Abstract
Objective
To gain insight in the expression profile of long non-coding RNAs (lncRNAs) in OA subchondral bone.
Methods
RNA sequencing data of macroscopically preserved and lesioned OA subchondral bone of patients that underwent joint replacement surgery due to OA (N = 22 pairs; 5 hips, 17 knees, Research osteoArthrits Articular Tissue (RAAK study) was run through an in-house pipeline to detect expression of lncRNAs. Differential expression analysis between preserved and lesioned bone was performed. Spearman correlations were calculated between differentially expressed lncRNAs and differentially expressed mRNAs identified previously in the same samples. Primary osteogenic cells were transfected with locked nucleic acid (LNA) GapmeRs targeting AC005165.1 lncRNA, to functionally investigate its potential mRNA targets.
Results
In total, 2816 lncRNAs were well-expressed in subchondral bone and we identified 233 lncRNAs exclusively expressed in knee and 307 lncRNAs exclusively in hip. Differential expression analysis, using all samples (N = 22 pairs; 5 hips, 17 knees), resulted in 21 differentially expressed lncRNAs [false discovery rate (FDR) < 0.05, fold change (FC) range 1.19–7.39], including long intergenic non-protein coding RNA (LINC) 1411 (LINC01411, FC = 7.39, FDR = 2.20 × 10−8), AC005165.1 (FC = 0.44, FDR = 2.37 × 10−6) and empty spiracles homeobox 2 opposite strand RNA (EMX2OS, FC = 0.41, FDR = 7.64 × 10−3). Among the differentially expressed lncRNAs, five were also differentially expressed in articular cartilage, including AC005165.1, showing similar direction of effect. Downregulation of AC005165.1 in primary osteogenic cells resulted in consistent downregulation of highly correlated frizzled related protein (FRZB).
Conclusion
The current study identified a novel lncRNA, AC005165.1, being dysregulated in OA articular cartilage and subchondral bone. Downregulation of AC005165.1 caused a decreased expression of OA risk gene FRZB, an important member of the wnt pathway, suggesting that AC005165.1 could be an attractive potential therapeutic target with effects in articular cartilage and subchondral bone.
Keywords: OA, long non-coding RNA, subchondral bone, articular cartilage
Rheumatology key messages.
Epigenetic differences between hip and knee OA subchondral bone were identified based on long non-coding RNAs (lncRNAs).
Twenty-one lncRNAs were identified as being differentially expressed between preserved and lesioned OA subchondral bone.
OA-related deregulation of FRZB might be caused by deregulation of lncRNA AC005165.1
Introduction
OA is a highly prevalent degenerative joint disease, characterized by articular cartilage degradation and subchondral bone remodelling [1–3]. Since OA is now considered a disease of the whole joint, recently focus has shifted towards characterization of gene expression profiles in OA synovium and subchondral bone [4, 5]. In this respect, we reported on mRNA expression profiling of OA subchondral bone of knee and hip joints [6]. We observed clustering of the samples based on joint site, suggesting distinct subchondral bone OA pathophysiological processes. This indicates that future therapeutic strategies particularly targeting bone should consider such differences between joint sites.
Different epigenetic mechanisms are described in OA, each of them modifying gene expression upon environmental cues such as mechanical stress or disease, without changing the genetic code. Among these, DNA methylation, histone modifications and miRNA expression are the most frequently studied in OA articular cartilage [1, 7–11]. In contrast, the role of long non-coding RNAs (lncRNAs) with OA pathophysiology is less explored as they show poor conservation between species [9]. lncRNAs are typically defined as RNAs >200 nucleotides in length, with little or no coding potential, and they are known to be involved in various transcriptional and (post-)translational processes, such as chromatin remodelling, mRNA/protein stabilization, production of short interfering RNAs and recruitment of scaffolding proteins, or they might act as pseudogenes [12, 13]. Moreover, the expression of lncRNAs can be highly tissue- and disease-specific [14, 15]. Due to the fact that OA is a disease of the whole joint, it is of added value to identify disease-specific lncRNAs that are expressed in various tissues involved in the OA pathophysiology, since these lncRNAs might serve as a potential druggable target with effects in several disease-relevant tissues.
Upon applying an in-house developed pipeline to reliably detect lncRNAs from RNA sequencing, we recently reported on the characterization of lncRNAs in OA cartilage. Notably, we identified prolyl 3-hydroxylase 2 antisense (P3H2-AS1) as being differentially expressed between macroscopically preserved and lesioned OA cartilage, and this was shown to regulate prolyl 3-hydroxylase 2 (P3H2) in cis [16]. Ajekigbe et al. [17] also reported on the expression levels of lncRNAs in OA cartilage, identifying among others LINC01411 and AC003090.1 as being differentially expressed between intact and damage OA cartilage from knees. Furthermore, Sun et al. [14] summarized the findings on the identification of lncRNAs involved in osteogenesis, such as maternally expressed 3 (MEG3), metastasis associated lung adenocarcinoma transcript 1 (MALAT1) and differentiation antagonizing non-protein coding RNA (DANCR). To our knowledge, however, there are no studies yet focussing on the characterization of lncRNA expression profiles with ongoing OA in subchondral bone.
