (500 samples with replacement), calculating bootstrap performance and applying same model in the original sample.
Average difference between model performance in bootstrap data and test performance in the original dataset.
Subtracting average optimism from apparent performance.
The Royston D statistic was 1.06 (95% CI 0.77, 1.35), corresponding to a HR of 2.89 (95% CI 2.16, 3.86) comparing the risk group above the median of the linear predictor to that below the median. The optimism-adjusted Royston D statistic was 0.73 (95% CI 0.44, 1.02), corresponding to a HR of 2.08 (95% CI 1.55, 2.77).
External validation
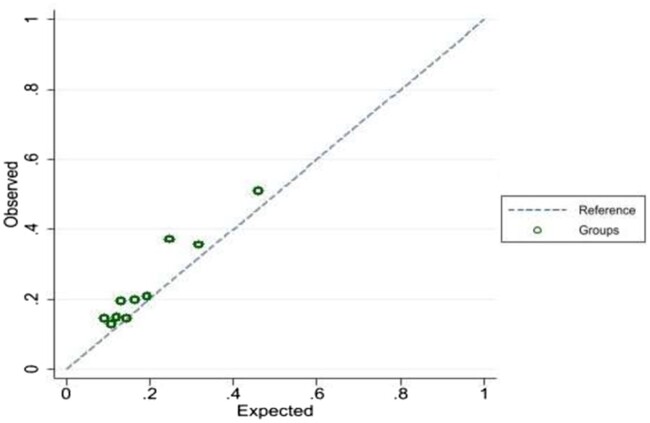
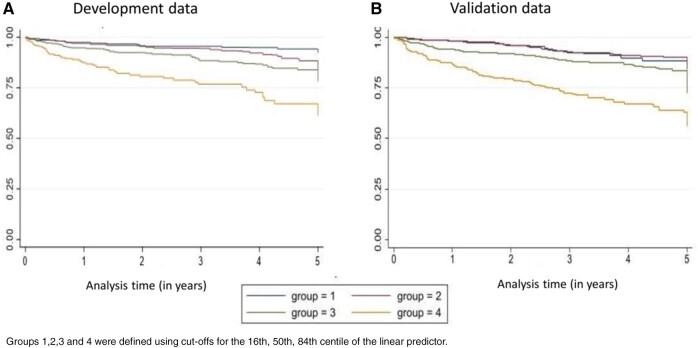
In the CPRD Aurum cohort there were 260 outcome events at a rate of 49.94/1000 person-years (95% CI 44.25, 56.37). Application of our final prognostic model to the independent population from CPRD Aurum yielded excellent calibration, with a calibration slope of 0.91 (95% CI 0.74, 1.07) (Fig. 1). The Royston D statistic in the validation data was 0.97 (95% CI 0.89, 1.05), corresponding to a HR of 2.64 (95% CI 2.44, 2.86), which suggests that our prediction model provided similar prognostic separation to that of the development dataset. Model discrimination in the derivation and validation data was broadly similar, but the model seemed less able to distinguish between the lowest two risk groups, particularly in the validation data (Fig. 2). The observed (and predicted) 5 year survival probabilities in validation data in these four risk groups were similar: 0.87 (0.90), 0.84 (0.87), 0.73 (0.79) and 0.56 (0.59), respectively.
Fig. 1.
Calibration plot in the validation dataset. C-slope 0.91 (95% CI 0.74–1.07)
Fig. 2.
Kaplan–Meier survival estimates in the model development and validation datasets
Groups 1,2,3 and 4 were defined using cut-offs for the 16th, 50th, 84th centile of the linear predictor.
Worked examples
A prognostic score to predict the absolute risk of LEF discontinuation after 6 months of primary care prescription and within the next 5 years may be calculated using the risk equation (Fig. 3, Supplementary Fig. S1, available at Rheumatology online). Participants with 16th centile and median linear predictor scores had 10.8% and 15.7% absolute risk of outcome event, respectively, over the 5 year follow-up period in the development datasets. The corresponding values were 10.9% and 15.3% in the validation dataset.
Fig. 3.