In the current study, we set out to characterize the lncRNA expression profile in subchondral bone using RNA sequencing data of patients that underwent joint replacement surgery due to OA (RAAK study). First, joint-specific lncRNAs expressed in OA subchondral bone were identified. Differential expression analysis comparing macroscopically preserved and lesioned OA bone (N = 22 paired samples) was then performed to identify robust differentially expressed lncRNAs. To investigate the role of the differentially expressed lncRNAs identified herein with OA pathophysiology, we correlated the expression levels of these lncRNAs with the expression levels of our previously identified differentially expressed mRNAs in subchondral bone of the same patients [6]. Finally, we functionally investigated the effect of a specific lncRNA on mRNA expression levels in primary osteogenic cells.
Methods
Sample description
The current study includes 41 participants of the RAAK study [2], who underwent a joint replacement surgery due to OA (supplementary Table S1, available at Rheumatology online). Macroscopically preserved and lesioned subchondral bone were collected from the joints of 37 of the 41 participants, for either RNA-sequencing (RNA-seq) (N = 22) or replication by means of reverse transcriptase-quantitative PCR (RT-qPCR) (N = 15) (supplementary Table S1A and B, available at Rheumatology online). Osteogenic cells were collected from 4 of the 41 participants (supplementary Table S1C, available at Rheumatology online). The classification of macroscopically preserved and lesioned OA subchondral bone was based on its preserved and lesioned classified overlying cartilage as described previously [2]. The results reported here were compared with our recently reported results on the expression of lncRNAs in OA articular cartilage [16], in which 98 samples were used (65 knees, 33 hips). Of these OA articular cartilage samples, 10 paired samples did overlap with the OA subchondral bone samples, i.e. of these 10 patients we had preserved and lesioned OA articular cartilage and OA subchondral bone. Written informed consent was obtained from all participants of the RAAK study and ethical approval for the RAAK study was given by the medical ethics committee of the Leiden University Medical Center (P08.239/P19.013).
RNA sequencing
Sequencing was performed on preserved and lesioned OA subchondral on the Illumina HiSeq4000 (San Francisco, California, USA). Detailed information on the RNA isolation, alignment, mapping and filtering on lncRNAs is available in the Supplementary Methods (available at Rheumatology online). To identify outliers, principal component analysis and hierarchical clustering on the samples was applied. Three extreme outliers were identified (supplementary Fig. S1, available at Rheumatology online) and upon performing sensitivity analysis, these outliers were removed from the dataset. Finally, non-paired samples were removed from the dataset resulting in 22 paired samples (N = 17 paired knee samples, N = 5 paired hip samples) for further analysis, of which 10 paired samples were overlapping with the cartilage samples of our previous study [16].
lncRNA expression
To identify the lncRNAs that are expressed in subchondral bone, we filtered the lncRNAs identified by our in-house pipeline on a minimal average read count of four and a minimal count of two in at least 80% of the samples, indicated as robustly expressed. Cluster analysis was based on Euclidean distance and a heatmap was created using the lncRNAs that were expressed in the total dataset, the knee dataset and the hip dataset.
Differential expression analysis
Prior to the differential expression analysis, the lncRNAs were filtered on a minimum average read counts of 4 to allow variation. Differential expression analysis was performed between preserved and lesioned OA subchondral bone. The results were validated and replicated by means of RT-qPCR. Additional information is available in the Supplementary Methods (available at Rheumatology online).
Correlation analysis
Correlation between the expression levels of previously identified differentially expressed mRNAs in subchondral bone [6] and the expression levels of the here identified differentially expressed lncRNAs in subchondral bone was calculated using a Spearman correlation. Additional information is available in the Supplementary Methods (available at Rheumatology online).
Functional validation of AC005165.1
Primary osteogenic cells were isolated from the OA joints (supplementary Table S1C, available at Rheumatology online), resulting in isolation of a mixture of bone cells, which was characterized by measuring osteogenic and chondrogenic markers (supplementary Fig. S2, available at Rheumatology online). Subsequently, osteogenic cells were transfected with antisense locked nucleic acid (LNA) GapmeRs (Qiagen, Hilden, Germany) targeting AC005165.1 or GapmeRs negative control. RT-qPCR was performed to measure gene expression levels. Additional information is available in the Supplementary Methods, available at Rheumatology online.
Data availability
The RNA-seq data are deposited at the European Genome-Phenome Archive (accession number: EGAS00001004476).
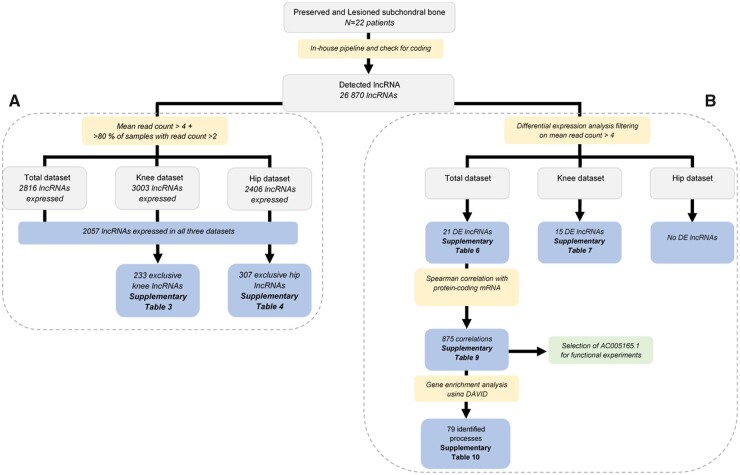
A complete overview of the approach applied to identify lncRNAs being expressed in subchondral bone is shown in Fig. 1A. An overview of the approach applied on identification of differential expressed lncRNAs with OA pathophysiology is shown in Fig. 1B.
Fig. 1.
Schematic overview of applied strategy
(A) Identification of expressed lncRNAs. (B) identification of lncRNAs differentially expressed between macroscopically preserved and lesioned OA subchondral bone. DE: differentially expressed; lncRNA: long non-coding RNA.
Results
Expression of lncRNAs in OA subchondral bone
Initially, we explored the expression profile of lncRNAs in OA subchondral bone (Fig. 1A). We applied our in-house pipeline [16] on an RNA sequencing dataset of 22 paired samples (5 hips, 17 knees; supplementary Table S1A, available at Rheumatology online) of macroscopically lesioned and preserved OA subchondral bone. Henceforth, we filtered on a minimal average read count of 4 and a minimal count of 2 in at least 80% of the samples, and we identified 2816 lncRNAs robustly expressed in OA subchondral bone.
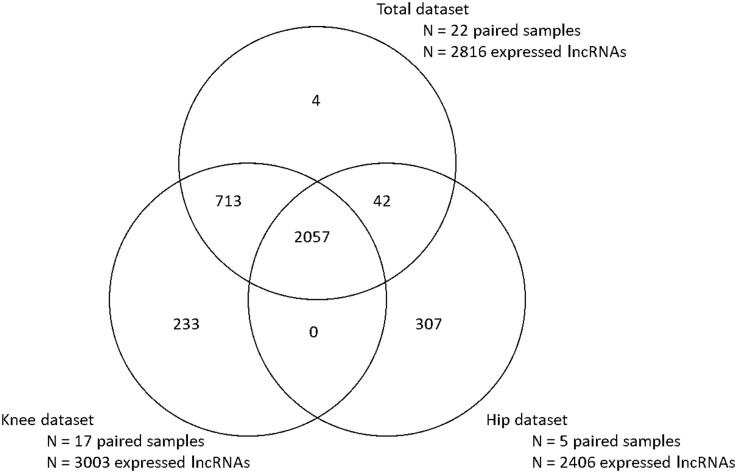
Since we observed major differences in mRNA expression levels between knee and hip OA subchondral bone in our previous study [6], we also explored lncRNA expression patterns in knee and hip subchondral bone separately, while including both preserved and lesioned samples. As shown in Fig. 2, we identified 2057 overlapping lncRNAs commonly expressed in the hip, knee and total datasets (mean counts between 4.02 and 3.40 × 105; supplementary Table S2, available at Rheumatology online). Moreover, we identified 233 exclusive knee lncRNAs (mean counts between 4.0 and 23; supplementary Table S3, available at Rheumatology online) and 307 exclusive hip lncRNAs (mean counts between 4.0 and 892; supplementary Table S4, available at Rheumatology online).
Fig. 2.
Venn diagram
Venn diagram of lncRNAs being expressed in the total, knee, and hip dataset of preserved and lesioned OA subchondral bone. lncRNA: long non-coding RNA.
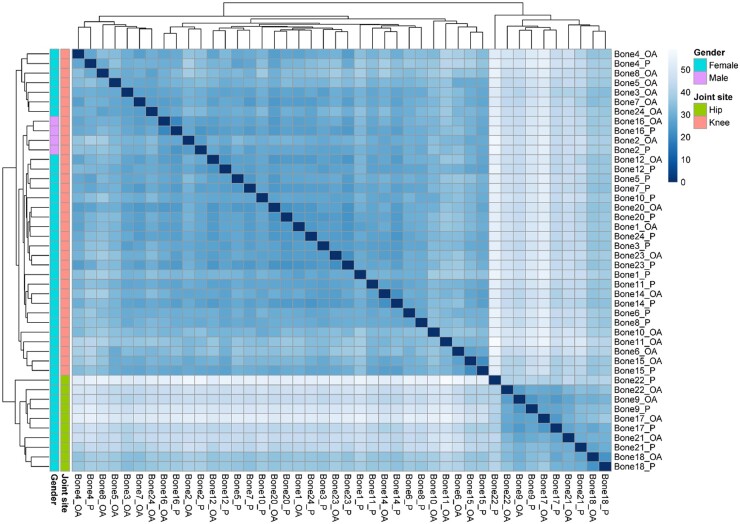
To investigate differences in expression levels of commonly expressed lncRNAs in knee and hip subchondral bone samples (N = 2057 lncRNAs; Fig. 2), we performed cluster analysis based on these commonly expressed lncRNAs using the Euclidian distance (Fig. 3). We observed, similar to the mRNA profile of subchondral bone, clustering of lncRNA expression profiles based on joint site. To investigate which lncRNAs are most contributing to this clustering, we performed differential expression analysis between the two clusters, with the hip cluster set as a reference. More specifically, we found 1069 lncRNAs being significantly differentially expressed between the two clusters (supplementary Table S5, available at Rheumatology online). The lncRNAs showing the highest fold difference (FD), i.e. lncRNAs highly expressed in knee samples, were AC068724.4 (FD = 158.87), AL034397.3 (FD = 157.82) and LINC02009 (FD = 89.21), while the lncRNAs with the lowest FD, i.e. highly expressed in hip samples, were AC105046.1 (FD = 0.15), transforming growth factor beta 2 overlapping transcript 1 (TGFB2-OT1, FD = 0.21) and LINC02328 (FD = 0.21).