Equation to predict the risk of LEF discontinuation after 6 months of primary care prescription and within the next 5 years
Discussion
This is the first study to develop and validate a prognostic model that predicts LEF discontinuation due to target organ damage. It includes routinely collected data and provides a readily applicable means of risk stratification. It has excellent calibration and good discrimination between higher- and lower-risk groups. It focused on patients successfully initiated on LEF and treated for >6 months, as this includes the majority of the burden of monitoring. Current guidelines recommend blood test monitoring every 3 months during long-term LEF treatment and more frequent monitoring in the context of polypharmacy or comorbidities [6, 7]. However, with the exception of concurrent MTX prescription, these factors are poorly understood [19]. Utilizing the results from this study, patients at high risk of LEF toxicity may be offered more careful monitoring or alternate treatments, while those at very low risk may undergo less frequent monitoring (e.g. 6 month testing). Additionally, this study reports that cytopenia and elevated liver enzymes, including those not sufficiently severe to withdraw treatment within the first 6 months of shared-care GP prescription, strongly predict target organ drug toxicity. This is a novel finding for LEF and is consistent with previous observations regarding MTX [12, 13]. Similarly, epilepsy and/or treatment with carbamazepine and sodium valproate were strong predictors of target organ drug toxicity. These data may help inform drug choices in these patients. Statins and paracetamol were also strong prognostic factors, while other DMARDs such as MTX and 5-aminosalicylates were weak prognostic factors.
We did not observe a statistically significant association between demographic and lifestyle factors, including alcohol excess, and AIRD type and outcomes of interest. There is weak evidence that alcohol consumption may be a risk factor for DILI due to specific drugs such as MTX, but not with other drugs [20]. Alcohol use in the preceding 12 months was a negative predictor of severe DILI in general [odds ratio 0.33 (95% CI 0.15, 0.76)] in a previous study [20]. These findings should be interpreted with caution as our study was not powered to detect these associations.
Overall, the prognostic model performed well in the external validation dataset with excellent calibration. It had low discriminant ability for those at very low and low predicted risk. This is unsurprising, as the absolute difference in risk over a 5 year horizon between these two groups was only 5%. Reassuringly, our model discriminated between low- and high-risk subsets, which it could be argued, is important for clinical application. In the future, discrimination may be improved by including variants associated with LEF transaminitis (e.g. C163A in the CYP1A2 gene and rs4244285 and rs12248560 in the CYP2C19 gene); reduced LEF metabolism (e.g. rs3213422 in DHODH gene) and excretion (rs2231137 in the ABCG2 gene, also linked with gout) [21–26].
Not all prognostic models change practice. To facilitate this, evidence from this study will be disseminated to the BSR DMARD monitoring guideline writing group and the monitoring strategy will be changed if the BSR recommendations are modified in light of the findings. The risk calculators will be available online and included in the in-practice software used by GPs.
Strengths of this study include adequate power, use of time-to-event methods, external validation in an independent dataset and the inclusion of prognostic factors that are simple to obtain during routine care and at no additional cost. We followed TRIPOD guidelines and used robust statistical methodology to develop and evaluate the prognostic model. The study included internal correction for optimism and missing data was estimated by multiple imputations. The generalizability of the model was enhanced by the use of a database with nationwide coverage. We used an exhaustive list of potential predictors using data-driven and theory-driven approaches.
However, there are several limitations of this study. First, dose reduction due to abnormal blood test results was not used to define the outcome, as 30% of data on LEF dose were missing in the CPRD, making it difficult to ascertain dose reductions [5]. Some outcomes may have been related to toxicity to other drugs. These two factors may have reduced our model’s performance due to misclassification bias. Second, it is possible that some outcome events may actually be due to a combination of lack of efficacy of LEF and a concurrent illness resulting in blood test abnormalities. However, our validation exercise revealed that 95% of outcome events were not explained by a concurrent illness [5]. Patients prescribed LEF from a rheumatology clinic were excluded from the study. However, this is unlikely to affect the generalizability of our findings, as the vast majority of long-term prescriptions in the UK are issued from primary care under a shared-care prescribing and monitoring agreement. Our development dataset had a high shrinkage factor, indicating a degree of overfitting.
In conclusion, we have developed and validated a risk prediction equation to quantify the absolute risk of LEF discontinuation due to abnormal monitoring blood test results over 5 years. We ascertained several strong risk factors that may be useful when choosing between DMARDs. Further research is warranted to validate the model in other populations and to evaluate the clinical outcomes using this model.
Funding: This work was funded by the National Institute for Health Research (NIHR) under its Research for Patient Benefit Programme (grant reference no. PB-PG-1217-20030).