Fig. 3.
Heatmap of sample distance
Heatmap is based on lncRNA expression levels of lncRNAs (N = 2057) expressed in all three datasets (i.e. total, hip and knee dataset of preserved and lesioned OA subchondral bone). lncRNA: long non-coding RNA.
Differential expression analysis of lncRNAs in OA subchondral bone
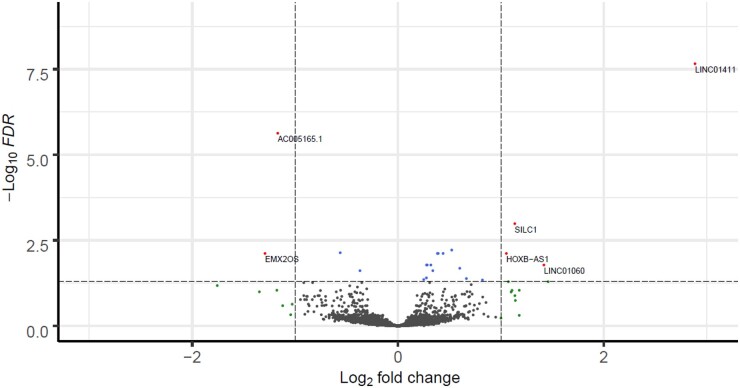
Next, we explored lncRNAs that change expression levels with OA pathophysiology, using a slightly different selection criteria to allow more variation (Fig. 1B). To identify robust lncRNAs that are associated with the OA pathophysiological process in subchondral bone, we filtered lncRNAs on a minimal average read count of 4 and we performed differential expression analyses between preserved and lesioned OA subchondral bone samples (knees and hips together). We identified 21 lncRNAs being false discovery rate (FDR) significantly differentially expressed between preserved and lesioned OA subchondral bone (Fig. 4; supplementary Table S6, available at Rheumatology online). Among these, LINC01411 [fold change (FC) = 7.39, FDR = 2.20 × 10−8] showed the highest and most significant upregulation, while AC005165.1 (FC = 0.44, FDR = 2.37 × 10−6) showed the most significant downregulation and EMX2OS (FC = 0.41, FDR = 7.64 × 10−3) the largest downregulation in lesioned compared with preserved OA subchondral bone. Differential expression analysis stratifying for joint site resulted in the identification of 15 lncRNAs being FDR significantly differentially expressed between preserved and lesioned knee samples (N = 17 paired samples; supplementary Fig. S3A, available at Rheumatology online), of which cancer susceptibility 15 (CASC15, FC = 1.48, FDR = 2.67 × 10−2) and AL135926.1 (FC = 1.70, FDR = 9.92 × 10−5) appeared to be exclusive knee lncRNAs, i.e. not significantly differentially expressed in the total nor the hip dataset (supplementary Table S7, available at Rheumatology online). We did not find any significantly differentially expressed lncRNAs between preserved and lesioned hip samples (N = 6 paired samples; supplementary Fig. S3B, available at Rheumatology online). To validate and replicate the results of the differential expression analysis by means of RT-qPCR, we included N = 9 paired samples for technical validation, i.e. samples overlapping with the RNA-seq dataset, and N = 15 paired samples for biological validation, i.e. additional preserved and lesioned OA subchondral bone samples (supplementary Table S1B, available at Rheumatology online). A selection of seven lncRNAs was measured in these samples: LINC01411, growth arrest specific 5 (GAS5), EMX2OS, PVT, LINC01060, sciatic injury induced lincRNA upregulator of SOX11 (SILC1) and AC005165.1. These lncRNAs showed similar directions of effect in the technical validation and the biological replication samples as compared with the direction of effect measured in the RNA-seq data, except for EMX2OS (supplementary Table S8, available at Rheumatology online).
Fig. 4.
Volcano plot
Volcano plot of differentially expressed lncRNAs in OA subchondral bone. The dots in the figure represent lncRNAs expressed in bone. Blue dots represent lncRNAs that are significantly differentially expressed, red dots represent lncRNAs that are significantly differentially expressed and have an absolute fold change of ≥2 and green dots represent the lncRNAs with an absolute fold change of ≥2 that are not significantly differentially expressed. FDR: false discovery rate; lncRNA: long non-coding RNA.
Correlation of mRNA and lncRNA in OA subchondral bone
To identify possible mRNA targets of the differentially expressed lncRNAs, i.e. lncRNAs regulating mRNAs with OA pathophysiology in subchondral bones, we filtered our recently reported differentially expressed mRNAs in subchondral bone [6] for protein-coding mRNAs (N = 1417 protein-coding differentially expressed mRNAs) and correlated them with expression levels of the differentially expressed lncRNAs (N = 21 lncRNAs) of the same patients (N = 22 paired samples). Upon prioritizing on high absolute correlations (−0.8 > ρ > 0.8) and significance (FDR < 0.05), we found 875 significant correlations between 16 lncRNAs and 378 mRNAs (supplementary Table S9, available at Rheumatology online). lncRNA small nucleolar RNA host gene 3 (SNHG3) showed the most interactions to mRNAs, with 174 significant correlations. In addition, the highest negative correlation was seen between SNHG3 and PTPRM (ρ = –0.92), encoding Protein Tyrosine Phosphatase Receptor Type M, whereas the highest positive correlation was seen between AC144548.1 and ILF2 (ρ = 0.92), encoding Interleukin Enhancer-binding Factor 2. Other notable interactions were those between AC005165.1 and FRZB (ρ = 0.85), encoding Frizzled Related Protein, and between SILC1 and POSTN (ρ = 0.81), encoding Periostin, which are both well-known OA genes.