Disclosure statement: This article presents independent research funded by the NIHR under its Research for Patient Benefit Programme (grant reference no. PB-PG-1217-20030). The views expressed are those of the authors and not necessarily those of the National Health Service, NIHR or the Department of Health and Social Care. C.D.M. is funded by the NIHR Applied Research Collaboration West Midlands, the NIHR School for Primary Care Research and an NIHR Research Professorship in General Practice (NIHR-RP-2014-04-026) for this research project. The study sponsor did not have any role in the conduct or reporting of this study. A.A. has received departmental research grants from AstraZeneca and Oxford Immunotec, speaker bureau fees from Menarini, scientific meeting support from Pfizer, consulting fees from Inflazome and author royalties from UpToDate and Springer, unrelated to this work. M.D. has received honoraria for attending ad hoc advisory boards on gout and osteoarthritis for Grunenthal, Mallinckrodt and Pfizer; author royalties from UpToDate; and was an investigator in an AstraZeneca-funded, investigator-led, non-drug study (the ‘Sons of Gout’ study), unrelated to this work. W.Z. has received honoraria from Regeneron and Eli Lilly for advice on treatment of OA. G.P.A. reports consulting fees from AstraZeneca, Amryt Pharma, Fractyl, Median Technologies and BerGenBio; advisory fees from Kandy Therapeutics, GlaxoSmithKline, Owlstone and Inventiva Pharma; research grant support from Preglem and Pfizer and meeting support from Roche Diagnostics unrelated to this work. The other authors have no conflicts of interest to declare. The study question was discussed at a PPI meeting in Nottingham and received support from all present. Study results were reported to the PPI group and modes of dissemination of study findings were also discussed and agreed to.
Data availability statement
This study used data from the CPRD. Due to the CPRD data-sharing policy, we are unable to share this study’s data. However, access to CPRD data can be directly requested from the CPRD.
Supplementary data
Supplementary data are available at Rheumatology online.
Supplementary Material
Contributor Information
Georgina Nakafero, Academic Rheumatology.
Matthew J Grainge, Population and Lifespan Sciences.
Tim Card, Population and Lifespan Sciences; Nottingham Digestive Diseases Centre, School of Medicine.
Maarten W Taal, Centre for Kidney Research and Innovation, University on Nottingham.
Guruprasad P Aithal, Nottingham Digestive Diseases Centre, School of Medicine; NIHR Nottingham BRC, Nottingham University Hospitals NHS Trust and the University of Nottingham.
Weiya Zhang, Academic Rheumatology.
Michael Doherty, Academic Rheumatology.
Christopher P Fox, Department of Haematology, Nottingham University Hospital NHS Trust, Nottingham.
Christian D Mallen, Primary Care Centre Versus Arthritis, School of Medicine, Keele University, Keele, UK.
Abhishek Abhishek, Academic Rheumatology; NIHR Nottingham BRC, Nottingham University Hospitals NHS Trust and the University of Nottingham.
References
- 1. Smolen JS, Landewé RBM, Bijlsma JWJ. et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis 2020;79:685–99. [DOI] [PubMed] [Google Scholar]
- 2. Strand V, Cohen S, Schiff M. et al. Treatment of active rheumatoid arthritis with leflunomide compared with placebo and methotrexate. Leflunomide Rheumatoid Arthritis Investigators Group. Arch Intern Med 1999;159:2542–50. [DOI] [PubMed] [Google Scholar]
- 3. Cohen S, Cannon GW, Schiff M. et al. Two-year, blinded, randomized, controlled trial of treatment of active rheumatoid arthritis with leflunomide compared with methotrexate. Utilization of Leflunomide in the Treatment of Rheumatoid Arthritis Trial Investigator Group. Arthritis Rheum 2001;44:1984–92. [DOI] [PubMed] [Google Scholar]
- 4. Emery P, Breedveld FC, Lemmel EM. et al. A comparison of the efficacy and safety of leflunomide and methotrexate for the treatment of rheumatoid arthritis. Rheumatology (Oxford) 2000;39:655–65. [DOI] [PubMed] [Google Scholar]
- 5. Nakafero G, Grainge MJ, Card T. et al. What is the incidence of methotrexate or leflunomide discontinuation related to cytopenia, liver enzyme elevation or kidney function decline? Rheumatology (Oxford) 2021;60:5785--94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ledingham J, Gullick N, Irving K. et al. BSR and BHPR guideline for the prescription and monitoring of non-biologic disease-modifying anti-rheumatic drugs. Rheumatology (Oxford) 2017;56:2257. [DOI] [PubMed] [Google Scholar]