To explore whether the differentially expressed lncRNAs are involved in certain processes or pathways, we performed gene enrichment analysis on their correlating mRNAs (supplementary Table S10, available at Rheumatology online). Genes correlated to 9 out of 16 lncRNAs showed significant enrichment. The genes correlated to AC006511.5 were enriched for Extracellular exosome [Gene Ontology (GO): 0070062, FDR = 3.67 × 10−4] and Myelin sheath (GO: 0043209, FDR = 3.67 × 10−4). Genes correlated to SILC1 were significantly enriched for the GO terms proteinaceous extracellular matrix (GO: 0005578, FDR = 1.07 × 10−4) and endoplasmic reticulum lumen (GO: 0005788, FDR = 4.62 × 10−2), while for example genes correlated to AC116533.1, AC245033.4 and GAS5 were all significantly enriched for transcriptional and translational processes such as translational initiation (GO: 0006413), poly(A) RNA binding (GO: 0044822) and viral transcription (GO: 0019083).
Functional investigation of AC005165.1
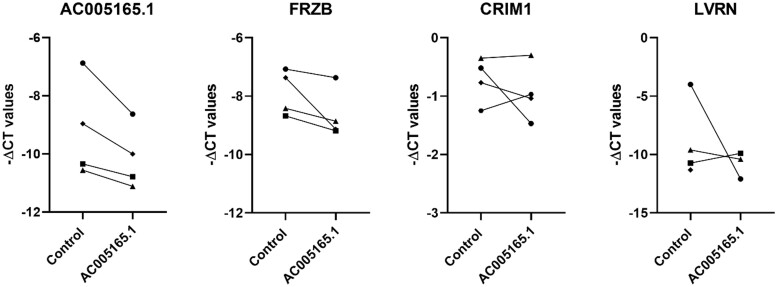
AC005165.1 was identified as the most significantly downregulated lncRNA (supplementary Table S6, available at Rheumatology online) and, among others, it showed high correlation with well-known OA gene FRZB (ρ = 0.85; supplementary Table S9, available at Rheumatology online). Therefore, we selected AC005165.1 to functionally investigate its possible mRNA targets in vitro. As shown in Fig. 5, upon downregulation of AC005165.1 (FC = 0.55, P = 0.51) by transfecting primary osteogenic cells (collected from N = 4 knees) with an LNA GapmeRs targeting AC005165.1, we observed consistent downregulation of FRZB (FC = 0.54), which was in line with the observed positive correlation (ρ = 0.85). However, the downregulation of FRZB did not reach statistical significance (P = 0.08). Other mRNAs highly correlating with AC0051651.1, such as cysteine rich transmembrane BMP regulator 1 (CRIM1, ρ = 0.82) and laeverin (LVRN, ρ = –0.84), showed more donor-dependent variation upon downregulation of AC005165.1.
Fig. 5.
Expression levels of AC005165.1, FRZB, CRIM1 and LVRN
Expression levels of AC005165.1, FRZB, CRIM1 and LVRN upon either transfecting primary osteogenic cells with LNA GapmeRs targeting AC005165.1 (indicated with AC005165.1) or transfecting primary osteogenic cells with a negative control (cells were collected from N = 4 knee joints).
Comparison of lncRNAs between subchondral bone and articular cartilage
Since subchondral bone and the articular cartilage are interacting tissues, we used our previously published results on lncRNAs in OA articular cartilage [16] to compare the identified differentially expressed lncRNAs between preserved and lesioned OA articular cartilage and preserved and lesioned subchondral bone. First, we selected the overlapping samples for which we had RNA-seq data of subchondral bone and articular cartilage (N = 10 paired samples; supplementary Table S1C, available at Rheumatology online). As shown in supplementary Fig. S4A (available at Rheumatology online), we found 1763 exclusive subchondral bone lncRNAs, 590 exclusive cartilage lncRNAs and 1090 lncRNAs that were expressed in both tissues (supplementary Table S11, available at Rheumatology online). Upon comparing the here identified differentially expressed lncRNAs in subchondral bone with our previously identified differentially expressed in articular cartilage [16], we found five lncRNAs to be differentially expressed in both tissues: AC005165.1, SILC1, LINC01411, AL590560.2 and AC079781.5 (supplementary Fig. S4B and Table S12, available at Rheumatology online). These five overlapping lncRNAs showed all similar directions of effect between preserved and lesioned samples in articular cartilage and subchondral bone.
Discussion
We set out to study lncRNAs in subchondral bone as function of joint site and OA pathophysiology. In doing so, we identified 2057 lncRNAs commonly expressed in subchondral bone of hip and knee joints, 233 exclusive knee lncRNAs and 307 exclusive hip lncRNAs. Moreover, we observed additionally clustering on joint site based on level of lncRNA expression (Fig. 3) among the commonly expressed lncRNA, signifying the difference between hip and knee OA subchondral bone pathophysiology. Differential expression analysis further identified 21 lncRNAs being differentially expressed between preserved and lesioned OA subchondral bone. Among the 21 differentially expressed lncRNAs we found AC005165.1, which was highly correlated to well-known OA gene FRZB (ρ = 0.86). Upon functional investigation of AC005165.1 in vitro by downregulating AC005165.1 using LNA GapmeRs, we observed a concurrent downregulation of FRZB. As lncRNAs tend to be highly tissue specific, lncRNAs, such as AC005165.1, could be attractive therapeutic OA targets with tissue specific effects.
Among the 21 differentially expressed lncRNAs, we identified LINC01411 (FC = 6.19, FDR = 2.20 × 10−8) as the most significantly and highest upregulated lncRNA, AC005165.1 (FC = 0.44, FDR = 2.37 × 10−6) as the most significantly downregulated lncRNA and EMX2OS as the most downregulated lncRNA (FC = 0.41, FDR = 7.64 × 10−3). The function of LINC0411 remains unknown, however in a recent study it was found to be differentially expressed between healthy and OA articular cartilage and between healthy and OA synovium, indicating its role in OA across multiple tissues [19]. According to biotype classification of Ensembl v97 [18], AC005165.1 was classified as a novel transcript and its function is still unknown. AC005165.1 is genomically located at chromosome 7, with no coding genes lying within a 200-kb window. EMX2OS is an antisense RNA to EMX2, encoding Empty Spiracles Homeobox 2, which is a transcription factor crucial for the CNS. Multiple differentially methylated sites between preserved and lesioned OA articular cartilage have been reported in both EMX2OS and its antisense gene EMX2 [20]. However, we did not find EMX2 among the differentially expressed genes in our cartilage dataset [1] nor among the differentially expressed genes in bone [6]. Notably, we were not able to either validate or replicate the differential expression of EMX2OS by means of RT-qPCR, which might be due to its low expression levels and its consistency across individuals. Other notable differentially expressed lncRNAs were GAS5 (FC = 1.21, FDR = 1.66 × 10−2) and PVT1 (FC = 1.52, FDR = 2.07 × 10−2), as they both have been previously associated with OA pathophysiology [14, 17, 21].
To explore the potential targets and interactions of the 21 differentially expressed lncRNAs identified here, we calculated Spearman correlations between these lncRNAs and the previously identified differentially expressed mRNAs in the same OA subchondral bone samples and gene enrichment analysis was performed (supplementary Tables S9 and S10, available at Rheumatology online). AC005165.1 was highly correlated with nine mRNAs, including FRZB, CRIM1 and LVRN. FRZB is a known OA gene and absence of FRZB in mice was previously shown to result in increased bone stiffness and increased cartilage degeneration [22]. CRIM1 is involved the TGF-β pathways by its binding to BMP-4 and BMP-7 [23], and LVRN is a metalloprotease that was previously linked to RA [24]. Despite the fact that LINC0411 showed a higher FC than AC005165.1, we selected AC005165.1 for functional investigation to determine the functional relation between AC005165.1 and the correlated mRNAs. Upon downregulation of AC005165.1 in primary osteogenic cells, we observed consistent downregulation of FRZB, while CRIM1 and LVRN expression levels did not change consistently. This suggests that AC005165.1 directly or indirectly targets FRZB gene expression, while CRIM1 and LVRN are functioning upstream of AC005165.1.
Similar to our mRNA expression profiling in OA subchondral bone [6], we here identified 233 exclusive knee, and 307 exclusive hip lncRNAs (supplementary Tables S3 and S4, available at Rheumatology online), indicating that lncRNA are not only tissue specific [14, 15], but also joint site specific. Additionally we showed (supplementary Fig. S3, available at Rheumatology online) that such differences are also captured by quantitative differences in expression levels. Consecutively, we showed knee joint specific differentially expressed lncRNAs between preserved and lesioned OA subchondral bone, such as CASC15 and AL135926.1 (supplementary Table S7, available at Rheumatology online). CASC15, which has not previously been associated to OA, is associated to cancer and involved in cell proliferation and migration [25]. AL135926.1 was classified as novel transcript by Ensembl v97 [18] and its exact function is still unknown. However, AL135926.1 is genomically located sense to protein-coding gene DPT, encoding dermatopontin, which was previously shown to inhibit BMP-2 activity in mice [26]. We did not find any FDR significantly differentially expressed lncRNAs when stratifying for hip samples, which is likely due to the low sample size. Together, the here detected tissue and joint site specificity of lncRNA’s qualifies them as eligible personalized therapeutic targets.
Although lncRNAs are known for their tissue specificity, we found a relatively large overlap of lncRNAs expressed in both articular cartilage and subchondral bone (N = 1090 lncRNAs), which might be due to their common origin. Among the overlapping differentially expressed between preserved and lesioned OA articular cartilage and subchondral bone, we found AC005165.1, making this lncRNA an attractive potential druggable target with effects in both tissues. The relative low number of differentially expressed lncRNAs identified in bone (N = 21) compared with those found in cartilage (N = 191) might be explained by the fact that cartilage is a single cell type tissue while subchondral bone multicellular and therefore more heterogeneous [27]. Moreover, the analysis on the subchondral bone included a lower sample size (N = 23 paired samples bone, N = 32 paired samples cartilage) and stricter threshold for including or excluding lncRNAs from the analysis.