- 7. Singh JA, Saag KG, Bridges SL Jr. et al. 2015 American College of Rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Rheumatol 2016;68:1–26. [DOI] [PubMed] [Google Scholar]
- 8. Herrett E, Gallagher AM, Bhaskaran K. et al. Data resource profile: Clinical Practice Research Datalink (CPRD). Int J Epidemiol 2015;44:827–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wolf A, Dedman D, Campbell J. et al. Data resource profile: Clinical Practice Research Datalink (CPRD) Aurum. Int J Epidemiol 2019;48:1740–1740g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chalasani N, Björnsson E.. Risk factors for idiosyncratic drug-induced liver injury. Gastroenterology 2010;138:2246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prakash A, Jarvis B.. Leflunomide: a review of its use in active rheumatoid arthritis. Drugs 1999;58:1137–64. [DOI] [PubMed] [Google Scholar]
- 12. Meijer B, Wilhelm AJ, Mulder CJJ. et al. Pharmacology of thiopurine therapy in inflammatory bowel disease and complete blood cell count outcomes: a 5-year database study. Ther Drug Monit 2017;39:399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dirven L, Klarenbeek NB, van den Broek M. et al. Risk of alanine transferase (ALT) elevation in patients with rheumatoid arthritis treated with methotrexate in a DAS-steered strategy. Clin Rheumatol 2013;32:585–90. [DOI] [PubMed] [Google Scholar]
- 14. Schafer JL. Multiple imputation: a primer. Stat Methods Med Res 1999;8:3–15. [DOI] [PubMed] [Google Scholar]
- 15. Steyerberg EW. Clinical prediction models: a practical approach to development, validation, and updating. Cham, Switzerland: Springer, 2019. [Google Scholar]
- 16. Royston P, Altman DG.. External validation of a Cox prognostic model: principles and methods. BMC Med Res Methodol 2013;13:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cox DR. Note on grouping. J Am Stat Assoc 1957;52:543–7. [Google Scholar]
- 18. Moons KG, Altman DG, Reitsma JB. et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015; 162:W1–73. [DOI] [PubMed] [Google Scholar]
- 19. Curtis JR, Beukelman T, Onofrei A. et al. Elevated liver enzyme tests among patients with rheumatoid arthritis or psoriatic arthritis treated with methotrexate and/or leflunomide. Ann Rheum Dis 2010;69:43–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.European Association for the Study of the Liver. EASL Clinical Practice Guidelines: drug-induced liver injury. J Hepatol 2019;70:1222–61. [DOI] [PubMed] [Google Scholar]
- 21. Grabar PB, Rozman B, Logar D, Praprotnik S, Dolzan V.. Dihydroorotate dehydrogenase polymorphism influences the toxicity of leflunomide treatment in patients with rheumatoid arthritis. Ann Rheum Dis 2009;68:1367–8. [DOI] [PubMed] [Google Scholar]
- 22. Sandoval-Plata G, Morgan K, Abhishek A.. Variants in urate transporters, ADH1B, GCKR and MEPE genes associate with transition from asymptomatic hyperuricaemia to gout: results of the first gout versus asymptomatic hyperuricaemia GWAS in Caucasians using data from the UK Biobank. Ann Rheum Dis 2021;80:1220–6. [DOI] [PubMed] [Google Scholar]
- 23. Bohanec Grabar P, Rozman B, Tomsic M. et al. Genetic polymorphism of CYP1A2 and the toxicity of leflunomide treatment in rheumatoid arthritis patients. Eur J Clin Pharmacol 2008;64:871–6. [DOI] [PubMed] [Google Scholar]
- 24. Hopkins AM, Wiese MD, Proudman SM. et al. Genetic polymorphism of CYP1A2 but not total or free teriflunomide concentrations is associated with leflunomide cessation in rheumatoid arthritis. Br J Clin Pharmacol 2016;81:113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wiese MD, Schnabl M, O’Doherty C. et al. Polymorphisms in cytochrome P450 2C19 enzyme and cessation of leflunomide in patients with rheumatoid arthritis. Arthritis Res Ther 2012;14:R163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim KA, Joo HJ, Park JY.. Effect of ABCG2 genotypes on the pharmacokinetics of A771726, an active metabolite of prodrug leflunomide, and association of A771726 exposure with serum uric acid level. Eur J Clin Pharmacol 2011;67:129–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study used data from the CPRD. Due to the CPRD data-sharing policy, we are unable to share this study’s data. However, access to CPRD data can be directly requested from the CPRD.