The RNA-seq dataset that we used in this study was primarily obtained for mRNA expression profiling. Nonetheless, by applying our in-house pipeline we were able to characterize robust lncRNA expression in the same samples. It should be noted, however, that the lncRNA that entered the analyses had relatively high expression levels, while lncRNAs generally tend to be expressed at low levels [28]. To this end, we used two different selection criteria. In our initial, descriptive analyses on the lncRNA being expressed in our (knee and hip) subchondral bone samples we used more stringent selection criteria than in our pairwise differential expression analysis. This because per definition differential pairwise expression analysis is less sensitive for confounding factors. However, in future research the identification of lncRNAs associated with OA pathophysiology might be improved by increasing the sequencing depth.
In conclusion, the current study identified differences between hip and knee OA subchondral bone based on robust lncRNA expression levels. Moreover, AC005165.1 was identified as an attractive potential therapeutic target, as it was here shown to be differentially expressed between preserved and lesioned OA subchondral bone and previously it was shown to be differentially expressed between preserved and lesioned OA articular cartilage. Furthermore, AC005165.1 was here shown to regulate well-known OA gene FRZB in vitro. Finally, AC005165.1 was not significantly differentially expressed between the hip and knee clusters, which could make AC005165.1 a suitable druggable target in OA articular cartilage and OA subchondral bone of both hips and knees. More research is still needed to further elucidate the role and mode of action of AC005165.1 in the OA pathophysiology. Together, this study shows that lncRNAs could bring new opportunities regarding joint tissue specific therapeutic strategies.
Supplementary Material
Acknowledgements
We thank all the participants of the RAAK study. The Leiden University Medical Center has and continues to support the RAAK study. We thank all the members of our group. We also thank Enrike van der Linden, Robert van der Wal, Anika Rabelink-Hoogenstraaten, Peter van Schie, Shaho Hasan, Maartje Meijer, Daisy Latijnhouwers and Geert Spierenburg for collecting the RAAK material. Moreover, data were generated within the scope of the Medical Delta programs Regenerative Medicine 4D: Generating complex tissues with stem cells and printing technology and Improving Mobility with Technology.
Funding: The study was funded by the Dutch Scientific Research council NWO/ZonMW VICI scheme (no. 91816631/528), Dutch Arthritis Society (DAA-10-1-402, DAF-16-1-405).
Disclosure statement: The authors have declared no conflicts of interest.
Data availability statement
The RNA-seq data are deposited at the European Genome-Phenome Archive (accession number: EGAS00001004476, https://ega-archive.org/). Data are available upon reasonable request by any qualified researchers who engage in rigorous, independent scientific research, and will be provided following review and approval of a research proposal and Statistical Analysis Plan (SAP) and execution of a Data Sharing Agreement (DSA). All other data relevant to the study are included in the article.
Supplementary data
Supplementary data are available at Rheumatology online.
Contributor Information
Margo Tuerlings, Department of Biomedical Data Sciences.
Marcella van Hoolwerff, Department of Biomedical Data Sciences.
Jessica M van Bokkum, Department of Biomedical Data Sciences.
H Eka D Suchiman, Department of Biomedical Data Sciences.
Nico Lakenberg, Department of Biomedical Data Sciences.
Demiën Broekhuis, Department of Orthopaedics, Leiden University Medical Center, Leiden, The Netherlands.
Rob G H H Nelissen, Department of Orthopaedics, Leiden University Medical Center, Leiden, The Netherlands.
Yolande F M Ramos, Department of Biomedical Data Sciences.
Hailiang Mei, Department of Biomedical Data Sciences.
Davy Cats, Department of Biomedical Data Sciences.
Rodrigo Coutinho de Almeida, Department of Biomedical Data Sciences.
Ingrid Meulenbelt, Department of Biomedical Data Sciences.
References
- 1. Coutinho de Almeida R, Ramos YFM, Mahfouz A. et al. RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage. Ann Rheum Dis 2019;78:270–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ramos YF, den Hollander W, Bovée JVMG. et al. Genes involved in the osteoarthritis process identified through genome wide expression analysis in articular cartilage; the RAAK study. PLoS One 2014;9:e103056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dunn SL, Soul J, Anand S. et al. Gene expression changes in damaged osteoarthritic cartilage identify a signature of non-chondrogenic and mechanical responses. Osteoarthritis Cartilage 2016;24:1431–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chou CH, Lee CH, Lu LS. et al. Direct assessment of articular cartilage and underlying subchondral bone reveals a progressive gene expression change in human osteoarthritic knees. Osteoarthritis Cartilage 2013;21:450–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kuttapitiya A, Assi L, Laing K. et al. Microarray analysis of bone marrow lesions in osteoarthritis demonstrates upregulation of genes implicated in osteochondral turnover, neurogenesis and inflammation. Ann Rheum Dis 2017;76:1764–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tuerlings M, Hoolwerff M, Houtman E. et al. RNA sequencing reveals interacting key determinants of osteoarthritis acting in subchondral bone and articular cartilage. Arthritis Rheumatol 2021;73:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coutinho de Almeida R, Ramos YFM, Meulenbelt I.. Involvement of epigenetics in osteoarthritis. Best Pract Res Clin Rheumatol 2017;31:634–48. [DOI] [PubMed] [Google Scholar]
- 8. Reynard LN, Barter MJ.. Osteoarthritis year in review 2019: genetics, genomics and epigenetics. Osteoarthritis Cartilage 2020;28:275–84. [DOI] [PubMed] [Google Scholar]
- 9. Rice SJ, Beier F, Young DA. et al. Interplay between genetics and epigenetics in osteoarthritis. Nat Rev Rheumatol 2020;16:268–81. [DOI] [PubMed] [Google Scholar]
- 10. Yang J, Qin S, Yi C. et al. MiR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett 2011;585:2992–7. [DOI] [PubMed] [Google Scholar]
- 11. Endisha H, Rockel J, Jurisica I. et al. The complex landscape of microRNAs in articular cartilage: biology, pathology, and therapeutic targets. JCI Insight 2018;3:e121630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frankish A, Diekhans M, Ferreira AM. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marchese FP, Raimondi I, Huarte M.. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 2017;18:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun H, Peng G, Ning X. et al. Emerging roles of long noncoding RNA in chondrogenesis, osteogenesis, and osteoarthritis. Am J Transl Res 2019;11:16–30. [PMC free article] [PubMed] [Google Scholar]
- 15. Quinn JJ, Chang HY.. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47–62. [DOI] [PubMed] [Google Scholar]
- 16. van Hoolwerff M, Metselaar PI, Tuerlings M. et al. Elucidating epigenetic regulation by identifying functional cis-acting long noncoding RNAs and their targets in osteoarthritic articular cartilage. Arthritis Rheumatol 2020;72:1845–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ajekigbe B, Cheung K, Xu Y. et al. Identification of long non-coding RNAs expressed in knee and hip osteoarthritic cartilage. Osteoarthritis Cartilage 2019;27:694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cunningham F, Achuthan P, Akanni W. et al. Ensembl 2019. Nucleic Acids Res 2019;47:D745–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li C, Zheng Z.. Identification of novel targets of knee osteoarthritis shared by cartilage and synovial tissue. Int J Mol Sci 2020;21:6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y, Fukui N, Yahata M. et al. Genome-wide DNA methylation profile implicates potential cartilage regeneration at the late stage of knee osteoarthritis. Osteoarthritis Cartilage 2016;24:835–43. [DOI] [PubMed] [Google Scholar]
- 21. Xing D, Liang JQ, Li Y. et al. Identification of long noncoding RNA associated with osteoarthritis in humans. Orthopaedic Surg 2014;6:288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lories RJ, Peeters J, Bakker A. et al. Articular cartilage and biomechanical properties of the long bones in Frzb-knockout mice. Arthritis Rheum 2007;56:4095–103. [DOI] [PubMed] [Google Scholar]
- 23. Wilkinson L, Kolle G, Wen D. et al. CRIM1 regulates the rate of processing and delivery of bone morphogenetic proteins to the cell surface. J Biol Chem 2003;278:34181–8. [DOI] [PubMed] [Google Scholar]
- 24. Haas CS, Creighton CJ, Pi X. et al. Identification of genes modulated in rheumatoid arthritis using complementary DNA microarray analysis of lymphoblastoid B cell lines from disease-discordant monozygotic twins. Arthritis Rheum 2006;54:2047–60. [DOI] [PubMed] [Google Scholar]
- 25. Sheng L, Wei R.. Long non-coding RNA-CASC15 promotes cell proliferation, migration, and invasion by activating Wnt/β-catenin signaling pathway in melanoma. Pathobiology 2020;87:20–9. [DOI] [PubMed] [Google Scholar]
- 26. Behnam K, Murray SS, Brochmann EJ.. BMP stimulation of alkaline phosphatase activity in pluripotent mouse C2C12 cells is inhibited by dermatopontin, one of the most abundant low molecular weight proteins in demineralized bone matrix. Connect Tissue Res 2006;47:271–7. [DOI] [PubMed] [Google Scholar]
- 27. Goldring MB, Goldring SR.. Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann N Y Acad Sci 2010;1192:230–7. [DOI] [PubMed] [Google Scholar]
- 28. Jarroux J, Morillon A, Pinskaya M.. History, discovery, and classification of lncRNAs. In: Rao MRS, ed. Long non coding RNA biology. Singapore: Springer Singapore, 2017: 1–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are deposited at the European Genome-Phenome Archive (accession number: EGAS00001004476).
A complete overview of the approach applied to identify lncRNAs being expressed in subchondral bone is shown in Fig. 1A. An overview of the approach applied on identification of differential expressed lncRNAs with OA pathophysiology is shown in Fig. 1B.
Fig. 1.
Schematic overview of applied strategy
(A) Identification of expressed lncRNAs. (B) identification of lncRNAs differentially expressed between macroscopically preserved and lesioned OA subchondral bone. DE: differentially expressed; lncRNA: long non-coding RNA.
The RNA-seq data are deposited at the European Genome-Phenome Archive (accession number: EGAS00001004476, https://ega-archive.org/). Data are available upon reasonable request by any qualified researchers who engage in rigorous, independent scientific research, and will be provided following review and approval of a research proposal and Statistical Analysis Plan (SAP) and execution of a Data Sharing Agreement (DSA). All other data relevant to the study are included in the article.